Abstract
We initially identified c-myc promoter binding protein 1 (MBP-1), which negatively regulates c-myc promoter activity, from a human cervical carcinoma cell expression library. Subsequent studies on the biological role of MBP-1 demonstrated induction of cell death in fibroblasts and loss of anchorage-independent growth, reduced invasive ability, and tumorigenicity of human breast carcinoma cells. To investigate the potential role of MBP-1 as a transcriptional regulator, a chimeric protein containing MBP-1 fused to the DNA binding domain of the yeast transactivator factor GAL4 was constructed. This fusion protein exhibited repressor activity on the herpes simplex virus thymidine kinase promoter via upstream GAL4 DNA binding sites. Structure-function analysis of mutant MBP-1 in the context of the GAL4 DNA binding domain revealed that MBP-1 transcriptional repressor domains are located in the N terminus (amino acids 1 to 47) and C terminus (amino acids 232 to 338), whereas the activation domain lies in the middle (amino acids 140 to 244). The N-terminal domain exhibited stronger transcriptional repressor activity than the C-terminal region. When the N-terminal repressor domain was transferred to a potent activator, transcription was strongly inhibited. Both of the repressor domains contained hydrophobic regions and had an LXVXL motif in common. Site-directed mutagenesis in the repressor domains indicated that the leucine residues in the LXVXL motif are required for transcriptional repression. Mutation of the leucine residues in the common motif of MBP-1 also abrogated the repressor activity on the c-myc promoter. In addition, the leucine mutant forms of MBP-1 failed to suppress cell growth in fibroblasts like wild-type MBP-1. Taken together, our results indicate that MBP-1 is a complex cellular factor containing multiple transcriptional regulatory domains that play an important role in cell growth regulation.
c-myc promoter binding protein 1 (MBP-1), initially identified from a human cervical carcinoma (HeLa) cell expression library, binds to the TATA box sequences of the c-myc P2 promoter and negatively regulates both human and mouse c-myc promoter activities (6, 25, 26). The c-myc proto-oncogene can promote cell proliferation, differentiation, and oncogenic transformation (7, 36) or apoptosis under certain conditions (9, 35). Regulation of c-myc occurs at multiple levels, such as the initiation or termination of transcription and the attenuation of transcription (20, 25). Recent studies have shown that MBP-1 and TATA binding protein bind simultaneously in the minor groove of the c-myc P2 promoter (6). It is possible that MBP-1 negatively regulates c-myc expression by preventing formation of a transcription initiation complex with a general transcriptional factor(s).
MBP-1 is expressed ubiquitously in normal human tissues (27) and localized at human chromosome 1p35-ter (38). Ectopic expression of MBP-1 in murine fibroblasts (NIH 3T3 cells) induces massive cell death, DNA fragmentation, and reduction of c-myc expression (26). Bcl2, a cell survival gene, protects against MBP-1-mediated cell death. Complementation of exogenous deregulated c-myc (without an MBP-1 binding site) also prevents MBP-1-induced cell death. Since MBP-1 negatively regulates c-myc transcription, downregulation of endogenous c-myc expression, which is compensated for by exogenous deregulated c-myc, may be a possible mechanism of protection from apoptotic cell death (26). However, the protective role of Bcl2 in MBP-1-mediated cell death suggests the involvement of another cell regulatory factor(s) in the mediation of biological activity of MBP-1. Thus, in addition to c-myc regulation, MBP-1 appears to exert a regulatory effect on cell growth through another, unknown, mechanism. Exogenous expression of MBP-1 in human breast carcinoma cells results in reduced invasiveness, loss of anchorage-independent growth, and suppression of tumor formation in athymic nude mice (28). Recent studies suggest that the C-terminal half of MBP-1 does not bind to the c-myc promoter (29). However, the C-terminal half of MBP-1 suppressed c-myc transcription and reduced cell growth. The mechanism by which MBP-1 exerts its biological activity is unknown. However, one reasonable explanation is that MBP-1 directly or indirectly modulates the expression of other genes necessary for cell proliferation. In this study, we embarked on a detailed analysis of MBP-1-related functional activities. We have used a number of MBP-1 deletion mutant proteins fused to the DNA binding domain of GAL4 to map its transcriptional regulatory activity. The repressor domains identified in the context of the GAL4 system correlated with the biological activities of MBP-1.
MATERIALS AND METHODS
Cells.
NIH Swiss mouse embryo (NIH 3T3) cells, human cervical carcinoma (HeLa) cells, and African monkey kidney (COS7) cells were obtained from the American Type Culture Collection. Cells were grown in Dulbecco’s modified Eagle medium supplemented with 10% heat-inactivated fetal bovine serum.
Plasmid constructs.
GAL4TK CAT, TK CAT (kindly provided by Y. Shi, Harvard Medical School, Boston, Mass.), G5E1BCAT (kindly provided by D. Dean, Washington University, St. Louis, Mo.), and c-myc CAT (25) plasmids were used as the reporter constructs in this study. The expression vector CMVGAL4 construct was prepared by substituting the cytomegalovirus (CMV) promoter for the simian virus 40 early promoter of pSG424 (32) containing the GAL4 DNA binding domain (amino acids 1 to 147). Plasmid GALMBP1-338 was constructed by PCR amplification of MBP-1 cDNA (25) and cloned in frame with the GAL4 DNA binding domain into CMVGAL4 plasmid DNA. For MBP-1 deletion mutant proteins, desired fragments were generated by PCR amplification using sense and antisense oligonucleotides (Table 1). Amplified fragments were digested with BamHI (5′ end) and XbaI (3′ end) and cloned in frame downstream of the GAL4 DNA binding domain of the CMVGAL4 vector. The mutant plasmids were analyzed by restriction enzyme digestion and DNA sequencing. pM3/3CGln (kindly provided by C. Sample, St. Jude Children’s Research Hospital, Nashville, Tenn.) containing a DNA fragment encompassing the glutamine-rich activation domain from Epstein-Barr virus transcription factor EBNA3C (21) was inserted in frame into the GAL4 amino acid 1 to 147 sequence in the pM3 vector (33). 3CGln(MBP-1) was derived by in frame ligation of the DNA fragment encoding MBP1-47 to the downstream portion of the GAL4-3CGln sequence in pM3/3CGln at the SalI (5′ end) and XbaI (3′ end) sites. The resulting double-stranded plasmid DNAs were transformed into Escherichia coli DH5α, and purified plasmid DNAs were used for in vitro transient expression assay.
TABLE 1.
Oligonucleotides used to construct mutant forms of GALMBP-1
| Primer sequencea | Initial amino acid of mutant MBP-1 |
|---|---|
| Sense | |
| 5′-AACTGAGGATCCAGATGGA-3′ | 1 |
| 5′-AAAGCTGGATCCCTGATAAG-3′ | 140 |
| 5′-CTGTACGGATCCTTCATCAA-3′ | 188 |
| 5′-CCAAAGAGGATCCCCAAGGCC-3′ | 232 |
| Antisense | |
| 5′-ACTTTGATCTAGAGGCAGTTG-3′ | 244 |
| 5′-CCAGACCTCTAGAACTCGGA-3′ | 155 |
| 5′-GATGACTCTAGAGTTGCCAG-3′ | 47 |
| 5′-CAGCAGCTCTAGACCTTCTT-3′ | 130 |
| 5′-GCCTGCCCTCTAGATTACTTG-3′ | 338 |
Underlined sequences indicate the first codon of the deletion mutant form of MBP-1. Boldface sequences indicate the restriction enzyme sites used for undirectional cloning.
CAT assay.
Cells (5 × 105) were cotransfected with 5 μg of effector plasmid DNA and 5 μg of reporter plasmid DNA (unless specified differently), and cell extracts were prepared after 48 h of transfection. Chloramphenicol acetyltransferase (CAT) assays were performed as described previously (26). The acetylated and nonacetylated forms of chloramphenicol were separated by thin-layer chromatography and scanned with a Molecular Dynamics PhosphorImager. The level of CAT activity was calculated as the percentage of the two acetylated forms of chloramphenicol relative to the total amount of [14C]chloramphenicol. Transfection efficiencies were normalized to an internal β-galactosidase control. Experiments were repeated at least three times for reproducibility.
Western blot analysis.
NIH 3T3 cells (2 × 105) were transfected with different GALMBP-1 constructs. Cell lysates were prepared after 48 h of transfection and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. GAL4 fusion proteins were detected by Western blot analysis using a monoclonal antibody to the GAL4 DNA binding domain (Santa Cruz) and the ECL system (Amersham Corporation).
Colony formation assay.
NIH 3T3 cells were transfected with 3 μg of wild-type or mutant MBP-1. Cells were split at a 1:9 ratio after 24 h of transfection and treated with 400-μg/ml G418 as described earlier (26). Antibiotic selection was continued for 3 weeks, and colonies were scored following crystal violet staining.
RESULTS
MBP-1 can function as a transcriptional repressor.
We have previously demonstrated that MBP-1 directly binds to the c-myc promoter and negatively regulates c-myc transcription (25, 26). Furthermore, ectopic expression of full-length MBP-1 and the carboxy-terminal half of MBP-1 suppresses the growth of human breast carcinoma cells (28, 29). To investigate whether MBP-1 could function as a general transcriptional repressor, we fused full-length MBP-1 to the DNA binding domain of the yeast GAL4 transcription factor (GALMBP1-338). This chimeric gene fusion construct allowed us to investigate the transcriptional regulatory role of MBP-1 under well-defined conditions. We performed in vitro transient-transfection assays with increasing amounts of GALMBP1-338 and a fixed amount (5 μg) of GAL4TK CAT in NIH 3T3 cells. GAL4TK CAT contains five GAL4 DNA binding sites upstream of the herpesvirus thymidine kinase (TK) promoter driving expression of the cat gene. Results from this experiment suggested that GALMBP1-338 represses TK promoter activity in a dose-dependent manner (Fig. 1). The MBP-1 expression plasmid, when used under control of the CMV promoter (without the GAL4 DNA binding domain) in an in vitro transient-transfection assay, did not repress transcription from the GAL4TK CAT reporter (Fig. 2), whereas GALMBP1-338 showed moderate suppression of the TK promoter (without the GAL4 DNA binding sites). Repression of transcription mediated by GALMBP-1 required both MBP-1 fused to the GAL4 DNA binding domain and GAL4 binding sites to be present in the reporter plasmid. Some promoter-specific transcription factors are active only in certain cell types (2). To determine whether MBP-1-mediated inhibition is cell type specific, in vitro transient-transfection assays were performed with HeLa cells and GAL4TK CAT and GALMBP1-338 constructs. GAL4TK CAT activity was inhibited in a dose-dependent manner by GALMBP1-338 in HeLa cells (data not shown).
FIG. 1.
Typical transcriptional activity of GALMBP1-338 in NIH 3T3 cells. Effector plasmid DNA was cotransfected at the indicated concentrations with 5 μg of the GAL4TK CAT reporter plasmid. The total amount of plasmid DNA (10 μg) was kept constant by addition of an empty vector (CMVGAL4) to each transfection mixture. Cell extracts were prepared 48 h posttransfection and assayed for CAT activity. The result indicates that the GALMBP1-338 fusion protein repressed TK promoter activity in a dose-dependent manner.
FIG. 2.
MBP-1 represses transcription when it is brought to the promoter through the GAL4 DNA binding domain. A 5-μg sample of a GAL4TK CAT or TK CAT reporter plasmid was cotransfected with MBP-1 or an empty vector used as a control. The results shown are average results from four independent assays. A relative CAT activity of 100% was arbitrarily assigned to the vector control.
Identification of MBP-1 regulatory domains.
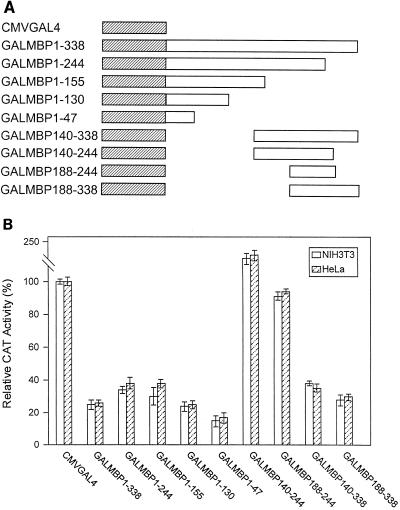
We constructed a number of effector plasmids consisting of deletion mutant forms of MBP-1 linked to the GAL4 DNA binding domain (Fig. 3A) to determine the region responsible for transcriptional repression. These constructs were tested for the ability to activate GAL4TK CAT in HeLa and NIH 3T3 cells. Our initial experiments indicated that MBP-1 possesses two repressor domains and one activation domain (Fig. 3B). The N-terminal (amino acids 1 to 47) and C-terminal (amino acids 188 to 338) regions of MBP-1 exhibited repressor activity on GAL4TK CAT. On the other hand, the middle region (amino acids 140 to 244) appeared to be an activator of CAT transcription, with activity ranging from 150 to 210% of the basal level. We also performed an in vitro transient-transfection assay using HeLa cells, the MBP-1 activation domain, and a different reporter construct, G5E1B CAT. This construct contains five GAL4 DNA binding sites upstream of the adenovirus E1B promoter driving expression of the cat gene. The GALMBP140-244 mutant construct demonstrated a strong activation effect on the E1B promoter (data not shown).
FIG. 3.
(A) Schematic representation of deletion mutant GALMBP-1 used in the analysis for MBP-1 transregulatory domains. The hatched box represents the GAL4 1-147 DNA binding domain, and the open box represents MBP-1 sequences. The numbers following MBP are amino acid positions. (B) Transcriptional activity of deletion mutant GALMBP-1 in NIH 3T3 and HeLa cells. Cells were cotransfected with equal amounts of deletion mutant GAL4TK CAT and GALMBP-1. The CAT assay was performed as described in Materials and Methods. The results suggest that MBP-1 possesses two repressor domains (at the N and C termini) and one activation domain (in the middle).
NIH 3T3 cells were transfected with the plasmid constructs to determine protein expression from the various deletion mutants. Cell lysates were analyzed by Western blotting using a monoclonal antibody to the GAL4 DNA binding domain. Expression of the fusion proteins in transfected NIH 3T3 cells was observed (Fig. 4). GALMBP188-338 appeared as a polypeptide with a lower molecular weight than that calculated (lane 11). Proteolytic cleavage of the fusion protein is a possible reason for this difference, as it also appeared with some other gene products showing multiple polypeptides.
FIG. 4.
Expression of deletion mutant MBP-1 by Western blot analysis using a monoclonal antibody to the GAL4 DNA binding domain. NIH 3T3 cells were transfected with 2 μg of each expression plasmid, and Western blot analysis was performed as described in Materials and Methods. Polypeptides detected by transfection of CMVGAL4 (lane 1), GALMBP1-338 (lane 2), GALMBP1-244 (lane 3), GALMBP1-155 (lane 4), and GALMBP1-130 (lane 5); by mock-transfected cell extract (lane 6); and by transfection of GALMBP140-244 (lane 7), GALMBP188-244 (lane 8), GALMBP140-338 (lane 9), GALMBP232-338 (lane 10), and GALMBP188-338 (lane 11) are shown. Molecular masses are shown on the right in kilodaltons.
Characterization of the repressor domains.
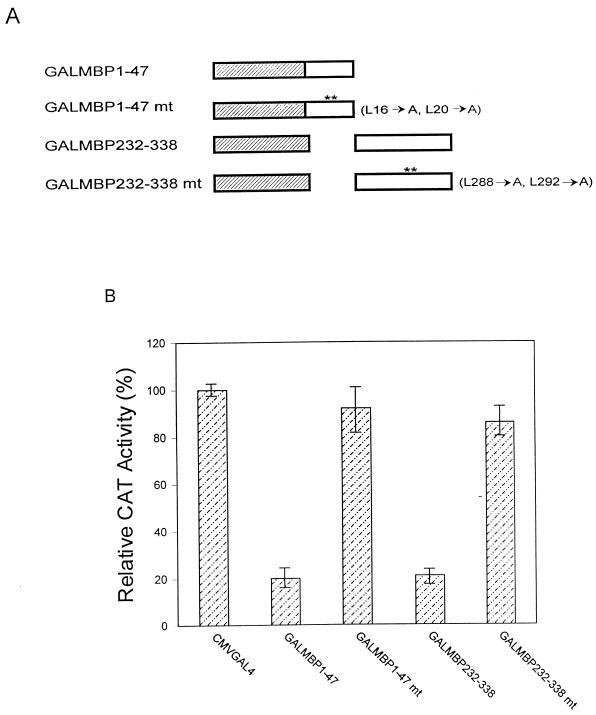
Initial experiments with GALMBP-1 fusion proteins demonstrated that MBP-1 possesses two repressor domains at the N and C termini. To further define the domain within the C-terminal repressor sequences, an additional construct, GALMBP232-338, was generated. An in vitro cotransfection assay suggested that the GALMBP232-338 construct represses CAT activity 80% (data not shown). Thus, the results described earlier and the present data suggest that the trans-repressor domains of MBP-1 are located between amino acids 1 to 47 and amino acids 232 to 338. Both of the repressor domains are highly hydrophobic in nature and have an LXVXL motif in common. To determine if the leucine residues in these regions were, in fact, responsible for repressor activity, point mutations were made by changing the Leu16, Leu20, Leu288, and Leu292 residues to alanine (Fig. 5A). The two leucine-to-alanine mutations in each of these repressor domains failed to inhibit CAT activity (Fig. 5B), suggesting that these regions are responsible for repressor activity. Similar results were obtained when N- or C-terminal leucine mutant forms of full-length MBP-1 were used (data not shown).
FIG. 5.
(A) Schematic representation of leucine-to-alanine point mutations in the LXVXL motif of MBP-1 repressor domains. (B) Transcriptional activities of the repressor domains and respective mutant constructs in NIH 3T3 cells are presented as means of three independent experiments. A relative CAT activity of 100% was arbitrarily assigned to the vector control.
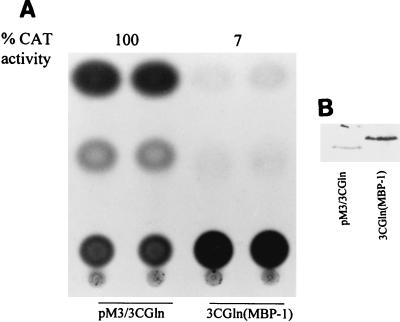
To determine whether the repressor sequence of MBP-1 is a transferable inhibitory domain, we constructed fusion proteins in which the MBP-1 repressor domain (amino acids 1 to 47) was attached to the C terminus of the glutamine-rich activator domain of EBNA3C. The activity was compared with that of its counterpart, which lacks the MBP1-47 sequences. The activity was inhibited when the activation domain (3CGln) of EBNA3C was fused to MBP1-47 (Fig. 6A), indicating that the repressor domain of MBP-1 can inhibit the transcriptional activity of a heterologous activation domain. Western blot analysis using a GAL4 monoclonal antibody exhibited expression of the MBP-1 repressor domain as a chimeric protein (Fig. 6B). As the N-terminal repressor domain demonstrated stronger suppression than the C-terminal domain, the MBP1-47 construct was only tested with a heterologous protein. However, either the N- or C-terminal repressor domain fused to the MBP-1 activation domain inhibited the transcriptional activity of the TK promoter (Fig. 3B) or the E1B promoter. This result further explains the dominant nature of the repressor domains and the overall suppressor activity of MBP-1.
FIG. 6.
The N-terminal region of MBP-1 (amino acids 1 to 47) acts as a transferable repressor domain. (A) The N-terminal repressor domain of MBP-1 was cloned in frame with a heterologous activation domain (3CGln) of Epstein-Barr virus transcription factor EBNA3C. CAT activities from pM3/3CGln were arbitrarily assigned a value of 100%, and CAT activity is shown at the top. (B) Western blot analysis for expression of pM3/3CGln and 3CGln(MBP-1) fusion proteins.
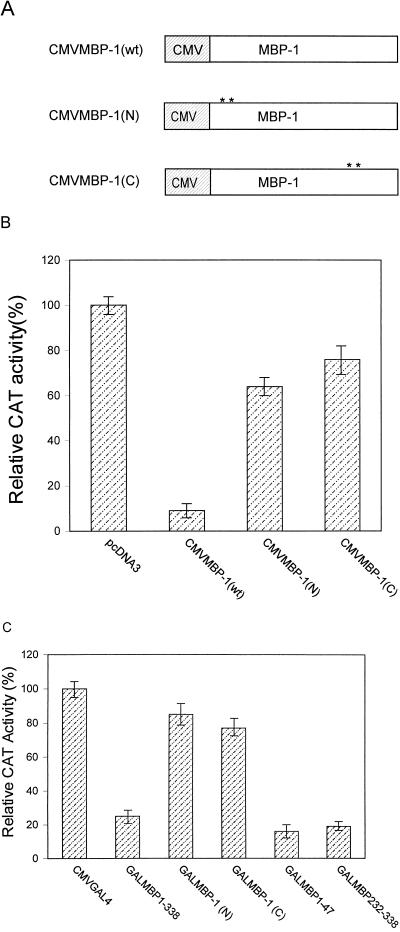
In order to investigate whether mutation of the leucine motif of the repressor domain has an effect on c-myc promoter activity, we mutated the leucine residues to alanine (Fig. 5A) in the N- and C-terminal repressor domains of full-length MBP-1. These mutant constructs were cloned into a pcDNA3 mammalian expression vector (Fig. 7A). An in vitro transient-transfection assay was performed by using the c-myc CAT reporter (25) and wild-type or mutant MBP-1 effector constructs. Results from this experiment suggested that both the N- and C-terminal mutant forms inhibited the repressor activity on the c-myc promoter (Fig. 7B). To further verify this finding, an in vitro transient-transfection assay was performed by using GAL4MBP-1 constructs and the c-myc cat reporter gene (does not contain GAL4 DNA binding sites). Our results suggested that both of the domains in the GAL4 chimera exhibit repressor activity on the c-myc promoter (Fig. 7C). On the other hand, the N- and C-terminal mutant forms of the repressor domain in MBP-1 as GAL4 fusion proteins lacked repressor activity on the c-myc promoter. Expression of the MBP-1 protein and its mutant forms in NIH 3T3 cells was determined by Western blot analysis using a GAL4 DNA binding domain-specific monoclonal antibody (Fig. 8). The ∼49-kDa polypeptide band corresponded to the calculated fusion protein. Additional polypeptides with smaller molecular sizes probably represent proteolytically degraded fusion proteins. Protein expression from the CMVMBP-1(wt), CMVMBP-1(N), and CMVMBP-1(C) constructs in NIH 3T3 cells were indistinguishable because of endogenous MBP-1 expression. However, in vitro-translated products from the MBP-1 mutant constructs confirmed the authenticity of the constructs (data not shown).
FIG. 7.
Transcriptional regulation of the c-myc promoter by wild-type and mutant MBP-1. (A) Schematic diagram of wild-type and mutant MBP-1 under control of the CMV promoter in a pcDNA3 expression vector. CMVMBP-1(wt) represents wild-type MBP-1, and CMVMBP-1(N) and CMVMBP-1(C) represent MBP-1 constructs with leucine-to-alanine mutations in the amino- and carboxy-terminal repressor domains, respectively. (B) Wild-type and mutant MBP-1 cotransfected with a c-myc CAT reporter construct in NIH 3T3 cells. Results are shown as averages of four independent assays. (C) GALMBP1-338, its mutant forms and repressor domains cotransfected with a c-myc CAT construct in NIH 3T3 cells. Results are shown as averages of three independent assays. In all cases, the relative CAT activity of the vector control was arbitrarily assigned a value of 100%.
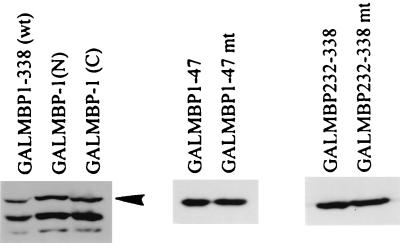
FIG. 8.
Western blot analysis of expression of wild-type and mutant MBP-1 as fusion proteins. The arrowhead corresponds to the calculated molecular weight of MBP-1.
Role of leucine motifs in MBP-1-mediated cell growth regulation.
We have shown previously that the growth of NIH 3T3 cells is suppressed when wild-type MBP-1 is overexpressed (26, 29). To investigate whether the leucine motif (LXVXL) in the repressor domains is required for MBP-1-mediated cell growth regulation, NIH 3T3 cell were transfected with wild-type or mutant MBP-1 (Fig. 7A). Cells were treated with G418 for 3 weeks, and the antibiotic-resistant colonies were counted. The mutations in the leucine motifs of repressor domains altered the growth suppression activity of MBP-1 (Table 2). These results suggested that the leucine motif in the repressor domains of MBP-1 separately exerts its effect on cell growth regulation.
TABLE 2.
Growth regulation in NIH 3T3 cells by wild-type or mutant MBP-1
| DNA transfected | Total no. of coloniesa in transfection:
|
||
|---|---|---|---|
| I | II | III | |
| pcDNA3 | 150 | 180 | NDb |
| CMVMBP-1 | 2 | 12 | ND |
| CMVMBP-1(N) | 92 | 87 | 93 |
| CMVMBP-1(C) | 105 | 110 | 108 |
Each primary plate of NIH 3T3 cells was transfected with 3 μg of plasmid DNA, and the total number of colonies was calculated from 6 × 105 cells. Colonies were counted after 3 weeks of transfection.
ND, not determined.
DISCUSSION
We have previously shown that MBP-1 represses c-myc transcription by direct interaction with the promoter sequences (6, 25, 26). Results from the present study demonstrate that MBP-1, when brought to the promoter by a GAL4 DNA binding domain, can significantly repress transcriptional activity. We propose that besides blocking the transcription of c-myc, MBP-1 can modulate cellular gene transcription through an alternative mechanism. For a long time, research has focused mainly on activator and coactivator proteins (11, 12), but it recently became clear that repressor and corepressor proteins also play an important role in the regulation of gene expression (8, 11, 12, 14, 16). Transcriptional repression can occur by several general mechanisms, such as competition for DNA binding sites, squelching, quenching, and direct or indirect (using a corepressor) interaction with transcription machinery (31). Interestingly, several of the transcription factors investigated act as both activators or repressors, depending on the target promoter and the cellular context (31). Thus, it appears that besides direct binding to the c-myc promoter, MBP-1 may function like an adapter which can regulate transcription through protein-protein interaction, bridging a specific transcription factor. Alternatively, MBP-1 may directly interact with other factors involved in transcription. Indeed, further studies are necessary to elucidate the mechanism of transcriptional regulation by MBP-1.
We have also used progressive deletion mutant forms of MBP-1 to characterize its functional domains. The deletion analysis suggested that the repressor activity resides within strongly hydrophobic domains encompassing amino acids 1 to 47 and amino acids 232 to 338 of MBP-1. Interestingly, both the repressor domains have an LXVXL motif and replacement of the leucine residues with alanine abrogated the repressor activity. This motif does not match any other repressor domain in the GenBank database (searched by the BLAST program), and the biological significance of this motif is unknown. We have demonstrated that mutation of the leucine residues in the LXVXL motif abrogated the repressor activity not only on the GAL4-TK promoter but also on the native c-myc promoter and altered the cell growth-regulatory function of MBP-1. The repressor domains are termed “portable” or “transferable” because they function in the context of heterologous activation and the DNA binding domains. The N-terminal repressor domain of MBP-1 can function independently, inhibiting transcription when attached to the activation domain of EBNA3C, like Oct-2A (10) and Mad (1).
Gene fusion experiments also demonstrated that a chimeric GAL4 MBP-1 protein containing amino acids 140 to 244 functions as a transcriptional activator when bound to a promoter bearing multiple GAL4 DNA binding sites. These sequences contain highly charged amino acid residues. The data further indicate that this may represent a bifunctional nature of the protein, but alternatively, the two repressor domains may mask the activator function for a predominant repressor activity of MBP-1. A similar phenomenon has been demonstrated for hMTF-1, which is a heavy metal-responsive transcription regulator (24), and activating transcription factor 2 (17). Transcription factors that activate in one circumstance and repress in another have been documented, and the molecular bases of these transitions are quite diverse (31). For instances, transcription factor Kruppel converts from an activator to a repressor in a dose-dependent manner (34). Sp3 is a dual-function regulator whose predominant activity depends upon the number of DNA-binding sites present in the promoter and the molecular basis of this transition is unknown (18). The transcriptional activity of Ets-1 is modified upon DNA binding, and allosteric changes have been suggested to alter the structure of a transcription factor as a result of interaction with DNA (22, 23). A recent study suggested that the T-cell oncogene RBTN-2 is a complex transcription factor possessing multiple trans-regulatory domains (19), similar to other transcription regulators, such as p53, c-fos, SRF, IRF, YY1, the visna virus Tat protein, Oct-2A, and activating transcription factor 2 (3, 4, 5, 10, 15, 17, 37, 39).
The presence of multiple trans-regulatory domains in MBP-1 suggests that the overall activity of this protein depends on the interplay among these repressor and activator regions, possibly interacting through a cellular factor(s). We have also demonstrated that MBP-1 downregulates the human immunodeficiency virus long terminal repeat (30). On the other hand, our recent observation suggests that MBP-1 transactivates the proliferating cell nuclear antigen promoter (unpublished data). How and under what conditions MBP-1 switches from a repressor to an activator is of general interest and requires further investigation. Results from previous studies and the present observations prompt us to speculate that besides regulating c-myc, MBP-1 may have an impact on the fine tuning of other cellular gene regulation. Some repressors might employ more than one operating mechanism, depending on the promoter context. Each type of repression mode could evoke particular biological regulatory properties of the repressor mediating developmental, differentiation, and cell growth strategies (13). The ability of the MBP-1 repressor domains to block c-myc promoter activity may provide a role for apoptosis and inhibition of tumorigenicity (26, 28). In fact, identification of target genes will contribute significantly to our understanding of the complex regulatory function of MBP-1 and its biological role in cell growth regulation.
ACKNOWLEDGMENTS
We thank D. Dean, Y. Shi, and C. Sample for providing research materials and SuzAnn Price for preparation of the manuscript.
This research work was supported by PHS grant CA-52799 from the National Cancer Institute.
REFERENCES
- 1.Ayer D E, Laherty C A, Lawrence Q A, Armstrong A P, Eisenman R E. Mad proteins contain a dominant transcription repression domain. Mol Cell Biol. 1996;16:5772–5781. doi: 10.1128/mcb.16.10.5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baichwal V R, Tjian R. Control of c-Jun activity by interaction of a cell-specific inhibitor with regulatory domain delta: differences between v- and c-Jun. Cell. 1990;63:815–825. doi: 10.1016/0092-8674(90)90147-7. [DOI] [PubMed] [Google Scholar]
- 3.Brown H J, Sutherland J A, Cook A, Bannister A J, Kouzarides T. An inhibitor domain in c-fos regulates activation domains containing HOB1 motif. EMBO J. 1995;14:124–131. doi: 10.1002/j.1460-2075.1995.tb06982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bushmeyer S, Park K, Atchison M L. Characterization of functional domains within the multifunctional transcription factor YY1. J Biol Chem. 1995;270:30213–30220. doi: 10.1074/jbc.270.50.30213. [DOI] [PubMed] [Google Scholar]
- 5.Carruth L M, Hardwick J M, Morse B A, Clements J E. Visna virus Tat protein: a potent transcription factor with both activator and suppressor domains. J Virol. 1994;68:6137–6146. doi: 10.1128/jvi.68.10.6137-6146.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaudhary D, Miller D M. The c-myc promoter binding protein (MBP-1) and TBP bind simultaneously in the minor groove of the c-myc P2 promoter. Biochemistry. 1995;34:3438–3445. doi: 10.1021/bi00010a036. [DOI] [PubMed] [Google Scholar]
- 7.Cole M D. The c-myc oncogene: its role in transformation and differentiation. Annu Rev Genet. 1986;20:361–384. doi: 10.1146/annurev.ge.20.120186.002045. [DOI] [PubMed] [Google Scholar]
- 8.Cowell I G. Repression versus activation in the control of gene transcription. Trends Biochem Sci. 1994;19:38–42. doi: 10.1016/0968-0004(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 9.Evan G I, Littlewood T D. The role of c-myc in cell growth. Curr Opin Genet Dev. 1993;3:44–49. doi: 10.1016/s0959-437x(05)80339-9. [DOI] [PubMed] [Google Scholar]
- 10.Friedl E M, Matthias P. Mapping of the transcriptional repression domain of the lymphoid-specific transcription factor Oct-2A. J Biol Chem. 1996;271:13927–13930. doi: 10.1074/jbc.271.24.13927. [DOI] [PubMed] [Google Scholar]
- 11.Goodrich J A, Tjian R. TBP-TAF complexes: selectivity factors for eukaryotic transcription. Curr Opin Cell Biol. 1994;6:403–409. doi: 10.1016/0955-0674(94)90033-7. [DOI] [PubMed] [Google Scholar]
- 12.Guarente L. Transcriptional coactivators in yeast and beyond. Trends Biochem Sci. 1995;20:517–521. doi: 10.1016/s0968-0004(00)89120-3. [DOI] [PubMed] [Google Scholar]
- 13.Hanna-Rose W, Hansen U. Active repression mechanisms of eukaryotic transcription repressors. Trends Genet. 1996;12:229–234. doi: 10.1016/0168-9525(96)10022-6. [DOI] [PubMed] [Google Scholar]
- 14.Herschbach B, Johnson A D. Transcriptional repression in eukaryotes. Annu Rev Cell Biol. 1993;9:479–509. doi: 10.1146/annurev.cb.09.110193.002403. [DOI] [PubMed] [Google Scholar]
- 15.Johansen F-E, Prywes R. Identification of transcriptional activation and inhibitory domains in serum response factor (SRF) by using GAL4-SRF constructs. Mol Cell Biol. 1993;13:4640–4647. doi: 10.1128/mcb.13.8.4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson A D. The price of repression. Cell. 1995;81:655–658. doi: 10.1016/0092-8674(95)90524-3. [DOI] [PubMed] [Google Scholar]
- 17.Li X-Y, Green M R. Intramolecular inhibition of activating transcription factor-2 function by its DNA-binding domain. Genes Dev. 1996;10:517–527. doi: 10.1101/gad.10.5.517. [DOI] [PubMed] [Google Scholar]
- 18.Majello B, De Luca P, Lania L. Sp3 is a bifunctional transcription regulator with modular independent activation and repression domains. J Biol Chem. 1997;272:4201–4026. doi: 10.1074/jbc.272.7.4021. [DOI] [PubMed] [Google Scholar]
- 19.Mao S, Neale G A M, Goorha R M. T-cell proto-oncogene rhombotin-2 is a complex transcription regulator containing multiple activation and repression domains. J Biol Chem. 1997;272:5594–5599. doi: 10.1074/jbc.272.9.5594. [DOI] [PubMed] [Google Scholar]
- 20.Marcu K B, Bossone S A, Patel A J. Myc function and regulation. Annu Rev Biochem. 1992;61:809–860. doi: 10.1146/annurev.bi.61.070192.004113. [DOI] [PubMed] [Google Scholar]
- 21.Marshall D, Sample C. Epstein-Barr virus nuclear antigen 3C is a transcriptional regulator. J Virol. 1995;69:3624–3630. doi: 10.1128/jvi.69.6.3624-3630.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKnight S L. Transcription revisited: a commentary on the 1995 Cold Spring Harbor Laboratory Meeting, “Mechanisms of eukaryotic transcription.”. Genes Dev. 1996;10:367–381. doi: 10.1101/gad.10.4.367. [DOI] [PubMed] [Google Scholar]
- 23.Patersen J M, Skalicky J J, Donaldson L W, McIntosh L P, Alber T, Graves B J. Modulation of transcription factor Ets-1 DNA binding: DNA-induced unfolding of an alpha helix. Science. 1995;269:1866–1869. doi: 10.1126/science.7569926. [DOI] [PubMed] [Google Scholar]
- 24.Radtke F, Georgiev O, Muller H-P, Brugnera E, Schaffner W. Functional domains of the heavy metal-responsive transcription regulator. Nucleic Acids Res. 1995;23:2277–2286. doi: 10.1093/nar/23.12.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ray R, Miller D M. Cloning and characterization of a human c-myc promoter-binding protein. Mol Cell Biol. 1991;11:2154–2161. doi: 10.1128/mcb.11.4.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ray R. Induction of cell death in murine fibroblasts by a c-myc promoter binding protein. Cell Growth Differ. 1995;6:1089–1096. [PubMed] [Google Scholar]
- 27.Ray R B, Sheikh M S, Fontana J F, Miller D M. Human breast carcinoma cells show correlation in expression of c-myc oncogene and the c-myc binding protein (MBP-1) Int J Oncol. 1994;5:1433–1436. doi: 10.3892/ijo.5.6.1433. [DOI] [PubMed] [Google Scholar]
- 28.Ray R B, Steele R, Seftor E, Hendrix M. Human breast carcinoma cells transfected with the gene encoding a c-myc promoter-binding protein (MBP-1) inhibits tumors in nude mice. Cancer Res. 1995;55:3747–3751. [PubMed] [Google Scholar]
- 29.Ray R B, Steele R. Separate domains of MBP-1 involved in c-myc promoter binding and growth suppressive activity. Gene. 1997;186:175–180. doi: 10.1016/s0378-1119(96)00693-2. [DOI] [PubMed] [Google Scholar]
- 30.Ray R B, Srinivas R V. Inhibition of human immunodeficiency virus type 1 replication by a cellular transcriptional factor MBP-1. J Cell Biochem. 1997;64:565–572. doi: 10.1002/(sici)1097-4644(19970315)64:4<565::aid-jcb4>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 31.Roberts S G E, Green M R. Transcription. Dichotomous regulators. Nature. 1995;375:105–106. doi: 10.1038/375105a0. [DOI] [PubMed] [Google Scholar]
- 32.Sadowski I, Ptashne M. A vector expressing CAL4 (1-147) fusions in mammalian cells. Nucleic Acids Res. 1989;17:7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sadowski I, Bell B, Broad P, Hollis M. Gal4 fusion vectors for expression in yeast or mammalian cells. Gene. 1992;118:137–141. doi: 10.1016/0378-1119(92)90261-m. [DOI] [PubMed] [Google Scholar]
- 34.Sauer F, Fondell J D, Ohkuma Y, Roeder R G, Jackle H. Control of transcription by Kruppel through interactions with TFIIB and TFIIE beta. Nature. 1995;375:162–164. doi: 10.1038/375162a0. [DOI] [PubMed] [Google Scholar]
- 35.Shi Y, Glynn J M, Guilbert L J, Cotter T G, Bissonnette R P, Green D R. Role for c-myc in activation-induced apoptotic cell death in T-cell hybridomas. Science. 1992;257:212–214. doi: 10.1126/science.1378649. [DOI] [PubMed] [Google Scholar]
- 36.Spencer C A, Gourdine M. Control of c-myc regulation in normal and neoplastic cells. Adv Cancer Res. 1991;53:1–48. doi: 10.1016/s0065-230x(08)60476-5. [DOI] [PubMed] [Google Scholar]
- 37.Subler M A, Martin D W, Deb S. Overlapping domains on the p53 protein regulate its transcriptional activation and repression functions. Oncogene. 1994;9:1351–1359. [PubMed] [Google Scholar]
- 38.White R A, Adkinson L R, Dowler L L, Ray R B. Chromosomal localization of the human gene encoding c-myc promoter-binding protein (MBP-1) to chromosome 1p35-pter. Genomics. 1997;39:406–408. doi: 10.1006/geno.1996.4499. [DOI] [PubMed] [Google Scholar]
- 39.Yamamoto H, Lampier M S, Fujita T, Taniguchi T, Harada H. The oncogenic transcription factor IRF-2 possesses a transcriptional repression and a latent activation domain. Oncogene. 1994;9:1423–1428. [PubMed] [Google Scholar]