Abstract
Chemotherapy-induced peripheral neuropathy (CIPN) is a frequent and potentially irreversible adverse event of cytotoxic chemotherapy. We evaluate whether sensory neurons derived from induced pluripotent stem cells (iPSC-DSN) can serve as human disease model system for chemotherapy induced neurotoxicity. Sensory neurons differentiated from two established induced pluripotent stem cell lines were used (s.c. BIHi005-A https://hpscreg.eu/cell-line/BIHi005-A and BIHi004-B https://hpscreg.eu/cell-line/BIHi004-B, Berlin Institute of Health Stem Cell Core Facility). Cell viability and cytotoxicity assays were performed, comparing susceptibility to four neurotoxic and two non-neurotoxic drugs. RNA sequencing analyses in paclitaxel vs. vehicle (DMSO)-treated sensory neurons were performed. Treatment of iPSC-DSN for 24 h with the neurotoxic drugs paclitaxel, bortezomib, vincristine and cisplatin led to a dose dependent decline of cell viability in clinically relevant IC50 ranges, which was not the case for the non-neurotoxic compounds doxorubicin and 5-fluorouracil. RNA sequencing analyses at 24 h, i.e. before paclitaxel-induced cell death occurred, revealed the differential expression of genes of neuronal injury, cellular stress response, and sterol pathways in response to 1 µM paclitaxel. Neuroprotective effects of lithium chloride co-incubation, which were previously shown in rodent dorsal root ganglia, could be replicated in human iPSC-DSN. Cell lines from the two different donors BIHi005-A and BIHi004-B showed different responses to the neurotoxic treatment in cell viability and cytotoxicity assays.
Keywords: Induced pluripotent stem cell derived sensory neurons (iPSC-DSN), Chemotherapy induced neuropathy, 3R, Replacement, Transcriptome, Lithium
Specifications Table
| Subject | Neuroscience: Cellular and Molecular |
| Specific subject area | Experimental neurology and disease modeling |
| Type of data | Table Chart Graph Figure Transcriptome data |
| How data were acquired | MTT-based cell viability and proteases-based cytotoxicity assays. RNA sequencing analyses. |
| Data format | Raw data Analyzed |
| Parameters for data collection | Data were collected from experiments using induced pluripotent stem cell derived sensory neurons. |
| Description of data collection | Data were collected from different cell viability and cytotoxicity assays based on signal detection by fluorescence, luminescence and absorbance readers. RNA sequencing analyses were performed as described in the manuscript [1]. |
| Data source location | Institution: Charité - Universitätsmedizin Berlin City/Town/Region: Berlin Country: Germany |
| Data accessibility | With the article |
| Related research article | Co-submission: Schinke et al. Modeling chemotherapy induced neurotoxicity with human induced pluripotent stem cell (iPSC) -derived sensory neurons. Neurobiology of disease 2021. https://doi.org/10.1016/j.nbd.2021.105391[1]. |
Value of the Data
-
•
These data give insight into the pathomechanisms of chemotherapy induced neurotoxicity modeled with human induced pluripotent stem cell derived sensory neurons of two healthy donors, applying cell viability and cytotoxicity assays and differential RNA sequencing analyses.
-
•
Basic scientists, clinician scientists, biostatisticians and toxicologists will benefit from these data.
-
•
The shared data give insight into transcriptional changes of sensory neurons in response to paclitaxel treatment. They may help scientists in the field plan and analyze data on cell viability, cytotoxicity or RNA sequencing and compare results with the data provided here.
1. Data Description
01_Viability_Cytotoxicity_Figures provides the graphs on the live/dead ratios of iPSC-DSN from two healthy donors exposed to six different chemotherapeutic drugs. The live/dead ratios normalized to vehicle and the respective curves are provided in this file. All graphs are numbered from 1 to 81 which allow reference to the raw data, which are provided below.
02_Viability_Cytotoxicity_Assays provides the calculated live/dead ratios of all cell viability and cytotoxicity assays that were performed in [1]. This Excel-file provides 17 sheets: The first sheet includes a table that briefly lists all assays performed, how many wells had to be excluded from further analyses and the reason for exclusion. The following 15 sheets provide the live/dead ratios from all assays that were shown in the figures of the file above and in the Supplementary Figs. 5–19 of [1]. This file also provides pictures of the cell culture plates and the viability curves. The raw data from which the live/dead ratios were calculated and the precise steps for their calculation are provided in the folder below (03_Viability_Cytotoxicity_Assays_Raw_Data), with excel files of which the respective numbers match the graphs in 01_Viability_Cytotoxicity_Figures and the assays in 02_Viability_Cytotoxicity_Assays.
03_Viability_Cytotoxicity_Assays_Raw_Data is a folder which contains 81 excel files with the raw and analyzed data of all assays performed in [1]. Each of these excel files consists of at least three sheets which are called MTT, Cytotox and Combined, and in in some cases also Combined_corrected. In the upper part of each sheet, the comma-separated values extracted from the .csv files of the assay readers are shown. Below, these values are put into a table in which the percentage relative to vehicle are calculated. The sheet Combined_Corrected shows the data after exclusion of the wells that did not pass microscopic quality control: e.g., if contamination with fibroblasts or fungus were to be found (usually marked with a red square and a dot in the middle on the plate) or if the cells were detached (usually marked with an x, see respective documentation in the foto of the plate). Statistical outliers (≥ 2 SDs in comparison to the average of the other wells in the same row) are marked red in the sheet Combined and excluded from further analyses in the sheet Combined_corrected. The latter also shows the graphs which are found in the file 01_Viability_Cytotoxicity_Figures. The exact calculation steps can be followed by clicking on the line with the respective result in the excel file. The graphical and statistical analyses are provided in the Graphpad Prism File 04_All _MTT_Cytotoxicity_Assays.
04_All _MTT_Cytotoxicity_Assays provides the Graphpad Prism file on how the calculations of all 81 assays were performed. By clicking on the graphs in the Layouts section, the respective figures can be found, and their graphical calculation and the respective raw data are linked to these layout files. Here, it is also referenced which data are pooled and to be found in the rectangles in 01_Viability_Cytotoxicity_Figures or in the Suppl. Figs. 5–19 in [1].
05_MTT_Cytotoxicity_Assays_Manuscript is the Graphpad Prism file, which includes data and figures presented in the manuscript of [1] and their respective calculation, linked in the Layout section. With these s.c. layout files, the live/dead ratios of the cell viability/cytotoxicity assays are linked, as well as the statistical analyses for the comparison of the cell types of the different healthy donors. All data, which are included here, are based on the raw data which are provided in the files above. To make it easier to track the data for the reader, the Graphpad layouts on which the figures in [1] are based have a * in front of their file name.
06_Diff_Gene_expression provides the table with the differentially expressed genes of iPSC-DSN treated with vehicle (DMSO 1/6000) or paclitaxel 1 µM for 24 h. Furthermore, the TPM file (Transcripts Per Kilobase Million) is included which allows the reader to search 58.000 genes for their mRNA expression in iPSC-DSN treated with vehicle or with paclitaxel.
07_Diff_Gene_Expression_HTML provides the HTML file with the analyzed comparative data of iPSC-DSN treated with vehicle (DMSO 1/6000) or paclitaxel 1 µM for 24 h.
08_RNA_seq_DNA_GEO. This file contains the GEO submission form for the RNA sequencing data.
09_RNA_sequencing_GEO_Link. This file provides access to the raw and analyzed transcriptome data.
2. Experimental Design, Materials and Methods
The experimental design, materials and methods are also described in detail in the manuscript “Modeling chemotherapy induced neurotoxicity with human induced pluripotent stem cell (iPSC) -derived sensory neurons” [1] and its supplemental methods.
2.1. iPSC lines
iPSC-derived sensory neurons were differentiated from the established stem cell lines BIHi005-A (https://hpscreg.eu/cell-line/BIHi005-A) and BIHi004-B (https://hpscreg.eu/cell-line/BIHi004-B, Berlin Institute of Health Stem Cell Core Facility), obtained by reprogramming of human dermal fibroblasts using Sendai viral vectors as previously reported [2,3]. Cells were cultured on growth factor reduced Geltrex (Gibco) in E8 media with daily media exchange. Cells were enzymatically clump-passaged when >70% confluency was reached, usually every 2–4 days using EDTA (UltraPure™ 0.5 M EDTA, Thermo Fisher).
2.2. Sensory neuron differentiation and maintenance
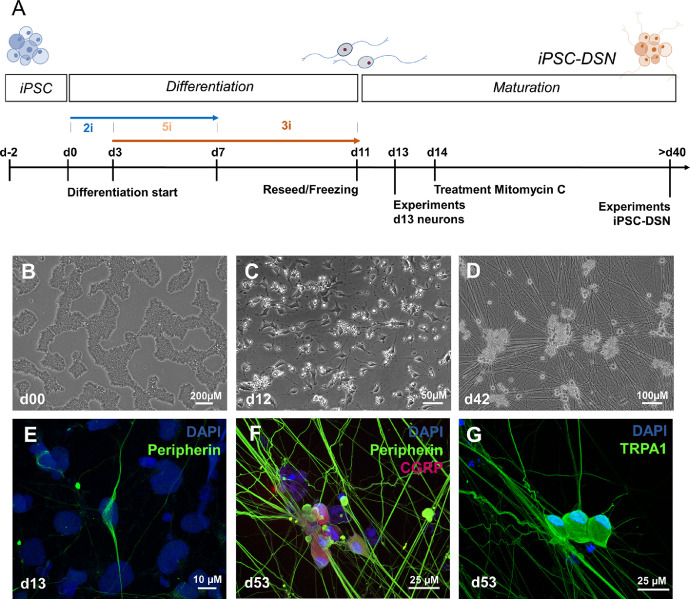
We applied the s.c. P2 protocol by Schwartzentruber et al. (2018) [4] for human iPSC differentiation. In brief, confluent iPSC were single cell seeded at a density between 200.000 and 400.000/well into 6-well plates (Falcon) on growth factor reduced Geltrex (Gibco) and maintained in E8 media until 70–80% confluency was achieved (usually after 48 to 72 h). On day 0, E8 media was exchanged for Knockout Serum Replacement Media (500 ml DMEM-KO [Gibco] supplemented with 130 ml CTS KnockOut SR XenoFree Medium (Gibco), 1 × MEM-Non-essential Amino Acid Solution (Sigma), 1 × Glutamax (Gibco) and 0.01 mM β-Mercapto-ethanol (Gibco) including the two small molecule inhibitors ‘2i’: 100 nM LDN-193189 (Sigma) and 10 µM SB-431542 (Peprotech) for neuroectoderm differentiation. On day 3, ‘3i’ consisting of 3 µM CHIR99021 (Sigma), 10 µM DAPT (Sigma) and 10 µM SU5402 (Sigma) were also added for neural crest specification. On day 4, N2B27 media (500 ml Neurobasal-Medium [Gibco], 5 ml of N-2 (100 ×) supplement [Gibco], 10 ml of B27-supplement (50 ×) without Vitamin A [Gibco], 1 × Glutamax and 0.01 mM β-Mercapto-ethanol [Sigma]) was progressively phased in. From day 7 onwards, ‘2i’ addition was ceased while ‘3i’ continued to be added until day 11 (Fig. 1 A). On day 11, iPSC-DSN were either reseeded or cryopreserved according to the protocols of Schwartzentruber et al. [4] and Stacey et al. [5] (also see detailed Suppl. Methods in [1]). On day 14, the few dividing non-neuronal cells were eliminated applying 1 µg/ml Mitomycin C (Sigma) for 2 h. iPSC-DSN were maturated for at least four more weeks, with half media changes every 3–4 days using N2B27 supplemented with BDNF, GDNF, βNGF and NT3 (all at 25 ng/ml, Peprotech). For the present study, we performed 12 independent differentiations, carried out by the same experimenter.
Fig. 1.
Differentiation and characterization of iPSC-DSN. (A). Cells were maintained in E8 media until differentiation was started with LDN19318 and SB431542 (s.c. 2i, dual SMAD inhibition), followed by the activation of WNT using CHIR99021, Notch inhibition with DAPT and inhibition of VEGF/FGF/PDGF applying SU5402 (s.c. 3i, when applied together with 2i referenced as 5i). After reseeding on d11, cells matured for at least 30 more days (maturation phase). (B–D) show representative phase contrast images of the differentiation phase, from stem cell colonies (B) to sensory neuron ganglia like structures >40d (D). (E–G) depict typical markers of the peripheral nervous system, which is Peripherin in d13 iPSC-DSN (E), or Peripherin and CGRP (F) and TRPA1 (G) in iPSC-DSN maturated 52d. Fig. 1 A was modified from[1].
2.3. Compound preparation
Paclitaxel (Adipogen), bortezomib (Cayman Chemical) and 5-fluorouracil (Sigma) were dissolved in DMSO. Cisplatin (Sigma) was solved in sterile 0.9% NaCl (all at 6 mM). Vincristine (Cayman Chemical), doxorubicin hydrochloride (Tocris) and lithium chloride (Sigma) were prepared in aqueous solutions using sterile water (stock solutions: 600 mM lithium chloride, vincristine and doxorubicin 1 mM). Final concentrations of 1 nM to 100 µM (cisplatin) or 100 pM to 10 µM (all other substances but lithium, which was applied in 10 to 1000 µM concentrations) were administered for 24 h for cell viability and cytotoxicity experiments, unless otherwise stated (e.g., the lithium experiments were carried out in a 72 h specimen, see [1]). Vehicle concentrations were 1/600 DMSO (for experiments with paclitaxel, bortezomib and 5-fluorouracil), 1/100 NaCl 0.9% or water (for experiments with vinristine, doxorubicin) and 1/60 (NaCl 0.9%, experiments with cisplatin). As only 1 µM paclitaxel was administered in the RNA sequencing experiments, the DMSO vehicle was applied in a 1/6000 concentration.
2.4. Cell viability and cytotoxicity assays
Chemotherapeutic drugs were added to iPSC-DSN maintained in 96-well plates and incubated for 24 h if not otherwise stated. Cell viability and cytotoxicity were assessed using MTT and proteases assays (Promega CytoTox-Fluor Assay and CytoTox-Glo Assay), according to the manufacturers' instructions. Live/dead ratios of MTT/proteases assays were calculated and normalized to vehicle (compare [1,6]). All raw data of these assays, their calculation to live/dead ratios and their graphical analyses are provided in the following files which are attached to this article: 01_Viability_Cytotoxicity_Figures (graphical overview; assays are numbered and allow reference to the raw files), 02_Viability_Cytotoxicity_Assays (calculated live/dead ratios), 03_Viability_Cytotoxicity_Assays_Raw_Data (Folder with excel files with MTT and cytotoxicity assays separately and the live/dead ratio normalized to vehicle), 04_All _MTT_Cytotoxicity_Assays (Graphpad File with the calculation of the graphs of all assays) and 05_MTT_Cytotoxicity_Assays_Manuscript (Graphpad file with the grahps shown in the main figures of [1]).
The z-factor was calculated applying digitonin (200 µg/ml, sigma) for 24 h on iPSC-DSN d13, resulting in a mean value of 0.76 ± 0.13, indicating a large separation band [7] (5 experiments pooled; see the last sheet of the file 02_Viability_Cytotoxicity_Assays). Experiments to evaluate lithium chloride as potential neuroprotectant were performed with the same setup, but in a 72 h treatment challenge (see 02_Viability_Cytotoxicity_Assays and the folder 03_Viability_Cytotoxicity_Assays_Raw_Data).
2.5. Transcriptome sequencing analyses
iPSC-DSN were cultured in Geltrex coated 6-well plates at 106 cells/well as described above. After maturation until d44 - d49, three wells were treated either with vehicle (DMSO at 1/6000) or paclitaxel (1 µM for 24 h), and RNA was harvested with the Aurum™ Total RNA Mini Kit according to manufacturers' instructions, with a total RNA amount between 1.49 and 2.97 µg. RNA sequencing was conducted by Brooks Life Sciences Genewiz®, applying PolyA selection for RNA removal, 2 × 150 bp sequencing configuration and 20–30 million reads per sample. 5 samples were analyzed (n = 3 treated with DMSO vs. n = 2 treated with paclitaxel).
Computational Methods. RNA Seq reads were mapped to human genome (GRCh38.p7) with STAR-version 2.7.3a [8] applying the default parameters. We obtained an average of 80.2% uniquely mapped reads per sample. Reads were assigned to genes with featureCounts [9] with the following parameters: -t exon -g gene_id, gene annotation - Gencode GRCh38/v25. The differential expression analysis was carried out with DESeq2-version 1.22.1 [10] using the default parameters. We kept genes that have at least 5 reads in at least 2 samples. Gene set enrichment analysis was carried out with R/tmod package [11].
2.6. Statistical analyses
Data were analyzed using Prism v8 (Graphpad Software, San Diego, CA). Data were checked for Gaussian distribution applying histograms and the Shapiro-Wilk normality test. Normally distributed data were analyzed with two-sided t-tests (2 groups). Data with low sample sizes or data which were not normally-distributed were analyzed with the Mann-Whitney-U test (2 groups). Cell viabilities were compared with one-way ANOVA and corrected for multiple comparison using Holm-Sidak's multiple comparison test (normally distributed data) or the Kruskal-Wallis test and corrected using the Dunn's multiple comparison test (data that were not normally distributed). Dose-response curves were calculated with non-linear regression models using GraphPad Prism (s.c. Log inhibitor vs. response, three parameters). The Graphpad files which include these calculations in detail are provided in this article (see the files 04_All _MTT_Cytotoxicity_Assays and 05_MTT_Cytotoxicity_Assays_Manuscript). The calculations can be reproduced by clicking on the respective layout file in the Graphpad file which then allows reference to the raw data and their further processing. All raw data of the assays are shown in the file 03_Viability_Cytotoxicity_Assays_Raw_Data, with the respective figures of all assays in the file 01_Viability_Cytotoxicity_Figures.
All experiments with >d40 iPSC-DSN were replicated at least 3 times from at least 2 independently differentiated or maturated iPSC-DSN cultures (biological replicates) with 4 technical replicates for each condition (see detailed description in Suppl. Material 1, Table 1 and Suppl. Figs. 5–19 in [1]). Data points with more than two standard deviations from the sample mean were treated as statistical outliers and excluded from analysis, which was the case in 1.66% of the analyzed wells (See Suppl. Mat. 1, Table 1 in [1] or the files provided with this article: 02_Viability_Cytotoxicity_Assays, 03_Viability_Cytotoxicity_Assays_Raw_Data). Normally distributed data are presented as mean ± SD. Not normally distributed data are shown as median with interquartile ranges. Results for all 81 analyzed assays are shown in Suppl. Mat. 1, Figs. 5–19 of [1] or as Figs. 1–15 attached to this Data-in-Brief article (see 01_Viability_Cytotoxicity_Figures).
2.7. Data availability
Open data publishing guidelines were followed. Raw data of cell viability and cytotoxicity assays are attached to this Data-in-Brief article. RNA sequencing data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus [12] and are accessible through GEO Series accession number GSE173610 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE173610). The respective download sources are provided in GEO under the link above: Raw data are available in SRA, and processed data are available on Series record.
Ethics Statement
The human induced pluripotent stem cell (hiPSC) lines used in this study are established stem cell lines generated by the Berlin Institute of Health (BIH). The detailed ethical statement including the approval for the use of the cell lines BIHi005-A and BIHi004-B is deposited under the following links: https://hpscreg.eu/cell-line/BIHi005-A and https://hpscreg.eu/cell-line/BIHi004-B.
CRediT authorship contribution statement
Christian Schinke: Conceptualization, Formal analysis, Funding acquisition, Investigation, Writing – original draft, Writing – review & editing. Valeria Fernandez Vallone: Conceptualization, Funding acquisition, Investigation, Supervision, Writing – review & editing. Andranik Ivanov: Investigation, Writing – review & editing. Yangfan Peng: Investigation, Writing – review & editing. Péter Körtvelyessy: Conceptualization, Resources, Writing – review & editing. Luca Nolte: Investigation, Writing – review & editing. Petra Huehnchen: Conceptualization, Formal analysis, Writing – original draft, Writing – review & editing. Dieter Beule: Writing – review & editing. Harald Stachelscheid: Conceptualization, Resources, Writing – review & editing. Wolfgang Boehmerle: Conceptualization, Funding acquisition, Formal analysis, Supervision, Writing – original draft, Writing – review & editing. Matthias Endres: Conceptualization, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors report no conflict of interest.
Acknowledgments
We thank Petra Loge for her excellent technical assistance, and Kristin Fischer, Judit Küchler and Tanja Fisch of the BIH Stem Cell Core Facility for their invaluable help in stem cell culture and maintenance. We thank Dr. Dorette Freyer for sharing her extensive cell culture expertise, Dr. Richard Kovács and Dr. Gisela Laettig for their support optimizing Ca2+ imaging experiments, and Smilla Maierhof for her support in cell culture maintenance and intellectual input in the final phase of the study. This research was supported by the Deutsche Forschungsgemeinschaft (NeuroCure Excellence Cluster to ME) and AnimalFreeResearch Switzerland to CS, WB and ME. This work was supported by Charité 3R - Replace - Reduce – Refine. ME receives funding from the Bundesministerium für Bildung und Forschung (Center for Stroke Research Berlin), the German Center for Neurodegenerative Diseases (DZNE), the German Center for Cardiovascular Research (DZHK) and the Corona Foundation. CS, PH and WB are members of the Clinician Scientist program funded by the Charité Universitätsmedizin Berlin and Berlin Institute of Health. PH is the recipient of a Rahel-Hirsch fellowship funded by the Charité Universitätsmedizin Berlin.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.dib.2021.107320.
Appendix. Supplementary Materials
References
- 1.Schinke C., Fernandez Vallone V., Ivanov A., Peng Y., Körtvelyessy P., Nolte L., Huehnchen P., Beule D., Stachelscheid H., Boehmerle W. Modeling chemotherapy induced neurotoxicity with human induced pluripotent stem cell (iPSC) -derived sensory neurons. Neurobiol. Dis. 2021;155 doi: 10.1016/j.nbd.2021.105391. [DOI] [PubMed] [Google Scholar]
- 2.Hennig A.F., Rössler U., Boiti F., Von Der Hagen M., Gossen M., Kornak U., Stachelscheid H. Generation of a human induced pluripotent stem cell line (BIHi002-A) from a patient with CLCN7-related infantile malignant autosomal recessive osteopetrosis. Stem Cell Res. 2019;35 doi: 10.1016/j.scr.2018.101367. [DOI] [PubMed] [Google Scholar]
- 3.Fusaki N., Ban H., Nishiyama A., Saeki K., Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc. Jpn. Acad. Ser. B. 2009;85(8):348–362. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwartzentruber J., Foskolou S., Kilpinen H., Rodrigues J., Alasoo K., Knights A.J., Patel M., Goncalves A., Ferreira R., Benn C.L. Molecular and functional variation in iPSC-derived sensory neurons. Nat. Genet. 2018;50(1):54–61. doi: 10.1038/s41588-017-0005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stacey P., Wassermann A.M., Kammonen L., Impey E., Wilbrey A., Cawkill D. Plate-based phenotypic screening for pain using human iPSC-derived sensory neurons. SLAS Discov. Adv. Life Sci. R&D. 2018;23(6):585–596. doi: 10.1177/2472555218764678. [DOI] [PubMed] [Google Scholar]
- 6.Von Der Ahe D., Huehnchen P., Balkaya M., Peruzzaro S., Endres M., Boehmerle W. Suramin-induced neurotoxicity: preclinical models and neuroprotective strategies. Molecules. 2018;23(2):346. doi: 10.3390/molecules23020346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J.H., Chung T.D.Y., Oldenburg K.R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999;4(2):67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 8.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 10.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;(12):15. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zyla J., Marczyk M., Domaszewska T., Kaufmann S.H.E., Polanska J., Weiner J. Gene set enrichment for reproducible science: comparison of CERNO and eight other algorithms. Bioinformatics. 2019;35(24):5146–5154. doi: 10.1093/bioinformatics/btz447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edgar R. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Open data publishing guidelines were followed. Raw data of cell viability and cytotoxicity assays are attached to this Data-in-Brief article. RNA sequencing data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus [12] and are accessible through GEO Series accession number GSE173610 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE173610). The respective download sources are provided in GEO under the link above: Raw data are available in SRA, and processed data are available on Series record.