Abstract
Phase contrast techniques, such as differential interference contrast (DIC) microscopy, are widely used to provide morphological images of unstained biological samples. The trans-illumination geometry required for these techniques restricts their application to thin samples. We introduce oblique back-illumination microscopy (OBM), a method of collecting en face phase gradient images of thick scattering samples, enabling near video-rate in vivo phase imaging with a miniaturized probe suitable for endoscopy.
Phase contrast microscopy techniques are widely used in biological research because they can provide high resolution images of unlabeled samples even when these are nearly transparent. For example, differential interference contrast (DIC), which reveals lateral phase gradients, is one of the more popular techniques because it provides apparent 3D sample relief using a standard widefield microscope equipped with a lamp and camera1. Simpler techniques based on oblique illumination can also be used with widefield microscopes and provide similar imaging as DIC2–5. However, in order to reveal lateral phase gradients, all these techniques must be operated in trans-illumination configurations, limiting their use to thin samples. Because one is often constrained to working instead with thick samples (e.g. endoscopy or in vivo applications), there is a need for a method that can provide DIC-like imaging in thick samples. We introduce such a method here.
Our technique is called oblique back-illumination microscopy (OBM). As its name suggests, this technique is based on a similar principle as oblique illumination microscopy, with the notable difference that illumination source and detection optics both reside on the same side of the sample (i.e. in a reflection geometry), allowing its application to samples of arbitrary thickness. Unlabeled samples can, of course, be imaged with microscopes based on direct light reflection. The most successful of these for tissue imaging is optical coherence tomography (OCT), which, like OBM, can also be operated in a widefield en face configuration6. However, microscopes based on light reflection intrinsically reveal only sample structure that varies rapidly in the axial direction, such as sharp interfaces or particles much smaller than the illumination wavelength7. In contrast, microscopes based on light transmission are not subject to this constraint, and can reveal even slowly varying sample structure in the lateral direction, thus providing images of subtle sample features impossible to see in reflection mode. An important characteristic of OBM is that, even though it is configured in a reflection geometry, it is in fact a transmission microscope in disguise. In effect, OBM uses multiple scattering in tissue to convert epi-illumination into trans-illumination. Because the illumination source is offset from the detection optical axis, the trans-illumination is oblique (Fig. 1). While illumination obliquity directly leads to phase gradient contrast, image intensity is also influenced by sample absorption. The use of two off-axis sources diametrically opposed to one another permits the acquisition of two raw images with similar absorption contrast but with phase gradient contrast of opposing sign (Supplementary Fig. 1). The subtraction of these raw images enhances phase gradient contrast while canceling absorption contrast; addition of the raw images has the opposite effect, revealing only absorption contrast while canceling phase gradient contrast. By this method, the sequential acquisition of two raw images using alternating illumination sources decouples absorption and phase gradient contrast5.
Figure 1.
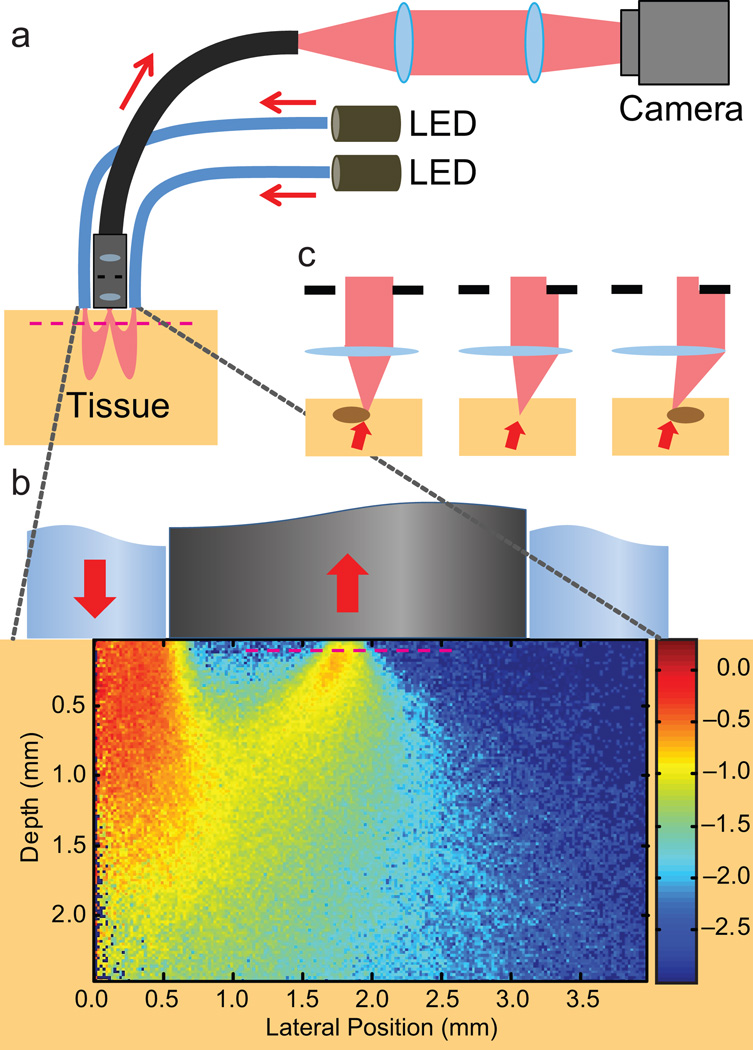
An OBM setup with a contact-mode endomicroscope probe. (a) Illumination from two LEDs is sequentially delivered into a thick sample by diametrically opposed optical fibers attached to the probe housing. Multiple scattering in the sample redirects the light so that it trans-illuminates the focal plane of the probe micro-objective (magenta dashed line). An image from the focal plane is then relayed by a flexible fiber bundle and projected onto a digital camera. (b) Close-up of the probe distal end, onto which is superposed a density map obtained by Monte Carlo simulation of the light energy in the sample that was injected by a single fiber (from left) and collected by the micro-objective (Online Methods). Note the obliqueness of the light distribution through the focal plane. (c) Oblique trans-illumination leads to phase gradient contrast. With no phase gradient, oblique illumination is partially blocked by the aperture in the micro-objective (center panel). Phase gradients due to index of refraction variations at the focal plane tilt the light more or less (right/left) depending on the slope of the variations. As a result, more or less light is blocked by the micro-objective aperture, leading to intensity variations in the recorded image and hence phase gradient contrast4,5.
We present results obtained from a miniaturized OBM built using a flexible endomicroscope probe comprising a distal micro-objective attached to an imaging fiber bundle (Online Methods). Two optical fibers attached opposite one another to the micro-objective housing deliver light to the sample from two independently controlled light emitting diodes (LEDs). Our endomicroscope probe was designed to operate in contact mode, meaning the micro-objective does not collect light reflected directly from the sample surface. Instead, the micro-objective only collects light that has been multiply scattered in the sample and redirected upwards through the focal plane, located here at a depth of 60 µm (the working distance of the micro-objective). Unless stated otherwise, all images presented here are individual frames from movies acquired, processed and displayed at a net rate of 17.5 Hz using a double-shutter camera that reads images pairwise. The exposure time per raw image was typically 1–5 ms.
To begin, we demonstrate OBM by imaging a 45 µm polystyrene bead embedded in a scattering tissue phantom consisting of 2 µm beads mixed in agarose (Fig. 2a). The phase gradient induced by the bead is observed to be approximately linear (Fig. 2b). Note that since the phase gradient image is derived from a difference of raw images, it contains both positive and negative values (zero is represented as an intermediate gray level). Because phase gradients must, by definition, arise from apparent sample structure, they must also arise from the vicinity of the focal plane (objects out of focus are blurred and exhibit little structure). Thus, phase gradient imaging exhibits apparent out-of-focus background rejection. This is manifest in Supplementary Video 1, in which only the in-focus 2 µm beads are visible. An axial profile of these beads shows a full width at half maximum (FWHM) axial resolution of 6 µm (Supplementary Fig. 2).
Figure 2.
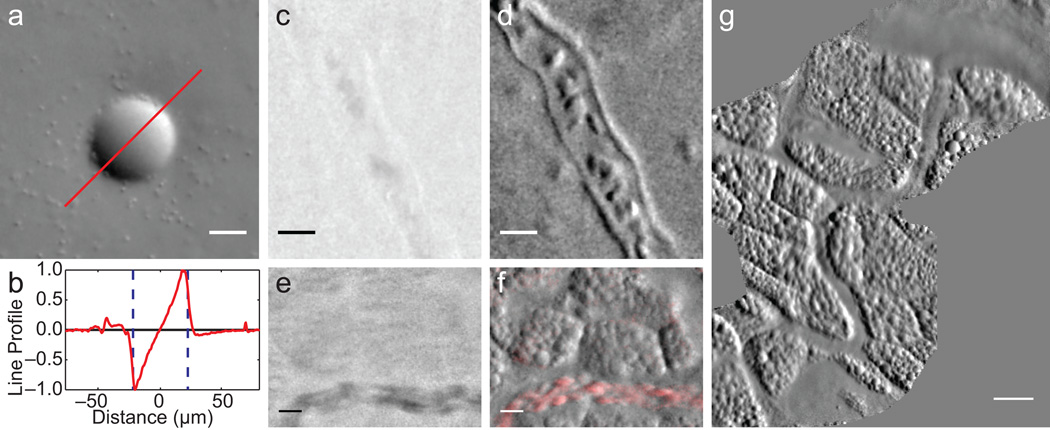
Demonstration of OBM in a tissue phantom and in vivo. (a) Phase gradient image of a 45 µm polystyrene bead in scattering medium (2 µm beads mixed in agarose, Supplementary Video 1). Note 2 µm beads are readily visible. (b) Corresponding phase gradient profile. (c-f) Simultaneously acquired absorption (c,e) and phase gradient (d,f) images of the CAM vascular system of day 11 chick embryo in ovo. Individual red blood cells (RBCs) and vessel walls are clearly visible (Supplementary Video 2). (f) Moving RBCs are highlighted in red using a sliding 3-frame temporal variance filter (Supplementary Video 3). (g) A CAM vasculature mosaic reconstructed from Supplementary Videos 4,5. Scale bars are (a,c-f) 20 µm, and (g) 50 µm.
We performed simultaneous absorption and phase gradient imaging of the chorioallantoic membrane (CAM) in a day 11 chick embryo in vivo (Fig. 2c–g, Supplementary Videos 2–5). Note the low contrast of the absorption images compared to the phase gradient images (Supplementary Video 4). Because the absorption images are derived from the sum of raw images, their values are positive definite (zero is black), and they do not exhibit out-of-focus background rejection (such imaging is similar to orthogonal polarization spectral imaging8). Compared to images obtained from OCT, OBM images are speckle-free. Moreover, owing to the large photon fluxes involved, they are relatively shot-noise free. Intensity noise arises, in our case, mostly from inhomogeneous image sampling due to an uneven distribution of fiber cores in the imaging fiber bundle (Online Methods).
Our endomicroscope probe was designed for gastrointestinal imaging. Accordingly, we demonstrate simultaneous absorption and phase gradient imaging of excised, unstained mouse intestinal epithelium (Figs. 3a-h). The absorption images are essentially featureless in this case, whereas the phase gradient images are information rich. For example, crypts of Lieberkühn and ileal villi are readily visible in Figures 3g (Supplementary Video 6) and 3h (Supplementary Video 7), respectively, illustrating the potential of OBM for in situ histopathology and “optical biopsies”.
Figure 3.
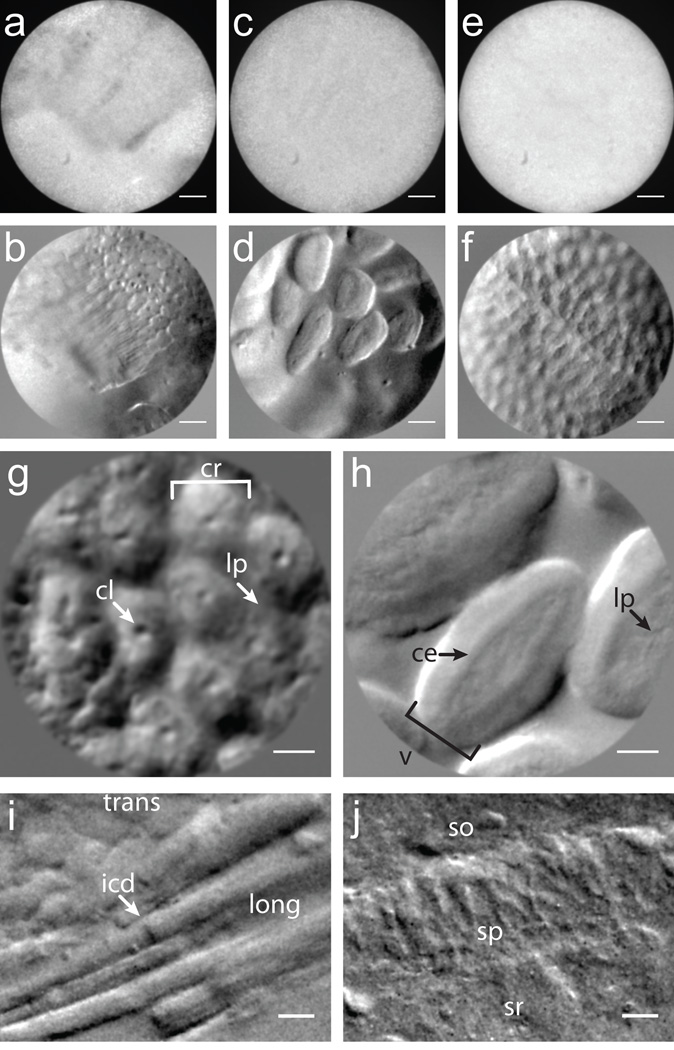
Demonstration of OBM in excised mouse tissue. (a-f) Simultaneously acquired amplitude (a,c,e) and phase gradient (b,d,f) images of intestinal epithelium taken with a 1× micro-objective (field of view (FOV) 600 µm). (g,h) Higher magnification phase gradient images of the epithelium in the distal colon (g) and small intestine (h) were taken with a 2.5× micro-objective (FOV 240 µm). Crypts of Lieberkühn (cr), crypt lumens (cl) and lamina propia (lp) are indicated with arrows (g, Supplementary Video 6); ileal villi (v), columnar epithelium (ce) and lamina propia (lp) are indicated with arrows (h, Supplementary Video 7). (i) Transverse (trans) and longitudinal (long) aspects of mouse cardiac muscle tissue; intercalated discs (icd) are readily distinguished. (j, Supplementary Video 8) Mouse brain slice reveals pyramidal neurons in the CA1 region of the hippocampus. Stratum oriens (so), stratum pyramidale (sp) and stratum radiatum (sr) are apparent. Scale bars are (a-f) 75 µm, (g-h) 30 µm and (i-j) 20 µm.
Finally, we image other types of tissue such as mouse cardiac muscle and brain tissue (Figs. 3i,j and Supplementary Video 8). These images are manifestly DIC-like in appearance, while being acquired from tissue sections many millimeters thick. Again, our imaging depth was limited by the 60 µm working distance of our micro-objective. Somewhat deeper imaging can be achieved with a longer working distance objective, which we demonstrate with a standard microscope setup (Supplementary Figs. 3,4).
In summary, we have presented an apparatus that, to our knowledge, is the first to provide sub-surface, DIC-like (i.e. transmission-like) phase gradient imaging from thick scattering tissue in a widefield reflection geometry. The apparatus is simple, fast, robust, and inexpensive, making it broadly appealing to biological and clinical researchers alike.
ONLINE METHODS
Hardware setup
Aspheric condenser lenses (Thorlabs ACL5040-A) coupled white light from two LEDs (Luxeon Star MR-WC310-20s) into optical fibers (Thorlabs BFL48-1000; 0.48 NA; 1,000 µm core). The fibers launched the illumination into the sample (25 mW per channel at the fiber output), where multiple scattering redirected it through the focal plane and into a micro-objective (Mauna-Kea Technologies; 2.6 mm diameter; 1× or 2.5× magnification; 60 µm working distance; water-immersion; 0.8 NA) coupled to a imaging fiber bundle (30,000 cores; 600 µm active area). The separation distance between the fiber axis and the micro-objective probe axis was approximately 1.8 mm. Standard microscope optics (Olympus Plan 10× 0.25 NA air objective, Linos AC f = 200 mm tube lens; 4f configuration) imaged the proximal face of the fiber bundle onto a digital camera (PCO Pixelfly USB; 14-bit; 2 × 2 binning; 35 fps; 1–5 ms exposure time per illumination direction) which recorded the images. Motion artifacts were minimized by operating the camera in double shutter mode, which reduced the inter-frame delay between exposures (200 µs) 9. Illumination power delivered by the left and right optical fibers was triggered (Thorlabs LEDD1B) to overlap with the first and second frame in the each image pair, respectively. Camera readout time limited the frame rate. In a separate experiment, an epi-illumination path was added to compare OBM with traditional reflection imaging. The strong specular reflection from the proximal fiber bundle surface was largely rejected by using a polarizing beamsplitter (Supplementary Fig. 5). Custom written software (National Instruments LabVIEW 11.0) performed the image acquisition and display. A data acquisition card (National Instruments PCI-6221) synchronized illumination gating and camera exposure.
Image processing
A preprocessing routine described previously9 first corrected for the quasiperiodic sampling pattern imparted by the fiber bundle cores. Each raw image was then normalized by its respective low-pass filtered version (Gaussian filter kernel with σ = 80 pixels) to correct for non-uniform illumination profiles and thus “flatten” the images. The two normalized images were then added or subtracted to produce absorption-only or phase-gradient-only images, respectively (Supplementary Fig. 1). Unless otherwise stated, positive definite absorption images are displayed with a linear grayscale mapping the value 0 to black and the maximum value of the image to white. Phase gradient images are displayed with a linear grayscale mapping such that 0 is represented by the middle gray level and the image is scaled to fill the dynamic range of the display (Supplementary Fig. 5). A graphics processing unit (GPU, NVIDIA GTX280) running custom-written software written in CUDA-C10 performed the image processing.
Monte Carlo simulations
We performed simulations using CUDAMCML11, a modification of MCML12 enabling execution on GPUs. We further modified CUDAMCML to execute on a cluster of CUDA-enabled workstations13. We modeled the tissue with a semi-infinite slab geometry and tissue optical parameters ntissue = 1.37, ls = 150 µm, ls* = 3,000 µm and g = 0.95 (n is index of refraction, ls and ls* are the scattering and transport mean free path lengths, respectively, and g = 1 – ls / ls* is the anisotropy factor). Illumination fiber parameters were nfiber = 1.37, diameter = 1,000 µm and numerical aperture, NA = 0.48. Micro-objective probe parameters were nprobe = 1.37, diameter = 240 µm and NA = 0.8. Fiber-probe separation was d = 1,818 µm. A Henyey-Greenstein phase function characterized photon scattering events14. We simulated 105 photons to estimate photon path density as a function of lateral position and depth, revealing the so-called photon banana (displayed in arbitrary units in log scale, Fig. 1b). We simulated 108 photons to estimate the distribution of exit angles of the detected photons as a function of fiber-probe separation (Supplementary Figs. 6,7).
Tissue phantom preparation
We prepared the scattering tissue phantom by heating a 30 mL solution of 2% (w/v) agarose (Sigma A5093-100G), 5% 2 µm diameter polystyrene beads (Polysciences 19814-15), and 0.1% 45 µm diameter polystyrene beads (Polysciences 07314-5) in H2O to 75 °C on a hotplate, followed by pouring the mixture into a 60 × 15 mm cell culture dish (Corning 430166). We covered the phantom with paraffin film and left it to cool to room temperature before imaging. Mie theory estimated the optical properties of the bulk medium to be ls = 74 µm, ls* = 1,040 µm and g = 0.93. The indices of refraction of hydrated agarose gel and polystyrene beads were n = 1.35 and n = 1.59, respectively15. We performed the imaging through water.
Chick embryo preparation
Fertilized Gallus gallus eggs (Carolina Biological Supply Co. 139290) were stored in an incubator at 37 °C and 50% humidity, being turned every 7 h to prevent fusion of the chorioallantoic membrane (CAM) with the shell membrane. Imaging was performed at embryonic day 11. A 1 cm diameter region of the shell and shell membrane was removed exposing the embryo and CAM. A layer of 37 °C saline was dripped over the preparation before imaging in ovo with the OBM probe. Following imaging, the embryos were euthanized by hypothermia by storing the eggs at −15 °C.
Mouse tissue preparation
Gastrointestinal tissue: Six week old C57 black 6 mice were euthanized by CO2 inhalation and the gastrointestinal tracts were immediately excised and washed with 4% paraformaldehyde. The colon and small intestine were cut longitudinally, unrolled and cleared of fecal matter. The preparations were incubated in 4% paraformaldehyde for 72 h. Before ex vivo imaging, the tissues were pinned to a silicone elastomer slab (Sylgard® 184, Corning) to expose the apical surfaces. Residual mucus and fecal matter were gently washed away with saline before imaging. Cardiac tissue: The intact heart was removed from the same mice as above and incubated in 4% paraformaldehyde for 72 h. Upon ex vivo imaging a transverse cut exposing the left ventricular cavity was made, exposing cardiac muscle fibers in both transverse and longitudinal aspects. Brain tissue: Eight week old Swiss Webster mice were anesthetized and decapitated, and the brain was incubated in 30% sucrose for 72 h. A 4.3 mm thick coronal slice was made to expose the CA1 region of the hippocampus. Skin tissue: Shaved, 3 × 3 cm area sections of ventral skin were removed from the same mice as above and incubated in 4% paraformaldehyde for 24 h. Upon ex vivo imaging the tissue was pinned to a silicon elastomer slab. The animals used in this study were treated in accordance with the guidelines of the Institutional Animal Care and Use Committee of Boston University.
Supplementary Material
Acknowledgments
We thank S. Singh (Boston University) and J. Ritt (Boston University) for supplying mouse gastrointestinal samples and M. Baum (Boston University) for supplying skin and brain tissue samples. We thank K. Calabro (Boston University) for helping develop the Monte Carlo simulation code. We thank R. Wu (Boston University) for help with the microscope setup used for Supplementary Figs. 3,4. We thank all the members of the Biomicroscopy Lab for their helpful conversations and careful review of this manuscript. This work was supported by National Institutes of Health grant R01-EB010059.
Footnotes
Author Contributions
T.N.F., K.K.C. and J.M. conceived and developed the technique. T.N.F. built the setup and acquired the data. T.N.F. and J.M. wrote the manuscript. J.M. supervised the project.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Nomarski G. J. Phys. Radium. 1955;16:S9. [Google Scholar]
- 2.Saylor CF. J. Res. Natl. Bur. Std. (US) 1935;15:277. [Google Scholar]
- 3.Axelrod D. Cell Biophys. 1981;3:167173. doi: 10.1007/BF02788132. [DOI] [PubMed] [Google Scholar]
- 4.Yi R, Chu KK, Mertz J. Opt. Express. 2006;14:5191–5200. doi: 10.1364/oe.14.005191. [DOI] [PubMed] [Google Scholar]
- 5.Mehta SB, Sheppard CJR. Opt. Lett. 2009;34:1924–1926. doi: 10.1364/ol.34.001924. [DOI] [PubMed] [Google Scholar]
- 6.Dubois A, Boccara AC. Chapter 19. In: Drexler W, Fujimoto JG, editors. Optical Coherence Tomography. Springer; 2009. pp. 565–591. [Google Scholar]
- 7.Mertz J. Introduction to Optical Microscopy. Roberts & Co.; 2009. [Google Scholar]
- 8.Groner W, et al. Nat. Med. 1999;5:1209–1212. doi: 10.1038/13529. [DOI] [PubMed] [Google Scholar]
- 9.Ford TN, Lim D, Mertz J. J. Biomed. Opt. 2012;17:021105. doi: 10.1117/1.JBO.17.2.021105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.NVIDIA CUDA Compute Unified Device Architecture Programming Guide, Version 2.0. 2008 [Google Scholar]
- 11.Alerstam E, Svensson T, Andersson-Engels S. CUDAMCML User manual and implementation notes. 2009 http://www.atomic.physics.lu.se/biophotonics/
- 12.Wang LH, Jacques SL, Zheng LQ. Comput. Meth. Prog. Bio. 1995;47:131–146. doi: 10.1016/0169-2607(95)01640-f. [DOI] [PubMed] [Google Scholar]
- 13.Calabro KW, Aizenberg E, Bigio IJ. Proc. SPIE. 2012;8230:82300H. [Google Scholar]
- 14.Henyey LG, Greenstein JL. Astrophys. J. 1941;93:70–83. [Google Scholar]
- 15.Pogue BW, Patterson MS. J. Biomed. Opt. 2006;11:041102. doi: 10.1117/1.2335429. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.