Abstract
Cotton, one of the most important crops in the world, produces natural fiber materials for the textile industry. WRKY transcription factors play important roles in plant development and stress responses. However, little is known about whether and how WRKY transcription factors regulate fiber development of cotton so far. In this study, we show that a fiber-preferential WRKY transcription factor, GhWRKY16, positively regulates fiber initiation and elongation. GhWRKY16-silenced transgenic cotton displayed a remarkably reduced number of fiber protrusions on the ovule and shorter fibers compared to the wild-type. During early fiber development, GhWRKY16 directly binds to the promoters of GhHOX3, GhMYB109, GhCesA6D-D11, and GhMYB25 to induce their expression, thereby promoting fiber initiation and elongation. Moreover, GhWRKY16 is phosphorylated by the mitogen-activated protein kinase GhMPK3-1 at residues T-130 and S-260. Phosphorylated GhWRKY16 directly activates the transcription of GhMYB25, GhHOX3, GhMYB109, and GhCesA6D-D11 for early fiber development. Thus, our data demonstrate that GhWRKY16 plays a crucial role in fiber initiation and elongation, and that GhWRKY16 phosphorylation by GhMPK3-1 is essential for the transcriptional activation on downstream genes during the fiber development of cotton.
GhWRKY16 is a crucial regulator promoting fiber initiation and elongation in cotton, whose phosphorylation by GhMPK3-1 is essential for the transcriptional activation of downstream genes.
Introduction
Upland cotton (Gossypium hirsutum) is the most important cotton cultivar planted in over 50 countries across the world, and accounts for 90% of the world cotton production (Wendel and Cronn, 2003). Cotton fiber is a specialized and elongated single epidermal cell that is derived from the seed coat. Fiber development is a delicate and complex process with cell differentiation lasting about 50 days and goes through four distinct but overlapping periods: initiation, elongation, secondary cell wall thickening, and maturation. From 3 days before to 1 day post anthesis (DPA), approximately 20%–30% of the ovule epidermal cells begin to differentiate into spinnable fibers. Fiber cells then enter a rapid elongation period, with a growth rate of more than 2 mm/day up to 20 DPA. The elongation period determines the final length of fiber cells. At around 16 DPA, cellulose biosynthesis begins in large quantity and is deposited on the secondary cell wall. This period lasts until 40 DPA, followed by the dehydration and maturation of cotton fibers (Kim and Triplett, 2001; Gou et al., 2007; Haigler et al., 2012). A number of factors affecting the development of cotton fibers have been identified: for example, ethylene biosynthesis plays a significant role during fiber elongation (Shi et al., 2006), and very-long-chain fatty acids may be involved in cotton fiber development by activating ethylene biosynthesis (Qin et al., 2007). In addition, ascorbate peroxidase also participates in cotton fiber cell development by modulating hydrogen peroxide homeostasis (Li et al., 2007; Qin et al., 2008). However, the regulatory mechanism of fiber development is still largely unknown.
Transcription factors (TFs) play essential regulatory roles by controlling the transcription rates of downstream genes during plant growth and development (Yang et al., 2004). In recent years, an increasing number of TFs has been reported to function in fiber development of cotton. For example, the MYB transcription factor GhMYB25 was shown to be involved in fiber cell differentiation (Machado et al., 2009). Overexpression of GhMYB25 in cotton increased the number of fiber initials, while silencing of GhMYB25 resulted in fewer initials as well as shorter fibers compared to the wild-type. The related TF GhMYB25-like shares 69% sequence identity with GhMYB25 and is also required for fiber cell differentiation. Silencing of GhMYB25-like in cotton resulted in fibreless seeds (Walford et al., 2011). Similarly, the downregulation of GhMYB109 expression produced seeds with shorter fibers, indicating that GhMYB109 plays an important role in fiber elongation of cotton (Pu et al., 2008). The cotton homeodomain leucine zipper (HD-ZIP) TF, GhHOX3, promotes fiber elongation by directly regulating the expression levels of cell wall loosening protein genes GhRDL1 (RESPONSIVE TO DESICCATION 22 [RD22]-like1) and EXPANSIN A1 (GhEXPA1; Shan et al., 2014). PACLOBUTRAZOL RESISTANCE 1 (GhPRE1), a basic helix–loop–helix (bHLH) protein, is a positive regulator of fiber elongation (Zhao et al., 2018). TEOSINTE BRANCHED, CYCLOIDEA AND PCF 14 (GhTCP14) participates in auxin-mediated fiber elongation by directly inducing the expression of the auxin response gene INDOLE-3-ACETIC ACID INDUCIBLE 3 (IAA3) and the auxin transporter genes PIN-FORMED 2 (PIN2) and AUXIN 1 (AUX1; Wang et al., 2013). We recently revealed that GhFSN1 (fiber secondary cell wall-related NAC1), a NAC domain TF, positively regulates fiber secondary cell wall biosynthesis (Zhang et al., 2018), and GhFP1 (Fibre-related Protein 1), a bHLH TF, promotes fiber elongation by modulating brassinosteroid (BR) biosynthesis and signaling in cotton (Liu et al., 2020). However, no WRKY TFs have been reported in cotton fiber development to date.
WRKY proteins are a class of plant-specific TFs that regulate various plant developmental and physiological processes. The first WRKY gene (SWEET POTATO FACTOR1, SPF1) was identified in sweet potato (Ipomoea batatas L.; Ishiguro and Nakamura, 1994), followed by the other WRKY genes ABSCISIC ACID RESPONSE ELEMENT BINDING FACTOR 1 (ABF1) and ABF2 in common wild oat (Avena fatua), PcWRKY1, PcWRKY2, and PcWRKY3 in parsley (Petroselinum crispum) and ZINC-DEPENDENT ACTIVATOR PROTEIN-1 (ZAP1) in Arabidopsis (Arabidopsis thaliana); all encoded proteins can bind to the DNA sequence “(T)(T)TGAC(C/T)” in promoters, known as the W-box (Rushton et al., 1995, 1996; de Pater et al., 1996). WRKY proteins contains at least one conserved 60-amino acid residues termed as the WRKY domain that consists of a highly conserved WRKYGQK polypeptide in the N terminus and a zinc finger motif in the C-terminus (Rushton et al., 2010). Based on the number of WRKY domains and the pattern of the C-terminal zinc finger motif, WRKY TFs are classified into three subfamilies: groups I, II, and III. Group I WRKYs are characterized by two WRKY domains with a C2H2-type zinc finger motif (C–X4–5–C–X22–23–H–X1–H), while groups II and III WRKYs only contain one WRKY domain, with C2H2-type and C2HC-type (C–X7–C–X23–H–X1–C) zinc finger motifs, respectively. Furthermore, group II WRKYs may be divided into subgroups IIa, IIb, IIc, IId, and IIe according to their conserved motifs (Eulgem et al., 2000). At least 72 WRKY members have been identified in Arabidopsis, 109 in rice (Oryza sativa), 136 in maize (Zea mays), 112 in Gossypium raimondii, and 109 in Gossypium arboreum (Wu et al., 2005; Wei et al., 2012; Ding et al., 2015a).
WRKY TFs have been shown to regulate plant growth and development. For example, Arabidopsis WRKY46 regulates the transcription of abscisic acid (ABA)-related and auxin-related genes to modulate the development of lateral roots (Ding et al., 2015b). WRKY71 accelerates Arabidopsis flowering by inducing the expression of FLOWERING LOCUS T (FT) and LEAFY (LFY) (Yu et al., 2016). The glandular trichome-specific transcription factor WRKY1 raises the contents of artemisinin and dihydroartemisinic acid in sweet wormwood (Artemisia annua), thereby exerting a positive regulator role in the artemisinin biosynthetic pathway (Chen et al., 2017). Besides, MdWRKY9 reduces BR production in apple (Malus domestica) by directly repressing the expression of DWARF4 (MdDWF4), encoding the rate-limiting enzyme for BR biosynthesis, leading to dwarf plants (Zheng et al., 2018). Wheat (Triticum aestivum) WRKY51 promotes lateral root formation by blocking the expression of 1-aminocyclopropane-1-carboxylate Synthase (ACS) genes, which are involved in ethylene biosynthesis, by binding to the W-box within their promoters (Hu et al., 2018). Arabidopsis WRKY36 promotes hypocotyl elongation by directly inducing the expression of LONG HYPOCOTYL 5 (HY5; Yang et al., 2018). However, little is known about how WRKY TFs function in fiber development of cotton so far. Here, we report that the WRKY TF GhWRKY16 positively regulates fiber development in cotton. We provide evidence that GhWRKY16 promotes fiber initiation and elongation by regulating the expression of its downstream target genes, and phosphorylation of GhWRKY16 may be crucial for its transcriptional activity during fiber development of cotton.
Results
GhWRKY16 promotes fiber initiation and elongation
In our previous study, we identified 26 WRKY genes in upland cotton (Zhou et al., 2014). Among them, a gene encoding a subgroup IId WRKY TF (GhWRKY16, Gh_D06G0175) was preferentially expressed in very early developing ovules and fibers, and reached its peak expression in 9 DPA fibers, suggesting that GhWRKY16 may contribute to fiber initiation and elongation. Since the genome sequence of tetraploid upland cotton (G. hirsutum) was released after this initial survey, we used 74 known Arabidopsis WRKY proteins as query to conduct an exhaustive Basic Local Alignment Search Tool search against the cotton protein database (https://cottonfgd.org/blast/, G. hirsutum, NAU) and identified 230 WRKY genes (Supplemental Data Set S1). Moreover, the homoeologs GhWRKY16-A (Gh_A06G0179) and GhWRKY16-D (i.e. GhWRKY16 hereafter, Gh_D06G0175) shared nearly 99% identity (Supplemental Figure S1A), with GhWRKY16-A maintaining a high expression level during fiber initiation and elongation, showing a peak in expression in 0 DPA ovules (Supplemental Figure S1B).
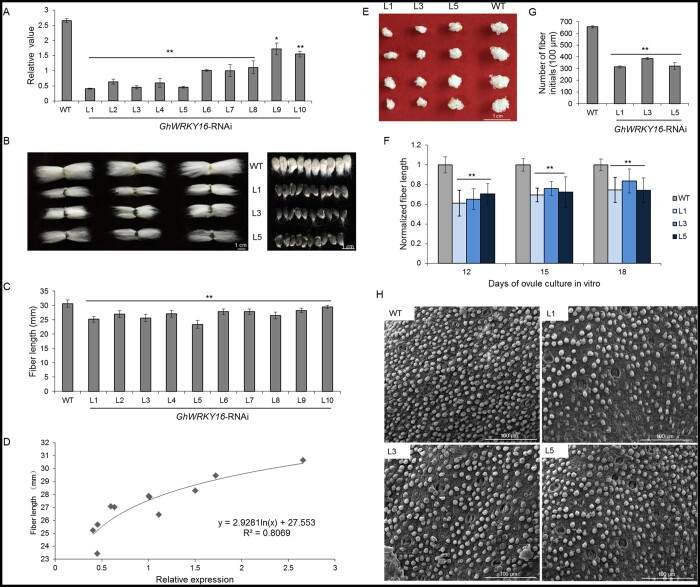
To explore the function of GhWRKY16 in fiber development, we generated transgenic cotton plants downregulated for GhWRKY16 transcript levels through RNA interference (RNAi). We regenerated more than 50 transgenic seedlings from 10 independent transformation events (T0 generation) and transplanted them to soil for growing to maturation. We extracted total RNA from 9 DPA fibers of GhWRKY16-RNAi cotton lines (T1–T4 generation) and determined GhWRKY16 transcript levels in the fibers of the transgenic lines, which were reduced to varying degrees (from 20% to 80%) compared to the wild-type (Figure 1A). We selected three transgenic lines (L1, L3, and L5) with much lower GhWRKY16 transcript levels for subsequent characterization. We also examined relative transcript levels for the other WRRKY genes GhWRKY16-A, GhWRKY11, GhWRKY15, GhWRKY26, GhWRKY29, GhWRKY31, and GhWRKY32 in fibers of RNAi plants, as they are highly homologous to GhWRKY16. As shown in Supplemental Figure S1C, only GhWRKY16-A transcripts also accumulated to lower levels in GhWRKY16-RNAi lines, indicating that the silencing of GhWRKY16 is specific.
Figure 1.
Silencing GhWRKY16 by RNA interference in cotton hinders fiber initiation and elongation. A, RT-qPCR analysis of GhWRKY16 (Gh_D06G0175) expression in 9 DPA fibers of the wild-type and GhWRKY16-RNAi lines. The cotton polyubiquitin gene GhUBI1 (EU604080) was used as reference; GhWRKY16 transcript levels in the wild-type were set to 1. B, Comparison of mature fiber length and seed phenotype with or without fuzz between GhWRKY16-RNAi lines (T2 generation) and the wild-type. C, Mean mature fiber length in GhWRKY16-RNAi lines and the wild-type. D, Fiber length and relative GhWRKY16 expression is positively correlated in the RNAi lines. E, In vitro cotton ovule culture. 1 DPA ovules of GhWRKY16-RNAi lines and the wild-type were cultured in liquid Beasley–Ting medium for 12 days. F, Mean fiber length of the cultured ovules (12–18 days) from GhWRKY16-RNAi lines and the wild-type (n > 30 ovules per line). Fiber length of wild-type ovules was set to 1. G, Mean number of fiber initials counted from the middle of 0 DPA ovules. H, Scanning electronic micrographs of ovule surface from GhWRKY16-RNAi lines and the wild-type. Error bars represent standard deviation (sd) of three biological replicates. **P <0.01 by Dunnett t test between the wild-type and GhWRKY16-RNAi lines. WT, wild-type; L1–L10, GhWRKY16-RNAi cotton lines. Scale bars: 1 cm (B, E), 100 µm (H)
We observed no significant differences in plant height, branch or flowering time between GhWRKY16-RNAi transgenic plants and the wild-type (Supplemental Figure S2A). However, mature fibers in the GhWRKY16-RNAi lines were shorter by 7%–26%, which was accompanied by the reduction or even the complete loss of fuzz relative to the wild-type (Figure 1, B and C; Supplemental Figure S2, B–G and Supplemental Table S1). Regression analysis showed that the reduction in GhWRKY16 transcript levels was strongly and positively correlated with fiber growth (Figure 1D). These results indicated that suppression of GhWRKY16 expression in cotton impedes fiber elongation, a phenotype that can be stably inherited. Additionally, we cultured 0 DPA ovules from the wild-type and GhWRKY16-RNAi transgenic lines in vitro for 12, 15, and 18 days. The elongation rate of transgenic fiber cells distinctly lagged behind that of the wild-type (Figure 1, E and F). Thus, the above data indicate that GhWRKY16 plays a positive role for fiber elongation in cotton.
As the fibers developing from GhWRKY16-RNAi ovules were sparser than those on wild-type ovules, we hypothesized that GhWRKY16 may also play an important role in fiber cell initiation. Scanning electron microscopy showed that the number of fiber protrusions on 0 DPA ovules from the transgenic lines is lower by about 40% compared to the wild-type (Figure 1, G and H). Furthermore, we observed hand-cut cross-sections of 0, 1, and 2 DPA ovules from GhWRKY16-RNAi lines and the wild-type, and determined that the fiber growth rate in GhWRKY16-RNAi lines is slower than that of the wild-type (Supplemental Figure S3A). Moreover, mature fiber weight from the same number of GhWRKY16-RNAi seeds was also remarkably lighter than that from wild-type seeds, although there was no significant difference in seed size or weight between the transgenic lines and the wild-type (Supplemental Figure S3, B and C). Thus, these results indicated that GhWRKY16 is involved in regulating fiber initiation of cotton.
GhWRKY16 regulates the expression of fiber elongation-related genes
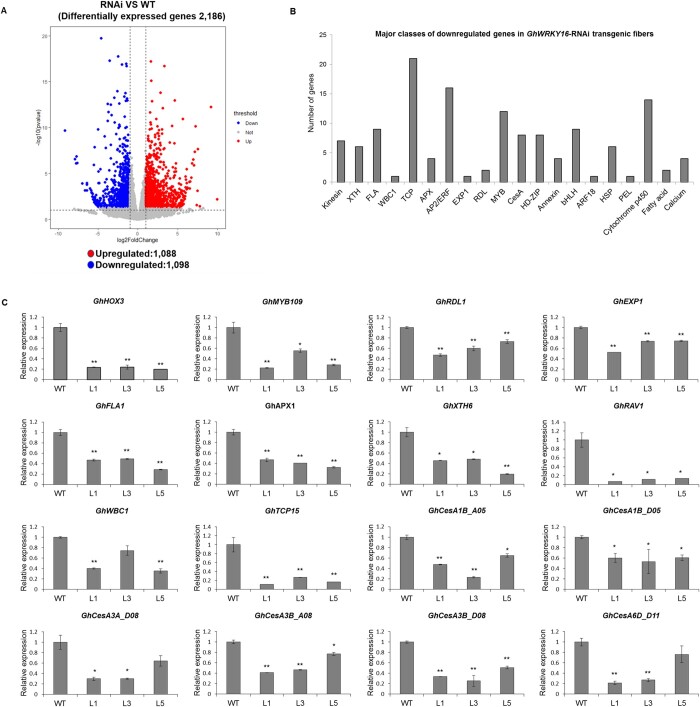
To explore the regulation mechanism of GhWRKY16 in fiber development, we performed a transcriptome deep sequencing (RNA-seq) analysis on 9 DPA fibers from the wild-type and GhWRKY16-RNAi seeds. We identified 2,186 differentially expressed genes (DEGs), consisting of 1,088 upregulated genes and 1,098 downregulated genes (Figure 2A; Supplemental Data Set S2). Furthermore, we validated the RNA-seq data by reverse transcription quantitative polymerase chain reaction (RT-qPCR) analysis (Supplemental Figure S4). Gene ontology cluster analysis showed that the DEGs regulated by GhWRKY16 are classified in diverse molecular functions, cellular components and biological processes (Supplemental Figure S5). Among downregulated genes, we noticed several TFs (such as HD-ZIP, bHLH, MYB, ETHYLENE RESPONSE FACTOR [ERF], and TCP) reported to participate in regulating cotton fiber development, as well as genes associated with cellulose biosynthesis, cell wall loosening, and cytoskeletal organization (Figure 2B). RT-qPCR analysis confirmed that these genes are significantly downregulated in the fibers the GhWRKY16-RNAi lines (Figure 2C). Thus, the above data indicated that GhWRKY16 promotes fiber initiation and elongation, possibly by regulating the transcription of its downstream genes involved in early fiber development of cotton.
Figure 2.
Transcriptome analysis of DEGs in fibers of GhWRKY16-RNAi transgenic cotton. A, Volcano of DEGs between the wild-type and GhWRKY16-RNAi lines. The red dots represent upregulated genes and the blue dots represent downregulated genes in 9 DPA fibers of the GhWRKY16-RNAi lines. B, Major classes of downregulated genes in GhWRKY16-RNAi fibers. C, RT-qPCR analysis of genes related to cotton fiber development in 9 DPA fibers of GhWRKY16 RNAi lines. Total RNA was isolated from 9 DPA fibers of the GhWRKY16 RNAi lines and wild-type controls. GhUBI1 (EU604080) was used as internal reference. Error bars represent sd of three biological replicates. *P <0.05; **P <0.01 by independent t-tests. WT, wild-type; L1, L3, and L5, GhWRKY16-RNAi cotton lines
GhWRKY16 is a typical WRKY transcriptional regulator
To localize GhWRKY16 in the cell, we transiently transfected Nicotiana benthamiana leaves with a GhWRKY16:eGFP (enhanced Green Fluorescent Protein) construct and then stained the leaves with the DNA dye DAPI. Confocal microscopy revealed that GhWRKY16-GFP fluorescence co-localizes with DAPI in nuclei (Supplemental Figure S6A), indicating that GhWRKY16 is a nucleus-localized protein.
WRKY TFs exert their function by binding preferentially to the cis-acting element W-box in the promoters of their downstream target genes (Eulgem et al., 2000). To assess whether GhWRKY16 can bind to the W-box, we cloned a sequence containing three W-box or mutated W-box (mW-box) tandem repeats into the pAbAi yeast one-hybrid vector and transformed the resulting constructs into the Y1HGold yeast strain (Supplemental Figure S6B). We separately cloned the GhWRKY16 coding sequence into the pGADT7 vector containing the GAL4 transcriptional activation domain. We then introduced the pGADT7 and pGADT7-GhWRKY16 vectors into the Y1HGold yeast strain carrying pAbAi-W-box or pAbAi-mW-box bait plasmids. All transformed yeast cells were plated on synthetic defined medium lacking leucine and uracil (SD–Leu–Ura) for selection and then tested on SD medium lacking leucine and containing the antibiotic aureobasidin (AbA). Only yeast cells harboring pAbAi-W-box and pGADT7-GhWRKY16 grew normally in the presence of AbA (Supplemental Figure S6C). Thus, the above results indicated that GhWRKY16 could bind to the W-box cis-acting elements for regulating the transcription of its downstream genes.
GhWRKY16 directly regulates its downstream genes for fiber elongation
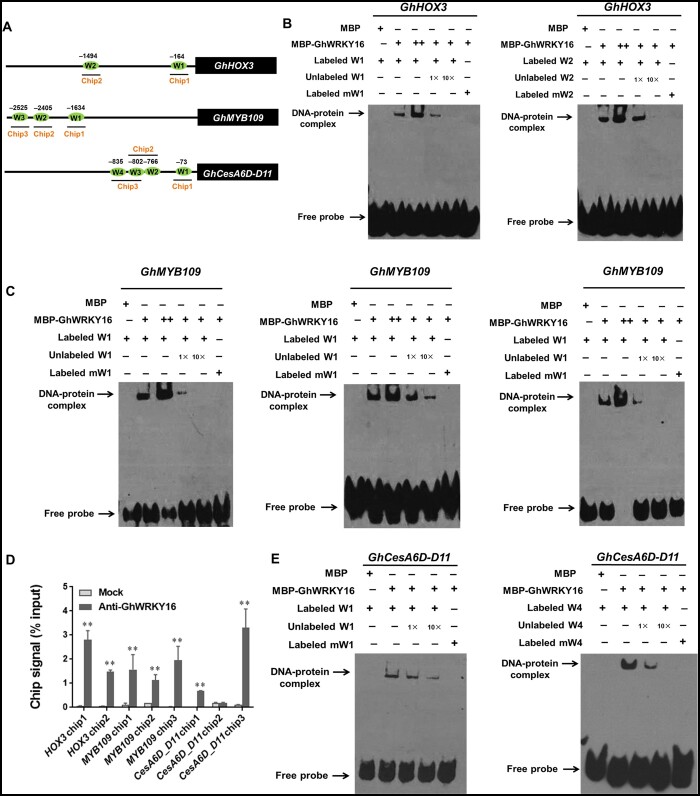
Since GhWRKY16 appeared to bind to W-boxes as a typical WRKY TF, we analyzed the promoters of those fiber development-related genes that are downregulated in GhWRKY16-RNAi transgenic lines for regulatory motifs, including W-boxes. As indicated in Supplemental Table S2, the promoters of predicted genes downstream of GhWRKY16 contained at least one W-box each. In agreement with the lower expression of the previously characterized fiber elongation-related genes GhHOX3 and GhMYB109 in GhWRKY16-RNAi fibers compared to the wild-type (Figure 2C), we identified two putative W-box elements in the GhHOX3 promoter and three in the GhMYB109 promoter (Figure 3A). We then performed electrophoretic mobility shift assays (EMSA) to test the binding of GhWRKY16 to these cis-elements. Indeed, we observed a band in gel when incubating recombinant maltose-binding protein (MBP)-tagged GhWRKY16 with biotin-labeled probes containing the W-box elements from the GhHOX3 and GhMYB109 promoters. The intensity of the binding complex increased in the gel with higher amounts of MBP-GhWRKY16 added. The signal was specific, as it was competed by excess unlabeled probes. Furthermore, we detected no DNA–protein complex when incubating recombinant MBP-GhWRKY16 with a mutated biotin-labeled probe (Figure 3, B and C). The above results demonstrated that GhWRKY16 can bind the W-box in the GhHOX3 and GhMYB109 promoters in vitro. To confirm these results in vivo, we performed chromatin immunoprecipitation followed by quantitative PCR (ChIP-qPCR) with an anti-GhWRKY16 antibody to detect the GhHOX3 and GhMYB109 promoter fragments, as illustrated in Figure 3A. Indeed, the GhHOX3 and GhMYB109 promoter fragments were highly enriched in the anti-GhWRKY16 precipitate (Figure 3D), indicating that GhWRKY16 directly binds to the GhHOX3 and GhMYB109 promoters in vivo. In contrast, ChIP-qPCR signals performed on GhWRKY16-RNAi lines were much lower than in the wild-type for the same promoter fragments (Supplemental Figure S7). Thus, we concluded that GhWRKY16 promotes fiber elongation by directly regulating the expression of GhHOX3 and GhMYB109.
Figure 3.
GhWRKY16 binds to W-box cis-elements in the promoters of its target genes in vitro and in vivo. A, Schematic representation of W-box in GhHOX3, GhMYB109, and GhCesA6D-D11 promoters. Horizontal lines represent the promoters, green ovals represent W-boxes, and the line underneath indicates the fragments detected by ChIP-qPCR. B,C,E, EMSA of GhWRKY16 binding to the W-box in the promoters of GhHOX3 (B), GhMYB109 (C), and GhCesA6D-D11 (E). Biotin-labeled probes were incubated with MBP-GhWRKY16 in vitro. Unlabeled probes were used for competition, and biotin-labeled mutated W-box cis-elements (TTGAC to AAAAC) were used as negative controls. D, ChIP-qPCR analysis of GhWRKY16 binding to the GhHOX3, GhMYB109, and GhCesA6D-D11 promoters. An anti-GhWRKY16 polyclonal antibody was used for ChIP, followed by qPCR analysis of bound chromatin from 9 DPA cotton fibers. The ChIP signal is expressed as the percentage of immunoprecipitated DNA in the total input DNA. Mock, ChIP without anti-GhWRKY16 antibody. Error bars represent sd of three biological replicates. **P <0.01 by t-test between mock and anti-GhWRKY16 antibody
In addition, cellulose synthase genes associated with primary cell wall biosynthesis were also significantly downregulated in GhWRKY16-RNAi fibers (Figure 2C), and their promoters contained W-box elements (Figure 3A). EMSA and CHIP-qPCR revealed that GhWRKY16 binds to the GhCesA6D_D11 promoter in vitro and in vivo (Figure 3, D and E), suggesting that GhWRKY16 may also directly induce the cellulose synthase gene to provide sufficient cellulose building blocks for the rapid elongation of cotton fiber cells. However, GhWRKY16 did not bind to the W-box elements in the GhCesA3D-D08 promoter (Supplemental Figure S8, A and B). Further analysis suggested that GhWRKY16 may preferably bind to W-box elements that are not surrounded by adjacent motifs, and the appearance of the motif GTACTGAARGAG near the W-box may exert a positive effect on GhWRKY16 binding to the W-box (Supplemental Figure S8C).
GhWRKY16 functions in fiber initiation by directly regulating GhMYB25 expression
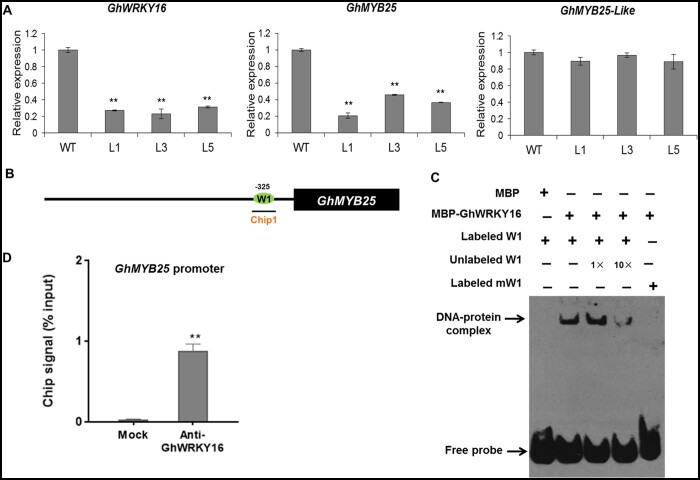
Fiber initiation was blocked in GhWRKY16-RNAi cotton plants. To explore the molecular mechanism underlying how GhWRKY16 affects fiber initiation, we measured transcript levels for fiber initiation-related genes (GhMYB25-like and GhMYB25) in ovules of RNAi lines and the wild-type at anthesis (0 DPA). As shown in Figure 4A, GhMYB25 expression was much lower in 0 DPA GhWRKY16-RNAi ovules compared to those of the wild-type, but not that of GhMYB25-like. Notably, the GhMYB25 promoter contained one putative W-box element (Figure 4B) that recombinant MBP-tagged GhWRKY16 bound to in EMSA. Binding to the probe was competed by incubation with excess unlabeled probe, and was abrogated when the recombinant protein was incubated with a biotin-labeled mutated probe (Figure 4C). CHIP-qPCR confirmed that GhWRKY16 directly binds to the GhMYB25 promoter in vivo (Figure 4D). Thus, the above data suggested that GhWRKY16 functions in fiber initiation by directly regulating the expression of GhMYB25.
Figure 4.
GhWRKY16 directly binds to the GhMYB25 promoter in vitro and in vivo. A, RT-qPCR analysis of GhMYB25-like and GhMYB25 relative transcript levels in 0 DPA ovules of GhWRKY16-RNAi transgenic lines. Total RNA was isolated from 0 DPA ovules from GhWRKY16-RNAi lines and the wild-type. GhUBI1 (EU604080) was used as reference; expression levels in the wild-type were set to 1. Error bars represent sd of three biological replicates. *P <0.05 and **P <0.01 by t-tests between GhWRKY16-RNAi lines and the wild-type. WT, wild-type; L1, L3, and L5, GhWRKY16-RNAi cotton lines. B, Schematic representation of W-box in the GhMYB25 promoter. Horizontal line represents the promoter, green oval represents the W-box, and the line underneath indicates the fragments detected by ChIP-qPCR. C, EMSA of GhWRKY16 binding to the W-box in the GhMYB25 promoter. Biotin-labeled probe containing the W-box element from GhMYB25 promoter was incubated with MBP-GhWRKY16 in vitro. Unlabeled probe was used for competition, and a biotin-labeled mutated W-box cis-element (TTGAC to AAAAC) was used as negative control. D, ChIP-qPCR analysis of GhWRKY16 binding to the GhMYB25 promoter. An anti-GhWRKY16 polyclonal antibody was used for ChIP, followed by qPCR of the bound chromatin from 0 DPA ovules of cotton. The ChIP signal is expressed as the percentage of immunoprecipitated DNA in the total input DNA. Mock, ChIP signals without IP with GhWRKY16 antibody. Error bars represent sd of three biological replicates. **P <0.01 by t-test between mock and anti-GhWRKY16 antibody
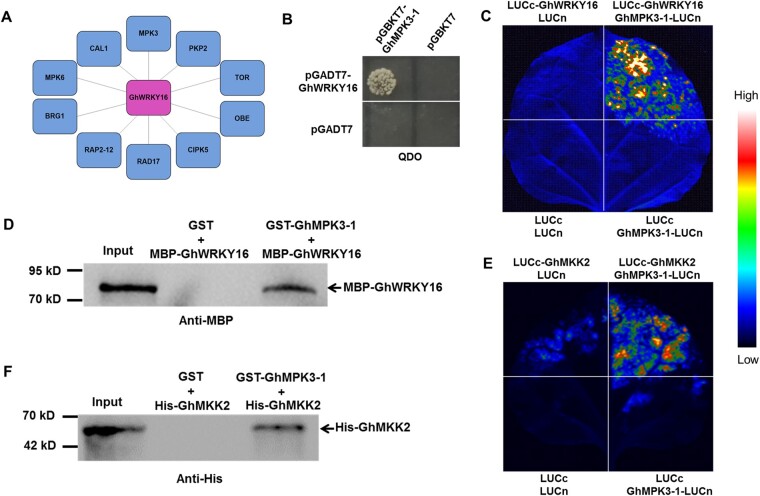
GhWRKY16 interacts with GhMPK3-1
Based on available gene co-expression networks for cotton (You et al., 2016), we identified several Mitogen Activated Protein Kinase (GhMPK) genes that are highly similar to Arabidopsis MPK3 and MPK6 and are co-expressed with GhWRKY16 in cotton fibers (Figure 5A; Supplemental Table S3). Furthermore, bioinformatics analysis revealed that GhWRKY16 contains a D motif (KKRKSRVKRVIRV), which has been reported to be present in all MPK-interacting proteins (Takuji et al., 2000). We first determined the expression patterns of these MPK genes in cotton fibers by RT-qPCR analysis. As shown in Supplemental Figure S9, GhMPK3-1, GhMPK3-2, GhMPK6-1, GhMPK6-2, and GhMPK6-3 were preferentially expressed in fibers at the initiation and elongation stages, thus following the same expression pattern as GhWRKY16. We then conducted a yeast two-hybrid assay to test whether GhWRKY16 interacts with these GhMPKs. As shown in Figure 5B and Supplemental Figure S10A, only colonies harboring the constructs pGADT7-GhWRKY16 with pBGKT7-GhMPK3-1 grew on selective medium, indicating that GhWRKY16 interacts with GhMPK3-1 in yeast cells. In addition, we performed luciferase (LUC) complementation imaging (LCI) assays to validate this interaction in plants cells. We fused GhMPK3-1 to the N-terminal half of LUC to form GhMPK3-1-LUCn, while we tagged GhWRKY16 with the C-terminal half of LUC to form LUCc-GhWRKY16. We detected strong LUC activity in N. benthamiana leaves co-infiltrated with GhMPK3-1-LUCn and LUCc-GhWRKY16 constructs (Figure 5C), indicating that GhWRKY16 can interact with GhMPK3-1 in plants. We confirmed the interaction between GhWRKY16 and GhMPK3-1 with pull-down assays: recombinant MBP-GhWRKY16 was able to bind to recombinant glutathione S-transferase (GST)-GhMPK3-1, but not to GST alone (control) in vitro (Figure 5D). Since MPKs are part of a kinase cascade, we also tested interaction between MPK3-1 and the MPK Kinases GhMKK1, GhMKK2, and GhMKK4: only GhMKK2 (and not GhMKK1 or GhMKK4) interacted with GhMPK3-1 in vivo and in vitro through both LCI and pull-down assays (Figure 5, E and F; Supplemental Figure S10B), indicating that GhWRKY16 likely functions downstream of the GhMKK2-GhMPK3-1 module.
Figure 5.
Assay of interaction between GhWRKY16 and GhMPK3-1, and GhMKK2 and GhMPK3-1 in vitro and in vivo. A, Co-expression network of GhWRKY16, as determined from ccNET (http://structuralbiology.cau.edu.cn/gossypium/). B, Yeast two-hybrid assay of the interaction between GhWRKY16 and GhMPK3-1. Yeast transformants containing pGADT7-GhWRKY16 or pGBKT7-GhMPK3-1 were spotted onto SD–Leu–Trp after yeast mating, and interaction was tested on SD–Leu–Trp–His–Ade, using pGBKT7 and pGADT7 empty vectors as negative controls. C, LCI assay of the interaction between GhWRKY16 and GhMPK3-1. Constructs expressing GhMPK3-1-LUCn and LUCc-GhWRKY16 were co-infiltrated in N. benthamiana leaves, using LUCn and LUCc as negative controls. D, Pull-down assay of GhWRKY16 and GhMPK3-1. Recombinant MBP-GhWRKY16 was incubated with GST-GhMPK3-1 in vitro, using GST as negative control. Pulled-down proteins were analyzed by immunoblotting with anti-MBP antibody. E, LCI assay of the interaction between GhMPK3-1 and GhMKK2. Constructs expressing GhMPK3-1-LUCn and LUCc-GhMKK2 were co-infiltrated in N. benthamiana leaves, using LUCn and LUCc as negative controls. F, Pull-down assay of GhMPK3-1 and GhMKK2. Recombinant GST-GhMPK3-1 was incubated with His-GhMKK2 in vitro, using GST as negative control. Pulled-down proteins were analyzed by immunoblotting with anti-His antibody
Phosphorylation of GhWRKY16 by GhMPK3-1 is essential for enhancing its transcriptional activity
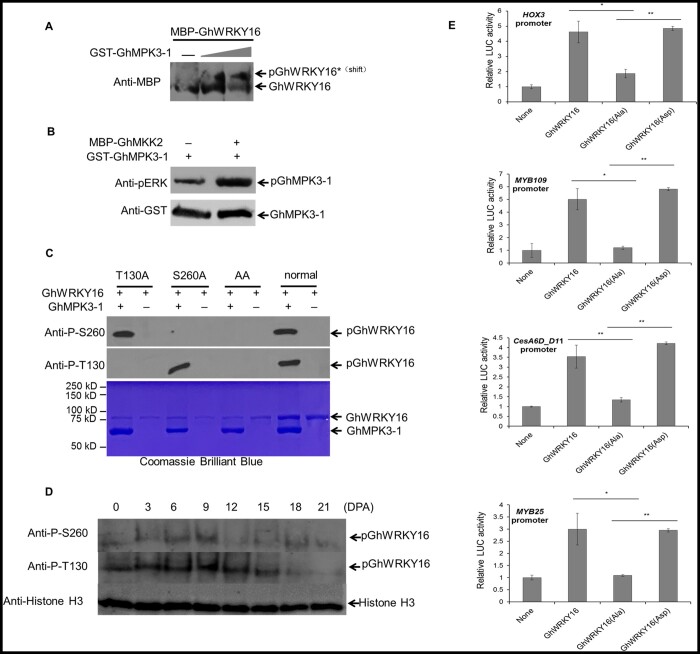
To test whether GhMPK3-1 can phosphorylate GhWRKY16, we conducted a Phos-tag mobility shift assay, in which phosphorylated proteins bind to Phos-tag, which then slows the migration of the phosphorylated protein during sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE), hence separating them from nonphosphorylated proteins (Mao et al., 2011). We detected recombinant MBP-GhWRKY16 as a single, faster migrating band in the absence of GST-GhMPK3-1, but we noticed the gradual appearance of a second, slower migrating band with increasing amounts of GST-GhMPK3-1, indicating that GhWRKY16 is phosphorylated by GhMPK3-1 in vitro (Figure 6A). Furthermore, an in vitro kinase activity assay revealed that GhMPK3-1 undergoes autophosphorylation, as well as phosphorylation by GhMKK2 (Figure 6B). The sites phosphorylated by MPKs are usually on a serine or threonine residue followed by a proline (S/T-P motif). We identified two predicted MPK phosphorylation sites in GhWRKY16: Thr-130 (EVSTFKPLCST130PSYK) and Ser-260 (KPIKGS260PHPRGYYKC). We prepared antibodies specifically recognizing the phosphorylated sites were prepared, and tested them on recombinant MBP-GhWRKY16 with the two phosphorylation sites intact (MBP-GhWRKY16), either site mutated (MBP-GhWRKY16T130A, MBP-GhWRKY16S260A) or both sites mutated (MBP-GhWRKY16T130A/S260A) incubated with GST-GhMPK3-1 for in vitro kinase activity assays. Both antibodies recognized a band of the appropriate size when MBP-GhWRKY16 was incubated with GST-GhMPK3-1 (Figure 6C). Each phospho-specific antibody recognized their cognate residues, demonstrating that Thr130 and Ser260 in GhWRKY16 are phosphorylated by GhMPK3-1 (Figure 6C). The nuclear localization of GhWRKY16 was not affected by the loss of phosphorylation (Supplemental Figure S11).
Figure 6.
Phosphorylation of GhWRKY16 by GhMPK3-1 enhances its transcriptional activation of downstream target genes. A, Immunoblotting analysis of in vitro phosphorylation of GhWRKY16 by recombinant GhMPK3-1 with SuperSep Phos-tagTM SDS–PAGE. Phosphorylated GhWRKY16 (pWRKY16) migrates more slowly in the gel. Gray triangle, increasing amount of MBP-GhWRKY16. B, In vitro kinase activity assay of GhMPK3-1 and GhMKK2. Phosphorylation of GhMPK3-1 by recombinant GhMKK2 was detected by immunoblotting with the anti-pERK antibody (Cell Signaling, Danvers, MA, USA), recognizing phosphorylated Extracellular Regulated protein Kinases (top), and anti-GST antibody to quantify GhMPK3-1 (bottom). C, Identification of phosphorylation sites in GhWRKY16 by in vitro kinase activity assay. T130A, GhWRKY16T130A; S260A, GhWRKY16S260A; AA, GhWRKY16T130A/S260A. Upper, immunoblot analysis; lower, SDS–PAGE gel stained by Coomassie Brilliant Blue for loading. Anti-P-T130, anti-phosphorylated WRKY16 Thr130 antibody; Anti-P-S260, anti-phosphorylated WRKY16 Ser260 antibody. D, Phosphorylation levels of GhWRKY16 in cotton fibers at different developmental stages. E, Effects of GhWRKY16 phosphorylation on the transcription of downstream target genes by dual LUC reporter assay. LUC activity was normalized to REN activity, with LUC/REN activity from the control without effector (None) set to 1. GhWRKY16, wild-type GhWRKY16; GhWRKY16(Ala), GhWRKY16T130A/S260A mutant; GhWRKY16(Asp), GhWRKY16 phospho-mimic. Error bars represent sd of three biological replicates. **P <0.01 by t test between phosphorylated GhWRKY16 and nonphosphorylated GhWRKY16(Ala)
We then explored the phosphorylation pattern of GhWRKY16 during fiber development. GhWRKY16 maintained a high phosphorylation level in initiating and elongating fibers (Figure 6D), suggesting that phosphorylation of GhWRKY16 is of great significance for its function in cotton fiber initiation and elongation. To test the significance of GhWRKY16 phosphorylation on its function as a TF, we employed the dual LUC system to measure the transcriptional activation potential of GhWRKY16 phospho-mutants on its downstream target genes. When constructs expressing GhWRKY16 or the phosphomimic GhWRKY16S260D together with GhMPK3-1 were co-infiltrated in N. benthamiana leaves, LUC activity derived from the GhHOX3pro:LUC, GhMYB109pro:LUC, GhCesA6D_D11pro:LUC, and GhMYB25pro:LUC reporters increased 2- to 4-fold over the reporter without effector. In contrast, LUC activity derived from N. benthamiana leaves co-infiltrated with constructs expressing the double mutant GhWRKY16T130A/S260A and GhMPK3-1 was comparable to the reporters without effector (Figure 6E). The above results demonstrate that phosphorylation of GhWRKY16 by GhMPK3-1 is essential for regulating the expression of its downstream target genes during fiber development in cotton.
Discussion
Cotton is one of the most important economic crops and is widely cultivated, providing natural raw materials for the textile industry, but high cotton planting costs and the poor quality of cotton fibers are bottlenecks to the development of a cotton-based economy. Meeting the increasing demand for higher quality and yield of cotton fibers will require new directions for cotton molecular breeding in the future. Thus, it is a pressing need to understand the molecular mechanism behind cotton fiber development. In this study, we demonstrated that the group II d WRKY TF GhWRKY16 functions in fiber initiation and elongation by regulating the expression of downstream target genes, such as GhHOX3, GhMYB109, GhCesA6D_D11, and GhMYB25. In addition, the phosphorylation of GhWRKY16 by the GhMKK2-GhMPK3-1 module is essential for its transcriptional activity during early fiber development of cotton.
WRKY transcription factors have been reported to participate in various plant physiological processes, such as abiotic and biotic stress responses, epidermal differentiation, leaf senescence, lateral roots development, and flowering (Ishida et al., 2007; Besseau et al., 2012; Yu et al., 2012; Ding et al., 2015b; Li et al., 2016; Wang et al., 2019). Here, we established that silencing of GhWRKY16 expression in cotton by RNAi leads to shorter mature fibers, with fewer or even the complete loss of fuzz on seeds, indicating that GhWRKY16 positively regulates fiber initiation and elongation. WRKY TFs play regulatory roles by binding to the W-box in the promoters of their target genes. In this study, we identified at least one W-box element in the promoters of genes predicted to act downstream of GhWRKY16, suggesting that GhWRKY16 may directly regulate their transcription for fiber elongation. However, GhWRKY16 did not affect the transcription of all genes with W-boxes, suggesting that the binding of GhWRKY16 to the W-box may be influenced by the sequence flanking the W-box. Indeed, GhWRKY16 tends to bind to W-box elements that are not surrounded by adjacent motifs, although the motif GTACTGAARGAG near a W-box may exert a positive effect on the binding of GhWRKY16 (Supplemental Figure S8). However, the specificity of such binding will require an in-depth investigation.
The fasciclin-like arabinogalactan protein GhFLA1 is involved in fiber initiation and elongation by affecting arabinogalactan protein composition and primary cell wall in cotton (Huang et al., 2013). The cotton cytosolic ascorbate peroxidase GhAPX1 functions in fiber elongation through regulating reactive oxygen species homeostasis during the fast fiber-cell elongation period (Li et al., 2007). A cotton TCP TF with high sequence identity to Arabidopsis TCP15 was shown to promote fiber elongation (Hao et al., 2012). The loss of cellulose synthases result in the reduction of primary cellulose production and a defect in cell elongation and cell wall integrity (Hu et al., 2019). In this study, transcriptome analysis revealed that the expression levels of numerous genes (encoding MYB, AP2/ERF, and TCP type TFs, auxin response factors, L-ascorbate peroxidases) with important roles in phytohormone biosynthetic and signaling pathways during cotton fiber initiation and elongation are prominently altered in GhWRKY16-RNAi fibers compared to the wild-type (Supplemental Data Set S1), suggesting that GhWRKY16 regulates cotton fiber development possibly through more than one pathway.
As the fuzz is too short to be collected by the cotton gin (mechanized cotton picker), a fuzz-free phenotype is typically regarded as a characteristic of better-quality cotton cultivars. However, the fuzz-free phenotype is often accompanied by lint loss or worse lint quality in cotton (Wan et al., 2016). Here, we determined that the suppression of GhWRKY16 expression results in shorter fibers and loss of fuzz on cotton seeds. GhHOX3 responds to gibberellin and affects cotton fiber elongation by regulating the expression levels of downstream genes GhRDL1 and GhEXPA1 (Shan et al., 2014). Our data showed that the expression level of GhHOX3 is lower in GhWRKY16-RNAi fibers, and that GhWRKY16 directly binds to the W-box elements in the GhHOX3 promoter, suggesting that GhWRKY16 functions directly upstream of GhHOX3 in early fiber development. The expression of the other important TF gene GhMYB109 was also significantly downregulated in GhWRKY16-RNAi fibers and may directly act downstream of GhWRKY16. Although GhMYB109-silenced cotton displayed a shorter fiber phenotype (Pu et al., 2008), the regulatory mechanism of MYB109-mediated fiber development is still unclear. In consequence, the identity of the fiber elongation-related genes downstream of GhMYB109 that are regulated by GhWRKY16 remain unknown. GhWRKY16 also regulated the expression of cellulose synthase genes associated with primary wall biosynthesis to meet the requirement of the rapidly elongating cotton fiber cells. Previous studies reported that GhMYB25 and GhMYB25-like are essential for fiber initiation and GhMYB25-like is upstream of GhMYB25 and GhMYB109 (Machado et al., 2009; Walford et al., 2011). At the initial stage of fiber development, GhWRKY16 also directly regulates the expression of GhMYB25 to promote fiber cell initiation. However, the expression of GhMYB25-like was unaltered in GhWRKY16-RNAi lines, and there was no interaction between GhWRKY16 and GhMYB25-like, suggesting that GhMYB25-like may function upstream from GhWRKY16 in fiber initiation of cotton.
MAPK signaling cascades are important in the regulation of plant growth and development as well as stress responses. A few WRKY TFs have been reported to be phosphorylated by MAPKs, thus affecting their transcriptional regulation activity. For instance, Arabidopsis WRKY34 is phosphorylated by MPK3 and MPK6 to maintain pollen vitality and promote pollen tube growth (Guan et al., 2014). During plant development, Arabidopsis MPK3 and MPK6 also phosphorylate WRKY2 to increase the transcription level of its downstream target gene WUSCHEL-RELATED HOMEOBOX 8 (WOX8), thereby affecting embryo developmental patterns (Ueda et al., 2017). The MAPK kinase cascade (GhMAPK3K15–GhMKK4–GhMAPK6) can directly phosphorylate GhWRKY59 to enhance the expression of the downstream drought-related gene DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2 (GhDREB2), hence improving cotton resistance to drought stress (Li et al., 2017). However, little is known about the phosphorylation of WRKY TFs by MPKs during cotton fiber development. In this study, our data revealed that GhWRKY16 is phosphorylated at Thr-130 and Ser-260 by GhMPK3-1, whose encoding gene is co-expressed with GhWRKY16 in fast elongating fibers of cotton. The phosphorylation of GhWRKY16 enhanced its transcriptional activation of the downstream fiber elongation-related genes to promote fiber elongation, suggesting that phosphorylation of GhWRKY16 is required for its transcriptional activation.
Phosphorylated proteins may exhibit altered functions, subcellular localization, or DNA binding affinity. For example, the phosphorylation of the cotton cytosolic pyruvate kinase GhPK6 inhibits its enzymatic activity and promotes its degradation during fiber elongation (Zhang and Liu, 2017). Similarly, cotton ACS2 is phosphorylated by the calcium-dependent protein kinase GhCPK1, resulting in increased ACS activity and ethylene production during fiber development (Wang et al., 2011). Phosphorylation of BRASSINAZOLE-RESISTANT 1 (GhBZR1) modulates its nuclear localization and directly affects the expression levels of XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 1 (GhXTH1) and GhEXP so as to modulate BR signaling in fiber initiation and elongation (Zhou et al., 2015). The phosphorylation of Arabidopsis MYB15 by MPK6 reduces its DNA binding affinity to its target genes (Kim et al., 2017). On the contrary, our data indicated that the nuclear localization of GhWRKY16 is not affected by phosphorylation (Supplemental Figure S11).
The conserved WRKY domain containing the WRKYGQK heptapeptide and the zinc-finger-like motif are essential for WRKY TFs binding to the W-box. In this study, we found that GhMPK3-1 phosphorylates GhWRKY16 at Thr-130 and Ser-260, while the WRKY domain is located between amino acids 245 and 306. We therefore speculated that phosphorylation at Ser-260 may affect the DNA binding affinity of GhWRKY16, thereby modulating the transcriptional regulation of GhWRKY16 on its downstream target genes. Additionally, we investigated expression levels of GhWRKY16 and GhMPK3-1 in fibers of four wild cotton species (TX2094, TX2090, TX2095, and TX665) and five domesticated cotton cultivars (MAXXA, TM1, Cascot L-7, Coker315, and CRB252) based on the data from Yoo and Wendel (2014). We discovered that, aside from higher expression levels in wild cotton TX665 and domesticated cotton MAXXA, GhWRKY16 showed consistent expression levels in early developing fibers of the other species, suggesting that the function of GhWRKY16 is conserved across cotton species during cotton fiber development. In contrast, GhMPK3-1 was induced in elongating fibers in domesticated cotton, suggesting that GhMPK3-1 has likely been selected during cotton domestication and may be potentially used for genetic improvement on cotton fibers (Supplemental Table S4).
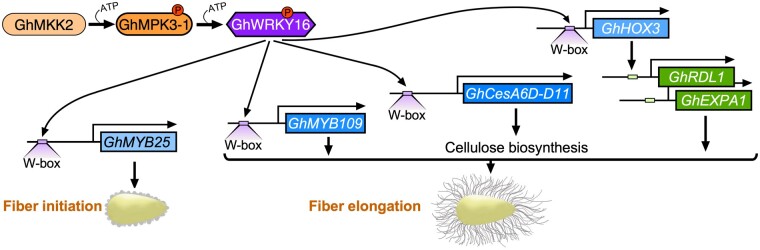
In summary, our data reveal a novel GhWRKY16-mediated regulatory mechanism for promoting fiber initiation and elongation in cotton (Figure 7). During early fiber development, GhMPK3-1 is phosphorylated by GhMKK2, and phosphorylated GhMPK3-1 then phosphorylates GhWRKY16 at Thr-130 and Ser-260 to activate the transcriptional activity of GhWRKY16. Subsequently, activated GhWRKY16 binds to the W-box cis-acting elements in the promoters of downstream target genes (such as GhMYB25, GhHOX3, GhMYB109, and GhCesA6D_D11) to activate their expression and promote fiber initiation and elongation. During the fiber initiation stage, GhWRKY16 directly regulates the expression of GhMYB25, a key regulator of early fiber development. At the fiber elongation stage, GhWRKY16 promotes fiber cell elongation through directly regulating the expression of GhHOX3 and GhMYB109, which are of vital importance for fiber elongation. GhHOX3 may also regulate the expression of the genes encoding the cell wall loosening proteins GhEXP1 and GhRDL1 to modulate fiber elongation (Shan et al., 2014). In parallel, the expression of the cellulose synthase gene GhCesA6D_D11 is also induce by GhWRKY16 to provide sufficient cellulose materials for primary cell wall biosynthesis in rapidly elongating fibers. Thus, this study provides a new understanding of the molecular mechanism governing GhWRKY16-regulated fiber development, and thereby offers a potential target to improve the yield and quality of cotton fibers.
Figure 7.
A model showing the molecular mechanism of GhWRKY16 regulating fiber initiation and elongation
Materials and methods
Plant materials
Seeds of upland cotton (Gossypium hirsutum cv. Cocker 312) were surface sterilized with 75% (v/v) ethanol for 1 min, and then 10% hydrogen peroxide for 2 h, and washed with sterile distilled water three to five times. After soaking in sterile distilled water overnight, the seeds were sown onto half-strength Murashige and Skoog medium and allowed to germinate for 5–6 days under 16-h light (provided by 12w LED light bulb; 5,000 lux light intensity [100 µE/m2/s])/8-h dark cycles at 28°C. The seedlings were transplanted to soil for further growth to maturation. The hypocotyls from these seedlings were used for cotton transformation, following the protocol described by Li et al. (2002). Fibers and other cotton tissues were collected for DNA, RNA, and protein extraction.
RT-qPCR analysis
Total RNA was extracted from 0 DPA ovules, and 3, 6, 9, 12, 15, 18, and 21 DPA fibers with the RNAprep Pure Polysaccharide Polyphenol Plant Total RNA Extraction Kit (Tiangen, China) according to manufacturer’s instructions. About 1.5–2 µg total RNA from cotton fibers was reverse-transcribed into cDNA using M-MLV reverse transcriptase (Promega, USA) according to the manufacturer’s manual before quantitative PCR using SYBR Green real-time PCR master mix (Toyobo, Japan) in a DNA Engine Opticon 2 detection system (MJ Research, Canada). The cDNAs were used as templates in real-time PCR reactions with gene-specific primers, and a cotton polyubiquitin gene GhUBI1 (EU604080) was used as reference. The relative expression of each target gene was calculated by the equation Y = 10(CtGhUBI-CtGhgene)/3×100%, the detailed experimental method refers to Li et al. (2005). Each RT-qPCR reaction was performed in triplicates, and results are reported as means and standard deviation. Gene-specific primers used in this analysis are listed in Supplemental Data Set S3.
Construction of vectors
To construct the GhWRKY16 RNAi vector, a 250-bp fragment specific for GhWRKY16 was cloned by PCR using Pyrococcus furiosus Pfu DNA polymerase (Takara, Japan) into the modified pBluescript SK with a Tubulin intron to generate an inverted repeat before cloning into the pBI121 vector. To construct vectors for recombinant protein production, the coding sequences of GhWRKY16 and GhMPK3-1 were cloned into pMAL vector (New England Biolabs, Ipswich, MA, USA) and pGEX-4T vector (GE Healthcare, Pittsburgh, PA, USA), respectively.
Microscopy of ovules and fiber morphology
For observations by optical microscopy, ovules at 0, 1, and 2 DPA were collected from the wild-type and GhWRKY16-RNAi lines grown in the field, cross-sectioned to 20–30 µm in thickness and observed under an optical microscope.
To observe the protuberances of fiber cells at very early developmental stages, cotton ovules at 0 and 1 DPA were fixed in 2.5% (v/v) glutaraldehyde under vacuum, and then sprayed with gold. Scanning electron microscopy was used to observe the surface of ovules at the School of Medicine, Wuhan University (Wuhan, China). The number of fiber protrusions was scored on the ovules at 0 DPA.
In vitro cotton ovule culture
Cotton bolls at 1 DPA were collected and sterilized with 75% ethanol for 1 min, and then washed two to three times with sterile distilled water. The ovules were excised from the bolls with a scalpel and placed into liquid Beasley–Ting medium containing 0.5-μM gibberellic acid and 5-μM indole-3-acetic acid. The ovules were cultured in the dark at 30°C for 12–18 days. Photographs were then taken and the length of the fibers on the ovule surface was measured.
EMSA
Biotin-labeled probes containing W-box from the GhHOX3, GhMYB109, GhCesA6D-D11, and GhMYB25 promoters were incubated with purified recombinant MBP-GhWRKY16 in binding buffer (10-mM Tris, pH 7.5, 1-mM EDTA, 50-mM NaCl, and 1-mM dithiothreitol) for 20 min at room temperature. Then the protein–DNA complex was separated by 6% non-denaturing polyacrylamide gel in 0.5×Tris Buffered EDTA buffer, pH 8.0. A 10-fold excess of unlabeled probes was used for competition assay. Biotin-labeled probes with a mutated W-box (TTGAC to AAAAC) were used as negative control. The DNA in the gel was electroblotted onto nitrocellulose membrane and detected by Chemiluminescent Nucleic Acid Detection Module Kit (Thermo Scientific).
ChIP assay
The ChIP assay was performed as previously described (Haring et al., 2007). Briefly, ∼2 g of fibers at 9 DPA from wild-type cotton was cross-linked with 1% (v/v) formaldehyde for 10 min at room temperature. Chromatin was then sheared with a sonicator and immunoprecipitated with an anti-GhWRKY16 antibody. The DNA was extracted and used as template for qPCR analysis. All primer sequences are listed in Supplemental Data Set S3.
Dual LUC assay
The dual LUC assay followed the protocol described previously (Liu et al., 2008). Briefly, the promoters of GhHOX3, GhMYB109, GhCesA6D_D11, and GhMYB25 were cloned into the pGreenII0800-LUC vector, while the coding sequence of the effector GhWRKY16 was cloned into the vector p2300. All resulting constructs were then introduced into Agrobacterium (Agrobacterium tumefaciens) strain GV3101 (with the helper PSoup+P19 plasmid). The Agrobacteria containing each reporter or effector were co-infiltrated into N. benthamiana leaves. After incubation for 48–64 h with bagging in the light, the infiltrated areas of N. benthamiana leaves were collected for total protein extraction. The firefly LUC activity, derived from the reporter constructs, and Renilla luciferase (REN) activity, derived from the constitutive cauliflower mosaic virus 35S promoter, were determined according to the operating instruction of the Dual-Luciferase Reporter Assay System kit (Promega). LUC activity was normalized to that of REN. The mean value and standard derivation were calculated from three independent biological replicate experiments.
Yeast two-hybrid assay
To analyze the interaction between GhWRKY16 and GhMPKs, the coding sequence of GhWRKY16 was cloned into the pGADT7 vector, and then introduced into yeast strain AH109. The coding sequences for GhMPK3-1, GhMPK3-2, GhMPK6-1, GhMPK6-2, and GhMPK6-3 were cloned into the pGBKT7 vector, and introduced individually into yeast strain Y187. After mating reactions between the two haploid strains, diploid colonies were selected on synthetic dextrose medium lacking leucine and tryptophan (SD–Leu–Trp). Positive interactions were detected on selective medium (SD–Leu–Trp–His–Ade; Zhang et al., 2010).
LCI assay
The coding sequence of GhMPK3-1 was cloned into the JW771 vector, while the coding sequences of GhWRKY16 and GhMKK1, GhMKK2, and GhMKK4 were cloned into the JW772 vector, as previously described (Gou et al., 2011). In the constructs, GhWRKY16 and GhMKK1/2/4 were fused to the C-terminal half of luciferase (LUCc) to generate LUCc-GhWRKY16, LUCc-GhMKK1/2/4 fusions, and GhMPK3-1 was fused to the N-terminal half of LUC (LUCn) to generate GhMPKs–LUCn fusion, using LUCc and LUCn as negative controls. The constructs were introduced into Agrobacterium strain GV3101 (with the helper pSoup+P19 plasmid). Agrobacteria containing the LUCc-GhWRKY16, LUCc-GhMKK1/2/4, or GhMPK3-1-LUCn constructs were resuspended in infiltration buffer (10-mM MgCl2, 10-mM MES [2-(N-morpholino) ethanesulfonic acid], pH 5.7, 150-μM acetosyringone), and infiltrated into N. benthamiana leaves. After incubation for 48–64 h with bagging in the light, the 0.8-mM D-luciferin was sprayed onto the infiltrated N. benthamiana leaves and the LUC signal was observed with a chemiluminescence imaging system (tanon-5200 multi).
Pull-down assay
Recombinant GST-GhMPK3-1, MBP-GhWRKY16, and His-GhMKK2 were produced in Escherichia coli strain BL21 (DE3) with the addition of 0.5-mM IPTG (isopropyl β-d-1-thiogalactopyranoside) at 37°C and OD600 =0.5. The supernatant containing the GST–GhMPK3-1 fusion protein was incubated with an appropriate amount of glutathione–agarose resin for 30 min, and the unbound proteins were washed away with 10 volumes of phosphate buffered saline. Then, purified MBP-GhWRKY16 or purified His-GhMKK2 was mixed with the GST-GhMPK3-1 resin suspension and incubated at 4°C for 2 h. Bound GST-GhMPK3-1 and any interacting protein were eluted with elution buffer (50-mM Tris–HCl, 10-mM glutathione, pH 8.0). The eluted proteins were subjected to immunoblot analysis with an anti-MBP antibody or anti-His antibody (working dilution 1:3,000; Abcam, UK).
Preparation of antibody against phosphorylated GhWRKY16 peptides
The phosphorylated antibodies specific for phosphorylated GhWRKY16 were prepared by GL Biochem Ltd (Shanghai, China). First, phosphorylated peptides and nonphosphorylated peptides were synthesized to a purity of over 90%. The phosphorylated peptides and nonphosphorylated peptides were coupled with keyhole limpet hemocyanin (KLH) to immunize rabbits. After the sixth immunization, an antiserum sample was taken to test the serum titer, and after the eighth immunization, the antiserum samples were collected. Second, a nonphosphorylated antigen affinity purification column was used to purify the antiserum to obtain antibodies recognizing the nonphosphorylated peptides until the peak of antibodies reached a certain low level. After all these antibodies were removed, a phosphorylated antigen affinity purification column was used to purify the antiserum to obtain the antibody recognizing the phosphorylated peptides. Enzyme-linked immunosorbent assay estimated the antibody titer to be above 1:32,000. Specificity of each phospho-specific antibody was tested by dot blotting assay; the phospho-specific antibodies did not cross-react with nonphosphorylated peptides (Supplemental Figure S12).
In vitro kinase activity assay
In vitro kinase activity assays were performed as described previously (Ding et al., 2018). Briefly, 3 mg of recombinant WRKY16 and 10 mg of recombinant MPK3-1 proteins were mixed in a 50-μL kinase reaction (25-mM Tris–HCl, pH 7.5, 1-mM EGTA, 20-mM MgCl2, 1-mM DTT, 200-μM ATP, and 1×phosphatase inhibitor) and incubated at room temperature for 1 h. The reaction was stopped by adding 5× SDS loading buffer and incubating at 100°C for 5 min. Samples were separated on SuperSep Phos-tag SDS–PAGE gels (Wako 193-16711) or 12% SDS–PAGE gels, and then analyzed by immunoblotting with an anti-MBP antibody (working dilution 1:3,000; Abcam, UK).
Accession numbers
Sequence data from this article can be found at The National Center for Biotechnological Information (NCBI) under the following accession numbers: GhHOX3 (Gh_A12G2462), GhMYB109 (Gh_A05G3123), GhRDL1 (Gh_A05G0391), GhEXP1 (Gh_D04G1924), GhFLA1 (Gh_D01G0733), GhAPX1 (Gh_D08G2094), GhXTH6 (Gh_D11G2065), GhRAV1 (Gh_D02G1153), GhWBC1 (Gh_D07G0603), GhTCP15 (Gh_D13G2529), GhCesA1B_A05 (Gh_A05G3967), GhCesA1B_D05 (Gh_D05G0077), GhCesA3A_D08 (Gh_D03G0611), GhCesA3B_A08 (Gh_A08G1305), GhCesA3B_D08 (Gh_D08G1597), GhCesA6D-D11 (Gh_D11G2235), GhMYB25 (Gh_D04G1901), GhMYB25-like (Gh_D12G1628), GhWRKY16 (Gh_D06G0175), GhMPK3-1 (Gh_D03G1283), GhMKK2 (Gh_D06G0748), GhMPK3-2 (Gh_D05G3876), GhMPK6-1 (Gh_D02G0105), GhMPK6-2 (Gh_D02G1080), GhMPK6-3 (Gh_D03G1422), GhMKK1 (Gh_D05G0323), GhMKK4 (Gh_D06G1960).
All sequences obtained by deep sequencing datasets were deposited at the Gene Expression Omnibus at NCBI under project GSE174452.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Data Set S1 . Characteristics of 230 WRKY genes in upland cotton (G. hirsutum).
Supplemental Data Set S2 . DEGs identified in 9 DPA fibers of GhWRKY16-RNAi cotton lines by RNA-seq analysis.
Supplemental Data Set S3 . Primers and probes used in this study.
Supplemental Data Set S4 . Summary of statistical analyses.
Supplemental Figure S1 . Expression profiles of selected GhWRKY genes in fibers of GhWRKY16-RNAi cotton.
Supplemental Figure S2 . Phenotypic analysis of GhWRKY16-RNAi transgenic cotton plants (T4 generation).
Supplemental Figure S3 . Observation of fiber initiation and assay of thousand-seed weight and fiber weight in GhWRKY16-RNAi transgenic cotton.
Supplemental Figure S4 . RT-qPCR analysis of DEGs in fibers of GhWRKY16-RNAi transgenic cotton lines.
Supplemental Figure S5 . Gene ontology (GO) functional cluster analysis of RNA-seq data from 9 DPA fibers of GhWRKY16-RNAi transgenic cotton lines.
Supplemental Figure S6 . GhWRKY16 functions as a typical WRKY transcriptional regulator.
Supplemental Figure S7 . ChIP-qPCR analysis of GhWRKY16 binding to the promoters of GhHOX3 and GhMYB109 in fibers of GhWRKY16-RNAi cotton lines.
Supplemental Figure S8 . Assay of the specificity of GhWRKY16 protein binding to the W-box element.
Supplemental Figure S9 . Expression profiles of MAPK genes in ovules and fibers of cotton at different developmental stages.
Supplemental Figure S10 . Interaction analysis between GhWRKY16 and GhMPKs, and GhMPK3-1 and MKK1/4 in planta.
Supplemental Figure S11 . Subcellular localization of mutated GhWRKY16(T130A/S260A).
Supplemental Figure S12 . Dot blotting assay of the specificity of antibodies against phosphorylated GhWRKY16.
Supplemental Table S1 . Comparison of fiber quality parameters between the wild-type and GhWRKY16-RNAi transgenic cotton.
Supplemental Table S2 . W-box elements in the promoters of predicted target genes downstream of GhWRKY16.
Supplemental Table S3 . Proteins interacting with GhWRKY16, as predicted by co-expression network analysis.
Supplemental Table S4 . Expression analysis of GhWRKY16 and GhMPK3-1 in domesticated cotton cultivars and wild cotton species.
Funding
This work was supported by the National Key R&D Program of China (Grant No. 2016YFD0100505), the Project of Transgenic Research from the Ministry of Agriculture of China (Grant No. 2016ZX08009003), and the National Natural Science Foundation of China (Grant No. 31500987).
Conflict of interest statement. The authors declare no competing interests.
Supplementary Material
X.B.L. and N.N.W. conceived and designed the research; N.N.W., Y.L., Y.H.C., R.L., L.Z., and Y.W. performed the experiments; N.N.W., Y.Z., and X.B.L. analyzed the data; N.N.W. and X.B.L. wrote the paper.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Xue-Bao Li (xbli@mail.ccnu.edu.cn).
References
- Besseau S, Li J, Palva ET (2012) WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J Exp Bot 63: 2667–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Yan T, Shen Q, Lu X, Pan Q, Huang Y, Tang Y, Fu X, Liu M, Jiang W, et al. (2017) GLANDULAR TRICHOME-SPECIFIC WRKY1 promotes artemisinin biosynthesis in Artemisia annua. New Phytol 214: 304–316 [DOI] [PubMed] [Google Scholar]
- de Pater S, Greco V, Pham K, Memelink J, Kijne J (1996) Characterization of a zinc-dependent transcriptional activator from Arabidopsis. Nucleic Acids Res 24: 4624–4631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H, He J, Wu Y, Wu X, Ge C, Wang Y, Zhong S, Peiter E, Liang J, Xu W (2018) The tomato mitogen-activated protein kinase SlMPK1 is as a negative regulator of the high-temperature stress response. Plant Physiol 177: 633–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding M, Chen J, Jiang Y, Lin L, Cao Y, Wang M, Zhang Y, Rong J, Ye W (2015a) Genome-wide investigation and transcriptome analysis of the WRKY gene family in Gossypium. Mol Genet Genom 290: 151–171 [DOI] [PubMed] [Google Scholar]
- Ding ZJ, Yan JY, Li CX, Li GX, Wu YR, Zheng SJ (2015b) Transcription factor WRKY 46 modulates the development of Arabidopsis lateral roots in osmotic/salt stress conditions via regulation of ABA signaling and auxin homeostasis. Plant J 84: 56–69 [DOI] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5: 199–206 [DOI] [PubMed] [Google Scholar]
- Gou JY, Wang LJ, Chen SP, Hu WL, Chen XY (2007) Gene expression and metabolite profiles of cotton fiber during cell elongation and secondary cell wall synthesis. Cell Res 17: 422–434 [DOI] [PubMed] [Google Scholar]
- Gou JY, Felippes FF, Liu CJ, Weigel D, Wang JW (2011) Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23: 1512–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Meng X, Khanna R, LaMontagne E, Liu Y, Zhang S (2014) Phosphorylation of a WRKY transcription factor by MAPKs is required for pollen development and function in Arabidopsis. PLoS Genet 10: e1004384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigler CH, Betancur L, Stiff MR, Tuttle JR (2012) Cotton fiber: a powerful single-cell model for cell wall and cellulose research. Front Plant Sci 3: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao J, Tu L, Hu H, Tan J, Deng F, Tang W, Nie Y, Zhang X (2012) GbTCP, a cotton TCP transcription factor, confers fibre elongation and root hair development by a complex regulating system. J Exp Bot 63: 6267–6281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haring M, Offermann S, Danker T, Horst I, Peterhansel C, Stam M (2007) Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods 3: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Wang R, Zheng M, Liu X, Meng F, Wu H, Yao Y, Xin M, Peng H, Ni Z, et al. (2018) TaWRKY51 promotes lateral root formation through negative regulation of ethylene biosynthesis in wheat (Triticum aestivum L.). Plant J 96: 372–388 [DOI] [PubMed] [Google Scholar]
- Hu H, Zhang R, Tang Y, Peng C, Wu L, Feng S, Chen P, Wang Y, Du X, Peng L (2019) Cotton CSLD3 restores cell elongation and cell wall integrity mainly by enhancing primary cellulose production in the Arabidopsis cesa6 mutant. Plant Mol Biol 101: 389–401 [DOI] [PubMed] [Google Scholar]
- Huang GQ, Gong SY, Xu WL, Li W, Li P, Zhang CJ, Li DD, Zheng Y, Li FG, Li XB (2013) A fasciclin-like arabinogalactan protein, GhFLA1, is involved in fiber initiation and elongation of cotton. Plant Physiol 161: 1278–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T, Hattori S, Sano R, Inoue K, Shirano Y, Hayashi H, Shibata D, Sato S, Kato T, Tabata S, et al. (2007) Arabidopsis TRANSPARENT TESTA GLABRA2 is directly regulated by R2R3 MYB transcription factors and is involved in regulation of GLABRA2 transcription in epidermal differentiation. Plant Cell 19: 2531–2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro S, Nakamura K (1994) Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5’ upstream regions of genes coding for sporamin and β-amylase from sweet potato. Mol Gen Genet 244: 563–571 [DOI] [PubMed] [Google Scholar]
- Kim HJ, Triplett BA (2001) Cotton fiber growth in planta and in vitro. Models for plant cell elongation and cell wall biogenesis. Plant Physiol 127: 1361–1366 [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Kim HS, Bahk S, An J, Yoo Y, Kim JY, Chung WS (2017) Phosphorylation of the transcriptional repressor MYB15 by mitogen-activated protein kinase 6 is required for freezing tolerance in Arabidopsis. Nucleic Acids Res 45: 6613–6627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XB, , Fan XP, , Wang XL, , Cai L, , Yang WC ( 2005) The cotton ACTIN1 gene is functionally expressed in fibers and participates in fiber elongation. Plant Cell 17: 859–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Li M, Wang P, Cox KL Jr, Duan L, Dever JK, Shan L, Li Z, He P (2017) Regulation of cotton (Gossypium hirsutum) drought responses by mitogen-activated protein (MAP) kinase cascade-mediated phosphorylation of GhWRKY59. New Phytol 215: 1462–1475 [DOI] [PubMed] [Google Scholar]
- Li HB, Qin YM, Pang Y, Song WQ, Mei WQ, Zhu YX (2007) A cotton ascorbate peroxidase is involved in hydrogen peroxide homeostasis during fibre cell development. New Phytol 175: 462–471 [DOI] [PubMed] [Google Scholar]
- Li W, Wang H, Yu D (2016) Arabidopsis WRKY transcription factors WRKY12 and WRKY13 oppositely regulate flowering under short-day conditions. Mol Plant 9: 1492–1503 [DOI] [PubMed] [Google Scholar]
- Li XB, Cai L, Cheng NH, Liu JW (2002) Molecular characterization of the cotton GhTUB1 gene that is preferentially expressed in fiber. Plant Physiol 130: 666–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Yu X, Li K, Klejnot J, Yang H, Lisiero D, Lin C (2008) Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322: 1535–1539 [DOI] [PubMed] [Google Scholar]
- Liu ZH, Chen Y, Wang NN, Chen YH, Wei N, Lu R, Li Y, Li XB (2020) A basic helix-loop-helix (bHLH) protein (GhFP1) promotes fiber elongation of cotton (Gossypium hirsutum) via modulating BR biosynthesis and signaling. New Phytol 225: 2439–2452 [DOI] [PubMed] [Google Scholar]
- Mao G, Meng X, Liu Y, Zheng Z, Chen Z, Zhang S (2011) Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23: 1639–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado A, Wu Y, Yang Y, Llewellyn DJ, Dennis ES (2009) The MYB transcription factor GhMYB25 regulates early fibre and trichome development. Plant J 59: 52–62 [DOI] [PubMed] [Google Scholar]
- Pu L, Li Q, Fan XP, Yang WC, Xue YB (2008) The R2R3 MYB transcription factor GhMYB109 is required for cotton fiber development. Genetics 180: 811–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin YM, Hu CY, Pang Y, Kastaniotis AJ, Hiltunen JK, Zhu YX (2007) Saturated very-long-chain fatty acids promote cotton fiber and Arabidopsis cell elongation by activating ethylene biosynthesis. Plant Cell 19: 3692–3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin YM, Hu CY, Zhu YX (2008) The ascorbate peroxidase regulated by H2O2 and ethylene is involved in cotton fiber cell elongation by modulating ROS homeostasis. Plant Signal Behav 3: 194–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Macdonald H, Huttly AK, Lazarus CM, Hooley R (1995) Members of a new family of DNA-binding proteins bind to a conserved cis-element in the promoters of a-Amy2 genes. Plant Mol Biol 29: 691–702 [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ (2010) WRKY transcription factors. Trends Plant Sci 15: 247–258 [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Torres JT, Parniske M, Wernert P, Hahlbrock K, Somissich IE (1996) Interaction of elicitor-induced DNA binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J 15: 5690–5700 [PMC free article] [PubMed] [Google Scholar]
- Shan CM, Shangguan XX, Zhao B, Zhang XF, Chao LM, Yang CQ, Wang LJ, Zhu HY, Zeng YD, Guo WZ, et al. (2014) Control of cotton fibre elongation by a homeodomain transcription factor GhHOX3. Nat Commun 5: 5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi YH, Zhu SW, Mao XZ, Feng JX, Qin YM, Zhang L, Cheng J, Wei LP, Wang ZY, Zhu YX (2006) Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell 18: 651–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takuji T, Adachi M, Moriguchi T, Nishida EA (2000) Conserved docking motif in MAP kinases common 972 to substrates, activators and regulators. Nat Cell Biol 2: 110–116 [DOI] [PubMed] [Google Scholar]
- Ueda M, Aichinger E, Gong W, Groot E, Verstraeten I, Vu LD, De Smet I, Higashiyama T, Umeda M, Laux T (2017) Transcriptional integration of paternal and maternal factors in the Arabidopsis zygote. Genes Dev 31: 617–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walford SA, Wu YR, Llewellyn DJ, Dennis ES (2011) GhMYB25-like: a key factor in early cotton fibre development. Plant J 65: 785–797 [DOI] [PubMed] [Google Scholar]
- Wan Q, Guan X, Yang N, Wu H, Pan M, Liu B, Fang L, Yang S, Hu Y, Ye W, et al. (2016) Small interfering RNAs from bidirectional transcripts of GhMML3_A12 regulate cotton fiber development. New Phytol 210: 1298–1310 [DOI] [PubMed] [Google Scholar]
- Wang H, Mei W, Qin Y, Zhu YX (2011) 1-Aminocyclopropane-1-carboxylic acid synthase 2 is phosphorylated by calcium-dependent protein kinase 1 during cotton fiber elongation. Acta Biochim Biophys Sin 43: 654–661 [DOI] [PubMed] [Google Scholar]
- Wang MY, Zhao PM, Cheng HQ, Han LB, Wu XM, Gao P, Wang HY, Yang CL, Zhong NQ, Zuo JR, et al. (2013) The cotton transcription factor TCP14 functions in auxin-mediated epidermal cell differentiation and elongation. Plant Physiol 162: 1669–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang NN, Xu SW, Sun YL, Liu D, Zhou L, Li Y, Li XB (2019) The cotton WRKY transcription factor (GhWRKY33) reduces transgenic Arabidopsis resistance to drought stress. Sci Rep 9: 724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei KF, Chen J, Chen YF, Wu LJ, Xie DX (2012) Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in maize. DNA Res 19: 153–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel JF, Cronn RC (2003) Polyploidy and the evolutionary history of cotton. Adv Agron 78: 139–186 [Google Scholar]
- Wu KL, Guo ZJ, Wang HH, Li J (2005) The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res 12: 9–26 [DOI] [PubMed] [Google Scholar]
- Yang Y, Liang T, Zhang L, Shao K, Gu X, Shang R, Shi N, Li X, Zhang P, Liu H (2018) UVR8 interacts with WRKY36 to regulate HY5 transcription and hypocotyl elongation in Arabidopsis. Nat Plants 4: 98–107 [DOI] [PubMed] [Google Scholar]
- Yang Z, Wang X, Li X, Yang C (2004) Advance on the study of transcription factors in higher plants. Hereditas 26: 403–408 [PubMed] [Google Scholar]
- Yoo MJ, Wendel JF (2014) Comparative evolutionary and developmental dynamics of the cotton (Gossypium hirsutum) fiber transcriptome. PLoS Genet 10: e1004073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Q, Xu WY, Zhang K, Zhang LW, Yi X, Yao DX, Wang CC, Zhang XY, Zhao XH, Provart N, et al. (2016) ccNET: database of co-expression networks with functional modules for diploid and polyploid Gossypium. Nucleic Acids Res 45: D1090–D1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Huaxia Y, Lu W, Wu C, Cao X, Guo X (2012) GhWRKY15, a member of the WRKY transcription factor family identified from cotton (Gossypium hirsutum L.), is involved in disease resistance and plant development. BMC Plant Biol 12: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Liu Z, Wang L, Kim SG, Seo PJ, Qiao M, Wang N, Li S, Cao X, Park CM, et al. (2016) WRKY71 accelerates flowering via the direct activation of FLOWERING LOCUS T and LEAFY in Arabidopsis thaliana. Plant J 85: 96–106 [DOI] [PubMed] [Google Scholar]
- Zhao B, Cao JF, Hu G, Chen ZW, Wang LY, Shangguan XX, Wang LJ, Mao YB, Zhang TZ, Wendel JF, et al. (2018) Core cis-element variation confers subgenome-biased expression of a transcription factor that functions in cotton fiber elongation. New Phytol 218: 1061–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Liu JY (2017) Serine phosphorylation of the cotton cytosolic pyruvate kinase GhPK6 decreases its stability and activity. FEBS Open Bio 7: 358–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Huang GQ, Zou D, Yan JQ, Li Y, Hu S, Li XB (2018) The cotton (Gossypium hirsutum) NAC transcription factor (FSN1) as a positive regulator participates in controlling secondary cell wall biosynthesis and modification of fibers. New Phytol 217: 625–640 [DOI] [PubMed] [Google Scholar]
- Zhang ZT, Zhou Y, Li Y, Shao SQ, Li BY, Shi HY, Li XB (2010) Interactome analysis of the six cotton 14-3-3s that are preferentially expressed in fibres and involved in cell elongation. J Exp Bot 61: 3331–3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Zhao Y, Shan D, Shi K, Wang L, Li Q, Wang N, Zhou J, Yao J, Xue Y, et al. (2018) MdWRKY9 overexpression confers intensive dwarfing in the M26 rootstock of apple by directly inhibiting brassinosteroid synthetase MdDWF4 expression. New Phytol 217: 1086–1098 [DOI] [PubMed] [Google Scholar]
- Zhou L, Wang NN, Kong L, Gong SY, Li Y, Li XB (2014) Molecular characterization of 26 cotton WRKY genes that are expressed differentially in tissues and are induced in seedlings under high salinity and osmotic stress. Plant Cell Tiss Organ Cult 119: 141–156 [Google Scholar]
- Zhou Y, Zhang ZT, Li M, Wei XZ, Li XJ, Li BY, Li XB (2015) Cotton (Gossypium hirsutum) 14‐3‐3 proteins participate in regulation of fibre initiation and elongation by modulating brassinosteroid signalling. Plant Biotechnol J 13: 269–280 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.