Abstract
Reactive oxygen species (ROS) are unstable reactive molecules that are toxic to cells. Regulation of ROS homeostasis is crucial to protect cells from dysfunction, senescence, and death. In plant leaves, ROS are mainly generated from chloroplasts and are tightly temporally restricted by the circadian clock. However, little is known about how ROS homeostasis is regulated in nonphotosynthetic organs, such as petals. Here, we showed that hydrogen peroxide (H2O2) levels exhibit typical circadian rhythmicity in rose (Rosa hybrida) petals, consistent with the measured respiratory rate. RNA-seq and functional screening identified a B-box gene, RhBBX28, whose expression was associated with H2O2 rhythms. Silencing RhBBX28 accelerated flower senescence and promoted H2O2 accumulation at night in petals, while overexpression of RhBBX28 had the opposite effects. RhBBX28 influenced the expression of various genes related to respiratory metabolism, including the TCA cycle and glycolysis, and directly repressed the expression of SUCCINATE DEHYDROGENASE 1, which plays a central role in mitochondrial ROS (mtROS) homeostasis. We also found that PHYTOCHROME-INTERACTING FACTOR8 (RhPIF8) could activate RhBBX28 expression to control H2O2 levels in petals and thus flower senescence. Our results indicate that the circadian-controlled RhPIF8–RhBBX28 module is a critical player that controls flower senescence by governing mtROS homeostasis in rose.
A circadian-dependent transcription factor module constrains Succinate Dehydrogenase1 expression and the TCA cycle to avoid excessive mitochondrial reactive oxygen species production in petals.
Introduction
Senescence is the final stage in the development of a plant organ and usually leads to programmed death of all constituent cells. Accordingly, senescence is a tightly controlled process during which plants remobilize nutrients from senescing leaves or petals to other developing organs, such as seeds, stems, or roots (van Doorn and Woltering, 2004; Rogers, 2006; Lim et al., 2007; Schippers et al., 2015; Rogers and Munné-Bosch, 2016; Kim et al., 2017). This important and unique process is coordinately regulated by various internal and external factors, including developmental status, reactive oxygen species (ROS) signaling, phytohormones, the circadian clock, and light (Gan and Amasino, 1995; Lim et al., 2007; van Doorn and Woltering, 2008; Rogers, 2013; Liebsch and Keech, 2016; Rogers and Munné-Bosch, 2016; Kim et al., 2017; Ma et al., 2018).
ROS play a key role in regulating plant organ senescence (Apel and Hirt, 2004; van Doorn and Woltering, 2008; Khanna-Chopra, 2012; Guo et al., 2017). Precocious leaf senescence is a typical phenotype seen in the Arabidopsis (Arabidopsis thaliana) constitutive expression of pr genes5 and jungbrunnen1 mutants, which produce excessive amounts of hydrogen peroxide (H2O2) (Jing et al., 2008; Wu et al., 2012). Conversely, age-dependent leaf senescence is delayed in nac with transmembrane motif 1-like4, ARABIDOPSIS A-FIFTEEN knockout, and wrky75 mutants that generate lower than normal amounts of H2O2 (Olsen et al., 2005; Chen et al., 2012; Lee et al., 2012; Guo et al., 2017).
ROS are generated as byproducts of various metabolic processes that occur in organelles with a highly oxidizing metabolic activity or an intense rate of electron flow, such as chloroplasts, mitochondria, peroxisomes, and microbodies in plant cells (Foyer et al., 1994; Apel and Hirt, 2004; Mittler et al., 2004; Navrot et al., 2007). ROS are commonly toxic to cells, although some, such as H2O2, are important regulatory molecules that are broadly involved in multiple cell signaling pathways (Cezary et al., 2018). Therefore, cellular ROS homeostasis must be tightly controlled to protect cells from dysfunction and death.
In leaves, chloroplasts are the main source of ROS, which are generated in chloroplast thylakoids through electron transfer from photosynthetic chain components (Asada, 2006; Foyer and Noctor, 2013; Ishida et al., 2014). Accumulating evidence shows that ROS homeostasis is governed by the circadian clock. The circadian clock is an endogenous timekeeping mechanism that controls a wide range of developmental and metabolic processes in most organisms (McClung, 2006; Edgar et al., 2012). In Arabidopsis leaves, ROS homeostasis and expression of ROS-responsive genes are circadian-regulated (Covington et al., 2008; Lai et al., 2012). For example, their H2O2 levels exhibit a typical diurnal oscillation, with a peak in the afternoon and a trough at midnight, consistent with the expression dynamics of photosynthetic genes (Lai et al., 2012). Mutation in the key circadian clock component gene CIRCADIAN CLOCK ASSOCIATED1 (CCA1) abolishes the rhythmicity of both H2O2 level and the expression of catalase genes, which encode the key enzyme for H2O2 scavenging, indicating that ROS homeostasis is tightly linked to the fluctuation of light-harvesting capacity in leaves (Harmer et al., 2000; Lai et al., 2012). Moreover, leaf senescence is also significantly accelerated in various mutants of circadian clock component genes, including cca1, late elongated hypocotyl, early-flowering 3 (elf3), elf4, and lux arrhythmo, suggesting that disrupting ROS rhythmicity may lead to senescence (Song et al., 2018; Zhang et al., 2018).
Generally, petals are considered to share common evolutionary origins with leaves, although they have very different functions (Friedman et al., 2004; Rogers and Munné-Bosch, 2016). Petals are a specialized form of leaves that have different colors and shapes to attract pollinators while lacking photosynthetic ability (Glover and Martin, 1998). Petal senescence is tightly regulated by various developmental signals and is less affected by environmental stimuli than leaf senescence (Rogers, 2006; van Doorn and Woltering, 2008). As for leaves, a rise in ROS levels and an imbalance in redox status are typical features of petal senescence. Since petals are nonphotosynthetic organs, they generally contain few active chloroplasts (Stubbs and Francis, 1971; Ljubesić et al., 1991; Marano et al., 1993; Juneau et al., 2010; Ohmiya et al., 2014). The main sources of ROS generation in petals are most likely the mitochondria, mainly through electron transfer from components of the respiratory chain (Rhoads et al., 2006; Rogers, 2013), representing a process fundamentally different from that in leaves. Respiration rate continues to increase until flowers wilt (Siegelman et al., 1958), implying that respiration-produced ROS might be involved in petal senescence in rose.
Although ROS is considered to play an important role in petal senescence (Rogers, 2005, 2006, 2012), the regulatory mechanism behind ROS homeostasis in petals has been largely unknown. Here, we show that H2O2 levels in rose (Rosa hybrida) petals follow a circadian rhythm and peak at night, which is consistent with the pattern of respiration rate in petals. Our combined physiological, biochemical, and genetic approaches revealed that a circadian regulatory module consisting of a phytochrome-interacting factor (PIF; RhPIF8) and a B-box gene (RhBBX28) plays critical roles in H2O2 homeostasis in petals and consequent petal senescence. Furthermore, we demonstrated that RhBBX28 directly repressed the expression of SUCCINATE DEHYDROGENASE 1 (RhSDH1), which encodes the central subunit of mitochondrial respiratory complex II. Together, our results establish a regulatory pathway for the circadian-regulated mitochondrial ROS (mtROS) homeostasis in petals, thus providing a better understanding of the underlying mechanism of ROS homeostasis in nonphotosynthetic organs.
Results
ROS accumulation in rose petals exhibits a circadian rhythm pattern
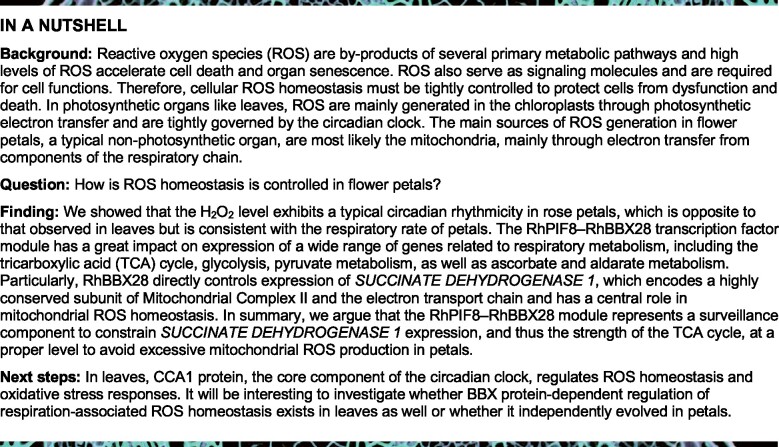
Chloroplasts are present in many petals, albeit in much smaller numbers than in leaves, and we hypothesized that the pattern of ROS production might differ from that in leaves. We, therefore, tested H2O2 levels in petals attached to flowers entrained to 16-h light/8-h dark (LD) photocycles. We discovered that H2O2 production peaked at dawn, Zeitgeber Time (ZT) 0, and dipped in the afternoon at ZT8 (Figure 1A), in a pattern opposite to that seen in leaves (Lai et al., 2012).
Figure 1.
ROS homeostasis exhibits circadian rhythmicity in rose petals. A–C, H2O2 levels in petals of rose flowers entrained under LD conditions (A), LL (B) or DD (C) for 3 days. For LL and DD treatments, flowers were initially grown under LD conditions and then transferred to LL or DD conditions. Experiments were performed independently twice, with similar results. One representative result is shown. Mean values ± sd are shown from three replicates (n = 3). White horizontal bars, day; black horizontal bars, night; gray horizontal bars, subjective day or night. D, Respiration rate of rose flowers in LD conditions. Mean values ± sd are shown (n = 3 for 0, 4, and 24 h, n = 4 for 8, 12, 16, and 20 h). E, Representative flower phenotype (left) and H2O2 accumulation (right) of petals under LL or DD conditions. H2O2 levels were determined by staining with DAB. Scale bars, 1 cm. F, Relative staining intensity of H2O2 accumulation (n = 4), (G) ion leakage (n = 4 for 0 days, n = 6 for 3 days), and (H) relative expression of RhSAG12 (n = 3) in fully opened (Stage 2) rose flower petals grown under LL or DD conditions. Mean values ± sd are shown. Experiments were performed independently 3 times, with similar results. Asterisks represent statistically significant differences (*P < 0.05; **P < 0.01), as determined by ANOVA (A–D) and Student’s t test (F–H).
We next investigated whether the observed pattern persisted in constant light (LL) and dark (DD) conditions by transferring rose plants grown in LD conditions to LL or DD conditions to monitor H2O2 levels in the absence of entraining cues. H2O2 production rhythms in petals persisted in LL and DD conditions as in LD conditions, but were higher in DD than in LL conditions (Figure 1, A–C). In addition, we monitored the respiration dynamic in petals. Notably, the respiration rate of petals also exhibited a circadian rhythm, with the same phase as the pattern of H2O2 production (Figure 1D).
It has been reported that respiration rate of rose petals increases during flower opening and senescence (Siegelman et al., 1958). Our results also demonstrated that H2O2 accumulation continued increasing after the flower began to open (Stage 2; Supplemental Figure S1), consistent with the respiration curve (Siegelman et al., 1958), thus indicating that respiration-produced ROS accumulate as flowers open.
In Arabidopsis, extended darkness triggers leaf senescence, which involves chlorophyll degradation and increases in ROS levels (Song et al., 2014; Liebsch and Keech, 2016). Interestingly, although petals have no chlorophyll, an extended dark treatment of 3 days also promoted petal senescence compared to plants maintained in LL, and this was accompanied by fading of petal color (Figure 1E, left). 3,3′-diaminobenzidine (DAB) staining showed a clear elevation of H2O2 levels in the petals of flowers maintained in DD relative to those of flowers experiencing LL conditions (Figure 1, E [right] and F). Electrolyte leakage from petals significantly increased in response to DD treatment, indicative of damage to biological membranes (Figure 1G). In addition, expression of the rose homolog of SENESCENCE-ASSOCIATED GENE12 (RhSAG12), a typical indicator of senescence (Lv et al., 2014), was also significantly enhanced in DD (Figure 1H), confirming that petal senescence is accelerated in darkness.
RhBBX28 is a key regulator of petal senescence
Since H2O2 levels in petals displayed a circadian rhythm, we speculated that the circadian clock might be involved in regulating H2O2 homeostasis in petals. Transcription of the B-box (BBX) gene family is tightly regulated by light and the circadian clock (Kumagai et al., 2008), and BBX genes, in turn, govern multiple clock-controlled development processes, such as photoperiodic flowering (Andrés and Coupland, 2012; Romera-Branchat et al., 2014; Song et al., 2015). To understand whether BBX genes contribute to H2O2 homeostasis in petals, we first searched our rose transcriptome database (for R. hybrida cv Samantha) (http://bioinfo.bti.cornell.edu/cgi-bin/ rose_454/index.cgi) and identified 17 BBX genes expressed in petals (Supplemental Figure S2A). We noticed that the expression of two BBX genes (RU06829 and RU18177) increased during the late opening stages (Stages 3 to 5) (Supplemental Figure S2A). Therefore, we selected these two genes for further study.
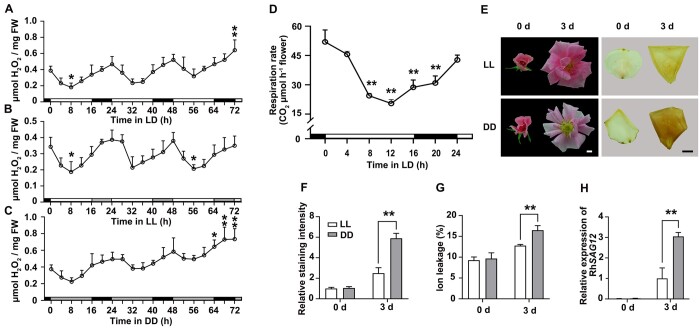
The full-length coding sequence of RU06829 was 989 bp, with a 726-bp open reading frame (ORF), and it was 2,378 bp, with a 1,128-bp ORF for RU18177. Phylogenetic analysis indicated that the predicted proteins of RU06829 and RU18177 show high sequence similarity with Arabidopsis BBX28/29 and BBX7, respectively (Figure 2A; Khanna et al., 2009), and thus we designated RU06829 and RU18177 as RhBBX28 and RhBBX7, respectively.
Reverse transcription-quantitative PCR (RT-qPCR) analysis confirmed that expression of RhBBX28 and RhBBX7 increases as flowers open, and both displayed a diurnal expression pattern in petals. Expression of RhBBX28 reached its peak at dawn (ZT24) and its trough (the lowest point in the oscillation) in the afternoon (ZT8), while RhBBX7 expression peaked at ZT12 and was lowest at ZT4 (Figure 2C; Supplemental Figure S2B). The expression pattern of RhBBX28 in LL and DD conditions was similar to that in LD conditions, although with a generally elevated expression level in DD, further confirming that RhBBX28 expression is regulated by the circadian clock (Figure 2C).
Figure 2.
Characterization of RhBBX28. A, Phylogenetic analysis of RhBBX28, RhBBX7, and BBX genes from Arabidopsis. The phylogenetic tree was constructed using MEGA software version 5.05 with the neighbor-joining algorithm. The scale bar indicates the branch length. RhBBX28 and RhBBX7 were highlighted. B, Effects of silencing of RhBBX28 on petal senescence. RT-qPCR analyses of RhBBX28 transcript levels in petals (upper, left), length of the senescence phase (from fully open flowers to senescence) (upper, right), and flower phenotype (lower) of RhBBX28-silenced and TRV (empty vector)-transformed control plants. Flower phenotypes were recorded starting once flowers were fully opened. Experiments were performed independently 3 times, with similar results. Mean values ± sd are shown from 10 replicates (n = 10) for phenotype and five replicates (n = 5) for gene expression test. Scale bar, 1 cm. C, Relative expression levels of RhBBX28 in petals of rose flowers entrained under LD, LL, or DD conditions for 3 days. For LL and DD treatments, flowers were initially grown under LD conditions and then transferred to LL or DD conditions. Experiments were performed independently twice, with similar results. One representative result is shown. Mean values ± sd are shown from five replicates (n = 5). White horizontal bars, day; black horizontal bars, night; gray horizontal bars, subjective day or night. D, Subcellular localization of RhBBX28-GFP in N. benthamiana leaves. The plasmid ProSuper:RhBBX28-GFP was co-infiltrated with the nuclear marker ProSuper:NF-YA4-mCherry. Green and red fluorescence were visualized by confocal microscopy 3 days after infiltration. Scale bar, 50 μm. E, Transcriptional repressor activity of RhBBX28 in N. benthamiana leaves. Upper, schematic representation of Reporter and Effector constructs. The coding sequence with the stop codon was inserted into the pBD effector vector and driven by the 35S promoter. pBD-VP16 was used as a positive control. Middle and bottom, live imaging (middle) and quantitative analysis (bottom) of transcriptional repression activity of the RhBBX28 protein. Mean values ± sd are shown from three replicates (n = 3). The experiments were performed independently twice, with similar results. Asterisks represent statistically significant differences (*P < 0.05; **P < 0.01), as determined by Student’s t test (B and E) and ANOVA (C).
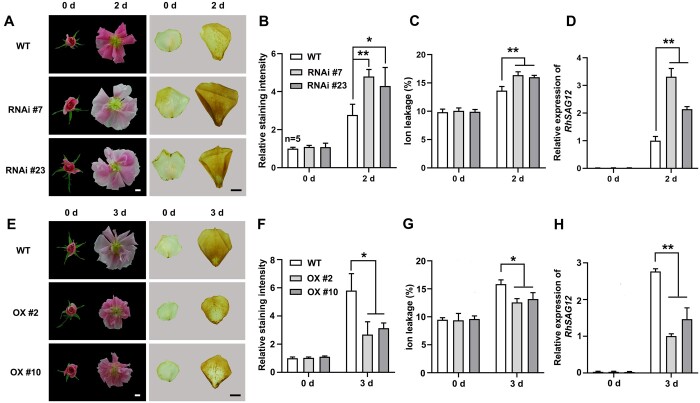
Next, we silenced RhBBX28 or RhBBX7 using a virus-induced gene silencing (VIGS) approach (Liu et al., 2002; Tian et al., 2014) in rose plants. In rose flowers, color fading and wilting of petals are two visible and typical indicators of petal senescence; we, therefore, defined the flower senescence phase as the period from the development of fully open flowers to the emergence of these senescence symptoms (Wu et al., 2017). Silencing of RhBBX28, but not RhBBX7, significantly accelerated petal senescence (Figure 2B; Supplemental Figure S3). Compared to control plants transformed with the Tobacco Rattle Virus (TRV) empty vector (Liu et al., 2002), DD-induced color fading of petals was more severe in RhBBX28-silenced flowers (Supplemental Figure S4, A and B). Similarly, ion leakage and RhSAG12 expression were also significantly higher in RhBBX28-silenced flowers relative to control flowers (Supplemental Figure S4, C and D). These results suggested that RhBBX28 might be involved in regulation of H2O2 accumulation and petal senescence.
A Basic Local Alignment Search Tool for Protein search revealed that the deduced polypeptide of RhBBX28 shares similarity to BBX proteins from a range of plant species. The putative RhBBX28 protein showed structure features of group-V members of the BBX family: a highly conserved single B-box domain in the N-terminus but no CONSTANS (CO), CO-like, and TOC1 (CCT) domain in the C terminus (Khanna et al., 2009; Gangappa and Botto, 2014; Supplemental Figure S5).
Protein structure analysis predicted a nuclear localization signal site in the C-terminus of RhBBX28 (Supplemental Figure S5). Transient co-infiltration of an RhBBX28-GFP construct with a nucleus marker construct (NUCLEAR FACTOR Y ALPHA4 [NF-YA4]-mCherry) in Nicotiana benthamiana leaves demonstrated that RhBBX28 localizes to the nucleus (Figure 2D). To test the transcriptional activity of RhBBX28, we constructed the effector plasmid pBD-RhBBX28 and used pBD-VP16 as a positive control. The dual-luciferase (LUC) reporter harbored the yeast (Saccharomyces cerevisiae) GAL4 DNA-binding element and a minimal cauliflower mosaic virus (35S promoter driving the expression of the firefly LUC reporter gene, with a Renilla (REN) reniformis LUC reporter under the control of the 35S promoter as an internal control (Figure 2E, upper). When co-infiltrated into N. benthamiana leaves with the reporter constructs, pBD-RhBBX28 resulted in significantly lower firefly LUC activity as compared to pBD alone (Figure 2E, lower), suggesting that RhBBX28 is a transcriptional repressor.
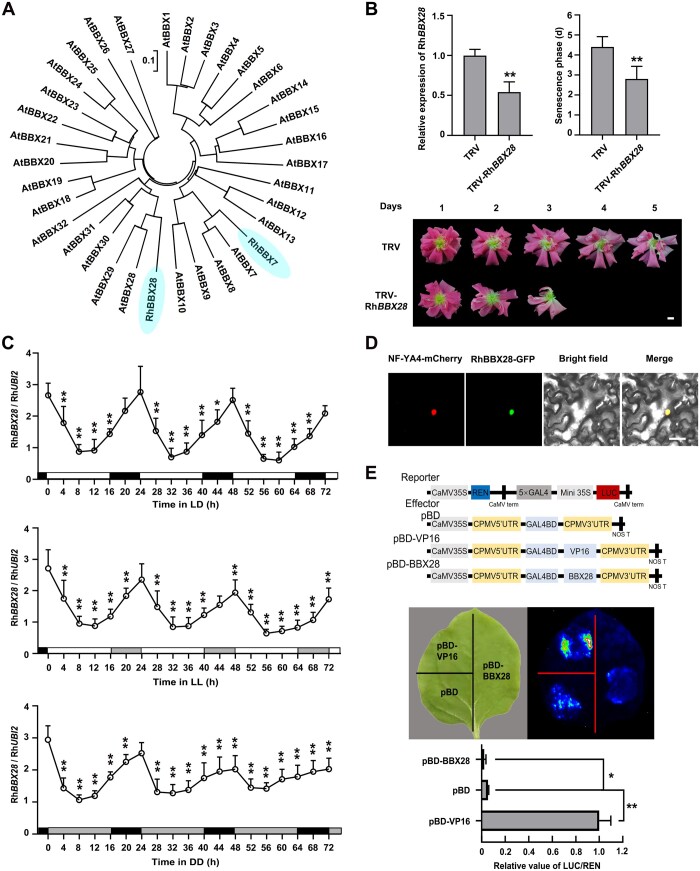
To further explore the function of RhBBX28 in petal senescence, we generated transgenic rose lines silenced for RhBBX28 by RNA interference (RhBBX28-RNAi) or overexpressing RhBBX28 (RhBBX28-OX) through Agrobacterium tumefaciens-mediated transformation of somatic embryos (Liu et al., 2021). RT-qPCR analysis confirmed that RhBBX28 transcript levels were significantly lower in the petals of RhBBX28-RNAi lines, and significantly higher in RhBBX28-OX lines, than in wild-type (WT) plants (Supplemental Figure S4E).
We determined that overexpressing RhBBX28 significantly prolonged the senescence phase (7.00 ± 0.53 day after flower, DAF is fully open for OX #2 and 6.29 ± 0.70 DAF for OX #10) compared to WT plants (4.21 ± 0.43 DAF). On the other hand, the senescence phase was much shorter in RhBBX28-RNAi transgenic lines (2.88 ± 0.33 DAF for RNAi #7 and 3.29 ± 0.45 DAF for RNAi #23) than in WT plants (Figure 3, A and B). In addition, overexpression of RhBBX28 produced smaller flowers with lighter color than in WT, while RhBBX28-RNAi plants had slightly larger flowers (Figure 3B).
Figure 3.
RhBBX28 is a key regulator of petal senescence. A and B, Flower phenotypes (A) and length of the senescence phase (B) of WT, RhBBX28-RNAi, and RhBBX28-OX plants. Flower phenotypes were recorded once flowers were fully opened. Experiments were performed independently 3 times, with similar results. C, D, Relative expression levels of RhBBX28 (C) and H2O2 levels (D) in petals of WT, RhBBX28-OX #10, and RhBBX28-RNAi #23 rose flowers entrained under DD conditions for 3 days. For DD treatments, flowers were initially grown under LD conditions and then transferred to DD conditions. Experiments were performed independently twice, with similar results. One representative result is shown. Mean values ± sd are shown from three replicates (n = 3). White horizontal bars, day; black horizontal bars, night; gray horizontal bars, subjective day or night. Mean values ± sd are shown from seven replicates (n = 7) for each line. RhUBI2 was used as an internal control. Asterisks represent statistically significant differences (*P < 0.05; **P < 0.01), as determined by ANOVA. Scale bar, 1 cm.
The rhythmicity of RhBBX28 was lost in the RhBBX28-OX line, while persisted in the RhBBX28-RNAi with a weakened amplitude relative to the WT (Figure 3C). To test whether RhBBX28 influences H2O2 accumulation, we monitored H2O2 levels in the petals of RhBBX28-RNAi and RhBBX28-OX plants. Notably, the level of H2O2 was elevated in the RhBBX28-RNAi line, especially in the dark period, while it was reduced in the RhBBX28-OX line (Figure 3D).
When WT and RhBBX28-RNAi plants were subjected to DD treatment for 2 days, we observed only slight color fading in the petals of WT flowers, while color fading was much more severe in RhBBX28-RNAi plants (Figure 4A, left). Likewise, DAB-staining revealed significantly higher H2O2 levels in the petals of RhBBX28-RNAi plants than in WT plants (Figure 4, A [right] and B). Electrolyte leakage and RhSAG12 expression in the petals of RhBBX28-RNAi plants were also significantly higher than in WT (Figure 4, C and D). In contrast, as compared to that in WT plants, dark-induced color fading was very mild and restricted to the petal margin in RhBBX28-OX lines (Figure 4E, left). Thus, H2O2 accumulation, electrolyte leakage and RhSAG12 transcript levels in RhBBX28-OX lines were significantly lower than in WT plants (Figure 4, E [right], F–H). Notably, although RhBBX28-OX lines exhibited lighter petal color than RhBBX28-RNAi lines, dark-induced color fading is slighter in RhBBX28-OX lines than in RhBBX28-RNAi lines, suggesting that RhBBX28 was important in dark-induced petal senescence. Moreover, petal senescence was delayed under LL conditions in WT, RhBBX28 OX, and RNAi lines, while senescence of RhBBX28 RNAi lines was still quicker than WT and RhBBX28 OX lines (Supplemental Figure S6). These results supported the hypothesis that RhBBX28 plays an important role in rose flower senescence via governing dark-induced ROS generation.
Figure 4.
RhBBX28 modulates dark-induced petal senescence and H2O2 accumulation. A, Flower phenotypes (left) and H2O2 accumulation (right) in petals of RhBBX28-RNAi and WT plants after 2 days in DD conditions. B, Relative staining intensity of H2O2 accumulation (n = 5 for WT in 0 days, n = 4 for the rest), (C) ion leakage (n = 4), and (D) relative expression of RhSAG12 (n = 3) in petals of RhBBX28-RNAi and WT plants in DD condition. For DD treatments, flowers were initially grown under LD conditions and then transferred to DD conditions. Experiments were performed independently 3 times, with similar results. Mean values ± sd are shown. E, Flower phenotypes (left) and H2O2 accumulation (right) in petals of RhBBX28-OX and WT plants after 3 days in DD conditions. Experiments were performed independently 3 times, with similar results. F, Relative staining intensity of H2O2 accumulation (n = 3), (G) ion leakage (n = 4 for WT and OX #2 in 0 days, and OX #10 in 3 days, n = 3 for the rest), (H) and relative expression of RhSAG12 (n = 3) in petals of RhBBX28-OX and WT plants in DD conditions. H2O2 levels were determined by staining with DAB. RhUBI2 was used as an internal control. Experiments were performed independently 3 times, with similar results. Mean values ± sd are shown. Asterisks represent statistically significant differences (*P < 0.05; **P < 0.01), as determined by Student’s t test. Scale bars, 1 cm.
RhBBX28 governs petal senescence by regulating RhSDH1 expression and consequent H2O2 homeostasis
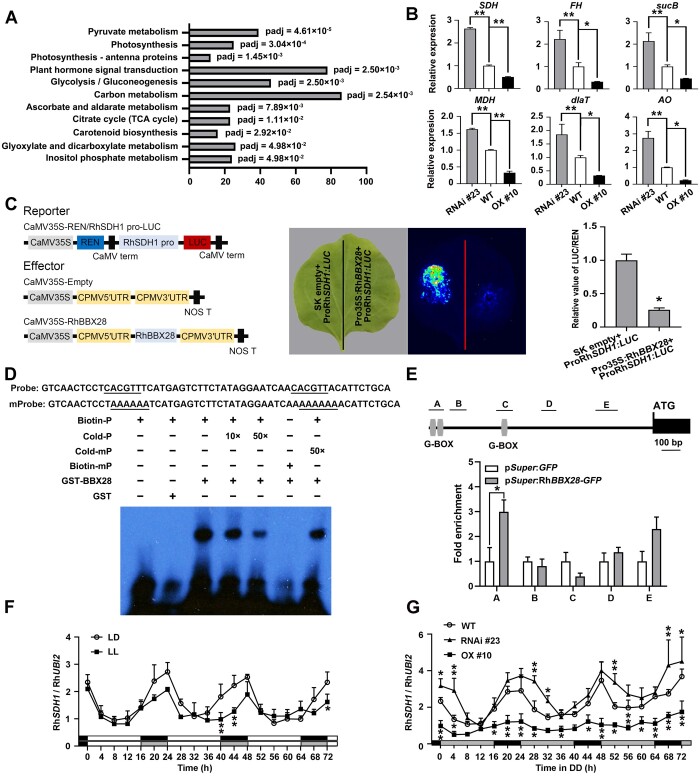
To investigate how RhBBX28 might influence H2O2 homeostasis, we conducted a transcriptome deep sequencing (RNA-seq) analysis to explore the possible regulatory network downstream of RhBBX28 using WT and RhBBX28-OX and -silencing lines. The differentially expressed genes (DEGs) (WT versus OX, fold change ≥2, adjusted P ≤ 0.05) were subjected toKyoto Encyclopedia of Genes and Genomes (KEGG) analysis. Remarkably, we noticed that several terms associated with respiratory metabolisms, such as citrate cycle (TCA cycle), glycolysis, pyruvate metabolism, and ascorbate and aldarate metabolism, were significantly enriched (Figure 5A). We independently confirmed that by RT-qPCR analysis the transcript levels of genes involved in respiratory metabolic pathways and H2O2 homeostasis were elevated in the petals of RhBBX28-RNAi lines, and repressed in RhBBX28-OX lines, including the genes encoding succinate dehydrogenase (RhSDH1), fumarate hydratase (RhFH), dihydrolipoamide succinyltransferase, malate dehydrogenase (RhMDH), dihydrolipoyllysine-residue acetyltransferase, and L-ascorbate oxidase (Figure 5B).
Figure 5.
RhBBX28 influences expression of genes related to respiratory metabolism and ROS homeostasis in rose petals. A, Significantly enriched KEGG pathways in RhBBX28-OX lines based on gene expression analysis. Padj, adjusted P-value of each enriched pathway. B, Expression of genes related to respiratory metabolism and ROS homeostasis in petals of WT, RhBBX28-OX #10, and RhBBX28-RNAi #23 plants. RhUBI2 was used as an internal control. Data are for the genes encoding succinate dehydrogenase (ubiquinone) flavoprotein subunit 1 (SDH, RchiOBHm_Chr2g0101771, and AT5G66760.1), fumarate hydratase 1 (FH, RchiOBHm_Chr6g0294521, and AT2G47510.2), dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex 2 (sucB, RchiOBHm_Chr2g0158941, and AT5G55070.1), malate dehydrogenase (MDH, RchiOBHm_Chr4g0393011, and AT5G43330.1), dihydrolipoyllysine-residue acetyltransferase component 3 of pyruvate dehydrogenase complex (dlaT, RchiOBHm_Chr2g0144451, and AT3G13930.1), and L-ascorbate oxidase homolog (AO, RchiOBHm_Chr6g0279531, and AT1G76160.1). Samples were collected 4 h after lights on (ZT0). Experiments were performed independently twice, with similar results. Mean values ± sd are shown from three biological replicates (n = 3). C, Interaction between RhBBX28 and the RhSDH1 promoter, as shown by a dual LUC reporter system. Left, schematic representation of the constructed dual LUC reporter system. Live imaging (middle) and quantitative analysis (right) of transcriptional repression of the RhSDH1 promoter by RhBBX28. The ProRhSDH1:LUC construct was co-infiltrated with Pro35S:RhBBX28 or SK empty vector into N. benthamiana leaves. Experiments were independently repeated 3 times, with similar results. A representative image of an N. benthamiana leaf 3 days after infiltration is shown. Mean values ± sd are shown from three replicates (n = 3). D, EMSA analysis of RhBBX28 binding to the RhSDH1promoter. The sequence of the region from –1,172 to –1,117 bp of the RhSDH1 promoter was used as a probe. As indicated, RhBBX28-dependent mobility shifts were detected and were competed by unlabeled WT probe in a dose-dependent manner, but not by unlabeled mutated probe. Experiments were performed independently twice, with similar results. For EMSA, the experiments were performed using 1 μg recombinant GST–RhBBX28 proteins mixing with 2 nmol biotin-labeled RhSDH1 probe. E, ChIP-qPCR assay showing RhBBX28 binding to the RhSDH1 promoter in planta. Upper, schematic representation of the RhSDH1 promoter. Black box, RhSDH1 ORF; Gray vertical lines, the putative G-box (−1,162 to −1,157, −1,132 to −1,127, and −889 to −884 bp); Lines above, the fragments amplified in the ChIP-qPCR analysis. A: −1,182 to −1,102 bp, B: −1,076 to −986 bp, C: −939 to −852 bp, D: −591 to −512 bp, E: −297 to −180 bp, relative to the RhSDH1 translation initiation codon (ATG). Lower, ChIP-enrichment of the indicated RhSDH1 promoter fragments (A–E). Petals of RhBBX28 OX #2 line were used for ChIP. The experiment was performed independently twice with similar results and one representative result is shown. G, Relative expression levels of RhSDH1 in petals of WT rose flowers entrained under LD or LL conditions for 3 days. H, Relative expression levels of RhSDH1 in petals of WT, RhBBX28-OX #10 and RhBBX28-RNAi #23 rose flowers entrained under DD conditions for 3 days. For LL and DD treatments, flowers were initially grown under LD conditions and then transferred to LL or DD conditions. Experiments were performed independently twice, with similar results. One representative result is shown. Mean values ± sd are shown from three replicates (n = 3). White horizontal bars, day; black horizontal bars, night; gray horizontal bars, subjective day or night. Asterisks represent statistically significant differences (*P < 0.05, **P < 0.01), as determined by ANOVA (B, F, and G) and Student’s t test (C and E).
We, therefore, isolated the promoters for RhSDH1, RhMDH, and RhFH and constructed firefly LUC reporters (ProSDH1:LUC, ProMDH:LUC, and ProFH:LUC) from each to test whether RhBBX28 might influence the transactivation of these promoters. We co-infiltrated each reporter construct, along with an effector construct consisting of RhBBX28 driven by the 35S promoter (Pro35S:RhBBX28) and REN Luc under the control of the 35S promoter (Pro35S:REN, as an internal control), into N. benthamiana leaves (Figure 5C). Live imaging and quantitative analysis demonstrated that co-infiltration of RhBBX28 with ProSDH1:LUC, but not ProMDH:LUC or ProFH:LUC, significantly repressed the expression of the Luc reporter, as evidenced by lower Luc activity, indicating that RhBBX28 might repress the expression of RhSDH1 (Figure 5C; Supplemental Figure S7A).
To test whether RhBBX28 might directly bind to the RhSDH1 promoter, we first searched for the presence of possible G-box sites recognized by BBX proteins in the RhSDH1 promoter using the PlantCARE online tool (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). We identified three potential sites in the RhSDH1 promoter, located from –1,162 to –1,157 bp, –1,132 to –1,127 bp, and –889 to –884 bp relative to the translation start codon (Supplemental Figure S7B). Next, we generated two biotin-labeled probes corresponding to fragments containing each potential site (from –1,172 to –1,117 bp and –905 to –866 bp) and performed an electrophoretic mobility shift assay (EMSA). RhBBX28 failed to bind to the –905 to –866 bp fragment (Supplemental Figure S7C), but it did bind to the probe corresponding to –1,172 to –1,117 bp (Figure 5D). An unlabeled WT probe, but not an unlabeled mutated probe, competed for binding to RhBBX28 with the biotin-labeled probe, indicating that RhBBX28 specifically binds to this region of the RhSDH1 promoter (Figure 5D). Moreover, we generated truncated RhBBX28 for EMSA to test which part of RhBBX28 protein is responsible for DNA binding. The results supported that B-box domain, but not the C-terminus fragment, was able to bind to the RhSDH1 promoter directly (Supplemental Figure S7, D–F), consistent with a previous report (Song et al., 2020). Chromatin immuno-precipitation qPCR (ChIP-qPCR) confirmed that RhBBX28 was able to bind to –1,172 to –1,117 bp of RhSDH1 promoter in vivo (Figure 5E).
Sequence alignment showed that RhSDH1 contains a highly conserved domain associated with succinate and Flavin Adenine Dinucletide (FAD) binding (Figueroa et al., 2002; Supplemental Figure S8A). Phylogenetic analysis indicated that RhSDH1 encodes a homolog of Arabidopsis SDH1-1 (At5g66760) (Supplemental Figure S8B). Notably, SDH1 is highly conserved in eukaryotes (Huang and Millar, 2013; Huang et al., 2019). SDH1 is the central subunit of the succinate dehydrogenase complex, also known as mitochondrial complex II, the only enzyme to participate in both the TCA cycle and the electron transport chain. Moreover, mitochondrial complex II is a major resource of ROS in nonphotosynthetic organs (Huang et al., 2019).
We monitored the expression pattern of RhSDH1 in WT, RhBBX28 OX, and RNAi lines. The results showed that RhSDH1 also exhibited circadian rhythmicity in petals, and this rhythmicity was abolished RhBBX28 OX lines but persisted in RhBBX28 RNAi lines (Figure 5, F and G). Moreover, expression level of RhSDH1 in RhBBX28 RNAi lines was significantly higher than WT, but that in RhBBX28 OX lines was significantly higher than WT (Figure 5G).
These results suggested that RhBBX28 might influence petal mitochondrial respiration and consequent mtROS homeostasis by regulating the expression level of RhSDH1.
To extend these findings, we tested for a possible role of BBX28 in leaf and flower senescence in Arabidopsis. We did not find a difference in leaf and flower senescence among Col-0, BBX28-OX lines, and the bbx28 mutant (Supplemental Figure S9, A and B). Moreover, ChIP-qPCR failed to detect the in vivo binding of BBX28 to the SDH1-1 promoter (Supplemental Figure S9C). Therefore, when compared with BBX28, RhBBX28 might play a different role in rose.
RhPIF8 controls petal senescence by regulating RhBBX28 expression
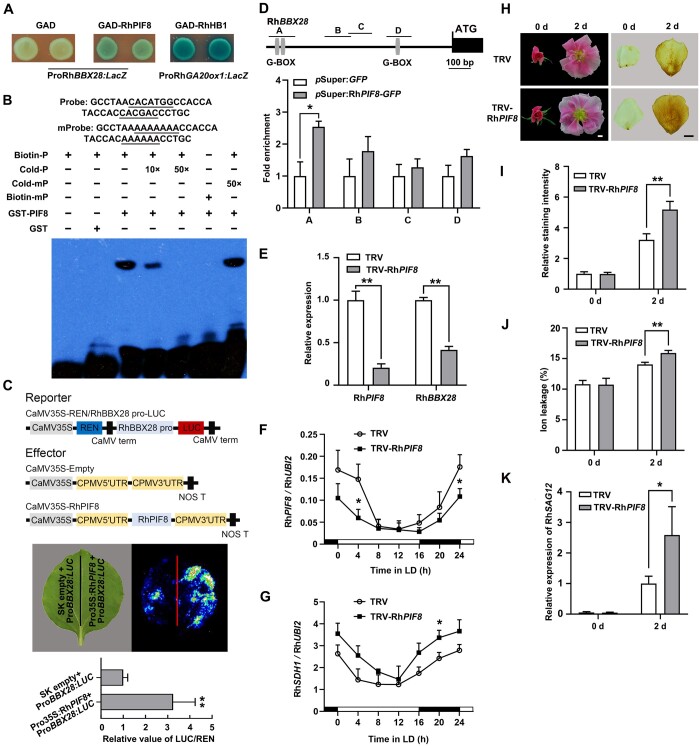
To better understand the regulatory mechanism of RhBBX28, we isolated an 834-bp promoter region upstream of the RhBBX28 coding sequence and looked for cis-regulatory elements using PlantCARE. The RhBBX28 promoter contained two G-box motifs that are specifically recognized by PIF proteins (Supplemental Figure S10A; Martínez-García et al., 2000; Oh et al., 2009; Hornitschek et al., 2012; Zhang et al., 2013; Sakuraba et al., 2014). By screening our rose transcriptomic database, we identified five putative PIF genes which were detectable in rose flowers, and we named RhPIF1, RhPIF3, RhPIF4, RhPIF7, and RhPIF8, based on phylogenetic analysis (Supplemental Figure S10B). Rose petals were collected on ZT4 from flowers at different opening stages, and RT-qPCR analysis revealed that the expression levels of RhPIF4 and RhPIF8 were relatively high among these five RhPIFs (Supplemental Figure S10C). We then investigated the expression pattern of RhPIF4 and RhPIF8 over a 24-h LD cycle. Interestingly, the diurnal expression pattern of RhPIF8, but not of RhPIF4, followed the same pattern seen with RhBBX28 (Supplemental Figure S10D). Therefore, we isolated the 834-bp promoter region upstream from the RhBBX28 coding sequence and tested whether RhPIF8 might bind and activate this promoter through a yeast one-hybrid assay. Indeed, RhPIF8 activated expression of the LacZ reporter when placed under the control of the RhBBX28 promoter, suggesting that RhPIF8 functions as a transactivator of RhBBX28 transcription (Figure 6A).
Figure 6.
RhPIF8 is a transcriptional activator of RhBBX28 and affects H2O2 accumulation in petals. A, Y1H analysis of the binding of RhPIF8 protein to the RhBBX28 promoter. GAD–RhPIF8, but not GAD itself, activates the expression of the lacZ reporter gene driven by the RhBBX28 promoter. GAD–RhHB1 with ProRhGA20ox1:LacZ was used as a positive control. Experiments were performed independently twice, with similar results. B, EMSA analysis of RhPIF8 binding to the G-box motifs in the RhBBX28 promoter. The sequence of the region from –764 to –729 bp of the RhBBX28 promoter was used as a probe. As indicated, RhPIF8-dependent mobility shifts were detected and were competed by unlabeled WT probe in a dose-dependent manner, but not by unlabeled mutated probe. For EMSA, the experiments were performed using 1 μg recombinant GST–RhPIF8 proteins mixing with 2 nmol biotin-labeled RhBBX28 probe. Experiments were performed independently twice, with similar results. C, Transactivation of RhBBX28 by RhPIF8. Upper, schematic representation of dual LUC reporter system. Middle and bottom, live imaging (middle), and quantitative analysis (bottom) of the transcriptional activation of the RhBBX28 promoter by RhPIF8. The ProRhBBX28:LUC construct was co-infiltrated with Pro35S:RhPIF8 or SK empty vector in N. benthamiana leaves. Experiments were independently repeated 3 times. A representative image of an N. benthamiana leaf is shown 3 days after infiltration. Mean values ± sd are shown from four replicates (n = 4). D, ChIP-qPCR assay showing RhPIF8 binding to the RhBBX28 promoter in planta. Upper, schematic representation of the RhBBX28 promoter. Black box, RhBBX28 ORF; Gray vertical lines, the putative G-box (−758 to −752, −739 to −734, and −271 to −266 bp); Lines above, the fragments amplified in the ChIP-qPCR analysis. A: −793 to −680 bp, B: −562 to −448 bp, C: −462 to −357 bp, D: −299 to −192 bp, relative to the RhBBX28 translation initiation codon (ATG). Lower, ChIP-enrichment of the indicated RhBBX28 promoter fragments (A–D). Rose petals expressing pSuper:RhPIF8-GFP were used for ChIP. The experiment was performed independently twice with similar results and one representative result is shown. E, RT-qPCR analysis of RhPIF8 and RhBBX28 transcript levels in petals of RhPIF8-silenced and TRV control plants. Mean values ± sd are shown from three replicates (n = 3). RhUBI2 was used as an internal control. F and G, Relative expression of RhPIF8 (F) and RhSDH1 (G) in petals of RhPIF8-silenced and TRV controls in LD conditions. Mean values ± sd are shown from at least three replicates (n = 4). RhUBI2 was used as an internal control. H, Flower phenotypes (left) and H2O2 accumulation (right) in petals of RhPIF8-silenced and TRV control plants in DD conditions for 2 days. For DD treatment, flowers were initially grown under LD conditions and then transferred to DD conditions. Experiments were performed independently twice, with similar results. One representative result is shown. Scale bars, 1 cm. I, Relative staining intensity of H2O2 accumulation (n = 6 for TRV-RhPIF8 in 0 days, n = 5 for TRV in 2 days, n = 7 for the rest), (J) ion leakage (n = 4 for TRV in 0 days, n = 3 for TRV-RhPIF8 in 0 days, n = 8 for 2 days), and (K) relative expression of RhSAG12 (n = 3) in petals of RhPIF8-silenced and TRV controls in DD conditions. H2O2 levels were determined by staining with DAB. Mean values ± sd are shown. RhUBI2 was used as an internal control. Asterisks represent statistically significant differences (*P < 0.05; **P < 0.01), as determined by Student’s t-test.
Next, we performed an EMSA to test whether RhPIF8 would directly bind to the G-box motif in the RhBBX28 promoter in vitro. We produced and purified recombinant RhPIF8 as a GST fusion protein (GST–RhPIF8) in Escherichia coli Rosetta strain. We detected shifted bands when recombinant GST–RhPIF8 protein was added to a biotin-labeled probe containing the predicted G-box cis-elements (from –764 to –729 bp in the RhBBX28 promoter). The intensity of the shifted band decreased upon addition of an unlabeled competitive probe but not of an unlabeled mutated probe (Figure 6B). These results supported the notion that RhPIF8 can directly bind to the G-box cis-element of the RhBBX28 promoter.
We further conducted dual-LUC reporter assay to test whether RhPIF8 can activate transcription from the RhBBX28 promoter in planta. Accordingly, we co-infiltrated a firefly LUC reporter driven by the RhBBX28 promoter (ProBBX28:LUC) with the effector construct harboring RhPIF8 expressed from the 35S promoter (Pro35S:RhPIF8) and Pro35S:REN, as an internal control in N. benthamiana leaves. Live imaging and measurement of the LUC:REN ratio, which reflects the transcriptional activity of the RhBBX28 promoter in vivo, demonstrated that co-infiltration of RhPIF8 with ProBBX28:LUC markedly activates the expression of the LUC reporter (Figure 6C), indicating that RhPIF8 acts as a transactivator of RhBBX28 transcription. ChIP-qPCR confirmed that RhPIF8 was able to bind to –821 to –708 bp of RhSDH1 promoter in vivo (Figure 6D).
To investigate the potential function of RhPIF8, we silenced RhPIF8 in rose plants by VIGS. RT-qPCR analysis showed that the expression of RhPIF1, RhPIF3, RhPIF4, and RhPIF7 was not affected in RhPIF8-silenced rose petals when compared to the TRV control, indicating that RhPIF8 was specifically silenced (Supplemental Figure S10E). Expression of RhBBX28 was significantly decreased in RhPIF8-silenced rose petals (Figure 6E), supporting the idea that RhPIF8 is a transactivator of RhBBX28. Moreover, expression level of RhSDH1 significantly increased in the RhPIF8-silenced lines (Figure 6, F and G). Upon DD treatment, RhPIF8-silenced plants exhibited a more severe color fading phenotype than TRV controls (Figure 6H, left). In addition, DAB staining indicated that H2O2 levels are significantly elevated in RhPIF8-silenced petals (Figure 6, H [right] and I). Electrolyte leakage and RhSAG12 expression were also significantly higher in the petals of RhPIF8-silenced plants than those of TRV control plants (Figure 6, J and K), which is in agreement with our hypothesis that RhPIF8 participates in the regulation of petal senescence and H2O2 homeostasis, probably through RhBBX28 and RhSDH1.
On the basis of these various data, we propose that the RhPIF8–RhBBX28 module plays a crucial role in petal senescence by regulating the expression of core genes involved in respiratory metabolism and consequent mtROS homeostasis at night (Figure 7).
Figure 7.
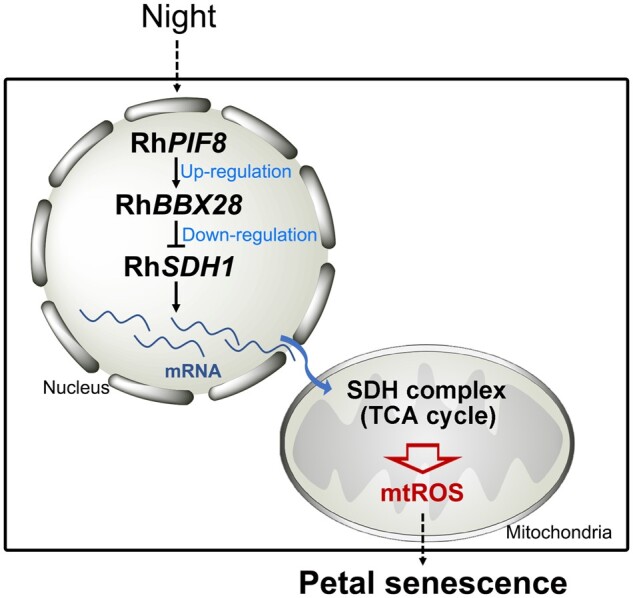
Schematic model of the role of the RhPIF8–RhBBX28 regulatory module in petal senescence. In rose petals, H2O2 levels follow a circadian rhythm, which is consistent with the respiratory rate of petals. The RhPIF8–RhBBX28 module represents a surveillance component to constrain RhSDH1 expression, and thus the strength of the TCA cycle, to a proper level to avoid excessive mtROS production in petals at night.
Discussion
H2O2 homeostasis exhibits a circadian rhythm paralleling respiratory rate changes in rose petals
Here we explored the potential link between circadian rhythm-dependent ROS homeostasis and petal senescence. Our results provide multiple lines of evidence that H2O2 levels follow the extent of mitochondrial respiration in petals and are controlled by a transcriptional cascade consisting of the clock-regulated transcription factors RhPIF8 and RhBBX28.
H2O2 levels in petals were well correlated with the strength of the respiratory rate, supporting the notion that H2O2 is produced mainly through electron transfer from respiratory chain components in rose. Interestingly, a previous report had shown that the growth rate of rose petals exhibits a typical diurnal rhythm. Indeed, when rose plants were entrained to 12-h light/12-h dark cycles, petal growth rate showed a rhythmic pattern, with a trough before the beginning of the dark period (ZT12) and a peak just before the light period (ZT0) (Horibe and Yamada, 2015). This growth pattern matches well with the respiratory rate curve we observed in our present work, indicating that the increase in respiratory rate is required to supply energy and metabolites for petal growth at night. Notably, leaf growth in dicotyledonous species, like petal growth, displays diurnal oscillations, with a maximal growth rate in the middle of the night (Poire et al., 2010). However, ROS levels in leaves remained much lower in darkness than in the light, likely due to the vast amounts of ROS generated from photosynthesis (Fryer et al., 2003).
The PIF8-BBX28 module controls mtROS homeostasis in petals by regulating the expression of genes related to respiratory metabolism
In Arabidopsis, PIF proteins are reported to be regulated by the circadian clock and to participate in various developmental processes regulated by light and the circadian clock, such as seed germination, photomorphogenesis, shading response, and leaf senescence (Yamashino et al., 2003; Nozue et al., 2007; Kidokoro et al., 2009; Niwa et al., 2009; Jeong and Choi, 2013; Leivar and Monte, 2014). PIF4 and PIF5 transcript levels exhibit a circadian rhythm, increasing from noon and reaching their highest levels at dawn (Nozue et al., 2007). LUX, the key component of the circadian clock, can directly bind to the PIF4 and PIF5 promoters (Helfer et al., 2011). Other circadian clock components, including CCA1, ELF3, ELF4, TIMING OF CAB2 EXPRESSION1, PSEUDO-RESPONSE REGULATOR5 (PRR5), and PRR7, also regulate the expression of PIF4 and PIF5 (Yamashino et al., 2003; Nozue et al., 2007; Niwa et al., 2009; Lu and Tobin, 2011; Herrero et al., 2012; Nusinow et al., 2012).
Several PIF genes are involved in ROS homeostasis and leaf senescence. Mutation of PIF1 and PIF3 results in excessive production of singlet oxygen during Arabidopsis de-etiolation, leading to extensive cell death. PIF1 and PIF3 physically interact with LONG HYPOCOTYL5 (HY5) and HY5 HOMOLOG to regulate the expression of several ROS-related genes, such as ASCORBATE PEROXIDASE2 and ZAT10 (Chen et al., 2013), by directly binding to their promoters. Mutation of PIF4 attenuates dark-induced accumulation of H2O2 and expression of ROS-responsive genes (Song et al., 2014). Our results indicate that in rose petals, RhPIF8 expression also displays diurnal oscillations. Here, we determined that silencing of RhPIF8 increases dark-induced H2O2 accumulation and accelerates petal senescence. In addition, RhPIF8 activates the transcription of RhBBX28 by binding directly to its promoter. In Arabidopsis, PIF1 and PIF3 can directly bind to the BBX23 promoter and activate BBX23 expression to modulate the expression of light-responsive and photomorphogenesis-related genes (Zhang et al., 2017). Whether more PIF-BBX transcription factor cascades play distinct functions in plant development should be explored in the future.
In Arabidopsis, BBX28 and BBX29, the two closest putative orthologs of RhBBX28, are reported to function in photomorphogenesis and flowering in a light-depended manner. In the dark, the E3 ubiquitin ligase CONSTITUTIVELY PHOTOMORPHOGENIC1 targets both BBX28 and BBX29 for degradation through the 26S proteasome. In the light, both proteins accumulate and interact with HY5, consequently compromising the binding and transcriptional activation potential of HY5 on its target genes, such as FAR-RED ELONGATED HYPOCOTYL1, CHALCONE SYNTHASE, HY5, BBX30, and BBX31 (Lin et al., 2018; Song et al., 2020). In addition, BBX28 can interact with CO and interferes with its recruitment to the FLOWERING LOCUS T locus; thus, BBX28 plays a negative role in flowering in LD conditions (Liu et al., 2020). Interestingly, BBX28 seems to barely influence leaf or flower senescence in Arabidopsis, implying BBX28 might play a different role in plant development when compared with RhBBX28.
In rose petals, RNA-seq analysis showed that RhBBX28 affects a wide range of biological processes that are responsible for ROS generation in mitochondria, such as the TCA cycle, glycolysis, and pyruvate metabolism, as well as ascorbate and aldarate metabolism. Notably, genes encoding several key enzymes in the TCA cycle (Isocitrate Dehydrogenase (IDH) and SDH) and glycolysis (pyruvate kinase), as well as mitochondrial proteins, have been reported to exhibit daily rhythms in Arabidopsis (Wijnen and Young, 2006; Lee et al., 2010). Moreover, diurnal rhythms of respiration have been identified in Chlamydomonas reinhardtii, a reference organism of unicellular green algae (Strenkert et al., 2019). Therefore, respiratory metabolism might be under diurnal and/or circadian control. Metabolite profiling also demonstrated that mutation of Arabidopsis PRR5, PRR7, and PRR9, core components of the circadian clock, results in a substantial increase of TCA cycle intermediates and antioxidant vitamins (ascorbate and α-tocopherol), indicating that the circadian clock maintains central metabolism in mitochondria (Fukushima et al., 2009). Our results support the idea that RhBBX28 directly binds to the promoter of RhSDH1, which encodes a highly conserved subunit of the SDH complex.
SDH, also known as mitochondrial complex II in eukaryotes, is a significant source of mtROS production in both mammalian and plant systems (Gleason et al., 2011; Quinlan et al., 2012; Jardim-Messeder et al., 2015). The SDH complex oxidizes succinate to fumarate and is the common only component of both the TCA cycle and the electron transport chain. Naturally, the SDH complex is indispensable for almost all multicellular organisms, and deficiency and/or mutation of SDH in mammals causes severe defects, such as Leigh syndrome, mitochondrial encephalopathy, optic atrophy, and even lethality at the embryonic stage (Cecchini, 2003; Yankovskaya et al., 2003; Rutter et al., 2010; Renkema et al., 2015; Moosavi et al., 2019). The eukaryotic SDH complex consists of four classical subunits: a flavoprotein, an iron–sulfur (Fe–S) protein, and two small integral membrane proteins. In flowering plants, four additional subunits have been identified (Eubel et al., 2003; Millar et al., 2004). The flavoprotein and the Fe–S protein are highly conserved across eukaryotes, while the integral membrane proteins are greatly diverged in plants, fungi, and mammals (Cecchini, 2003; Huang and Millar, 2013).
Interestingly, compared to other mitochondrial complexes such as complex I, SDH is more abundant in nonphotosynthetic tissues, such as roots and flowers. Mutation of complex I usually affects growth of both shoots and roots (Meyer et al., 2009; Koprivova et al., 2010; Poire et al., 2010), while sdh mutants exhibit normal or even better growth of shoots. In Arabidopsis, for instance, the flavoprotein is encoded by two highly conserved genes, SDH1-1 (At5g66760) and SDH1-2 (At2g18450). Heterozygous SDH1-1/sdh1-1 plants (with a ∼30% reduction in SDH activity) displayed enhanced CO2 assimilation rates and improved growth in nitrogen-limiting conditions, possibly through increased stomatal conductance (Fuentes et al., 2011). However, some level of SDH activity is necessary under other growth conditions, as a point mutation of SDH1-1 with reduced SDH activity, disrupted in stress responses1, abrogated salicylic acid-induced mitochondrial H2O2 production in roots and caused higher susceptibility to Pseudomonas syringae pv. tomato DC3000 and fungal pathogens (Alternaria brassicicola and Rhizoctonia solani) (Gleason et al., 2011).
In addition to roots, SDH1-1, SDH3, and SDH4, but not SDH1-2 and SDH2-3, are highly expressed in flowers (Figueroa et al., 2001, 2002; Elorza et al., 2004). Loss-of-function mutation of SDH1-1 leads to failure of both male and female gametophyte development as well as seed abortion, indicating that microspore SDH activity is essential for proper pollen development (León et al., 2007). This phenotype might be due to the very high energy requirement during pollen development for the biosynthesis of proteins and other macromolecules (Tadege and Kuhlemeier, 1997; McCormick, 2004). Here, we discovered that RhSDH1, a homolog of Arabidopsis SDH1-1, may be involved in petal senescence of roses. RhSDH1 is a direct target of RhBBX28 whose expression is repressed by the transcription factor. In agreement with this observation, RhSDH1 expression significantly increased in RhBBX28-RNAi lines and decreased in RhBBX28-OE lines. In Arabidopsis leaves, expression of SDH1-1 is much higher in the dark than in the light, which is consistent with the dynamics of mitochondrial respiration rate (Popov et al., 2007, 2010). In addition, the light-mediated reduction of SDH1-1 levels is specifically dependent on the red/far-red photoreceptor phytochrome A (Popov et al., 2010). Here, our results showed that RhPIF8, a PIF, affects RhSDH1 expression indirectly by activating RhBBX28 transcription. Considering that silencing of RhBBX28 shortened petal longevity and enhanced H2O2 accumulation in the dark, we speculate that RhBBX28 might represent a surveillance component acting to constrain RhSDH1 expression, and thus the strength of the TCA cycle, to an appropriate level to avoid excessive mtROS production at night.
Interestingly, the trade-off between energy expenditure and the production of free radicals in mitochondria has attracted much attention for the determination of lifespan (Harman, 1958; Balaban et al., 2005). In worms and mammals, accumulating evidence indicates that dietary caloric restriction and a moderate reduction of TCA-based metabolism will increase lifespan (Weir et al., 2017). In wheat (Triticum aestivum), application of bixafen, an inhibitor of SDH, also significantly delayed senescence (Berdugo et al., 2012, 2013). Additional studies will be required to clarify whether plants and animals share similar mechanisms to balance TCA metabolism and senescence (in plants) or lifespan (in animals).
Materials and methods
Plant materials and growth conditions
Rose (Rosa hybrid cv. Samantha) plantlets used in this study were propagated by in vitro culture as described previously (Wu et al., 2017; Zhang et al., 2019). Rose stems with at least one node were cultured on propagation medium (4.4 g L−1 Murashige and Skoog [MS] salts, 30 g L−1 sucrose, 1.0 mg L−1 6-BA, 0.05 mg L−1 1-Naphthylacetic Acid (NAA), 3.0 mg L−1 GA3, pH 6.0, and 6 g L−1 agar) for 30 days, and were then transferred to rooting medium (4.4 g L−1 MS salts, 0.1 mg L−1 NAA, pH 6.0, and 7 g L−1 agar) for 30 days. Rooted plants were transplanted into pots (9-cm diameter) containing a mixture of 1:1 (v/v) nutritive soil and vermiculite and grown at 22 ± 1°C, ∼50% relative humidity, and 100 μmol m−2 s−1 illumination with fluorescent lamps (SINOL, SN-T5, and 16 W) under a 16-h light/8-h dark cycle.
For long-day (LD, 16-h light/8-h dark) treatments, rose plantlets were grown on soil at 22 ± 1°C, ∼50% relative humidity, and 100 μmol m−2 s−1 illumination with fluorescent lamps. For LL or DD treatment, rose plantlets were grown in LD conditions until Stage 2 of flower development (Ma et al., 2005) and were then transferred to LL conditions. DD treatment was performed as described previously (Keech et al., 2007) with modifications, with flowers wrapped in aluminum foil for continuous dark treatment. For LL treatment, the flowers were also wrapped in aluminum foil, but the top was replaced with transparent plastic wrap to allow light to pass through.
Arabidopsis thaliana and N. benthamiana plants were grown in a mixture of vermiculite and nutritive soil (1:1) at 23 ± 1°C with ∼50% relative humidity, and grown under a LD condition (100 μmol m−2 s−1 with fluorescent lamps, 16-h light/8-h dark). For inducing senescence of detached leaves or inflorescences in darkness, the fourth and fifth rosette leaves from 3-week-old Arabidopsis plants or inflorescences from the primary bolts of 5-week-old were excised and placed in petri dishes for 3–5 days (Trivellini et al., 2012; Sakuraba et al., 2014).
DAB staining and H2O2 measurements
For in situ detection of H2O2, DAB was dissolved in deionized water and the pH of the solution was adjusted to pH 3.8 with HCl. Petals were infiltrated by vacuum in 1 mg mL−1 DAB staining solution and were then incubated in the dark for 8 h. Stained petals were transferred to bleaching solution (ethanol:acetic acid:glycerol [3:1:1, v/v/v]) for 3 days. The DAB staining picture was converted to a grayscale image with Photoshop CS6 software, and image intensity was then determined with ImageJ software (Wu et al., 2015).
Determination of H2O2 concentration was performed using the Amplex Red H2O2/Peroxidase Assay Kit (Invitrogen), according to the manufacturer’s instructions with minor modifications. Briefly, petals were frozen in liquid nitrogen and ground into a fine powder; 30 mg of each sample was fully suspended in 200 μL H2O2 extraction buffer (25 mM sodium phosphate buffer, pH 6.5). The extracts were centrifuged at 13,000 rpm for 15 min at 4°C, and the supernatant was prepared for quantification (Guo et al., 2017). Fluorescence was then measured with a Thermo Scientific Varioskan Flash reader using excitation at 530 nm and fluorescence detection at 590 nm.
Ion leakage measurements
Ion leakage was determined as described previously (Lv et al., 2014). Briefly, rose petal discs were collected at each time point. For each treatment, petal discs were placed in a 50-mL tube containing 20 mL of deionized water and incubated at 25°C for 30 min on an orbital shaker at 200 rpm. Initial conductivity of the fluid was measured with a conductivity meter (FiveGo F3; Mettler-Toledo International Inc., Columbus, OH, USA). Petal discs were then boiled for 15 min in deionized water and cooled to room temperature. Total conductivity was determined as above, and relative electrolyte leakage was calculated as the percentage of initial conductivity versus total conductivity.
RNA extraction and deep RNA-seq
Total RNA was extracted from rose (R. hybrida cv Samantha) petals using the hot borate method (Zhang et al., 2019). RNA integrity was assessed using the RNA Nano 6000 Assay Kit on a Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA). For cDNA library construction, 1 µg total RNA per sample was used. Libraries were sequenced on an Illumina platform by Beijing Novogene Bioinformatics Technology Co., Ltd (http://www.novogene.com/). RNA-seq data were processed, assembled, and annotated as previously described (Zhang et al., 2019). The DEGs (WT versus OX, fold change ≥2, adjusted P ≤ 0.05) were subjected to KEGG analysis.
RT-qPCR
Reverse transcription was performed with 1 μg total RNA using HiScript II Q Select RT SuperMix for qPCR (+gDNA wiper) (Vazyme, Nanjing, China). RT-qPCR reactions (20 μL volume containing 1 μL [20 ng] cDNA as the template) were performed using the StepOne Plus real-time Polymerase Chain Reaction (PCR) system (Applied Biosystems, Foster City, CA, USA) in standard mode with the KAPA SYBR FAST qPCR kit (Kapa Biosystems, Wilmington, MA, USA). RhUBI2 shows stable expression levels during rose flower opening and was used as an internal control (Meng et al., 2013). All experiments were performed independently 3 times. PCR primers are listed in Supplemental Table S1.
VIGS
For VIGS, the engineered vectors pTRV1 and pTRV2 originated from TRV were used (Liu et al., 2002). Gene-specific fragments of RhBBX28 (438 bp), RhBBX7 (295 bp), and RhPIF8 (492 bp) were separately inserted into the pTRV2 vector to construct the pTRV2-RhBBX28, pTRV2-RhBBX7, and pTRV2-RhPIF8 plasmids, respectively. VIGS-mediated silencing of RhBBX28, RhBBX7, and RhPIF8 was performed as described previously in rose plantlets (Wu et al., 2017; Zhang et al., 2019). The pTRV1, pTRV2, pTRV2-RhBBX28, pTRV2-RhBBX7, and pTRV2-RhPIF8 vectors were transformed individually into A. tumefaciens strain GV3101. Single colonies were cultured in Luria-Bertani medium containing 50 mg L−1 kanamycin and 50 mg L−1 rifampicin. Agrobacterium tumefaciens cells were harvested by centrifugation at 5,000 rpm for 10 min, and re-suspended in infiltration buffer (10 mM 2-(N-morpholino)-ethanesulfonic acid, 10 mM MgCl2, 0.2 mM acetosyringone, pH 5.6) to a final optical density at 600 nm of ∼1.0. Mixtures of A. tumefaciens cultures containing an equal ratio (v/v) of pTRV1 and pTRV2 (TRV control), pTRV1 and pTRV2-RhBBX28, pTRV1 and pTRV2-RhBBX7, or pTRV1 and pTRV2-RhPIF8 were placed in the dark at room temperature for 3 h before vacuum infiltration. Rose plantlets were submerged in infiltration buffer with A. tumefaciens colonies harboring pTRV1 and pTRV2, pTRV1 and pTRV2-RhBBX28, pTRV1 and pTRV2-RhBBX7, or pTRV1 and pTRV2-RhPIF8 (Optical Density (OD)600 = 0.8) and exposed to a vacuum of –25 KPa twice, each time for 60 s. After infiltration, plantlets were briefly washed with deionized water and planted in pots for subsequent experiments. Infiltrated plants were grown at 22 ± 1°C with relative humidity (∼60%) and 16-h light/8-h dark photoperiod. The flowers of all rose plantlets were observed and photographed when indicated. Three independent experiments were performed with at least 60 plantlets in each experiment. Primer sequences used for plasmid construction are listed in Supplemental Table S1.
Subcellular localization
The coding sequence of RhBBX28 was fused to GFP and inserted into the pCAMBIA1300 vector harboring a Super promoter to construct the pSuper:RhBBX28-GFP vector (Gong et al., 2002). pSuper:NF-YA4-mCherry was used as a nuclear marker (Zhang et al., 2019). The vectors were transformed into A. tumefaciens strain GV3101, respectively, and co-infiltrated into N. benthamiana leaves. After 3 days, the leaves epidermal cells were monitored with laser confocal fluorescence microscopy (Olympus FluoView FV1000). The primers used in subcellular localizations are listed in Supplemental Table S1.
Dual-LUC reporter assay
For transcription activity analysis, the coding sequence (with the stop codon) of RhBBX28 was inserted into pBD (Sainsbury et al., 2009). pBD-VP16 was used as a positive control and the empty vector pBD-empty was used as a negative control according to Han et al. (2016).
For the transactivation assay of RhPIF8 to the RhBBX28 promoter, the 834-bp promoter sequence of RhBBX28 was inserted into pGreenII 0800-LUC vector upstream to the LUC gene to construct ProRhBBX28:LUC reporter plasmid. Similarly, the 1,182 bp of RhSDH1 promoter, 747 bp of RhMDH promoter, and 1,218 bp of RhFH promoter were used to construct ProRhSDH1:LUC, ProRhMDH:LUC, and ProRhFH:LUC reporter plasmids, respectively.
The coding sequence of RhPIF8 and RhBBX28 was inserted into the pGreenII 62-SK vector to construct Pro35S:RhPIF8 and Pro35S:RhBBX28 effector plasmids, respectively. The constructs were transformed into A. tumefaciens strain GV3101 harboring the pSoup plasmid, respectively. The transformed A. tumefaciens lines were cultured in Luria-Bertani medium with selection antibiotics. The cultures were harvested by centrifugation at 5,000 rpm for 10 min, and re-suspended in infiltration buffer (10 mM 2-(N-Morpholino)-ethanesulfonic acid, 10 mM MgCl2, 0.2 mM acetosyringone, pH 5.6) to a final optical density at 600 nm of ∼1.0. Nicotiana benthamiana plants with 4–5 young leaves were used for co-infiltration with Pro35S:RhPIF8 and ProRhBBX28:LUC, Pro35S:RhBBX28 and ProRhSDH1:LUC, Pro35S:RhBBX28 and ProRhMDH:LUC, Pro35S:RhBBX28 and ProRhFH:LUC, respectively. At 3 days after infiltration, the ratios of LUC to REN were measured using the dual-LUC assay reagents (Promega, Madison, WI, USA) on a GLO-MAX 20/20 luminometer (Promega; Guo et al., 2017; Wei et al., 2017). Images of LUC signals were captured by a CDD camera (CHEMIPROHT 1300B/LND, 16 bits; Roper Scientific, Sarasota, FL, USA). Primer sequences used for DLR are listed in Supplemental Table S1.
Stable transformation of roses
Agrobacterium tumefaciens of strain EHA105 harboring the pSuper:RhBBX28-GFP or pFGC1008-RhBBX28 vector were used to transform rose plants (R. hybrida cv. Samantha) by A. tumefaciens-mediated transformation with modifications (Liu et al., 2021). Leaflets with petioles of tissue-cultured rose plants were used as explants and were placed with their abaxial side up on Somatic embryo Induction Medium (MS salts + 3.0 mg L−1 2,4-D + 0.05 mg L−1 kinetin + 60 g L−1 glucose + 20 g L−1 mannitol + 2.5 g L−1 gelrite) in the dark at room temperature to induce somatic embryos. For A. tumefaciens-mediated transformation, the pSuper:RhBBX28-GFP or pFGC1008-RhBBX28 vectors were introduced into A. tumefaciens strain EHA105. Agrobacteria were grown overnight with appropriate antibiotics and then collected by centrifugation. Cell pellets were re-suspended in infiltration solution (MS salts + 45 g L−1 glucose + 100 µmol L−1 acetosyringone) and the absorbance was adjusted to OD600 = 0.5. The bacterial suspension was incubated for 2 h at 28°C with shaking. Infiltration was performed by immersing somatic embryos in the bacterial suspension followed by incubation for 40 min at 28°C with shaking (180 rpm). Somatic embryos were then briefly washed and dried on sterilized paper towels to remove excess bacteria. Somatic embryos were subsequently grown on Co-cultivation Medium (MS salts + 1.0 mg L−1 2,4-D + 0.05 mg L−1 6-BA + 60 g L−1 glucose + 2.5 g L−1 gelrite + 100 mg L−1 casein hydrolysate + 100 μmol L−1 acetosyringone) in the dark at 22 ± 1°C for 4 days. After co-cultivation, somatic embryos were transferred to Selection and Germination Medium (half-strength MS salts + 1.0 mg L−1 6-BA + 0.01 mg L−1 NAA + 0.5 mg L−1 GA3 + 300 mg L−1 cefotaxime + 30 mg L−1 hygromycin B + 100 mg L−1 casein hydrolysate +30 g L−1 glucose + 2.5 g L−1 gelrite + 20 g L−1 mannitol) and incubated at 22 ± 1°C under a 16-h light/8-h dark photoperiod. Resistant buds were transferred to Rooting Medium (half-strength MS salts +1.0 mg L−1 NAA + 30 mg L−1 hygromycin B + 30 g L−1 sucrose + 7.5 g L−1 gelrite) to induce roots. Rooted plants were transferred to pots containing a 1:1 volume of peat moss and vermiculite and grown at 22 ± 1°C with relative humidity ∼60% under a 16-h light/8-h dark photoperiod. Hygromycin-resistant transformants were screened by PCR and RT-qPCR to validate authentic transgenic plants. Primer sequences used for plasmid construction are listed in Supplemental Table S1.
EMSAs
Oligonucleotide probes were synthesized and labeled with biotin, while unlabeled DNA of the same sequence was used as a competitor. EMSA was performed using the Light Shift Chemiluminescent EMSA Kit (Thermo Fisher, Waltham, MA, USA) according to the manufacturer’s instructions with minor modifications. Briefly, GST–RhPIF8, GST–RhBBX28, GST–RhBBX28-BBOX, or GST–RhBBX28-CEND (C-terminus) was introduced into E. coli strain Rosetta. The bacteria were grown to OD600 = 0.6 at 37°C and then incubated at 16°C for 8 h. Protein expression was induced with isopropyl thio-β-d-galactoside (Sigma, St Louis, MO, USA) to a final concentration of 0.2 mM. Recombinant GST–RhPIF8, GST–RhBBX28, GST–RhBBX28-BBOX, or GST–RhBBX28–CEND fusion proteins were purified with glutathione Sepharose 4B beads (GE Healthcare, Chicago, IL, USA) following the manufacturer’s instructions, then 1-μg fusion proteins was incubated with 2 nM biotin-labeled probe in binding buffer (2.5% glycerol, 50 mM KCl, 5 mM MgCl2, and 10 mM EDTA) at 25°C for 25 min. Competition experiments were performed by adding unlabeled DNA probes. Samples were then loaded onto a 5% polyacrylamide gel and subjected to electrophoresis in 0.5 × Tris borateEthylene Diamine Tetraacetic Acid (EDTA) buffer until the bromophenol blue dye had migrated approximately three-quarters of the way down the length of the gel. Primer and probe sequences used for EMSA are listed in Supplemental Table S1.
ChIP-qPCR assay
The ChIP-qPCR assay was performed as previously described (Cheng et al., 2021). For binding of RhBBX28 to the RhSDH1 promoter, the petals of RhBBX28-OX # 2 line were used. For binding of BBX28 to the SDH1 promoter, the leaves of 2-week-old YFP-BBX28 OX Arabidopsis were used. For binding of RhPIF8 to the RhBBX28 promoter, pSuper:RhPIF8-GFP construct was introduced into A. tumefaciens of strain GV3101, and the transformed A. tumefaciens were incubated to OD600 = 1.5. The A. tumefaciens were collected by centrifugation and were re-suspended in infiltration solution (10 mM 2-(N-morpholino)-ethanesulfonic acid, 10 mM MgCl2, 0.2 mM acetosyringone, pH 5.6). The A. tumefaciens suspension was adjusted with infiltration solution to an OD600 = 1.0 and was incubated for 3 h at room temperature in dark. Then the A. tumefaciens suspension was infiltrated into rose petals. The infiltrated petals were incubated on wet filter paper for 3 days and then were used. For ChIP, 2 g of rose petals or Arabidopsis leaves were cross-linked using 1% (v/v) formaldehyde. The chromatin complexes were fragmented by sonication and incubated with anti-GFP (AE012; ABclonal, Wuhan, China) for 6 h. After washing, the eluted samples were incubated at 65°C for 6 h to reverse the cross-linking. The co-precipitated DNA was purified and analyzed using qPCR. The primers used are listed in Supplemental Table S1.
Yeast one-hybrid assay
Yeast one-hybrid assays were performed as previously described with minor modifications (Pei et al., 2013; Wu et al., 2017). Briefly, pGAD–RhPIF8 was co-transformed with ProRhBBX28:LacZ reporter into yeast strain EGY48 according to the Yeast Protocols Handbook (Clontech). Transformants were grown on synthetic defined media minus Ura and Trp plates containing X–gal (5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside) for blue color development. GAD-RhHB1 with ProRhGA20ox1:LacZ was used as a positive control (Lv et al., 2014). Primer sequences used for yeast one-hybrid are listed in Supplemental Table S1.
Sequence analysis and phylogenetic analysis
The amino acid sequences were aligned using the ClustalW (https://www.genome.jp/tools-bin/clustalw) with default parameters. The phylogenetic tree was constructed based on the alignment result using the Neighbor-joining method in MEGA version 5.05 software (Tamura et al., 2011) using 1,000 bootstrap replicates and the following parameters: Poisson correction, pairwise deletion, and uniform rates. The alignments used to construct the phylogenetic tree are provided as Supplemental Files S1–S3.
Statistical analyses
The statistical significance of the data was determined by GraphPad Prism version 8.0 (GraphPad Software Inc., San Diego, CA, USA: http://www.graphpad.com/). Comparisons between two groups of data were calculated by Student’s t test orAnalysis Of Variance (ANOVA) (*P < 0.05; **P < 0.01). Statistical data are provided in Supplemental File S4.
Accession numbers
Rose gene sequences from this article can be found in GenBank (http://www.ncbi.nlm.nih.gov) under the following accession numbers: RhBBX28 (MT813030) and RhPIF8 (MT813031).
Data availability
Transcriptome raw sequence data were deposited in the NCBI Sequence Read Archive database under accession number SRR12329579-SRR12329584 and SRR12329600-SRR12329605.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. H2O2 levels in petals during rose flower opening and senescence.
Supplemental Figure S2. Expression pattern of the BBX gene family in rose petals.
Supplemental Figure S3. Effects of silencing of RhBBX7 on flower senescence.
Supplemental Figure S4. Silencing of RhBBX28 promotes dark-induced petal senescence and H2O2 accumulation in roses.
Supplemental Figure S5. Alignment analysis of deduced amino acid sequences of RhBBX28 with BBX proteins of subfamily I from Arabidopsis thaliana.
Supplemental Figure S6. Knockdown of RhBBX28 accelerates petal senescence and H2O2 accumulation even in LL conditions.
Supplemental Figure S7. Analysis of direct targets of RhBBX28.
Supplemental Figure S8. Alignment analysis and phylogenetic tree of the deduced amino acid sequences of RhSDH1.
Supplemental Figure S9. BBX28 barely affects leaf and flower senescence and fails to bind to the promoter of SDH1-1 in Arabidopsis.
Supplemental Figure S10. Phylogenetic analysis and expression pattern of RhPIF genes.
Supplemental Table S1. List of primers used in this study
Supplemental Files S1–S3. Alignments used to generate phylogenetic trees.
Supplemental File S4. Summary of statistical tests.
Supplementary Material
Acknowledgments
We thank Dr Jiren Chen (Hunan Agricultural University), Dr Guogui Ning (Huazhong Agricultural University) for his generous help with generation of transgenic rose plants. We thank Dr Fang Lin (Lanzhou University) for providing the seeds of bbx28 mutant and BBX28-OX lines. We thank Dr Wangjin Lu (South China Agricultural University) for providing the pBD, pBD-VP16, and dual-reporter vector.
Funding
This work was supported by the National Key Research and Development Program (Grant 2018YFD1000400), the National Natural Science Foundation of China (grant numbers 31730079 and 31522049), the Construction of Beijing Science and Technology Innovation and Service Capacity in Top Subjects (CEFF-PXM2019_014207_000032), and the 111 Project (B17043).
Conflict of interest statement. The authors declare no conflict of interest.
Y.Z., Z.W., M.F., J.C., M.Q., W.W., Y.B., Q.X., and Y.Y. performed the experiments. N.M. and J.G. designed the research. C.M, C.J., S.G., H.Z., Y.C., B.H., and N.M. provided technical support, conceptual advice, and data analysis. Y.Z., Z.W., N.M., and J.G. wrote the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Nan Ma (ma_nan@cau.edu.cn).
References
- Andrés F, Coupland G (2012) The genetic basis of flowering responses to seasonal cues. Nat Rev Genet 13: 627. [DOI] [PubMed] [Google Scholar]
- Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Asada K (2006) Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol 141: 391–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban RS, Nemoto S, Finkel T (2005) Mitochondria, oxidants, and aging. Cell 120: 483–495 [DOI] [PubMed] [Google Scholar]
- Berdugo CA, Mahlein AK, Steiner U, Dehne HW, Oerke EC (2013) Sensors and imaging techniques for the assessment of the delay of wheat senescence induced by fungicides. Funct Plant Biol 40: 677–689 [DOI] [PubMed] [Google Scholar]
- Berdugo CA, Steiner U, Dehne HW, Oerke EC (2012) Effect of bixafen on senescence and yield formation of wheat. Pestic Biochem Physiol 104: 171–177 [Google Scholar]
- Cecchini G (2003) Function and structure of complex II of the respiratory chain. Annu Rev Biochem 72: 77–109 [DOI] [PubMed] [Google Scholar]
- Cezary W, Melanie C, Jaakko K (2018) Reactive oxygen species in plant signaling. Annu Rev Plant Biol 69: 209–236 [DOI] [PubMed] [Google Scholar]
- Chen D, Xu G, Tang W, Jing Y, Ji Q, Lin FR (2013) Antagonistic basic helix-loop-helix/bZIP transcription factors form transcriptional modules that integrate light and reactive oxygen species signaling in Arabidopsis. Plant Cell 25: 1657–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GH, Liu CP, Chen SCG, Wang LC (2012) Role of ARABIDOPSIS A-FIFTEEN in regulating leaf senescence involves response to reactive oxygen species and is dependent on ETHYLENE INSENSITIVE2. J Exp Bot 63: 275–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Yu Q, Wang Y, Wang H, Dong Y, Ji Y, Zhou X, Li Y, Jiang C, Gan S, et al. (2021) Ethylene-regulated asymmetric growth of the petal base promotes flower opening in rose (Rosa hybrida). Plant Cell 33: 1229–1251 [DOI] [PubMed] [Google Scholar]
- Covington MF, Maloof JN, Straume M, Kay SA, Harmer SL (2008) Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol 9: R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RS, Green EW, Zhao YW, van Ooijen G, Olmedo M, Qin XM, Xu Y, Pan M, Valekunja UK, Feeney KA, et al. (2012) Peroxiredoxins are conserved markers of circadian rhythms. Nature 485: 459–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elorza A, León G, Gómez I, Mouras A, Holuigue L, Araya A, Jordana X (2004) Nuclear SDH2-1 and SDH2-2 genes, encoding the iron-sulfur subunit of mitochondrial complex II in Arabidopsis, have distinct cell specific expression patterns and promoter activities. Plant Physiol 136: 4072–4087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eubel H, Jansch L, Braun HP (2003) New insights into the respiratory chain of plant mitochondria. Supercomplexes and a unique composition of complex II. Plant Physiol 133: 274–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa P, Léon G, Elorza A, Holuigue L, Araya A, Jordana X (2002) The four subunits of mitochondrial respiratory complex ii are encoded by multiple nuclear genes and targeted to mitochondria in Arabidopsis thaliana. Plant Mol Biol 50: 725–734 [DOI] [PubMed] [Google Scholar]
- Figueroa P, Léon G, Elorza A, Holuigue L, Jordana X (2001) Three different genes encode the iron-sulfur subunit of succinate dehydrogenase in Arabidopsis thaliana. Plant Mol Biol 46: 241–250 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G (2013) Redox signaling in plants. Antioxid Redox Signal 18: 2087–2090 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Lelandais M, Kunert KJ (1994) Photooxidative stress in plants. Plant Physiol 92: 696–717 [Google Scholar]
- Friedman WE, Moore RC, Purugganan MD (2004) The evolution of plant development. Am J Bot 91: 1726–1741 [DOI] [PubMed] [Google Scholar]
- Fryer MJ, Ball L, Oxborough K, Karpinski S, Mullineaux PM, Baker NR (2003) Control of Ascorbate Peroxidase 2 expression by hydrogen peroxide and leaf water status during excess light stress reveals a functional organisation of Arabidopsis leaves. Plant J 33: 691–705 [DOI] [PubMed] [Google Scholar]
- Fuentes D, Meneses M, Nunes-Nesi A, Araujo WL, Tapia R, Gomez I, Holuigue L, Gutierrez RA, Fernie AR, Jordana X (2011) A deficiency in the flavoprotein of arabidopsis mitochondrial complex II results in elevated photosynthesis and better growth in nitrogen-limiting conditions. Plant Physiol 157: 1114 1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima A, Kusano M, Nakamichi N, Kobayashi M, Hayashi N, Sakakibara H, Mizuno T, Saito K (2009) Impact of clock-associated Arabidopsis pseudo-response regulators in metabolic coordination. Proc Natl Acad Sci U S A 106: 7251–7256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan S, Amasino R (1995) M. Inhibition of leaf senescence by autoregulated production of cytokinin. Science 270: 1986–1988 [DOI] [PubMed] [Google Scholar]
- Gangappa SN, Botto JF (2014) The BBX family of plant transcription factors. Trends Plant Sci 19: 460–470 [DOI] [PubMed] [Google Scholar]
- Gleason C, Huang S, Thatcher LF, Foley RC, Anderson CR, Carroll AJ, Millar AH, Singh KB (2011) Mitochondrial complex II has a key role in mitochondrial-derived reactive oxygen species influence on plant stress gene regulation and defense. Proc Natl Acad Sci U S A 108: 10768–10773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover BJ, Martin C (1998) The role of petal cell shape and pigmentation in pollination success in Antirrhinum majus. Heredity 80: 778–784 [Google Scholar]
- Gong Z, Lee H, Xiong L, Jagendorf A, Stevenson B, Zhu J (2002) RNA helicase-like protein as an early regulator of transcription factors for plant chilling and freezing tolerance. Proc Natl Acad Sci U S A 99: 11507–11512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo PG, Li ZH, Huang PX, Li BS, Fang S, Chu JF, Guo HW (2017) A tripartite amplification loop involving the transcription factor WRKY75, salicylic acid, and reactive oxygen species accelerates leaf senescence. Plant Cell 29: 2854–2870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Kuang J, Chen J, Liu X, Xiao Y, Fu C, Wang J, Wu K, Lu W (2016) Banana transcription factor MaERF11 recruits histone deacetylase MaHDA1 and represses the expression of MaACO1 and expansins during fruit ripening. Plant Physiol 171: 1070–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harman JW ( 1958) Cytochondrial aspects of cellular pathology. J Clin Path 11: 495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang SH, Han B, Zhu T, Wang X, Kreps JA, Kay SA (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290: 2110–2113 [DOI] [PubMed] [Google Scholar]
- Helfer A, Nusinow DA, Chow BY, Gehrke AR, Bulyk ML, Kay SA (2011) LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr Biol 21: 126–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero E, Kolmos E, Bujdoso N, Yuan Y, Wang MM, Berns MC, Uhlworm H, Coupland G, Saini R, Jaskolski M, et al. (2012) Early flowering 4 recruitment of early flowering 3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 24: 428–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horibe T, Yamada K (2015) Diurnal rhythm of petal growth in cut rose flowers. Acta Hortic 1064: 241–245 [Google Scholar]
- Hornitschek P, Kohnen MV, Lorrain S, Rougemont J, Ljung K, López-Vidriero I, Franco-Zorrilla JM, Solano R, Trevisan M, Pradervand S, et al. (2012) Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J 71: 699–711 [DOI] [PubMed] [Google Scholar]
- Huang S, Braun HP, Gawryluk RMR, Millar AH (2019) Mitochondrial complex ii of plants: subunit composition, assembly, and function in respiration and signaling. Plant J 98: 405–417 [DOI] [PubMed] [Google Scholar]
- Huang S, Millar AH (2013) Succinate dehydrogenase: the complex roles of a simple enzyme. Curr Opin Plant Biol 16: 344–349 [DOI] [PubMed] [Google Scholar]
- Ishida H, Izumi M, Wada S, Makino A (2014) Roles of autophagy in chloroplast recycling. Biochim Biophys Acta 1837: 512. [DOI] [PubMed] [Google Scholar]
- Jardim-Messeder D, Caverzan A, Rauber R, de Souza Ferreira E, Margis-Pinheiro M, Galina A (2015) Succinate dehydrogenase (mitochondrial complex II) is a source of reactive oxygen species in plants and regulates development and stress responses. New Phytol 208: 776–789 [DOI] [PubMed] [Google Scholar]
- Jeong J, Choi G (2013) Phytochrome-interacting factors have both shared and distinct biological roles. Mol Cells 35: 371–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing HC, Hebeler R, Oeljeklaus S, Sitek B, Stühler K, Meyer HE, Sturre MJG, Hille J, Warscheid B, Dijkwel PP (2008) Early leaf senescence is associated with an altered cellular redox balance in Arabidopsis cpr5/old1 mutants. Plant Biol 10: 85–98 [DOI] [PubMed] [Google Scholar]
- Juneau P, Lay PL, Béla B, Samson G, Popovic R (2010) Relationship between the structural and functional changes of the photosynthetic apparatus during chloroplast–chromoplast transition in flower bud of Lilium longiflorum. Photochem Photobiol 75: 377–381 [DOI] [PubMed] [Google Scholar]
- Keech O, Pesquet E, Ahad A, Askne A, Gardestrm P (2007) The different fates of mitochondria and chloroplasts during dark-induced senescence in Arabidopsis leaves. Plant Cell Environ 30: 1523–1534 [DOI] [PubMed] [Google Scholar]
- Khanna R, Kronmiller B, Maszle RD, Coupland G, Holm M, Mizuno T, Wu SH (2009) The Arabidopsis B-Box zinc finger family. Plant Cell 21: 3416–3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna-Chopra R (2012) Leaf senescence and abiotic stresses share reactive oxygen species-mediated chloroplast degradation. Protoplasma 249: 469–481 [DOI] [PubMed] [Google Scholar]
- Kidokoro S, Maruyama K, Nakashima K, Imura I, Narusaka Y, Shinwari ZK, Osakabe Y, Fujita Y, Mizoi Y, Shinozaki K, et al. (2009) The phytochrome-interacting factor PIF7 negatively regulates DREB1 expression under circadian control in Arabidopsis. Plant Physiol 151: 2046–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kim JH, Lyu JI, Woo HR, Lim PO (2017) New insights into the regulation of leaf senescence in Arabidopsis. J Exp Bot 4: 787–799 [DOI] [PubMed] [Google Scholar]
- Koprivova A, Mugford ST, Kopriva S (2010) Arabidopsis root growth dependence on glutathione is linked to auxin transport. Plant Cell Rep 29: 1157–1167 [DOI] [PubMed] [Google Scholar]
- Kumagai T, Ito S, Nakamichi N, Niwa Y, Murakami M, Yamashino T, Mizumo T (2008) The common function of a novel subfamily of B-Box zinc finger proteins with reference to circadian-associated events in Arabidopsis thaliana. Biosci Biotechnol Biochem 72: 1539–1549 [DOI] [PubMed] [Google Scholar]
- Lai AG, Doherty CJ, Mueller-Roeber B, Kay SA, Schippers JHM, Dijkwel PP (2012) CIRCADIAN CLOCK-ASSOCIATED 1 regulates ROS homeostasis and oxidative stress responses. Proc Natl Acad Sci U S A 109: 17129–17134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CP, Eubel H, Millar AH (2010) Diurnal changes in mitochondrial function reveal daily optimization of light and dark respiratory metabolism in Arabidopsis. Mol Cell Proteomics 9: 2125–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Seo PJ, Lee HJ, Park CM (2012) A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant J 70: 831–844 [DOI] [PubMed] [Google Scholar]
- Leivar P, Monte E (2014) PIFs: systems integrators in plant development. Plant Cell 26: 56–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- León G, Holuigue L, Jordana X (2007) Mitochondrial complex II is essential for gametophyte development in Arabidopsis. Plant Physiol 143: 1534–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebsch D, Keech O (2016) Dark-induced leaf senescence: new insights into a complex light-dependent regulatory pathway. New Phytol 212: 563–570 [DOI] [PubMed] [Google Scholar]
- Lim PO, Kim HJ, Nam HG (2007) Leaf senescence. Annu Rev Plant Biol 58: 115–136 [DOI] [PubMed] [Google Scholar]
- Lin F, Jiang Y, Li J, Yan TT, Fan LM, Liang JS, Chen JF, Xu DQ, Deng XW (2018) B-BOX DOMAIN PROTEIN28 negatively regulates photomorphogenesis by repressing the activity of transcription factor HY5 and undergoes COP1-mediated degradation. Plant Cell 30: 2006–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Yuan Y, Jiang H, Ning G, Zhao L, Zhou X, Zhou H, Gao J, Ma N (2021) Agrobacterium tumefaciens-mediated transformation of modern rose (Rosa hybrida) using leaf-derived embryogenic callus. Hortic Plant J 10.1016/j.hpj.2021.02.001 [DOI] [Google Scholar]
- Liu Y, Lin G, Yin C, Fang Y (2020) B-box transcription factor 28 regulates flowering by interacting with CONSTANS Sci Rep 10: 17789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP (2002) Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J 30: 415–429 [DOI] [PubMed] [Google Scholar]
- Ljubesić N, Wrischer M, Devidé Z (1991) Chromoplasts: the last stages in plastid development. Int J Dev Biol 35: 251–258 [PubMed] [Google Scholar]
- Lu SX, Tobin EM (2011) Chromatin remodeling and the circadian clock: Jumonji C-domain containing proteins. Plant Signal Behav 6: 810–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv PT, Zhang CQ, Liu JT, Liu XW, Jiang GM, Jiang XQ, Khan AM, Wang LS, Hong B, Gao JP (2014) RhHB1 mediates the antagonism of gibberellins to ABA and ethylene during rose (Rosa hybrida) petal senescence. Plant J 78: 578–590 [DOI] [PubMed] [Google Scholar]
- Ma N, Ma C, Liu Y, Shahid MO, Wang CP, Gao JP (2018) Petal senescence: a hormone view. J Exp Bot 69: 719–732 [DOI] [PubMed] [Google Scholar]
- Ma N, Cai L, Lu W, Tan H, Gao J (2005) Exogenous ethylene influences flower opening of cut roses (Rosa hybrida) by regulating the genes encoding ethylene biosynthesis enzymes. Sci Chin C Life Sci 48: 434–444 [DOI] [PubMed] [Google Scholar]
- Marano MR, Serra EC, Orellano EG, Carrrillo N (1993) The path of chromoplast development in fruits and flowers. Plant Sci 94: 1–17 [Google Scholar]
- Martínez-García JF, Huq E, Quail PH (2000) Direct targeting of light signals to a promoter element-bound transcription factor. Science 288: 859–863 [DOI] [PubMed] [Google Scholar]
- McClung CR (2006) Plant circadian rhythms. Plant Cell 18: 792–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick S. (2004) Control of male gametophyte development. Plant Cell 16 (Suppl): S142–S153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y, Li N, Tian J, Gao J, Zhang C (2013) Identification and validation of reference genes for gene expression studies in postharvest rose flower (Rosa hybrida). Sci Hortic 158: 16–21 [Google Scholar]
- Meyer EH, Tomaz T, Carroll AJ, Estavillo G, Delannoy E, Tanz SK, Small ID, Pogson BJ, Millar AH (2009) Remodeled respiration in ndufs4 with low phosphorylation efficiency suppresses Arabidopsis germination and growth and alters control of metabolism at night. Plant Physiol 151: 603–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Breusegem FV (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9: 490–498 [DOI] [PubMed] [Google Scholar]
- Millar AH, Eubel H, Jansch L, Kruft V, Heazlewood JL, Braun HP (2004) Mitochondrial cytochrome c oxidase and succinate dehydrogenase complexes contain plant specific subunits. Plant Mol Biol 56: 77–90 [DOI] [PubMed] [Google Scholar]
- Moosavi B, Berry EA, Zhu XL, Yang WC, Yang GF (2019) The assembly of succinate dehydrogenase: a key enzyme in bioenergetics. Cell Mol Life Sci. 76: 4023–4042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navrot N, Rouhier N, Gelhaye E, Jacquot JP (2007) Reactive oxygen species generation and antioxidant systems in plant mitochondria. Plant Physiol 129: 185–195 [Google Scholar]
- Niwa Y, Yamashino T, Mizuno T (2009) The circadian clock regulates the photoperiodic response of hypocotyl elongation through a coincidence mechanism in Arabidopsis thaliana. Plant Cell Physiol 50: 838–854 [DOI] [PubMed] [Google Scholar]
- Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL, Maloof JM (2007) Rhythmic growth explained by coincidence between internal and external cues. Nature 448: 358–361 [DOI] [PubMed] [Google Scholar]
- Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farré EM, Kay SA (2012) The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Kang H, Yamaguchi S, Park J, Lee D, Kamiya Y, Choi G (2009) Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell 21: 403–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmiya A, Hirashima M, Yagi M, Tanase K, Yamamizo C (2014) Identification of genes associated with chlorophyll accumulation in flower petals. PLoS One 9: e113738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen AN, Ernst HA, Leggio LL, Skriver K (2005) NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 10: 79–87 [DOI] [PubMed] [Google Scholar]
- Pei H, Ma N, Tian J, Luo J, Chen W, Li J, Zheng Y, Chen X, Fei Z, Gao J (2013) An NAC transcription factor controls ethylene-regulated cell expansion in flower petals. Plant Physiol 163: 775–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poire R, Schneider H, Thorpe MR, Kuhn AJ, Schurr U, Walter A (2010) Root cooling strongly affects diel leaf growth dynamics, water and carbohydrate relations in Ricinus communis. Plant Cell Environ 33: 408–417 [DOI] [PubMed] [Google Scholar]
- Popov VN, Eprintsev AT, Fedorin DN, Igamberdiev AU (2010) Succinate dehydrogenase in Arabidopsis thaliana is regulated by light via phytochrome A. FEBS Lett 584: 199–202 [DOI] [PubMed] [Google Scholar]
- Popov VN, Fedorin DN, Eprintsev AT (2007) Light regulation of succinate dehydrogenase expression in Arabidopsis thaliana leaves. Russ J Plant Physiol 54: 360–365 [Google Scholar]
- Quinlan CL, Orr AL, Perevoshchikova IV, Treberg JR, Ackrell BA, Brand MD (2012) Mitochondrial complex II can generate reactive oxygen species at high rates in both the forward and reverse reactions. J Biol Chem 287: 27255–27264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renkema GH, Wortmann SB, Smeets RJ, Venselaar H, Antoine M, Visser G, Ben-Omran T, van den Heuvel LP, Timmers HJ, Smeitink JA (2015) SDHA mutations causing a multisystem mitochondrial disease: novel mutations and genetic overlap with hereditary tumors. Eur J Hum Genet 23: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads DM, Umbach AL, Subbaiah CC, Siedow JN (2006) Mitochondrial reactive oxygen species: contribution to oxidative stress and interorganellar signaling. Plant Physiol 141: 357–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers H, Munné-Bosch S (2016) Production and scavenging of reactive oxygen species and redox signaling during leaf and flower senescence: similar but different. Plant Physiol 171: 1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers HJ (2005) Cell death and organ development in plants. Curr Top Dev Biol 71: 225–261 [DOI] [PubMed] [Google Scholar]
- Rogers H.J. (2013) From models to ornamentals: how is flower senescence regulated? Plant Mol Biol 82: 563–574 [DOI] [PubMed] [Google Scholar]
- Rogers HJ (2012) Is there an important role for reactive oxygen species and redox regulation during floral senescence? Plant Cell Environ 35: 217–233 [DOI] [PubMed] [Google Scholar]
- Rogers HJ (2006) Programmed cell death in floral organs: how and why do flowers die? Ann Bot 97: 309–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romera-Branchat M, Andrés F, Coupland G (2014) Flowering responses to seasonal cues: what's new? Curr Opin Plant Biol 21: 120–127 [DOI] [PubMed] [Google Scholar]
- Rutter J, Winge DR, Schiffman JD (2010) Succinate dehydrogenase—assembly, regulation and role in human disease. Mitochondrion 10: 393–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuraba Y, Jeong J, Kang MY, Kim J, Paek NC, Choi G (2014) Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat Commun 5: 4636. [DOI] [PubMed] [Google Scholar]
- Sainsbury F, Thuenemann EC, Lomonossoff GP (2009) pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol J 7: 682–693 [DOI] [PubMed] [Google Scholar]
- Schippers JH, Schmidt R, Wagstaff C, Jing HC (2015) Living to die and dying to live: the survival strategy behind leaf senescence. Plant Physiol 169: 914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegelman HW, Chow CT, Biale JB (1958) Respiration of developing rose petals. Plant Physiol 33: 403–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Yang CW, Gao S, Zhang W, Li L, Kuai B (2014) Age-triggered and dark-induced leaf senescence require the bHLH transcription factors PIF3, 4, and 5. Mol Plant 7: 1776–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Shim JS, Kinmonthschultz HA, Imaizumi T (2015) Photoperiodic flowering: time measurement mechanisms in leaves. Annu Rev Plant Biol 66: 441–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Jiang Y, Kuai B, Li L (2018) CIRCADIAN CLOCK-ASSOCIATED 1 inhibits leaf senescence in Arabidopsis. Front Plant Sci 9: 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, Yan T, Liu J, Bian Y, Heng Y, Lin F, Jiang Y, Deng XW, Xu D (2020) BBX28/BBX29, HY5 and BBX30/31 form a feedback loop to fine-tune photomorphogenic development. Plant J 104: 377–390 [DOI] [PubMed] [Google Scholar]
- Strenkert D, Schmollinger S, Gallaher SD, Salomé PA, Purvine SO, Nicora CD, Mettler-Altmann T, Soubeyrand E, Weber APM, Lipton MS, et al. (2019) Multiomics resolution of molecular events during a day in the life of Chlamydomonas. Proc Natl Acad Sci U S A 116: 2374–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs JM, Francis MJ (1971) Electron microscopical studies of rose petal cells during flower maturation. Planta Med 20: 211–218 [DOI] [PubMed] [Google Scholar]
- Tadege M, Kuhlemeier C (1997) Aerobic fermentation during tobacco pollen development. Plant Mol Biol 35: 343–354 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J, Pei H, Zhang S, Chen J, Chen W, Yang R, Meng Y, You J, Gao J, Ma N (2014) TRV-GFP: a modified Tobacco rattle virus vector for efficient and visualizable analysis of gene function. J Exp Bot 65: 311–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivellini A, Jibran R, Watson LM, O’Donoghue EM, Ferrante A, Sullivan KL, Dijkwel PP, Hunter DA (2012) Carbon deprivation-driven transcriptome reprogramming in detached developmentally arresting Arabidopsis inflorescences. Plant Physiol 160: 1357–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn WG, Woltering EJ (2008) Physiology and molecular biology of petal senescence. J Exp Bot 59: 453–480 [DOI] [PubMed] [Google Scholar]
- van Doorn WG, Woltering EJ (2004) Senescence and programmed cell death: substance or semantics? J Exp Bot 55: 2147–2153 [DOI] [PubMed] [Google Scholar]
- Wei Q, Ma C, Xu Y, Wang T, Chen Y, Lü J, Zhang L, Jiang CZ, Hong B, Gao J (2017) Control of chrysanthemum flowering through integration with an aging pathway. Nat Commun 8: 829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir HJ, Yao P, Huynh FK, Escoubas CC, Goncalves RL, Burkewitz K, Laboy R, Hirschey MD, Mair WB (2017) Dietary restriction and AMPK increase lifespan via mitochondrial network and peroxisome remodeling. Cell Metab 26: 884–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnen H, Young MW (2006) Interplay of circadian clocks and metabolic rhythms. Annu Rev Genet 40: 409–448 [DOI] [PubMed] [Google Scholar]
- Wu A, Allu AD, Garapati P, Siddiqui H, Dortay H, Zanor MI, Asensi-Fabado MA, Munné-Bosch S, Antonio C, Tohge T (2012) JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 24: 482–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Sun YF, Zhao YN, Zhang J, Luo LL, Li M, Wang JL, Yu H, Liu GF, Yang LS (2015) Deficient plastidic fatty acid synthesis triggers cell death by modulating mitochondrial reactive oxygen species. Cell Res 25: 621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Ma N, Jia Y, Zhang Y, Feng M, Jiang CZ, Ma C, Gao J (2017) An ethylene-induced regulatory module delays flower senescence by regulating cytokinin content. Plant Physiol 173: 853–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashino T, Akinori M, Toru F, Shusei S, Tomohiko K, Satoshi T, Mizuno M (2003) A link between circadian-controlled bHLH factors and the APRR1/TOC1 quintet in Arabidopsis thaliana. Plant Cell Physiol 44: 619–629 [DOI] [PubMed] [Google Scholar]
- Yankovskaya V, Horsefield R, Törnroth S, Luna-Chavez C, Miyoshi H, Léger C, Byrne B, Cecchini G, Iwata S (2003) Architecture of succinate dehydrogenase and reactive oxygen species generation. Science 299: 700–704 [DOI] [PubMed] [Google Scholar]
- Zhang S, Feng M, Chen W, Zhou X, Lu J, Wang Y, Li Y, Jiang CZ, Gan SS, Ma N, et al. (2019) In rose, transcription factor PTM balances growth and drought survival via PIP2;1 aquaporin. Nat Plants 5: 290–299 [DOI] [PubMed] [Google Scholar]
- Zhang X, Huai J, Shang F, Xu G, Tang W, Jing Y, Lin R (2017) A PIF1/PIF3-HY5-BBX23 transcription factor cascade affects photomorphogenesis. Plant Physiol 174: 2487–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Mayba O, Pfeiffer A, Shi H, Tepperman JM, Speed TP, Quail PH (2013) A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet 9: e1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YY, Wang Y, Wei H, Li N, Tian W, Chong K, Wang L (2018) Circadian evening complex represses jasmonate-induced leaf senescence in Arabidopsis. Mol Plant 11: 326–337 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Transcriptome raw sequence data were deposited in the NCBI Sequence Read Archive database under accession number SRR12329579-SRR12329584 and SRR12329600-SRR12329605.