Abstract
Human rRNA synthesis by RNA polymerase I requires at least two auxiliary factors, upstream binding factor (UBF) and SL1. UBF is a DNA binding protein with multiple HMG domains that binds directly to the CORE and UCE elements of the ribosomal DNA promoter. The carboxy-terminal region of UBF is necessary for transcription activation and has been shown to be extensively phosphorylated. SL1, which consists of TATA-binding protein (TBP) and three associated factors (TAFIs), does not have any sequence-specific DNA binding activity, and its recruitment to the promoter is mediated by specific protein interactions with UBF. Once on the promoter, the SL1 complex makes direct contact with the DNA promoter and directs promoter-specific initiation of transcription. To investigate the mechanism of UBF-dependent transcriptional activation, we first performed protein-protein interaction assays between SL1 and a series of UBF deletion mutants. This analysis indicated that the carboxy-terminal domain of UBF, which is necessary for transcriptional activation, makes direct contact with the TBP-TAFI complex SL1. Since this region of UBF can be phosphorylated, we then tested whether this modification plays a functional role in the interaction with SL1. Alkaline phosphatase treatment of UBF completely abolished the ability of UBF to interact with SL1; moreover, incubation of the dephosphorylated UBF with nuclear extracts from exponentially growing cells was able to restore the UBF-SL1 interaction. In addition, DNase I footprinting analysis and in vitro-reconstituted transcription assays with phosphatase-treated UBF provided further evidence that UBF phosphorylation plays a critical role in the regulation of the recruitment of SL1 to the ribosomal DNA promoter and stimulation of UBF-dependent transcription.
RNA polymerase I (Pol I) directs RNA synthesis from a single class of genes, the rRNA genes, which are found in multiple tandem arrayed copies in the nucleoli of eukaryotic cells (16, 25, 29). Fractionation of human nuclear extracts indicates that, in addition to RNA Pol I, at least two auxiliary factors are necessary to direct accurate and promoter-specific initiation of transcription, upstream binding factor (UBF) and selectivity factor 1 (SL1) (2, 20, 21). Human UBF has been purified to homogeneity and has been found to be a 94- to 97-kDa polypeptide that recognizes both the CORE and UCE elements of the human rRNA promoter (2, 17). The cloning of cDNA encoding human UBF revealed that it has an amino-terminal region that mediates dimerization and four domains termed HMG boxes, with high homology to the nonhistone chromosomal high-mobility group proteins, HMG 1 and 2 (17, 23). The first HMG box of UBF is necessary and sufficient for DNA binding specificity, while HMG boxes 2, 3, and 4 appear to modulate DNA binding efficiency (18). Another feature of UBF is the carboxy-terminal tail, rich in acidic amino acids, which is required for transcriptional activation (18, 37). Intriguingly, while UBF has been cloned from organisms such as mice, rats, and Xenopus laevis, a UBF-like activity has not been identified in Saccharomyces cerevisiae and Acanthamoeba castellanii (25, 29).
The second essential factor necessary for accurate RNA Pol I transcription is the selectivity factor, SL1. SL1 is analogous to TFIID and TFIIIB, which are involved in RNA Pol II and III transcription, respectively, in that it is a multisubunit complex composed of the TATA-binding protein (TBP) and three TAFs (TBP-associated factors) (6, 7). SL1 is required to direct initiation from the rRNA promoter and plays a crucial role in the promoter recognition properties of the rRNA transcriptional apparatus. While the SL1 complex alone does not bind specifically to the rRNA promoter, in the presence of UBF, SL1 makes contact with the DNA template and extends the DNase I footprint at both the CORE and the UCE promoter elements (2). Mutations in the promoter sequence that affect either the binding of UBF to the DNA template or the interaction of UBF with SL1 result in a drastic reduction of transcriptional activity (11). These findings strongly suggest that the network of protein-protein and protein-DNA interactions among UBF, SL1, and the promoter elements plays a major role in Pol I transcription. Recent studies indicate that UBF binds to SL1 and that this interaction is mediated by protein-protein contacts between UBF and two subunits of the SL1 complex, namely, TBP and TAFI48 (1, 13). Moreover, at least two of the TAFIs appear to make direct contact with the DNA, upon recruitment of SL1 to the promoter. The published data for this finding are still controversial, with either TAFI48/TAFI110 or TAFI63/TAFI110 being reported to make contact with the DNA (1, 32).
In eukaryotic cells, RNA Pol I activity is tightly linked to the signals that control cell growth (25, 29, 30). A number of extracellular stimuli, such as serum deprivation, glucocorticoids, insulin, and phorbol esters, affect the rate of RNA Pol I transcription (4, 5, 9, 12, 24). In addition, Pol I transcription is regulated during the progression of the cell cycle and is repressed at prometaphase and anaphase during mitosis (25, 29, 31). Interestingly, when transcription is arrested during mitosis, UBF appears to remain associated with the DNA (8). Although it is presently unclear how Pol I transcription can be modulated, it has been proposed that posttranslational modifications of UBF and/or SL1 may affect the transcriptional activities of these two factors. For example, phosphorylation of UBF is modulated during muscle cell activation or serum deprivation, and several studies have indicated, at least in vitro, that the phosphorylated form of UBF is severalfold more transcriptionally active than the dephosphorylated moiety (27).
Since the recruitment of SL1 to the promoter occurs primarily through protein-protein contacts with UBF, this interaction represents an important step in the UBF-mediated transcriptional activation of the rRNA promoter. Indeed, a great deal of experimental evidence indicates that the functional cooperativity among UBF, SL1, and the human rRNA promoter is crucial for promoter function and rRNA synthesis. To better understand the mechanism of Pol I transcriptional activation by UBF, we have addressed the requirement for the UBF-mediated recruitment of SL1 to the ribosomal DNA (rDNA) promoter. To begin with, we have dissected the region of UBF involved in the interaction with SL1 by an in vitro protein-protein interaction assay. These studies reveal that the carboxy-terminal region of UBF makes direct contact with SL1, implying that an important role of the transcription activation domain is to mediate interactions with the TBP-TAFI complex SL1. Interestingly, the carboxy-terminal tail of UBF has been shown to be extensively phosphorylated, and the phosphorylation-dephosphorylation of this region has been linked to transcriptional activity. Thus, we have analyzed the effect of phosphorylation on UBF-SL1 interaction. We have shown, through in vitro protein-protein interaction and DNase I footprinting assays, that the phosphorylation state of UBF plays a crucial role in the recruitment of SL1 to the UCE and CORE elements of the rRNA promoter. In conclusion, our experimental data demonstrate that the carboxy-terminal activation domain of UBF directly interacts with SL1 and that UBF phosphorylation plays a critical role in the assembly of a stable and productive initiation complex at the rRNA promoter.
MATERIALS AND METHODS
Plasmid constructs and recombinant proteins.
Flag-tagged baculovirus expression vectors were constructed by removing UBF deletion mutants from pTβ STOP vector (generous gift of M.-H. Jantzen) with NdeI-EcoRI and inserting the fragments downstream of flag epitope engineered into pVL 1392 HAX (HAX was removed) at NdeI-EcoRI (7). Restriction analysis and DNA sequencing confirmed the identity of the clones. The synthesis of recombinant baculoviruses and infection of Sf9 insect cells were performed as previously described (38). Cells were harvested at 36 to 48 h and lysed with an ultrasonic disrupter in TM buffer as described in references 7 and 38.
In vitro protein-protein interaction assays.
Flag-tagged UBF full-length protein (FL), deletion mutants, and hepatitis C virus Pol (HCV Pol) were affinity purified on anti-Flag M2 resin (Kodak) by nutating at 4°C for 1 h and then washing three times in TM 10++ (50 mM Tris [pH 7.9], 12.5 mM MgCl2, 1 mM EDTA, 10% glycerol, 0.1% Nonidet P-40 [NP-40])–0.4 M KCl and two times in TM 10++–0.1 M KCl. Alternatively, proteins used in the protein-protein interaction assays were immobilized on anti-Flag M2 resin (Kodak) and washed three times in dissociation buffer (50 mM Tris [pH 7.9], 1% sodium dodecyl sulfate [SDS], 250 mM LiCl, 0.5% NP-40) and two times in TM 10++–0.1 M KCl. An aliquot of each protein was resolved on SDS-polyacrylamide gel electrophoresis (PAGE) and silver stained, and equivalent amounts of each protein were used per reaction. Alkaline phosphatase (AP)-treated UBF was equilibrated in 1× AP reaction buffer and then incubated with 0.5 to 1 U of either shrimp or calf intestine AP for 15 min at 30°C. Immobilized proteins were then washed twice in radioimmunoprecipitation assay buffer (50 mM Tris [pH 7.9], 150 mM KCl, 0.5% deoxycholate [DOC], 0.1% SDS, 1% NP-40) and twice in TM 10 (no NP-40)–0.1 M KCl. Reaction mixtures treated with nuclear extracts were incubated with 300 μg of nuclear extracts prepared from exponentially growing HeLa cells (6) for 10 min at 30°C in the presence of 1 μM okadaic acid and 1 μM ATP. Reaction mixtures were then washed three times in TM 10++–0.1 M KCl. Ten micrograms of partially purified SL1 from HeLa cells (prepared as described in reference 6) was then added to the immobilized proteins and nutated at 4°C overnight. The resulting complex was washed four times in TM 10++–0.1 M KCl, eluted with 0.05 ml of BCO buffer (20 mM Tris [pH 8.0], 0.5 mM EDTA, 20% glycerol, 1 M KCl, 1% DOC) for 30 min at 4°C, and precipitated with a 1/4 volume of 100% trichloroacetic acid–4 mg of DOC per ml at 4°C for 20 min (as described in reference 10). The pellet was washed with 100% acetone, air dried, resuspended in SDS sample buffer, and heated at 95°C for 3 min. Complexes were separated by SDS–8% PAGE and transferred to nitrocellulose membranes for Western blot analysis. SL1 was detected with anti-TAFI110 and anti-TBP polyclonal antibody. All washes and elution buffers contained a cocktail of protease inhibitors (1 mM dithiothreitol, 1 mM sodium metabisulfite, 0.1 mM phenylmethylsulfonyl fluoride, 100 μg of aprotinin per ml, 10 μg of leupeptin per ml).
Protein purification.
Recombinant flag epitope-tagged UBF FL and UBF 670C deletion mutant were expressed and purified from Sf9 insect cells infected with recombinant baculoviruses by either one of the following protocols. (i) Forty-eight hours after infection, the cells were collected, washed twice with ice-cold phosphate-buffered saline, and lysed in lysis buffer (20 mM Tris [pH 7.5], 500 mM NaCl, 10% glycerol). The lysate was then brought to 55% saturation with ammonium sulfate, and proteins were precipitated by centrifugation at 26,000 × g for 15 min at 4°C. The pellet was resuspended in TM 10–0.25 M KCl, dialyzed against TM 10–0.25 M KCl, and centrifuged at 100,000 × g for 45 min at 4°C prior to being loaded onto a DEAE (Pharmacia Biotech) column preequilibrated to TM 10–0.25 M KCl. The column was washed with TM 10–0.25 M KCl, and the peak flowthrough fraction, as determined by protein concentration, was loaded onto a heparin-agarose column (Poros HE1). The column was washed with TM 10–0.25 M KCl, and the proteins were eluted by a linear salt gradient from 0.25 to 1 M KCl in TM 10 buffer. Fractions containing UBF were pooled and dialyzed against TM 10–0.1 M KCl. After this purification step, UBF was about 99% pure, as determined by SDS-PAGE and silver-stained gel. All buffers contained a cocktail of protease inhibitors (1 mM dithiothreitol, 0.1 mM phenylmethylsulfonyl fluoride, and 1 mM metabisulfite). Dephosphorylation of UBF was achieved by incubating purified UBF with agarose-bound AP (Sigma) for 30 min at 30°C. The bound AP was then separated from soluble UBF by low-speed centrifugation. (ii) Alternatively, flag epitope-tagged UBF and UBF 670C expressed in Sf9 cells were purified by using anti-Flag M2 affinity resin (Kodak). Cells were lysed in TM–0.5 M KCl and incubated with anti-Flag M2 resin for 1 h at 4°C on a nutator. After extensive washing, the bound material was eluted from the affinity resin by treatment with elution buffer (50 mM glycine, 150 mM NaCl, pH 5) and neutralized with 0.05 volume of 2 M Tris (pH 7.9). Eluted proteins were dialyzed to TM–0.1 M KCl and quantitated by SDS-PAGE and silver staining. AP-treated UBF was affinity purified as described above and treated with AP prior to elution and dialysis. The complete removal of AP from the UBF samples was confirmed by incubating an aliquot of the treated proteins with a 32P-end-labeled DNA probe. After absorption to Whatman DE-81 filters, no loss of 32P label above the background was observed for the AP-treated UBF sample.
RNA Pol I used in the reconstituted transcription reaction was prepared as follows. Nuclear extracts from HeLa cells were chromatographed on a heparin agarose column with a salt gradient from 0.1 to 0.7 M KCl. Fractions eluted at 250 mM KCl were pooled, dialyzed against TM buffer containing 0.1 M KCl, and fractionated on a Sepharose 300 (Pharmacia Biotech) gel filtration column. Active fractions were then loaded onto a Q-Sepharose column (Poros HQ) equilibrated against TM 10–0.1 M KCl. Proteins were eluted with a salt gradient from 0.1 to 0.7 M KCl in TM 10 buffer. The active fractions were pooled, dialyzed to 0.125 M KCl, aliquoted, and stored at −80°C. This RNA Pol I preparation contained no detectable UBF and SL1 activity. SL1 was purified from HeLa cells as previously described (3, 6). Protein concentration was determined by using a Bradford assay kit (Bio-Rad).
DNase I footprinting analysis.
DNase I footprinting analysis was performed as previously described (3, 18) with pSBr24 (−500 to +24 of the human rRNA promoter cloned into pUC18) as the template. The addition of 200 mM sodium orthovanadate to the reaction mixture was the only modification.
In vitro transcription assays.
In vitro-reconstituted transcription assays were performed as previously described (6, 38) with the following modification: each transcription reaction was performed with 30 ng of rRNA template prHu3. RNA products were detected by S1 analysis with a single-stranded oligonucleotide overlapping the transcription initiation site (from −20 to +40) (2).
RESULTS
The carboxy-terminal acidic tail of UBF is necessary for protein-protein interaction with SL1.
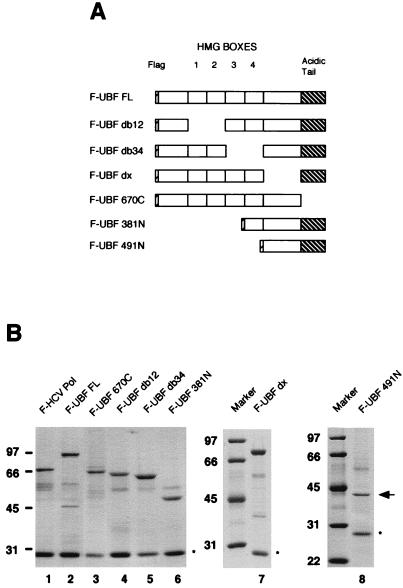
To identify the regions of UBF that interact with SL1, we constructed vectors containing a series of flag-tagged UBF deletion mutants for expression in the baculovirus expression system. Experimental results from our laboratory show unequivocally that the recombinant UBF is functionally indistinguishable from the UBF isolated and purified from human cells. To begin our studies, we generated UBF mutants lacking the carboxy-terminal region (F-UBF 670C), lacking the amino-terminal region (F-UBF 381N and F-UBF 491N), having deletions of certain HMG boxes (F-UBF db12 and F-UBF db34), or having deletions of the region between HMG box 4 and the acidic tail (F-UBF dx) as schematically represented in Fig. 1A. Each protein was expressed in Sf9 insect cells and purified by affinity chromatography on anti-Flag M2 resin, under high-stringency conditions (see Materials and Methods for details). The purity and the amount of wild-type and mutant UBF proteins were assessed by resolving the proteins on SDS-PAGE and subsequently staining the gel with Coomassie blue (Fig. 1B). In addition to the series of UBF proteins, recombinant flag-tagged HCV Pol (F-HCV Pol) was expressed and purified from Sf9 cells and used as a negative control in the protein-protein interaction assays.
FIG. 1.
UBF deletion mutants. (A) Schematic representation of UBF FL and deletion mutations. HMG boxes 1 to 4 and the carboxy-terminal acidic tail are indicated in the diagram. The carboxy-terminal deletion (UBF 670C), internal deletions (UBF db12, UBF db34, and UBF dx), and amino-terminal deletions (UBF 381N and UBF 491N) of flag-tagged UBF are shown. (B) Recombinant proteins expressed in Sf9 cells were purified on anti-Flag M2 resin, resolved on SDS-PAGE, and stained with Coomassie blue. The proteins were F-HCV Pol (lane 1), F-UBF FL (lane 2), F-UBF 670C (lane 3), F-UBF db12 (lane 4), F-UBF db34 (lane 5), F-UBF 381N (lane 6), F-UBF dx (lane 7), and F-UBF 491N (lane 8). The arrow indicates the position of F-UBF 491N. The asterisks indicate immunoglobulin G light chain. Markers at the left of each panel show molecular mass in kilodaltons.
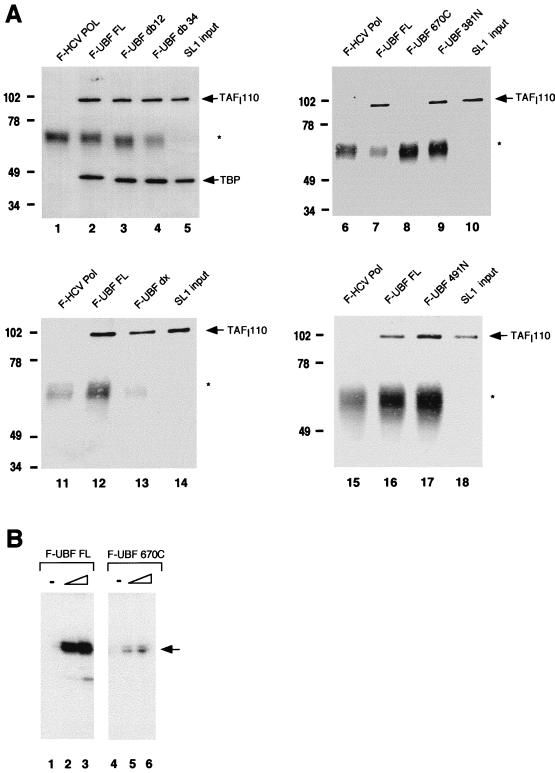
Each of the flag-tagged UBF deletion mutants was then tested in a series of in vitro protein-protein interaction studies. Equal molar amounts of full-length and mutant UBF proteins and HCV Pol, as judged by silver stained SDS-PAGE, were immobilized on anti-Flag M2 affinity resin and incubated with human SL1. Although two subunits of SL1, TBP and TAFI48, can interact directly with UBF (1, 19), we reasoned that it was more relevant to perform these studies with the intact SL1 complex, since we cannot exclude the possibility that interactions observed with isolated subunits may involve contact surfaces that are not accessible in the context of the intact SL1. Thus, the SL1 fraction used in the protein-protein interaction assays was partially purified from HeLa nuclear extracts and was depleted of any UBF and RNA Pol I activity. Each reaction mixture was then extensively washed, and the bound complexes were eluted from the affinity beads by treatment with high salts and detergents. The eluted proteins were precipitated with trichloroacetic acid, dissolved in SDS sample buffer, and then resolved by SDS-PAGE. After transfer to nitrocellulose, the blots were probed with antibody raised against TAFI110 and/or TBP to detect the bound SL1. As shown in Fig. 2A, SL1 interacts efficiently with UBF (lanes 2, 7, 12, and 16), UBF deletion mutants missing HMG boxes 1 and 2 or 3 and 4 (lanes 3 and 4), or mutants missing the region between HMG box 4 and the acidic tail (lane 13). Moreover, SL1 efficiently binds to amino-terminal deletions of UBF containing the region from HMG box 4 to the carboxy-terminal tail (lane 9) and the 274 carboxy-terminal amino acids (lane 17). On the other hand, the removal of the carboxy-terminal acidic tail completely abolishes SL1 binding (lane 8). This same mutant has been shown by us and others to be transcriptionally inactive (Fig. 2B) (18). Taken together, these results indicate that the carboxy-terminal tail of UBF mediates the interaction between UBF and SL1 and reinforces the concept that functional cooperativity between the transcription factors UBF and SL1 is required for the activation of rRNA transcription. Moreover, our results indicate that the HMG boxes, some of which were postulated to have been involved in binding to SL1, are not essential for this protein-protein interaction.
FIG. 2.
SL1 interacts with the carboxy-terminal domain of UBF. (A) Recombinant flag-tagged proteins were immobilized on anti-Flag M2 resin as bait and incubated with human SL1. The resulting complex was eluted, resolved on SDS-PAGE, and transferred to nitrocellulose for Western analysis to detect coimmunoprecipitated SL1 (see Materials and Methods). The bait proteins used in each reaction are as indicated above each panel. F-HCV Pol is the negative control, and the input is 10% of the SL1 used per reaction. Nitrocellulose was probed with polyclonal anti-TAFI110 antibody and reprobed with polyclonal anti-TBP antibody (lanes 1 to 5) or probed with anti-TAFI110 antibody alone (lanes 6 to 18). The asterisks denote immunoglobulin G heavy chain. Markers to the left of each gel show molecular mass in kilodaltons. (B) Increasing amounts (1 and 2 ng) of affinity-purified recombinant UBF FL (lanes 2 and 3) and UBF 670C (lanes 5 and 6) were used with partially purified RNA Pol I (2 μl; 5 mg/ml) and SL1 (1 μl; 0.8 mg/ml) in reconstituted transcription reactions. UBF amounts were estimated by SDS-PAGE and silver staining. In vitro-synthesized transcripts were detected by S1 nuclease protection assay. The arrow indicates the protected oligonucleotide fragment.
UBF phosphorylation plays an important role in the regulation of the protein interactions between UBF and SL1.
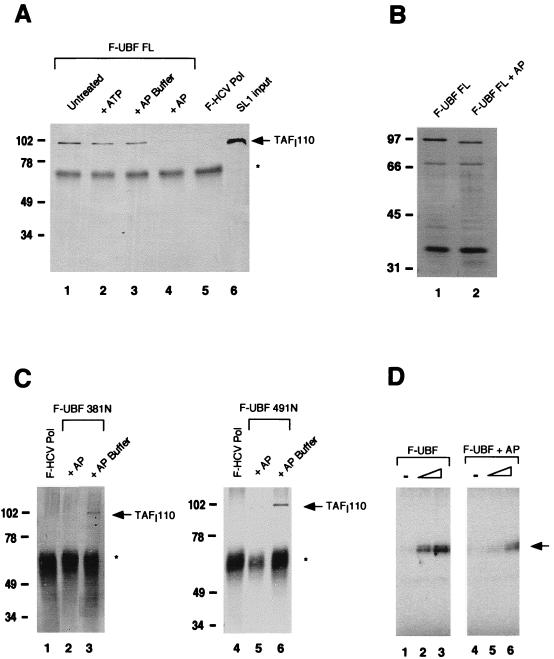
The carboxy-terminal region of UBF has also been shown to be heavily phosphorylated, and the phosphorylation state of UBF appears to be important for its transcriptional activity (14, 37) (see also below). To determine if phosphorylation plays a role in the regulation of the SL1-UBF interaction, additional in vitro interaction assays were performed with UBF that was dephosphorylated by treatment with AP. Flag-tagged UBF immobilized on anti-Flag M2 affinity resin was incubated in the presence of either shrimp or calf intestine AP for 15 min at 30°C. Dephosphorylation of UBF correlated with the appearance of a faster-migrating form of UBF on SDS-PAGE (Fig. 3B). Then, the reaction mixture was extensively washed to remove the phosphatase prior to incubation of the immunocomplex with SL1. The results of this experiment, shown in Fig. 3A, reveal that UBF treated with AP fails to interact with SL1 (lane 4), as determined by the absence of TAFI110 in the immunoprecipitated product. On the other hand, when UBF is incubated either with ATP (lane 2) or with the buffer used for the AP reaction (lane 3), it binds to SL1 as well as does the untreated wild-type UBF (lane 1). Similarly, two UBF amino-terminal deletion mutants, UBF 381N and UBF 491N, which can normally associate with SL1 (Fig. 3C, lanes 3 and 6), did not bind to SL1 once treated with AP (lanes 2 and 5). The finding that a posttranslational modification of UBF is possibly involved in the regulation of SL1 binding was further confirmed by the observation that Escherichia coli-expressed UBF mutant 381N, which contains the carboxy-terminal tail, failed to bind to SL1 (data not shown). E. coli-expressed full-length UBF could not be tested in this assay because it is predominantly synthesized as a truncated mutant missing the carboxy-terminal tail (37a). Thus, our results indicate that dephosphorylation by AP treatment strongly affects the ability of UBF to interact with SL1 and suggest that this posttranslational modification plays an important role in the regulation of protein-protein interactions between UBF and SL1. Moreover, in vitro-reconstituted transcription assays show that AP treatment of UBF sharply decreases its transcriptional activity (Fig. 3D). These results provide further evidence that there is a tight link between UBF-dependent activation and UBF-SL1 binding.
FIG. 3.
Role of UBF phosphorylation in SL1 binding. (A) F-UBF FL was immobilized on flag antibody beads and either untreated (lane 1) or treated with 5 mM ATP (lane 2), AP buffer alone (lane 3), or with buffer plus AP (lane 4). The binding assay was then performed as previously described. Coimmunoprecipitated SL1 was detected by Western blot analysis with anti-TAFI110 antibody. Lanes 5 and 6 are negative control and SL1 input, respectively. (B) Silver-stained SDS-PAGE of untreated UBF (lane 1) and AP-treated UBF (lane 2) after immunoprecipitation shows faster migration of dephosphorylated UBF. (C) F-UBF 381N and F-UBF 491N were immobilized on flag antibody beads and treated either with AP buffer alone (lanes 3 and 6) or with buffer plus AP (lanes 2 and 5). SL1 binding assays were done as previously described. (D) In vitro transcription assays containing partially purified RNA Pol I (10 μg), SL1 (0.8 μg), and increasing amounts (0.25 and 1.25 ng) of purified recombinant UBF (lanes 2 and 3) or dephosphorylated UBF (lanes 5 and 6) were performed as described in Materials and Methods. The transcription assays with UBF and AP-treated UBF were quantified with a phosphorimager. The mean fold activation in the presence of UBF or AP-treated UBF, calculated from two independent experiments, is 8.5- and 2.0-fold, respectively. Asterisks in panels A and C indicate immunoglobulin G heavy chain. Markers in panels A to C show molecular mass in kilodaltons. The arrow in panel D indicates the protected oligonucleotide fragment.
Incubation of AP-treated UBF with HeLa nuclear extract rescues the binding to SL1.
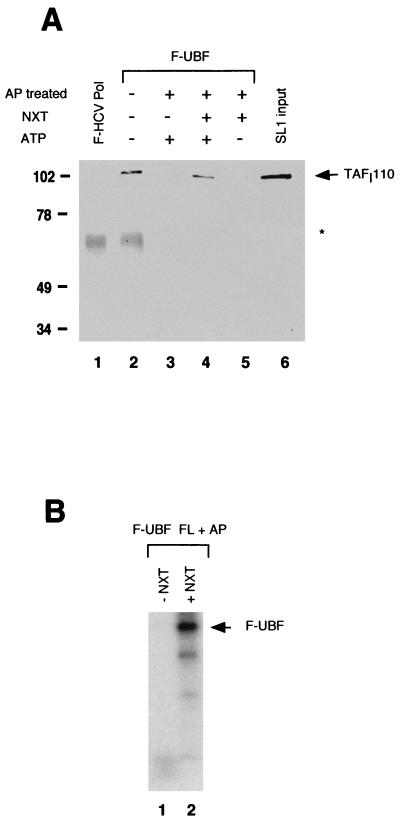
To determine if the SL1 binding activity of dephosphorylated UBF could be restored, AP-treated flag-tagged UBF bound to affinity resin was incubated with nuclear extracts prepared from exponentially growing HeLa cells, in the presence of ATP. After incubation at 30°C, the reaction mixture was washed extensively and incubated with human SL1, and the resulting complex was detected by immunoblotting with anti-TAFI110, as previously described. As shown in Fig. 4A, while the AP-treated UBF fails to interact with SL1 (lane 3), preincubation of AP-treated UBF with HeLa nuclear extracts (lane 4) reestablishes a stable complex formation between UBF and SL1 to levels similar to that of the untreated UBF (lane 2). Reactivation of SL1 binding is dependent on ATP, since the incubation of dephosphorylated UBF with nuclear extracts in the absence of ATP fails to yield a stable UBF-SL1 complex (compare lane 4 with lane 5). Finally, we show that UBF can be readily radiolabeled in the presence of a small amount of [γ-32P]ATP during the incubation with the nuclear extracts (Fig. 4B, lane 2), further suggesting a functional link between UBF and cellular kinases. Importantly, the restored protein interaction is dependent on the carboxy-terminal tail, since a UBF deletion mutant missing the carboxy-terminal tail (UBF 670C) which has been preincubated with nuclear extracts does not bind to SL1 (data not shown).
FIG. 4.
Reconstitution of SL1 binding with dephosphorylated UBF. (A) The protein interaction assay was performed as described in Materials and Methods with untreated UBF (lane 2) and AP-treated UBF. Prior to the addition of SL1, dephosphorylated UBF was incubated in TM buffer plus 1 μM ATP (lane 3) or with nuclear extracts (NXT) prepared from exponentially growing HeLa cells in the presence (lane 4) or absence (lane 5) of 1 μM ATP. Western blotting was performed with anti-TAFI110 antibody. The asterisk indicates immunoglobulin G heavy chain. Markers show molecular mass in kilodaltons. (B) Immobilized flag-tagged UBF was treated with AP before incubation with 10 μCi of [γ-32P]ATP in the presence (lane 2) or absence (lane 1) of 300 μg of nuclear extracts (NXT) from exponentially growing HeLa cells. Following separation on SDS-PAGE, phosphorylation was detected by autoradiography.
Taken together, our data indicate that the protein-protein interaction between UBF and SL1 is mediated by the carboxy-terminal tail of UBF and, more importantly, that this interaction is regulated by a phosphorylation-dephosphorylation mechanism.
UBF phosphorylation regulates the recruitment of SL1 to the rDNA promoter.
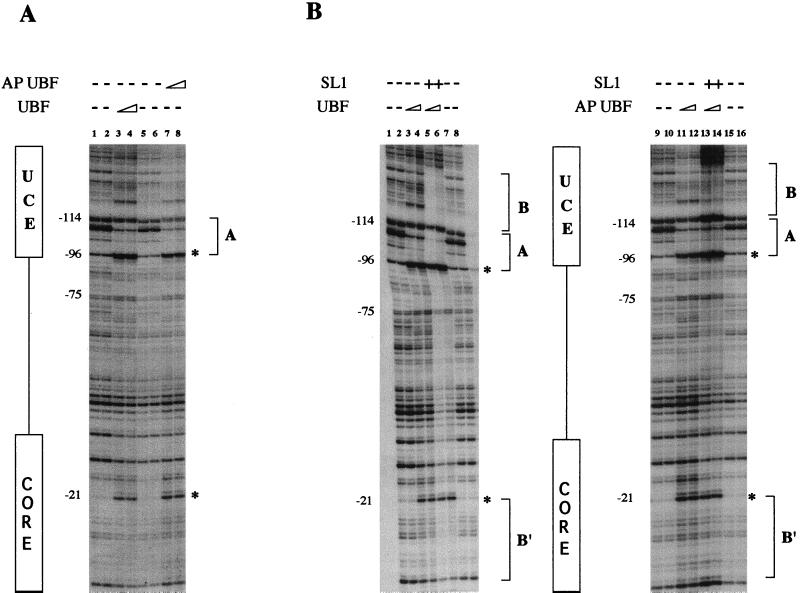
The experiments presented so far suggest that the weak transcriptional activity of dephosphorylated UBF is at least in part due to its inability to recruit SL1 to the promoter. To establish unambiguously the requirement of UBF phosphorylation in the formation of a stable preinitiation complex at the human rDNA promoter, we performed DNase I protection assays with either phosphorylated or dephosphorylated UBF. As shown in Fig. 5A, the protection pattern of phosphorylated (lanes 3 and 4) or dephosphorylated (lanes 7 and 8) UBF does not reveal any significant difference. The DNase I footprinting shows that both forms of UBF protect a region between −75 and −114, overlapping the UCE (site A). In addition, a weaker interaction with the CORE element results in an enhanced cleavage at position −21 (3). Thus, phosphorylated and AP-treated UBF bind with equal affinities to the human rDNA promoter. On the other hand, comparison of the footprinting pattern obtained with phosphorylated (Fig. 5B, lanes 5 and 6) and AP-treated (lanes 13 and 14) UBF in the presence of SL1 shows a substantial difference of the protected region in both the CORE and the UCE elements. The enhanced protection pattern of UBF over the UCE (site B) promoter element in the presence of SL1 is sharply reduced in the presence of the dephosphorylated form of UBF. An even more dramatic effect is seen in the CORE region, where the SL1 protection over the promoter (site B′) is completely abolished. In summary, our results indicate that UBF phosphorylation-dephosphorylation does not affect the ability of UBF to recognize and bind to the rRNA promoter but rather regulates the formation of a strong and stable initiation complex with SL1, as indicated by the formation of new DNA-protein contacts at the promoter in the presence of phosphorylated UBF.
FIG. 5.
UBF phosphorylation mediates SL1 recruitment to the rDNA promoter. (A) DNase I digestion of UBF and hypophosphorylated UBF on the coding strand of the human rDNA promoter with UCE and CORE region as indicated on the left. Shown are protection patterns with no protein added (lanes 1, 2, 5, and 6) or with increasing amounts of either UBF (lanes 3 and 4) or dephosphorylated UBF (lanes 7 and 8). (B) Footprinting analysis was performed as described for panel A with both forms of UBF in the presence of 1 μg of SL1. Shown are results with naked DNA only (lanes 1, 2, 7, 8, 9, 10, 15, and 16), UBF (lanes 3 and 4), AP-treated UBF (lanes 11 and 12), increasing amounts of UBF with SL1 (lanes 5 and 6), and increasing amounts of AP-treated UBF with SL1 (lanes 13 and 14). The region of DNA protected by UBF is indicated by bracket A, while SL1 extended footprinting is indicated by bracket B (UCE) and bracket B′ (CORE). Hypersensitive sites at positions −96 and −21 are indicated by asterisks.
DISCUSSION
In this report, we have examined the cooperative interaction between SL1 and UBF and its relationship to RNA Pol I transcriptional activation. Transcription of rRNA by RNA Pol I requires the cooperative interaction of at least two auxiliary factors, UBF and SL1. UBF binds to the minor groove of the rRNA promoter, primarily through HMG box 1, and induces a bend in the DNA (25). Once bound to the promoter, UBF recruits the selectivity and species-specific factor SL1. Human SL1 does not have any specific or nonspecific DNA binding activity; therefore, its recruitment to the DNA promoter region occurs via protein-protein interactions with UBF.
Using purified human SL1 and recombinant human UBF, purified from baculovirus-infected insect cells, we have characterized the interaction between UBF and SL1 with the aim of better understanding the process of transcription initiation by RNA Pol I. Our data demonstrate for the first time that the interaction between UBF and SL1 is mediated by direct interaction of SL1 with the carboxy-terminal domain of UBF. Since this domain is required for transcriptional activation, our results establish a functional link between the transactivation function of UBF and its ability to bind to SL1. This notion provides strong support for the concept that a key role of the transcription activation domain of UBF is to mediate the interaction with the TBP-TAFI complex SL1. This is reminiscent of many Pol II-transcribed genes, where the TBP-TAF complex appears to function as a bridge between the transcription activation domains and the RNA Pol holoenzyme-basal transcriptional machinery (22, 35).
Jantzen et al. (18), based on indirect evidence from DNase I footprinting analysis, postulated that HMG boxes 3 and 4 might also be involved in the interaction with SL1. Our data indicate that this is unlikely, since deletion mutants containing these domains can associate quite well with SL1 in the protein interaction assay. Rather, we interpret the inability of the UBF HMG box 3 and 4 deletion mutant to produce an SL1 footprinting pattern or to activate transcription as a conformational defect of these UBF mutants which does not allow SL1 to make the correct contacts with the promoter and consequently fails to promote efficient initiation of transcription. In this regard, it has been shown that the topology of the initiation complex on the DNA is rather important, and for example, mutations that affect the spacing between the promoter elements have a significant effect on Pol I activity (28). It is also possible that interactions between UBF and other components of the Pol I transcriptional machinery (i.e., RNA Pol I), possibly mediated by one or more of the HMG boxes, may be important for the formation of a productive initiation complex (33).
The presence of multiple phosphorylation sites at serine residues in the carboxy-terminal domain of UBF prompted us to test whether this posttranslational modification might play a role in the regulation of UBF-SL1 interaction. To our surprise, dephosphorylation of UBF by AP completely abolishes the binding of SL1 to UBF. Importantly, the binding can be rescued by preincubation of dephosphorylated UBF with a nuclear extract prepared from exponentially growing HeLa cells. Moreover, our footprinting analysis shows that in the presence of AP-treated UBF most of the SL1-specific contacts within the UCE and CORE elements of the promoter are lost, thus providing further evidence of the inability of dephosphorylated UBF to form a productive preinitiation complex. These results have two major implications. First, they demonstrate for the first time that the activation domain of a transactivating protein regulates its interaction with a basal component of the transcription complex through phosphorylation and dephosphorylation. Second, they suggest a mechanism of rRNA synthesis regulation by physiological stimuli, which involves one or more cellular kinases acting through signal transduction pathways. Our results are in agreement with studies which indicate that UBF is hypophosphorylated and transcriptionally inactive in quiescent or serum-deprived cells (26). All of these results point to a key role for UBF phosphorylation in the control of growth-dependent rRNA transcription. Interestingly, in the last few years it has become apparent that posttranslational modifications, such as phosphorylation, play an important function in the regulation of the interaction between a variety of transcription factors and, ultimately, in the modulation of gene expression (15, 34). For RNA Pol I transcription, this mechanism of regulation offers a very simple process which enables the cell to rapidly regulate ribosome biosynthesis in response to a variety of extracellular stimuli.
Recent experimental data indicate that UBF can be found bound to the DNA even in the absence of RNA Pol I transcription (8). The authors proposed that modification of the transcriptional machinery might be involved in the inactivation of transcription. In view of our results, we could speculate that the absence of Pol I transcription could be the consequence of UBF dephosphorylation. However, we cannot exclude that modifications in one or more of the SL1 subunits may also be important in modulation of RNA Pol I transcription during the cell cycle or in growth-dependent rRNA synthesis.
The results of our experiments also show that nuclear extracts from exponentially growing cells contain factors that can phosphorylate recombinant UBF and, by doing so, facilitate the binding of SL1. However, this UBF preparation was only 1.5- to 2.0-fold more active than dephosphorylated UBF in transcription assays (data not shown). It is possible that since the kinase reaction with nuclear extracts is not very efficient, the dephosphorylated UBF present in the reaction, which can bind to the promoter as well as the phosphorylated form, may negatively affect the transcription reaction. Nevertheless, our observation will certainly be useful for future studies on the biochemical purification and characterization of cellular kinase(s) that can regulate UBF-SL1 interaction and Pol I transcriptional activity. The protein kinases involved in this process are currently unknown, and previous work has suggested that a hierarchic series of phosphorylation events, mediated most likely by several cellular kinases, modulates UBF activity (36). Ultimately, because of the known correlation between UBF phosphorylation and cell growth, the identification and biochemical characterization of this kinase(s) may provide an important tool for understanding the biological role of cellular kinases during growth and cell proliferation.
ACKNOWLEDGMENTS
We are grateful to the members of the Gene Expression Group at USC for helpful advice and discussions. We thank H.-M. Jantzen for sharing constructs and Tiffany Bui for technical support.
W.Z. is partially supported by the Heidelberger Predoctoral Scholarship Award in Cancer Research. This work was funded by grant RPG-97-058-01-NP from the American Cancer Society.
REFERENCES
- 1.Beckmann H, Chen J L, O’Brien T, Tijan R. Coactivator and promoter-selective properties of RNA polymerase I TAFs. Science. 1995;270:1506–1509. doi: 10.1126/science.270.5241.1506. [DOI] [PubMed] [Google Scholar]
- 2.Bell S P, Learned R M, Jantzen H-M, Tijan R. Functional cooperativity between transcription factors UBF1 and SL1 mediates human ribosomal RNA synthesis. Science. 1988;241:1192–1197. doi: 10.1126/science.3413483. [DOI] [PubMed] [Google Scholar]
- 3.Bell S P, Pikaard C S, Reeder R H, Tijan R. Molecular mechanisms governing species-specific transcription of ribosomal RNA. Cell. 1989;59:489–497. doi: 10.1016/0092-8674(89)90032-9. [DOI] [PubMed] [Google Scholar]
- 4.Cavanaugh A H, Hempel W M, Taylor L J, Rogalsky V, Todorov G, Rothblum L I. Activity of RNA polymerase I transcription factor UBF blocked by Rb gene product. Nature. 1995;374:177–180. doi: 10.1038/374177a0. [DOI] [PubMed] [Google Scholar]
- 5.Cavanaugh A H, Thompson E A. Hormonal regulation of transcription of rDNA. J Biol Chem. 1986;261:12738–12744. [PubMed] [Google Scholar]
- 6.Comai L, Tanese N, Tijan R. The TATA-binding protein and associated factors are integral components of the RNA polymerase I transcription factor, SL1. Cell. 1992;68:965–976. doi: 10.1016/0092-8674(92)90039-f. [DOI] [PubMed] [Google Scholar]
- 7.Comai L, Zomerdijk J C B M, Beckmann H, Zhou S, Admon A, Tijan R. Reconstitution of transcription factor SL1: exclusive binding of TBP by SL1 or TFIID subunits. Science. 1994;266:1966–1972. doi: 10.1126/science.7801123. [DOI] [PubMed] [Google Scholar]
- 8.Gebrane-Younes J, Fomproix N, Hernandez-Verdun D. When rDNA transcription is arrested during mitosis, UBF is still associated with non-condensed rDNA. J Cell Sci. 1997;110:2429–2440. doi: 10.1242/jcs.110.19.2429. [DOI] [PubMed] [Google Scholar]
- 9.Grummt I, Smith A V, Grummt F. Amino acid starvation affects the initiation frequency of nucleolar RNA polymerase. Cell. 1976;7:439–445. doi: 10.1016/0092-8674(76)90174-4. [DOI] [PubMed] [Google Scholar]
- 10.Gu W, Shi X-L, Roeder R. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–822. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 11.Haltiner M M, Smale S T, Tijan R. Two distinct promoter elements in the human rRNA gene identified by linker scanning mutagenesis. Mol Cell Biol. 1986;6:227–235. doi: 10.1128/mcb.6.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hammond L M, Bowman H L. Insulin stimulates the translation of ribosomal proteins and the transcription of rRNA in mouse myoblast. J Biol Chem. 1988;263:17785–17791. [PubMed] [Google Scholar]
- 13.Hempel W H, Cavanaugh A H, Hannan R D, Taylor L, Rothblum L I. The species-specific RNA polymerase I transcription factor SL-1 binds to upstream binding factor. Mol Cell Biol. 1996;16:557–563. doi: 10.1128/mcb.16.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hershey J C, Hautmann M, Thompson M M, Rothblum L I, Haystead T A J, Owens G K. Angiotensin II-induced hypertrophy of rat vascular smooth muscle is associated with increased 18S rRNA synthesis and phosphorylation of the rRNA transcription factor, upstream binding factor. J Biol Chem. 1995;270:25096–25101. doi: 10.1074/jbc.270.42.25096. [DOI] [PubMed] [Google Scholar]
- 15.Hunter T, Karin M. The regulation of transcription through phosphorylation. Cell. 1992;70:375–387. doi: 10.1016/0092-8674(92)90162-6. [DOI] [PubMed] [Google Scholar]
- 16.Jacob S T. Regulation of ribosomal gene transcription. Biochem J. 1995;306:617–626. doi: 10.1042/bj3060617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jantzen H-M, Admon A, Bell S P, Tijan R. Nucleolar transcription factor hUBF contains a DNA-binding motif with homology to HMG proteins. Nature. 1990;344:830–836. doi: 10.1038/344830a0. [DOI] [PubMed] [Google Scholar]
- 18.Jantzen H-M, Chow A M, King D S, Tijan R. Multiple domains of the RNA polymerase I activator hUBF interact with the TATA-binding protein complex hSL1 to mediate transcription. Genes Dev. 1992;6:1950–1963. doi: 10.1101/gad.6.10.1950. [DOI] [PubMed] [Google Scholar]
- 19.Kwon H, Green M. The RNA polymerase I transcription factor, upstream binding factor, interacts directly with the TATA box-binding protein. J Biol Chem. 1994;269:30140–30146. [PubMed] [Google Scholar]
- 20.Learned R M, Cordes S, Tijan R. Purification and characterization of a transcription factor that confers promoter specificity to human RNA polymerase I. Mol Cell Biol. 1985;5:1358–1369. doi: 10.1128/mcb.5.6.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Learned R M, Learned T K, Haltiner M M, Tijan R. Human rRNA transcription is modulated by the coordinate binding of two factors to an upstream control element. Cell. 1986;45:847–857. doi: 10.1016/0092-8674(86)90559-3. [DOI] [PubMed] [Google Scholar]
- 22.Lee T I, Young R A. Regulation of gene expression by TBP-associated proteins. Genes Dev. 1998;12:1398–1408. doi: 10.1101/gad.12.10.1398. [DOI] [PubMed] [Google Scholar]
- 23.McStay B, Frazier M W, Reeder R H. xUBF contains a novel dimerization domain essential for RNA polymerase I transcription. Genes Dev. 1991;5:1957–1968. doi: 10.1101/gad.5.11.1957. [DOI] [PubMed] [Google Scholar]
- 24.Mishima Y M, Matsui T, Muramatsu M. The mechanism of decrease in nucleolar RNA synthesis by protein synthesis inhibition. J Biochem. 1979;85:807–818. [PubMed] [Google Scholar]
- 25.Moss T, Stefanovsky V Y. Promotion and regulation of ribosomal transcription in eukaryotes by RNA polymerase I. Prog Nucleic Acids Res Mol Biol. 1995;50:25–66. doi: 10.1016/s0079-6603(08)60810-7. [DOI] [PubMed] [Google Scholar]
- 26.O’Mahony D J, Smith S D, Xie W, Rothblum L I. Analysis of the phosphorylation, DNA-binding and dimerization properties of the RNA polymerase I transcription factors UBF1 and UBF2. Nucleic Acids Res. 1992;20:1301–1308. doi: 10.1093/nar/20.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Mahony D J, Xie W, Smith S D, Singer H A, Rothblum L I. Differential phosphorylation and localization of the transcription factor UBF in vivo in response to serum deprivation. J Biol Chem. 1992;267:35–38. [PubMed] [Google Scholar]
- 28.Pape L K, Windle J J, Sollner-Webb B. Half helical turn spacing changes convert a frog into a mouse rDNA promoter: a distant upstream domain determines the helix face of the initiation site. Genes Dev. 1990;4:52–62. doi: 10.1101/gad.4.1.52. [DOI] [PubMed] [Google Scholar]
- 29.Paule M R. Transcription of ribosomal RNA genes by eukaryotic RNA polymerase I. New York, N.Y: Springer-Verlag, Inc.; 1998. [Google Scholar]
- 30.Paule M R, Iida C T, Perna P J, Harris G H, Knoll D A, D’Alessio J M. In vitro evidence that eukaryotic ribosomal RNA transcription is regulated by modification of RNA polymerase I. Nucleic Acids Res. 1984;12:8161–8180. doi: 10.1093/nar/12.21.8161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roussel P, Andre C, Comai L, Hernandez-Verdun D. The rDNA transcription machinery is stabilized as initiation complex during mitosis in active NORs and absent in inactive NORs. J Cell Biol. 1996;133:235–246. doi: 10.1083/jcb.133.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rudloff U, Eberhard D, Tora L, Stunnenberg H, Grummt I. TBP associated factors interact with DNA and govern species specificity of RNA polymerase I transcription. EMBO J. 1994;13:2611–2616. doi: 10.1002/j.1460-2075.1994.tb06551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schnapp G, Santori F, Carles C, Riva M, Grummt I. The HMG box-containing nucleolar transcription factor UBF interacts with a specific subunit of RNA polymerase I. EMBO J. 1994;13:190–199. doi: 10.1002/j.1460-2075.1994.tb06248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Treisman R. Regulation of transcription by MAP kinase cascade. Curr Opin Cell Biol. 1996;8:205–215. doi: 10.1016/s0955-0674(96)80067-6. [DOI] [PubMed] [Google Scholar]
- 35.Verrijzer C P, Tijan R. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem Sci. 1996;21:338–342. [PubMed] [Google Scholar]
- 36.Voit R, Kuhn A, Sander E E, Grummt I. Activation of mammalian ribosomal gene transcription requires phosphorylation of the nucleolar transcription factor UBF. Nucleic Acids Res. 1995;23:2593–2599. doi: 10.1093/nar/23.14.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voit R, Schnapp A, Kuhn A, Rosenbauer H, Hirschmann P, Stunnenberg H G, Grummt I. The nucleolar transcription factor mUBF is phosphorylated by casein kinase II in the C-terminal hyperacidic tail which is essential for transactivation. EMBO J. 1992;11:2211–2218. doi: 10.1002/j.1460-2075.1992.tb05280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37a.Zhai, W., and L. Comai. Unpublished observation.
- 38.Zhai W, Tuan J, Comai L. SV40 large T antigen binds to the TBP-TAFI complex SL1 and coactivates ribosomal RNA transcription. Genes Dev. 1997;11:1605–1617. doi: 10.1101/gad.11.12.1605. [DOI] [PubMed] [Google Scholar]