Abstract
The ataxia-telangiectasia mutated (ATM) and ATM and Rad3-related (ATR) kinases coordinate the DNA damage response. The roles described for Arabidopsis thaliana ATR and ATM are assumed to be conserved over other plant species, but molecular evidence is scarce. Here, we demonstrate that the functions of ATR and ATM are only partially conserved between Arabidopsis and maize (Zea mays). In both species, ATR and ATM play a key role in DNA repair and cell cycle checkpoint activation, but whereas Arabidopsis plants do not suffer from the absence of ATR under control growth conditions, maize mutant plants accumulate replication defects, likely due to their large genome size. Moreover, contrarily to Arabidopsis, maize ATM deficiency does not trigger meiotic defects, whereas the ATR kinase appears to be crucial for the maternal fertility. Strikingly, ATR is required to repress premature endocycle onset and cell death in the maize endosperm. Its absence results in a reduction of kernel size, protein and starch content, and a stochastic death of kernels, a process being counteracted by ATM. Additionally, while Arabidopsis atr atm double mutants are viable, no such mutants could be obtained for maize. Therefore, our data highlight that the mechanisms maintaining genome integrity may be more important for vegetative and reproductive development than previously anticipated.
Differently from Arabidopsis, ATR activity in maize plays an essential role under control growth conditions, ensuring genome stability during kernel Development.
Introduction
Due to their sessile lifestyle, plants can be exposed to severe adverse environmental stresses, resulting in aberrant DNA replication and loss of genome integrity, both potentially affecting growth and plant survival (Hu et al., 2016; Nisa et al., 2019). To cope with such aberrations, eukaryotic cells activate highly coordinated cellular networks, collectively termed as the DNA damage response (DDR), which are crucial for maintaining genome stability (Hu et al., 2016; Nisa et al., 2019). DDR activation relies on two phosphatidylinositol 3 kinase-like protein kinases, called ataxia-telangiectasia mutated (ATM) and ATM and Rad3-related (ATR) (Hu et al., 2016). In general, DNA double-strand breaks (DSBs) activate the ATM kinase, whereas ATR is predominantly activated by single-stranded DNA (ssDNA) and defects in replication fork progression (Maréchal and Zou, 2013). ATM is activated by the MRN complex (MRE11, RAD50, and NBS1) that recognizes DSBs (Nisa et al., 2019); ATR responds to the accumulation of the replication protein A at ssDNA sites that are generated in response to any lesion that perturbs replication (Iyer and Rhind, 2017).
Remarkably, whereas the upstream kinases ATM and ATR and the proteins that participate directly in the DNA repair mechanisms appear to be largely conserved across eukaryotes, the factors that transduce the DNA damage signal appear to have diverged (Hu et al., 2016; Nikitaki et al., 2018). For instance, plants lack orthologous genes for the canonical signal transducer kinases CHK1/2 and the transcription factor p53 that act downstream of ATM and ATR in mammals (Nikitaki et al., 2018). Instead, Arabidopsis possesses a set of plant-specific proteins, including the SOG1 transcription factor (Yoshiyama et al., 2009; 2013a) and the SIAMESE/SIAMESE-RELATED (SMR) family of cyclin-dependent kinase inhibitors (Yi et al., 2014). SOG1 is phosphorylated by ATM in response to DSBs (Yoshiyama et al., 2013b) and can be phosphorylated in vitro by ATR (Sjogren et al., 2015). This phosphorylation is crucial for SOG1 activation in response to DSBs (Yoshiyama et al., 2017), representing a direct and immediate connection between DNA damage and the transcriptional activation of DDR genes (Yoshiyama et al., 2009; Bourbousse et al., 2018; Ogita et al., 2018).
In contrast to mammals, where disruption of ATR induces embryonic death (Brown and Baltimore, 2000; Maréchal and Zou, 2013; Menolfi and Zha, 2020) and ATM mutations lead to developmental defects (Menolfi and Zha, 2020), Arabidopsis atr and atm knockout mutants do not show any vegetative developmental defect under nonstressed conditions (Garcia et al., 2003; Culligan et al., 2004), although atm mutant plants display partial sterility (Garcia et al., 2003). Even the atr atm double mutant is viable, but fully infertile due to severe defects during meiosis (Culligan and Britt, 2008). In spite of these discrepancies at the organismal scale, the respective roles of ATM and ATR in DNA damage signaling appear to be conserved between plants and other eukaryotes. Indeed, in Arabidopsis, loss of ATM results in hypersensitivity to genotoxic agents that induce DSBs either directly (e.g. exposure to γ-irradiation) or indirectly during the replication process (e.g. crosslinking agents), as observed in all eukaryotes (Garcia et al., 2003; Culligan et al., 2006; Menolfi and Zha, 2020). Likewise, the Arabidopsis atr mutants are hypersensitive to replication-blocking agents (Culligan et al., 2004), as other eukaryotes (Friedel et al., 2009; Segurado and Tercero, 2009; Jossen and Bermejo, 2013). However, ATR deficiency leads to high sensitivity to DSBs in yeast and mammals (Segurado and Tercero, 2009; Jossen and Bermejo, 2013), whereas Arabidopsis atr mutant plants show only a slight sensitivity to ionizing irradiation, suggesting only a minor role of this kinase in the response to DSBs in plants (Culligan et al., 2004, 2006).
The described roles for ATR and ATM in Arabidopsis have been assumed to be conserved in other plants such as crops, but molecular evidence is scarce (Manova and Gruszka, 2015; Szurman-Zubrzycka et al., 2019; Zhang et al., 2020), and some differences may exist between plant species. Indeed, recently, the characterization of both kinases in Physcomitrium patens revealed differences with their Arabidopsis orthologs, with PpATR playing a predominant role in the transcriptional response to DSBs, rather than PpATM (Martens et al., 2020) and triggering the formation of stem cells from differentiated tissue in response to DSBs (Gu et al., 2020).
So far, the roles of ATR and ATM have not been studied during the development of crop seeds. The maize seed consists of three main compartments: the seed coat or pericarp, of maternal origin, and the embryo and endosperm, the two products of double fertilization (Doll et al., 2017). Endosperm development begins with a triploid (3C) cell formed from the two polar nuclei of the ovule and one sperm cell (Domínguez and Cejudo, 2014). After fertilization, mitotic cell proliferation begins, and at ∼10–12 days after pollination (DAP), the endosperm cells transition from a mitotic cell cycle to endoreduplication, resulting in a dramatic increase in nuclear volume and DNA content that peaks at 16–20 DAP. Grain filling begins simultaneously through the accumulation of storage compounds, such as starch and proteins, necessary to support seedling growth following germination. During the final stages of seed development, the starchy endosperm undergoes programmed cell death (PCD), but these cells and their contents remain intact in the mature grain. The cell death of the starchy endosperm is critical to facilitate the rapid mobilization of the storage reserves for the growth of the germinated embryo. Premature induction of PCD would limit reserve deposition and thus jeopardize germination (Young et al., 1997; Young and Gallie, 2000; Domínguez and Cejudo, 2014).
Here, we demonstrate that the functional roles of the ATR and ATM kinases are only partially conserved between Arabidopsis and maize, as exemplified by the accumulation of replication defects in maize ATR-deficient plants and differences in fertility defects between the two species. Moreover, our data reveal a crucial role for the ATR kinase in proper kernel development, playing a pivotal role in repressing premature endocycle onset and cell death of the endosperm.
Results
Generation of CRISPR/Cas9 mutants for the maize ATR and ATM genes
Maize orthologs of the Arabidopsis ATR and ATM genes were initially identified using the PLAZA platform (Van Bel et al., 2018). For the AtATR gene (AT5G40820), this resulted in a one-on-one relationship with the Zm00001d014813 gene, encoding a protein with 67.2% sequence similarity. The ATM gene (AT3G48190) showed a one-on-one orthology with the maize Zm00001d040166 gene, although the encoded protein shares only a 47.7% sequence similarity. While the AtATM gene contains 63 exons and is spread over 23 kb, the ZmATM gene has 78 exons and is spread over >123 kb.
With both orthologous genes identified, we designed two single guide RNAs to induce mutations in the coding sequence of the maize ATR and ATM genes using CRISPR/Cas9. We introduced these constructs into immature maize embryos via Agrobacterium-mediated transformation to generate knockouts of the maize ATR and ATM genes. For each construct, T0 mutant regenerated plants were crossed with B104 wild-type (WT) plants. Within the offspring, plants were selected for heterozygosity for the mutation and absence of the Cas9 transgenes. These plants were subsequently self-pollinated to obtain isogenic WT and homozygous mutant plants. We selected two independent CRISPR/Cas9-induced mutant lines for both genes, named ATR-A and ATR-B, representing Zmatr mutants, and ATM-A and ATM-B, representing Zmatm mutants. While the ATR-B and ATM-A mutant alleles were caused by an indel that generates a frameshift, ATR-A and ATM-B were deletion mutants of hundreds of base pairs (Supplemental Figure S1).
Conserved role of ZmATR in the response to replicative stress
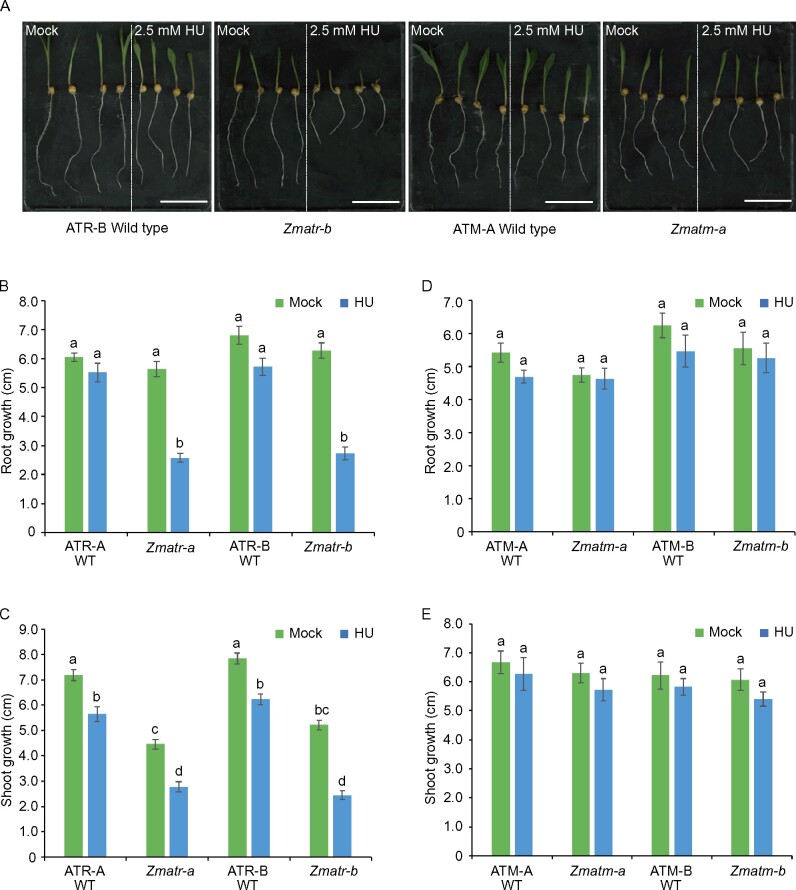
In all eukaryotes studied so far, ATR is required for responses to replicative stress such as fork stalling, which is associated with hypersensitivity to replication-blocking agents (Friedel et al., 2009; Segurado and Tercero, 2009; Jossen and Bermejo, 2013). The main characteristic of Arabidopsis atr mutants is a rapid loss of meristem activity due to the inability to slow down the replication process in the presence of stalled replication forks (Culligan et al., 2004). To confirm that maize ATR shares this conserved role, we challenged the WT and mutant seedlings with hydroxyurea (HU), which is an inhibitor of ribonucleotide reductase. HU treatment depletes cellular deoxyribonucleotide pools, and thereby induces stalling of replication forks. The sensitivity to DNA stress was initially determined by measuring Zmatr-b root length during 4 days of growth within medium supplemented with HU (2.5 and 5.0 mM). The mutant seedlings displayed a dramatic decrease in root growth at both concentrations while the WT plants were only sensitive at 5.0 mM (Supplemental Figure S2A). Therefore, for further phenotypic analysis using both independent mutants, the lowest HU concentration was used. Although both the WT and Zmatr mutant roots grew at a very similar rate in medium without genotoxin (Figure 1, A and B), the shoots of the Zmatr mutant lines were 40%–50% smaller in comparison with their corresponding WT plants (Figure 1, A and C). When treated with HU, the WT roots were not affected, whereas the Zmatr mutant roots were reduced by ∼60% in comparison with untreated plants (Figure 1, A and B). A similar effect was observed for the Zmatr mutant shoot length with a decrease of ∼40% compared with approximately 15%–20% in WT plants (Figure 1, A and C). As expected, ZmATM is not involved in the stabilization of stalled forks, as both the Zmatm mutant lines did not display any inhibition of growth under treatment nor under control conditions (Figure 1, A, D, and E).
Figure 1.
Zmatr plants are hypersensitive to replicative stress induced by HU. A, Representative WT and mutant seedlings following growth for 3 days under control conditions (Mock) or in the presence of 2.5 mM HU. Scale bar = 5.5 cm. B and D, Quantification of root growth of ATR (B) and ATM (D) WT and mutant seedlings treated as in (A). C and E, Quantification of shoot growth of ATR (C) and ATM (E) WT and mutant seedlings treated as in (A). Values are average ± standard error (se); n = 8–10. Letters on the bars indicate statistically different means (P < 0.05, ANOVA mixed model analysis, Tukey’s correction for multiple testing). See also Supplemental Data Set S1.
In Arabidopsis, the ATR and ATM kinases transduce the DNA stress signal to the SOG1 transcription factor, which controls the transcriptional induction of DNA repair genes (Yoshiyama et al., 2013a; Hu et al., 2016; Yoshiyama et al., 2017). To determine whether the ZmATR and ZmATM kinases control the transcriptional response under replicative stress, we monitored the expression-level changes of several DDR-related genes, including the putative maize orthologous genes for RAD51A, RAD51B, BRCA1, XRI, and RAD54 involved in homologous recombination (HR), RAD7A that participates in nucleotide excision repair, and the RNR1 and TSO2 ribonucleotide reductase subunits that function in the synthesis of nucleotides (Roa et al., 2009; Sánchez-Pons et al., 2011; Lahari et al., 2018). To map the rapid transcriptional changes, plants were treated with 5 mM HU for 90 min. With exception of the RAD54 gene, the early transcriptional response toward HU was dependent on ZmATR, but not ZmATM (Supplemental Figure S3). These results show that ZmATR plays a conserved role in the replicative stress response.
ZmATR plays a predominant role in the response to DSBs, whereas ZmATM shows a minor role
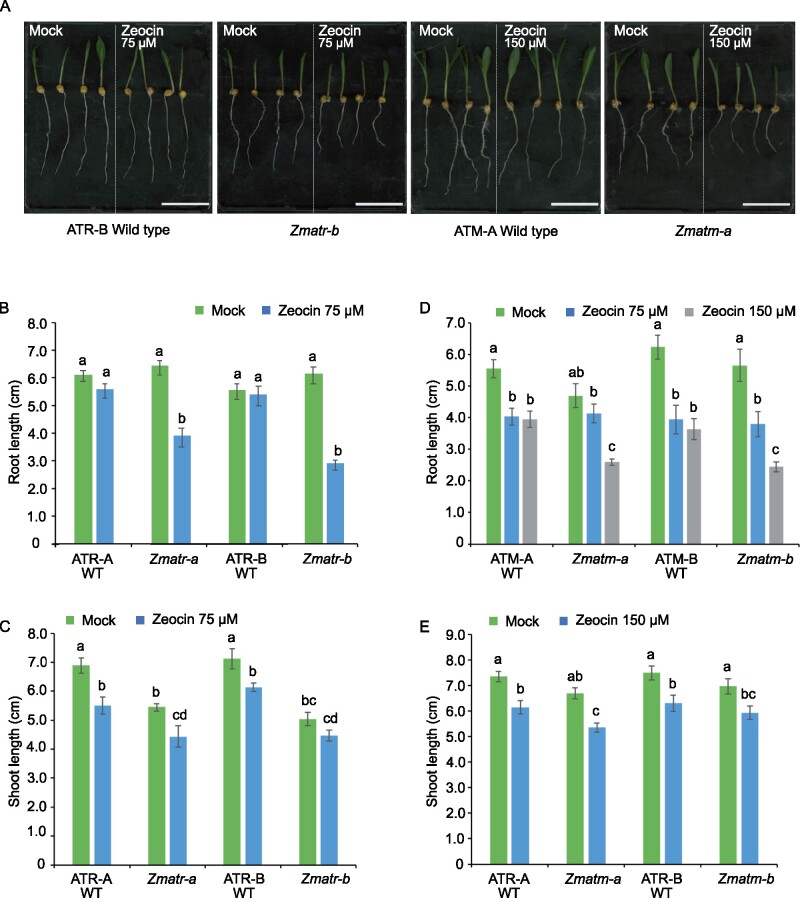
To explore the roles of ZmATR and ZmATM in response to DSBs, we evaluated the sensitivity of the corresponding mutants to zeocin, which is a radiomimetic agent that generates mainly DSBs (Chankova et al., 2007). Initially, Zmatr-b and Zmatm-b seedlings were treated with zeocin (75 and 150 µM). The Zmatr-b mutant displayed a pronounced root growth sensitivity in response to both concentrations (Supplemental Figure S2B) compared with the control plants. Contrastingly, Zmatm-b only displayed a slight sensitivity to zeocin at 150 µM in comparison to its WT line (Supplemental Figure S2C). To validate these observations, we challenged the two independent lines for Zmatr and Zmatm and their corresponding WT plants with 75 µM zeocin. Both Zmatr lines showed a reduction of root length between 40% and 50% compared with the untreated plants, whereas WT plants did not show a significant reduction of root growth (Figure 2, A and B). Again, with 75 µM zeocin, no increased sensitivity was observed for the Zmatm mutants compared with WT plants, which did show a ∼10% reduction in root length (Figure 2D). Nevertheless, with the 150-µM zeocin treatment, a clear difference of reduction in root growth was observed in Zmatm mutants compared with WT roots. Moreover, under all conditions, we observed a slight inhibition of shoot growth, which was similar between all genotypes examined (Figure 2, C and E).
Figure 2.
Both maize ATR and ATM kinases are necessary for resistance to zeocin. A, Representative WT and mutant seedlings following growth for 3 days under control conditions (Mock) or in the presence of 75 or 150 µM zeocin. Scale bar = 5.5 cm. B and D, Quantification of root growth of ATR (B) and ATM (D) WT and mutant seedlings treated as in (A). C and E, Quantification of shoot growth of ATR (C) and ATM (E) WT and mutant seedlings treated as in (A). Letters indicate statistically different growth for each genotype (P < 0.0001, mixed model analysis, Tukey’s correction for multiple testing). Values are average ± se; n = 8–11. Letters on the bars indicate statistically different means (P < 0.05, ANOVA mixed model analysis, Tukey’s correction for multiple testing). See also Supplemental Data Set S1.
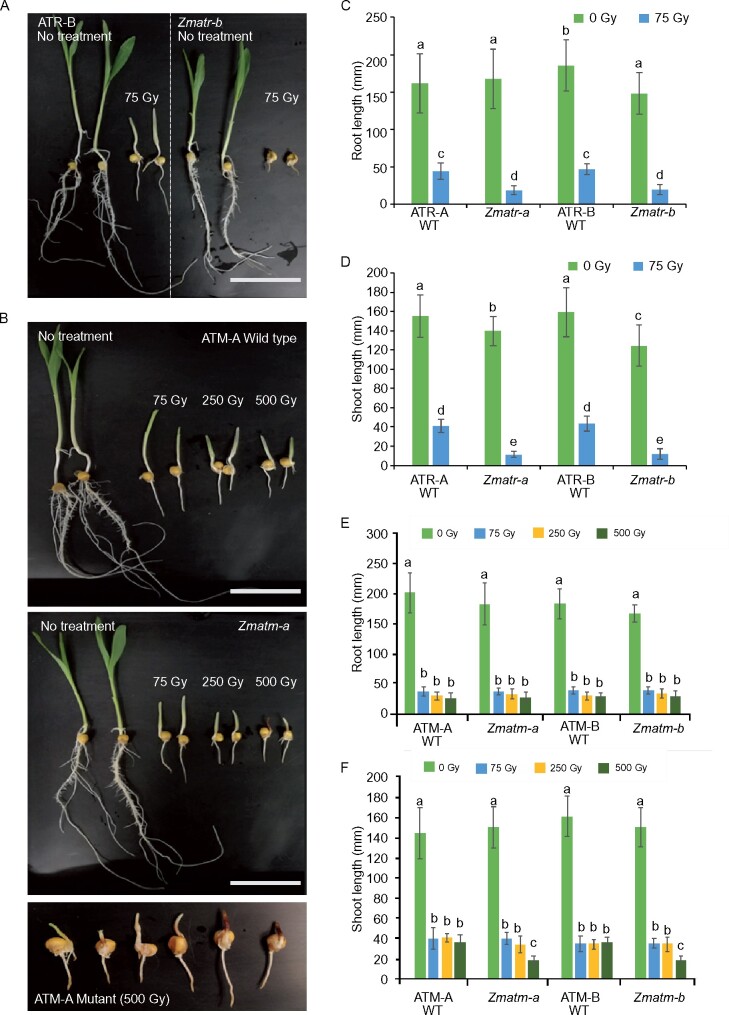
To validate these results on DSB sensitivity, we exposed the seedlings to γ-irradiation, which is known to also induce DSBs. Two-day-old seedlings were treated with a dose of 75 Gy. Root and shoot lengths were measured after 4 days of recovery. Under these treatment conditions, the Zmatr mutant lines clearly were more sensitive to the treatment compared with WT plants (Figure 3, A, C, and D), but no differences were observed for the Zmatm mutant (Figure 3, B, E, and F). To examine whether ZmATM is required to respond to higher levels of DNA damage as observed with zeocin treatment, we irradiated with doses of 250 and 500 Gy. Only at 500 Gy, shoot growth in Zmatm was severely affected (Figure 3, B, E, and F) and most of the shoots showed accumulation of anthocyanins, revealed by purple coloring (Figure 3B, lowest part). In summary, our results show that ZmATR plays a crucial role in the DDR triggered by DSBs in maize, as described for yeast and mammals (Cimprich and Cortez, 2008; Blackford and Jackson, 2017), whereas ZmATM contributes to the response under severe DNA-damaging conditions only.
Figure 3.
Maize ATR plays a predominant role in the response to γ-irradiation-induced DNA damage. A and B, Representative 7-day-old ATR (A) and ATM (B) WT and mutant seedlings grown under control conditions (no treatment) or γ‐irradiated at doses of 75, 250, or 500 Gy on day 2. Scale bar = 6.5 cm. C and D, Average root (C) and shoot (D) length of ATR WT and mutant seedlings. E and F, Average root (E) and shoot (F) length in ATM WT and mutant seedlings. Values are average ± se; n = 25–31 (C-D); n = 24–32 (E-F). Letters on the bars indicate statistically different means (P < 0.05, ANOVA mixed model analysis, Tukey’s correction for multiple testing). See also Supplemental Data Set S1.
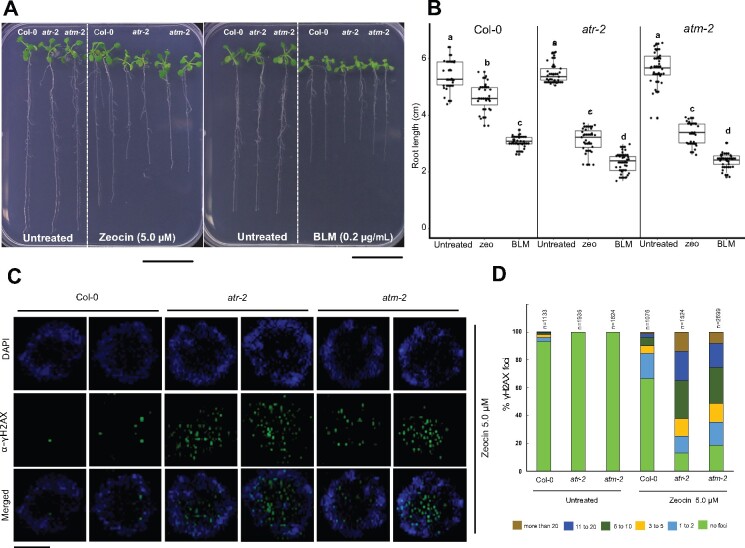
Previous studies in Arabidopsis suggested that in comparison with ATM, ATR plays only a minor role in the response to DSBs, because Arabidopsis atm mutant plants display an enhanced sensitivity to γ-irradiation, whereas atr mutants show only slight sensitivity in comparison with the WT (Culligan et al., 2004, 2006). To test this hypothesis, we challenged Arabidopsis atr and atm mutant plants to different concentrations of zeocin and the related drug bleomycin (Supplemental Figure S4). Notably, the concentration of zeocin needed to trigger in Arabidopsis a root growth inhibition similar to that observed in maize was remarkedly lower, at 5 µM. We found that both atr and atm mutants displayed an increased sensitivity to both DSB-inducing agents in comparison with the WT (Figure 4, A and B). These results suggest that in Arabidopsis ATR is also crucial for resistance to DSBs, next to ATM.
Figure 4.
Atatr and Atatm mutants display similar sensitivity to zeocin and bleomycin, both DSB inducers. A, Representative seedlings after growth for 12 days under control conditions (untreated) or in the presence of 5.0-µM zeocin or 0.2 µg/mL bleomycin (BLM). Scale bar = 3.0 cm. B, Root length was measured after 12 days under control conditions (untreated) or in the presence of 5.0-µM zeocin or bleomycin 0.2 µg/mL (n ≥ 20 roots for each condition). Data are from two independent experiments. Letters on the data indicate statistically different means (P < 0.05, ANOVA mixed model analysis, Tukey’s correction for multiple testing). See also Supplemental Data Set S1. C, Immunostaining of γH2AX foci accumulation (green) in nuclei stained with DAPI (blue) in Arabidopsis root tips from Col-0, atr-2 and atm-2 plants after 24 h of treatment with 5.0-µM zeocin. Two representative nuclei are shown for each line. Scale bar = 3 µm. D, Quantification of the number of γH2AX foci per nucleus in Col-0, atr-2, and atm-2 root tips after 24 h under control conditions (untreated) or in the presence of 5.0 µM zeocin. The number of nuclei analyzed for each genotype and condition is shown on each bar. For each sample, the γH2AX foci of analyzed nuclei were counted and grouped into six categories: no focus, 1–2, 3–5, 6–10, 11–20, and >20 foci per nucleus.
Next, we examined whether the transcriptional response to DSBs in maize is affected in atr and atm mutants. Because γ-irradiation can generate reactive oxygen species and other types of stresses that lead to the generation of ssDNA (Kim et al., 2019), we only used zeocin to avoid such secondary effects. The same group of genes mentioned above for the response to HU was employed to evaluate the early transcriptional response to zeocin, by harvesting root tips for quantitative reverse transcription PCR (RT-qPCR) analysis after 3-h and 6-h treatments. WT maize plants presented a rapid and strong transcriptional induction to this DNA-damaging agent (Supplemental Figure S3), as previously reported for Arabidopsis (Adachi et al., 2011). The loss of either ATR or ATM impeded the early transcriptional response to DSBs for all the genes evaluated (Supplemental Figure S3). These observations appear contrasting with previous observations published for Arabidopsis, where ATR was reported to play only a minor role in the transcriptional response to DSBs (Culligan et al., 2006; Ricaud et al., 2007). A possible explanation is the source of material, being whole Arabidopsis seedlings compared with maize root tips. Therefore, to check whether the role of ATR in response to DSBs differs between maize and Arabidopsis, we decided to evaluate the transcriptional response in the Arabidopsis root tips. Three cyclin-dependent kinase inhibitors (SMR4, SMR5, and SMR7) that are transcriptionally activated by DNA damage (Yi et al., 2014) and DNA repair genes (RAD51A, RAD51B, BRCA1, XRI-1, RAD7A, CYCB1;1, and PARP2) were included in the analysis. In Atatr root tips, the early transcriptional response was dramatically attenuated by zeocin treatment for most of the genes tested, whereas Atatm displayed a full loss of transcriptional induction (Supplemental Figure S5). These results show that ATR helps to achieve a proper transcriptional induction in response to DSBs in proliferating tissues in both plant species.
Lack of ATR and ATM in maize leads to impaired DNA repair and activation of checkpoints in response to DNA stress
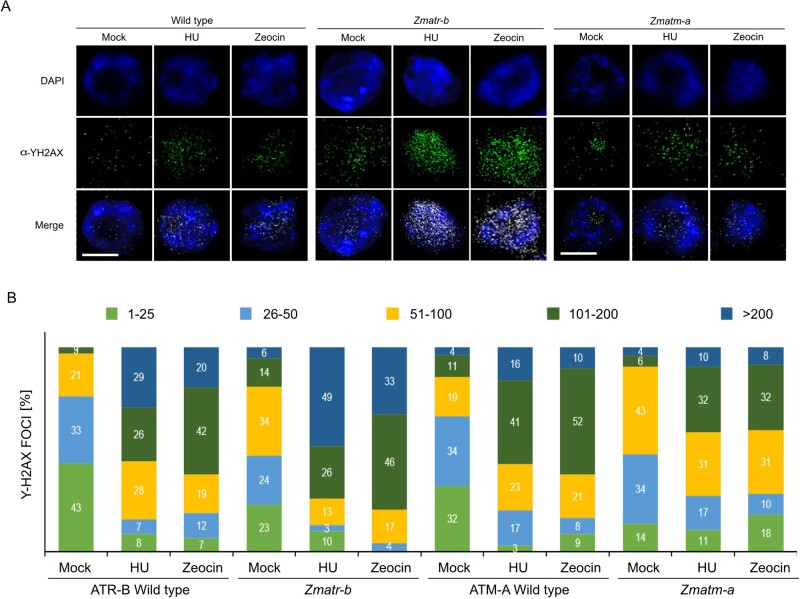
One essential mechanism to maintain genome integrity is the accumulation of γH2AX foci at DNA lesions in an ATM- and ATR-dependent manner (Friesner et al., 2005; Amiard et al., 2010; Maréchal and Zou, 2013; Waterworth et al., 2019). Upon DNA damage, both kinases phosphorylate the histone variant H2AX to generate γH2AX foci that recruit repair factors to the break sites (Turinetto and Giachino, 2015; Waterworth et al., 2019). To determine whether the observed sensitivity of the Zmatr and Zmatm mutants to DNA-damaging agents was associated with the loss of repair capacity and accumulation of DNA damage, we performed immunodetection of γH2AX foci on root tip nuclei of untreated and treated plants. The number of foci per nucleus varied widely. To visualize the results, we grouped the nucleus population into five categories, depending on the number of foci present per nucleus. Strikingly, all nuclei observed of untreated WT plants showed γH2AX foci (Figure 5), indicating the natural occurrence of DNA lesions. The Zmatr-b and Zmatm-a root tips showed a higher number of foci per nucleus compared with their respective WT line (Figure 5, A and B).
Figure 5.
Accumulation of γH2AX foci in Zmatr and Zmatm root tips in response to HU and zeocin. A, Immunostaining of γH2AX foci accumulation (green) in nuclei stained with DAPI (blue) in maize root tips of WT plants and Zmatr-b and Zmatm-a mutants after 24 h of treatment with 5.0 mM HU or 150-µM zeocin. Scale bar = 5 µm. B, Quantification of the number of γH2AX foci per nucleus in ATR-B WT, ATM-A WT, Zmatr-b and Zmatm-a root tips after 24 h of treatment with HU (5 mM), or zeocin (150 µM). For each sample, the γH2AX foci of 100 nuclei were counted and grouped into five categories: 1–25, 26–50, 51–100, 10–200, and >200 foci per nucleus.
In response to replication stress induced by 24 h exposure to HU (5 mM), the number of γH2AX foci per nucleus increased in the WT ATR and ATM lines, with around 80% of the nuclei showing ≥51 γH2AX foci. Zmatr-b mutant nuclei showed a hyperaccumulation of DNA damage foci, with around 49% nuclei presenting ≥200 foci, whereas in the isogenic WT line, this category represented only 29% (Figure 5B). Zmatm-a displayed a less dramatic accumulation of foci per nucleus, with only 42% nuclei showing ≥101 γH2AX foci, while in its WT line, around 57% nuclei were grouped in this category (Figure 5B).
We also treated the roots for 24 h with zeocin (150 µM). Strikingly, we observed a hyperaccumulation of γH2AX foci in Zmatr-b mutant nuclei, while the Zmatm-a mutant displayed only a slightly increased number of γH2AX foci (Figure 5A). Indeed, while the WT ATR-B and ATM-A nuclei showed around 62% nuclei with ≥101 γH2AX foci, the population of Zmatm-a mutant nuclei in this category was 40%. In the case of the Zmatr-b mutant, almost 80% of nuclei displayed this degree of accumulation of DNA damage foci (Figure 5B). These results are consistent with above-mentioned hypersensitivity of Zmatr mutants to zeocin. As comparison, we examined in Arabidopsis whether the formation of γH2AX foci in response to zeocin is affected in atr and atm mutants. Interestingly, both mutants showed an equal hyperaccumulation of γH2AX foci after treatment (Figure 4, C and D), while untreated plants did not display foci, as previously reported (Friesner et al., 2005; Amiard et al., 2010; Weimer et al., 2016).
To explore whether Zmatr and Zmatm mutants are impaired in the activation of cell cycle checkpoints, we examined the cell cycle progression by performing flow cytometry on root tips after 24 h of treatment with HU (5 mM) or zeocin (150 µM). In untreated root tips, no differences in the proportion of G1, S, and G2 nuclei were observed between the ATR-A WT and Zmatr-a mutant (Supplemental Figure S6A). After 24-h treatment with HU, in WT root tips, the population of nuclei in S-phase was enriched as was expected because HU triggers the activation of the intra S-phase checkpoint (Cools et al., 2011). However, in the Zmatr-a mutant, the nucleus population that arrested in the S-phase was smaller than in the WT, and there was an increase in the fraction of nuclei in G1/S. In contrast, the Zmatm-a mutant did not display differences from the WT in response to HU (Supplemental Figure S6A).
The profiles of flow cytometry in response to zeocin were less clear and any differences were nonsignificant (Supplemental Figure S6A). Therefore, because in other eukaryotes, ATM is required for adequate activation of a G2 checkpoint in response to DSBs (Blackford and Jackson, 2017), we determined the number of mitotic events per root tip in WT ATM-A and mutant Zmatm-a. No differences were observed between both lines in untreated root tips (Supplemental Figure S6B). However, in response to zeocin after 24 h, the WT roots showed a drastic decrease in the mitotic figures, from 175 on average per untreated root to 44 mitoses per treated root tip, whereas Zmatm-a mutant roots displayed an average of 94 per treated root tip (Supplemental Figure S6B). These results indicate that ZmATM is required for a proper cell cycle arrest in response to DSBs generated by zeocin.
The maize atr mutant displays defective kernel development
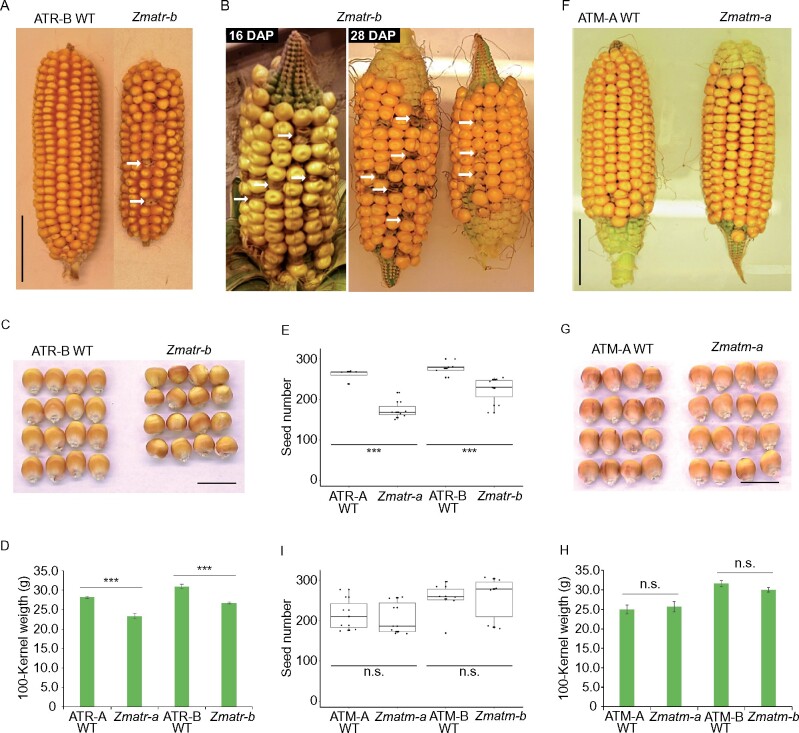
Similar to Atatr and Atatm mutants, their counterparts in maize were visually indistinguishable from WT plants at the adult stage when they are grown under optimal greenhouse conditions, despite the fact that Zmatr seedlings were smaller in the first days post-germination in vitro. In contrast to Atatr plants that are fully fertile (Culligan et al., 2004), Zmatr mutants displayed smaller cobs (Figure 6A; Supplemental Figure S7A), with a reduced kernel number per cob (Figure 6E), compared with WT plants. Intriguingly, although the Zmatr cobs appeared to be fully fertilized at early stages post-fertilization (16 and 28 DAP), a small population of kernels displayed an abnormal phenotype, showing a brown and wrinkled appearance (Figure 6B). These kernels died before maturity, resulting in gaps in the mature cobs (Figure 6A; Supplemental Figure S7A). The Zmatr kernels that did reach maturity were smaller in size and weight compared with WT kernels (Figure 6, C and D). The Zmatm mutant plants did not display defects on fertility (Figure 6F), contrary to what has been reported for Atatm mutants, which are semi-sterile (Garcia et al., 2003). The Zmatm mutant lines were indistinguishable from WT plants in the cob and kernel phenotypes (Figure 6, F and G; Supplemental Figure S7B), number of kernels per cob (Figure 6I), and mature kernel weight (Figure 6H).
Figure 6.
Phenotypic cob and kernel features of Zmatr and Zmatm mutant plants. A, Representative mature F2 ear of WT and Zmatr-b. Arrows indicate gaps left by early death of kernels. Scale bar = 4 cm. B, Representative F2 ears of Zmatr-b at 16 and 28 DAP. Arrows indicate kernels with abnormal phenotype that eventually die. C, Representative mature kernels of WT and Zmatr-b from a segregated F2 ear. Scale bar = 1.5 cm. D, Comparison of the 100-kernel weight of mature WT and Zmatr kernels of two independent lines. Weight was determined in a segregated F2 population. Values are means ± se; n = 8–12 (***P < 0.001, Student’s t-test). See also Supplemental Data Set S1. E, Number of kernels per cob in WT and Zmatr plants of two independent lines. Values are means ± se; n = 7–10 (***P < 0.001, Student’s t-test). See also Supplemental Data Set S1. F, Representative mature F2 ear of WT and Zmatm-a. Scale bar = 4 cm. G, Representative mature kernels of WT and Zmatm-a from a segregated F2 ear. Scale bar = 1.5 cm. H, Comparison of the 100-kernel weight of mature WT and Zmatm kernels of two independent lines. Weight was determined in a segregated F2 population. Values are means ± se; n = 8–9 (ns, not significant; Student’s t-test). See also Supplemental Data Set S1. I, Number of kernels per cob in WT and Zmatm plants of two independent lines. Values are means ± se; n = 6–7 (ns, not significant; Student’s t- test). See also Supplemental Data Set S1.
To gain better insight into the reasons for the apparent death shown for some Zmatr mutant kernels, we counted the percentage of abnormal kernels per cob. The abnormal kernel phenotype was observed between 5% and 16% (Supplemental Table S1), which is not consistent with the expected 25% Mendelian ratio for an embryo-lethal mutation, thus rather suggesting a stochastic frequency. Since Arabidopsis ATM is crucial for pollen viability (Garcia et al., 2003), we examined whether the abnormal kernel phenotype might be associated with a defective pollen phenotype through viability staining on pollen from WT and Zmatr mutant plants. Nearly all the pollen from Zmatr mutant plants were viable, similar to pollen of WT plants (Supplemental Figure S8A). To investigate the maternal effect, reciprocal backcrosses with B104 WT plants were performed. Pollen from WT plants was not able to rescue the Zmatr mutant cob phenotype. In contrast, Zmatr mutant plants used as pollen donor for the WT showed normal cob development (Supplemental Figure S8B and Supplemental Table S2). These reciprocal crosses hint to a maternal fertility defect in the Zmatr plants.
Lack of ZmATR and ZmATM results in endogenous DNA damage in the embryo
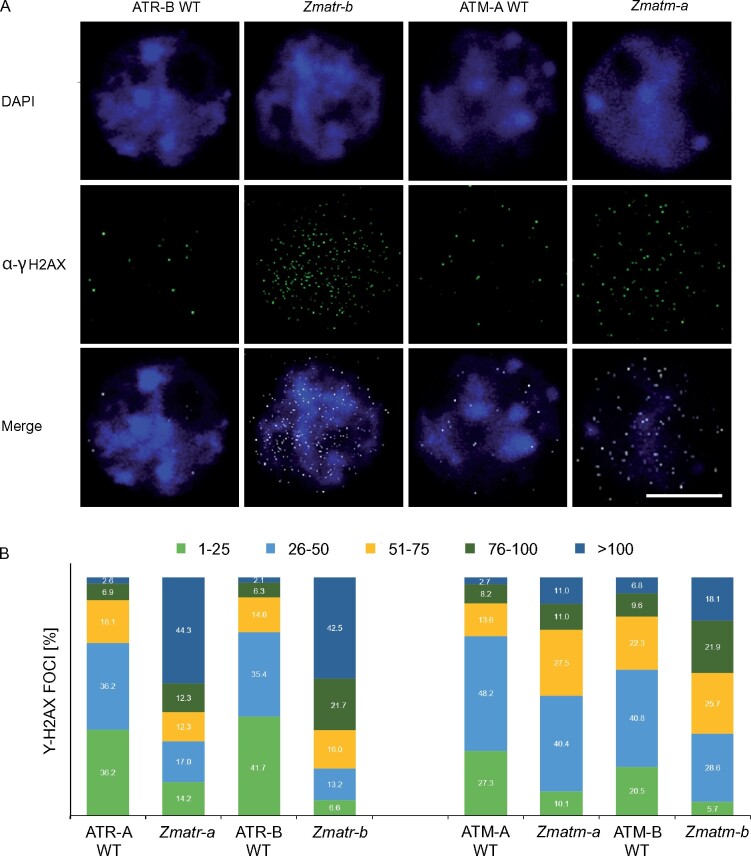
To determine whether lack of ZmATR or ZmATM may lead to the accumulation of DNA damage in the embryo, which is a developmental stage with a high rate of DNA replication and thus potentially sensitive to endogenous replicative stress, we analyzed γH2AX foci accumulation in embryos at 16 DAP. As observed in root tips, all nuclei of Zmatr-a and Zmatr-b embryos showed a higher number of foci per nucleus compared with their respective WT lines (Figure 7, A and B). In the Zmatr embryo, the majority of nuclei had ≥51 foci, being 68.9% for Zmatr-a and 80.2% in Zmatr-b, while for WT lines, the percentage of the nuclei with this foci number was 27.6% and 23.0%, respectively (Figure 7B). The Zmatm mutant embryo nuclei also displayed an increase of foci number although this was less outspoken compared with the Zmatr mutant embryos (Figure 7A). For instance, Zmatm-a and Zmatm-b nuclei with ≥51 γH2AX foci were 49.5% and 65.7% respectively, whereas their corresponding WT lines showed a percentage of 24.5% and 38.7%. The higher accumulation of DNA damage sites within the Zmatr embryonic nuclei was reflected by at least 40% of the nuclei analyzed showing ≥100 γH2AX foci compared with their WT lines, in which only 2.1%–2.6% of their nuclei displayed this amount of DNA damage foci. In the Zmatm mutant, only between 11.0% and 18.1% of the nuclei contained ≥100 γH2AX foci (Figure 7B). Thus, these results indicate that lack of ATR leads to a significant accumulation of DNA damage in the embryo. Moreover, although Zmatm mutant plants did not show macroscopic differences, they also showed increased endogenous DNA damage at least during the embryo development.
Figure 7.
Detection of γH2AX foci in Zmatr and Zmatm mutant embryos. A, Immunostaining of γH2AX foci accumulation (green) in nuclei stained with DAPI (blue) of ATR WT, Zmatr, ATM-WT, and Zmatm embryos at 16 DAP. A representative nucleus is shown for each line. Scale bar = 5 µm. B, Quantification of γH2AX foci in ATR-WT, Zmatr, ATM-WT, and Zmatm embryos at 16 DAP. For each sample, the γH2AX foci of 100 nuclei were counted and grouped into five categories: 1–25, 26–50, 51–75, 76–100, and >100 foci per nucleus. Two independent lines were analyzed.
Loss of ZmATR activity triggers premature endoreduplication in the endosperm
During maize kernel development, the endosperm switches from a mitotic cell program to endoreplication and eventually to PCD (Sabelli and Larkins, 2009). ATR and ATM are relatively strongly expressed in embryo and endosperm cells of maize, both in mitotic phase and throughout the endoreplication phase (Supplemental Figure S9), suggesting that both kinases might have a relevant role during endosperm development.
To test whether the mitotic cell cycle might be affected in the Zmatr and Zmatm mutant endosperm, expression levels of cell cycle-related genes were analyzed through RT-qPCR at 10, 12, 14, 16, and 19 DAP. The expression levels for almost all evaluated genes with exception of CDK2 were reduced in Zmatr endosperm at 19 DAP in comparison with corresponding WT endosperm, whereas at earlier time points, the significant differences were variable (Supplemental Figure S10A). These results indicate that at 19 DAP, most cells of Zmatr endosperm exited the mitotic cell cycle prematurely. In Zmatm endosperm, a slight decrease in the expression of cell cycle-related genes at 14 DAP was observed, but at later time points, expression levels recovered to those of WT endosperm.
Following the exit of the mitotic cell cycle, the endosperm cells undergo endoreplication (Sabelli and Larkins, 2009). To determine the effects of lack of ATR or ATM on the endoreplication process, developing endosperms of at least three independent cobs were analyzed by flow cytometry. Lack of ZmATM did not lead to changes in endoreplication level (Supplemental Figure S11). In contrast, the mean ploidy of Zmatr-a and Zmatr-b endosperm nuclei significantly increased at 14 DAP relative to WT endosperm, and this phenomenon was sustained for up to 19 DAP (Figure 8A). This increase was attributed to increased frequencies of 12C, 24C, and 48C endosperm nuclei in comparison with WT endosperm (Figure 8B): around 32% of the nuclei in the WT endosperm were between 12C and 48C, whereas these classes represented around 40% of nuclei in the Zmatr mutants (Figure 8B).
Figure 8.
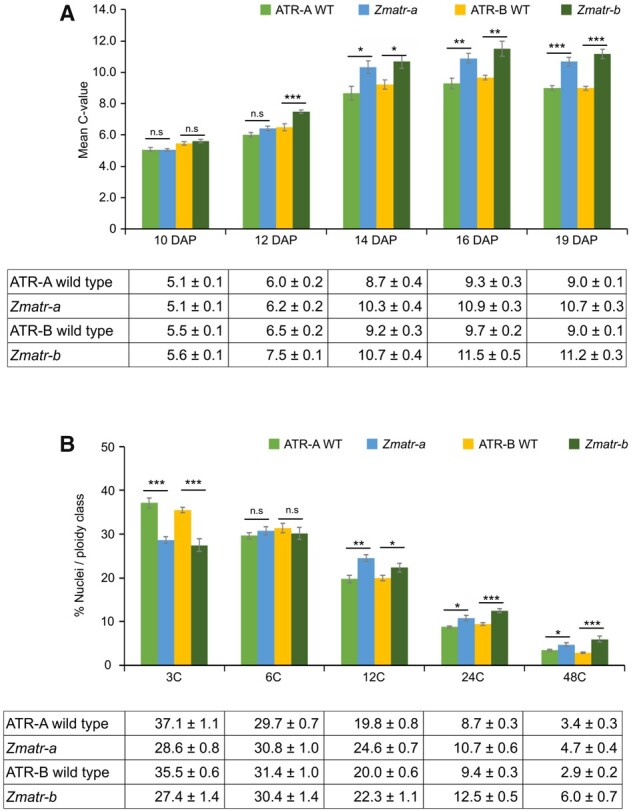
Premature endocycle onset in Zmatr endosperm. A, The mean C-values in ATR-WT and Zmatr endosperm at five developmental stages between 10 and 19 DAP. All analyses were carried out from at least three kernels from three independent cobs for each genotype. Values are means ± se; n = 9–11. Significantly different (ns, P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001, Student’s t-test). See also Supplemental Data Set S1. B, Distribution of endosperm nuclei (expressed as a percentage of the total number of nuclei) among different ploidy classes in 19-DAP ATR-WT and Zmatr endosperms. All analyses were carried out from at least three kernels from three independent cobs for each genotype. Values are means ± se; n = 9–11 (ns, not significant; P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001, Student’s t-test). See also Supplemental Data Set S1.
We next evaluated the expression levels of DNA replication-related genes such as MINICHROMOSOME 2-7 GENE FAMILY and PROLIFERATING CELL NUCLEAR ANTIGEN. The transcript levels were measured by RT-qPCR on RNA extracted from WT and Zmatr-b endosperms at 10, 12, 14, 16, and 19 DAP. Contrary to our expectations, reduced expression levels were observed for all evaluated genes at 19 DAP in Zmatr endosperms compared with WT plants (Supplemental Figure S10B).
Cell death is enhanced in ZmATR endosperm
Upon completion of endoreduplication, starchy endosperm cells undergo PCD, resulting in extensive DNA degradation (Young and Gallie, 2000). We evaluated whether the pattern and timing of cell death in Zmatr and Zmatm mutant endosperms were affected. First, the viability of endosperm cells during kernel development was examined by staining fresh sections with Evans blue, a dye that is excluded from living cells with intact plasma membranes, thereby staining only the cytoplasm of non-viable cells (Young et al., 1997). Between 16 and 22 DAP, staining within the central endosperm was more prominent and clearly more extended in Zmatr endosperm compared with WT endosperm (Figure 9A). To quantify these observations, we determined the cell death area (%) per endosperm of at least seven kernels from three different cobs. At the three time points evaluated (16, 19, and 22 DAP), the cell death area (%) was significantly higher in both Zmatr mutants in comparison with their respective WT lines (Figure 9B). In contrast, Zmatm mutants did not show differences with their WT plants, except for time point 22 DAP in the Zmatm-b mutant, for which a significant increase in the cell death area (%) was observed (Supplemental Figure S12). However, because this result was not reproducible for the other independent line, it might be caused by variability rather than the lack of ATM.
Figure 9.
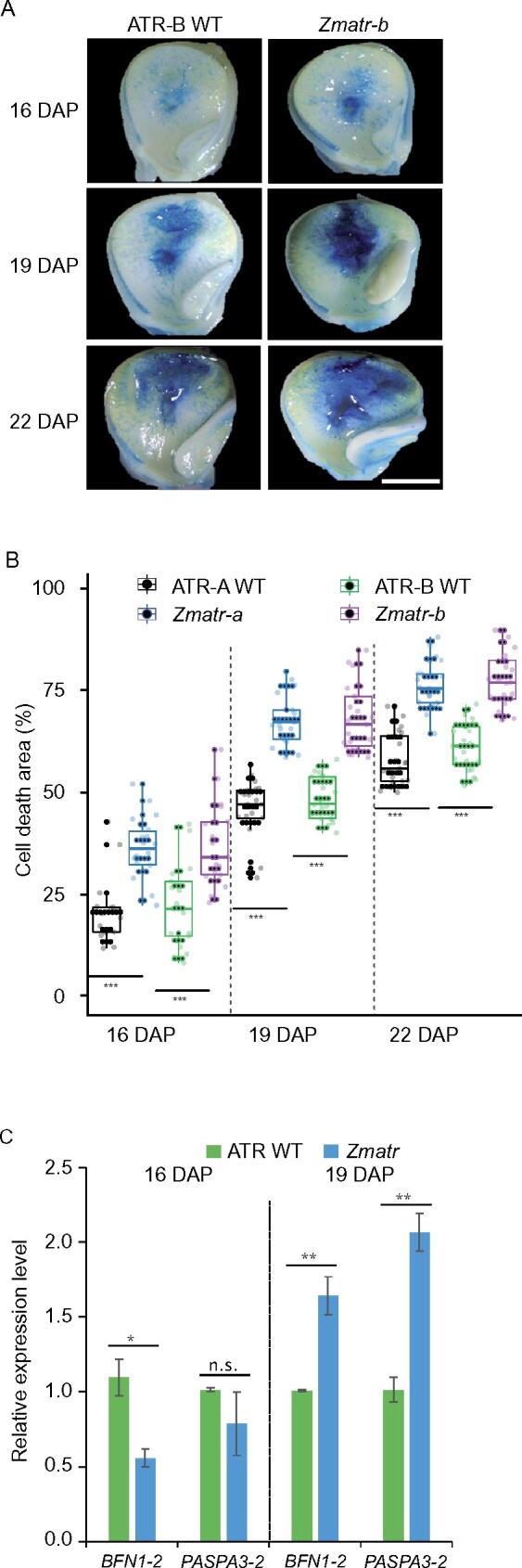
Lack of maize ATR results in enhanced cell death in endosperm. A, Progression of endosperm cell death in ATR-B WT and Zmatr-b kernels at indicated DAP, as indicated by Evans Blue staining. Dead cells are dark-stained. Scale bar = 3 mm. B, Quantification of the percentage of cell death within the endosperm area in ATR-A and ATR-B WT, and Zmatr-a and Zmatr-b mutants. All analyses were carried out from at least seven kernels of three independent cobs for each genotype. Values are means ± se; n = 21 (***P < 0.001, Student’s t-test). See also Supplemental Data Set S1. C, Expression levels of the BFN1-2 and PASPA3-2 transcripts in ATR-B WT and Zmatr-b endosperms at 16 and 19 DAP. RNA was extracted from an endosperm pool extracted from at least three kernels of three cobs for each genotype, and the expression levels were measure by quantitative RT-qPCR. Each bar shows the mean ± standard deviation (sd) sd of three biological replicates (**P < 0.05; **P < 0.01, Student’s t-test). See also Supplemental Data Set S1.
To study further the cell death within Zmatr endosperm, we determined the changes of expression of two lytic protein genes that accompany the progression of PCD in the root cap, the S1-P1 nuclease BFN1 and the aspartic protease PASPA3-2, both expressed in the endosperm of Arabidopsis (Farage-Barhom et al., 2008; Fendrych et al., 2014; Moussu et al., 2017). At 19 DAP, the expression of PASPA3-2 and BFN1-2 was 1.6- and 2-fold, respectively, higher in Zmatr than in WT endosperm (Figure 9C). Together, these results indicate that lack of ATR leads to more prominent and extended cell death in the endosperm, likely by an earlier transition to cell death. Although this phenomenon was observed in viable Zmatr kernels, abnormal Zmatr kernels exhibited massive cell death in the whole endosperm at 16 DAP (Supplemental Figure S13), which was consistent with the loss of viability of these kernels.
To determine whether the increased endoreduplication and early cell death affected starch and storage protein synthesis in the Zmatr kernels, we first evaluated the expression of genes involved in protein (19KDa zein) and starch synthesis (Opaque1, Opaque10, SS4, SSIIa, and Shrunken 2) (He et al., 2019). The expression levels of genes linked to starch synthesis displayed a significant increase of >2.5-fold at 19 DAP in Zmatr-b endosperm compared with WT endosperm (Supplemental Figure S14A). Next, we compared starch and protein (nonzein and zein) contents in Zmatr-b and WT endosperms. While no differences were observed at 19 DAP, mature endosperm of Zmatr-b kernels showed a significant reduction in the starch and zein protein content (Supplemental Figures S14, C and E), but not in nonzein protein content (Supplemental Figure S14, F and G). In Zmatm endosperm at 19 DAP, a lower accumulation of starch was found (Supplemental Figure S14B), but no difference was observed in mature endosperm (Supplemental Figure S14C). Likewise, although Zmatm endosperm displayed a significant reduction of starch and protein synthesis-related genes (Supplemental Figure S14A), these changes did not affect the final accumulation of these nutrients in mature kernels. Together, our results indicate that lack of ZmATR, but not of ZmATM, leads to early induction of endoreduplication and cell death in starchy endosperm, affecting starch and protein storage, resulting in the reduction of mature kernel size.
The presence of either ZmATR or ZmATM is essential for survival
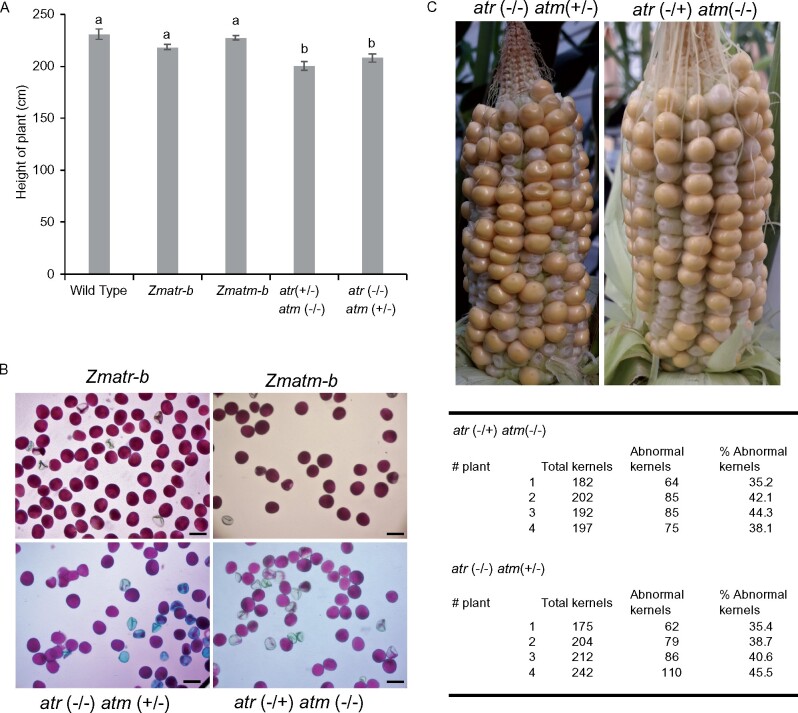
To determine whether the Zmatr Zmatm double mutant is viable, we crossed the Zmatr-b and Zmatm-b single mutants. No progeny with homozygous mutations in both genes were obtained after genotyping 200 seedlings from the F2 generation and 100 plants from the F3 generation. To gain more insight into the basis for the apparent lethality associated with the double mutation, the atr−/− atm+/− and atr+/− atm−/− sesquimutant lines, single mutants and WT plants were recovered for further analysis during the development. The sesquimutant plants displayed a slight but significant reduction in the plant height in comparison with the adult plants of single mutants and WT lines of adult plants that grew under optimal conditions in the greenhouse (Figure 10A).
Figure 10.
Vegetative and reproductive phenotypes of atm (+/−) atr (−/−) and atm (−/−) atr (+/−) sesquimutant plants. A, Height of plants of WT, Zmatr-b and Zmatm-b single mutants, and atm (+/−) atr (−/−) and atm (−/−) atr (+/−) sesquimutants. Height was measured from the soil up to the top tassel. Values are average ± se; n = 4–5. Letters on the bars indicate statistically different means (P < 0.05, ANOVA mixed model analysis, Tukey’s correction for multiple testing). See also Supplemental Data Set S1. B, Representative pollen of Zmatr-b and Zmatm-b single mutants, and atm (+/−) atr (−/−) and atm (−/−) atr (+/−) sesquimutants. Viability was visualized by Alexander staining. Scale bar = 50 µm. C, Cob phenotype of the atm (+/−) atr (−/−) and atm (−/−) atr (+/−) sesquimutants at 19 DAP (top). Percentage of abnormal kernels per cob of atm (+/−) atr (−/−) and atm (−/−) atr (+/−) sesquimutants (lower).
To better understand the basis for the apparent kernel death associated with the double mutation, we performed viability staining on pollen from the sesquimutant and single-mutant plants. Contrary to single mutants, where no differences in the pollen viability were observed compared with WT plants, some of the pollen of sesquimutant plants were non-viable (Figure 10B). Strikingly, despite the sesquimutants displaying a population of non-viable pollen, full cobs were obtained after self-pollination, although the percentage of abnormal kernels was dramatically increased in comparison with the Zmatr single mutant (Figure 10C). To evaluate whether the phenotype of abnormal kernels is consistent with a Mendelian ratio, we estimated the percentage of abnormal kernels per cob (Figure 10C). Interestingly, the kernels with a lethal phenotype were between 35% and 45%, rather than 25% that would be expected for a lethality associated uniquely to double mutant kernels; the early death of some Zmatr single-mutant or sesquimutant kernels likely contributed to this higher percentage.
Discussion
ATR and ATM signaling functions are largely conserved among Arabidopsis and maize upon exposure to DNA-damaging agents
Maize atr mutants show hypersensitivity toward HU, which initially induces stalling of replication forks by depleting cellular deoxyribonucleotide pools, but also results in DSB formation because of fork collapse (Singh and Xu, 2016). We found that the early transcriptional DDR induced by HU was ATR-dependent and that a lack of ZmATR failed to arrest cells in the S-phase, accompanied by a strong accumulation of γH2AX foci that are indicative of DNA break accumulation. These findings suggest that Zmatr mutant plants are not able to stabilize the HU-induced stalled replication forks, similarly to what is described for other species. Together, these results show that ZmATR has a conserved role in the response to replicative stress. Likewise, as observed in Arabidopsis, we found that ZmATM plays an important role in the induction of cell cycle arrest after DSB induction, as illustrated by the high frequency of mitotic events observed in Zmatm mutants exposed to zeocin.
On the other hand, we found that, in Arabidopsis and maize, the DSB-induced DDR depends on both ATR and ATM. In maize, both atr and atm mutants showed a higher accumulation of γH2AX foci under control conditions than WT plants do. However, only atr mutants showed an excessive increase of γH2AX foci following zeocin treatment, to a level similar to what was observed upon exposure to HU, indicating that under these conditions, loss of the ATR-dependent DDR leads to an accumulation of DNA damage, correlated with the observed inhibition of root growth. In contrast, maize atm mutants showed fewer foci per nucleus compared to WT plants after treatment with zeocin or HU. Since γH2AX foci generation is completely dependent on ATM and ATR, the constitutive γH2AX presence in each mutant must be dependent on the other kinase. Likely the lower number of γH2AX foci after treatment with zeocin in Zmatm compared to WT plants might be related with a predominant role of ZmATR in the response to DNA damage, where the constitutive activation of the ATR-dependent DDR might confer less accumulation of DNA damage by a pre-activation of DNA repair mechanisms. Nevertheless, Zmatm displayed sensitivity to zeocin at prolongated exposure times, likely by a failure in the activation of ATM-dependent checkpoints. Because γH2AX has been shown to facilitate HR in both mammals and Arabidopsis (Maréchal and Zou, 2013; Biedermann et al., 2017), we may hypothesize that in maize the absence of ATR leads to replication stress or DSBs that activate an ATM-dependent increase of γH2AX foci. Under normal growth conditions, the relatively low levels of DSBs can be repaired by HR, explaining the viability of the Zmatr mutant plants. However, when such plants are challenged by exogenous inducers of DSBs, the break repair machinery fails, leading to massive accumulation of DNA damage. Consistent with this idea, it has been observed that mammalian cells deficient for ATR display elevated chromosomal fragmentation after DSB induction due to defects in repair by HR (Wang et al., 2004).
Strikingly, in Arabidopsis, both atr and atm mutants show similar γH2AX foci accumulation levels in response to zeocin, which was consistent with an equal degree of zeocin or bleomycin (BLM) sensitivity. Similar results were obtained when the mutants were treated with cisplatin (Weimer et al., 2016) or following activation of DNA damage by aluminum (Chen et al., 2019), with both treatments resulting in a comparable percentage of nuclei with γH2AX foci in atr and atm mutants, but with atm mutants displaying a higher number of foci per nucleus. These data suggest that both Arabidopsis kinases play a role relevant in the generation of γH2AX foci during elevated and/or prolonged induction of DNA damage by different genotoxins. Nevertheless, in response to ionizing radiation, Arabidopsis ATM plays a more prominent role than ATR in the phosphorylation of γH2AX at least in mitotic cells (Friesner et al., 2005; Waterworth et al., 2019). Future research is necessary to better understand these differences.
ATR and ATM transduce the DNA stress signals to SOG1, whose phosphorylation is crucial for the transcriptional response to DNA damage, including the transcriptional induction of genes needed for the activation of a transient cell cycle arrest and PCD (Yoshiyama et al., 2009, 2017; Bourbousse et al., 2018; Ogita et al., 2018). We found that the early transcriptional response to DSBs in maize was dependent on ZmATM, as occurs in Arabidopsis (Culligan et al., 2006; Ricaud et al., 2007). Furthermore, in both Arabidopsis and maize, ATR participates in the early transcriptional response to DSBs in root tip meristems. This important role of ATR in the DSB response had probably been overlooked in Arabidopsis due to the use of whole plantlets for the analysis of changes in gene expression, rather than focusing on meristems as we did here. This would suggest that the respective roles of ATM and ATR vary according to the cell type, ATR playing a more crucial role in proliferating cells, in which DNA damage can interfere with the replication process, whereas ATM may be more prominent in differentiated cells. In addition, the relative contribution of ATR appears to be species specific. Indeed, in Arabidopsis atr mutants, the transcriptional activation of DDR genes is dramatically attenuated, but in Zmatr it is completely absent. Consistently, the relative importance of ATR and ATM for the regulation of the transcriptional response to DSBs has diverged between eukaryotes. In Saccharomyces cerevisiae, Tel1/ATM only marginally directs DSB repair and is of little importance in checkpoint control (Mantiero et al., 2007), playing only a secondary role in signaling pathways (Craven et al., 2002; Jaehnig et al., 2013), whereas these processes are dominated by Mec1/ATR (Gasch et al., 2001; Watson et al., 2004). Likewise, it has been shown for the moss P. patens that the early transcriptional response induced by BLM was largely dependent on ATR and only marginally on ATM (Martens et al., 2020). Note, however, that both in yeast and P. patens, DNA repair almost exclusively relies on HR and less on nonhomologous end-joining (Martens et al., 2020), which might account for the differential response in these organisms.
Although many functions can be compensated between ATR and ATM in diverse eukaryotes (Tomimatsu et al., 2009; Maréchal and Zou, 2013; Menolfi and Zha, 2020), including Arabidopsis (Friesner et al., 2005; Amiard et al., 2010; Roitinger et al., 2015; Waterworth et al., 2019), other roles are nonredundant or performed predominantly by one of the two kinases, and thus lack of any of them leads to developmental defects as a consequence of genetic instability (Murga et al., 2009; Maréchal and Zou, 2013; Menolfi and Zha, 2020). In maize, this partial functional redundancy between the two kinases appears to be conserved, as at the adult vegetative stage, the Zmatr and Zmatm single mutants are visually indistinguishable from WT plants, like their counterparts in Arabidopsis (Garcia et al., 2003; Culligan et al., 2004). However, the sesquimutant plants display a slight but significant reduction in stature and the double mutant is unviable, demonstrating that at least one of the kinases is required for an adequate development. This observation contrasts with Arabidopsis, where the atr atm double mutant is viable (Culligan and Britt, 2008).
Essential role of maize ATR in proliferating cells
As discussed above, contrary to Arabidopsis, where atr and atm mutant root nuclei do not show spontaneous appearance of γH2AX foci (Friesner et al., 2005; Amiard et al., 2010), we found that maize tissues with a high division rate, such as the developing embryo and root tips of atr mutants, display an increased presence of the DNA damage marker γH2AX in comparison with WT plants, suggesting that mutants suffer from replicative stress and/or DNA breakages in proliferative tissues. The maize genome is at least 17-fold bigger than the Arabidopsis genome, and contains much more complex repetitive regions (e.g. microsatellites and quasi-palindromic AT-rich repeats; Haberer et al., 2005; Schnable et al., 2009). These characteristics probably make its genome more prone to replicative stress and DNA breaks, implying that the DDR pathway plays a more essential role in species with large versus small genomes. Consistent with this hypothesis, atr mutant plants have been isolated in barley, whose genome size is of a same order of magnitude as maize, and around 60% of root meristem cells display endogenous DNA damage, resulting in plants with a reduced size (Szurman-Zubrzycka et al., 2019).
Contrasting requirements for ATM and ATR during meiosis and early stages of development in plants
Intriguingly, contrasting requirements in different species have been observed for plant ATM proteins during meiosis: ATM deficiency results in semi-sterility in Arabidopsis and full infertility in rice (Garcia et al., 2003; Zhang et al., 2020), a phenomenon that we did not observe in maize. In Arabidopsis, although ATM deficiency alone impacts meiotic DSB repair leading to partial sterility, the atr atm double mutant displays full sterility and accumulation of chromosomal fragmentation, indicating partial functional redundancy between the two proteins during meiosis (Culligan and Britt, 2008). Recently, it was demonstrated that Arabidopsis ATM is essential to limit the number of meiotic DSBs, acting at multiple steps during DSB formation and processing (Kurzbauer et al., 2021). Our results suggest that, in maize, ATR may function redundantly with ATM in meiotic cells. Consistent with this idea, the maize sesquimutants showed the appearance of non-viable pollen, whereas single mutants did not display differences compared with WT plants. In Arabidopsis, atm meiocytes display extensive chromosome fragmentation but do not undergo cell death before the end of the meiosis and formation of gametophytes, thus the high lethality in the pollen population is likely the result of an aberrant chromosomal content (Garcia et al., 2003). Such fragmentation is exacerbated in the atr atm double mutant (Culligan and Britt, 2008). Notably, the production of non-viable pollen in the sesquimutants could either be due to meiotic defects or to defects occurring later during the development of gametophytes, as occurring in Arabidopsis. However, how ZmATR and ZmATM precisely participate in meiosis and/or female gametophyte development remain to be revealed.
Conversely, whereas a lack of ATR does not affect the fertility in Arabidopsis and barley (Culligan et al., 2004; Szurman-Zubrzycka et al., 2019), in maize it resulted in a slight reduction of fertility, as reflected by a smaller cob size, decrease in the number of seeds per cob, and early abortion of some kernels. However, this effect likely relates to severe replicative stress during the early stages of embryo and endosperm development rather than to meiotic defects, since pollen viability was unaffected in Zmatr mutants.
ATR prevents early endosperm endoreplication and programmed cell death
As mentioned above, mutant maize atr cobs showed the stochastic appearance of abnormal kernels, which is characterized by an early death before maturity. As we observed that a lack of ATR activity results in an increase of replicative stress in embryos of viable kernels, we may hypothesize that the viable Zmatr kernels successfully cope with the endogenous DNA damage, while the abnormal kernels accumulate higher levels of breakages of DNA and genetic instability, eventually triggering early death of the kernel.
In the Arabidopsis root meristem, DSBs trigger cell differentiation, which includes the transition into the endocycle (Adachi et al., 2011). Furthermore, the excessive accumulation of DNA damage activates PCD in meristematic cells when the DNA damage cannot be repaired (Fulcher and Sablowski, 2009; Furukawa et al., 2010). In Arabidopsis, both endoreduplication and PCD are triggered by the perception of DNA damage through ATR and/or ATM (Fulcher and Sablowski, 2009; Furukawa et al., 2010; Adachi et al., 2011). The single mutants display a delay in the induction of PCD, but they do not lose the capacity to activate this process (Furukawa et al., 2010), while the endoduplication activation is not affected (Furukawa et al., 2010; Adachi et al., 2011). In contrast, induction of endoreduplication and PCD are fully compromised in double mutants (Furukawa et al., 2010; Adachi et al., 2011).
Considering that endoreduplication and PCD are an intrinsic part of maize endosperm development, and those processes are induced by DSBs in Arabidopsis through an ATM/ATR-dependent pathway, we may hypothesize that during the mitotic division of endosperm cells, lack of ZmATR leads to the accumulation of DSBs that act as a signal driving an early transition into the endocycle. During the subsequent events of DNA replication, even more DNA damage may accumulate, which would lead to a premature onset of PCD.
The early onset of endocycle and PCD in Zmatr kernels is reminiscent of the early senescence observed in Atatm mutants, in which the level of DSBs increases but the DNA repair efficiency decreases, because AtATM represses the DSB‐induced expression of genes associated with senescence (Li et al., 2020). Consistent with this idea, the endosperm of non-viable Zmatr kernels displayed an early cell death phenotype. Likewise, the Zmatr Zmatm double mutant kernels aborted early, indicating that in maize, the two kinases also have partially redundant functions in these mechanisms: ZmATM could be involved but has a minor role, which in its absence can be fully compensated by ZmATR. Furthermore, the elevated expression of both kinases during endosperm development supports the idea that ATM and ATR play a crucial role during the endosperm development of WT kernels.
The viable Zmatr kernels displayed a reduction in the weight and size of mature seeds, accompanied with a decrease in the starch and protein content as a consequence of an accelerated kernel maturity process in the endosperm triggered by early endoreduplication and PCD. This reduced accumulation of the storage nutrients may jeopardize the germination, which explains the early shoot growth phenotype of the Zmatr seedlings, when they still rely on nutrients supplied by the endosperm. However, this delay of growth may also be related with the importance of DDR during the germination of seeds (Waterworth et al., 2015, 2016).
In summary, we found that the necessity of the ATM and ATR kinases under nonstress conditions appears to be much higher in maize compared with Arabidopsis, with the absence of ATR resulting in both vegetative and reproductive phenotypes that are partially offset by ATM. Our data highlight that the mechanisms involved in the maintenance of genome integrity may be more important for plant development than previously anticipated and indirectly suggest that the presence of a nonoptimal DDR might severely affect the yield of field-grown crop plants.
Materials and methods
Plant medium and growth conditions
Maize seedlings were grown to maturity in soil in a 24°C growth chamber with a 16-h light/8-h dark regime, 55% relative humidity and light intensity (300 µmol m−2 s−1 photosynthetically active radiation at plant level) provided by a combination of high-pressure sodium vapor (RNP-T/LR/400W/S/230/E40; Radium) and metal halide lamps with quartz burners (HRI-BT/400W/D230/E40; Radium). For seedling analysis, the maize seeds were germinated using a paper roll system. Seeds were sterilized for 3 min in 100% ethanol, then submerged in 5% NaClO for 30 min, and finally the seeds were rinsed three times with sterilized water. Twelve seeds were spread out over the paper roll, and subsequently, the paper roll with seeds was transferred to a recipient with liquid 0.5 × Murashige and Skoog (MS) medium (Basalt Salt Mixture M0221; Duchefa, Haarlem, The Netherlands).
Arabidopsis plants were grown in vitro under long-day conditions (16-h light/8-h dark, Lumilux Cool White lm, 50–70 µmol m−2 s−1) at 21°C on solidified medium (half-strength MS [2.151 g/L], 10 g/L sucrose, and 0.5 g/L 2-(N-morpholino) ethanesulfonic acid, adjusted to pH 5.7 with 1 M KOH and 8 or 10 g/L agar). The T‐DNA insertion mutant lines atr-2 and atm-2 were previously described (Garcia et al., 2003; Culligan et al., 2004). Arabidopsis thaliana seeds were sterilized with 5% NaClO for 20 min and subsequent washing with sterile water. To obtain homogeneous germination, the seeds were vernalized for 2 days at 4°C.
Vector construction
The CRISPR construct was adapted from Xing et al. (2014). The pBUN411 plasmid was digested by HindIII, leading to two fragments of 2 kb (hereafter called fragment A) and 11.5 kb. The 11.5-kb fragment was further digested with SpeI, resulting in two fragments of 8.5 kb (called fragment B) and 3 kb. The pP+ plasmid was digested with XbaI and HindIII and was ligated to fragments A and B. This resulted in the whole backbone of pBUN411 being replaced by the backbone of pP+, containing a spectinomycin resistance cassette and a more efficient origin of replication. This new vector was called pBUN411-Sp. Using the CRISPR-P in silico gRNA design tool (Lei et al., 2014), two possible target sites were selected for both ATR and ATM. Designed primers with the target sites (listed in Supplemental Table S3) were used to perform PCR on the pCBC-MT1T2 plasmid (Xing et al., 2014), resulting in a fragment containing the desired target sites and the correct sites for ligation into pBUN411-Sp. This was done for both targeted genes. The vector contained the bar (bialaphos resistance) marker gene for selection of transformed callus in plants.
Maize transformation and selection of CRISPR/Cas9 lines
Immature embryos of the maize B104 inbred line were transformed using Agrobacterium tumefaciens EHA101 containing the pBUN411-Sp expression constructs. Transformed calli were selected on increasing concentrations of phosphinothricin, after which transformed T0 shoots were regenerated as described before (Coussens et al., 2012). For genotyping, DNA was extracted from transformed T0 shoots or leaf material in the next generations (∼2 cm2) using the Wizard® Genomic DNA purification kit (Promega, Madison, WI, USA ). We screened T0 transformants through PCR (primers are listed in Supplemental Table S3), followed by Sanger sequencing to identify CRISPR/Cas9-induced mutations in the targeted genes. The plants were screened for the absence of the bar marker by ammonium well assay (Coussens et al., 2012), followed by genotyping for the absence of the Cas9 gene construct. Two independent atm and atr mutants were selected and backcrossed with B104 WT plants. These lines were upscaled and screened in F2 to obtain WT and homozygous mutant plants (Zmatm or Zmatr) in the same genetic background. The characterization of the maize lines as well as all the experiments shown in this work was performed in F2 and F3 plants.
Genotoxic treatments
Maize seeds were germinated in a paper roll system for three days to ensure a straight root orientation. At this time point, seedlings of similar size (2.5–3.5 cm) were transferred to a Falcon tube (50 mL) with liquid 0.5 × MS medium (mock or supplemented with genotoxin) that was used like a hydroponic growth system. Treatments were performed with 2.5 mM HU (Sigma-Aldrich, MO, USA) or zeocin (75 µM or 150 µM). After three to four days of treatment, the plantlets were analyzed for root and shoot lengths.
For treatment with γ-irradiation, the maize seeds were grown hydroponically using the paper roll system. After two days, the seedlings were treated with γ‐irradiation using a 137Cs source (C.I.S. Bio, Gif Sur Yvette, France) at doses of 75, 250, or 500 Gy, and then returned for growth under hydroponic conditions. After five days, the root and shoot lengths were measured.
To evaluate the sensitivity to genotoxic agents in Arabidopsis, seedlings were grown for four days on 0.5 × MS and transferred to genotoxin-supplemented medium (5.0 µM zeocin or 0.2 µg/mL bleomycin). After ten days, the plantlets were analyzed for root length. All the measurements were performed using the ImageJ software package (https://imagej.net/ImageJ_1.x).
RNA extraction and RT-qPCR
The RNA extractions for maize were performed in pools of ten root tips (1–2 mm) or endosperms of nine independent kernels (three half endosperms per cob of three independent cobs were mixed to perform the RNA extraction). Total RNA was extracted using a Direct-zol RNA MiniPrep Plus kit (Zymo Research, Irvine, CA, USA). For Arabidopsis experiments, seeds were germinated on control medium on a nylon mesh and transferred five days after germination to control medium or medium supplemented with 20-µM zeocin for the indicated time, after which at least 200 root tips (1–2 mm) were harvested. RNA was isolated with the RNeasy Mini kit (Qiagen, Hilden, Germany) and was treated on-column with the RQ1 RNase-Free DNase (Promega, Madison, WI, USA).
RNA quality and concentration were determined using Nanodrop (Thermo Fisher Scientific, Waltham, MA, USA). The iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA) was used to prepare cDNA from 1 μg of RNA according to the manufacturer’s instructions. Quantitative real-time PCR was performed in a 384-well plate on a LightCycler 480 (Roche, Machelen, Belgium) with SYBR Green I Master mix (Roche, Belgium) in a final volume of 5 μL and 0.25 μM primer concentration. Each reaction was done with three technical repeats for each independent experiment. Three independent RNA extractions were done as three biological repeats. EF-α and 18S rRNA primers were used for normalization of the maize data, while for Arabidopsis three reference genes were used: EMB2386, PAC1, and RPS26E. Relative expression values were manually calculated using the 2−ΔΔCT method. Primers were designed using Primer3Plus (http://primer3plus.com/cgi-bin/dev/primer3plus.cgi). Primers used for RT-qPCR are listed in Supplemental Table S3.
Immunostaining of γH2AX
Maize root tips of 3-day-old seedlings or embryos of 16 DAP were fixed overnight in 4% paraformaldehyde in a solution of 1× PME (50 mM Pipes pH 6.9, 5 mM MgSO4, 1 mM EGTA) and then washed three times for 5 min in 1× PME. For one experimental replication, ten root meristems or six embryos were pooled, respectively, for nucleus isolation. The dissected tissues were chopped in Galbraith’s buffer (45 mM MgCl2, 20 mM MOPS, 30 mM sodium citrate, 0.1% [v/v] Triton X-100, adjusted to pH 7.0 using 1 M NaOH). After filtering through a 50-μm CellTrics filter and centrifugation under 200 g for 10 min at 4°C, the supernatant was removed. The nuclei were resuspended in 30 μL Galbraith’s buffer and spread on slides. The samples were left to dry for 1 h at room temperature.
Immunostaining for maize samples was performed as described (Amiard et al., 2010), with the following modifications. Each slide was incubated overnight at 4°C with 80 μL of a rabbit anti-plant γH2AX antibody (described in Amiard et al., 2010 and kindly provided by Charles White, CNRS, Clermont-Ferrand, France) in a 1:600 dilution in fresh blocking buffer (3% BSA in 1× PBS). Slides were washed three times for 5 min in 1× PBS solution and then incubated for 3 h at room temperature in 100-μL blocking buffer containing Alexa 488‐conjugated goat anti‐rabbit secondary antibody (Molecular Probes, Invitrogen, Carlsbad, CA, USA), diluted 1:1,000 in fresh blocking buffer. Finally, DNA was counterstained with 2 µg/mL DAPI for 30 min, after which slides were washed in 1× PBS and mounted in mounting medium.
Untreated or treated (5.0 µM zeocin) Arabidopsis 5-day-old seedlings from Col-0, atr-2, and atm-2 were fixed and immunostaining for γH2AX was performed as described by Amiard et al. (2010). Imaging of the nuclei was done using an Olympus FV1000 confocal microscope. Microscope configuration was the following: scanning mode: sequential unidirectional; excitation: 405 nm (DAPI) and 488 nm (Alexa Fluor 488); laser transmissivity: ˂1% and 5% were used for DAPI and Alexa Fluor 488, respectively. Microscope settings and camera detector exposure times were kept constant for each respective channel (Alexa Fluor 488 or DAPI) but were optimized for individual experiments.
Mitosis phase counting
The number of mitotic events per root tip was examined in untreated and treated root tips with 150 µM zeocin. Root tips were fixed overnight in 4% paraformaldehyde in a solution of 1× PME (50 mM Pipes pH 6.9, 5 mM MgSO4, 1 mM EGTA) and then washed three times for 5 min in 1× PME. Root apices were digested in enzyme mix (1% w/v cellulase, 0.5% w/v cytohelicase, and 1% w/v pectolyase in 1× PME) for 2 h at 37°C. After three washes with 1× PME, root apices were squashed gently between the slide and a coverslip, and frozen in liquid nitrogen. Afterward, the coverslip was removed, and the slides were left to dry for 1 h at room temperature. The slides were stained with 2-µg/mL DAPI for 30 min, after which slides were washed in 1× PBS and mounted in mounting medium. Imaging of the nuclei was done with an epifluorescence microscope (Axio Imager.Z2.). The microscope is equipped with an AxioCam Mrm camera and appropriate Zeiss filter sets for the fluorochromes used in this work (filter no. 49—DAPI). The number of mitoses was counted by eye.
Pollen staining
Fresh pollen from tassels was collected in the morning in Carnoy’s fixative (60% ethanol, 30% chloroform, and 10% acetic acid). Staining was done with a modified Alexander stain as reported by Peterson et al. (2010). The pollen was spread on slides, after which and then the staining solution was applied. After 10–15 min of incubation, samples were visualized under a microscope (Olympus BX51). Pollen from at least six independent plants was analyzed.
Flow cytometry
For maize ploidy analysis of roots, seeds were germinated using the paper roll system. Control and treated root tips (1–2 mm) of 3-day-old seedlings were analyzed. For one experimental replication, ten root meristems were analyzed and three replications per treatment were used. The root tips were chopped with a razor blade in 1 mL of Gif nucleus-isolation buffer (45 mM MgCl2, 30 mM sodium citrate, 60 mM MOPS, 1% (w/v) polyvinylpyrrolidone 10,000, pH 7.2) containing 0.1% (w/v) Triton X-100, supplemented with 5 mM sodium metabisulphite and RNAse (5 U/mL). Propidium iodide was added to the filtered supernatants to a final concentration of 50 μg/mL. The samples were analyzed using a Cyflow SL3 flow cytometer (Partec-Sysmex, Münster, Germany) with a 532-nm solid-state laser (30 mW) excitation and an emission collected after a 590-nm long-pass filter. For cell cycle analysis, we used the algorithm available in the Flomax software.
For endosperm analysis, tissue was obtained by removing the seed coat and embryos from each kernel. At least three kernels were analyzed for each plant. Three individual ears were used for flow cytometry analysis. The endosperm was rapidly chopped on a glass Petri dish using a razor blade in 1 mL ice-cold Galbraith’s buffer (45 mM MgCl2, 20 mM MOPS, 30 mM sodium citrate, 0.1% [v/v] Triton X-100, adjusted to pH 7.0 using 1 M NaOH). The homogenate was passed through two layers of a 100-µM nylon mesh and filtered through a 50-μm CellTrics filter. Then the nuclei were precipitated (200 g, 10 min, 4°C), and the supernatant was removed. The nuclei were suspended in 200-μL nucleus extraction buffer after which 1 mL staining buffer was added (Cystain UV Precise P, Sysmex Partec, Germany). The mix was filtered through a 50-μm CellTrics filter (Sysmex Partec, Görlitz, Germany) and analyzed by the Cyflow MB flow cytometer (Sysmex Partec, Germany). For each sample, at least 10,000 nuclei were collected and analyzed using a logarithmic scale display. The Cyflogic software was used for ploidy measurements. Mean C-values were calculated according to the following equation: ([n3 × 3] + [n6 × 6] + [n12 × 12] + ….)/total number of nuclei, where n3 = total number of nuclei in the 3C peak, and so on.
Evans blue staining
Fresh kernels were collected at 16, 19, and 22 DAP for each WT and mutant line. Near-median longitudinal hand sections (∼3 mm thick) were stained in 0.1% (w/v) Evans blue (C.I 23860) for 2 min as reported by Young et al. (1997). Stained sections were washed with water for 30 min and photographed. The analyses were carried out from at least seven kernels of three independent cobs for each genotype.
Analysis of total protein and starch contents
The total protein and starch contents of the kernels were analyzed as described (Zhang et al., 2019), with some modifications. Twenty endosperms of immature (19 DAP) or mature kernels from the same ear were pooled as a single replicate. Three biological replicates were used for the analysis. Flour was obtained by grinding dry endosperm under liquid nitrogen, and then lyophilized overnight.
Starch content determination was performed following the method described by instructions of the Total Starch Assay Kit (Catalog: K-TSTA-50A, Megazyme, Australia) based on the use of thermostable α-amylase and amyloglucosidase. Analyses were performed on 100 mg of flour.
For the measurement of protein content, ∼50 mg of flour was used for the content analysis of zein and nonzein proteins. The nonzein proteins were extracted by adding 1 mL of the buffer (12.5 mM sodium borate, 5% sodium dodecyl sulfate (SDS), 2% 2-mercaptoethanol, proteases inhibitors) to 50 mg flour and incubated overnight at 37°C with constant shaking. Subsequently, the samples were centrifugated at 12,000 g at room temperature for 15 min to precipitate debris. Supernatant was collected and absolute ethanol was added to a final concentration of 70%. The samples were incubated at 37°C for 2 h, then centrifugated at 12,000 g for 10 min at room temperature. The supernatant was transferred to a new tube (zein fraction), and then dried in a Speedvac and resuspended in water. The pellet was used to extract nonzein proteins with 1 mL buffer (12.5 mM sodium borate, 5% SDS, 2% 2-mercaptoethano, proteases inhibitors). The mix was incubated at 37°C for 2 h, and then centrifugated for 15 min at 12,000 g and room temperature. The pellet was washed twice with 70% ethanol, air-dried, and resuspended in 8 M Urea. Protein quantification was performed with the Compat-Able Protein Assay Preparation Reagent Kit (Catalog: 23215, Thermo Fisher Scientific, MA, USA) and the Pierce BCA Protein Assay Kit (Catalog: 23225, Thermo Fisher Scientific, MA, USA) following manufacturer’s manual.
Statistical analysis
All statistical analyses were performed using the R software (https://www.r-project.org/).
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: Zm00001d014813 (Zmatr), Zm00001d040166 (Zmatm), AT5G40820 (AtATR), and AT3G48190 (AtATM).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 . Map of CRISPR/Cas9 mutations.
Supplemental Figure S2 . Root growth kinetics of ATR-B WT, Zmatr-b, ATM-B, and Zmatm-b on medium supplemented with HU or zeocin.
Supplemental Figure S3 . Early transcriptional response in Zmatr-b and Zmatm-a root tips treated with HU or zeocin.
Supplemental Figure S4 . Root growth kinetics of arabidopsis Col-0, atr-2 and atm-2 Roots on medium supplemented with zeocin and bleomycin.
Supplemental Figure S5 . Early transcriptional response in Atatr and Atatm root tips treated with zeocin.
Supplemental Figure S6 . Zmatr and Zmatm root tips display impaired checkpoint activation in response to DNA stress.
Supplemental Figure S7 . Cob phenotype of WT and Zmatr and Zmatm mutant plants.
Supplemental Figure S8 . Pollen viability of Zmatr is not affected, but reciprocal crosses indicate a maternal defect.
Supplemental Figure S9 . Expression levels of Zmatr and Zmatm in different tissues and during the endosperm development in WT plants.
Supplemental Figure S10 . Expression levels of genes associated with cell cycle regulation and replication initiation during the development of Zmatm and/or Zmatr endosperms.
Supplemental Figure S11 . Endoreduplication is not affected in Zmatm endosperm.
Supplemental Figure S12 . Zmatm endosperm does not display differences in cell death timing or abundance.
Supplemental Figure S13 . Cell death in endosperm of Zmatr aborted kernels.
Supplemental Figure S14 . Starch and protein content in Zmatr and Zmatm endosperm.
Supplemental Table S1 . Percentage of abnormal kernels in Zmatr cobs
Supplemental Table S2 . Number of kernels in reciprocal crosses between ATR WT and mutant plants
Supplemental Table S3 . Primers used in this study
Supplemental Data set S1 . Statistical analysis.
Supplementary Material
Acknowledgments
The authors thank Annick Bleys for help in preparing the manuscript, Charles White (CNRS, Clermont-Ferrand, France) for the γH2AX antibody, Fernando Guzman-Chavez (University of Cambridge, Plant Sciences Department, UK) for help with the statistical analysis and Jeannine Drouin-Whabi (IPS2, France) for assistance with maize plant sowing and growth.
Funding
This work was supported by grants of the Research Foundation Flanders (G011420N and G010820N). Ignacio Achon was benefited from a doctoral fellowship “Don Carlos Antonio Lopez” by El Programa Nacional de Becas from Paraguay (BECAL #164/2017).
Conflict of interest statement. The authors declare no conflict of interest.
Contributor Information
Jose Antonio Pedroza-Garcia, Department of Plant Biotechnology and Bioinformatics, Ghent University, Ghent B-9052, Belgium; Center for Plant Systems Biology, VIB, Ghent B-9052, Belgium.
Thomas Eekhout, Department of Plant Biotechnology and Bioinformatics, Ghent University, Ghent B-9052, Belgium; Center for Plant Systems Biology, VIB, Ghent B-9052, Belgium.
Ignacio Achon, Department of Plant Biotechnology and Bioinformatics, Ghent University, Ghent B-9052, Belgium; Center for Plant Systems Biology, VIB, Ghent B-9052, Belgium.
Maher-Un Nisa, Institute of Plant Sciences Paris-Saclay (IPS2), CNRS, INRA, University Paris-Sud, University of Evry, Paris University, Sorbonne Paris-Cite, University of Paris-Saclay, 91405, Orsay, France.
Griet Coussens, Department of Plant Biotechnology and Bioinformatics, Ghent University, Ghent B-9052, Belgium; Center for Plant Systems Biology, VIB, Ghent B-9052, Belgium.
Ilse Vercauteren, Department of Plant Biotechnology and Bioinformatics, Ghent University, Ghent B-9052, Belgium; Center for Plant Systems Biology, VIB, Ghent B-9052, Belgium.
Hilde Van den Daele, Department of Plant Biotechnology and Bioinformatics, Ghent University, Ghent B-9052, Belgium; Center for Plant Systems Biology, VIB, Ghent B-9052, Belgium.
Laurens Pauwels, Department of Plant Biotechnology and Bioinformatics, Ghent University, Ghent B-9052, Belgium; Center for Plant Systems Biology, VIB, Ghent B-9052, Belgium.
Mieke Van Lijsebettens, Department of Plant Biotechnology and Bioinformatics, Ghent University, Ghent B-9052, Belgium; Center for Plant Systems Biology, VIB, Ghent B-9052, Belgium.
Cécile Raynaud, Institute of Plant Sciences Paris-Saclay (IPS2), CNRS, INRA, University Paris-Sud, University of Evry, Paris University, Sorbonne Paris-Cite, University of Paris-Saclay, 91405, Orsay, France.
Lieven De Veylder, Department of Plant Biotechnology and Bioinformatics, Ghent University, Ghent B-9052, Belgium; Center for Plant Systems Biology, VIB, Ghent B-9052, Belgium.
J.A.P.G., T.E., and L.D.V. conceived and designed the research. J.A.P.G., T.E., I.A., M-U.N., I.V., H.V.d.D., and C.R. performed the experiments. G.C., M.V.L., and L.P. did the maize transformations. J.A.P.G., T.E., I.A., C.R., and L.D.V analyzed data. J.A.P.G. and L.D.V. wrote the article. All authors read, revised, and approved the article.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Lieven De Veylder (lieven.deveylder@psb.vib-ugent.be).
References
- Adachi S, Minamisawa K, Okushima Y, Inagaki S, Yoshiyama K, Kondou Y, Kaminuma E, Kawashima M, Toyoda T, Matsui M, et al. (2011) Programmed induction of endoreduplication by DNA double-strand breaks in Arabidopsis. Proc Natl Acad Sci USA 108: 10004–10009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiard S, Charbonnel C, Allain E, Depeiges A, White CI, Gallego ME (2010) Distinct roles of the ATR kinase and the Mre11-Rad50-Nbs1 complex in the maintenance of chromosomal stability in Arabidopsis. Plant Cell 22: 3020–3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedermann S, Harashima H, Chen P, Heese M, Bouyer D, Sofroni K, Schnittger A (2017) The retinoblastoma homolog RBR1 mediates localization of the repair protein RAD51 to DNA lesions in Arabidopsis. EMBO J 36: 1279–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackford AN, Jackson SP (2017) ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol Cell 66: 801–817 [DOI] [PubMed] [Google Scholar]
- Bourbousse C, Vegesna N, Law JA (2018) SOG1 activator and MYB3R repressors regulate a complex DNA damage network in Arabidopsis. Proc Natl Acad Sci USA 115: E12453–E12462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EJ, Baltimore D (2000) ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev 14: 397–402 [PMC free article] [PubMed] [Google Scholar]
- Chankova SG, Dimova E, Dimitrova M, Bryant PE (2007) Induction of DNA double-strand breaks by zeocin in Chlamydomonas reinhardtii and the role of increased DNA double-strand breaks rejoining in the formation of an adaptive response. Radiat Environ Biophys 46: 409–416 [DOI] [PubMed] [Google Scholar]
- Chen P, Sjogren CA, Larsen PB, Schnittger A (2019) A multi-level response to DNA damage induced by aluminium. Plant J 98: 479–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D (2008) ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol 9: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cools T, Iantcheva A, Weimer AK, Boens S, Takahashi N, Maes S, Van den Daele H, Van Isterdael G, Schnittger A, De Veylder L (2011) The Arabidopsis thaliana checkpoint kinase WEE1 protects against premature vascular differentiation during replication stress. Plant Cell 23: 1435–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens G, Aesaert S, Verelst W, Demeulenaere M, De Buck S, Njuguna E, Inzé D, Van Lijsebettens M (2012) Brachypodium distachyon promoters as efficient building blocks for transgenic research in maize. J Exp Bot 63: 4263–4273 [DOI] [PubMed] [Google Scholar]
- Craven RJ, Greenwell PW, Dominska M, Petes TD (2002) Regulation of genome stability by TEL1 and MEC1, yeast homologs of the mammalian ATM and ATR genes. Genetics 161: 493–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culligan K, Tissier A, Britt A (2004) ATR regulates a G2-phase cell-cycle checkpoint in Arabidopsis thaliana. Plant Cell 16: 1091–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culligan KM, Britt AB (2008) Both ATM and ATR promote the efficient and accurate processing of programmed meiotic double-strand breaks. Plant J 55: 629–638 [DOI] [PubMed] [Google Scholar]
- Culligan KM, Robertson CE, Foreman J, Doerner P, Britt AB (2006) ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J 48: 947–961 [DOI] [PubMed] [Google Scholar]
- Doll NM, Depège-Fargeix N, Rogowsky PM, Widiez T (2017) Signaling in early maize kernel development. Mol Plant 10: 375–388 [DOI] [PubMed] [Google Scholar]
- Domínguez F, Cejudo FJ (2014) Programmed cell death (PCD): an essential process of cereal seed development and germination. Front Plant Sci 5: 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farage-Barhom S, Burd S, Sonego L, Perl-Treves R, Lers A (2008) Expression analysis of the BFN1 nuclease gene promoter during senescence, abscission, and programmed cell death-related processes. J Exp Bot 59: 3247–3258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendrych M, Van Hautegem T, Van Durme M, Olvera-Carrillo Y, Huysmans M, Karimi M, Lippens S, Guérin CJ, Krebs M, Schumacher K, et al. (2014) Programmed cell death controlled by ANAC033/SOMBRERO determines root cap organ size in Arabidopsis. Curr Biol 24: 931–940 [DOI] [PubMed] [Google Scholar]
- Friedel AM, Pike BL, Gasser SM (2009) ATR/Mec1: coordinating fork stability and repair. Curr Opin Cell Biol 21: 237–244 [DOI] [PubMed] [Google Scholar]
- Friesner JD, Liu B, Culligan K, Britt AB (2005) Ionizing radiation-dependent γ-H2AX focus formation requires ataxia telangiectasia mutated and ataxia telangiectasia mutated and Rad3-related. Mol Biol Cell 16: 2566–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulcher N, Sablowski R (2009) Hypersensitivity to DNA damage in plant stem cell niches. Proc Natl Acad Sci USA 106: 20984–20988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa T, Curtis MJ, Tominey CM, Duong YH, Wilcox BWL, Aggoune D, Hays JB, Britt AB (2010) A shared DNA-damage-response pathway for induction of stem-cell death by UVB and by gamma irradiation. DNA Repair 9: 940–948 [DOI] [PubMed] [Google Scholar]
- Garcia V, Bruchet H, Camescasse D, Granier F, Bouchez D, Tissier A (2003) AtATM is essential for meiosis and the somatic response to DNA damage in plants. Plant Cell 15: 119–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Huang M, Metzner S, Botstein D, Elledge SJ, Brown PO (2001) Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR homolog Mec1p. Mol Biol Cell 12: 2987–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu N, Tamada Y, Imai A, Palfalvi G, Kabeya Y, Shigenobu S, Ishikawa M, Angelis KJ, Chen C, Hasebe M (2020) DNA damage triggers reprogramming of differentiated cells into stem cells in Physcomitrella. Nat Plants 6: 1098–1105 [DOI] [PubMed] [Google Scholar]
- Haberer G, Young S, Bharti AK, Gundlach H, Raymond C, Fuks G, Butler E, Wing RA, Rounsley S, Birren B, et al. (2005) Structure and architecture of the maize genome. Plant Physiol 139: 1612–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Wang J, Qi W, Song R (2019) Maize Dek15 encodes the cohesin-loading complex subunit SCC4 and is essential for chromosome segregation and kernel development. Plant Cell 31: 465–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Cools T, De Veylder L (2016) Mechanisms used by plants to cope with DNA damage. Annu Rev Plant Biol 67: 439–462 [DOI] [PubMed] [Google Scholar]
- Iyer DR, Rhind N (2017) The intra-S checkpoint responses to DNA damage. Genes 8: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaehnig EJ, Kuo D, Hombauer H, Ideker TG, Kolodner RD (2013) Checkpoint kinases regulate a global network of transcription factors in response to DNA damage. Cell Rep 4: 174–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jossen R, Bermejo R (2013) The DNA damage checkpoint response to replication stress: a game of forks. Front Genet 4: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Ryu TH, Lee SS, Lee S, Chung BY (2019) Ionizing radiation manifesting DNA damage response in plants: an overview of DNA damage signaling and repair mechanisms in plants. Plant Sci 278: 44–53 [DOI] [PubMed] [Google Scholar]
- Kurzbauer MT, Janisiw MP, Paulin LF, Prusen Mota I, Tomanov K, Krsicka O, Haeseler AV, Schubert V, Schlogelhofer P (2021) ATM controls meiotic DNA double-strand break formation and recombination and affects synaptonemal complex organization in plants. Plant Cell 33: 1633--1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahari T, Lazaro J, Marcus JM, Schroeder DF (2018) RAD7 homologues contribute to Arabidopsis UV tolerance. Plant Sci 277: 267–277 [DOI] [PubMed] [Google Scholar]
- Lei Y, Lu L, Liu HY, Li S, Xing F, Chen LL (2014) CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol Plant 7: 1494–1496 [DOI] [PubMed] [Google Scholar]
- Li Z, Kim JH, Kim J, Lyu JI, Zhang Y, Guo H, Nam HG, Woo HR (2020) ATM suppresses leaf senescence triggered by DNA double-strand break through epigenetic control of senescence-associated genes in Arabidopsis. New Phytol 227: 473–484 [DOI] [PubMed] [Google Scholar]
- Manova V, Gruszka D (2015) DNA damage and repair in plants – from models to crops. Front Plant Sci 6: 885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantiero D, Clerici M, Lucchini G, Longhese MP (2007) Dual role for Saccharomyces cerevisiae Tel1 in the checkpoint response to double-strand breaks. EMBO Rep 8: 380–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal A, Zou L (2013) DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb Perspect Biol 5: a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens M, Horres R, Wendeler E, Reiss B (2020) The importance of ATM and ATR in Physcomitrella patens DNA damage repair, development, and gene targeting. Genes 11: 752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menolfi D, Zha S (2020) ATM, ATR and DNA-PKcs kinases — the lessons from the mouse models: inhibition ≠ deletion. Cell Biosci 10: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussu S, Doll NM, Chamot S, Brocard L, Creff A, Fourquin C, Widiez T, Nimchuk ZL, Ingram G (2017) ZHOUPI and KERBEROS mediate embryo/endosperm separation by promoting the formation of an extracuticular sheath at the embryo surface. Plant Cell 29: 1642–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murga M, Bunting S, Montaña MF, Soria R, Mulero F, Cañamero M, Lee Y, McKinnon PJ, Nussenzweig A, Fernandez-Capetillo O (2009) A mouse model of ATR-Seckel shows embryonic replicative stress and accelerated aging. Nat Genet 41: 891–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitaki Z, Holá M, Donà M, Pavlopoulou A, Michalopoulos I, Angelis KJ, Georgakilas AG, Macovei A, Balestrazzi A (2018) Integrating plant and animal biology for the search of novel DNA damage biomarkers. Mutat Res 775: 21–38 [DOI] [PubMed] [Google Scholar]
- Nisa MU, Huang Y, Benhamed M, Raynaud C (2019) The plant DNA damage response: signaling pathways leading to growth inhibition and putative role in response to stress conditions. Front Plant Sci 10: 653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogita N, Okushima Y, Tokizawa M, Yamamoto YY, Tanaka M, Seki M, Makita Y, Matsui M, Okamoto-Yoshiyama K, Sakamoto T, et al. (2018) Identifying the target genes of SUPPRESSOR OF GAMMA RESPONSE 1, a master transcription factor controlling DNA damage response in Arabidopsis. Plant J 94: 439–453 [DOI] [PubMed] [Google Scholar]
- Peterson R, Slovin JP, Chen C (2010) A simplified method for differential staining of aborted and non-aborted pollen grains. Int J Plant Biol 1: e13 [Google Scholar]
- Ricaud L, Proux C, Renou JP, Pichon O, Fochesato S, Ortet P, Montané MH (2007) ATM-mediated transcriptional and developmental responses to γ-rays in Arabidopsis. PLoS One 2: e430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roa H, Lang J, Culligan KM, Keller M, Holec S, Cognat V, Montané MH, Houlné G, Chabouté ME (2009) Ribonucleotide reductase regulation in response to genotoxic stress in Arabidopsis. Plant Physiol 151: 461–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roitinger E, Hofer M, Köcher T, Pichler P, Novatchkova M, Yang J, Schlögelhofer P, Mechtler K (2015) Quantitative phosphoproteomics of the ataxia telangiectasia-mutated (ATM) and ataxia telangiectasia-mutated and rad3-related (ATR) dependent DNA damage response in Arabidopsis thaliana. Mol Cell Proteomics 14: 556–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabelli PA, Larkins BA (2009) The contribution of cell cycle regulation to endosperm development. Sex Plant Reprod 22: 207–219 [DOI] [PubMed] [Google Scholar]
- Sánchez-Pons N, Irar S, García-Muniz N, Vicient CM (2011) Transcriptomic and proteomic profiling of maize embryos exposed to camptothecin. BMC Plant Biol 11: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Segurado M, Tercero JA (2009) The S-phase checkpoint: targeting the replication fork. Biol Cell 101: 617–627 [DOI] [PubMed] [Google Scholar]
- Singh A, Xu YJ (2016) The cell killing mechanisms of hydroxyurea. Genes 7: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjogren CA, Bolaris SC, Larsen PB (2015) Aluminum-dependent terminal differentiation of the Arabidopsis root tip is mediated through an ATR-, ALT2-, and SOG1-regulated transcriptional response. Plant Cell 27: 2501–2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelpflug SC, Sekhon RS, Vaillancourt B, Hirsch CN, Buell CR, de Leon N, Kaeppler SM (2016) An expanded maize gene expression atlas based on RNA sequencing and its use to explore root development. Plant Genome 9: 1–16 [DOI] [PubMed] [Google Scholar]
- Szurman-Zubrzycka ME, Nawrot M, Jelonek J, Dziekanowski M, Kwasniewska J, Szarejko I (2019) ATR, a DNA damage signaling kinase, is involved in aluminum response in barley. Front. Plant Sci 10: 1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomimatsu N, Mukherjee B, Burma S (2009) Distinct roles of ATR and DNA-PKcs in triggering DNA damage responses in ATM-deficient cells. EMBO Rep 10: 629–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turinetto V, Giachino C (2015) Multiple facets of histone variant H2AX: a DNA double-strand-break marker with several biological functions. Nucleic Acids Res 43: 2489–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bel M, Diels T, Vancaester E, Kreft L, Botzki A, Van de Peer Y, Coppens F, Vandepoele K (2018) PLAZA 4.0: an integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Res 46: D1190–D1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang H, Powell SN, Iliakis G, Wang Y (2004) ATR affecting cell radiosensitivity is dependent on homologous recombination repair but independent of nonhomologous end joining. Cancer Res 64: 7139–7143 [DOI] [PubMed] [Google Scholar]
- Waterworth WM, Bray CM, West CE (2015) The importance of safeguarding genome integrity in germination and seed longevity. J Exp Bot 66: 3549–3558 [DOI] [PubMed] [Google Scholar]
- Waterworth WM, Footitt S, Bray CM, Finch-Savage WE, West CE (2016) DNA damage checkpoint kinase ATM regulates germination and maintains genome stability in seeds. Proc Natl Acad Sci USA 113: 9647–9652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterworth WM, Wilson M, Wang D, Nuhse T, Warward S, Selley J, West CE (2019) Phosphoproteomic analysis reveals plant DNA damage signalling pathways with a functional role for histone H2 AX phosphorylation in plant growth under genotoxic stress. Plant J 100: 1007–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson A, Mata J, Bähler J, Carr A, Humphrey T (2004) Global gene expression responses of fission yeast to ionizing radiation. Mol Biol Cell 15: 851–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimer AK, Biedermann S, Harashima H, Roodbarkelari F, Takahashi N, Foreman J, Guan Y, Pochon G, Heese M, Van Damme D, et al. (2016) The plant-specific CDKB 1-CYCB 1 complex mediates homologous recombination repair in Arabidopsis. EMBO J 35: 2068–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14: 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi D, Lessa Alvim Kamei C, Cools T, Vanderauwera S, Takahashi N, Okushima Y, Eekhout T, Yoshiyama KO, Larkin J, Van den Daele H, et al. (2014) The Arabidopsis SIAMESE-RELATED cyclin-dependent kinase inhibitors SMR5 and SMR7 regulate the DNA damage checkpoint in response to reactive oxygen species. Plant Cell 26: 296–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiyama K, Conklin PA, Huefner ND, Britt AB (2009) Suppressor of gamma response 1 (SOG1) encodes a putative transcription factor governing multiple responses to DNA damage. Proc Natl Acad Sci USA 106: 12843–12848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiyama KO, Sakaguchi K, Kimura S (2013a) DNA damage response in plants: conserved and variable response compared to animals. Biology 2: 1338–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiyama KO, Kaminoyama K, Sakamoto T, Kimura S (2017) Increased phosphorylation of Ser-Gln sites on SUPPRESSOR OF GAMMA RESPONSE1 strengthens the DNA damage response in Arabidopsis thaliana. Plant Cell 29: 3255–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiyama KO, Kobayashi J, Ogita N, Ueda M, Kimura S, Maki H, Umeda M (2013b) ATM-mediated phosphorylation of SOG1 is essential for the DNA damage response in Arabidopsis. EMBO Rep 14: 817–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young TE, Gallie DR (2000) Regulation of programmed cell death in maize endosperm by abscisic acid. Plant Mol Biol Rep 42: 397–414 [DOI] [PubMed] [Google Scholar]
- Young TE, Gallie DR, DeMason DA (1997) Ethylene-mediated programmed cell death during maize endosperm development of wild-type and shrunken2 genotypes. Plant Physiol 115: 737–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhang F, Cheng X, Liu K, Tang J, Li Y, Tang D, Cheng Z, Yu H (2020) OsATM safeguards accurate repair of meiotic double-strand breaks in rice. Plant Physiol 183: 1047–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Dong J, Ji C, Wu Y, Messing J (2019) NAC-type transcription factors regulate accumulation of starch and protein in maize seeds. Proc Natl Acad Sci USA 116: 11223–11228 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.