Abstract
Centipedegrass [Eremochloa ophiuroides (Munro) Hack.], a member of the Panicoideae subfamily, is one of the most important warm-season turfgrasses originating from China. This grass has an extremely developed prostrate growth habit and has been widely used in transitional and warm climatic regions. To better understand the genetic basis of important biological characteristics, such as prostrate growth and seed yield, in warm-season turfgrasses, we present a high-quality reference genome for centipedegrass and use PacBio, BioNano, and Hi-C technologies to anchor the 867.43 Mb genome assembly into nine pseudochromosomes, with a scaffold N50 of 86.05 Mb and 36,572 annotated genes. Centipedegrass was most closely related to sorghum and diverged from their common ancestor ~16.8 Mya. We detected a novel chromosome reshuffling event in centipedegrass, namely, the nest chromosome fusion event in which fusion of chromosomes 8 and 10 of sorghum into chromosome 3 of centipedegrass likely occurred after the divergence of centipedegrass from sorghum. The typical prostrate growth trait in centipedegrass may be linked to the expansion of candidate PROSTRATE GROWTH 1 (PROG1) genes on chromosome 2. Two orthologous genes of OsPROG1, EoPROG1, and EoPROG2, were confirmed to increase the stem number and decrease the stem angle in Arabidopsis. Collectively, our assembled reference genome of centipedegrass offers new knowledge and resources to dissect the genome evolution of Panicoideae and accelerate genome-assisted breeding and improvement of plant architecture in turf plants.
Subject terms: Evolutionary biology, Plant development
Introduction
Centipedegrass [Eremochloa ophiuroides (Munro) Hack.] is an indigenous, perennial warm-season (C4) grass species in China that is also well adapted for use as turfgrass in transitional and warm climatic regions (Fig. 1a, b). It is now widely used in the southern and eastern USA, Southeast Asia and tropical northern and eastern parts of Australia, and the southern Yangtze River region of China1,2. E. ophiuroides is well known for good adaptation to infertile and acidic soils and a wide range of climatic conditions3. It possesses great potential for use in the turf industry due to its lower management and fertilization requirements compared with other turfgrasses. However, the genomic features that underlie these important biological characteristics are still unclear. Although a low-quality draft genome assembly of Zoysia has been released4, no high-quality reference genomes representing all warm-season turfgrasses, including important species such as bermudagrass, zoysiagrass, seashore paspalum, and centipedegrass, have been reported to date.
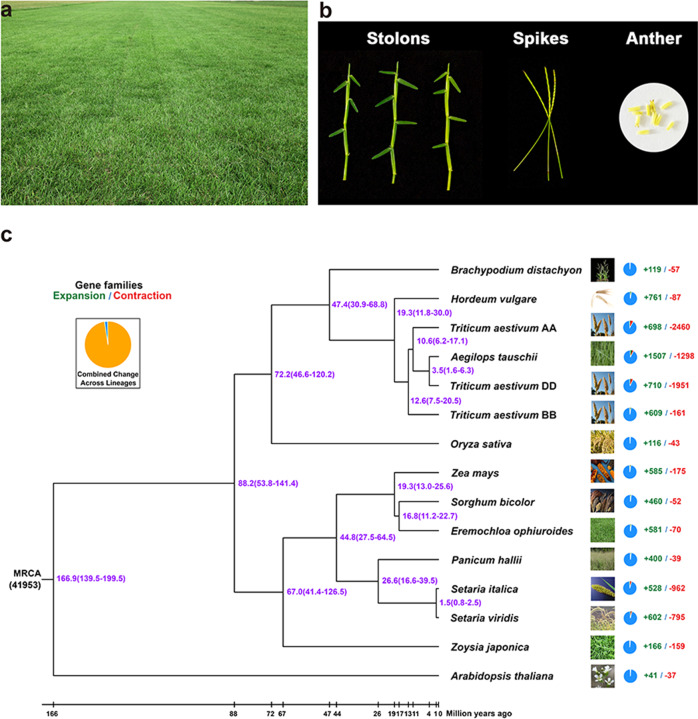
Fig. 1. Photographs and comparative genomic analysis of E. ophiuroides.
a Turf performance of E. ophiuroides. b Stolons, spikes, and anthers of ‘Ganbei’ centipedegrass. c Phylogenetic analysis, divergence time, and gene family expansions and contractions among 13 plant species. The phylogenetic tree was constructed on the basis of 1114 single-copy orthologous genes and A. thaliana as the outgroup. Divergence times (Mya) are indicated by the purple numbers beside the branch nodes. Gene family expansions and contractions are indicated by green and red numbers, respectively
E. ophiuroides belongs to the genus Eremochloa in the family Poaceae and subfamily Panicoideae5. It is a diploid species with a somatic chromosome number of 2n = 2x = 186,7 and a genome size of ~800 Mb8. E. ophiuroides is closely related to several main crop species with complex genomes, such as maize (Zea mays), sorghum (Sorghum bicolor), and foxtail millet (Setaria italica). Among these species that also belong to Panicoideae, maize underwent a separate whole-genome duplication (WGD) event, with a chromosome number 2n = 2x = 20 and a size of ~2.3 Gb9,10; the sorghum genome is ~730 Mb in size, with a chromosome number of 2n = 2x = 20, and includes large transposon structures and substantial genomic rearrangements11; and the foxtail millet genome also underwent chromosome reshuffling events, with a genome size of ~423 Mb and chromosome number of 2n = 2x = 1812. Although E. ophiuroides presents a comparable genome size to sorghum, its chromosome number is closer to that of foxtail millet. This implies that the E. ophiuroides genome most likely formed through a series of genomic events that remain to be determined.
E. ophiuroides is the only species in the Eremochloa genus that can be used as turfgrass due to its typical prostrate growth habit in the field. During crop domestication, plant architecture was modified in a direction more conducive to increasing crop yield. The transition from prostrate growth to erect growth is one of the most critical events during this process, and genes controlling growth habits play a decisive role in this transition. Previous studies demonstrated that the PROSTRATE GROWTH 1 (PROG1) gene is highly associated with plant architecture changes during domestication in rice and that dysfunction and/or alterations in the expression patterns of PROG1 have led to erect growth in both Asian and African cultivated rice13–15. PROG1 encodes a C2H2 zinc-finger domain transcription factor located on the short arm of rice chromosome 7 that controls the transition of plant architecture from prostrate growth to erect growth during rice domestication13,14. Recently, Wu et al.16 found that deletions on rice chromosome 7 eliminating a tandem repeat of zinc-finger genes are closely linked to the role of the PROG1 gene in plant architecture changes during domestication in both Asian and African rice; they also noted a similar tandem repeat of zinc-finger protein-coding genes and a complex structural variation at the RICE PLANT ARCHITECTURE DOMESTICATION (RPAD) locus in foxtail millet (S. italica) and its presumed progenitor, green foxtail (Setaria viridis). While erect growth is a more beneficial architecture for most crop plants, prostrate growth is an adaptation to disturbed habitats in wild crop plants and is also the preferred plant architecture for turfgrasses. Considering the extremely developed prostrate growth habit of E. ophiuroides, ascertaining the structural characteristics and genome distribution of the PROG1 gene as well as its expression status in different tissues will contribute to revealing the prostrate growth mechanism of centipedegrass and wild species of crop plants, which will be of great significance for promoting the breeding of turfgrasses and some important crops.
Here, we present a high-quality chromosome-level genome assembly for E. ophiuroides obtained via next-generation Illumina, Pacific Biosciences (PacBio), and BioNano sequencing combined with 10× genomic and high-throughput/resolution chromosome conformation capture (Hi-C) technologies. We systematically analyzed the evolutionary position of E. ophiuroides in Poaceae, investigated the expansion/contraction of gene families in the E. ophiuroides genome assembly through comparative genomic analysis, and proposed a model for the structural evolution of E. ophiuroides. The key genes controlling prostrate growth were further screened and analyzed to reveal the typical prostrate growth characteristics of E. ophiuroides. Thus, the E. ophiuroides reference genome will be a valuable resource for genetic studies and breeding programs in turf plants, both for exploring the genome evolution of Poaceae and the genome-assisted breeding of novel cultivars with desired traits, such as favorable plant architecture traits.
Materials and methods
Plant material and genome sequencing and assembly
The sequenced centipedegrass material was a ‘Ganbei’ accession collected from Mount Lushan in Jiangxi Province, China (28°36′N, 116°00′E); this variety has been largely applied in water conservation projects in the lower reaches of the Yangtze River and is preserved in the Grass Research Center of the Institute of Botany, Jiangsu Province and Chinese Academy of Science, China. Leaf tissue of E. ophiuroides ‘Ganbei’ was used for Illumina, PacBio, BioNano, and Hi-C library construction. The BioNano library was sequenced on the Saphyr platform. For 10× Genomics sequencing, DNA samples were sequenced on the Illumina HiSeq X Ten platform. The Hi-C library was sequenced on the Illumina NovaSeq PE150 platform. All DNA extraction and sequencing procedures were performed by the Novogene Company (Tianjin, China) (http://www.novogene.com/).
The PacBio reads were used for de novo assembly and further polished with Illumina data using Pilon v1.2217 to correct indel errors associated with homopolymer repeats in the PacBio data. The sequence consistency and quality of the assembled genome were evaluated using the Burrows-Wheeler Aligner (BWA)18 and Benchmarking Universal Single-Copy Orthologs (BUSCO)19. To anchor scaffolds onto pseudochromosomes, HiCUP v0.6.1 was used to map and process the reads from the Hi-C library20. The genome was divided into bins of equal sizes (500 k), and a contact map plotted with HiCPlotter confirmed the genome structure and quality21. The detailed methods are provided in the Supplementary Methods.
RNA-seq
Root, stem, leaf, node, and spike tissues of the same E. ophiuroides ‘Ganbei’ individual were sampled, frozen in liquid nitrogen, and subjected to extraction22. Thereafter, cDNA libraries were constructed, and the transcriptomes were sequenced on the Illumina HiSeq 2500 platform by Novogene (Tianjin, China) (http://www.novogene.com/). In addition, these five RNA samples were mixed, and full-length transcriptome sequencing was performed on the PacBio Sequel platform.
Genome prediction and annotation
Genome prediction and annotation mainly included repeat sequence prediction, gene annotation, and noncoding RNA (ncRNA) prediction. Repeat sequences were predicted through homology searches in the Repbase database (http://www.girinst.org/repbase) by using RepeatMaster and RepeatProteinMask software (http://www.repeatmasker.org/)23. The insertion times of the long terminal repeats (LTRs) were estimated using the formula K/2r (r = 1.3 × 10−8), considering a higher substitution rate in intergenic regions than in coding regions.
Augustus (http://bioinf.uni-greifswald.de/augustus/), GlimmerHMM (http://ccb.jhu.edu/software/glimmerhmm/) and SNAP (http://homepage.mac.com/iankorf/) were used for de novo gene structure prediction. RNA-seq data from roots, stems, leaves, nodes and spikes were also used to identify gene structures with BLAT (http://genome.ucsc.edu/cgi-bin/hgBlat). In total, 45,472 primitive gene models predicted from the above three prediction methods were corrected with RNA-seq data and integrated into a nonredundant and more complete gene set24. Finally, a gene set containing 36,572 gene models was obtained. Furthermore, gene function annotation was performed via BLASTP (E value ≤ 1e−5) searches against the NR (https://www.ncbi.nlm.nih.gov/), SwissProt (http://www.UniProt.org/), KEGG (http://www.genome.jp/kegg/) and InterPro (https://www.ebi.ac.uk/interpro/) protein databases.
The noncoding RNA annotations included tRNAs, rRNAs, miRNAs, and snRNAs. tRNAscan-SE (http://lowelab.ucsc.edu/tRNAscan-SE/) was used to search the tRNA sequences in the genome assembly of E. ophiuroides. The rRNAs of E. ophiuroides were identified by BLAST searches in related species, and the miRNA and snRNA sequences were predicted by applying INFERNAL (http://infernal.janelia.org/) against the Rfam database (v12.0). The detailed methods are provided in the Supplementary Methods.
Comparative genomic analysis
Genes of thirteen plant species (E. ophiuroides, Aegilops tauschii, Brachypodium distachyon, Hordeum vulgare, Oryza sativa, Panicum hallii, S. italic, S. viridis, Z. mays, S. bicolor, Zoysia japonica, Triticum aestivum and Arabidopsis thaliana) were used to analyze gene families. First, the gene sets of each species were filtered by retaining the longest transcript in the coding region and excluding genes encoding proteins with <50 amino acids. Thereafter, OrthoMCL v2.0.925 was used to cluster the filtered gene sets of the thirteen plant species. Through this analysis, we obtained single-copy orthologous groups for phylogenetic analysis and divergence time estimation. A phylogenetic tree was constructed using RAxML with the maximum likelihood-based method and the GTRGAMMA model26. Divergence times were predicted using the MCMCtree module of PAML v4.7 (http://abacus.gene.ucl.ac.uk/software/paml.html)27 with the following process parameter settings: burn-in = 10,000, sample-number = 100,000 and sample frequency = 2. Furthermore, CAFE (v1.6) (http://sourceforge.net/projects/cafehahnlab/) was used to analyze the expansion and contraction of gene families28. The unique, expanded, and contracted gene families of E. ophiuroides were annotated via GO and KEGG analyses.
Resistance (R) gene identification
To identify R genes in the E. ophiuroides and S. bicolor genomes, HMMER v3.1 with the hidden Markov model (HMM) profile was used to scan for the NB-ARC domain in the Pfam protein family (NB-ARC: PF00931)29. The candidate genes containing the nucleotide-binding site (NBS) domain were confirmed in the NCBI Conserved Domain Database (CDD)30 and Pfam database31, and the genes that did not contain the NBS domain were removed. Each of the candidate genes was checked manually by using available annotations in GenBank to confirm that they encoded the corresponding NBS candidate proteins.
EoPROG gene expression assays and protein structure prediction
Total RNA was extracted from root, stem, leaf, node, and spike tissues using RNA extraction reagent (Vazyme, Nanjing, China). Six EoPROG genes, presenting close evolutionary relationships with OsPROG1 in rice, were subjected to expression assays with quantitative RT-PCR in different tissues. The primers for these genes were designed using Primer 5.0 software, and they are listed in Supplementary Table 1. EoActin was used as a housekeeping gene32. Furthermore, the protein structures of EoPROG1, EoPROG2, OsPROG1, and Sobic.002G036600.2 were predicted using SWISS-MODEL (https://swissmodel.expasy.org/).
Ectopic expression of EoPROG1 and EoPROG2 in Arabidopsis
The coding sequences (CDSs) of EoPROG1 and EoPROG2 were first cloned into the pMD19-T vector and then introduced into the pCAMBIA1305.1 vector by LR recombination. The pCAMBIA1305.1 vector contains the hygromycin resistance gene Hyg. The pCAMBIA1305.1-EoPROG plasmids were introduced into Agrobacterium tumefaciens strain EHA105 and then transformed into the Col-0 ecotype33. The transgenic plants were screened by 50 μg mL−1 hygromycin and identified using RT-PCR with the primer pair Hyg-F/R. The reference sequence was actin 2 (AtAct2, tair: AT3G18780). The primers were listed in Supplementary Table 1. The significant difference analysis was performed by SPSS Statistics v.18.0 (Duncan’s test) (SPSS Inc., Chicago, IL, United States).
Results
Assembly and annotation
The E. ophiuroides genome was assessed by survey analysis, with an estimated genome size, heterozygosity level, and repeat frequency of 856.12 Mb, 0.76% and 59.87%, respectively (Supplementary Table 2, Supplementary Fig. 1a). The assembled genome size was confirmed to be 852.31 Mb, including 640 scaffolds with an N50 of up to 3.67 Mb via next-generation Illumina, Pacific Biosciences (PacBio), and Nanopore sequencing combined with 10× Genomics technologies (Table 1, Supplementary Table 3). In the sequence consistency assessment, the mapping rate of the transcriptome reads in the assembled genome was ~98.87%, and the coverage was ~99.78% (Supplementary Table 4). After quality assessment, the completeness of the gene set in the assembled genome was 95.4% (Supplementary Table 5). These results showed that the assembled E. ophiuroides genome presented high sequence consistency and completeness.
Table 1.
Statistics of assembled E. ophiuroides
| Assembly | BioNano assembly | Hi-C assembly | |
|---|---|---|---|
| Total length | 852.31 Mb | 867.40 Mb | 867.43 Mb |
| Contig N50 | 1.86 Mb | 1.86 Mb | 1.76 Mb |
| Scaffold number | 640 | 363 | 341 |
| Scaffold N50 | 3.67 Mb | 6.10 Mb | 86.05 Mb |
| Scaffold N90 | 0.91 Mb | 1.51 Mb | 65.49 Mb |
| Longest scaffolds | 15.67 Mb | 18.07 Mb | 114.81 Mb |
After further improvement of the BioNano assembly, the genome size was confirmed to be 867.40 Mb, including 363 scaffolds, with a scaffold N50 of up to 6.10 Mb (Table 1, Supplementary Table 3). The contigs of the Hi-C library were clustered into 9 pseudochromosomes (Supplementary Table 6, Supplementary Fig. 1b), which was consistent with the results of previous studies. A total of 92.61% of the contigs were assembled into 9 pseudochromosomes, and the scaffold N50 reached 86.05 Mb (Supplementary Table 6, Table 1). The longest and shortest pseudochromosomes were chromosomes 1 and 9, respectively, with sizes of 114.81 and 65.49 Mb (Supplementary Table 6). According to the above analyses, the final assembly of the E. ophiuroides genome was 867.43 Mb in size and included 341 scaffolds, with a scaffold N50 of 86.05 Mb (Table 1). The E. ophiuroides genome assembly was improved to the chromosome level, with a 92.61% mapping rate on 9 pseudochromosomes.
We identified a total of ~536.07 Mb of repetitive elements, which occupied 61.80% of the genome (Supplementary Table 7). In total, ~528.49 Mb of transposon elements (TEs), which occupied 60.93% of the genome, were annotated (Supplementary Table 8). LTR retrotransposons were the most abundant class of transposons, accounting for 54.26% (~470.69 Mb) of the E. ophiuroides genome (Supplementary Table 8). Among these elements, Ty1/copia and Ty3/gypsy occupied 10.50% and 41.67% of the assembled genome, respectively (Supplementary Table 8). DNA transposons, long interspersed nuclear elements (LINEs) and short interspersed nuclear elements (SINEs) accounted for 4.15, 1.98, and 0.003% of the assembled genome (Supplementary Table 8).
In total, 36,572 genes were identified, with an average length of 3.28 kb and a mean CDS length of 1.14 kb (Supplementary Table 9). The average number of exons per gene and the exon length in the E. ophiuroides genome were similar to those in other reported Poaceae plant genomes (Supplementary Table 9). However, the average intron length in the E. ophiuroides genome was obviously longer than that in other Poaceae plants (Supplementary Table 9). In addition to protein-coding genes, 2856 miRNA, 616 tRNA, 231 rRNA, and 5174 snRNA genes were identified, accounting for a total of 0.13% of the centipedegrass genome (Supplementary Table 10). The 36,572 identified genes were compared with protein sequences in the NR, SwissProt, KEGG, and InterPro protein databases, and 34,161 (93.4%) genes were annotated in at least one of the protein databases (Supplementary Table 11, Supplementary Fig. 2a). The annotated genes were subjected to BUSCO analysis, and 91.6% completion was achieved.
Comparative genomic analysis
To reveal the gene family contraction and expansion of E. ophiuroides, we compared the gene families of A. tauschii, B. distachyon, H. vulgare, O. sativa, P. hallii, S. italica, S. viridis, Z. mays, S. bicolor, Z. japonica, T. aestivum (T. aestivum AA, T. aestivum BB and T. aestivum DD), E. ophiuroides and A. thaliana, which served as an outgroup (Supplementary Table 12). All the genes from these species were clustered into 41,956 gene families, among which 9313 gene families were shared by all Poaceae species. A total of 643 gene families containing 1650 genes appeared to be unique to E. ophiuroides. The GO enrichment analysis of unique E. ophiuroides genes showed that these genes were associated with 50 GO terms, and the most enriched GO terms were “transferase activity” and “zinc ion binding” (Supplementary Fig. 2b). In addition, KEGG pathway analysis of the unique E. ophiuroides genes showed that the most enriched pathways were “base excision repair” and “plant-pathogen interaction” (Supplementary Fig. 2c).
Altogether, 1114 single-copy genes were identified from E. ophiuroides and 13 other Poaceae species for phylogenetic analysis (Supplementary Fig. 3a). Our results indicated that E. ophiuroides and S. bicolor evolved as sister groups that diverged from their common ancestor ~16.8 Mya, and Z. mays was identified as sister to the Eremochloa-Sorghum lineage (Fig. 1c). This phylogenetic topology was consistent with the traditional classification in which E. ophiuroides and S. bicolor belong to Andropogoneae and Z. mays belong to Maydeae in Panicoideae. To further study the genome evolution of E. ophiuroides, we investigated the intracollinearity of E. ophiuroides and the collinearity between E. ophiuroides and other Poaceae species. By analyzing the paralogous relationships among the nine chromosomes of E. ophiuroides, we identified seven major duplications between chromosomes 7 and 5, 3 and 4, 1 and 2, 9 and 2, 3 and 1, 6 and 3, and 8 and 6 and one major duplications within chromosome 1 (Supplementary Fig. 3b).
Furthermore, an analysis of gene family expansion/contraction during the evolution of those 13 plant species was performed based on the phylogenetic relationships. A total of 581 gene families were found to be expanded in centipedegrass, and 70 gene families were contracted (Fig. 1c). The KEGG pathway analysis of the 581 expanded gene families containing 3100 genes showed that these expanded gene families were involved in pathogen resistance-related pathways, among which the most enriched pathway was “plant-pathogen interaction” (Supplementary Fig. 4a). In addition, the GO enrichment analysis showed that these genes were associated with disease resistance, such as “defense response”, “response to stress” and “response to stimulus” (Supplementary Fig. 4b).
After that, we analyzed the shared gene families between S. bicolor and E. ophiuroides (Fig. 1c). A total of 338 gene families were shared between E. ophiuroides and S. bicolor only, and 643 gene families appeared to be unique to E. ophiuroides (Supplementary Fig. 4c). In addition, the comparison of gene families between E. ophiuroides and S. bicolor revealed 83 expanded gene families and 56 contracted gene families in E. ophiuroides. The above results showed that “plant-pathogen interaction” was the most enriched pathway in unique genes and expanded gene families in E. ophiuroides.
Reactive oxygen species (ROS)-related genes, flavonoid-related genes, and plant disease-related R genes play significant roles in detecting pathogen attack and activating the defensive response to pathogens34–36. Two upstream regulatory gene families of ROS (https://www.kegg.jp/pathway/map04626) were identified in E. ophiuroides and S. bicolor, and the gene numbers were similar between these two species (Supplementary Table 13). Moreover, 14 flavonoid-related gene families were identified in E. ophiuroides and S. bicolor, and the gene numbers of the cinnamate 4-hydroxylase (C4H), flavonoid 3’-hydroxylase (F3’H) and flavonoid 3-O-glucosyltransferase (UFGT) families in E. ophiuroides were greater than those in S. bicolor (Supplementary Table 13). The best-characterized R genes encode products that contain an NBS domain and a series of leucine-rich repeats (LRRs)37. Interestingly, the NBS gene family nearly doubled in E. ophiuroides in comparison to S. bicolor (597 vs. 337) (Supplementary Table 13, Table 2). The greatest number of NBS genes in E. ophiuroides was found on chromosome 4, and their distribution was dispersive (Table 2, Supplementary Fig. 4d). However, the greatest number of NBS genes in S. bicolor was found on chromosome 5 (Table 2), and most NBS gene clusters were located at the distal ends of the chromosomes38.
Table 2.
NBS gene numbers in E. ophiuroides and S. bicolor
| E. ophiuroides | S. bicolor | ||
|---|---|---|---|
| Chromosome | NBS gene | Chromosome | NBS gene |
| Chr1 | 30 | Chr1 | 18 |
| Chr2 | 62 | Chr2 | 51 |
| Chr3 | 71 | Chr3 | 15 |
| Chr4 | 210 | Chr4 | 8 |
| Chr5 | 29 | Chr5 | 101 |
| Chr6 | 14 | Chr6 | 18 |
| Chr7 | 39 | Chr7 | 31 |
| Chr8 | 18 | Chr8 | 47 |
| Chr9 | 60 | Chr9 | 21 |
| – | – | Chr10 | 26 |
| Other | 64 | Other | 1 |
| Total | 597 | 337 | |
Differences in the distribution of genes and repeat sequences between E. ophiuroides and S. bicolor
The E. ophiuroides and S. bicolor genomes exhibited similar retrotransposon contents (56.24% and 54.52%, respectively)39, but the distribution of LTRs on the chromosomes differed between the two species. The LTR density in distal chromosome regions was higher in E. ophiuroides than in S. bicolor, and the gene density in distal chromosome regions was lower in E. ophiuroides (Supplementary Fig. 5). The results indicated that more LTRs were retained in intergenic regions in E. ophiuroides, which might be caused by LTR family expansion or a low LTR turnover rate. To reveal the evolution of LTRs in E. ophiuroides and S. bicolor, we identified full-length LTRs and calculated their insertion times. A total of 21,326 and 19,968 full-length LTRs were identified in E. ophiuroides and S. bicolor, respectively, and LTRs underwent expansion in E. ophiuroides ~2 million years ago, while most LTR insertions in S. bicolor occurred <1 million years ago (Supplementary Fig. 6a).
The athila, del and tat subfamilies of the Ty3/gypsy elements and the sire subfamily of the Ty1/copia elements were the most enriched retrotransposon classes in E. ophiuroides and S. bicolor (Supplementary Table 14). The insertion times of the del elements were similar between E. ophiuroides and S. bicolor, while the other three subfamilies all showed earlier insertions in E. ophiuroides (Supplementary Fig. 6b). We also illustrated the distribution of athila and del elements on the chromosomes. del elements distributed in distal and proximal chromosome regions in E. ophiuroides and S. bicolor. athila elements were rarely found in gene-rich regions in S. bicolor but were distributed in distal regions of the E. ophiuroides chromosomes (Supplementary Fig. 5). These results implied that athila elements have been inserted and maintained in the E. ophiuroides genome for ~2 million years. The formation of solo-LTRs through unequal homologous recombination counterbalances the amplification of LTR retrotransposons40. The retention of athila elements in E. ophiuroides is likely due to their perennation and vegetative propagation characteristics, which reduce the recombination rate.
Reshuffling and structural evolution of E. ophiuroides chromosomes
It has been reported that the chromosomes of grasses undergo extensive rearrangements despite the close evolutionary relationships among grasses12,41. Previous studies have revealed that the ancestor of grasses experienced one WGD and two nest chromosome fusion events, producing 12 common chromosomes12,41,42. To investigate the evolution of the E. ophiuroides chromosomes, the genomes of E. ophiuroides, B. distachyon, O. sativa, S. bicolor, Z. mays, and S. italica, belonging to the three subfamilies of grasses (Pooideae, Ehrhartoideae, and Panicoideae), were chosen for comparative analyses. Massive collinear blocks were identified among these grasses, which indicated well-preserved genome structures in this family. A total of 45.10% (15,620 gene pairs), 46.39% (16,066 gene pairs), 58.36% (20,212 gene pairs), 51.72% (17,911), and 50.33% (17.430) of the E. ophiuroides genome was syntenic with B. distachyon, O. sativa, S. bicolor, Z. mays, and S. italica, respectively (Fig. 2, Supplementary Table 15).
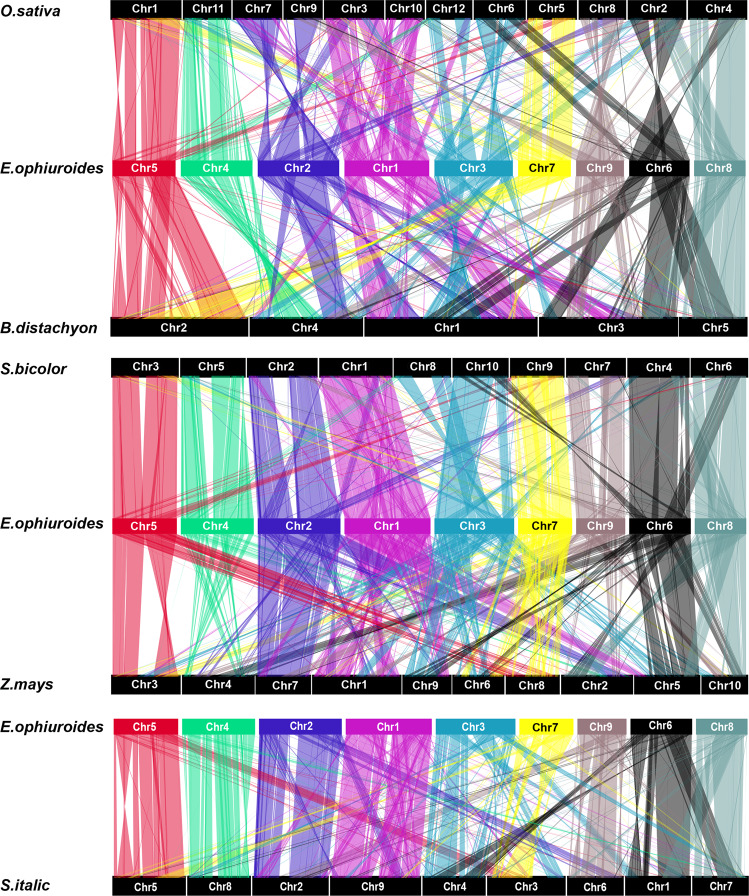
Fig. 2. Chromosome analyses of E. ophiuroides.
Syntenic blocks between E. ophiuroides and other sequenced grass genomes, including O. sativa, B. distachyon, S. bicolor, Z. mays, and S. italica
Using the rice genome as the reference comparison strain genome because of its 12 retained chromosomes, we found that E. ophiuroides chromosome 1 is largely collinear with rice chromosomes 3 and 10, E. ophiuroides chromosome 2 is collinear with rice chromosomes 7 and 9, and E. ophiuroides chromosome 3 is collinear with rice chromosomes 3 and 10 (Fig. 2). In addition, large regions of collinearity were identified between E. ophiuroides chromosome 6 and rice chromosome 2 and between E. ophiuroides chromosome 9 and rice chromosome 8, while small regions of collinearity were detected between E. ophiuroides chromosome 6 and rice chromosomes 6 and 4 and between E. ophiuroides chromosome 9 and rice chromosomes 9 and 4 (Fig. 2). This result demonstrated that complex chromosome rearrangements as well as chromosome fusions have occurred among the E. ophiuroides chromosomes. In addition to the three nest chromosome fusions that occurred in E. ophiuroides (chromosomes 1, 2, and 3), two and three nest chromosome fusions were identified in the S. bicolor and S. italica genomes, respectively (Fig. 2). Considering the close relationship between S. bicolor and E. ophiuroides, a further comparative analysis of collinearity was performed between them. We noted that E. ophiuroides chromosome 3 was highly collinear with S. bicolor chromosomes 8 and 10 (Fig. 2), indicating that S. bicolor chromosomes 8 and 10 might have fused during their evolution, leading to the difference in chromosome number between S. bicolor and E. ophiuroides. A similar chromosome fusion event associated with high collinearity between chromosome 3 of S. italica and chromosomes 8 and 9 of S. bicolor was also detected12.
The phylogenetic relationships among the grasses were refined and showed that E. ophiuroides split from S. bicolor, which occurred after the divergence of S. italica (Fig. 3). By combining the data obtained in the current study with previously reported genome data from rice41, sorghum43, and foxtail millet12, we performed a detailed analysis of the common and lineage-specific patterns of conservation between those different genomes. Accordingly, we proposed a model (Fig. 3) of the structural evolution of E. ophiuroides and three other grass species from a common ancestor with a basic number of five chromosomes (A5, A7, A11, A8, and A4), including orthologous chromosomes that exhibit shared ancestral duplications. In rice, sorghum, and foxtail millet, the shared duplications corresponding to ancestral chromosomes A5, A7, A11, A8, and A4 were congruent with those reported previously by Salse et al.41 and Zhang et al.12. In E. ophiuroides, a duplication corresponding to ancestral chromosome A5 was found between chromosomes 5 and 7; a duplication corresponding to A7 was found on chromosomes 1 and 2; ancestral chromosome A11 corresponded to conserved chromosomes 4 and 3; a duplication corresponding to A8 was detected between chromosomes 9 and 2; and a duplication corresponding to A4 was observed on orthologous chromosomes 3, 8, and 6 (Fig. 3).
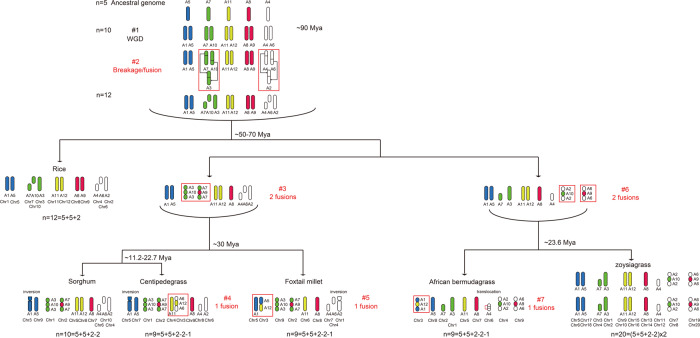
Fig. 3. A model of chromosome structural evolution of grasses.
A model of chromosome structural evolution in the centipedegrass, rice, sorghum, foxtail millet, African bermudagrass, and zoysiagrass genomes starting from a common ancestor with n = 5 chromosomes
As described in the previous studies12,41,42,44, an intermediate ancestor with n = 12 (5 + 5 + 2) chromosomes evolved from a common ancestor with a genome including five chromosomes (A5, A7, A11, A8, and A4) after a single WGD (event #1) and two nest chromosome fusion events (event #2) observed in grass species (Fig. 3). Subsequently, rice retained the basic structure of 12 chromosomes, although additional segmental duplications occurred in its genome. In the ancestral genome with 12 chromosomes, the occurrence of two chromosomal fusions (between A3 and A10 and between A7 and A9) resulted in another intermediate ancestor of sorghum, foxtail millet, and centipedegrass with n=10 chromosomes (5 + 5 + 2 − 2) (event #3) (Fig. 3). Thereafter, the sorghum genome structure remained similar to that of the ancestral genome with ten chromosomes (5 + 5 + 2 − 2), while the centipedegrass genome underwent an additional chromosome fusion (between A6 and A12) (event #4), as did foxtail millet (between A5 and A12) (event #5) (Fig. 3). Thus, similar to foxtail millet, centipedegrass evolved independently from the ancestor with n = 10 chromosomes and finally developed a genome structure with 9 chromosomes (n = 9 = 5 + 5 + 2 − 2 − 1) (Fig. 3). Compared with sorghum, foxtail millet, and centipedegrass, two chromosomal fusions occurred in the intermediate ancestor of African bermudagrass and zoysiagrass (between A2 and A10 and between A6 and A9) (event #6) (Fig. 3). After that, the African bermudagrass genome underwent additional chromosome fusion (between A1 and A12) (event #7) and chromosome translocation in A4 (Fig. 3).
The prostrate growth habit of E. ophiuroides
Compared with domesticated grain crops, E. ophiuroides retains its prostrate growth habit. It has been reported that the prostrate growth of wild rice from Yuanjiang County in China is controlled by a semidominant gene, OsPROG1 (Os07g0153600), on chromosome 7, which encodes a single Cys(2)-His(2) zinc-finger protein13. Inactivating OsPROG1 function can lead to erect growth and an increased grain number13. The NCBI database shows that the preferred name of the OsPROG1 gene of rice is zinc finger protein GIS3 (GLABROUS INFLORESCENCE STEMS 3), and there are 5 GIS3 genes in O. sativa. According to these sequences, we identified six and 14 GIS3 homologs in S. bicolor and E. ophiuroides, respectively (Supplementary Table 16). Furthermore, three GIS3 homologs in S. bicolor were located on chromosome 2, and eight GIS3 homologs in E. ophiuroides were also located on chromosome 2 (Supplementary Table 16). Chromosome 2 of S. bicolor was highly collinear with chromosome 2 of E. ophiuroides (Fig. 2). Previous studies have shown that chromosome 2 of S. bicolor is collinear with chromosomes 7 and 9 of O. sativa and that the terminal sequences of chromosome 2 of S. bicolor aligned with those of chromosome 7 of O. sativa39. The collinearity analysis of chromosome segments containing OsPROG1 genes in O. sativa and PROG homologs in S. bicolor and E. ophiuroides showed an obvious series of PROG homologs in E. ophiuroides, resulting in the formation of gene clusters (Fig. 4a). These results showed obvious expansion of PROG homologs on chromosome 2 in the E. ophiuroides genome.
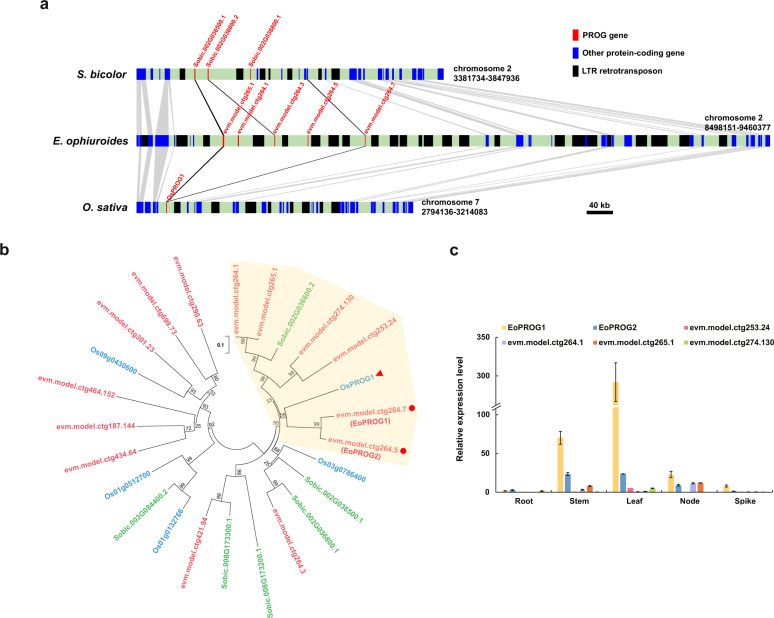
Fig. 4. Screening and analysis of candidate PROG genes for prostrate growth.
a Collinearity analysis of chromosome segments containing PROG homologs in O. sativa, S. bicolor, and E. ophiuroides. The red, blue, and black boxes indicate PROG genes, other protein-coding genes, and LTR retrotransposons, respectively. The gray region represents the shared collinear sequence. Black lines indicate the orthologous relationships of PROG genes. b Phylogenetic tree of PROG proteins. The gene IDs in blue represent the OsPROG1 gene (indicated by the red triangle) and its homologs in O. sativa. The gene IDs in green represent the homologs of the OsPROG1 gene in S. bicolor. The gene IDs in red represent the homologs of the OsPROG1 gene in E. ophiuroides. The red dots indicate the closest genetic relationships between candidate PROG genes and OsPROG1 in rice. The yellow area shows that one homologous gene in S. bicolor and six homologous genes in E. ophiuroides exhibited close genetic relationships with OsPROG1 in rice. c The expression patterns of six candidate PROG genes of E. ophiuroides in roots, stems, leaves, nodes, and spikes
The phylogenetic tree showed that 1 homolog (Sobic.002G036600.2) in S. bicolor and 6 homologs (evm.model.ctg264.7, evm.model.ctg264.5, evm.model.ctg264.1, evm.model.ctg265.1, evm.model.ctg274.130 and evm.model.ctg253.24) in E. ophiuroides showed close evolutionary relationships with OsPROG1 in rice (Fig. 4b). Among these genes, evm.model.ctg264.7 (EoPROG1) and evm.model.ctg264.5 (EoPROG2) exhibited the closest genetic relationship with OsPROG1 in rice (Fig. 4b). Amino acid sequence alignment showed that these genes shared a conserved C2H2-type zinc-finger domain (Supplementary Fig. 7a). According to quantitative RT-PCR data from various E. ophiuroides tissues (root, stem, leaf, node, and spike), EoPROG1 and EoPROG2 both showed high expression levels in stems and leaves (Fig. 4c). Protein structure prediction showed that these four proteins exhibited similar structures (Supplementary Fig. 7b).
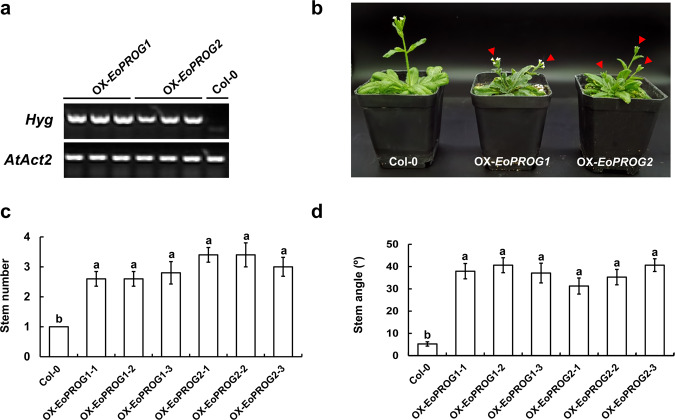
To investigate the function of EoPROGs, the CDSs of EoPROG1 and EoPROG2 were driven by the lac promoter and introduced into Arabidopsis. In total, eight hygromycin (Hyg)-resistant T1 transgenic lines of EoPROG1 and four T1 transgenic lines of EoPROG2 were obtained. Three T3 transgenic lines of EoPROG1 and EoPROG2 were identified with RT-PCR, and the EoPROG1 and EoPROG2 transgenic lines showed similar phenotypes (Fig. 5a, b). Compared with Col-0 plants, overexpressing EoPROG1 and EoPROG2 influenced the erect growth of Arabidopsis (Fig. 5b). The stem number of the OX-EoPROG transgenic lines was obviously greater than that of the Col-0 plants (Fig. 5c). Moreover, the stem angle of the OX-EoPROG transgenic lines was significantly higher than that of Col-0 plants (Fig. 5d). These results indicated that EoPROG1 and EoPROG2 might have significant functions in regulating the prostrate growth of E. ophiuroides.
Fig. 5. Ectopic expression of EoPROG1 and EoPROG2 in Col background Arabidopsis (Col-0).
a RT-PCR analyses of T3-overexpressing EoPROG1 and EoPROG2 transgenic lines. The reference gene is AtAct2. The resistance marker gene is Hyg. b Phenotypes of 48-day-old wild-type (Col-0) and T3 transgenic plants. c The stem number of wild-type (Col-0) and T3 transgenic plants. d The stem angle of wild-type (Col-0) and T3 transgenic plants. Data are the means (n = 10), with error bars showing the standard deviation. Letters above bars indicate significant differences between the respective values (p < 0.05)
Discussion
Chromosome structural evolution in E. ophiuroides
In our study, massive collinear blocks were detected by intergenic analyses between centipedegrass, brachypodium, rice, sorghum, maize, and foxtail millet, which revealed a close evolutionary relationship among these grass species. However, the apparent difference in genome size and chromosome numbers between centipedegrass and other grass species implied that centipedegrass chromosomes may have undergone different rearrangements from other species during its evolution. It is known that extensive chromosome rearrangements occurred in grass species despite their close evolutionary relationships12,41,45, and different grass species evolved from the first intermediate ancestor with 12 common chromosomes, which were derived from one WGD and two nest chromosome fusion events in the ancestor of the grasses12,41,42. Through a detailed comparative genomic analysis, we inferred that centipedegrass diverged from the second intermediate ancestor with n = 10 chromosomes, which was formed by two chromosomal fusions (between A3 and A10 A7 and A9) of the first intermediate ancestor. Interestingly, since Zhang et al.12 reported that foxtail millet chromosome 3 was collinear with ancestral chromosomes A5 and A12, we also found that centipedegrass chromosome 3 was homologous to ancestor chromosomes A6 and A12 in this study (Fig. 3). This finding indicated that two chromosomes (A6 and A12 or A5 and A12) of the ancestor might have fused into one chromosome corresponding to chromosome 3 in centipedegrass or foxtail millet. Since sorghum retains a similar chromosome constitution as the intermediate ancestor possessing n = 10 chromosomes, the occurrence of this extra chromosome fusion might be the major driving force for the formation of different genome structures in centipedegrass and foxtail millet. Furthermore, high collinearity between centipedegrass chromosome 3 and sorghum chromosomes 8 and 10 indicated that centipedegrass chromosome 3 resulted from the fusion of sorghum chromosomes 8 and 10, which most likely occurred after the divergence of centipedegrass from sorghum. A similar chromosome rearrangement was also discovered in foxtail millet12.
African bermudagrass and zoysiagrass are both warm-season turfgrasses classified in the Chloridoideae subfamily4,46; they differ from centipedegrass belonging to the Panicoideae subfamily. The chromosome rearrangement analysis showed that African bermudagrass and zoysiagrass had different intermediate ancestors with two chromosomal fusions (between A2 and A10 and between A6 and A9), while sorghum, centipedegrass and foxtail millet had the same intermediate ancestor as the other two chromosomal fusions (between A3 and A10 and between A7 and A9) (Fig. 3). These results showed that the different chromosomal fusion methods were related to the differentiation of different subfamilies. On that basis, the African bermudagrass genome underwent additional chromosome fusion (between A1 and A12) and chromosome translocation in A4 (Fig. 3), indicating that chromosome fusion events may have caused genomic variation and contributed to the speciation of grass plants.
Prostrate growth habit of E. ophiuroides
Among the three nest chromosome fusion events identified between E. ophiuroides and O. sativa, the fusion of chromosomes 7 and 9 of O. sativa to produce chromosome 2 of E. ophiuroides might have an important impact on the centipedegrass prostrate growth habit. In wild rice, the semidominant gene PROG1 on chromosome 7 has been reported to affect prostrate growth, and the inactivation of PROG1 function can lead to erect growth and more grains13. The PROG1 gene in rice encodes a single C2H2 zinc finger protein13 with the preferred name zinc finger protein GIS3 according to the NCBI database. In Asian cultivated rice O. sativa, a 110 kb deletion on the short arm of chromosome 7, which contains a tandem repeat of seven zinc finger genes, is closely linked to the OsPROG1 gene16. Deletions of the three zinc finger genes mentioned above have been verified to regulate the prostrate growth of wild rice16. In addition, a similar tandem repeat of a zinc finger protein-coding gene deletion is observed in African cultivated rice, indicating that the zinc finger gene cluster may exhibit the conserved function of regulating prostrate plant growth in the Poaceae family16. In African cultivated rice (Oryza glaberrima), the PROG7 gene, which is identical to OsPROG1, was verified to be necessary for the prostrate growth of African wild rice (Oryza barthii)47. The expression pattern showed that ObPROG7 had high expression levels in young leaves, leaf sheaths, and shoot apical meristems47. PROG homologs exhibited obvious expansion on chromosome 2 of the E. ophiuroides genome, and their locations were focused at the chromosome 2 termini (Fig. 4a). In addition, a large number of LTR retrotransposons appeared near PROG homologs, indicating that PROG gene expansion might be closely associated with LTR retrotransposon insertion (Fig. 4a). Therefore, the PROG homologs on chromosome 2 of the E. ophiuroides genome may serve significant functions in the prostrate growth of centipedegrass.
The phylogenetic tree showed that 1 PROG homolog (Sobic.002G036600.2) in sorghum and 6 PROG homologs (evm.model.ctg264.7, evm.model.ctg264.5, evm.model.ctg264.1, evm.model.ctg265.1, evm.model.ctg274.130 and evm.model.ctg253.24) in centipedegrass exhibited close genetic relationships with OsPROG1 in rice (Fig. 4b). Among these genes, EoPROG1 and EoPROG2 showed the closest genetic relationship with OsPROG1 (Fig. 4b). Both EoPROG1 and EoPROG2 exhibited high expression levels in the stems and leaves of centipedegrass, which is consistent with the expression pattern of ObPROG7 in African wild rice (Fig. 4c). Furthermore, EoPROG1 and EoPROG2 presented similar protein structures to OsPROG1 in rice (Supplementary Fig. 7b). The transgenic results showed that overexpressing EoPROG1 and EoPROG2 in Arabidopsis increased the number and angle of stems (Fig. 5c, d), which was consistent with previous research results in rice16. These results showed that EoPROG1 and EoPROG2 might be the most important genes in centipedegrass prostrate growth regulation.
The generation of a reference genome sequence of E. ophiuroides fills the sequencing gap in Eremochloa genus plants, enriches the available turfgrass genome sequences, and provides resources for centipedegrass molecular breeding. The comparative analysis of E. ophiuroides genomes allows us to better understand chromosome evolution in Poaceae and has great significance for our understanding of turfgrass prostrate growth.
Supplementary information
Acknowledgements
We thank Prof. Jun Wu and associate Prof. Jiayu Xue from Nanjing Agriculture University (China) for improving this manuscript and Prof. Kehua Wang and doctoral student Fengchao Cui from China Agricultural University for providing the genomic data of African bermudagrass. This work was funded by the National Natural Science Foundation of China [Grant No. 31902060, 31771870, 31902046, 32072608], the Natural Science Foundation of Jiangsu Province, China [Grant No. BK20180315, BK20200288], the Jiangsu Agricultural Science and Technology Innovation Fund [Grant No. CX(20)3141], and the Open Fund of Jiangsu Provincial Key Laboratory for the Research and Utilization of Plant Resource, China [Grant No. JSPKLB201840, JSPKLB201817, JSPKLB201927].
Author contributions
J.W., J.x.L., and J.Z. conceived and designed the study. R.W. and J.W. carried out the laboratory work. H.Z. supported data archiving. J.W., J.j.L., H.Z., R.W., H.W., R.C., H.G., J.C., and L.L. analyzed the data. H.Z., J.W., and J.j.L. contributed to data interpretation. J.W., H.Z., and J.j.L. wrote the paper.
Data availability
The genomic raw data in this study can be found in the NCBI repository http://www.ncbi.nlm.nih.gov/bioproject/PRJNA682293. The assembly and annotation data in this study can be found at https://figshare.com/s/8256acffdb73bb050045.
Conflict of interest
The authors declare no competing interests.
Footnotes
These authors contributed equally: Jingjing Wang, Hailing Zi
Contributor Information
Jianjian Li, Email: lijianjian2013@yahoo.com.
Junqin Zong, Email: zongjq1980@163.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41438-021-00636-6.
References
- 1.Li J, et al. Genetic diversity in centipedegrass [Eremochloa ophiuroides (Munro) Hack.] Hortic. Res. 2020;7:4. doi: 10.1038/s41438-019-0228-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Islam MA, Hirata M. Centipedegrass (Eremochloa ophiuroides (Munro) Hack.): growth behavior and multipurpose usages. Grassl. Sci. 2005;51:183–190. doi: 10.1111/j.1744-697X.2005.00014.x. [DOI] [Google Scholar]
- 3.Milla-Lewis SR, et al. Use of sequence-related amplified polymorphism (SRAP) markers for comparing levels of genetic diversity in centipedegrass (Eremochloa ophiuroides (Munro) hack.) germplasm. Genet. Resour. Crop Ev. 2012;59:1517–1526. doi: 10.1007/s10722-011-9780-8. [DOI] [Google Scholar]
- 4.Tanaka H, et al. Sequencing and comparative analyses of the genomes of zoysiagrasses. DNA Res. 2016;23:171–180. doi: 10.1093/dnares/dsw006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watson, L. & Dallwitz, M. J. The Grass Genera of the World (CAB International, 1992).
- 6.Brown WV. A cytological study of some texas gramineae. Bull. Torre Bot. Club. 1950;77:63–76. doi: 10.2307/2482267. [DOI] [Google Scholar]
- 7.Hanna WW, Burton GW. Cytology, reproductive behavior and fertility characteristics of centipedegrass. Crop Sci. 1978;18:835–837. doi: 10.2135/cropsci1978.0011183X001800050038x. [DOI] [Google Scholar]
- 8.Qu, R., Luo, H. & Meier V. D. Turfgrass. Compendium of Transgenic Crop Plants: Transgenic Plantation Crops, Ornamentals and Turf Grasses. (2008).
- 9.Schnable PS, et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 10.Tenaillon MI, Hufford MB, Gaut BS, Ross-Ibarra J. Genome size and transposable element content as determined by high-throughput sequencing in maize and Zea luxurians. Genome Biol. Evol. 2011;3:219–229. doi: 10.1093/gbe/evr008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deschamps S, et al. A chromosome-scale assembly of the sorghum genome using nanopore sequencing and optical mapping. Nat. Commun. 2018;9:4844. doi: 10.1038/s41467-018-07271-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang G, et al. Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nat. Biotechnol. 2012;30:549–554. doi: 10.1038/nbt.2195. [DOI] [PubMed] [Google Scholar]
- 13.Tan L, et al. Control of a key transition from prostrate to erect growth in rice domestication. Nat. Genet. 2008;40:1360–1364. doi: 10.1038/ng.197. [DOI] [PubMed] [Google Scholar]
- 14.Jin J, et al. Genetic control of rice plant architecture under domestication. Nat. Genet. 2008;40:1365–1369. doi: 10.1038/ng.247. [DOI] [PubMed] [Google Scholar]
- 15.Hua S, Cao B, Zheng B, Lia B, Sun C. Quantitative evaluation of influence of Prostrate Growth 1 gene on rice canopy structure based on three-dimensional structure model. Field Crops Res. 2016;194:65–74. doi: 10.1016/j.fcr.2016.05.004. [DOI] [Google Scholar]
- 16.Wu Y, et al. Deletions linked to PROG1 gene participate in plant architecture domestication in Asian and African rice. Nat. Commun. 2018;9:1–10. doi: 10.1038/s41467-017-02088-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walker BJ, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 20.Wingett, S. et al. HiCUP: pipeline for mapping and processing Hi-C data. F1000Res4 (2015). [DOI] [PMC free article] [PubMed]
- 21.Akdemir KC, Chin L. HiCPlotter integrates genomic data with interaction matrices. Genome Biol. 2015;16:198. doi: 10.1186/s13059-015-0767-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie Q, et al. De novo assembly of the Japanese lawngrass (Zoysia japonica Steud.) root transcriptome and identification of candidate unigenes related to early responses under salt stress. Front. Plant Sci. 2015;6:610. doi: 10.3389/fpls.2015.00610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bao W, Kojima KK, Kohany O. Repbase update, a database of repetitive elements in eukaryotic genomes. Mob. DNA-UK. 2015;6:11. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haas BJ, et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 2008;9:R7. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fischer S, et al. Using OrthoMCL to assign proteins to OrthoMCL-DB groups or to cluster proteomes into new Ortholog groups. Curr. Protoc. Bioinforma. 2011;35:6.12.1–6.12.19. doi: 10.1002/0471250953.bi0612s35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 27.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 28.Bie TD, Cristianini N, Demuth JP, Hahn MW. CAFE: a computational tool for the study of gene family evolution. Bioinformatics. 2006;22:1269–1271. doi: 10.1093/bioinformatics/btl097. [DOI] [PubMed] [Google Scholar]
- 29.Eddy SR. Accelerated profile HMM searches. PLoS Comput. Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marchler-Bauer A, et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015;43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finn RD, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chung MS, et al. Functional and genomic characterization of a wound-and methyl jasmonate-inducible chalcone isomerase in Eremochloa ophiuroides [Munro] Hack. Plant Physiol. Bioch. 2019;144:355–364. doi: 10.1016/j.plaphy.2019.10.008. [DOI] [PubMed] [Google Scholar]
- 33.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 34.Camejo D, Guzmán-Cedeño A, Vera-Macias L, Jiménez A. Oxidative post-translational modifications controlling plant-pathogen interaction. Plant Physiol. Bioch. 2019;144:110–117. doi: 10.1016/j.plaphy.2019.09.020. [DOI] [PubMed] [Google Scholar]
- 35.Camargo-Ramírez R, Val-Torregrosa B, San Segundo B. MiR858-mediated regulation of flavonoid-specific MYB transcription factor genes controls resistance to pathogen infection in Arabidopsis. Plant Cell Physiol. 2018;59:190–204. doi: 10.1093/pcp/pcx175. [DOI] [PubMed] [Google Scholar]
- 36.Gururani MA, et al. Plant disease resistance genes: current status and future directions. Physiol. Mol. Plant P. 2012;78:51–65. doi: 10.1016/j.pmpp.2012.01.002. [DOI] [Google Scholar]
- 37.Lupas A, Van DM, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 38.Cheng X, et al. A genomic analysis of disease-resistance genes encoding nucleotide binding sites in Sorghum bicolor. Genet. Mol. Biol. 2010;33:292–297. doi: 10.1590/S1415-47572010005000036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paterson AH, et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- 40.Sharma A, Schneider KL, Presting GG. Sustained retrotransposition is mediated by nucleotide deletions and interelement recombinations. Proc. Natl Acad. Sci. USA. 2008;105:15470–15474. doi: 10.1073/pnas.0805694105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salse J, et al. Identification and characterization of shared duplications between rice and wheat provide new insight into grass genome evolution. Plant Cell. 2008;20:11–24. doi: 10.1105/tpc.107.056309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murat F, et al. Ancestral grass karyotype reconstruction unravels new mechanisms of genome shuffling as a source of plant evolution. Genome Res. 2010;20:1545–1557. doi: 10.1101/gr.109744.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paterson AH, Bowers JE, Chapman BA. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl Acad. Sci. USA. 2004;101:9903–9908. doi: 10.1073/pnas.0307901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gaut BS. Evolutionary dynamics of grass genomes. N. Phytol. 2002;154:15–28. doi: 10.1046/j.1469-8137.2002.00352.x. [DOI] [Google Scholar]
- 45.Li G, et al. A high-quality genome assembly highlights rye genomic characteristics and agronomically important genes. Nat. Genet. 2021;53:574–584. doi: 10.1038/s41588-021-00808-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cui F, et al. The genome of the warm-season turfgrass African bermudagrass (Cynodon transvaalensis) Hortic. Res. 2021;8:1–16. doi: 10.1038/s41438-020-00428-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu M, et al. The domestication of plant architecture in African rice. Plant J. 2018;94:661–669. doi: 10.1111/tpj.13887. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genomic raw data in this study can be found in the NCBI repository http://www.ncbi.nlm.nih.gov/bioproject/PRJNA682293. The assembly and annotation data in this study can be found at https://figshare.com/s/8256acffdb73bb050045.