Abstract
Background: Collagen type 1 alpha 1 chain (COL1A1) is an extracellular matrix protein comprising two alpha 1 chains and one alpha 2 chain. Our previous study identified that COL1A1 is the key gene during the development and progression of lung adenocarcinoma by multi-omics analysis. However, the clinical significance of COL1A1 expression in lung cancer samples remains largely unknown. Here, we aimed to evaluate the level of COL1A1 in lung cancer samples and correlate its level with the clinical outcome.
Methods:COL1A1 gene expression in lung cancer samples was analyzed using the Oncomine database (www.oncomine.org). A total of 308 lung cancer samples (208 formalin-fixed paraffin-embedded tissues and 100 blood samples) were assessed for protein expression of COL1A1. Immunohistochemistry staining and enzyme-linked immunosorbent assay were used to detect COL1A1 expression in tissues and serum, respectively.
Results: We identified an elevation of COL1A1 in mRNA level and gene amplification in lung cancer tissues compared with normal lung tissues. High COL1A1 expression was observed in lung cancer tissues and serum (P < 0.05), it was significantly correlated with the peripheral type tumor, the larger diameter of the tumor, the occurrence of lymph node metastases and distant metastases, a higher TNM stage, and smoking (P < 0.05). High COL1A1 expression was associated with poor progression-free survival (PFS) and chemoresistance in lung cancer patients (P < 0.05). Multivariable Cox-regression analysis showed that COL1A1 expression was an independent prognostic factor (P < 0.05). Furthermore, the area under the receiver operating characteristic (AUC) curve was 0.909 for the combined COL1A1 and carcinoembryonic antigen (CEA) measurement.
Conclusion: Our findings revealed that COL1A1 could be used as a novel diagnostic, prognostic, and chemoresistance biomarker of human lung cancer, and these results provide a potential therapeutic strategy for lung cancer patients.
Keywords: Extracellular matrix protein, COL1A1, Chemoresistance, Metastatic, Lung cancer
Introduction
Lung cancer is one of the most common malignant tumors and the leading cause of cancer deaths worldwide 1. Genetic factors and environmental exposure impact the development and progression of this complex disease. Smoking and various susceptibility genes associated with lung cancer have been considered a possible mechanism for the development of lung cancer 2. However, the biological risk factors of lung cancer aggressiveness and chemoresistance remain elusive. In our previous research, we identified collagen type 1 alpha 1 chain (COL1A1) as the key gene during the development and progression of lung adenocarcinoma (LUAD) by multi-omics analysis 3.
COL1A1 is a component of the extracellular matrix (ECM), and its gene is located on chromosome 17 (17q21.23) and contains the myopia 5 (MYP517q21-22) locus 4, 5. This gene encodes the pro-alpha 1 chain of type I collagen, which has a triple helix comprising two alpha 1 chains and one alpha 2 chain 6. Specifically, COL1A1 mutations can lead to collagen-related diseases, including autosomal dominant osteogenesis imperfecta (OI) and the rare arthrochalasia subtype of Ehlers-Danlos syndrome (EDS) 7. Recently, it has been shown that a high level of COL1A1 is indicative of a more aggressive cellular behavior and poorer prognosis in patients with breast cancer, especially in those with ER+ breast cancer 8. Furthermore, COL1A1 expression was correlated with the key genes of the WNT/PCP signaling pathway, which may promote colorectal cancer cell migration 9. In mouse models, COL1A1 was frequently upregulated in gastric cancer tissues; it increased cell proliferation, colony-forming efficiency, migration ability, and invasion ability, while it promoted the development of grafted tumors in mice 10. However, little is known about the clinical role of COL1A1 in lung cancer.
For decades, despite advances in targeted therapies and immunotherapies, platinum-based chemotherapy has remained the standard first-line treatment for most advanced lung cancer 11. Intrinsic or acquired resistance to chemotherapy is the major cause of therapeutic failure 12. There is currently no available method to identify intrinsically or acquired chemoresistance in patients at baseline. Carcinoembryonic antigen (CEA) is a serum glycoprotein. However, it is widely used as a validated biomarker for disease monitoring and assessing the response and relapse in lung cancer; the results often lack specificity and sensitivity 13, 14. A previous study found that stellate cell-secreted COL1A1 can promote invasion and migration of pancreatic cancer cells, which is also associated with chemoresistance 15.
In the present study, we found that the COL1A1 level was significantly elevated in lung cancer tissues and serum samples, and it was closely associated with poor PFS and chemoresistance of lung cancer. These findings suggest that COL1A1 might be a candidate diagnostic, prognostic, and chemoresistance biomarker for lung cancer patients.
Materials and methods
Materials
Anti-Collagen I antibody (ab34710) was purchased from Abcam PLC (Cambridge, UK). Human COL1A1 enzyme-linked immunosorbent assay (ELISA) kit was purchased from Beijing Dogesce Biological Technology, Ltd. (Beijing, People's Republic of China).
Methods
Analysis of Oncomine data
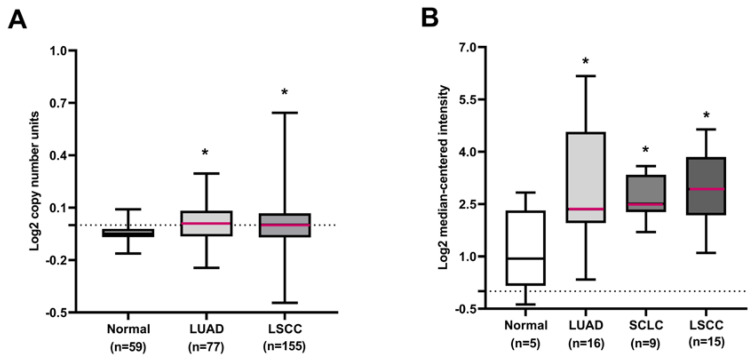
To determine the expression pattern of COL1A1 in lung cancer, we used the datasets in Oncomine (https://www.oncomine.org), a cancer microarray database, and an online data-mining platform, aimed at promoting discovery from genome-wide expression analyses 16. The data cohorts of Weiss et al. deposited in Oncomine.com, which includes 155 lung squamous cell carcinomas (LSCC), 77 LUAD, and 59 normal lung tissues, were analyzed for gene copy number 17. The study of Rohrbeck et al., which included 16 LUAD, 9 small cell lung cancer (SCLC), and 5 normal lung tissues, analyzed the mRNA level 18. The data were displayed using a Box chart.
Human specimens
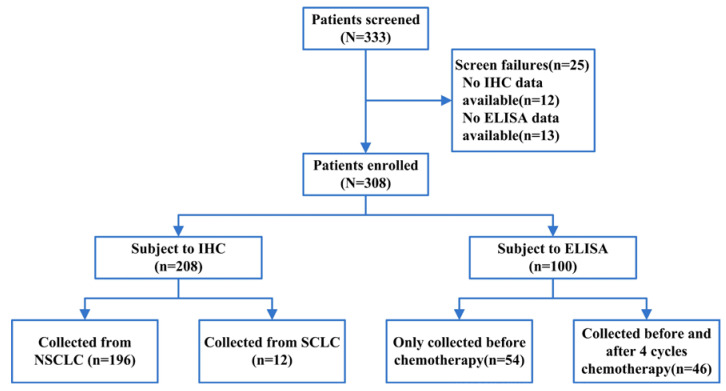
Gene expression analysis was successful in 308 biopsy specimens with full clinical data out of a total of 333 trial participants (Figure 1). A total of 208 formalin-fixed paraffin-embedded human lung cancer specimens were obtained from the Harbin Medical University Cancer Hospital. Human solid lung cancer specimens were collected from January 2018 to July 2019. All patients had histologically confirmed lung cancer with information on the histologic type and tumor stage (AJCC, The American Joint Committee on Cancer). This study, involving human lung tumor specimens, was approved by the Institutional Review Board of the Harbin Medical University Cancer Hospital.
Figure 1.
The flowchart of enrolled patients.
Blood samples were obtained from 100 patients with advanced lung cancer from January 2018 to July 2019. Serum samples of 54 patients were collected before chemotherapy, and the samples of 46 patients were collected before and after four cycles of chemotherapy. In addition, 40 serum samples donated voluntarily by healthy individuals were used as a control group during the study period. Fasting peripheral venous blood (5 ml) was collected from all participants in the morning.
Immunohistochemistry (IHC) and hematoxylin and eosin (H&E) staining
Paraffin sections were then stained with the avidin-biotin complex IHC stain following a standard staining protocol to detect the expression of COL1A1. After endogenous peroxidase was quenched with 3% hydrogen peroxide and blocked for 10 min, sections were incubated overnight with the primary antibody at 4°C. Rabbit primary monoclonal antibodies directed against COL1A1 (1:100 dilution; Abcam) were used in the experiment. Biotinylated immunoglobulin and peroxidase-conjugated streptavidin were then added. Finally, 3, 3'-diaminobenzidine was added for color development, and hematoxylin was used for counterstaining. The staining intensity and extent scored protein expression levels. Tissues with no staining were rated as 0, those with faint staining or moderate to strong staining in 10% of cells were rated as 1, those with moderate staining or strong staining in 10%-50% of cells were rated as 2, and those with strong staining in > 50% of cells were rated as 3. Cases that registered levels 0 and 1 were considered negative, and those that registered levels 2 and 3 were defined as positive 19. H&E staining was used to indicate the pathology.
ELISA
COL1A1 concentration was determined using an ELISA method. The assays were performed according to the manufacturer's protocols. Furthermore, 100 μl of HRP-conjugate reagent was added to the serum samples from cancer patients and healthy controls, covered with an adhesive strip, and incubated for 60 min at 37°C. Each well was aspirated and washed, and it was washed by filling with Wash Solution (400 μl) using a squirt bottle, manifold dispenser, or auto washer. Samples of 50 μl were added to each well. They were gently mixed and incubated for 15 min at 37°C, and they were protected from light. Finally, 50 μl Stop Solution was added to each well. All of the samples were examined three times, and the median values were used for statistical analysis.
Assessment and statistical analysis
All of the patients had a follow-up ≥ 12 months. PFS was defined as the interval from the initial chemotherapy to disease progression or death, without any evidence of progression. Serum samples were divided into the chemosensitive (partial or complete response) and chemoresistant (progressive disease) groups based on the Response Evaluation Criteria in Solid Tumors (RECIST Edition 1.1).
All of the data were analyzed with Statistical Package for the Social Sciences version 24 software (SPSS Inc., Chicago, IL, USA). The intragroup expression difference detected by the IHC assay was analyzed by the rank-sum test. We analyzed the association between COL1A1 and clinicopathologic parameters using the independent t-test, Mann-Whitney test, or Kruskal-Wallis test as appropriate. The paired t-test was used for ELISA group analysis. Receiver operator characteristic (ROC) curves were created to determine the predictive value of COL1A1 and CEA to distinguish between lung cancer patients and controls. Survival curves for PFS were estimated using the Kaplan-Meier method, and the differences were evaluated with the log-rank test. Univariate and multivariate analyses of the PFS were performed using Cox-regression analysis. Continuous variables were summarized using mean and standard deviation. Statistical significance was set at P < 0.05.
Results
COL1A1 gene amplification and elevation at mRNA levels in lung cancer
To assess the clinical relevance of COL1A1 in human primary lung cancers, the expression profile of COL1A1 was characterized by bioinformatics analysis of public datasets from Oncomine. The results showed that the COL1A1 gene copy number increased in lung cancer tissues compared to normal lung tissues in the Weiss lung dataset (Figure 2A). We also observed upregulation of the COL1A1 mRNA level in lung cancer tissues compared with normal lung tissues in the Rohrbeck Lung dataset (Figure 2B). Additionally, Merry et al. identified that a characteristic amplification of sequences from chromosome 17q, demarcated by the COL1A1 gene, was associated with elevated expression of the COL1A1 in dermatofibrosarcoma protuberans (DFSP) 20. These observations suggested that the COL1A1 protein may be highly expressed in lung cancer tissues.
Figure 2.
COL1A1 DNA or mRNA expression in human lung cancers using the Oncomine database. (A) The COL1A1 gene copy number in normal lung tissues and different histological subtypes of lung cancer. (B) COL1A1 mRNA expression in normal lung tissues and different histological subtypes of lung cancer. *P < 0.05 compared with the normal group.
COL1A1 expression in lung cancer tissues by IHC
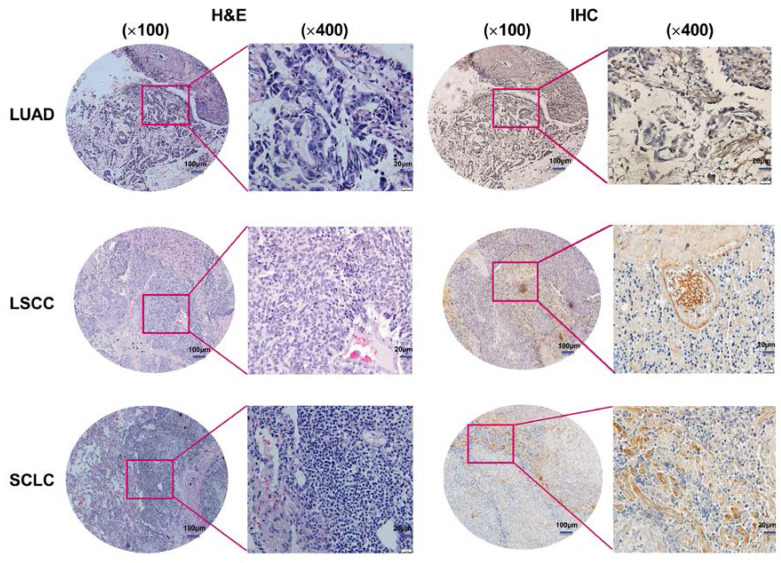
To examine the correlation between COL1A1 expression and clinicopathologic factors in lung cancer, we evaluated its expression in lung cancer tissues by IHC. The results showed that 110 of the 208 primary lesions exhibited positive staining for COL1A1, but there was no staining of the adjacent normal tissues (Figure 3). Statistical analysis revealed that higher COL1A1 expression were significantly correlated with the peripheral type tumor (P = 0.001), larger diameter of the tumor (P = 0.009), occurrence of lymph node metastases (P = 0.009) and distant metastases (P < 0.001), higher TNM stage (III, IV) (P = 0.006), and smoking (P < 0.001) (Table 1). However, no statistically significant correlations were identified between the expression of COL1A1 and other clinicopathologic parameters, including local invasion and pathologic type.
Figure 3.
COL1A1 expression in lung cancer tissues. Representative H&E and IHC staining of COL1A1 in LUAD, LSCC, and SCLC patients (magnification×100, ×400). Immunoreactivity was observed in the malignant cell-extracellular matrix.
Table 1.
Correlation between the COL1A1 expression and clinicopathologic parameters in lung cancer tissues.
| Cases (n) | COL1A1 average rank | Statistical test | P-value | |
|---|---|---|---|---|
| Location | ||||
| Central type | 108 | 93.15 | ||
| Peripheral type | 100 | 116.76 | Mann-Whitney test | 0.001 |
| Tumor diameter | ||||
| d≤3cm | 71 | 90.41 | ||
| 3cm<d≤7cm | 113 | 109.42 | ||
| d>7cm | 24 | 123.00 | Kruskal-Wallis test | 0.009 |
| Lymph node metastasis | ||||
| No | 66 | 90.70 | ||
| Yes | 142 | 110.92 | Independent t-test | 0.009 |
| Local invasion | ||||
| T1-T2 | 184 | 102.09 | ||
| T3-T4 | 24 | 123.00 | Mann-Whitney test | 0.062 |
| Distant metastasis | ||||
| No | 82 | 70.37 | ||
| Yes | 126 | 126.71 | Independent t-test | <0.001 |
| Pathologic type | ||||
| LSCC | 106 | 102.89 | ||
| LUAD | 90 | 108.56 | ||
| SCLC | 12 | 88.33 | Kruskal-Wallis test | 0.399 |
| Stage | ||||
| Ⅰ-Ⅱ | 61 | 89.33 | ||
| Ⅲ-IV | 147 | 110.80 | Mann-Whitney test | 0.006 |
| Smoking | ||||
| No | 110 | 80.93 | ||
| Yes | 98 | 130.96 | Independent t-test | <0.001 |
Elevated serum levels of COL1A1 in lung cancer patients
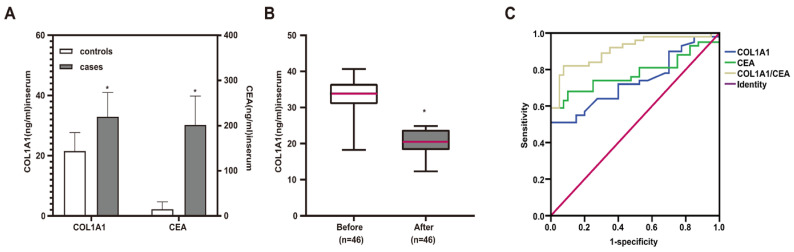
To investigate the potential of COL1A1 as a serological marker for lung cancer, we analyzed the level of COL1A1 in serum from lung cancer patients (n = 100) and healthy individuals (n = 40) by ELISA. The result showed that the serum levels of COL1A1 in lung cancer patients were significantly higher than those in the healthy group (32.90 ± 8.08 ng/ml vs. 21.51 ± 6.16 ng/ml, P < 0.001) (Figure 4A). In addition, the serum levels of COL1A1 in lung cancer patients before chemotherapy were significantly higher than in patients after four cycles of chemotherapy (32.67 ± 5.84 ng/ml vs. 19.98 ± 3.95 ng/ml, P < 0.001) (Figure 4B).
Figure 4.
Elevated COL1A1 levels in lung cancer serum samples. (A) Comparison of serum COL1A1 and CEA levels between lung cancer patients and healthy individuals. *P < 0.05 healthy individuals vs. lung cancer patients (independent sample t-test) (B) The serum levels of COL1A1 before and after four cycles of chemotherapy in lung cancer patients. *P < 0.05 before chemotherapy vs. after four cycles of chemotherapy (paired t-test) (C) ROC curves for the serum COL1A1, CEA, and the combination of COL1A1 and CEA in the diagnosis of lung cancer.
Then, we analyzed the association between the levels of COL1A1 in the serum of lung cancer patients and clinicopathologic parameters. As shown in Table 2, the higher concentrations of COL1A1 were significantly correlated with the peripheral type tumor (P < 0.001), larger diameter of the tumor (P = 0.028), occurrence of lymph node metastases (P < 0.001) and distant metastases (P = 0.001), higher TNM stage (III, IV) (P = 0.049), and smoking (P < 0.001).
Table 2.
Correlation between the COL1A1 level and clinicopathologic parameters in the serum of lung cancer patients.
| Cases (n) | COL1A1(ng/ml) | Statistical test | P-value | |
|---|---|---|---|---|
| Location | ||||
| Central type | 39 | 31.27±7.18 | ||
| Peripheral type | 61 | 33.78±8.19 | Mann-Whitney test | <0.001 |
| Tumor diameter | ||||
| d≤3cm | 45 | 30.55±6.95 | ||
| 3cm<d≤7cm | 43 | 34.49±10.53 | ||
| d>7cm | 12 | 35.40±5.76 | Kruskal-Wallis test | 0.028 |
| Lymph node metastasis | ||||
| No | 30 | 29.31±5.82 | ||
| Yes | 70 | 34.51±7.65 | Independent t-test | <0.001 |
| Local invasion | ||||
| T1-T2 | 82 | 33.89±6.67 | ||
| T3-T4 | 18 | 35.58±8.62 | Mann-Whitney test | 0.424 |
| Distant metastasis | ||||
| No | 24 | 30.63±8.04 | ||
| Yes | 76 | 33.16±7.55 | Mann-Whitney test | 0.001 |
| Pathologic type | ||||
| LSCC | 15 | 32.90±5.52 | ||
| LUAD | 64 | 34.27±7.41 | ||
| SCLC | 21 | 31.93±8.41 | Kruskal-Wallis test | 0.790 |
| Stage | ||||
| Ⅰ-Ⅱ | 12 | 28.46±6.63 | ||
| Ⅲ-IV | 88 | 34.07±4.61 | Independent t-test | 0.049 |
| Smoking | ||||
| No | 49 | 30.94±7.35 | ||
| Yes | 51 | 35.28±8.18 | Independent t-test | <0.001 |
The diagnostic value of COL1A1 as a serologic biomarker for lung cancer patients
To further evaluate the potential clinical utility of COL1A1 only or combined with CEA as the diagnostic serum marker, we employed the ROC curve analysis. CEA being a common serum marker, its serum levels in patients with lung cancer (30.19 ± 9.64 ng/ml) were higher than those in healthy volunteers (2.20 ± 2.47 ng/ml, P < 0.001) (Figure 4A). As shown in Figure 4C, the AUC of COL1A1 was 0.737 (95% CI: 0.656-0.817), although it did not outperform the serum CEA (AUC = 0.780, 95% CI: 0.706-0.854). The combined examination of COL1A1 and CEA had a better discriminating potential (AUC = 0.909, 95% CI: 0.861-0.957).
Overexpression of COL1A1 correlates with poor prognosis in lung cancer patients
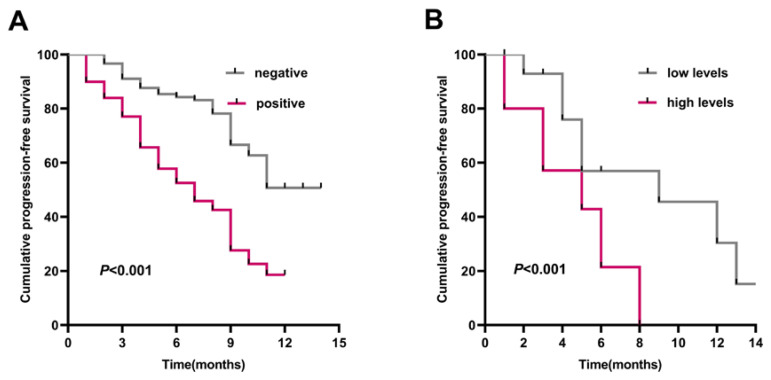
To determine the correlation between COL1A1 expression and clinical outcome, we analyzed the prognostic significance of COL1A1 using Kaplan-Meier analysis. In the IHC group, the PFS time of lung cancer patients with COL1A1-positive expression was markedly shorter than in the patients with COL1A1-negative expression (Figure 5A). In the ELISA group, a cutoff value corresponding to the median of COL1A1 was used to separate the patients into two groups: those with high serum levels (≥28.46 ng/ml) versus those with low serum levels (<28.46 ng/ml). Patients with low serum COL1A1 levels had a median PFS of 9 months compared to patients with high serum COL1A1 levels who had a PFS of 5 months (P < 0.001, log-rank test) (Figure 5B).
Figure 5.
The prognostic value of COL1A1 expression. Kaplan-Meier survival curves comparing lung cancer patients with high and low expression of COL1A1. (A) PFS survival curves of lung cancer patients in tissue samples. (B) PFS survival curves of lung cancer patients in blood samples.
Moreover, the subsequent univariate and multivariate Cox-regression model showed that the positive of COL1A1 (HR = 2.696; 95% CI: 1.827-3.980; P < 0.001), occurrence of lymph node metastases (HR = 4.586; 95% CI: 2.916-7.213; P < 0.001), higher local invasion at stage T3-T4 (HR = 3.483; 95% CI: 2.025-5.990; P < 0.001), and smoking (HR = 1.488; 95% CI: 1.039-2.132; P = 0.030) were statistically independent predictive factors of poorer prognosis for lung cancer patients (Table 3). The chemotherapy regimens for all patients with lung cancer are shown in Table 4.
Table 3.
Univariate and multivariate analyses of Cox proportional hazard regression model for PFS in lung cancer patients.
| Pathologic parameters | PFS | ||
|---|---|---|---|
| HR | 95% CI | P-value | |
| Univariate analysis | |||
| COL1A1 | |||
| Positive vs Negative | 2.732 | 1.863-4.006 | <0.001 |
| Location | |||
| Central type vs Peripheral type | 0.965 | 0.679-1.371 | 0.841 |
| Tumor diameter | |||
| d≤3cm vs 3cm<d≤7cm vs d>7cm | 1.849 | 1.380-2.476 | <0.001 |
| Lymph node metastasis | |||
| Yes vs No | 4.481 | 2.823-7.111 | <0.001 |
| Local invasion | |||
| T1-T2 vs T3-T4 | 2.470 | 1.476-4.136 | 0.001 |
| Distant metastasis | |||
| Yes vs No | 1.828 | 1.259-2.655 | 0.002 |
| Pathologic type | |||
| LSCC vs LUAD vs SCLC | 0.997 | 0.754-1.317 | 0.981 |
| Stage | |||
| Ⅰ-Ⅱ vs Ⅲ-IV | 4.340 | 2.691-6.998 | <0.001 |
| Smoking | |||
| Yes vs No | 1.434 | 1.007-2.042 | 0.046 |
| Multivariate analysis | |||
| COL1A1 | |||
| Positive vs Negative | 2.696 | 1.827-3.980 | <0.001 |
| Tumor diameter | |||
| d≤3cm vs 3cm<d≤7cm vs d>7cm | 1.400 | 0.913-2.145 | 0.205 |
| Lymph node metastasis | |||
| Yes vs No | 4.586 | 2.916-7.213 | <0.001 |
| Local invasion | |||
| T1-T2 vs T3-T4 | 3.483 | 2.025-5.990 | <0.001 |
| Distant metastasis | |||
| Yes vs No | 1.526 | 0.977-2.384 | 0.099 |
| Stage | |||
| Ⅰ-Ⅱ vs Ⅲ-IV | 0.536 | 0.211-1.361 | 0.293 |
| Smoking | |||
| Yes vs No | 1.488 | 1.039-2.132 | 0.030 |
Table 4.
Chemotherapy regimens for lung cancer patients.
| Type of lung cancer | Chemotherapy regimens |
|---|---|
| NSCLC | Pemetrexed + cisplatin |
| Gemcitabine + cisplatin | |
| Paclitaxel + cisplatin | |
| Vinorelbine + cisplatin | |
| Docetaxel + cisplatin | |
| Pemetrexed + carboplatin | |
| Gemcitabine + carboplatin | |
| Paclitaxel + carboplatin | |
| Gemcitabine | |
| Docetaxel | |
| SCLC | Etoposide + cisplatin |
| Etoposide + carboplatin | |
| Irinotecan + cisplatin | |
| Paclitaxel | |
| Docetaxel | |
| Gemctabine |
Discriminating potential of serum COL1A1 to identify lung cancer patients with intrinsic chemoresistance
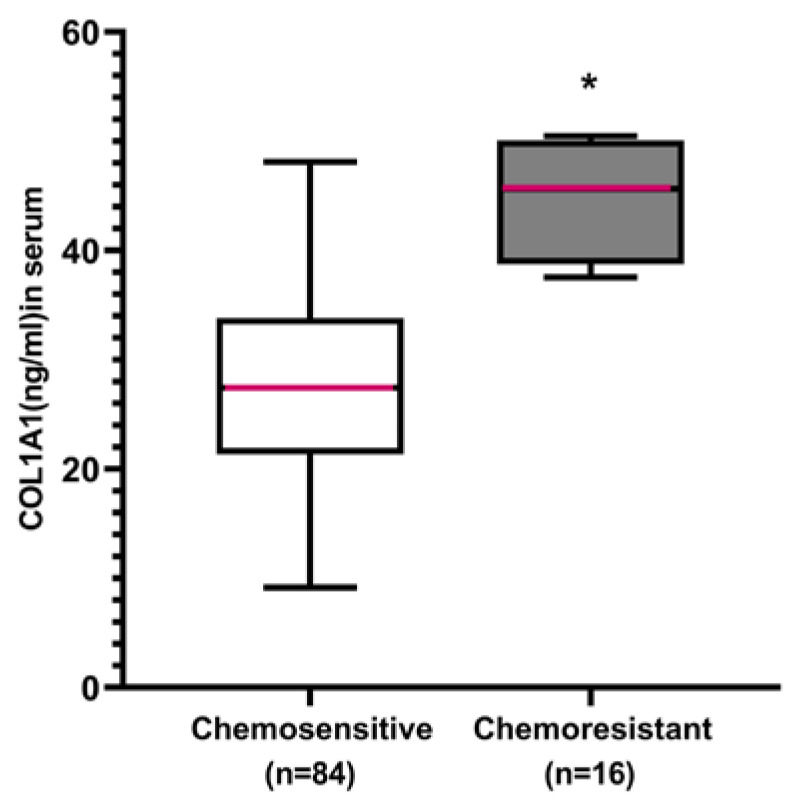
To further evaluate the clinical utility of COL1A1 as the intrinsic chemoresistance serum marker, we analyzed the serum COL1A1 levels in 100 lung cancer patients using ELISA. Of the 100 patients, 84 were chemosensitive and 16 were chemoresistant. Serum COL1A1 levels were significantly lower in the chemo-sensitive patients (28.68 ± 9.03 ng/ml) compared with the chemoresistant patients (44.82 ± 5.35 ng/ml, P < 0.001) (Figure 6).
Figure 6.
The potential of serum COL1A1 to identify lung cancer patients with intrinsic chemoresistance. Box plots representing the serum level of COL1A1 in lung cancer patients with chemoresistance and chemosensitivity to first-line chemotherapy. *P < 0.05 chemosensitive group vs. chemoresistance group (independent sample t-test). All experiments were repeated three times.
Discussion
It is challenging to explore a specific biomarker for the diagnostic, prognosis, and intrinsic chemoresistance of lung cancer. Based on our previous bioinformatics analysis, genetic alterations of the extracellular matrix protein COL1A1 are likely to be a frequent event in human lung cancers. Currently, in lung cancer, the clinical role of COL1A1 is not sufficiently understood. In our experimental data, COL1A1 was highly expressed in human lung cancer tissues and was correlated with some clinicopathologic parameters, such as the occurrence of lymph nodes and distant metastases. Our ELISA analysis revealed that the clinical significance of the serum level of COL1A1 in lung cancer patients was consistent with lung cancer tissues. Furthermore, our statistical analysis revealed that COL1A1 was associated with poor prognosis and chemoresistance of lung cancer. These data suggest that COL1A1 can be used as a potential biomarker in the diagnosing, prognosis, and chemoresistance of human lung cancer.
COL1A1 is one of the most abundant proteins in the human body. It is essential to maintain the structure of bone, skin, tendons, cornea, blood vessel walls, and other connective tissues 21. In addition, COL1A1 promotes embryonic stem cell proliferation and contributes to increased extracellular environment stiffness in the vertebrates 22. It is widely accepted that COL1A1 is produced by stromal fibroblasts 23. COL1A1 is associated with response to hypoxia and oxidative stress, which may help the cells adapt to oxidative stress and hypoxia, thus promoting tumor cell growth and metastasis 24. Currently, in lung cancer cells, the role of COL1A1 is not sufficiently understood. COL1A1 is implicated in the induction of epithelial-mesenchymal transition (EMT) through the transforming growth factor (TGF)-β-dependent pathway in non-SCLC cell lines 25, 26. Therefore, blocking TGF-β signaling may be an effective treatment to prevent cancer progression in patients with non-small-cell lung cancer (NSCLC). Our data showed that COL1A1 was highly expressed in lung cancer specimens (52.8%), which is in line with the previous reports on gastric cancer 27.
Moreover, COL1A1 was closely associated with the overall survival of cancer patients. Boguslawska et al. found that TGF-β1 induced the expression of COL1A1 in a renal cell carcinoma-derived cell line, and disturbed expression of COL1A1 occurred early during renal cell carcinoma carcinogenesis and correlated with poor survival 28. Likewise, COL1A1 has also been reported to be one component of hepatic fibrosis, and COL1A1 gene methylation might be correlated with the poor clinical outcomes of hepatocellular carcinoma 29. In our study, higher COL1A1 levels were significantly associated with shorter PFS, and overexpression of COL1A1 may predict a poor prognosis in advanced lung cancer patients.
However, because of chemoresistance, lung cancer has a low five-year survival rate and a high fatality rate, leading to chemotherapy failure 30. It is critical to identify the patients who would benefit from chemotherapy. A recent study revealed that activated fibroblasts play a prominent role in lung carcinogenesis, and cancer-associated fibroblasts are responsible for deposition of extracellular matrix 31. Consequently, proteins related to the extracellular matrix are released into the blood and could be considered potential circulating biomarkers 32. Cisplatin is a cell-cycle nonspecific agent widely used in the first-line chemotherapy of lung cancer patients 33. MiR-29b-3p can reverse cisplatin resistance by inhibiting the expression of the COL1A1 gene in NSCLC cell lines 34. In our study, COL1A1 could be used as a predictive biomarker for chemoresistant patients with advanced lung cancer in first-line chemotherapy.
In conclusion, COL1A1 can be used as a novel diagnostic, prognostic, and chemoresistance biomarker for lung cancer. These findings provide novel directions for studying the precise molecular mechanism of the COL1A1 gene and new strategies to develop potential COL1A1-associated anti-lung cancer drugs.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 81903070), the Natural Science Foundation of Heilongjiang Province (No. LH2019H039), the Haiyan Foundation of Harbin Medical University Cancer Hospital (No. JJZD2020-09), the Health Commission Foundation of Heilongjiang Province (2018247), the Jilin Science and Technology Agency funding (20190103099JH), and the Fundamental Research Funds for the Heilongjiang Provincial Universities (2019-KYYWF-0360).
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Yang D, Liu Y, Bai C, Wang X, Powell CA. Epidemiology of lung cancer and lung cancer screening programs in China and the United States. Cancer Lett. 2020;468:82–87. doi: 10.1016/j.canlet.2019.10.009. [DOI] [PubMed] [Google Scholar]
- 3.Yang Z, Liu B, Lin T, Zhang Y, Zhang L, Wang M. Multiomics analysis on DNA methylation and the expression of both messenger RNA and microRNA in lung adenocarcinoma. J Cell Physiol. 2019;234(5):7579–7586. doi: 10.1002/jcp.27520. [DOI] [PubMed] [Google Scholar]
- 4.Sun S, Wang Y, Wu Y, Gao Y, Li Q, Abdulrahman AA. et al. Identification of COL1A1 as an invasion-related gene in malignant astrocytoma. Int J Oncol. 2018;53(6):2542–2554. doi: 10.3892/ijo.2018.4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin GM, Zhao XJ, Chen AM, Chen YX, Li Q. Association of COL1A1 polymorphism with high myopia: a Meta-analysis. Int J Ophthalmol. 2016;9(4):604–9. doi: 10.18240/ijo.2016.04.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maasalu K, Nikopensius T, Kõks S, Nõukas M, Kals M, Prans E. et al. Whole-exome sequencing identifies de novo mutation in the COL1A1 gene to underlie the severe osteogenesis imperfecta. Hum Genomics. 2015;9(1):6. doi: 10.1186/s40246-015-0028-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin Z, Zeng J, Wang X. Compound phenotype of osteogenesis imperfecta and Ehlers-Danlos syndrome caused by combined mutations in COL1A1 and COL5A1. Biosci Rep. 2019;39(7):BSR20181409. doi: 10.1042/BSR20181409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J, Shen JX, Wu HT, Li XL, Wen XF, Du CW. et al. Collagen 1A1 (COL1A1) promotes metastasis of breast cancer and is a potential therapeutic target. Discov Med. 2018;25(139):211–223. [PubMed] [Google Scholar]
- 9.Zhang Z, Wang Y, Zhang J, Zhong J, Yang R. COL1A1 promotes metastasis in colorectal cancer by regulating the WNT/PCP pathway. Mol Med Rep. 2018;17(4):5037–5042. doi: 10.3892/mmr.2018.8533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong XZ, Zhao ZR, Hu Y, Lu YP, Liu P, Zhang L. LncRNA COL1A1-014 is involved in the progression of gastric cancer via regulating CXCL12-CXCR4 axis. Gastric Cancer. 2020;23(2):260–272. doi: 10.1007/s10120-019-01011-0. [DOI] [PubMed] [Google Scholar]
- 11.Rossi A, Di Maio M. Platinum-based chemotherapy in advanced non-small-cell lung cancer: optimal number of treatment cycles. Expert Rev Anticancer Ther. 2016;16(6):653–60. doi: 10.1586/14737140.2016.1170596. [DOI] [PubMed] [Google Scholar]
- 12.Tan LM, Li X, Qiu CF, Zhu T, Hu CP, Yin JY. et al. CLEC4M is associated with poor prognosis and promotes cisplatin resistance in NSCLC patients. J Cancer. 2019;10(25):6374–6383. doi: 10.7150/jca.30139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J, Chu Y, Li J, Zeng F, Wu M, Wang T. et al. Development of a prediction model with serum tumor markers to assess tumor metastasis in lung cancer. Cancer Med. 2020;9(15):5436–5445. doi: 10.1002/cam4.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, Tian X, Gao L, Jiang X, Fu R, Zhang T. et al. Clinical significance of circulating tumor cells and tumor markers in the diagnosis of lung cancer. Cancer Med. 2019;8(8):3782–3792. doi: 10.1002/cam4.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chakravarthy D, Muñoz AR, Su A, Hwang RF, Keppler BR, Chan DE. et al. Palmatine suppresses glutamine-mediated interaction between pancreatic cancer and stellate cells through simultaneous inhibition of survivin and COL1A1. Cancer Lett. 2018;419:103–115. doi: 10.1016/j.canlet.2018.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu W, Ouyang S, Zhou Z, Wang M, Wang T, Qi Y. et al. Identification of genes associated with cancer progression and prognosis in lung adenocarcinoma: Analyses based on microarray from Oncomine and The Cancer Genome Atlas databases. Mol Genet Genomic Med. 2019;7(2):e00528. doi: 10.1002/mgg3.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weiss J, Sos ML, Seidel D, Peifer M, Zander T, Heuckmann JM. et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med. 2010;2(62):62ra93. doi: 10.1126/scitranslmed.3001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rohrbeck A, Neukirchen J, Rosskopf M, Pardillos GG, Geddert H, Schwalen A. et al. Gene expression profiling for molecular distinction and characterization of laser captured primary lung cancers. J Transl Med. 2008;6:69. doi: 10.1186/1479-5876-6-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang M, Zhang G, Zhang Y, Cui X, Wang S, Gao S. et al. Fibrinogen Alpha Chain Knockout Promotes Tumor Growth and Metastasis through Integrin-AKT Signaling Pathway in Lung Cancer. Mol Cancer Res. 2020;18(7):943–954. doi: 10.1158/1541-7786.MCR-19-1033. [DOI] [PubMed] [Google Scholar]
- 20.Merry E, Thway K, Jones RL, Huang PH. Predictive and prognostic transcriptomic biomarkers in soft tissue sarcomas. NPJ Precis Oncol. 2021;5(1):17. doi: 10.1038/s41698-021-00157-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho Duy B, Zhytnik L, Maasalu K, Kändla I, Prans E, Reimann E, er al. Mutation analysis of the COL1A1 and COL1A2 genes in Vietnamese patients with osteogenesis imperfecta. Hum Genomics. 2016;10(1):27. doi: 10.1186/s40246-016-0083-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang VW. Collagen, stiffness, and adhesion: the evolutionary basis of vertebrate mechanobiology. Mol Biol Cell. 2020;31(17):1823–1834. doi: 10.1091/mbc.E19-12-0709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Winkler J, Abisoye-Ogunniyan A, Metcalf KJ, Werb Z. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat Commun. 2020;11(1):5120. doi: 10.1038/s41467-020-18794-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oleksiewicz U, Liloglou T, Tasopoulou KM, Daskoulidou N, Gosney JR, Field JK. et al. COL1A1, PRPF40A, and UCP2 correlate with hypoxia markers in non-small cell lung cancer. J Cancer Res Clin Oncol. 2017;143(7):1133–1141. doi: 10.1007/s00432-017-2381-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fang S, Dai Y, Mei Y, Yang M, Hu L, Yang H. et al. Clinical significance and biological role of cancer-derived Type I collagen in lung and esophageal cancers. Thorac Cancer. 2019;10(2):277–288. doi: 10.1111/1759-7714.12947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goswami MT, Reka AK, Kurapati H, Kaza V, Chen J, Standiford TJ. et al. Regulation of complement-dependent cytotoxicity by TGF-β-induced epithelial-mesenchymal transition. Oncogene. 2016;35(15):1888–98. doi: 10.1038/onc.2015.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, Ding Y, Li A. Identification of COL1A1 and COL1A2 as candidate prognostic factors in gastric cancer. World J Surg Oncol. 2016;14(1):297. doi: 10.1186/s12957-016-1056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boguslawska J, Kedzierska H, Poplawski P, Rybicka B, Tanski Z, Piekielko-Witkowska A. Expression of Genes Involved in Cellular Adhesion and Extracellular Matrix Remodeling Correlates with Poor Survival of Patients with Renal Cancer. J Urol. 2016;195(6):1892–902. doi: 10.1016/j.juro.2015.11.050. [DOI] [PubMed] [Google Scholar]
- 29.Jiang K, Liu H, Xie D, Xiao Q. Differentially expressed genes ASPN, COL1A1, FN1, VCAN and MUC5AC are potential prognostic biomarkers for gastric cancer. Oncol Lett. 2019;17(3):3191–3202. doi: 10.3892/ol.2019.9952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qi MM, Ge F, Chen XJ, Tang C, Ma J. MiR-124 changes the sensitivity of lung cancer cells to cisplatin through targeting STAT3. Eur Rev Med Pharmacol Sci. 2019;23(12):5242–5250. doi: 10.26355/eurrev_201906_18190. [DOI] [PubMed] [Google Scholar]
- 31.Chen FF, Lv X, Zhao QF, Xu YZ, Song SS, Yu W. et al. Inhibitor of DNA binding 3 reverses cisplatin resistance in human lung adenocarcinoma cells by regulating the PI3K/Akt pathway. Oncol Lett. 2018;16(2):1634–1640. doi: 10.3892/ol.2018.8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuchnio A, Moens S, Bruning U, Kuchnio K, Cruys B, Thienpont B. et al. The Cancer Cell Oxygen Sensor PHD2 Promotes Metastasis via Activation of Cancer-Associated Fibroblasts. Cell Rep. 2015;12(6):992–1005. doi: 10.1016/j.celrep.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 33.Bourgot I, Primac I, Louis T, Noël A, Maquoi E. Reciprocal Interplay Between Fibrillar Collagens and Collagen-Binding Integrins: Implications in Cancer Progression and Metastasis. Front Oncol. 2020;10:1488. doi: 10.3389/fonc.2020.01488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jia R, Wang C. MiR-29b-3p Reverses Cisplatin Resistance by Targeting COL1A1 in Non-Small-Cell Lung Cancer A549/DDP Cells. Cancer Manag Res. 2020;12:2559–2566. doi: 10.2147/CMAR.S246625. [DOI] [PMC free article] [PubMed] [Google Scholar]