Abstract
In the last few years, cellular metabolic reprogramming has been acknowledged as a hallmark of human cancer and evaluated for its crucial role in supporting the proliferation and survival of human cancer cells. In a variety of human tumours, including hepatocellular carcinoma (HCC), breast cancer and non-small-cell lung cancer (NSCLC), a large amount of carbon is reused in serine/glycine biosynthesis, accompanied by higher expression of the key glycine synthetic enzyme mitochondrial serine hydroxymethyltransferase 2 (SHMT2). This enzyme can convert serine into glycine and a tetrahydrofolate-bound one-carbon unit, ultimately supporting thymidine synthesis and purine synthesis and promoting tumour growth. In tumour samples, elevated expression of SHMT2 was found to be associated with poor prognosis. In this review, the pivotal roles of SHMT2 in human carcinogenesis are described, highlighting the underlying regulatory mechanisms through promotion of tumour progression. In conclusion, SHMT2 may serve as a prognostic marker and a target for anticancer therapies.
Keywords: Serine hydroxymethyltransferase 2 (SHMT2), human carcinogenesis, predictive biomarker, cell proliferation, tumour growth
Introduction
Cumulative evidence suggests that the serine and glycine biosynthetic pathways are linked to the biosynthesis of cellular components to sustain cell proliferation. Dysfunction of genes involved in the one-carbon unit biosynthetic pathway has been linked to increased risks of various human cancers 1, 2. SHMT2 is a key enzyme in one-carbon unit metabolism and catalyzes the conversion of serine and tetrahydrofolate (THF) to glycine and 5,10-methylenetetrahydrofolate (5,10-CH2-THF) 3. Specifically, SHMT2 expression exhibits a significant correlation with a wide variety of human phenotypes, including neural tube defects and childhood acute leukaemia. To assess the relationship between SHMT2 and human cancers, we analysed SHMT2 expression with public RNA-seq data from TCGA in multiple malignancies. As shown in Figure 1, SHMT2 was significantly elevated in more than 85% of cancer types compared with normal tissues. Similarly, a series of studies have reported that SHMT2 expression is increased in a subset of human cancers including colon cancer, breast cancer, lung cancer, ovarian cancer and prostate cancer. Recently, an advanced understanding of the mechanisms of SHMT2 in human carcinogenesis has provided novel opportunities for drug development, dietary intervention and therapeutic targets 4.
Figure 1.
Different expression levels of SHMT2 in human tumour types from TCGA database (*p < 0.05, **p < 0.01, ***p < 0.001).
Identification and Characterization of SHMT2
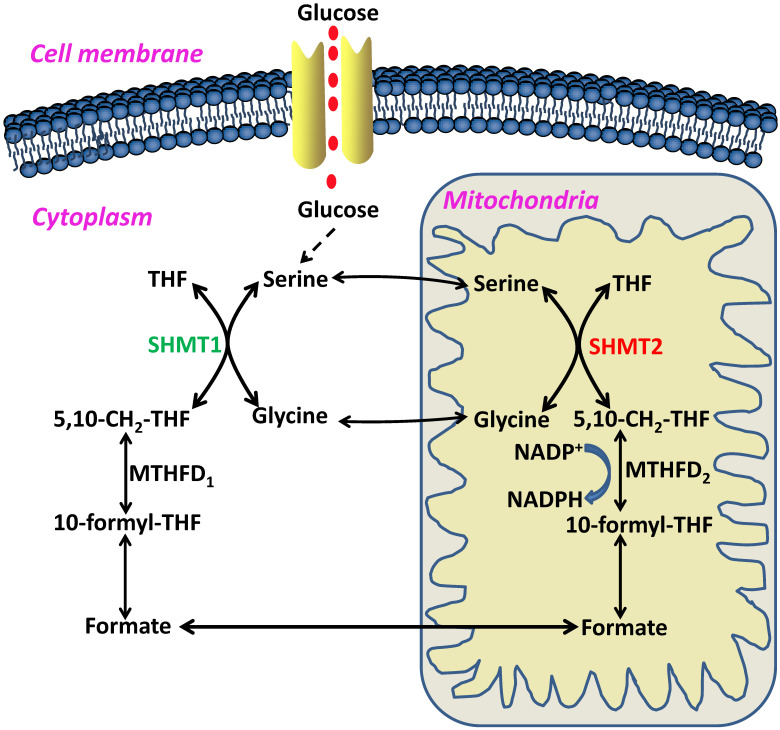
In 1981, Stauffer and his colleagues first cloned the glyA gene, which encodes the E. coli SHMT protein, a pyridoxal 5'-phosphate (PLP) dependent enzyme 5. The reverse reaction between glycine and serine catalyzed by SHMT was named the 'futile cycle', and critical roles of the reaction product 5-formylTHF have been found in maintaining one-carbon unit homeostasis 6, 7 (Figure 2). In humans, SHMT consists of two isoforms, SHMT1, which is located in the cytosol (cSHMT), and SHMT2, which is located in the mitochondria (mSHMT). These two isoforms display an amino acid sequence identity of approximately 66% (Figure 3). In addition, SHMT1 forms a tetramer in the cytoplasm, while SHMT2 exists in a dimer-to-tetramer transition depending on pyridoxal 5'-phosphate (PLP) binding 8, 9. In fact, SHMT2 can encode SHMT2α, a second transcript that lacks the import signal location to the mitochondria and is localized in the cytoplasm. Due to its important role in the cycle of one-carbon unit, SHMT2 is ubiquitous and highly conserved in all organisms.
Figure 2.
Schematic overview of serine/glycine metabolism in the cytoplasm and mitochondria. THF, tetrahydrofolate; 5,10-CH2-THF, 5,10-methylenetetrahydrofolate; MTHFD2, methylenetetrahydrofolate dehydrogenase 2; NADPH, nicotinamide adenine dinucleotide phosphate; 10-formyl-THF, 10-formyl-tetrahydrofolate; MTHFD1, methylenetetrahydrofolate dehydrogenase 1; SHMT1, serine hydroxymethyltransferase 1; SHMT2, serine hydroxymethyltransferase 2.
Figure 3.
Sequence of the protein SHMT2 aligned with SHMT1. Identical residues are highlighted in dark.
Deregulation of SHMT2 in Various Human Cancers
Many studies have demonstrated the crucial roles of SHMT2 in maintaining normal methylation patterns, DNA stability and genetic variation 10, 11. SHMT2 maps to 12q13 with all intron/exon splice junctions conforming to the GT/AG rule 6, 12. Several studies have suggested that the encoded product is expressed predominately in the mitochondria and primarily responsible for glycine synthesis in the cell 13. While SHMT1 maps to 17p11.2 and is expressed as multiple splice variants with 330 nucleotides of the 5'-untranslated region. There are several consensus DNA recognition sites for transcription factor binding in the 5'-promoter region of the mSHMT gene. Sequence analysis of the mSHMT gene revealed two Myc consensus binding sites in its promoter. Transcriptional activation of SHMT2 is regulated by the oncogenic transcription factor c-Myc 11. Moreover, transcriptional regulatory proteins, including Sp-1, AP-2, and PEA3, may control endogenous expression of SHMT2 for further investigation. Primer extension analysis and 5'-RACE analysis reveal that transcription initiation of cSHMT occurs at multiple sites. Northern blot analysis of multiple tissues suggests that cSHMT displays cell-specific splicing patterns. Multiple forms of cSHMT are present in MCF-7 cells, whereas a single form is expressed in SH_SY5Y cells 7. Although SHMT1 and SHMT2 catalyze the same biochemical reactions, they play different biological roles in tumors. Recent studies have more focused on single nucleotide polymorphism (SNP) of SHMT1 and indicated that the SNPs may associate with cancers 14, 15. However, meta-analysis by Wang Q et al about roles of SHMT1 gene in various cancer types indicated the inconsistent roles of SHMT1 in multiple tumor procession 16. Then in view of the consistent roles of SHMT2 and complex regulatory mechanisms in human tumour, we reviewed the current studies in multiple cancers for a comprehensive understanding of SHMT2.
Hepatocellular carcinoma (HCC)
Intracellular biosynthesis of glycine catalysed by SHMT2 is essential for hepatocyte growth and proliferation. Knockdown of SHMT2 expression worsened hepatic ischaemia-reperfusion and delayed liver regeneration after partial hepatectomy 17, 18. However, aberrant SHMT2 promoted changes in metabolism that enhanced the survival capacities of tumour cells to survive in the ischaemic microenvironment. Lee et al. found that SHMT2 was significantly upregulated at both the mRNA and protein levels in three human HCC cell lines (Hep3B, HepG2 and Huh-7) compared with an immortalized normal liver cell line (THLE2) 19. Inhibition of SHMT2 led to reduced cell proliferation and tumorigenicity in liver cancer cells. In addition, Huh-7 cells with downregulated SHMT2 expression failed to form tumours in a human tumour xenograft mouse model, whereas SHMT2 overexpression enhanced liver cancer cell survival and proliferation 19, 20. Moreover, an increasing number of studies have demonstrated that SHMT2 is overexpressed in the tissues of hepatocellular and intrahepatic biliary carcinoma patients and that a higher SHMT2 level is clinically related to a worse survival outcome than a lower level of SHMT2, suggesting that SHMT2 could be used as a potential prognostic biomarker in HCC 20, 21.
Gastrointestinal cancer
The role of the mitochondrial metabolic enzyme SHMT2 in gastrointestinal cancer has been reported in recent years for its function in providing one-carbon unit of serine-glycine conversion involved in one-carbon unit metabolism. SHMT2 acetylation was decreased in human colonic epithelial cell lines, which eventually promoted colorectal carcinogenesis 22. Later studies confirmed upregulated SHMT2 expression in colon cancer tissues 23 and showed that decreased invasion and inhibition of proliferation of colorectal carcinoma cells were accompanied by downregulation of SHMT2 expression in vitro 24. Clinically, SHMT2 was overexpressed in gastric cancer, oesophageal cancer and colorectal cancer (CRC) tissues at both the mRNA and protein levels, as measured by qRT-PCR and immunohistochemistry assays, compared to paired normal adjacent tissues. Furthermore, survival and correlation analysis revealed that SHMT2 expression was an independent prognostic factor for recurrence-free survival and disease-specific survival in gastric cancer, oesophageal cancer and CRC 25. Similarly, Shi et al. assessed SHMT2 expression in 130 gastric cancer tissues using immunohistochemistry and demonstrated that SHMT2 expression was enhanced compared to that in adjacent tissues. Patients with higher expression of SHMT2 showed significantly worse outcomes, which supported that SHMT2 acted as an attractive target for gastrointestinal cancer chemotherapy 26.
Glioma
SHMT2 expression was also associated with the survival of brain cancer cells within the ischaemic zones of gliomas 27. Overexpression of SHMT2, on the one hand, restricted the activity of pyruvate kinase and reinforced glycolytic progression, thus lessening oxygen demand and assisting cancer cells in enduring and surviving in an ischaemic tumour microenvironment. On the other hand, the enhanced glycine production also rendered these cells sensitive to inhibition by the glycine cleavage system. Intriguingly, SHMT2 expression can also be induced in glioblastoma cell lines (LN-308, LNT-229 and G55 cells); this molecule cooperates with phosphoglycerate dehydrogenase (PHGDH) to support glioblastoma cell adaption in hypoxic conditions 28, 29. In line with that, clinical detection of SHMT2 expression by immunohistochemistry revealed that it was highly upregulated in glioma tissues compared to that in the control group. In addition, a Kaplan-Meier analysis showed that higher SHMT2 expression was implicated in poorer progression-free survival (PFS) and overall survival (OS), although the data failed to link the expression of SHMT2 with drug resistance mechanisms in glioma patients 30, 31.
Breast cancer
Emerging studies have demonstrated the pivotal roles of SHMT2 protein in breast cancer progression 32. As shown in the work of Li et al., subclones of MDA-MB-231 cell lines with elevated one-carbon unit biosynthesis activity displayed increased metastatic potential 33. Knockdown of SHMT2 expression potently repressed the proliferation of metastatic subclones in vitro. Moreover, impaired cancer cell proliferation due to the inhibition of SHMT2-induced metabolic reprogramming was further validated in animal models. Compared to that in the matched noncancerous tissues, SHMT2 protein expression was also upregulated in clinical breast cancer tissues. Consistently, there was a significant correlation between the expression level of SHMT2 and breast cancer grade. More importantly, both primary and metastatic tumour growth was suppressed in the SHMT2 knockdown group. Similarly, another study showed that SHMT2 protein expression was detected in 128 breast cancer cases and associated with tumour aggressiveness (TNM staging and Elson grade) in a dose-dependent manner. Stratified analysis results indicated that SHMT2 was helpful in predicting outcomes, especially in oestrogen receptor (ER)-negative breast cancer patients. In cases involving stage IIb breast cancer, chemotherapy significantly extended survival time among patients with higher SHMT2 expression 34. Although this molecule is considered a prognostic biomarker of breast cancer, other studies have reported that abnormal expression of SHMT2 may predict negative prognosis 35, 36. Nevertheless, there was no association with age, sex, lymph node status, TNM stage or vascular invasion status 37, 38.
Notably, recent studies have linked SHMT2 overexpression with resistance to clinical therapy in breast cancer. Once exposed to lapatinib, cells with elevated SHMT2 could attenuate the toxicity of reactive oxygen species (ROS) and survive 39. In addition, though upregulating the expression of the antioxidant enzymes HMOX1, SHMT2 and SLC7A11, breast cancer cells exhibited resistance to paclitaxel through the EIF2AK3/EIF2AK4-pEIF2S1-ATF4 axis 40.
Collectively, these results indicated that SHMT2 may be a valuable prognostic biomarker in breast cancer. Furthermore, SHMT2 may serve as a potential therapeutic target for breast cancer treatment and drug discovery 37, 41.
Other cancers
The SHMT2 expression level was not only increased and implicated in poor prognosis in lung cancer 42, 43 but was also associated with idiopathic pulmonary fibrosis (IPF) 44. Based on application of western blotting and immunohistochemistry staining, the expression of SHMT2 was found to be increased in response to TGF-β in vitro and in lung tissues obtained from four patients with IPF compared with expression in healthy donors. Furthermore, upregulated SHMT2 expression increased the aggressiveness of prostate cancer cells 45. Additionally, a study of osteoarthritis showed that SHMT2 was overexpressed in osteoarthritis patients and involved in the pathogenesis of osteoarthritis by modulating one-carbon unit metabolism 46.
Altogether, SHMT2 acts as an important enzyme in one-carbon unit metabolism pathway. Different studies suggest the diverse functions of SHMT2 in the majority of tumours. A genome-wide oncogenic screening strategy and RNAi screen pioneered by Lee's group identified SHMT2 as a candidate oncogenic driver gene across multiple solid tumour types 47. Subsequent functional assays showed that SHMT2 was necessary for tumour cell survival across a panel of 32 cancer cell lines. A series of experimental and functional analyses revealed the oncogenic properties of SHMT2 in promoting cancer cell survival and tumour growth in vivo.
Mechanisms Underlying the Oncogenic Role of SHMT2 in Human Carcinogenesis
SHMT2 has a role in folate metabolism, as it provides active one-carbon unit contributing to the biosynthesis of nucleotides and proteins involved in tumour growth. Although the physiological function of the isozyme has been widely studied, the specific mechanisms of the oncogenic role of SHMT2 in human carcinogenesis remain to be further determined.
SHMT2 is primarily involved in the production of glycine and 5,10-CH2-THF 48. In mitochondria, SHMT2-dependent production of methylene-THF contributes to mitochondrial NADPH generation and redox balance during hypoxia 29. Hypoxia plays critical roles in metastasis and angiogenesis by upregulating the expression of glycolysis-related enzymes, including HK2, LDHA and PDK1. The upregulated expression of these enzymes suppresses entry of pyruvate into the TCA cycle, thus reducing ROS generation and promoting tumour growth. Previous studies have proposed that SHMT2 is a transcriptional target of Myc, illustrating an intrinsic link between tumorigenesis and cellular metabolism 11, 49. Myc amplification was shown to promote upregulation of SHMT2 expression in a hypoxia-inducible factor-1α (HIF-1α)-dependent manner, resulting in increased production of NADPH from NADP+, restrained cellular reactive oxygen species, and enhanced tumour cell survival 29. SHMT2 knockdown significantly inhibited xenograft tumour growth and aggravated ischaemia-reperfusion injury in mice through the ROS/JNK/P53 signaling pathway 17, 29. Moreover, SHMT2 can activate the Akt/mTOR signaling pathway by its metabolic product, which facilitates hepatocyte cell regeneration 18. SHMT2 expression can also be upregulated through the IL-6/JAK2/STAT3 regulatory pathway in human prostate carcinoma LNCaP cells, speeding up the cell transition towards more advanced stages 45.
Studies on noncoding RNAs in tumours have been increasing in recent years, and it was also reported that SHMT2 could be directly and negatively regulated by miR-370 in human articular chondrocytes and miR-615-5p in HCC cells 46. In NSCLC cells, long noncoding RNA (lncRNA) Gm15290 exerted a pro-oncogene effect by negatively regulating the expression of miR-615-5p and thus increasing the protein levels of miR-615-5p target genes, including SHMT2 50. In addition, the long noncoding RNA LINC01234 promoted colon cancer cell proliferation through the LINC01234-miR642a-5p-SHMT2 axis 24. Furthermore, it was found that circular RNA 0072995 directly targets miR-149-5p, thereby upregulating the expression of its downstream gene SHMT2 and enhancing breast tumour cell proliferation in vitro and in vivo 51.
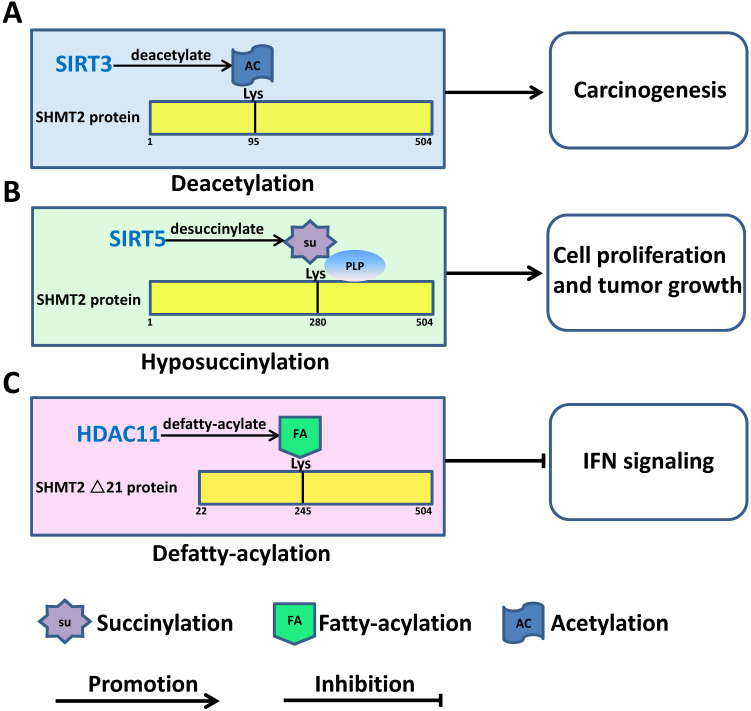
In addition to interaction with signaling pathways or modulation by miRNA, inadequate SHMT denaturalization can lead to human cancer progression via post-translational modification. SHMT2 could be acetylated at lysine K95 in different cancer cells, including CRC cells. K95 acetylation inhibits SHMT2 enzymatic activity and promotes its degradation via macroautophagy. SIRT3, a deacetylase localized in the mitochondria, deacetylated SHMT2 and eventually increased its activity to promote cell proliferation and colorectal carcinogenesis 22 (Figure 4A). SHMT2 interacts with nicotinamide adenine dinucleotide (NAD+)-dependent lysine desuccinylase SIRT5 directly. Under metabolic stress, desuccinylation at lysine 280 of SHMT2 by SIRT5 enhances SHMT2 activity to accelerate cell metabolism and facilitate cell proliferation and tumour growth in vivo and in vitro 52-54 (Figure 4B). HDAC11 has been proven to be an efficient lysine defatty-acylase that directly removes fatty acyl groups from SHMT2. In MCF-7 and A549 cells, knockdown of HDAC11 significantly increased the fatty acylation level of SHMT2. Although SHMT2 fatty acylation does not affect its enzymatic activity, K245 is the major fatty acylation site of HDAC11. Upon treatment with IFNα, fatty-acylated SHMT2 was recruited to late endosomes/lysosomes to deubiquitinate IFNαR1 and activate IFN signaling 55-57 (Figure 4C).
Figure 4.
Post-translational modification of SHMT2 regulates human cancer progression. A. Deacetylation of SHMT2 by SIRT3 at K95 promotes colorectal carcinogenesis. B. Desuccinylation of SHMT2 by SIRT5 at K280 promotes cancer cell proliferation and tumour growth. C. Defatty acylation of SHMT2 by HDAC11 at K95 regulates type I interferon signaling.
Perspectives
In cellular metabolism, glycine metabolism is a key step in the proliferation and survival of human cancer cells 58. SHMT2 is an oncogene, and thus, it may serve as a promising target for anticancer therapy. The identification of selective SHMT2 inhibitors could be an innovative and promising approach for the treatment of various cancers. Although SHMT2 is a difficult target for inhibition, as it is located in mitochondria, further drug discovery efforts are warranted. There are already preclinical studies exploring and evaluating the potential efficacy of inhibitors targeting SHMT2 and cytosolic C1 enzymes 59-61, among which AGF347 showed a satisfying and compelling anti-tumour effects in vivo in both early-stage and advanced stage MIA PaCa-2 pancreatic tumour xenografts62. Notably, anticancer models using SHMT2 knockout for treating human cancer may overestimate the anticancer effect of SHMT2 63, 64. The deletion of SHMT2 will inhibit translation in the mitochondria, whereas the effects of the drug may not achieve the depletion of one-carbon unit required to produce the same phenotype 65, 66. In summary, tumour cells become more aggressive under internal and external stresses. The prevention of tumour development, metastasis and recurrence by the perturbation of specific SHMT2 inhibitors requires further exploration.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 81802885, 81820108023); the National Key Research and Development Program of China (No. 2018YFC1705505) and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Author contributions
YZ wrote the draft manuscript and provided financial support; JZ contributed to figure preparation; MX, FC and RZ collected all the related articles and provided the data from TCGA database; JY and YZ revised the manuscript and participated in the whole work; NC contributed to editing of the manuscript and figures; YC was involved in providing guidance for this review. All authors have read and approved the final manuscript.
Abbreviations
- NSCLC
non-small-cell lung cancer
- HCC
hepatocellular carcinoma
- SHMT2
serine hydroxymethyltransferase 2
- THF
tetrahydrofolate
- 5,10-CH2-THF
5,10-methylenetetrahydrofolate
- PLP
pyridoxal 5'-phosphate
- CRC
colorectal cancer
- IPF
idiopathic pulmonary fibrosis
- HIF-1α
hypoxia-inducible factor-1α
References
- 1.Vander Heiden MG, DeBerardinis RJ. Understanding the Intersections between Metabolism and Cancer Biology. Cell. 2017;168:657–69. doi: 10.1016/j.cell.2016.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ducker GS, Rabinowitz JD. One-Carbon Metabolism in Health and Disease. Cell Metab. 2017;25:27–42. doi: 10.1016/j.cmet.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Minton DR, Nam M, McLaughlin DJ, Shin J, Bayraktar EC, Alvarez SW. et al. Serine Catabolism by SHMT2 Is Required for Proper Mitochondrial Translation Initiation and Maintenance of Formylmethionyl-tRNAs. Mol Cell. 2018;69:610–21. doi: 10.1016/j.molcel.2018.01.024. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parsa S, Ortega-Molina A, Ying HY, Jiang M, Teater M, Wang J. et al. The serine hydroxymethyltransferase-2 (SHMT2) initiates lymphoma development through epigenetic tumor suppressor silencing. Nat Cancer. 2020;1:653–64. doi: 10.1038/s43018-020-0080-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stauffer GV, Plamann MD, Stauffer LT. Construction and expression of hybrid plasmids containing the Escherichia coli glyA genes. Gene. 1981;14:63–72. doi: 10.1016/0378-1119(81)90148-7. [DOI] [PubMed] [Google Scholar]
- 6.Stover PJ, Chen LH, Suh JR, Stover DM, Keyomarsi K, Shane B. Molecular cloning, characterization, and regulation of the human mitochondrial serine hydroxymethyltransferase gene. J Biol Chem. 1997;272:1842–8. doi: 10.1074/jbc.272.3.1842. [DOI] [PubMed] [Google Scholar]
- 7.Girgis S, Nasrallah IM, Suh JR, Oppenheim E, Zanetti KA, Mastri MG. et al. Molecular cloning, characterization and alternative splicing of the human cytoplasmic serine hydroxymethyltransferase gene. Gene. 1998;210:315–24. doi: 10.1016/s0378-1119(98)00085-7. [DOI] [PubMed] [Google Scholar]
- 8.Giardina G, Brunotti P, Fiascarelli A, Cicalini A, Costa MG, Buckle AM. et al. How pyridoxal 5'-phosphate differentially regulates human cytosolic and mitochondrial serine hydroxymethyltransferase oligomeric state. FEBS J. 2015;282:1225–41. doi: 10.1111/febs.13211. [DOI] [PubMed] [Google Scholar]
- 9.Witschel MC, Rottmann M, Schwab A, Leartsakulpanich U, Chitnumsub P, Seet M. et al. Inhibitors of plasmodial serine hydroxymethyltransferase (SHMT): cocrystal structures of pyrazolopyrans with potent blood- and liver-stage activities. J Med Chem. 2015;58:3117–30. doi: 10.1021/jm501987h. [DOI] [PubMed] [Google Scholar]
- 10.Hebbring SJ, Chai Y, Ji Y, Abo RP, Jenkins GD, Fridley B. et al. Serine hydroxymethyltransferase 1 and 2: gene sequence variation and functional genomic characterization. J Neurochem. 2012;120:881–90. doi: 10.1111/j.1471-4159.2012.07646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nikiforov MA, Chandriani S, O'Connell B, Petrenko O, Kotenko I, Beavis A. et al. A functional screen for Myc-responsive genes reveals serine hydroxymethyltransferase, a major source of the one-carbon unit for cell metabolism. Mol Cell Biol. 2002;22:5793–800. doi: 10.1128/MCB.22.16.5793-5800.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garrow TA, Brenner AA, Whitehead VM, Chen XN, Duncan RG, Korenberg JR. et al. Cloning of human cDNAs encoding mitochondrial and cytosolic serine hydroxymethyltransferases and chromosomal localization. J Biol Chem. 1993;268:11910–6. [PubMed] [Google Scholar]
- 13.Pfendner W, Pizer LI. The metabolism of serine and glycine in mutant lines of Chinese hamster ovary cells. Arch Biochem Biophys. 1980;200:503–12. doi: 10.1016/0003-9861(80)90382-3. [DOI] [PubMed] [Google Scholar]
- 14.Wang YW, Zhang SD, Xue WJ, Zhu ML, Zheng LZ. SHMT1 C1420T polymorphism contributes to the risk of non-Hodgkin lymphoma: evidence from 7309 patients. Chin J Cancer. 2015;34:573–82. doi: 10.1186/s40880-015-0065-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bahari G, Hashemi M, Naderi M, Sadeghi-Bojd S, Taheri M. Association of SHMT1 gene polymorphisms with the risk of childhood acute lymphoblastic leukemia in a sample of Iranian population. Cell Mol Biol (Noisy-le-grand) 2016;62:45–51. [PubMed] [Google Scholar]
- 16.Wang Q, Lu K, Du H, Zhang Q, Chen T, Shu Y. et al. Association between cytosolic serine hydroxymethyltransferase (SHMT1) gene polymorphism and cancer risk: a meta-analysis. Biomed Pharmacother. 2014;68:757–62. doi: 10.1016/j.biopha.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 17.Wu H, Bai H, Duan S, Yuan F. Downregulating Serine Hydroxymethyltransferase 2 Deteriorates Hepatic Ischemia-Reperfusion Injury through ROS/JNK/P53 Signaling in Mice. Biomed Res Int. 2019;2019:2712185. doi: 10.1155/2019/2712185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang M, Yuan F, Bai H, Zhang J, Wu H, Zheng K. et al. SHMT2 Promotes Liver Regeneration Through Glycine-activated Akt/mTOR Pathway. Transplantation. 2019;103:e188–e97. doi: 10.1097/TP.0000000000002747. [DOI] [PubMed] [Google Scholar]
- 19.Woo CC, Chen WC, Teo XQ, Radda GK, Lee PT. Downregulating serine hydroxymethyltransferase 2 (SHMT2) suppresses tumorigenesis in human hepatocellular carcinoma. Oncotarget. 2016;7:53005–17. doi: 10.18632/oncotarget.10415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ji L, Tang Y, Pang X, Zhang Y. Increased Expression of Serine Hydroxymethyltransferase 2 (SHMT2) is a Negative Prognostic Marker in Patients with Hepatocellular Carcinoma and is Associated with Proliferation of HepG2 Cells. Med Sci Monit. 2019;25:5823–32. doi: 10.12659/MSM.915754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ning S, Ma S, Saleh AQ, Guo L, Zhao Z, Chen Y. SHMT2 Overexpression Predicts Poor Prognosis in Intrahepatic Cholangiocarcinoma. Gastroenterol Res Pract. 2018;2018:4369253. doi: 10.1155/2018/4369253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei Z, Song J, Wang G, Cui X, Zheng J, Tang Y. et al. Deacetylation of serine hydroxymethyl-transferase 2 by SIRT3 promotes colorectal carcinogenesis. Nat Commun. 2018;9:4468. doi: 10.1038/s41467-018-06812-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyo M, Konno M, Colvin H, Nishida N, Koseki J, Kawamoto K. et al. The importance of mitochondrial folate enzymes in human colorectal cancer. Oncol Rep. 2017;37:417–25. doi: 10.3892/or.2016.5264. [DOI] [PubMed] [Google Scholar]
- 24.Lin C, Zhang Y, Chen Y, Bai Y, Zhang Y. Long noncoding RNA LINC01234 promotes serine hydroxymethyltransferase 2 expression and proliferation by competitively binding miR-642a-5p in colon cancer. Cell Death Dis. 2019;10:137. doi: 10.1038/s41419-019-1352-4. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Liu Y, Yin C, Deng MM, Wang Q, He XQ, Li MT. et al. High expression of SHMT2 is correlated with tumor progression and predicts poor prognosis in gastrointestinal tumors. Eur Rev Med Pharmacol Sci. 2019;23:9379–92. doi: 10.26355/eurrev_201911_19431. [DOI] [PubMed] [Google Scholar]
- 26.Shi H, Fang X, Li Y, Zhang Y. High Expression of Serine Hydroxymethyltransferase 2 Indicates Poor Prognosis of Gastric Cancer Patients. Med Sci Monit. 2019;25:7430–8. doi: 10.12659/MSM.917435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim D, Fiske BP, Birsoy K, Freinkman E, Kami K, Possemato RL. et al. SHMT2 drives glioma cell survival in ischaemia but imposes a dependence on glycine clearance. Nature. 2015;520:363–7. doi: 10.1038/nature14363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Engel AL, Lorenz NI, Klann K, Munch C, Depner C, Steinbach JP. et al. Serine-dependent redox homeostasis regulates glioblastoma cell survival. Br J Cancer. 2020;122:1391–8. doi: 10.1038/s41416-020-0794-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye J, Fan J, Venneti S, Wan YW, Pawel BR, Zhang J. et al. Serine catabolism regulates mitochondrial redox control during hypoxia. Cancer Discov. 2014;4:1406–17. doi: 10.1158/2159-8290.CD-14-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang B, Wang W, Zhu Z, Zhang X, Tang F, Wang D. et al. Mitochondrial serine hydroxymethyltransferase 2 is a potential diagnostic and prognostic biomarker for human glioma. Clin Neurol Neurosurg. 2017;154:28–33. doi: 10.1016/j.clineuro.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 31.Wu M, Wanggou S, Li X, Liu Q, Xie Y. Overexpression of mitochondrial serine hydroxyl-methyltransferase 2 is associated with poor prognosis and promotes cell proliferation and invasion in gliomas. Onco Targets Ther. 2017;10:3781–8. doi: 10.2147/OTT.S130409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K. et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–50. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li AM, Ducker GS, Li Y, Seoane JA, Xiao Y, Melemenidis S. et al. Metabolic Profiling Reveals a Dependency of Human Metastatic Breast Cancer on Mitochondrial Serine and One-Carbon Unit Metabolism. Mol Cancer Res. 2020;18:599–611. doi: 10.1158/1541-7786.MCR-19-0606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang L, Chen Z, Xue D, Zhang Q, Liu X, Luh F. et al. Prognostic and therapeutic value of mitochondrial serine hydroxyl-methyltransferase 2 as a breast cancer biomarker. Oncol Rep. 2016;36:2489–500. doi: 10.3892/or.2016.5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Antonov A, Agostini M, Morello M, Minieri M, Melino G, Amelio I. Bioinformatics analysis of the serine and glycine pathway in cancer cells. Oncotarget. 2014;5:11004–13. doi: 10.18632/oncotarget.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernhardt S, Bayerlova M, Vetter M, Wachter A, Mitra D, Hanf V. et al. Proteomic profiling of breast cancer metabolism identifies SHMT2 and ASCT2 as prognostic factors. Breast Cancer Res. 2017;19:112. doi: 10.1186/s13058-017-0905-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL. et al. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336:1040–4. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yin K. Positive correlation between expression level of mitochondrial serine hydroxymethyltransferase and breast cancer grade. Onco Targets Ther. 2015;8:1069–74. doi: 10.2147/OTT.S82433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li X, Zhang K, Hu Y, Luo N. ERRalpha activates SHMT2 transcription to enhance the resistance of breast cancer to lapatinib via modulating the mitochondrial metabolic adaption. Biosci Rep. 2020. 40. [DOI] [PMC free article] [PubMed]
- 40.Chen L, He J, Zhou J, Xiao Z, Ding N, Duan Y. et al. EIF2A promotes cell survival during paclitaxel treatment in vitro and in vivo. J Cell Mol Med. 2019;23:6060–71. doi: 10.1111/jcmm.14469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Amelio I, Cutruzzola F, Antonov A, Agostini M, Melino G. Serine and glycine metabolism in cancer. Trends Biochem Sci. 2014;39:191–8. doi: 10.1016/j.tibs.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paone A, Marani M, Fiascarelli A, Rinaldo S, Giardina G, Contestabile R. et al. SHMT1 knockdown induces apoptosis in lung cancer cells by causing uracil misincorporation. Cell Death Dis. 2014;5:e1525. doi: 10.1038/cddis.2014.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DeNicola GM, Chen PH, Mullarky E, Sudderth JA, Hu Z, Wu D. et al. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat Genet. 2015;47:1475–81. doi: 10.1038/ng.3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nigdelioglu R, Hamanaka RB, Meliton AY, O'Leary E, Witt LJ, Cho T. et al. Transforming Growth Factor (TGF)-beta Promotes de Novo Serine Synthesis for Collagen Production. J Biol Chem. 2016;291:27239–51. doi: 10.1074/jbc.M116.756247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marrocco I, Altieri F, Rubini E, Paglia G, Chichiarelli S, Giamogante F, Shmt2: A Stat3 Signaling New Player in Prostate Cancer Energy Metabolism. Cells. 2019. 8. [DOI] [PMC free article] [PubMed]
- 46.Song J, Kim D, Chun CH, Jin EJ. miR-370 and miR-373 regulate the pathogenesis of osteoarthritis by modulating one-carbon metabolism via SHMT-2 and MECP-2, respectively. Aging Cell. 2015;14:826–37. doi: 10.1111/acel.12363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee GY, Haverty PM, Li L, Kljavin NM, Bourgon R, Lee J. et al. Comparative oncogenomics identifies PSMB4 and SHMT2 as potential cancer driver genes. Cancer Res. 2014;74:3114–26. doi: 10.1158/0008-5472.CAN-13-2683. [DOI] [PubMed] [Google Scholar]
- 48.Tong J, Krieger JR, Taylor P, Bagshaw R, Kang J, Jeedigunta S. et al. Cancer proteome and metabolite changes linked to SHMT2. PLoS One. 2020;15:e0237981. doi: 10.1371/journal.pone.0237981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haggerty TJ, Zeller KI, Osthus RC, Wonsey DR, Dang CV. A strategy for identifying transcription factor binding sites reveals two classes of genomic c-Myc target sites. Proc Natl Acad Sci U S A. 2003;100:5313–8. doi: 10.1073/pnas.0931346100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dong Y, Huo X, Sun R, Liu Z, Huang M, Yang S. lncRNA Gm15290 promotes cell proliferation and invasion in lung cancer through directly interacting with and suppressing the tumor suppressor miR-615-5p. Biosci Rep. 2018. 38. [DOI] [PMC free article] [PubMed]
- 51.Qi C, Qin X, Zhou Z, Wang Y, Yang Q, Liao T. Circ_0072995 Promotes Cell Carcinogenesis via Up-Regulating miR-149-5p-Mediated SHMT2 in Breast Cancer. Cancer Manag Res. 2020;12:11169–81. doi: 10.2147/CMAR.S272274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang X, Wang Z, Li X, Liu B, Liu M, Liu L. et al. SHMT2 Desuccinylation by SIRT5 Drives Cancer Cell Proliferation. Cancer Res. 2018;78:372–86. doi: 10.1158/0008-5472.CAN-17-1912. [DOI] [PubMed] [Google Scholar]
- 53.Sadhukhan S, Liu X, Ryu D, Nelson OD, Stupinski JA, Li Z. et al. Metabolomics-assisted proteomics identifies succinylation and SIRT5 as important regulators of cardiac function. Proc Natl Acad Sci U S A. 2016;113:4320–5. doi: 10.1073/pnas.1519858113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rardin MJ, He W, Nishida Y, Newman JC, Carrico C, Danielson SR. et al. SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell Metab. 2013;18:920–33. doi: 10.1016/j.cmet.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cao J, Sun L, Aramsangtienchai P, Spiegelman NA, Zhang X, Huang W. et al. HDAC11 regulates type I interferon signaling through defatty-acylation of SHMT2. Proc Natl Acad Sci U S A. 2019;116:5487–92. doi: 10.1073/pnas.1815365116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng H, Gupta V, Patterson-Fortin J, Bhattacharya S, Katlinski K, Wu J. et al. A BRISC-SHMT complex deubiquitinates IFNAR1 and regulates interferon responses. Cell Rep. 2013;5:180–93. doi: 10.1016/j.celrep.2013.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walden M, Tian L, Ross RL, Sykora UM, Byrne DP, Hesketh EL. et al. Metabolic control of BRISC-SHMT2 assembly regulates immune signalling. Nature. 2019;570:194–9. doi: 10.1038/s41586-019-1232-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang WC, Shyh-Chang N, Yang H, Rai A, Umashankar S, Ma S. et al. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell. 2012;148:259–72. doi: 10.1016/j.cell.2011.11.050. [DOI] [PubMed] [Google Scholar]
- 59.He L, Bao J, Yang Y, Dong S, Zhang L, Qi Y. et al. Study of SHMT2 Inhibitors and Their Binding Mechanism by Computational Alanine Scanning. J Chem Inf Model. 2019;59:3871–8. doi: 10.1021/acs.jcim.9b00370. [DOI] [PubMed] [Google Scholar]
- 60.Scaletti E, Jemth AS, Helleday T, Stenmark P. Structural basis of inhibition of the human serine hydroxymethyltransferase SHMT2 by antifolate drugs. FEBS Lett. 2019;593:1863–73. doi: 10.1002/1873-3468.13455. [DOI] [PubMed] [Google Scholar]
- 61.Paiardini A, Tramonti A, Schirch D, Guiducci G, di Salvo ML, Fiascarelli A. et al. Differential 3-bromopyruvate inhibition of cytosolic and mitochondrial human serine hydroxymethyltransferase isoforms, key enzymes in cancer metabolic reprogramming. Biochim Biophys Acta. 2016;1864:1506–17. doi: 10.1016/j.bbapap.2016.08.010. [DOI] [PubMed] [Google Scholar]
- 62.Dekhne AS, Shah K, Ducker GS, Katinas JM, Wong-Roushar J, Nayeen MJ. et al. Novel Pyrrolo[3,2-d]pyrimidine Compounds Target Mitochondrial and Cytosolic One-carbon Metabolism with Broad-spectrum Antitumor Efficacy. Mol Cancer Ther. 2019;18:1787–99. doi: 10.1158/1535-7163.MCT-19-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Calvo SE, Clauser KR, Mootha VK. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2016;44:D1251–7. doi: 10.1093/nar/gkv1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rath S, Sharma R, Gupta R, Ast T, Chan C, Durham TJ. et al. MitoCarta3.0: an updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res. 2021;49:D1541–D7. doi: 10.1093/nar/gkaa1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ducker GS, Ghergurovich JM, Mainolfi N, Suri V, Jeong SK, Hsin-Jung Li S. et al. Human SHMT inhibitors reveal defective glycine import as a targetable metabolic vulnerability of diffuse large B-cell lymphoma. Proc Natl Acad Sci U S A. 2017;114:11404–9. doi: 10.1073/pnas.1706617114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]