Abstract
Pancreatic cancer is a highly aggressive cancer with an exceedingly low rate of response to treatments, which calls for comprehensive molecular characterization of pancreatic cancer cell lines (PCCLs). We screened multi-layer molecular data of 36 PCCLs, including gene mutation, gene expression, microRNA (miRNA) expression, and protein profiles. Our comparative analysis of genomic mutations found that PCCLs recapitulated genomic alterations of the primary tumor and suggested potential therapeutic strategies for clinical interventions. The panel of 36 PCCLs was classified into 2 subgroups based on transcriptomic mRNA expression, wherein the C1 subgroup was characterized with differentiation, whereas C2 cell lines were featured with immunity, angiogenesis, epidermis, and proliferation. Transcriptomic classification was further recapitulated by miRNA and protein expression. Additionally, the differential proteins between C1 and C2 subgroups were prominently involved in epidermal growth factor receptor (EGFR) signaling, phosphatidylinositol 3-kinase (PI3K) signaling, and mitogen-activated protein kinase (MAPK) signaling pathways. Tumor samples from different subgroups exhibited distinct infiltration of CD4 naive cells and monocytes. Remarkably, patients in subgroups C1 showed longer survival, whereas those in C2 had worse clinical outcome. Further integrative analysis revealed that temozolomide and NVP-TAE684 showed higher sensitivity in the C1 subgroup, whereas the C2 cell lines were more sensitive to SR1001 and SRT-1720. Our results also showed that PCCLs with mutations in CDKN2A, TP53, and SMAD4 were more sensitive to certain anti-cancer drugs. Our integrative analysis identified molecular features of pancreatic cancer that were associated with clinical significance and drug sensitivity, providing potentially effective strategies for precision treatments of patients with pancreatic cancer.
Keywords: pancreatic cancer, cancer cell lines, drug response, multi-omics data, integrative analysis, pancreatic cancer subgroups
Graphical abstract
This study provides a comprehensive characterization of multiple pancreatic cancer cell lines, which are divided into 2 subgroups that showed therapeutic differences.
Introduction
Pancreatic cancer is a major cause of cancer-related mortality worldwide, which is associated with extremely poor prognosis.1, 2, 3, 4 Patients with pancreatic cancer are typically diagnosed in advanced stages and refractory to most clinical treatments of tumor.5 One of the major obstacles impeding the improvement of diagnosis and therapy for pancreatic cancer is that the whole scene of molecular alterations in pancreatic cancer has been remaining obscure. Therefore, it’s urgent to characterize the molecular landscape of pancreatic cancer to facilitate the discovery of biomarkers for early diagnosis and potential targets for clinical interventions.
Immortalized cell lines derived from human cancers have been widely used in exploring tumor biology and discovery of anti-cancer drugs.6,7 Characterization of molecular alterations in human cancer cell lines has shown relevant contributions to understanding the sophisticated mechanisms underlying tumor development.8,9 The project of Cancer Cell Line Encyclopedia (CCLE) generated multi-layer, high-throughput data of more than 1,000 cancer cell lines,6,10,11 including genome (whole-exome sequencing [WES] for 326 cell lines; whole-genome sequencing [WGS] for 329 cell lines), transcriptome (RNA sequencing [RNA-seq] for 1,019 cell lines; NanoString microRNA [miRNA] profiling for 954 cell lines), proteome (reverse-phase protein array [RPPA] for 213 cell lines), epigenome (reduced representation bisulfite sequencing [RRBS] for 843 cell lines; global profiling of 42 histone modifications for 897 cell lines), and metabolome (metabolic profiling of 225 metabolites for 928 cell lines). Furthermore, the Cancer Dependency Map Project12 (DepMap) provided a cancer-dependency profile generated from high-throughput screening of loss-of-function experiments, such as CRISPR and RNAi. DepMap also cataloged valuable resources for systematically identifying biomarkers of cancer vulnerabilities and drug response, including omics data from CCLE and drug sensitivities from the Genomics of Drug Sensitivity in Cancer (GDSC)13 and the Cancer Therapeutics Response Portal (CTRP).14 These collaborative international efforts have offered the cancer research community a great opportunity of exploring the molecular features, cancer vulnerability, and drug response for human cancers. Furthermore, the examination of the omics data of large panels of cancer cell lines derived from individual cancer types would benefit the precision treatment for tumor patients. For example, Caruso et al.9 presented a molecular landscape of liver cancer by screening genetic, mRNA, miRNA, and protein profiles in 34 liver cancer cell lines. They identified genetic alternations and gene-expression patterns associated with the sensitivity of 31 anti-cancer agents, which will benefit the precision treatments for patients with liver cancer. However, an encyclopedic depiction of pancreatic cancer cell lines (PCCLs) has been lacking.
In the present study, we performed integrative analysis of multi-omics data derived from 36 PCCLs. Our analysis found that PCCLs remained the major genomic alterations of the primary pancreatic tumor. We classified the PCCLs into 2 subgroups based on mRNA expression, which was further recapitulated by miRNA and protein expression. The tumor samples were further classified into 3 subgroups according to their expression patterns associated with PCCL subgroups. By integrating the drug sensitivity data, we identified drugs that exhibited variant sensitivity in different subgroups. Our results will contribute to the understanding of molecular alterations in pancreatic cancer and propel the clinical practice of treating pancreatic cancer patients by proposing promising candidates for targeted therapy.
Results
PCCLs share genomic mutations with primary tumor
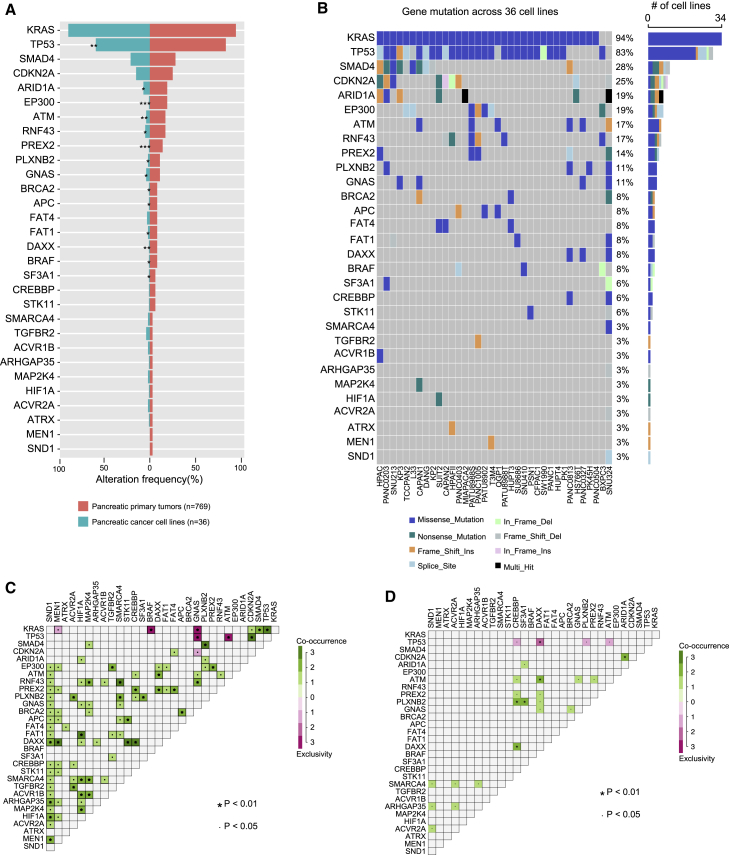
To systematically examine whether PCCLs retain the major genomic alterations that occurred in primary pancreatic tumors, we first analyzed gene mutations across 36 PCCLs (Table S1). The mutation frequency of cancer driver genes was compared between 36 PCCLs and 769 primary pancreatic tumor samples from The Cancer Genome Atlas (TCGA) pancreatic cancer cohort5 (Figure 1A; Table S2). More specifically, 53.3% of cancer driver genes showed no difference of mutation frequency between PCCLs and the primary pancreatic tumor, such as KRAS, SMAD4, and CDKN2A. However, some cancer driver genes exhibited significant discrepancy in PCCLs, such as TP53, ARID1A, and EP300. Additionally, the majority of these cancer driver genes harbored a missense mutation (Figure 1B; Table S3). Many causing genes in cancer show co-occurring or mutually exclusive mutation patterns.15 We next examined all possible gene pairs to detect the co-occurrence and mutual exclusivity of gene mutations in both PCCLs and primary pancreatic cancer samples (see Materials and methods). Consequently, 87 gene pairs of co-occurrence and 6 gene pairs of mutual exclusivity were identified in TCGA pancreatic cancer patient cohort (Figure 1C). In the pancreatic cancer patient cohort, KRAS and BRAF were significantly mutually exclusive presented, which may be explained by the senescence induced by co-expression of mutant KRAS and BRAF.16 This observation proposed a potential treatment that pharmacologically targeting KRAS in BRAF mutated patients or targeting BRAF in patients with KRAS mutation might be an effective clinical intervention. However, much fewer of such events were observed in PCCLs, wherein 20 gene pairs of co-occurrence and 4 gene pairs of mutual exclusivity were detected (Figure 1D). This may be due to the relatively small amount of PCCLs, which only covers a portion of pancreatic cancer subtypes. Our analysis revealed that our curated panel of PCCLs recapitulated the major genomic dysregulation in primary pancreatic cancer samples. Furthermore, our analysis of co-occurrence and mutual exclusivity shed lights on therapeutic strategies for pancreatic cancer patients.
Figure 1.
Gene mutation across pancreatic cancer cell lines (PCCLs)
(A) Mutation of cancer driver genes in pancreatic primary tumors from TCGA (right panel) and PCCLs (left panel). (B) The mutation frequency and mutation types of cancer driver genes across 36 PCCLs. The mutation co-occurrence and exclusivity of cancer driver genes in pancreatic primary tumors (C) and PCCLs (D). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
PCCLs were classified into two subgroups by transcriptomic mRNA
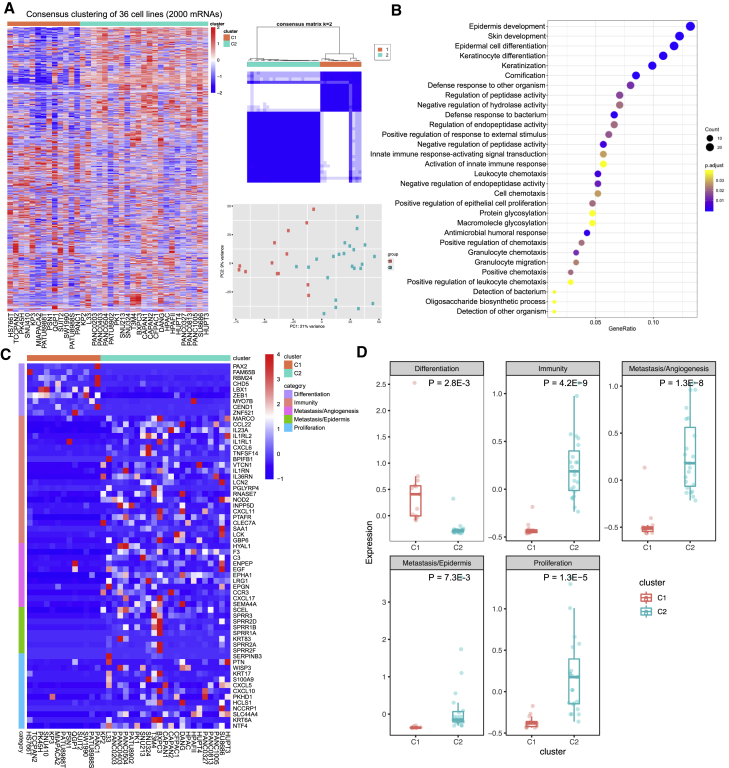
Samples derived from different cancer subtypes are often characterized with various molecular features. We first explored the transcriptomic patterns of mRNA in PCCLs. The algorithm of unsupervised consensus clustering was employed to classify 36 PCCLs (see Materials and methods). The clustering obtained the best performance at K = 2, and PCCLs were classified into 2 subgroups, that is, the C1 and C2 subgroups (Figure 2A). Sample classification was further confirmed by the principal component analysis (PCA) using mRNA expression, where samples in one subgroup were closer to each other than those in the other subgroup. To further examine the biological discrepancies between these 2 subgroups, functional enrichment analysis was performed on differentially expressed genes between these 2 subgroups. Consequently, our analysis revealed multiple biological processes that these 2 subgroups are different, including epidermis biology, such as “epidermis development,” “epidermal cell differentiation,” and “positive regulation of epithelial cell proliferation” (Figure 2B). Furthermore, the C1 subgroup is mainly associated with “differentiation,” indicating that the C1 cell lines are more differentiated with epithelial features, which is characterized with high expression of differentiation-related marker genes, such as FAM65B and RBM24 (Figure 2C). However, PCCLs of the C2 subgroup exhibited other molecular features, such as “immunity,” “metastasis/angiogenesis,” “metastasis/epidermis,” and “proliferation.” As expected, the C2 subgroup cell lines showed high abundance of relevant marker genes, such as immunity of interleukin (IL)-23A and IL-1RL2, angiogenesis of ENPEP and EPHA1, epidermis of SPRR3 and SPRR2D, and proliferation of NTF4 and KRT6A (Figure 2D). In summary, the PCCLs were classified into 2 subgroups with remarkable biological discrepancies.
Figure 2.
mRNA transcriptomic analysis of 36 PCCLs
(A) Consensus clustering and principal component analysis (PCA) of mRNA profiles in PCCLs for the optimal number of clusters at K = 2. (B) Functional enrichment of differentially expressed genes between Cluster1 and Cluster2. (C) Expression pattern of gene-related differentiation, immunity, angiogenesis, epidermis, and proliferation that showed differential expression between 2 subgroups of pancreatic cancer. (D) Comparison of mean expression levels of genes in biological features between 2 subgroups of PCCLs.
miRNA expression is highly associated with PCCL transcriptomic subgroups
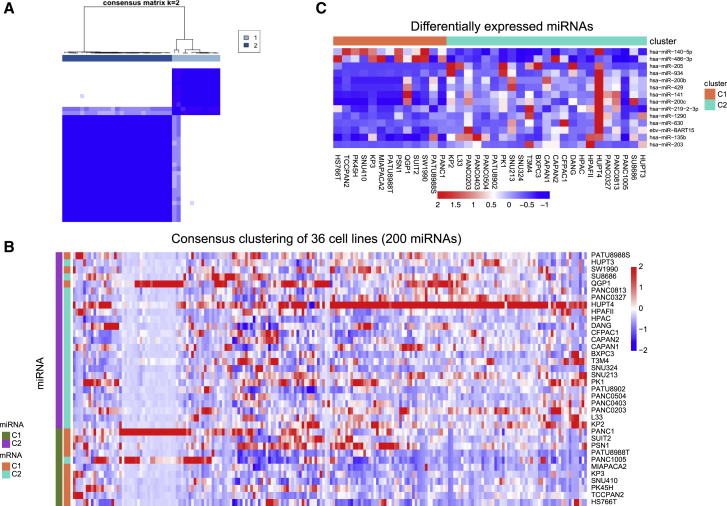
To further examine the PCCL transcriptomic subgroups, we analyzed the expression profile of 734 miRNAs across 36 PCCLs. The top 200 miRNAs with the largest variance of expression levels were adopted to perform the unsupervised consensus clustering (see Materials and methods). In particular, the consensus clustering showed the optimal performance at K = 2, where 36 PCCLs were classified into 2 subgroups (Figure 3A). PCCLs in miRNA subgroups were then assigned to mRNA subgroups. Notably, the miRNA expression-based classification was highly consistent with the mRNA transcriptomic subgroups, with minor exception of 4 cell lines (Figure 3B). The miRNA expression levels were further compared between the 2 transcriptomic subgroups, revealing 2 upregulated and 12 downregulated miRNAs in the C1 subgroup over the C2 subgroup (Figure 3C). Surprisingly, one Epstein-Barr virus (EBV)-related miRNA, EBV-miR-BART15, showed higher abundance in the C2 subgroup compared with those in the C1 subgroup. These results showed the close association between miRNA and mRNA expression patterns in PCCLs.
Figure 3.
miRNA profile analysis in 36 PCCLs
(A) Consensus matrix at the optimal number K = 2. (B) Consensus clustering of miRNA profiles in PCCLs at K = 2. (C) Differentially expressed miRNAs between 2 subgroups of PCCLs.
Protein profiling recapitulates PCCL transcriptomic subgroups
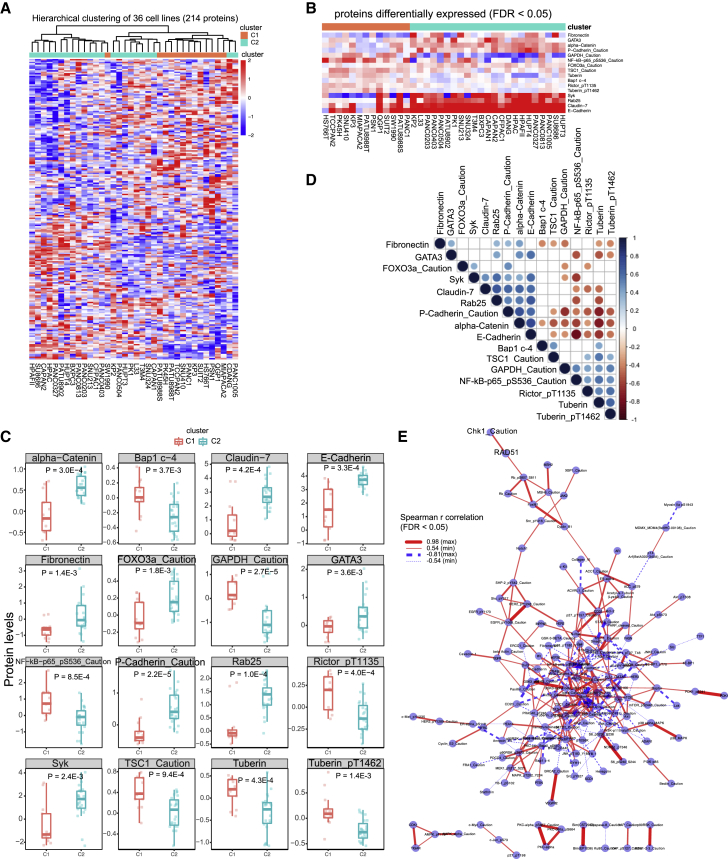
The expression profile of 214 proteins across 36 PCCLs was further analyzed to characterize the protein features. Of note, the PCCLs were classified into 2 subgroups based on protein levels, where only 3 cell lines showed discrepancy with the transcriptomic classification (Figure 4A). Differential analysis disclosed 16 proteins that exhibited a remarkable difference (9 upregulation and 7 downregulation in the C2 subgroup) between the C1 and C2 subgroups of PCCLs (Figure 4B). For example, Rab25, P-Cadherin_Caution, and alpha-Catenin showed significantly higher levels in the C2 subgroup than those in the C1 subgroup, whereas Rictor_pT1135, Tuberin, and TSC1_Caution were downregulated in the C2 subgroup (Figure 4C). Among these differential proteins, 9 proteins exhibited significant correlations with their host mRNAs (Table S4). Moreover, the correlation analysis was performed to examine the co-regulation of these differential proteins across PCCLs. Intriguingly, the majority of differential proteins exhibited remarkably positive or negative correlations across 36 PCCLs (Figure 4D). More specifically, GATA3 and E-Cadherin were highly co-expressed across various PCCLs, whereas Tuberin and P-Cadherin_Caution showed remarkably exclusive expression. The protein interaction network was further constructed based on the co-regulation relations between protein pairs. Network analysis revealed the discrepancy of biological properties among different subgroups, which was reflected by relevant pathways, such as epidermal growth factor receptor (EGFR) signaling, phosphatidylinositol 3-kinase (PI3K) signaling, and mitogen-activated protein kinase (MAPK) signaling pathways (Figure 4E). Our analysis revealed that different subgroups harbored distinct molecular features of pancreatic cancer, which might benefit the clinical practice of classification management and precision treatments for patients with pancreatic cancer.
Figure 4.
Proteomics analysis in 36 PCCLs
(A) Hierarchical clustering of pancreatic cancer proteome profiles and association with mRNA subgroups. (B) Heatmap showed the differential proteins between 2 subgroups. (C) Associations among differential proteins. (D) Boxplots show 19 differential proteins between 2 subgroups. (E) Protein interaction network showed the significantly positive (red solid lines) and negative (blue dashed lines) protein correlations across 36 PCCLs.
PCCL subgroups showed clinical significance
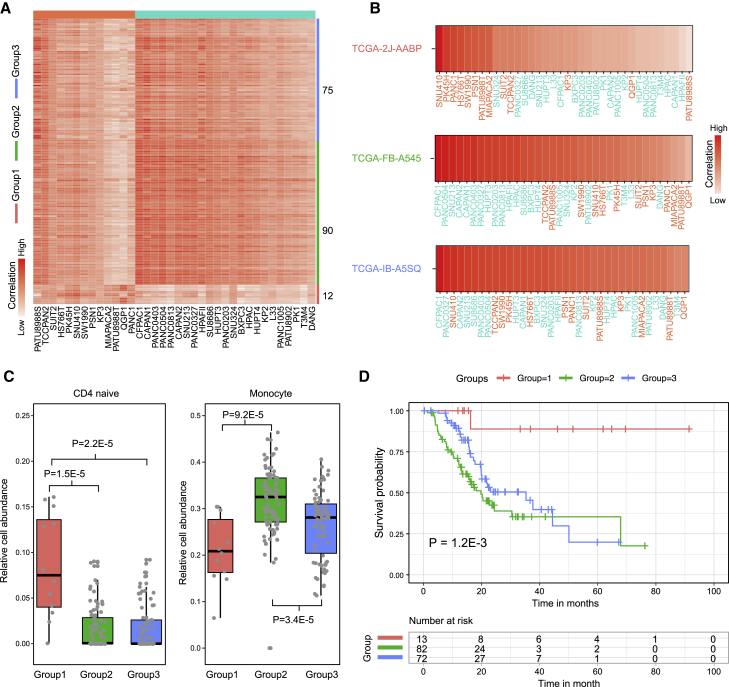
To further explore the clinical significance of PCCL subgroups, pancreatic cancer samples derived from TCGA pancreatic adenocarcinoma (PAAD) cohort were mapped to corresponding subgroups (see Materials and methods). PAAD tumor samples were classified into three subgroups according to their expression correlations with PCCLs (Figure 5A). In particular, 12 tumor samples showed the most similar expression profiles with PCCLs in subgroup C1, whereas 90 samples were highly correlated with those in subgroup C2. Additionally, a portion of tumor samples (n = 75) exhibited mixed expression patterns of subgroups C1 and C2, which were assigned as subgroup C3. Tumor samples from subgroup C1 (such as TCGA-2J-AABP) displayed top correlations with cell lines in the C1 subgroup, whereas those in subgroup C2 (such as TCGA-FB-A545) have higher similarity with C2 cell lines (Figure 5B). The C3 tumor samples (such as TCGA-IB-A5SQ) were highly correlated with cell lines from both C1 and C2 subgroups. Histologically, most samples in C2 and C3 subgroups were a ductal adenocarcinoma subtype, whereas the majority in C1 subgroups were the other adenocarcinoma subtype (Table S5). The abundance of infiltrated immune cells in each tumor sample was estimated to investigate the immune infiltration among different subgroups. Different subgroups showed extensive discrepancy in abundance of infiltrated immune cells (Table S6). In particular, our analysis revealed that tumor samples from the C1 subgroup had significantly higher infiltration of CD4 naive cells than those from C2 and C3 subgroups (p = 1.5E−5 for C2, and p = 2.2E−5 for C3; Figure 5C). In addition, tumor samples in the C2 subgroup were more infiltrated by monocytes than those in C1 and C3 subgroups (p = 9.2E−5 for C1, and p = 3.4E−4 for C3). Kaplan-Meier analysis revealed that the survival time among different subgroups showed significant difference (p = 1.2E−3; Figure 5D). This result is consistent with our observation that the C1 subgroup shows more differentiated features than the C2 subgroup (Figure 2D). These observations suggested that our subgroup classification has remarkable clinical significance and may promote more efficient clinical management for patients with pancreatic cancer.
Figure 5.
PCCL subgroups showed clinical significance
(A) Heatmap showed the correlations between tumor samples and PCCLs in different subgroups. (B) The correlations between PCCLs and example samples from different subgroups. (C) Comparisons of immune cell abundance among samples from different subgroups. (D) Kaplan-Meier curve showed the survival differences among patients from different subgroups.
Integrated analysis identified drug sensitivity-associated molecular features in PCCLs
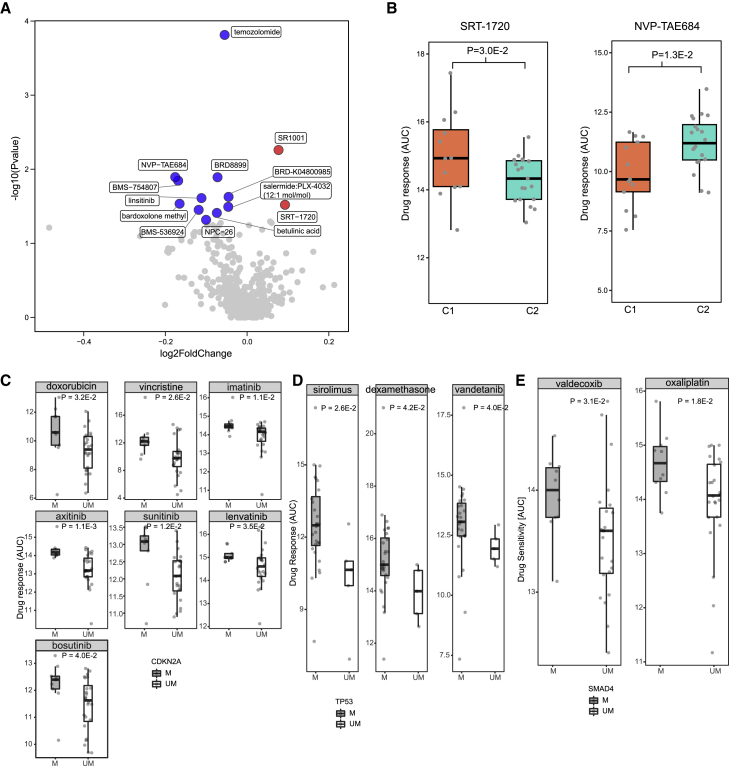
To maximize the clinical utility of multi-omics data in PCCLs, we further integrated the drug sensitivity data to explore the applications of molecular features in precision treatments of pancreatic cancer. By comparing the sensitivity to 497 anti-tumor agents between cell lines in two subgroups, we identified 13 agents with distinct sensitivity in different PCCL subgroups (Figure 6A). In total, 2 agents showed higher sensitivity in the C2 subgroup, whereas C1 PCCLs were more sensitive to 11 agents. Specifically, the small molecule activator of the sirtuin subtype SIRT1, SRT-1720, exhibited significantly higher sensitivity in C2 PCCLs (p = 3.0E−2; Figure 6B). Pancreatic cells in C1 subgroups were more sensitive to another small molecule anaplastic lymphoma kinase (ALK) inhibitor, NVP-TAE684 (p = 1.3E−2). Furthermore, we explored the associations between genomic mutations and drug sensitivity, which were often used to distinguish cells with different sensitivity to anti-tumor drugs.17,18 Among all 61,789 gene-drug comparison pairs, 3,028 were found significantly associated (p < 0.05; Table S7). In particular, pancreatic cancer cells with a CDKN2A mutation showed much more sensitivity of such drugs as doxorubicin, vincristine, imatinib, axitinib, sunitinib, lenvatinib, and bosutinib (Figure 6C). TP53-mutated PCCLs were more sensitive than those with wild-type TP53 to sirolimus, dexamethasone, and vandetanib (Figure 6D). Additionally, valdecoxib and oxaliplatin were highly sensitive in SMAD4-mutated PCCLs (Figure 6E). Our analysis identified molecular features associated with drug sensitivity, offering potential strategy for precision treatments of pancreatic cancer.
Figure 6.
Drug response and associated molecular features of 36 PCCLs
(A) Volcano plot showed the differences of drug response among pancreatic cancer subgroups. (B) Boxplots represented 2 drugs in which their sensitivity showed significant variance between 2 pancreatic cancer subgroups. (C) Boxplots represented 7 drugs in which their sensitivity showed significant variance between CDKN2A-mutated and -non-mutated PCCLs. (D) Boxplots represented 3 drugs in which their sensitivity showed significant variance between TP53-mutated and -non-mutated pancreatic cell lines. (E) Boxplots represented 2 drugs in which their sensitivity showed significant variance between SMAD4-mutated and -non-mutated PCCLs.
Discussion
In this study, we presented a comprehensive molecular characterization of 36 PCCLs. Our analysis of gene mutation showed that pancreatic cancer cells remained the major genomic alterations of the primary pancreatic tumor. The PCCLs were classified into 2 subgroups exhibiting different biological properties based on transcriptomic mRNA profiling, which was also recapitulated by miRNA expression and protein levels. Although the transcriptomic subgroups showed no overall variance of drug sensitivity, the C1 cell lines were more sensitive to such drugs as sildenafil and nelarabine. Additionally, our analysis also found that PCCLs with genomic mutations of CDKN2A, TP53, and SMAD4 exhibited remarkably more sensitive to some drugs, such as doxorubicin, sirolimus, and valdecoxib, respectively.
We included 36 PCCLs in the current study, which might not be sufficient to reflect the whole heterogeneity of primary pancreatic cancer. Covering more PCCLs will offer a relatively more comprehensive characterization. Up to date, our study provided the broadest picture of molecular alterations of pancreatic cancer from the aspect of cell lines, which contributes much to the understanding of pancreatic cancer. We classified the PCCLs by using mRNA, miRNA, and protein expression, respectively. The subgroups based on miRNA or protein expression exhibited high consistence with those based on mRNA expression. The result indicated that mRNA expression is sufficient to catch the major biological discrepancies among different subgroups. Furthermore, mRNA expression has been used to classify subtypes in many malignancies, such as breast cancer,19 lymphoma,20 colorectal cancer,21 and pancreatic cancer22 as well. Our classification in PCCLs will facilitate the development of precision therapeutics.
In the analysis of miRNA profiling, we identified that an EBV-derived miRNA, EBV-miR-BART15 was highly expressed in the C2 subgroup. EBV-miR-BART15 is an EBV-derived miRNA. Multiple studies have shown that EBV-miR-BART15 is strongly associated with apoptosis and epithelial-mesenchymal transition (EMT).23,24 EBV-miR-BART15 has been proven to play regulatory roles in human cancer, including gastric cancer.25 Although there has been no direct evidence demonstrating that EBV-miR-BART15 plays a role in pancreatic cancer, our results implicate the potential relevance of EBV-miR-BART15 in pancreatic cancer cells. Further efforts are needed to validate the role of EBV-miR-BART15 in pancreatic cancer.
Our subgroup classification, which is different from other studies, is based on cellular activities. The classification in our study will be helpful for biological function and pre-clinical precision meditation research. We further mapped pancreatic cancer samples to PCCLs and divided tumor samples into 3 different subgroups. These patients from different subgroups showed distinct immune infiltration and clinical outcomes, which suggests potential strategies of more efficient clinical management for patients in different subgroups. Larger pancreatic cancer cohorts from multiple medical centers were needed to explore the latent clinical utility of our subgroup classification.
In addition, we performed integrated analysis to explore the variance of drug sensitivity across different subgroups of PCCLs. We identified cell lines that showed significantly different sensitivity to anti-cancer drugs. These findings paved the way for exploring precision patient management and treatments for pancreatic cancer.
In summary, our study recapitulated molecular features of pancreatic cancer patients through PCCLs and will accelerate the development of precision treatments for pancreatic cancer patients.
Materials and methods
Omics data of PCCLs
Multi-omics data across 36 PCCLs were retrieved from DepMap (https://depmap.org/portal/), which aims to systematically catalog and identify biomarkers of genetic vulnerabilities and drug sensitivities in hundreds of cancer models, further to aid the precision treatments of cancer patients.26 Our study includes gene mutation, mRNA expression, miRNA expression, and protein levels. All downloaded data were preprocessed. Specifically, the gene mutation was annotated in MAF format, mRNA expression was normalized in fragments per kilobase per million (FPKM) mapped reads, miRNA expression was normalized in reads per million (RPM) mapped reads, and protein levels derived from RPPA were normalized by following the pipeline of the MD Anderson Cell Lines Project.27
Manual curation of cancer driver genes in pancreatic cancer
We manually curated cancer driver genes from public databases, including Catalogue of Somatic Mutations in Cancer (COSMIC),28 Integrative Onco Genomics (IntOGen),29 DriverDB,30 and cBio Cancer Genomics Portal.31 Totally, 30 cancer driver genes were curated for pancreatic cancer. The mutation frequencies of these cancer driver genes were calculated in 36 PCCLs and 769 primary pancreatic cancer samples, respectively.
Evaluation of co-occurrence and exclusivity of mutated genes
The somaticInteractions function in maftools, which is a powerful R package for genomic analysis,32 was employed to identify mutual exclusivity or a co-occurring set of genes. More specifically, pairwise Fisher’s Exact test was performed between all potential pairs of genes in PCCLs and primary pancreatic tumor, respectively.
Classification of PCCLs using an mRNA expression profile
The unsupervised consensus clustering was performed by using the top 2,000 mRNAs with the highest variance across 36 PCCLs by employing the ConsensusClusterPlus package.33 In particular, the consensus partitions of 2,000 mRNA expression profiles were established in K clusters (for K = 2, 3, …, 10). For each partition of K clusters, 1,000 re-sampling interactions of hierarchical clustering was performed with Ward’s linkage method and Euclidean distance as the distance metric. The optimal number of clusters was determined as 2 by adopting the cumulative distribution functions (CDFs) of the consensus matrices,34 where both the CDF area under the curves (AUCs) and the shape of the functions were considered.
Differential expression analysis
The read counts of mRNAs were adopted to perform differential expression analysis by using DESeq2.35 The differential analysis of miRNAs and proteins was performed by using Student’s t test. All of the differential analyses were performed among different clustering groups. Genes, miRNAs, and proteins with a p value <0.05 were considered as significantly differentially expressed.
Functional enrichment of gene sets
Gene set enrichment analysis was performed on the significantly differential genes by utilizing the clusterProfiler package.36 The biological processes annotated in the Gene Ontology (GO) database37 were adopted in the enrichment analysis. Biological processes with a p value <0.05 were considered as significantly enriched by the differential genes.
Construction of protein interaction network
We computed the correlations between each potential pair of proteins by utilizing the method of Spearman’s correlation. The protein pairs with a false discovery rate (FDR) < 0.05 were considered significantly interacted protein pairs. The protein interaction network was further constructed and visualized by Cytoscape software (version 3.7.2).38
Mapping tumor samples to PCCL subgroups
The gene expression matrix of pancreatic cancer samples in TCGA PAAD cohort was retrieved from the Genomic Data Commons (GDC) data portal (https://portal.gdc.cancer.gov/). Customized python scripts were utilized to process the data for download analysis. The 2,000 genes used in consensus clustering were adapted to calculate the correlations between individual PCCLs and tumor samples. For each PAAD sample, PCCLs were ranked by Spearman coefficients, and the top 3 PCCLs were used to define subgroups. In particular, PAAD samples were defined as the C1 or C2 subgroup when top 3 PCCLs are all from the C1 or C2 subgroup. If the top 3 PCCLs include both C1 and C2 subgroups, then PAAD samples were assigned as a mix subgroup, the C3 subgroup.
Estimation of immune cell abundance in tumor samples
The Immune Cell Abundance Identifier (ImmuCellAI) method (http://bioinfo.life.hust.edu.cn/ImmuCellAI#!/)39 was employed to estimate the immune cell abundance for each tumor sample from gene expression profiles. Specifically, ImmuCellAI is a gene set signature-based method to precisely calculate the abundance of various immune cell types by comparing with the reference expression profiles. In total, 24 different types of immune cells were included, covering 18 T cell subsets: “CD4+,” “CD8+,” “CD4+ naive,” “CD8+ naive,” “central memory T,” “effector memory T,” “Tr1” (T regulatory [Treg] type 1), “iTreg” (induced Treg), “nTreg” (natural Treg), “Th1” (T helper [Th]1), “Th2,” “Th17,” “Tfh” (T follicular helper), “Tc” (T cytotoxic), “MAIT” (mucosal-associated invariant T), “Tex,” (exhausted T cells) “gamma delta T,” and “natural killer (NK) T cells” and 6 other important immune cell types: “B cells,” “macrophages,” “monocytes,” “neutrophils,” “DC” (dendritic cell), and “NK cells.”
Drug response analysis in PCCLs
Drug response data used in this study were retrieved from the CTRP (https://portals.broadinstitute.org/ctrp.v2.1/)14 and the GDSC (https://www.cancerrxgene.org/),13 which cover 497 anti-cancer agents across 36 PCCLs. Specifically, the drug sensitivity was measured by the fitted dose response AUC values. Additionally, the differences of drug response among distinct subgroups of PCCLs were tested using chi-square test for each anti-cancer agent. Drugs with a p value <0.05 were considered to have significantly different sensitivity among corresponding different subgroups.
Statistical analysis
In this study, statistical analysis and data visualization were all performed in R software (The R Foundation, Vienna, Austria; https://www.R-project.org). Unless specific statements, all tests were two tailed, and p < 0.05 was considered to be statistically significant.
Availability of data and materials
All accession codes, unique identifiers, or web links for publicly available datasets are described in the paper. All data supporting the findings of the current study are listed in the Supplemental information.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (number [no.] 8187140429), Natural Science Foundation of Shanghai, China (no. 19ZR1411400), and Shanghai General Hospital Startup Funding (02.06.01.20.06).
Author contributions
S.L. conceived the project. S.L., J.Q., and L.L. supervised the project. L.S., Wei Hu, S.Q., and Z.F. downloaded the data and performed the computational analysis. D.Y., T.L., J.W., Y.W., and A.W. performed the statistical analysis and assisted in plotting. L.F., J.X., B.Z., Wenguang He, and Z.N. interpreted the results. S.L. and L.S. wrote the manuscript with comments from all authors. All listed authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.06.015.
Contributor Information
Luming Liu, Email: sh9147@outlook.com.
Jiang-Jiang Qin, Email: jqin@zcmu.edu.cn.
Shengli Li, Email: shengli.li@shsmu.edu.cn.
Supplemental information
References
- 1.Kleeff J., Korc M., Apte M., La Vecchia C., Johnson C.D., Biankin A.V., Neale R.E., Tempero M., Tuveson D.A., Hruban R.H., Neoptolemos J.P. Pancreatic cancer. Nat. Rev. Dis. Primers. 2016;2:16022. doi: 10.1038/nrdp.2016.22. [DOI] [PubMed] [Google Scholar]
- 2.Kamisawa T., Wood L.D., Itoi T., Takaori K. Pancreatic cancer. Lancet. 2016;388:73–85. doi: 10.1016/S0140-6736(16)00141-0. [DOI] [PubMed] [Google Scholar]
- 3.Neoptolemos J.P., Kleeff J., Michl P., Costello E., Greenhalf W., Palmer D.H. Therapeutic developments in pancreatic cancer: current and future perspectives. Nat. Rev. Gastroenterol. Hepatol. 2018;15:333–348. doi: 10.1038/s41575-018-0005-x. [DOI] [PubMed] [Google Scholar]
- 4.Strobel O., Neoptolemos J., Jäger D., Büchler M.W. Optimizing the outcomes of pancreatic cancer surgery. Nat. Rev. Clin. Oncol. 2019;16:11–26. doi: 10.1038/s41571-018-0112-1. [DOI] [PubMed] [Google Scholar]
- 5.Raphael B.J., Hruban R.H., Aguirre A.J., Moffitt R.A., Yeh J.J., Stewart C., Robertson A.G., Cherniack A.D., Gupta M., Getz G., et al. Cancer Genome Atlas Research Network. Electronic address: andrew_aguirre@dfci.harvard.edu. Cancer Genome Atlas Research Network Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell. 2017;32:185–203.e13. doi: 10.1016/j.ccell.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghandi M., Huang F.W., Jané-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R., 3rd, Barretina J., Gelfand E.T., Bielski C.M., Li H., et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature. 2019;569:503–508. doi: 10.1038/s41586-019-1186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li S., Zhang Z., Han L. Molecular Treasures of Cancer Cell Lines. Trends Mol. Med. 2019;25:657–659. doi: 10.1016/j.molmed.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 8.Li S., Hu Z., Zhao Y., Huang S., He X. Transcriptome-Wide Analysis Reveals the Landscape of Aberrant Alternative Splicing Events in Liver Cancer. Hepatology. 2019;69:359–375. doi: 10.1002/hep.30158. [DOI] [PubMed] [Google Scholar]
- 9.Caruso S., Calatayud A.L., Pilet J., La Bella T., Rekik S., Imbeaud S., Letouzé E., Meunier L., Bayard Q., Rohr-Udilova N., et al. Analysis of Liver Cancer Cell Lines Identifies Agents With Likely Efficacy Against Hepatocellular Carcinoma and Markers of Response. Gastroenterology. 2019;157:760–776. doi: 10.1053/j.gastro.2019.05.001. [DOI] [PubMed] [Google Scholar]
- 10.Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehár J., Kryukov G.V., Sonkin D., et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H., Ning S., Ghandi M., Kryukov G.V., Gopal S., Deik A., Souza A., Pierce K., Keskula P., Hernandez D., et al. The landscape of cancer cell line metabolism. Nat. Med. 2019;25:850–860. doi: 10.1038/s41591-019-0404-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boehm J.S., Golub T.R. An ecosystem of cancer cell line factories to support a cancer dependency map. Nat. Rev. Genet. 2015;16:373–374. doi: 10.1038/nrg3967. [DOI] [PubMed] [Google Scholar]
- 13.Iorio F., Knijnenburg T.A., Vis D.J., Bignell G.R., Menden M.P., Schubert M., Aben N., Gonçalves E., Barthorpe S., Lightfoot H., et al. A Landscape of Pharmacogenomic Interactions in Cancer. Cell. 2016;166:740–754. doi: 10.1016/j.cell.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Basu A., Bodycombe N.E., Cheah J.H., Price E.V., Liu K., Schaefer G.I., Ebright R.Y., Stewart M.L., Ito D., Wang S., et al. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell. 2013;154:1151–1161. doi: 10.1016/j.cell.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szczurek E., Beerenwinkel N. Modeling mutual exclusivity of cancer mutations. PLoS Comput. Biol. 2014;10:e1003503. doi: 10.1371/journal.pcbi.1003503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cisowski J., Sayin V.I., Liu M., Karlsson C., Bergo M.O. Oncogene-induced senescence underlies the mutual exclusive nature of oncogenic KRAS and BRAF. Oncogene. 2016;35:1328–1333. doi: 10.1038/onc.2015.186. [DOI] [PubMed] [Google Scholar]
- 17.Hauser A.S., Chavali S., Masuho I., Jahn L.J., Martemyanov K.A., Gloriam D.E., Babu M.M. Pharmacogenomics of GPCR Drug Targets. Cell. 2018;172:41–54.e19. doi: 10.1016/j.cell.2017.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haverty P.M., Lin E., Tan J., Yu Y., Lam B., Lianoglou S., Neve R.M., Martin S., Settleman J., Yauch R.L., Bourgon R. Reproducible pharmacogenomic profiling of cancer cell line panels. Nature. 2016;533:333–337. doi: 10.1038/nature17987. [DOI] [PubMed] [Google Scholar]
- 19.Parker J.S., Mullins M., Cheang M.C., Leung S., Voduc D., Vickery T., Davies S., Fauron C., He X., Hu Z., et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alizadeh A.A., Eisen M.B., Davis R.E., Ma C., Lossos I.S., Rosenwald A., Boldrick J.C., Sabet H., Tran T., Yu X., et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 21.Sadanandam A., Lyssiotis C.A., Homicsko K., Collisson E.A., Gibb W.J., Wullschleger S., Ostos L.C.G., Lannon W.A., Grotzinger C., Del Rio M., et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat. Med. 2013;19:619–625. doi: 10.1038/nm.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collisson E.A., Bailey P., Chang D.K., Biankin A.V. Molecular subtypes of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 2019;16:207–220. doi: 10.1038/s41575-019-0109-y. [DOI] [PubMed] [Google Scholar]
- 23.Choi H., Lee H., Kim S.R., Gho Y.S., Lee S.K. Epstein-Barr virus-encoded microRNA BART15-3p promotes cell apoptosis partially by targeting BRUCE. J. Virol. 2013;87:8135–8144. doi: 10.1128/JVI.03159-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Torres K., Landeros N., Wichmann I.A., Polakovicova I., Aguayo F., Corvalan A.H. EBV miR-BARTs and human lncRNAs: Shifting the balance in competing endogenous RNA networks in EBV-associated gastric cancer. Biochim. Biophys. Acta Mol. Basis Dis. 2021;1867:166049. doi: 10.1016/j.bbadis.2020.166049. [DOI] [PubMed] [Google Scholar]
- 25.Choi H., Lee S.K. TAX1BP1 downregulation by EBV-miR-BART15-3p enhances chemosensitivity of gastric cancer cells to 5-FU. Arch. Virol. 2017;162:369–377. doi: 10.1007/s00705-016-3109-z. [DOI] [PubMed] [Google Scholar]
- 26.Tsherniak A., Vazquez F., Montgomery P.G., Weir B.A., Kryukov G., Cowley G.S., Gill S., Harrington W.F., Pantel S., Krill-Burger J.M., et al. Defining a Cancer Dependency Map. Cell. 2017;170:564–576.e16. doi: 10.1016/j.cell.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J., Zhao W., Akbani R., Liu W., Ju Z., Ling S., Vellano C.P., Roebuck P., Yu Q., Eterovic A.K., et al. Characterization of Human Cancer Cell Lines by Reverse-phase Protein Arrays. Cancer Cell. 2017;31:225–239. doi: 10.1016/j.ccell.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tate J.G., Bamford S., Jubb H.C., Sondka Z., Beare D.M., Bindal N., Boutselakis H., Cole C.G., Creatore C., Dawson E., et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019;47(D1):D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gonzalez-Perez A., Perez-Llamas C., Deu-Pons J., Tamborero D., Schroeder M.P., Jene-Sanz A., Santos A., Lopez-Bigas N. IntOGen-mutations identifies cancer drivers across tumor types. Nat. Methods. 2013;10:1081–1082. doi: 10.1038/nmeth.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng W.C., Chung I.F., Chen C.Y., Sun H.J., Fen J.J., Tang W.C., Chang T.Y., Wong T.T., Wang H.W. DriverDB: an exome sequencing database for cancer driver gene identification. Nucleic Acids Res. 2014;42:D1048–D1054. doi: 10.1093/nar/gkt1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mayakonda A., Lin D.C., Assenov Y., Plass C., Koeffler H.P. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28:1747–1756. doi: 10.1101/gr.239244.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilkerson M.D., Hayes D.N. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu T., Sung S.Y. Consensus clustering. Intell. Data Anal. 2005;9:551–565. [Google Scholar]
- 35.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. The Gene Ontology Consortium Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miao Y.R., Zhang Q., Lei Q., Luo M., Xie G.Y., Wang H., Guo A.Y. ImmuCellAI: A Unique Method for Comprehensive T-Cell Subsets Abundance Prediction and its Application in Cancer Immunotherapy. Adv. Sci. (Weinh.) 2020;7:1902880. doi: 10.1002/advs.201902880. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All accession codes, unique identifiers, or web links for publicly available datasets are described in the paper. All data supporting the findings of the current study are listed in the Supplemental information.