Abstract
PURPOSE
Although NRG1 fusions are oncogenic drivers across multiple tumor types including lung cancers, these are difficult to study because of their rarity. The global eNRGy1 registry was thus established to characterize NRG1 fusion–positive lung cancers in the largest and most diverse series to date.
METHODS
From June 2018 to February 2020, a consortium of 22 centers from nine countries in Europe, Asia, and the United States contributed data from patients with pathologically confirmed NRG1 fusion–positive lung cancers. Profiling included DNA-based and/or RNA-based next-generation sequencing and fluorescence in situ hybridization. Anonymized clinical, pathologic, molecular, and response (RECIST v1.1) data were centrally curated and analyzed.
RESULTS
Although the typified never smoking (57%), mucinous adenocarcinoma (57%), and nonmetastatic (71%) phenotype predominated in 110 patients with NRG1 fusion–positive lung cancer, further diversity, including in smoking history (43%) and histology (43% nonmucinous and 6% nonadenocarcinoma), was elucidated. RNA-based testing identified most fusions (74%). Molecularly, six (of 18) novel 5′ partners, 20 unique epidermal growth factor domain–inclusive chimeric events, and heterogeneous 5′/3′ breakpoints were found. Platinum-doublet and taxane-based (post–platinum-doublet) chemotherapy achieved low objective response rates (ORRs 13% and 14%, respectively) and modest progression-free survival medians (PFS 5.8 and 4.0 months, respectively). Consistent with a low programmed death ligand-1 expressing (28%) and low tumor mutational burden (median: 0.9 mutations/megabase) immunophenotype, the activity of chemoimmunotherapy and single-agent immunotherapy was poor (ORR 0%/PFS 3.3 months and ORR 20%/PFS 3.6 months, respectively). Afatinib achieved an ORR of 25%, not contingent on fusion type, and a 2.8-month median PFS.
CONCLUSION
NRG1 fusion–positive lung cancers were molecularly, pathologically, and clinically more heterogeneous than previously recognized. The activity of cytotoxic, immune, and targeted therapies was disappointing. Further research examining NRG1-rearranged tumor biology is needed to develop new therapeutic strategies.
INTRODUCTION
Gene fusions are enriched in non–small-cell lung cancers (NSCLCs). Many of these fusions encode chimeric oncoproteins that drive cancer growth.1-4 Activating fusions involving ALK,5-7 ROS1,8-11 RET,12-15 NTRK1, NTRK2, or NTRK32,16,17 result in constitutive kinase domain activation that drives downstream pathway signaling, promoting lung cancer cell proliferation and survival. Most importantly, the identification of these fusions matches patients to highly active targeted therapies that are approved by one or more regulatory agencies around the world.2-4
CONTEXT
Key Objective
The goals of the eNRGy1 global multicenter registry are to characterize the features of NRG1 fusion–positive lung cancers and elucidate the clinical activity of systemic therapy in a centrally curated real-world database of patients with these rare cancers.
Knowledge Generated
NRG1 fusion–positive lung cancers are pathologically, clinically, and molecularly more diverse than previously recognized. Many fusions are first detected by RNA-based sequencing. A variety of unique chimeric events are identified. Most tumors are characterized by no or low programmed death ligand-1 expression and low tumor mutational burden. The activity of a variety of cytotoxic, immunotherapy, and targeted therapy regimens is modest at best.
Relevance
Comprehensive sequencing to identify NRG1 fusions should capture molecularly heterogeneous events and not be biased toward particular clinical or pathologic features. To develop novel therapeutic strategies, stakeholders should prioritize research into the underexplored biology of NRG1 fusion–positive tumors and the development of rationally designed drugs.
NRG1 fusions are a relatively recent addition to this list of fusion oncogenes.18-21 Structurally, these alterations are distinct from the aforementioned fusions. The transmembrane chimeric oncoprotein contains the epidermal growth factor or epidermal growth factor–like binding domain of NRG1, a known ligand of ERBB3. Binding of the oncoprotein to ERBB3 results in the formation of heterodimers between ERBB3 and other ERBB family members, thereby activating oncogenic signaling and cancer growth. Of these heterodimers, ERBB3-ERBB2 is the most transforming. Therapeutic targeting of these fusions has thus centered on the inhibition of ERBB3 and/or ERBB2.22-29 For example, individual case reports or small series have noted clinical benefit with the pan-ERBB1/2/4 tyrosine kinase inhibitor afatinib in selected patients.22,23,25,26,30
Although NRG1 fusions were first discovered in lung cancers in 2014,19 the clinical, pathologic, and molecular features of these cancers are yet to be comprehensively characterized in a large series.23,31 In addition, the activity of many systemic therapies in this molecularly enriched cohort of patients has not been well-described. To address this unmet need, we formed the eNRGy1 global multicenter consortium of thoracic oncology investigators to contribute data on patients with NRG1 fusion–positive lung cancers to a central registry.
METHODS
eNRGy1 Global Multicenter Registry
Investigators taking part in the consortium were initially identified on the basis of their contributions to existing registries for other molecularly defined lung cancer subtypes, including those with RET rearrangements,14 BRAF mutations,32 HER2 mutations,33,34 and ROS1 rearrangements.35 All investigators were certified in good clinical practice and obtained ethics review board approval through their individual institutions.
Eligible Patients
Patients were considered eligible for registry inclusion if they had a pathologically confirmed diagnosis of lung cancer with an NRG1 fusion as determined by testing in an accredited laboratory. Acceptable testing methods for NRG1 fusion detection included fluorescence in situ hybridization using the Agilent, Clinisciences, or ZytoVision assays (fusion-positive tumors were defined as those with split signals or isolated red [3′] signals in ≥ 15% of enumerated tumor cells)36; DNA-based and/or RNA-based next-generation sequencing (NGS) using MSK-IMPACT, FoundationOne, Caris NGS, ION Ampliseq, Oncomine, StrataNGS, or Archer; reverse transcription-polymerase chain reaction (PCR); or through detection of imbalanced gene expression via nCounter gene fusion panels (NanoString Technologies, Seattle, WA).
Clinicopathologic Data
Investigators were asked to record data on patient demographics (including sex, age at diagnosis, smoking habits, and ethnicity) and tumor pathologic features (including stage, histology as determined by a local pathologist, and NRG1 fusion partner). Treatment history, including the date of diagnosis, treatments received, dates of progression, and survival status were documented. For survival analysis, patients were followed through February 2020. Best overall response to treatment was determined according to RECIST version 1.1, which was assessed locally at each institution.
Immunophenotype
Programmed death ligand-1 (PD-L1) expression in tumor cells was determined by immunohistochemistry.37 Because of the variability in measures of tumor mutational burden (TMB) using different sequencing assays,38-40 TMB was only collected for those patients whose tissue underwent sequencing using a single NGS assay, MSK-IMPACT. MSK-IMPACT is a targeted, hybrid capture-based NGS DNA assay that covers up to 468 cancer-related genes.41 This assay has been extensively validated for the assessment of TMB.40,42,43 The TMB of NRG1 fusion–positive tumors was compared with that recorded for all other lung cancers that underwent sequencing using MSK-IMPACT.
Data Collection and Analysis
Investigators from the global consortium submitted anonymized data to a database maintained at one institution between June 2018 and February 2020. Categorical variables were compared using Fisher's exact tests. Continuous variables were compared using Mann-Whitney testing. Progression-free survival (PFS) was assessed from therapy initiation until radiologic progression (by RECIST v1.1) or death. Overall survival (OS) was assessed from the date of initial diagnosis through death. Survival analyses were carried out according to the Kaplan-Meier method, with surviving patients censored at the date of last follow-up.
Statistical analyses were performed using GraphPad Prism version 8.4.2 (San Diego) or R version 3.4.0 (R Project for Statistical Computing, Vienna, Austria). STATA (version 16, College Station, TX) was used to calculate confidence intervals for Kaplan-Meier curves. The results were considered statistically significant if they fell below the P = .05 threshold.
RESULTS
Clinicopathologic Features
Data from 110 patients with NRG1 fusion–positive lung cancers were contributed by a total of 22 different centers from nine countries in Europe, Asia, and the United States. Demographics are summarized in Table 1. The median age was 64 years. The majority of patients were either Asian (52%) or White (46%). Most patients (57%) were never smokers. In patients with a prior or current history of smoking (n = 36), the median pack-year history was 37.
TABLE 1.
Clinicopathologic Characteristics

At the time of diagnosis, most (71%, n = 58/82) patients had nonmetastatic (stages I-III) disease. In patients with metastatic disease diagnosed at any time during their disease course (n = 44), the most common sites of metastasis were the lung (71%, n = 31/44), bone (34%, n = 15/44), and lymph nodes (23%, n = 10/44). Intrathoracic metastases (involving the mediastinum [2%, n = 1/44], the pleura [16%, n = 7/44], the contralateral lung [71%, n = 31/44], and lymph nodes [23%, n = 10/44]) were frequent (77%, n = 34/44). Extrathoracic metastases were found in 43% (n = 19/44) of patients. The frequency of metastases and their sites are shown in Figure 1A and the Data Supplement (online only). Adenocarcinoma was the most common histology, found in 94% (n = 103/110) of patients. In adenocarcinomas, invasive mucinous adenocarcinoma (IMA) was the most frequent (57%) subtype as shown in Figure 1B.
FIG 1.
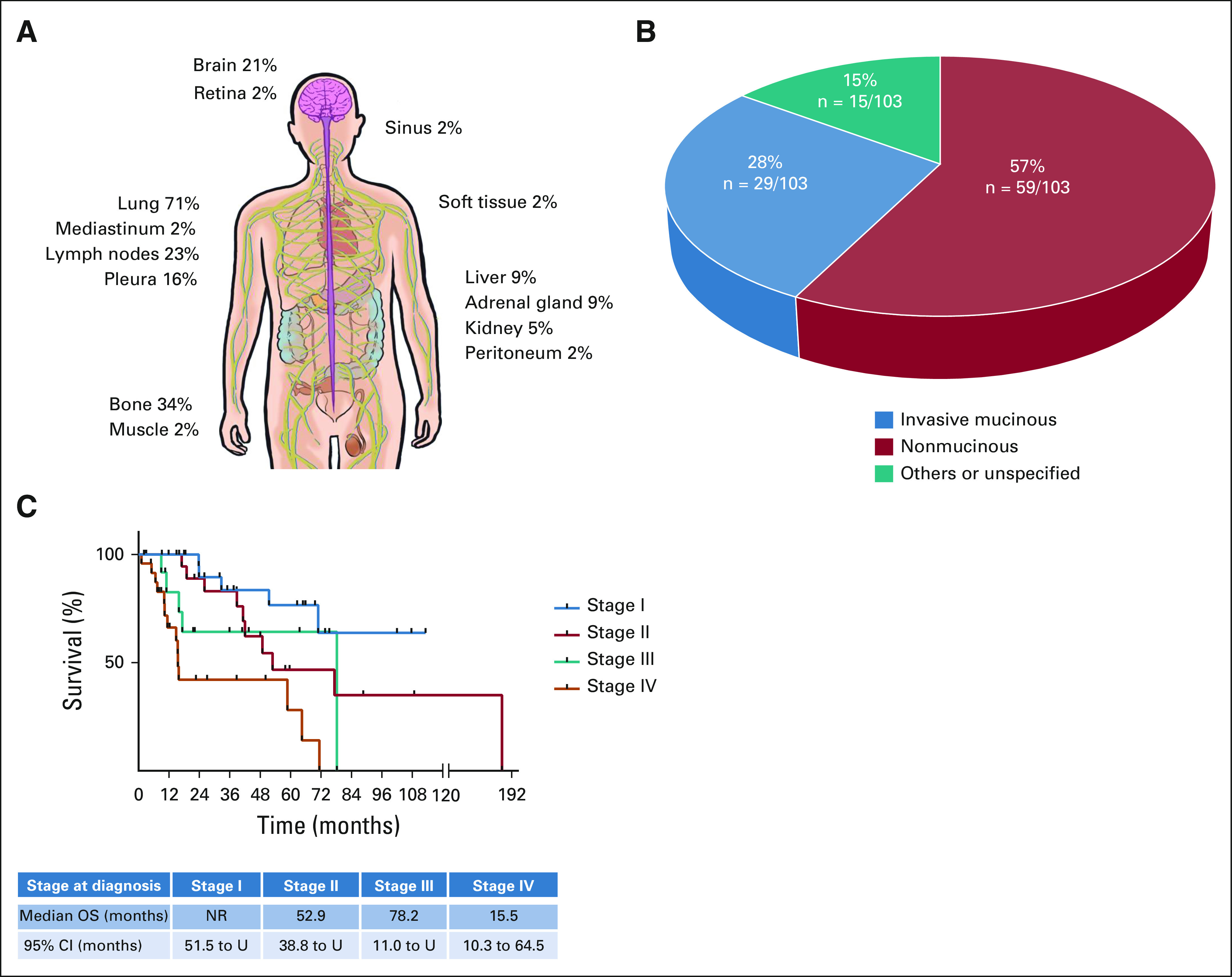
Clinicopathologic features. (A) The frequency of metastasis to selected anatomic sites is shown for patients with NRG1 fusion–positive lung cancers. (B) The histologic subtypes of 103 NRG1 fusion–positive adenocarcinomas are shown. These are divided into invasive mucinous adenocarcinomas, noninvasive mucinous adenocarcinomas, and other subtypes. (C) Kaplan-Meier curves of OS are shown for stage I (blue), stage II (red), stage III (green), and stage IV (orange) disease at diagnosis. The median duration of follow-up was 32 months (range, 1-179 months). NR, not reached; OS, overall survival; U, undefined.
Kaplan-Meier plots of OS are shown in Figure 1C and the Data Supplement by stage at diagnosis. The median OS by stage was not reached (95% CI, 51.5 to undefined) for stage I (n = 26) and was 52.9 months (95% CI, 38.8 to undefined) for stage II (n = 19), 78.2 months (95% CI, 11.0 to undefined) for stage III (n = 13), and 15.5 months (95% CI, 10.3 to 64.5) for stage IV (n = 24).
Fusion Diagnosis
RNA-based testing was the most common method that identified NRG1 fusions: 74% were detected by RNA-based assays and 26% were detected by DNA-based assays (Fig 2A). Of the RNA-based assays, NRG1 fusions were most commonly identified by amplicon-based RNA sequencing using anchored multiplex PCR (62%, n = 50/81) followed by reverse transcription-PCR (27%, n = 22/81) and expression analysis using nCounter (11%, n = 9/81).
FIG 2.
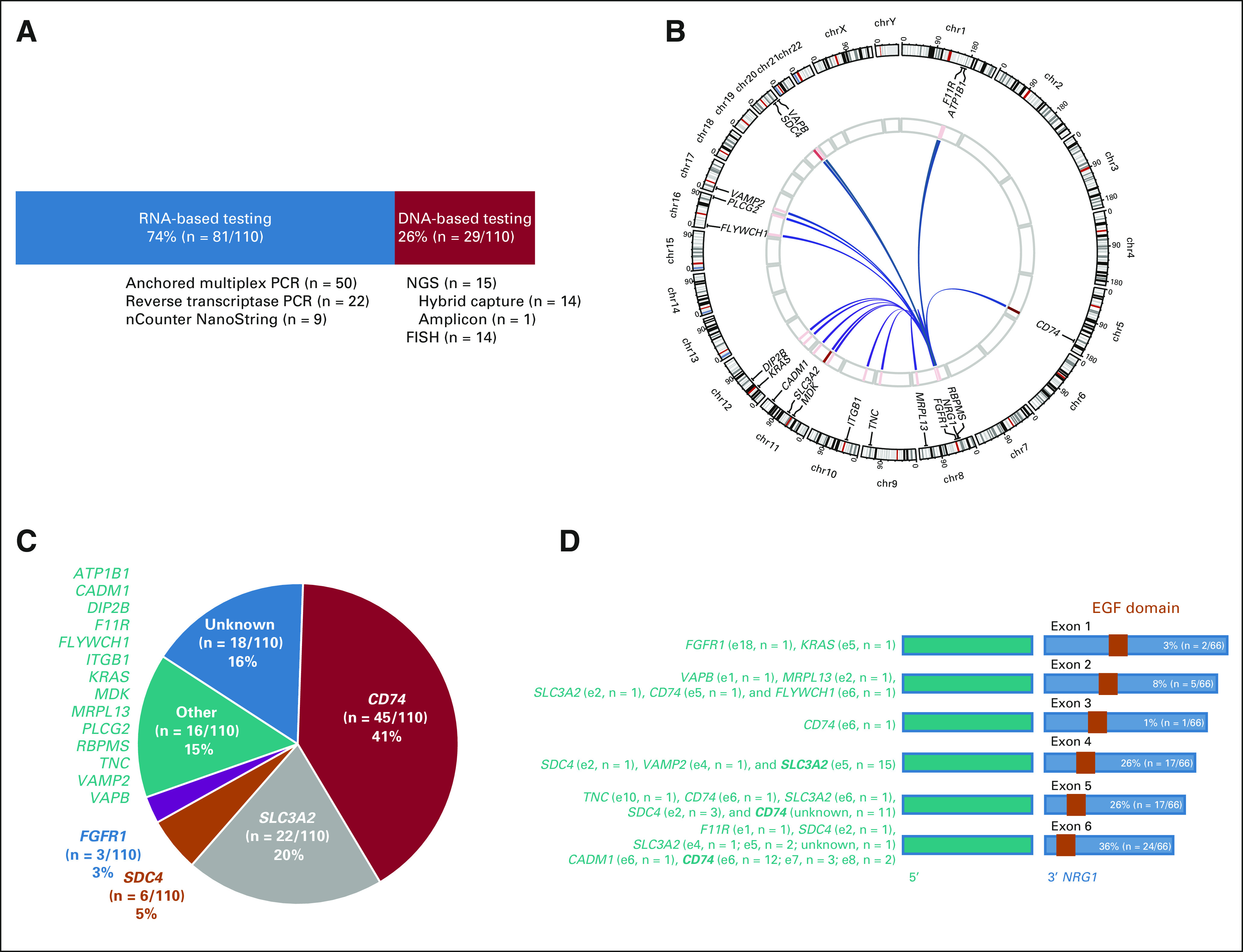
Molecular features. (A) The primary assay that identified the NRG1 fusion in cancers from 110 patients in this registry is divided into RNA-based (blue) and DNA-based (red) assays. Below each corresponding bar, a list and number of the individual assays are shown. (B) A Circos plot of the various NRG1 fusions detected and their corresponding upstream partners is shown. The intensity of the red bars in the inner circle represents the frequency of each fusion event, with darker bars representing more common fusions and lighter bars representing less common fusions. (C) The frequency of upstream partners is shown. The most common 5′ partners—CD74, SLC3A2, SDC4, and FGFR1—are shown individually, whereas less common partners are aggregated into other partners (green). (D) When known, the exon that precedes the 5′ breakpoint is shown in green along with the frequency of each event. Exons and exon numbers are abbreviated as eX (e for exon and X for exon number), and events that occur in more than 10 fusions in aggregate are in boldface. The structure of the corresponding 3′ NRG1 gene is shown, with the first exon shown after the breakpoint noted above each blue bar. EGF domains are depicted as orange boxes. EGF, epidermal growth factor; FISH, fluorescence in situ hybridization; NGS, next-generation sequencing; PCR, polymerase chain reaction.
Using DNA-based assays, NRG1 fusions were almost equally detected by NGS (52%, n = 15/29) and fluorescence in situ hybridization (48%, n = 14/29). When detected by NGS, the majority of NRG1 fusions were detected using hybrid capture-based testing (93%, n = 14/15) compared with amplicon-based testing (7%, n = 1/15).
Molecular Features
A plot of the various NRG1 fusions identified is shown in Figure 2B and summarized in the Data Supplement. Upstream gene partners were identified in 92 fusions (84%), and breakpoints in 67 fusions (61%). Eighteen unique upstream gene partners were identified, and 13 with known exonic breakpoints are depicted in Figure 2C. The most common upstream partners were CD74 (41%) and SLC3A2 (20%). Less common partners were SDC4, FGFR1, ATP1B1, CADM1, DIP2B, F11R, FLYWCH1, ITGB1, KRAS, MDK, MRPL13, PLCG2, RBPMS, TNC, VAMP2, and VAPB.
The various breakpoints as reported by local molecular testing assays are shown in Figure 2D for 20 unique chimeric events. For CD74, the breakpoint occurred most commonly after exon 6, followed by exon 8 and exon 7. For SLC3A2, the breakpoint occurred most commonly after exon 5. For NRG1, the breakpoint most commonly involved exon 6, followed by exons 4 and 5, followed by exon 2. All NRG1 fusions included the EGF domain that binds ERBB3.
NRG1 fusions were mutually exclusive with other known oncogenic drivers in the majority of patients (94%, n = 103/110). In the remaining seven patients (Data Supplement), a concurrent driver was identified. Four had hotspot KRAS mutations (KRAS G12C, n = 1; KRAS G12V, n = 1; KRAS G12D, n = 2), all of which are drivers known to occur in IMAs. Three had either an EGFR mutation (EGFR L858R, n = 2) or an ALK fusion (EML4-ALK variant 3, n = 1). In three patients (Data Supplement), the concurrent driver was clearly present de novo. Two patients with surgically resected stage IB/IIB NRG1 fusion–positive IMAs had a concurrent KRAS G12D substitution found at the time of surgery (with no preceding neoadjuvant therapy). One patient had NRG1 and ALK fusions that were both found in the same sample acquired at the diagnosis of metastatic disease before any systemic therapy. This patient responded to crizotinib for 13 months, followed by ceritinib for 18 months.
Immunophenotype
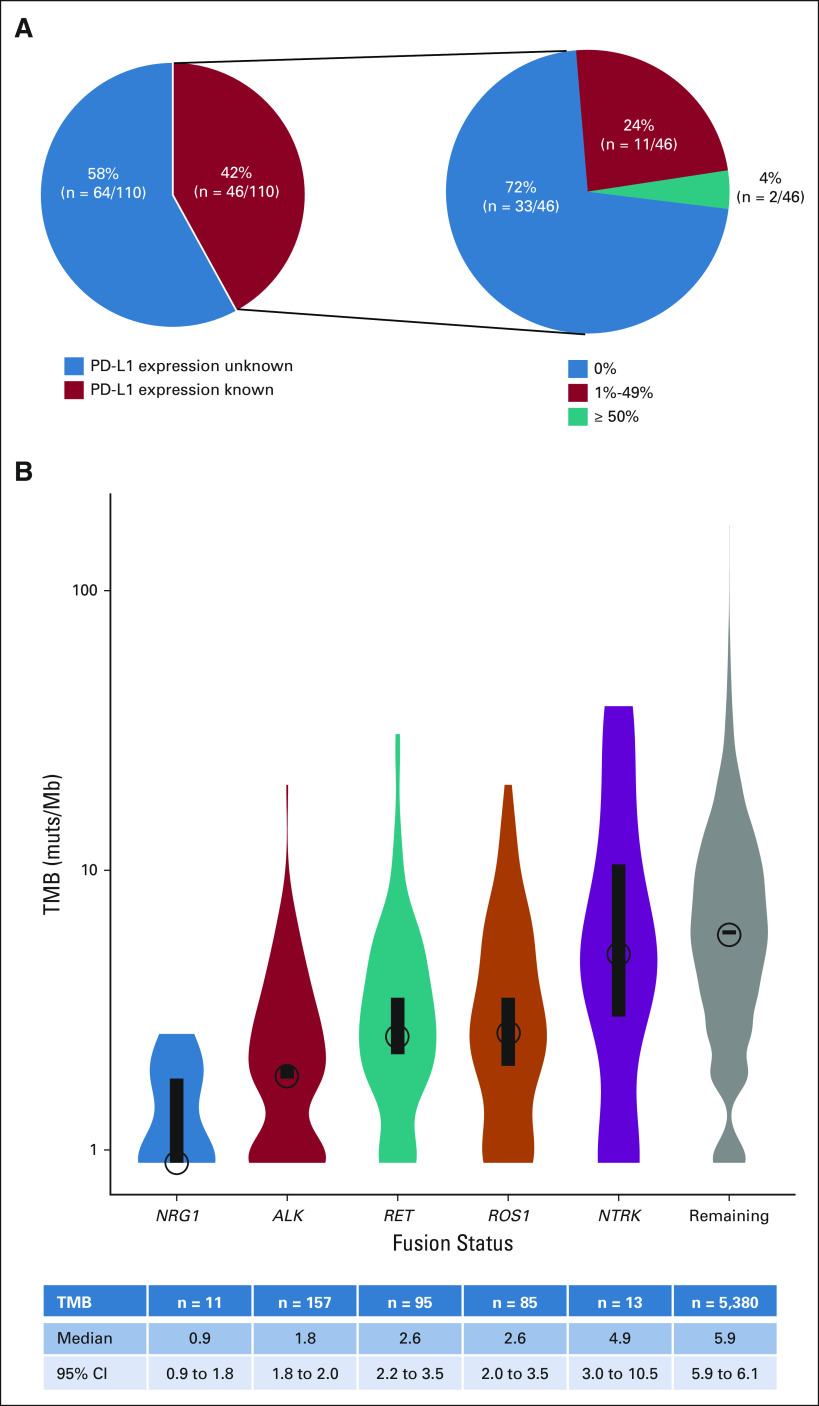
Tumor PD-L1 status was known for 46 of the 110 patients (42%) and is shown in Figure 3A. The antibodies used for PD-L1 testing were 22C3 (n = 26), E1L3N (n = 14), and QR1 (n = 4), with testing on two tumors carried out using an unspecified assay. High PD-L1 expression (50% or greater) was rare (4%, n = 2/46). The majority of tumors had either no expression of PD-L1 in 72% (n = 33/46) of tumors or PD-L1 expression of 1%-49% in 24% (n = 11/46) of tumors.
FIG 3.
Immunophenotype. (A) Of the 110 patients with NRG1 fusion–positive lung cancers, PD-L1 status was known in 46 patients as shown in the pie chart on the left. Of these 46 patients, PD-L1 expression is divided into 0%, 1%-49%, and ≥ 50%, the frequencies of which are shown in the pie chart on the right. The size of the pie graphs relative to each other is not scaled to the total size of the corresponding populations. (B) Violin plots of TMB in mutations per megabase are shown for patients with NRG1 fusion–positive lung cancers compared with those that harbor ALK, ROS1, RET, or NTRK1/2/3 fusions and those whose lung cancers do not harbor these alterations (gray). The circles and black bars indicate the median and 95% CIs, respectively. PD-L1, programmed death ligand-1; TMB, tumor mutational burden.
NRG1 fusions were also characterized by low TMB as shown in Figure 3B. As measured by MSK-IMPACT, the median TMB of NRG1 fusion–positive lung cancers was 0.9 mutations/megabase (range, 0-2.6; n = 11). This was lower than that in patients with ALK (1.8 mutations/megabase, P = .03; n = 157), ROS1 (2.6 mutations/megabase, P = .0008; n = 85), RET (2.6 mutations/megabase, P = .0006; n = 95), and NTRK1/2/3 (4.9 mutations/megabase, P = .003; n = 13) fusion–positive lung cancers. Similarly, the median TMB of NRG1 fusion–positive was lower (P < .0001) than that of 5,380 lung cancers (5.9 mutations/megabase) that did not harbor fusions involving ALK, ROS1, RET, or NTRK.
Chemotherapy and Immunotherapy Activity
The activity of systemic therapy was assessed in patients either diagnosed with or who developed metastatic disease during the course of their disease (Data Supplement). Outcomes are summarized in Table 2. In evaluable patients who received platinum-doublet–based chemotherapy, many of whom received pemetrexed, only 13% (n = 2/15) had a response; the disease control rate was 60% (n = 9/15). The median PFS was 5.8 months (95% CI, 2.2 to 9.8; range, 0.7-12.1 months; Fig 4A and Data Supplement). In patients who received taxane-based chemotherapy in the post–platinum-doublet setting, one response (14%, n = 1/7) was observed and the most common outcome was progressive disease (71%, n = 5/7). The median PFS was 4.0 months (95% CI, 0.8 to 5.3; range, 0.8-5.5 months; Fig 4B).
TABLE 2.
Activity of Systemic Therapy
FIG 4.
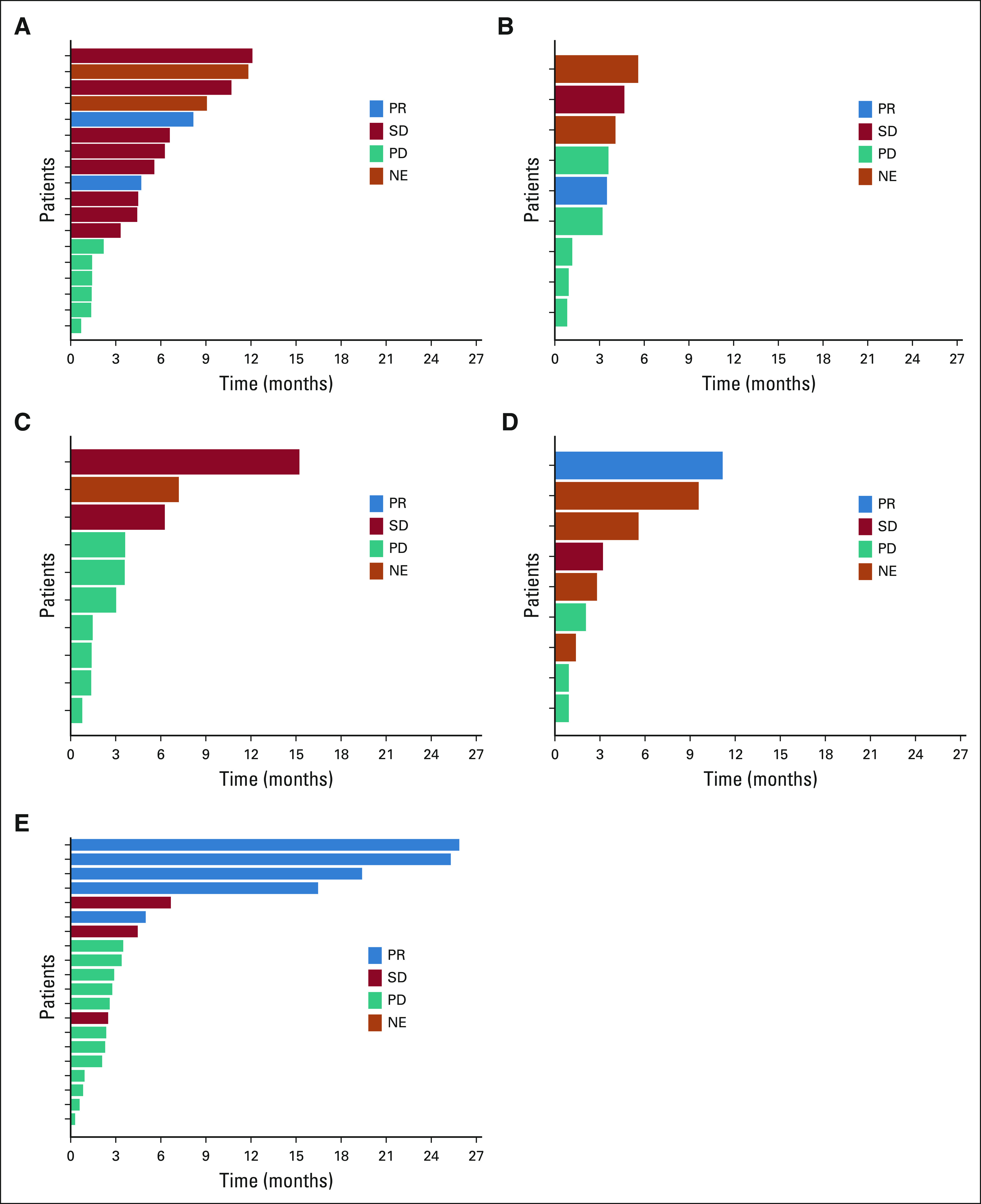
Systemic therapy activity. Swimmer plots of the duration of therapy are shown. Best response to therapy is indicated by the blue (PR), red (SD), and teal (PD) bars. Note that no patients had complete responses. The duration of treatment for patients for whom best response could not be evaluated (such as those with nonmeasurable disease) is shown in orange. Plots are separated into patients who received (A) platinum-doublet chemotherapy (n = 18), (B) taxane-based chemotherapy after prior platinum-doublet chemotherapy (n = 9), (C) combination immune checkpoint inhibition and chemotherapy (n = 10), (D) single-agent immune checkpoint inhibition (n = 9), and (E) targeted therapy with the pan-ERBB family inhibitor, afatinib (n = 20). NE, could not be evaluated; PD, progressive disease; PR, partial response; SD, stable disease.
Consistent with the immunophenotype of these cancers, the activity of single-agent immune checkpoint inhibition was modest (Data Supplement, Figs 4C and 4D). In patients evaluable for response, the most common outcome was progressive disease (60%, n = 3/5). Only one patient had a partial response that lasted more than 11 months. The median PFS was 3.6 months (95% CI, 0.9 to undefined; range, 0.9-11.2 months; Data Supplement). No responses (0%, n = 0/9) were observed in patients treated with chemoimmunotherapy (most of whom received carboplatin, pemetrexed, and pembrolizumab), for whom progressive disease occurred in more than half of patients (56%, n = 5/9). The median PFS was 3.3 months (95% CI, 1.4 to 6.3; range, 1.4-15.2 months; Fig 4D).
Afatinib Activity
As NRG1 fusions are dependent on ERBB signaling, several investigators have explored the use of afatinib, a pan-ERBB inhibitor, in patients with these cancers.22,25,26,30,44 A response was achieved in 25% (n = 5/20, all partial responses) of evaluable patients treated with afatinib (Table 2 and Data Supplement). The fusion partners were known in four of five patients, including CD74 (n = 2), SLC3A2 (n = 1), and SDC4 (n = 1). Stable disease occurred in another 15% (n = 3/20) of patients. The most common response to afatinib was progressive disease, which occurred in 60% (n = 12/20) of patients. The response rate in cancers with CD74-NRG1 and non-CD74-NRG1 fusions was 22% (n = 2/9) and 27% (n = 3/11), respectively.
In addition, the duration of clinical benefit with afatinib was limited. The swimmer's plot of afatinib monotherapy is shown in Figure 4E. The median PFS with afatinib was 2.8 months (95% CI, 1.9 to 4.3; range, 0.3-25.3 months; Data Supplement). PFS did not differ for patients with tumors harboring CD74 fusion partners versus other fusion types (Data Supplement). There was no significant difference (P > .05) in OS when patients who received afatinib were compared with patients who did not receive afatinib.
DISCUSSION
This global registry represents the largest series of patients with NRG1 fusion–positive lung cancers reported to date. As a testament to the utility of multinational consortia such as this one, the number of patients featured here is several fold higher than the number identified through analysis of data from single institutions, large-scale sequencing laboratories, or even The Cancer Genome Atlas.23,31 Despite the retrospective nature of the study and its inherent limitations such as reporting bias and the lack of prospective sequencing data, complete clinical annotation for every patient, and central radiologic assessment, this underscores the utility of such cooperative endeavors to generate meaningful real-world data, particularly in rare genotype-driven cancers.
Although the data generated here confirm preliminary observations reported in prior smaller series or case reports, including publications from members of this registry,23,30,36 several new clinicopathologic observations emerged. More than half of patients were initially diagnosed with stage I or II disease, although many subsequently developed metastatic disease. Whereas intrathoracic metastases predominated, consistent with the natural history of many IMAs,45,46 extrathoracic metastases were observed in more than 40% of patients. Additionally, although NRG1 fusions were strongly associated with IMAs in prior series,24,46,47 nonmucinous adenocarcinomas represented more than a quarter of cases. These fusions were also found in nonadenocarcinoma histologies, including squamous cell and large cell neuroendocrine cancers, suggesting that screening for this driver should not be biased solely toward IMAs.
From a diagnostic perspective, NRG1 fusions can be difficult to detect using DNA sequencing alone.23,31 In our series, only 27% of patients with NRG1 fusion–positive tumors were identified through DNA-based testing, whereas 73% of patients primarily required RNA-based testing to identify these alterations. The design of this registry did not allow a diagnostic performance evaluation of DNA-based and/or RNA-based testing for NRG1 detection. Specifically, a denominator of prospectively sequenced samples was not available to determine the true frequency of NRG1 fusion detection by DNA versus RNA sequencing, and a proportion of samples were sequenced after DNA-based NGS returned negative for MAPK pathway drivers. Nevertheless, RNA-based assays appear to be the best molecular testing method to identify NRG1 fusions. NRG1 fusion breakpoints, while highly heterogenous as demonstrated here, convergently occur in large intronic regions that are challenging to tile and capture by DNA-based assays.23 This observation is consistent with previous reports showing that even comprehensive contemporary DNA-based hybrid capture NGS can fail to identify selected fusions.48 In contrast, RNA-based assays overcome common difficulties associated with DNA-based assays. In particular, assays such as anchored multiplex PCR are preferred over those that assess expression imbalance as some fusions may have high expression of both 3′ and 5′ ends. Furthermore, recognizing that NRG1 fusions were found de novo with other drivers at low frequencies,36 screening algorithms should consider NRG1 fusion interrogation in KRAS-mutant disease and in other driver-positive cancers, particularly after progression on a prior matched TKI.
In our study, all 20 unique chimeric events retained the EGF domain of NRG1, which is known to bind ERBB3 and activate oncogenesis. Notably, 18 unique upstream partners were identified. The most common fusion was CD74-NRG1,23 which was identified in 41% of tumors, with SLC3A2-NRG1 being the second most common, found in 20% of tumors. Importantly, we identified previously unreported upstream partners, including FGFR1, CADM1, F11R, FLYWCH1, KRAS, and PLCG2; this highlights the molecular diversity of NRG1 fusions and the need to screen for these oncogenes with a comprehensive assay poised to detect all possible rearrangements. Furthermore, NRG2α fusions have been identified in cancers, including NSCLCs.49,50 These clinical observations are informative for preclinical experiments that explore fusion diversity and their ability to localize subcellularly and operate differentially in tumor cells.19,31,48,51-53
The most striking observation in this series is the limited lack of activity of systemic therapy in advanced NRG1 fusion–positive lung cancers, acknowledging the small number of patients treated with selected regimens. Response to platinum-based or taxane-based post–platinum-doublet chemotherapy was poor relative to the historic activity of these agents in previously published registrational data sets that treated unselected NSCLCs. It is thus unsurprising that the median OS for patients with stage IV disease was 15.5 months. The lack of response to platinum-based chemotherapy is of interest, given that other fusion-positive tumors, such as those involving ALK, ROS1, and RET, are known to be sensitive to first-line chemotherapy, particularly regimens that include pemetrexed.14,54-57
As with other fusion-positive NSCLCs, NRG1 fusion–positive lung cancers derived limited benefit from immunotherapy.58-62 Response was rare and only observed in one patient of those who received either single-agent immune checkpoint inhibition or immunotherapy combined with chemotherapy. This was most surprising in the latter group of patients for whom progressive disease was observed in more than half of patients. The lack of efficacy was consistent, however, with the immunophenotypic profile of NRG1 fusion–positive tumors in our registry. TMB was not only lower than that in unselected NSCLCs but also interestingly lower in comparison with ALK, ROS1, RET, and NTRK1/2/3 fusion–positive lung cancers. The biologic reasons that underlie such an observation remain unknown and will need to be explored. On top of this, most tumors did not express PD-L1, and only a minority (4%) of cancers had PD-L1 expression of 50% or greater, similar to NRG2α fusions.50
Disappointingly, the activity of targeted therapy with afatinib was also modest. The response rate of 25% and the median PFS of 2.8 months were substantially less than those observed with highly active contemporary targeted therapeutics for ALK, ROS1, RET, and NTRK1/2/3 fusion–positive cancers.4 Although the multicenter Targeted Agent and Profiling Utilization Registry trial (ClinicalTrials.gov identifiers: NCT02925234, NCT02693535)63 will help confirm the prospective activity of afatinib in this setting, novel therapeutics should continue to be explored for these tumors.22,23,25-29,64 For example, promising preclinical and/or clinical activity has been seen with ERBB3 (seribantumab, NCT04383210) or ERBB3/ERBB2 (zenocutuzumab, NCT02912949) monoclonal antibody–based therapy and pan-ERBB covalent TKI therapy (tarloxotinib, NCT03805841) in NRG1 fusion–positive tumors.4,22,23,25,26,44,53 Targeting NRG2α fusion–positive cancers may, in contrast, require strategies that take into account that these fusions may preferentially bind ERBB4.50
In conclusion, NRG1 fusions have a diversity of fusion partners and an EGF binding domain that binds ERBB3. Detection should focus on the inclusion of RNA-based sequencing, which maximizes the likelihood of fusion identification. NRG1 fusion–positive cancers typically do not express high levels of PD-L1 and have a low TMB, consistent with their poor response to immunotherapy. Furthermore, responses to chemotherapy or targeted therapy with afatinib are underwhelming. The development of novel therapeutics for these cancers is thus an unmet need.
Alexander Drilon
Honoraria: Medscape, OncLive, PeerVoice, Physicans' Education Resource, Targeted Oncology, MORE Health, Research to Practice, Foundation Medicine, Peerview
Consulting or Advisory Role: Ignyta, Loxo, TP Therapeutics, AstraZeneca, Pfizer, Blueprint Medicines, Genentech/Roche, Helsinn Therapeutics, BeiGene, Hengrui Therapeutics, Exelixis, Bayer, Tyra Biosciences, Verastem, Takeda/Millennium, BerGenBio, MORE Health, Lilly, Verastem, AbbVie, 14ner Oncology/Elevation Oncology, Remedica, Archer, Monopteros Therapeutics, Novartis, EMD Serono/Merck, Melendi, Repare Therapeutics
Research Funding: Foundation Medicine
Patents, Royalties, Other Intellectual Property: Wolters Kluwer (royalties for Pocket Oncology)
Other Relationship: Merck, GlaxoSmithKline, Teva, Taiho Pharmaceutical, Pfizer, PharmaMar, Puma Biotechnology
Michael Duruisseaux
Consulting or Advisory Role: AstraZeneca, MSD Oncology, BMS, Pfizer, Roche, Takeda, Boehringer Ingelheim, Janssen Oncology, Amgen, AbbVie
Travel, Accommodations, Expenses: Boehringer Ingelheim, Merck Sharp & Dohme
Ji-Youn Han
Honoraria: Roche, AstraZeneca, Bristol Myers Squibb, Takeda
Consulting or Advisory Role: MSD Oncology, AstraZeneca, Bristol Myers Squibb, Lilly, Novartis, Takeda, Pfizer
Research Funding: Roche, Pfizer, Ono Pharmaceutical, Takeda
Soo-Ryum Yang
Consulting or Advisory Role: Invitae
Yonina R. Murciano-Goroff
Travel, Accommodations, Expenses: AstraZeneca
Morihito Okada
Speakers' Bureau: Taiho Pharmaceutical, Johnson & Johnson, Covidien, Lilly, Chugai Pharma, AstraZeneca, Ono Pharmaceutical, CSL Behring
Research Funding: Taiho Pharmaceutical, Nippon Kayaku, Chugai Pharma, Covidien, Johnson & Johnson, Daiichi Sankyo, Yakult Honsha, Lilly Japan, Nihon Medi-Physics, Pfizer, Mochida Pharmaceutical Co Ltd, Shionogi, Ono Pharmaceutical, Kyowa Hakko Kirin
Miguel Angel Molina
Employment: Pangaea Oncology
Research Funding: AstraZeneca, Merck Serono, In3Bio
Marie Wislez
Consulting or Advisory Role: Boehringer Ingelheim, Roche, MSD Oncology, Bristol Myers Squibb, AstraZeneca, Amgen
Speakers' Bureau: Boehringer Ingelheim, Amgen, Roche, MSD Oncology, Bristol Myers Squibb, AstraZeneca
Travel, Accommodations, Expenses: Roche, MSD Oncology
Alexa Schrock
Employment: Foundation Medicine
Stock and Other Ownership Interests: Foundation Medicine, Roche
Siraj Ali
Employment: EQRX (I), Foundation Medicine, EQRX
Leadership: Incysus Inc, Elevation Oncology, Pillar Biosciences
Stock and Other Ownership Interests: Exelixis, Merus NV, Pfizer
Consulting or Advisory Role: Azitra (I), Princepx Tx (I)
Patents, Royalties, and Other Intellectual Property:Patents via Seres Health on microbiome in non neoplastic disease (I), Foundation Medicine
Valérie Gounant
Consulting or Advisory Role: Bristol Myers Squibb, Takeda, Roche, AstraZeneca, Boehringer Ingelheim, Novartis, Chugai Pharma Europe
Travel, Accommodations, Expenses: Takeda, Roche, Pfizer
Alison M. Schram
Research Funding: Merus, Kura Oncology, Surface Oncology, AstraZeneca, Lilly, Northern Biologics, Pfizer, Black Diamond Therapeutics, BeiGene, Relay Therapeutics
Isabelle Monnet
Travel, Accommodations, Expenses: MSD Oncology, Roche, Takeda, Pfizer
Jin-Yuan Shih
Honoraria: AstraZeneca, Boehringer Ingelheim, Pfizer, Novartis, Merck Sharp & Dohme, Ono Pharmaceutical, Bristol Myers Squibb, Roche, Chugai Pharma, Lilly
Consulting or Advisory Role: Chugai Pharma, Boehringer Ingelheim, Bristol Myers Squibb, Roche, AstraZeneca, Lilly, Takeda, CStone Pharmaceuticals, Janssen Oncology, Novartis, Pfizer, Ono Pharmaceutical, Merck Sharp & Dohme
Research Funding: Roche
Travel, Accommodations, Expenses: Bristol Myers Squibb, Pfizer, Chugai Pharma, Roche
Joshua Sabari
Consulting or Advisory Role: AstraZeneca, Janssen Oncology, Navire, Pfizer, Regeneron, Medscape, Takeda
Maurice Pérol
Consulting or Advisory Role: Lilly, Roche/Genentech, Pfizer, AstraZeneca, Boehringer Ingelheim, Merck Sharp & Dohme, Bristol Myers Squibb, Novartis, Amgen, Takeda, Chugai Pharma, Gritstone Oncology
Research Funding: AstraZeneca, Roche, Takeda, Boehringer Ingelheim
Travel, Accommodations, Expenses: AstraZeneca, Roche, Bristol Myers Squibb, Merck Sharp & Dohme, Pfizer, Takeda, Chugai Pharma
Viola W. Zhu
Stock and Other Ownership Interests: TP Therapeutics
Honoraria: AstraZeneca, Roche/Genentech, Takeda, Blueprint Medicines, Xcovery
Consulting or Advisory Role: AstraZeneca, Takeda, TP Therapeutics, Roche/Genentech, Xcovery
Speakers' Bureau: AstraZeneca, Roche/Genentech, Takeda, Blueprint Medicines
Travel, Accommodations, Expenses: AstraZeneca, Roche/Genentech, Takeda, TP Therapeutics
Misako Nagasaka
Consulting or Advisory Role: AstraZeneca, Caris Life Sciences, Daiichi Sankyo, Takeda, Novartis, EMD Serono
Speakers' Bureau: Blueprint Medicines
Research Funding: Tempus
Travel, Accommodations, Expenses: Anheart Therapeutics
Robert Doebele
Employment: Rain Therapeutics
Leadership: Rain Therapeutics
Stock and Other Ownership: Rain Therapeutics
Consulting or Advisory Role: GreenPeptide, AstraZeneca, Roche/Genentech, Bayer, Takeda, Rain Therapeutics, Anchiano, Blueprint Medicines, Foundation Medicine, Guardant Health
Patents, Royalties, and Other Intellectual Property: Abbott Molecular for Patent PCT/US2013/057495, Rain Therapeutics, Genentech (Inst), Foundation Medicine (Inst), Black Diamond (Inst), Pearl River (Inst), Voronoi (Inst)
Travel, Accommodations, Expenses: Rain Therapeutics, Roche/Genentech
D. Ross Camidge
Honoraria: Roche, Takeda, AstraZeneca, Daiichi Sankyo, Bio-Thera, Ribon Therapeutics, Bristol Myers Squibb, Inivata, AbbVie, Apollomics, Elevation Oncology, EMD Serono, Helsinn Therapeutics, Lilly, Nuvalent Inc, Seattle Genetics, Turning Point Therapeutics, Kestrel Labs, Amgen Astellas BioPharma, Anchiano, Eisai, GlaxoSmithKline, Janssen, OnKure, Mersana, Pfizer, QiLu Pharmaceutical, Sanofi
Research Funding: Takeda
Maria Arcila
Honoraria: Invivoscribe, Biocartis
Consulting or Advisory Role: AstraZeneca
Travel, Accommodations, Expenses: AstraZeneca, Invivoscribe, Raindance Technologies
Sai-Hong Ignatius Ou
Stock and Other Ownership Interests: Turning Point Therapeutics, Elevation Oncology
Honoraria: Pfizer, Roche Pharma AG, Genentech/Roche, ARIAD/Takeda, AstraZeneca
Consulting or Advisory Role: Pfizer, Roche/Genentech, AstraZeneca, Takeda, Jassen/JNJ
Speakers' Bureau: AstraZeneca, Genentech/Roche
Research Funding: Pfizer, Roche Pharma AG, AstraZeneca/MedImmune, AstraZeneca, ARIAD, Revolution Medicines, Mirati Therapeutics, Jassen/JNJ
Denis Moro-Sibilot
Consulting or Advisory Role: Roche/Genentech, Boehringer Ingelheim, Lilly/ImClone, Sanofi, Novartis, Amgen, Pfizer, AstraZeneca, Clovis Oncology, MSD Oncology, ARIAD, Bristol-Myers Squibb, Takeda, Abbvie
Research Funding: Abbvie (Inst), Boehr (Inst), Roche/Genentech (Inst), Bristol-Myers Squibb (Inst)
Expert Testimony: MSD Oncology
Travel, Accommodations, Expenses: Roche/Genentech, Lilly/ImClone, Pfizer, MSD Oncology, Bristol-Myers Squibb
Lucia Anna Muscarella
Travel, Accommodations, Expenses: Boehringer Ingelheim
Stephen V. Liu
Consulting or Advisory Role: Genentech, Pfizer, Lilly, Bristol Myers Squibb, AstraZeneca, Takeda, Regeneron, G1 Therapeutics, Guardant Health, Janssen Oncology, MSD Oncology, Jazz Pharmaceuticals, Blueprint Medicines, Inivata, PharmaMar, Daiichi Sankyo/UCB Japan, BeiGene, Amgen
Research Funding: Genentech/Roche, Pfizer, Corvus Pharmaceuticals, Bayer, Merck, Lycera, AstraZeneca, Molecular Partners, Blueprint Medicines, Lilly, Rain Therapeutics, Alkermes, Bristol Myers Squibb, Turning Point Therapeutics, RAPT Therapeutics, Merus, Debiopharm Group, Elevation Oncology
Travel, Accommodations, Expenses: AstraZeneca, Roche/Genentech, MSD Oncology
Jacques Cadranel
Honoraria: AstraZeneca/MedImmune, Bristol Myers Squibb, Roche/Genentech, Merck Sharp & Dohme, Boehringer Ingelheim
Consulting or Advisory Role: AstraZeneca/MedImmune, Roche/Genentech, Boehringer Ingelheim, Bristol Myers Squibb, Takeda, Merck Sharp & Dohme, Pfizer, Lilly, Novartis
Research Funding: Pfizer, Novartis, AstraZeneca/MedImmune
No other potential conflicts of interest were reported.
SUPPORT
Supported by the National Cancer Institute at the National Institutes of Health (Grant No. P30-CA-008748).
A.D., M.D., S.V.L., and J.C. contributed equally to this work.
AUTHOR CONTRIBUTIONS
Conception and design: Alexander Drilon, Michael Duruisseaux, Haiquan Chen, Fanny Magne, Maria Arcila, Rafael Rosell, Stephen V. Liu, Jacques Cadranel
Administrative support: Christina Falcon, Rafael Rosell
Provision of study materials or patients: Michael Duruisseaux, Ji-Youn Han, Masaoki Ito, Miguel Angel Molina, Marie Wislez, Clarisse Dupont, Valérie Gounant, Alison M. Schram, Joshua Sabari, Robert Doebele, D. Ross Camidge, D. Ross Camidge, Sai-Hong Ignatius Ou, Denis Moro-Sibilot, Stephen V. Liu, Jacques Cadranel
Collection and assembly of data: Alexander Drilon, Michael Duruisseaux, Ji-Youn Han, Masaoki Ito, Christina Falcon, Morihito Okada, Miguel Angel Molina, Marie Wislez, Philippe Brun, Clarisse Dupont, Eva Branden, Giulio Rossi, Alexa Schrock, Siraj Ali, Valérie Gounant, Fanny Magne, Torsten Gerriet Blum, Alison M. Schram, Isabelle Monnet, Jin-Yuan Shih, Joshua Sabari, Maurice Pérol, Misako Nagasaka, Robert Doebele, D. Ross Camidge, Maria Arcila, Sai-Hong Ignatius Ou, Denis Moro-Sibilot, Lucia Anna Muscarella, Stephen V. Liu, Jacques Cadranel
Data analysis and interpretation: Alexander Drilon, Michael Duruisseaux, Ji-Youn Han, Christina Falcon, Soo-Ryum Yang, Yonina R. Murciano-Goroff, Miguel Angel Molina, Fanny Magne, Alison M. Schram, Jin-Yuan Shih, Joshua Sabari, Maurice Pérol, Viola W. Zhu, Misako Nagasaka, Robert Doebele, Maria Arcila, Sai-Hong Ignatius Ou, Stephen V. Liu, Jacques Cadranel
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Clinicopathologic Features and Response to Therapy of NRG1 Fusion–Driven Lung Cancers: The eNRGy1 Global Multicenter Registry
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/authors/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Alexander Drilon
Honoraria: Medscape, OncLive, PeerVoice, Physicans' Education Resource, Targeted Oncology, MORE Health, Research to Practice, Foundation Medicine, Peerview
Consulting or Advisory Role: Ignyta, Loxo, TP Therapeutics, AstraZeneca, Pfizer, Blueprint Medicines, Genentech/Roche, Helsinn Therapeutics, BeiGene, Hengrui Therapeutics, Exelixis, Bayer, Tyra Biosciences, Verastem, Takeda/Millennium, BerGenBio, MORE Health, Lilly, Verastem, AbbVie, 14ner Oncology/Elevation Oncology, Remedica, Archer, Monopteros Therapeutics, Novartis, EMD Serono/Merck, Melendi, Repare Therapeutics
Research Funding: Foundation Medicine
Patents, Royalties, Other Intellectual Property: Wolters Kluwer (royalties for Pocket Oncology)
Other Relationship: Merck, GlaxoSmithKline, Teva, Taiho Pharmaceutical, Pfizer, PharmaMar, Puma Biotechnology
Michael Duruisseaux
Consulting or Advisory Role: AstraZeneca, MSD Oncology, BMS, Pfizer, Roche, Takeda, Boehringer Ingelheim, Janssen Oncology, Amgen, AbbVie
Travel, Accommodations, Expenses: Boehringer Ingelheim, Merck Sharp & Dohme
Ji-Youn Han
Honoraria: Roche, AstraZeneca, Bristol Myers Squibb, Takeda
Consulting or Advisory Role: MSD Oncology, AstraZeneca, Bristol Myers Squibb, Lilly, Novartis, Takeda, Pfizer
Research Funding: Roche, Pfizer, Ono Pharmaceutical, Takeda
Soo-Ryum Yang
Consulting or Advisory Role: Invitae
Yonina R. Murciano-Goroff
Travel, Accommodations, Expenses: AstraZeneca
Morihito Okada
Speakers' Bureau: Taiho Pharmaceutical, Johnson & Johnson, Covidien, Lilly, Chugai Pharma, AstraZeneca, Ono Pharmaceutical, CSL Behring
Research Funding: Taiho Pharmaceutical, Nippon Kayaku, Chugai Pharma, Covidien, Johnson & Johnson, Daiichi Sankyo, Yakult Honsha, Lilly Japan, Nihon Medi-Physics, Pfizer, Mochida Pharmaceutical Co Ltd, Shionogi, Ono Pharmaceutical, Kyowa Hakko Kirin
Miguel Angel Molina
Employment: Pangaea Oncology
Research Funding: AstraZeneca, Merck Serono, In3Bio
Marie Wislez
Consulting or Advisory Role: Boehringer Ingelheim, Roche, MSD Oncology, Bristol Myers Squibb, AstraZeneca, Amgen
Speakers' Bureau: Boehringer Ingelheim, Amgen, Roche, MSD Oncology, Bristol Myers Squibb, AstraZeneca
Travel, Accommodations, Expenses: Roche, MSD Oncology
Alexa Schrock
Employment: Foundation Medicine
Stock and Other Ownership Interests: Foundation Medicine, Roche
Siraj Ali
Employment: EQRX (I), Foundation Medicine, EQRX
Leadership: Incysus Inc, Elevation Oncology, Pillar Biosciences
Stock and Other Ownership Interests: Exelixis, Merus NV, Pfizer
Consulting or Advisory Role: Azitra (I), Princepx Tx (I)
Patents, Royalties, and Other Intellectual Property:Patents via Seres Health on microbiome in non neoplastic disease (I), Foundation Medicine
Valérie Gounant
Consulting or Advisory Role: Bristol Myers Squibb, Takeda, Roche, AstraZeneca, Boehringer Ingelheim, Novartis, Chugai Pharma Europe
Travel, Accommodations, Expenses: Takeda, Roche, Pfizer
Alison M. Schram
Research Funding: Merus, Kura Oncology, Surface Oncology, AstraZeneca, Lilly, Northern Biologics, Pfizer, Black Diamond Therapeutics, BeiGene, Relay Therapeutics
Isabelle Monnet
Travel, Accommodations, Expenses: MSD Oncology, Roche, Takeda, Pfizer
Jin-Yuan Shih
Honoraria: AstraZeneca, Boehringer Ingelheim, Pfizer, Novartis, Merck Sharp & Dohme, Ono Pharmaceutical, Bristol Myers Squibb, Roche, Chugai Pharma, Lilly
Consulting or Advisory Role: Chugai Pharma, Boehringer Ingelheim, Bristol Myers Squibb, Roche, AstraZeneca, Lilly, Takeda, CStone Pharmaceuticals, Janssen Oncology, Novartis, Pfizer, Ono Pharmaceutical, Merck Sharp & Dohme
Research Funding: Roche
Travel, Accommodations, Expenses: Bristol Myers Squibb, Pfizer, Chugai Pharma, Roche
Joshua Sabari
Consulting or Advisory Role: AstraZeneca, Janssen Oncology, Navire, Pfizer, Regeneron, Medscape, Takeda
Maurice Pérol
Consulting or Advisory Role: Lilly, Roche/Genentech, Pfizer, AstraZeneca, Boehringer Ingelheim, Merck Sharp & Dohme, Bristol Myers Squibb, Novartis, Amgen, Takeda, Chugai Pharma, Gritstone Oncology
Research Funding: AstraZeneca, Roche, Takeda, Boehringer Ingelheim
Travel, Accommodations, Expenses: AstraZeneca, Roche, Bristol Myers Squibb, Merck Sharp & Dohme, Pfizer, Takeda, Chugai Pharma
Viola W. Zhu
Stock and Other Ownership Interests: TP Therapeutics
Honoraria: AstraZeneca, Roche/Genentech, Takeda, Blueprint Medicines, Xcovery
Consulting or Advisory Role: AstraZeneca, Takeda, TP Therapeutics, Roche/Genentech, Xcovery
Speakers' Bureau: AstraZeneca, Roche/Genentech, Takeda, Blueprint Medicines
Travel, Accommodations, Expenses: AstraZeneca, Roche/Genentech, Takeda, TP Therapeutics
Misako Nagasaka
Consulting or Advisory Role: AstraZeneca, Caris Life Sciences, Daiichi Sankyo, Takeda, Novartis, EMD Serono
Speakers' Bureau: Blueprint Medicines
Research Funding: Tempus
Travel, Accommodations, Expenses: Anheart Therapeutics
Robert Doebele
Employment: Rain Therapeutics
Leadership: Rain Therapeutics
Stock and Other Ownership: Rain Therapeutics
Consulting or Advisory Role: GreenPeptide, AstraZeneca, Roche/Genentech, Bayer, Takeda, Rain Therapeutics, Anchiano, Blueprint Medicines, Foundation Medicine, Guardant Health
Patents, Royalties, and Other Intellectual Property: Abbott Molecular for Patent PCT/US2013/057495, Rain Therapeutics, Genentech (Inst), Foundation Medicine (Inst), Black Diamond (Inst), Pearl River (Inst), Voronoi (Inst)
Travel, Accommodations, Expenses: Rain Therapeutics, Roche/Genentech
D. Ross Camidge
Honoraria: Roche, Takeda, AstraZeneca, Daiichi Sankyo, Bio-Thera, Ribon Therapeutics, Bristol Myers Squibb, Inivata, AbbVie, Apollomics, Elevation Oncology, EMD Serono, Helsinn Therapeutics, Lilly, Nuvalent Inc, Seattle Genetics, Turning Point Therapeutics, Kestrel Labs, Amgen Astellas BioPharma, Anchiano, Eisai, GlaxoSmithKline, Janssen, OnKure, Mersana, Pfizer, QiLu Pharmaceutical, Sanofi
Research Funding: Takeda
Maria Arcila
Honoraria: Invivoscribe, Biocartis
Consulting or Advisory Role: AstraZeneca
Travel, Accommodations, Expenses: AstraZeneca, Invivoscribe, Raindance Technologies
Sai-Hong Ignatius Ou
Stock and Other Ownership Interests: Turning Point Therapeutics, Elevation Oncology
Honoraria: Pfizer, Roche Pharma AG, Genentech/Roche, ARIAD/Takeda, AstraZeneca
Consulting or Advisory Role: Pfizer, Roche/Genentech, AstraZeneca, Takeda, Jassen/JNJ
Speakers' Bureau: AstraZeneca, Genentech/Roche
Research Funding: Pfizer, Roche Pharma AG, AstraZeneca/MedImmune, AstraZeneca, ARIAD, Revolution Medicines, Mirati Therapeutics, Jassen/JNJ
Denis Moro-Sibilot
Consulting or Advisory Role: Roche/Genentech, Boehringer Ingelheim, Lilly/ImClone, Sanofi, Novartis, Amgen, Pfizer, AstraZeneca, Clovis Oncology, MSD Oncology, ARIAD, Bristol-Myers Squibb, Takeda, Abbvie
Research Funding: Abbvie (Inst), Boehr (Inst), Roche/Genentech (Inst), Bristol-Myers Squibb (Inst)
Expert Testimony: MSD Oncology
Travel, Accommodations, Expenses: Roche/Genentech, Lilly/ImClone, Pfizer, MSD Oncology, Bristol-Myers Squibb
Lucia Anna Muscarella
Travel, Accommodations, Expenses: Boehringer Ingelheim
Stephen V. Liu
Consulting or Advisory Role: Genentech, Pfizer, Lilly, Bristol Myers Squibb, AstraZeneca, Takeda, Regeneron, G1 Therapeutics, Guardant Health, Janssen Oncology, MSD Oncology, Jazz Pharmaceuticals, Blueprint Medicines, Inivata, PharmaMar, Daiichi Sankyo/UCB Japan, BeiGene, Amgen
Research Funding: Genentech/Roche, Pfizer, Corvus Pharmaceuticals, Bayer, Merck, Lycera, AstraZeneca, Molecular Partners, Blueprint Medicines, Lilly, Rain Therapeutics, Alkermes, Bristol Myers Squibb, Turning Point Therapeutics, RAPT Therapeutics, Merus, Debiopharm Group, Elevation Oncology
Travel, Accommodations, Expenses: AstraZeneca, Roche/Genentech, MSD Oncology
Jacques Cadranel
Honoraria: AstraZeneca/MedImmune, Bristol Myers Squibb, Roche/Genentech, Merck Sharp & Dohme, Boehringer Ingelheim
Consulting or Advisory Role: AstraZeneca/MedImmune, Roche/Genentech, Boehringer Ingelheim, Bristol Myers Squibb, Takeda, Merck Sharp & Dohme, Pfizer, Lilly, Novartis
Research Funding: Pfizer, Novartis, AstraZeneca/MedImmune
No other potential conflicts of interest were reported.
REFERENCES
- 1.Benayed R, Offin M, Mullaney K, et al. High yield of RNA sequencing for targetable kinase fusions in lung adenocarcinomas with no mitogenic driver alteration detected by DNA sequencing and low tumor mutation burden Clin Cancer Res 254712–47222019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farago AF, Azzoli CG.Beyond ALK and ROS1: RET, NTRK, EGFR and BRAF gene rearrangements in non-small cell lung cancer Transl Lung Cancer Res 6550–5592017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kohno T, Nakaoku T, Tsuta K, et al. Beyond ALK-RET, ROS1 and other oncogene fusions in lung cancer Transl Lung Cancer Res 4156–1642015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schram AM, Chang MT, Jonsson P, et al. Fusions in solid tumours: Diagnostic strategies, targeted therapy, and acquired resistance Nat Rev Clin Oncol 14735–7482017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shaw AT, Engelman JA.ALK in lung cancer: Past, present, and future J Clin Oncol 311105–11112013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peters S, Camidge DR, Shaw AT, et al. Alectinib versus crizotinib in untreated ALK-positive non-small-cell lung cancer N Engl J Med 377829–8382017 [DOI] [PubMed] [Google Scholar]
- 7.Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer N Engl J Med 3682385–23942013 [DOI] [PubMed] [Google Scholar]
- 8.Drilon A, Jenkins C, Iyer S, et al. ROS1-dependent cancers—Biology, diagnostics and therapeutics Nat Rev Clin Oncol 1835–552021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drilon A, Ou SI, Cho BC, et al. Repotrectinib (TPX-0005) is a next-generation ROS1/TRK/ALK inhibitor that potently inhibits ROS1/TRK/ALK solvent-front mutations Cancer Discov 81227–12362018 [DOI] [PubMed] [Google Scholar]
- 10.Drilon A, Siena S, Dziadziuszko R, et al. Entrectinib in ROS1 fusion-positive non-small-cell lung cancer: Integrated analysis of three phase 1-2 trials Lancet Oncol 21261–2702020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drilon A, Siena S, Ou SI, et al. Safety and antitumor activity of the multitargeted pan-TRK, ROS1, and ALK inhibitor entrectinib: Combined results from two phase I trials (ALKA-372-001 and STARTRK-1) Cancer Discov 7400–4092017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drilon A, Hu ZI, Lai GGY, et al. Targeting RET-driven cancers: Lessons from evolving preclinical and clinical landscapes Nat Rev Clin Oncol 15151–1672018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drilon A, Wang L, Hasanovic A, et al. Response to Cabozantinib in patients with RET fusion-positive lung adenocarcinomas Cancer Discov 3630–6352013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gautschi O, Milia J, Filleron T, et al. Targeting RET in patients with RET-rearranged lung cancers: Results from the global, multicenter RET registry J Clin Oncol 351403–14102017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Subbiah V, Velcheti V, Tuch BB, et al. Selective RET kinase inhibition for patients with RET-altered cancers Ann Oncol 291869–18762018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cocco E, Scaltriti M, Drilon A.NTRK fusion-positive cancers and TRK inhibitor therapy Nat Rev Clin Oncol 15731–7472018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drilon A, Laetsch TW, Kummar S, et al. Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children N Engl J Med 378731–7392018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dimou A, Camidge DR.Detection of NRG1 fusions in solid tumors: Rare gold? Clin Cancer Res 254865–48672019 [DOI] [PubMed] [Google Scholar]
- 19.Fernandez-Cuesta L, Plenker D, Osada H, et al. CD74-NRG1 fusions in lung adenocarcinoma Cancer Discov 4415–4222014 [DOI] [PubMed] [Google Scholar]
- 20.Muscarella LA, Rossi A.NRG1: A cinderella fusion in lung cancer? Lung Cancer Manag 6121–1232017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trombetta D, Rossi A, Fabrizio FP, et al. NRG1-ErbB lost in translation: A new paradigm for lung cancer? Curr Med Chem 244213–42282017 [DOI] [PubMed] [Google Scholar]
- 22.Cheema PK, Doherty M, Tsao MS.A case of invasive mucinous pulmonary adenocarcinoma with a CD74-NRG1 fusion protein targeted with afatinib J Thorac Oncol 12e200–e2022017 [DOI] [PubMed] [Google Scholar]
- 23.Drilon A, Somwar R, Mangatt BP, et al. Response to ERBB3-directed targeted therapy in NRG1-rearranged cancers Cancer Discov 8686–6952018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandez-Cuesta L, Thomas RK.Molecular pathways: Targeting NRG1 fusions in lung cancer Clin Cancer Res 211989–19942015 [DOI] [PubMed] [Google Scholar]
- 25.Gay ND, Wang Y, Beadling C, et al. Durable response to afatinib in lung adenocarcinoma harboring NRG1 gene fusions J Thorac Oncol 12e107–e1102017 [DOI] [PubMed] [Google Scholar]
- 26.Jones MR, Lim H, Shen Y, et al. Successful targeting of the NRG1 pathway indicates novel treatment strategy for metastatic cancer Ann Oncol 283092–30972017 [DOI] [PubMed] [Google Scholar]
- 27.Jones MR, Williamson LM, Topham JT, et al. NRG1 gene fusions are recurrent, clinically actionable gene rearrangements in KRAS wild-type pancreatic ductal adenocarcinoma Clin Cancer Res 254674–46812019 [DOI] [PubMed] [Google Scholar]
- 28.Schaefer G, Fitzpatrick VD, Sliwkowski MX.Gamma-heregulin: A novel heregulin isoform that is an autocrine growth factor for the human breast cancer cell line, MDA-MB-175 Oncogene 151385–13941997 [DOI] [PubMed] [Google Scholar]
- 29.Shin DH, Jo JY, Han JY.Dual targeting of ERBB2/ERBB3 for the treatment of SLC3A2-NRG1-mediated lung cancer Mol Cancer Ther 172024–20332018 [DOI] [PubMed] [Google Scholar]
- 30.Cadranel J, Liu SV, Duruisseaux M, et al. Therapeutic potential of afatinib in NRG1 fusion-driven solid tumors: A case series Oncologist 267–162020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jonna S, Feldman RA, Swensen J, et al. Detection of NRG1 gene fusions in solid tumors Clin Cancer Res 254966–49722019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gautschi O, Milia J, Cabarrou B, et al. Targeted therapy for patients with BRAF-mutant lung cancer: Results from the European EURAF cohort J Thorac Oncol 101451–14572015 [DOI] [PubMed] [Google Scholar]
- 33.Mazieres J, Barlesi F, Filleron T, et al. Lung cancer patients with HER2 mutations treated with chemotherapy and HER2-targeted drugs: Results from the European EUHER2 cohort Ann Oncol 27281–2862016 [DOI] [PubMed] [Google Scholar]
- 34.Mazieres J, Peters S, Lepage B, et al. Lung cancer that harbors an HER2 mutation: Epidemiologic characteristics and therapeutic perspectives J Clin Oncol 311997–20032013 [DOI] [PubMed] [Google Scholar]
- 35.Mazieres J, Zalcman G, Crino L, et al. Crizotinib therapy for advanced lung adenocarcinoma and a ROS1 rearrangement: Results from the EUROS1 cohort J Clin Oncol 33992–9992015 [DOI] [PubMed] [Google Scholar]
- 36.Muscarella LA, Trombetta D, Fabrizio FP, et al. ALK and NRG1 fusions coexist in a patient with signet ring cell lung adenocarcinoma J Thorac Oncol 12e161–e1632017 [DOI] [PubMed] [Google Scholar]
- 37.Ancevski Hunter K, Socinski MA, Villaruz LC.PD-L1 testing in guiding patient selection for PD-1/PD-L1 inhibitor therapy in lung cancer Mol Diagn Ther 221–102018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hendriks LE, Rouleau E, Besse B.Clinical utility of tumor mutational burden in patients with non-small cell lung cancer treated with immunotherapy Transl Lung Cancer Res 7647–6602018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Melendez B, Van Campenhout C, Rorive S, et al. Methods of measurement for tumor mutational burden in tumor tissue Transl Lung Cancer Res 7661–6672018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vokes NI, Liu D, Ricciuti B, et al. Harmonization of tumor mutational burden quantification and association with response to immune checkpoint blockade in non-small-cell lung cancer. JCO Precis Oncol. 2019;3 doi: 10.1200/PO.19.00171. 10.1200/PO.19.00171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients Nat Med 23703–7132017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rizvi H, Sanchez-Vega F, La K, et al. Molecular determinants of response to anti-programmed cell death (PD)-1 and anti-programmed death-ligand 1 (PD-L1) blockade in patients with non-small-cell lung cancer profiled with targeted next-generation sequencing J Clin Oncol 36633–6412018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samstein RM, Lee CH, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types Nat Genet 51202–2062019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goto Y, Cadranel J, Weinberg BA, et al. 63O—NRG1-fusion-driven solid tumours: A case series indicating the therapeutic potential of afatinib Ann Oncol 30ix23–ix242019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cha YJ, Kim HR, Lee HJ, et al. Clinical course of stage IV invasive mucinous adenocarcinoma of the lung Lung Cancer 10282–882016 [DOI] [PubMed] [Google Scholar]
- 46.Shim HS, Kenudson M, Zheng Z, et al. Unique genetic and survival characteristics of invasive mucinous adenocarcinoma of the lung J Thorac Oncol 101156–11622015 [DOI] [PubMed] [Google Scholar]
- 47.Trombetta D, Graziano P, Scarpa A, et al. Frequent NRG1 fusions in Caucasian pulmonary mucinous adenocarcinoma predicted by Phospho-ErbB3 expression Oncotarget 99661–96712018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nagasaka M, Ou SI.Neuregulin 1 fusion-positive NSCLC J Thorac Oncol 141354–13592019 [DOI] [PubMed] [Google Scholar]
- 49.Kohsaka S, Hayashi T, Nagano M, et al. Identification of novel CD74-NRG2α fusion from comprehensive profiling of lung adenocarcinoma in Japanese never or light smokers J Thorac Oncol 15948–9612020 [DOI] [PubMed] [Google Scholar]
- 50.Ou S-HI, Xiu J, Nagasaka M, et al. Identification of novel CDH1-NRG2 and F11R-NRG2 fusions in NSCLC plus additional novel fusions in other solid tumors by whole transcriptome sequencing. JTO Clin Res Rep. 2021;2:100132. doi: 10.1016/j.jtocrr.2020.100132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dhanasekaran SM, Balbin OA, Chen G, et al. Transcriptome meta-analysis of lung cancer reveals recurrent aberrations in NRG1 and Hippo pathway genes. Nat Commun. 2014;5:5893. doi: 10.1038/ncomms6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shin DH, Lee D, Hong DW, et al. Oncogenic function and clinical implications of SLC3A2-NRG1 fusion in invasive mucinous adenocarcinoma of the lung Oncotarget 769450–694652016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakaoku T, Tsuta K, Ichikawa H, et al. Druggable oncogene fusions in invasive mucinous lung adenocarcinoma Clin Cancer Res 203087–30932014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Camidge DR, Kono SA, Lu X, et al. Anaplastic lymphoma kinase gene rearrangements in non-small cell lung cancer are associated with prolonged progression-free survival on pemetrexed J Thorac Oncol 6774–7802011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Drilon A, Bergagnini I, Delasos L, et al. Clinical outcomes with pemetrexed-based systemic therapies in RET-rearranged lung cancers Ann Oncol 271286–12912016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen YF, Hsieh MS, Wu SG, et al. Efficacy of pemetrexed-based chemotherapy in patients with ROS1 fusion-positive lung adenocarcinoma compared with in patients harboring other driver mutations in east asian populations J Thorac Oncol 111140–11522016 [DOI] [PubMed] [Google Scholar]
- 57.Shaw AT, Varghese AM, Solomon BJ, et al. Pemetrexed-based chemotherapy in patients with advanced, ALK-positive non-small cell lung cancer Ann Oncol 2459–662013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Berghoff AS, Bellosillo B, Caux C, et al. Immune checkpoint inhibitor treatment in patients with oncogene- addicted non-small cell lung cancer (NSCLC): Summary of a multidisciplinary round-table discussion. ESMO Open. 2019;4:e000498. doi: 10.1136/esmoopen-2019-000498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gainor JF, Shaw AT, Sequist LV, et al. EGFR mutations and ALK rearrangements are associated with low response rates to PD-1 pathway blockade in non-small cell lung cancer: A retrospective analysis Clin Cancer Res 224585–45932016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee CK, Man J, Lord S, et al. Checkpoint inhibitors in metastatic EGFR-mutated non-small cell lung cancer—A meta-analysis J Thorac Oncol 12403–4072017 [DOI] [PubMed] [Google Scholar]
- 61.Mazieres J, Drilon A, Lusque A, et al. Immune checkpoint inhibitors for patients with advanced lung cancer and oncogenic driver alterations: Results from the IMMUNOTARGET registry Ann Oncol 301321–13282019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Offin M, Rizvi H, Tenet M, et al. Tumor mutation burden and efficacy of EGFR-tyrosine kinase inhibitors in patients with EGFR-mutant lung cancers Clin Cancer Res 251063–10692019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Laskin JJ, Cadranel J, Renouf DJ, et al. Afatinib as a novel potential treatment option for NRG1 fusion-positive tumors. J Glob Oncol. 2019;5:110. [Google Scholar]
- 64.Schram AM, Drilon A, Mercade TM, et al. 685TiP—A phase II basket study of MCLA-128, a bispecific antibody targeting the HER3 pathway, in NRG1 fusion-positive advanced solid tumours. Ann Oncol. 2019;30:v317. [Google Scholar]