Abstract
Background and Objectives:
Bacteriocins are considered alternative non-conventional antimicrobials produced by certain bacteria with activity against closely related species. The present study focuses on screening, characterization, and partial purification of bacteriocins produced by Staphylococcus sp. isolated from different clinical sources such as pus and blood.
Materials and Methods:
A total of 100 Staphylococcus isolates were screened for bacteriocin production using spot on lawn assay and agar diffusion method against five indicator bacteria. Bacteriocins from five selected highly active isolates were subjected to proteinase-K enzyme, different pH, and heating at different temperatures, and investigated the stabilities of their antimicrobials. Two selected isolates, MK65 and MK88, were molecularly identified by 16S rRNA gene sequencing, explored for the presence of 18 bacteriocin genes, and liquid chromatography-high resolution electrospray ionization mass spectrometry (LC-HRESIMS) was used to identify their different metabolites.
Results:
Twenty isolates exhibited inhibitory effect against at least one indicator bacteria. Micrococcus luteus ATCC 4698 showed the highest sensitivity to such bacteriocins. Proteinase K, acidic pH, and heating at 100°C triggered marked activity inhibition. However, amylase enzyme, alkaline pH, and heating at 80°C caused trivial effects. Four out of eighteen bacteriocin genes were detected using PCR. Fermentation, partial purification, and LC-HRESIMS of total protein extracts of two selected isolates, MK65 and MK88, revealed the production of different antimicrobial peptides.
Conclusion:
To the best of our knowledge, this is the first study to report the production of micrococcin and α-circulocin from Staphylococcus aureus MK65 and the production of amonabactin from Staphylococcus epidermidis MK88.
Keywords: Staphylococcus sp., Bacteriocins, Amonabactin, Micrococcin, Micrococcus luteus
INTRODUCTION
The continuing appearance of multi-resistant bacterial infections reinforces the urgent need to find and develop new antibiotics. Bacteriocins are agents that can display good activity against various human pathogens and help combat multi-resistant bacteria (1). Bacteriocins are ribosomally synthesized, heat-stable antimicrobial peptides produced by bacteria, and they can exhibit broad and narrow range inhibition spectra. These bacteriocins have been grouped into three different classes. Briefly, Class I consists of the small, heat-stable, post-translationally modified peptides known as lantibiotics due to the presence of the modified residues lanthionine and methyllanthionine. Class II consists of small, heat-stable peptides without modified residues. Class III consists of abundant, heat-labile proteins (2).
The human body commensal microbiota helps the host’s wellbeing and is intended to protect the host from various infections. One such defensive mechanism is bacteriocin production. These antimicrobial peptides may be seen as healthy as they can easily be degraded by proteolytic enzymes of the mammalian gastrointestinal tract. Bacteriocins are fascinating alternatives to chemical preservatives in milk products, particularly their purified forms, because they do not raise any health concerns. Furthermore, Staphylococcus bacteriocins demonstrated antagonistic activity against different pathogens, including Listeria monocytogenes, and thus showed applicability for food preservation (3).
The prospect of transforming bacteriocins into the next generation of antibiotics, in addition to the rapid genetic and nanotechnological advances, opens up the opportunity to more interesting applications (3).
Clinically isolated bacteria from wounds produce bacteriocins with broad or narrow range inhibition spectra. For instance, these bacteriocins display inhibitory effect against various Gram-positive bacteria, notably some opportunistic skin pathogens such as Propionibacterium acnes, Staphylococcus epidermidis, and methicillin-resistant Staphylococcus aureus (MRSA) (4). It was also shown that 80% of Staphylococcus nasal isolates released bacteriocins with a broad range of activity, especially during stressful conditions, such as iron deficiency and hydrogen peroxide exposure. Knockout of bacteriocin genes decreased the competitiveness of the producers, showing that bacteriocins may be a key driver for interactions of microbiota in poor ecosystems (5).
In the present study, we targeted the isolation of Staphylococcal bacteria from different clinical specimens and investigated the presence of promising biologically active metabolites in addition to their inhibitory activity against various pathogens through bacteriocin production.
MATERIALS AND METHODS
Collection of samples.
A total of 100 Staphylococcus sp. isolates were recovered from different pus and blood samples from children. These bacteria were isolated based on the streaking technique using mannitol salt agar and incubated for 24 hours at 37°C. Gram stain and biochemical tests such as catalase, gelatinase, and caseinase tests were used to identify suspected Staphylococcus isolates. Maintenance of these isolates was carried out in 20% glycerol and stored at −80°C.
Screening for bacteriocin production using the spot on lawn assay.
A number of 100 Staphylococcus isolates were allowed to grow in brain heart infusion broth for 24 hours at 37°C. Ten microliters from each growing isolate were spotted on brain heart agar, and plates were incubated for 24 hours at 37°C. Growing Staphylococcus isolates spots were overlaid with 5 ml soft agar containing 70 μl of indicator strain and incubated overnight. Five different indicator bacteria were included; Enterococcus faecalis V583 ATCC 700802, Lactobacillus sakei ATCC 15521, Micrococcus luteus ATCC 4698, Bacillus subtilis ATCC 6633, and Escherichia coli ATCC 8739. The antimicrobial activities due to bacteriocin were observed as clear inhibition zones around colonies.
Antibiotic-resistant pattern and typing of bacteriocinogenic Staphylococcus sp.
A five selected bacteriocin producing Staphylococcus isolates were biotypically compared and characterized for their sensitivity to 26 different antibiotics. CLSI disc diffusion method (M02-A10) was used to determine the antibiotic resistance phenotype of Staphylococcus isolates. Mueller-Hinton agar (Oxoid) was used, and the test was proceeding, as described (6). The plates were incubated at 37°C for 24 hr before measuring and recording zones of inhibition surrounding the discs as described (7).
Fermentation and preparation of crude bacteriocin with confirmatory antimicrobial activities using agar well diffusion method (cup technique).
Staphylococcus isolates, showing significant antimicrobial activities, were undergoing small-batch fermentation. Briefly, each Staphylococcus isolate was sub-cultured in 10 mL brain heart infusion broth (BHI) for 24 hr at 37°C, and later on 1% of starting inoculum was used to inoculate 1 L of brain heart infusion broth (BHI) in a 2 L Erlenmeyer flask and then incubated on a rotary shaker incubator 160 RPM at 37°C for 24 hr (8). The cell-free supernatant from each flask was collected after centrifugation at 8,000 g for 20 min at 4°C. Total proteins were partially purified and precipitated for each isolate using 40% ammonium sulfate precipitation technique. Brown protein pellets were collected after cooling centrifugation at 4°C for 40 minutes at 14,000 g. The total precipitated protein for each isolate was dissolved in 10 mL distilled water to achieve 100× final concentration.
Confirmatory antimicrobial activity was conducted by applying the agar diffusion method (cup technique) using the cell-free culture supernatant and precipitated protein, adjusted to pH 7. Aliquot of 50 μl of the pH adjusted supernatant or protein solution was added into 0.5 cm wells, made in inoculated agar plates with the indicator strain Micrococcus luteus ATCC 4698. These plates were incubated overnight, and the formation of growth inhibition zones was checked (9).
Physicochemical properties of crude bacteriocin.
The cell-free culture supernatant of bacteriocinogenic Staphylococcus isolates was subjected to different treatment conditions and investigated for the stability of their antimicrobial activities. Thermal stability was tested by heating the cell-free culture supernatant at 55°C, 80°C, 100°C, and autoclaving at 121°C for 30 min. Sensitivity towards different degrading enzymes as proteinase K, α-chymotrypsin, trypsin, α-amylase, and RNAse, was done at a final concentration of 1 mg/mL by incubating cell-free culture supernatant with each specified enzyme at 37°C for 1 h. Sensitivity to a surface-active agent as DTT at a final concentration of 1% (v/v) was estimated by incubating with the crude bacteriocin solution at 37°C for 1 h (10). The antimicrobial activity was determined compared to the non-treated cell-free culture supernatant by agar well diffusion technique using Micrococcus luteus ATCC 4698 as an indicator strain and expressed as a relative activity.
Bacterial DNA extraction, amplification, and sequencing of 16S rRNA gene.
Genomic DNA isolation was done with some modifications of a previously described protocol (11). Briefly, 1.5 ml of the overnight culture was centrifuged for 10 min at 3,000 g, the supernatant was discarded, and pellets were resuspended in 200 μL spheroplast buffer (10% sucrose, 25 mM Tris pH 8.4, 25 mM EDTA pH 8.0, 2 mg/mL lysozyme and 0.4 mg/mL RNase A), vortexed and incubated at 37°C for 10 minutes until cell lysis occurred. Then, 50 μL of each of 5% SDS (lysis buffer 1) and 5 M NaCl (lysis buffer 2) was added, mixed, and incubated at 65°C for 5 minutes. 100 μL neutralizing buffer (60 mL 5M potassium acetate, 11.5 mL glacial acetic acid, and 28.5 mL dH2O) was then added, vortexed, and put in ice for 5 min before centrifugation at 18,000 g at 4°C for 15 minutes. The supernatant (approximately 400 μL) was transferred into a new tube, mixed with an equal volume of isopropanol, left 5 minutes at room temperature, and centrifuged at 18,000 g at room temperature for 15 minutes to precipitate DNA. The resulting pellet was washed with 70% ethanol by centrifugation at 18,000 g at room temperature for 5 minutes. The final pellets were air-dried and resuspended in 50 μL 1× TE buffer pH 8 and stored in a refrigerator at 4°C.
PCR amplification technique of the 16S rRNA gene was performed using universal forward primer (27F 5′-AGAGTTTGATCCTGGCTCAG-3′) and universal reverse primer (1492R 5′-GGTTACCTTGTTAC-GACTT-3′). The PCR mixture was prepared in a 50 μL of PCR reaction volume using 10 μL 5× buffer, 500 ng genomic DNA, 10 mM dNTPs mixture, 2.5 units of Taq DNA polymerase enzyme, and 1 μL of both forward and reverse primers. The PCR program, product purification, and sequencing using the ABI Prism BigDye terminator kit version 3.1 with the ABI Prism 3100 generic analyzer were accomplished as previously described (12).
Sequence manipulation and phylogenetic analysis.
The Blast tool in the National Center for Biotechnology Information (NCBI) was used to compare the good quality sequences to the GenBank database to identify the closest related species with highly similar sequences to the amplified ones. Finally, the multiple sequence alignment of the amplified sequences and closely related sequences from NCBI was carried out, followed by phylogenetic analysis using MEGA7 software (13).
PCR screening for bacteriocin genes.
The presence of genes encoding for 18 known bacteriocins in Staphylococcus sp. was investigated by PCR using specific primers purchased from Invitrogen (Table 1). The primers were designed using the Primer3 website https://primer3plus.com. The PCR product of interest was detected by agarose gel electrophoreses as described before (14), using 1.5% (w/v) agarose gels with reference to 100 bp DNA ladder (Fermentas, Finland).
Table 1.
PCR primers of 18 known bacteriocins structural genes used in this study
| Gene | Ref. Gene Accession no. | Primer | Sequence (5′–3′) | Annealing temp (°C) | Amplicon size (bp) | Primers Ref. |
|---|---|---|---|---|---|---|
| Lactococcocin-972 | NC_021670 | F | TAGCTGGTGGTGTTTGGAGT | 51 | 177 | This study |
| R | TTTTGTTTCCACCCCAAGCC | |||||
| BsaA1 | NC_003923 | F | ACCCGTCTTTTCACAACCAA | 50 | 111 | This study |
| R | GTTCTTGATTTAGACGTGCAAGT | |||||
| BsaA2 | NC_009641 | F | CCTTTCAGGCTCTATTTGCTGT | 50 | 161 | This study |
| R | GTTCTTGATTTAGACGTGCAAGT | |||||
| Nukacin–ISK-1 nuKA | NC_005207 | F | AGGAGGTAACAAACATGG | 45 | 195 | (35) |
| R | CGGAGGGATATATTATGG | |||||
| Lactacin F | NC_018952 | F | GAGCAGGTTAGGAGGAGCT | 50 | 113 | This study |
| R | CCTCCTCTAAAATGCTCCGG | |||||
| Aureocin A53 | NC_025194 | F | GAAGTTGTGAAAACTATTA | 45 | 322 | (36) |
| R | CATAAAACAAAGAGCCAAAGT | |||||
| Aureocin A70 | NC_020227 | F | CCTTATAACTTCGAATGCT | 45 | 525 | (36) |
| R | AAATATTAACAAGAGAAA | |||||
| Epidermin A | NC_007795 | F | GGAGTGTTTAAAATGGAAGC | 45 | 431 | (37) |
| R | CCTTTTCCCAGTCTATTTTG | |||||
| Gallidermin | NC_017351 | F | GGAGTGTTTAAAATGGAAGCAGT | 45 | 153 | This study |
| R | GAAGCTACCTGTTTTGGCACA | |||||
| Epicidin 280 | Y14023.1 | F | CGGAGGGATATATTATGG | 45 | 195 | (36) |
| R | CAATCACTACTATTGACAATCAC | |||||
| Epilancin K7 elKA | U20348 | F | CTCAAAAGAGTGATTTAAGTCCGC | 50 | 115 | (35) |
| R | CCACCAGTAATATTGCAACCGC | |||||
| Pep5 pepA | L23967 | F | AGAGGAGGTGGTTATATATG | 45 | 427 | (35) |
| R | TGAGTTCCATGCCCAGTG | |||||
| SAP057A_040 | NC_013334 | F | AGCGTGGTGATTCTTATG | 45 | 499 | (37) |
| R | TCTGATTTATTTAGTTCTGGAT | |||||
| Epidermicin n101 | JQ025382 | F | GCTTTTGGGTTGATGGGCAT | 50 | 193 | This study |
| R | TGGATTAACGCCGGTCAAAG | |||||
| BacCH91 | NC_020227 | F | TTAGTGAAAATAAATAGTA | 45 | 399 | (20) |
| R | CATTTGTAAGCACCTCAC | |||||
| Delta-lysinl | JX901171 | F | AGGAGTGATTTCAATGGCAGC | 50 | 297 | This study |
| R | TGGGATGGCTTAATAACTCACC | |||||
| Warnericin_RC | JX901171 | F | GGTATGCTTTAGGGCCATAGG | 50 | 229 | This study |
| R | AACTACACCTTGGGAAATACGT | |||||
| Epilancin_15x | JQ979180 | F | GGATTATAGGAGGTGAGATTTA | 50 | 180 | This study |
| R | CCAGTAAAGTGACATCCACAAGTT |
Partial purification of bacteriocin and LC-HRESIMS profiling analysis.
Total proteins were analyzed by liquid chromatography/mass spectrometry using a Thermo Instruments MS system (LTQ XL/LTQ Orbitrap Discovery) coupled to a Thermo Instruments HPLC system (Accela PDA detector, Accela PDA autosampler, and Accela pump) TUV Rheinland of North America Inc with Waters Sun Fire C18 RP analytical HPLC column (5 μm, 4.6 × 150 mm) as described (11, 15). Metabolites molecular formulas were deducted by exact mass of eluted peaks, and identified in Dictionary of Natural Products, CRC press, online version for matching expected compounds.
Statistical analysis.
R statistical platform (https://www.r-project.org) and GraphPad Prism (La Jolla, CA, USA) programs and software packages were used for data visualization.
RESULTS
Isolation, characterization, and screening for bacteriocin production.
A total of 100 Staphylococcus isolates were recovered from different clinical samples, at which 64% were isolated from blood samples, and 36% were recovered from pus samples (Fig. 1A). After growing in mannitol salt agar, the color of colonies and morphology varied between opaque golden yellow, lemon-yellow, or white color. All Staphylococcus isolates showed a Gram-positive violet bunches appearance. A spot on lawn technique to screen for bacteriocin production revealed that only 20 isolates showed possible bacteriocin production against at least one indicator strain. The five most active bacteriocin producing isolates were selected for further work steps. Micrococcus luteus ATCC 4698 was chosen as indicator bacteria during the rest of experimental work because it showed the best inhibitiory zones in the case of the 5 selected isolates (Fig. 1B).
Fig. 1.
Resistance phenotypes of 5 selected bacteriocin producing Staphylococcus isolates to 26 different antibiotics. a) A transposed heatmap is representing the resistance phenotypes of each isolate according to the color code shown in the figure key. Isolates are shown on the X-axis and antibiotics on the Y-axis. Both isolates and antibiotics are ordered by hierarchical clustering, reflected by the horizontal and vertical trees, respectively. The heatmap was created in R by the basic heatmap function. b) A stacked bar plot summarizing the disc diffusion resistance patterns.
Phenotypic characterization of antibiotics resistant pattern.
Five selected bacteriocin producing isolates were tested for their sensitivity against 26 different antibiotics. These isolates were susceptible to vancomycin, linezolid, ciprofloxacin, gatifloxacin, moxifloxacin, ofloxacin, chloramphenicol, clindamycin, quinupristin/dalfopristin, gentamicin, and sulfamethoxazole/trimethoprim. However, 4 isolates showed oxacillin/methithlin resistance (MRSA) with a positive coagulase test. Also, some isolates were resistant to cephalosporins, tetracyclines, and azithromycin (Table 1). We also attempted to cluster the 5 selected isolates based on their resistance pattern, and the clustering pattern was presented as a heatmap (Fig. 1). The heatmap has the advantage of showing the overall trends and patterns within isolates.
Production and physicochemical properties of crude bacteriocin.
The five selected bacteriocin producing Staphylococcus isolates were allowed to grow in a shaking incubator at 160 rpm for 24 hr at 37°C. Antimicrobial activity of crude bacteriocin in the supernatant or concentrated precipitated protein was tested against Micrococcus luteus ATCC 4698 indicator strain. The zone of inhibition was measured and recorded in millimeters (mm). Optimization of incubation time required for fermentation and production of bacteriocin was conducted and revealed that the maximum production was achieved after 10 hr incubation in shaking incubator.
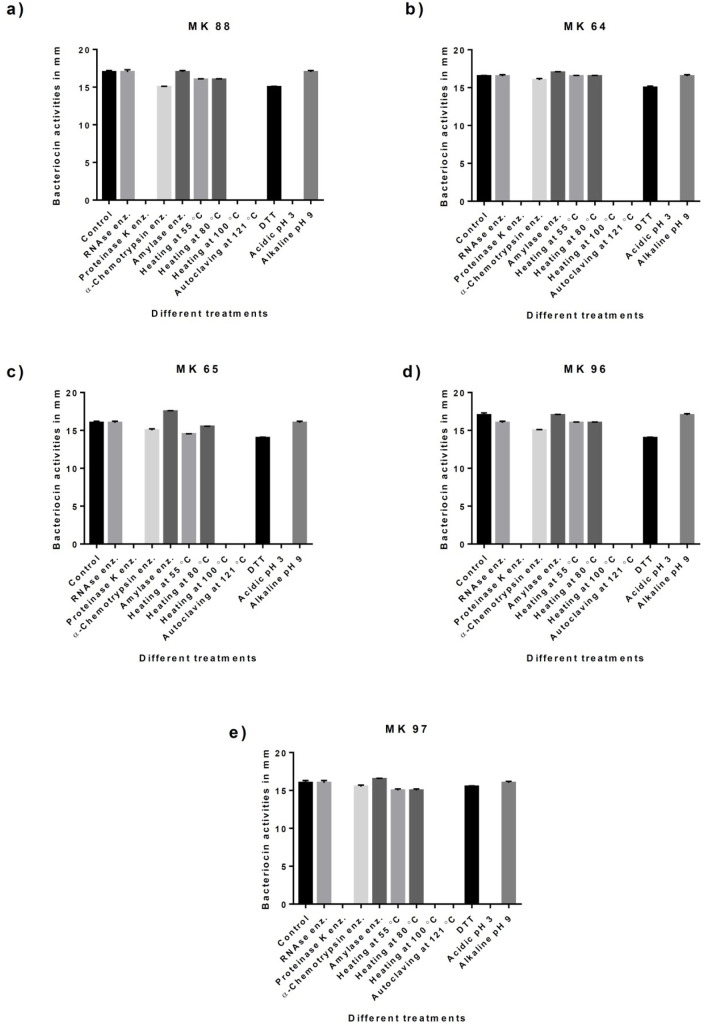
The physicochemical properties, including sensitivity towards different degrading enzymes; proteinase K, α-chymotrypsin, α-amylase and RNAse, sensitivity to DTT, various heat stresses, and varying pH, were investigated. All bacteriocins lost their activity when treated with proteinase K enzyme, pH 3, heating at 100°C, and autoclaving, as seen in Table 2 and Fig. 2.
Table 2.
LC-HRESIMS analysis for total protein extracts of isolate MK65
| Retention time | Accurate mass | Molecular formula | Tentative identification |
|---|---|---|---|
| 5.18 | 481.1304 | C17H24N2O14 | Staphyloferrin A |
| 7.29 | 533.3294 | C23H44N6O8 | IC-1202Bsiderophore |
| 7.85 | 681.3413 | C26H48N8O13 | Ornibactin C4 |
| 9.18 | 758.4320 | C38H63NO14 | Lactenocin macrolide |
| 10.45 | 599.3188 | C30H42N6O7 | BE-18257-A cyclic peptide |
| 11.14 | 652.4029 | C31H53N7O8 | L-Valyl-L-leucyl-L-prolyl-L-valyl-L-prolyl-L-glutamine |
| 11.84 | 683.2546 | C32H42O16 | Bruceoside B |
| 13.51 | 565.4208 | C32H52O8 | AB-023b polyene |
| 13.60 | 609.4470 | C33H52O10 | Strevertene-D polyene |
| 15.18 | 651.1458 | C34H18O14 | No hits |
| 15.87 | 674.2821 | C35H39N5O9 | Vulnibactinsiderophore |
| 16.28 | 751.3298 | C37H46N6O11 | Amonabactin-P-750siderophore |
| 17.39 | 633.3171 | C36H44N2O8 | SDZ-220-542 polyketide |
| 17.62 | 679.4641 | C36H62N4O8 | No hits |
| 18.51 | 655.2760 | C36H38N4O8 | Coproporphyin Isiderophore |
| 20.15 | 671.4104 | C38H54O10 | Milbemycin B |
| 20.69 | 873.3446 | C39H52N8O13S | Peptidthiollacton |
| 22.93 | 747.3124 | C40H46N2O12 | No hits |
| 23.93 | 752.4214 | C39H61NO13 | L-687795polyketide |
| 24.38 | 860.5337 | C40H73N7O13 | Amphibactin H |
| 25.41 | 930.6020 | C47H79N9O10 | α-Circulocin |
| 26.21 | 1144.2175 | C48H49N13O9S6 | Micrococcin |
| 26.96 | 739.5657 | C50H74O4 | Haloxanthin |
Fig. 2.
Response of crude bacteriocins from Staphylococcus isolates to different treatment conditions.
Bacteriocins of Staphylococcus sp. were slightly sensitive to heat, and during a gradual increase in temperature, the activity was decreased until completely diminished at 100°C. They showed stable activity in alkaline media at pH 9; however, complete loss of activity was observed in acidic media at pH 3. They did not affect by RNAse enzyme; however, the effects of proteolytic enzymes proved the proteinaceous nature of bacteriocins at which their activities were decreased by the effect of alpha chymotrypsin and entirely lost by the effect of proteinase K enzyme. The effect of surfactants was observed as a slight decrease in activity by DTT.
DNA sequencing for selected isolates and phylogenetic analysis.
The partial 16S rRNA gene sequencing of both isolates MK65 and MK88 revealed similarity as 98.6% with Staphylococcus aureus and 98.1% with Staphylococcus epidermidis, respectively, according to the BLAST tool at NCBI GenBank as shown in (Table 3).
Table 3.
LC-HRESIMS analysis for total protein extracts of isolate MK88
| Retention time | Accurate mass | Molecular formula | Tentative identification |
|---|---|---|---|
| 7.52 | 353.1952 | C13H28N4O7 | Fortimicin-KL1aminosuger |
| 9.18 | 758.4320 | C38H63NO14 | Lactenocin macrolide |
| 11.18 | 381.1769 | C17H24N4O6 | Cutinostatin B |
| 12.91 | 331.2843 | C19H38O4 | Aggreceride B |
| 14.16 | 359.2793 | C20H38O5 | Persicomycin 1 |
| 15.93 | 561.2975 | C31H45O7P | Oxydifficidin |
| 16.29 | 751.3298 | C37H46N6O11 | Amonabactin-P-750siderophore |
| 17.40 | 546.3652 | C29H47N5O5 | Stacopin P-1 |
| 17.94 | 562.3601 | C29H47N5O6 | Stacopin P-2 |
| 20.26 | 679. 4640 | C36H62N4O8 | No hits |
| 23.92 | 752.4214 | C39H61NO13 | L-687795polyketide |
| 24.39 | 581.4354 | C41H56O2 | No hits |
| 25.37 | 851.4997 | C42H74O17 | CP 64537 polyketide |
| 26.91 | 1100.5696 | C45H77N15O17 | Epidermidin |
| 27.05 | 889.4944 | C48H72O15 | No hits |
The resulting sequence was aligned to 20 of the closely related Staphylococcus sp. by retrieving their sequences from the NCBI GenBank database and assembled in MEGA7 software for phylogenetic analysis using the Neighbor-Joining tree method. The evolutionary distances were computed using the Kimura 2-parameter method. The obtained phylogenetic tree (Fig. 3) confirmed the similarity of both MK65 and MK88 isolates to S. aureus and S. epidermidis with a similarity matrix bootstrap value of 100 and 99, respectively. The resulted nucleotide sequences were deposited in Genbank under accession numbers MN611706 and MN611707.
Fig. 3.
The evolutionary history was inferred using the Neighbor-Joining method (38). The optimal tree with the sum of branch length = 0.19041993 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Kimura 2-parameter method and are in the units of the number of base substitutions per site. The analysis involved 22 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 1346 positions in the final dataset. Evolutionary analyses were conducted in MEGA7 (39).
PCR screening for bacteriocin structural genes and LC-HRESIMS analysis.
Isolates MK65 and MK88 were explored for the presence of genes encoding 18 known bacteriocins in Staphylococcus sp. by PCR amplification using specific primers. Isolate MK88 showed positive amplification of bsaA1, bsaA2, Nukacin–ISK-1, and Lactococcocin-972. However, isolate MK65 showed only positive results in the case of bsaA1 and bsaA2 genes.
LC-HRESIMS screening for total protein extracts of isolates MK65 and MK88 showed the presence of different compounds, including bacteriocins and antimicrobial agents (Tables 2 and 3) (Fig. 4). It showed the presence of α-circulocin and micrococcin in extracts of isolate MK65 and confirmed the presence of epidermedin in extracts of isolate MK88.
Fig. 4.
HPLC chromatogram of total protein extracts related to isolate MK65 (a) and isolate MK88 (b), and all their relative retention times and molecular masses are presented in Table 2 and Table 3, respectively.
DISCUSSION
The emerging cases and threats posed to public health by various multidrug-resistant bacteria due to the overuse and misuse of antibiotics have enforced the discovery and the use of new antimicrobials to combat infections.
The current study aimed at screening 100 human-derived Staphylococcus sp. for bacteriocin production, suggesting that clinically isolated Staphylococcus sp. has the potentiality to produce bacteriocins that can be used as an alternative therapeutic agent in the prevention and treatment of many infections. Previously, it has been shown that 60% of bacteriocin producers came from clinical materials (16). Agar-based spot on lawn assays revealed that only 20% (20/100) of our isolates reproducibly exhibited inhibitory effect against at least one of the five tested indicator strains. Previous studies supported this finding that showed a similar percentage of bacteriocin production from clinical isolates and effective against different groups of bacteria (17). A study by O’ Sullivan and coworker (4) showed that only 0.112% (101/90,000) of their isolates could exhibit antimicrobial activity. However, Janek and his colleagues (5) performed a similar spot on the lawn screen method to determine bacteriocin production by Staphylococcus sp. and they found that 86.5% (77/89) exhibited antimicrobial activity. Also, Lynch and colleagues (18) reported that 94% of their isolated Staphylococcus sp. could show antagonistic bacteriocin production. These findings suggest that bacteriocin production is a common phenotype among Staphylococcus isolates. The difference in the percentage of bacteriocin producers between our study and the other studies may be due to the difference in indicator strains used in each study. For example, Sullivan et al. used only one indicator microorganism (Lactobacillus delbrueckii subsp. Bulgaricus), while different indicator microorganisms were used in the other two studies. In the current study, staphylococcal bacteriocins showed activities against Enterococcus faecalis V583 ATCC 700802, Lactobacillus sakei ATCC 15521, Micrococcus luteus ATCC 4698, Bacillus subtilis ATCC 6633, and Escherichia coli ATCC 8739. This finding agrees with the results reported by Klaenhammer (17), which recognized that 99% of all bacteriocin-producing bacteria could have antimicrobial activities to at least one indicator bacteria if they were checked against suitable indicators. The inhibitory spectrum was more pronounced against most of the Gram-positive organisms when compared to Gram-negative bacteria. This might be due to the presence of more receptors for bacteriocin adsorption in peptidoglycan based cell wall of Gram-positive bacteria (19).
The twenty bacteriocinogenic isolates were short-listed to only 5 isolates based on their higher activities in cell-free supernatants. Only isolates MK65 and MK88 were undergoing genotyping and further analysis of their total protein extracts. Micrococcus luteus ATCC 4698 was selected for further investigation as it was considered the most sensitive indicator bacteria. The optimal initial pH for higher production of the inhibitory factor was 7.5, with an incubation period of 10 hours in a shaking incubator. Partial purification steps achieved bacteriocin recovery based on ammonium sulfate precipitation followed by dissolving in distilled water (20). The activity of bacteriocin was retained and increased after successive purification steps. These inhibitory activities of bacteriocins were completely inactivated after heat treatment at 100°C for 15 minutes, indicating their heat-labile proteinaceous nature. The inhibitory activity of bacteriocins was stable at pH 4–7, but it was either decreased or inactivated after exposure to pH 3, which was reported in a previous study (20). The antagonistic effects produced by Staphylococcus sp. were wholly or partially inactivated when preparations were treated with proteolytic enzymes suggesting their proteinaceous nature. This came in contact with other previous studies at which AI-type epidermin-like and lantibiotics were reported to be typically cleaved by trypsin after prolonged incubation (21). Concerning the application of bacteriocins biological preservatives in foods and feeds, sensitivity to proteolytic enzymes suggests that their ingestion will not affect the microbial flora of gastrointestinal tract.
In this study, the detection of 4 out of 18 genes responsible for bacteriocin production in Staphylococcus sp. could confirm the capabilities of clinically isolated Staphylococci sp. to produce bacteriocin (22). bsaA1 and bsaA2 were detected in both tested isolates (MK65 and MK88), while nukacin–ISK-1 and Lactococcocin-972 were found in isolate MK88. Previously, bsaA1 or bsaA2 genes were reported to be produced from clinically isolated Staphylococcus with significant bactericidal effects (23). From our investigations and other studies, it was apparent that there could be a particular association between the bactericidal effect of Staphylococcus sp. and the presence of the bsa genes. Nukacin–ISK-1 was previously reported as bacteriocin derived from Staphylococcus warneri (24), and Lactococccin-972 was reported as a lactococcal bacteriocin that has a bactericidal effect on sensitive Lactococci (25).
Total proteins were analyzed by liquid chromatography/high-resolution mass spectrometry (LC-HRESIMS profiling analysis). HRESIMS allows the detection of proteins and metabolites to the nearest 0.001 atomic mass units. In this study, micrococcin was reported for the first time to be produced by S. aureus MK65. Micrococcin was only reported before to be produced by Staphylococcus equorum WS 2733 (26). Also, epidermidin was detected in Staphylococcus epidermidis MK88, and it was previously reported to be secreted by Staphylococcus epidermidis (27). The α-Circulocin is an antibacterial lipopeptides from Bacillus circulans (28), however in this study and for the first time, it has been produced by S. aureus MK65. Also, siderophore compounds were detected in the two analyzed samples, which are iron-chelating compounds secreted by staphylococci and serving to transport iron across cell membranes. In isolate MK65, different siderophores were found as staphyloferrin A, IC-1202B, vulnibactin, ornibactin C4, amonabactin-P-750 and amphibactin H. These siderophores were shown in previous studies to be derived from Staphylococcus sp. (29). In this study and for the first time, isolate MK 88 showed production of amonabactin that was not detected before in Staphylococcus sp. and it was only reported to be produced from Aeromonas hydrophila (30). Aggreceride B was also detected in isolate MK65 with an accurate mass of 331.2843 dalton, which is a platelet aggregation inhibitor derived before from Streptomyces sp. (31). Also, stacopin P-1 and stacopin P-2 were detected in this study, which are proteinase inhibitors produced by Staphylococcus sp. (32). Oxydifficidin was also recognized, which is macrocyclic polyene lactone phosphate esters and had activity against aerobic, anaerobic bacteria, and lethal bacteremia caused by Klebsiella pneumoniae (33). Fortimicin-KL1 aminosuger, which is actinomyces antibiotic, was also detected (34).
In conclusion, these results show the potential usefulness of bacteriocins produced by clinically isolated Staphylococcus sp. to be used in further applications to combat multi-resistant microorganisms. To the best of our knowledge, this is the first study to report the production of micrococcin and α-circulocin by S. aureus MK65, and the production of amonabactin, by Staphylococcus epidermidis MK88. Further work is however required to understand thoroughly the molecular dynamics, structural-functional relationships and mechanisms of bacteriocin action.
REFERENCES
- 1.Fahim HA, Rouby W, El-Gendy AO, Khairalla AS, Naguib IA, Farghali AA. Enhancement of the productivity of the potent bacteriocin avicin A and improvement of its stability using nanotechnology approaches. Sci Rep 2017; 7: 10604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nes IF, Diep DB, Holo H. Bacteriocin diversity in Streptococcus and Enterococcus. J Bacteriol 2007; 189: 1189–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chikindas ML, Weeks R, Drider D, Chistyakov VA, Dicks LM. Functions and emerging applications of bacteriocins. Curr Opin Biotechnol 2018; 49: 23–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Sullivan JN, Rea MC, O’Connor PM, Hill C, Ross RP. Human skin microbiota is a rich source of bacteriocin-producing staphylococci that kill human pathogens. FEMS Microbiol Ecol 2019; 95:fiy241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janek D, Zipperer A, Kulik A, Krismer B, Peschel A. High frequency and diversity of antimicrobial activities produced by nasal Staphylococcus strains against bacterial competitors. PLoS Pathog 2016; 12(8): e1005812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sandiford S, Upton M. Identification, characterization, and recombinant expression of epidermicin NI01, a novel unmodified bacteriocin produced by Staphylococcus epidermidis that displays potent activity against Staphylococci. Antimicrob Agents Chemother 2012; 56: 1539–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Molham F, Khairalla AS, Azmy AF, El-Gebaly E, El-Gendy AO, AbdelGhani S. Anti-proliferative and anti-biofilm potentials of bacteriocins produced by non-pathogenic Enterococcus sp. Probiotics Antimicrob Proteins 2020: 10.1007/s12602-020-09711-1. [DOI] [PubMed] [Google Scholar]
- 8.Dündar H, Atakay M, Çelikbıçak Ö, Salih B, Bozoğlu F. Comparison of two methods for purification of enterocin B, a bacteriocin produced by Enterococcus faecium W3. Prep Biochem Biotechnol 2015; 45: 796–809. [DOI] [PubMed] [Google Scholar]
- 9.Ansari A, Zohra RR, Tarar OM, Qader SAU, Aman A. Screening, purification and characterization of thermostable, protease resistant Bacteriocin active against methicillin resistant Staphylococcus aureus (MRSA). BMC Microbiol 2018; 18: 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu Y, Yan H, Li X, Gu Y, Wang X, Yi Y, et al. Physicochemical properties and mode of action of a novel bacteriocin BM1122 with broad antibacterial spectrum produced by Lactobacillus crustorum MN047. J Food Sci 2020; 85: 1523–1535. [DOI] [PubMed] [Google Scholar]
- 11.Lotfy MM, Hassan HM, Mohammed R, Hetta M, El-Gendy AO, Rateb ME, et al. Chemical profiling and biological screening of some river Nile derived-microorganisms. Front Microbiol 2019; 10: 787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahmad MS, El-Gendy AO, Ahmed RR, Hassan HM, El-Kabbany HM, Merdash AG. Exploring the antimicrobial and antitumor potentials of Streptomyces sp. AGM12-1 isolated from Egyptian soil. Front Microbiol 2017; 8:438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 2007; 24: 1596–1599. [DOI] [PubMed] [Google Scholar]
- 14.Naguib MM, Khairalla AS, El-Gendy AO, Elkhatib WF. Isolation and characterization of mercury-resistant bacteria from wastewater sources in Egypt. Can J Microbiol 2019; 65: 308–321. [DOI] [PubMed] [Google Scholar]
- 15.El-Helw NO, El-Gendy AO, El-Gebaly E, Hassan HM, Rateb ME, El-Nesr KA. Characterization of natural bioactive compounds produced by isolated bacteria from compost of aromatic plants. J Appl Microbiol 2019; 126: 443–451. [DOI] [PubMed] [Google Scholar]
- 16.Blinkova LP, Dorofeeva ES, Baturo AP, Romanenko EE, Katosova l K, Polikarpova SV, et al. [The detection of bacteriogenic causes of opportunistic infections]. Vestn Ross Akad Med Nauk 2008; (4): 14–18. [PubMed] [Google Scholar]
- 17.James R, Penfold CN, Moore GR, Kleanthous C. Killing of E. coli cells by E group nuclease colicins. Biochimie 2002; 84: 381–389. [DOI] [PubMed] [Google Scholar]
- 18.Lynch D, O’Connor PM, Cotter PD, Hill C, Field D, Begley M. Identification and characterisation of capidermicin, a novel bacteriocin produced by Staphylococcus capitis. PLoS One 2019; 14(10): e0223541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Padilla C, Lobos O, Brevis P, Abaca P, Hubert E. Effects of the bacteriocin PsVP-10 produced by Pseudomonas sp. on sensitive bacterial strains. Rev Latinoam Microbiol 2002; 44: 19–23. [PubMed] [Google Scholar]
- 20.Wladyka B, Wielebska K, Wloka M, Bochenska O, Dubin G, Dubin A, et al. Isolation, biochemical characterization, and cloning of a bacteriocin from the poultry-associated Staphylococcus aureus strain CH-91. Appl Microbiol Biotechnol 2013; 97: 7229–7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuipers OP, Rollema HS, de Vos WM, Siezen RJ. Biosynthesis and secretion of a precursor of nisin Z by Lactococcus lactis, directed by the leader peptide of the homologous lantibiotic subtilin from Bacillus subtilis. FEBS Lett 1993; 330: 23–27. [DOI] [PubMed] [Google Scholar]
- 22.Bierbaum G, Götz F, Peschel A, Kupke T, van de Kamp M, Sahl HG. The biosynthesis of the lantibiotics epidermin, gallidermin, Pep5 and epilancin K7. Antonie Van Leeuwenhoek 1996; 69: 119–127. [DOI] [PubMed] [Google Scholar]
- 23.Daly KM, Upton M, Sandiford SK, Draper LA, Wescombe PA, Jack RW, et al. Production of the Bsa lantibiotic by community-acquired Staphylococcus aureus strains. J Bacteriol 2010; 192: 1131–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sashihara T, Kimura H, Higuchi T, Adachi A, Matsusaki H, Sonomoto K, et al. A novel lantibiotic, nukacin ISK-1, of Staphylococcus warneri ISK-1: cloning of the structural gene and identification of the structure. Biosci Biotechnol Biochem 2000; 64: 2420–2428. [DOI] [PubMed] [Google Scholar]
- 25.Martínez B, Suárez JE, Rodríguez A. Lactococcin 972: a homodimeric lactococcal bacteriocin whose primary target is not the plasma membrane. Microbiology (Reading) 1996; 142: 2393–2398. [DOI] [PubMed] [Google Scholar]
- 26.Carnio MC, Höltzel A, Rudolf M, Henle T, Jung G, Scherer S. The macrocyclic peptide antibiotic micrococcin P(1) is secreted by the food-borne bacterium Staphylococcus equorum WS 2733 and inhibits Listeria monocytogenes on soft cheese. Appl Environ Microbiol 2000; 66: 2378–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu C, Wiseman GM. The nature of epidermidins, new antibiotics from staphylococci. Can J Microbiol 1972; 18: 121–125. [DOI] [PubMed] [Google Scholar]
- 28.He H, Shen B, Korshalla J, Carter GT. Circulocins, new antibacterial lipopeptides from Bacillus circulans, J2154. Tetrahedron 2001; 57: 1189–1195. [Google Scholar]
- 29.Konetschny-Rapp S, Jung G, Meiwes J, Zähner H. Staphyloferrin A: a structurally new siderophore from staphylococci. Eur J Biochem 1990; 191: 65–74. [DOI] [PubMed] [Google Scholar]
- 30.Barghouthi S, Young R, Olson MO, Arceneaux JE, Clem LW, Byers BR. Amonabactin, a novel tryptophan- or phenylalanine-containing phenolate siderophore in Aeromonas hydrophila. J Bacteriol 1989; 171: 1811–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akeda Y, Shibata K, Ping X, Tanaka T, Taniguchi M. AKD-2A, B, C and D, new antibiotics from Streptomyces sp. OCU-42815. Taxonomy, fermentation, isolation, structure elucidation and biological activity. J Antibiot (Tokyo) 1995; 48: 363–368. [DOI] [PubMed] [Google Scholar]
- 32.Saito M, Kawaguchi N, Hashimoto M, Kodama T, Higuchi N, Tanaka T, et al. Purification and structure of novel cysteine proteinase inhibitors, staccopins pi and p2, from staphylococcus tanabeensis. Agric Biol Chem 1987; 51: 861–868. [Google Scholar]
- 33.Zimmerman SB, Schwartz CD, Monaghan RL, Pelak BA, Weissberger B, Gilfillan EC, et al. Difficidin and oxydifficidin: novel broad spectrum antibacterial antibiotics produced by Bacillus subtilis. I. Production, taxonomy and antibacterial activity. J Antibiot (Tokyo) 1987; 40: 1677–1681. [DOI] [PubMed] [Google Scholar]
- 34.Dairi T, Hasegawa M. Common biosynthetic feature of fortimicin-group antibiotics. J Antibiot (Tokyo) 1989; 42: 934–943. [DOI] [PubMed] [Google Scholar]
- 35.Rahmdel S, Shekarforoush SS, Hosseinzadeh S, Torriani S, Gatto V. Antimicrobial spectrum activity of bacteriocinogenic Staphylococcus strains isolated from goat and sheep milk. J Dairy Sci 2019; 102: 2928–2940. [DOI] [PubMed] [Google Scholar]
- 36.dos Santos Nascimento J, Fagundes PC, de Paiva Brito MA, dos Santos KR, do Carmo de Freire Bastos M. Production of bacteriocins by coagulase-negative staphylococci involved in bovine mastitis. Vet Microbiol 2005; 106: 61–71. [DOI] [PubMed] [Google Scholar]
- 37.Ceotto H, Nascimento Jdos S, Brito MA, Bastos Mdo C. Bacteriocin production by Staphylococcus aureus involved in bovine mastitis in Brazil. Res Microbiol 2009; 160: 592–599. [DOI] [PubMed] [Google Scholar]
- 38.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 1987; 4: 406–425. [DOI] [PubMed] [Google Scholar]
- 39.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 2016; 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]