Abstract
The core plant circadian oscillator is composed of multiple interlocked transcriptional–translational feedback loops, which synchronize endogenous diel physiological rhythms to the cyclic changes of environmental cues. PSEUDO-RESPONSE REGULATORS (PRRs) have been identified as negative components in the circadian clock, though their underlying molecular mechanisms remain largely unknown. Here, we found that a subfamily of zinc finger transcription factors, B-box (BBX)-containing proteins, have a critical role in fine-tuning circadian rhythm. We demonstrated that overexpressing Arabidopsis thaliana BBX19 and BBX18 significantly lengthened the circadian period, while the null mutation of BBX19 accelerated the circadian speed. Moreover, BBX19 and BBX18, which are expressed during the day, physically interacted with PRR9, PRR7, and PRR5 in the nucleus in precise temporal ordering from dawn to dusk, consistent with the respective protein accumulation pattern of PRRs. Our transcriptomic and genetic analysis indicated that BBX19 and PRR9, PRR7, and PRR5 cooperatively inhibited the expression of morning-phased clock genes. PRR proteins affected BBX19 recruitment to the CCA1, LHY, and RVE8 promoters. Collectively, our findings show that BBX19 interacts with PRRs to orchestrate circadian rhythms, and suggest the indispensable role of transcriptional regulators in fine-tuning the circadian clock.
BBX19 functions as a regulator of circadian rhythm by complexing with PRR proteins to enhance their repressive effect on CCA1 transcription.
Introduction
The circadian clock is a timekeeping mechanism synchronizing self-sustained physiological rhythms to the 24-h environmental cycles. In land plants, the clock is composed of multiple interconnected transcriptional feedback loops (Creux and Harmer, 2019; McClung, 2019), in which sequentially expressed circadian core components allow plants to predict daily changes of zeitgebers by fine-tuning circadian parameters of the rhythmic expression of their target genes. In the Arabidopsis thaliana circadian clock, the morning loop consists of two MYB-like transcription factors CIRCADIAN CLOCK ASSOCIATED (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), and their homologs, REVEILLE8 (RVE8/LHY-CCA1-LIKE5/LCL5) and RVE4, as well as PSEUDO-RESPONSE REGULATOR (PRR7 and PRR9). CCA1 and LHY inhibit transcription of evening-phased PRR5, TOC1/PRR1, and LUX ARRHYTHMO (LUX; Lau et al., 2011; Nagel et al., 2015; Kamioka et al., 2016). In contrast, RVE8 and RVE4 dynamically interact with transcriptional coactivators, NIGHT LIGHT-INDUCIBLE AND CLOCK-REGULATED1 (LNK1) and LNK2, in the morning to positively regulate expression of PRR5 and TOC1 (Rugnone et al., 2013; Xie et al., 2014). In turn, PRRs function as transcriptional repressors of morning-phased clock genes (Nakamichi et al., 2005, 2010; Gendron et al., 2012; Huang et al., 2012). TOC1 interacts with TCP transcription factor CCA1 HIKING EXPEDITION (CHE) to prevent the activation of CCA1 at night (Pruneda-Paz et al., 2009). PRR5, PRR7, and PRR9 interact with the plant Groucho/TUP1 corepressor, TOPLESS (TPL), and binds to the CCA1 promoter to inhibit its expression, thereby regulating the circadian period length (Wang et al., 2013). This highly wired transcriptional network ensures the stability and robustness of the circadian clock.
The B-box zinc-finger subfamily BBX IV in Arabidopsis consists of eight members, BBX18–BBX25, all of which have two B-box motifs in their N terminus but lack one C-terminal CONSTANS, CONSTANS-like (CO-like), and TOC1 (CCT) domain (Khanna et al., 2009). BBX18 and BBX23 are critical for thermomorphogenesis, as they interact with EARLY FLOWERING3 (ELF3) to regulate the PIF4-dependent gene expression and participate in modulating hypocotyl elongation under warm temperature conditions (Ding et al., 2018). ELF3 is required for the formation of the ELF3–ELF4–LUX evening complex (EC) of the circadian clock, and functions in clock gating, photoperiod sensing, and hypocotyl growth (Covington et al., 2001; Liu et al., 2001; Yu et al., 2008; Nusinow et al., 2011; Chow et al., 2012; Anwer et al., 2020). BBX19 overexpression caused photoperiodic late flowering, in which BBX19 interacted with CONSTANS to inhibit the transcription of FLOWERING LOCUS T (Wang et al., 2014). BBX19 was therefore considered to function in clock output pathways. In addition, ELF3 is recruited by BBX19 and then degraded by COP1 to regulate the formation of EC, which inhibits PIF4 and PIF5 expression, thus promoting evening hypocotyl growth (Wang et al., 2015).
Recently, BBX IV components were reported to modulate photomorphogenesis via their DNA binding ability. BBX21 (STH2) is required for anthocyanin accumulation through direct binding to the HY5 promoter under light conditions (Datta et al., 2006, 2007; Xu et al., 2016, 2018). BBX21 binds to MYB12 and F3H promoter regions, and this process depends on HY5 (Bursch et al., 2020). HY5 is also required for the binding of BBX20 and BBX23 to the promoter regions of target genes (Zhang et al., 2017; Bursch et al., 2020). BBX21, BBX22 (LZF1/STH3), BBX24 (STO), and BBX25 physically interacts with COP1, suggesting that light signaling regulates BBX proteins via COP1-mediated ubiquitination and proteasomal degradation (Datta et al., 2008; Jiang et al., 2012; Xu et al., 2016; Song et al., 2020). BBX24 and BBX25 interact with HY5, potentially to form inactive heterodimers, direct inhibiting binding of HY5 to the BBX22 promoter during early seedling development (Gangappa et al., 2013).
BBX20 (DBB2/BZS1) is regulated by light and COP1-mediated ubiquitination, and acts as negative regulator in brassinosteroid pathway to mediate crosstalk between hormone and light signaling (Kumagai et al., 2008; Khanna et al., 2009; Fan et al., 2012; Wei et al., 2016). In addition, BBX32, a member of the BBX V family, is regulated by the circadian clock, and its overexpression resulted in a lengthened period of circadian rhythm and late flowering (Tripathi et al., 2017). The ectopic expression of Arabidopsis BBX32 in soybeans affects the transcription pattern of soybean clock genes, thereby increasing grain yield (Preuss et al., 2012). However, due to the divergence of BBX family functions, the roles of BBX family members in plant growth, and especially in the circadian system are largely unknown.
In this study, we found that BBX19 and BBX18 proteins dynamically interact with PRR9, PRR7, and PRR5 from the early morning onward in the nucleus to regulate circadian periodicity. Temporal transcriptome and genetic analysis showed that BBX19 and PRR9, PRR7, and PRR5 jointly repressed the expression of morning-phased clock genes CCA1, LHY, and RVE8. BBX19 interacted with PRR9 and PRR7 to bind to CCA1, LHY, and RVE8 promoters to modulate their transcription. These findings demonstrated that BBX19–PRRs complexes function directly in transcriptional regulation of the circadian clock, further bridging the feedback inhibition of morning circadian genes by sequentially expressed PRRs.
Results
Mutation of BBX19 shortens the circadian period
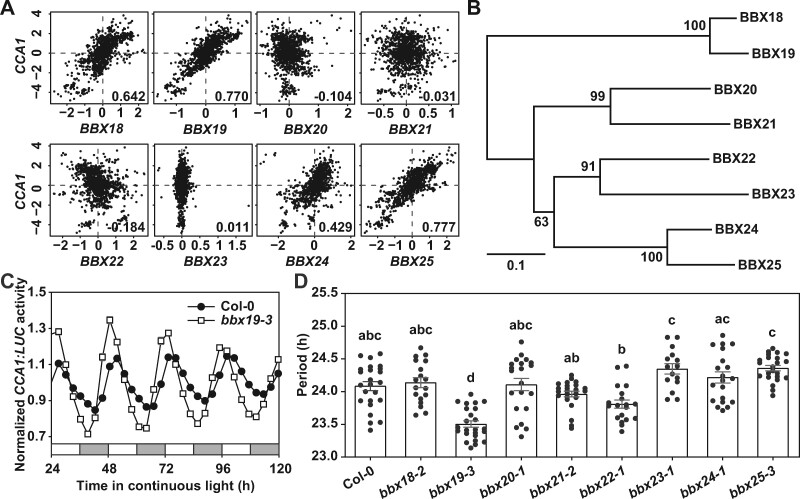
To expand the known molecular architecture of the circadian clock, we checked multiple microarray- and RNAseq-based coexpression data sets in ATTED-II (http://atted.jp) and retained the top 50 genes highly co-expressed with CCA1 or LHY. After alignment, 32 genes associated with both CCA1 and LHY were obtained (Supplemental Table S1), including RVE8 and RVE4, which encode MYB-like transcription factors similar to CCA1 and LHY (Farinas and Mas, 2011; Rawat et al., 2011). LNK2, LNK3, and LNK4 also showed high correlation coefficients, and LNK2 was reported to physically interact with CCA1, LHY, RVE4, and RVE8 (Xie et al., 2014). In addition, five genes BBX18, BBX19, BBX25, COL1, and COL2 were identified that belong to the functionally diverse BBX family (Figure 1, A and B; Supplemental Table S1 and Supplemental Data Set S1). Overexpression of COL1 and COL2 (belonging to BBX structural group I) accelerates the circadian clock, thereby generating a shortened circadian rhythm (Ledger et al., 2001), but no further studies have revealed how they are involved in the circadian clock.
Figure 1.
Dysfunction of BBX19 leads to the accelerated circadian pace. A, Estimation of correlation between CCA1 and BBX subfamily IV genes in co-expression analysis using the multiple microarray- and RNAseq-based coexpression data sets in ATTED-II (http://atted.jp/top_draw.shtml#CoexViewer). The Pearson’s correlation coefficient (r-value) was listed in the lower right corner of each panel, which is used to represent the linear association between CCA1 and BBX subfamily genes. The r-value of 0 indicates that there is no association, while values of −1 or +1 indicates that there is a strongest linear correlation. B, The phylogenetic radiant tree of eight full-length orthologs of BBX subfamily IV in Arabidopsis. The evolutionary distance was inferred using the neighbor-joining method, and phylogenetic tree was constructed using the Jukes–Cantor genetic distance model in Geneious Tree Builder. C, D, Circadian rhythms of CCA1:LUC in the bbx18-bbx25 mutants were monitored under free-running conditions. Data showing mean ± se for three independent experiments. At least 15 individual seedlings were used for each analysis. Open bars indicate subjective day, and gray bars indicate subjective night (C). Dots indicate individual samples and bars mean period ± se (D). Multiple groups were analyzed with one-way ANOVA followed by Tukey’s multiple comparison test, P < 0.05
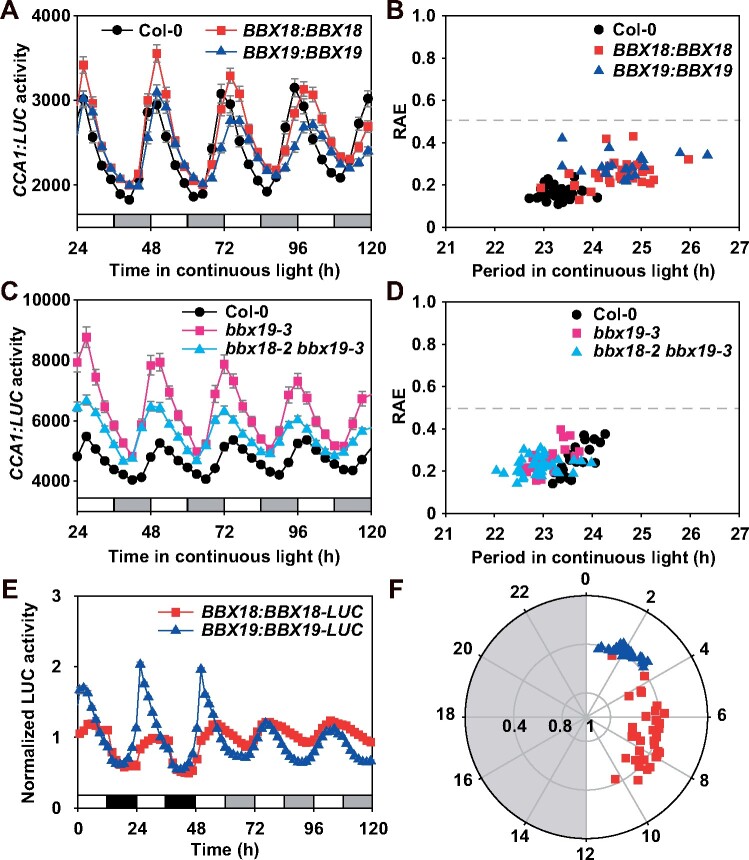
Circadian rhythms were therefore monitored in BBX subfamily IV gene mutants. A bioluminescence rhythms assay under free-running conditions (constant light, LL) indicated that only the null mutation of BBX19 (bbx19-1, bbx19-2, and bbx19-3) significantly shortened the period of self-sustained CCA1:LUC expression (23.5 h in bbx19-3 versus 24.1 h in the wild-type; Figure 1, C and D; Supplemental Figures S1 and S2, and Supplemental Tables S2 and S3). The genome sequence of BBX19:BBX19 complemented the circadian phenotype of bbx19 T-DNA insertion mutant lines (Supplemental Figure S2C and Supplemental Table S3). In addition, bbx19-4, a CRISPR/Cas9-mediated genome editing mutation line, was created and also showed a shortened circadian period (Supplemental Figure S2, D–F and Supplemental Table S3). BBX19 and BBX18 shared ∼69.4% identity at the amino acid level in evolutionary analyses (Supplemental Figure S3 and Supplemental Data Set S2). Moreover, increased expression of BBX19 and BBX18 showed a significant lengthening of the circadian period (24.5 h in BBX18:BBX18/Col-0, 24.7 h in BBX19:BBX19/Col-0 versus 23.3 h in Col-0), indicating that they function in maintaining the circadian clock (Figure 2, A and B;Supplemental Table S4). In addition, we found that the bbx18-2 bbx19-3 double mutant had similarly shortened periodicity to that of the bbx19-3 mutant alone (Figure 2, C and D; Supplemental Table S4). Furthermore, BBX19–GFP and BBX18–GFP fusion proteins were evident in the nucleus (Supplemental Figure S4). The accumulation of BBX19 and BBX18 proteins showed robust oscillations in both light/dark diurnal cycle and LL conditions, in which the BBX19 peak phase occurred around dawn while the peak of BBX18 occurred in the early afternoon (Figure 2, E and F). In summary, BBX19 and BBX18 are involved in adjusting the circadian rhythm.
Figure 2.
Morning-phased BBX19 and BBX18 are involved in regulating self-sustained circadian period. A, B, Increased expression of BBX18 or BBX19 lengthened the circadian period length. The full-length gene constructs of BBX18:BBX18 and BBX19:BBX19 were transformed into the wild-type plants to generate the overexpression transgenic lines. Period estimation for of individual CCA1:LUC rhythm (A) is plotted against their relative amplitude errors (RAEs) (B). RAE is used to define the limit of rhythmicity, a complete sine-fitting wave is defined as 0, and a value of 1 defines the weakest rhythm. Data represent mean ± se from three independent experiments. At least 24 individual seedlings were used for each analysis. Open bars indicate subjective day, and gray bars indicate subjective night. C, D, Circadian rhythm (C) and period estimate (D) of the bbx18 bbx19 double mutant under free-running conditions. The bbx18-2 bbx19-3, together with Col-0 and bbx19-3 seedlings were entrained under 12-h:12-h LD cycles for 2 weeks and then released to constant light (LL) at 22°C for 5 days. E, F, The daily expression of BBX18 and BBX19 proteins were regulated by the circadian clock, with a peak phase appeared in the morning. The CT phase angles for individual seedlings were plotted against their RAE values to indicate the peak position and the robustness of rhythmicity, respectively (F)
BBX19 and BBX18 sequentially interact with PRR9, PRR7, and PRR5 proteins
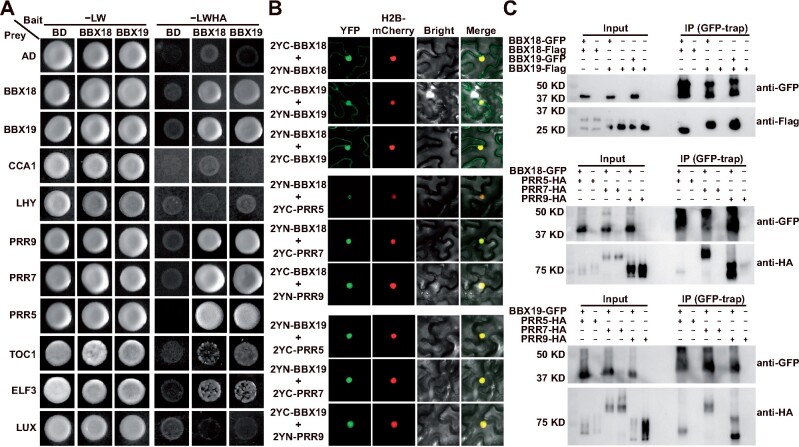
To unravel the underlying mechanism of BBX family genes in circadian regulation, we first used a yeast two-hybrid system to identify whether core oscillators are direct partners of BBX19 and BBX18 (Figure 3A). The results demonstrated that BBX19, BBX18 interacted with PRR9, PRR7, and PRR5, but not CCA1, LHY, and LUX. In addition to complexing with clock proteins, BBX19 and BBX18 proteins also interact with themselves or each other to form homodimers and heterodimers, respectively. BiFC assays were also used to verify that the BBX19, BBX18 dimers, BBX19–BBX18–PRR9, PRR7–PRR5 interactions occur in the nucleus in epidermis cells of the co-infiltrated leaves of Nicotiana benthamiana (Figure 3B;Supplemental Figure S5). Moreover, co-immunoprecipitations were further performed with protein extracts from infiltrated N. benthamiana leaves using anti-GFP antibody (Figure 3C). Together, BBX19 and BBX18 were characterized as forming protein complexes with PRR proteins.
Figure 3.
BBX19 and BBX18 physically interact with PRR proteins in vitro and in vivo. A, Yeast two-hybrid system to screen the interacting proteins of BBX18 and BBX19 among the known clock proteins. AD, activating domain; BD, binding domain; -LW, synthetic dropout medium without leucine and tryptophan; -LWHA, selective medium without leucine, tryptophan, histidine, and adenine. B, BiFC assay showing the interaction between BBX18/19 and PRR proteins predominantly occurred in nucleus. Each protein was tagged with either the N- or C-terminal fragment of YFP as indicated. The fluorescent signal in N. benthamiana epidermal cells was imaged at 48 h after A. tumefaciens-mediated infiltration. C, Co-immunoprecipitation analysis of BBX18, BBX19, and PRRs with transiently expressed proteins in N. benthamiana. Anti-GFP antibody was used for performing immunoprecipitation. The proteins were detected with anti-Flag and anti-HA for immunoblotting as indicated
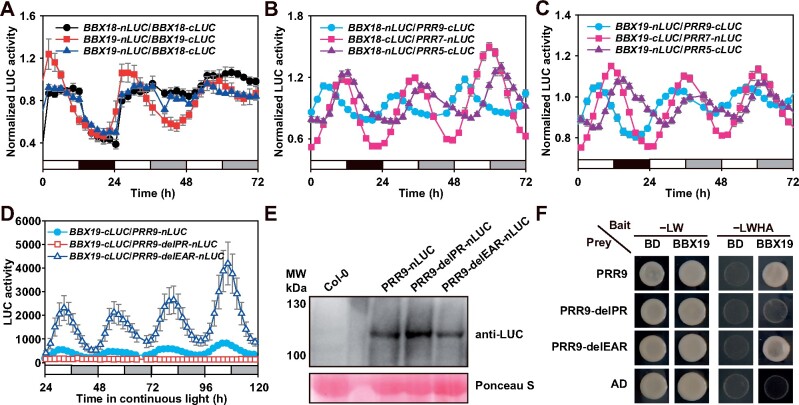
Furthermore, a luciferase complementation analysis was used to check the dynamic interactions of PRRs with BBX19 or BBX18 under both 12-h:12-h light: dark (LD) cycle and LL conditions (Figure 4;Supplemental Figure S6). The expression of the fusion proteins was driven by their own promoters. The formation of BBX19 and BBX18 homodimers and heterodimers all displayed oscillation patterns, and the dynamic interaction of BBX19 homodimer showed good robustness (Figure 4A;Supplemental Figure S6A). In the LD cycle, the BBX19 dimer peaked in the early morning, while the BBX18 dimer lagged slightly. Overall, the dynamic pattern of BBX19–BBX18 interaction is similar to that of the BBX18 homodimer. The protein–protein interactions of BBX19 and BBX18 with PRR9, PRR7, and PRR5 also displayed robust circadian oscillations in LL conditions (Figure 4, B and C; Supplemental Figure S6, B–D), and the interaction peak of each pair occurred at different times of the day, including a BBX–PRR9 peak in the morning, a BBX–PRR7 peak around late afternoon, and a BBX–PRR5 peak in the evening. Also, from the Y2H analysis and recombinant LUC activity, BBX19 showed very weak interactions with TOC1 and ELF3 (Figure 3A;Supplemental Figure S7). Collectively, our findings suggested that BBX19 and BBX18 likely act as partners of sequentially expressed PRR9, PRR7, and PRR5 in the circadian clock.
Figure 4.
Dynamic protein–protein interactions between BBX19/18 and PRR proteins. A–C, The diurnal and circadian oscillations of the formation of each protein pair. The fusion proteins driven by their own promoters were fused to C-terminal domain of nLUC or cLUC, then the transgenic Arabidopsis plants were generated by genetic cross. The recombined LUC activity in F1 generation was continuously monitored for 72 h with a TopCount luminometer. Data represent mean ± se for three independent experiments. D, Deletion analysis showed that the PR domain of PRR9 is essential for its interaction with BBX19. E, Immunoblot analysis showed the expression of PRR9 in PRR9-nLUC, PRR9-delPR-nLUC, and PRR9-delEAR-nLUC plants. The seedlings were grown under 12-h:12-h LD cycles for 10 days and then sampled at ZT5. Total proteins were separated by 10% sodium dodecyl sulphate–polyacrylamide gel electrophoresis and PRR9 proteins were confirmed by immunoblotting with anti-LUC (AS163691A, from Agrisera). The molecular weight of the PRR9–nLUC fusion protein is expected to be about 99 kDa; PRR9-delPR-nLUC to be about 86 kDa; PRR9-delEAR-nLUC to be about 97 kDa. F, Yeast two-hybrid analysis of BBX19 and PRR9 protein interaction domains
Ethylene-responsive element binding factor-associated amphiphilic repression (EAR) is a conserved repression motif in plant transcriptional regulators (Kagale and Rozwadowski, 2011), which is necessary for PRR9, PRR7, and PRR5 to interact with TPL family proteins and inhibit CCA1 and LHY expression (Wang et al., 2013). The PR domain is similar to the conserved signal receiver domain of response regulators (Farré and Liu, 2013). To identify which motif mediates protein–protein interactions, we examined the function of N-terminal EAR and PR domains of the PRR9 protein (Figure 4D;Supplemental Figure S6D). The results suggested that deleting EAR caused more robust protein–protein interactions between PRR9 and BBX19. However, the lack of a PR domain resulted in a complete loss of the dynamic protein–protein interactions between PRR9 and BBX19 (Figure 4D). To examine whether deleting the PR or EAR domain affects stability of the PRR9 protein, we analyzed the protein accumulation of PRR9 using immunoblotting (Figure 4E). The levels of PRR9 protein in the wild-type PRR9-nLUC, PRR9-delPR-nLUC, and PRR9-delEAR-nLUC were similar. Also, yeast two-hybrid analysis confirmed that the interaction between BBX19 and PRR9 depends on its PR domain (Figure 4F). In summary, the above results suggested that the PR domain of PRR9 protein is essential for interacting with BBX19 protein, and the EAR domain probably hinders their interaction.
PRR genes are genetically required to regulate BBX19 in the circadian period
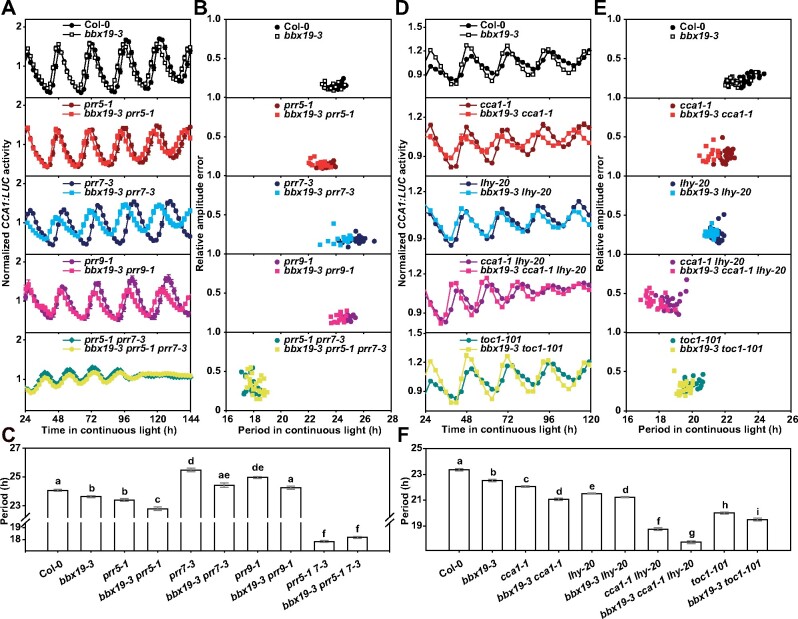
To clarify the genetic relationship between BBX and PRR genes, we generated the bbx19-3 prr5-1, bbx19-3 prr7-3, bbx19-3 prr9-1, and bbx19-3 prr5-1 prr7-3 mutant lines. Circadian rhythms of CCA1:LUC reporter in the mutants were monitored under LL conditions, and variance of circadian period length was compared within groups (Figure 5, A–C; Supplemental Table S5). We found that bbx19-3 prr7-3 and bbx19-3 prr9-1 double mutants exhibited a relatively short period, compared with the long period in the prr7-3 and prr9-1 single mutants. In addition, both bbx19-3 and prr5-1 displayed a shortened period (23.6 h and 23.4 h, respectively), while the bbx19-3 prr5-1 double mutant had a shorter period length (22.8 h). The consistently shortened phenotype in the bbx19 prr double null mutant indicated that BBX19 potentially imposes a brake on the circadian rhythm. However, the periods in bbx19-3 prr7-3 and bbx19-3 prr9-1 were still longer than that in bbx19-3. The bbx19-3 prr5-1 prr7-3 triple mutant (18.2 h) showed a slightly longer period than the prr5-1 prr7-3 line (17.8 h). In summary, the results suggested an epistatic effect of prr5, prr7, and prr9 over bbx19.
Figure 5.
PRRs are genetically required for the regulation of BBX19 on circadian period. A, B, Circadian rhythm of CCA1:LUC was measured in the bbx19-3 prr5-1, bbx19-3 prr7-3, bbx19-3 prr9-1, and bbx19-3 prr5-1 prr7-3 knockout mutants. Arabidopsis seedlings were grown under 12:12 LD cycles, 22°C, for 7 days before transferred to LL for luminescence measurement. The circadian parameters analysis was performed using the fast Fourier transform-nonlinear least squares based on LL24-120 rhythmic traces (A). Period estimation for individual seedlings is plotted against their relative amplitude errors (RAEs value the robustness of rhythmicity) (B). C, Period length estimation of CCA1:LUC circadian rhythm (B). Multiple groups were analyzed with one-way ANOVA followed by Tukey’s multiple comparison test, P < 0.05. D–F, Circadian rhythm of CCA1:LUC was measured in the bbx19-3 cca1-1, bbx19-3 lhy-20, bbx19-3 cca1-1 lhy-20, and bbx19-3 toc1-101 mutants
Furthermore, we found that the period length in the double mutants bbx19-3 cca1-1 and bbx19-3 lhy-20 were similar (21.1 h and 21.2 h, respectively), and slightly shorter than single mutants of cca1-1 and lhy-20 (Figure 5, D–F; Supplemental Table S6). The period length in the bbx19-3 cca1-1 lhy-20 triple mutant was about 17.7 h, which is much shorter than cca1-1 lhy-20 (18.8 h), indicating that BBX19 acts independently with CCA1 and LHY. In addition, the period of bbx19-3 toc1-101 was about 20 h, which is slightly shorter than toc1-101 by about half an hour (Figure 5, D–F; Supplemental Table S6). The data suggested that bbx19-3 also produces an additive effect to the short period displayed by prr5-1, cca1-1, lhy-20, and toc1-101. Given the physical interactions with PRRs and the epistasis of the prr null mutant over bbx19, our results showed that BBX19 likely regulates the circadian period through the interaction with PRRs.
Temporal transcriptome analysis of the BBX19-regulated circadian process
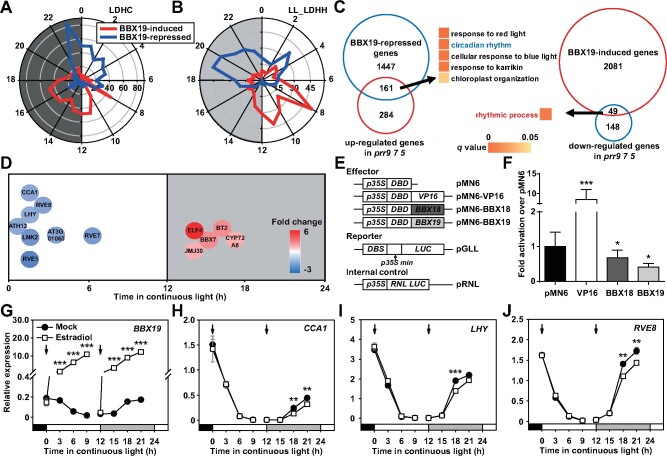
To further investigate the potential mechanism of BBX19 in regulating the circadian clock, we used RNA sequencing to profile the circadian transcriptome from BBX19 inducible overexpression lines (Figure 6;Supplemental Data Set S3). We cloned BBX19 into the pER8 vector system to check for inducible expression in transgenic plants (Zuo et al., 2000). Estradiol was applied to pER8-BBX19 transgenic seedlings at ZT12 to induce excessive accumulation of BBX19 in the next morning. Analyzing samples taken from ZT2 and oscillated differentially expressed genes (DEGs) using the microarray data (http://diurnal.mocklerlab.org/), we identified several hundred transcripts whose accumulation oscillated with a 24-h period in either LD diurnal cycles or LL conditions (Figure 6, A and B). There were 1,608 genes specifically inhibited by BBX19 (fold change >1.5), 34% of which exhibited diurnal rhythms and 27% of which exhibited circadian rhythms, with peaks appearing around dawn (ZT19-ZT4). Among the genes with increased expression promoted by BBX19, 26% in LD and 20% in LL showed enrichment of rhythmic transcripts, but their peaks appeared from the afternoon to late evening.
Figure 6.
BBX19 inhibits the expression of morning-phased circadian core components. A, B, Radial plots with number of BBX19-controlled genes on the radius and circadian phase (peak phase) on the circumference. For RNA-sequencing, the Arabidopsis seedlings carrying a pER8-BBX19-YFP-HA transgene were grown under 12:12 LD cycles for 10 days before BBX19 were induced with β-estradiol at ZT12. Samples were harvested at ZT2 of the next day for RNA extraction and the subsequent RNA seq experiments. Analysis of DEGs (P < 0.05 and fold change >1.5) using the microarray data (http://diurnal.mocklerlab.org/) identified circadian-regulated genes (rhythmic expression under LD and LL conditions). Light and shading represent day and night, respectively. C, GO analysis of the overlapping genes between BBX19-controlled genes and DEGs in the d975 triple mutant of PRR9, 7 and 5 (Nakamichi et al., 2009). D, A plot showing circadian phase of the genes co-regulated by BBX19 and PRR9, PRR7, and PRR5 over the course of a 24-h day. The background color of the letters represents the changes of the genes in the inducible BBX19 expression lines. E, F, Identifying the transcriptional repressive activity of BBX19 and BBX18 in Arabidopsis protoplasts. Schematic diagrams of the effectors and LUC reporter constructs used for transient dual-luciferase transactivation assays in Arabidopsis protoplasts (E). DBD, GAL4 DNA binding domain; DBS, GAL4 DNA binding site; RNL LUC, Renilla luciferase. 35S:RLUC, internal control. BBX19 and BBX18 inhibited the expression of the LUC reporter gene (F). The transcriptional activation is indicated by the ratio of LUC/RLUC. Data showing mean ± se for three independent experiments (*P < 0.05; ***P < 0.001 compared to the negative control using Student’s t test). G–J, Estradiol-induced BBX19 expression at subjective night inhibited the transcript accumulation of CCA1, LHY, and RVE8 (**P < 0.01; ***P < 0.001; Student’s t test). Data show mean ± se of three technical replicates from one of the three independent biological experiments (also shown in Supplemental Figure S8); IPP2 was used as a normalization control; all experiments yielded congruent results
Comparing the transcriptome in BBX19-inducible overexpression material with the transcriptome in the prr975 triple mutant (Nakamichi et al., 2009), we found that 36% of the genes up-regulated in prr975 (161 genes) were inhibited by BBX19 (Figure 6C). Gene ontology enrichment analysis indicated that these 161 genes participated in diverse biological processes including the circadian rhythm and those closely related to the function of the circadian system such as responses to light (Figure 6C, left). Correspondingly, 25% of the genes downregulated in prr975 (49 genes) were promoted by BBX19, and they were also mainly involved in the circadian processes (Figure 6C, right). We further analyzed the acrophase (peak phase) of genes related to clock regulation and found a few morning-phased genes, including CCA1, LHY, RVE8, and RVE1, whose expression was negatively regulated by BBX19; evening-phased genes, including ELF4 and JMJ30, were positively regulated by BBX19 (Figure 6D).
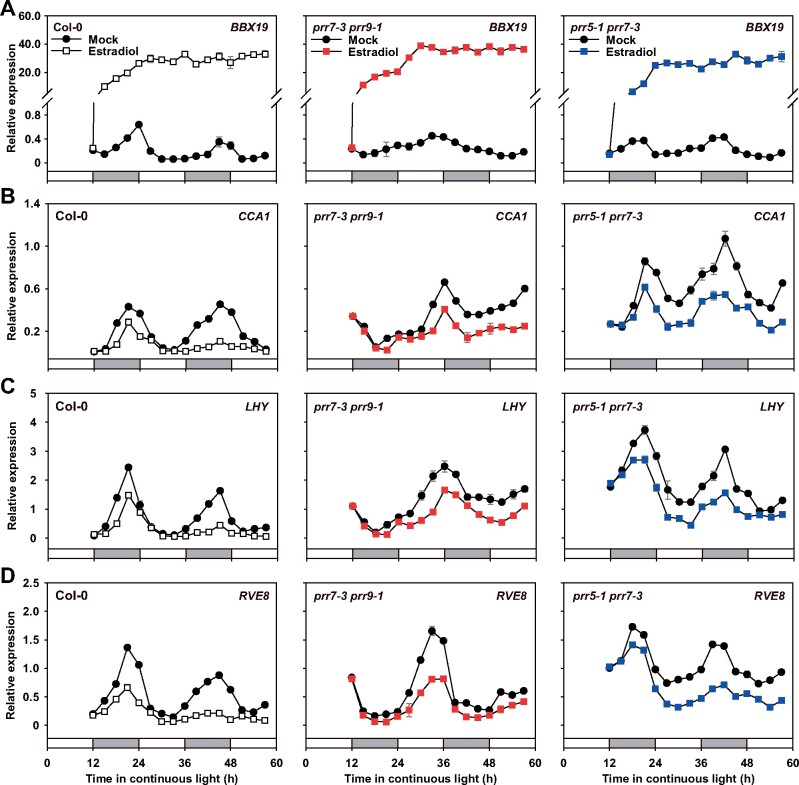
Moreover, the transient gene expression system using Arabidopsis mesophyll protoplasts indicated that BBX19 alone had no transcriptional activation activity, and instead slightly repressed the expression of LUC reporter compared to the negative control (Figure 6, E and F). To substantiate the effect of BBX19 on inhibiting gene transcription, we induced the expression of BBX19 during the day or night, and examined the transcript levels of CCA1, LHY, and RVE8 (Figure 6, G–J; Supplemental Figure S8). Estradiol was applied to pER8-BBX19 materials at ZT0 and ZT12. BBX19 was significantly overexpressed at ZT3 or ZT15 (i.e. 3 h after estradiol treatment), and with the time extension, the transcript levels of BBX19 were very similar between the two independent treatments (Figure 6G;Supplemental Figure S8A). The overexpression of BBX19 inhibited the transcript accumulation of CCA1, LHY, and RVE8 before dawn, but not during the daytime (Figure 6, H–J; Supplemental Figure S8, B–D).
After BBX19 overexpression, transcript accumulation was monitored under LL conditions for 48 h (Figure 7;Supplemental Figure S9). The results showed that accumulation of CCA1, LHY, and RVE8 transcripts began to decline in Col-0 within 12 h after treatment with estradiol. After that, the level of transcripts for each gene was extremely low. Hence, we proposed that, after dawn, the transcription of CCA1, LHY, and RVE8 is already declining or at a trough, and the effect of overexpressing BBX19 is not significant during the day. However, from evening to dawn, when the transcripts of CCA1, LHY, and RVE8 would be rising, overexpressing BBX19 will significantly inhibit those target genes. In addition, we further analyzed the function of BBX19 overexpression on morning-phased genes in the prr7-3 prr9-1 and prr5-1 7-3 mutants. The results showed that the inhibitory effect of BBX19 on CCA1, LHY, or RVE8 expression in the mutants was significantly weakened compared to the wild-type (Col-0), especially on the second day after inducing BBX19, when the transcription peaks of CCA1, LHY, and RVE8 in the wild-type were strongly suppressed (Figure 7;Supplemental Figure S9). The data predicted that PRR9, PRR7, and PRR5 are required for BBX19 to negatively regulate the expression of CCA1, LHY, or RVE8. Therefore, combined with transcriptome analysis, we proposed that BBX19 maintains the endogenous circadian rhythm by modulating the expression of morning-phased clock components such as CCA1, LHY, and RVE8.
Figure 7.
BBX19 inhibits the accumulation of CCA1, LHY, and RVE8 transcripts. The wild-type (Col-0), prr7-3 prr9-1, and prr5-1 prr7-3 mutants containing pER8-BBX19 were grown under 12:12 LD cycles for 10 days before BBX19 were induced at ZT12 with β-estradiol (A). quantitative reverse transcription polymerase chain reaction analysis of the transcript accumulation of CCA1 (B), LHY (C), and RVE8 (D) in the Col-0, prr7-3 prr9-1, and prr5-1 prr7-3 mutants. Data show mean ± se of three technical replicates from one of three independent biological experiments (also shown in Supplemental Figure S9); IPP2 was used as a normalization control; all experiments yielded congruent results. White or gray bars represent subjective day or subjective night, respectively
PRR9, PRR7, and PRR5 are involved in the binding of BBX19 to the CCA1 promoter and inhibit its transcription
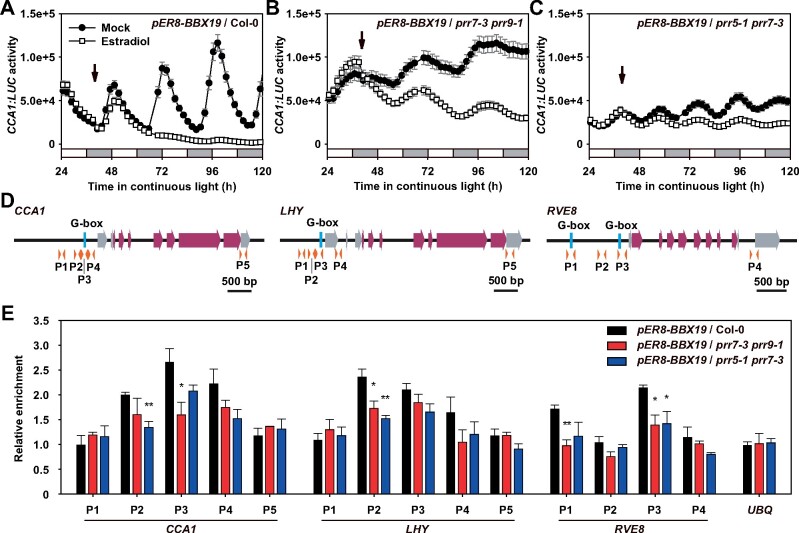
To investigate the effect of BBX19 in clock gene expression in real time, we examined the promoter activity of CCA1 in pER8-BBX19 materials. We found that estradiol treatment did not affect CCA1:LUC activity in the wild-type (Supplemental Figure S10A), but overexpressing BBX19 significantly inhibited CCA1:LUC activity (Figure 8A), indicating its inhibitory function on transcription of CCA1. The inhibitory effect of BBX19 overexpression on CCA1:LUC activity was markedly blocked in the prr7-3 prr9-1 and prr5-1 prr7-3 mutants (Figure 8, B and C). In addition, overexpressing BBX19 caused a lengthened period and slightly reduced the circadian amplitude of TOC1:LUC in free-running conditions (Supplemental Figure S10B), consistent with the circadian phenotype of CCA1:LUC in BBX19:BBX19/Col-0 plants (Figure 2, A and B). Collectively, these results indicated that BBX19 and its interacting proteins, PRR9, PRR7, and PRR5, jointly modulated morning clock gene expression.
Figure 8.
PRR9, PRR7, and PRR5 are required for the association of BBX19 with CCA1 promoter and inhibit its transcription. A–C, Measurement of CCA1:LUC activity in the prr7-3 prr9-1 and prr5-1 prr7-3 mutant with or without the induced expression of BBX19. Arabidopsis seedlings carrying pER8-BBX19 were grown under 12:12 LD cycles for 7 days before transferred into LL and treated with β-estradiol at CT39. LUC activity was measured in LL using a TopCount luminometer. D, Schematic diagram of CCA1, LHY, and RVE8 gene structure including the upstream region. G-box elements in the promoter region (blue vertical bar), exon (purple box with arrow), 5′ and 3′ untranslated region (gray box with arrow), and orange arrow heads below represent the location of primers used in chromatin immunoprecipitation quantitative real-time polymerase chain reaction (ChIP-qPCR) assay. E, ChIP-qPCR assay of BBX19-YFP-HA protein in Col-0, prr7-3 prr9-1, and prr5-1 prr7-3 mutants with promoters of CCA1, LHY, and RVE8. Seedlings were grown under 12:12 LD cycles for 14 days before BBX19 were induced at ZT12 with β-estradiol. Sampling was performed at ZT3 when BBX19 expression reached a significant peak. Anti-HA antibody was used for precipitating of BBX19 protein, followed by qPCR detection. For relative enrichment of DNA fragments, the ratios between the levels of immuno-precipitated DNA in signal samples (using anti-HA antibody) and in reference samples (no antibody) were calculated. Data represent mean ± se of three biological replicates (**P < 0.01; *P < 0.05; Student’s t test)
Given the physical interactions between BBX19 and PRR9, PRR7, and PRR5, together with the genetic requirement for PRR9, PRR7, and PRR5 in regulating the circadian period, chromatin immunoprecipitation was used to compare the relative abundance of BBX19 protein within the promoter regions of its putative target genes, CCA1, LHY, and RVE8 (Figure 8, D and E). Chromatin was isolated from BBX19-YFP-HA/Col-0, BBX19-YFP-HA/prr7-3 prr9-1, and BBX19-YFP-HA/prr5-1 7-3 seedlings, which were harvested at ZT3 to match the peak expression of BBX19 in a 24-h day. The results showed a significant association of BBX19 in Col-0 plants with the CCA1 promoter, and the regions around the G-box were necessary to mediate the transcriptional regulation (Figure 8E). In the prr7-3 prr9-1 or prr5-1 7-3 mutants, the associations of BBX19 with CCA1, LHY, and RVE8 promoter regions were weakened (Figure 8E), and the prr7-3 prr9-1 and prr5-1 7-3 mutations did not affect the accumulation of BBX19 protein (Supplemental Figure S11). Thus, the data suggested that protein complexes formed by BBX19 and PRR9, PRR7, and PRR5 might facilitate their binding to common target genes. Together, our data demonstrated that BBX19 negatively regulates morning-phased clock gene expression by forming protein complexes with PRRs.
Discussion
The transcript and protein accumulation of CCA1 exhibited a robust 24-h rhythm, reaching a peak immediately after dawn, and then its expression was continuously suppressed until the night, when the CCA1 transcript level reached a trough and then began to be enriched again (Yakir et al., 2009); the mechanism of this is unclear. PRR9, PRR7, PRR5, and TOC1 are expressed sequentially throughout the day, and act as inhibitors to regulate the expression of CCA1 and LHY (Nakamichi et al., 2010; Gendron et al., 2012; Huang et al., 2012). Previous results of Chromatin immunoprecipitation (ChIP)-sequencing show that PRR proteins, including PRR9, PRR7, and PRR5 associate to chromatin regions rich in G-box-like motifs, and distinct PRR-targeted genes include the morning-phased clock genes, CCA1, LHY, RVE1, RVE2, RVE7, RVE8, and the transcriptional cofactor genes LNK1, LNK2, LNK3, and LNK4 (Liu et al., 2016). Here, we identified a member of BBX subfamily IV with DNA binding activity, BBX19, which acted on the self-sustained circadian rhythm (Figures 1 and 2). Chromatin immunoprecipitation analysis showed that BBX19 preferentially associated to the chromatin region containing a G-box element (Figure 8) and negatively regulated the expression of morning-phased core clock genes, including CCA1, LHY, and RVE8. In the prr9-1 prr7-3 and prr5-1 7-3 mutants, the binding ability of BBX19 with CCA1, LHY, and RVE8 promoters was weakened, together with the physical interaction between BBX19 and PRR proteins, indicating that BBX19 regulates the transcription process by interacting with PRR proteins.
In this study, it was noteworthy that the protein–protein interactions between PRR9, PRR7, PRR5 and BBX19 displayed robust circadian oscillations over a 24-h day, with the BBX19-PRR9 protein pair peak appearing at noon, BBX19-PRR7 peaking in late afternoon, and BBX19-PRR5 peaking in the evening (Figures 3 and 4). Our results hence revealed a dynamic molecular mechanism in which BBX19, a zinc-finger transcription factor, interacts with PRR9, PRR7, and PRR5 sequentially from early morning to evening, to directly inhibit CCA1, LHY, and RVE8 expression (Figure 9). Previously, BBX19 was also reported to interact with ELF3 and then be degraded by COP1 to participate in the formation of clock ELF3–ELF4–LUX evening complex (Wang et al., 2015). Regarding how PRRs regulate the transcription of target genes, there are two possible mechanisms based on previous studies. Early studies shown that TOC1 and PRR5 can directly bind to the promoter through the CCT domain, and the latest studies have shown that PRRs can also be recruited by PIFs and indirectly bind to G-box cis-elements on the promoters of target genes (Gendron et al., 2012; Nakamichi et al., 2012; Zhu et al., 2016; Zhang et al., 2020). Studies have also shown that TPL can interact with the EAR motif of PRR and contribute to the inhibitory effect of PRRs (Wang et al., 2013).
Figure 9.
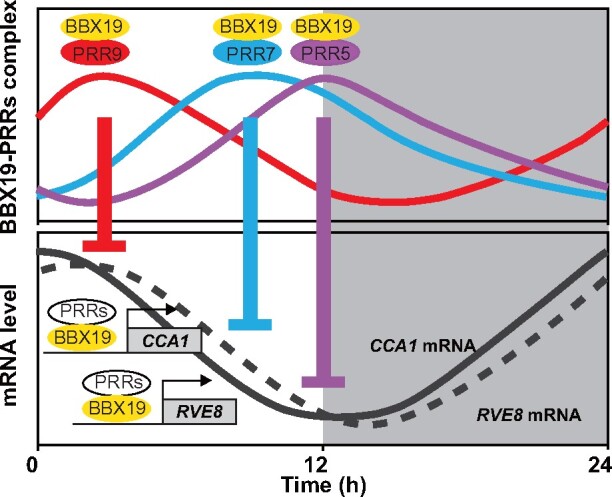
A proposed working model for the dynamic formation of BBX19–PRRs complex over a 24 h in regulating the CCA1 and RVE8 expression. Zinc finger transcription factor, BBX19 protein, is expressed during the daytime. Sequentially expressed PRR9, PRR7, and PRR5 interact with BBX19 in precise temporal ordering from dawn to dusk. PRR proteins affect BBX19 recruitment to the CCA1 and RVE8 promoters. BBX19–PRRs complexes function directly in transcriptional regulation of the circadian clock to orchestrate circadian rhythms
The plant Groucho/TUP1 family component has been identified as transcriptional corepressor of the circadian clock (Wang et al., 2013). TPL physically interacts with PRR9, PRR7, and PRR5 separately, and jointly bound to the promoters of CCA1 and LHY in the ChIP assay. Dysfunction of TPL causes increased levels of CCA1 and LHY transcripts, as well as a lengthened circadian period. As the common interacting protein of TPL and BBX19, the working model for PRR9, PRR7, and PRR5 sequential expression on CCA1 transcriptional regulation has become more complicated. Notably, the peak of TPL transcript and protein enrichment occurs around dawn of a 24-h day (Wang et al., 2013), which is quite different from the peak expression of BBX19 in the morning (Figure 2, E and F). TPL interacts with the EAR motif of PRRs. However, we found that BBX19 interacted with the PR domain, but the interaction between BBX19 and PRR9 was even augmented when EAR is missing (Figure 4D), implying that the regulatory mechanism for PRRs, BBX19, and TPL needs to be further investigated. In addition, BBX19 was previously reported to have particularly high expression in the vasculature (Wang et al., 2014). Therefore, it would be helpful to analyze the genetic relationship between TPL and BBX19 in the circadian system, and to examine the spatial and temporal organization of TPL and BBX19 in the circadian clock and clock outputs. Nonetheless, our findings provided new insights into how the circadian clock finely regulates growth and development.
Previously, BBX19 was shown to act similarly to BBX21 of the BBX IV family in mediating photomorphogenesis: BBX21 specifically binds to the T/G-box (CACGTT) element in the HY5 promoter but activates its expression (Xu et al., 2016), while PRRs have inhibitory roles in the transcriptional regulation of circadian oscillators. Here, we found that BBX19 significantly inhibited CCA1 promoter activity through interacting with PRR proteins (Figure 8, A and C). In the prr7-3 prr9-1 or prr5-1 prr7-3 mutants, the amplitude of CCA1:LUC rhythmic expression was significantly rescued compared to the wild-type material, indicating that PRRs are necessary for the inhibitory effect of BBX19 on CCA1 expression. The expression pattern of BBX19 is very similar to that of CCA1. The transcription and translation of both start around midnight and peak in the morning. Based on our results, the inhibitory effect of BBX19 on the accumulation of CCA1, LHY, and RVE8 is likely to start at midnight. This implied that BBX19–PRRs worked as a transcriptional repressor complex involved in regulating transcription initiation of morning-phased circadian oscillators.
In view of this, the other BBX IV transcription factors may form a transcription repressive complex with certain components in a similar way as BBX19–PRRs and function directly in the temporal and spatial expression of their target genes. The overexpression of COL1 and COL2, which belongs to the BBX subfamily, leads to a short-period phenotype (Ledger et al., 2001). PRRs interact with the CO protein of the BBX family to stabilize CO, thereby regulating photoperiod-dependent flowering. Also, the results of chromatin immunoprecipitation quantitative real-time polymerase chain reaction indicate that CO in the prr975 toc1 quadrant cannot bind to the FT promoter region (Hayama et al., 2017). In addition to BBX19, there are a few members from different BBX subfamilies that participate in circadian clock-related transcriptional regulation, and there may be synergy or antagonism among them.
Circadian core components—such as CCA1, PRR7, and ELF3—regulate multiple physiological outputs, such as hypocotyl elongation, in response to photoperiodic zeitgebers (Harmer, 2009; Lu et al., 2012; Martin et al., 2018; Zheng et al., 2018). We further investigated whether BBX19 also responds to light. Although the lack of BBX19 altered circadian periodicity (Figure 1), we found that the trend of the phase response curve to light pulses and the fluence-rate response curve were consistent with that of the wild-type, indicating that the responsiveness of the circadian clock in the bbx19-3 to external light signals was not affected (Supplemental Figure S12). We speculate that there may be zeitgebers other than light that reset the circadian clock via BBX19. Our results provide a molecular mechanism enhancing in-depth understanding of the fine regulation mechanism of PRRs, which may help elucidate how the circadian clock regulates growth and development in the future.
Materials and methods
Plant materials and growth conditions
The following T-DNA insertion lines were obtained from the Arabidopsis Biological Resource Center (ABRC, Ohio State University): bbx18-2 (SALK_061956), bbx19-1 (SALK_088902), bbx19-2 (SALK_087493), bbx19-3 (SALK_032997), bbx20-1 (CS878932), bbx21-2 (SALK_105390), bbx22-1 (SALK_105367), bbx23-1 (SALK_053389), bbx24-1 (SALK_067473), and bbx25-3 (CS2103310). The cca1-1 lhy-20 double mutant, in which cca1-1 is in a Ws background (Green and Tobin, 1999), was created by backcrossing six times with lhy-20 in a Col-0 background (Michael et al., 2003). toc1-101 was a gift from Peter Quail (Kikis et al., 2005). Arabidopsis seeds were sterilized in 20% bleach before being placed on 1/2 Murashige and Skoog (MS) medium (M524, PhytoTechnology Laboratories) plus 2% sucrose, and then stratified for 3 days at 4°C in the dark. Plants were grown under a 12:12 LD cycle (white light, 70 μmol m−2 s−1) at 22°C in a growth chamber (Percival CU-36L5).
Constructs
For the split luciferase complementation assays, constructs were produced following the method described previously (Li et al., 2020). Full-length BBX18, BBX19, PRR9, PRR7, and PRR5 genomic DNAs were amplified from Col-0 genomic DNA with primer pairs BBX18-F/BBX18-R and BBX19-F/BBX19-R (Supplemental Table S7), then PCR products were cloned into the pENTR 1A vector. Two SfiI sites were inserted just before the stop codons of BBX18, BBX19, PRR9, PRR7, and PRR5 through PCR amplification with primer pairs BBX18-SfiI-F/BBX18-SfiI-R and BBX19-SfiI-F/BBX19-SfiI-R (Supplemental Table S7). PCR products of either LUC, nLUC or cLUC with SfiI sites at both ends were amplified and then cloned to create in-frame translational fusions. The donor vectors with BBX18, BBX19, PRR9, PRR7, and PRR5 were finally recombined into binary vectors. Constructs consisting of PRR9 lacking the PR domain (PRR9-delPR, 118 amino acids, positions 38–156) or EAR domain (PRR9-delEAR, 20 amino acids, positions 250–269) were fused to the N-terminal domain of LUC (nLUC) before being transformed into BBX19-cLUC/Col-0 plants. To generate an estradiol-inducible pER8 expression vector, the BBX19 CDS sequences were amplified by PCR before they were inserted into pENTR/SD/D-TOPO (Invitrogen), and were then recombined by LR reaction Gateway technology into destination vector pER8-GW (Papdi et al., 2008).
The bbx19-4 Cas9-free mutant was generated using a CRISPR/Cas9 approach according to the previously published paper (Gao et al., 2016). The target sequence was cloned into the U6-gRNA unit, then the U6-gRNA unit was assembled into the pHDE-35SCas9-mCherry vector though the PmeI site. The bbx19-4 CRISPR/Cas9 constructs were transformed into Arabidopsis using the floral dip method. T1 plants were screened on MS medium with hygromycin, genomic DNA samples extracted from T1 plants were used as templates for PCR, and bbx19-4-F(PCR) and bbx19-4-R(PCR) primers were used to amplify the fragment containing the target site for Sanger sequencing. Cas9-free T2 seeds were separated by a fluorescence microscope according to the mCherry signals and the Cas9-free T2 plants were sequenced to obtain homozygous genome-editing plants. All primer sequences are listed in Supplemental Table S7.
Circadian rhythm measurement
The luciferase reporter gene fusion CCA1:LUC was introduced into the wild-type and bbx18-bbx25 mutant lines. Transgenic seedlings were entrained under 12:12 LD cycles for 7 days before they were grown in constant light (LL) at 22°C for 5 days. Circadian rhythms of LUC activity were captured using a back-illuminated CCD sensor from e2v (CCD47-40) and normalized to the mean value over the time series. Fast Fourier transform-nonlinear least squares analysis of circadian parameters were conducted on a data window of ZT24-120. The bioluminescence activity of BBX18:BBX18-LUC and BBX19:BBX19-LUC fusion proteins were measured on a Packard TopCountTM luminometer and used as a read-out of the state of BBX18 and BBX19 under LD (ZT0-48) and LL (ZT48-120) conditions.
Temporal transcriptome (RNA-seq) analysis
Seedlings of pER8:BBX19-YFP-HA/Col-0 were grown under 12:12 LD cycles at 22°C for 10 days, and then were treated by 30-μM β-estradiol or mock at ZT12. The materials were collected at ZT2 of the next morning and were immediately frozen in liquid nitrogen. RNA-seq libraries were prepared using the Illumina Directional mRNA-Seq Library Preparation Kit and sequenced on an Illumina HiSeq 2000, resulting in single-end 50-bp reads in each sample. RNA sequencing produced an average of 10.9 million reads for the mock sample and an average of 11.2 million reads for the estradiol sample. Sequence reads were aligned to the TAIR10 genome and analyzed using CLC Genomics Workbench 11 software (Qiagen). The ratio of reads mapped to the reference genome in the two groups was 99.59% and 98.86%, respectively. DEGs between the estradiol- and mock-treated pER8-BBX19-YFP-HA/Col-0 transgenic plants were identified by a significance analysis when the change was more than 1.5-fold with P < 0.05. The diurnal rhythm and circadian rhythm of DEGs were identified using microarray data (http://diurnal.mocklerlab.org/). Gene ontology (GO) term enrichment analysis for the DEGs was performed using PANTHER (http://www.pantherdb.org;Mi et al., 2013).
Co-immunoprecipitation assays
To generate pCsVMV:PRRs-HA-1300 constructs, full-length PRR9, PRR7, and PRR5 coding sequence were amplified and inserted into the vector of pCsVMV:HA-1300. Fragments containing the ORFs of BBX18 and BBX19 were separately inserted into pCsVMV:GFP-1300 and 2 × 35S:FLAG-1307 vectors. All primer sequences are listed in Supplemental Table S7. The combinations of Agrobacterium carrying the indicated vectors were co-infiltrated into the leaves of 5-week-old N. benthamiana, and the samples were collected after 3 days of infiltration. Protein extraction and immunoprecipitation assays were performed following a method described previously (Wang et al., 2013) using GFP-Trap (GTMA-20, ChromoTek) magnetic beads. The incubation was about 1 h at 4°C followed by washing four times with protein extraction buffer using a magnetic stand. For immunoblot detection, GFP antibody (Cat#ab6556, Abcam), HA antibody (Cat#11867423001, Roche), and FLAG antibody (Cat#M20008M, Abmart) were used to detect the tagged proteins.
ChIP assays
ChIP assays were performed following a previously described method (Saleh et al., 2008). Seedlings of pER8-BBX19-YFP-HA/Col-0, pER8-BBX19-YFP-HA/prr7-3 prr9-1, and pER8-BBX19-YFP-HA/prr5-1 prr7-3 were grown under 12:12 LD cycles at 22°C for 2 weeks, and then treated with 30-μM β-estradiol at ZT12. The materials were harvested and cross-linked with 1% formaldehyde at ZT3 of the next morning. Protein G-Agarose beads (Roche, Cat. # 11243233001) and an anti-HA antibody (Sigma-Aldrich, Cat. #H3663) were used for ChIP analysis. Primers amplifying a fragment in UBQ were used for the negative control. All primer sequences are listed in Supplemental Table S7.
Phylogenetic analysis
For the phylogenetic tree, sequence information on different plants was retrieved via a BLASTP search of Phytozome 12 (https://phytozome.jgi.doe.gov/pz/portal.html). Sequence alignments and evolutionary analyses were performed with the software MEGA 7 (Kumar et al., 2016). Multiple sequence alignments were performed using ClustalW and phylogenic trees were generated using the neighbor-joining method (Saitou and Nei, 1987). Statistical support of the nodes was calculated with the bootstrap method with 1,000 replicates (Felsenstein, 1985).
Accession numbers
Sequence data for the genes described in this article can be found in the GenBank/EMBL databases under the following accession numbers: BBX18 (AT2G21320), BBX19 (AT4G38960), BBX20 (AT4G39070), BBX21 (AT1G75540), BBX22 (AT1G78600), BBX23 (AT4G10240), BBX24 (AT1G06040), BBX25 (AT2G31380), CCA1 (AT2G46830), LHY (AT1G01060), RVE8 (AT3G09600), TOC1 (AT5G61380), PRR5 (AT5G24470), PRR7 (AT5G02810), PRR9 (AT2G46790), ELF3 (AT2G25930), LUX (AT3G46640), IPP2 (AT3G02780), and UBQ (AT4G05320).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Circadian rhythms of CCA1:LUC in BBX subfamily IV gene mutation lines under free-running conditions.
Supplemental Figure S2. Circadian rhythms in the BBX19 mutation and complementation lines.
Supplemental Figure S3. Phylogenetic assessment of AtBBX18 and AtBBX19 orthologs in land plants.
Supplemental Figure S4. Subcellular localization of BBX18 and BBX19.
Supplemental Figure S5. Negative controls for BiFC assays.
Supplemental Figure S6. LUC bioluminescence analysis showed dynamic protein–protein interactions between BBX19/18 and PRR proteins.
Supplemental Figure S7. Dynamic protein–protein interactions between BBX19 and TOC1, ELF3 proteins.
Supplemental Figure S8. Estradiol-induced BBX19 expression at subjective night inhibited the transcript accumulation of CCA1, LHY, and RVE8.
Supplemental Figure S9. BBX19 inhibits the accumulation of CCA1, LHY, and RVE8 transcripts.
Supplemental Figure S10. BBX19 overexpression leads to the reduced amplitude and lengthened period of TOC1:LUC.
Supplemental Figure S11. Inducible expression of BBX19 protein in the BBX19-YFP-HA transgenic lines.
Supplemental Figure S12. Characteristics of circadian rhythms in response to environmental light cues.
Supplemental Table S1. Five genes of BBX subfamily IV, co-expressed with CCA1 and LHY in multiple microarray- and RNAseq-based coexpression data sets in ATTED-II (http://atted.jp), were highly ranked in the co-expression list.
Supplemental Table S2. Period length of CCA1:LUC circadian rhythms shown in Figure 1, C and D.
Supplemental Table S3. Period length of circadian rhythms shown in Figure S2.
Supplemental Table S4. Period length of CCA1:LUC circadian rhythms shown in Figure 2, A–D.
Supplemental Table S5. Period length of CCA1:LUC circadian rhythms shown in Figure 5, A–C.
Supplemental Table S6. Period length of CCA1:LUC circadian rhythms shown in Figure 5, D–F.
Supplemental Table S7. Oligonucleotides (shown 5′ to 3′) used in this study.
Supplemental Data Set S1. Text file of the alignment used for the phylogenetic analysis shown in Figure 1B.
Supplemental Data Set S2. Text file of the alignment used for the phylogenetic analysis shown in Supplemental Figure S3.
Supplemental Data Set S3. RNA sequencing of the circadian transcriptome from BBX19 inducible overexpression lines shown in Figure 6, A–D.
Supplementary Material
Acknowledgment
We thank Jun-Xian He for sharing the pMN6 transient assay system.
Funding
This work was supported by the National Natural Science Foundation of China to X.X. (U1904202, 31570285) and Q.X. (31670285), the Natural Science Foundation of Hebei (17966304D) and the Hebei Hundred Talents Program (E2016100018) to Q.X., National Natural Science Foundation of China (31570292) and Strategic Priority Research Program of the Chinese Academy of Sciences (XDB27030206) to L.W.
Conflict of Interest statement: The authors declare no conflicts of interest.
These authors contributed equally to this work (L.Y., Y.Y.).
X.X., Q.X., and L.W. conceived the project and wrote the article. L.Y. and Y.Y. analyzed circadian rhythm with BBXs gene mutants. L.Y. constructed pER8-BBX19 expression vector, generated genetic materials, performed temporal transcriptome, RT-PCR, dynamic protein–protein interactions, and ChIP analysis. Y.Y. performed confocal imaging, completed Y2H, BiFC, and Co-IP analysis and related constructs. M.L. performed the WB assay, Y.S. performed RT-PCR assay, H.L. completed partial Y2H construct. Q.X. and Q.W. completed the constructs of PRRs for LCA analysis. X.X., Q.X., L.Y., Y.Y. and L.W. analyzed the data.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) are: Xiaodong Xu (xiaodong.xu@henu.edu.cn), Lei Wang (wanglei@ibcas.ac.cn), and Qiguang Xie (qiguang.xie@henu.edu.cn).
References
- Anwer MU, Davis A, Davis SJ, Quint M (2020) Photoperiod sensing of the circadian clock is controlled by EARLY FLOWERING 3 and GIGANTEA. Plant J 101: 1397–1410 [DOI] [PubMed] [Google Scholar]
- Bursch K, Toledo-Ortiz G, Pireyre M, Lohr M, Braatz C, Johansson H (2020) Identification of BBX proteins as rate-limiting cofactors of HY5. Nat Plants 6: 921–928 [DOI] [PubMed] [Google Scholar]
- Chow BY, Helfer A, Nusinow DA, Kay SA (2012) ELF3 recruitment to the PRR9 promoter requires other Evening Complex members in the Arabidopsis circadian clock. Plant Signal Behav 7: 1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington MF, Panda S, Liu XL, Strayer CA, Wagner DR, Kay SA (2001) ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell 13: 1305–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creux N, Harmer S (2019) Circadian rhythms in plants. Cold Spring Harb Perspect Biol 11: a034611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Hettiarachchi GHCM, Deng XW, Holm M (2006) Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth. Plant Cell 18: 70–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Hettiarachchi C, Johansson H, Holm M (2007) SALT TOLERANCE HOMOLOG2, a B-Box protein in Arabidopsis that activates transcription and positively regulates light-mediated development. Plant Cell 19: 3242–3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Johansson H, Hettiarachchi C, Irigoyen ML, Desai M, Rubio V, Holm M (2008) LZF1/SALT TOLERANCE HOMOLOG3, an Arabidopsis B-box protein involved in light-dependent development and gene expression, undergoes COP1-mediated ubiquitination. Plant Cell 20: 2324–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Wang S, Song ZT, Jiang Y, Han JJ, Lu SJ, Li L, Liu JX (2018) Two B-box domain proteins, BBX18 and BBX23, interact with ELF3 and regulate thermomorphogenesis in Arabidopsis. Cell Rep 25: 1718–1728 [DOI] [PubMed] [Google Scholar]
- Fan XY, Sun Y, Cao DM, Bai MY, Luo XM, Yang HJ, Wei CQ, Zhu SW, Sun Y, Chong K, et al. (2012). BZS1, a B-box protein, promotes photomorphogenesis downstream of both brassinosteroid and light signaling pathways. Mol Plant 5: 591–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farinas B, Mas P (2011) Functional implication of the MYB transcription factor RVE8/LCL5 in the circadian control of histone acetylation. Plant J 66: 318–329 [DOI] [PubMed] [Google Scholar]
- Farré EM, Liu T (2013) The PRR family of transcriptional regulators reflects the complexity and evolution of plant circadian clocks. Curr Opin Plant Biol 16: 621–629 [DOI] [PubMed] [Google Scholar]
- Felsenstein J (1985) Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39: 783–791 [DOI] [PubMed] [Google Scholar]
- Gangappa SN, Crocco CD, Johansson H, Datta S, Hettiarachchi C, Holm M, Botto JF (2013) The Arabidopsis B-BOX protein BBX25 interacts with HY5, negatively regulating BBX22 expression to suppress seedling photomorphogenesis. Plant Cell 25: 1243–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Chen J, Dai X, Zhang D, Zhao Y (2016) An effective strategy for reliably isolating heritable and Cas9-free Arabidopsis mutants generated by CRISPR/Cas9-mediated genome editing. Plant Physiol 171: 1794–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron JM, Pruneda-Paz JL, Doherty CJ, Gross AM, Kang SE, Kay SA (2012) Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc Natl Acad Sci USA 109: 3167–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RM, Tobin EM (1999) Loss of the circadian clock-associated protein 1 in Arabidopsis results in altered clock-regulated gene expression. Proc Natl Acad Sci USA 96: 4176–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL (2009) The circadian system in higher plants. Annu Rev Plant Biol 60: 357–377 [DOI] [PubMed] [Google Scholar]
- Hayama R, Sarid-Krebs L, Richter R, Fernandez V, Jang S, Coupland G (2017) PSEUDO RESPONSE REGULATORs stabilize CONSTANS protein to promote flowering in response to day length. EMBO J 36: 904–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Pérez-García P, Pokhilko A, Millar AJ, Antoshechkin I, Riechmann JL, Mas P (2012) Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336: 75–79 [DOI] [PubMed] [Google Scholar]
- Jiang L, Wang Y, Li QF, Bjorn LO, He JX, Li SS (2012) Arabidopsis STO/BBX24 negatively regulates UV-B signaling by interacting with COP1 and repressing HY5 transcriptional activity. Cell Res 22: 1046–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S, Rozwadowski K (2011) EAR motif-mediated transcriptional repression in plants: an underlying mechanism for epigenetic regulation of gene expression. Epigenetics 6: 141–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamioka M, Takao S, Suzuki T, Taki K, Higashiyama T, Kinoshita T, Nakamichi N (2016) Direct repression of evening genes by CIRCADIAN CLOCK-ASSOCIATED1 in the Arabidopsis circadian clock. Plant Cell 28: 696–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R, Kronmiller B, Maszle DR, Coupland G, Holm M, Mizuno T, Wu SH (2009) The Arabidopsis B-box zinc finger family. Plant Cell 21: 3416–3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikis EA, Khanna R, Quail PH (2005) ELF4 is a phytochrome-regulated component of a negative-feedback loop involving the central oscillator components CCA1 and LHY. Plant J 44: 300–313 [DOI] [PubMed] [Google Scholar]
- Kumagai T, Ito S, Nakamichi N, Niwa Y, Murakami M, Yamashino T, Mizuno T (2008) The common function of a novel subfamily of B-box zinc finger proteins with reference to circadian-associated events in Arabidopsis thaliana. Biosci Biotechnol Biochem 72: 1539–1549 [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau OS, Huang X, Charron J-B, Lee J-H, Li G, Deng XW (2011) Interaction of Arabidopsis DET1 with CCA1 and LHY in mediating transcriptional repression in the plant circadian clock. Mol Cell 43: 703–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledger S, Strayer C, Ashton F, Kay SA, Putterill J (2001) Analysis of the function of two circadian-regulated CONSTANS-LIKE genes. Plant J 26: 15–22 [DOI] [PubMed] [Google Scholar]
- Li Y, Wang L, Yuan L, Song Y, Sun J, Jia Q, Xie Q, Xu X (2020) Molecular investigation of organ-autonomous expression of Arabidopsis circadian oscillators. Plant Cell Environ 43: 1501–1512 [DOI] [PubMed] [Google Scholar]
- Liu TL, Newton L, Liu MJ, Shiu SH, Farre EM (2016) A G-box-like motif is necessary for transcriptional regulation by circadian pseudo-response regulators in Arabidopsis. Plant Physiol 170: 528–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XL, Covington MF, Fankhauser C, Chory J, Wagner DR (2001) ELF3 encodes a circadian clock–regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell 13: 1293–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SX, Webb CJ, Knowles SM, Kim SHJ, Wang Z, Tobin EM (2012) CCA1 and ELF3 interact in the control of hypocotyl length and flowering time in Arabidopsis. Plant Physiol 158: 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Rovira A, Veciana N, Soy J, Toledo-Ortiz G, Gommers CMM, Boix M, Henriques R, Minguet EG, Alabadi D, et al. (2018) Circadian waves of transcriptional repression shape PIF-regulated photoperiod-responsive growth in Arabidopsis. Curr Biol 28: 311–318 [DOI] [PubMed] [Google Scholar]
- McClung CR (2019) The plant circadian oscillator. Biology (Basel) 8: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Thomas PD (2013) PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res 41: D377–D386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Salomé PA, Yu HJ, Spencer TR, Sharp EL, Alonso JM, Ecker JR, McClung CR (2003) Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science 302: 1049–1053 [DOI] [PubMed] [Google Scholar]
- Nagel DH, Doherty CJ, Pruneda-Paz JL, Schmitz RJ, Ecker JR, Kay SA (2015) Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc Natl Acad Sci USA 112: E4802–4810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kita M, Ito S, Sato E, Yamashino T, Mizuno T (2005). PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol 46: 686–698 [DOI] [PubMed] [Google Scholar]
- Nakamichi N, Kiba T, Henriques R, Mizuno T, Chua NH, Sakakibara H (2010) PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22: 594–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kiba T, Kamioka M, Suzuki T, Yamashino T, Higashiyama T, Sakakibara H, Mizuno T (2012) Transcriptional repressor PRR5 directly regulates clock-output pathways. Proce Natl Acad Sci USA 109: 17123–17128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kusano M, Fukushima A, Kita M, Ito S, Yamashino T, Saito K, Sakakibara H, Mizuno T (2009) Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol 50: 447–462 [DOI] [PubMed] [Google Scholar]
- Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farre EM, Kay SA (2011) The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papdi C, Ábrahám E, Joseph MP, Popescu C, Koncz C, Szabados L (2008) Functional identification of Arabidopsis stress regulatory genes using the controlled cDNA overexpression system. Plant Physiol 147: 528–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss SB, Meister R, Xu Q, Urwin CP, Tripodi FA, Screen SE, Anil VS, Zhu S, Morrell JA, Liu G, et al. (2012). Expression of the Arabidopsis thaliana BBX32 gene in soybean increases grain yield. PLoS One 7: e30717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda-Paz JL, Breton G, Para A, Kay SA (2009) A functional genomics approach reveals CHE as a novel component of the Arabidopsis circadian clock Science 323: 1481–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat R, Takahashi N, Hsu PY, Jones MA, Schwartz J, Salemi MR, Phinney BS, Harmer SL (2011) REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock. PLoS Genet 7: e1001350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rugnone ML, Faigón Soverna A, Sanchez SE, Schlaen RG, Hernando CE, Seymour DK, Mancini E, Chernomoretz A, Weigel D, Más P, et al. (2013) LNK genes integrate light and clock signaling networks at the core of the Arabidopsis oscillator. Proc Natl Acad Sci USA 110: 12120–12125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Saleh A, Alvarez-Venegas R, Avramova Z (2008) An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nature Protoc 3: 1018–1025 [DOI] [PubMed] [Google Scholar]
- Song Z, Bian Y, Liu J, Sun Y, Xu D (2020) B-box proteins: pivotal players in light-mediated development in plants. J Integr Plant Biol 62: 1293–1309 [DOI] [PubMed] [Google Scholar]
- Tripathi P, Carvallo M, Hamilton EE, Preuss S, Kay SA (2017) Arabidopsis B-BOX32 interacts with CONSTANS-LIKE3 to regulate flowering. Proc Natl Acad Sci USA 114: 172–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CQ, Guthrie C, Sarmast MK, Dehesh K (2014) BBX19 interacts with CONSTANS to repress FLOWERING LOCUS T transcription, defining a flowering time checkpoint in Arabidopsis. Plant Cell 26: 3589–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CQ, Sarmast MK, Jiang J, Dehesh K (2015) The transcriptional regulator BBX19 promotes hypocotyl growth by facilitating COP1-mediated EARLY FLOWERING3 degradation in Arabidopsis. Plant Cell 27: 1128–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Kim J, Somers DE (2013) Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc Natl Acad Sci USA 110: 761–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CQ, Chien CW, Ai LF, Zhao J, Zhang Z, Li KH, Burlingame AL, Sun Y, Wang ZY (2016) The Arabidopsis B-box protein BZS1/BBX20 interacts with HY5 and mediates strigolactone regulation of photomorphogenesis. J Genet Genomics 43: 555–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Wang P, Liu X, Yuan L, Wang L, Zhang C, Li Y, Xing H, Zhi L, Yue Z, et al. (2014) LNK1 and LNK2 are transcriptional coactivators in the Arabidopsis circadian oscillator. Plant Cell 26: 2843–2857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Jiang Y, Li J, Holm M, Deng XW (2018) The B-box domain protein BBX21 promotes photomorphogenesis. Plant Physiol 176: 2365–2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Jiang Y, Li J, Lin F, Holm M, Deng XW (2016) BBX21, an Arabidopsis B-box protein, directly activates HY5 and is targeted by COP1 for 26S proteasome-mediated degradation. Proc Natl Acad Sci USA 113: 7655–7660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakir E, Hilman D, Kron I, Hassidim M, Melamed-Book N, Green RM (2009) Posttranslational regulation of CIRCADIAN CLOCK ASSOCIATED1 in the circadian oscillator of Arabidopsis. Plant Physiol 150: 844–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JW, Rubio V, Lee NY, Bai S, Lee SY, Kim SS, Liu L, Zhang Y, Irigoyen ML, Sullivan JA, et al. (2008). COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol Cell 32: 617–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Huai J, Shang F, Xu G, Tang W, Jing Y, Lin R (2017) A PIF1/PIF3-HY5-BBX23 transcription factor cascade affects photomorphogenesis. Plant Physiol 174: 2487–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Pfeiffer A, Tepperman JM, Dalton-Roesler J, Leivar P, Gonzalez Grandio E, Quail PH (2020) Central clock components modulate plant shade avoidance by directly repressing transcriptional activation activity of PIF proteins. Proc Natl Acad Sci USA 117: 3261–3269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Zhang F, Wang S, Su Y, Ji X, Jiang P, Chen R, Hou S, Ding Y (2018) MLK1 and MLK2 coordinate RGA and CCA1 activity to regulate hypocotyl elongation in Arabidopsis thaliana. Plant Cell 30: 67–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JY, Oh E, Wang T, Wang ZY (2016) TOC1-PIF4 interaction mediates the circadian gating of thermoresponsive growth in Arabidopsis. Nat Commun 7: 13692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Niu Q-W, Chua N-H (2000) An estrogen-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J 24: 265–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.