Abstract
Skeletal muscle gene expression is dependent on combinatorial associations between members of the MyoD family of basic helix-loop-helix (bHLH) transcription factors and the myocyte enhancer factor 2 (MEF2) family of MADS-box transcription factors. The transmembrane receptor Notch interferes with the muscle-inducing activity of myogenic bHLH proteins, and it has been suggested that this inhibitory activity of Notch is directed at an essential cofactor that recognizes the DNA binding domains of the myogenic bHLH proteins. Given that MEF2 proteins interact with the DNA binding domains of myogenic bHLH factors to cooperatively regulate myogenesis, we investigated whether members of the MEF2 family might serve as targets for the inhibitory effects of Notch on myogenesis. We show that a constitutively activated form of Notch specifically blocks DNA binding by MEF2C, as well as its ability to cooperate with MyoD and myogenin to activate myogenesis. Responsiveness to Notch requires a 12-amino-acid region of MEF2C immediately adjacent to the DNA binding domain that is unique to this MEF2 isoform. Two-hybrid assays and coimmunoprecipitations show that this region of MEF2C interacts directly with the ankyrin repeat region of Notch. These findings reveal a novel mechanism for Notch-mediated inhibition of myogenesis and demonstrate that the Notch signaling pathway can discriminate between different members of the MEF2 family.
Members of the basic helix-loop-helix (bHLH) family of transcription factors control development and differentiationof many cell types, including muscle, neural, and hematopoietic cells. Skeletal muscle differentiation is regulated by four bHLH factors—MyoD, myogenin, Myf-5, and MRF4 (49, 74), each of which can initiate myogenesis when expressed in nonmuscle cells. These factors dimerize with the ubiquitous bHLH proteins E12, E47, and HEB, known as E proteins, and activate muscle transcription by binding E boxes (CANNTG) in the control regions of skeletal muscle genes.
Biochemical and genetic evidence indicates that the myogenic bHLH factors activate myogenesis in collaboration with members of the myocyte enhancer factor 2 (MEF2) family of MADS (MCM1, agamous, deficiens, and serum response factor)-box transcription factors (reviewed in references 7, 53, and 64). The MADS box, located at the N termini of MEF2 factors, is a conserved 57-amino-acid domain responsible for DNA binding, dimerization, and interaction with myogenic and neurogenic bHLH factors. Immediately adjacent to the MADS box of the MEF2 factors is a 29-amino-acid region known as the MEF2 domain which influences DNA binding site specificity. There are four MEF2 genes in vertebrates, MEF2A, -B, -C, and -D (10, 12, 35, 43, 44, 45, 57, 73), and a single MEF2 gene in fruit flies (37, 51). MEF2 factors bind as homo- and heterodimers to the consensus sequence CTA(A/T)4TAG/A in the control regions of muscle genes and act as transcriptional activators (12, 24, 57). The different MEF2 factors exhibit similar activities in transfection assays, but there is evidence from gene knockout experiments in mice that they play different roles in vivo (40, 50).
During embryogenesis, the MEF2 genes are expressed throughout developing skeletal and cardiac muscle lineages, as well as in the nervous system (19, 42, 65, 68). MEF2C is the first member of the family to be expressed in developing muscle cell lineages, with transcripts appearing in precardiac cells by about embryonic day 7.75 and in skeletal muscle precursor cells within the myotome of the developing somites by embryonic day 8.5. Soon thereafter, the other MEF2 genes are expressed in overlapping patterns (19). After birth, the expression of MEF2A, -B, and -D becomes ubiquitous, whereas the expression of MEF2C becomes restricted to skeletal muscle, brain, and spleen (43).
Mutational analyses of the myogenic bHLH factors have shown that their basic regions play a dual role in muscle gene activation by mediating DNA binding and interactions with a myogenic cofactor (11, 16). Members of the MEF2 family appear to fit the criteria for such a cofactor. Myogenic bHLH factors interact with MEF2 factors, resulting in cooperative activation of muscle-specific transcription (31, 47). This interaction enables either factor bound to DNA to recruit the other through protein-protein interactions without the necessity of both factors binding DNA (6, 47). In cells expressing a dominant negative form of MEF2A, MyoD and myogenin are unable to activate myogenesis (54) and in Drosophila mutants lacking MEF2, the myogenic bHLH gene nautilus is expressed in skeletal myoblasts, but it is devoid of myogenic activity (9, 38). Thus, activation of the skeletal muscle program appears to require the combined activities of myogenic bHLH and MEF2 factors.
A variety of extracellular signals inhibit skeletal myoblast differentiation by interfering with the activity of myogenic bHLH proteins. The transmembrane receptor Notch and its cell surface-associated ligand Delta have been shown to prevent myogenesis in tissue culture, as well as in Xenopus and Drosophila embryos (1, 4, 32, 39, 52, 63). Notch proteins contain an extracellular domain consisting of 34 to 36 epidermal growth factor (EGF) repeats, a cysteine-rich domain, and three Notch/lin-12 repeats (reviewed in references 2 and 3) and an intracellular domain composed of six tandem ankyrin/cdc10 repeats, flanked by putative nuclear localization signals, followed by a PEST sequence that mediates protein degradation. Activation of Notch signalling normally requires binding to transmembrane ligands on adjacent cells. The Notch receptor is processed by proteolytic cleavage in the trans-Golgi network to generate two fragments, one containing the extracellular domain, and the other, the transmembrane and intracellular domains. These two fragments are tethered at the cell surface and form the signalling-competent heterodimeric receptor (8, 56). There is evidence of a second ligand-dependent cleavage event of the intracellular fragment which leads to its nuclear translocation (25, 29, 32, 33, 36, 62). The receptor can also be activated by deletion of the transmembrane and extracellular regions (reviewed in reference 2).
When Notch is activated by ligand binding, the intracellular domain, which is released by proteolytic cleavage, interacts with the transcription factors Suppressor of hairless [Su(H)] proteins in Drosophila and their vertebrate homologs CBF1/KBF2/RBP-Jk (25, 28, 29, 41). The resulting complex upregulates genes of the Drosophila enhancer-of-split complex [E(spl)] and their mammalian homolog HES-1, respectively, which encode bHLH proteins that inhibit the activities of other bHLH proteins. HES-1 has been reported to inhibit the activity of MyoD (61), and this has been proposed as a mechanism whereby Notch inhibits myogenesis. However, a mutant form of Notch that lacks the CBF1-binding domain and cannot induce HES-1 retains the ability to inhibit myogenesis (63), suggesting the existence of a CBF1/HES-1-independent pathway through which Notch inhibits myogenesis.
Activated Notch has been reported to inhibit the myogenic activity of MyoD without affecting its DNA binding activity (32). The inhibitory signal from Notch is directed at the DNA binding domain of MyoD and appears to occur through interference with the expression or activity of an essential MyoD cofactor. Here we investigated the possibility that members of the MEF2 family might be targets for negative regulation by Notch. We demonstrate that activated Notch can specifically inhibit the ability of MEF2C to activate myogenesis in cooperation with the myogenic bHLH factors. However, there must also be other mechanisms for Notch-mediated repression of myogenesis because other members of the MEF2 family, which also cooperate with myogenic bHLH factors to control muscle gene expression, are refractory to the effects of Notch. Thus, Notch signaling provides a mechanism for selective regulation of MEF2C functions, as well as for the inhibition of myogenesis through MEF2-independent mechanisms.
MATERIALS AND METHODS
Plasmids.
Expression vectors encoding wild-type and mutant forms of mouse MEF2C, myogenin, and MyoD have been described (48). To delete the exon of MEF2C that encodes that Notch-interacting domain, cDNAs encoding amino acids 1 to 86 and 134 to 465 were synthesized by PCR such that a SacII site was introduced at the internal junction of the two fragments. The resulting clone, called alternate MEF2C, encodes amino acids 1 to 86, with the addition of Pro-Arg at positions 87 and 88, followed by Ala-134 to the carboxyl terminus of MEF2C. The CMV-NotchIC and CMV-NotchΔ clones, which were derived from human Notch2, were the gift of T. Kadesch (University of Pennsylvania) and are described in Blaumueller et al. (8). The GAL4-Notch clones, derived from mouse Notch1, were the gift of S. D. Hayward (Johns Hopkins University) and are described in Hsieh et al. (28).
The reporter plasmids 4R-tk-CAT and MEF2x2-CAT have been described previously (48). 4R-tk-CAT contains four tandem copies of the right E box from the MCK enhancer linked to the thymidine kinase (tk) basal promoter and MEF2x2-CAT contains two tandem copies of the MEF2 site from the MCK enhancer linked to the β-myosin heavy-chain (MHC) promoter. RSV-CAT and pSV2CAT contain the Rous sarcoma virus or the simian virus 40 promoters and enhancers, respectively. PG5E1b-CAT contains five tandem copies of the GAL4 binding site linked to the E1b promoter upstream of CAT and was used as a reporter for experiments with GAL4-Notch fusions.
Cells and transfections.
10T1/2 mouse fibroblasts were grown in Dulbecco modified Eagle medium (DMEM) (Gibco-BRL, Gaithersburg, Md.) supplemented with 10% fetal calf serum (FCS). Cells were transfected by the calcium phosphate method. The total amount of plasmid DNA was equivalent in all transfections. Methods used for mammalian two-hybrid assays and assays for synergy between myogenic bHLH and MEF2 factors were as described previously (6).
For CAT assays, 10T1/2 fibroblasts were seeded into 60-mm-diameter dishes and transfected for 16 h, after which the cells were rinsed in phosphate-buffered saline (PBS) to remove excess precipitate, and cells were maintained in growth medium. The ratios and amounts of plasmid DNA are indicated in the figure legends. At 48 h posttransfection, cells were harvested, and freeze-thaw cell lysates were made in 0.25 M Tris (pH 7.5). The amount of protein in each sample was determined by protein assay (Bio-Rad, Hercules, Calif.), and chloramphenicol acetyltransferase (CAT) assays were performed with an equal amount of total protein.
For differentiation assays, 10T1/2 fibroblasts were seeded into 35-mm-diameter dishes that had been coated in 0.1% (wt/vol) gelatin (Sigma, St. Louis, Mo.) and then were transfected with 3 μg of EMSV-myobHLH, EMSV-myogenin, or EMSV-MyoD; 1 μg of CMV-MEF2 expression vectors; and 3 μg of CMV-NotchIC or NotchΔ. The amount of total DNA added in each transfection was constant and was maintained by the addition of pcDNAI or EMSV. Cells were transfected for 20 h, excess precipitate was rinsed off, and cells were maintained for an additional 24 h in growth medium. Cells were then transferred to differentiation medium (DMEM supplemented with 2% horse serum [Gibco-BRL]) and maintained for 5 more days, with daily changes of medium. For immunostaining, cells were fixed in cold methanol at −20°C for 8 min, followed by rehydration in PBS and incubation with an α-myosin heavy-chain (MHC) antibody, MY-32 (1:400 dilution; Sigma) for 1 h. MHC-positive cells were detected by peroxidase staining by using the HistoStain SP kit (Zymed, San Francisco, Calif.).
Gel mobility shift assays.
Coupled in vitro transcription-translation reactions were performed with 0.5 μg of plasmid DNA and TNT reticulocyte lysates with T7 polymerase (Promega, Madison, Wis.). The efficiency of translation was determined by performing duplicate translation reactions in the presence of Trans-[35S] (DuPont-NEN). As a probe, we used a double-stranded oligonucleotide corresponding to the muscle creatine kinase (MCK) MEF2 site (24) that was end labeled with [γ-32P]ATP (Dupont-NEN) and T4 polynucleotide kinase (New England Biolabs, Beverly, Mass.). For each DNA binding reaction, 2 μg of the total translation products was added to a 20-μl total reaction mixture along with 40,000 cpm of probe in binding buffer (40 mM KCl; 15 mM HEPES, pH 7.9; 1 mM EDTA; 0.4 mM dithiothreitol, 50% [vol/vol] glycerol), and 2 μg of poly(dI-dC) (Pharmacia Biotech, Piscataway, N.J.). Binding reactions were carried out for 20 min at room temperature, and protein-DNA complexes were analyzed on 5% 0.5× TBE polyacrylamide gels.
Immunoprecipitations and Western blots.
In vitro translations were performed with the TNT T7 translation kit from Promega. Translation products were labeled with Trans-[35S] (DuPont-NEN).
For immunoprecipitations, COS cells were plated at 2 × 105 cells/ml in 10-cm dishes. Cells were transfected by calcium phosphate precipitation by using a total of 10 μg of DNA per plate. At 48 h after transfection, plates were rinsed twice in ice-cold PBS, and then 1 ml of ice-cold NTT lysis buffer (140 mM NaCl; 50 mM Tris, pH 7.5; 1 mM EDTA; 0.1% Triton X-100; 10% glycerol) containing the complete protease inhibitor cocktail (Boehringer Mannheim, Indianapolis, Ind.) was added to each plate. Cells were scraped from plates into 1.5-ml tubes and incubated for 15 min on ice to lyse them. Lysed cells were vortexed and centrifuged for 15 min in a microfuge at 4°C to remove cellular debris. The lysate was transferred to a new tube for immunoprecipitation. Each lysate received 2 μl of M2 anti-FLAG antibody (Eastman Kodak) and 25 μl of protein A/G-Plus agarose (Santa Cruz Biotechnology). Lysates were then incubated for 4 h at 4°C with shaking, followed by centrifugation and then three washes in NTT buffer. Immunoprecipitates were then separated by sodium dodecyl sulfate (SDS)–12% polyacrylamide gel electrophoresis (PAGE).
Gels were transferred to Immobilon-P membrane in BS-N transfer buffer (48 mM Tris, 39 mM glycine; pH 9.2) by using the Bio-Rad semidry transfer apparatus. Membranes were blocked by incubation for 1 h at room temperature in Tris-buffered saline (TBS) containing 5% Carnation nonfat dry milk. The membrane was rinsed in TBS with 0.05% Tween 20, followed by incubation for 1 h in TBS–0.05% Tween 20–1% bovine serum albumin (BSA) with a 1:100 dilution of anti-hemagglutinin (anti-HA) antibody coupled to horseradish peroxidase (HRP) (Boehringer Mannheim). The membrane was then washed four times for 10 min each in TBS–0.05% Tween 20–1% BSA and exposed to Renaissance chemiluminescence reagents (DuPont-NEN) for 1 min and exposed to hyperfilm.
AnkR-HA encompasses the ankyrin repeat domain and intracellular amino acids immediately N terminal to this domain of human Notch2 cloned into pCDNA1. The hemagglutinin epitope of influenza virus (YPVDVPDYA) was added by using a double-stranded oligonucleotide at the C terminus of the protein. MEF2C-FLAG was cloned into pCDNA1 and the FLAG epitope (DYKDDDDK) was added in frame at the EcoRI site such that the C-terminal 28 amino acid residues are missing and replaced with the FLAG epitope.
For immunoprecipitation, proteins were diluted in NTT buffer (0.14 M NaCl, 0.05 M Tris [pH 7.5], 0.1% Triton X-100, 1 mM EDTA, 10% glycerol) containing a complete protease inhibitor cocktail (Boehringer Mannheim) and were immunoprecipitated for 4 h at 4°C with M2 α-FLAG antibody (Eastman Kodak Co., Rochester, N.Y.). Immunoprecipitated proteins were recovered with protein A/G-Plus agarose (Santa Cruz Biotechnology), and the agarose beads were washed in NTT buffer containing inhibitors. Proteins were analyzed on SDS–10% PAGE with Benchmark prestained molecular-weight markers (Gibco-BRL).
RESULTS
Notch blocks cooperative activation of myogenesis by myogenin and MEF2C.
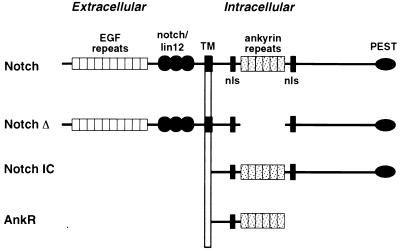
Previous studies suggested that activated Notch blocked the ability of myogenic bHLH factors to activate myogenesis by interfering with an essential cofactor (32). Because members of the MEF2 family act as cofactors for myogenic bHLH proteins, our initial interest was to determine whether MEF2 factors could be targets for the inhibitory activity of Notch. To examine the effect of Notch on myogenesis, we used several mutant forms of Notch. NotchIC comprises the intracellular domain of Notch (Fig. 1) that is localized to the nucleus and signals constitutively (8). NotchΔ represents the entire receptor with the ankyrin repeats deleted (Fig. 1) and has been shown to function as a dominant negative inhibitor of ligand-dependent Notch signalling (58). However, this mutant protein retains the intracellular region required for CBF1 activation (28, 67). Previous studies have demonstrated that these Notch mutants are stable in transfected cells.
FIG. 1.
Schematic diagrams of Notch proteins. Structures of full-length Notch and the various deletion mutants used in this study are shown. The extracellular region of Notch consists of 34 to 36 EGF repeats followed by three novel Notch/lin12 domains. The transmembrane domain (TM) is shown with the orientation relative to the plasma membrane. The intracellular region of Notch contains six ankyrin repeats flanked by putative nuclear localization signals (nls), and a PEST sequence is located at the C terminus. NotchΔ lacks the ankyrin repeats and NotchIC contains only the intracellular region. AnkR consists of the ankyrin repeats and the N-terminal intracellular residues, along with an HA epitope tag at the C terminus. Unless otherwise specified, most experiments were performed with wild-type or mutant forms of human Notch2.
Myogenic conversion of transiently transfected 10T1/2 cells was assayed by immunostaining for MHC-positive cells. MEF2 factors lack myogenic activity alone but augment the myogenic activity of myogenin or MyoD (31, 47). We reported previously that the synergistic activation of muscle gene expression by MEF2 and myogenic bHLH factors was especially apparent when only the bHLH region of myogenin was used (47). This region of the protein can dimerize with E proteins and bind DNA, but it is devoid of myogenic activity because it lacks the transcription activation domains at the N and C termini of the wild-type protein. The myogenic activity of this myogenin deletion mutant, referred to as Myo-bHLH, can be restored by expression together with MEF2 factors (47). We therefore asked whether activated Notch could inhibit this type of cooperative interaction. As shown in Table 1, transfection of 10T1/2 ells with expression vectors encoding Myo-bHLH and MEF2A, -C, or -D resulted in efficient activation of myogenesis. However, when NotchIC was expressed with Myo-bHLH and MEF2C, there was a dramatic reduction in the number of MHC-positive cells. In contrast, NotchIC only marginally inhibited myogenic conversion in the presence of Myo-bHLH and MEF2A or -D (Table 1). Thus, NotchIC specifically inhibited the ability of MEF2C to synergize with myogenin and MyoD to induce myogenesis. NotchΔ also exhibited weak inhibition of differentiation induced by Myo-bHLH plus MEF2C (Table 1).
TABLE 1.
Inhibition of myogenic conversion by Notcha
| Plasmid | Myogenic conversion (%) |
|---|---|
| MEF2C | 0 |
| MEF2A | 0 |
| MEF2D | 0 |
| Myo-bHLH | 0 |
| Myo-bHLH + MEF2C | 100 ± 20 |
| Myo-bHLH + MEF2C + Notch IC | 13 ± 1 |
| Myo-bHLH + MEF2C + NotchΔ | 66 ± 1 |
| Myo-bHLH + MEF2A | 50 ± 2 |
| Myo-bHLH + MEF2A + Notch IC | 76 ± 4 |
| Myo-bHLH + MEF2D | 36 ± 14 |
| Myo-bHLH + MEF2D + Notch IC | 50 ± 28 |
10T1/2 cells were transiently transfected with the indicated plasmids and later stained for MHC expression as described in Materials and Methods. With Myo-bHLH plus MEF2C, 113 MHC-positive cells were observed per field, which was assigned a value of 100%. All other values are expressed relative to Myo-bHLH plus MEF2C.
Activated Notch blocks cooperative activation of transcription by MEF2 and myogenic bHLH proteins.
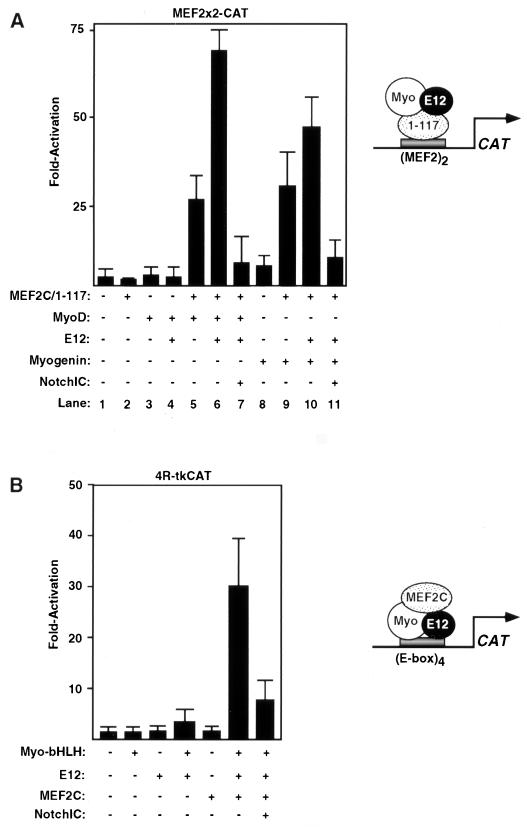
Myogenic bHLH factors and MEF2C can cooperate to activate transcription of reporter genes containing binding sites for only one factor or the other. This type of cooperativity is dependent on interactions between the bHLH and MADS/MEF2 domains (47). To further define the mechanism whereby activated Notch interfered with the functions of MyoD and myogenin, we tested whether Notch could inhibit the ability of MyoD or myogenin to activate a MEF2-dependent reporter gene in the presence of the MEF2C mutant, 1–117, which contains the MADS and MEF2 domains but lacks the C-terminal activational domains. The MEF2-dependent reporter used in this assay, MEF2x2CAT, contains two MEF2 sites upstream of a basal promoter and can be activated by wild-type MEF2 protein but not by the MEF2/1–117 mutant. Because this reporter does not contain an E box, it can only be activated by myogenic bHLH proteins through protein-protein interactions with the DNA binding domain of MEF2 bound to its target sites. In this assay, transcriptional activation is greatest in the presence of three factors; MyoD or myogenin, E12, and MEF2C/1–117 (Fig. 2A, lane 6). NotchIC inhibited this type of transcriptional synergy (Fig. 2A, compare lanes 6 and 7 and lanes 10 and 11).
FIG. 2.
NotchIC inhibits cooperative activation of E-box- and MEF2-dependent reporters by myogenic bHLH proteins and MEF2C. (A) 10T1/2 cells were transfected with 3 μg of each reporter, 1 μg of each activator, and 3 μg of NotchIC, and CAT assays were performed as described in Materials and Methods. The data is presented as the fold activity versus that observed with the reporter gene alone and represents the mean ± the standard error of the mean for three experiments performed with at least two different preparations of the plasmids. (A) MEF2x2-CAT was used as the reporter. A schematic of the putative protein-protein interactions required for reporter gene activation is shown to the right. (B) 4R-tkCAT was used as the reporter. A schematic of the putative protein-protein interactions required for reporter gene activation is shown on the right.
In a converse series of experiments, we tested whether NotchIC could inhibit the ability of full-length MEF2C to activate an E-box-dependent CAT reporter in the presence of the bHLH regions of myogenin and E12. The reporter, 4R-tkCAT, contains four tandem copies of the right E box from the MCK enhancer upstream of the thymidine kinase promoter and does not respond to MEF2C alone in transiently transfected 10T1/2 fibroblasts, since this reporter lacks a MEF2 binding site (Fig. 2B). Myo-bHLH, which lacks transcriptional activity on its own, was also unable to activate this CAT reporter in the presence of E12. However, if MEF2C was expressed in 10T1/2 cells together with the bHLH region of myogenin and E12, there was a 25-fold activation of the CAT reporter, which reflects recruitment of MEF2 to the protein-DNA complex via interaction with the bHLH heterodimer bound to the E box. NotchIC inhibited synergistic activation of the E-box-dependent reporter by the three transcription factors (Fig. 2B). Thus, the data presented in Fig. 2 show that activated Notch inhibits the ability of MEF2C and myogenic bHLH factors to cooperatively activate transcription irrespective of which factor is bound to DNA.
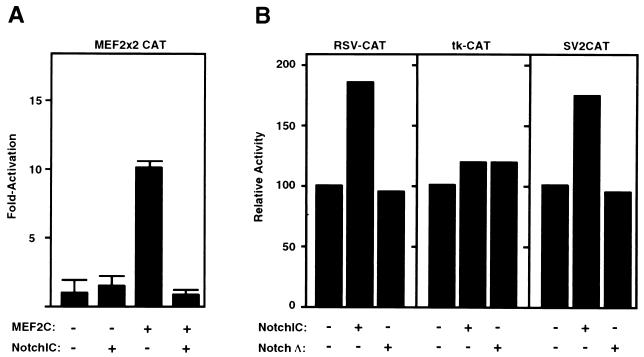
Activated Notch blocks MEF2-dependent transcription.
We next tested whether activated Notch was able to interfere with the ability of full-length MEF2 factors to transactivate MEF2x2CAT. This reporter is efficiently transactivated by MEF2C, -A, and -D (Fig. 3A). Consistent with the preferential inhibition of MEF2C’s ability to cooperate with myogenic bHLH factors to induce myogenesis, transactivation by MEF2C was strongly inhibited by Notch (Fig. 3A). The transcriptional activity of MEF2A was only slightly inhibited and MEF2D was unaffected by activated Notch (data not shown). Activated Notch did not inhibit RSV-CAT, tk-CAT, or SV2CAT, indicating that it did not act as a general inhibitor of transcription (Fig. 3B). Similarly, previous studies showed that activated Notch did not block activity of the transactivation domain of MyoD in the absence of MEF2C (32).
FIG. 3.
NotchIC inhibits the ability of MEF2C to transactivate a MEF2-dependent reporter gene. 10T1/2 cells were transfected with 2 μg of each reporter, 2 μg of each MEF2 plasmid, and 2 μg of NotchIC, and CAT assays were performed as described in Materials and Methods. (A) MEF2x2CAT was used as the reporter. The ratio of plasmids used was based on data from titration experiments (data not shown). The data is presented as the fold activity versus that observed with the reporter gene alone and represents the mean ± the standard error of the mean for three experiments performed with at least two different preparations of the plasmids. (B) RSV-CAT, tk-CAT, and SV2CAT were used as reporters. Values represent the results of a representative experiment and are expressed as CAT activity relative to that with each reporter gene alone, which was assigned a value of 100.
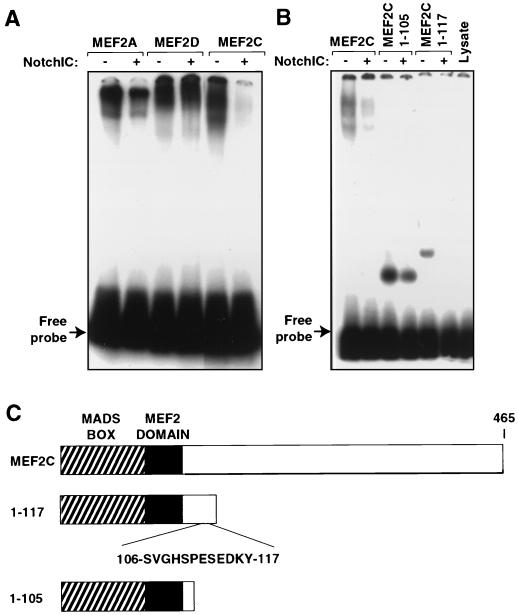
Activated Notch inhibits DNA binding by MEF2C.
To further characterize the mechanism for Notch-mediated repression of MEF2C function, we investigated whether the DNA binding activity of MEF2C was inhibited by NotchIC. Indeed, when NotchIC was translated together with MEF2C in a rabbit reticulocyte lysate and MEF2 binding activity was measured in a gel shift assay with a labeled probe corresponding to the MCK MEF2 site, DNA binding activity was dramatically reduced (Fig. 4A). In agreement with the apparent selectivity of Notch for inhibition of MEF2C function, DNA binding activity of MEF2A and -D was not substantially affected by NotchIC (Fig. 4A). Translation efficiencies, monitored by duplicate Trans-[35S]-labeled translations, were comparable in the presence and absence of Notch (data not shown).
FIG. 4.
NotchIC inhibits the ability of MEF2C to bind DNA. MEF2 and NotchIC proteins were translated from plasmid templates by using TNT reticulocyte lysates and T7 polymerase. The efficiency of translation was determined with duplicate Trans-[35S]-labeled reaction mixtures (data not shown). A 32P-end-labeled double-stranded oligonucleotide representing the MCK MEF2 site was used as the probe. DNA binding reactions were carried out as described in Materials and Methods, and protein-DNA complexes were analyzed on 5% 0.5× TBE polyacrylamide gels. (A) The ability of MEF2C to bind DNA was specifically inhibited when cotranslated with NotchIC. (B) Wild-type MEF2C or C-terminal truncation mutants 1–105 or 1–117 were translated in the presence or absence of NotchIC and tested for DNA binding activity. MEF2C/1–117 was inhibited by NotchIC, whereas MEF2C/1–105 was not. Lysate alone is shown at the right. (C) Schematic diagrams of the MEF2C proteins used in panel B.
The MADS and MEF2 domains, which are highly homologous among the four vertebrate MEF2 proteins, are encoded by the first 86 amino acids of the proteins. Outside of this region, the sequences of these proteins diverge. Since there is conservation of the amino acid sequences in the N termini of the different MEF2 gene products, it seemed most likely that the selective responsiveness of MEF2C to Notch was mediated by another unique region of the protein. We therefore used a series of C-terminal truncation mutations to map the region of MEF2C that was the target of Notch inhibition. Carboxyl-terminal deletion mutants of MEF2C that retained the first 117 amino acids of the protein were inhibited from binding DNA in the presence of activated Notch, whereas a mutant containing only residues 1 to 105 was not inhibited (Fig. 4B). Several longer deletion mutants that extended further toward the C terminus were also inhibited from binding DNA in the presence of NotchIC (data not shown). These results suggested that the minimal region of MEF2C required for Notch-mediated repression lay between amino acids 105 and 117.
Interaction of activated Notch and MEF2C in a mammalian two-hybrid assay.
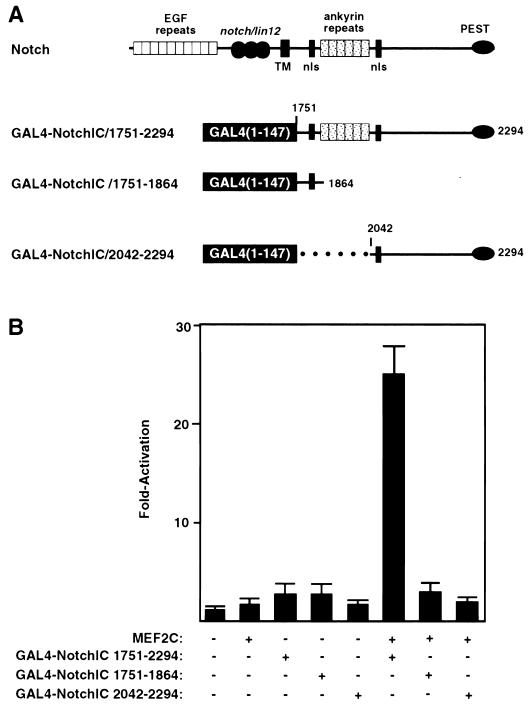
To determine whether activated Notch could interact directly with MEF2C, we used a mammalian two-hybrid assay. In this assay, we tested whether a series of GAL4 DNA binding domain-Notch fusion proteins (Fig. 5A) could recruit MEF2C to activate the GAL4-dependent reporter, pG5E1bCAT, which contains five GAL4 binding sites upstream of the E1b promoter linked to CAT. Because GAL4-Notch fusion proteins do not activate transcription alone, activation of the GAL4 dependent reporter gene would require interaction of Notch with MEF2C and the resulting recruitment of the MEF2C transcription activation domain to the promoter. As shown in Fig. 5B, NotchIC fused to GAL4 (GAL4-NotchIC1751–2294) had no transcriptional activity on its own. However, in the presence of full length MEF2C, GAL4-NotchIC1751–2294 was able to activate the GAL4-dependent reporter gene, indicating that it interacted with MEF2C and thereby recruited the MEF2C transcription activation domain to the reporter. None of the other vertebrate MEF2 proteins demonstrated activation in this assay (data not shown). This interaction was dependent on the ankyrin repeats in Notch. Coexpression of MEF2C and the GAL4 DNA binding domain fused to residues of Notch that are N terminal of the ankyrin repeat domain (GAL4-NotchIC1751–1864) demonstrated no activation. Similarly, the region of Notch C terminal to the ankyrin repeat region, when fused to GAL4 (GAL4-NotchIC2042–2294), failed to interact with MEF2C to activate the GAL4-dependent reporter gene (Fig. 5A and 5B). These results suggested that MEF2C could form a complex with the intracellular domain of Notch and that this interaction required the ankyrin repeats.
FIG. 5.
Detection of MEF2-Notch interaction by using a two-hybrid assay. (A) Schematic diagrams of Notch and the GAL4 fusion proteins used in two-hybrid assays. The GAL4 DNA binding domain was fused to the entire intracellular portion of mouse Notch1 (GAL4/Notch1751–2294) or the residues N terminal (GAL4/Notch1751–1869) or C terminal (GAL4/Notch2042–2294) to the ankyrin repeats. TM, transmembrane domain; nls, putative nuclear localization signal. (B) 10T1/2 cells were transfected with 2 μg of the pG5E1bCAT reporter, 2 μg of MEF2C, and 2 μg of each GAL4/Notch plasmid, and the CAT activity was determined as described in Materials and Methods. The data are presented as the fold activity versus that observed with the reporter gene alone and represents the average ± the standard error of four experiments performed with at least two different preparations of the plasmids. Significant reporter gene activation was seen only with MEF2C and Gal4/NotchIC1751–2294.
It is important to emphasize that in this two-hybrid assay, interaction between MEF2C and Notch results in the activation of transcription, whereas in the types of two-hybrid assays shown in Fig. 2, interaction of MEF2C and Notch results in the inhibition of transcription by blocking formation of a functional multiprotein transcriptional complex among MEF2C, myogenin or MyoD, and E12 (see Discussion). Both types of assays lead to the same conclusion: that Notch interacts directly with MEF2C.
Coimmunoprecipitation of Notch and MEF2C in vivo.
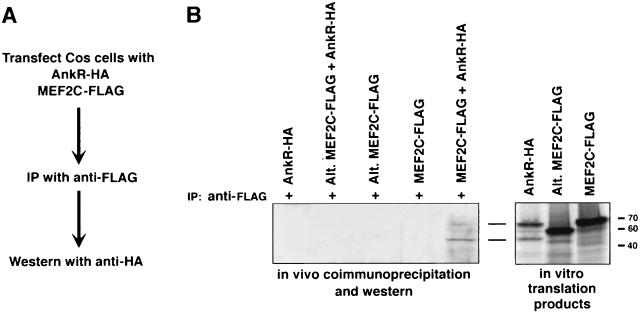
To further validate the interaction between Notch and MEF2C, we examined whether the proteins could be coimmunoprecipitated from cell lysates. COS cells were transfected with expression vectors encoding the ankyrin repeat region (AnkR) of Notch with an HA epitope and encoding MEF2C with a FLAG epitope. Extracts were then immunoprecipitated with anti-FLAG antibodies, and immunoprecipitates were resolved by SDS-PAGE and subjected to Western blot analysis with anti-HA antibody. AnkR-HA, which migrates as a doublet of approximately 45 and 65 kDa, was selectively immunoprecipitated with MEF2C (Fig. 6B, lane 5). In contrast, an alternatively spliced variant of MEF2C (43), which lacks residues 87 to 135 including the Notch-binding domain, was not coimmunoprecipitated with MEF2C (lane 3). In vitro-translated proteins were run on separate lanes as markers.
FIG. 6.
Coimmunoprecipitation of Notch and MEF2C from cell extracts. COS cells were transfected with expression vectors encoding the ankyrin repeat region of Notch fused to an HA epitope tag (AnkR-HA) and MEF2C fused to a FLAG tag. Cell lysates were immunoprecipitated with anti-FLAG antibody, immunoprecipitates were separated by SDS–12% PAGE, followed by Western blot with anti-HA antibody and anti-HRP. (A) Schematic of the experiment. (B) Western blot of extracts from cells transfected with the indicated expression vectors and 35S-labeled in vitro translation products in adjacent lanes as markers. AnkR-HA, which migrates as a doublet of approximately 45 and 64 kDa, coimmunoprecipitates with wild-type MEF2C-FLAG (lane 5), but not with the alternate isoform of MEF2C lacking the Notch-binding domain (lane 3).
These results confirm the transfection and two-hybrid assays which demonstrate direct interaction between Notch and MEF2C and show that the unique amino acids immediately C terminal to the MEF2 domain mediate this interaction.
DISCUSSION
On the basis of previous studies which suggested that the inhibitory effects of Notch were directed at a cofactor that recognized the MyoD basic region and on evidence that members of the MEF2 family potentiate the myogenic activity of MyoD by interacting with its basic region, we examined whether MEF2 factors might be the targets for Notch-dependent inhibition of myogenesis. Our results lead to the following conclusions about the inhibition of myogenesis by Notch. (i) Activated Notch selectively inhibits the DNA binding and myogenic activities of MEF2C but not of other MEF2 isoforms. (ii) Notch prevents formation of a “functional” multiprotein transcription complex between MEF2C and heterodimers of myogenic bHLH proteins and E12. (iii) Inhibition of MEF2C activity appears to be mediated by direct interaction of the ankyrin repeat region of Notch with residues 105 to 117 of MEF2C. (iv) Inhibition of MEF2C function by Notch is one of multiple pathways for Notch-dependent inhibition of myogenesis.
Signaling from Notch to MEF2C.
The different MEF2 gene products show similar activities when tested in transfection assays for their abilities to transactivate MEF2-dependent reporter genes and to cooperate with myogenic bHLH proteins to activate muscle transcription. However, our results indicate that Notch can specifically interact with, and inhibit the activity of, MEF2C. The region of MEF2C that appears to be minimally required for interaction with Notch, residues 105 to 117, is encoded by an alternatively spliced exon in MEF2C that is present in transcripts from skeletal muscle and brain and is not conserved in other MEF2 family members (43).
Notch receptors are activated in response to binding the cell surface ligand delta on adjacent cells. After ligand-dependent activation of Notch, the cytoplasmic domain of the receptor is clipped by an intracellular protease, enabling it to migrate to the nucleus (25, 29, 33, 36, 62). We do not know where within the cell activated Notch interacts with MEF2C, though we presume this occurs in the nucleus. The region of MEF2C that is required for interaction with Notch lies C terminal to the minimal DNA binding and dimerization domain and has not previously been assigned a specific function. Our results show that interaction of Notch with this region prevents binding of MEF2C to DNA. In addition, Notch blocks the formation of a transcriptionally active complex between MEF2C and MyoD-E12 or myogenin-E12 heterodimers (Fig. 2A). The ability of Notch to block this type of synergy between MEF2C and myogenic bHLH proteins could reflect an inhibition of the interactions between MEF2C and myogenic bHLH factors or a block to transmission of transcription activation signals from the multiprotein complex (7). Since the binding site for activated Notch on MEF2C lies immediately adjacent to the region required for these activities, Notch may interfere with these activities of MEF2C by steric hindrance.
Multiple pathways for Notch-mediated inhibition of myogenesis.
Activated Notch proteins have been shown to interact with the transcription factor CBF1 in vertebrates and its Drosophila homolog Su(H) to form a complex that activates transcription of the bHLH genes HES-1 and E(spl), respectively (28–30, 41). This interaction requires the region between the transmembrane domain and the ankyrin-repeat region of Notch (28, 67). HES proteins form inactive heterodimers with myogenic bHLH proteins and inhibit myogenesis (61). While it has been proposed that HES-1 mediates the inhibitory effects of Notch on myogenesis (61), it has also been demonstrated that the ankyrin repeats alone are sufficient for the inhibition of myogenesis by Notch (32, 63). Since this region of Notch does not induce HES-1, there must also be other mechanisms for the inhibition of myogenesis by Notch.
The inhibitory effects of NotchIC on myogenesis have been explained by the inhibition of expression or activity of a cofactor that recognized the basic region of MyoD (32). Consistent with this conclusion, NotchIC did not affect DNA binding by MyoD, and inhibition could not be reversed by overexpression of E12, suggesting that E proteins are not the target for inhibition (32). Because members of the MEF2 family function as cofactors for myogenic bHLH proteins (31, 47), they are potential candidates for the inhibitory targets of Notch signalling. While our results show that Notch can specifically interfere with the ability of myogenin and MyoD to cooperate with MEF2C to induce myogenesis, this cannot account for all of the inhibitory effects of Notch on myogenesis. Activation of myogenesis in 10T1/2 cells transfected with MyoD or myogenin alone, for example, is unlikely to require MEF2C, since this member of the MEF2 family is expressed relatively late in the myogenic program in muscle cells in tissue culture (43, 45). Thus, there must be other Notch-dependent mechanisms for repression that are likely to be mediated by different myogenic cofactors.
The coactivator CBP/p300 has been shown to be a cofactor for myogenic bHLH proteins, as well as MEF2 (60), but recent studies have shown that p300 activity is not affected by Notch (55). In addition, the E protein E47 has been shown to be inactivated in the presence of Notch (55). Suppression of E47 activity also seems unlikely to account for the inhibitory effects of Notch on myogenesis or the synergistic activation of muscle transcription by myogenic bHLH factors and MEF2C because excess E12 or E47 does not alleviate myogenic suppression (32).
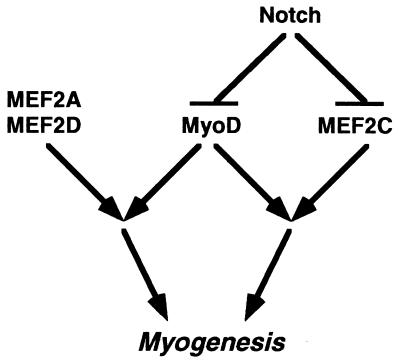
Previous studies showed that point mutations in the fourth ankyrin repeat of truncated Notch abolish the ability of NotchIC to inhibit myogenesis (32). Our results also point to the importance of the ankyrin repeats of Notch for the suppression of MEF2C activity. However, we also observed partial inhibition of myogenesis by NotchΔ, which lacks the ankyrin repeats but contains the region known to interact with CBF1. This is consistent with the conclusion that Notch acts through multiple mechanisms to block the myogenic program. A simplified model of the regulatory relationship between Notch and myogenic transcription factors is shown in Fig. 7. According to this model, Notch inhibits MEF2C activity, which prevents muscle gene activation by myogenic bHLH factors and MEF2C. Notch also acts through other MEF2C-independent mechanisms to inhibit the functions of myogenic bHLH factors. MEF2A and MEF2D are not responsive to Notch, but when the activity of myogenic bHLH factors is blocked by Notch, they have no effect on muscle gene expression. In addition to the effects of Notch on the transcriptional activity of myogenic bHLH factors, activated Notch has also been shown to inhibit the expression of these factors (63). Thus, multiple steps in the myogenic pathway are blocked by Notch. Indeed, recent studies in Drosophila cells have revealed at least three potential steps in the myogenic pathway perturbed by Notch (23).
FIG. 7.
Schematic of the influence of Notch on MEF2C and myogenic bHLH factor interactions. Notch signaling interferes with the myogenic activity of MEF2C. In addition, the activity of MyoD and other myogenic bHLH factors is blocked by Notch through a MEF2C-independent pathway.
Regulation of cell fate decisions by Notch.
Notch signalling regulates cell fate decisions in numerous cell types in addition to muscle. In Drosophila, Notch signalling inhibits ectodermal cells from entering a neurogenic pathway and, instead, leads them to adopt an epidermal cell fate (20, 25, 27). Suppression of neurogenesis by Notch has also been observed in vertebrate embryos, as well as in cultured cells (17, 36, 52, 56). Notch also controls the development of T lymphocytes and early hematopoietic myeloid cells and influences somite formation during vertebrate embryogenesis (14, 46, 59, 69).
Under what conditions might Notch normally inhibit MEF2C activity? During embryogenesis, MEF2C is expressed in the somite myotome beginning at about embryonic day 8.5 (19, 65). There are four Notch genes in vertebrates, which show overlapping but distinct expression patterns (23, 34, 70–72). Notch1 and Notch2 are coexpressed with MEF2C in the somites, and the Notch ligand delta is expressed on adjacent cells. Thus, Notch could play a role in modulating the early stages of myogenesis in the embryo. The observation that mice bearing null mutations in the different Notch alleles do not show defects in muscle development (14, 66) probably reflects the functional overlap among the different family members.
It is also possible that negative regulation of MEF2C function by Notch plays a role in other cell types during development. For example, MEF2C is expressed specifically in developing neurons in different regions of the brain, as well as in the early heart, spleen, and in monocytes (19, 42, 43). In light of the role of Notch in the control of neurogenesis in Drosophila sp. (25–27) and vertebrates (13, 52), it is possible that Notch signaling is important for preventing the activation of MEF2C-dependent genes in these cell types at certain stages of development. In this regard, we have shown previously that in neural cells, MEF2C can cooperate with MASH1 to activate transcription (5). Thus, the form of Notch-mediated repression described here in skeletal muscle cells may also be operative in the developing nervous system.
ACKNOWLEDGMENTS
We are grateful to the following individuals for reagents: S. Hayward, T. Kadesch, and G. Weinmaster. We also thank A. Tizenor for assistance with graphics.
This work was supported by grants from The NIH and the Muscular Dystrophy Association to E.N.O.
REFERENCES
- 1.Anant S, Roy S, VijayRaghavan K. Twist and Notch negatively regulate adult muscle differentiation in Drosophila. Development. 1998;125:1361–1369. doi: 10.1242/dev.125.8.1361. [DOI] [PubMed] [Google Scholar]
- 2.Artavanis-Tsakonis S, Matsuno K, Fortini M E. Notch signaling. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- 3.Artavanis-Tsakonis S, Simpson P. Choosing a cell fate: a view from the Notch locus. Trends Genet. 1991;7:403–408. doi: 10.1016/0168-9525(91)90264-q. [DOI] [PubMed] [Google Scholar]
- 4.Baker R, Schubiger G. Autonomous and nonautonomous Notch functions for embryonic muscle and epidermis development in Drosophila. Development. 1996;122:617–626. doi: 10.1242/dev.122.2.617. [DOI] [PubMed] [Google Scholar]
- 5.Black B L, Ligon K L, Zhang Y, Olson E N. Cooperative transcriptional activation by the neurogenic basic helix-loop-helix protein MASH1 and members of the myocyte enhancer factor-2 (MEF2) family. J Biol Chem. 1996;271:26659–26663. doi: 10.1074/jbc.271.43.26659. [DOI] [PubMed] [Google Scholar]
- 6.Black B L, Molkentin J D, Olson E N. Multiple roles for the MyoD basic region in transmission of transcriptional activation signals and interaction with MEF2. Mol Cell Biol. 1998;18:69–77. doi: 10.1128/mcb.18.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Black B L, Olson E N. Transcriptional control of muscle development by myocyte enhancer factor 2 (MEF2) proteins. Annu Rev Cell Dev Biol. 1998;14:167–196. doi: 10.1146/annurev.cellbio.14.1.167. [DOI] [PubMed] [Google Scholar]
- 8.Blaumueller C M, Qi H, Zagouras P, Artavanis-Tsakonis S. Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell. 1997;90:281–291. doi: 10.1016/s0092-8674(00)80336-0. [DOI] [PubMed] [Google Scholar]
- 9.Bour B A, O’Brien M A, Lockwood W L, Goldstein E S, Bodmer R, Taghert P H, Abmayr S M, Nguyen H T. Drosophila MEF2, a transcription factor that is essential for myogenesis. Genes Dev. 1995;9:730–741. doi: 10.1101/gad.9.6.730. [DOI] [PubMed] [Google Scholar]
- 10.Breitbart R, Liang C, Smoot L, Laheru D, Mahdavi V, Nadal-Ginard B. A fourth human MEF-2 transcription factor, hMEF2D, is an early marker of the myogenic lineage. Development. 1993;118:1095–1106. doi: 10.1242/dev.118.4.1095. [DOI] [PubMed] [Google Scholar]
- 11.Brennan T J, Chakraborty T, Olson E N. Mutagenesis of the myogenin basic region identifies an ancient protein motif critical for activation of myogenesis. Proc Natl Acad Sci USA. 1991;88:5676–5679. doi: 10.1073/pnas.88.13.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chambers A E, Kotecha S, Towers N, Mohun T J. Muscle-specific expression of SRF-related genes in the early embryo of Xenopus laevis. EMBO J. 1992;11:4981–4991. doi: 10.1002/j.1460-2075.1992.tb05605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chitnis A, Henrique D, Lewis J, Ish-Horowicz D, Kintner C. Primary neurogenesis in Xenopus embryos regulated by a homologue of the Drosophila neurogenic gene Delta. Nature. 1995;375:761–766. doi: 10.1038/375761a0. [DOI] [PubMed] [Google Scholar]
- 14.Conlon R A, Reaume A G, Rossant J. Notch1 is required for the coordinate segmentation of somites. Development. 1995;121:1533–1545. doi: 10.1242/dev.121.5.1533. [DOI] [PubMed] [Google Scholar]
- 15.Cserjesi P, Olson E N. Myogenin induces myocyte-specific enhancer binding factor MEF-2 independently of other muscle-specific gene products. Mol Cell Biol. 1991;11:4854–4862. doi: 10.1128/mcb.11.10.4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis R L, Weintraub H. Acquisition of myogenic specificity by replacement of three amino acid residues from MyoD into E12. Science. 1992;256:1027–1030. doi: 10.1126/science.1317057. [DOI] [PubMed] [Google Scholar]
- 17.de la Pompa J L, Wakeham A, Correia K M, Samper E, Brown S, Aguilera R J, Nakano T, Honjo T, Mak T W, Rossant J, Conlon R A. Conservation of the Notch signalling pathway in mammalian neurogenesis. Development. 1997;124:1139–1148. doi: 10.1242/dev.124.6.1139. [DOI] [PubMed] [Google Scholar]
- 18.del Amo F F, Smith D E, Swiatek P J, Gendron-Maguire M, Greenspan R J, McMahon A P, Gridley T. Expression pattern of Motch, a mouse homolog of Drosophila Notch, suggests an important role in early postimplantation mouse development. Development. 1992;115:737–744. doi: 10.1242/dev.115.3.737. [DOI] [PubMed] [Google Scholar]
- 19.Edmondson D G, Lyons G E, Martin J F, Olson E N. Mef2 gene expression marks the cardiac and skeletal muscle lineages during mouse embryogenesis. Development. 1994;120:1251–1263. doi: 10.1242/dev.120.5.1251. [DOI] [PubMed] [Google Scholar]
- 20.Fehon R G, Koon P J, Rebay R, Regan C L, Xu T, Muskavitch M A T, Artavanis-Tsakonis S. Molecular interactions between the protein products of the neurogenic loci Notch and Delta, two EGF-homologous genes in Drosophila. Cell. 1990;61:523–534. doi: 10.1016/0092-8674(90)90534-l. [DOI] [PubMed] [Google Scholar]
- 21.Fortini M E, Rebay I, Caron L A, Artavanis-Tsakonis S. An activated Notch receptor blocks cell-fate commitment in the developing Drosophila eye. Nature. 1993;365:555–557. doi: 10.1038/365555a0. [DOI] [PubMed] [Google Scholar]
- 22.Fortini M E, Artavanis-Tsakonis S. The Suppressor of hairless protein participates in Notch receptor signaling. Cell. 1994;79:273–282. doi: 10.1016/0092-8674(94)90196-1. [DOI] [PubMed] [Google Scholar]
- 23.Fuerstenberg, S., and E. Ginger. Multiple roles for Notch in Drosophila myogenesis. Dev. Biol., in press. [DOI] [PubMed]
- 24.Gossett L A, Kelvin D J, Sternberg E A, Olson E N. A new myocyte-specific enhancer binding factor that recognizes a coserved element associated with multiple muscle-specific genes. Mol Cell Biol. 1989;9:5022–5033. doi: 10.1128/mcb.9.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heitzler P, Simpson P. The choice of cell fate in the epidermis of Drosophila. Cell. 1991;64:1083–1092. doi: 10.1016/0092-8674(91)90263-x. [DOI] [PubMed] [Google Scholar]
- 26.Heitzler P, Simpson P. Altered epidermal growth factor-like sequences provide evidence for a role of Notch as receptor for epidermal development. Development. 1993;117:1113–1123. doi: 10.1242/dev.117.3.1113. [DOI] [PubMed] [Google Scholar]
- 27.Hoppe P E, Greenspan R J. The Notch locus of Drosophila is required in epidermal cells for epidermal development. Development. 1990;109:875–885. doi: 10.1242/dev.109.4.875. [DOI] [PubMed] [Google Scholar]
- 28.Hsieh J J-D, Henkel T, Salmon P, Robey E, Peterson M G, Hayward S D. Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol Cell Biol. 1996;16:952–959. doi: 10.1128/mcb.16.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jarriault S, Brou C, Logeat F, Schroeter E H, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 30.Jennings B, de Celis J, Delidakis C, Preiss A, Bray S. Role of Notch and achaete-scute complex in the expression of Enhancer of split bHLH proteins. Development. 1995;121:3745–3752. [Google Scholar]
- 31.Kaushal S, Schneider J W, Nadal-Ginard B, Mahdavi V. Activation of the myogenic lineage by MEF2A, a factor that induces and cooperates with MyoD. Science. 1994;266:1236–1240. doi: 10.1126/science.7973707. [DOI] [PubMed] [Google Scholar]
- 32.Kopan R, Nye J S, Weintraub H. The intracellular domain of mouse Notch: a constitutively activated repressor of myogenesis directed at the basic helix-loop-helix region of MyoD. Development. 1994;120:2385–2396. doi: 10.1242/dev.120.9.2385. [DOI] [PubMed] [Google Scholar]
- 33.Kopan R, Schroeter E H, Nye J S, Weintraub H. Signal transduction by activated mNotch: importance of proteolytic processing and its regulation by the extracellular domain. Proc Natl Acad Sci USA. 1996;93:1683–1688. doi: 10.1073/pnas.93.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lardelli M, Lendahl U. MotchA and MotchB-Two mouse Notch homologues coexpressed in a wide variety of tissues. Exp Cell Res. 1993;204:364–372. doi: 10.1006/excr.1993.1044. [DOI] [PubMed] [Google Scholar]
- 35.Leifer D, Krainc D, Yu Y-T, McDermott J, Breitbart R E, Heng J, Neve R L, Kosofsky B, Nadal-Ginard B, Lipton S A. MEF2C, a MADS/MEF2-family transcription factor expressed in a laminar distribution in cerebral cortex. Proc Natl Acad Sci USA. 1993;90:1546–1550. doi: 10.1073/pnas.90.4.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lieber T, Kidd S, Alcamo E, Corbin V, Young M W. Antineurogenic phenotypes induced by truncated Notch proteins indicate a role in signal transduction and may point to a novel function for Notch in nuclei. Genes Dev. 1993;7:1949–1965. doi: 10.1101/gad.7.10.1949. [DOI] [PubMed] [Google Scholar]
- 37.Lilly G, Galewsky S, Firulli A B, Schulz R A, Olson E N. D-MEF2: a MADS box transcription factor expressed in differentiating mesoderm and muscle cell lineages during Drosophila embryogenesis. Proc Natl Acad Sci USA. 1994;91:5662–5666. doi: 10.1073/pnas.91.12.5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lilly B, Zhao B, Ranganayakulu G, Paterson B M, Schulz R A, Olson E N. Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science. 1995;267:688–693. doi: 10.1126/science.7839146. [DOI] [PubMed] [Google Scholar]
- 39.Lindsell C E, Shawber C J, Boulter J, Weinmaster G. Jagged: a mammalian ligand that activates Notch1. Cell. 1995;80:909–917. doi: 10.1016/0092-8674(95)90294-5. [DOI] [PubMed] [Google Scholar]
- 40.Lin Q, Schwarz J, Bucana C, Olson E N. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science. 1997;276:1404–1407. doi: 10.1126/science.276.5317.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu F, Lux S E. Constitutively active human Notch1 binds to the transcription factor CBF1 and stimulates transcription through a promoter containing a CBF1-responsive element. Proc Natl Acad Sci USA. 1996;93:5663–5667. doi: 10.1073/pnas.93.11.5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lyons G E, Micales B K, Schwarz J, Martin J F, Olson E N. Expression of Mef2 genes in the mouse central nervous system suggests a role in neuronal maturation. J Neurosci. 1995;15:5727–5738. doi: 10.1523/JNEUROSCI.15-08-05727.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin J F, Schwarz J J, Olson E N. Myocyte enhancer factor (MEF) 2C: a tissue-restricted member of the MEF-2 family of transcription factors. Proc Natl Acad Sci USA. 1993;90:5282–5286. doi: 10.1073/pnas.90.11.5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin J F, Miano J M, Hustad C M, Copeland N G, Jenkins N A, Olson E N. A Mef2 gene that generates a muscle-specific isoform via alternative mRNA splicing. Mol Cell Biol. 1994;14:1647–1656. doi: 10.1128/mcb.14.3.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McDermott J C, Cardoso M C, Yu Y-T, Andres V, Leifer D, Krainc D, Lipton S A, Nadal-Ginard B. hMEF2C gene encodes skeletal muscle- and brain-specific transcription factors. Mol Cell Biol. 1993;13:2564–2577. doi: 10.1128/mcb.13.4.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Milner L A, Bigas A, Kopan R, Brashem-Stein C, Bernstein I D, Martin D I K. Inhibition of granulocytic differentiation by mNotch1. Proc Natl Acad Sci USA. 1996;93:13014–13019. doi: 10.1073/pnas.93.23.13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Molkentin J D, Black B L, Martin J F, Olson E N. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995;83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- 48.Molkentin J D, Black B L, Martin J F, Olson E N. Mutational analysis of the DNA binding, dimerization, and transcriptional activation domains of MEF2C. Mol Cell Biol. 1996;16:2627–2636. doi: 10.1128/mcb.16.6.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Molkentin J D, Olson E N. Combintorial control of muscle development by basic helix-loop-helix and MADS-box transcription factors. Proc Natl Acad Sci USA. 1996;93:9366–9373. doi: 10.1073/pnas.93.18.9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Molkentin, J. D., and E. N. Olson. Unpublished data.
- 51.Nguyen H T, Bodmer R, Abmayr S M, McDermott J C, Spoerel N A. D-mef2: a Drosophila mesoderm-specific MADS box-containing gene with a biphasic expression profile during embryogenesis. Proc Natl Acad Sci USA. 1994;91:7520–7524. doi: 10.1073/pnas.91.16.7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nye J S, Kopan R, Axel R. An activated Notch suppresses neurogenesis and myogenesis but not gliogenesis in mammalian cells. Development. 1994;120:2421–2430. doi: 10.1242/dev.120.9.2421. [DOI] [PubMed] [Google Scholar]
- 53.Olson E N, Perry M, Schulz R A. Regulation of muscle differentiation by the MEF2 family of MADS box transcription factors. Dev Biol. 1995;172:2–14. doi: 10.1006/dbio.1995.0002. [DOI] [PubMed] [Google Scholar]
- 54.Ornatsky O L, Andreucci J J, McDermott J C. A dominant-negative form of transcription factor MEF2 inhibits myogenesis. J Biol Chem. 1997;272:33271–33278. doi: 10.1074/jbc.272.52.33271. [DOI] [PubMed] [Google Scholar]
- 55.Ordentlich P, Lin A, Shen C-P, Blaumueller C, Matsuno K, Artavanis-Tsakonas S, Kadesch T. Notch inhibition of E47 supports the existence of a novel signaling pathway. Mol Cell Biol. 1998;18:2230–2239. doi: 10.1128/mcb.18.4.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pan D, Rubin G M. Kuzbanian controls proteolytic processing of Notch and mediates lateral inhibition during Drosophila and vertebrate neurogenesis. Cell. 1997;90:217–280. doi: 10.1016/s0092-8674(00)80335-9. [DOI] [PubMed] [Google Scholar]
- 57.Pollock R, Treisman R. Human SRF-related proteins: DNA binding properties and potential regulatory targets. Genes Dev. 1995;5:2327–2341. doi: 10.1101/gad.5.12a.2327. [DOI] [PubMed] [Google Scholar]
- 58.Rebay I, Fehon R G, Artavanis-Tsakonis S. Specific truncations of Drosophila Notch define dominant activated and dominant negative forms of the receptor. Cell. 1993;74:319–329. doi: 10.1016/0092-8674(93)90423-n. [DOI] [PubMed] [Google Scholar]
- 59.Robey E, Chang D, Itano A, Cado D, Alexander H, Lans D, Weinmaster G, Salmon P. An activated form of Notch influences the choice between CD4 and CD8 T cell lineages. Cell. 1996;87:483–492. doi: 10.1016/s0092-8674(00)81368-9. [DOI] [PubMed] [Google Scholar]
- 60.Sartorelli V, Huang J, Hamamori Y, Kedes L. Molecular mechanisms of myogenic coactivation by p300: direct interaction with the activation domain of MyoD and with the MADS box of MEF2C. Mol Cell Biol. 1997;17:1010–1026. doi: 10.1128/mcb.17.2.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sasai Y, Kagayama R, Tagawa Y, Shigemoto R, Nakanishi S. Two mammalian helix-loop-helix factors structurally related to Drosophila hairy and Enhancer of split. Genes Dev. 1992;6:2620–2634. doi: 10.1101/gad.6.12b.2620. [DOI] [PubMed] [Google Scholar]
- 62.Schroeter E H, Kisslinger J A, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 63.Shawber C, Nofziger D, Hsieh J J-D, Lindsell C, Bogler O, Hayward D, Weinmaster G. Notch signaling inhibits muscle cell differentiation through a CBF-1 independent pathway. Development. 1996;122:3765–3773. doi: 10.1242/dev.122.12.3765. [DOI] [PubMed] [Google Scholar]
- 64.Shore P, Sharrocks A D. The MADS-box family of transcription factors. Eur J Biochem. 1995;229:1–13. doi: 10.1111/j.1432-1033.1995.tb20430.x. [DOI] [PubMed] [Google Scholar]
- 65.Subramanian S V, Nadal-Ginard B. Early expression of the different isoforms of the myocyte enhancer factor-2 (MEF2) protein in myogenic as well as non-myogenic cell lineages during mouse embryogenesis. Mech Dev. 1996;57:103–112. doi: 10.1016/0925-4773(96)00542-4. [DOI] [PubMed] [Google Scholar]
- 66.Swiatek P J, Lindsell C E, del Amo F F, Weinmaster G, Gridley T. Notch1 is essential for postimplantation development in mice. Genes Dev. 1994;8:707–719. doi: 10.1101/gad.8.6.707. [DOI] [PubMed] [Google Scholar]
- 67.Tamura K, Taniguchi Y, Minoguchi S, Sakai T, Tun T, Furukawa T, Honjo T. Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-Jκ/Su(H) Curr Biol. 1995;5:1416–1423. doi: 10.1016/s0960-9822(95)00279-x. [DOI] [PubMed] [Google Scholar]
- 68.Ticho B S, Stainier D Y, Fishman M C, Breitbart R E. Three zebrafish MEF2 genes delineate somitic and cardiac muscle development in wild-type and mutant embryos. Mech Dev. 1996;59:205–218. doi: 10.1016/0925-4773(96)00601-6. [DOI] [PubMed] [Google Scholar]
- 69.Washburn T, Schweighoffer E, Gridley T, Chang D, Fowlkes B J, Cado D, Robey E. Notch activity influences the αβ versus γδ T cell lineage decision. Cell. 1997;88:833–843. doi: 10.1016/s0092-8674(00)81929-7. [DOI] [PubMed] [Google Scholar]
- 70.Weinmaster G, Roberts V J, Lemke G. A homolog of Drosophila Notch expressed during mammalian development. Development. 1991;113:199–205. doi: 10.1242/dev.113.1.199. [DOI] [PubMed] [Google Scholar]
- 71.Weinmaster G, Roberts V J, Lemke G. Notch2: a second mammalian Notch gene. Development. 1992;116:931–941. doi: 10.1242/dev.116.4.931. [DOI] [PubMed] [Google Scholar]
- 72.Williams R, Lendahl U, Lardelli M. Complementary and combinatorial patterns of Notch gene family expression during early mouse development. Mech Dev. 1995;53:357–368. doi: 10.1016/0925-4773(95)00451-3. [DOI] [PubMed] [Google Scholar]
- 73.Yu Y-T, Breitbart R E, Smoot L B, Lee Y, Mahdavi V, Nadal-Ginard B. Human myocyte-specific enhancer factor 2 comprises a group of tissue-restricted MADS box transcription factors. Genes Dev. 1992;6:1783–1798. doi: 10.1101/gad.6.9.1783. [DOI] [PubMed] [Google Scholar]
- 74.Yun K, Wold B. Skeletal muscle determination and differentiation: story of a core regulatory network and its context. Curr Opin Cell Biol. 1996;8:877–889. doi: 10.1016/s0955-0674(96)80091-3. [DOI] [PubMed] [Google Scholar]