Abstract
Global conservation policy and action have largely neglected protecting and monitoring genetic diversity—one of the three main pillars of biodiversity. Genetic diversity (diversity within species) underlies species’ adaptation and survival, ecosystem resilience, and societal innovation. The low priority given to genetic diversity has largely been due to knowledge gaps in key areas, including the importance of genetic diversity and the trends in genetic diversity change; the perceived high expense and low availability and the scattered nature of genetic data; and complicated concepts and information that are inaccessible to policymakers. However, numerous recent advances in knowledge, technology, databases, practice, and capacity have now set the stage for better integration of genetic diversity in policy instruments and conservation efforts. We review these developments and explore how they can support improved consideration of genetic diversity in global conservation policy commitments and enable countries to monitor, report on, and take action to maintain or restore genetic diversity.
Keywords: genetic diversity, policy, adaptation, monitoring, indicators
Now is a critical time to embrace transformational change in society's relationship with nature, to conserve biodiversity, and to support resilient ecosystems. This will require appropriate targets and goals, coordinated and ambitious action, and ongoing monitoring with scalable, relevant, and reliable measures of progress, for all levels of biodiversity (Díaz et al. 2020, WWF 2020). Numerous biodiversity initiatives are facing this challenge. The foremost global commitment—the Convention on Biological Diversity (CBD)—has had five components since its inception in 1992: to conserve the three levels of biodiversity (genetic, taxonomic, and ecosystem), to achieve sustainable use of nature's contributions to people, and to ensure shared and equitable benefits from such use (www.cbd.int). The CBD Secretariat, Parties, and experts are currently drafting priorities, goals, targets, and indicators for 2020–2030 and beyond; a final agreement is expected in 2021. Other relevant initiatives making biodiversity and societal commitments include the Sustainable Development Goals (SDG) 2020–2030 Decade of Action, the Global Strategy for Plant Conservation (GSPC) post-2020 goals, the UN Decade of Ocean Science, the EU Biodiversity Strategy for 2030, the IPBES work plan for 2030, and the International Union for Conservation of Nature's (IUCN) Key Biodiversity Areas. In addition to existing policy, ambitious proposals exist for safeguarding species’ biodiversity, such as reducing extinctions to near zero (Rounsevell et al. 2020), and for protecting ecosystems, such as preserving 30%–50% of land and sea area.
However, ambitious, quantitative goals for genetic diversity are lacking (Díaz et al. 2020, Hoban et al. 2020a). Genetic erosion is ongoing, in rare and common species (Stoffel et al. 2018, Leigh et al. 2019, CBD 2020a). This loss is serious; genetic diversity (heritable variation among individuals and populations, encoded in DNA, e.g., intraspecific diversity) is necessary for species to adapt to the degree and pace of modern environmental change, including climate change and diseases. Long-term species survival critically depends on it. It is also important for short-term survival to avoid inbreeding depression. The mortality rate for the offspring of related parents is often 30%– 40% higher than when the parents are unrelated, and the surviving inbred offspring often have lower health, growth rates, or fertility (Ralls et al. 1979, Charpentier et al. 2007). Genetic diversity also underlies resilient and diverse ecosystems and is a resource for innovation and a margin of safety to protect the welfare of society in a changing world (Des Roches et al. 2021).
Because international biodiversity conservation instruments are currently being revised, there is a unique opportunity to commit to action to avert genetic diversity loss and to advance monitoring of genetic change over time (Díaz et al. 2020, Hoban et al. 2020a, Des Roches et al. 2021). A strong commitment to ambitious genetic diversity conservation is needed and is feasible, especially as genetics gains more importance in ecology and natural resource management (McCallen et al. 2019). However, recent drafts for the CBD post-2020 framework have a vague commitment to genetic diversity conservation, a single biodiversity goal rather than a goal for each of the three levels (CBD 2020a, 2020b), and proposed genetic indicators that are weak and primarily restricted to agricultural species (Hoban et al. 2020a).
In this article, we critically examine the enabling conditions for global agreements to explicitly and comprehensively commit to strong goals, concerted action, and monitoring for genetic diversity (figure 1). We first explain recent findings showing that nature and society need the resilience provided by genetic diversity more than ever, along with findings that document rapid, alarming loss of genetic diversity. We then review monitoring tools to effectively incorporate genetic diversity under global policy frameworks, reliable indicators based on proxies, and infrastructure and technology to affordably collect, share, and interpret genetic data. Then we highlight successful regional policies protecting genetic diversity and adaptive processes and capacity building through international partnerships. This breadth of topics and the inclusion of recent and practical solutions to longstanding issues in genetic monitoring complement and build on previous reviews in conservation genetics (e.g., Smith et al. 2014, Shafer et al. 2015). We conclude that commitment to genetic diversity conservation, supported by monitoring using a combination of DNA based studies and available proxies, is now both possible and necessary.
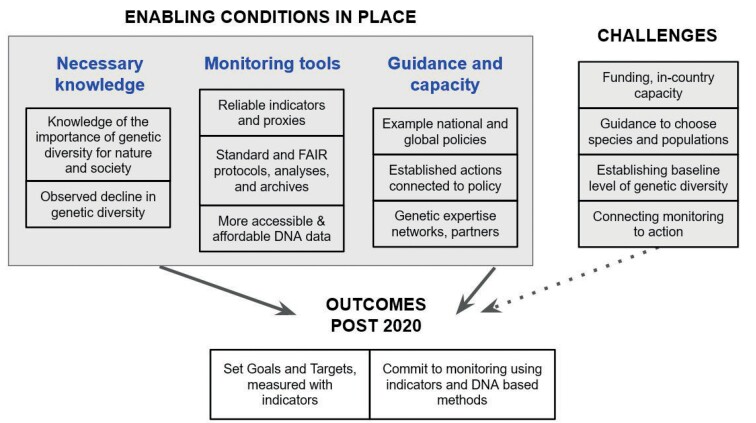
Figure 1.
Summary of the enabling conditions, and the issues or challenges to overcome, that will enable positive global policies to set goals, targets and indicators, and help countries to commit to monitoring and reporting (Outcomes post 2020). Abbreviation: FAIR, findable, accessible, interoperable, and reusable.
First set of conditions: Necessary knowledge
The first elements needed to motivate and guide genetic diversity monitoring are knowledge regarding the importance of genetic diversity and the current rate of loss of genetic diversity (figure 1). This knowledge tells policy makers that genetic diversity is critical, and genetic diversity is declining rapidly.
Genetic diversity is the foundation of resilience in nature. Genetic diversity provides the capacity for all species to adapt; without it, species cannot survive changing environments, climate change, and new pest and disease impacts. Genetic diversity is also important in unchanging environments. When populations are reduced to small numbers of individuals (i.e., a few hundred), negative consequences of inbreeding can occur, lowering fitness and fecundity, which threatens species’ survival and makes recovery more expensive and difficult (Blomqvist et al. 2010). Moreover, the dependence of ecosystem resilience and recovery on genetic diversity has become increasingly clear, particularly for surviving extreme weather events and warmer temperatures—for example, in corals, kelp, and seagrass ecosystems (Reusch et al. 2005, Wernberg et al. 2018, Morikawa and Palumbi 2019). Genetic diversity within species also enables greater species diversity by affecting niche space and competition, as in forest trees and in insect communities (Keith et al. 2017, Clark 2010). Resilience in ecosystems supports services such as coastal protection, water management, pest and disease management, carbon sequestration, and fisheries (for a review, see Stange et al. 2020, Des Roches et al. 2021). In fact, genetic diversity underpins ecosystem function and community structure as fundamentally as species diversity does (Prieto et al. 2015, Raffard et al. 2019).
Genetic diversity enables sustainable development and innovations in agriculture and semimanaged systems, supporting stable societies. Naturally occurring genetic diversity has been incorporated within numerous crops, mitigating major production losses due to flooding, high salinity, and toxic soils—challenges that are increasing with climate change (for a review, see Mickelbart et al. 2015). The genetic diversity in major tree genera that are harvested for timber and biofuels (such as Pinus, Fagus, Acacia, and Fraxinus) is allowing forest restoration and recovery after recent devastating pathogen and pest attacks (Sniezko and Koch 2017). The genetic diversity within wild species that can be bred with major crops (i.e., crop wild relatives) to provide pest and disease resilience, increased yield, and add other novel traits was valued in 1997 at US$115 billion per year globally. For a specific example, the contribution of wild relatives to sunflower is at least US$267 million (Pimentel et al. 1997, Hein and Gatzweiler 2006, Seiler et al. 2017). Such valuation was not inclusive of the direct contributions of thousands of wild species used by humans for food, fiber, fuel, and medicine, or their contribution to ecosystem services, including pollination and multiuse grasslands (Hajjar et al. 2008). The importance of genetic diversity for nature and society under increasing climate change, habitat modification, and new diseases mean that protecting, assessing, and monitoring genetic diversity are essential.
Specific anthropogenic drivers are causing rapid, alarming loss of genetic diversity. Researchers have worked for decades to measure genetic diversity patterns and understand how environmental change and human impacts cause genetic diversity change. Early work identified lower genetic diversity in rare and threatened species (Frankham 1996, Garner et al. 2005). Subsequent studies further documented how population reductions and habitat fragmentation rapidly affect populations’ genetic composition. A recent meta-analysis showed 6% loss of genetic diversity in populations of 91 species over the past century (Leigh et al. 2019). A 6% decline is greater than estimated losses in species’ diversity via extinctions (e.g., 1% of mammals extinct in the twentieth century; Ceballos et al. 2015). Genetic erosion on island systems is even worse—a striking 28% (Leigh et al. 2019). Such losses are likely in other island-like systems (lakes, mountain peaks, isolated habitats, etc.). The impact of harvesting is also concerning, with genetic diversity in harvested fish 12% lower than unharvested counterparts (Pinsky and Palumbi 2014). Moreover, the extirpation of populations causes the loss of unique genetic adaptations to local environments (Ceballos et al. 2020). The recently reported 68% decline in the Living Planet Index, which is based on vertebrate population sizes (WWF 2020), will result in dramatic losses of genetic diversity. Table 1 portrays the potential magnitude of such loss, by using population genetic theory to predict the amount of genetic diversity (using two metrics for genetic diversity—alleles and heterozygosity—that change at different rates) for populations with a given effective population size (Ne; for a definition, see table 1). On the basis of this analysis, assuming global 68% population reductions, many species are predicted to experience more than 50% loss of genetic diversity if no protective actions are taken (table 1, supplemental table S1).
Second set of conditions: Monitoring tools
Genetic diversity monitoring will require three pillars that complement and enhance each other: globally applicable indicators based on reasonable proxies of genetic diversity, improved standards and infrastructure, and technically feasible and affordable DNA-based monitoring (figure 1).
Reliable indicators exist to effectively incorporate genetic diversity into global policy frameworks. Indicators are measurements that are used to provide insight into a system; they are used to monitor the state of a system, track overall trends, assess which policy interventions have strong impact, and aid decision-makers in prioritizing resources. The lack of effective indicators for tracking genetic diversity status and change has been recognized (CBD 2020b). Specifically, existing indicators such as the Red List Index, the size of seed banks, and the number of threatened livestock breeds poorly reflect the status, threats, or trends in genetic diversity of species (Hoban et al. 2020a).
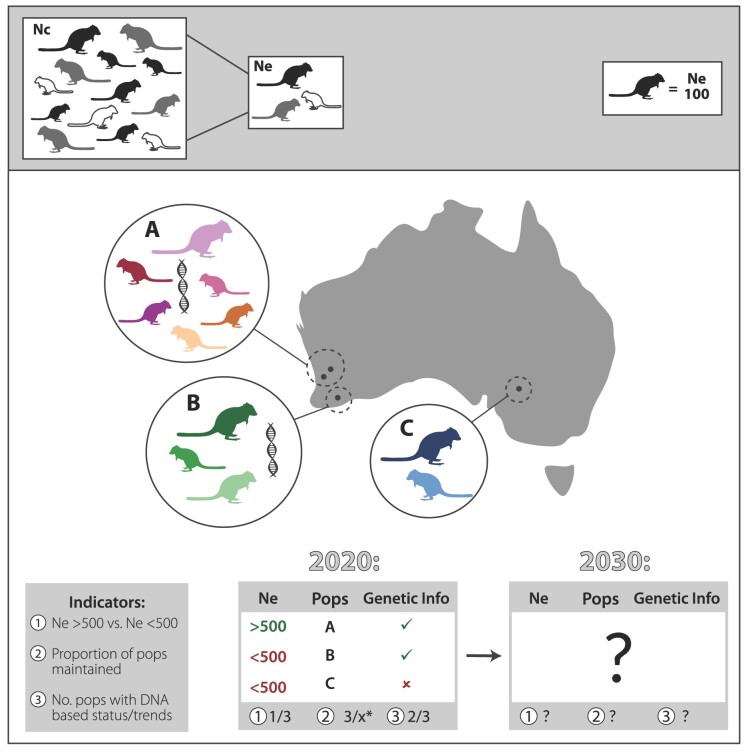
Recent research shows that sometimes environmental, demographic, and geographic data can help assess, monitor, and conserve adaptive potential and genetic resilience of populations in situ and ex situ (Beckman et al. 2019, Hollingsworth et al. 2020, Hoban et al. 2020b, Di Santo and Hamilton 2021). On the basis of this knowledge, (Hoban et al. 2020a) proposed three new pragmatic indicators for genetic diversity status and trends for the CBD (figure 2):
Figure 2.
Hypothetical example demonstrating how indicators can be used to establish baseline and current levels of genetic diversity, and monitor this into the future to guide management decisions. Indicators 1 and 2 can be based on genetic data (effective population size [Ne] and genetically defined populations), or proxies (10% of census size [Nc] or geographic location). The top bar reminds the reader that Ne is much smaller than Nc and shows that each organism in the map represents an Ne of 100. *x is the historical baseline number of populations.
First is the proportion of all populations that are sufficiently large to maintain genetic diversity. Populations maintaining an effective population size (Ne)—a metric reflecting the rate of genetic erosion—greater than 500 should retain substantial adaptive variation for many generations (Franklin 1980, Allendorf et al. 2013, Ryman et al. 2019). Contemporary Ne can often be estimated from genetic or demographic data, using a number of available methods, which has been done in hundreds of cases (see Hoban et al. 2020a). Recognizing the assumptions and limitations of each method and appropriate sampling methods is crucial; fortunately, guidance on these areas is available (Gilbert and Whitlock 2015, Frankham et al. 2019, Ryman et al. 2019). Alternatively, Ne can be estimated by a general rule where Ne is approximately 10% of census size (Nc), which translates to Nc of 5000 (Hoban et al. 2020a). The inclusion of population size as a component of Favorable Conservation Status, a designation under the EU Habitats Directive (Epstein et al. 2016), suggests that Ne > 500 or Nc > 5000 can be a feasible genetic indicator for reporting.
Second is the number of populations conserved overall. Preventing population losses can help maintain unique and locally adapted genetic variants, which will allow species greater evolutionary potential for the future. Multiple efforts documenting population losses (Ceballos et al. 2020, WWF 2020) globally demonstrate this as a feasible indicator. One Australian State, New South Wales, has also used this population-based metric to estimate that 9%–21% of genetic diversity has likely been lost over all species, and an alarming 51% may have been lost in highly threatened species (DPIE 2020).
Third is the number of populations with monitoring of genetic diversity. Gathering knowledge about genetic diversity in situ and ex situ can continue to inform conservation action as it has in hundreds of cases (e.g., identifying vulnerable or especially valuable populations, optimizing translocations, resolving species taxonomy, and estimating connectivity between populations; see Allendorf et al. 2013).
These indicators are SMART (specific, measurable, achievable, realistic, and timely; see table 2); they represent pragmatic ways of monitoring genetic change at national and global scales. They also complement the indicator known as “comprehensiveness of the conservation of useful wild plants,” which estimates the amount of a species’ geographic range in protected areas or conserved ex situ—another vital measure of genetic diversity conservation (Khoury et al. 2019). As DNA sequencing technology, regional genetic diversity maps (e.g., Schmidt et al. 2020), and analytical models continue to improve, these indicators can be compared with and enhanced by genetic data collected in the future and with other indicators (Hollingsworth et al. 2020, Lefevre et al. 2020).
Table 2.
The SMART characteristics of three recently proposed indicators for genetic diversity that rely on proxies and DNA based studies.
| Indicator | Index for maintaining genetic diversity | Index for maintaining populations and their adaptations | Index for monitoring populations’ genetic diversity |
|---|---|---|---|
| Specific—how are indicators quantitative | Quantifies the number of populations with Ne > 500 versus Ne < 500. | Quantifies the number or proportion of populations maintained. | Quantifies the number of populations with DNA based status and trends. |
| Measurable—where does data exist (including baseline from the past) | 10% of census size (Nc) is a reasonable proxy of Ne. Nc estimates exist for thousands of populations in the LPI, and RL assessments. Also, agencies, NGOs, and museums often hold data on Nc estimates. Scientific literature also has numerous Nc and Ne estimates. Ne estimates will increase rapidly as genomic data becomes widely affordable and accessible. | Data on the number of populations within species or proportion of populations remaining is available in existing databases (LPI, GBIF, museums, RL assessments, stock assessments and government reports). | Information on genetic status publications is available on scholarly databases or Google Scholar. Information on genetic data sets is available on data portals (GenBank, GEOME, Dryad; see Supplemental S2). Genetic experts can offer guidance on suitable data availability. |
| Achievable—what are feasible actions or policy levers | Support many large populations (Ne > 500) and baseline connectivity among populations. Action is taken to increase size or connectivity among populations above 500. | Protect all populations that remain now. It is achievable. Restore populations to baseline when possible. | Invest in research and knowledge sharing. The number of populations investigated genetically is rapidly increasing. |
| Relevant—what is the connection to genetic change and biodiversity conservation | With small Ne, rapid genetic erosion occurs and remaining diversity is at risk. | Populations often contain adaptations to local environments. Maintaining populations maintains genetic diversity. | Knowledge of genetic diversity informs conservation action, and supports indicators 1 and 2. |
| Time bound—deadlines | Stability by 2030, increasing by 2050. | Aim for no loss by 2030, restoration by 2050. | Capacity in place by 2030. |
Abbreviations: Ne, effective population size; LPI, Living Planet Index; RL, Red List.
Table 1.
Predicted loss of two metrics of genetic diversity, heterozygosity and allelic diversity, after a 68% decline from a baseline effective population size (Ne).
| Baseline Ne population size | Ne population size after 68% decline, as is reported in the Living Planet Index | Predicted loss of genetic variation of individuals (heterozygositya; as a percentage) | Predicted loss of genetic variation that helps populations adapt (allelesb; as a percentage) |
|---|---|---|---|
| 1000 | 320 | 60.3 | 19.4 |
| 10000 | 3200 | 29.8 | 54.4 |
| 100000 | 32000 | 4.9 | 66.30 |
| 1000000 | 320000 | 0.5 | 67.8 |
Note: Effective population size is a genetic parameter used to measure loss of genetic diversity; it is often one-tenth of the census size. The 68% decline is the average vertebrate population decline reported by the Living Planet Index (LPI). We use (Kimura and Crow 1964) to predict expected heterozygosity, He, from effective population size and we use (Jost 2010) to predict allelic diversity, AD, from He. A decline in Ne causes a loss of He, with the largest proportional losses in small populations because of unequal family size and mating among relatives. A decline in Ne causes a loss of AD, with the largest proportional losses in large populations because large populations harbor many rare alleles that can be easily lost. The smaller proportional loss of AD in small populations is because they have fewer alleles to begin with. Note that the percentage loss shown will not precisely equate to a species’ ability to adapt; in particular, even though fewer alleles are lost in small populations, this should not be interpreted as unimportant. These figures suggest that genetic diversity losses are very large if populations decline in abundance similar to declines in the LPI. Full table and details on methodology and caveats in supplement S1. aExpected heterozygosity, the genetic variation most useful for immediate recovery. bAllelic diversity, the genetic variation most important for long term response to environmental change.
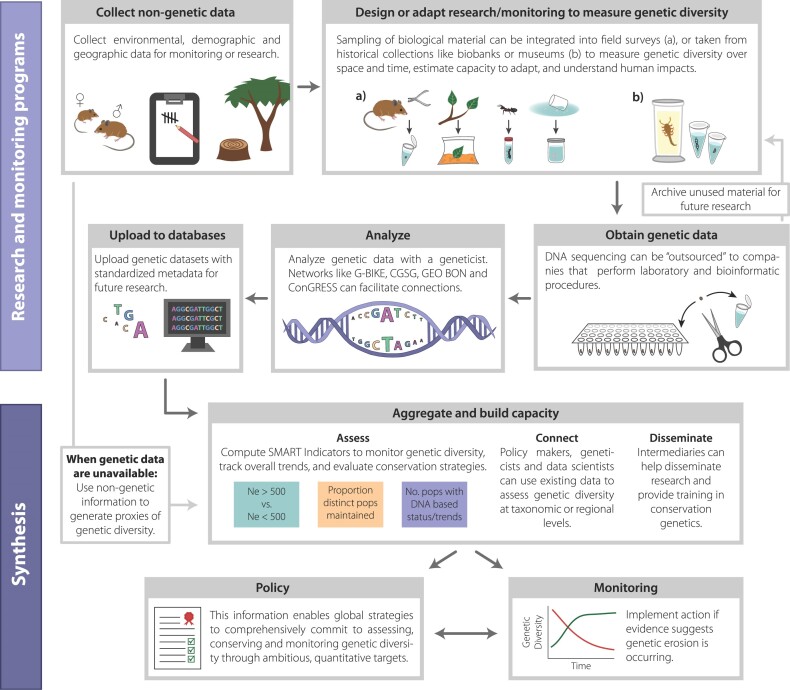
Practices, standards, and infrastructure are established to assess relevant genetic change through more accessible, affordable DNA-based monitoring. Indicators one and two should be complemented by genetic monitoring; directly analyzing DNA from individuals and populations within species over time is the most accurate way to assess genetic diversity status (figure 3). This is because losses of genetic diversity are not always apparent. Populations that underwent strong declines (e.g., because of overhunting) and lost genetic diversity in the past but recovered in numbers may still have low genetic diversity today (such as numerous seals; see Stoffel et al. 2018). This makes such species vulnerable to environmental change and increasing pathogen pressure, as in musk ox (Hansen et al. 2018). Extreme weather or other pressures can also reduce genetic diversity even as census size remains high. After an unprecedented heat wave in Western Australia, little impact was seen on the spatial extent and abundance of two seaweed species, but diversity loss was massive; over half of genetic diversity in one species was lost (Gurgel et al. 2020). These documented losses highlight that genetic diversity erodes long before species disappear and suggest that many systems are highly vulnerable to future extreme events. Monitoring genetic diversity directly can help detect this vulnerability. Although DNA-based monitoring will not be globally routine for some time, numerous countries already assess and monitor genetic change directly, in an increasingly standardized fashion, and are making data available for global use (Bruford et al. 2017, Posledovich et al. 2021).
Figure 3.
Schematic of including genetic data in monitoring and policy, from collecting genetic and nongenetic data, to sequencing, analysis and archiving, to aggregation and indicator calculation, to policy and implementation of actions.
Since the 1980s, geneticists have contributed more than 2 billion DNA sequences to the searchable database GenBank (www.ncbi.nlm.nih.gov/genbank/statistics), and the cost of DNA sequencing continues to decline. DNA sequencing can now often be outsourced by sending biological samples to companies who efficiently perform laboratory and bioinformatic procedures (when sending samples internationally, adherence to legislation and consideration of access and benefits is necessary). This approach is becoming highly affordable and rapidly replacing older technologies such as microsatellites, although the choice of appropriate marker system depends on existing local infrastructure. In any case, obtaining data on genetic diversity no longer requires significant infrastructure or laboratory personnel, although expert guidance on interpreting data remains essential (and is often lacking; see the “Guidance and capacity” section below). Countries can also work in partnership with other countries and international organizations such as the IUCN, zoo and botanic garden organizations, and GEO BON (the Group on Earth Observations Biodiversity Observation Network).
Genetic data are increasingly FAIR—or findable, accessible, interoperable, and reproducible. Laboratory protocols and data storage formats are increasingly standardized, and data analysis and modeling is often performed using open-source code and established workflows (Holderegger et al. 2020). There is, of course, room for improvement, and large-scale projects (genome-scale data across many populations of dozens to hundreds of species), such as the California Conservation Genomics Project and FORGENIUS, will further demonstrate the importance of documented, standardized workflows. Metadata standards—for instance, those based on Darwin Core (Wieczorek et al. 2012)—applied to data repositories such as the Genomics Observatory Metadatabase (GEOME; see also supplement S2) mean that genetic data sets can be searched, aggregated, filtered, and analyzed across space and time. Also, the 2011 Joint Data Archiving Policy mandated publicly available, archived genetic data sets for numerous major journals (Fairbairn 2011, Whitlock 2011), whereas numerous groups have created searchable archives with thousands of DNA data sets, sometimes with environmental or trait data (see supplement S2). These advances can make genetic monitoring routine, in the way that large databases of species’ occurrence (e.g., the Global Biodiversity Information Facility, GBIF) help monitor and predict change in species distributions (figure 3). Geneticists and data scientists can and do use these resources to provide information on genetic diversity to policymakers (Hollingsworth et al. 2020) or identify potential partnerships nationally or internationally for generating new data. Meanwhile, the issue of access and benefits of genetic diversity data use are increasingly recognized, including in the policies of major journals (Marden et al. 2021).
The diversity of sources of DNA samples, which are required for genetic monitoring, is also rapidly expanding. Biomaterial collections (e.g., the European Association of Zoos and Aquariums Biobank, CryoArks, the Millennium Seed Bank) can provide preserved tissue to analyze genetic status (including comparing archival with modern populations or in situ and ex situ populations), helping fill spatial and temporal gaps in monitoring programs, establish baselines for calculating change, and provide reference points for management. DNA from fossil, museum, herbaria, or archived specimens can also help document genetic diversity baseline levels (Lord et al. 2020). Also, genetic variation has been increasingly characterized using hair, feces, and other noninvasive sources of small amounts of DNA (Chiou and Bergey 2018). This has made genetic monitoring possible, routine, and affordable for rare, dangerous, nocturnal, or elusive species. (Environmental DNA from soil, water, or air is commonly employed for determining species’ occurrence or counting species but has likely limitations in monitoring genetic diversity within populations.) Novel technologies such as handheld sequencers are starting to allow scientists to collect near-real-time data in the field (Pomerantz et al. 2018). Institutions are also collaborating across borders to share samples and data to document genetic diversity across species with wide geographic ranges (e.g., trees, wolves, wildcats, bears, raptors). Together, these endeavors can help monitor and predict genetic change affordably, in standard fashion, for most species.
Third set of conditions: Guidance and capacity
Protecting and monitoring genetic diversity at international scales requires successful model policies at local and national scales, evidence of successful interventions, and an increase in capacity building and networks that can cross science-policy divides (figure 1).
Conservation policy at regional levels increasingly recognizes and protects genetic diversity: Providing models for global efforts. Although CBD Aichi target 13, SDG 2.5 and GSPC target 9 acknowledge and commit to conserving genetic diversity, they are vague and focus strongly on agricultural and other socioeconomically important species. However, at subglobal scales, numerous governments, agricultural sectors, and conservation groups already measure and protect genetic diversity more explicitly for wild species (Santamaría and Méndez 2012). Such examples demonstrate the will and competency to develop genetic diversity conservation policy, which global initiatives can learn from and follow.
The first legal protection of genetic diversity was likely the US Endangered Species Act (USFWS 1973), which recognized distinct population segments (genetically or geographically isolated) as units of protection equivalent to species. Subsequently, genetic data has frequently been used in decision-making because of the act's mandate to use the best available science (Kelly 2010). Numerous countries have followed in protecting genetic diversity (e.g., Canada and Australia). Legal policies based on conserving genetic diversity and managing genetic issues are also common in forestry and fisheries sectors. Hundreds of forested areas to support genetic conservation are recognized across Europe (www.euforgen.org). Large-scale monitoring of genetic diversity of salmonids occurs, such as monitoring genetic diversity in Baltic Sea salmon by the Helsinki Commission since 1998 (Laikre et al. 2016) and required hatchery genetic management plans for monitoring and maintaining genetic diversity in North America (National Marine Fisheries Service 2005). In addition, national guidelines exist for minimum population sizes for producing genetically viable, appropriate seed for forestry restoration (www.euforgen.org). Meanwhile, favorable population status under Natura 2000 in Europe, to secure long-term viability, requires large “effective population sizes” (Epstein et al. 2016) and effective gene exchange among populations. Several nations, including Sweden and Switzerland, have committed to formal genetic monitoring programs to track genetic diversity change over time (Black-Samuelsson et al. 2020, KORA Foundation 2020, Posledovich et al. 2021), whereas Scotland assesses genetic status and threats in 26 species (Hollingsworth et al. 2020).
The IUCN also increasingly recognizes the importance of genetic diversity. The designation of Key Biodiversity Areas (KBAs), a worldwide system of sites “contributing significantly to the global persistence of biodiversity,” includes a genetic distinctiveness criterion (IUCN 2016). IUCN (2020) mentions genetic exchange as one of the key objectives of corridors (Hilty et al. 2020). At the 2016 World Conservation Congress, IUCN resolution 104 asked its member states to recognize, protect, and manage forest genetic units, whereas the 2020 IUCN resolution 109 calls for increased integration of genetic diversity into all IUCN planning activities, including protected area planning and natural capital. On the other hand, although the IUCN Red List, the globally most important species’ assessment tool, recognizes that the degree of genetic exchange is an important criterion for delimiting populations, it does not systematically incorporate genetic concepts or data into its threat assessment methodology, although proposals have been made for this (Willoughby et al. 2015, Vitorino et al. 2019, Garner et al. 2020).
Actions based on policies can and do improve the status of genetic diversity. First, maintaining genetic diversity requires maintaining populations throughout a species range, not just in small areas. Australian mammals, for example, show higher genetic diversity in remnant mainland populations, even after drastic declines, than in offshore island populations (Eldridge et al. 2004); each population contributes to future resilience. Second, populations must also be large enough to maintain genetic diversity. Maintaining genetic diversity may require more, larger, and better connected habitat areas than the area needed for maintaining species diversity (Struebig et al. 2011). Genetic diversity erodes long before species become extinct—like trees hollowed out before they fall. Third, strategic actions such as translocations can increase genetic diversity or fitness of small, inbred populations (genetic rescue; Weeks et al. 2011, Whiteley et al. 2015), for which recent risk-analysis (e.g., outbreeding, disease, invasiveness) and decision-making frameworks (when to translocate and how much) have become available (Ralls et al. 2018, Van Rossum and Hardy 2020). Retaining and restoring genetic connectivity, such as through habitat corridors, can allow exchange of genetic variants between populations and slow the loss of diversity. It is possible to calculate the population sizes, minimum habitat area, or protected area configuration needed to maintain genetic diversity (Méndez et al. 2014). Genetic data can provide crucial knowledge on how landscape structure impedes or supports gene flow and genetic diversity and informs large landscape planning. Finally, for many species, ex situ collections in seed banks, zoos, and botanic gardens help forestall the loss of genetic diversity and sometimes conserve genetic variation no longer extant in the wild, although most are currently not sufficiently large, representative, or replicated (Khoury et al. 2019, Hoban et al. 2020b, Wei and Jiang 2021). Genetic data collected before and after any of these actions can determine whether they have succeeded (also see the “Looking ahead: Challenges” section below).
Decades of genetic monitoring in zoo and captive breeding populations have helped to establish genetic methods and appropriate goals for guiding action. Agronomists have long sought to conserve more than 95% of genetic diversity within populations in seed banks. In zoos, meanwhile, pedigree-based management typically aims to retain more than 90% of genetic diversity over 100 years (Ballou et al. 2010). Zoos and botanic gardens increasingly incorporate molecular genetic data to carefully select breeding individuals (Wood et al. 2020), whereas other programs aim to conserve genetic variation through rapid population size increase (Wildt et al. 2019). Ideally genetic diversity may be best conserved via integrated management of wild and captive populations, informed by genetic data of in situ and ex situ populations, following the One Plan approach (Byers et al. 2013, Ogden et al. 2020).
New initiatives connect policymakers to genetic expertise, guidance and tools, and help obtain and apply genetic data. Intermediaries between scientists and government agencies are working to raise awareness of and ability to conserve genetic diversity, including in cases in which the data are sparse or controversial (see figure 3). Networks housed within the Society for Conservation Biology, the Group on Earth Observations, the IUCN, and other groups are assisting in capacity building and the application of genetic indicators and monitoring (see supplemental S2, and also see Hoban et al. 2020a). These organizations have produced policy briefs, white papers, and webinars on indicators; key concepts in conservation genetics; and genetic technologies (for examples, see supplements S3 and S4). This work is informed by foundational efforts that identified legislation for which genetic diversity is relevant (Laikre et al. 2010, Santamaría and Méndez 2012). Genetic diversity assessment capacity is also emerging in lower-income, high-biodiversity regions (Torres-Florez et al. 2018), including through international collaborations (Blanco et al. 2020). Efforts are also being made to identify barriers preventing practitioners from using genetic data (Taylor et al. 2017), synthesize the evidence base to guide decision-making (Cook and Sgrò 2017), determine how frequently genetic research findings are used by nature management agencies (Bowman et al. 2016), improve dissemination and accessibility of genetic findings (Hoban et al. 2013), and increase the relevance of research to practice and policy (Taft et al. 2020).
Looking ahead: Challenges
Geneticists and conservationists continue to make progress in generating genetic data and making it accessible and useful to decision-makers, but several issues remain (figure 1).
One major challenge is that many parts of the world have scarce capacity to gather and interpret genetic data (because of a lack of resources, or conflict zones). This causes genetic knowledge gaps, mirroring gaps in species observations and biodiversity monitoring broadly, often in regions of high threats to biodiversity. It is necessary to urgently build local assessment and monitoring capacity and establish international research partnerships (Holderegger et al. 2020, Marden et al. 2021), while ensuring fair attribution and adherence to international protocols, including the Nagoya Protocol on Access and Benefit-Sharing. Efforts include regional groups of the IUCN Conservation Genetics Specialist Group in the global south (Africa, South America, South Asia) and initiatives for education, training, and biodiversity monitoring in central Africa (Anthony et al. 2015). Detailed guidance on using genetic diversity indicators and other tools in diverse situations is also needed. The G-BiKE (Genomic Biodiversity Knowledge for Resilient Ecosystems) network is extending such training and collaboration across 39 countries of Europe, North Africa, and the Middle East (www.cost.eu/actions/CA18134).
Although the indicators presented above are comparable across many kinds of species, another challenge is choosing which species to actually include in genetic monitoring programs. Broad issues should first be addressed: Who are the potential stakeholders? What are the program timeframe, the costs, and the long-term funding availability? How will data and samples be stored? What currently available resources can be leveraged (personnel, etc.)? Is coordination across national borders feasible? Choosing species involves numerous additional considerations, including national values and interests (for more detail, see Laikre et al. 2008, Posledovich et al. 2021), and we mention three as short illustrations of the process: the ability to detect change if it occurs, a representation of species in a region, and technical feasibility. First, because genetic change happens over generations, lifespan must be considered; genetic change will be detected more quickly if the species’ generation time is moderate (1–5 years, for example). Still, genetic monitoring can occur for long-lived organisms by sampling the offspring of successive cohorts (as with tree seedlings). Second, programs should include diverse taxonomic groups, life history traits (e.g., size, dispersal ability), ecosystems or biomes, and degrees of commonness (Laikre et al. 2008, Bruford et al. 2017). For instance (Hollingsworth et al. 2020) chose species from different groups (fungi, bryophyte, vascular plant, mollusk, insect, and vertebrates) and value systems (conservation, cultural value, ecosystem services, food or medicinal foraging, game species). Chosen species may include flagship or umbrella species to attract public and policy attention to genetic diversity. Third, feasibility is higher for species that are fairly accessible or that leave remnants (e.g., feathers, scat) and that have some genomic resources (sequenced genomes or exomes or an SNP chip), allowing a faster start-up and increasing informativeness and affordability. We do not advocate particular DNA markers, because the field is changing rapidly and most of today's markers will be compatible with the future of whole genome sequencing. Species should be chosen in consultation with managers to ensure that genetic data can address their needs and to coordinate with existing species and habitat monitoring programs. Finally, these considerations apply whether a program is choosing species to monitor with genetic data or proxies, although if the program uses proxies, species choice may also consider how much data exists in national or global occurrence record databases (e.g., GBIF, the European Forest Genetic Resources Programme, forestry inventories).
Establishing a “baseline” for temporal comparison is another challenge (Black-Samuelsson et al. 2020), common to all monitoring. There are unique opportunities with genetic data, because DNA can be obtained from museum samples, other archives, seed banks, or herbaria, sometimes hundreds of years old. Therefore genetic diversity loss over long time periods can be quantified (Hoban et al. 2014, Bruford et al. 2017). This historic DNA provides both a direct window into the genetic diversity levels preceding anthropogenic impacts, as well as knowledge on variability, which can help define thresholds of change that are cause for alarm. Still, such samples are relatively scarce, and statistical analysis of this data remains challenging (Gauthier et al. 2020). To serve as a resource for the researchers of tomorrow, contemporary genetic data, metadata, and DNA samples should be stored securely and shared globally, and analyses should be modified as new technologies and knowledge develop.
A final challenge is to better connect monitoring data to action, especially to determine which actions to take when particular genetic change thresholds have been crossed. For instance, in populations showing declining trends in genetic variation, when should translocations be considered? In a population showing increasing, detrimental hybridization, when should hybrids be culled? There are some clear warning signals, as well as tools available to help. For most organisms, populations with an Ne lower than 50 are subject to near-term inbreeding and severe genetic erosion and are principle candidates for translocations, but a decision should be made using existing risk assessment frameworks for outbreeding depression, maladaptations, and disease (Ralls et al. 2018). A new approach has also been developed to detect genomic maladaptation, which can identify populations lacking certain genetic variants needed to help survive environmental change, such as predicted warming (Bay et al. 2017, Razgour et al. 2019, Gougherty et al. 2021). Therefore insight from both neutral and adaptive genetic data can help prevent the loss of genetic variation (Flanagan et al. 2018). In addition, simulation models are increasingly used to make decisions about translocations, acceptable harvest levels (e.g., fisheries, forestry, hunting), captive breeding programs, the removal of hybrids, habitat connectivity, or the design of protected areas to safeguard genetic diversity across many species (van Wyk et al. 2017, Grueber et al. 2019). Simulations are also useful to predict impacts of management, after which monitoring can validate and improve models in the future (Hoban 2014, Wright et al. 2021). The specific thresholds and management actions for genetic diversity will vary depending on the species and region (e.g., biological traits, land or seascape structure, abundance, human influences) and should include risk assessments.
Conclusions
In the past, genetic diversity concepts have been considered complicated, impractical, or inaccessible for policymakers and genetic data viewed as too expensive and sparse for global conservation. This led to neglect of genetic diversity concerns and monitoring (Vernesi et al. 2008, Laikre et al. 2010, 2020, Hoban et al. 2020a). Several challenges must be overcome, but in general the necessary enabling conditions are in place for documenting and monitoring genetic diversity, as are the knowledge and indicators for ensuring its conservation. The CBD post-2020 framework and other policies that aim for transformative change must fully acknowledge the crucial role of genetic diversity for nature and society and commit to effectively conserve, assess, and monitor it in selected species that represent all life, not just economically important species (Díaz et al. 2020, Des Roches et al. 2021). There must be a CBD post-2020 framework with a clear, measurable, and numerical genetic diversity goal, of the same standing as species and ecosystems, as well as associated action targets (Hoban et al. 2020a); inclusion and implementation of practical genetic diversity indicators in the CBD and other global biodiversity commitments (e.g., IPBES, SDG); and increased establishment and scaling up of genetic monitoring programs, with those actors having sufficient resources assisting others.
Genetic diversity should be conserved with the same urgency as species diversity, to support food security, wellbeing, culture, and adaptation. Strong genetic diversity policy goals, targets, and indicators are possible, and monitoring can be linked to action. Researchers and conservation professionals must work with policymakers to draft frameworks protecting Earth's remaining biodiversity. With bold action, genetic diversity can be measured, tracked, and maintained for a resilient future.
Supplementary Material
Acknowledgments
We thank anonymous reviewers and the editor for helpful comments. This article is based on work from COST Action G-BiKE CA 18134, supported by COST (European Cooperation in Science and Technology). We acknowledge logistical support from the Group on Earth Observations Biodiversity Observation Network. CK was supported by grant no. 2019–67012–29733 and project accession no. 1019405 from the USDA National Institute of Food and Agriculture. PG acknowledges the structural support of the Flemish government. LL was funded by the Swedish Research Council and SRC Formas. SH and MPG were supported by the Institute of Museum and Library Services (grants no. MG-30–16–0085–16 and no. MA-30–18–0273–18). This article does not represent the official finding or policy of any government or institution. This article has been peer reviewed and approved for publication consistent with USGS Fundamental Science Practices (https://pubs.usgs.gov/circ/1367). Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the US government or any government.
Author Biographical
Sean Hoban is affiliated (shoban@mortonarb.org) with The Morton Arboretum, Center for Tree Science, in Lisle, Illinois, in the United States. Michael W. Bruford is affiliated with the School of BioSciences at Cardiff University, in Cardiff, Wales, in the United Kingdom, W. Chris Funk is affiliated with the Department of Biology in the Graduate Degree Program in Ecology, at Colorado State University, in Fort Collins, Colorado, in the United States. Peter Galbusera is affiliated with the Royal Zoological Society of Antwerp, Centre for Research and Conservation, in Antwerp, Belgium. M. Patrick Griffith is affiliated with the Montgomery Botanical Center, in Coral Gables, Florida, in the United States. Catherine E. Grueber is affiliated with the University of Sydney's School of Life and Environmental Sciences, Faculty of Science, in Sydney, New South Wales, in Australia. Myriam Heuertz is affiliated with INRAE, and the University of Bordeaux, Biogeco, in Cestas, France. Margaret E. Hunter is affiliated with the US Geological Survey's Wetland and Aquatic Research Center, in Gainesville, Florida, in the United States. Christina Hvilsom is affiliated with the Copenhagen Zoo, in Frederiksberg, Denmark. Belma Kalamujic Stroil is affiliated with the University of Sarajevo Institute for Genetic Engineering and Biotechnology, Laboratory for Molecular Genetics of Natural Resources, in Sarajevo, Bosnia and Herzegovina. Francine Kershaw is affiliated with the Natural Resources Defense Council, in New York, New York, in the United States. Colin K. Khoury is affiliated with the International Center for Tropical Agriculture, in Cali, Colombia. Colin K. Khoury is affiliated with Saint Louis University's Department of Biology, in St. Louis, Missouri, in the United States. Linda Laikre is affiliated the Department of Zoology, Division of Population Genetics, at Stockholm University, in Stockholm, Sweden. Margarida Lopes-Fernandes is affiliated with the Instituto da Conservação da Natureza e das Florestas, in Lisbon, Portugal. Anna J. MacDonald is affiliated with the Australian National University, John Curtin School of Medical Research and Research School of Biology, in Canberra, Australia. Joachim Mergeay is affiliated with the Research Institute for Nature and Forest, in Geraardsbergen, Belgium. Joachim Mergeay is affiliated with the Laboratory of Aquatic Ecology, Evolution, and Conservation, at KULeuven, in Leuven, Belgium. Mariah Meek is affiliated with the Michigan State University Department of Integrative Biology, AgBio Research, and with the Ecology, Evolution, and Behavior Program, in East Lansing, Michigan, in the United States. Cinnamon Mittan is affiliated Cornell University's Department of Ecology and Evolutionary Biology, in Ithaca, New York, in the United States. Tarek A. Mukassabi is affiliated with the University of Benghazi Department of Botany, Faculty of Sciences, in Benghazi, Libya. David O'Brien is affiliated with NatureScot, in Inverness, Scotland, in the United Kingdom. Rob Ogden is affiliated with the Royal (Dick) School of Veterinary Studies and with the Roslin Institute, at the University of Edinburgh, Easter Bush Campus, in Edinburgh, Scotland, in the United Kingdom. Clarisse Palma-Silva is affiliated with the Universidade de Campinas, in Campinas, Brazil. Uma Ramakrishnan is affiliated with the Department of Ecology and Evolution at the National Centre for Biological Sciences, in Bangalore, India. Gernot Segelbacher is the chair of wildlife ecology and management at the University Freiburg, in Freiburg, Germany. Robyn E. Shaw is affiliated with the Department of Environmental and Conservation Sciences at Murdoch University, in Perth, Australia. Per Sjögren-Gulve is affiliated with the Wildlife Analysis Unit at the Swedish Environmental Protection Agency, in Stockholm, Sweden. Nevena Veličković is affiliated with the University of Novi Sad's Faculty of Sciences, Department of Biology and Ecology, in Novi Sad, Serbia. Cristiano Vernesi is affiliated with the Forest Ecology and Biogeochemical Fluxes Unit, Research and Innovation Centre, at the Fondazione Edmund Mach, in San Michele all’ Adige, Italy.
Contributor Information
Sean Hoban, The Morton Arboretum, Center for Tree Science, Lisle, Illinois, United States.
Michael W Bruford, Cardiff University, Cardiff, Wales, United Kingdom.
W Chris Funk, Department of Biology, Graduate Degree Program in Ecology, Colorado State University, Fort Collins, Colorado, United States.
Peter Galbusera, Royal Zoological Society of Antwerp, Centre for Research and Conservation, Antwerp, Belgium.
M Patrick Griffith, Montgomery Botanical Center, Coral Gables, Florida, United States.
Catherine E Grueber, University of Sydney's School of Life and Environmental Sciences, Faculty of Science, Sydney, New South Wales, Australia.
Myriam Heuertz, INRAE, and the University of Bordeaux, Biogeco, Cestas, France.
Margaret E Hunter, US Geological Survey's Wetland and Aquatic Research Center, Gainesville, Florida, United States.
Christina Hvilsom, Copenhagen Zoo, Frederiksberg, Denmark.
Belma Kalamujic Stroil, University of Sarajevo Institute for Genetic Engineering and Biotechnology, Laboratory for Molecular Genetics of Natural Resources, Sarajevo, Bosnia and Herzegovina.
Francine Kershaw, Natural Resources Defense Council, New York, New York, United States.
Colin K Khoury, International Center for Tropical Agriculture, Cali, Colombia; Saint Louis University's Department of Biology, St. Louis, Missouri, United States.
Linda Laikre, Department of Zoology, Division of Population Genetics, Stockholm University, Stockholm, Sweden.
Margarida Lopes-Fernandes, Instituto da Conservação da Natureza e das Florestas, Lisbon, Portugal.
Anna J MacDonald, Australian National University, John Curtin School of Medical Research and Research School of Biology, Canberra, Australia.
Joachim Mergeay, Research Institute for Nature and Forest, Geraardsbergen, Belgium; Laboratory of Aquatic Ecology, Evolution, and Conservation, KULeuven, Leuven, Belgium.
Mariah Meek, Michigan State University Department of Integrative Biology, AgBio Research, Ecology, Evolution, and Behavior Program, East Lansing, Michigan, United States.
Cinnamon Mittan, Cornell University's Department of Ecology and Evolutionary Biology, Ithaca, New York, United States.
Tarek A Mukassabi, University of Benghazi Department of Botany, Faculty of Sciences, Benghazi, Libya.
David O'Brien, NatureScot, Inverness, Scotland, United Kingdom.
Rob Ogden, Royal (Dick) School of Veterinary Studies and with the Roslin Institute, University of Edinburgh, Easter Bush Campus, Edinburgh, Scotland, United Kingdom.
Clarisse PALMA-SILVA, Universidade de Campinas, Campinas, Brazil.
Uma Ramakrishnan, Department of Ecology and Evolution, National Centre for Biological Sciences, Bangalore, India.
Gernot Segelbacher, Chair of wildlife ecology and management, University Freiburg, Freiburg, Germany.
Robyn E Shaw, Department of Environmental and Conservation Sciences, Murdoch University, Perth, Australia.
Per Sjögren-Gulve, Wildlife Analysis Unit, Swedish Environmental Protection Agency, Stockholm, Sweden.
Nevena Veličković, University of Novi Sad's Faculty of Sciences, Department of Biology and Ecology, Novi Sad, Serbia.
Cristiano Vernesi, Forest Ecology and Biogeochemical Fluxes Unit, Research and Innovation Centre, Fondazione Edmund Mach, San Michele all’ Adige, Italy.
References
- Allendorf FW, Luikart GH, Aitken SN. 2013. Conservation and the Genetics of Populations, 2nd ed.Wiley-Blackwell. [Google Scholar]
- Anthony NM, et al. 2015. Evolution and conservation of central African biodiversity: Priorities for future research and education in the Congo basin and gulf of guinea. Biotropica 47: 6–17. [Google Scholar]
- Ballou JD, Lees C, Faust LJ, Long S, Lynch C, Bingaman Lackey L, Foose TJ. 2010. Demographic and genetic management of captive populations. Pages 219–252 in Kleiman DG, Thompson KV, Baer CK, eds. Wild Mammals in Captivity: Principles and Techniques for Zoo Management. University of Chicago Press. [Google Scholar]
- Bay RA, Rose N, Barrett R, Bernatchez L, Ghalambor CK, Lasky JR, Brem RB, Palumbi SR, Ralph P. 2017. Predicting responses to contemporary environmental change using evolutionary response architectures. American Naturalist 189: 463–473. [DOI] [PubMed] [Google Scholar]
- Beckman E, Meyer A, Denvir A, Gill D, Man G, Pivorunas D, Shaw K, Westwood M. 2019. Conservation Gap Analysis of Native U.S. Oaks. The Morton Arboretum. [Google Scholar]
- Black-Samuelsson S, Eriksson A, Bergqvist J. 2020. The Second Report on The State of the World's Forest Genetic Resources Sweden. Skogsstyrelsen. Report no. 2020/3. [Google Scholar]
- Blanco MB, Greene LK, Rasambainarivo F, Toomey E, Williams RC, Andrianandrasana L, Larsen PA, Yoder AD. 2020. Next-generation technologies applied to age-old challenges in Madagascar. Conservation genetics 21: 785–793. [Google Scholar]
- Blomqvist D, Pauliny A, Larsson M, Flodin L-Å. 2010. Trapped in the extinction vortex? Strong genetic effects in a declining vertebrate population. BMC Evolutionary Biology 10: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman J, Greenhorn JE, Marrotte RR, McKay MM, Morris KY, Prentice MB, Wehtje M. 2016. On applications of landscape genetics. Conservation Genetics 17: 753–760. [Google Scholar]
- Bruford MW, Davies N, Dulloo ME, Faith DP, Walters M. 2017. Monitoring changes in genetic diversity. Pages 107–128 in Walters M, Scholes RJ, eds. The GEO Handbook on Biodiversity Observation Networks. Springer. [Google Scholar]
- Byers O, Lees C, Wilcken J, Schwitzer C. 2013. The One Plan approach: The philosophy and implementation of CBSG's approach to integrated species conservation planning. WAZA Magazine 14: 2–5. [Google Scholar]
- [CBD] Convention on Biological Diversity . 2020a. Global Biodiversity Outlook 5. CBD. [Google Scholar]
- [CBD] Convention on Biological Diversity . 2020b. Zero Draft of the post-2020 global biodiversity framework. CBD. [Google Scholar]
- Ceballos G, Ehrlich PR, Barnosky AD, García A, Pringle RM, Palmer TM. 2015. Accelerated modern human–induced species losses: Entering the sixth mass extinction. Science Advances 1: e1400253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceballos G, Ehrlich PR, Raven PH. 2020. Vertebrates on the brink as indicators of biological annihilation and the sixth mass extinction. Proceedings of the National Academy of Sciences 117: 13596–13602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier MJE, Widdig A, Alberts SC. 2007. Inbreeding depression in non-human primates: A historical review of methods used and empirical data. American Journal of Primatology 69: 1370–1386. [DOI] [PubMed] [Google Scholar]
- Chiou KL, Bergey CM. 2018. Methylation-based enrichment facilitates low-cost, noninvasive genomic scale sequencing of populations from feces. Scientific Reports 8: 1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark JS. 2010. Individuals and the variation needed for high species diversity in forest trees. Science 327: 1129–1132. [DOI] [PubMed] [Google Scholar]
- Cook CN, Sgrò CM. 2017. Aligning science and policy to achieve evolutionarily enlightened conservation. Conservation Biology 31: 501–512. [DOI] [PubMed] [Google Scholar]
- Des Roches S, Pendleton LH, Shapiro B, Palkovacs EP. 2021. Conserving intraspecific variation for nature's contributions to people. Nature Ecology Evolution. 10.1038/s41559-021-01403-5 [DOI] [PubMed] [Google Scholar]
- Díaz S, et al. 2020. Set ambitious goals for biodiversity and sustainability. Science 370: 411–413. [DOI] [PubMed] [Google Scholar]
- Di Santo LN, Hamilton JA. 2021. Using environmental and geographic data to optimize ex situ collections and preserve evolutionary potential. Conservation Biology 35: 733–744. [DOI] [PubMed] [Google Scholar]
- [DPIE] Department of Planning, Industry and Environment . 2020. NSW Biodiversity Outlook Report: Results from the Biodiversity Indicator Program, First Assessment. DPIE. [Google Scholar]
- Eldridge MDB, Kinnear JE, Zenger KR, McKenzie LM, Spencer PBS. 2004. Genetic diversity in remnant mainland and ‘pristine’ island populations of three endemic Australian macropodids (Marsupialia): Macropus eugenii, Lagorchestes hirsutus, and Petrogale lateralis. Conservation Genetics 5: 325–338. [Google Scholar]
- Epstein Y, López-Bao JV, Chapron G. 2016. A legal-ecological understanding of favorable conservation status for species in Europe. Conservation Letters 9: 81–88. [Google Scholar]
- Fairbairn DJ. 2011. The advent of mandatory data archiving. Evolution 65: 1–2. [DOI] [PubMed] [Google Scholar]
- Flanagan SP, Forester BR, Latch EK, Aitken SN, Hoban S. 2018. Guidelines for planning genomic assessment and monitoring of locally adaptive variation to inform species conservation. Evolutionary Applications 11: 1035–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankham R. 1996. Relationship of genetic variation to population size in wildlife. Conservation Biology 10: 1500–1508. [Google Scholar]
- Frankham R, Ballou JD, Ralls K, Eldridge M, Dudash MR, Fenster CB, Lacy RC, Sunnucks P. 2019. A Practical Guide for Genetic Management of Fragmented Animal and Plant Populations. Oxford University Press. [Google Scholar]
- Franklin IR. 1980. Evolutionary change in small populations. Pages 135–149 in Soule ME, Wilcox BA, eds. Conservation Biology: An Evolutionary-Ecological Perspective. Sinauer Associates. [Google Scholar]
- Garner A, Rachlow JL, Hicks JF. 2005. Patterns of genetic diversity and its loss in mammalian populations. Conservation Biology 19: 1215–1221. [Google Scholar]
- Garner BA, Hoban S, Luikart G. 2020. IUCN Red List and the value of integrating genetics. Conservation Genetics 21: 795–801. [Google Scholar]
- Gauthier J, Pajkovic M, Neuenschwander S, Kaila L, Schmid S, Orlando L, Alvarez N. 2020. Museomics identifies genetic erosion in two butterfly species across the 20th century in Finland. Molecular Ecology Resources 20: 1191–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert KJ, Whitlock MC. 2015. Evaluating methods for estimating local effective population size with and without migration. International Journal of Organic Evolution 69: 2154–2166. [DOI] [PubMed] [Google Scholar]
- Gougherty AV, Keller SR, Fitzpatrick MC. 2021. Maladaptation, migration and extirpation fuel climate change risk in a forest tree species. Nature Climate Change 11: 166–171. [Google Scholar]
- Grueber CE, Fox S, McLennan EA, Gooley RM, Pemberton D, Hogg CJ, Belov K. 2019. Complex problems need detailed solutions: Harnessing multiple data types to inform genetic management in the wild. Evolutionary Applications 12: 280–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurgel CFD, Camacho O, Minne AJP, Wernberg T, Coleman MA. 2020. Marine heatwave drives cryptic loss of genetic diversity in underwater forests. Current Biology 30: 1199–1206.e2. [DOI] [PubMed] [Google Scholar]
- Hajjar R, Jarvis DI, Gemmill-Herren B. 2008. The utility of crop genetic diversity in maintaining ecosystem services. Agriculture, Ecosystems, and Environment 123: 261–270. [Google Scholar]
- Hansen CCR, Hvilsom C, Schmidt NM, Aastrup P, Van Coeverden de Groot PJ, Siegismund HR, Heller R. 2018. The Muskox lost a substantial part of its genetic diversity on its long road to greenland. Current Biology 28: 4022–4028.e5. [DOI] [PubMed] [Google Scholar]
- Hein L, Gatzweiler F. 2006. The economic value of coffee (Coffea arabica) genetic resources. Ecological Economics 60: 176–185. [Google Scholar]
- Hilty J, Worboys G, Keeley A, Woodley S, Lausche B, Locke H, Carr M, Pulsford I, Pittock J, White JW, Others. 2020. Guidelines for Conserving Connectivity through Ecological Networks and Corridors. International Union for Conservation of Nature World Commission on Protected Areas. [Google Scholar]
- Hoban S. 2014. An overview of the utility of population simulation software in molecular ecology. Molecular Ecology 23: 2383–2401. [DOI] [PubMed] [Google Scholar]
- Hoban S, et al. 2020a. Genetic diversity targets and indicators in the CBD post-2020 Global Biodiversity Framework must be improved. Biological Conservation 248: 108654. [Google Scholar]
- Hoban S, Arntzen JA, Bruford MW, Godoy JA, Rus Hoelzel A, Segelbacher G, Vilà C, Bertorelle G. 2014. Comparative evaluation of potential indicators and temporal sampling protocols for monitoring genetic erosion. Evolutionary Applications 7: 984–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoban S, et al. 2020b. Taxonomic similarity does not predict necessary sample size for ex situ conservation: A comparison among five genera. Proceedings of the Royal Society B 287: 20200102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoban SM, et al. 2013. Bringing genetic diversity to the forefront of conservation policy and management. Conservation Genetics Resources 5: 593–598. [Google Scholar]
- Holderegger R, Schmidt BR, Grünig C, Meier R, Csencsics D, Gassner M, Rellstab C, Stapfer A. 2020. Ready-to-use workflows for the implementation of genetic tools in conservation management. Conservation Genetics Resources 12: 691–700. [Google Scholar]
- Hollingsworth PM, et al. 2020. Scotland's Biodiversity Progress to 2020 Aichi Targets: Conserving Genetic Diversity: Development of a National Approach for Addressing Aichi Biodiversity Target 13 that Includes Wild Species. Scottish Natural Heritage. [Google Scholar]
- [IUCN] International Union for Conservation of Nature . 2016. A global standard for the identification of key biodiversity areas. IUCN. https://portals.iucn.org/library/sites/library/files/documents/2016-048.pdf. [Google Scholar]
- [IUCN] International Union for Conservation of Nature . 2020. Guidelines for Conserving Connectivity through Ecological Networks and Corridors. IUCN. [Google Scholar]
- Jost L. 2010. The relation between evenness and diversity. Diversity 2: 207–232. [Google Scholar]
- Keith AR, Bailey JK, Lau MK, Whitham TG. 2017. Genetics-based interactions of foundation species affect community diversity, stability and network structure. Proceedings of the Royal Society B 284: 2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly RP. 2010. The use of population genetics in Endangered Species Act listing decisions. Ecology Law Quarterly 37: 1107–1158. [Google Scholar]
- Khoury CK, et al. 2019. Comprehensiveness of conservation of useful wild plants: An operational indicator for biodiversity and sustainable development targets. Ecological Indicators 98: 420–429. [Google Scholar]
- Kimura M, Crow JF. 1964. The number of alleles that can be maintained in a finite population. Genetics 49: 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KORA Foundation . 2020. Jahresbericht 2019. KORA Foundation. [Google Scholar]
- Laikre L, et al. 2010. Neglect of genetic diversity in implementation of the convention on biological diversity. Conservation Biology 24: 86–88. [DOI] [PubMed] [Google Scholar]
- Laikre L, Larsson LC, Palmé A, Charlier J, Josefsson M, Ryman N. 2008. Potentials for monitoring gene level biodiversity: Using Sweden as an example. Biodiversity and Conservation 17: 893–910. [Google Scholar]
- Laikre L, Lundmark C, Jansson E, Wennerström L, Edman M, Sandström A. 2016. Lack of recognition of genetic biodiversity: International policy and its implementation in Baltic Sea marine protected areas. Ambio 45: 661–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laikre L, et al. 2020. Post-2020 goals overlook genetic diversity. Science 367: 1083–1085. [DOI] [PubMed] [Google Scholar]
- Leigh DM, Hendry AP, Vázquez-Domínguez E, Friesen VL. 2019. Estimated six per cent loss of genetic variation in wild populations since the industrial revolution. Evolutionary Applications 12: 1505–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefèvre F, Alía R, Bakkebo Fjellstad K, Oggioni S, Rusanen M, Vendramin GG, Bozzano M. 2020. Dynamic Conservation and Utilization of Forest Tree Genetic Resources: Indicators for in Situ and ex Situ Genetic Conservation and Forest Reproductive Material. European Forest Genetic Resources Programme, European Forest Institute. [Google Scholar]
- Lord E, et al. 2020. Pre-extinction demographic stability and genomic signatures of adaptation in the woolly Rhinoceros. Current Biology 30: 3871–3879. [DOI] [PubMed] [Google Scholar]
- Marden E, et al. 2021. Sharing and reporting benefits from biodiversity research. Molecular Ecology 30: 1103–1107. [DOI] [PubMed] [Google Scholar]
- McCallen E, Knott J, Nunez-Mir G, Taylor B, Jo I, Fei S. 2019. Trends in ecology: Shifts in ecological research themes over the past four decades. Frontiers in Ecology and the Environment 17: 109–116. [Google Scholar]
- Méndez M, Vögeli M, Tella JL, Godoy JA. 2014. Joint effects of population size and isolation on genetic erosion in fragmented populations: Finding fragmentation thresholds for management. Evolutionary Applications 7: 506–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickelbart MV, Hasegawa PM, Bailey-Serres J. 2015. Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nature Reviews Genetics 16: 237–251. [DOI] [PubMed] [Google Scholar]
- Morikawa MK, Palumbi SR. 2019. Using naturally occurring climate resilient corals to construct bleaching-resistant nurseries. Proceedings of the National Academy of Sciences 116: 10586–10591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Service National Marine Fisheries. 2005. Policy on the Consideration of Hatchery-Origin Fish in Endangered Species Act Listing Determinations for Pacific Salmon and Steelhead. National Marine Fisheries Service, Protected Resources Division, National Oceanic and Atmospheric Administration, Department of Commerce, Portland, Oregon. [Google Scholar]
- Ogden R, Chuven J, Gilbert T, Hosking C, Gharbi K, Craig M, Al Dhaheri SS, Senn H. 2020. Benefits and pitfalls of captive conservation genetic management: Evaluating diversity in scimitar-horned oryx to support reintroduction planning. Biological Conservation 241: 108244. [Google Scholar]
- Pimentel D, Wilson C, McCullum C, Huang R, Dwen P, Flack J, Tran Q, Saltman T, Cliff B. 1997. Economic and environmental benefits of biodiversity. BioScience 47: 747–757. [Google Scholar]
- Pinsky ML, Palumbi SR. 2014. Meta-analysis reveals lower genetic diversity in overfished populations. Molecular Ecology 23: 29–39. [DOI] [PubMed] [Google Scholar]
- Pomerantz A, Peñafiel N, Arteaga A, Bustamante L, Pichardo F, Coloma LA, Barrio-Amorós CL, Salazar-Valenzuela D, Prost S. 2018. Real-time DNA barcoding in a rainforest using nanopore sequencing: Opportunities for rapid biodiversity assessments and local capacity building. GigaScience 7. 10.1093/gigascience/giy033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posledovich D, Ekblom R, Laikre L. 2021. Mapping and Monitoring of Genetic Diversity in Sweden: A Proposal for Program to Start 2020. Swedish Environmental Protection Agency. [Google Scholar]
- Prieto I, Violle C, Barre P, Durand J-L, Ghesquiere M, Litrico I. 2015. Complementary effects of species and genetic diversity on productivity and stability of sown grasslands. Nature Plants 1: 15033. [DOI] [PubMed] [Google Scholar]
- Raffard A, Santoul F, Cucherousset J, Blanchet S. 2019. The community and ecosystem consequences of intraspecific diversity: A meta-analysis. Biological Reviews of the Cambridge Philosophical Society 94: 648–661. [DOI] [PubMed] [Google Scholar]
- Ralls K, Ballou JD, Dudash MR, Eldridge MDB, Fenster CB, Lacy RC, Sunnucks P, Frankham R. 2018. Call for a paradigm shift in the genetic management of fragmented populations. Conservation Letters 11: e12412. [Google Scholar]
- Ralls K, Brugger K, Ballou J. 1979. Inbreeding and juvenile mortality in small populations of ungulates. Science 206: 1101–1103. [DOI] [PubMed] [Google Scholar]
- Razgour O, Forester B, Taggart JB, Bekaert M, Juste J, Ibáñez C, Puechmaille SJ, Novella-Fernandez R, Alberdi A, Manel S. 2019. Considering adaptive genetic variation in climate change vulnerability assessment reduces species range loss projections. Proceedings of the National Academy of Sciences 116: 10418–10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusch TBH, Ehlers A, Hämmerli A, Worm B. 2005. Ecosystem recovery after climatic extremes enhanced by genotypic diversity. Proceedings of the National Academy of Sciences 102: 2826–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rounsevell MDA, Harfoot M, Harrison PA, Newbold T, Gregory RD, Mace GM. 2020. A biodiversity target based on species extinctions. Science 368: 1193–1195. [DOI] [PubMed] [Google Scholar]
- Ryman N, Laikre L, Hössjer O. 2019. Do estimates of contemporary effective population size tell us what we want to know? Molecular Ecology 28: 1904–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaría L, Méndez PF. 2012. Evolution in biodiversity policy–current gaps and future needs. Evolutionary Applications 5: 202–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler GJ, Qi LL, Marek LF. 2017. Use of Sunflower Crop Wild Relatives for Cultivated Sunflower Improvement. Crop Science 57: 1083–1101. [Google Scholar]
- Shafer ABA, et al. 2015. Genomics and the challenging translation into conservation practice. Trends in Ecology and Evolution 30: 78–87. [DOI] [PubMed] [Google Scholar]
- Smith TB, Kinnison MT, Strauss SY, Fuller TL, Carroll SP. 2014. Prescriptive evolution to conserve and manage biodiversity. Annual Review of Ecology, Evolution, and Systematics 45: 1–22. [Google Scholar]
- Sniezko RA, Koch J. 2017. Breeding trees resistant to insects and diseases: Putting theory into application. Biological Invasions 19: 3377–3400. [Google Scholar]
- Stange M, Barrett RDH, Hendry AP. 2020. The importance of genomic variation for biodiversity, ecosystems and people. Nature Reviews Genetics 22: 89–105. [DOI] [PubMed] [Google Scholar]
- Stoffel MA, et al. 2018. Demographic histories and genetic diversity across pinnipeds are shaped by human exploitation, ecology and life-history. Nature Communications 9: 4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struebig MJ, Kingston T, Petit EJ, Le Comber SC, Zubaid A, Mohd-Adnan A, Rossiter SJ. 2011. Parallel declines in species and genetic diversity in tropical forest fragments. Ecology Letters 14: 582–590. [DOI] [PubMed] [Google Scholar]
- Taft HR, McCoskey DN, Miller JM, Pearson SK, Coleman MA, Fletcher NK, Mittan CS, Meek MH, Barbosa S. 2020. Research–management partnerships: An opportunity to integrate genetics in conservation actions. Conservation Science and Practice 2: 53. [Google Scholar]
- Taylor BL, Perrin WF, Reeves RR, Rosel PE, Wang JY, Cipriano F, Scott Baker C, Brownell RL Jr. 2017. Why we should develop guidelines and quantitative standards for using genetic data to delimit subspecies for data-poor organisms like cetaceans. Marine Mammal Science 33: 12–26. [Google Scholar]
- Torres-Florez JP, Johnson WE, Nery MF, Eizirik E, Oliveira-Miranda MA, Galetti PM. 2018. The coming of age of conservation genetics in Latin America: What has been achieved and what needs to be done. Conservation Genetics 19: 1–15. [Google Scholar]
- [USFWS} US Fish and Wildlife Service . 1973. Endangered species act of 1973. USFWS. [Google Scholar]
- Van Rossum F, Hardy OJ. 2020. Guidelines for genetic monitoring of translocated plant populations. Conservation Biology. [DOI] [PubMed] [Google Scholar]
- van Wyk AM, Dalton DL, Hoban S, Bruford MW, Russo I-RM, Birss C, Grobler P, van Vuuren BJ, Kotzé A. 2017. Quantitative evaluation of hybridization and the impact on biodiversity conservation. Ecology and Evolution 7: 320–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernesi C, Bruford MW, Bertorelle G, Pecchioli E, Rizzoli A, Hauffe HC. 2008. Where's the conservation in conservation genetics? Conservation Biology 22: 802–804. [DOI] [PubMed] [Google Scholar]
- Vitorino LC, Borges Souza UJ, TP F, Ballesteros–Mejia L. 2019. Towards inclusion of genetic diversity measures into IUCN assessments: A case study on birds. Animal Biodiversity and Conservation 42: 317–335. [Google Scholar]
- Weeks AR, et al. 2011. Assessing the benefits and risks of translocations in changing environments: A genetic perspective. Evolutionary Applications 4: 709–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Jiang M. 2021. Meta-analysis of genetic representativeness of plant populations under ex situ conservation in contrast to wild source populations. Conservation Biology 35: 12–23. [DOI] [PubMed] [Google Scholar]
- Wernberg T, Coleman MA, Bennett S, Thomsen MS, Tuya F, Kelaher BP. 2018. Genetic diversity and kelp forest vulnerability to climatic stress. Scientific Reports 8: 1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley AR, Fitzpatrick SW, Funk WC, Tallmon DA. 2015. Genetic rescue to the rescue. Trends in Ecology and Evolution 30: 42–49. [DOI] [PubMed] [Google Scholar]
- Whitlock MC. 2011. Data archiving in ecology and evolution: Best practices. Trends in Ecology and Evolution 26: 61–65. [DOI] [PubMed] [Google Scholar]
- Wieczorek J, Bloom D, Guralnick R, Blum S, Döring M, Giovanni R, Robertson T, Vieglais D. 2012. Darwin Core: An evolving community-developed biodiversity data standard. PLOS ONE 7: e29715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildt D, et al. 2019. Breeding Centers, Private Ranches, and Genomics for Creating Sustainable Wildlife Populations. BioScience 69: 928–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willoughby JR, Sundaram M, Wijayawardena BK, Kimble SJA, Ji Y, Fernandez NB, Antonides JD, Lamb MC, Marra NJ, DeWoody JA. 2015. The reduction of genetic diversity in threatened vertebrates and new recommendations regarding IUCN conservation rankings. Biological Conservation 191: 495–503. [Google Scholar]
- Wood J, et al. 2020. Applying the zoo model to conservation of threatened exceptional plant species. Conservation Biology 34: 1416–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright BR, Hogg CJ, McLennan EA, Belov K, Grueber CE. 2021. Assessing evolutionary processes over time in a conservation breeding program: A combined approach using molecular data, simulations and pedigree analysis. Biodiversity and Conservation 30: 1011–1029. [Google Scholar]
- [WWF] World Wildlife Fund . 2020. Living Planet Report 2020: Bending the Curve of Biodiversity Loss. WWF. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.