Abstract
PURPOSE
MET exon 14 (METex14) skipping alterations are oncogenic drivers in non–small-cell lung cancer (NSCLC). We present a comprehensive overview of METex14 samples from 1,592 patients with NSCLC, associated clinicogenomic characteristics, potential mechanisms of acquired resistance, treatment patterns, and outcomes to MET inhibitors.
METHODS
Hybrid capture–based comprehensive genomic profiling (CGP) was performed on samples from 69,219 patients with NSCLC. For treatment patterns and outcomes analysis, patients with advanced METex14-altered NSCLC were selected from the Flatiron Health-Foundation Medicine clinicogenomic database, a nationwide deidentified electronic health record–derived database linked to Foundation Medicine CGP for patients treated between January 2011 and March 2020.
RESULTS
A total of 1,592 patients with NSCLC (2.3%) were identified with 1,599 METex14 alterations spanning multiple functional sites (1,458 of 60,244 tissue samples and 134 of 8,975 liquid samples). Low tumor mutational burden and high programmed death ligand 1 expression were enriched in METex14-altered samples. MDM2, CDK4, and MET coamplifications and TP53 mutations were present in 34%, 19%, 11%, and 42% of tissue samples, respectively. Comparing tissue and liquid cohorts, coalteration frequency and acquired resistance mechanisms, including multiple MET mutations, EGFR, ERBB2, KRAS, and PI3K pathway alterations, were generally similar. Positive percent agreement with the tissue was 100% for METex14 pairs collected within 1 year (n = 7). Treatment patterns showed increasing adoption of MET inhibitors in METex14-altered NSCLC after receipt of CGP results; the real-world response rate to MET inhibitors was 45%, and time to treatment discontinuation was 4.4 months.
CONCLUSION
Diverse METex14 alterations were present in 2%-3% of NSCLC cases. Tissue and liquid comparisons showed high concordance and similar coalteration profiles. Characterizing common co-occurring alterations and immunotherapy biomarkers, including those present before or acquired after treatment, may be critical for predicting responses to MET inhibitors and informing rational combination strategies.
INTRODUCTION
MET exon 14 skipping alterations are established drivers of oncogenesis and occur in approximately 3% of patients with non–small-cell lung cancer (NSCLC).1-3 Exon 14 encodes the juxtamembrane domain of the MET receptor tyrosine kinase, including the Y1003 residue required for efficient recruitment of the CBL E3 ubiquitin ligase, which targets MET for ubiquitin-mediated degradation.4 Heterogenous alterations in exon 14 and its adjacent introns can interfere with splicing, resulting in exon 14 skipping, or directly alter or delete the CBL binding site, leading to increased MET stability and oncogenic potential.2,4-8
CONTEXT
Key Objective
This study examined more than 1,500 individual patients with METex14-altered non–small-cell lung cancer allowing granular description of diverse alterations activating MET, as well as exploring coalterations and signatures, which may predict resistance and sensitivity to combination therapies.
Knowledge Generated
METex14 skipping alterations were identified via tissue and liquid biopsy, as well as a smaller subset of alterations predicted to disrupt the exon 14 CBL binding site or activate MET via other mechanisms. Coalteration observations from previous reports were confirmed in this larger data set, and targetable mechanisms of resistance to MET inhibitors were identified. Treatment patterns and outcomes using a real-world data set were also described.
Relevance
METex14 skipping alterations are targetable drivers in non–small-cell lung cancer. This study confirms the diversity of these alterations and highlights the need for rigorous methods of detection and standardization of biomarker definitions. Genomic profiling to identify coalterations and mechanisms of acquired resistance is warranted to further tailor personalized treatments.
MET exon 14 skipping alterations confer sensitivity to the multityrosine kinase inhibitor (TKI) crizotinib, and initial data suggest that response rates are similar across METex14 skipping alteration subtypes.9 Multiple other TKIs, including capmatinib and tepotinib, have received US Food and Drug Administration (FDA) approval for NSCLC with MET exon 14 skipping alterations.10-12 Because of the numerous sites within and around exon 14 that bind the spliceosome complex, many diverse genomic variations can result in deleterious alterations that cause exon 14 skipping and ultimately disrupt CBL binding and MET downregulation.2,4 This complexity introduces challenges related to comprehensive detection of all classes of MET exon 14 skipping alterations and consistency across biomarker definitions. Given the accelerated progress of targeted therapy development, broad detection of these alterations is essential as it is the elucidation of the coalteration and immune biomarker landscape that may drive resistance to MET inhibitors or predict responsiveness to immunotherapies.
We performed a large-scale analysis of MET exon 14 skipping alterations detected using comprehensive genomic profiling (CGP) of tissue and liquid biopsies in the routine clinical care from patients with NSCLC, including characterization of coalterations, which may contribute to sensitivity or resistance to MET inhibitors.13-16 We also report tumor mutational burden (TMB) and programmed death ligand 1 (PD-L1) expression across MET exon 14-altered tissue samples as previous reports have suggested that despite elevated PD-L1 expression, patients with NSCLC harboring MET exon 14 skipping alterations may not respond well to immunotherapy (IO).17 Finally, we describe the demographics and evolution of treatment patterns of patients with MET exon 14-altered NSCLC captured in a real-world clinicogenomic database (CGDB).
METHODS
Foundation Medicine CGP
Hybrid capture–based CGP was performed on formalin-fixed paraffin-embedded tumor tissue or circulating tumor DNA (ctDNA) samples collected from 69,219 patients with primarily advanced NSCLC during routine clinical care. Testing was performed in a Clinical Laboratory Improvement Amendments-certified, College of American Pathologists-accredited, and New York State-regulated reference laboratory (Foundation Medicine Inc, Cambridge, MA). Approval for this study, including a waiver of informed consent and a Health Insurance Portability and Accountability Act waiver of authorization, was obtained from the Western Institutional Review Board (Protocol No. 20152817). For additional details, see Appendix.
Flatiron Health-Foundation Medicine Clinicogenomic Database
This study used the nationwide deidentified advanced NSCLC Flatiron Health-Foundation Medicine CGDB (FH-FMI CGDB). Retrospective longitudinal clinical data were derived from electronic health records (EHR), comprising patient-level structured and unstructured data, curated via technology-enabled abstraction, and were linked to genomic data derived from FMI CGP tests by deidentified, deterministic matching.18 During the study period, the deidentified data originated from approximately 280 cancer clinics (approximately 800 sites of care). This study included 8,614 patients who had a diagnosis of advanced NSCLC, received care within the FH network between January 2011 and March 2020, and underwent tissue CGP (FoundationOne or FoundationOne CDx) between August 2012 and March 2020. Institutional Review Board approval with waiver of informed consent was obtained before conducting the study. For additional details, see Appendix 1.
RESULTS
Foundation Medicine Genomic Database
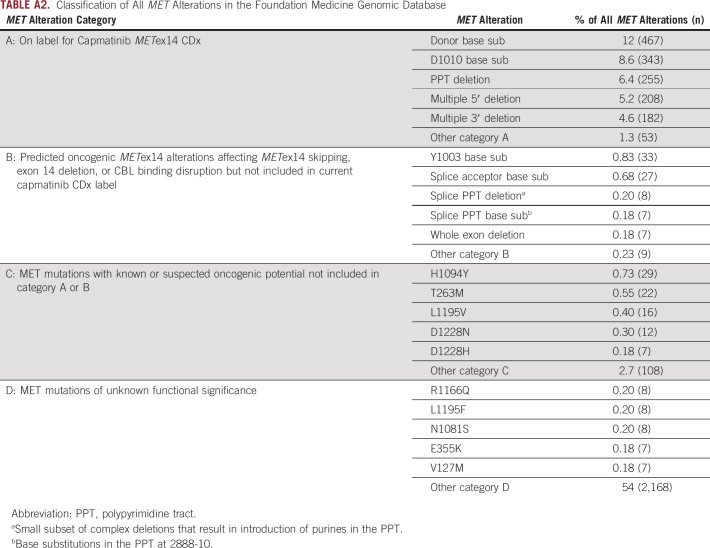
Analysis of the FMI genomic database of 69,219 NSCLC samples identified 1,599 alterations predicted to affect MET exon 14 skipping or CBL binding, herein referred to as METex14 alterations, in samples from 1,592 unique patients (2.3%). METex14 alterations were identified in 2.4% (1,458 of 60,244) of tissue specimens and 1.8% (134 of 7,468) of liquid specimens with detectable ctDNA. The median age of patients with METex14 alterations was 75 years, with 85% being ≥ 65 years, compared with a median age of 67 years in patients with NSCLC wild type (WT) for METex14 alterations (P < .0001; Table 1). Subanalysis comparing METex14 patients age ≥ 65 and < 65 years did not reveal any significant differences among sex, disease histology, or genomic characteristics (Appendix Table A1). The majority of events classified as METex14 alterations and included in this analysis are also included in the capmatinib CDx label (Appendix Fig A1, Appendix Table A2, category A, n = 1,508). However, additional variants expected to have the same functional effect but not included (eg, Y1003 CBL binding mutation or whole METex14 deletion) or observed (eg, certain rare METex14 splice variants) in the GEOMETRY trial, and therefore not covered under the CDx label, were also included in this study (category B, n = 91). MET short variant alterations (point mutations or indels) outside METex14 or within METex14 but with unresolved functional implications were excluded from this analysis (category C, n = 194). Variants of unknown significance (category D, n = 2,206) were also excluded. Overall, alterations approved under the current capmatinib CDx label represent 94% (1,508 of 1,599) of alterations predicted to activate MET through disruption of exon 14, and more generally, 84% (1,508 of 1,793) of all MET alterations with known or suspected oncogenic potential.
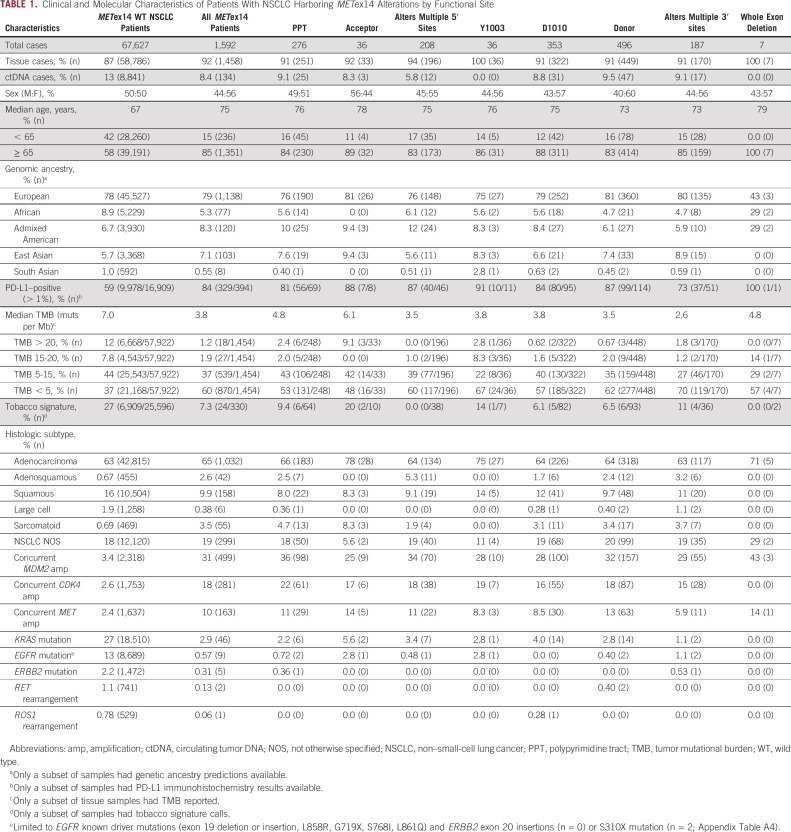
TABLE 1.
Clinical and Molecular Characteristics of Patients With NSCLC Harboring METex14 Alterations by Functional Site
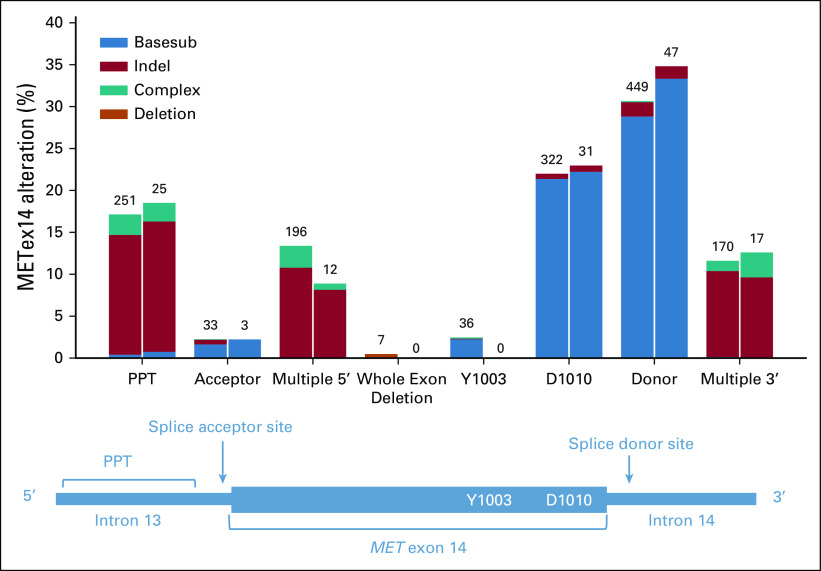
METex14 alterations were observed across exon 14 splice functional sites: donor (31%), D1010 (22%), polypyrimidine tract (PPT, 17%), multiple 5′ sites (13%), multiple 3′ sites (12%), acceptor (2.3%), at Y1003 (2.3%), and whole exon deletions (0.44%). Alterations at the donor and acceptor sites were primarily base substitutions (94% and 75%), whereas indels were more likely to occur within the PPT and multiple functional sites at the 3′ or 5′ ends of exon 14. Similar rates of detection were observed in tissue and liquid cohorts (Fig 1). Median TMB of METex14-altered NSCLC tumor samples was 3.8 mutations per megabase (muts per Mb) compared with 7.0 muts per Mb for NSCLC WT for METex14 alterations (P < .001; Fig 2A). PD-L1 expression was available for 394 (25%) METex14-altered and 16,909 (25%) METex14 WT NSCLC tumor specimens. Of these, 84% METex14-altered had a tumor proportion score (TPS) of > 1% compared with 59% of WT specimens (P < .001). Notably, the fraction of cases with low positive (1%-49%) TPS was relatively similar between WT and METex14 cohorts (29% v 23%; P = .08), whereas high positive (≥ 50%) was significantly enriched in the METex14-altered subset (30% v 60%; P < .001; Fig 2B). Neither median TMB nor PD-L1 expression significantly differed across alteration functional sites.
FIG 1.
Distribution of MET exon 14 alterations by functional site and event type. Functional site categories are shown on the x-axis aligned with the corresponding schematic showing exon 14 and flanking introns. Colored bars indicate METex14 alteration type. Deletion events (including short variants and rearrangements) resulting in complete deletion of exon 14 are indicated in orange. Left bars: METex14 events detected in tissue. Right bars: METex14 events detected in liquid. PPT, polypyrimidine tract.
FIG 2.
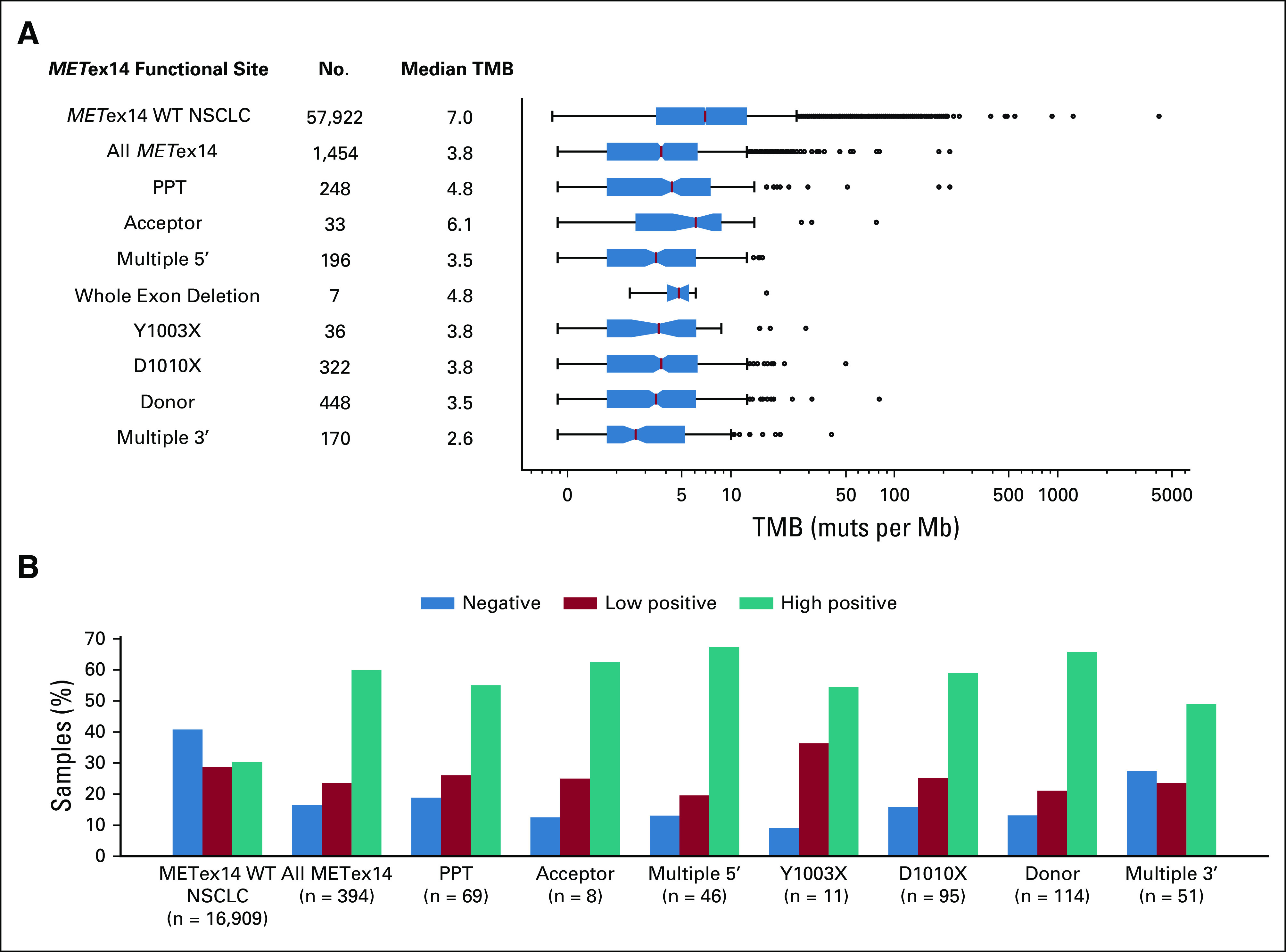
Tumor mutational burden and PD-L1 expression across METex14 alteration subtypes. (A) Distribution and median TMB (muts per Mb) and (B) PD-L1 expression by immunohistochemistry by METex14 functional site compared with NSCLC WT for METex14 alterations. PD-L1 expression was reported using the TPS: negative if the percentage of tumor cells staining with ≥ 1 + intensity was < 1%, low positive if 1%-49%, or high positive if ≥ 50%. mut, mutations; NSCLC, non–small-cell lung cancer; PD-L1, XXX; PPT, polypyrimidine tract; TMB, tumor mutational burden; TPS, tumor proportion score; WT, wild type.
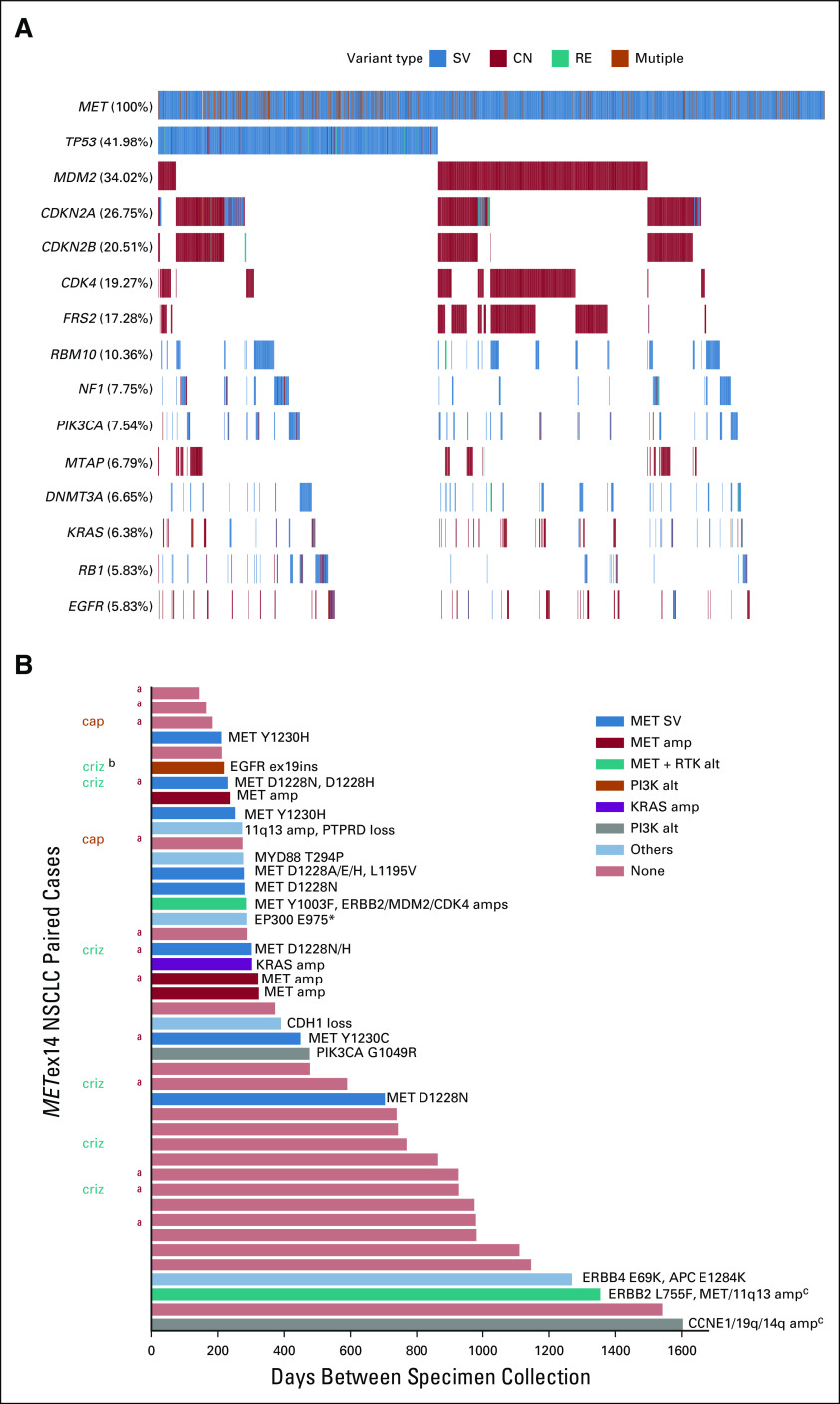
The most frequent coalterations in METex14-altered tissue specimens were TP53 alterations (42%) and MDM2 amplification (34%); co-occurring CDK4 and MET amplifications were observed in 19% and 11% of cases (Fig 3A). MDM2 amplification and TP53 alterations were largely mutually exclusive, co-occurring in just 2.7% of cases. MDM2 and CDK4, both on chromosome 12q, were coamplified in 17% of METex14-altered specimens; 90% of CDK4-amplified METex14-altered specimens had MDM2 coamplification.
FIG 3.
Co-occurring alterations in NSCLC tissue samples positive for METex14 alterations. (A) Oncoprint of co-occurring alterations in > 5% of cases. (B) Potential acquired resistance alterations in 44 paired samples. Acquired alterations are listed at the top of each bar, and bars are color coded by the category of acquired alteration. aIndicates that second paired sample was liquid biopsy. bOriginal METex14 alteration was not detected in the second sample; in one of these cases, a different METex14 alteration was acquired, but similar coalterations were observed between samples; in the other case, an EGFRex19ins was acquired postcrizotinib with different somatic coalterations suggesting potential detection of DNA from a second primary tumor in the lymph node biopsy. cGenes included on 11q13 amp include CCND1, FGF19, FGF3, and FGF4. Genes on 19q amp include CCNE1, AKT2, and AXL. Genes on 14q amp include BCL2L2, NFKBIA, and NKX2-1. amp, amplification; cap, known interim treatment with capmatinib; CN, copy number alteration; criz, known interim treatment with crizotinib; del, homozygous deletion; ins, insertion; NSCLC, non–small-cell lung cancer; RE, gene rearrangement; RTK, receptor tyrosine kinase; SV, short variant mutation.
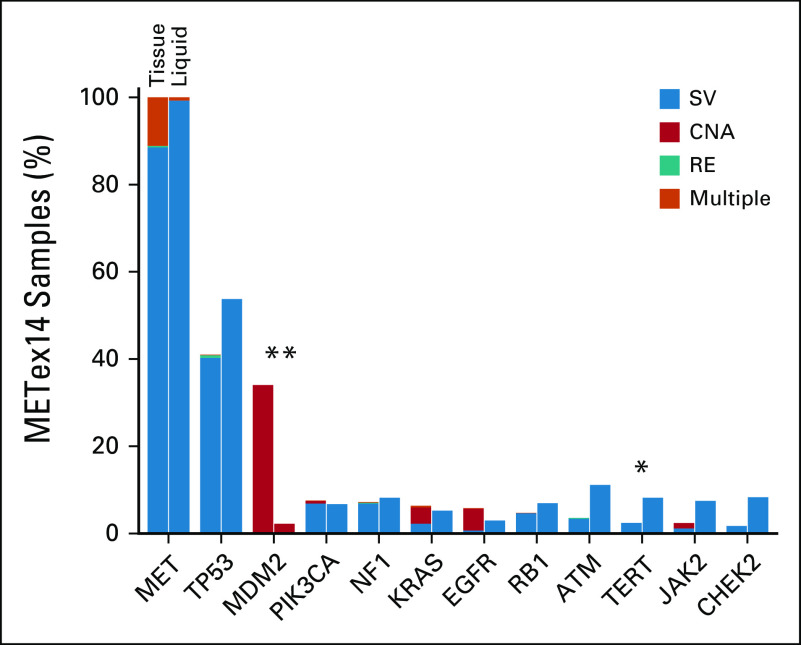
In METex14-altered NSCLC ctDNA samples, co-occurrence patterns were generally similar to tissue for genes and alterations baited across both assays; however, MDM2 amplification was less commonly detected in liquid specimens (P < .001). MET coamplification was also significantly less common in liquid versus tissue METex14-altered cases (0.75% v 11%; P = .0004; Appendix Fig A2).
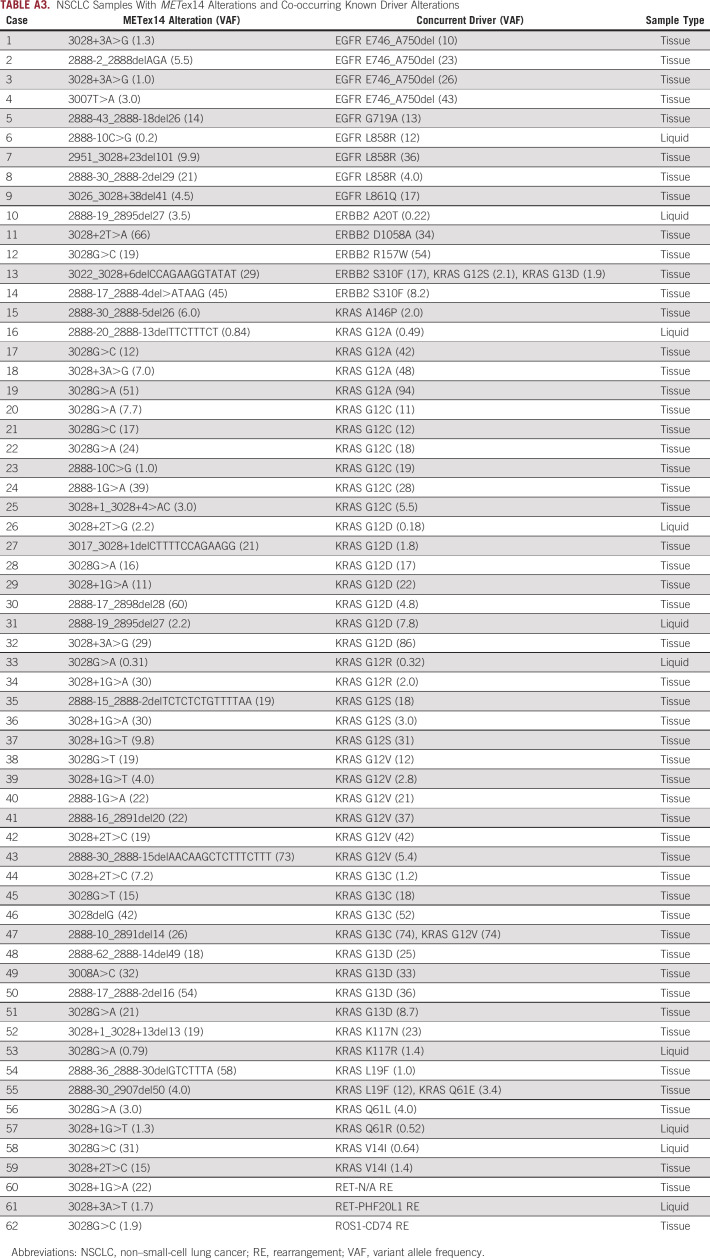
Assessing all METex14-altered samples, KRAS mutations occurred in 46 (2.9%), including 13% G12C, 52% other changes at codon G12, 20% changes at codon G13, and 15% other. Activating EGFR and ERBB2 mutations co-occurred in nine (0.57%) and five (0.31%) samples, respectively. Two samples harbored RET intron 11 rearrangements, and one harbored a CD74-ROS1 fusion (Appendix Table A3). Overall, codriver alterations were enriched in ctDNA (7.5%, 10 of 134) versus tissue (3.6% 52 of 1,458; P = .03), and the presence of a second driver may represent acquired resistance (AR) in a subset of cases. Concurrent BRAF V600E, ALK rearrangements, and NTRK fusions were not observed.
Samples from the same patient collected ≥ 60 days apart (median 372 days, range 144-1,603) with a METex14 alteration in the primary sample were available for 43 patients including tissue-tissue (n = 29), tissue-ctDNA (n = 9), ctDNA-tissue (n = 1), and ctDNA-ctDNA pairs (n = 4). In total, 42 of 43 patients (98%) retained the primary METex14 alteration, and 22 of 43 patients (51%) had reportable acquired alteration(s) detected. At least one secondary MET mutation (Y1230C/H, D1228A/E/N/H, L1195V, and Y1003F) was acquired in nine specimens, with three specimens (one tissue and two ctDNA) acquiring ≥ 2 MET mutations. Four samples had acquired MET amplification (7-13 copies). Other acquired alterations included known oncogenic alterations in EGFR, ERBB2, KRAS, and AKT2 as well as alterations in other genes with likely significance in cancer, but with unestablished roles in AR. Acquired alterations, including secondary MET mutations, were observed across METex14 alteration subtypes. Although treatment history was not available for most cases, eight patients with multiple samples were confirmed to have had interim treatment with crizotinib or capmatinib (Fig 3B).
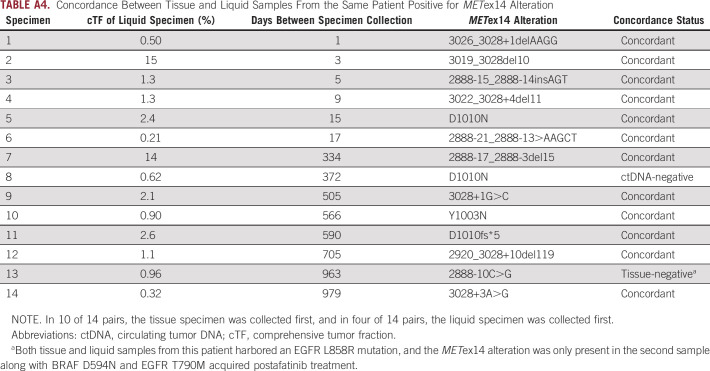
Fourteen patients with METex14-alterations had both tissue and ctDNA samples available for concordance analysis (median 353 days between specimen collection, range 1-979; Appendix Table A4). Positive percent agreement with the tissue biopsy was 92% over all pairs and 100% for pairs collected within 1 year. In 2 of 14 discordant pairs (14%), one lacked METex14 alteration in a ctDNA specimen with a low estimated composite tumor fraction (0.62%), and in the other, the primary tissue specimen harbored EGFR L858R while the ctDNA specimen collected post-afatinib treatment harbored multiple heterogenous AR mechanisms including a METex14 skipping alteration.
Cases With Multiple METex14 Alterations
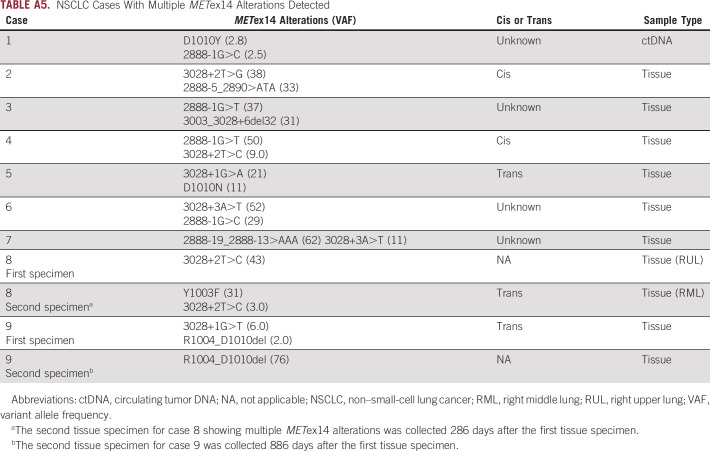
Among 1,592 METex14-altered NSCLC samples, nine samples (eight tissue samples and one ctDNA sample) harbored multiple METex14 alterations (Appendix Table A5). In five cases, on the basis of proximity, the alterations were able to be assessed for cis and trans status and two were in cis and three were in trans. In one case, a single METex14 alteration (43% variant allele frequency [VAF]) was detected in a primary lung sample, and then a second lung biopsy collected 286 days later revealed the initial donor site alteration (3% VAF) and acquired MET Y1003F (31% VAF) in trans.
CGDB Treatment Patterns and Outcomes Analysis
Of 6,439 patients with advanced NSCLC in the CGDB meeting criteria for assessment, METex14 alterations were detected in 148 (2.3%) cases (Appendix Fig A3). Clinical characteristics, genomic characteristics, and therapeutic regimens are listed in Table 2. Fifty-three patients began first-line therapy after the CGP report: 24 (45%) on an IO-containing regimen without MET TKI, 13 (25%) on the chemotherapy-containing regimen without IO or MET TKI, 11 (21%) on MET TKI (crizotinib or cabozantinib)-containing regimen, and five (9.4%) with an unspecified clinical study drug that may have included a MET TKI (Fig 4A). For comparison, 61 patients began first-line therapy before the CGP report: 35 (57%) on the chemo-containing regimen, 20 (33%) on the IO-containing regimen, three (4.9%) on the MET TKI-regimen, two (3.3%) on erlotinib (EGFR TKI), and one (1.6%) on an unspecified clinical study drug (Fig 4A). For the three patients who started MET TKI before CGP, therapy was initiated less than a month before the report, so we suspect that a preliminary report may have influenced the treatment decision. Similar patterns were observed for second-line therapy pre- and post-CGP reports.
TABLE 2.
Treatment Patterns and Clinicogenomic Characteristics of Patients With NSCLC Harboring METex14 Alterations' Patients in the FMI-FH CGDB
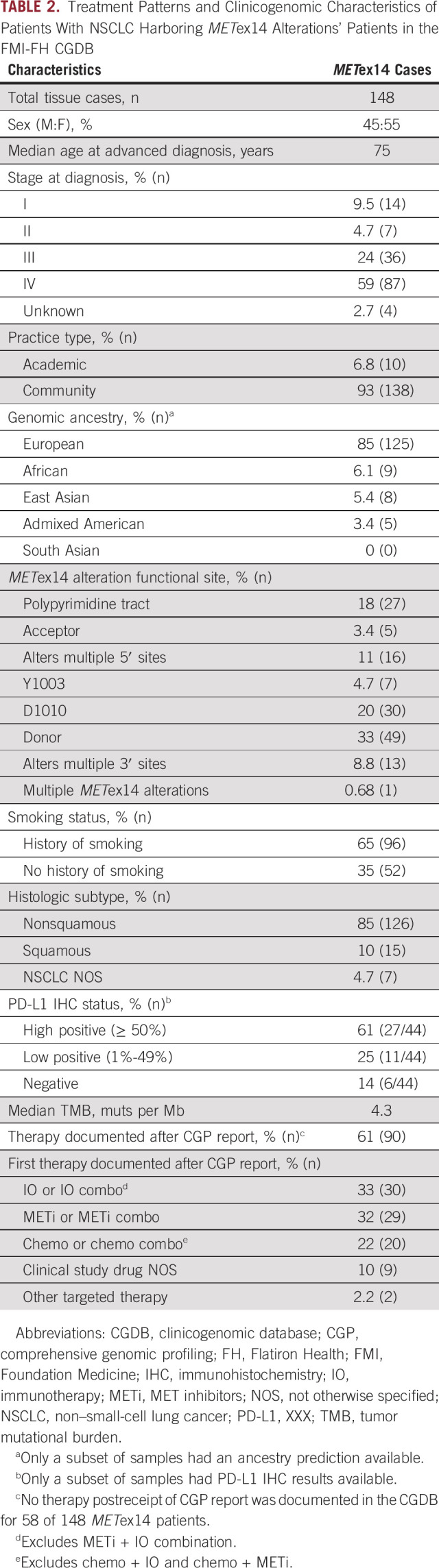
FIG 4.
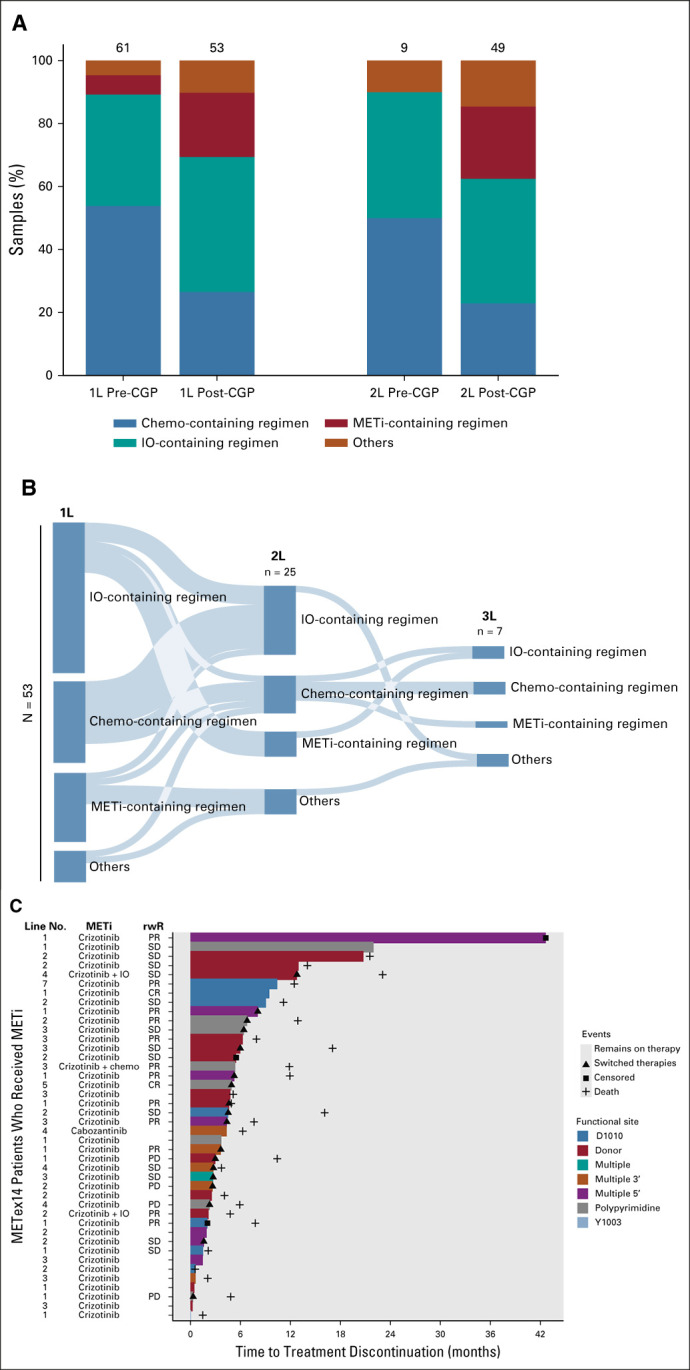
Real-world treatment patterns of patients with METex14-altered NSCLC in the CGDB. (A) Distribution of 1L and 2L therapies before and after CGP for 114 METex14-altered NSCLC who started treatment after CGP biopsy. All patients captured started 1L therapy between 2012 and 2020. (B) Sankey diagram of 53 patients with METex14-altered NSCLC with documented receipt of 1L therapy in the CGDB after report of CGP results. All patients captured started 1L therapy post-CGP report between 2015 and 2020. Categorical groupings include (1) METi-containing regimen: MET TKI monotherapy, MET TKI + chemotherapy, and MET TKI + IO, (2) IO-containing regimen: IO monotherapy, IO + chemotherapy, IO + clinical study drug, or IO + other targeted therapy, (3) chemo-containing regimen: chemotherapy monotherapy, chemotherapy + clinical study drug, or chemotherapy + other targeted therapy, and (4) other: non-MET targeted therapy (erlotinib, aftatinib, ceretinib, or bevacizumab) or unspecified clinical study drug, which may have included a MET TKI. Six patients who received MET TKI in the fourth to seventh lines are not captured here but are shown in (C). (C) Swimmer plot displaying time to treatment discontinuation for patients with METex14-altered NSCLC who received MET TKI. rwR was available for a subset of patients. 1L, first line; 2L, second line; CGP, comprehensive genomic profiling; CR, complete response; IO, immunotherapy; NSCLC, non–small-cell lung cancer; METi, MET inhibitors; PD, progressive disease; PR, partial response; rwR, real-world response; SD, stable disease; TKI, tyrosine kinase inhibitor.
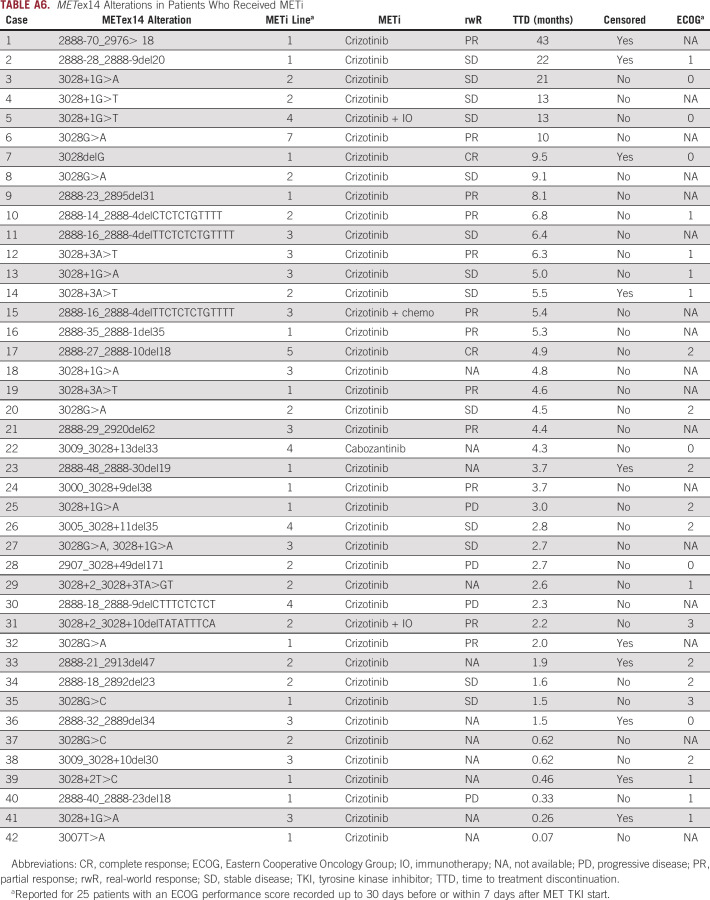
Of patients who received a MET TKI first-line post-CGP report, five of 11 patients (45%) went on to receive second-line therapy; however, four of 11 patients remained on first-line therapy at the time of analysis (Fig 4B). Overall, 42 of 148 METex14 patients had documented receipt of MET TKI post-CGP biopsy (primarily crizotinib monotherapy) at some point during their treatment history, distributed across first-seventh lines. For 31 patients with available data, the real-world response (rwR) rate was 45% (2 complete response and 12 partial response) and median time to therapy discontinuation (TTD) on MET TKI was 4.4 months, with seven patients still on therapy at the time of analysis (Fig 4C, Appendix Table A6).
DISCUSSION
We assessed the largest cohort to date of 1,592 METex14-altered NSCLC samples. The METex14 alteration frequency observed here (2.3%) is similar to previously published studies and shows a diverse spectrum of alterations.1-3 The large size of this landmark analysis allowed for high-resolution confirmation of frequencies of coalterations and IO biomarkers reported in previous smaller studies. We identified rare cases with co-occurring drivers, including 1.9% with KRAS mutations at G12, which may represent AR. In available paired cases, a subset of whom had documented interim treatment with crizotinib or capmatinib, the majority had reportable acquired alterations detected, including secondary MET resistance mutations, MET amplification, and alterations in EGFR, ERBB2, KRAS, and the PI3K pathway, as well as other acquired alterations potentially relevant in cancer but not previously described as AR mechanisms to MET inhibitors.13-16 In 49% of paired cases, no acquired alterations were reported, suggesting either a yet to be identified mechanism driving resistance, an AR mechanism present but below the limit of detection, or that the second sample may not have been collected at the point of AR to MET inhibition. Interestingly, identification of acquired alterations was less common when the samples were collected > 2 years apart. Multiple heterogenous AR mechanisms were observed in both tissue and liquid samples and were not more frequent in ctDNA in our cohort of paired samples, although in nonpaired samples, codrivers were more commonly detected with METex14 in ctDNA vs tissue. Cases with multiple drivers may be sensitive to combination treatment with approved and investigational therapies now available targeting receptor tyrosine kinases and KRAS G12C in NSCLC.
Assessment of IO biomarkers confirmed that PD-L1 expression is elevated in METex14-altered samples; however, clinical data have suggested that despite elevated PD-L1 expression, patients with METex14-altered NSCLC have low response rates to IO and responses are not correlated with PD-L1 expression, which is consistent with low median TMB observed in METex14-altered samples.17,19
METex14 skipping alterations are currently the only MET alterations associated with FDA-approved therapies and a companion diagnostic in NSCLC.11 However, METex14 skipping alterations that are on-label for capmatinib represent 84% (Appendix Fig A1) of MET short variant alterations with known or suspected oncogenic potential in the FMI database, and 94% of METex14 variants predicted to result in activation through disruption of exon 14 or CBL binding.20 It is worth noting that mutations surrounding the CBL binding site at positions D1002 and R1004-E1009 may affect CBL binding and confer sensitivity to MET TKIs similar to Y1003 mutation; however, these alterations are still relatively uncharacterized and thus excluded from primary analysis in this study.21 Ultimately, variation in how METex14 biomarkers are defined may lead to confusion in terms of actionability, highlighting the need for standardization.
Given the diversity of METex14 alterations, distinguishing accurate methods for detection can be challenging. A variety of DNA- and RNA-based methods are clinically available for METex14 alteration detection, but there is only one FDA-approved CDx. Some DNA-based platforms have incomplete coverage of regions where alteration is known to alter METex14 skipping, such as the splice acceptor site, so critical assessment of available diagnostics is essential for comprehensive detection.22 RNA profiling is also an emerging methodology to detect exclusion of exon 14 from the mRNA. In rare cases, RNA profiling may detect variants not detected via DNA sequencing; however, the FoundationOne CDx DNA assay was shown in the GEOMETRY trial to be highly concordant with RNA sequencing methods.11 Furthermore, RNA profiling would not detect mutations at the CBL binding site and would not distinguish distinct METex14 alterations in DNA, which we report to occur in the same patient in rare instances.
Comparing tissue and liquid cohorts, METex14 alterations were less commonly detected in ctDNA, which may be due in part to practice testing patterns. We also saw decreased frequency of coamplifications in the ctDNA cohort, which is generally known regarding amplification detection in ctDNA,23 and should be taken into consideration as the functional and therapeutic significance of these coamplifications is further elucidated. In a small cohort of 14 cases with tissue and liquid samples collected from the same patient, concordance was high for METex14 detection (100% positive percent agreement for samples collected < 1 year apart). In the VISION trial, similar outcomes to tepotinib were reported for cases with METex14 alterations detected in tissue or ctDNA, further supporting the utility of either method for detecting these alterations.12
In very rare cases, we saw multiple distinct METex14 skipping alterations in the same tissue (n = 8) or ctDNA (n = 1) sample. Both cis and trans relationships between the two METex14 events were observed, and VAF relationships for METex14 events were also variable. Unfortunately, clinical histories were not available for most of these cases, and it remains unclear whether these cases typically represent multiple primaries, AR, or another phenomenon. However, in a separate NSCLC case, two tissue samples determined to have been collected from multiple primary tumors in the same patient each harbored a distinct METex14 alteration (Mark Awad, ASCO 2020 Oral Presentation).
Using the CGDB, we observed higher MET inhibitor usage post-CGP report compared with before CGP was performed (39% v 4.9%), although off-label use of MET inhibitors was still a minority presumably due to lack of physician confidence in crizotinib efficacy, barriers to access, or preference for IO on the basis of positive PD-L1 staining. We plan to investigate timing of receipt of PD-L1 IHC results and METex14 CGP results, and potential impact on therapy selection in follow-up studies. Compared with results reported for the PROFILE 1001 trial,9 the rwR rate of 45% seen here was somewhat higher than the 32% ORR reported, although limitations to interpretation of rwR are noted below. Conversely, PFS was not available in the CGDB, but the median TTD of 4.4 months seen here was shorter than the 7.3 month median PFS reported on trial. This could be due to a number of reasons including (1) for our TTD analysis, seven patients still remained on MET TKI at the time of data cutoff, (2) patients treated in the RW setting likely had worse performance status on average, and (3) differences between TTD and PFS as noted below.
Limitations of this study include lack of clinical history and treatment information for most patients in the FMI genomic database. For the CGDB cohort, clinical data were derived from EHR and data not documented in the EHR may be incomplete or missing, particularly for events occurring outside of the FH network. In particular, rwR and TTD were retrospectively captured from EHR, which differs from prospective collection of progression data within the context of a clinical trial. TTD is also used as a proxy for PFS, and patients may have discontinued drug for reasons other than disease progression. Furthermore, all patients in this study received CGP, which may introduce selection bias for patients treated by physicians with distinct practice patterns. Data were collected over a period where the MET inhibitor treatment landscape was rapidly evolving, and outcomes data are biased toward off-label crizotinib, which was available before newer specific MET inhibitors were developed.
Overall, we assessed available characteristics for more than 1,500 METex14-altered NSCLC cases, including the landscape of coalterations that may modulate response to targeted and immunotherapies. We also compared genomics of tissue and liquid cohorts and observed high concordance for METex14 alteration detection in a small cohort of cases, suggesting that both tissue and liquid CGP have utility in detecting METex14 alterations, although coamplifications may be better assessed in tissue. These data and continued use of CGP to characterize this patient population will be useful to predict response to MET inhibitors and may be imperative for the selection of effective combination therapies.
APPENDIX 1. SUPPLEMENTAL MATERIALS
Foundation Medicine Comprehensive Genomic Profiling
As previously described, for 60,244 tumor tissue specimens, DNA (> 50 ng) was extracted from formalin-fixed paraffin-embedded specimens and next-generation sequencing was performed by hybridization-captured, adapter ligation-based libraries to high, uniform coverage (> 500×) for all coding exons of 187-324 cancer-related genes plus selected introns.17 For 8,975 liquid samples, plasma was isolated from 20 mL of peripheral whole blood and ≥ 20 ng of circulating tumor DNA (ctDNA) was extracted to create adapted sequencing libraries for coding exons of 60 or 70 genes before hybrid capture and sample-multiplexed sequencing.24 Results were analyzed for base substitutions (subs), short insertions and deletions (indels), copy number gains or losses, and rearrangements; copy losses were not reported for liquid samples included in this study. For all assay versions, all coding exons of MET were baited, but there was no dedicated baiting of MET introns.
Tumor mutational burden was defined as the number of somatic mutations per megabase (mb) postfiltering to remove known somatic and deleterious mutations. The total number of mutations was divided by the coding region target territory of the assay to calculate the mutation burden per mb.18
PD-L1 expression was determined by immunohistochemistry performed on tissue sections using the 22C3 (Dako) PD-L1 antibody. PD-L1 expression was reported using the tumor proportion score (TPS), where TPS = No. of PD-L1–positive tumor cells/(total No. of PD-L1–positive + PD-L1–negative tumor cells). The TPS result was stratified into negative (< 1%), low positive (1%-49%), and high positive (≥ 50%).
For liquid biopsy specimens, composite tumor fraction (cTF) was used to estimate the ctDNA fraction. cTF leverages two complementary methods, tumor fraction (TF) estimate and maximum somatic allele frequency (MSAF). When ctDNA fraction is higher, a TF estimate is calculated on the basis of a measure of tumor aneuploidy that incorporates observed deviations in coverage across the genome for a given sample. Calculated values for this metric are calibrated against a training set on the basis of samples with well-defined tumor fractions to generate an estimate of TF. When ctDNA content is lower, MSAF is determined by calculating the allele fraction for all known somatic, likely somatic, and variant of unknown significance base substitutions, excluding certain common and rare germline variants. The cTF is based on the TF estimate when available and is generated from MSAF when lack of tumor aneuploidy limits the ability to return an informative estimate of TF.
Genomic ancestry was determined for each individual. Single-nucleotide polymorphisms targeted by each of our comprehensive genomic profiling (CGP) tissue tests were superimposed with phase III 1000 genomes' data. Using an established approach, we projected the single-nucleotide polymorphisms down to the top five principal components and used random forest ensemble learning to train a classifier on each profiling test. Classifiers were trained to recognize five general ancestries: African (AFR), admixed American (AMR), East Asian (EAS), European (EUR), and South Asian (SAS).24
Flatiron Health-Foundation Medicine Clinicogenomic Database
Patients who were diagnosed with metastatic disease > 90 days before their first visit within the Flatiron Health (FH) network or who received their Foundation Medicine report > 60 days after their last FH visit date were excluded to ensure all therapies received before CGP were captured and to exclude patients who left the FH network before CGP. This left 6,439 unique patients eligible for this study. Clinical characteristics and treatment information were obtained via technology-enabled abstraction of clinical notes and radiology and pathology reports and linked to CGP data.
Statistical Analysis and Real-World End Points
The Fisher exact test was used to assess significance of categorical relationships, and the Kruskal-Wallis test was used to assess significance of continuous variables. False discovery rate correction was performed by the Benjamini-Hochberg procedure to correct P values for multiple tests.
FIG A1.
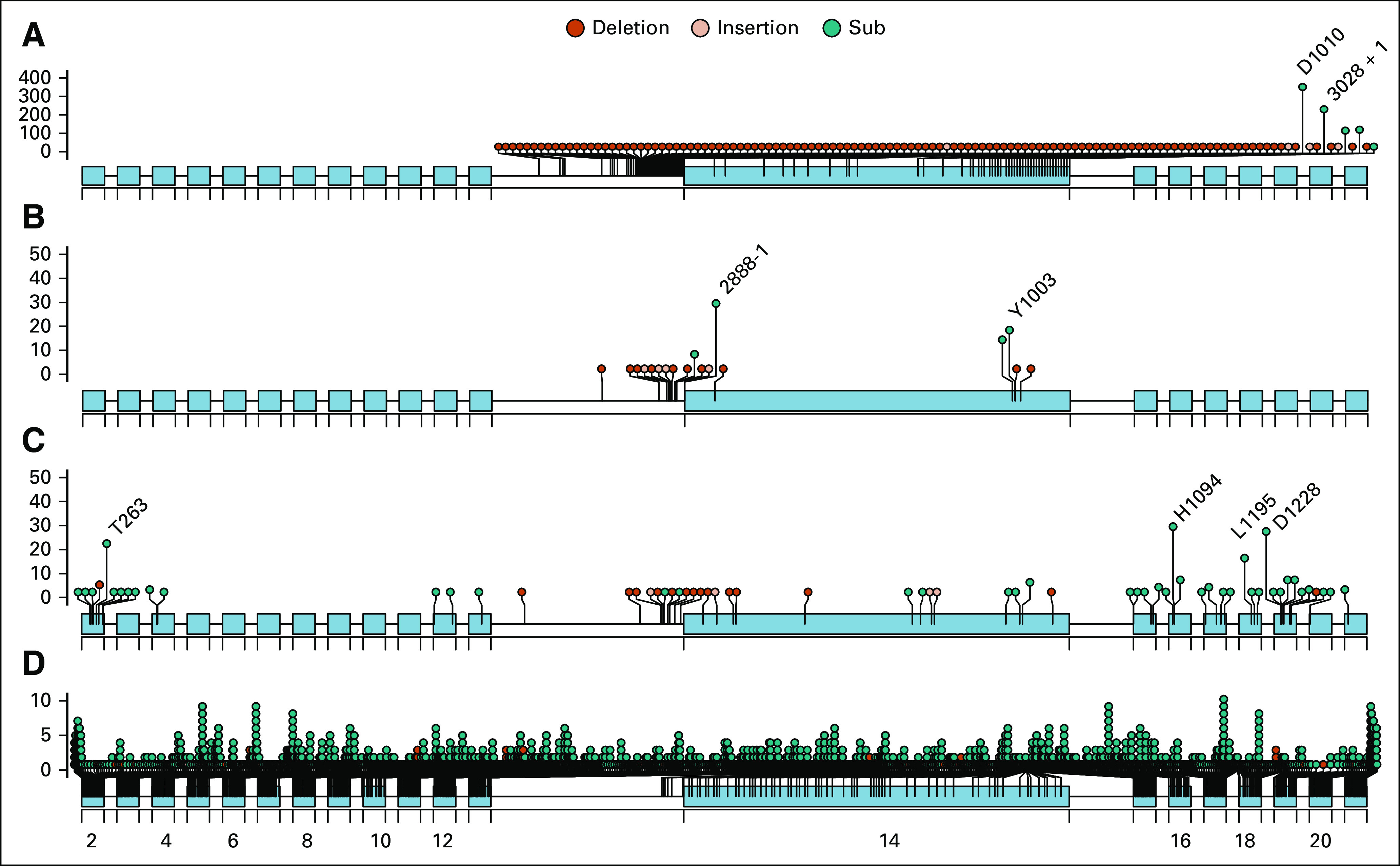
Lollipop diagram of all MET alterations identified in the Foundation Medicine genomic database. (A) On label for capmatinib METex14 CDx. (B) Alteration affecting METex14 skipping, deletion, or CBL binding disruption but not included in current capmatinib CDx label. (C) MET SV (base substitution or indel) alteration with known or likely functional significance not included in category A or B. (D) MET SV of unknown functional significance. SV, short variant.
FIG A2.
Co-occurring alterations in METex14-positive tissue and liquid specimens. Analysis is limited to genes and alterations baited across both assays. Genes altered in > 5% of samples in either cohort are shown. Homozygous losses were not reported in the majority of liquid specimens and were excluded. Cases with multiple MET alterations were primarily those with METex14 skipping alteration and co-MET amplification. *q < 0.05; **q < 0.001. CNA, copy number alteration; RE, rearrangement; SV, short variant.
FIG A3.
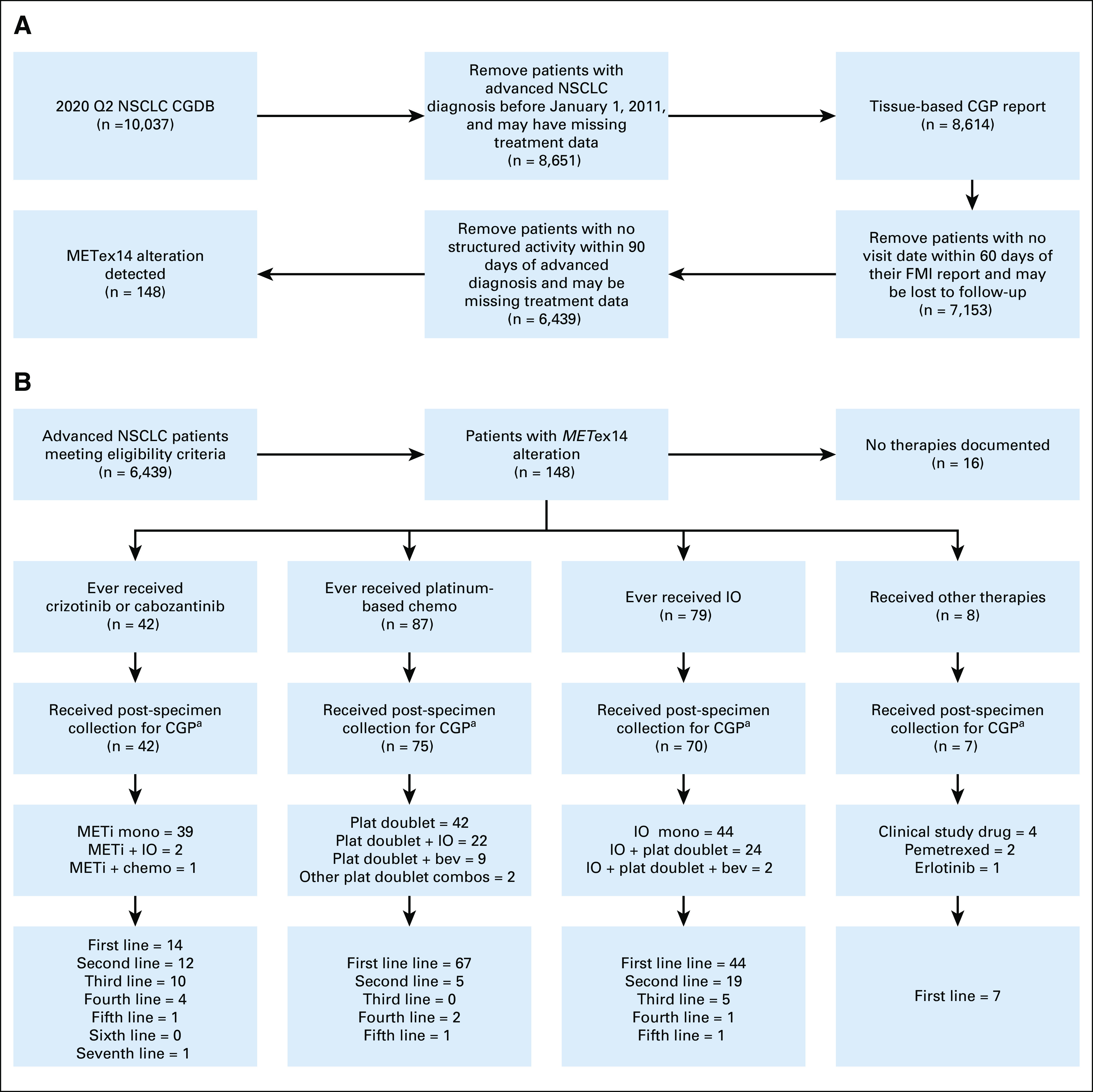
Cohort selection and treatment patterns for patients with METex14-altered NSCLC available for assessment in the CGDB: (A) cohort eligibility diagram and (B) treatment patterns diagram. aOf patients who received therapy postspecimen collection, 37 of 42 METi-treated, 24 of 75 platinum chemotreated, and 40 of 70 IO-treated patients began therapy post-CGP report receipt. CGDB, clinicogenomic database; CGP, comprehensive genomic profiling; FMI, Foundation Medicine; IO, immunotherapy; METi, MET inhibitors; NSCLC, non–small-cell lung cancer.
TABLE A1.
Genomic Comparison Between Patients Age < 65 and ≥ 65 Years With METex14-Altered NSCLC
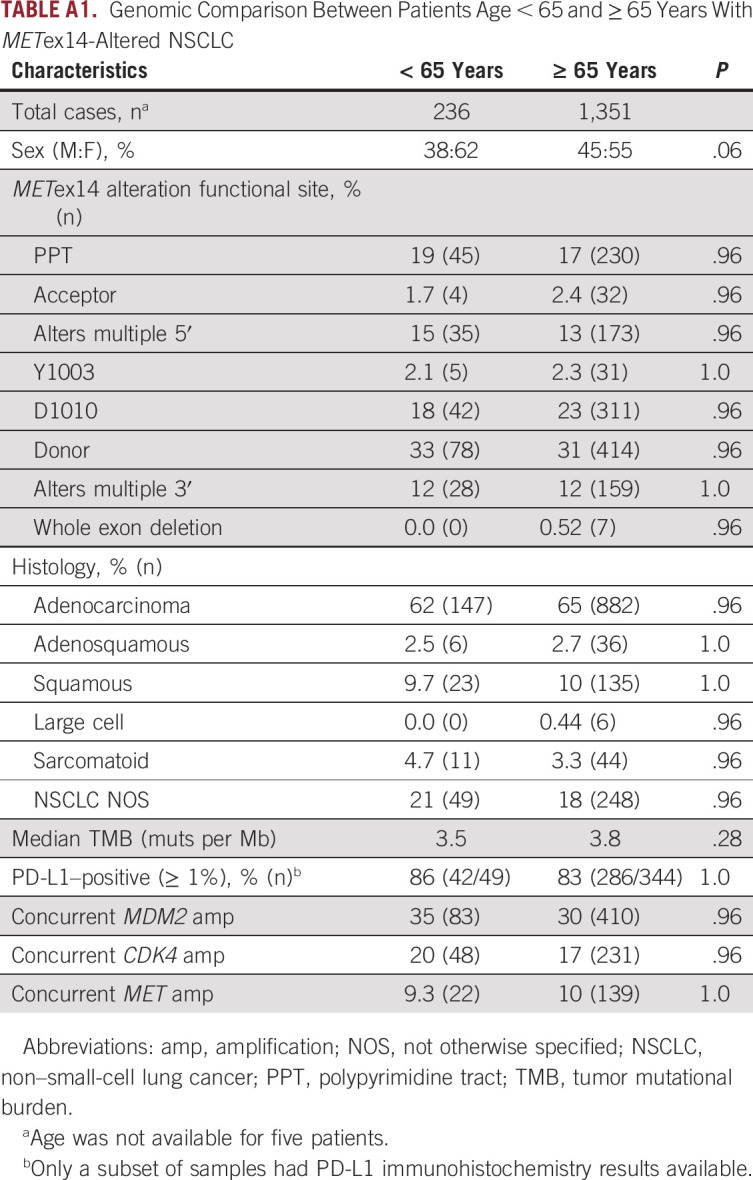
TABLE A2.
Classification of All MET Alterations in the Foundation Medicine Genomic Database
TABLE A3.
NSCLC Samples With METex14 Alterations and Co-occurring Known Driver Alterations
TABLE A4.
Concordance Between Tissue and Liquid Samples From the Same Patient Positive for METex14 Alteration
TABLE A5.
NSCLC Cases With Multiple METex14 Alterations Detected
TABLE A6.
METex14 Alterations in Patients Who Received METi
Jessica K. Lee
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Russell Madison
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Anthony Classon
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Ole Gjoerup
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Mark Rosenzweig
Employment: Foundation Medicine
Stock and Other Ownership Interests: Foundation Medicine
Garrett M. Frampton
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Brian M. Alexander
Employment: Foundation Medicine
Leadership: Foundation Medicine
Stock and Other Ownership Interests: Roche
Research Funding: Lilly, Puma Biotechnology, Celgene
Open Payments Link: https://openpaymentsdata.cms.gov/physician/854258/summary
Geoffrey R. Oxnard
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Honoraria: Guardant Health, Foundation Medicine
Consulting or Advisory Role: AstraZeneca, Inivata, Takeda, LOXO, DropWorks, GRAIL, Janssen, Sysmex, Illumina, AbbVie, Merck
Patents, Royalties, Other Intellectual Property: DFCI has a patent pending titled “Non-invasive blood-based monitoring of genomic alterations in cancer,” on which I am a coauthor
Jeffrey M. Venstrom
Employment: Foundation Medicine, Genentech/Roche
Leadership: Foundation Medicine
Stock and Other Ownership Interests: Roche
Consulting or Advisory Role: Synkrino Biotherapeutics
Travel, Accommodations, Expenses: Foundation Medicine, Genentech/Roche
Mark M. Awad
Consulting or Advisory Role: Genentech, Merck, Pfizer, Boehringer Ingelheim, AbbVie, AstraZeneca/MedImmune, Clovis Oncology, Nektar, Bristol Myers Squibb, ARIAD, Foundation Medicine, Syndax, Novartis, Blueprint Medicines, Maverick Therapeutics, Achilles Therapeutics, Neon Therapeutics, Hengrui Therapeutics, Gritstone Oncology, Archer, Mirati Therapeutics, NextCure, EMD Serono, AstraZeneca, Panasonic, Instil Bio
Research Funding: Genentech/Roche, Lilly, AstraZeneca, Bristol Myers Squibb
Alexa B. Schrock
Employment: Foundation Medicine
Stock and Other Ownership Interests: Foundation Medicine, Roche
No other potential conflicts of interest were reported.
PRIOR PRESENTATION
Presented in part at ASCO 2020 oral presentation (Awad et al, abstr 9511).
AUTHOR CONTRIBUTIONS
Conception and design: Jessica K. Lee, Garrett M. Frampton, Alexa B. Schrock
Financial support: Brian M. Alexander
Administrative support: Brian M. Alexander
Collection and assembly of data: Garrett M. Frampton, Alexa B. Schrock
Data analysis and interpretation: All authors
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by the authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Jessica K. Lee
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Russell Madison
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Anthony Classon
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Ole Gjoerup
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Mark Rosenzweig
Employment: Foundation Medicine
Stock and Other Ownership Interests: Foundation Medicine
Garrett M. Frampton
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Brian M. Alexander
Employment: Foundation Medicine
Leadership: Foundation Medicine
Stock and Other Ownership Interests: Roche
Research Funding: Lilly, Puma Biotechnology, Celgene
Open Payments Link: https://openpaymentsdata.cms.gov/physician/854258/summary
Geoffrey R. Oxnard
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Honoraria: Guardant Health, Foundation Medicine
Consulting or Advisory Role: AstraZeneca, Inivata, Takeda, LOXO, DropWorks, GRAIL, Janssen, Sysmex, Illumina, AbbVie, Merck
Patents, Royalties, Other Intellectual Property: DFCI has a patent pending titled “Non-invasive blood-based monitoring of genomic alterations in cancer,” on which I am a coauthor
Jeffrey M. Venstrom
Employment: Foundation Medicine, Genentech/Roche
Leadership: Foundation Medicine
Stock and Other Ownership Interests: Roche
Consulting or Advisory Role: Synkrino Biotherapeutics
Travel, Accommodations, Expenses: Foundation Medicine, Genentech/Roche
Mark M. Awad
Consulting or Advisory Role: Genentech, Merck, Pfizer, Boehringer Ingelheim, AbbVie, AstraZeneca/MedImmune, Clovis Oncology, Nektar, Bristol Myers Squibb, ARIAD, Foundation Medicine, Syndax, Novartis, Blueprint Medicines, Maverick Therapeutics, Achilles Therapeutics, Neon Therapeutics, Hengrui Therapeutics, Gritstone Oncology, Archer, Mirati Therapeutics, NextCure, EMD Serono, AstraZeneca, Panasonic, Instil Bio
Research Funding: Genentech/Roche, Lilly, AstraZeneca, Bristol Myers Squibb
Alexa B. Schrock
Employment: Foundation Medicine
Stock and Other Ownership Interests: Foundation Medicine, Roche
No other potential conflicts of interest were reported.
REFERENCES
- 1.Schrock AB, Frampton GM, Suh J, et al. Characterization of 298 patients with lung cancer harboring MET Exon 14 skipping alterations J Thorac Oncol 111493–15022016 [DOI] [PubMed] [Google Scholar]
- 2.Frampton GM, Ali SM, Rosenzweig M, et al. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors Cancer Discov 5850–8602015 [DOI] [PubMed] [Google Scholar]
- 3.Awad MM, Oxnard GR, Jackman DM, et al. MET exon 14 mutations in non-small-cell lung cancer are associated with advanced age and stage-dependent MET genomic amplification and c-Met overexpression J Clin Oncol 34721–7302016 [DOI] [PubMed] [Google Scholar]
- 4.Kong-Beltran M, Seshagiri S, Zha J, et al. Somatic mutations lead to an oncogenic deletion of Met in lung cancer Cancer Res 66283–2892006 [DOI] [PubMed] [Google Scholar]
- 5.Onozato R, Kosaka T, Kuwano H, et al. Activation of MET by gene amplification or by splice mutations deleting the juxtamembrane domain in primary resected lung cancers J Thorac Oncol 45–112009 [DOI] [PubMed] [Google Scholar]
- 6.Comoglio PM, Trusolino L, Boccaccio C.Known and novel roles of the MET oncogene in cancer: A coherent approach to targeted therapy Nat Rev Cancer 18341–3582018 [DOI] [PubMed] [Google Scholar]
- 7.Peschard P, Fournier TM, Lamorte L, et al. Mutation of the c-Cbl TKB domain binding site on the Met receptor tyrosine kinase converts it into a transforming protein Mol Cel 8995–10042001 [DOI] [PubMed] [Google Scholar]
- 8.van der Steen N, Giovannetti E, Pauwels P, et al. CMET exon 14 skipping: From the structure to the clinic J Thorac Oncol 111423–14322016 [DOI] [PubMed] [Google Scholar]
- 9.Drilon A, Clark JW, Weiss J, et al. Antitumor activity of crizotinib in lung cancers harboring a MET exon 14 alteration Nat Med 2647–512020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu S, Fang J, Cao L, et al. Preliminary efficacy and safety results of savolitinib treating patients with pulmonary sarcomatoid carcinoma (PSC) and other types of non-small cell lung cancer (NSCLC) harboring MET exon 14 skipping mutations. Cancer Res. 2019;79 abstr CT031. [Google Scholar]
- 11.Wolf J, Seto T, Han JY, et al. Capmatinib in MET exon 14-mutated or MET-amplified non-small-cell lung cancer N Engl J Med 383944–9572020 [DOI] [PubMed] [Google Scholar]
- 12.Paik PK, Felip E, Veillon R, et al. Tepotinib in non–small-cell lung cancer with MET exon 14 skipping mutations N Engl J Med 383931–9432020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzawa K, Offin M, Lu D, et al. Activation of KRAS mediates resistance to targeted therapy in MET exon 14–mutant non–small cell lung cancer Clin Cancer Res 251248–12602019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ou SHI, Young L, Schrock AB, et al. Emergence of preexisting MET Y1230C mutation as a resistance mechanism to crizotinib in NSCLC with MET exon 14 skipping J Thorac Oncol 12137–1402017 [DOI] [PubMed] [Google Scholar]
- 15.Recondo G, Bahcall M, Spurr LF, et al. Molecular mechanisms of acquired resistance to MET tyrosine kinase inhibitors in patients with MET exon 14–mutant NSCLC Clin Cancer Res 262615–26252020 [DOI] [PubMed] [Google Scholar]
- 16.Rotow JK, Gui P, Wu W, et al. Co-occurring alterations in the RAS–MAPK pathway limit response to MET inhibitor treatment in MET exon 14 skipping mutation-positive lung cancer Clin Cancer Res 26439–4492020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sabari JK, Leonardi GC, Shu CA, et al. PD-L1 expression, tumor mutational burden, and response to immunotherapy in patients with MET exon 14 altered lung cancers Ann Oncol 292085–20912018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Birnbaum B, Nussbaum N, Seidl-Rathkopf K, et al. Model-assisted cohort selection with bias analysis for generating large-scale cohorts from the EHR for oncology research. 2020 http://arxiv.org/abs/2001.09765
- 19.Calles A, Riess JW, Brahmer JR.Checkpoint blockade in lung cancer with driver mutation: Choose the road wisely Am Soc Clin Oncol Ed Book 40372–3842020 [DOI] [PubMed] [Google Scholar]
- 20.Miao YL, Xu QQ.MET Y1003S point mutation shows sensitivity to crizotinib in a patient with lung adenocarcinoma Lung Cancer 13084–862019 [DOI] [PubMed] [Google Scholar]
- 21.Chu LP, Franck D, Parachoniak CA, et al. MET genomic alterations in head and neck squamous cell carcinoma (HNSCC): Rapid response to crizotinib in a patient with HNSCC with a novel MET R1004G mutation Oncologist 241305–13082019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jurkiewicz M, Saqi A, Mansukhani MM, et al. Efficacy of DNA versus RNA NGS-based Methods in MET Exon 14 skipping mutation detection. J Clin Oncol. 2020;38 suppl; abstr 9036. [Google Scholar]
- 23.Schrock AB, Welsh A, Chung JH, et al. Hybrid capture–based genomic profiling of circulating tumor DNA from patients with advanced non–small cell lung cancer J Thorac Oncol 14255–2642019 [DOI] [PubMed] [Google Scholar]
- 24.Newberg J, Connelly C, Frampton G. Determining patient ancestry based on targeted tumor comprehensive genomic profiling. Cancer Res. 2019;79 suppl; abstr 1599. [Google Scholar]