Abstract
Objective:
We present the Clinical Trial Knowledge Base, a regularly updated knowledge base of discrete clinical trial eligibility criteria equipped with a web-based user interface for querying and aggregate analysis of common eligibility criteria.
Materials and methods:
We used a natural language processing (NLP) tool named Criteria2Query (Yuan et al., 2019) to transform free text clinical trial eligibility criteria from ClinicalTrials.gov into discrete criteria concepts and attributes encoded using the widely adopted Observational Medical Outcomes Partnership (OMOP) Common Data Model (CDM) and stored in a relational SQL database. A web application accessible via RESTful APIs was implemented to enable queries and visual aggregate analyses. We demonstrate CTKB’s potential role in EHR phenotype knowledge engineering using ten validated phenotyping algorithms.
Results:
At the time of writing, CTKB contained 87,504 distinctive OMOP CDM standard concepts, including Condition (47.82%), Drug (23.01%), Procedure (13.73%), Measurement (24.70%) and Observation (5.28%), with 34.78% for inclusion criteria and 65.22% for exclusion criteria, extracted from 352,110 clinical trials. The average hit rate of criteria concepts in eMERGE phenotype algorithms is 77.56%.
Conclusion:
CTKB is a novel comprehensive knowledge base of discrete eligibility criteria concepts with the potential to enable knowledge engineering for clinical trial cohort definition, clinical trial population representativeness assessment, electronical phenotyping, and data gap analyses for using electronic health records to support clinical trial recruitment.
Keywords: Clinical Trial, Eligibility Criteria, Knowledge Base, Natural Language Processing
1. Introduction
Learning from past Randomized Clinical Trials (RCTs) promises to improve the design of future RCTs. One of the barriers to successful RCTs is insufficient patient recruitment [2–5], which often results from restrictive eligibility criteria that specify the characteristics of qualifying participants. It remains challenging to optimize the feasibility and recruitment efficiency for eligibility criteria, whose definition process remains opaque, unscalable and insufficiently inclusive [6]. Clinical study designers often reuse and adapt eligibility criteria from existing protocols [7]. They rarely use data standards such as clinical terminologies or provide explicit rationale [7,8]. Thus, eligibility criteria are often excessively complex or restrictive. Butler et al. [9] compared clinical trial eligibility criteria to electronic health records (EHR) data elements of a cohort of Alzheimer’s Disease patients in order to assess the data gap for eligibility screening. The authors found many criteria do not have corresponding EHR data elements.
Efforts have been made to enable knowledge reuse of eligibility criteria. Daniel et al. developed a knowledge base called PICASSO for clinical trials [10], aiming at evaluating and critiquing clinical trial protocol design. Ravid et al. [11], proposed an ontological framework Epoch that supports the knowledge-based reasoning for the clinical trials of the immune tolerance therapies. The TrialBank project captured the information of 19 randomized controlled trials into a structured electronic knowledge base [12]. Speedie et al. proposed an information model PCRO to support the development of clinical trial management system for the community-based primary care research [13]. The collaborative database “OpenTrials” tries to integrate all available information on all clinical trials together via the donations of structured data, but the datasets contained in OpenTrials so far is very limited [14]. Sun and Loparo [15] used external medical knowledge base to improve the information extraction from clinical trials, but the extracted results are not organized and stored for further applications [16]. A relational database based on the EHR data standard of OMOP Common Data Model (CDM) v5 is created by Si et al. [17] to manage the parsed results of Alzheimer’s clinical trials, including the medical terms and their relations.
Prior knowledge bases designed for the sharing and reuse of eligibility criteria, such as Trial Bank [5], Epoch [11], SysBank [18] lack scalability. Manual annotation of eligibility criteria entails high costs for knowledge engineering and timely updates. Small scale knowledge bases with manually annotated eligibility criteria are often domain-biased and have inadequate information to support data-driven analyses. Therefore, they are underpowered for meaningful generalization and compromise the goal of population representativeness. Another challenge in creating such a knowledge base is the lack of a widely accepted representation standards for eligibility criteria. Multiple proposals for such standards have been put forth by Musen [19], Tu [20], Weng [21], Sim [22], and others since the 1980s, but none achieved acceptable scalability and adoption yet.
To remedy the aforementioned defects in existing knowledge bases for clinical trial eligibility criteria including inefficiency in knowledge engineering, insufficient extensibility, not data-driven but primarily expert-driven, and inadequate interoperability with EHR data, we aim to develop a new knowledge base that is standards-based and can be automatically populated via text knowledge engineering using advanced natural language processing methods. This study contributes an open-source Clinical Trial Knowledge Base (CTKB) that includes all discrete eligibility concepts and their frequently used attributes extracted from ClinicalTrials.gov and represents them using clinical data standards to improve their EHR interoperability.
2. Methods
2.1. The CTKB architecture
The organization for CTKB with its foundation and external applications are depicted in Fig. 1. To translate free-text criteria to structured data representations, an end-to-end data flow pipeline (Fig. 1) is developed with the following six layers: (1) Data layer with eligibility criteria collection and decomposition; (2) Criteria2Query-based NLP layer with information extraction and normalization; (3) Clinical trial metadata layer with data deposition into MySQL database; (4) Domain knowledge layer with categorization of computable eligibility criteria into domains; (5) Web components layer with eligibility criteria query, analysis, and visualization; (6) External applications layer with criteria translation into cohort definitions through ATLAS (an OHDSI open source cohort identification tool available at https://github.com/OHDSI/Atlas), Cohort representativeness (GIST) [23], electronic patient phenotyping, patient-centric trials search service (DQueST) [24], etc. The design and development of CTKB follow the goal that the stored structured eligibility criteria with their associated attributes can be easily translated into computer-interpretable standardized queries into EHR repositories. The core of CTKB is composed of three layers, i.e.: the data storage layer (Clinical trial metadata), the domain knowledge layer, and the data access layer (Web components). Thus, besides a data storage layer and a data access layer, we introduced an intermediate layer to store domain-specific data after each criterion is processed and normalized. The bottom storage layer of CTKB preserves meta information of clinical trials and eligibility rules. Moreover, the domain knowledge layer is distilled after eligibility rules are segmented into entities and standardized to concepts of different domains. Concepts, obtained from standardizing eligibility criteria entities, with the same domain (along with the metadata of a concept, such as trial id, temporal attributes, or value attributes) are grouped and saved in the same table in our database. The data access layer supports user queries and visualizations from various web components in addition to application programming interfaces (APIs) to enable external extensions of CTKB.
Fig. 1.
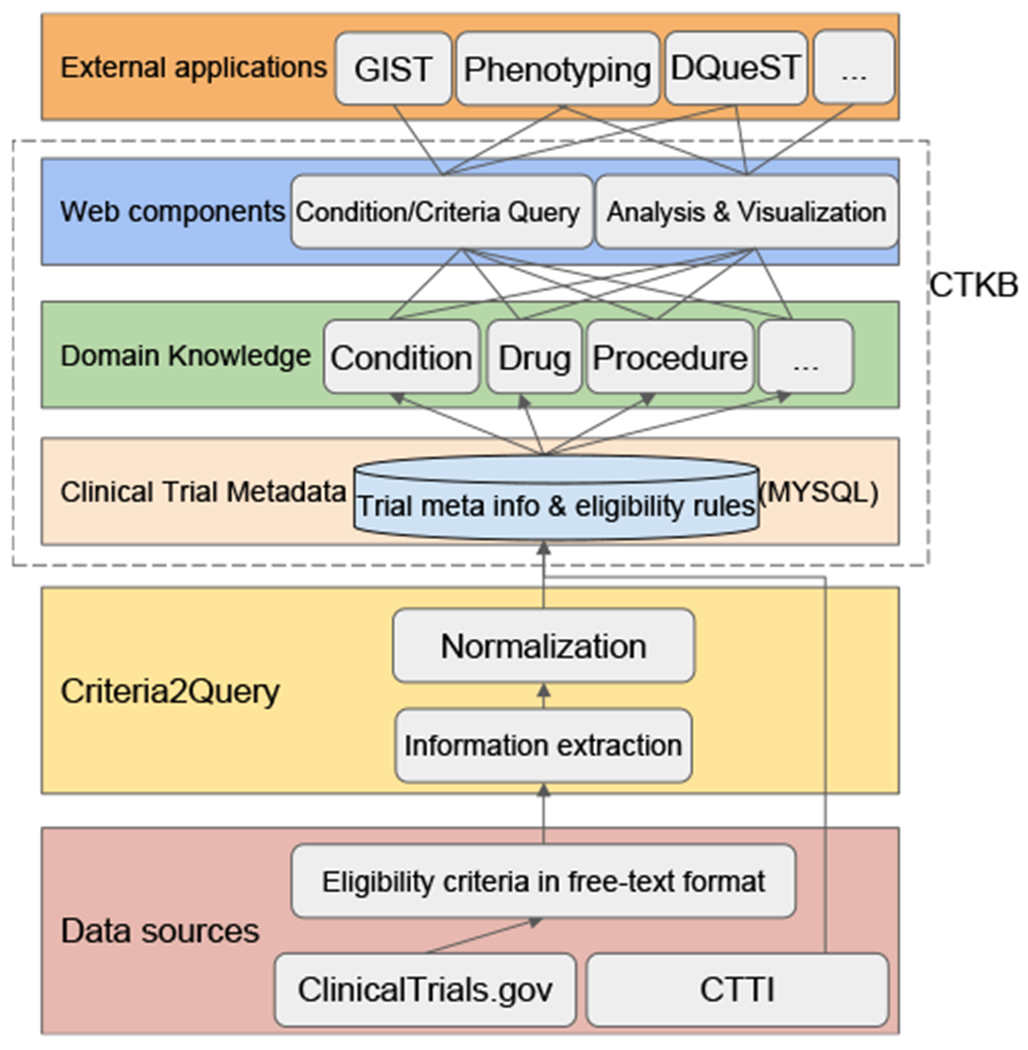
CTKB architecture with three layers including relational databases, domain knowledge, and web components supporting condition and criteria queries and statistical analysis and visualization of the selected criteria. CTTI (Clinical Trials Transformation Initiative).
All the components along the pipeline are modularized and automated to facilitate comprehensive maintenance and updates of the whole system. This arrangement also ensures that CTKB remains scalable and maintainable as it can periodically incorporate the latest increment from its data resources, i.e., ClinicalTrials.gov [25], enhancing CTKB’s sustainability.
2.2. Data source and extraction
We collected 352,110 clinical trials registered on ClinicalTrials.gov by October 1st, 2020. These trials were downloaded into plain text files and each trial was named by its National Clinical Trial ID (NCTID) as its unique identifier. All the information of a trial, including both structured and unstructured data, are retained and recorded in its corresponding text file. The structured data of a clinical trial is stored as one record in a table with the trial ID as its primary key. The columns of this table are the structured feature fields of a trial, including study type, title, status, phase, etc. For the unstructured data, we are particularly interested in eligibility criteria which are manually curated in free text. Thus, we relied on an NLP tool, Criteria2Query, to further process such data.
2.3. Criteria2Query
To parse the eligibility criteria of a clinical trial into standard data elements, we employed an open-sourced NLP tool named Criteria2Query, for its promising results on identifying entities and extracting relations from free-text eligibility criteria. Hence, we employed Criteria2Query as an information extraction tool in this study to convert free-text eligibility criteria to a machine-readable format. The data preparation is comprised of three steps, (1) recognizing named entities and attributes from eligibility criteria; (2) extracting the relations between entities and attributes; and (3) normalizing the entities and attributes. We illustrate step 1 and step 2 with an excerpt of a clinical trial on Type 2 Diabetes Mellitus (NCT01640873, available at https://clinicaltrials.gov/ct2/show/NCT01640873) in Fig. 2. For instance, from the description of inclusion criteria (upper half of Fig. 2), Body Mass Index, Type 2 Diabetes, and Metformin are recognized as Measurement, Condition, and Drug entities, respectively. The numerical expression “≤40 kg/m2” is recognized as a Value attribute, whose association with measurement Body Mass Index is implicitly identified. Similarly, from the description of exclusion criteria (lower half of Fig. 2), History of and in the past 3 months are identified as Observation entity and a Temporal attribute, respectively. Each of these recognized entities, along with its assigned OMOP CDM categories, will be standardized using OMOP vocabulary (Section 2.4) as Step 3 and then saved in our CTKB database.
Fig. 2.
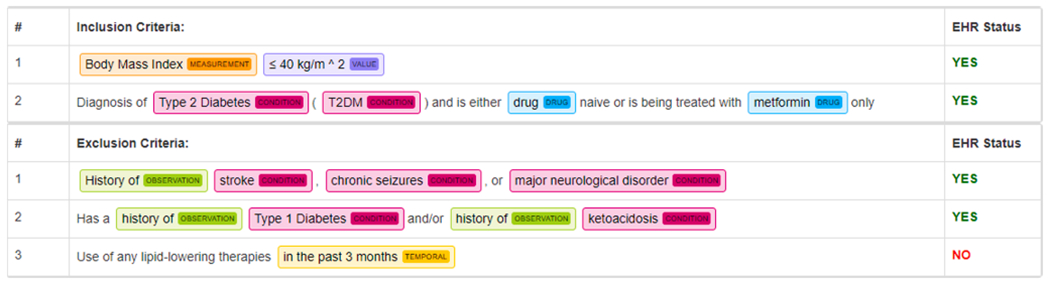
Example of named entity recognition results of Criteria2Query on excerpt of eligibility criteria of a Type 2 Diabetes Mellitus clinical trial (NCT01640873). Condition entity is denoted in red, Drug entity is denoted in blue, Measurement entity is denoted in orange, Observation entity is denoted in green, Value (Numerical) entity is denoted in purple, and Temporal entity is denoted in yellow. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.4. Concept standardization
Prior to standardization, entities recognized by Criteria2query are text snippets containing potential medical concepts or measured numerical attributes. More standardization details are described in the work of Criteri2Query [1]. The underlying vocabulary used for standardizing concepts is adapted from the OMOP CDM, which consolidates 81 vocabularies frequently utilized for different aspects of documenting health care information. By introducing a few of commonly used structural components, OMOP CDM harmonizes variability originated from those different vocabularies designed with disparate formats, quality, and comprehensiveness. Entities are mapped to standard concepts in the OMOP CDM standard vocabulary through concept normalization. For example, in Fig. 2, the recognized entity “Type 2 diabetes” is standardized to a condition concept with ID 201826 and concept name of Type 2 diabetes mellitus (originated from SNOMED CT), while “Metformin” is standardized to a drug concept with ID 1503297 and name Metformin (originated from RxNorm). Temporal attributes and numeric attributes are standardized by concept normalization and linked with corresponding concepts.
The same entity may be mapped to different concepts. For example, entity “Neck pain” can be mapped to a Condition concept Neck pain in SNOMED CT, or an Observation concept Neck or shoulder pain in UK Biobank terminology. Hence, the clinical category that an entity belongs to can help determine its proper standardization to a concept. In OMOP, more than 20 clinical entity categories (e.g., Condition, Drug, Race, Specimen, etc.) are defined and each concept is assigned with at least one domain in the Standardized Vocabulary. For our concept standardization, we adopted five domains including Condition, Drug, Measurement, Observation, and Procedure. These five domains align with the domain labels predicted by Criteria2Query in addition to its task of recognizing entities.
2.5. Database design
All data are stored and managed by a relational database. For each clinical trial, we used its NCTID as its unique identifier across all tables in the database. Table 1 lists the name and record description of 13 data tables in CTKB’s database. The 13 data tables are named according to the types of information stored in them. For instance, a record in tables with the prefix “ctgov” (short for ClinicalTrials.gov) stores the summary information of one clinical trial including its title, status, phrase, start date, etc. Tables with the prefix “ec” (short for Eligibility Criteria) store information to the granularity of one eligibility criterion rule parsed by Criteria2Query. Concepts with the same domain (along with the metadata of a concept, such as trial id, temporal attributes, or value attributes) are saved in the same table in our database. For example, the ec_condition table that holds all eligibility criterion rules specified with a condition concept after standardization. Similarly, the ec_drug table holds eligibility criterion rules specified with a drug concept after standardization. In addition, there are tables host useful statistical knowledge computed from data in other tables. For instance, the rank of criterion usage frequency (ec_criterion_rank table) is derived from ec_all_criteria table, the prevalence of a criterion used with one condition (ec_common_condition table) are derived from ctgov_trial_condition and ec_condition tables.
Table 1.
Names and descriptions of the 13 data tables in CTKB.
| Table name | Record Description |
|---|---|
| ctgov_trial_info | A trial’s title, phase, status, start date, etc. |
| ctgov_trial_condition | A trial’s target condition |
| ctgov_trial_intervention | A trial’s target intervention |
| ec_all_criteria | An eligibility criterion rule of a trial |
| ec_condition | An eligibility criterion rule specified with a condition |
| ec_drug | An eligibility criterion rule specified with a drug |
| ec_measurement | An eligibility criterion rule specified with a measurement |
| ec_observation | An eligibility criterion rule specified with an observation |
| ec_procedure | An eligibility criterion rule specified with a procedure |
| ec_common_condition | Eligibility criteria grouped by the trial’s condition types |
| ec_common_intervention | Eligibility criteria grouped by the trial’s intervention types |
| ec_common_criteria_stats | Phase count of eligibility criteria grouped by the trial’s condition types |
| ec_criterion_rank | Frequency rank of a criterion |
2.6. APIs
As an open-source database, CTKB currently supports free access to all records. The eligibility information of all clinical trials in our database may be accessed programmatically with REST API, which contains 15 types of queries. Detailed documentation and examples for each query will be available at http://ctkb.io/api.
2.7. Implementation
CTKB’s implementation is composed of four major components: a data server, a RESTful back-end web server, a front-end web server, and web-based user interface (UI). MySQL (http://www.mysql.org) is used as the database engine of the data server. Spring MVC is employed as the back-end web framework to follow the three-level modeling (Model–View–Controller, MVC). The technologies used to implement the front-end web server and UI include JSP (Java Server Pages), AJAX (Asynchronous JavaScript and XML), and Bootstrap (https://getbootstrap.com) which provides a series of web page templates. In addition, data visualization was powered by ECharts (a comprehensive charting library https://www.echartsjs.com) to add interactive charts on the web pages.
3. Results
3.1. Descriptive statistics
CTKB is deployed at http://ctkb.io. The summary statistics of CTKB can be found in Table 2. A total number of 3,647,567 eligibility criteria rules are parsed and standardized using OMOP CDM among 352,110 clinical trials. These trials are targeting to 3,844 unique conditions and 3,106 unique interventions. The total number of processed criteria entities is 8,695,529, which were mapped to 87,504 unique medical concepts in standard medical vocabularies. Among the unique 87,504 entities, 47.82% are Condition entities, 23.01% are Drug entities; 13.73% are Procedure entities; 24.70% are Measurement entities; and 5.28% are Observation entities. To breakdown the standardized 8,695,529 entities as their usage in inclusion criteria versus exclusion criteria, we observed that 34.78% of entities are used as inclusion criteria, while 65.22% of entities are used in exclusion criteria.
Table 2.
Data content of CTKB data release 2020-10-01. A total number of 3,647,567 eligibility criteria rules are processed from 352,110 clinical trials. The target of these trials covers 3,844 unique conditions and 3,106 unique interventions. A total number of 8,695,529 entities are standardized, mapping into 87,504 unique medical concepts. The distribution of 87,504 unique standardized entities among five OMOP domains. Note that the percentage sum of five domains exceeds 100% due to a concept can have multiple domains. The ratios of entities used as inclusion and exclusion criteria are reported.
| Statistics | ||
|---|---|---|
| Trials processed | 352,110 | |
| Individual criterion sentences or rules | 3,647,567 | |
| Unique conditions | 3,844 | |
| Unique interventions | 3,106 | |
| Total number of eligibility criteria entities | 8,695,529 | |
| Unique medical concepts as eligibility criteria entities | 87,504 | |
| Condition | 47.82% | |
| Drug | 23.01% | |
| Procedure | 13.73% | |
| Measurement | 24.70% | |
| Observation | 5.28% | |
| Inclusion criteria ratio | 34.78% | |
| Exclusion criteria ratio | 65.22% | |
In Fig. 3(a) we listed the top ten most frequently used criteria in CTKB. For example, Pregnant is used 159,974 times in all the trials and is the second most used criteria. To discover which diseases or topics are studied most frequently, we display the top ten most frequently studied conditions or interventions in Fig. 3(b). For example, there exists 8,238 trials targeting Depression, which is the most studied topic. Similarly, 5,643 trials were registered for Type 2 Diabetes Mellitus, which is the fifth most studied topic. Fig. 3(c) shows the number of clinical trials registered on each year. For example, the number of clinical trials was increasing from 17,194 in the year of 2010 to 25,658 in the year of 2018. The phase distribution of all the clinical trials in CTKB is calculated in Fig. 2(d), including 42,818 Phase 2 trials and 25,591 Phase 3 trials.
Fig. 3.
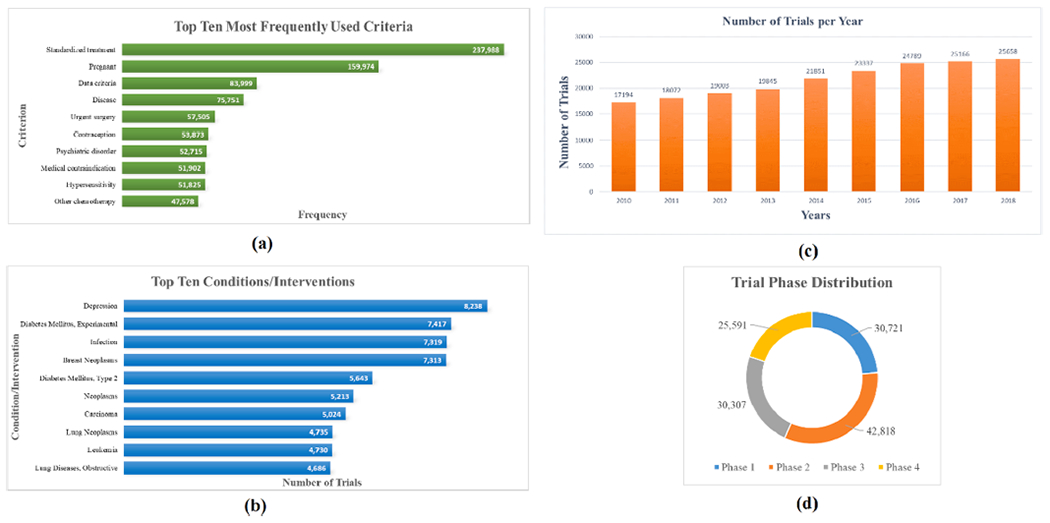
Statistics of clinical trials and criteria in CTKB. (a) The top ten most frequently used criteria. (b) The top ten conditions/interventions that are studied. (c) The number of clinical trials registered each year. (d) The phase distribution of clinical trials in CTKB.
CTKB can provide condition-centric statistical analysis in various aspects, given that all the clinical trial level and criterion level data are preserved and integrated in CTKB. We illustrate this with 3,255 clinical trials on COVID-19 that are included into CTKB.
Table 3 displays the most frequently used 10 criteria for each of the five domains (Condition, Drug, Measurement, Procedure, Observation) for COVID-19 clinical trials. For example, the most frequently used condition criterion is Disease caused by severe acute respiratory syndrome coronavirus (4,126 times), while most frequently used drug is Immunosuppressants (313 times) and the second most frequently used drug is Hydroxychloroquine (302 times).
Table 3.
The 10 most frequently used criteria (regardless inclusion or exclusion) for Condition, Drug, Measurement, Procedure, Observation domains in 3,255 COVID-19 clinical trials.
| Domain | Concept Names | Frequency |
|---|---|---|
| Condition | Disease caused by severe acute respiratory syndrome coronavirus | 4126 |
| Pregnant | 1063 | |
| Severe acute respiratory syndrome | 720 | |
| Post-term pregnancy | 689 | |
| ENT symptoms | 634 | |
| Infection | 632 | |
| Medical contraindication | 532 | |
| Condition in fetus originating in the perinatal period | 479 | |
| Fever | 474 | |
| Breastfeeding painful | 427 | |
| Drug | Immunosuppressants | 313 |
| hydroxychloroquine | 302 | |
| Anti-inhibitor coagulant complex 1 UNT | 234 | |
| phenylbutazone | 234 | |
| Serum | 201 | |
| remdesivir 100 MG Injection | 165 | |
| chloroquine | 155 | |
| Alanine | 137 | |
| coumarin | 115 | |
| Azithromycin | 109 | |
| Measurement | Centor criteria | 763 |
| Polymerase chain reaction analysis | 508 | |
| Inspired oxygen tension | 354 | |
| Respiratory rate | 345 | |
| Measurement of oxygen saturation at periphery | 331 | |
| Oxygen measurement, partial pressure, arterial | 328 | |
| RAST | 284 | |
| Malt RAST | 273 | |
| Standard pregnancy test | 233 | |
| Measurement of 2019 novel coronavirus antigen | 161 | |
| Procedure | General treatment | 1673 |
| Checking blood and blood products | 553 | |
| Mechanical ventilation | 456 | |
| Active immunization | 414 | |
| Education about hospitalization | 299 | |
| Analysis using meta-PCR | 269 | |
| Viscosupplementation | 214 | |
| Management of drug regimen | 202 | |
| Emergency surgery | 195 | |
| Intubation | 184 | |
| Observation | Reason for hospitalization | 369 |
| Contraception | 344 | |
| Home | 162 | |
| Alcohol | 147 | |
| Survival time | 116 | |
| Normal breast feeding | 92 | |
| Support | 82 | |
| Life expectancy [Time] Estimated | 78 | |
| Activity | 64 | |
| Antibody to hepatitis C virus | 62 |
3.2. User interface I — Criteria summary
From CTKB’s criteria search portal (http://ctkb.io/#/criteria), users can access a pre-defined criteria summary template page that provides insight into a specific criterion on the scale of hundreds of thousands of clinical trials as well as some aggregate analysis. A user can search a criterion by its name in a search box which is located at the top of the page (Fig. 4(a)). Once a user selects one criterion, we will retrieve and aggregate the information from CTKB database, to fill up a template which is designed from the perspective of clinical researchers. This template contains four fixed sections for criteria from all domains and one additional value distribution section for criteria from the measurement domain. The first section is a frozen row with four columns including a criterion’s usage frequency among all clinical trials processed in CTKB, its frequency used as inclusion criterion, its frequency used as exclusion criterion, and its rank among all criteria in our knowledge base. Below the first section is the Disease Concept Distribution section (Fig. 4(b)). This section shows the counts of a criterion used as inclusion criterion (red bar) and exclusion criterion (black bar) binned by target disease. The diseases are sorted by the total count of this criterion used in the clinical trials. Users can use the seamless horizontal scrolling bar to navigate to different groups of disease and use the mouse wheel to zoom in or out to view various numbers of diseases. The data and picture for this figure are downloadable through the buttons on the right top corner of the figure.
Fig. 4.
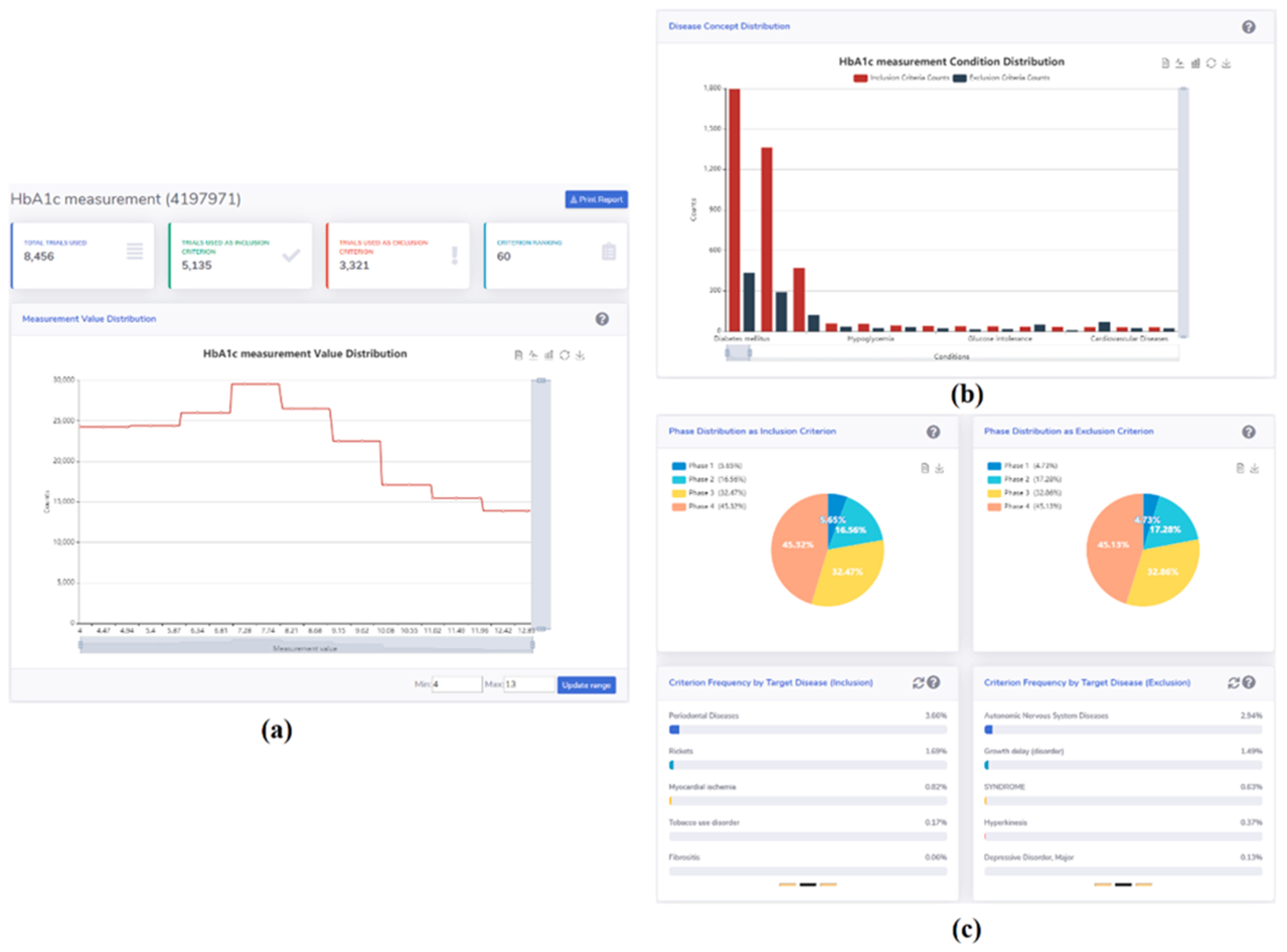
The summary page for the HbA1c criterion.
The third section is the Phase Distribution of a criterion (Fig. 4(c) top part). We use two pie charts to represent the phase distributions of a criterion used as inclusion criteria and exclusion criteria, respectively. We display the percentage of a criterion used in four phases in each pie chart.
The fourth section is the Criteria Frequency by Target Disease (Fig. 4(c) bottom part). In this section, we show that the frequency percentage of a criterion among all the criteria that are used for a target disease. The left carousel randomly displays the frequency percentage of such criterion used as an inclusion criterion for 15 diseases, with 5 diseases per group. The right carousel randomly displays 15 diseases for which this criterion is used as an exclusion criterion. The refresh button on the right top corner for each carousel can be used to randomly generate another group of 15 diseases.
Another section, called Measurement Value Distribution (bottom part of Fig. 4(a)), is added on top of the second section if the searched criterion is from the measurement domain. This section shows, among all clinical trials, what is the value range of a measurement criterion and how the frequency of these values are distributed within the range. A user can customize the range by specifying the lower and upper limits in the “Min” and “Max” input boxes, respectively, and click on the “Update range” button to update the drawing. To view the counts of values in a specific range, users can use the mouse wheel to zoom in or out. Note that the description of each section will be displayed by clicking on the question mark on the right side of the section name row. The results can be downloaded as a pdf file.
3.3. User interface II — Cohort definition
We integrated a cohort definition template (http://ctkb.io/#/condition) for users who are interested in defining cohorts for clinical trial recruitment: i.e., a user can search a condition or intervention and select relevant criteria from the returned results, then use these criteria to create a cohort definition. The three steps supported in this template are: (1) query, (2) selection, and (3) projection. The query capability of step 1 consists of a single search-box which lies on top of the page (Fig. 5(a)). A user’s input into this search box will trigger the retrieval of all criteria related to the input. The returned inclusion and exclusion criteria, ranked by their frequency with the searched condition/intervention, are shown in two tables separately. In step 2, users are free to select one or multiple criteria to be included in patient cohort definition, a user can toggle the checkbox associated with this criterion. After a group of criteria is selected, a user can view the additional information of the selected criteria by clicking on the button “Begin Building Patient Cohort” to transit to the detail page (Fig. 5(b)). On this detail page, a user can review the selected criteria and make modifications, as necessary. For example, for a criterion from measurement domain, a user can customize the value range of such numerical criterion. In step 3 (Fig. 5(c)), the selected criteria can be used to build cohort definition through three options for different types of users: (1) automatically map these criteria into ATLAS for building a queryable cohort definition; (2) download the JSON format of the selected criteria; (3) generate a human readable text for these criteria. These three options address the varying needs around eligibility criteria among different stakeholders, including experienced ATLAS researchers, data analysts, and lay persons (potential participants and IRB personnel).
Fig. 5.
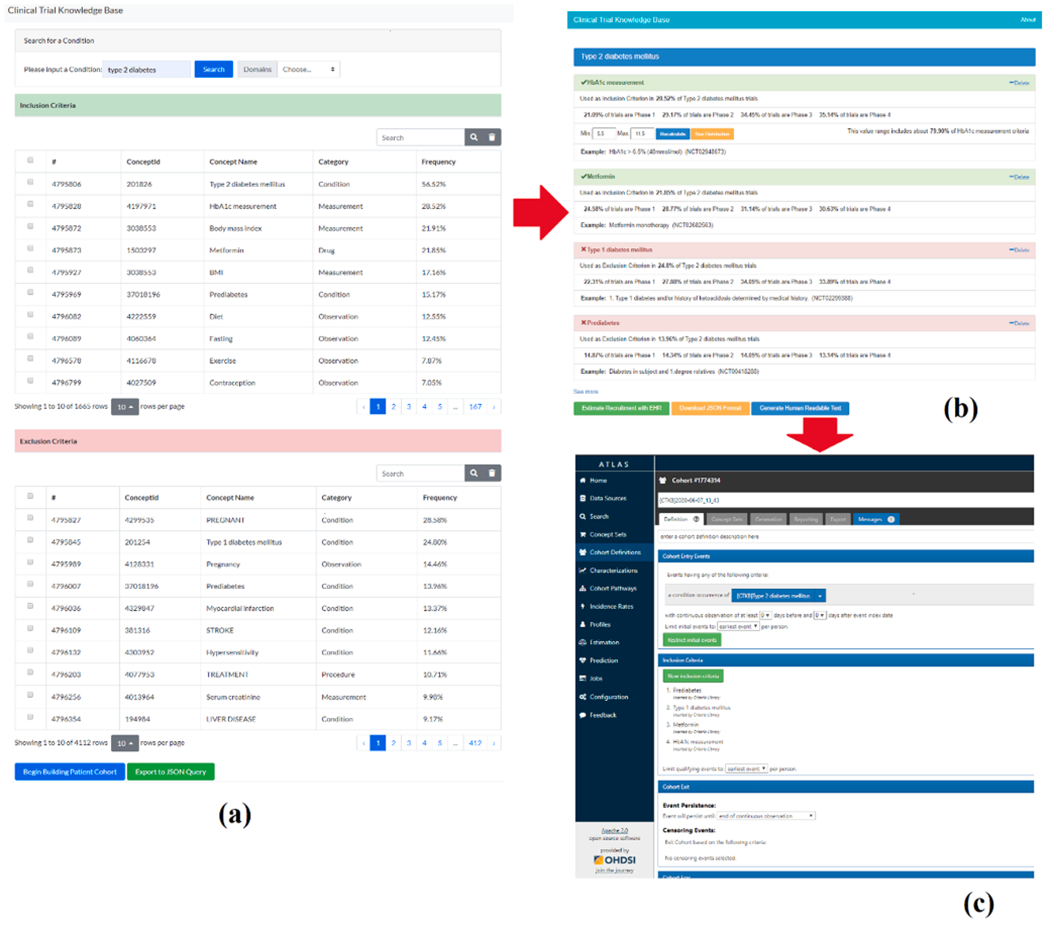
Example of using CTKB’s cohort definition template to define criteria for Type 2 diabetes patient cohort. (a) Search criteria by Type 2 diabetes, (b) select and modify criteria concept encoding, and (c) convert selected criteria concept codes to an ATLAS cohort definition for querying synthetic patient EHR database for protocol feasibility assessment.
3.4. CTKB for electronic patient phenotyping
We demonstrate an example of using CTKB for Electronic Phenotyping (mining phenotyping knowledge from clinical trial eligibility criteria).
Despite its widely recognized importance, generating accurate and sharable clinical phenotypes is labor intensive and time-consuming. The eMERGE Network is a multi-site collaborative network combining biorepositories with electronic health record systems for genomic discovery and electrical phenotyping research [26]. eMERGE phenotype algorithms were developed, validated, and implemented by a consortium of phenotyping researchers. Phenotypes implemented by eMERGE are represented by narrative definitions, codes for clinical concepts, and logic flowcharts. Hence eMERGE phenotypes are a reliable knowledge source for providing solid phenotyping variables. One of the major bottlenecks in eMERGE’s research is the efficiency and accuracy in generating and sharing phenotypes [27]. Mining phenotyping knowledge from other sources, such as clinical trial eligibility criteria, can facilitate this process and even potentially validate the results. Given the similarity between eligibility criteria rules and eMERGE phenotyping rules, we hypothesize that CTKB can be a potential knowledge source for phenotyping.
To verify our hypothesis that CTKB can serve as a source of valuable and reusable phenotype knowledge, we compared the top eligibility criteria queried from CTKB with manually annotated criteria used for phenotyping by the eMERGE community. Ten diseases were randomly selected among 53 diseases annotated by eMERGE researchers. To ensure reasonable comparison, all variables without eligibility value (e. g., Has visit or Note count) were excluded from this analysis. For each variable from the eMERGE phenotype, we verified if this variable exists in the top 25 inclusion or exclusion criteria for the appropriate disease that are automatically extracted from CTKB. This is denoted as Hit@25, i.e., if a variable is found among the top 25 criteria, it is a hit. Otherwise, it is not hit. The Hit rate is then calculated as the percentage of all variables found in the top 25 criteria, highlighting overlap between automatically extracted criteria in CTKB and human generated concepts from eMERGE.
The ten selected diseases for analysis are shown in Table 4 along with number of trials in the CTKB, number of eMERGE variables, the Hit@25 count, and the hit rate for each disease. Three diseases have hit rates of 100% and the lowest agreement rate found in Colorectal Cancer is 42.86%. On average, out of 156 eMERGE variables, 121 are identified in CTKB, result in an 77.56% hit rate. The Hit@25 scores were also generated according to eMERGE variable domains (Table 5). Three domains had hit rates of 100% and the lowest hit rate was 50% found in the Note variables. Averaged across all variable domains, the hit rate was 82.48%. Overall, this analysis shows a high recall of automatically identified eligibility concepts against manually curated variable concepts from the eMERGE community and underscores the utility of reusing criteria knowledge from CTKB for future phenotyping research.
Table 4.
Ten target diseases with CTKB trial count, eMERGE variable count, Hit@25 count, and hit rate.
| Phenotype | Trial Count | Variable Count | Hit@25 | Hit Rate |
|---|---|---|---|---|
| Type 2 Diabetes Mellitus | 5,643 | 20 | 20 | 100.00% |
| Rheumatoid Arthritis | 2,021 | 6 | 6 | 100.00% |
| Diverticulosis and Diverticulitis | 145 | 8 | 8 | 100.00% |
| Benign Prostatic Hyperplasia | 436 | 15 | 14 | 93.33% |
| Chronic Obstructive Lung Disease | 2,618 | 13 | 12 | 92.31% |
| Bipolar Disorder | 1,148 | 9 | 8 | 88.89% |
| Hypothyroidism | 133 | 17 | 15 | 88.24% |
| Venous Thromboembolism | 422 | 12 | 10 | 83.33% |
| Chronic Kidney Disease | 1,436 | 28 | 16 | 57.14% |
| Colorectal Cancer | 3,033 | 28 | 12 | 42.86% |
| Total | 17,035 | 156 | 121 | 77.56% |
Table 5.
eMERGE variable domains with variable count, Hit@25 count and hit rate.
| Variable Domain | Variable Count | Hit@25 | Hit Rate |
|---|---|---|---|
| Disease | 48 | 40 | 83.33% |
| Lab Tests | 40 | 21 | 52.5% |
| Demographic | 26 | 23 | 88.46% |
| Medication | 17 | 16 | 94.12% |
| Procedure | 9 | 8 | 88.89% |
| Observation | 6 | 5 | 83.33% |
| Report | 3 | 3 | 100% |
| Phenotype | 3 | 2 | 66.67% |
| Note | 2 | 1 | 50% |
| Family History | 1 | 1 | 100% |
| Problem List | 1 | 1 | 100% |
| Average | 82.48% |
4. Discussion
ClinicalTrials.gov is a valuable and reusable source for clinical trial designs, Tasneem et al. developed the Aggregate Analysis of ClinicalTrials.gov (AACT) database as a publicly accessible analysis dataset [28]. AACT allows filtering and aggregation of trials by study descriptors, such as study phase, intervention type, recruiting status, etc. However, the free-text eligibility rules are not included into AACT to support patient cohort definition and characteristic analyses in a programmatic and scalable way. CTKB leveraged NLP tools to automatically convert unstructured eligibility criteria into computable entities and attributes, to optimize reusing eligibility criteria for future clinical studies based on past studies. He et al. developed a tool, named VITTA [29], to visualize aggregated clinical trial study populations using numeric expressions for some medical conditions in ClinicalTrials.gov. VITTA was built upon a database called COMPACT (Commonalities in Target Populations of Clinical Trials [30]), which extracted prefixed numerical and categorical eligibility features (e.g., HbA1c or BMI for Type 2 diabetes studies) from clinical trials registered on ClinicalTrials.gov. While VITTA was built upon traditional text mining techniques, CTKB leveraged the advanced NLP tool Criteria2Query for better accuracy in named entity recognition and relation extraction. In addition, while VITTA profiles target populations with one condition using only one quantitative eligibility criterion each time, CTKB overcome this limitation by analyzing all eligibility criteria associated with one condition, and further enable correlation analyses of eligibility criteria and conditions to the scale of tens of thousands of clinical trials.
The National Institutes of Health (NIH) advocates the use of Common Data Elements (CDEs) to facilitate comparing and combining data across studies, including data elements derived from electronic health records [31]. The use of CDEs can facilitate the integration of patient clinical data from diverse sources and allows interoperability between databases. In our case, if clinical trials, particularly eligibility criteria used for patient recruitment, are created using the same data elements and measures, then researchers can compare data across studies more easily and accurately. CTKB’s content are mapped to data elements using OMOP CDM, which is a framework to accommodate CDEs for organizing and standardizing observational databases. Such a framework allows seamlessly execution of a number of standard queries and analytic methods built on the basis of OMOP structure. The effective conversion of a source database into a CDM is generally assessed whether an acceptable proportion of terms and database records can be mapped using the common vocabularies. For example, Overhage et al. [32] transformed data from five different observational databases (a mix of US claims databases and EHR data) into separate CDM instances and concluded that a range of 93.2–99.7% for conditions and 88.8–97.6% for medications are mapped. Matcho et al. [33] transformed the Clinical Practice Research Datalink (CPRD) to the OMOP CDM, with 99.9% of database condition records and 89.7% of database drug records were mapped. To evaluate the effective conversion of data elements in CTKB, we calculated ratios of entities that are recognized by Critiera2Query but not standardized using OMOP CDM for both Condition and Drug domains. We found that only 1.2% of eligibility criteria with condition entities and 0.28% of drug entities were not mapped into OMOP standardized vocabularies.
4.1. CTKB as backend knowledge source for other systems
CTKB can potentially interact with other information extraction or analytics tools for mining meaningful knowledge from clinical trials. We propose the linking of CTKB with two external tools: GIST (calculating a trial’s representativeness of real-world patients in the granularity of criteria) and DQueST (providing patient-centered trial search services).
4.1.1. GIST
To assess the population representativeness of a trial, its eligibility criteria will be compared to the profile of the target patient population. For example, a trial testing a new drug in Type 2 Diabetes Mellitus should represent the overall Type 2 Diabetes Mellitus patient population. To address this, a tool was published by Sen et al. called Generalizability Index for Study Traits (GIST) [23]. This tool uses normalized eligibility criteria stored in OMOP format and a subset of de-identified patient data in the trial’s target population to statistically assess the degree to which the criteria represent real-world populations, both at the trial level and the criterion level. Further, using this statistic on 16 sepsis trials, they found a significant correlation with reported serious adverse events (SAEs) such that trials with more inclusive/less restrictive eligibility criteria experienced fewer SAEs [23]. GIST could potentially take advantage of the comprehensive distribution of normalized eligibility criteria provided by CTKB, to statistically prioritize each individual criterion’s impact to patients filtering. This enables GIST to rigorously calculate the clinical trial representativeness or generalizability, and allow trial designers and researchers alike to identify restrictive criteria and adjust accordingly.
4.1.2. DQueST
Conventional patient-facing questionnaires are often designed for a specific trial and tend to be long (up to a few hundred questions) due to their static nature and hence involve tedious efforts (up to hours) for patients to answer [34]. The adoption of EHRs has made it possible to use e-screening to identify eligible patients. Many trial search engines such as ClinicalTrials.gov, findMeCure (www.findmecure.com), and trialstoday (www.trialstoday.org), provide patient-centered trial search services and while these tools use keyword-based methods to retrieve relevant trials, this often leads to information overload for the patient. DQueST [24] provides a novel dynamic questionnaire that prompts clinical question in real-time based on a patient’s answers to previous questions and common eligibility criteria in the disease they are searching for, drastically reducing questionnaire length and information overload for the trial seeker. DQueST can be powered by CTKB so that DQueST can access eligibility criteria concepts and relations for any disease from all trials on ClinicalTrials.gov, and further rigorously select the best clinical questions by ranking relevant criteria with their corresponding information gains derived from aggregate analysis results of clinical trial eligibility criteria.
4.2. Limitations
One major limitation of this work is the lack of comprehensive evaluation of CTKB. A comprehensive assessment consists of three general categories: content-quality assessment (e.g., normalization accuracy); task-based assessment (e.g., feasibility for external usage); and user-centered assessment (e.g., user interface friendliness). Regarding data quality, CTKB inherited from Criteria2Query the accuracy of converting free-text eligibility criteria into standardized entities and attributes. Therefore, we can quantify the normalization quality of CTKB’s products to some extent by examining the performance of Criteria2Query. Criteria2Query achieved 0.795 and 0.805 F1 score for entity recognition and relation extraction, respectively, in an experiment that involves 125 sentences of free-text eligibility criteria extracted from 10 clinical trials covering different disease domains, encompassing 215 entities and 34 relations. There was no overlap between this test data and the training data. Evaluation was based on end-to-end results of Criteria2Query. Two domain experts generated the gold standards based on inter-rater agreement by reviewing all cohort definitions independently. To assess CTKB’s feasibility for extension, we reported CTKB’s high recall of overlapping with eMERGE phenotyping variables and added our rationale on how CTKB can support GIST and DQueST calculation: CTKB enables the queries of common eligibility criteria variables and their uses in GIST and DQueST, both using discrete common eligibility criteria variables to either match with electronic health records variables or to optimize clinical trial search. The technical details and evaluations for GIST and DQueST were provided elsewhere [23,24,35]. The usability of CTKB remains untested. We plan to invite diverse users to gain their experience and feedback on using our search modules and information templates.
4.3. Future work
The external use cases of CTKB demonstrate the importance and potential of clinical research driven by the clinical trial eligibility criteria. Our effort to establish CTKB was prompted by the great need of researchers to access a comprehensive dataset of standardized eligibility criteria. To accommodate the increasing demand for large-scale analysis, we will implement a process to minimize the data gap between CTKB and ClinicalTrials.gov by periodically processing new clinical trials that are registered on ClinicalTrials.gov.
Since CTKB’s content is fully automated using underlying machine learning natural language processing tools, the errors reside in entity parsing and concept mapping will inevitably propagate to CTKB. Thus, to strive for higher level of data accuracy and reusability, we will invite experienced reviewers to uncover data inconsistency, concept normalization error, and Boolean logics of the eligibility rules. One major issue we are working on is normalizing value (range) for all the measurement concept. For example, the value for Aspartate transaminase (AST) measurement are described with flexible free text such as “<40 mL/min”, “≤2.5 times upper limit of normal (ULN)”, “<5% below the lower limit of normal”, and “<35%”. The freedom of using different unit or existing normal limits proposes a serious challenge for normalizing measurement concept with consistent and comprehensive values.
Furthermore, we will collaborate with clinicians and physicians to survey their requirements and research interests for providing further advanced and meaningful analysis. In addition, we will conduct usability tests on CTKB to evaluate its feasibility in various clinical research practice. Users will be invited to validate the free-text parsing and mapping results, and their feedback will be recorded and used to further improve Criteria2Query’s accuracy on extracting eligibility criteria entities and attributes.
To enrich CTKB’s APIs for advanced external usage, we also plan to support more complicated searches such as criteria combination with Boolean operations. In the future version of CTKB, users are allowed to manually set the priority of the search criteria by changing the positions of brackets in the query string.
5. Conclusions
CTKB is a comprehensive knowledge base. It enables explorative analyses of clinical trial eligibility criteria. Comparing to existing clinical trial knowledge bases, the content of CTKB is more dynamic and current through its use of advanced NLP tools. Our experiment with eMERGE phenotypes shows the potential of reusing criteria knowledge from CTKB for phenotyping knowledge engineering. It also promises to ease cohort definitions and to facilitate protocol feasibility assessment at scale.
Acknowledgements
This project was sponsored by National Library of Medicine grant R01LM009886-11 and National Center for Advancing Clinical and Translational Science grants UL1TR001873 and 1OT2TR003434-01.
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].Yuan C, Ryan PB, Ta C, Guo Y, Li Z, Hardin J, et al. , Criteria2Query: a natural language interface to clinical databases for cohort definition, J. Am. Med. Inform. Assoc 26 (4) (2019) 294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Lovato LC, Hill K, Hertert S, Hunninghake DB, Probstfield JL, Recruitment for controlled clinical trials: literature summary and annotated bibliography, Control. Clin. Trials 18 (4) (1997) 328–352. [DOI] [PubMed] [Google Scholar]
- [3].Fogel DB, Factors associated with clinical trials that fail and opportunities for improving the likelihood of success: a review, Contemp. Clin. Trials Commun 11 (2018) 156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Jenkins V, Farewell V, Farewell D, Darmanin J, Wagstaff J, Langridge C, et al. , Drivers and barriers to patient participation in RCTs, Br. J. Cancer 108 (7) (2013) 1402–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Mills EJ, Seely D, Rachlis B, Griffith L, Wu P, Wilson K, et al. , Barriers to participation in clinical trials of cancer: a meta-analysis and systematic review of patient-reported factors, Lancet Oncol. 7 (2) (2006) 141–148. [DOI] [PubMed] [Google Scholar]
- [6].Richesson RL, Rusincovitch SA, Wixted D, Batch BC, Feinglos MN, Miranda ML, et al. , A comparison of phenotype definitions for diabetes mellitus, J. Am. Med. Inform. Assoc 20 (e2) (2013) e319–e326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hao T, Rusanov A, Boland MR, Weng C, Clustering clinical trials with similar eligibility criteria features, J. Biomed. Inform 52 (2014) 112–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Yaman A, Chakrabarti S, Sen A, Weng C, How have cancer clinical trial eligibility criteria evolved over time?, in: AMIA Summits on Translational Science Proceedings, 2016, 2016, p. 269. [PMC free article] [PubMed] [Google Scholar]
- [9].Butler A, Wei W, Yuan C, Kang T, Si Y, Weng C, The data gap in the EHR for clinical research eligibility screening, in: AMIA Summits on Translational Science Proceedings, 2018, 2018, p. 320. [PMC free article] [PubMed] [Google Scholar]
- [10].Rubin DL, Gennari J, Musen MA, Knowledge representation and tool support for critiquing clinical trial protocols. Proceedings of the AMIA Symposium, American Medical Informatics Association, 2000. [PMC free article] [PubMed] [Google Scholar]
- [11].Shankar RD, Martins SB, O’Connor MJ, Parrish DB, Das AK, Epoch: an ontological framework to support clinical trials management. Proceedings of the International Workshop on Healthcare Information and Knowledge Management, 2006. [Google Scholar]
- [12].Sim I, Olasov B, Carini S, The Trial Bank system: capturing randomized trials for evidence-based medicine. AMIA Annual Symposium proceedings AMIA Symposium, American Medical Informatics Association, 2003. [PMC free article] [PubMed] [Google Scholar]
- [13].Speedie SM, Taweel A, Sim I, Arvanitis TN, Delaney B, Peterson KA, The Primary Care Research Object Model (PCROM): a computable information model for practice-based primary care research, J. Am. Med. Inform. Assoc 15 (5) (2008) 661–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Goldacre B, Gray J, OpenTrials: towards a collaborative open database of all available information on all clinical trials, Trials 17 (1) (2016) 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Sun Y, Loparo K, Information extraction from free text in clinical trials with knowledge-based distant supervision. 2019 IEEE 43rd Annual Computer Software and Applications Conference (COMPSAC), IEEE, 2019. [Google Scholar]
- [16].Sun Y, Loparo K, Knowledge-guided text structuring in clinical trials, arXiv preprint arXiv:191212380, 2019.
- [17].Si Y, Weng C, An OMOP CDM-based relational database of clinical research eligibility criteria, Stud. Health Technol. Inform 245 (2017) 950. [PMC free article] [PubMed] [Google Scholar]
- [18].Carini S, Sim I, SysBank: a knowledge base for systematic reviews of randomized clinical trials. AMIA Annual Symposium Proceedings, American Medical Informatics Association, 2003. [PMC free article] [PubMed] [Google Scholar]
- [19].Musen M, Rohn J, Fagan L, Shortliffe E, Knowledge engineering for a clinical trial advice system: uncovering errors in protocol specification, Bull. Cancer 74 (3) (1987) 291–296. [PubMed] [Google Scholar]
- [20].Tu SW, Kemper CA, Lane NM, Carlson RW, Musen MA, A methodology for determining patients’ eligibility for clinical trials, Methods Inf. Med 32 (04) (1993) 317–325. [PubMed] [Google Scholar]
- [21].Weng C, Gennari JH, McDonald DW, A collaborative clinical trial protocol writing system, Medinfo (2004). [PubMed] [Google Scholar]
- [22].Sim I, Detmer DE, Beyond trial registration: a global trial bank for clinical trial reporting, PLoS Med. 2 (11) (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sen A, Chakrabarti S, Goldstein A, Wang S, Ryan PB, Weng CGIST, 2.0: A scalable multi-trait metric for quantifying population representativeness of individual clinical studies, J. Biomed. Inform 63 (2016) 325–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Liu C, Yuan C, Butler AM, Carvajal RD, Li ZR, Ta CN, et al. , DQueST: dynamic questionnaire for search of clinical trials, J. Am. Med. Inform. Assoc 26 (11) (2019) 1333–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zarin DA, Tse T, Williams RJ, Califf RM, Ide NC, The ClinicalTrials.gov results database—update and key issues, N. Engl. J. Med 364 (9) (2011) 852–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].McCarty CA, Chisholm RL, Chute CG, Kullo IJ, Jarvik GP, Larson EB, et al. , The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies, BMC Med. Genomics 4 (1) (2011) 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Gottesman O, Kuivaniemi H, Tromp G, Faucett WA, Li R, Manolio TA, et al. , The electronic medical records and genomics (eMERGE) network: past, present, and future, Genet. Med 15 (10) (2013) 761–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Tasneem A, Aberle L, Ananth H, Chakraborty S, Chiswell K, McCourt BJ, et al. , The database for aggregate analysis of ClinicalTrials.gov (AACT) and subsequent regrouping by clinical specialty, PloS One 7 (3) (2012) e33677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].He Z, Carini S, Sim I, Weng C, Visual aggregate analysis of eligibility features of clinical trials, J. Biomed. Inform 54 (2015) 241–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].He Z, Carini S, Hao T, Sim I, Weng C, A method for analyzing commonalities in clinical trial target populations. AMIA Annual Symposium Proceedings, American Medical Informatics Association, 2014. [PMC free article] [PubMed] [Google Scholar]
- [31].Cohen MZ, Thompson CB, Yates B, Zimmerman L, Pullen CH, Implementing common data elements across studies to advance research, Nurs. Outlook 63 (2) (2015) 181–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Overhage JM, Ryan PB, Reich CG, Hartzema AG, Stang PE, Validation of a common data model for active safety surveillance research, J. Am. Med. Inform. Assoc 19 (1) (2012) 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Matcho A, Ryan P, Fife D, Reich C, Fidelity assessment of a clinical practice research datalink conversion to the OMOP common data model, Drug Saf 37 (11) (2014) 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Rothman KJ, Mikkelsen EM, Riis A, Sørensen HT, Wise LA, Hatch EE, Randomized trial of questionnaire length, Epidemiology 20 (1) (2009) 154. [DOI] [PubMed] [Google Scholar]
- [35].Weng C, Li Y, Ryan P, Zhang Y, Liu F, Gao J, et al. , A distribution-based method for assessing the differences between clinical trial target populations and patient populations in electronic health records, Appl. Clin. Inform 5 (2) (2014) 463. [DOI] [PMC free article] [PubMed] [Google Scholar]


