Abstract
MOT1 is an ATPase which can dissociate TATA binding protein (TBP)-DNA complexes in a reaction requiring ATP hydrolysis. Consistent with this observation, MOT1 can repress basal transcription in vitro. Paradoxically, however, some genes, such as HIS4, appear to require MOT1 as an activator of transcription in vivo. To further investigate the function of MOT1 in basal transcription, we performed in vitro transcription reactions using yeast nuclear extracts depleted of MOT1. Quantitation of MOT1 revealed that it is an abundant protein, with nuclear extracts from wild-type cells containing a molar excess of MOT1 over TBP. Surprisingly, MOT1 can weakly activate basal transcription in vitro. This activation by MOT1 is detectable with amounts of MOT1 that are approximately stoichiometric to TBP. With amounts of MOT1 similar to those present in wild-type nuclear extracts, MOT1 behaves as a weak repressor of basal transcription. These results suggest that MOT1 might activate transcription via an indirect mechanism in which limiting TBP can be liberated from nonpromoter sites for use at promoters. In support of this idea, excess nonpromoter DNA sequesters TBP and represses transcription, but this effect can be reversed by addition of MOT1. These results help to reconcile previous in vitro and in vivo results and expand the repertoire of transcriptional control strategies to include factor-assisted redistribution of TBP between promoter and nonpromoter sites.
MOT1 is an essential Saccharomyces cerevisiae protein implicated in the regulation of a diverse set of genes (8, 9, 15). MOT1 was originally identified by a mutation, mot1-1, that resulted in elevated levels of reporter gene expression driven by a weak promoter (9). Subsequently, MOT1 was uncovered in several other similar genetic screens (12, 13, 16, 19). In unrelated lines of work, MOT1 was identified as a TATA binding protein (TBP)-associated factor (TAF) (17) and as a protein with ATP-dependent TBP-DNA dissociating activity in vitro (2). Consistent with these biochemical activities, MOT1 can repress basal transcription in vitro (1, 2). The activity of MOT1 in vitro can also be at least partially overcome by factors such as TFIIA that bind to TBP and block the interaction of TBP with MOT1 and/or stabilize the binding of TBP to DNA (1). Additionally, the transcriptional activator GAL4-VP16 can overcome MOT1-mediated repression of basal transcription in vitro (2). These combined biochemical and genetic observations lead to the suggestion that MOT1 functions in vivo as a global repressor of basal transcription. This simple picture of MOT1 function was challenged, however, by the surprising observation that mutation of MOT1 can have no detectable effect on expression of many genes or actually lead to a decrease in gene expression of certain genes in vivo (8, 15, 19). The HIS4 gene is particularly interesting in this regard because, while in vitro experiments supported the model that MOT1 can repress HIS4 basal transcription (2), in vivo, MOT1 appears to activate HIS4 transcription (8, 15).
How can the known TBP-DNA dissociating activity of MOT1 in vitro be reconciled with its apparent function as an activator at some promoters in vivo? One possibility is that MOT1 affects HIS4 expression in vivo by an indirect mechanism. For instance, MOT1 might regulate the expression of a protein that itself regulates the expression of many genes. Alternatively, MOT1 might function as an activator of transcription by disassembling TBP (or TBP-containing complexes) bound to some promoters. Certain promoters might direct the assembly of stable but kinetically “dead” transcription complexes. If transcriptionally competent complexes can form on these promoters only at a low frequency, then MOT1-catalyzed TBP-DNA dissociation at these sites might increase the levels of steady-state mRNA by providing an opportunity for multiple attempts to form a competent preinitiation complex.
Another possibility is that MOT1 is targeted to TBP-DNA complexes formed on high-affinity sites that are not present in promoters. TBP binds with high affinity to a variety of AT-rich sequences (7, 11, 22), and it is likely that many spurious TBP-binding sites are fortuitously present in the genome. Binding of TBP to bona fide TATA boxes could then be stabilized from MOT1 action at promoters by the association of TBP with other general transcription factors (1). In this study, in vitro transcription experiments were performed to test the idea that MOT1 might function to redistribute TBP among promoter and nonpromoter sites. The results indicate that low levels of MOT1 (stoichiometric to TBP) function to activate basal transcription. In contrast, a molar excess of MOT1 which approximates the amount of MOT1 found in nuclear extracts from wild-type cells leads to repression of basal transcription. Furthermore, the behavior of MOT1 can be switched in vitro from that of a weak activator to that of a weak repressor by adjusting the amount of nonpromoter DNA present in the reaction mixture. These results support a model in which one function of MOT1 in vivo is to facilitate the distribution of a limiting pool of TBP between promoter and nonpromoter sites.
MATERIALS AND METHODS
Nuclear extracts.
Nuclear extracts were prepared from 4-liter cultures of either wild-type (JD194 [9]) or mot1-1 (JD215b [9]) yeast grown in yeast extract-peptone-dextrose (YPD) at 30°C. Cells were pelleted in 500-ml centrifuge bottles in a GS3 rotor spun at 4,000 rpm for 9 min in a Sorvall RC 5B centrifuge. All subsequent centrifugations were performed under these conditions unless otherwise noted. Cell pellets were then resuspended in a total volume of 270 ml of 50 mM Tris-HCl (pH 7.5) containing 30 mM dithiothreitol (DTT) and incubated at 30°C for 15 min with gentle shaking. Cells were then pelleted and resuspended in a total volume of 40 ml of YPD-S (10 g of yeast extract, 20 g of peptone, 20 g of glucose, and 182.2 g of sorbitol per liter) containing 60 mg of Zymolyase 100T (ICN), 2 μM pepstatin, 0.6 μM leupeptin, chymostatin (2 μg/ml), 1 mM phenylmethylsulfonyl fluoride, and 2 mM benzamidine. Cell suspensions were then incubated for up to 2 h at 30°C, and spheroplast formation was monitored by measuring the optical density at 600 nm of 10 μl of cell suspension in 1 ml of 1% sodium dodecyl sulfate and by visualization, under a light microscope, of spheroplast ghosts formed in 1% sodium dodecyl sulfate. The reaction was terminated by the addition of 540 ml of YPD-S when approximately 70 to 80% of the cells had been converted to spheroplasts. Cells were then pelleted and resuspended in 1 liter of YPD-S. Following a 30-min incubation at 30°C with gentle shaking, cells were again pelleted and washed twice with 540 ml of YPD-S, followed by one wash with 540 ml of 1 M sorbitol at 4°C. All subsequent steps were performed at 4°C. The cell pellets were resuspended in approximately 250 ml of buffer A (18% [wt/vol] polysucrose 400 [Sigma], 10 mM Tris-Cl [pH 7.5], 20 mM KCl, 5 mM MgCl2, 1 mM EDTA, 0.5 mM spermidine, 0.15 mM spermine, 3 mM DTT, and protease inhibitors as described above). Spheroplasts were then homogenized by two passes through a Yamada LH21 homogenizer at 1,000 rpm. Large cellular debris and unlysed cells were removed by five centrifugations in 250-ml bottles spun in a GSA rotor at 5,400 rpm for 5 min each. The nuclei were then pelleted by spinning the extract at 13,000 rpm for 30 min in an SS34 rotor, and the nuclei were then resuspended in 10 to 20 ml of buffer B (100 mM Tris-acetate [pH 7.9], 50 mM potassium acetate, 10 mM MgSO4, 20% glycerol, 2 mM EDTA, 3 mM DTT, and protease inhibitors as described above) by using a Dounce homogenizer. To lyse the nuclei, 3 M ammonium sulfate (pH 7.6) was added to the nuclear suspension to achieve a final concentration of 0.5 M. Following stirring for 30 min, the debris was pelleted by spinning the lysate at 28,000 rpm for 75 min in an SW28 rotor. Ammonium sulfate (0.35 g/ml of nuclear extract) was then added, the extract was stirred for 30 min, and then nuclear proteins were pelleted by centrifugation at 20,000 rpm for 30 min in an SW28 rotor. Nuclear proteins were then resuspended in approximately 0.5 ml of buffer C (20 mM HEPES-KOH [pH 7.6], 10 mM MgSO4, 10 mM EGTA, 20% glycerol, 5 mM DTT, and protease inhibitors as described above) and dialyzed against 1 liter of buffer C for 4 h. Protein concentrations were determined by Bradford assay using bovine serum albumin as a standard; protein concentrations were typically 30 to 40 mg/ml. The experiments described in this paper were performed with two independently prepared batches of nuclear extract from mot1-1 cells, and the results were indistinguishable.
Recombinant proteins.
Recombinant yeast TBP was obtained from an Escherichia coli overexpression strain as previously described (21). MOT1 was obtained from a yeast overexpression strain and purified with antibody-coupled beads as previously described (3). One unit of MOT1 activity is defined as the amount of MOT1 required to completely dissociate all of the TBP-DNA complexes formed under the conditions previously described (1); MOT1 activity was assayed by gel mobility shift assay as previously described (1). Under these conditions, 1 U of MOT1 represents approximately 35 ng of MOT1 polypeptide, but it is important to note that a precise conversion between mass and activity is difficult since MOT1 specific activity is batch dependent and susceptible to degradation even in optimal storage buffers and as a result of freeze-thaw cycles.
Recombinant MOT1 and MOT1 (K1303A) mutant protein were obtained by using a baculovirus expression system. MOT1 (K1303A) contains a single amino acid change at lysine 1303, which destroys the ATPase activity of MOT1 without affecting the ability of the protein to bind to TBP-DNA complexes (3). Site-directed mutagenesis was used to insert a BglII restriction site in place of the MOT1 ATG at position +250. The resulting ∼6-kb BglII fragment was subcloned into the baculovirus vector pACHLT-A. Insect cells were infected according to the standard protocol (Pharmingen). Hi5 cells infected with the MOT1-containing virus were harvested and lysed by sonication in buffer A (sodium phosphate [pH 8], 250 mM NaCl, and 0.05% Triton X-100, plus the protease inhibitors phenylmethylsulfonyl fluoride benzamidine, TPCK [N-tosyl-l-phenylalanine chloromethyl ketone], TLCK [Nα-p-tosyl-l-lysine chloromethyl ketone], leupeptin, pepstatin, and aprotinin). Cell lysates were then incubated with Qiagen Ni-nitrilotriaacetic acid resin for 2 h at 4°C. MOT1 protein bound to the resin was washed extensively with buffer A containing 10 mM imidazole (pH 6) and finally eluted with buffer A plus 500 mM imidazole (pH 6). The recombinant MOT1 protein used in these studies was >90% pure.
In vitro transcription.
In vitro transcription was performed essentially as described previously (2, 6), with plasmids containing the HIS4 (pSH387), CYC1 (−52 TATA element; pCZGal3), or ACT1 (pSH385) core promoters (2). Transcription reactions were also performed with a plasmid (pTM04) containing a 542-bp fragment of the HIS4 gene obtained by PCR using the primers 5′-GGCTCGAGATTTGAGCAAGGAACTATTTTTGA-3′ and 5′-CCGGATCCGGTCATTATTCAGAAAAAAAATTTTGT-3′. This fragment was cloned into the XhoI and BamHI sites of pSH387 to generate an in vitro transcription template which directs the synthesis of RNA, which can be quantitated, and whose ends can be mapped by using the same primer as was used for the other templates. All of the transcription reactions except those shown in Fig. 3 (lanes 8 to 14) were performed in buffer containing 10 mM HEPES-KOH (pH 7.6), 100 mM potassium glutamate, 10 mM magnesium acetate, 5 mM EGTA, and 3.5% glycerol. The reactions in Fig. 3 (lanes 8 to 14) were performed in buffer containing 4 mM Tris-acetate (pH 8), 60 mM potassium acetate, 5 mM magnesium acetate, and 4% glycerol; for unknown reasons, under these conditions MOT1’s ability to bind to TBP-DNA is similar to that seen in glutamate-containing buffer but its ATP-dependent TBP-DNA dissociating activity is much greater than that detected in glutamate-containing buffer (not shown). Transcription reaction mixtures contained approximately 150 μg of nuclear extract protein, and specifically initiated RNA was detected by primer extension as previously described (20). The oligonucleotide duplexes used in Fig. 6 were obtained by combining 5′-CCCCGAC CGGGTGTTCCTGAAGGGGGGCTATAAAAGGGGGTGGGGGCGCG-3′ and 5′-CGCGCCCCCACCCCCTTTTATAGCCCCCCTTCAGGAACACCCGGTCGGGG-3′ to obtain a wild-type TATA-containing DNA or by combining 5′-CCCCGACCGGGTGTTCCTGAAGGGGGGCTGTAAAAGGGGGTGG GGGCGCG-3′ and 5′-CGCGCCCCCACCCCCTTTTACAGCCCCCCTTCAGGAACACCCGGTCGGGG-3′ to obtain a DNA duplex containing the sequence TGTAAAAG in the TATA box. The DNAs were mixed in a solution containing 10 mM Tris-Cl (pH 8), 1 mM EDTA, and 0.1 M NaCl; boiled; and then slowly cooled to room temperature for 30 min before use.
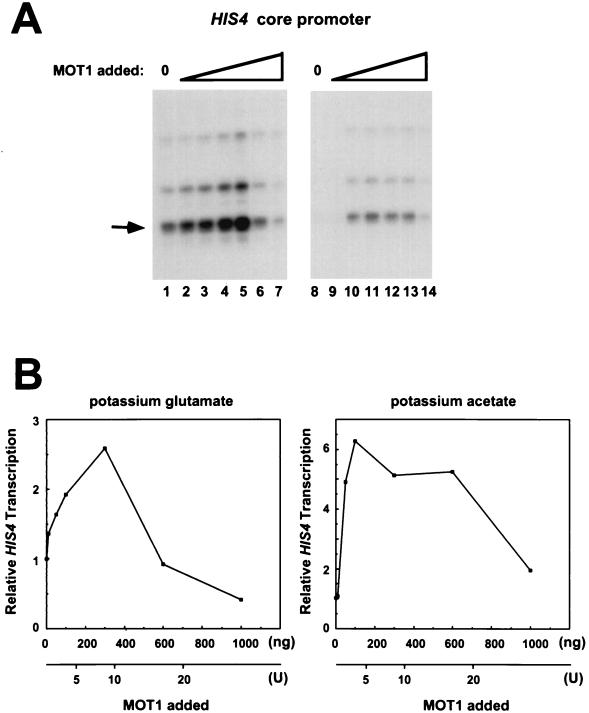
FIG. 3.
Recombinant MOT1 can activate or repress basal transcription driven by the HIS4 core promoter. (A) Experiments were performed as described in the legend to Fig. 2, but with increasing amounts of baculovirus-expressed MOT1 (rMOT1). Transcription reactions were performed in standard potassium glutamate-containing buffer (lanes 1 to 7) or in buffer containing potassium acetate (lanes 8 to 14) (see Materials and Methods). The reaction mixtures contained 10 ng (lanes 2 and 9), 50 ng (lanes 3 and 10), 100 ng (lanes 4 and 11), 300 ng (lanes 5 and 12), 600 ng (lanes 6 and 13), or 1,000 ng (lanes 7 and 14) of MOT1. The bands that were quantitated are indicated by the arrow. (B) Quantitation by phosphorimager analysis of the data shown in panel A. The graph on the left is of data from lanes 1 to 7, and the graph on the right is of data obtained from lanes 8 to 14. For comparison with the results obtained with purified MOT1 from yeast (yMOT1), the x axes also indicate the approximate units of MOT1 activity added.
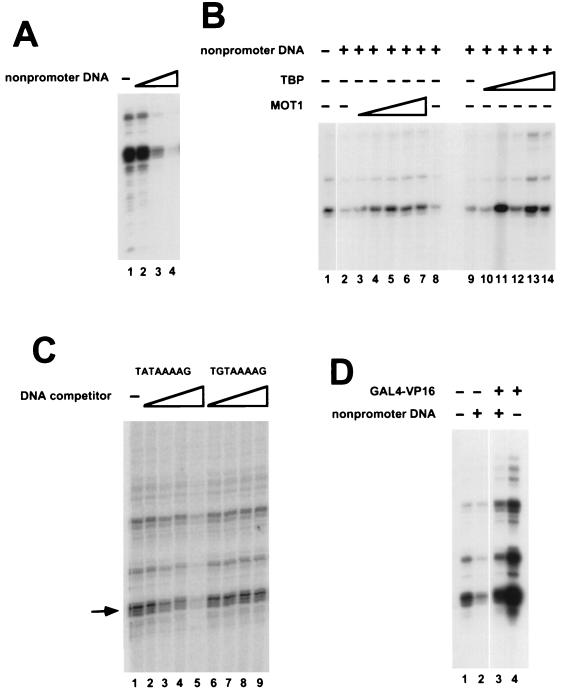
FIG. 6.
Repression of HIS4 basal transcription by nonpromoter DNA and effects of addition of TBP or GAL4-VP16. (A) Transcription reactions were performed with nuclear extract from mot1-1 cells. The reaction mixtures contained 0 μg (lane 1), 0.24 μg (lane 2), 1.2 μg (lane 3), or 2.4 μg (lane 4) of pKS II+ plasmid in addition to 0.25 μg of HIS4-containing plasmid template (see Materials and Methods). (B) Transcription reaction mixtures contained either 0 μg (lane 1) or 0.8 μg (lanes 2 to 14) of pKS II+. MOT1 purified from yeast was added to lanes 3 to 7 (0.5, 1.25, 2.5, 5, or 7.5 U, respectively); lane 8 contains a volume of mock-purified MOT1 equivalent to the volume of MOT1 added to lane 7. In addition to the TBP already present in the extract, recombinant yeast TBP was added to reaction mixtures in lanes 10 to 14 (1, 3, 10, 20, or 30 ng, respectively). The minus symbols in other lanes indicate that reaction mixtures contained TBP present in the nuclear extract but no additional recombinant TBP was added to the reaction. (C) HIS4 core promoter activity was assayed in the absence (lane 1) or presence of 50-mer oligonucleotide duplexes containing a wild-type TATA sequence (lanes 2 to 5) or a mutant TATA sequence, TGTAAAAG, which does not bind TBP (lanes 6 to 9). The reaction mixtures contained 3 ng (lanes 2 and 6), 10 ng (lanes 3 and 7), 30 ng (lanes 4 and 8), or 100 ng (lanes 5 and 9) of the indicated competitor oligonucleotides. (D) Transcription reaction mixtures contained pKS II+ (lanes 2 and 3) and/or GAL4-VP16 which was preincubated with DNA prior to the addition of mot1-1 nuclear extract. GAL4-VP16 activated transcription fivefold in the absence of pKS II+ and fourfold in the presence of pKS II+.
Western blot analysis.
Nuclear extract protein (10 μg) or recombinant MOT1 or TBP was fractionated on 10% protein gels and transferred to Immobilon P membranes (Millipore). The blots were probed with rabbit polyclonal anti-TBP antiserum (21) or rabbit polyclonal anti-MOT1 antiserum. The MOT1 antiserum was raised by using a bacterially expressed fragment of MOT1 encoding the C-terminal 67 amino acids. Immunoreactive bands were detected by enhanced chemiluminescence (Amersham ECL+), and because the relationship between band intensity and amount of protein is nonlinear, for purposes of quantitation, only bands of similar intensity were compared.
RESULTS AND DISCUSSION
Quantitation of MOT1 in wild-type and mot1-1 nuclear extracts.
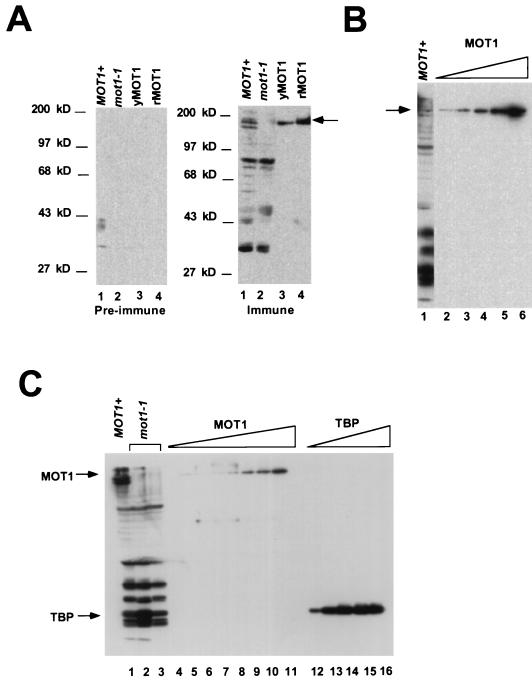
The amount of MOT1 in nuclear extracts from wild-type and mot1-1 cells was determined by Western blotting using rabbit polyclonal antiserum raised against the C-terminal 67 amino acids of MOT1. Previous results showed that this C-terminal tail of MOT1 is essential for function both in vitro and in vivo (3). Consequently, any MOT1 molecules lacking an intact C terminus which are not detectable with this antiserum are inactive. As shown in Fig. 1A, MOT1 antiserum specifically detects a protein in wild-type nuclear extracts migrating with an apparent molecular mass of 175 kDa. This polypeptide comigrates with MOT1 purified from a yeast overexpression strain (yMOT1) as well as MOT1 purified from a baculovirus overexpression system (rMOT1) (Fig. 1A, right-hand panel). Both purified preparations of MOT1 dissociate TBP-DNA complexes in an ATP-dependent manner, and both of these preparations have similar specific activities (not shown; see Materials and Methods). The immune antiserum also detects a series of smaller proteins in yeast nuclear extract, which may be degraded forms of MOT1. Since an intact N terminus was also shown to be essential for MOT1’s activity both in vitro and in vivo (3), N-terminally degraded forms of MOT1 are all presumed to be inactive. Extract from mot1-1 cells contains no detectable full-length MOT1 protein, although similar levels of smaller immunoreactive species are present in both wild-type and mot1-1 extracts (Fig. 1A, right-hand panel, lanes 1 and 2; Figure 1C, lanes 1 to 3). The amount of MOT1 in wild-type nuclear extract was estimated by comparing the signal obtained in nuclear extract with that from various amounts of purified baculovirus-expressed MOT1 (Fig. 1B). Based on this and other blots (not shown), there is approximately 90 ng of full-length MOT1 polypeptide in 10 μg of wild-type nuclear extract. Quantitation of TBP in these same nuclear extracts indicates that there is approximately 0.5 ng of TBP per 10 μg of nuclear extract protein (Fig. 1C). Additionally, the amount of TBP polypeptide is unchanged by the mot1-1 mutation. Remarkably, this indicates that MOT1 is present in vast excess (>20-fold molar excess) over TBP in our nuclear extracts.
FIG. 1.
Western blot analysis of wild-type and mot1-1 nuclear extracts. (A) Specificity of antiserum and comparison of native and recombinant MOT1. The two panels were loaded with identical protein samples; the left-hand panel was probed with preimmune serum, and the right-hand panel was probed with anti-MOT1 antiserum. Lanes 1 and 2 contain 10 μg of nuclear extract from wild-type and mot1-1 yeast cells, respectively. Lane 4 contains 250 ng of rMOT1; lane 3 contains a comparable amount of yMOT1 whose activity was not determined. The arrow indicates the position of full-length immunoreactive MOT1. (B) Quantitation of MOT1 in wild-type nuclear extract. Lane 1 contains 10 μg of total nuclear extract protein. Lanes 2 to 6 contain 90, 120, 210, 240, or 300 ng, respectively, of recombinant MOT1. The blot was probed as described for panel A, with anti-MOT1 antiserum. (C) Quantitation of MOT1 and TBP in wild-type or mot1-1 nuclear extract. Lane 1 contains 20 μg of wild-type nuclear extract protein. Lanes 2 and 3 contain 20 μg of total protein from two independently prepared samples of mot1-1 nuclear extract. Lanes 4 to 11 contain MOT1 purified from a yeast overexpression strain (0.5, 1.0, 1.5, 2.0, 2.5, 3.0, 4.0, or 5.0 U, respectively). Lanes 12 to 16 contain 1, 3, 9, 15, or 20 ng, respectively, of recombinant yeast TBP. The blot was probed with anti-MOT1 (top half) or anti-TBP (bottom half) rabbit polyclonal antiserum.
The molar excess of MOT1 over TBP in these nuclear extracts is likely due to loss of TBP (and/or TBP-containing complexes) during the extract preparation. Lysis of a known number of yeast cells followed by direct analysis of the lysate by Western blotting indicates that TBP is present in at least twofold molar excess over any single RNA polymerase II- or III-specific TAF, including MOT1 (4a). Additionally, the MOT1 human homologue, TAF172, was found to be present in amounts roughly equimolar to TBP (5) and not in the large excess observed for MOT1 in our nuclear extracts. This large concentration of MOT1 can readily explain why basal transcription is difficult to detect in nuclear extracts prepared from wild-type cells by this procedure (2).
Determination of the amount of MOT1 in these extracts was important to establish a range of MOT1 to add back to the in vitro transcription reaction mixtures described below. While the absolute level of MOT1 in wild-type nuclear extracts probably does not reflect the in vivo stoichiometry, it is important to note that activation of basal transcription by MOT1 (described below) occurs with amounts of MOT1 that reflect the probable ratio of MOT1 to TBP in vivo. Furthermore, reconstitution of mot1-1 nuclear extracts with levels of MOT1 present in wild-type nuclear extract supports the idea that MOT1 can repress basal transcription when present at high levels. Affinity-purified yMOT1 is obtained in amounts too low to accurately quantitate by protein assay, but the relative amounts of yMOT1 were determined by immunoblotting as shown in Fig. 1, and the activities of yMOT1 and rMOT1 in the in vitro transcription assay are discussed below. To provide a more accurate assessment of the effects of different small-scale preparations of yMOT1 on basal transcription, in subsequent experiments the amounts of yMOT1 added are expressed as units of MOT1 activity (described in Materials and Methods and reference 1).
Activation and repression of basal transcription by MOT1 in vitro.
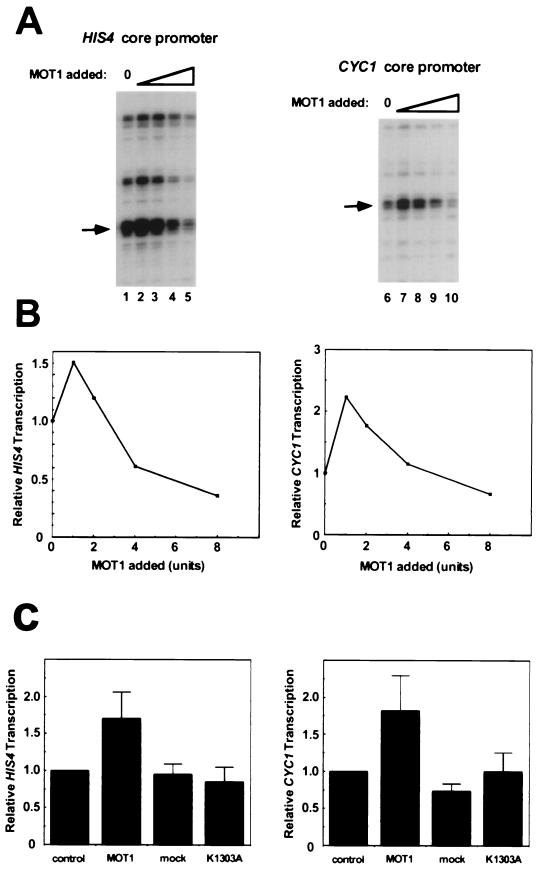
To determine the effects of exogenously added MOT1 on basal transcription, increasing amounts of yMOT1 were added to transcription reaction mixtures containing a yeast promoter and mot1-1 nuclear extract. Specifically initiated RNAs were quantitated by primer extension as described elsewhere (20). Results obtained with the HIS4 and CYC1 promoters are shown in Fig. 2A, and quantitation of the band intensities obtained with a PhosphorImager are shown in Fig. 2B. The HIS4 promoter is of particular interest, since MOT1 was shown to repress HIS4 basal transcription in vitro but activate HIS4 transcription in vivo (2, 8, 15). Basal transcription is almost undetectable in wild-type nuclear extract (2), but the levels of basal transcription obtained with mot1-1 nuclear extract are readily detected (Fig. 2A, lanes 1 and 6) (2). The addition of low levels of MOT1 (1 to 2 U) results in weak activation of transcription. Since these reaction mixtures contain approximately 7.5 ng of TBP, this amount of MOT1 would be insufficient to disrupt all of the TBP-DNA complexes formed under our standard gel mobility shift or footprinting conditions with this level of purified TBP. When 8 U of MOT1 are added to an otherwise identical reaction mixture, basal transcription is weakly repressed (Fig. 2A, lanes 5 and 10). In our standard assays, using purified components, for MOT1’s ATP-dependent TBP-DNA dissociating activity, 8 U of activity is just sufficient to disrupt all of the TBP-DNA complexes formed in a reaction mixture containing 7.5 ng TBP (not shown). Multiple replicates of the experiment shown in Fig. 2A established that the weak activation seen with low levels of yMOT1 is reproducible and statistically significant (Fig. 2C). This activation depends on the presence of the MOT1 polypeptide in the yMOT1 preparation and depends on a catalytically active MOT1 ATPase (Fig. 2C). The activity of ATPase-defective MOT1 is described more fully below.
FIG. 2.
Purified MOT1 can weakly activate or repress basal transcription in vitro. (A) MOT1 purified from a yeast overexpression strain was titrated into transcription reaction mixtures containing nuclear extract from mot1-1 cells and either the HIS4 core promoter or the CYC1 core promoter as indicated. The amount of MOT1 added is expressed as units of activity, with 1 U being the amount of MOT1 which can completely dissociate all of the TBP-DNA complexes formed under the conditions previously described (1). No MOT1 was added to the reaction mixtures in lanes 1 and 6; the amounts of MOT1 added to the other reaction mixtures were 1 U (lanes 2 and 7), 2 U (lanes 3 and 8), 4 U (lanes 4 and 9), and 8 U (lanes 5 and 10). Transcripts were detected by primer extension. The major start sites of transcription are identical to those previously observed both in vitro and in vivo (2). The bands that were quantitated are indicated by the arrows. (B) The relative amounts of specifically initiated RNA obtained in panel A are plotted versus the amount of MOT1 added. (C) Statistical analysis of the effect of exogenously added MOT1. The maximum stimulation of basal transcription by MOT1 is compared to the effects of adding mock-affinity-purified eluate obtained with whole-cell extract from cells containing MOT1 without an epitope tag (mock) or an equivalent amount of purified MOT1 harboring a point mutation which destroys ATPase activity (K1303A). Relative transcription levels are normalized to those in reactions which contained no additional eluate (control). The peak of activation by MOT1 varied slightly from experiment to experiment and also varied with different preparations of yMOT1, but the maximal activation was generally seen with 1 to 4 U of yMOT1 (see text). The error bars represent the standard deviation obtained by averaging the results of at least three independent experiments.
Given the well-established activity of MOT1 as both a repressor of basal transcription and an ATP-dependent TBP-dissociating enzyme (2), we were initially surprised that repression of basal transcription by wild-type yMOT1 was difficult to observe. This appears to be due in part to both the high levels of MOT1 in wild-type nuclear extracts and the low concentration of yMOT1 in our small-scale preparations, which make it difficult to reconstitute reactions with as much yMOT1 as is present in wild-type extracts. Therefore, the effects of rMOT1 on basal transcription were tested next, since rMOT1 was obtained in good yield and in high concentration. As shown in Fig. 3A (lanes 1 to 7) and Fig. 3B (left-hand graph), titration of rMOT1 led to a biphasic response, with activation of HIS4 basal transcription seen at low levels of MOT1 and repression of basal transcription seen at more elevated levels of MOT1. The peak of HIS4 activation was seen when 300 ng of rMOT1 was added to the reaction mixture; this corresponds to an approximately fivefold molar excess of MOT1 over TBP. The amount of MOT1 in an equivalent amount of extract from wild-type cells is approximately 1,300 ng, and amounts of rMOT1 in the 600 to 1,000 ng range generally gave rise to weak repression of basal transcription. Thus, these data (Fig. 3) are consistent with the observation that our standard nuclear extracts generate exceedingly low basal transcription signals with this HIS4 template (reference 2 and data not shown).
The standard in vitro transcription reaction mixtures contain glutamate in a buffer in which MOT1 can bind to TBP-DNA complexes but ATP-dependent TBP-DNA dissociation is inefficient (not shown). To determine if solution conditions more favorable for the TBP-DNA dissociation reaction might affect basal transcription differently, conditions were established for assaying basal transcription in which the TBP-DNA dissociation reaction can be readily detected independently by DNase I footprinting (see Materials and Methods). As shown in Fig. 3A (lanes 8 to 14) and in Fig. 3B (right-hand graph), addition of rMOT1 to in vitro transcription reaction mixtures leads to a result qualitatively similar to that seen in glutamate-containing buffer: basal HIS4 transcription is activated at low levels of rMOT1, and this activation is not seen with high levels of rMOT1. That no large differences in MOT1’s effects on basal transcription were observed under these two sets of conditions implies that the rate-limiting step in determining the magnitude of MOT1’s transcriptional activity involves a different step than its ATP-dependent TBP-DNA dissociation activity. Since TBP can stably interact with many other TAFs (18), and MOT1 associates with TBP in a complex distinct from other TBP-TAF complexes (17) one possibility is that the effects of MOT1 in vitro are limited by the rate at which TBP dissociates from TAFs that prevent interaction between TBP and MOT1 (see below).
In view of MOT1’s TBP-DNA dissociating activity and its previously described activity as a transcriptional repressor (2), it is remarkable that MOT1 can behave as an activator of basal transcription in vitro. Importantly, activation is only seen at levels of MOT1 that are roughly stoichiometric with TBP. Two models can explain these results. Since the promoters are contained on plasmid DNA, these results are consistent with a model in which low levels of MOT1 liberate limiting TBP from nonpromoter sites on the template-containing plasmid for use at promoters. An alternative model is that inactive, kinetically trapped, TBP-containing complexes can form on promoters and that MOT1 functions to clear such complexes to provide for additional opportunities for functional preinitiation complex formation. These results also suggest that the effects of MOT1 mutation on transcription in vivo depend on the allele of MOT1 used, since different mutant alleles which give rise to different residual levels of MOT1 activity would lead to qualitatively and quantitatively different effects on individual promoters.
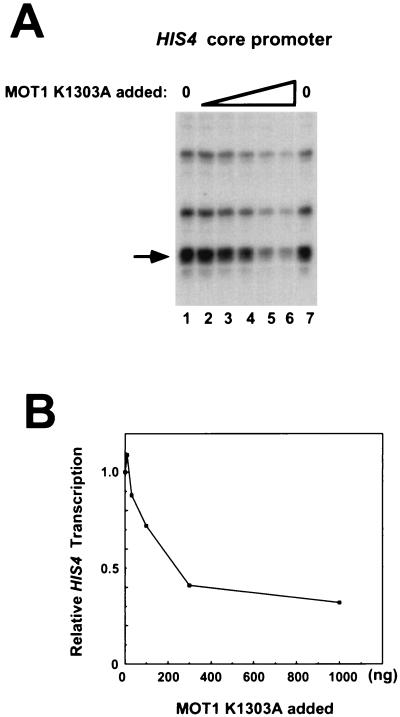
The effects of recombinant ATPase-defective MOT1 K1303A on basal transcription were tested next (Fig. 4). Titration of rMOT1 K1303A into transcription reaction mixtures containing the HIS4 core promoter revealed that in addition to the failure of ATPase-defective MOT1 to support activation, elevated levels of this protein repressed basal transcription (Fig. 4B). Since this protein is strongly dominant negative in vivo (2) and the dominant negativity can be fully suppressed by overexpression of TBP (2, 3), we infer that the ATPase-defective MOT1 protein interferes with transcription by forming an inactive complex with TBP either on or off DNA.
FIG. 4.
ATPase-defective MOT1 represses basal transcription. (A) Basal transcription reactions were performed as described in the legends to Fig. 2 and 3 except that reaction mixtures contained 10 ng (lane 2), 30 ng (lane 3), 100 ng (lane 4), 300 ng (lane 5), or 1,000 ng (lane 6) of MOT1 K1303A. The reaction mixture in lane 1 contained no added MOT1 K1303A. The bands that were quantitated are indicated by the arrows. (B) Quantitation of the data shown in panel A.
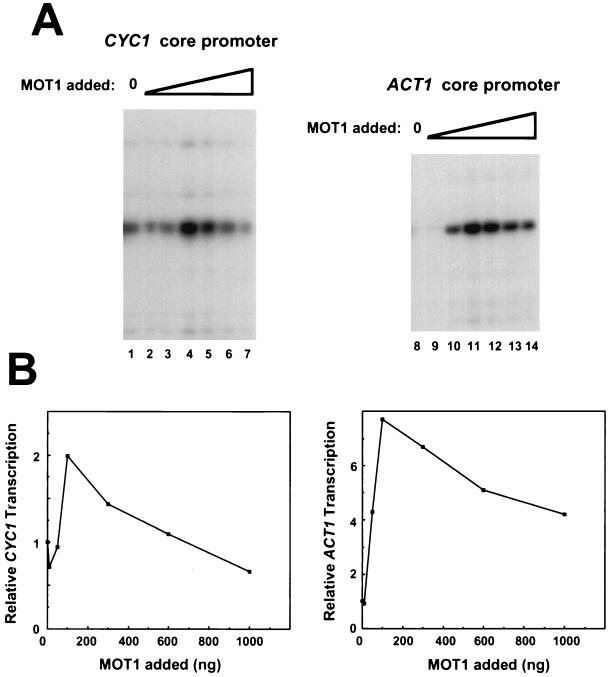
The effects of rMOT1 on basal transcription driven by the CYC1 and ACT1 promoters are shown in Fig. 5. Effects seen with each of these promoters are similar to those seen with HIS4: basal transcription is activated by rMOT1 when added at low levels and is unchanged or repressed when rMOT1 is added at levels comparable to those in wild-type extracts. The maximum amount of activation for all of the promoters varied by about a factor of 2, and the amount of rMOT1 required to achieve maximal activation varied slightly from experiment to experiment. Overall, maximum activation required rMOT1 in the reaction mixture at a one- to fivefold molar excess over TBP, and reconstitution of the extract to MOT1 levels seen in wild-type extracts resulted in weak repression of transcription. Whereas HIS4 expression is repressed by mutation of MOT1 in vivo (8, 15), CYC1-driven expression is activated by a mutation in MOT1 (2). As described above, based on the biphasic response of the transcription apparatus to addition of MOT1 reported here, we anticipate that in vivo, promoters will respond in complex ways to mutations in MOT1 since different alleles of MOT1 and different growth conditions would presumably give rise to different residual levels of MOT1 activity in vivo.
FIG. 5.
Recombinant MOT1 can activate or repress basal transcription driven by either the CYC1 or ACT1 promoters. (A) Basal transcription reactions were performed as described in the legends to Fig. 2 and 3, with 10 ng (lanes 2 and 9), 50 ng (lanes 3 and 10), 100 ng (lanes 4 and 11), 300 ng (lanes 5 and 12), 600 ng (lanes 6 and 13), or 1,000 ng (lanes 7 and 14) of MOT1. (B) Quantitation by phosphoimager analysis of the data shown in panel A.
The similar behavior of MOT1 when assayed with each of the three unrelated templates might be due to an indirect mechanism in which the activation function of MOT1 is due to dissociation of TBP from nonpromoter sites present in the plasmid vector. The idea that MOT1 might function by liberating TBP from other DNA sites for use at promoters is consistent with the observation that mutations in MOT1 have an Spt phenotype (15), suggesting that MOT1 mutations can alter start sites of transcription. At some spurious sites, TBP binding which is unchecked by MOT1 might lead to the formation of preinitiation complexes which are capable of initiating the synthesis of aberrant RNA. In vitro, TBP binding to fortuitous sites can lead to the assembly of transcription complexes capable of initiating RNA synthesis (7). Additionally, the similar behaviors of these promoters in vitro with respect to MOT1 might also reflect the fact that these templates have similar promoter strengths in these assays.
In reactions containing wild-type levels of MOT1, basal transcription is weakly repressed. This is likely due to the direct action of MOT1 on TBP-containing complexes bound to the promoter. Given the clear-cut effects of MOT1 both on TBP-DNA binding and in wild-type versus mot1-1 nuclear extracts, why is the repressive effect of MOT1 so modest in these reactions? As discussed above, the modest effects of MOT1 on repression of basal transcription probably result from ineffective competition between MOT1 and other TAFs which have formed stable complexes with TBP in the extract prior to the addition of MOT1. It is not known if MOT1 can dissociate TFIID-DNA complexes, but the ability of TFIIA to block MOT1 action (2) suggests that other TAFs might interfere with MOT1 action. For example, the inhibitory domain of yeast TAF130/145 (4, 14) interacts with amino acids on the convex surface of TBP (14) which are critical for interaction with both TFIIA (14) and MOT1 (1), suggesting that MOT1 and TAF130/145 interactions with TBP are mutually exclusive. Additionally, transcription experiments suggest that TAF172, a human homologue of MOT1, is targeted to TAF-free TBP in vitro (5).
Roles of MOT1, TBP, and GAL4-VP16 in overcoming repression by nonpromoter DNA.
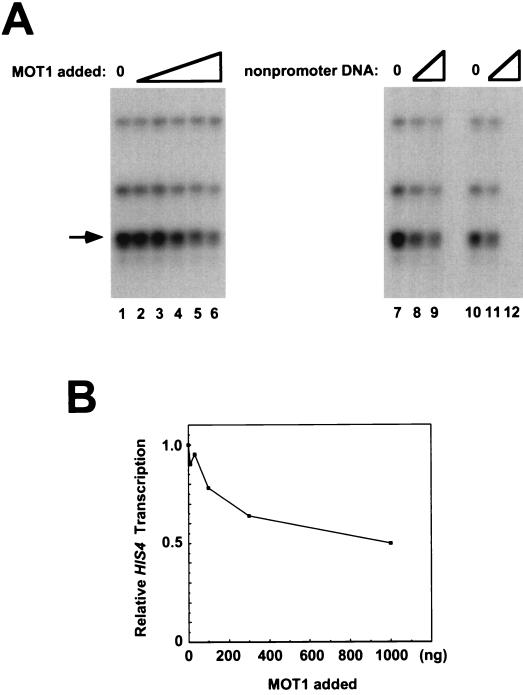
To test the idea that the effect of MOT1 on basal transcription depends on competition for TBP binding between different TBP binding sites, we first tested the effect of exogenously added nonpromoter DNA on basal transcription driven by the HIS4 promoter. In these experiments, increasing amounts of plasmid DNA (lacking a yeast promoter) were added to basal transcription reaction mixtures otherwise carried out as described above. As shown in Fig. 6A, addition of nonpromoter DNA led to a three- to fivefold decrease in basal transcription. This decrease appears to be due to the binding of TBP (or a TBP-containing complex) to nonpromoter sites on the plasmid, because the addition of increasing amounts of TBP rescues HIS4 basal transcription in reaction mixtures containing nonpromoter DNA (Fig. 6B, lanes 9 to 14). The idea that repression of basal transcription is due to binding of TBP to nonpromoter DNA is also supported by the observation that basal transcription can be repressed by addition of a TATA box-containing oligonucleotide but not by addition of an equimolar amount of an oligonucleotide duplex containing a single point mutation in the TATA box which prevents TBP binding (Fig. 6C). Basal transcription driven by the HIS4 core promoter could also be rescued by the addition of yMOT1 (Fig. 6B, lanes 2 to 8), suggesting that under these conditions, MOT1 might activate transcription by facilitating the distribution of TBP between promoter and nonpromoter sites. As an aside, the effects of the transcriptional activator GAL4-VP16 were compared under conditions under which excess nonpromoter DNA was present or absent. The results in Fig. 6D demonstrate that GAL4-VP16 activates HIS4 basal transcription fivefold in the absence of exogenous nonpromoter DNA and fourfold in the presence of nonpromoter DNA. Since the fold activation is similar but the activator does not reconstitute transcription to similar levels under these two conditions, we conclude that the activation seen here does not reflect recruitment of TBP (or TFIID) to the HIS4 promoter. In contrast, GAL4-VP16 activates transcription from the HIS4 basal promoter to similar levels in extracts from wild-type or mot1-1 cells (2), consistent with the idea that this activator functions in at least two steps in the process of preinitiation complex formation.
Differential effect of MOT1 on HIS4 basal transcription in reactions with different amounts of nonpromoter DNA.
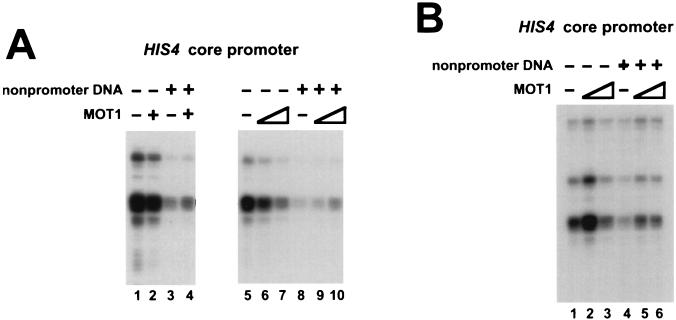
The observation that MOT1 can rescue basal transcription under conditions of excess nonpromoter DNA suggests that an equivalent amount of MOT1 has different effects on transcription, depending on the amount of nonpromoter DNA present in the reaction mixture. To test this, MOT1 was added to transcription reaction mixtures which contained or did not contain excess nonpromoter DNA. As shown in Fig. 7, an amount of MOT1 that leads to weak repression of transcription under standard conditions (Fig. 7A lanes 1, 2, and 5 to 7) caused weak activation when additional nonpromoter DNA was present in the reaction mixture (Fig. 7A lanes 3, 4, and 8 to 10). Another example of these differential effects is shown in Fig. 7B in which low levels of MOT1 activate basal transcription whether or not excess nonpromoter DNA is present (lanes 1, 2, 4, and 5), whereas increasing the concentration of MOT1 leads to either weak repression of transcription (lanes 1 and 3) or weak activation of transcription (lanes 4 and 6), depending on the amount of nonpromoter DNA which is present in the reaction mixture.
FIG. 7.
Comparison of the effects of MOT1 on HIS4 basal transcription in the presence and absence of nonpromoter DNA. Transcription reaction mixtures contained 3.4 μg of pKS II+ nonpromoter DNA as indicated and/or MOT1. (A) Transcription reactions were performed with the HIS4 core promoter with (+) or without (−) MOT1 purified from yeast. The reaction mixtures in lanes 2 and 3 contained 3 U of MOT1, those in lanes 6 and 9 contained 2 U of MOT1, and those in lanes 7 and 10 contained 8 U of MOT1. (B) Reactions were performed as described for panel A but with 250 ng (lanes 2 and 5) or 800 ng (lanes 3 and 6) of recombinant MOT1 from baculovirus.
Each of the three core promoters tested responds similarly to exogenously added MOT1. Thus, while the in vitro system described here defines a weak activation function for MOT1, the in vitro data do not recapitulate the differential response of promoters to MOT1 seen in vivo. One possibility is that larger fragments of promoter DNA respond differently to MOT1 than the core promoters tested as described above. To test this, transcription driven by a 542-bp fragment of HIS4 upstream DNA was compared to transcription driven by the 149-bp HIS4 core promoter. As shown in Fig. 8A, lanes 1 to 6, the larger HIS4 promoter fragment drives transcription which is repressed by high levels of MOT1, but weak activation like that seen with the HIS4 core promoter is not observed (Fig. 8C). This suggests that factors bound to the larger HIS4 promoter facilitate recruitment of TBP and associated factors such that this promoter can now more effectively compete with limiting TBP in the reaction mixture. If this is the case, repression of transcription might still be observed when high levels of MOT1 out-compete upstream activation factors for interaction with TBP and/or TAFs. In support of this suggestion is the observation that transcription driven by the HIS4 core promoter can be repressed by the addition of exogenous nonpromoter DNA, whereas transcription driven by the larger fragment of HIS4 upstream DNA is resistant to competition for TBP binding by nonpromoter DNA (Fig. 8A, lanes 7 to 12).
FIG. 8.
Transcription driven by a larger (542-bp) fragment of the HIS4 promoter is repressed but not activated by MOT1. (A) Transcription reactions were performed with the plasmid containing the larger 542-bp fragment of the HIS4 promoter (lanes 1 to 9) or the HIS4 core promoter (lanes 10 to 12). The reaction mixtures in lanes 1 and 7 to 12 contained no added MOT1, whereas rMOT1 was added to the reaction mixtures in lane 2 (10 ng), lane 3 (30 ng), lane 4 (100 ng), lane 5 (300 ng), and lane 6 (1,000 ng). The reaction mixtures in lanes 8 and 11 contained 1.2 μg of pKS II+ nonpromoter DNA, and the reaction mixtures in lanes 9 and 12 contained 2.6 μg pKS II+. The bands that were quantitated are indicated by the arrow. (B) Quantitation of the data shown in panel A, lanes 1 to 6.
These results support a model in which one function of MOT1 is to liberate limiting TBP from nonpromoter sites for utilization at promoters. It is important to point out, however, that there are promoters which might be targeted directly for repression by MOT1 in vivo (9). It seems reasonable, therefore, that the function of MOT1 in vivo probably involves dissociating TBP from a complicated array of sites, including fortuitous high-affinity TBP binding sites in nonpromoter DNA as well as certain promoters which are susceptible to MOT1-mediated repression. While the present work defines an activation function for MOT1, this in vitro system cannot explain all of the promoter-specific effects of MOT1 observed in vivo. For instance, the in vitro system does not explain the apparent requirement of HIS4 for MOT1 in vivo (8, 15). This might be explained in part by the ability of MOT1 to regulate the expression of transcriptional regulators. It is also obvious that our in vitro system cannot mimic the complex competition between promoters and other sites for TBP binding which occurs in vivo and which we suggest is crucial in determining the consequences of MOT1 function. Unraveling the factors that determine the effects of MOT1 on specific promoters is likely to yield insight into the complexities of how preinitiation complexes are formed in vivo at specific promoters.
The similar behaviors of three different core promoters could also mean that MOT1 does not activate transcription in vitro by disassembling inactive TBP-containing complexes formed on promoters. On the other hand, there may be templates which respond in this manner to MOT1 which have not so far been tested. One possibility is that very weak promoters are activated by MOT1 by such a direct mechanism. To test this idea, the MOT1 response of basal promoters with mutated TATA boxes was examined. Unfortunately, these promoters did not drive detectable RNA synthesis in either the presence or absence of MOT1 (3a). It would be interesting to compare the in vitro response to MOT1 of a naturally occurring TATA-less promoter to the response of one of the templates described here, but a system for assaying TATA-less transcription in vitro by using wild-type or MOT1-depleted extracts is not currently available. A large difference in the response of two such templates in vitro would provide a biochemical strategy for understanding the molecular basis of the complex effects of MOT1 on transcription, and this will be the subject of future work. Given the evolutionary conservation of MOT1 (5, 10, 23), the analysis of yeast MOT1 function is likely to provide additional general insights into the global regulation of eukaryotic transcription.
ACKNOWLEDGMENTS
We thank Fred Winston, John Chicca, Frank Pugh, Joe Reese, members of the Auble and Weil laboratories, and the University of Virginia Sixes and Sevens Research Discussion Group for insightful discussions.
This work was supported by grants from the National Institutes of Health (GM55763 to D.T.A. and GM52461 to P.A.W.).
REFERENCES
- 1.Auble D T, Hahn S. An ATP-dependent inhibitor of TBP binding to DNA. Genes Dev. 1993;7:844–856. doi: 10.1101/gad.7.5.844. [DOI] [PubMed] [Google Scholar]
- 2.Auble D T, Hansen K E, Mueller C G F, Lane W S, Thorner J, Hahn S. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 1994;8:1920–1934. doi: 10.1101/gad.8.16.1920. [DOI] [PubMed] [Google Scholar]
- 3.Auble D T, Wang D, Post K W, Hahn S. Molecular analysis of the SNF2/SWI2 protein family member MOT1, an ATP-driven enzyme that dissociates TATA-binding protein from DNA. Mol Cell Biol. 1997;17:4842–4851. doi: 10.1128/mcb.17.8.4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Auble, D. T. Unpublished observations.
- 4.Bai Y, Perez G M, Beechem J M, Weil P A. Structure-function analysis of TAF130: identification and characterization of a high-affinity TATA-binding protein interaction domain in the N terminus of yeast TAFII130. Mol Cell Biol. 1997;17:3081–3093. doi: 10.1128/mcb.17.6.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Campbell, A. M., and P. A. Weil. Unpublished observations.
- 5.Chicca J J I, Auble D T, Pugh B F. Cloning and biochemical characterization of TAF-172, a human homolog of yeast MOT1. Mol Cell Biol. 1998;18:1701–1710. doi: 10.1128/mcb.18.3.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colbert T, Hahn S. A yeast TFIIB-related factor involved in RNA polymerase III transcription. Genes Dev. 1992;6:1940–1949. doi: 10.1101/gad.6.10.1940. [DOI] [PubMed] [Google Scholar]
- 7.Coleman R A, Pugh B F. Evidence for functional binding and stable sliding of the TATA binding protein on nonspecific DNA. J Biol Chem. 1995;270:13850–13859. doi: 10.1074/jbc.270.23.13850. [DOI] [PubMed] [Google Scholar]
- 8.Collart M A. The NOT, SPT3, and MOT1 genes functionally interact to regulate transcription at core promoters. Mol Cell Biol. 1996;16:6668–6676. doi: 10.1128/mcb.16.12.6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis J L, Kunisawa R, Thorner J. A presumptive helicase (MOT1 gene product) affects gene expression and is required for viability in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:1879–1892. doi: 10.1128/mcb.12.4.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldman-Levi R, Miller C, Bogoch J, Zak N B. Expanding the Mot1 subfamily: 89B helicase encodes a new Drosophila melanogaster SNF2-related protein which binds to multiple sites on polytene chromosomes. Nucleic Acids Res. 1996;24:3121–3129. doi: 10.1093/nar/24.16.3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hahn S, Buratowski S, Sharp P A, Guarente L. Yeast TATA-binding protein TFIID binds to TATA elements with both consensus and nonconsensus DNA sequences. Proc Natl Acad Sci USA. 1989;86:5718–5722. doi: 10.1073/pnas.86.15.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang Y W, Stillman D J. Epigenetic effects on yeast transcription caused by mutations in an actin-related protein present in the nucleus. Genes Dev. 1996;10:604–619. doi: 10.1101/gad.10.5.604. [DOI] [PubMed] [Google Scholar]
- 13.Karnitz L, Morrison M, Young E T. Identification and characterization of three genes that affect expression of ADH2 in Saccharomyces cerevisiae. Genetics. 1992;132:351–359. doi: 10.1093/genetics/132.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kokubo T, Swanson M J, Nishikawa J-I, Hinnebusch A G, Nakatani Y. The yeast TAF145 inhibitory domain and TFIIA competitively bind to TATA-binding protein. Mol Cell Biol. 1998;18:1003–1012. doi: 10.1128/mcb.18.2.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Madison J M, Winston F. Evidence that Spt3 functionally interacts with Mot1, TFIIA, and TBP to confer promoter-specific transcriptional control in Saccharomyces cerevisiae. Mol Cell Biol. 1996;17:287–295. doi: 10.1128/mcb.17.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piatti S, Tazzi R, Pizzagalli A, Plevani P, Lucchini G. Control of DNA synthesis genes in budding yeast: involvement of the transcriptional modulator MOT1 in the expression of the DNA polymerase alpha gene. Chromosoma. 1992;102:S107–S113. doi: 10.1007/BF02451793. [DOI] [PubMed] [Google Scholar]
- 17.Poon D, Campbell A M, Bai Y, Weil P A. Yeast Taf170 is encoded by MOT1 and exists in a TBP-TAF complex distinct from TFIID. J Biol Chem. 1994;269:23135–23140. [PubMed] [Google Scholar]
- 18.Poon D, Weil P A. Immunopurification of yeast TATA-binding protein and associated factors. J Biol Chem. 1993;268:15325–15328. [PubMed] [Google Scholar]
- 19.Prelich G. Saccharomyces cerevisiae BUR6 encodes a DRAP1/NC2α homolog that has both positive and negative roles in transcription in vivo. Mol Cell Biol. 1997;17:2057–2065. doi: 10.1128/mcb.17.4.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ranish J A, Hahn S. The yeast general transcription factor TFIIA is composed of two polypeptide subunits. J Biol Chem. 1991;266:19320–19327. [PubMed] [Google Scholar]
- 21.Reddy P, Hahn S. Dominant negative mutations in yeast TFIID define a bipartite DNA-binding region. Cell. 1991;65:349–357. doi: 10.1016/0092-8674(91)90168-x. [DOI] [PubMed] [Google Scholar]
- 22.Schroeder S C, Wang C K, Weil P A. Identification of the cis-acting DNA sequence elements regulating the transcription of the Saccharomyces cerevisiae gene encoding TBP, the TATA box binding protein. J Biol Chem. 1994;269:28335–28346. [PubMed] [Google Scholar]
- 23.van der Knaap J A, Borst J W, van der Vliet P C, Gentz R, Timmers H T M. Cloning of the cDNA for the TATA-binding protein-associated factorII170 subunit of transcription factor B-TFIID reveals homology to global transcription regulators in yeast and drosophila. Proc Natl Acad Sci USA. 1997;94:11827–11832. doi: 10.1073/pnas.94.22.11827. [DOI] [PMC free article] [PubMed] [Google Scholar]