Abstract
Telomeric DNA consists of short, tandemly repeated sequences at the ends of chromosomes. Telomeric DNA in the ciliate Paramecium tetraurelia is synthesized by an error-prone telomerase with an RNA template specific for GGGGTT repeats. We have previously shown that misincorporation of TTP residues at the telomerase RNA templating nucleotide C52 accounts for the 30% GGGTTT repeats randomly distributed in wild-type telomeres. To more completely characterize variable repeat synthesis in P. tetraurelia, telomerase RNA genes mutated at C52 (A, U, and G) were expressed in vivo. De novo telomeric repeats from transformants indicate that the predominant TTP misincorporation error seen in the wild-type telomerase is dependent on the presence of a C residue at template position 52. Paradoxically, the effects of various other telomerase RNA template and alignment region mutations on de novo telomeres include significant changes in fidelity, as well as the synthesis of aberrant, 5-nucleotide telomeric repeats. The occurrence of deletion errors and the altered fidelity of mutated P. tetraurelia telomerase, in conjunction with misincorporation by the wild-type enzyme, suggest that the telomerase RNA template domain may be analogous to homopolymeric mutational hot spots that lead to similar errors by the human immunodeficiency virus proofreading-deficient reverse transcriptase.
Telomeres, the specialized DNA-protein structures found at the ends of eukaryotic chromosomes, are necessary for chromosome stability and to facilitate the complete replication of chromosomal termini. In the absence of a proper telomere “cap,” chromosomes may be recognized as damaged DNA, leading to cell cycle arrest (50, 22), as well as illegitimate fusions, degradation, and aneuploidy (reviewed in reference 7).
Telomeric DNA from most eukaryotes consists of a variable number of short, tandemly repeated sequences. The sequences invariably have a strand bias, with a typically G-rich strand oriented 5′ to 3′ toward the chromosome terminus. The telomeric G-rich strand is synthesized by the ribonucleoprotein enzyme telomerase. Telomerase RNA (TER) subunits, which have been isolated from a number of ciliates, yeast, and mammals, all share the characteristic feature of a templating domain that dictates the species-specific telomeric repeat added to the 3′ end of the chromosome (reviewed in references 17 and 46).
Protein components associated with telomerase from ciliates, yeasts, and mammals have been isolated. A 123-kDa protein from the ciliate Euplotes aediculatus (35) and its homologs from Tetrahymena thermophila, Oxytricha trifallax, Saccharomyces cerevisiae, Schizosaccharomyces pombe, and humans (9, 13, 43, 44) all have the sequence and functional characteristics of a reverse transcriptase related to retrotransposon and retroviral reverse transcriptases (reviewed in reference 10). Mutational studies of the S. cerevisiae gene EST2 (34, 36) and in vitro reconstitution experiments with the human and Tetrahymena recombinant proteins (3, 13, 53) have confirmed a catalytic function for these Ea p123 homologs. Two other proteins that copurify with Tetrahymena thermophila telomerase, p80 and p95 (15), have also been described. It has been postulated that a p80-p95 complex functions to position the DNA substrate at the RNA template for extension by the telomerase holoenzyme (21). Mammalian homologs to T. thermophila p80 have also been identified (45, 27).
A nucleolytic activity associated with T. thermophila and Euplotes crassus telomerase cleaves the 3′ termini of primers in vitro (14, 42). This activity either is an intrinsic property of telomerase or is catalyzed by a factor that remains tightly associated with telomerase through extensive purification. It has been postulated that the in vitro endonucleolytic removal of nontelomeric sequences from primers by telomerase may be part of a proofreading system in which mismatched nucleotides are removed prior to elongation (25). Such a proofreading mechanism may have evolved to ensure high fidelity for telomerase, since deviation from species-specific telomeric repeats can have severe consequences on cell viability and nuclear division (31, 38, 41, 48, 54). Alternatively, the associated nucleolytic activity may be involved in de novo telomere addition to restructured chromosomes during macronuclear development in the late stages of conjugation (reviewed in reference 16).
Naturally occurring variable telomeres have been documented for the yeast Saccharomyces spp., the malarial protozoan Plasmodium falciparum (8), and many species of the ciliate Paramecium (2, 12, 20, 40). Irregular repeat synthesis in Saccharomyces is a consequence of partial translocation and stuttering along the RNA template, which results in duplicate copying of one or more nucleotides during each round of polymerization (11). Despite a single telomerase RNA consistent with G4T2 repeat synthesis, wild-type telomeres in most Paramecium spp. consist of a random mixture of G4T2 and G3T3 repeats at 70 and 30%, respectively (40). This variability in Paramecium telomeres is due to a stereotypical misincorporation of TTP at templating nucleotide C52 (39). Paradoxically, Paramecium caudatum telomerase faithfully synthesizes invariant G4T2 repeats as dictated by its RNA template. The high fidelity exhibited by the P. caudatum enzyme is not solely a property of the telomerase RNA, since expression and utilization of the P. caudatum telomerase RNA in P. tetraurelia transformants do not impart high fidelity to the telomerase of that species in vivo (39).
To further characterize variable telomeric repeat synthesis by P. tetraurelia telomerase, we have extended our mutational analysis of the P. tetraurelia telomerase RNA template and alignment domains. Mutated genes were introduced into P. tetraurelia macronuclei by microinjection, and de novo telomeric repeats from transformant clonal lines were analyzed. We show that misincorporation of TTP by the enzyme at templating nucleotide 52 is dependent on a C residue at that position, part of a homopolymeric tract of four C residues. The effects of various other mutations on de novo telomere synthesis include significant increased and decreased fidelity, as well as the occurrence of aberrant, 5-nucleotide telomeric repeats. The patterns of altered fidelity and the appearance of novel, high-frequency deletion errors by mutated P. tetraurelia telomerase suggest that the telomerase RNA template domain may represent a homopolymeric mutational hot spot, similar to those documented for the human immunodeficiency virus type 1 proofreading-deficient reverse transcriptase.
MATERIALS AND METHODS
Materials.
P. tetraurelia (nd6/nd6 and cam2/cam2 [29, 33]) was maintained at room temperature in a monoxenic wheat grass infusion, containing Enterobacter aerogenes (51). Restriction enzymes and other molecular biology reagents were purchased from New England Biolabs (Beverly, Mass.). The antibiotic G-418 was purchased from Sigma (St. Louis, Mo.).
General methods.
Conventional PCR methods and molecular techniques (49) were used for plasmid constructions. Genomic DNA was isolated from P. tetraurelia, and radiolabeled telomerase RNA gene probe was generated by a PCR strategy that was described previously (40). Oligonucleotides were radiolabeled at the 5′ end with T4 polynucleotide kinase and [γ-32P]ATP (7,000 Ci/mmol; ICN). DNA sequencing was performed with either Sequenase (U.S. Biochemical Corp., Cleveland, Ohio) or the CircumVent thermal cycle DNA-sequencing kit (New England Biolabs).
Transformation vector modification.
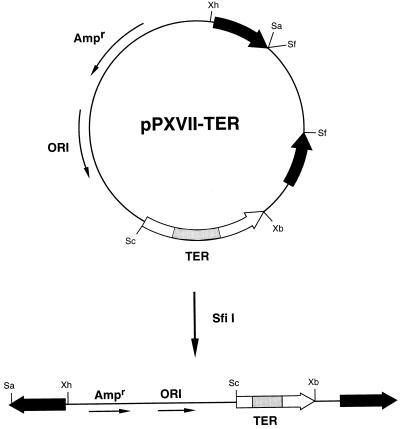
To facilitate the efficient sequencing of de novo telomeric repeats from transformants, unique XhoI and SalI restriction sites flanking one of the “seed” telomeres were introduced into plasmid pPXVI (39). A 1.2-kb BsaI-BssHII fragment from pPXVI, which includes one of the seed telomeres and a portion of the β-lactamase gene, was subcloned into a modified version of pLITMUS 28 (New England Biolabs). The modified pLITMUS 28 had the M13 intergenic region cloned in a clockwise orientation, opposite that of the commercially available phagemid (47a). The XhoI site was introduced by oligonucleotide site-directed mutagenesis (32). This construct was linearized at a unique SfiI site, the termini were blunted with T4 DNA polymerase, and the following linker sequence introduced by ligation with T4 DNA ligase: 5′-GGG TCG ACG GGG TTG GGG TTG GGG CTG CAG GCC TAC GTG GCC CG-3′. This linker includes unique SalI and PstI sites, separated by 16 bp of telomeric sequence. The SfiI cloning site, destroyed as part of the cloning strategy, was reintroduced at the 3′ end of this linker. The modified 1.2-kb BsaI-BssHII fragment was subcloned back into pPXVI, to yield the transformation vector pPXVII (see Fig. 2).
FIG. 2.
Plasmid for P. tetraurelia transformation. Plasmid pPXVII-TER was constructed as described in Materials and Methods. Wild-type and mutated TER genes, including nucleotides −240 through +325, were cloned so that the direction of transcription is toward the terminus of linearized plasmids. Shaded regions are indicative of transcribed nucleotides. A bacterial origin of replication (ORI) and ampicillin resistance marker (Ampr) are as indicated. Solid arrows represent tandem G4T2 telomeric repeats, located at the termini when plasmids are linearized by SfiI digestion. Restriction sites: Sa, SalI; Sc, SacI; Sf, SfiI; Xb, XbaI (Xb); Xh, XhoI. The approximate size of the pPXVII-TER construct is 4.3 kb (figure not drawn to scale).
P. tetraurelia telomerase RNA mutagenesis.
Oligonucleotide site-directed mutagenesis of the TER gene in plasmid pPTER (39) was performed by the method of Kunkel et al. (32). The TER genes were cloned into pPXVII by virtue of unique SacI and XbaI sites. The sequence of the modified genes in the resultant pPXVII-TER constructs (see Fig. 2) were confirmed by double-stranded DNA sequencing.
Transformation of P. tetraurelia by microinjection.
Plasmids suitable for microinjection were prepared with the Wizard DNA purification system (Promega, Madison, Wis.). Plasmids pPXV-NEO (28), pPXVII, and various pPXVII-TER constructs were linearized by SfiI digestion, and microinjection of P. tetraurelia was performed precisely as previously described (39). Transformant clonal lines were maintained at a density of 200 cells/ml and expanded for 20 to 22 fissions following microinjection.
Southern blot hybridizations.
Preparation of total DNA from transformant clonal lines, DNA transfer from agarose gels onto Nytran membranes, hybridization of radiolabeled DNA probes, and blot-washing conditions were all as previously described (40). A Molecular Dynamics PhosphorImager was used to quantitate the relative ratio of the plasmid-encoded to endogenous TER genes present in total DNA from transformants.
Cloning de novo telomeres from P. tetraurelia transformants.
Total DNA (5 μg) extracted from transformants 20 to 22 fissions after microinjection was digested with XbaI in a 50-μl reaction mixture, effectively removing one telomeric end from the microinjected pPXVII-TER plasmids. The XbaI-digested DNA was treated with 6 U of T4 DNA polymerase (New England Biolabs) in a standard reaction buffer (0.1 mM deoxynucleoside triphosphates [dNTPs]) at 12°C for 15 min to generate blunt ends suitable for ligation. Following phenol-chloroform (1:1) extraction and ethanol precipitation, 0.5 μg of the DNA was incubated overnight at 16°C with T4 DNA ligase (100-μl reaction volume). Competent Escherichia coli DH10B was transformed by electroporation with the ligation products. DNA prepared from ampicillin-resistant clones was screened for the TER gene by Southern blot hybridization.
Positive clones were screened for the presence of a unique SalI site situated between the remaining seed telomere and any de novo telomeric repeats added by telomerase in transformants. The seed telomere was removed from rescued plasmids by a restriction digestion with XhoI and SalI, followed by a ligation of the resultant compatible, cohesive ends with T4 DNA ligase. The secondary-rescue ligation products were used to transform competent E. coli (DH10B) by electroporation. The C strands of cloned, de novo telomeric repeats were sequenced by using a primer complementary to plasmid vector sequence (5′-TAA CTT TTA CTC AAT GTC AAA G-3′) that is adjacent to the XhoI-SalI site. The identities of TER genes on rescued plasmids were also confirmed by sequencing through the plasmid-encoded genes with the minus strand primer (5′-GCG TCT AGA AAT AAC TAT TTA GAG C-3′), complementary to nucleotides +209 to +191, inclusive, of the TER gene.
RESULTS
Telomerase RNA mutations.
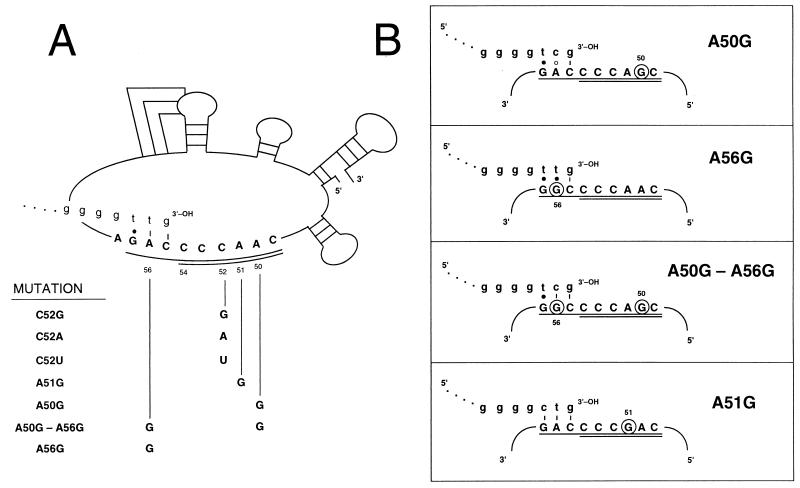
We have previously reported that the naturally occurring variability in P. tetraurelia telomeres is due to misincorporation of TTP at telomerase RNA templating nucleotide C52. Expression of a C52A TER template mutation in P. tetraurelia transformants leads to a dramatic increase in the number of de novo G3T3 repeats (39). To address whether the stereotypical misincorporation observed for the wild-type telomerase is dependent on a novel rC · dT base pair during the polymerization cycle, we have included single C52U and C52G substitutions in a parallel analysis (Fig. 1A).
FIG. 1.
(A) P. tetraurelia telomerase RNA nucleotide substitutions. A schematic of the Paramecium telomerase RNA secondary structure and nucleotide positions is taken from reference 40. Alignment nucleotides (1) are underlined and templating nucleotides are double-underlined. The 3′ end of a de novo telomere is shown (lowercase letters) base paired with the alignment nucleotides prior to elongation by telomerase. The various mutations analyzed are identified by the wild-type telomerase RNA position followed by the altered nucleotide. (B) Base-pairing potential of telomeric repeats with mutated telomerase RNAs. Only the telomerase RNA alignment (underlined) and templating (doubly underlined) nucleotides are shown. Substituted nucleotides are circled, and the 3′ end of the predicted de novo telomere is shown in lowercase letters, base paired with the alignment region prior to elongation. Dashes represent Watson-Crick base pairs, solid circles represent rG · dT pairs, and open circles represent mismatched nucleotides.
We had also observed inefficient utilization of an A50G template mutation by transformants. Only two G4TC telomeric repeats (the predicted sequence for an A50G template) were detected in 10 de novo telomeres isolated from this class of transformants (39). One consequence of G4TC repeat synthesis from the A50G template is a diminished base-pairing potential of de novo repeats with the telomerase RNA alignment region (Fig. 1B). Therefore, we have introduced an alignment nucleotide mutation, A56G, both singly and in combination with the A50G template substitution. Introduction of this alignment mutation tests whether A56G can compensate for the A50G template, the net effect being efficient G4TC repeat synthesis in A50G A56G transformants. We have also constructed an A51G template mutation, predicted to give rise to G4CT repeats. In direct contrast to the A50G mutation, de novo repeats from the A51G template should have the potential to form three Watson-Crick base pairs with the alignment region (Fig. 1B).
P. tetraurelia transformation by microinjection.
Mutated telomerase RNA genes were cloned into pPXVII, a plasmid suitable for Paramecium transformation (39). Prior to comicroinjection, both pPXVII-TER (Fig. 2) and pPXV-NEO (28) were digested with SfiI. The resultant linearized molecules have tracts of approximately 45 G4T2 telomeric repeats at their termini (Fig. 2), which increases the efficiency of transformation. Plasmid pPXV-NEO provides resistance to the antibiotic G-418, a selectable marker for transformants. Approximately 2 × 106 copies of each linearized plasmid were comicroinjected into the macronuclei of postautogamous P. tetraurelia.
Following microinjection, single-cell isolates were fed and allowed to expand to approximately 100 cells over 3 to 4 days at room temperature, at which time they were tested for G-418 resistance. Resistant clonal lines were maintained at 200 cells/ml for an additional 7 to 10 days, and total DNA and RNA were isolated approximately 20 to 22 fissions after microinjection. There were no obvious growth or morphological phenotypes for any of the various pPXVII-TER transformants compared to uninjected controls or cells transformed with pPXV-NEO alone (data not shown).
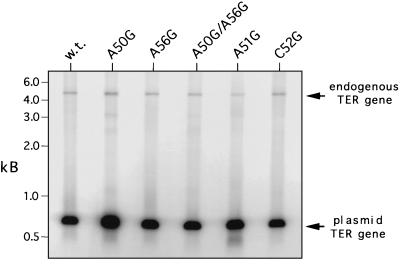
Total DNA from transformants was digested with SacI and XbaI to determine the relative amounts of the plasmid-borne TER gene (0.7-kb fragment) and the endogenous gene (4.5-kb fragment) present in clonal cell lines. Quantitation of Southern blots probed with the TER gene (Fig. 3) by PhosphorImager analysis indicates that the plasmid-borne gene accounts for no less than 98% of the telomerase RNA gene present in transformants (data not shown). Northern blot analyses indicate that the amount of TER transcribed from the plasmid-borne gene relative to that from the endogenous gene is directly proportional to the gene dosage (reference 39 and data not shown).
FIG. 3.
Endogenous and plasmid-encoded telomerase RNA genes in transformants. Southern blots of total DNA (1 μg), isolated from transformants and digested with SacI plus XbaI, were probed with the radiolabeled P. tetraurelia TER gene. The endogenous and plasmid-encoded gene fragments migrate to 4.4 and 0.7 kb, respectively. All transformants analyzed included the TER gene as indicated. Analyses of C52A and C52U transformants are not shown. w.t., wild type.
De novo telomere synthesis in transformants.
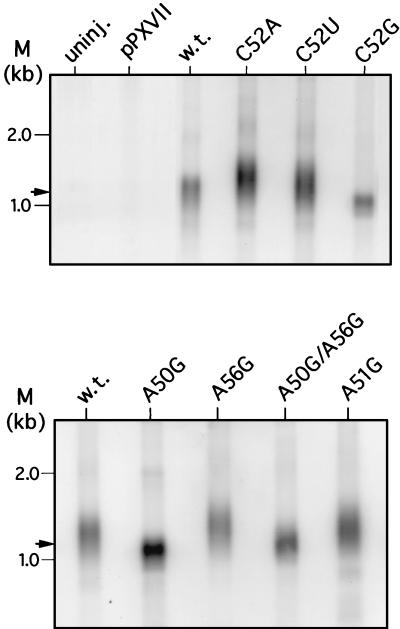
The fixed length of microinjected plasmid termini represent a convenient reference point in determining de novo repeat synthesis by transformant telomerase (39). In vivo extension of a SacI telomeric fragment that contains the plasmid-encoded TER gene (1.2 kb; Fig. 2) was measured for all classes of transformants 20 to 22 cell divisions after microinjection (Fig. 4). De novo telomeres in C52A and C52U transformants were on average 200 to 250 bp longer than those in cells transformed with the wild-type telomerase RNA gene. In contrast, the same telomeric fragment was consistently shorter than the 1.2-kb input length in cells expressing the C52G template mutation.
FIG. 4.
Southern blot analyses of a defined telomeric fragment from P. tetraurelia transformants. Digestion of the pPXVII-TER series of linearized, microinjected plasmids with SacI generates a telomeric fragment of approximately 1.2 kb that includes the telomerase RNA gene (arrow; see Fig. 2). Southern blots of total DNA (1 μg) from transformants digested with SacI and probed with the radiolabeled P. tetraurelia telomerase RNA gene are shown. uninj., DNA from uninjected cells; PXVII, DNA from clones coinjected with pPXVII (without TER) and pPXV-NEO. All other samples were from clones coinjected with pPXV-NEO and pPXVII-TER, with the telomerase RNA substitutions as indicated. w.t., wild type.
De novo telomere synthesis in A50G transformants was similar to that of the C52G mutants, with minimal telomere extension in these cells. The relative efficiency of telomere extension was greater in the A50G A56G double mutation, approaching that of the wild-type control (Fig. 4). Finally, the average lengths of de novo telomeres in A56G and A51G transformants exceeded those of wild-type transformants, although not to the same degree as those from cells expressing the C52A and C52U template mutations.
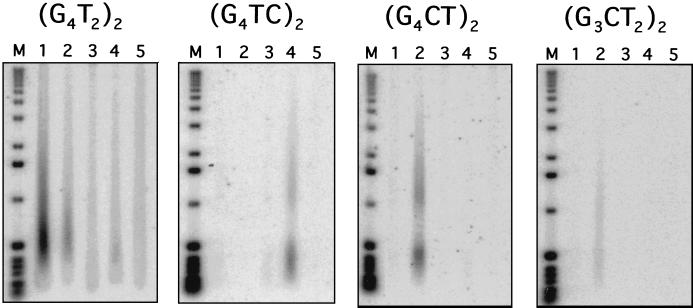
Telomere extension in five different transformants was also gauged by Southern blot hybridization of total DNA to various telomeric oligonucleotides (Fig. 5). There was a very low level of (G4TC)2 hybridization to A50G telomeres (lane 3), in contrast to the strong hybridization of this probe to A50G A56G transformant DNA (lane 4). Telomeres from the A51G transformant (lane 2) hybridized strongly to the (G4CT)2 probe and somewhat weakly to (G3CT2)2 (see Discussion). The C52G transformant DNA hybridized to the wild-type telomeric repeat (G4T2)2, but not at all to the sequence G3CT2, which was predicted for this template mutation (lane 5).
FIG. 5.
Southern blots of telomeres from transformants. Total DNA (8 μg) was digested with DraI and separated on 0.8% agarose gels (in quadruplicate) prior to capillary transfer of the DNA to Nytran filters. The radiolabeled oligonucleotides used as probes are as indicated. Telomerase RNAs expressed in transformants are as follows: lane 1, wild type; lane 2, A51G; lane 3, A50G; lane 4, A50G A56G; lane 5, C52G.
Telomere addition to the microinjected plasmid termini has made it possible to routinely clone and sequence the de novo telomeric repeats shown in Fig. 4 from P. tetraurelia cells expressing mutated telomerase RNA. Briefly, total DNA from transformants is digested to completion with XbaI, effectively removing one telomere from the linearized pPXVII-TER plasmids (Fig. 2). The XbaI-treated DNA is incubated with T4 DNA polymerase and dNTPs, resulting in blunt ends that facilitate the circularization of molecules with T4 DNA ligase. Transformation of competent E. coli with the ligation products and selection for ampicillin resistance leads to the rescue of circular plasmids from transformant DNA containing de novo telomeric repeats. Due to the exonucleolytic activity associated with T4 DNA polymerase, the most distal telomeric repeats may be lost during blunt end formation.
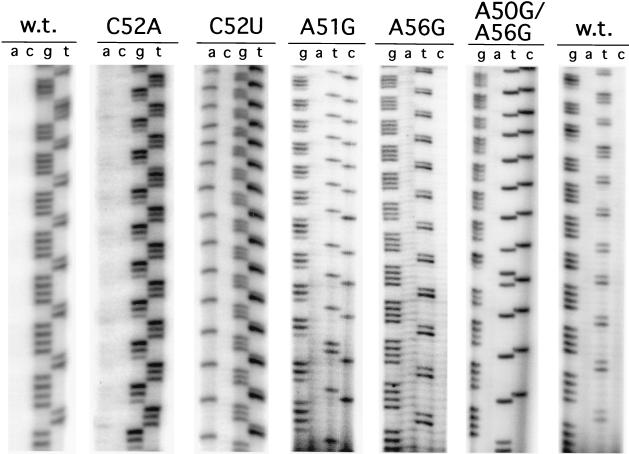
Cloned telomeric repeats were sequenced as described in Materials and Methods; representative examples from various transformants are shown in Fig. 6 and a compilation of de novo repeats is presented in Table 1. Wild-type transformants produced a mixture of G4T2 and G3T3 repeats, consistent with the composition of wild-type Paramecium telomeres (2, 20). As previously reported (39), the vast majority of de novo telomeric DNAs dictated by the C52A template mutation consist of G3T3 repeats. The C52U substitution leads to an equally high percentage of G3AT2 repeats, indicative of an enzyme that does not misincorporate TTP at template position 52. Finally, there was no extension of seed telomeres on the microinjected plasmids recovered from cells expressing the C52G template. This was somewhat anticipated, given the lack of de novo telomere extension in these cells (Fig. 4 and 5). There was also the apparent loss of distal repeats from the microinjected plasmid telomeres in C52G transformants, since none of the rescued plasmids retained a unique SalI restriction site 30 bp proximal to the telomere end (see Materials and Methods) (Fig. 2). Further evidence of telomere attrition in C52G transformants is that the number of repeats per seed telomere from rescued plasmids was smaller than that initially present during transformation (data not shown).
FIG. 6.
Representative telomere sequences from P. tetraurelia transformants. The C-rich strand of de novo telomeres cloned from transformants were sequenced as described in the text. The mutated telomerase RNAs expressed in transformants are as indicated. w.t., wild type.
TABLE 1.
Sequence of de novo telomeric repeats from transformants expressing mutated telomerase RNAsa
| Construct | % of repeats
|
No. of repeats | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| G4T2 | G3T3 | G3T2 | G3AT2 | G4CT | G3CT | G4TC | G3TC | Other | ||
| Wild type | 70 | 26 | 2 | 2 | 110 | |||||
| C52A | 7 | 93 | 210 | |||||||
| C52U | 5 | 1 | 1 | 93 | 224 | |||||
| C52G | 0 | |||||||||
| A50G | 0 | |||||||||
| A51G | 3 | 1 | 31 | 62 | 3 | 113 | ||||
| A56G | 85 | 10 | 5 | 197 | ||||||
| A50G/A56G | 2 | 1 | 66 | 24 | 7 | 80 | ||||
Telomeric repeats were cloned and sequenced from transformant DNA approximately 20 to 22 generations after microinjection, as described in Methods. The percentages of the various telomeric repeats are as indicated, and the total numbers of de novo repeats sequenced for each transformant class are shown on the right. The percentages of repeat sequences predicted for a given telomerase RNA template are shown in boldface.
De novo telomeric repeats were not detected in plasmids rescued from cells expressing the A50G template mutation. In contrast, cells expressing the A50G A56G double mutation efficiently synthesized G4TC repeats, as well as novel G3TC repeats (66 and 24%, respectively). Five-nucleotide repeats (G3T2) have been documented for wild type Paramecium telomeres, albeit at less than 3% (39) (Table 1). There was an apparent increase in telomerase fidelity in cells expressing telomerase RNA with the A56G mutation alone. The percentage of de novo G4T2 repeats increased from 72% in wild type transformants to 85% in cells with the A56G mutation (Table 1). Finally, the A51G template substitution leads to a surprisingly high percentage of 5-nucleotide G3CT repeats (62%). The 6-nucleotide repeat predicted for this template mutation, G4CT, accounts for 31% of the de novo telomeric DNA sequenced.
DISCUSSION
Error discrimination by DNA polymerases can occur at a number of steps along the polymerization cycle (reviewed in reference 19). Binding of the correct dNTP to a template-primer complex is favored over that of the incorrect dNTPs, due to hydrogen bonding and the geometry of the base-pair and base-stacking interactions at the catalytic site. The rate of phosphodiester bond formation is presumably higher for the cognate dNTP, and subsequent extension is more efficient with a correctly paired 3′-hydroxyl terminus. Crystallographic studies of Bacillus stearothermophilus and bacteriophage T7 DNA polymerases complexed with their substrates reveal that the basis for error discrimination is the steric complementarity between the enzyme and a correctly formed Watson-Crick base pair at the active site (18, 30). Mismatched nucleotides cannot bind with the same geometry, resulting in misalignment of the primer 3′-OH terminus, which effectively prevents misincorporation.
DNA polymerases, with the exception of retroviral reverse transcriptases, can also preferentially remove misincorporated nucleotides with an associated nucleolytic activity. Subsequently, error rates for proofreading DNA polymerases, which range from 10−7 to 10−11 error per nucleotide, are orders of magnitude lower than those for reverse transcriptase (37). Misincorporation error frequencies as high as 10−2 have been documented for spleen necrosis virus reverse transcriptase (47). Studies with avian myeloblastosis virus and human immunodeficiency virus type 1 reverse transcriptases have also shown that certain template sequences constitute mutational hot spots, where misincorporation, insertion, and deletion errors occur more frequently (reviewed in reference 6). These hot spots all have a common feature: they are homopolymeric nucleotide sequences (4). Furthermore, single-nucleotide substitutions within and adjacent to the hot spots can significantly decrease polymerization error frequencies (5).
Our in vivo data on both wild-type and mutated P. tetraurelia telomerase suggest that the telomerase RNA residues C52 to C55 comprise a homopolymeric mutational hot spot, similar to those described for retroviral reverse transcriptases. Substitutions within and adjacent to these template and alignment nucleotides have a direct effect on telomerase fidelity in vivo. A net effect of the P. tetraurelia telomerase RNA templating nucleotide C52 substitutions was to reduce the homopolymeric tract of C residues from four to three. Coincident with two of these mutations (C52A and C52U) was a decrease in misincorporation of TTP at this templating position. As anticipated, the C52A substitution simply changes the template to specify G3T3 repeats, with greater than 90% of the de novo repeats synthesized the cognate G3T3 (39) (Fig. 6 and Table 1). The importance of C52 in TTP misincorporation was clearly demonstrated by telomere sequences from C52U transformants. The composition of de novo telomeres from cells expressing the C52U template was 93% G3AT2 repeats. If TTP addition were independent of an rC residue at this template position, a substantial percentage of de novo G3T3 repeats would have been detected in telomeres from C52U transformants.
In contrast to the C52A and C52U transformants, plasmids rescued from cells expressing the C52G mutation lacked de novo telomeric repeats. There was a cumulative net loss from the input seed telomere over the approximately 26 fissions following microinjection (Fig. 4), and the predicted G3CT2 repeats from this template mutation were not detected (Fig. 5 and data not shown). Although telomere attrition was also seen with the A50G mutation (Fig. 4), two cognate G4TC repeats dictated by this template were detected in a previous study (39) and are present at low levels in transformants (Fig. 5). Whereas the A50G template mutation results in an inefficient telomerase, the effect of the C52G substitution appears to be somewhat more severe. There is efficient incorporation of dC residues in other telomeric repeats in vivo (see below), and so the inclusion of dC in the 3′ strand of a Paramecium telomere per se cannot be the sole reason for a block of activity. Some Tetrahymena telomerase RNA template mutations are also apparently nonfunctional in vivo (54). It is formally possible that telomeres from C52G transformants include G3CT2 repeats that are not protected by telomeric proteins from degradation. The exact nature of the apparent block in telomere maintenance by C52G transformants cannot be determined at this level of analysis.
A single nucleotide substitution in the TER alignment region (G57A) increases the incidence of G3T3 errors from 30 to 50% in P. tetraurelia transformants (39). Paradoxically, an A56G substitution that decreases the base-pairing potential between de novo repeats and the alignment region (5′-GGGTTG-3′ base paired to 3′-GGC-5′, where boldface type indicates the base-paired nucleotides [Fig. 1]) increases telomerase fidelity, leading to a decrease in G3T3 misincorporation that is statistically significant (χ2 = 12.4; P < 0.005). This phenotype is reminiscent of that previously characterized for P. tetraurelia expressing a C49A template substitution. The predominant telomeric repeat synthesized by C49A transformants was G4T3. There was a complete absence of G3T4 repeats detected in these cells, although it was the predicted sequence if there had been continued TTP misincorporation at C52 (39).
Inefficient maintenance of telomeres by A50G transformants is not due to the failure of telomeric proteins to recognize and protect the mutated telomere. The A50G template mutation, coupled with a compensatory A56G substitution in the alignment region, results in efficient extension of telomeres with G4TC repeats (Fig. 5 and 6; Table 1). The Watson-Crick base-pairing potential between the 3′ end of the de novo telomeric repeat (5′-GGGTCG-3′) and the mutated alignment region (3′-GGC-5′) is restored in these transformants and approximates that of the wild-type interaction (5′-GGGTTG-3′ base pairing with 3′-GAC-5′). Similarly, there is perfect base-pairing potential of de novo repeats from A51G transformants with the wild-type alignment region (5′-GGGCTG-3′ base-paired to 3′-GAC-5′); the cognate G4CT repeats were easily detected in telomeres from transformants (Fig. 5 and Table 1). These data support the conclusion of an in vitro study with Tetrahymena telomerase that correct positioning and efficient elongation of primers is largely dependent on an analogous interaction with the alignment domain (23).
Telomeres from P. tetraurelia expressing wild-type telomerase RNA include a small percentage of 5-nucleotide repeats, G3T2 (39) (Table 1). Surprisingly, both A50G A56G and A51G transformants synthesized high percentages of G3TC and G3CT de novo repeats (24 and 62%, respectively). This high incidence of G3CT repeats accounts for the observed hybridization of a (G3CT2)2 probe to DNA from A51G transformant DNA under low-stringency wash conditions (Fig. 5): only 2 of 12 bp are mismatched between this probe and the 12-nucleotide telomeric sequence (G3CT)(G4CT)G.
Taken together, our results demonstrate that substitutions within or adjacent to the homopolymeric tract from C52 to C55 of the Paramecium telomerase RNA template can either increase or decrease TTP misincorporation in vivo or can promote stereotypical deletion errors at greater than 50%. Errors by retroviral reverse transcriptase at homopolymeric mutational hot spots are also affected by changes at neighboring nucleotides, with error rates at particular hot spots never being greater than 2% (5). Experiments with Tetrahymena telomerase also indicate that altered TERs can promote misincorporation and premature disassociation of primers, although error frequencies for mutated telomerase were not monitored in these in vitro studies (23, 24). Together, these data support the paradigm of a complex catalytic site for telomerase, with a role for the telomerase RNA beyond that of a passive template. In the case of wild-type and mutated P. tetraurelia telomerase, templating nucleotide C52 appears to be at the center of both misincorporation and deletion errors that can occur at a high frequency.
Based on the sequence of cloned telomeres and the analysis of reaction products from activity assays, the error frequency of Tetrahymena telomerase has been estimated at no greater than 10−2 (24). Nucleolytic activities associated with both the Tetrahymena and the equally precise Euplotes enzyme (14, 42) have been postulated to be part of a proofreading system, in which misincorporated nucleotides are removed from de novo repeats prior to elongation (25). It is conceivable that telomerase from most Paramecium species lack an efficient mechanism to prevent or correct TTP misincorporation, resulting in a high percentage of G3T3 errors. A notable exception may be the P. caudatum telomerase, whose telomeres consist solely of G4T2 repeats (40). A comparison of a precise P. caudatum telomerase with an imprecise P. tetraurelia enzyme in vitro may determine whether a proofreading nucleolytic activity contributes to ciliate telomerase fidelity. Interestingly, telomerase from the malarial parasite Plasmodium falciparum, whose telomeres consist of G3T3A and G3T2CA repeats (52), lacks an associated nucleolytic activity (8).
The current model for telomere length regulation is that of a competitive relationship between telomere binding proteins and telomerase for access to the telomere end (reviewed in reference 26). There is an apparent loss of length regulation in P. tetraurelia transformants that do not synthesize high percentages of both G4T2 and G3T3 (39) (Fig. 4 and Table 1). When the vast majority of repeats are G3T3 (C52A) or when G4T2 repeats predominate (A56G), there is a concomitant increase in telomere length. However, telomere extension did not differ from that in the wild type when the ratio of the two naturally occurring repeats was shifted from 70:30 to 50:50 in G57A transformants (39). It is possible that the juxtaposition of G4T2 and G3T3 repeats at regular intervals is necessary for efficient binding by P. tetraurelia telomere binding proteins. It follows that the affinity of the P. tetraurelia telomere binding protein for (G4T2G3T3)n would be greater than that for either (G4T2)n or (G3T3)n. Conversely, a P. caudatum telomere binding protein would be predicted to have a higher affinity for (G4T2)n. A more complete comparative analysis of the various factors that dictate telomere synthesis and regulation in P. tetraurelia and P. caudatum may shed light on the determinants that dictate telomerase precision.
ACKNOWLEDGMENTS
We thank Thomas Marsh for critical reading of and helpful comments on the manuscript.
This research was supported by Public Health Service grant GM-50861 from the National Institutes of Health (to D.P.R.).
REFERENCES
- 1.Autexier C, Greider C W. Boundary elements of the Tetrahymena telomerase RNA template and alignment domains. Genes Dev. 1995;9:2227–2239. doi: 10.1101/gad.9.18.2227. [DOI] [PubMed] [Google Scholar]
- 2.Baroin A, Prat A, Caron F. Telomeric site position heterogeneity in macronuclear DNA of Paramecium primaurelia. Nucleic Acids Res. 1987;15:1717–1728. doi: 10.1093/nar/15.4.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beattie T L, Zhou W, Robinson M O, Harrington L. Reconstitution of human telomerase activity in vitro. Curr Biol. 1998;8:177–180. doi: 10.1016/s0960-9822(98)70067-3. [DOI] [PubMed] [Google Scholar]
- 4.Bebenek K, Abbotts J, Roberts J D, Wilson S H, Kunkel T A. Specificity and mechanism of error-prone replication by human immunodeficiency virus-1 reverse transcriptase. J Biol Chem. 1989;264:16948–16956. [PubMed] [Google Scholar]
- 5.Bebenek K, Abbotts J, Wilson S H, Kunkel T A. Error-prone polymerization by HIV-1 reverse transcriptase. Contribution of template-primer misalignment, miscoding, and termination probability to mutational hot spots. J Biol Chem. 1993;268:10324–10334. [PubMed] [Google Scholar]
- 6.Bebenek K, Kunkel T A. The fidelity of retroviral reverse transcriptases. In: Skalka A M, Goff S P, editors. Reverse transcriptase. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 85–102. [Google Scholar]
- 7.Blackburn E H, Greider C W, editors. Telomeres. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1995. [Google Scholar]
- 8.Bottius E, Bakhsis N, Scherf A. Plasmodium falciparum telomerase—de novo telomere addition to telomeric and nontelomeric sequences and role in chromosome healing. Mol Cell Biol. 1998;18:919–925. doi: 10.1128/mcb.18.2.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bryan T M, Sperger J M, Chapman K B, Cech T R. Telomerase reverse transcriptase genes identified in Tetrahymena thermophila and Oxytricha trifallax. Proc Natl Acad Sci USA. 1998;95:8479–8484. doi: 10.1073/pnas.95.15.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cech T R, Nakamura T M, Lingner J. Telomerase is a true reverse transcriptase—a review. Biochemistry (Moscow) 1997;62:1202–1205. [PubMed] [Google Scholar]
- 11.Cohn M, Blackburn E H. Telomerase in yeast. Science. 1995;269:396–400. doi: 10.1126/science.7618104. [DOI] [PubMed] [Google Scholar]
- 12.Cohn M, McEachern M J, Blackburn E H. Telomeric sequence diversity within the genus Saccharomyces. Curr Genet. 1998;33:83–91. doi: 10.1007/s002940050312. [DOI] [PubMed] [Google Scholar]
- 13.Collins K, Gandhi L. The reverse transcriptase component of the Tetrahymena telomerase ribonucleoprotein complex. Proc Natl Acad Sci USA. 1998;95:8485–8490. doi: 10.1073/pnas.95.15.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collins K, Greider C W. Tetrahymena telomerase catalyzes nucleolytic cleavage and nonprocessive elongation. Genes Dev. 1993;7:1364–1376. doi: 10.1101/gad.7.7b.1364. [DOI] [PubMed] [Google Scholar]
- 15.Collins K, Kobayashi R, Greider C W. Purification of Tetrahymena telomerase and cloning of genes encoding the two protein components of the enzyme. Cell. 1995;81:677–686. doi: 10.1016/0092-8674(95)90529-4. [DOI] [PubMed] [Google Scholar]
- 16.Coyne R S, Chalker D L, Yao M C. Genome downsizing during ciliate development—nuclear division of labor through chromosome restructuring. Annu Rev Genet. 1996;30:557–578. doi: 10.1146/annurev.genet.30.1.557. [DOI] [PubMed] [Google Scholar]
- 17.Dokudovskaya S S, Petrov A V, Dontsova O A, Bogdanov A A. Telomerase is an unusual RNA-containing enzyme—a review. Biochemistry (Moscow) 1997;62:1206–1215. [PubMed] [Google Scholar]
- 18.Doublie S, Tabor S, Long A M, Richardson C C, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 19.Echols H, Goodman M F. Fidelity mechanisms in DNA replication. Annu Rev Biochem. 1991;60:477–511. doi: 10.1146/annurev.bi.60.070191.002401. [DOI] [PubMed] [Google Scholar]
- 20.Forney J D, Blackburn E H. Developmentally controlled telomere addition in wild-type and mutant Paramecia. Mol Cell Biol. 1988;8:251–258. doi: 10.1128/mcb.8.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gandhi L, Collins K. Interaction of recombinant Tetrahymena telomerase proteins p80 and p95 with telomerase RNA and telomeric DNA substrates. Genes Dev. 1998;12:721–733. doi: 10.1101/gad.12.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garvik B, Carson M, Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol Cell Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilley D, Blackburn E H. Specific RNA residue interactions required for enzymatic functions of Tetrahymena telomerase. Mol Cell Biol. 1996;16:66–75. doi: 10.1128/mcb.16.1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilley D, Lee M S, Blackburn E H. Altering specific telomerase RNA template residues affects active site function. Genes Dev. 1995;9:2214–2226. doi: 10.1101/gad.9.18.2214. [DOI] [PubMed] [Google Scholar]
- 25.Greene E C, Bednenko J, Shippen D E. Flexible positioning of the telomerase-associated nuclease leads to preferential elimination of nontelomeric DNA. Mol Cell Biol. 1998;18:1544–1552. doi: 10.1128/mcb.18.3.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greider C W. Telomere length regulation. Annu Rev Biochem. 1996;65:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- 27.Harrington L, Mcphail T, Mar V, Zhou W, Oulton R, Bass M B, Arruda I, Robinson M O. A mammalian telomerase-associated protein. Science. 1997;275:973–977. doi: 10.1126/science.275.5302.973. [DOI] [PubMed] [Google Scholar]
- 28.Haynes W J, Ling K Y, Saimi Y, Kung C. Induction of antibiotic resistance in Paramecium tetraurelia by the bacterial gene APH-3′-II. J Eukaryot Microbiol. 1995;42:83–91. doi: 10.1111/j.1550-7408.1995.tb01545.x. [DOI] [PubMed] [Google Scholar]
- 29.Kanabrocki J A, Saimi Y, Preston R R, Haynes W J, Kung C. Efficient transformation of cam2, a behavioral mutant of Paramecium tetraurelia, with the calmodulin gene. Proc Natl Acad Sci USA. 1991;88:10845–10849. doi: 10.1073/pnas.88.23.10845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiefer J R, Mao C, Braman J C, Beese L S. Visualizing DNA replication in a catalytically active Bacillus DNA polymerase crystal. Nature. 1998;391:304–307. doi: 10.1038/34693. [DOI] [PubMed] [Google Scholar]
- 31.Kirk K E, Harmon B P, Reichardt I K, Sedat J W, Blackburn E H. Block in anaphase chromosome separation caused by a telomerase template mutation. Science. 1997;275:1478–1481. doi: 10.1126/science.275.5305.1478. [DOI] [PubMed] [Google Scholar]
- 32.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-directed mutagenesis without phenotypic selection. Methods Enzymol. 1987;155:166–178. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 33.Lefort-Tran M, Aufderheide K, Pouphile M, Rossignol M, Beisson J. Control of exocytotic processes: cytological and physiological studies of trichocyst mutants in Paramecium tetraurelia. J Cell Biol. 1981;88:301–311. doi: 10.1083/jcb.88.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lendvay T S, Morris D K, Sah J, Balasubramanian B, Lundblad V. Senescence mutants of saccharomyces cerevisiae with a defect in telomere replication identify three additional est genes. Genetics. 1996;144:1399–1412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lingner J, Cech T R. Purification of telomerase from Euplotes aediculatus—requirement of a primer 3′ overhang. Proc Natl Acad Sci USA. 1996;93:10712–10717. doi: 10.1073/pnas.93.20.10712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lingner J, Hughes T R, Shevchenko A, Mann M, Lundblad V, Cech T R. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 37.Litvak S. Retroviral reverse transcriptases. New York, N.Y: Chapman & Hall; 1996. [Google Scholar]
- 38.Marusic L, Anton M, Tidy A, Wang P, Villeponteau B, Bacchetti S. Reprogramming of telomerase by expression of mutant telomerase RNA template in human cells leads to altered telomeres that correlate with reduced cell viability. Mol Cell Biol. 1997;17:6394–6401. doi: 10.1128/mcb.17.11.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCormick-Graham M, Haynes W J, Romero D P. Variable telomeric repeat synthesis in Paramecium tetraurelia is consistent with misincorporation by telomerase. EMBO J. 1997;16:3233–3242. doi: 10.1093/emboj/16.11.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCormick-Graham M, Romero D P. A single telomerase RNA is sufficient for the synthesis of variable telomeric DNA repeats in ciliates of the genus Paramecium. Mol Cell Biol. 1996;16:1871–1879. doi: 10.1128/mcb.16.4.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McEachern M J, Blackburn E H. Runaway telomere elongation caused by telomerase RNA gene mutations. Nature. 1995;376:403–409. doi: 10.1038/376403a0. [DOI] [PubMed] [Google Scholar]
- 42.Melek M, Greene E C, Skippen D E. Processing of nontelomeric 3′ ends by telomerase—default template alignment and endonucleolytic cleavage. Mol Cell Biol. 1996;16:3437–3445. doi: 10.1128/mcb.16.7.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meyerson M, Counter C M, Eaton E N, Ellisen L W, Steiner P, Caddle S D, Ziaugra L, Beijersbergen R L, Davidoff M J, Liu Q Y, Bacchetti S, Haber D A, Weinberg R A. Hest2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- 44.Nakamura T M, Morin G B, Chapman K B, Weinrich S L, Andrews W H, Lingner J, Harley C B, Cech T R. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 45.Nakayama J, Saito M, Nakamura H, Matsuura A, Ishikawa F. TLP1: a gene encoding a protein component of mammalian telomerase is a novel member of WD repeats family. Cell. 1997;88:875–884. doi: 10.1016/s0092-8674(00)81933-9. [DOI] [PubMed] [Google Scholar]
- 46.Nugent C I, Lundblad V. The telomerase reverse transcriptase—components and regulation. Genes Dev. 1998;12:1073–1085. doi: 10.1101/gad.12.8.1073. [DOI] [PubMed] [Google Scholar]
- 47.Pathak V K, Temin H M. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: substitutions, frameshifts, and hypermutations. Proc Natl Acad Sci USA. 1990;87:6019–6023. doi: 10.1073/pnas.87.16.6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47a.Romero, D. Unpublished data.
- 48.Romero D P, Blackburn E H. Circular rDNA replicons persist in Tetrahymena thermophila transformants synthesizing GGGGTC telomeric repeats. J Eukaryot Microbiol. 1995;42:32–43. doi: 10.1111/j.1550-7408.1995.tb01537.x. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 50.Sandell L L, Zakian V A. Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- 51.Sonneborn T M. Methods in Paramecium research. Methods Cell Physiol. 1970;4:242–339. [Google Scholar]
- 52.Vernick K D, McCutchan T F. Sequence and structure of a Plasmodium falciparum telomere. Mol Biochem Parasitol. 1988;28:85–94. doi: 10.1016/0166-6851(88)90055-2. [DOI] [PubMed] [Google Scholar]
- 53.Weinrich S L, Pruzan R, Ma L B, Ouellette M, Tesmer V M, Holt S E, Bodnar A G, Lichtsteiner S, Kim N W, Trager J B, Taylor R D, Carlos R, Andrews W H, Wright W E, Shay J W, Harley C B, Morin G B. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- 54.Yu G-L, Bradley J D, Attardi L D, Blackburn E H. In vivo alteration of telomere sequences and senescence caused by mutated Tetrahymena telomerase RNAs. Nature. 1990;344:126–132. doi: 10.1038/344126a0. [DOI] [PubMed] [Google Scholar]