Abstract
Malate oxidation by plant mitochondria enables the generation of both oxaloacetate and pyruvate for tricarboxylic acid (TCA) cycle function, potentially eliminating the need for pyruvate transport into mitochondria in plants. Here, we show that the absence of the mitochondrial pyruvate carrier 1 (MPC1) causes the co-commitment loss of its putative orthologs, MPC3/MPC4, and eliminates pyruvate transport into Arabidopsis thaliana mitochondria, proving it is essential for MPC complex function. While the loss of either MPC or mitochondrial pyruvate-generating NAD-malic enzyme (NAD-ME) did not cause vegetative phenotypes, the lack of both reduced plant growth and caused an increase in cellular pyruvate levels, indicating a block in respiratory metabolism, and elevated the levels of branched-chain amino acids at night, a sign of alterative substrate provision for respiration. 13C-pyruvate feeding of leaves lacking MPC showed metabolic homeostasis was largely maintained except for alanine and glutamate, indicating that transamination contributes to the restoration of the metabolic network to an operating equilibrium by delivering pyruvate independently of MPC into the matrix. Inhibition of alanine aminotransferases when MPC1 is absent resulted in extremely retarded phenotypes in Arabidopsis, suggesting all pyruvate-supplying enzymes work synergistically to support the TCA cycle for sustained plant growth.
Pyruvate is supplied by three independent processes that act synergistically to maintain metabolic flux and support plant respiration in a variety of circumstances.
Introduction
Pyruvate occupies a pivotal node in the regulation of carbon metabolism as the end product of glycolysis and a major substrate for mitochondrial respiration. Mitochondrial pyruvate oxidation is catalyzed by the pyruvate dehydrogenase complex (PDC) to provide carbon skeletons to the tricarboxylic acid (TCA) cycle, and its depletion leads to extensive changes in metabolism, abnormal organ development, and severe growth retardation in plants (Yu et al., 2012; Ohbayashi et al., 2019). PDC and malate dehydrogenase (MDH) work cooperatively to provide substrates, acetyl-CoA, and oxaloacetate (OAA) respectively, for citrate synthesis (Selinski and Scheibe, 2019). In terms of kinetics, malate production by MDH is favored over OAA formation (Hüdig et al., 2015); therefore, in order to drive the reaction toward OAA and maintain the continuity of respiration, OAA needs to be continuously removed by NAD-malic enzyme (NAD-ME; Tronconi et al., 2010a, 2010b), citrate synthase (Schmidtmann et al., 2014), and a putative OAA exporter (Haferkamp and Schmitz-Esser, 2012). Malate oxidation by plant mitochondria enables the generation of both OAA (via MDH) and pyruvate (via NAD-ME) for TCA cycle function, and in isolated plant mitochondria malate-feeding enables a much faster respiratory rate than pyruvate alone. This indicates that pyruvate import may be slower than malate import, and based on this it has long been thought that the need for pyruvate transport across the inner mitochondrial membrane in plants may be minimal (Day and Hanson, 1977). In the following decades, this concept has been further tested by various in vitro biochemical approaches (Edwards et al., 1998; Jenner et al., 2001) and in vivo 13C-labeling studies (Tcherkez et al., 2005, 2008, 2009; Lehmann et al., 2016), but genetic evidence to prove it had been lacking.
The availability of full genome information and insertional mutants in Arabidopsis enabled significant advances in our understanding the fate of malate in plant mitochondria. Insertional loss-of-function of both mitochondrial MDH enzymes resulted in a slow growth phenotype and an elevated leaf respiration rate (Tomaz et al., 2010). Insertional loss-of-function of both mitochondrial NAD-ME subunits in Arabidopsis caused a significant diversion of excess malate to amino acid synthesis at night without affecting vegetative growth (Tronconi et al., 2008). Gene insertion-based reduction in PDC activity caused a greatly retarded vegetative phenotype in Arabidopsis (Ohbayashi et al., 2019). This suggests that mitochondrial pyruvate supply and oxidation are both important for optimal plant growth.
The transport of glycolytically derived pyruvate into the mitochondrial matrix is carried out by the mitochondrial pyruvate carrier (MPC). Its existence was first proposed in the 1970s based on the specific inhibition of mitochondrial pyruvate uptake by cinnamate analogs, such as α-cyano-4-hydroxycinnamate and UK-5099 (Halestrap and Denton, 1974; Halestrap, 1975). Attempts to purify and identify the carrier proteins by hydroxyapatite chromatography or affinity with an immobilized inhibitor were not successful due to numerous technical challenges at the time (Thomas and Halestrap, 1981; Brailsford et al., 1986; Nałecz et al., 1986; Capuano et al., 1990). Screening of yeast mutants identified a candidate in the mitochondrial carrier family that influences pyruvate uptake and is sensitive to UK-5099, but it was later shown to be a NAD+ carrier (Hildyard and Halestrap, 2003; Todisco et al., 2006). It was not until 2012 that the proteins responsible for UK-5099-sensitive MPC activity were identified by two independent research groups (Bricker et al., 2012; Herzig et al., 2012). MPCs are 12–16-kDa proteins that share limited homology with the semi-SWEET family transporters (Bender and Martinou, 2016). In yeast, a functional MPC complex, composed of a heterodimer of a core subunit (MPC1) and a regulatory unit (MPC2 or MPC3), facilitates the co-import of pyruvate with a proton (Tavoulari et al., 2019). Yeast without MPC1 grows slowly in medium without amino acids and has a lower pyruvate transport (Bricker et al., 2012; Herzig et al., 2012). MPC mutation in Drosophila is lethal with a sugar-only diet and caused impaired pyruvate oxidation-linked metabolism under a standard diet. Human MPC1 harboring a single point mutation resulted in lactic acidosis and hyperpyruvatemia due to impaired pyruvate oxidation, while knockout of MPC1 in mice led to embryonic lethality (Brivet et al., 2003). From these reports, it can be concluded that MPC1 is an essential component for supplying pyruvate to mitochondria in yeast and mammals.
In Arabidopsis, four MPC isoforms are present, with Arabidopsis MPC1 (At5g20090) showing the highest homology with yeast MPC1 (Shen et al., 2017). The role of Arabidopsis MPC in pyruvate transport has been proven indirectly through complementation of a yeast mutant (Li et al., 2014). In Arabidopsis, MPC1 has been shown to interact with MPC3 or MPC4, but it remains uncertain if an interaction exists between MPC3 and MPC4 (Shen et al., 2017; He et al., 2019). In plants, lack of MPC has only been linked to phenotypes of cadmium sensitivity and drought tolerance (Li et al., 2014; Shen et al., 2017; He et al., 2019). Single or higher-order knockouts of MPC isoforms do not cause any obvious growth defect under normal conditions (He et al., 2019), indicating that unlike in other eukaryotes, pyruvate transport is probably not essential for respiratory pyruvate supply in Arabidopsis. Apart from the direct transport of glycolysis-derived pyruvate into mitochondria and malate oxidation, pyruvate can also be supplied from alanine through the combined actions of cytosolic and mitochondrial alanine aminotransferases (AlaATs; Miyashita et al., 2007) and a yet-to-be characterized mitochondrial alanine carrier (Passarella et al., 2003; Bender and Martinou, 2016). However, the role of AlaATs in reversibly converting pyruvate to alanine has only been reported during hypoxia and recovery from hypoxia (Ricoult et al., 2006; Miyashita and Good, 2008; Diab and Limami, 2016).
MPC in yeast and mammalian systems can only facilitate pyruvate transport from the cytosol to the mitochondrial matrix and not vice versa (Bricker et al., 2012; Herzig et al., 2012). Pyruvate export from mitochondria in cancer cells is mediated by a different class of transporter called the monocarboxylate carrier MCT1 (Hong et al., 2016). Under some circumstances in plants, the export of pyruvate from mitochondria is needed for phosphoenolpyruvate synthesis in the cytosol by pyruvate orthophosphate dikinase to respond to water stress in rice (Oryza sativa) seedlings (Netting, 2002) or to support the recycling of carbon intermediates and CO2 fixation in Crassulacean acid metabolism (Hartwell et al., 2016) and C4 plants (Rao and Dixon, 2016). However, the identity of the plant mitochondrial pyruvate exporter is currently unknown. It remains unclear which MPC isoforms facilitate pyruvate transport in plants, which direction of transport they facilitate, and how pyruvate transport activity via MPC is co-regulated with other pyruvate-supplying pathway in contributing to the plant mitochondrial pyruvate pool.
In this study, we show the primary role of MPC1 in pyruvate import into Arabidopsis mitochondria using a reverse genetic approach in combination with direct evidence from in vitro, in organello, and in vivo analyses. We obtained evidence that plant mitochondrial pyruvate import is mainly mediated by MPC1, which is needed to maintain pyruvate supply for Arabidopsis respiratory metabolism. NAD-ME can independently supply pyruvate to the TCA cycle in Arabidopsis mitochondria, but our data indicate that it has a relatively small contribution to pyruvate-linked metabolism in vivo. Instead, the action of AlaATs provides an important functional backup for MPC complex and these enzymes most likely work in a cooperative manner to supply mitochondrial pyruvate needed for sustaining respiration at night in plants. These data provide insight into the flexibility of pyruvate-supplying pathways in plants to efficiently use and extract energy from the cellular carbon repertoire in different environments.
Results
MPC1 is essential for mitochondrial pyruvate carrier complex accumulation in Arabidopsis and its absence causes the co-commitment loss of MPC3 and MPC4
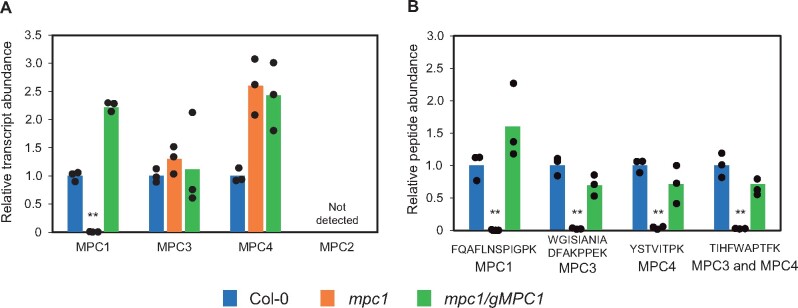
In Arabidopsis, MPC1, MPC3, and MPC4 (At5g20090, At4g22310, and At4g05590) are ubiquitously expressed in all tissue types, while MPC2 (At4g14695) is expressed exclusively in flowers (Waese et al., 2017). MPC2, MPC3, and MPC4 share about 60%–80% amino acid sequence identity, whereas MPC1 is the least similar to these isoforms (<33%) but shares the highest sequence identity with yeast MPC1 (Schwacke et al., 2003; Li et al., 2014). Thus, MPC1 is the most promising candidate as a nonredundant core component of the plant MPC complex. A T-DNA insertion line, mpc1, was previously confirmed to be a homozygous mutant that contained an insertion 400-bp upstream of the start codon (Shen et al., 2017; Supplemental Figure S1A). Under long-day conditions, mpc1 does not exhibit an obvious aberrant vegetative phenotype (Supplemental Figure S1B). Quantitative real-time polymerase chain reaction (qPCR) analysis of whole leaf and quantitation of selected peptides from isolated mitochondria by liquid chromatography–selective reaction monitoring–mass spectrometry (LC–SRM–MS) revealed that mpc1 is unable to produce full length MCP1 transcript or accumulate MPC1 protein, confirming that it is a MPC1 knockout mutant (Figure 1). Even though no obvious change in MPC3 and MPC4 transcript abundance was observed in mpc1, MPC3 and MPC4 proteins were completely absent in isolated mitochondria (Figure 1B). It is possible that MPC3 and MPC4 are transcribed but their protein products may be unstable, leading to their degradation. When a MPC1 transgene was expressed under the control of its native promoter in mpc1 plants (mpc1/gMPC1), both MPC3 and MPC4 abundances were restored in mitochondria (Figure 1A). Thus, MPC1 is required for the assembly of the MPC complex.
Figure 1.
MPC1 is required for the assembly of mitochondrial pyruvate carrier complex in Arabidopsis. A, Relative transcript abundance of MPCs in Col-0, mpc1, and mpc1/gMPC1. Transcript abundance was measured using qPCR of MPC1, MPC2, MPC3, MPC4 inter-exon sequences. Primers are provided in Supplemental Table S1. Elongation factor 1 alpha EF-1α was used as a housekeeping control. B, Relative protein abundance of MPC isoforms in Col-0, mpc1, and mpc1/gMPC1. Protein abundances were determined by quantifying peak area of unique peptides using LC–SRM–MS. FQAFLNSPIGPK was unique for MPC1, WGISIANIADFAKPPEK was unique for MPC3, YSTVITPK was unique for MPC4. The abundance of a mitochondrial voltage-dependent anion channel 1 (VDAC1) peptide was used as a housekeeping control. Each data point represents an averaged value from three biological replicates with error bars indicating standard error. Significant differences among mpc1, Col-0, and mpc1/gMPC1 are denoted by asterisks based on Student’s t test (**P < 0.01)
MPC1 is required for pyruvate-dependent respiration and is a target of the pyruvate transport inhibitor UK-5099
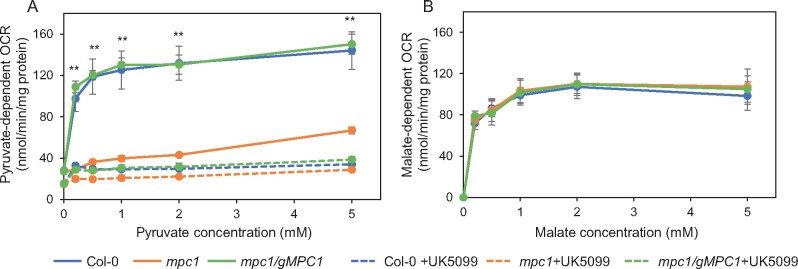
Evidence that plant MPC1 functions as a mitochondrial pyruvate importer to date is based on yeast complementation analysis (Li et al., 2014) and sequence conservation with yeast and mammal homologs. To provide direct experimental proof for its role in respiration and pyruvate transport, we used isolated mitochondria as an in vitro model system to monitor MPC function by manipulating respiratory substrate supply. We first measured the ability of isolated mitochondria to oxidize pyruvate in the presence of ADP and cofactors. Pyruvate as the sole substrate was unable to drive significant oxygen consumption, probably due to the lack of OAA replenishment. Respiratory assays at pH 7.2 have been shown previously to minimize malic enzyme conversion of exogenous malate to pyruvate in isolated mitochondria (Willeford and Wedding, 1987). When malate alone was supplied at pH 7.2 in our experiments, only 20% of the oxygen consumption rate in the presence of malate and saturating pyruvate was measured (27 ± 1.6 nmol·min–1·mg protein–1, compared to 149 ± 23 nmol·min–1·mg–1). Titrations with different nonsaturating concentrations of pyruvate revealed a ∼70% reduction in oxygen consumption rate (called pyruvate-dependent OCR) by mpc1 mitochondria compared to the wild-type (Figure 2), suggesting that mpc1 has a diminished ability to oxidize pyruvate for mitochondrial respiration. This could be the result of (1) altered PDC abundance and/or activity, (2) defects in TCA cycle and/or electron transport chain (ETC) functions, and/or (3) inability to import pyruvate. We found that the relative abundance and activity of PDC and the relative abundance of TCA cycle enzymes showed no difference between Col-0 and mpc1 (Supplemental Figure S2, A and C). In addition, the relative abundance of ETC components was unaffected by the loss of MPC1 (Supplemental Figure S2B); thus, the possibility of a defect in mitochondrial function other than pyruvate import was unlikely. As an additional control, we measured malate-dependent respiration at pH 6.4 to promote NAD-ME activity and found no difference among wild-type, mutant, and complemented line (Figure 2B).
Figure 2.
MPC1 is required for pyruvate-dependent respiration of Arabidopsis mitochondria. Mitochondrial respiration in the presence of ADP measured as OCR was assayed at varying concentrations (A) of pyruvate at pH 7.2 supplemented with 0.5-mM malate or (B) of malate at pH 6.4. UK-5099 was used as a pyruvate carrier inhibitor where shown. Each data point represents an averaged value from four biological replicates with error bars indicating standard error. Significant differences among mpc1, Col-0, and mpc1/gMPC1 are denoted by double asterisks based on Student’s t test (**P < 0.01)
When we measured mitochondrial pyruvate-dependent respiration in the presence of UK-5099, a known inhibitor for MPC in mammals (Halestrap, 1975), we found mpc1 mitochondria were insensitive to the UK-5099 treatment (Figure 2A). In contrast, mitochondria isolated from the wild-type and mpc1/gMPC1 were sensitive to UK-5099 treatment, showing pyruvate dependence of oxygen consumption diminished to a rate similar to untreated mpc1 mitochondria. This confirmed that the presence of MPC1 was required for UK-5099-sensitive pyruvate-dependent respiration in Arabidopsis mitochondria.
MPC1 is required for pyruvate import into plant mitochondria
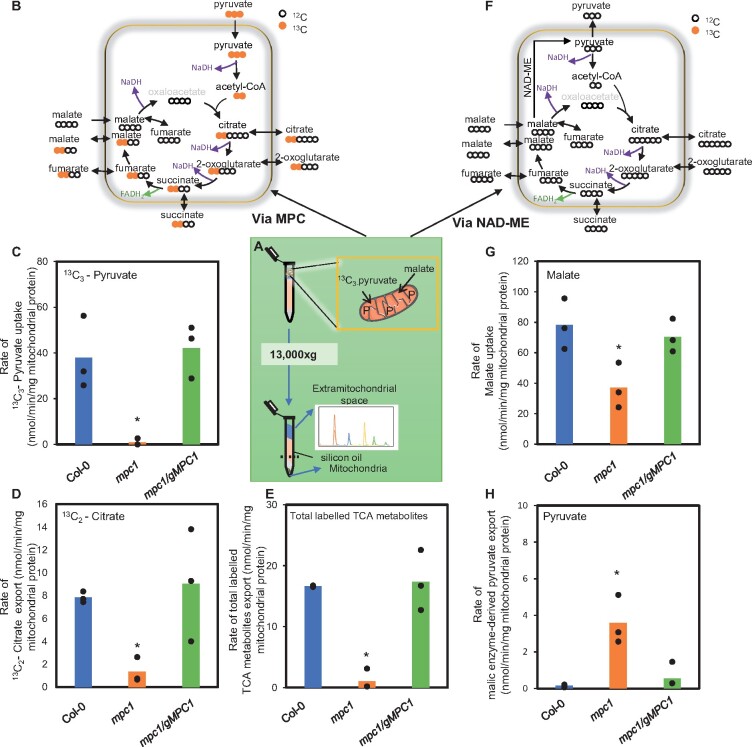
To directly demonstrate whether pyruvate uptake is defective in mpc1 mitochondria, we provided isolated mitochondria with both unlabeled malate and 13C3-pyruvate in the presence of ADP and other cofactors (see “Methods” section) at pH 7.2, followed by SRM–MS analysis to monitor the depletion of these metabolites in the extra-mitochondrial space (Figure 3). Figure 3, A, B, and F provides a detailed explanation of the experimental setup and the 13C incorporation patterns from the provided substrates into TCA cycle intermediates. The incorporation of 13C3-pyruvate into TCA cycle intermediates by freshly isolated mitochondria was tracked by measuring the level of metabolites in the extra-mitochondrial space (Figure 3, B–E). The amount of 13C3-pyruvate decreased linearly in the extra-mitochondrial space of the wild-type and mpc1/gMPC1 mitochondria, while pyruvate uptake rate was negligible in mpc1 mitochondria, resulting in a significant difference in net pyruvate uptake (Figure 3C; Supplemental Figure S3A). This showed that the absence of MPC1 effectively abolished pyruvate import into the mitochondrial matrix.
Figure 3.
MPC1 is required for the uptake and usage of pyruvate by Arabidopsis mitochondria. Experimental design of combined malate and 13C3-pyruvate feeding of isolated mitochondria at pH 7.2 (A) showing the isotopic incorporation patterns of labeled pyruvate into TCA cycle intermediates along with the steps that generate NADH for consumption by the ETC via MPC (B) and NAD-ME (F). Movements of organic acids across mitochondrial membranes are also shown. Bar graphs show the (C) uptake rate of 13C3-pyruvate, (D) export rate of labeled citrate, (E) export rate of total labeled TCA metabolites made from imported pyruvate (G) uptake rate of unlabeled malate, and (H) rate of unlabeled pyruvate made via NAD-ME. Quantification was carried out using SRM–MS to directly assess substrate consumption and product generation of substrate-fed mitochondria after separating mitochondria from the extra-mitochondrial space by centrifugation through a single silicon oil layer. The rates were calculated from time course values of metabolite concentration recorded in the extra-mitochondrial space after varying incubation periods. Each data point represents averaged value from three or more replicates with error bars indicating standard error. Significant differences among mpc1, Col-0, and mpc1/gMPC1 are denoted by asterisks based on Student’s t test (*P < 0.05)
Mitochondria fed with substrates and cofactors to drive full TCA cycle operation have been shown to generate TCA intermediates in excess which are rapidly exported to the extra-mitochondrial medium (Brailsford et al., 1986). We have recently demonstrated that it is possible to assess the function of mitochondrial carriers in isolated mitochondria by following such conversion rates of supplied respiratory substrate(s) using SRM–MS analysis (Lee et al., 2020). 13C2-citrate accumulated linearly in the extra-mitochondrial space of the wild-type and mpc1/gMPC1 mitochondria while in mpc1 only 15% of that rate was recorded (Figure 3D). This residual rate was most likely due to non-MPC-facilitated diffusion of pyruvate into mitochondria and/or a low level of free citrate synthase activity released from damaged mitochondria during the procedure. Treatment of mitochondria from all three genotypes with UK-5099 resulted in a drastic reduction in labeled citrate production, similar to that observed in mpc1 (Supplemental Figure S3B). The total labeled carbon incorporated into measured TCA cycle intermediates from 13C3-pyruvate was significantly higher in the wild-type compared to mpc1 (Figure 3E), which confirmed mpc1 could not sustain the same rate of supply of pyruvate in the matrix for TCA cycle-driven respiration (Figure 2). These results could be reproduced in a label swap experiment using unlabeled pyruvate and 13C4-malate as substrates, generating completely independent isotopic patterns but the same conclusions with regards to the rate of pyruvate transport in mpc1 (Supplemental Figures S4 and S5). Taken together, these data support the claim that MPC1 is an essential subunit of the MPC complex in Arabidopsis responsible for pyruvate uptake into the matrix of plant mitochondria.
Internal pyruvate synthesis by mitochondrial NAD-dependent malic enzyme is a compensation pathway for the loss of MPC1
In the same combined malate and 13C3-pyruvate feeding of isolated mitochondria experiment, we were also able to trace the amount of malate that was imported into the mitochondria and how it was consumed (Figure 3, F–H). The amount of malate taken up by mpc1 mitochondria was found to be half of that by the wild-type and mpc1/gMPC1 mitochondria (Figure 3G). This could be due to a lack of pyruvate in mpc1 mitochondria to remove OAA, which could then sequentially result in a feedback inhibition of MDH, an increased malate concentration in the matrix and a reduced demand for malate uptake.
Unlike yeast and mammals where MPC1 loss caused a range of growth defects (Bricker et al., 2012; Herzig et al., 2012), the absence of MPC1 did not result in growth delay in Arabidopsis (Shen et al., 2017). It is therefore very likely that alternative pyruvate-supplying pathways exist in plants, with at least one of them being more active in plants than in yeast or mammals. Apart from feeding carbon through OAA via malate dehydrogenase, malate can also be converted into pyruvate to contribute to the matrix pyruvate pool (Figure 3F). The mitochondrial NAD-ME pathway (encoded by AtNAD-ME1–At2g13560 and AtNAD-ME2–At4g00570) is the most likely candidate given its close association with the TCA cycle, its use of malate as primary substrate, its ability to provide pyruvate in the matrix without a transport step, and evidence that its loss results in a readjustment in TCA cycle flux at night (Tronconi et al., 2008). Malate oxidation by NAD-ME in intact isolated mitochondria can generate a significant amount of pyruvate (Day, 1980), which can be assayed in the extra-mitochondrial medium. We found the rate at which pyruvate was synthesized from malate and exported to the extra-mitochondrial space was significantly higher in mpc1 compared to the wild-type and mpc1/gMPC1 (Figure 3H), even though they have similar maximal NAD-ME activities (Supplemental Figure S2D). Similar results were observed using unlabeled pyruvate and 13C4-malate as substrates in the label swap experiment (Supplemental Figures S4 and S5). These data indicate that NAD-ME substantively provided an alternative internal pyruvate source when MPC1 was absent.
The absence of MPC in NAD-ME knockout plants results in a vegetative phenotype due to limited pyruvate supply as a respiratory substrate
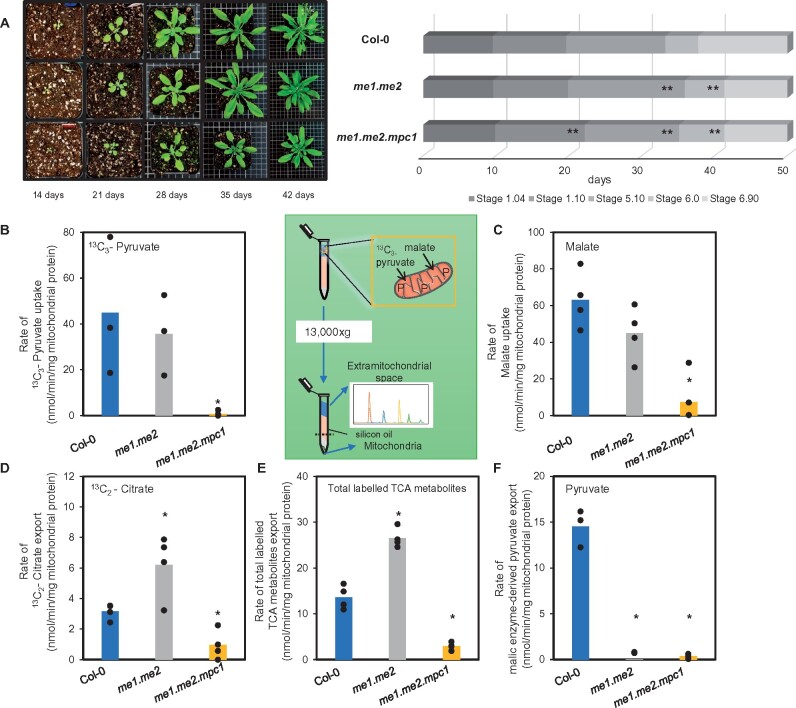
The elevation of NAD-ME-derived pyruvate in mpc1 mitochondria suggested the role of MPC1 could be enhanced in NAD-ME knockout plants. To investigate this, we generated a triple knockout by crossing mpc1 and nad.me1.1 x nad.me2.1 (me1.me2) plants to assess their combined effect on growth and metabolic phenotype in planta. Plants homozygous for all three mutations nad.me1.1 x nad.me2.1 x mpc1 (me1.me2.mpc1; Supplemental Figure S2A) displayed significantly decreased vegetative growth rate compared to the wild-type and other genotypes (Figure 4; Supplemental Figure S6). me1.me2.mpc1 plants were noticeably smaller at the four-leaf stage (Stage 1.04) onwards even though the number of leaves were identical. At later developmental stages (Stage 1.10—10 rosette leaves > 1 mm; Stage 5.10—first flower bud visible; Stage 6.00—first flower opened), me1.me2.mpc1 plants were about 2 to 6 days delayed. The mutant leaves did not reach the full diameter of Col-0 leaves after 1 week of bolting. The retarded phenotype was observed in both long-day and short-day growth conditions (Supplemental Figure S6C); however, it could be restored to a wild-type-like phenotype upon re-introduction of the MPC1 transgene under the control of its native promoter (me1.me2.mpc1/gMPC1; Supplemental Figure S6D). These results showed that MPC is an important component for plant growth and development when NAD-ME activity is impaired.
Figure 4.
Vegetative and respiratory phenotypes of NAD-ME knockout and MPC x NAD-ME knockouts. A, Quantitative phenotyping of me1.me2 and me1.me2 x mpc1 plants in a soil-based experiment (n = 15) in long-day conditions (16-h light/8-h dark, 60% humidity). Photos were taken at 14 days, 21 days, 28 days, 35 days, and 42 days after sowing to track plant growth. Representative top views of various genotype rosettes are shown. Each gradient bar represents distinct growth stages: 1.04—four rosette leaves > 1 mm; 1.10—ten rosette leaves > 1 mm; 5.10—first flower bud visible; 6.00—first flower opened; 6.90—flowering complete. Significant differences between tested genotypes and Col-0 are indicated based on Student’s t test and denoted by asterisks (**P < 0.01). Bar graphs show the uptake rate of 13C3-pyruvate (B); uptake rate of malate (C); export rate of citrate (D) and export rate of total labeled TCA metabolites synthesized via MPC (E) and export rate of pyruvate synthesized via NAD-ME (F, including 13C2-citrate, 13C2-2-OG, 13C2-succinate, and 13C2-malate). Mitochondria were incubated in 500-µM 13C3-pyruvate and 500-µM unlabeled malate in the presence of ADP at pH 6.4. Metabolic reaction was stopped by centrifugation through a single silicon oil layer in which the mitochondrial pellet was separated from the extra-mitochondrial medium. The rates were calculated from time courses of metabolite concentration recorded in the extra-mitochondrial space that were quantified using LC–SRM–MS as outlined in the “Methods” section. Each data point represents an averaged value from three or more replicates with error bars indicating standard error. Significant differences between mutants and Col-0 are indicated based on Student’s t test and denoted by asterisks (*P < 0.05)
The functional importance of MPC is also highlighted by how mitochondria isolated from NAD-ME double knockout plants consumed externally fed malate and pyruvate. To show this, isolated mitochondria were fed with both 13C3-pyruvate and malate at pH 6.4 to maximize the rate of NAD-ME activity. At pH 6.4, MPC activity of Col-0 appeared to be similar to that at pH 7.2 (Figures 3C and 4B). me1.me2 plants did not have a vegetative phenotype (Figure 4A) and therefore were expected to have wild-type pyruvate transport activity using MPC. Indeed, malate and 13C3-pyruvate were taken up by the wild-type and me1.me2 mitochondria at a similar rate (Figure 4, B and C). However, while wild-type mitochondria converted imported malate to pyruvate via NAD-ME, no rate of pyruvate export could be measured in me1.me2. Instead, me1.me2 mitochondria showed an even higher consumption rate of 13C3-pyruvate than the wild-type, as evidenced by an increased rate of 13C2-citrate synthesis and export (Figure 4D). This enhanced conversion into downstream labeled TCA intermediates, such as succinate and malate, was consistent with me1.me2 having a rate of imported pyruvate consumption higher than the wild-type (Figure 4E; Supplemental Figure S6). In contrast, the loss of both MPC and NAD-ME in me1.me2.mpc1 resulted in the inability of isolated mitochondria to import and oxidize externally fed pyruvate and malate, presumably due to the accumulation of OAA in the matrix. Mitochondria from these triple knockout plants displayed nearly no U-13C-pyruvate transport and relatively low malate transport, 0.75 ± 0.58 and 7.4 ± 7.6 nmol·min–1·mg–1 respectively, compared to 33.7 ± 17.4 and 63.1 ± 7.6 nmol·min–1·mg–1 protein in wild-type mitochondria (Figure 4B). Reduced incorporation of the supplied substrates into me1.me2.mpc1 mitochondria was confirmed by the limited rate of conversion into subsequent TCA intermediates (Supplemental Figure S7).
Alanine is a third source of mitochondrial pyruvate in Arabidopsis
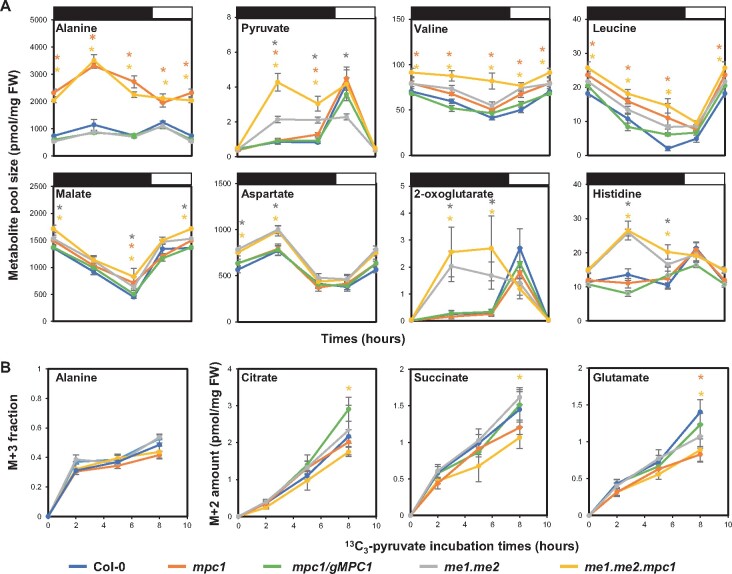
Compared to the substantial growth phenotype caused by a decrease in PDC activity (Ohbayashi et al., 2019), the phenotype of me1.me2.mpc1 plants is comparatively mild. This suggests that there could be a third source of mitochondrial pyruvate to support the development of mutant plants lacking both NAD-ME and MPC. To explore which metabolite could be responsible, we measured pool sizes of primary metabolites over a light/dark cycle. Metabolomics revealed that mpc1 leaves accumulated four- to six-fold higher levels of alanine throughout the diurnal cycle compared to the wild-type (Figure 5). An increase in the abundances of valine, leucine, and isoleucine was also observed in mpc1. These amino acids are most likely synthesized as a consequence of glycolytic pyruvate that cannot enter into the mitochondrial matrix (Singh and Shaner, 1995). Pyruvate and malate were slightly increased in abundance in mpc1 at night but no overall abundance change in TCA cycle intermediate was observed (Supplemental Figure S8). In contrast, knockout of NAD-ME led to elevated pyruvate and 2-oxoglutarate (2-OG) levels at night. me1.me2.mpc1 appeared to exhibit the combined effect of mpc1 and me1.me2 on metabolite abundances (Figure 5A). In addition, knockout of both MPC1 and NAD-ME magnified the pyruvate accumulation to four-fold compared to the wild-type, which was consistent with a block in pyruvate-dependent mitochondrial metabolism (Figure 5A).
Figure 5.
Alanine accumulation and usage helps supply pyruvate in the absence of MPC and NAD-ME. A, Diurnal changes of representative metabolites of Col-0 (blue), mpc1 (orange), mpc1/gMPC1 (green), me1.me2 (gray), and me1.me2.mpc1 (yellow). Leaf discs were collected from 6-week-old plants grown under short-day conditions. Samples were collected at 1, 8, 15 h after the switch to darkness and 4 h after the switch to light. The length of dark and light periods is indicated by black and white bars, respectively. The unit of metabolite absolute amount (vertical axis) is pmol·mg−1 fresh weight. Each data point represents an averaged value from at least six replicates with error bars indicating the standard error. Significant differences between tested genotypes versus Col-0 are indicated based on Student’s t test and denoted by corresponding colored asterisks. B, Time courses of 13C-labelling into metabolites using 13C3-pyruvate in darkened leaf discs. Leaf discs were collected from 6-week-old plants grown under short-day conditions and incubated in 20-mM 13C3-pyruvate solution for 2, 5, and 8 h, respectively. Incorporation patterns of 13C3-pyruvate into TCA cycle intermediates via PDC are similar to information shown in Figure 3A. [M + 3] alanine fraction and absolute abundances of [M + 2] citrate, [M + 2] succinate and [M + 2] glutamate are shown. Means ± se (n = 4). Significant differences between mutants and Col-0 are indicated based on Student’s t test and denoted by corresponding colored asterisks (*P < 0.05)
To probe the rate of pyruvate incorporation into primary metabolism in intact tissue, we fed 13C3- pyruvate to leaf discs in the dark. Amongst five genotypes, only me1.me2.mpc1 showed a slightly lower rate of 13C-incorporation into citrate and succinate, which could be attributed to the deficiency in mitochondrial pyruvate utilization due to the lack of NAD-ME and MPC activities (Figure 5B). The M + 3 alanine fraction (directly produced from amination of 13C3-pyruvate) showed no significant change in relative distribution (Figure 5B) while the M + 3 amount increased more significantly in mpc1 and me1.me2.mpc1 than that in the wild-type (Supplemental Figure S8), which indicates that the increased alanine pool size was due to a higher demand for this amino acid rather than blockage in alanine-requiring enzymatic reactions. Similar results were also observed following 13C6-glucose labeling of leaf discs (Supplemental Figure S9). These data provide evidence that alanine-2-OG transamination is a third pathway for providing pyruvate to sustain mitochondrial TCA cycle metabolism and respiration. Mitochondrial AlaAT abundance measured at the wild-type level in mpc1 and me1.me2.mpc1 (Supplemental Figure S2), suggesting that the increase in alanine use was not driven by enzyme concentration. Indeed, the significantly lower amount of labeled glutamate (Figure 5B) and the elevated level of 2-OG pool size at night (Figure 5A) in me1.me2.mpc1 plants indicate an altered equilibrium in total cellular transamination activities compared to the wild-type. Such a change in 2-OG:glutamate in plants lacking MPC1 could be a consequence of increased demand for alanine transamination reactions in mitochondria and cytosol. This pathway via AlaAT appeared to carry sufficient pyruvate flux for plant metabolism as label incorporation into most TCA cycle metabolites was maintained at wild-type levels, except for citrate and succinate (Figure 5B; Supplemental Figure S8).
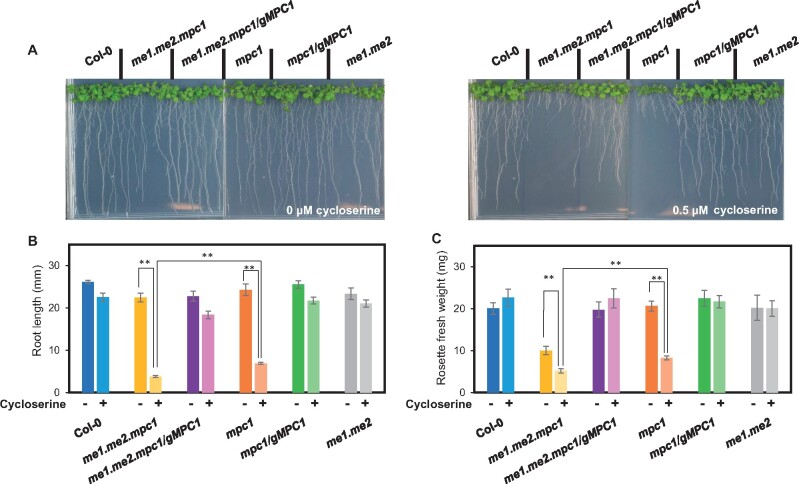
Plants lacking MPC are hypersensitive to inhibition of alanine aminotransferases by cycloserine
The existence of a yet-to-be identified mitochondrial alanine carrier (Passarella et al., 2003) could enable alanine to be imported from the cytosol into mitochondria. As AlaATs in mitochondria and the cytosol can inter-convert pyruvate and alanine, it would be possible to bypass MPC by replacing pyruvate with alanine as a primary entry point for glycolytic products to enter mitochondrial metabolism. Using the established AlaAT inhibitor cycloserine (Wong et al., 1973; Cornell et al., 1984; Duff et al., 2012), we subjected mpc1, me1.me2 and me1.me2.mpc1 and wild-type plants to chemical inhibition of AlaATs to assess the contribution of different pyruvate sources to seedling development (Figure 6). The me1.me2.mpc1 plants displayed only a moderate difference in rosette size under normal conditions, suggesting that AlaAT alone can largely support seedling growth. In the presence of 0.5-µM cycloserine; however, the rosette size and root length of me1.me2.mpc1 and mpc1 was significantly reduced compared to that of wild-type plants. These phenotypes could be complemented by reintroducing the MPC1 transgene into the mpc1-carrying mutants (Figure 6). me1.me2.mpc1 was more sensitive than mpc1 to this inhibitor, as shown by the significant difference in their rosette size and root length. This could be interpreted as evidence that NAD-ME activity in mpc1 accounts for a small percentage of pyruvate generation in vivo. In contrast, me1.me2 seedlings were insensitive to cycloserine treatment, indicating that MPC alone was sufficient to supply the bulk of pyruvate needed for sustaining seedling growth.
Figure 6.
Phenotypes of MPC1 and/or NAD-ME mutants in response to cycloserine treatment. Seeds were sterilized and placed on 1/2 Murashige-Skoog (MS) plates in the absence (left) or presence (right) of 0.5-µM cycloserine and grown under long-day conditions (16-h light/8-h dark, 60% humidity) (A). Representative photos were taken after 2 weeks. Bar graphs showing root length (B) and rosette fresh weight (C) of the six genotypes measured after 2 weeks. Each data point represents an averaged value from 10 replicates with error bars indicating the standard error. Significant differences between genotypes are indicated based on Student’s t test and denoted by asterisks (**P < 0.01)
Taken together, our data show that MPC and NAD-ME can both independently supply pyruvate to the TCA cycle in Arabidopsis mitochondria. However, when both these pyruvate sources are absent, metabolite pool sizes are still largely maintained (Supplemental Figure S7) with the exception of alanine utilization, and a mild change in plant development is observed. Blocking AlaATs severely affects growth in me1.me2.mpc1 and mpc1 while it has little effect on wild-type plants. This suggests pyruvate–alanine cycling via cytosolic and mitochondrial AlaATs is another important pathway that works cooperatively with MPC and NAD-ME to maintain mitochondrial pyruvate supply for dark respiration in plants.
Discussion
Mitochondrial pyruvate transport has not been considered as the dominant pathway for supplying oxidative substrates to respiration in plants for decades (Day and Hanson, 1977; Tcherkez et al., 2005; Araújo et al., 2012; Lehmann et al., 2016), since malate alone can provide pyruvate and OAA concomitantly within the matrix to drive the TCA cycle operation (Macrae and Moorhouse, 1970, Selinski and Scheibe, 2019). Although MPC1 has been recently shown to impact cadmium tolerance and stomatal opening in plants (Shen et al., 2017; He et al., 2019), no studies have been conducted in plants to prove that the MPC complex transports pyruvate into mitochondria or demonstrate its roles in mitochondrial respiratory metabolism. In this study, we provided genetic and biochemical evidence for MPC1 being the core carrier isoform responsible for the assembly and function of the mitochondrial pyruvate import complex in plants. In yeast, the presence of yeast MPC1 stabilizes yeast MPC2 and MPC3 through physical interaction as functional heterodimers (Tavoulari et al., 2019). In a similar manner, the loss of MPC1 in Arabidopsis resulted in the absence of both MPC3 and MPC4 (Figure 1B). This explains why mpc1 and higher-order mutants carrying the mpc1 allele produce the same cadmium-dependent short-root phenotypes as mpc1 (He et al., 2019). Our results show the MPC acts only as a mitochondrial pyruvate importer, as mitochondrial matrix generated pyruvate could still be readily exported to the extra-mitochondrial space by mpc1 mitochondria (Figure 3G;Supplemental Figures S3 and S4). This suggests the existence of other mitochondrial pyruvate transporter(s) that export pyruvate from the mitochondrial matrix, which is consistent with evidence that carriers other than MPC are responsible for mitochondrial pyruvate export in mammals (Hong et al., 2016; Chinopoulos, 2020). More crucially, our study also provides strong evidence that mitochondrial NAD-ME and cytosolic and mitochondrial AlaATs operate cooperatively with MPC to maintain the mitochondrial pyruvate pool in vivo in Arabidopsis (Figure 7).
Figure 7.
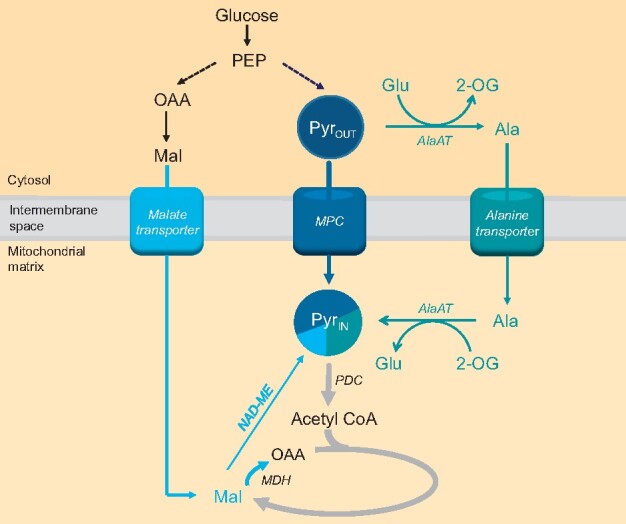
The mitochondrial pyruvate pool is contributed to by multiple supply pathways. A combination of in vitro, in organello, and in vivo analyses suggest the supply of mitochondrial pyruvate is facilitated mainly by MPC and AlaAT pathways; inhibition of one is compensated for by the other (e.g. increased alanine level is the most prominent feature of mpc1 mutants). The majority of malate imported into the matrix under physiological pH conditions is likely to be oxidized into OAA; thus, NAD-ME would synthesize a relatively small amount of pyruvate compared to the two major pathways, which was confirmed by analysis of me1.me2. MPC was the only pathway able to fully supply the pyruvate pool on its own, evidenced by the fact that cycloserine-treated me1.me2 seedlings can maintain normal growth, while ALT alone in me1.me2.mpc1 had a growth deficiency phenotype. Dashed lines indicate unknown relative fluxes. Bolder arrows indicate larger flux compared to other pathways using or generating the same metabolite. PEP, phosphoenolpyruvate; Pyrout, pyruvate in the cytosol, Pyrin, pyruvate inside the mitochondrial matrix; Ala, alanine; Glu, glutamate
The degree to which MPC1 affects growth of different organisms reflects the flexibility of pyruvate metabolism and the proportion of oxidative phosphorylation generated by MPC-dependent pyruvate flux in different organisms. In mice, MPC is the primary carrier of oxidative substrates for energy production in mitochondria and catalyzes most of the pyruvate flux, which cannot be sufficiently compensated for by other potential pyruvate-supplying pathways, leading to embryonic lethality when MPC is absent (Vanderperre et al., 2016). In yeast, the role of MPC is less important as the existence of a PDC bypass involving cytosolic pyruvate decarboxylation enables mitochondrial transport of substrates independently of MPC function (Boubekeur et al., 1999). Such flexibility of pyruvate utilization also exists in Arabidopsis as seen by a lack of an obvious phenotype under normal growth conditions in mutant lacking the MPC complex (Supplemental Figure S1B). Our findings indicate that instead, plants utilize a mixture of MPC and MPC bypasses to supply mitochondria with pyruvate. NAD-ME was more active in converting malate to pyruvate in isolated mpc1 mitochondria compared to the wild-type (Figures 2, B and 3, G). Conversely, knocking out NAD-ME resulted in an increase in MPC activity in isolated mitochondria as evidenced by the 50% increase in 13C incorporation into TCA intermediates (Figure 4E). The effect of impaired pyruvate supply on plant growth becomes visually apparent when MPC1 was knocked out in the me1.me2 background. This triple mutant was not only unable to import pyruvate and convert malate to pyruvate but also showed a reduced the rate of mitochondrial malate transport, likely due to feedback inhibition by OAA accumulation in the matrix. Consistent with our results, knocking out both MPC and mitochondrial NAD-ME in yeast magnified the slow-growth phenotype under nonstresses conditions which was not observed in the single MPC1 knockout (Bricker et al., 2012). Taken together, the loss of MPC1 appears to trigger the promotion of NAD-ME flux to pyruvate and vice versa, at least in isolated mitochondria. This complementary regulation of MPC and mitochondrial NAD-ME activities is not as clear in single knockouts in planta due to metabolic flexibility, which ensures that the overall metabolic and energy homeostasis are maintained without plant growth consequences.
Metabolomics and 13C-pyruvate tracing in vivo only showed slight changes in metabolite levels in all genotypes examined, highlighting the metabolic flexibility of Arabidopsis leaves to maintain metabolite pool size in plants lacking MPC1. The only exceptions were alanine and 2-OG abundances, which were higher in mpc1 and me1.me2, respectively. The phenotype of me1.me2.mpc1 is most likely caused by the combined metabolic effects of these mutations, and the change in 13C labeling into glutamate (Figure 6B), which could be an indicator of an altered flux through AlaAT (Zhang et al., 2020). The hypersensitivity of the me1.me2.mpc1 and mpc1 plants to cycloserine treatment indicates that pyruvate–alanine cycling between mitochondria and cytosol can become a major pathway under certain conditions, such as when MPC1 is absent (Figure 7). However, there appears to be an energy cost associated with pyruvate–alanine cycling as the main mitochondrial pyruvate supply. The priority of diverting glutamate and 2-OG pools into inter-conversion of pyruvate and alanine in mitochondria and cytosol makes these metabolites less available for other aminotransferases, which are critical for modulating cellular nitrogen metabolism and mediating nitrogen-dependent stress response (McCommis et al., 2015). Decreased cadmium sensitivity of mpc1 (He et al., 2019) is an example of the consequences of increased flux through the pyruvate–alanine cycling pathway, as glutamate is not as readily available to produce glutathione to scavenge cadmium ions (Sytar et al., 2013; Tompkins et al., 2019) and to adjust amino acid profiles, which have been shown to be important for heavy metal stress response (Blasco and Puppo, 1999; Sharma and Dietz, 2006; Zhu et al., 2018). Moreover, just as in MPC and NAD-ME single knockout mutants, mutant lacking the major AlaAT isoform (alt1) does not cause a vegetative phenotype (Miyashita et al., 2007), suggesting that at least two out of three pyruvate-supplying pathways are needed to support mitochondrial respiration and prevent an impact on plant growth.
Metabolic flexibility has long been considered a hallmark of what makes plant mitochondria unique (Douce and Neuburger, 1989). Much of the research on this flexibility has focused on the branching of the ETC (Vanlerberghe and McIntosh, 1997; Rasmusson et al., 2004; Millar et al., 2011) and the range of substrates that contribute reducing potential to the ETC (Schertl and Braun, 2014). Metabolic flexibility has been shown for TCA cycle components as well, given that these enzymes are mostly not exclusively targeted to mitochondria (Schnarrenberger and Martin, 2002; Araújo et al., 2012). Reduction in the activity of TCA cycle components such as mitochondrial citrate synthase (Sienkiewicz-Porzucek et al., 2008), NAD-dependent isocitrate dehydrogenase (Sienkiewicz-Porzucek et al., 2010), aconitase (Carrari et al., 2003), malate dehydrogenase (Nunes-Nesi et al., 2005), and fumarase (Nunes-Nesi et al., 2007) mostly resulted in normal phenotypes and minor changes in respiratory and photosynthetic performances in tomato (Solanum lycopersicum) plants due to metabolic re-adjustment through their counterparts in the cytosol, peroxisome, and/or plastids. Alternatively, the catabolism of amino acids, some of which accumulated in mpc1 mutants (Supplemental Figure S8), can bypass pyruvate import to supply mitochondria with TCA cycle substrates (Araújo et al., 2011; Kochevenko et al., 2012; Cavalcanti et al., 2017). Furthermore, mitochondrial components often play an important role in a larger, highly flexible metabolic network linking carbon metabolism and nitrogen metabolism. For example, 2-OG as the main checkpoint of both processes can be produced by the concerted action of isocitrate dehydrogenases, aminotransaminases, and glutamate dehydrogenase either in the mitochondrial matrix or cytosol (Weber and Flügge, 2002; Foyer et al., 2003). The appropriate allocation of 2-OG to TCA cycle and amino acid biosynthesis determines metabolic homeostasis. Also, the gamma-aminobutyric acid (GABA) shunt can bypass two steps of TCA cycle by allowing the synthesis of GABA in the cytosol during stresses including salinity, drought, heat, and hypoxia (Bouché and Fromm, 2004) and transport GABA back to the mitochondria to make succinate and re-enter the TCA cycle (Michaeli et al., 2011). In a similar manner, this work introduces AlaAT as a major flux contributor to the canonical pyruvate supply pathways by efficiently bypassing pyruvate import from the cytosol to the mitochondrial matrix rather than just being considered a stress response pathway (Ricoult et al., 2006; Miyashita and Good, 2008; Diab and Limami, 2016). In this context, it is notable that a positive correlation of respiratory rates in Arabidopsis leaves was more apparent for alanine than for pyruvate, sugars, or other organic and amino acids (O'Leary et al., 2017) and alanine is a strong stimulator of respiratory rate in vivo in Arabidopsis (O'Leary et al., 2020).
Mitochondrial pyruvate supply requires flexibility as pyruvate is an essential starting material for carbon metabolism in mitochondria, greatly affecting all downstream reactions and eventually energy production and plant development. Given that MPC is the major mitochondrial pyruvate supplier in autotropic tissues, its activity must match to changes in demand due to regulation of PDC by post-translational modification. Consistent with reports of a night-time increase in PDC activity (Budde and Randall, 1990; Tovar-Méndez et al., 2003), the absence of MPC1 resulted in the accumulation of pyruvate and related amino acids only in the dark (Supplemental Figure S8). Since MPC1 is ubiquitously expressed in both heterotrophic and autotrophic tissues (Shen et al., 2017), the need for MPC activity in different tissue types is most likely dependent on PDC regulation and the requirement for TCA cycle initiation. However, the quantitative involvement of three pathways under changing internal and external environments in plants contributes to a remarkable plasticity in pyruvate supply in plants. While NAD-ME alone failed to compensate for the loss of both MPC and AlaAT, AlaAT alone (in me1.me2.mpc1) only resulted in a moderate vegetative phenotype while MPC alone (in cycloserine treated me1.me2) was able to maintain a normal plant growth (Figure 6). This suggests their relative contribution to the mitochondrial pyruvate pool in vivo and as a result their roles in central metabolism. Future experiments could investigate the activity of different MPC complexes or the contribution of MPC to total pyruvate flux into mitochondria in different tissues such as stems, roots, siliques, or flowers.
In conclusion, our data suggest plants possess at least three pathways that all synergistically contribute to mitochondrial pyruvate supply and usage for respiration. These pathways are coordinated in such a way that, when one is compromised the other two increase in flux to maintain metabolic and redox homeostasis for optimal growth. This allows metabolic flexibility to cope with changes in cellular substrate availability, particularly during environmental changes such as nutrient and metal stresses, hypoxia, cold and drought. NAD-ME appears to contribute less to pyruvate supply than MPC and AlaAT pathways as AlaAT and MPC alone can largely support normal plant growth, but not NAD-ME alone (Figure 7). Meanwhile, AlaAT is an efficient alternative pathway to divert pyruvate flux from MPC especially when MPC is defective (Figure 7). A challenge for the future will be to accurately quantify the relative fluxes of all three pyruvate-contributing pathways to leaf respiration under different nutrient and stress conditions. This will be important to further understand one of the most important mitochondrial processes, and from there, the exploitation of plant energy generation and usage in plants.
Materials and methods
Plant material, growth conditions, and quantitative RNA analysis
The MPC1 T-DNA insertion line SALK008945 was obtained from the Arabidopsis Biological Resource Center (https://abrc.osu.edu/). me1.me2 seeds were previously characterized and published (Tronconi et al., 2008) and were obtained from Professor Verónica G. Maurino (University of Bonn). me1.me2.mpc1 was created from crossing between me1.me2 and mpc1. The complemented lines mpc1/gMPC1 and me1.me2.mpc1/gMPC1 were generated by introducing the genomic version of MPC1 with its endogenous promoter into the corresponding mutants using pCambia1380 as the vector. Specifically, genomic MPC1 was amplified using gMPC1_F and gMPC1_R primers (Supplemental Table S1) and cloned into the plasmid pCambia1380, which was then transformed into Escherichia coli to replicate the plasmid. The mutants were floral dipped in the culture of Agrobacterium transformed with pCambia1380-gMPC1. Seeds were collected and selected using 15 μg·mL–1 hygromycin B plates for three generations before being utilized for experiments to ensure genetic stability.
Seeds were stratified for 3 days in the dark at 4°C before being transferred to either long-day or short-day conditions. Phenotype analysis was performed on plants grown under long-day conditions of 16-h light, 8-h dark, and 60% humidity (110 µmol s−1 m−1 light intensity with tubular fluorescent lighting). For cycloserine treatments, Arabidopsis seedlings were grown on vertical one-half strength Murashige and Skoog agar (10 g·L–1 agar, 0.4 g·L–1 MES) plates with 0.5-µM cycloserine added after autoclaving.
Plants used in metabolic and stable isotope labeling analysis were grown under short-day conditions of 8-h light, 16-h dark, and 60% humidity (110 µmol s−1 m−1 light intensity with tubular fluorescent lighting).
RNA was extracted from about 20-mg leaf tissue of three biological replicates per genotype using the Spectrum Plant Total RNA Kit (Sigma-Aldrich) according to the manufacturer’s instructions. The RNA quality and quantity were assessed using a nanodrop ND-1000 spectrophotometer (ThermoFisher). RNA was mixed with primers and qPCR reaction components provided in QuantiNova SYBR Green RT-PCR Kit in 96-well plate. Quantitative transcript analysis was done using the LightCycler 480 System (Roche). A list of qPCR primers is provided in Supplemental Table S1.
Isolation of mitochondria, O2 electrode measurements, and enzyme activity assays
Arabidopsis seeds were surface sterilized and dispensed into one-half strength Murashige and Skoog liquid media (1.1 g·L–1 agar, 0.4 g·L–1 MES, 10 g·L–1 sucrose) within enclosed, sterilized 100-mL polypropylene containers. The containers were seated on a rotating shaker in the long-day conditions as described above and seedlings were harvested after 2 weeks.
Mitochondria were isolated from 2-week-old Arabidopsis seedlings as described previously (Millar et al., 2007). O2 electrode measurements were carried out as previously described (Lee et al., 2010), except pyruvate was added at 0, 0.5, 1, 2, or 5 mM and malate was added at 0.5 mM for pyruvate-dependent OCR measurement. PDH, NAD-ME in vitro activity assays were carried out as described previously (Huang et al., 2015).
Analyses of metabolites by LC–SRM–MS
Leaf discs (∼20 mg) were collected from 6-week-old short-day grown (8-h light/16-h dark) plants at 1, 8, and 15 h after dark switch and 4 h after light switch. Samples were snap-frozen in liquid nitrogen and stored at –80°C. Frozen samples were ground using Retsch mixer mill before metabolites were extracted using 90% methanol containing 13C-leucine and adipic acid as internal standards. After incubation at 75°C for 20 min and centrifugation at room temperature at 18,000g for 3 min, supernatants were aliquoted and dried using a SpeedVac concentrator.
For quantitation of organic acids, dried samples were re-suspended in 100-µL methanol and derivatized as described previously (Han et al., 2013). Samples were analyzed by an Agilent 1100 HPLC system coupled to an Agilent 6430 Triple Quadrupole (QQQ) mass spectrometer equipped with an electrospray ion source as described previously (Lee et al., 2020). Chromatographic separation was performed on a Kinetex C18 column, using 0.1% formic acid in water (solvent A) and 0.1% formic acid in methanol (solvent B) as the mobile phase for binary gradient elution (Supplemental Table S2). Data were acquired using Agilent MassHunter Workstation Data Acquisition software. The column flow rate was 0.3 mL·min–1; the column temperature is 40°C, and the autosampler was kept at 10°C. SRM transitions for each targeted analyte are shown in Supplemental Table S3. Optimizations for metabolite transitions for SRM–MS were described in detail elsewhere (Le et al., 2021).
For amino acid quantification, dried samples were re-suspended in 50 µL water. Samples were analyzed using the same LC–MS system as described above. Chromatographic separation was performed using Agilent Poroshell 120 HILIC-Z column, using mobile phases of 20-mM ammonium formate in water (solvent A) and 20-mM ammonium formate in acetonitrile (solvent B, Supplemental Table S4). The column flow rate was 0.4 mL·min–1; the column temperature was 35°C, and the autosampler was kept at 10°C. SRM transitions for each targeted analyte are listed in Supplemental Table S5.
Substrate feeding of isolated mitochondria
The detailed methods and materials for MS-based mitochondria feeding assays were described previously (Le et al., 2021). In short, 100-µg isolated mitochondria were mixed with substrates (0.5-mM pyruvate and 0.5-mM malate), cofactors (2-mM NAD+, 0.2-mM TPP, and 0.012-mM CoA), and 1-mM ADP (for ATP synthesis) in a final volume of 200 µL. At specified times, this reaction mixture was layered on top of silicon oil (AR200, 90 µL), which was layered above the stopping sucrose solution (0.5 M sucrose, pH 1.0). Substrate transport was stopped by rapid centrifugation (12,000g at 20°C for 3 min) to harvest the mitochondria at the bottom of the tube. Five microliters of the extra-mitochondrial medium (the top layer) was collected and extracted for quantitative analysis by LC–SRM–MS.
13C-pyruvate and 13C-glucose labeling of Arabidopsis leaf discs and analysis of labeled metabolites
Leaf discs (∼50 mg) were prepared from 6-week-old short-day grown (8-h light/16-h dark) plants 1 h before the end of a normal light photoperiod. They were floated on leaf respiratory buffer containing 20-mM 13C3-pyruvate (99% purity, Sigma Aldrich), 20-mM 13C6-glucose (99% purity, Sigma Aldrich), or an unlabeled counterpart. At the specified incubation time, leaf discs were briefly washed with respiratory buffer to remove excess labeled pyruvate and frozen in liquid nitrogen for metabolite extraction as stated above. Analyses of total, untargeted metabolites were performed using an Agilent 1100 HPLC system coupled to an Agilent 6520 Quadrupole/Time-of-Flight mass spectrometer equipped with an electrospray ion source. Chromatographic separation was performed on an Agilent Poroshell 120 HILIC-Z column, using 10-mM ammonium acetate in water (solvent A) and 10-mM ammonium acetate in acetonitrile (solvent B) as mobile phases (Supplemental Table S6). Solvents A and B were supplied with 0.1% (v/v) Infinity Lab deactivator additive (Agilent) to improve peak shapes. The column flow rate was 0.25 mL·min–1; the column temperature was 40°C, and the autosampler was kept at 10°C. The scanning range was from 70 m/z to 230 m/z.
Peak extraction was done using Agilent Mass Hunter Quantitative analysis software based on known m/z values and retention time obtained from authentic standards. Raw peak was then isotopically corrected for natural 12C abundances using IsoCor software (Millard et al., 2019). Percentage of each isotopolog of a compound of interest was calculated as the ratio of the peak area of the isotopolog over the total area of both labeled and unlabeled fractions. Absolute labeled amount of each isotopolog of a compound was calculated through multiplying the percentage of labeled isotopolog by the absolute amount of that compound obtained from the analysis of unlabeled sample by LC–SRM–MS as described above.
Relative quantification of mitochondrial protein abundances by LC–SRM–MS
One hundred micrograms frozen mitochondrial proteins were precipitated in 100% acetone for 24 h at −20°C and the pellets were washed with acetone for three times. Samples were alkylated, trypsin digested, desalted and cleaned as previously described (Petereit et al., 2020). Samples were loaded onto an AdvanceBio Peptide Map column (2.1 × 250 mm, 2.7-μm particle size; part number 651750-902, Agilent), using an Agilent 1290 Infinity II LC System coupled to an Agilent 6495 Triple Quadrupole MS. The column was heated to 60°C and the column flow rate was 0.4 mL·min-1. The binary elution gradient for HPLC and the list of peptide transitions used for SRM–MS are provided in Supplemental Table S7 and Supplemental Data Set S1, respectively. Peak area of targeted peptides was determined using the Skyline software package. Peptide abundances from each sample were normalized against voltage-dependent anion channel (VDAC) since its abundance was similar amongst all samples.
Statistical analysis
All statistical analyses were performed using the two-sided t test function built in Excel 2010. Statistical tests and the number of biological replicates are indicated in figure legends. Biological replicates indicate samples that were collected from different batches of plants grown under the same conditions except biological replicates for transcript analysis and metabolite analysis were samples collected from different plants grown at the same time.
Accession numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AtMPC1 (At5g20090), AtMPC4 (At4g05590), AtMPC3 (At4g22310), AtMPC2 (At4g14695), AtNAD-ME1 (At2g13560), and AtNAD-ME2 (At4g00570).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 . Gene model and phenotypes of mpc1.
Supplemental Figure S2 . Changes in the abundance and activity of respiratory enzymes in tested mutants.
Supplemental Figure S3 . 13C3-Pyruvate and malate feeding to isolated mitochondria of Col-0, mpc1 and mpc1/gMPC1.
Supplemental Figure S4 . Swap label experiment of pyruvate and 13C4-malate feeding into isolated mitochondria of Col-0, mpc1 and mpc1/gMPC1.
Supplemental Figure S5 . Calculated import and export rates from the swap label time course experiment of pyruvate and 13C4-malate feeding into isolated mitochondria of Col-0, mpc1 and mpc1/gMPC1.
Supplemental Figure S6 . Soil-based phenotypes of mpc1, me1.me2 and complementary genotypes.
Supplemental Figure S7 . Time courses of metabolite concentrations in the extra-mitochondrial space of 13C3-pyruvate and malate feeding into Col-0, me1.me2, and me1.me2.mpc1 mitochondria
Supplemental Figure S8 . Diurnal changes of representative metabolites in different genotypes.
Supplemental Figure S9 . Incorporation of 13C3-pyruvate and 13C-glucose into alanine, TCA cycle intermediates, and related amino acids of leaf discs from different genotypes in the dark.
Supplemental Table S1 . A list of qPCR primers used.
Supplemental Table S2 . The binary elution gradient of solvents A and B for HPLC used for organic acid quantification.
Supplemental Table S3 . The SRM transitions used as quantifier and qualifier for derivatized labeled and unlabeled TCA cycle metabolites of interest.
Supplemental Table S4 . The binary elution gradient of solvents A and B for HPLC used for amino acid quantification.
Supplemental Table S5 . The SRM transitions used as quantifier and qualifier for amino acid measurement.
Supplemental Table S6 . The binary elution gradient of solvents A and B for HPLC used for untargeted metabolite analysis.
Supplemental Table S7 . The binary elution gradient of solvents A and B for HPLC used for mitochondrial protein abundance measurement.
Supplemental Data Set S1 . The SRM transitions used to measure relative abundance of mitochondrial proteins.
Supplemental Data Set S2 . All figure statistical tests.
Supplementary Material
Acknowledgments
We thank Ricarda Fenske for optimizing method and running LC–SRM–MS for the mitochondrial protein abundance measurements. We thank Brendan O’Leary for a critical reading of the manuscript.
Funding
This work was supported by the Australian Research Council Centre of Excellence in Plant Energy Biology (CE140100008) and X.H.L. is a Forrest Scholar supported by the Forrest Research Foundation and a receiver of Research Training Program scholarships from the Department of Education, Skills and Employment in the Australian Government.
Conflict of interest statement. The authors declare that they have no conflict of interest.
X.H.L., C.P.L., and A.H.M. designed the research. X.H.L. performed most of the experiments and data analysis, C.P.L. assisted with some of the mass spectrometry and data analysis. X.H.L., C.P.L., and A.H.M. wrote the paper.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: A. Harvey Millar (harvey.millar@uwa.edu.au).
References
- Araújo WL, Tohge T, Ishizaki K, Leaver CJ, Fernie AR (2011) Protein degradation – an alternative respiratory substrate for stressed plants. Trends Plant Sci 16: 489–498 [DOI] [PubMed] [Google Scholar]
- Araújo WL, Nunes-Nesi A, Nikoloski Z, Sweetlove LJ, Fernie AR (2012) Metabolic control and regulation of the tricarboxylic acid cycle in photosynthetic and heterotrophic plant tissues. Plant Cell Environ 35: 1–21 [DOI] [PubMed] [Google Scholar]
- Bender T, Martinou JC (2016) The mitochondrial pyruvate carrier in health and disease: to carry or not to carry? Biochim Biophys Acta 1863: 2436–2442 [DOI] [PubMed] [Google Scholar]
- Blasco J, Puppo J (1999) Effect of heavy metals (Cu, Cd and Pb) on aspartate and alanine aminotransferase in Ruditapes philippinarum (Mollusca: Bivalvia). Comp Biochem Physiol C Pharmacol Toxicol Endocrinol 122: 253–263 [DOI] [PubMed] [Google Scholar]
- Boubekeur S, Bunoust O, Camougrand N, Castroviejo M, Rigoulet M, Guérin B (1999) A mitochondrial pyruvate dehydrogenase bypass in the yeast Saccharomyces cerevisiae. J Biol Chem 274: 21044–21048 [DOI] [PubMed] [Google Scholar]
- Bouché N, Fromm H (2004) GABA in plants: just a metabolite? Trends Plant Sci 9: 110–115 [DOI] [PubMed] [Google Scholar]
- Brailsford MA, Thompson AG, Kaderbhai N, Beechey RB (1986) Pyruvate metabolism in castor-bean mitochondria. Biochem J 239: 355–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricker DK, Taylor EB, Schell JC, Orsak T, Boutron A, Chen YC, Cox JE, Cardon CM, Van Vranken JG, Dephoure N, et al. (2012). A mitochondrial pyruvate carrier required for pyruvate uptake in yeast, Drosophila, and humans. Science 337: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brivet M, Garcia-Cazorla A, Lyonnet S, Dumez Y, Nassogne MC, Slama A, Boutron A, Touati G, Legrand A, Saudubray JM (2003) Impaired mitochondrial pyruvate importation in a patient and a fetus at risk. Mol Genet Metab 78: 186–192 [DOI] [PubMed] [Google Scholar]
- Budde RJ, Randall DD (1990) Pea leaf mitochondrial pyruvate dehydrogenase complex is inactivated in vivo in a light-dependent manner. Proc Natl Acad Sci USA 87: 673–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capuano F, Di Paola M, Azzi A, Papa S (1990) The monocarboxylate carrier from rat liver mitochondria. Purification and kinetic characterization in a reconstituted system. FEBS Lett 261: 39–42 [DOI] [PubMed] [Google Scholar]
- Carrari F, Nunes-Nesi A, Gibon Y, Lytovchenko A, Loureiro ME, Fernie AR (2003) Reduced expression of aconitase results in an enhanced rate of photosynthesis and marked shifts in carbon partitioning in illuminated leaves of wild species tomato. Plant Physiol 133: 1322–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalcanti JHF, Quinhones CGS, Schertl P, Brito DS, Eubel H, Hildebrandt T, Nunes-Nesi A, Braun H-P, Araújo WL (2017) Differential impact of amino acids on OXPHOS system activity following carbohydrate starvation in Arabidopsis cell suspensions. Physiologia Plantarum 161: 451–467 [DOI] [PubMed] [Google Scholar]
- Chinopoulos C (2020) From glucose to lactate and transiting intermediates through mitochondria, bypassing pyruvate kinase: considerations for cells exhibiting dimeric PKM2 or otherwise inhibited kinase activity. Front Physiol 11: 543564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell NW, Zuurendonk PF, Kerich MJ, Straight CB (1984) Selective inhibition of alanine aminotransferase and aspartate aminotransferase in rat hepatocytes. Biochem J 220: 707–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day DA (1980) Malate decarboxylation by Kalanchoë daigremontiana mitochondria and its role in Crassulacean acid metabolism. Plant Physiol 65: 675–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day DA, Hanson JB (1977) Pyruvate and malate transport and oxidation in corn mitochondria. Plant Physiol 59: 630–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diab H, Limami AM (2016) Reconfiguration of N metabolism upon hypoxia stress and recovery: roles of alanine aminotransferase (AlaAT) and glutamate dehydrogenase (GDH). Plants (Basel) 5 :25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douce R, Neuburger M (1989) The uniqueness of plant mitochondria. Annu Rev of Plant Physiol Plant Mol Biol 40: 371–414 [Google Scholar]
- Duff SMG, Rydel TJ, McClerren AL, Zhang W, Li JY, Sturman EJ, Halls C, Chen S, Zeng J, Peng J, et al. (2012) The enzymology of alanine aminotransferase (AlaAT) isoforms from Hordeum vulgare and other organisms, and the HvAlaAT crystal structure. Arch of Biochem and Biophys 528: 90–101 [DOI] [PubMed] [Google Scholar]
- Edwards S, Nguyen BT, Do B, Roberts JKM (1998) Contribution of malic enzyme, pyruvate kinase, phosphoenolpyruvate carboxylase, and the Krebs cycle to respiration and biosynthesis and to intracellular pH regulation during hypoxia in maize root tips observed by nuclear magnetic resonance imaging and gas chromatography-mass spectrometry. Plant Physiol 116: 1073–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Parry M, Noctor G (2003) Markers and signals associated with nitrogen assimilation in higher plants. J Exp Bot 54: 585–593 [DOI] [PubMed] [Google Scholar]
- Haferkamp I, Schmitz-Esser S (2012) The plant mitochondrial carrier family: functional and evolutionary aspects. Front Plant Sci 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halestrap AP (1975) The mitochondrial pyruvate carrier. Kinetics and specificity for substrates and inhibitors. Biochem J 148: 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halestrap AP, Denton RM (1974) Specific inhibition of pyruvate transport in rat liver mitochondria and human erythrocytes by α-cyano-4-hydroxycinnamate (Short Communication). Biochem J 138: 313–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Gagnon S, Eckle T, Borchers CH (2013) Metabolomic analysis of key central carbon metabolism carboxylic acids as their 3-nitrophenylhydrazones by UPLC/ESI-MS. Electrophoresis 34: 2891–2900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell J, Dever LV, Boxall SF (2016) Emerging model systems for functional genomics analysis of Crassulacean acid metabolism. Curr Opin Plant Biol 31: 100–108 [DOI] [PubMed] [Google Scholar]
- He L, Jing Y, Shen J, Li X, Liu H, Geng Z, Wang M, Li Y, Chen D, Gao J, Zhang W (2019) Mitochondrial pyruvate carriers prevent cadmium toxicity by sustaining the TCA cycle and glutathione synthesis. Plant Physiol 180: 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzig S, Raemy E, Montessuit S, Veuthey J-L, Zamboni N, Westermann B, Kunji ERS, Martinou JC (2012) Identification and functional expression of the mitochondrial pyruvate carrier. Science 337: 93. [DOI] [PubMed] [Google Scholar]
- Hildyard JCW, Halestrap AP (2003) Identification of the mitochondrial pyruvate carrier in Saccharomyces cerevisiae. Biochem J 374: 607–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong CS, Graham NA, Gu W, Espindola Camacho C, Mah V, Maresh EL, Alavi M, Bagryanova L, Krotee PAL, Gardner BK, et al. (2016) MCT1 modulates cancer cell pyruvate export and growth of tumors that co-express MCT1 and MCT4. Cell Rep 14: 1590–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Lee CP, Millar AH (2015) Activity assay for plant mitochondrial enzymes. Methods Mol Biol 1305: 139–149 [DOI] [PubMed] [Google Scholar]
- Hüdig M, Maier A, Scherrers I, Seidel L, Jansen EEW, Mettler-Altmann T, Engqvist MKM, Maurino VG (2015) Plants possess a cyclic mitochondrial metabolic pathway similar to the mammalian metabolic repair mechanism involving malate dehydrogenase and l-2-hydroxyglutarate dehydrogenase. Plant Cell Physiol 56: 1820–1830 [DOI] [PubMed] [Google Scholar]
- Jenner HL, Winning BM, Millar AH, Tomlinson KL, Leaver CJ, Hill SA (2001) NAD malic enzyme and the control of carbohydrate metabolism in potato tubers. Plant Physiol 126: 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochevenko A, Araújo WL, Maloney GS, Tieman DM, Do PT, Taylor MG, Klee HJ, Fernie AR (2012) Catabolism of branched chain amino acids supports respiration but not volatile synthesis in tomato fruits. Mol Plant 5: 366–375 [DOI] [PubMed] [Google Scholar]
- Le X, Millar AH, Lee CP (2021) Assessing the kinetics of metabolite uptake and utilisation by isolated mitochondria using selective reaction monitoring-mass spectrometry (SRM-MS). Methods Mol Biol 2363: 10.1007/978-1-0716-1653-6_8 [DOI] [PubMed] [Google Scholar]
- Lee CP, Eubel H, Millar AH (2010) Diurnal changes in mitochondrial function reveal daily optimization of light and dark respiratory metabolism in Arabidopsis. Mol Cell Proteomics 9: 2125–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CP, Elsässer M, Fuchs P, Fenske R, Schwarzländer M, Millar AH (2020) The Arabidopsis mitochondrial dicarboxylate carrier 2 maintains leaf metabolic homeostasis by uniting malate import and citrate export. bioRxiv, doi: 10.1101/2020.04.28.065441
- Lehmann MM, Wegener F, Barthel M, Maurino VG, Siegwolf RTW, Buchmann N, Werner C, Werner RA (2016) Metabolic fate of the carboxyl groups of malate and pyruvate and their influence on δ13C of leaf-respired CO2 during light enhanced dark respiration. Front Plant Sci 7: 739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CL, Wang M, Ma XY, Zhang W (2014) NRGA1, a putative mitochondrial pyruvate carrier, mediates ABA regulation of guard cell ion channels and drought stress responses in Arabidopsis. Mol Plant 7: 1508–1521 [DOI] [PubMed] [Google Scholar]
- Macrae AR, Moorhouse R (1970) The oxidation of malate by mitochondria isolated from cauliflower buds. Eur J Biochem 16: 96–102 [DOI] [PubMed] [Google Scholar]
- McCommis KS, Chen Z, Fu X, McDonald WG, Colca JR, Kletzien RF, Burgess SC, Finck BN (2015) Loss of mitochondrial pyruvate carrier 2 in the liver leads to defects in gluconeogenesis and compensation via pyruvate-alanine cycling. Cell Metab 22: 682–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaeli S, Fait A, Lagor K, Nunes-Nesi A, Grillich N, Yellin A, Bar D, Khan M, Fernie AR, Turano FJ, et al. (2011) A mitochondrial GABA permease connects the GABA shunt and the TCA cycle, and is essential for normal carbon metabolism. Plant J 67: 485–498 [DOI] [PubMed] [Google Scholar]
- Millar AH, Liddell A, Leaver CJ (2007) Isolation and subfractionation of mitochondria from plants. Methods Cell Biol 80: 65–90 [DOI] [PubMed] [Google Scholar]
- Millar AH, Whelan J, Soole KL, Day DA (2011) Organization and regulation of mitochondrial respiration in plants. Annu Rev Plant Biol 62: 79–104 [DOI] [PubMed] [Google Scholar]
- Millard P, Delépine B, Guionnet M, Heuillet M, Bellvert F, Létisse F (2019) IsoCor: isotope correction for high-resolution MS labeling experiments. Bioinformatics 35: 4484–4487 [DOI] [PubMed] [Google Scholar]
- Miyashita Y, Good AG (2008) Contribution of the GABA shunt to hypoxia-induced alanine accumulation in roots of Arabidopsis thaliana. Plant Cell Physiol 49: 92–102 [DOI] [PubMed] [Google Scholar]
- Miyashita Y, Dolferus R, Ismond KP, Good AG (2007) Alanine aminotransferase catalyses the breakdown of alanine after hypoxia in Arabidopsis thaliana. Plant J 49: 1108–1121 [DOI] [PubMed] [Google Scholar]
- Nałecz MJ, Nałecz KA, Azzi A (1991) Purification and functional characterisation of the pyruvate (monocarboxylate) carrier from baker’s yeast mitochondria (Saccharomyces cerevisiae). Biochim Biophys Acta 1079: 87–95 [DOI] [PubMed] [Google Scholar]
- Nałecz MJ, Nałecz KA, Broger C, Bolli R, Wojtczak L, Azzi A (1986) Extraction, partial purification and functional reconstitution of two mitochondrial carriers transporting keto acids: 2-oxoglutarate and pyruvate. FEBS Lett 196: 331–336 [DOI] [PubMed] [Google Scholar]
- Netting AG (2002) pH, abscisic acid and the integration of metabolism in plants under stressed and non‐stressed conditions. II. Modifications in modes of metabolism induced by variation in the tension on the water column and by stress. J Exp Bot 53: 151–173 [PubMed] [Google Scholar]
- Nunes-Nesi A, Carrari F, Lytovchenko A, Smith AMO, Ehlers Loureiro M, Ratcliffe RG, Sweetlove LJ, Fernie AR (2005) Enhanced photosynthetic performance and growth as a consequence of decreasing mitochondrial malate dehydrogenase activity in transgenic tomato plants. Plant Physiol 137:611–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Nesi A, Carrari F, Gibon Y, Sulpice R, Lytovchenko A, Fisahn J, Graham J, Ratcliffe RG, Sweetlove LJ, Fernie AR (2007) Deficiency of mitochondrial fumarase activity in tomato plants impairs photosynthesis via an effect on stomatal function. Plant J 50: 1093–1106 [DOI] [PubMed] [Google Scholar]
- O'Leary BM, Oh GGK, Lee CP, Millar AH (2020) Metabolite regulatory interactions control plant respiratory metabolism via target of rapamycin (TOR) kinase activation. Plant Cell 32: 666–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary BM, Lee CP, Atkin OK, Cheng R, Brown TB, Millar AH (2017) Variation in leaf respiration rates at night correlates with carbohydrate and amino acid supply. Plant Physiol 174: 2261–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohbayashi I, Huang S, Fukaki H, Song X, Sun S, Morita MT, Tasaka M, Millar AH, Furutani M (2019) Mitochondrial pyruvate dehydrogenase contributes to auxin-regulated organ development. Plant Physiol 180: 896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passarella S, Atlante A, Valenti D, de Bari L (2003) The role of mitochondrial transport in energy metabolism. Mitochondrion 2: 319–343 [DOI] [PubMed] [Google Scholar]
- Petereit J, Duncan O, Murcha MW, Fenske R, Cincu E, Cahn J, Pružinská A, Ivanova A, Kollipara L, Wortelkamp S, et al. (2020). Mitochondrial CLPP2 assists coordination and homeostasis of respiratory complexes. Plant Physiol 184: 148–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao X, Dixon RA (2016) The differences between NAD-ME and NADP-ME subtypes of C4 photosynthesis: more than decarboxylating enzymes. Front Plant Sci 7: 1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmusson AG, Soole KL, Elthon TE (2004) Alternative NAD(P)H dehydrogenases of plant mitochondria. Annu Rev Plant Biol 55: 23–39 [DOI] [PubMed] [Google Scholar]
- Ricoult C, Echeverria LO, Cliquet JB, Limami AM (2006) Characterization of alanine aminotransferase (AlaAT) multigene family and hypoxic response in young seedlings of the model legume Medicago truncatula. J Exp Bot 57: 3079–3089 [DOI] [PubMed] [Google Scholar]
- Schertl P, Braun H-P (2014) Respiratory electron transfer pathways in plant mitochondria. Front Plant Sci 5: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidtmann E, König A-C, Orwat A, Leister D, Hartl M, Finkemeier I (2014) Redox regulation of Arabidopsis mitochondrial citrate synthase. Mol Plant 7: 156–169 [DOI] [PubMed] [Google Scholar]
- Schnarrenberger C, Martin W (2002) Evolution of the enzymes of the citric acid cycle and the glyoxylate cycle of higher plants. Eur J Biochem 269: 868–883 [DOI] [PubMed] [Google Scholar]
- Schwacke R, Schneider A, van der Graaff E, Fischer K, Catoni E, Desimone M, Frommer WB, Flügge U-I, Kunze R (2003) ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol 131: 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selinski J, Scheibe R (2019) Malate valves: old shuttles with new perspectives. Plant Biol 21: 21–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SS, Dietz KJ (2006) The significance of amino acids and amino acid-derived molecules in plant responses and adaptation to heavy metal stress. J Exp Bot 57: 711–726 [DOI] [PubMed] [Google Scholar]
- Shen JL, Li CL, Wang M, He LL, Lin MY, Chen DH, Zhang W (2017) Mitochondrial pyruvate carrier 1 mediates abscisic acid-regulated stomatal closure and the drought response by affecting cellular pyruvate content in Arabidopsis thaliana. BMC Plant Biol 17: 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienkiewicz-Porzucek A, Nunes-Nesi A, Sulpice R, Lisec J, Centeno DC, Carillo P, Leisse A, Urbanczyk-Wochniak E, Fernie AR (2008) Mild reductions in mitochondrial citrate synthase activity result in a compromised nitrate assimilation and reduced leaf pigmentation but have no effect on photosynthetic performance or growth. Plant Physiol 147: 115–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienkiewicz-Porzucek A, Sulpice R, Osorio S, Krahnert I, Leisse A, Urbanczyk-Wochniak E, Hodges M, Fernie AR, Nunes-Nesi A (2010) Mild reductions in mitochondrial NAD-dependent isocitrate dehydrogenase activity result in altered nitrate assimilation and pigmentation but do not impact growth. Mol Plant 3: 156–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh BK, Shaner DL (1995) Biosynthesis of branched chain amino acids: from test tube to field. Plant Cell 7: 935–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sytar O, Kumar A, Latowski D, Kuczynska P, Strzałka K, Prasad MNV (2013) Heavy metal-induced oxidative damage, defense reactions, and detoxification mechanisms in plants. Acta Physiol Plant 35: 985–999 [Google Scholar]
- Tavoulari S, Thangaratnarajah C, Mavridou V, Harbour ME, Martinou JC, Kunji ER (2019) The yeast mitochondrial pyruvate carrier is a hetero-dimer in its functional state. EMBO J 38: e100785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherkez G, Cornic G, Bligny R, Gout E, Ghashghaie J (2005) In vivo respiratory metabolism of illuminated leaves. Plant Physiol 138: 1596–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherkez G, Bligny R, Gout E, Mahé A, Hodges M, Cornic G (2008) Respiratory metabolism of illuminated leaves depends on CO2 and O2 conditions. Proc Natl Acad Sci USA 105:797–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherkez G, Mahé A, Gauthier P, Mauve C, Gout E, Bligny R, Cornic G, Hodges M (2009) In folio respiratory fluxomics revealed by 13C isotopic labeling and H/D isotope effects highlight the noncyclic nature of the tricarboxylic acid “cycle” in illuminated leaves. Plant Physiol 151: 620–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas AP, Halestrap AP (1981) Identification of the protein responsible for pyruvate transport into rat liver and heart mitochondria by specific labelling with [3H]N-phenylmaleimide. Biochem J 196: 471–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todisco S, Agrimi G, Castegna A, Palmieri F (2006) Identification of the mitochondrial NAD+ transporter in Saccharomyces cerevisiae. J Biol Chem 281: 1524–1531 [DOI] [PubMed] [Google Scholar]
- Tomaz T, Bagard M, Pracharoenwattana I, Lindén P, Lee CP, Carroll AJ, Ströher E, Smith SM, Gardeström P, Millar AH (2010) Mitochondrial malate dehydrogenase lowers leaf respiration and alters photorespiration and plant growth in Arabidopsis. Plant Physiol 154: 1143–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompkins SC, Sheldon RD, Rauckhorst AJ, Noterman MF, Solst SR, Buchanan JL, Mapuskar KA, Pewa AD, Gray LR, Oonthonpan L, et al. (2019) Disrupting mitochondrial pyruvate uptake directs glutamine into the TCA cycle away from glutathione synthesis and impairs hepatocellular tumorigenesis. Cell Rep 28: 2608–2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar-Méndez A, Miernyk JA, Randall DD (2003) Regulation of pyruvate dehydrogenase complex activity in plant cells. Eur J Biochem 270: 1043–1049 [DOI] [PubMed] [Google Scholar]
- Tronconi MA, Maurino VG, Andreo CS, Drincovich MF (2010a) Three different and tissue-specific NAD-malic enzymes generated by alternative subunit association in Arabidopsis thaliana. J Biol Chem 285: 11870–11879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronconi MA, Gerrard Wheeler MC, Maurino VG, Drincovich MF, Andreo CS (2010b) NAD-malic enzymes of Arabidopsis thaliana display distinct kinetic mechanisms that support differences in physiological control. Biochem J 430: 295–303 [DOI] [PubMed] [Google Scholar]
- Tronconi MA, Fahnenstich H, Gerrard Weehler MC, Andreo CS, Flügge UI, Drincovich MF, Maurino VG (2008) Arabidopsis NAD-malic enzyme functions as a homodimer and heterodimer and has a major impact on nocturnal metabolism. Plant Physiol 146: 1540–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderperre B, Herzig S, Krznar P, Hörl M, Ammar Z, Montessuit S, Pierredon S, Zamboni N, Martinou JC (2016) Embryonic lethality of mitochondrial pyruvate carrier 1 deficient mouse can be rescued by a ketogenic diet. PLoS Genet 12: e1006056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe GC, McIntosh L (1997) Alternative oxidase: from gene to function. Annu Rev Plant Physiol Plant Mol Biol 48: 703–734 [DOI] [PubMed] [Google Scholar]
- Waese J, Fan J, Pasha A, Yu H, Fucile G, Shi R, Cumming M, Kelley LA, Sternberg MJ, Krishnakumar V, et al. (2017) ePlant: visualizing and exploring multiple levels of data for hypothesis generation in plant biology. Plant Cell 29: 1806–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A, Flügge UI (2002) Interaction of cytosolic and plastidic nitrogen metabolism in plants. J Exp Bot 53: 865–874 [DOI] [PubMed] [Google Scholar]
- Willeford K, Wedding R (1987) pH effects on the activity and regulation of the NAD malic enzyme. Plant Physiol 84: 1084–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong DT, Fuller RW, Molloy BB (1973) Inhibition of amino acid transaminases by L-cycloserine. Adv Enzyme Regul 11: 139–154 [DOI] [PubMed] [Google Scholar]
- Yu H, , Du X, , Zhang F, , Zhang F, , Hu Y, , Liu S, , Jiang X, , Wang G, , Liu D ( 2012) A mutation in the E2 subunit of the mitochondrial pyruvate dehydrogenase complex in Arabidopsis reduces plant organ size and enhances the accumulation of amino acids and intermediate products of the TCA Cycle. Planta 236: 387–399 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Taufalele PV, Cochran JD, Robillard-Frayne I, Marx JM, Soto J, Rauckhorst AJ, Tayyari F, Pewa AD, Gray LR, et al. (2020) Mitochondrial pyruvate carriers are required for myocardial stress adaptation. Nat Metab 2: 1248–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Xiao H, Guo Q, Zhang Z, Zhao J, Yang D (2018) Effects of cadmium stress on growth and amino acid metabolism in two Compositae plants. Ecotoxicol Environ Saf 158: 300–308 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.