Abstract
Endoplasmic reticulum-associated degradation (ERAD) is known to regulate plant responses to diverse stresses, yet its underlying molecular mechanisms and links to various stress signaling pathways are poorly understood. Here, we show that the ERAD component ubiquitin-conjugating enzyme UBC32 positively regulates drought tolerance in Arabidopsis thaliana by targeting the aquaporins PIP2;1 and PIP2;2 for degradation. Furthermore, we demonstrate that the RING-type ligase Rma1 acts together with UBC32 and that the E2 activity of UBC32 is essential for the ubiquitination of Rma1. This complex ubiquitinates a phosphorylated form of PIP2;1 at Lys276 to promote its degradation, thereby enhancing plant drought tolerance. Extending these molecular insights into crops, we show that overexpression of Arabidopsis UBC32 also improves drought tolerance in rice (Oryza sativa). Thus, beyond uncovering the molecular basis of an ERAD-regulated stress response, our study suggests multiple potential strategies for engineering crops with improved drought tolerance.
The ubiquitin-conjugating enzyme UBC32 enhances drought tolerance by cooperating with the RING-type E3 ligase Rma1 to ubiquitinate a phosphorylated form of PIP2;1 at Lys276 to promote its degradation.
Introduction
Drought is a severe environmental stress with a direct impact on plant growth and plant yield. The plant hormone abscisic acid (ABA), a major internal signal that regulates stress-adaptive responses, functions in the drought response by regulating transcription factors or promoting stomatal closure. Under stress conditions, these transcription factors regulate the expression of stress-related genes by binding to ABA-responsive elements (ABREs) in their promoters (Kang et al., 2002). In addition, drought also induces ABA-independent events. Dehydration-responsive element-binding 1 (DREB1)/C-repeat binding factor (CBF) transcription factors recognize the cis-elements DRE/(C-repeat)CRT, which participate in the ABA-independent pathway, and their overexpression increases plant drought tolerance (Seki et al., 2001). Almost all drought-related genes identified to date function in ABA-dependent and ABA-independent pathways. For example, the U-box E3 ligases U-box containing protein (PUB)18/19 function in the ABA-dependent pathway, and PUB22/23 function in the ABA-independent pathway (Cho et al., 2008; Seo et al., 2012). The U-box N-terminal domain (UND) in PUB18/19 determines this difference (Seo et al., 2016). Both PUB18/19 and PUB22/23 are negative regulators in drought stress responses.
Ubiquitination appears in almost all plant development events, including growth, hormonal responses, and stress responses. This process is catalyzed by three types of enzymes: Ubiquitin (Ub)-activating enzymes (E1), Ub-conjugating enzymes (E2), and Ub ligases (E3; Kraft et al., 2005; Stone et al., 2005). Several E3 ligases negatively or positively participate in the drought stress response. The RING-type E3 ligase SALT- AND DROUGHT-INDUCED RING FINGER 1 (SDIR1) positively regulates the drought stress response in an ABA-dependent manner (Zhang et al., 2007). Mutants of the Arabidopsis thaliana U-box E3 ligase genes PUB12/13 show enhanced sensitivity to drought stress due to the accumulation of the ABA co-receptor ABI1 (Kong et al., 2015). The U-box E3 ligase genes PUB18/19 and PUB22/23 are negative regulators of drought stress responses in an ABA-dependent or ABA-independent manner, respectively (Cho et al., 2008; Seo et al., 2012). However, few E2s and E2/E3 pairs have been reported to confer drought tolerance in plants.
Aquaporins are also involved in the drought response. Aquaporin genes are downregulated by several abiotic stresses, including drought stress (Jang et al., 2004; Alexandersson et al., 2005). Plasma membrane intrinsic proteins (PIPs), the most abundant aquaporins, appear to function in intercellular water transport. An Arabidopsis mutant of PIP2;2, which is expressed in roots, showed decreased hydraulic conductivity in roots (Javot et al., 2003). Moreover, Nicotiana benthamiana overexpressing PIP1b showed hypersensitivity to drought treatment (Aharon et al., 2003). In addition, overexpressing Rma1H1 increased drought tolerance in Arabidopsis. Rma1H1 functions in drought tolerance by mediating the downregulation of PIP2;1 protein levels and inhibiting its trafficking from the endoplasmic reticulum (ER) to the plasma membrane (Lee et al., 2009). Based on these findings, plasma membrane-localized aquaporins are thought to play crucial roles during water deficit stress, although the underlying mechanism is unknown.
We previously identified UBC32, a ubiquitin-conjugating enzyme, as an ER-associated degradation (ERAD) component (Cui et al., 2012). Stress treatment increases the expression and stability of UBC32 (Cui et al., 2012; Chen et al., 2016). However, the detailed mechanism of how UBC32 participates in the stress response is unclear. In this study, using immunoprecipitation–mass spectrometry (IP–MS), we determined that the aquaporins PIP2;1 and PIP2;2 interact with UBC32. The interaction was confirmed by luciferase complementation imaging (LCI), bimolecular fluorescence complementation (BiFC), and pull-down assays. Under drought stress treatment, the Arabidopsis ubc32 mutant showed increased sensitivity to drought, while pip2;1/2;2 exhibited reduced drought sensitivity compared to the wild-type (WT). The ubiquitin-conjugating enzyme UBC32 interacts with the RING-type E3 ligase Rma1. This Rma1–UBC32 complex associates with the Ser280/283-phosphorylated version of PIP2;1 and then ligates ubiquitin to Lys276 of PIP2;1 to promotes its degradation, thereby increasing drought tolerance in Arabidopsis. Expanding this discovery to crops, we demonstrate that UBC32 improves drought tolerance in rice (Oryza sativa) by downregulating PIP2;1 protein levels. Thus, we uncovered a direct link between ERAD and drought stress tolerance.
Results
UBC32 physically interacts with the aquaporin proteins PIP2;1 and PIP2;2
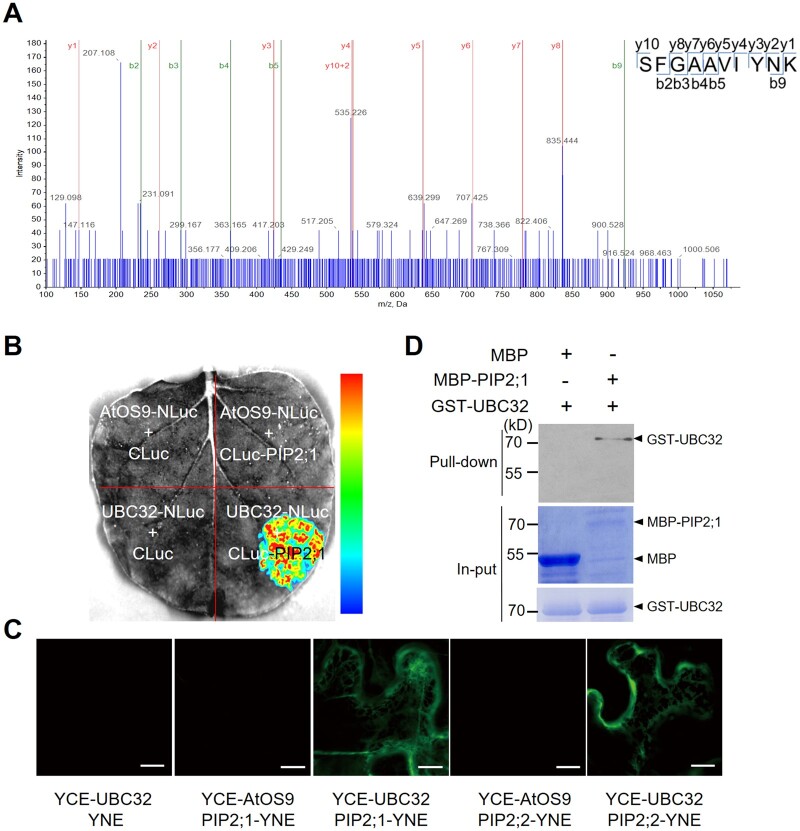
To explore the roles of UBC32 in plant stress responses, we performed immunoprecipitation experiments with a UBC32–GFP fusion protein in transgenic Arabidopsis plants with the aim of identifying proteins that associate with UBC32. Liquid chromatography–tandem mass spectrometry (LC–MS/MS) analysis of the immunoprecipitated proteins identified two aquaporin proteins: PIP2;1 and PIP2;2 (Figure 1A;Supplemental Data Set S1). Both PIP2;1 and PIP2;2 are known as stress-related proteins with characterized functions in water transport. Therefore, it was reasonable to speculate that these two proteins somehow contribute to the role of UBC32 in abiotic stress responses.
Figure 1.
UBC32 directly interacts with PIP2;1 and PIP2;2. A, PIP2;1 and PIP2;2 were found to interact with UBC32 by mass spectrometry. Total proteins were extracted from UBC32–GFP overexpression plants and immunoprecipitated for mass spectrometry. The SFGAAVIYNK peptide of PIP2;1 and PIP2;2 was identified in the UBC32–GFP immunoprecipitated complex, as illustrated on the right. B, PIP2;1 binds to UBC32 in planta based on LCI experiments. An interaction was detected between UBC32-NLuc and PIP2;1-CLuc, but not in negative controls including AtOS9-NLuc and PIP2;1-CLuc. The pseudocolor bar on the right shows the range of luminescence intensity. C, PIP2;1 interacts with UBC32, as revealed by BiFC assay. The full-length coding sequence of UBC32 was cloned into pSPYCE (M) and the full-length coding sequences of PIP2s were inserted into pSPYNE (R) 173. AtOS9 was used as a negative control in this BiFC assay, bar = 10 μm. D, PIP2;1 interacts with UBC32 in a pull-down assay. E. coli-expressed MBP, MBP-PIP2;1, and GST-UBC32 were used in the pull-down assay. Equal amounts of MBP and MBP-PIP2;1 bound to amylose resin were incubated with 2 μg GST-UBC32. Pull-down products were detected using anti-GST antibody. The arrowheads on the right indicate the positions of full-length GST/MBP-tagged proteins.
To confirm the interaction between UBC32 and PIP2;1 (and PIP2;2), we performed LCI assays using the pUBC32-NLuc and pCLuc-PIP2;1 plasmids. OS9, a lectin that recognizes misfolded proteins in the ER, was used as a negative control in this assay. pUBC32-NLuc and pCLuc-PIP2;1 and the control vectors were co-expressed in N. benthamiana leaves. Only samples expressing the combination of CLuc-PIP2;1 and UBC32-NLuc showed strong LUC complementation signals, but samples expressing CLuc-PIP2;1 with AtOS9-NLuc did not (Figure 1B). We measured the levels of CLuc-PIP2;1, UBC32-NLuc, and control proteins using anti-Luc antibody (Supplemental Figure S1A). Similar results were obtained using plasmids for UBC32 and PIP2;2 (Supplemental Figure S1, B and C). We then performed a BiFC assay to further confirm the UBC32–PIP2;1 interaction. Specifically, PIP2s were fused with the N-terminal half of the yellow fluorescent protein (YFP) gene and UBC32 was fused into the C-terminal half of the YFP gene. Confocal microscopy revealed that UBC32 binds to PIP2;1 and PIP2;2 (Figure 1C). The proteins used in the BiFC assays were well expressed in plants (Supplemental Figure S1D).
Furthermore, we tested the direct interactions between UBC32 and PIP2;1 and PIP2;2 using a pull-down approach. Only MBP-PIP2;1 pulled down the GST-UBC32 proteins but not the MBP control (Figure 1D). The same result was obtained for the interaction between UBC32 and PIP2;2 (Supplemental Figure S1E). UBC32 also bound to PIP2;1 in a split ubiquitin yeast two-hybrid assay (Supplemental Figure S2). We further determined the region of UBC32 that associated with PIP2;1 by performing a split ubiquitin yeast two-hybrid assay. Our results showed that the C-terminal region from amino acid 253–309 of UBC32 containing the transmembrane domain was responsible for its interaction with PIP2;1 (Supplemental Figure S2). Taken together, these results indicate that UBC32 physically interacts with PIP2;1 (and PIP2;2) both in vivo and in vitro.
UBC32 functions as a positive regulator of drought tolerance and the ABA response
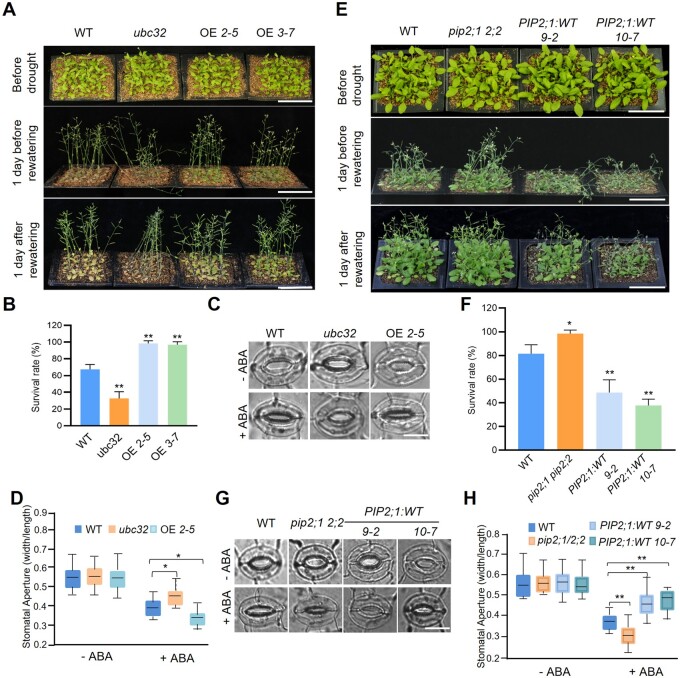
Given our evidence that UBC32 binds to PIP2;1 directly, we next investigated how UBC32 and PIP2;1/PIP2;2 function together in the drought stress response. We previously showed that the Arabidopsis ubc32 mutant exhibited reduced sensitivity to salt and ABA treatment and that UBC32 is a drought-induced gene at the transcriptional level (Cui et al., 2012). We first examined the drought tolerance of the WT, ubc32, and two independent UBC32 overexpression lines generated in our previous study (OE2-5 and OE3-7). Under normal conditions, WT, ubc32, and UBC32 overexpression seedlings all grew well, exhibiting no obvious differences. However, upon withholding water for 20 days, only 33% of ubc32 seedlings survived, whereas 67% of WT seedlings, 98% of OE 2–5 and 97% of OE 3–7 seedlings remained alive (Figure 2, A and B). These results suggest that the ubiquitin-conjugating enzyme UBC32 functions in the drought stress response.
Figure 2.
UBC32 functions as a positive regulator while PIP2;1/PIP2;2 are negative regulators of ABA-related drought stress tolerance. A, Drought tolerance test of WT, ubc32 mutant, and 35S:UBC32 transgenic plants. Two-week-old seedlings were subjected to drought stress for ∼20 days and rewatered when significant differences in wilting were observed. Representative photographs obtained from three independent experiments are shown, bar = 5 cm. B, Statistical analysis of the seedling survival rate in (A). Values represent the mean ± sd (five biological replicates; n = 80). Statistical significance was analyzed by one-way ANOVA (**P <0.01). Detailed experiment replicates and statistical analysis are described in the “Method” section. C, Comparison of ABA-induced stomatal closure in WT, ubc32 mutant and 35S:UBC32 transgenic plants. Leaves from 4-week-old plants incubated in stomatal opening buffer were exposed to the light for 3 h. ABA (0 or 10 μM) was added to the samples and stomatal closure was observed after 3 h of treatment. Representative photographs are shown, bar = 10 µm. D, Quantitative analysis of stomatal aperture. The data were obtained from approximately bout 60 stomata. The box plots show the median (central line), the lower and upper quartiles (box) and the minimum and maximum values (whiskers). Significant differences between WT and ubc32 mutant or 35S:UBC32 transgenic plants were determined by one-way ANOVA (*P <0.05). E, Drought tolerance test of the pip2;1 pip2;2 mutant and two independent 35S:PIP2;1 transgenic plants lines. Two-week-old seedlings were subjected to drought stress for ∼20 days and rewatered when significant differences in wilting were observed. Representative photographs obtained from three independent experiments are shown, bar = 5 cm. F, Statistical analysis of the seedling survival rate in (E). Values represent the mean ± sd (five biological replicates; n = 80). Statistical significance was analyzed by one-way ANOVA (*P <0.05, **P <0.01). Detailed experiment replicates and statistical analysis are described in the “Method” section. G, Comparison of ABA-induced stomatal closure in the WT, pip2;1 pip2;2, and two independent 35S:PIP2;1 transgenic plants lines. ABA treatment was performed as in (C). More than 40 stomata were measured, and representative photographs are shown, bar = 10 μm. H, Quantitative analysis of stomatal aperture. The data were obtained from approximately 40 stomata. The box plots show the median (central line), the lower and upper quartiles (box) and the minimum and maximum values (whiskers). Significant differences between WT and pip2;1 pip2;2 or 35S:PIP2;1 transgenic plants were determined by one-way ANOVA (**P <0.01).
Plant responses to drought stress are complex but frequently involve regulatory activity by ABA (Zhu, 2002). To determine whether the drought stress responses of ubc32 and the UBC32 overexpression lines are ABA-related, we examined the stomata behaviors of WT, ubc32, and the OE2-5 plants under ABA treatment. The stomatal apertures (the ratio of width to length) of the different plants were not significantly different under 1/2 MS treatment (Figure 2, C and D). After being treated with 1/2 MS containing 10-μM ABA for 3 h, the stomatal aperture of the UBC32 overexpression plants was much smaller while that of ubc32 was much larger compared to the WT (Figure 2, C and D). Furthermore, we analyzed the expression of various ABA and drought-induced genes in WT, ubc32, and UBC32 overexpression seedlings via reverse transcription polymerase chain reaction. Following 4 h of drought treatment, the expression levels of almost all genes examined showed a significant increase in the UBC32 overexpression line, while the expression levels in ubc32 were not significantly different from that of the WT (Supplemental Figure S3). Taken together, these results indicate that UBC32 functions as a positive regulator of plant responses to drought and ABA.
Overexpressing AcPIP2 from chamiso (Atriplex canescens) in Arabidopsis led to reduced drought tolerance, and drought stress reduces the stability of PIP2;1 via 26S proteasome-mediated degradation (Lee et al., 2009; Li et al., 2015). We obtained PIP2;1 or PIP2;2 overexpression transgenic plants in the Col-0 (WT) background (Supplemental Figure S4, A and 4B). The pip2;1 pip2;2 double mutant was generated as previously reported (Qing et al., 2016). After drought treatment in soil, the pip2;1/2/2 seedlings were more tolerant to drought stress, while PIP2;1 (and PIP2;2) overexpression transgenic plants were more sensitive to drought than the WT, which is consistent with previous results (Figure 2, E and F;Supplemental Figure S5, A and B). Given that UBC32 functions in ABA-related drought tolerance, we examined whether PIP2;1/2;2 regulates ABA-induced stomatal closure using isolated epidermal peels. ABA-induced stomatal closure was enhanced in the pip2;1 pip2;2 mutants and impaired in 35S:PIP2;1 overexpression plants compared to the WT (Figure 2, G and H). A similar stomatal response to ABA was observed in 35S:PIP2;2 transgenic plants (Supplemental Figure S5, C and D). Collectively, these data indicate that PIP2;1 and PIP2;2 are involved in ABA-related stomatal responses.
PIP2;1 is essential for the drought-stress-related functions of UBC32
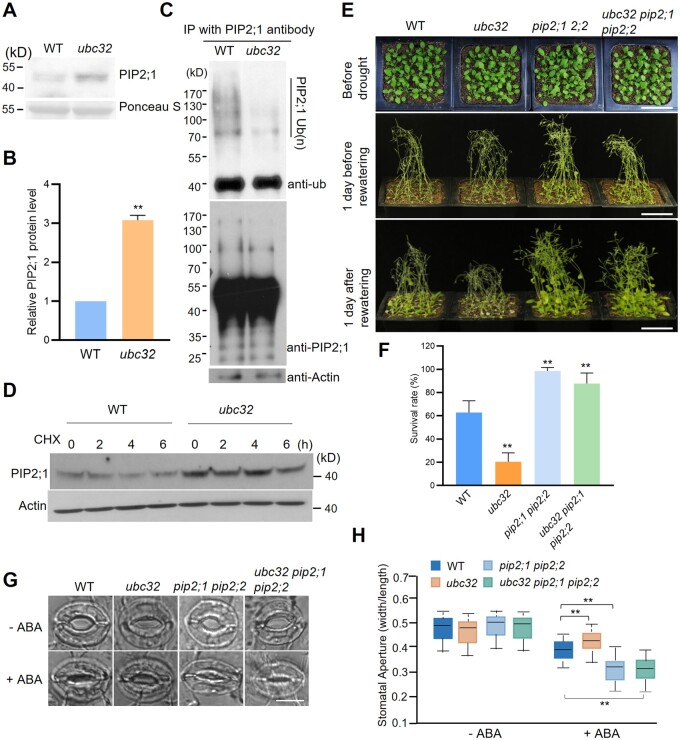
UBC32 and PIP2;1 (and PIP2;2) exhibited an opposite response upon drought stress. Because UBC32 is a ubiquitin-conjugating enzyme, we analyzed the protein levels of PIP2;1 in the WT and ubc32 seedlings using anti-PIP2;1 antibody (Qing et al., 2016). PIP2;1 protein levels were higher in ubc32 compared with the WT (Figure 3, A and B), while PIP2;1 transcript levels in WT and ubc32 did not differ (Supplemental Figure S6A). These results demonstrate that the protein level of PIP2;1 is negatively regulated by UBC32. Based on the finding that PIP2;1 can be ubiquitinated in plants (Lee et al., 2009), we investigated whether the ubiquitination of PIP2;1 is mediated by UBC32. We purified PIP2;1 from WT and ubc32 using anti-PIP2;1 antibody. Using ubiquitin and PIP2;1 antibodies, we obtained the same result—fewer modified bands in the immune complex from the ubc32 background compared with WT (Figure 3C). This result suggests that UBC32 promotes the ubiquitination modification process of PIP2;1. Consistent with the ubiquitination results, PIP2;1 exhibited reduced degradation in ubc32 compared with the WT background (Figure 3D). PIP2;2 protein levels decreased gradually with increasing amounts of UBC32, whereas the transcription of PIP2;2 was not changed by UBC32 (Supplemental Figure S6, A and B). Taken together, these findings suggest that UBC32 mediates the ubiquitination and degradation of PIP2;1 and PIP2;2.
Figure 3.
PIP2;1 and PIP2;2 are negatively regulated by UBC32 at the biochemical and genetic levels. A, The protein level of PIP2;1 in WT and ubc32 mutant. PIP2;1 protein level in 2-week-old WT and ubc32 seedlings was detected using anti-PIP2;1 antibody. Ponceau S represents the loading control. B, Quantification of PIP2;1 protein levels in WT and ubc32 in (A). Values represent the mean ± sd. Significantly different PIP2;1 protein levels in WT and ubc32 were determined by Student’s t test, **P < 0.01. C, In vivo ubiquitination level of PIP2;1 in WT and ubc32. Crude protein extracted from WT and ubc32 seedlings was immunoprecipitated by anti-PIP2;1 antibody and detected using anti-ubiquitin antibody and anti-PIP2;1 antibody. D, The in vivo degradation rate of PIP2;1 in WT and ubc32. WT and ubc32 seedlings were treated with 50-µΜ cycloheximide for the indicated time points. The PIP2;1 protein was checked using anti-PIP2;1 antibody. Actin represents the loading control. E, Drought tolerance test of WT, ubc32, pip2;1 pip2;2, and ubc32 pip2;1 pip2;2. The phenotype was analyzed in the same way as in Figure 2A. Representative photographs obtained from three independent experiments are shown, bar = 5 cm. F, Statistical analysis of the seedling survival rate in (E). Values represent the mean ± sd (five biological replicates; n = 80). Statistical significance was analyzed by one-way ANOVA (**P <0.01). Detailed experiment replicates and statistical analysis are described in the “Method” section. G, Comparison of ABA-induced stomatal closure in the WT, ubc32, pip2;1 pip2;2, and ubc32 pip2;1 pip2;2. Leaves from 4-week-old plants incubated in stomatal opening buffer were exposed to the light for 3 h. ABA (0 or 10 µM) was added to the samples and stomatal closure was observed after 3 h of treatment. More than 60 stomata were measured, and representative photographs are shown, bar = 10 µm. H, Statistics of the stomatal aperture in (G). The data were obtained from approximately 60 stomata. The box plots show the median (central line), the lower and upper quartiles (box) and the minimum and maximum values (whiskers). Significant differences between WT and ubc32, pip2;1 pip2;2 or ubc32 pip2;1 pip2;2 were determined by one-way ANOVA (**P <0.01).
Next we determined the genetic relationship between UBC32 and PIP2;1/2;2 in plants under drought stress. Under drought stress treatment, the ubc32 pip2;1 pip2;2 triple mutant exhibited a similar drought tolerance phenotype to that of the pip2;1 pip2;2 double mutant, demonstrating that PIP2;1 (and PIP2;2) is epistatic to UBC32 at the genetic level (Figure 3, E and F). We also examined the stomatal aperture of these seedlings under ABA treatment. The stomata of the ubc32 pip2;1 pip2;2 triple mutant showed similar movement to that of the pip2;1 pip2;2 double mutant after 3 h of ABA treatment (Figure 3, G and H). We also observed the phenotypes of plants with different genotypes under ABA treatment. In the presence of 0.5-μM ABA, the ubc32 mutant grew better than the WT, which is consistent with a previous report (Cui et al., 2012). The ubc32 pip2;1 pip2;2 triple mutant show enhanced sensitivity to ABA, and it recovered the less sensitive phenotype of ubc32 compared with WT (Supplemental Figure S6, C and D). These results indicate that PIP2;1 and PIP2;2 are essential for the drought-stress-related functions of UBC32.
The E2–E3 pair UBC32 and Rma1 regulate the stability of PIP2;1 by ubiquitinating its Lys276
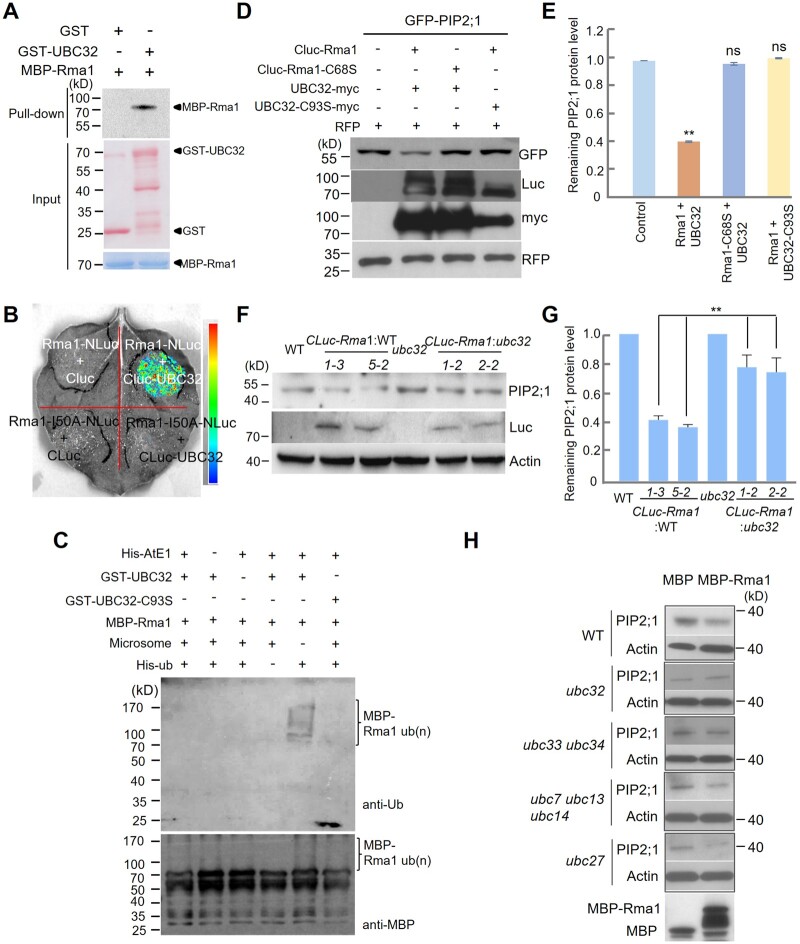
E3 ligases are usually required for the ubiquitination process. Rma1H1, a drought stress-induced ER membrane-anchored E3 ligase, negatively regulates PIP2;1 levels via ubiquitination, as observed in transgenic Arabidopsis plants (Lee et al., 2009). This finding suggests that Rma1 might function with UBC32 as an E3–E2 pair to regulate PIP2 protein accumulation in Arabidopsis. First, we examined their physical interaction. GST-UBC32 and MBP-Rma1 proteins were prepared from Escherichia coli for the pull-down assay. Compared with the GST control, only GST-UBC32 bound to MBP-Rma1 in the assay (Figure 4A). We also detected the in vivo interaction by LCI and obtained a positive result (Figure 4B). Rma1-I50A, in which the isoleucine residue involved in the E2-RING interaction was mutated, was used as negative control. We examined protein levels by immunoblot analysis (Supplemental Figure S7A). We conclude that UBC32 is associated with Rma1 in vitro and in vivo.
Figure 4.
UBC32 and Rma1 function as an E2–E3 pair to ubiquitinate and degrade PIP2;1/PIP2;2. A, Rma1 interacts with UBC32 based on a pull-down assay. GST, GST-UBC32, and MBP-Rma1 expressed in E. coli were used in this pull-down assay. Equal amounts of GST and GST-UBC32 bound to anti-GST resins were incubated with 2-μg MBP-Rma1. Pull-down products were detected using anti-MBP antibody. The arrowheads on the right indicate the positions of full-length GST/MBP-tagged proteins. B, Rma1 binds to UBC32 in planta based on an LCI experiment. CLuc-UBC32 interacts with Rma1-NLuc, but not the mutated RING domain form (Rma1-I50A-NLuc). The pseudocolor bar on the right shows the range of luminescence intensity. C, In vitro autoubiquitination assay of Rma1. MBP-Rma1, His-AtE1, Ub, and 200 ng microsome were combined with GST-32 or GST-UBC32-C93S, respectively. The reaction was carried at 30°C for 90 min, and immunoblot analysis was performed using anti-MBP and anti-ub antibodies. D, The E3 ligase activity of Rma1 and E2 activity of UBC32 are necessary for the degradation of PIP2;1. Single amino acid mutations Rma1-C68S and UBC32-C93S, which lost E3 ligase activity or E2 activity, respectively, were used in this assay. Agrobacterium containing the GFP-PIP2;1 plasmid was co-injected into N. benthamiana with Agrobacterium containing the corresponding plasmids as indicated. Total protein was extracted from N. benthamiana leaves at 3 days post infiltration and analyzed by immunoblot analysis. RFP was used as the co-expressed control in this assay. E, Quantification of GFP-PIP2;1 protein levels in (D). Values represent the mean ± sd (n = 3, n means biological replicate). Statistical significance was determined by one-way ANOVA (**P <0.01; ns, no significant difference). F, The regulation of PIP2;1 by UBC32 is Rma1-dependent. PIP2;1 protein level was detected in WT, CLuc-Rma1:WT transgenic plants, ubc32 and CLuc-Rma1:ubc32 transgenic plants using anti-PIP2;1 antibody. G, Quantification of PIP2;1 protein levels in (F). Values represent the mean ± sd. Statistical significance was determined by one-way ANOVA (**P <0.01). H, PIP2;1 stability is not affected by other E2s in the ERAD system. Crude extracts of WT and different E2 mutants were incubated with MBP and MBP-Rma1 at 22°C and PIP2;1 protein was checked using anti-PIP2;1 antibody.
We then explored whether UBC32 is the E2 enzyme responsible for the E3 ligase activity of Rma1. Using GST-UBC32 expressed in E. coli as the E2, no E3 ligase activity signal was detected. This result is consistent with a previous study in which the E3 ligase activity of Rma1 was not detected with UBC32-△TM protein expressed in E. coli (Zhao et al., 2013). In an animal ERAD study, the microsome (containing the ER membrane and other factors potentially needed for ERAD) was needed to reconstitute the ERAD in a cell-free ubiquitin assay system (Nakatsukasa et al., 2008). After adding microsomes from ubc32 plants to the E3 ligase activity detection assay, MBP-Rma1 showed E3 ligase activity using GST-UBC32 as the E2 enzyme, but not the mutant form GST-UBC32-C93S (Figure 4C;Supplemental Figure S7B). This result was further confirmed by an enhanced E3 ligase activity using WT crude protein as an E2 source compared with ubc32 crude protein as an E2 source (Supplemental Figure S7C). Taken together, these results demonstrate that the active UBC domain of UBC32 is essential for the E3 ligase activity of Rma1.
Next, we analyzed whether Rma1 and UBC32 function jointly to enhance PIP2;1/2;2 degradation by co-expressing different combinations of proteins in N. benthamiana leaves. In this experiment, the RING domain mutant Rma1-C68S and the UBC domain mutant UBC32-C93S were used in parallel. Compared with the control, PIP2;1 accumulation was reduced only when co-expressed with Rma1 and UBC32 together but not with combined Rma1-C68S/UBC32 or Rma1/UBC32-C93S (Figure 4, D and E). Thus, Rma1 and UBC32 appear to cooperate in a RING domain- and UBC domain-dependent manner to degrade PIP2;1; a similar effect occurred for PIP2;2 (Supplemental Figure S8, A and B). We further confirmed the partnership between Rma1 and UBC32 in transgenic Arabidopsis plants. Compared with WT, overexpressing Rma1 in the WT background indeed reduced the protein level of PIP2; 1 (Figure 4F). However, when the same experiment was performed in the ubc32 mutant background, the reduction in PIP2;1 protein levels in Rma1:ubc32 versus ubc32 was much smaller than in the WT background (Figure 4, F and G). These results further demonstrate that the regulatory effect of Rma1 on PIP2;1 is UBC32-dependent. To examine the role of Rma1 and UBC32 in regulating the stability of PIP2;1, we added MBP and MBP-Rma1 into cell lysates of different mutants of E2 in the ERAD compartment. In the presence of Rma1, PIP2;1 protein levels were reduced in the cell lysates of WT and almost all E2 mutants except ubc32 (Figure 4H). Taken together, these results demonstrate that the degradation of PIP2;1 depends on the E3 ligase Rma1 and the ubiquitin-conjugating enzyme UBC32 complex.
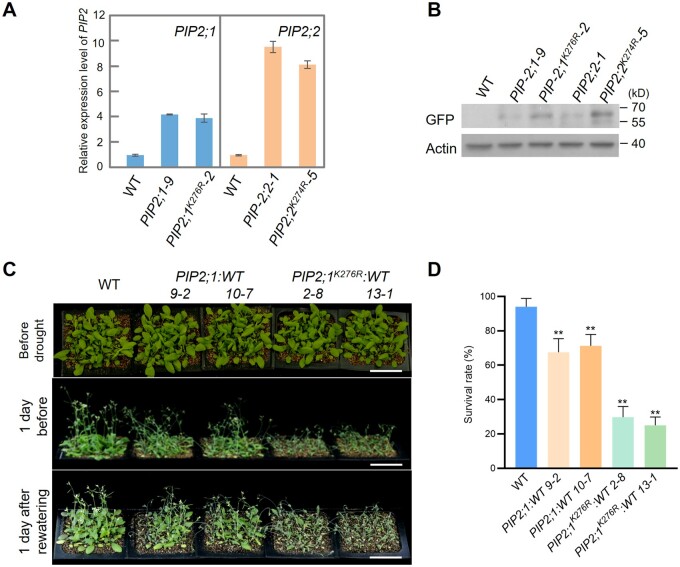
Next, we sought to identify possible ubiquitination sites in PIP2;1. GFP-PIP2;1 was enriched and purified from total protein extracts from MG132-treated PIP2;1 transgenic seedlings using GFP-trap magnetic agarose. LC–MS/MS analysis suggested that Lys276 of PIP2;1 might be the ubiquitination site (Supplemental Figure S9A). To experimentally verify that this residue is a ubiquitination site, we generated GFP-PIP2;1K276R transgenic plants. Although they had similar RNA expression levels, GFP-PIP2;1K276R plants accumulated more PIP2;1 protein compared to GFP-PIP2;1 plants (Figure 5, A and B). Following immunoprecipitation using anti-GFP antibody, we measured the ubiquitination levels of GFP-PIP2;1 and GFP-PIP2;1K276R using anti-ubiquitin antibody. A lower ubiquitination level was detected in GFP-PIP2;1K276R, demonstrating that this K276R (Lys–Arg) mutation reduced the ubiquitination of GFP-PIP2;1 (Supplemental Figure S9B). These results suggest that this Lys276 is an active ubiquitination site. Because this peptide is conserved between PIP2;1 and PIP2;2, we further confirmed that the Lys274 of PIP2;2 is also an active ubiquitination site (Figure 5, A and B).
Figure 5.
Lys276 and Lys274 are the ubiquitination sites of PIP2;1 and PIP2;2, respectively. A, The RNA level of PIP2;1/2;2 determined by qPCR in WT, GFP-PIP2;1 transgenic plants (PIP2;1-9 and PIP2;1K276R-2) or GFP-PIP2;2 transgenic plants (PIP2;2-1 and PIP2;2K274R-5). ACTIN2 was used as an internal control. B, GFP-PIP2;1/2;2 protein level was determined in WT, GFP-PIP2;1 transgenic plants (PIP2;1-9 and PIP2;1K276R-2) or GFP-PIP2;2 transgenic plants (PIP2;2-1 and PIP2;2K274R-5) using anti-GFP antibody. Actin was used as an internal control. C, Drought tolerance test of PIP2;1 and PIP2;1K276R transgenic plants. Two independent lines were used for each gene. The phenotype was analyzed as in Figure 2a, bar = 5 cm. D, Statistical analysis of the seedling survival rate in (C). Values represent the mean ± sd (five biological replicates; n = 80). Statistical significance was analyzed by one-way ANOVA (**P <0.01). Detailed experiment replicates and statistical analysis are described in the “Method” section.
To examine the role of the Lys276 in the drought stress response, we subjected the GFP-PIP2;1K276R lines to drought treatment. Compared with PIP2;1 overexpression plants, PIP2;1K276R plants exhibited a more drought-sensitive phenotype (Figure 5, C and D;Supplemental Figure S9C). Similar results were obtained for PIP2;2K274R transgenic plants (Supplemental Figure S9, D–F). These results confirm the notion that Lys276 is an active ubiquitination site in PIP2;1 (Lys274 in PIP2;2) that contributes to the drought stress response in Arabidopsis.
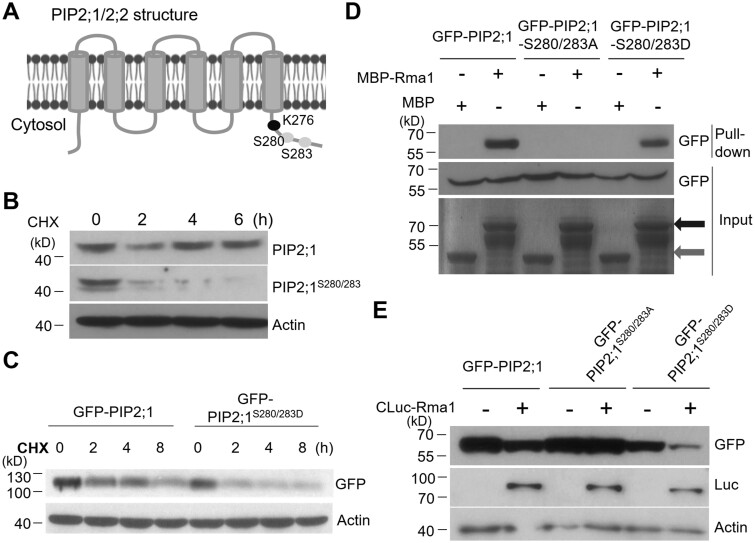
Phosphorylation of PIP2;1 promotes its degradation
The ubiquitination site Lys276 is located in the C-terminus of PIP2;1 (Figure 6A). The post-translational modification of PIP2;1, especially the phosphorylation modification at the C-terminus, which is localized to the cytosol, has been well studied (Maurel et al., 2015). The C-terminal (Ser280 and Ser283, termed S280/283 below) phosphorylation form of PIP2;1 is the active form, which exhibits increased water transport (Qing et al., 2016). Using anti-PIP2;1 antibody and anti-pS280/283 antibody, we examined the protein stability of PIP2:1 with or without phosphorylation of S280/283 under mannitol treatment (Qing et al., 2016). Both the levels of WT PIP2;1 and S280/S283-phosphorylated PIP2;1 were reduced under mannitol treatment, and the levels of phosphorylated PIP2;1 showed a greater reduction than PIP2;1 under these conditions (Supplemental Figure S10A). To test whether this result is due to a difference in protein stability between PIP2;1 and phosphorylated PIP2;1, we measured protein levels under treatment with the protein synthesis inhibitor cycloheximide. As expected, S280/283-phosphorylated PIP2;1 protein exhibited a shorter half-life compared to PIP2;1 protein (Figure 6B). We also measured the degradation rates of GFP-PIP2;1 and GFP-PIP2;1-SD (PIP2;1S280D/S283D, Ser280/283 phospho-mimic variant) after cycloheximide treatment. GFP-PIP2;1-SD degraded much more rapidly than GFP-PIP2;1, which is consistent with the in vivo results determined using anti-PIP2:1 antibody (Figure 6C).
Figure 6.
Phosphorylated PIP2;1 shows a faster degradation rate than the nonphosphorylated form. A, Schematic diagram of modification sites in the C-terminus of PIP2;1. Lys276, Ser280, and Ser283 are indicated. B, Phosphorylated PIP2;1 shows a faster turnover rate than the non-phosphorylated form. WT seedlings were treated with 50-µΜ cycloheximide (CHX) for 0, 2, 4, 6 h and total proteins were examined using anti-PIP2;1 antibody and anti-pS280/283 antibody, respectively. C, GFP-PIP2;1S280/283D degrades much more rapidly than GFP-PIP2;1. Total proteins were extracted from N. benthamiana expressing GFP-PIP2;1 and GFP-PIP2;1S280/283D and incubated at 22°C for different time points. GFP-PIP2;1 and GFP-PIP2;1S280/283D protein levels were measured using anti-GFP antibody. D, Rma1 interacts with PIP2;1 and PIP2;1S280/283D but not PIP2;1S280/283A. MBP and MBP-Rma1 were expressed and purified from E. coli, while GFP-PIP2;1, GFP-PIP2;1S280/283D, and PIP2;1S280/283A were expressed in N. benthamiana. MBP and MBP-Rma1 were bound to amylose resins, and the resins were incubated with equal amounts of PIP2;1 or PIP2;1 variant proteins. The dark gray arrow indicates input of MBP-Rma1, and the light gray arrow indicates input of MBP. MBP was used as a negative control. E, CLuc-Rma1 accelerates the turnover of GFP-PIP2;1 and GFP-PIP2;1S280/283D but not GFP-PIP2;1S280/283A. Different PIP2;1 variants were co-expressed with Rma1 in N. benthamiana and PIP2;1 protein levels were detected using anti-GFP antibody.
We then tested whether the increased degradation of S280/S283-phosphorylated PIP2;1 is dependent on the Rma1–UBC32 complex. First, we examined the effects of phosphorylation on the Rma1–PIP2;1 association. We generated phospho-dead PIP2;1S280A/S283A (PIP2;1-SA) and phospho-mimic PIP2;1S280D/S283D (PIP2;1-SD) variants. A pull-down assay showed that, compared to the MBP control, PIP2;1 and PIP2;1-SD were associated with MBP-Rma1, whereas PIP2;1-SA failed to interact with MBP-Rma1 (Figure 6D). An LCI assay showed that, when all proteins were well expressed, UBC32 interacted with PIP2;1 and PIP2;1-SD, but not with PIP2;1-SA in planta (Supplemental Figure S10B and 10C). We also detected in vivo ubiquitination levels in PIP2;1 and its variants. Compared with GFP–PIP2;1, GFP–PIP2;1-SD exhibited increased ubiquitin levels, while GFP–PIP2;1-SA showed reduced levels (Supplemental Figure S10D). Consistent with the results of interaction and ubiquitination assays, when coexpressed in N. benthamiana, PIP2;1 and PIP2;1-SD were destabilized by Rma1, whereas PIP2;1-SA was largely unaffected (Figure 6E). We also examined the effects of the C-terminal phosphorylation of PIP2;1 on drought tolerance. PIP2;1-SD-overexpressing plants were more tolerant to drought than PIP2;1-overexpressing plants (Supplemental Figure S11, A and B). Together, these results demonstrate that Rma1 associates with S280/S283-phosphorylated PIP2;1 and subsequently mediates its ubiquitination and degradation under drought stress. Our data suggest that the phosphorylation of PIP2;1 has a biological function besides increasing the water channel activity of this protein.
Overexpressing UBC32 improves the growth of rice under drought stress
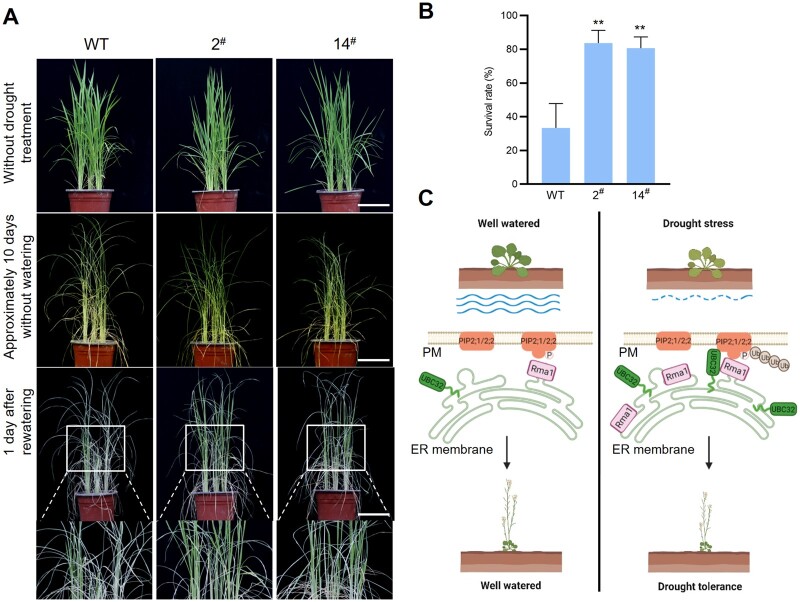
Amino acid sequence alignment revealed that the UBC32 homolog in rice is highly similar to UBC32 from Arabidopsis (Supplemental Figure S12A). To investigate whether UBC32 also positively regulates the drought response in crops, we examined the phenotypes of transgenic rice plants overexpressing Arabidopsis UBC32 driven by the maize Ubiquitin promoter (Ub:UBC32) in the Nipponbare background. The UBC32 expression level of two independent lines was confirmed by qPCR (Supplemental Figure S12B). Under sufficient water conditions, the overexpression lines exhibited similar growth to the WT control (Nipponbare). Under drought conditions, the overexpression lines showed better growth compared with WT. At the same time, <40% of WT seedlings and >80% of Ub:UBC32 transgenic seedlings were alive (Figure 7, A and B). These results demonstrate that UBC32 plays a positive role in drought tolerance in rice.
Figure 7.
Arabidopsis UBC32 enhances drought tolerance in rice. A, Drought tolerance test of WT (Nipponbare) and Ub:UBC32 transgenic plants (2# and 14#). B, Statistical analysis of the seedling survival rate in (A). Values represent the mean ± sd (four biological replicates; n = 48). Statistical significance was analyzed by one-way ANOVA (**P <0.01). C, A proposed working model for the role of UBC32 and Rma1 in the plant response to drought stress. Under drought stress, the expression of UBC32 and Rma1 is induced. UBC32 cooperates with Rma1 to recognize phosphorylated PIP2;1 and PIP2;2 and target them for ubiquitin-dependent degradation to enhance drought tolerance in plants. PM, plasma membrane.
In conclusion, we provided evidence that UBC32–Rma1 function together to target phosphorylated PIP2;1 and PIP2;2 for ubiquitin-dependent degradation to enhance drought tolerance in plants (Figure 7C). We also demonstrated that overexpressing UBC32 increased drought tolerance in rice.
Discussion
Degradation mediated by the ubiquitin proteasome system participates in a series of physiological processes in plants, such as plant growth, hormonal signaling, and biotic and abiotic stress responses. Here, we propose a working model for how the ERAD ubiquitin-conjugating enzyme UBC32 and the RING-type E3 ligase Rma1 function together to regulate plant drought stress responses by modifying the stability of two aquaporin proteins: PIP2;1 and PIP2;2. Drought stress induces the expression of both UBC32 and Rma1 (Lee et al., 2009; Cui et al., 2012). Rma1 binds to the phosphorylated form of PIP2;1 to initiate its proteasome-mediated degradation. UBC32 overexpression in transgenic Arabidopsis resulted in reduced PIP2;1 protein levels and increased tolerance to drought stress.
Much research about the functional impacts of ubiquitination on plant stress responses has focused on E3 ligases. In Arabidopsis, many E3 ligases were shown to be involved in plant stress responses. For example, PUB12/13 plays a role in the drought stress response, SDIR1 functions in the salt and drought stress responses and HOS1 functions in the cold stress response (Dong et al., 2006; Zhang et al., 2007; Kong et al., 2015; Zhang et al., 2015). E2 ubiquitin-conjugating enzymes have received relatively little attention, although there are examples of E2/E3 pairs that together regulate development and stress responses in plants. For example, in Arabidopsis, UBC1 and UBC2 interact with the E3 ligases HUB1 and HUB2 to mediate histone H2B monoubiquitination, which in turn increases cellular levels of FLC, a central flowering repressor (Cao et al., 2008; Gu et al., 2009). PHOSPHATE2 (PHO2), which encodes the ubiquitin-conjugating E2 enzyme UBC24, is critical for Pi homeostasis. PHO2 knockout plants suffered Pi toxicity due to the enhanced uptake and root-to-shoot translocation of Pi (Aung et al., 2006). The role of UBC24 in Pi homeostasis has been characterized in Arabidopsis, rice, and wheat (Liu et al., 2012; Park et al., 2014; Ouyang et al., 2016; Ying et al., 2017).
In general, E3 enzymes recruit substrates directly, while E2 enzymes function by binding to E3 (Ye and Rape, 2009). However, there are several examples of E2 enzymes directly interacting with their targets. For example, the E2-conjugating enzyme UBC24 in Arabidopsis directly associates with and modulates the stability of the phosphate transporter PHO1 (Liu et al., 2012). UBC27 also directly interacts with ABI1 and promotes its ubiquitination and degradation (Pan et al., 2020). As a core ERAD component, the ubiquitin conjugase UBC32 might have many substrates that it does not recognize and physically interact with directly. Instead, it might interact with these substrates through other ERAD components, such as E3 ligases or the adaptor protein AtOS9. However, in this study, we demonstrated that UBC32 directly interacts with its targets PIP2;1 and PIP2;2 in pull-down assays. These findings support the notion that some ubiquitin-conjugating enzymes not only mediate the ubiquitination of their targets by binding to E3 ligases, but they also participate in recognizing the targets themselves.
We previously determined that abiotic stresses including salt and drought stress induce UBC32 expression (Cui et al., 2012). In the present study, we identified the role of UBC32 in the drought stress response. Specifically, we showed that UBC32 functions in drought stress tolerance by targeting aquaporins (PIP2;1 and PIP2;2 here) for degradation. UBC32, UBC33, and UBC34 interact with PUB18, a U-box E3 ligase that functions as a negative regulator of plant responses to drought stress (Ahn et al., 2018). Knocking out UBC32, UBC33, and UBC34 improved the growth of Arabidopsis under drought stress conditions (Ahn et al., 2018). In contrast, we observed improved growth under drought stress conditions in our gene overexpression lines. Thus, it is not obvious how to reconcile our finding that the UBC32 knockout mutant exhibited decreased growth under drought stress conditions. Many factors could potentially explain the different results from these two studies. For example, compared to the previous study, we observed plant phenotypes during different growth stages, the plants were grown under different environmental conditions, or different knockout mutant lines were used in the drought stress assay.
The ER membrane-anchored E3 ligase Rma1H1 positively regulates drought tolerance by negatively regulating PIP2;1 protein levels via ubiquitination, as demonstrated in transgenic Arabidopsis plants (Lee et al., 2009). Furthermore, research in mammals has shown that the Rma1 homolog (RMA1) cooperates with UBC32 homolog (Ubc6e) as an E3–E2 pair to mediate the turnover of CFTRΔF508 (Younger et al., 2006). Therefore, we investigated the possibility that Rma1 cooperates with UBC32 to monitor the degradation of PIP2;1/2;2. Rma1 was associated with UBC32 in vivo and in vitro (Figure 4). The degradation of PIP2;1/2;2 is dependent on the RING-domain of Rma1 and the UBC-domain of UBC32.
The phosphorylation of RhPIP2;1 on S273 in rose is enhanced by dehydration stress and is sufficient to cause nuclear accumulation of the C-terminus of RhPTM, a membrane-tethered MYB-like transcription factor (Zhang et al., 2019). PIP2;1 contributes to ABA-triggered stomatal closure through OPEN STOMATA1 (OST1)-mediated phosphorylation at S121 of PIP2;1 (Grondin et al., 2015). Our result shows that S280/S283-phosphorylated PIP2;1 has a shorter half-life compared to PIP2;1. Rma1 interacts with and accelerates the degradation of PIP2;1 and PIP2;1-SD, but not PIP2;1-SA. Based on these results, different phosphorylation sites in PIP2;1 might have distinct functions. In the stomatal movement experiments with pip2;1 pip2;2, the difference results between these two studies might be due to the use of different mutants and/or different experimental protocols to measure stomatal aperture. pip2;1 was used in Grondin et al. (2015), while the pip2;1 pip2;2 double mutant was used in the current study. In the ABA-induced stomatal closure assay, we used intact rosette leaves to observe stomatal movement. Six to eight rosette leaves were treated with ABA for 3 h, and epidermal peels were collected and used to measure stomatal aperture. Grondin et al. treated epidermal peels from leaves, but not whole leaves, with ABA directly. Perhaps the signaling response is distinct between whole leaves and epidermal peels. The multiple phosphorylation-based regulatory mechanisms associated with PIP2;1 highlight its response to multiple stress signaling pathways and emphasize the centrality of this protein’s function in plant water relations.
In addition to the model plant Arabidopsis, crops are facing serious threats from drought. Drought is the greatest limiting factor to rice production worldwide. Studying the drought response mechanism and breeding drought-tolerant rice are essential for maintaining yield gains and ensuring food security. We overexpressed UBC32 in Nipponbare and found that, compared with WT, Ub:UBC32 transgenic plants showed better growth under water deficiency conditions. Thus, Arabidopsis UBC32 increases drought tolerance in both Arabidopsis and rice. These results provide a possible target for breeding and/or engineering drought tolerant rice and other crops in the future.
Materials and methods
Plant materials and growth conditions
Arabidopsis thaliana ecotype Columbia-0 was used as the WT, and all mutants used in this study were in the Col-0 background. The ubc32 and pip2;1/2;2 mutants were generated as previously reported (Chen et al., 2016; Qing et al., 2016). After surface sterilization, seeds were sown on Murashige and Skoog (MS) medium and vernalized for 3 days at 4°C. Ten days later, seedlings grown on MS medium were transferred to soil in a greenhouse with a 16-h light/8-h dark photoperiod at 22°C; LED light at an intensity of 100 μmol m−2 s−1. To observe seedling phenotypes, 0.5-μM ABA was added to the MS medium. The drought stress test was performed as described previously (Qin et al., 2008; Ding et al., 2015). Plants were grown for ∼2 weeks under normal watering conditions and subjected to water stress by withholding watering for ∼2 to 3 weeks. In each experiment, 16 plants were grown in a small pot under a 16-h light/8-h dark photoperiod at 22°C. At least five independent experiments were performed. The plants were rewatered when significant differences in wilting were observed. Twenty-four hours after rewatering, surviving plants were counted. For the rice drought stress test, in each experiment, 24 plants were grown in a small pot under a 12-h light/12-h dark photoperiod at 28°C. At least five independent experiments were performed. Ten days later, the plants were rewatered when significant differences in wilting were observed.
Constructs
All constructs for UBC32 were described previously (Cui et al., 2012; Chen et al., 2016). To generate constructs for PIP2;1 and PIP2;2, the full-length coding sequences of PIP2;1 and PIP2;2 were inserted into pMalC2 in the EcoRI and SalI sites, pCambia-CLuc in the KpnI and SalI sites, pCambia1300-221-GFP vectors in the KpnI and BamHI sites, and pVYNE (R) in the SpeI and SalI sites. GFP-PIP2 plasmids were introduced into Agrobacterium tumefaciens strain GV3101 and transformed into plants by the floral dip method (Clough and Bent, 1998). All primers used in this study are listed in Supplemental Table S1. To generate constructs for Rma1, the Rma1 coding sequence was inserted into pMalC2 in the EcoRI and SalI sites and pCambia-CLuc in the KpnI and SalI sites. pMalC2 and pCambia1300-221-GFP are as previous described (Chen et al., 2016), pCambia-CLuc and Cambia-NLuc were described in Chen et al. (2008), pSPYNE (R) 173 and pSPYCE (M) were described in Waadt et al. (2008).
IP–MS assay
To identify UBC32-interacting proteins, immunoprecipitation was performed using UBC32–GFP transgenic plants. Total proteins were extracted from the plants the native buffer (50-mM Tris–MES, pH 8.0, 0.5 M sucrose, 1-mM MgCl2, 10-mM EDTA, 5-mM DTT, protease inhibitor cocktail Complete Mini tablets (Roche)) and purified using GFP-trap magnetic agarose. The enriched proteins were shown in Supplemental Data Set S1. To identify the ubiquitination sites of PIP2;1, GFP-PIP2;1 was enriched and purified using GFP-trap magnetic agarose from the total protein extracts from 50-µM MG132-treated PIP2;1 transgenic seedlings. After purification, samples were separated on a 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS–PAGE) gel and the gel was stained with Coomassie brilliant blue.
The gel containing samples was cut into 2–3 mm2 pieces and de-colored in 25-mM ammonium bicarbonate containing 50% acetonitrile buffer. The proteins were reduced with 10-mM DTT at 56°C for 1 h and alkylated by 55-mM iodoacetamide in the dark for 45 min. The products were further digested with trypsin (Sigma T1426; enzyme-to-substrate ratio 1:50) at 37°C overnight. Tryptic peptides were extracted from the gel with buffer containing 5% trifluoroacetic acid and 50% acetonitirile by ultrasonic extraction twice. The liquids were freeze-dried in a SpeedVac, and peptides were resolubilized in 0.1% formic acid and filtered through a 0.45-μm centrifugal filter. The peptides were identified on a TripleTOF 5600 mass spectrometer (AB Sciex, Canada) coupled to an Eksigent nanoLC 1D platform assembled in Information Dependent Mode. A 90 min LC gradient (A = 0.1% formic acid in H2O, B = 0.1% formic acid in acetonitrile) was used to separate the peptides at a flow rate of 300 nL·min-1. The peptides were identified from MS/MS spectra using ProteinPilotTM software 4.2 by searching against the Arabidopsis database downloaded from UniProt. The fixed modification was carbamidomethylation of cysteine residues. Trypsin was specified as the proteolytic enzyme with two missed cleavages allowed. Mass tolerance was set to 0.05 Da, and the maximum false discovery rate for proteins and peptides was 1%.
Pull-down assays
MBP, MBP-PIP2;1/2;2, GST-UBC32, and MBP-Rma1 were expressed in E. coli strain BL21 and purified with buffer containing 0.5% NP-40. After incubating with 2-μg purified MBP and MBP-PIP2;1/2;2 for 2 h at 4°C, the amylose resin was washed three times with PBS and blocked by 5% BSA for 1 h at 4°C. The beads were washed three times again with PBS and incubated with 2 μg of purified GST-UBC32 for 2 h at 4°C. Finally, the beads were washed six times with PBS containing 150-mM NaCl and analyzed by SDS–PAGE. To examine the interaction of MBP-Rma1 with GFP-fused PIP2;1 variants, GFP-fused proteins were expressed in N. benthamiana. The proteins were extracted and filtrated as described in the immunoprecipitation section before being used in the pull-down assay.
qPCR assay
Two-weeks-old Arabidopsis seedlings grown on 1/2 MS medium plates were harvested for RNA extraction. For drought treatment, seedlings were pretreated with 1/2 MS liquid medium for ∼8 h and grown without liquid medium for 4 h before being harvested. Total RNA was isolated from the seedlings using an Ultrapure RNA Kit (Cwbio). The total RNA (2 μg) was denatured and used for first-strand cDNA synthesis using a Fast Quant RT Kit (Tiangen). Quantitative polymerase chain reaction (qPCR) was performed using a Quant One Step qRT-PCR Kit (SYBR Green) (Tiangen) and the CFX96 Touch Real-Time PCR Detection System (Bio-Rad). The primer sequences used for qPCR are shown in Supplemental Table S1.
Split ubiquitin yeast two-hybrid
The vectors pPR3N and pBT3-STE were used in this assay. The full-length UBC32 coding sequence or a truncated sequence was cloned into pPR3N, while the full-length PIP2;1 coding sequence was cloned into pBT3-STE. The corresponding plasmids were co-transformed into yeast (Saccharomyces cerevisiae) NMY51 cells as described previously (Thaminy et al., 2004). After 3 days, yeast colonies that grew on yeast synthetic dropout medium lacking Leu and Trp were suspended in water to optical density (OD)600 = 1. The suspended cells were then diluted to 0.1, 0.01, and 0.001 and plated on yeast synthetic dropout medium lacking His, Leu, Trp, and Ade to test protein interactions.
LCI assays
LCI assays were performed as described previously (Chen et al., 2008). The assays were performed to examine the interactions between UBC32 fused with NLuc and PIP2;1 (PIP2;1-SA and PIP2;1-SD) fused with CLuc; E3 ligase Rma1 fused with NLuc; and UBC32 fused with CLuc. Equal volumes of Agrobacterium tumefaciens containing pCambia-NLuc, pCambia-CLuc (or their derivative constructs), at a final concentration of OD600 = 1.5, were mixed with A. tumefaciens harboring 35S-p19, which expresses the tombusvirus silencing suppressor p19 to ensure efficient expression of the fusion proteins. Different combinations of constructs were infiltrated into different positions of the same leaf of N. benthamiana. Three days later, 1-mM luciferin was sprayed onto the leaves, which were then kept in the dark for 5 min. A low-light cooled charge-coupled device imaging apparatus (NightOWL LB 983 in vivo imaging system) was used to take the LUC images for 10 min.
BiFC assays
UBC32 and PIP2;1/2;2 were fused to the C- and N-termini of Venus to produce YCE-UBC32 and YNE-PIP2;1/2;2 respectively (Waadt et al., 2008). YCE and YNE were used as negative controls. A. tumefaciens strain EHA105 cells carrying BiFC vectors YCE-UBC32 and YNE-PIP2;1/2;2 or their negative controls were mixed with A. tumefaciens harboring 35S-p19 before being infiltrated into N. benthamiana leaves. A Leica TCS SP5 confocal laser scanning microscope was used to capture the fluorescence images of the infiltrated leaves at 3 days post infiltration.
ABA-related stomatal closure assay
In this assay, fresh leaves at similar developmental stages were harvested. These leaves were incubated in stomatal opening buffer (5-mM KCl, 50-mM CaCl2, and 10-mM MES–Tris, pH 6.1) at 22°C under high-light conditions (light intensity of 300 μmol m−2 s−1) for at least 3 h to ensure the opening of all stomata. ABA was then added to the buffer to a final concentration of 10 µM. After 3 h of incubation, photographs were taken, and stomatal aperture was measured using ImageJ. At least 60 stomata were measured for each material.
Immunoprecipitation, immunoblot analysis, and antibodies
The co-infiltrated parts of N. benthamiana leaves were used to extract total proteins with native protein extraction buffer (50-mM Tris–MES, pH 8.0, 0.5 M sucrose, 1-mM MgCl2, 10-mM EDTA, 5-mM DTT, protease inhibitor cocktail Complete Mini tablets (Roche)) and centrifuged at 4°C for 6 min at 16,000g; this process was repeated three times. Anti-PIP2;1 antibody coupled to IgG agarose was added to the crude protein, incubated at 4°C for 2 h, and washed at least five times with PBS. The agarose was suspended in protein loading buffer and boiled at 95°C for 5 min before being analyzed by SDS–PAGE. GFP-PIP2;1 (and its variants) extracted from overexpression plants or N. benthamiana leaves was incubated with GFP-trap magnetic agarose and the agarose was washed at least five times with PBS buffer. The samples were used for semi-in vivo co-immunoprecipitation assays or the identification of ubiquitination sites.
The antibodies used in this assay were as follows: anti-LUC (1:5,000 diluted, Sigma, Cat# L0159), anti-myc (1:5,000 diluted, EASY-BIO, Cat# BE2073), anti-MBP (1:5,000 diluted, EASY-BIO, Cat# BE2021), anti-GST (1:2,000 diluted, EASY-BIO, Cat# BE2013), anti-RFP (1:2,000 diluted, EASY-BIO, Cat# BE2023), anti-Actin (1:5,000 diluted, EASY-BIO, Cat# BE0027), anti-GFP (1:1,500 diluted, Roche, Cat#11814460001), goat anti-mouse (1:5,000 diluted, Proteintech, Cat# SA00001-1), goat anti-rabbit (1:5,000 diluted, Proteintech, Cat# SA00001-2). Anti-ubiquitin, anti-PIP2;1 and anti-pS280/283 were used as previously described (Zhao et al., 2013, Qing et al., 2016).
Protein degradation assays
For the in vivo degradation assay in plants, 2-week-old WT and ubc32 seedlings were treated with cycloheximide (75 μM) for the indicated time and collected for protein extraction in protein extraction buffer (50-mM Tris–MES, pH 8.0, 0.5 M sucrose, 1-mM MgCl2, 10-mM EDTA, 5-mM DTT, protease inhibitor cocktail Complete Mini tablets (Roche)) on ice. Protein samples were analyzed using SDS–PAGE. For the combined degradation assays of PIP2;1/2;2, different A. tumefaciens cultures harboring UBC32 (or Rma1) were mixed with equal volumes of A. tumefaciens harboring PIP2;1/2;2, and co-expressed in N. benthamiana. Three days later, total proteins were extracted and detected using the corresponding antibodies.
Plant microsomal preparation
Microsomes were extracted from plants using a Minute Plant Microsomal Membrane Extraction Kit (Invent Biotech, catalog MM-018). Buffers and the filter cartridge in a collection tube were pre-chilled on ice. A total of 200 mg freshly ground plant tissue was placed in the filter with 300-µL buffer A. The extracts were centrifuged at 14,000g at 4°C for 20 min. The supernatant was completely removed, and the pellet was resuspended in 300-μL buffer B, followed by centrifugation at 11,000g for 10 min at 4°C. The supernatants were transferred to 1 mL 1× PBS and centrifuged at 14,000g for 30 min at 4°C. The pellet (microsomal fraction) was dissolved in 50–200 µL detergent-containing buffer.
In vitro ubiquitination assay
For the in vitro ubiquitination assay, His-AtE1 (50 ng), His-ub (UBQ14; 2 μg), GST-UBC32 (500 ng), GST-UBC32-C93S (500 ng), and MBP-Rma1 (1 μg) purified from E. coli (BL21) and microsomal extracts from seedlings were used. Reactions were performed at 30°C for 90 min and were stopped by the addition of 4× protein loading sample buffer (258-mM Tris–HCl, pH 6.8, 8% SDS, 40% glycerol, 0.4% Coomassie Brilliant Blue, and 0.4 M β-mercaptoethanol). Samples were detected using anti-ub and anti-MBP antibodies.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 8.0 and one-way analysis of variance (ANOVA). The ANOVA table is shown in Supplemental File S1. In the statistical analysis of the survival rate under drought stress, values represent the mean ± sd of five biological replicates from at least two independent experiments. In one independent experiment, three or four pots (16 seedlings per pot) were treated with drought stress. * represents P < 0.05 and ** represents P < 0.01. In the statistical analysis of the ABA-related stomatal closure assay, stomatal aperture was measured using ImageJ. Two or three independent experiments were performed, and 30 stomata were measured in one experiment.
Accession numbers
Sequence information from this study can be found in the GenBank/EMBL databases under the following accession numbers: UBC32 (At3g17000), Rma1 (At4g03510), PIP2;1 (At3g53420), PIP2;2 (At2g37170), ABF3 (At4g34000), ABF4 (At3g19290), RD22 (At5g25610), RD29A (At5g52310), RD29B (At5g52300), ACTIN7 (At5g09810).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 . PIP2;2 is associated with UBC32 in vivo and in vitro.
Supplemental Figure S2 . The C-terminus of UBC32 containing the transmembrane domain is required for its interaction with PIP2;1.
Supplemental Figure S3 . UBC32 plays a positive role in drought tolerance and the ABA response.
Supplemental Figure S4 . GFP-PIP2;1/2;2 protein levels in 35S:PIP2;1/2;2 transgenic plants.
Supplemental Figure S5 . PIP2;2 is a negative regulator of ABA-related drought tolerance.
Supplemental Figure S6 . PIP2;1 and PIP2;2 act downstream of UBC32.
Supplemental Figure S7 . The E2 activity of UBC32 is important for the E3 ligase activity of Rma1.
Supplemental Figure S8 . Rma1 functions with UBC32 to regulate PIP2;2.
Supplemental Figure S9 . 35S:PIP2;1 K276R/PIP2;2 K274R transgenic plants show enhanced drought sensitivity compared to PIP2;1/PIP2;2 transgenic plants.
Supplemental Figure S10 . The phosphorylated form of PIP2;1 increases its binding to UBC32.
Supplemental Figure S11 . The phosphorylated form of PIP2;1 increases drought tolerance.
Supplemental Figure S12 . UBC32 enhances drought tolerance in rice.
Supplemental Table S1 . Primers used in this study.
Supplemental Data Set S1 . IP-MS identification of proteins associated with UBC32.
Supplemental File S1 . ANOVA and t test tables.
Supplementary Material
Acknowledgments
We are grateful to Prof. Ning Li from The Hong Kong University of Science and Technology for providing the PIP2;1 antibodies and pip2;1 pip2;2 double mutants.
Funding
This work was supported by grant no. 2016YFA0500500 from the National Key Research and Development Program of China and grant no. 31700234 from the National Natural Science Foundation of China.
Conflict of interest statement. The authors declare no conflict of interests.
Q.X. and Q.C. designed the research. Q.C. performed most of the experiments. R.L., Q.W., and Y.Z. performed some immunoblot analysis. S.W., R.X., and X.S. helped grow rice and analyzed the drought stress phenotypes of rice. X.Y., X.H., and Y.W. performed IP–MS and analyzed the results. Q.C. and Q.X. wrote the manuscript. Y.W., F.Y., and L.L. participated in the discussion of the results and modification of the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Qi Xie (qxie@genetics.ac.cn).
References
- Aharon R, Shahak Y, Wininger S, Bendov R, Kapulnik Y, Galili G (2003) Overexpression of a plasma membrane aquaporin in transgenic tobacco improves plant vigor under favorable growth conditions but not under drought or salt stress. Plant Cell 15: 439–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn MY, Oh TR, Seo DH, Kim JH, Cho NH, Kim WT (2018) Arabidopsis group XIV ubiquitin-conjugating enzymes AtUBC32, AtUBC33, and AtUBC34 play negative roles in drought stress response. J Plant Physiol 230: 73–79 [DOI] [PubMed] [Google Scholar]
- Alexandersson E, Fraysse L, Sjovall-Larsen S, Gustavsson S, Fellert M, Karlsson M, Johanson U, Kjellbom P (2005) Whole gene family expression and drought stress regulation of aquaporins. Plant Mol Biol 59: 469–484 [DOI] [PubMed] [Google Scholar]
- Aung K, Lin SI, Wu CC, Huang YT, Su CL, Chiou TJ (2006) pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol 141: 1000–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Dai Y, Cui S, Ma L (2008) Histone H2B monoubiquitination in the chromatin of FLOWERING LOCUS C regulates flowering time in Arabidopsis. Plant Cell 20: 2586–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM (2008) Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Zhong Y, Wu Y, Liu L, Wang P, Liu R, Cui F, Li Q, Yang X, Fang S, et al. (2016). HRD1-mediated ERAD tuning of ER-bound E2 is conserved between plants and mammals. Nat Plants 2: 16094. [DOI] [PubMed] [Google Scholar]
- Cho SK, Ryu MY, Song C, Kwak JM, Kim WT (2008) Arabidopsis PUB22 and PUB23 are homologous U-box E3 ubiquitin ligases that play combinatory roles in response to drought stress. Plant Cell 20: 1899–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana .Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cui F, Liu LJ, Zhao QZ, Zhang ZH, Li QL, Lin BY, Wu YR, Tang SY, Xie Q (2012) Arabidopsis ubiquitin conjugase UBC32 is an ERAD component that functions in brassinosteroid-mediated salt stress tolerance. Plant Cell 24: 233–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding SC, Zhang B, Qin F (2015) Arabidopsis RZFP34/CHYR1, a ubiquitin E3 ligase, regulates stomatal movement and drought tolerance via SnRK2.6-mediated phosphorylation. Plant Cell 27: 3228–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong CH, Agarwal M, Zhang Y, Xie Q, Zhu JK (2006) The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc Natl Acad Sci U S A 103: 8281–8286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grondin A, Rodrigues O, Verdoucq L, Merlot S, Leonhardt N, Maurel C (2015) Aquaporins contribute to ABA-triggered stomatal closure through OST1-mediated phosphorylation. Plant Cell 27: 1945–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Jiang D, Wang Y, Bachmair A, He Y (2009) Repression of the floral transition via histone H2B monoubiquitination. Plant J. 57: 522–533 [DOI] [PubMed] [Google Scholar]
- Jang JY, Kim DG, Kim YO, Kim JS, Kang HS (2004) An expression analysis of a gene family encoding plasma membrane aquaporins in response to abiotic stresses in Arabidopsis thaliana. Plant Mol Biol 54: 713–725 [DOI] [PubMed] [Google Scholar]
- Javot H, Lauvergeat V, Santoni V, Martin-Laurent F, Guclu J, Vinh J, Heyes J, Franck KI, Schaffner AR, Bouchez D, et al. (2003) Role of a single aquaporin isoform in root water uptake. Plant Cell 15: 509–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JY, Choi HI, Im MY, Kim SY (2002) Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14: 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Cheng J, Zhu Y, Ding Y, Meng J, Chen Z, Xie Q, Guo Y, Li J, Yang S, et al. (2015). Degradation of the ABA co-receptor ABI1 by PUB12/13 U-box E3 ligases. Nat Commun 6: 8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft E, Stone SL, Ma LG, Su N, Gao Y, Lau OS, Deng XW, Callis J (2005) Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol 139: 1597–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HK, Cho SK, Son O, Xu Z, Hwang I, Kim WT (2009) Drought stress-induced Rma1H1, a RING membrane-anchor E3 ubiquitin ligase homolog, regulates aquaporin levels via ubiquitination in transgenic Arabidopsis plants. Plant Cell 21: 622–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yu G, Sun X, Liu Y, Liu J, Zhang X, Jia C, Pan H (2015) AcPIP2, a plasma membrane intrinsic protein from halophyte Atriplex canescens, enhances plant growth rate and abiotic stress tolerance when overexpressed in Arabidopsis thaliana. Plant Cell Rep 34: 1401–1415 [DOI] [PubMed] [Google Scholar]
- Liu TY, Huang TK, Tseng CY, Lai YS, Lin SI, Lin WY, Chen JW, Chiou TJ (2012) PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. Plant Cell 24: 2168–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel C, Boursiac Y, Luu DT, Santoni V, Shahzad Z, Verdoucq L (2015) Aquaporins in Plants. Physiol Rev 95: 1321–1358 [DOI] [PubMed] [Google Scholar]
- Nakatsukasa K, Huyer G, Michaelis S, Brodsky JL (2008) Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell 132: 101–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang X, Hong X, Zhao X, Zhang W, He X, Ma W, Teng W, Tong Y (2016) Knock out of the PHOSPHATE 2 gene TaPHO2-A1 improves phosphorus uptake and grain yield under low phosphorus conditions in common wheat. Sci Rep 6: 29850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan WB, Lin BY, Yang XY, Liu LJ, Xia R, Li JG, Wu YR, Xie Q (2020). The UBC27-AIRP3 ubiquitination complex modulates ABA signaling by promoting the degradation of ABI1 in Arabidopsis. Proc Natl Acad Sci U S A 117: 27694–27702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BS, Seo JS, Chua NH (2014) NITROGEN LIMITATION ADAPTATION recruits PHOSPHATE2 to target the phosphate transporter PT2 for degradation during the regulation of Arabidopsis phosphate homeostasis. Plant Cell 26: 454–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin F, Sakuma Y, Tran PLS, Maruyama K, Kidokoro S, Fujita Y, Fujita M, Umezawa T, Sawano Y, Miyazono KI, et al. (2008). Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 20: 1693–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing D, Yang Z, Li M, Wong WS, Guo G, Liu S, Guo H, Li N (2016) Quantitative and functional phosphoproteomic analysis reveals that ethylene regulates water transport via the C-terminal phosphorylation of aquaporin PIP2;1 in Arabidopsis. Mol. Plant 9: 158–174 [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Abe H, Kasuga M, Yamaguchi-Shinozaki K, Carninci P, Hayashizaki Y, Shinozaki K (2001) Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell 13: 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo DH, Ahn MY, Park KY, Kim EY, Kim WT (2016) The N-terminal UND motif of the Arabidopsis U-box E3 ligase PUB18 is critical for the negative regulation of ABA-mediated stomatal movement and determines its ubiquitination specificity for exocyst subunit Exo70B1. Plant Cell 28: 2952–2973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo DH, Ryu MY, Jammes F, Hwang JH, Turek M, Kang BG, Kwak JM, Kim WT (2012) Roles of four Arabidopsis U-box E3 ubiquitin ligases in negative regulation of abscisic acid-mediated drought stress responses. Plant Physiol 160: 556–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Hauksdottir H, Troy A, Herschleb J, Kraft E, Callis J (2005) Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol 137: 13–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaminy S, Miller J, Stagljar I (2004) The split-ubiquitin membrane-based yeast two-hybrid system. Methods Mol Biol 261: 297–312 [DOI] [PubMed] [Google Scholar]
- Waadt R, Schmidt LK, Lohse M, Hashimoto K, Bock R, Kudla J (2008) Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J 56: 505–516 [DOI] [PubMed] [Google Scholar]
- Ye Y, Rape M (2009)Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol 10: 755–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Y, Yue W, Wang S, Li S, Wang M, Zhao Y, Wang C, Mao C, Whelan J, Shou H (2017) Two h-type thioredoxins interact with the E2 ubiquitin conjugase PHO2 to fine-tune phosphate homeostasis in rice. Plant Physiol 173: 812–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younger JM, Chen L, Ren HY, Rosser MF, Turnbull EL, Fan CY, Patterson C, Cyr DM (2006) Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell 126: 571–582 [DOI] [PubMed] [Google Scholar]
- Zhang H, Cui F, Wu Y, Lou L, Liu L, Tian M, Ning Y, Shu K, Tang S, Xie Q (2015) The RING finger ubiquitin E3 ligase SDIR1 targets SDIR1-INTERACTING PROTEIN1 for degradation to modulate the salt stress response and ABA signaling in Arabidopsis. Plant Cell 27: 214–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Feng M, Chen W, Zhou X, Lu J, Wang Y, Li Y, Jiang CZ, Gan SS, Ma N, et al. (2019). In rose, transcription factor PTM balances growth and drought survival via PIP2;1 aquaporin. Nat Plants 5: 290–299 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yang C, Li Y, Zheng N, Chen H, Zhao Q, Gao T, Guo H, Xie Q (2007). SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell 19: 1912–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Tian M, Li Q, Cui F, Liu L, Yin B, Xie Q (2013). A plant-specific in vitro ubiquitination analysis system. Plant J 74: 524–533 [DOI] [PubMed] [Google Scholar]
- Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.