Abstract
The removal of oxidative damage from Saccharomyces cerevisiae DNA is thought to be conducted primarily through the base excision repair pathway. The Escherichia coli endonuclease III homologs Ntg1p and Ntg2p are S. cerevisiae N-glycosylase-associated apurinic/apyrimidinic (AP) lyases that recognize a wide variety of damaged pyrimidines (H. J. You, R. L. Swanson, and P. W. Doetsch, Biochemistry 37:6033–6040, 1998). The biological relevance of the N-glycosylase-associated AP lyase activity in the repair of abasic sites is not well understood, and the majority of AP sites in vivo are thought to be processed by Apn1p, the major AP endonuclease in yeast. We have found that yeast cells simultaneously lacking Ntg1p, Ntg2p, and Apn1p are hyperrecombinogenic (hyper-rec) and exhibit a mutator phenotype but are not sensitive to the oxidizing agents H2O2 and menadione. The additional disruption of the RAD52 gene in the ntg1 ntg2 apn1 triple mutant confers a high degree of sensitivity to these agents. The hyper-rec and mutator phenotypes of the ntg1 ntg2 apn1 triple mutant are further enhanced by the elimination of the nucleotide excision repair pathway. In addition, removal of either the lesion bypass (Rev3p-dependent) or recombination (Rad52p-dependent) pathway specifically enhances the hyper-rec or mutator phenotype, respectively. These data suggest that multiple pathways with overlapping specificities are involved in the removal of, or tolerance to, spontaneous DNA damage in S. cerevisiae. In addition, the fact that these responses to induced and spontaneous damage depend upon the simultaneous loss of Ntg1p, Ntg2p, and Apn1p suggests a physiological role for the AP lyase activity of Ntg1p and Ntg2p in vivo.
Reactive oxygen species generated by normal cellular metabolism or produced by exogenous agents can induce several types of DNA damage, including DNA base damage and the formation of apurinic/apyrimidinic (AP) sites. These types of DNA damage are thought to be processed primarily through the base excision repair (BER) pathway. According to the classic model of the BER pathway, a damaged base is removed by a specific N-glycosylase, and the resulting AP site is cleaved by an AP endonuclease. Following the processing of the 5′ terminus by deoxyribose phosphodiesterase, DNA polymerase fills in the gap, and DNA ligase seals the ends together. Several DNA N-glycosylases possess an associated AP lyase activity that mediates strand scission at the abasic sites generated by the removal of damaged bases (13). Whether such AP lyases are capable of functioning in vivo in the repair of abasic sites which are generated independently of N-glycosylase activity is currently unknown. The Saccharomyces cerevisiae Ntg1 and Ntg2 proteins are homologs of Escherichia coli endonuclease III (endo III) (40). Ntg1p and Ntg2p are N-glycosylase-associated AP lyases with similar substrate specificities directed primarily against oxidatively damaged pyrimidines (32, 40). However, unlike Ntg2p and all other known endo III homologs, Ntg1p does not possess a C-terminal Fe-S cluster (40). In addition, Ntg1p has a putative mitochondrial targeting sequence, while Ntg2p does not (2, 40). Finally, although recent studies have demonstrated that the expression of NTG1 is induced by H2O2 (11, 40), expression of NTG2 does not appear to be H2O2 inducible (40). The possible functional overlap of Ntg1p and Ntg2p in vivo has not been examined and may serve as an important model of the response to oxidative DNA damage in other eukaryotes.
The nucleotide excision repair (NER) pathway generally removes bulky DNA lesions, but recent studies have also implicated NER in the repair of oxidative damage (19). The NER pathway removes damaged DNA bases by introducing nicks 5′ and 3′ to the damage. In S. cerevisiae, the 3′ incision is produced by Rad2p (17), whereas the 5′ incision is produced by the Rad1-Rad10 protein complex (10). After the oligonucleotide including the damaged DNA is removed, DNA polymerase fills in the gap, and DNA ligase joins the ends (13). The E. coli NER complex (i.e., UvrABC) has been shown to introduce nicks 3′ and 5′ to an AP site in vitro (34), but the relevance of this activity in vivo is currently unknown.
Recombination is involved in the repair of single- or double-strand breaks. S. cerevisiae cells that are deficient in RAD52 epistasis group proteins are highly sensitive to the killing effects of agents that produce strand breaks (e.g., ionizing radiation) (27). The involvement of recombination in the processing of certain types of DNA damage has also been examined. DNA lesions that block replication, such as cyclobutane pyrimidine dimers, appear to induce recombination (39). In addition, recombination rates increase upon exposure to H2O2, suggesting a role for recombination in the response in yeast to oxidative DNA damage (5). It should be noted that with some types of lesions, recombination may constitute a damage tolerance rather than a damage removal mechanism, since the damaged bases remain in the genome. Such recombinational bypass of damage would allow the cell to progress through mitosis, and the remaining DNA damage would presumably be removed at later times by other repair pathways.
A second mechanism of damage tolerance involves translesion synthesis (TLS). In certain environments, DNA polymerase must bypass DNA lesions in order for a cell to survive. Under these conditions, it is possible that certain lesions are not recognized by a particular repair pathway due to a blocked polymerase or that the damage is too extensive to be removed efficiently (13). In S. cerevisiae, TLS involves DNA polymerase ζ (Pol ζ), a complex of two proteins (Rev3p and Rev7p) which is able to bypass several types of DNA lesions, including cyclobutane pyrimidine dimers and AP sites (21, 25). Consequently, cells utilizing TLS are able to proceed through the cell cycle but with a corresponding increase in mutation rate.
In order to determine the in vivo roles of Ntg1p and Ntg2p in BER and to examine the overlap between different DNA repair pathways, we constructed a series of yeast mutants lacking DNA repair proteins from the BER, NER, recombination, and/or TLS pathway. The sensitivities of these mutants to a variety of DNA-damaging agents, as well as their spontaneous recombination and mutation rates, were examined. Our results indicate unexpected overlaps between different DNA repair and DNA damage tolerance pathways in the processing of oxidative and spontaneous DNA damage.
MATERIALS AND METHODS
Media and growth conditions.
Yeast strains were grown nonselectively on YEPD medium (1% yeast extract–2% Bacto-peptone–2% dextrose–2.5% agar for plates). Synthetic complete (SC) medium (33) lacking lysine and containing 2% dextrose was used for selective growth of Lys+ recombinants and revertants. SC medium lacking arginine and containing 60 mg of canavanine per liter was used for the determination of spontaneous-mutation frequency, and SC medium containing 1 g of 5-fluoroorotic acid per liter (4) was used to select Ura− yeast segregants. Luria-Bertani medium (1% yeast extract–0.5% Bacto-tryptone–1% NaCl–1.5% agar for plates) was used for the growth of E. coli strains. Ampicillin was added at 100 μg/ml to Luria-Bertani medium for the growth of plasmid-containing strains. Yeast and bacterial strains were grown at 30 and 37°C, respectively.
Strain construction.
Yeast transformations were carried out according to the method of Gietz et al. (14) with modifications as noted. All strains used in this study are isogenic derivatives of SJR751, a Leu− derivative of SJR357 (MATα ade2-101ochis3Δ200 ura3ΔNco lys2ΔBgl CAN1S) (9). Wild-type alleles in SJR751 or in its isogenic derivatives were replaced with disruption alleles by one-step gene disruption (30). ntg1Δ::LEU2 was introduced by transformation with NcoI-NdeI-digested pLF298 (2), rad52Δ::URA3 was introduced by transformation with EcoRI-SalI-digested pBRΔHSURA3 (22), apn1Δ::HIS3 was introduced by transformation with EcoRI-BamHI-digested pSCP19A (28), ntg2Δ::hisG-URA3-hisG was introduced by transformation with XhoI-SacI-digested pGEM-ntg2::hisG-URA3-hisG (40), and rad1Δ::hisG-URA3-hisG was introduced by transformation with SalI-EcoRI-digested pR1.6 (31). When the hisG-URA3-hisG cassette was used for disruption, Ura− segregants were isolated on 5-fluoroorotic acid.
A PCR-generated rev3Δ::kan disruption fragment was used to delete REV3. Primers 5′-ATGTCGAGGGAGTCGAACGACACAATACAGAGCGATACGGTTAGATCATCCTCTAAATCACAGCTGAAGCTTCGTACG-3′ (forward) and 5′-TTACCAATCATTTAGAGATATTAATGCTTCTTCCCTTTGAACA GATTGATTATCTCTCAAAGGCCACTAGTGATCTG-3′ (reverse) were used to amplify an approximately 1-kb disruption fragment with pFA6-kanMX2 (37) as a template. The first 60 bases of each primer are complementary to REV3, and the 3′ ends are complementary to the kanamycin resistance cassette. The PCR product was precipitated and resuspended in water and used directly for transformation. Following transformation, cells were grown for 3 h in 2 ml of YEPD before selective plating on YEPD containing 200 mg of Geneticin (Sigma) per liter. After 2 days of incubation at 30°C, the colonies were replica plated onto fresh Geneticin-containing medium. All gene disruptions were confirmed by Southern blot or PCR analysis. Loss of Ntg1p and Ntg2p activities was enzymatically confirmed as previously described (40). Loss of Apn1p activity was confirmed by incubating cell extracts with a 3′-end-labeled AP-containing substrate by using a previously described method (1) (data not shown). Loss of Rad1p and Rad52p activities was confirmed by gauging the sensitivity of mutants to UV irradiation or X-irradiation, respectively (data not shown).
Sensitivity of strains to DNA-damaging agents.
Yeast cells grown in 5 ml of YEPD overnight were pelleted, washed twice in sterile H2O, and resuspended in 1× phosphate-buffered saline (equal in volume to initial culture). Aliquots of cells were then subjected to various concentrations of menadione (see Fig. 1) for 30 min at 30°C with shaking. Sensitivity to H2O2 was monitored by a method described by Ramotar et al. (28). Exponential-phase cultures (approximately 2 × 107 cells/ml) were pelleted, washed twice in sterile H2O, and resuspended in 1× phosphate-buffered saline (equal in volume to initial culture). The cells were treated with various concentrations of H2O2 for 1 h at 30°C with shaking. After treatment, the cells were diluted and plated on YEPD medium. Colonies were counted after 2 to 4 days of growth.
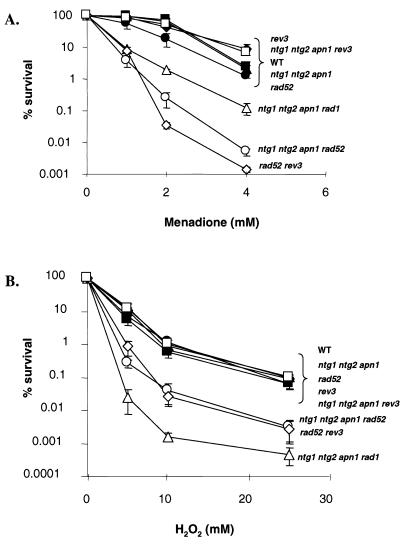
FIG. 1.
Sensitivity of mutant strains to menadione and H2O2. (A) Cells were exposed to increasing concentrations of menadione (0 to 4 mM). (B) Cells were exposed to increasing concentrations of H2O2 (0 to 25 mM). Error bars represent the standard deviations for three separate cultures.
Measurement of recombination and mutation rates.
Rates of prototroph formation were determined by the method of the median (23). Independent 2-day-old colonies were inoculated into 5 ml of YEPD liquid medium and grown nonselectively to 2 × 108 cells/ml. Cells were harvested by centrifugation, washed once with sterile H2O, and resuspended in 1 ml of water. One-hundred-microliter aliquots of appropriate dilutions were plated onto SC medium lacking lysine and containing 2% dextrose to select recombinants and lys2ΔBgl revertants, onto canavanine-containing medium to identify forward mutations in CAN1, and onto YEPD to determine viable cell numbers. Canr colonies were counted on day 2, and Lys+ colonies were counted on day 3 after selective plating. The data from a minimum of 10 cultures were used for each rate determination.
RESULTS
Sensitivity to oxidizing agents.
In vitro assays have demonstrated that endo III and its homologs, Ntg1p and Ntg2p, remove oxidative base damage produced by H2O2, menadione, gamma rays, and UV light (18, 40). To examine the potential biological role(s) of Ntg1p and Ntg2p in yeast, mutants lacking one or both of these proteins were assessed for their survival following exposures to a variety of DNA-damaging agents, including H2O2, menadione, UV light, and ionizing radiation. The single (ntg1 or ntg2) and double (ntg1 ntg2) mutants showed no increase in sensitivity over the wild type to any of the agents tested (data not shown). These results suggested that other BER N-glycosylases or other DNA repair pathways were repairing lethal lesions normally processed by Ntg1p and/or Ntg2p. To determine if other N-glycosylases were removing the induced damages, the S. cerevisiae major AP endonuclease (Apn1p) was eliminated in combination with Ntg1p and Ntg2p to significantly decrease the cells’ ability to repair oxidative DNA damage via BER. No increase in sensitivity to H2O2, menadione, or ionizing radiation was observed for any of the strains (Fig. 1 and data not shown).
The insensitivity of the ntg1 ntg2 apn1 triple mutant to oxidizing agents suggested the involvement of other DNA repair pathways in the repair of, or tolerance to, DNA damage processed by Ntg1p, Ntg2p, and Apn1p. To investigate the role of NER in the repair of oxidative DNA damage, RAD1 was disrupted singly and in combination with ntg1, ntg2, or apn1, but no further increase in sensitivity to H2O2 or menadione was observed in the rad1 single mutant, the ntg1 ntg2 rad1 triple mutant, or the apn1 rad1 double mutant relative to the wild-type strain (data not shown). However, when rad1 was disrupted in the ntg1 ntg2 apn1 triple-mutant background, the cells became highly sensitive to both menadione and H2O2 (Fig. 1).
To test the possible involvement of recombination in the tolerance of oxidative DNA damage, RAD52 was disrupted in wild-type, ntg1, ntg2, apn1, ntg1 apn1, ntg2 apn1, and ntg1 ntg2 apn1 strains. Only the ntg1 ntg2 apn1 rad52 quadruple mutant exhibited increased sensitivity to cell killing by either H2O2 or menadione (Fig. 1). This effect was dependent upon the loss of Ntg1p, Ntg2p, Apn1p, and Rad52p, since all triple-mutant combinations, in which only one of these four proteins was present, retained wild-type sensitivity (Fig. 1 and data not shown).
To investigate the role of TLS in the bypass of oxidative DNA damage, rev3 was disrupted in wild-type, ntg1 ntg2 apn1, and rad52 strains. The ntg1 ntg2 apn1 rev3 quadruple mutant, which is deficient in both BER and TLS, remained resistant to the killing effects of H2O2 and menadione (Fig. 1). However, the rad52 rev3 double mutant, which was deficient in TLS and recombination, was highly sensitive to both menadione and H2O2 (Fig. 1). This demonstrates the importance of both recombination and TLS in the tolerance of the cell to substantial levels of DNA damage induced by high concentrations of these damaging agents. Whereas the ntg1 ntg2 apn1 rad52 and rad52 rev3 mutants were highly sensitive to oxidizing agents, the ntg1 ntg2 apn1 rev3 mutant was not, suggesting that recombination is a major pathway for dealing with oxidative DNA damage. Interestingly, the ntg1 ntg2 apn1 rad1 quadruple mutant was more sensitive to H2O2 than the ntg1 ntg2 apn1 rad52 and rad52 rev3 mutants (Fig. 1B), while the ntg1 ntg2 apn1 rad52 quadruple mutant and rad52 rev3 double mutant were more sensitive to menadione than the ntg1 ntg2 apn1 rad1 mutant (Fig. 1A). This difference is likely due to the different types of lesions produced by menadione and H2O2. Although both H2O2 and menadione are oxidizing agents, it has been suggested that menadione produces strand breaks in DNA (26), which may explain the increased sensitivity to menadione of the rad52 mutant combinations.
Spontaneous mutator and hyperrecombinogenic (hyper-rec) phenotypes.
In situations where multiple pathways process a common DNA lesion, elimination of one pathway can often be compensated for by shuttling the damage into alternative pathways. The elimination of an error-free pathway, for example, may lead to an increase in either mutation or recombination rate, and the elimination of an error-prone pathway may reduce mutation rates but stimulate recombination. The rates of spontaneous frameshift mutations were assessed in various mutant strains by measuring the reversion rates of the lys2ΔBgl frameshift allele (16), and the rates of spontaneous base substitutions were estimated by measuring the forward mutation rates of the CAN1 locus (36). In addition, to investigate the role of recombination in the processing of DNA damage, a second lys2 allele (lys2Δ3500) was introduced into the relevant strains (20). Recombination between the lys2ΔBgl allele located on chromosome II and the lys2Δ3500 allele located on chromosome V was monitored by measuring the rate of Lys+ prototroph production. The spontaneous recombination rates and spontaneous mutation rates for various repair mutant combinations are presented in Table 1. The spontaneous mutation and recombination rates for the ntg1, ntg2, ntg1 ntg2, ntg1 apn1, and ntg2 apn1 strains were not increased relative to those of the wild-type parental strain (Table 1 and data not shown). For the ntg1 ntg2 apn1 triple mutant, however, an 18-fold increase in the recombination rate was observed, indicating that recombination is involved in the processing of lesions normally repaired by the BER pathway. The ntg1 ntg2 apn1 mutant, but not the single or double mutants, also had an elevated spontaneous-mutation rate with a 2.6-fold increase in frameshift mutations and an 11-fold increase in forward mutations at CAN1.
TABLE 1.
Spontaneous recombination and mutation ratesa
| Relevant genotype | Recombination rate (10−8) | lys2ΔBgl reversion rate (10−9) | Canr rate (10−7) |
|---|---|---|---|
| WT | 5.3 (1.0) | 3.2 (1.0) | 1.2 (1.0) |
| ntg1 ntg2 | 5.5 (1.1) | 2.2 (0.8) | 1.5 (1.3) |
| apn1 | 10 (1.9) | 3.8 (1.2) | 2.1 (1.8) |
| ntg1 ntg2 apn1 | 93 (18) | 8.2 (2.6) | 13 (11) |
| rad1 | 6.8 (1.3) | 5.9 (1.8) | 2.1 (1.8) |
| ntg1 ntg2 rad1 | 8.2 (1.5) | 11 (3.4) | 5.5 (4.6) |
| apn1 rad1 | 25 (4.7) | 23 (7.2) | 9.8 (8.2) |
| ntg1 ntg2 apn1 rad1 | 890 (170) | 350 (110) | 74 (62) |
| rad52 | ND | 22 (6.9) | 8.5 (7.1) |
| ntg1 ntg2 apn1 rad52 | ND | 120 (38) | 180 (150) |
| rev3 | 9.1 (1.7) | 1.5 (0.5) | 0.81 (0.7) |
| ntg1 ntg2 apn1 rev3 | 310 (58) | 2.2 (0.7) | 2.4 (2.0) |
Rates are expressed as recombinants or mutants per cell per generation. Numbers in parentheses indicate the fold increase over wild type values. WT, wild type; ND, not determined.
The disruption of NER in combination with BER (ntg1 ntg2 apn1 rad1 quadruple mutant) resulted in a synergistic (i.e., greater than additive) increase in mutation and recombination rates relative to the ntg1 ntg2 apn1 and rad1 mutants (Table 1): frameshifts increased 110-fold, base substitutions increased 62-fold, and recombination increased 170-fold. This suggests that in the ntg1 ntg2 apn1 triple mutant, NER is active in the removal of AP sites, and in the rad1 mutant, BER is responsible for the removal of AP sites. However, if both BER and NER are compromised simultaneously, the DNA damage tolerance pathways (recombination and TLS) must deal with the damage, suggesting that BER and NER are competing pathways. In agreement with a previous report (38), an apn1 rad1 double mutant does exhibit a weak mutator phenotype, but the strong synergism is observed only in the ntg1 ntg2 apn1 rad1 quadruple mutant.
Elimination of recombination alone (rad52 mutant) resulted in a moderate mutator phenotype with a 6.9-fold increase in frameshift mutations and a 7.1-fold increase in forward mutations at CAN1 relative to wild type. Disruption of both the BER and recombination pathways (ntg1 ntg2 apn1 rad52 quadruple mutant) resulted in a synergistic increase in mutation rates: a 38-fold increase in frameshift mutations and a 150-fold increase in forward mutations at CAN1 relative to the wild type. Similarly, when both the TLS and BER pathways were eliminated (ntg1 ntg2 apn1 rev3 quadruple mutant), the recombination rate increased 58-fold. The mutation rates in this particular background were similar to levels in a wild-type strain, confirming that the majority of mutations accumulating in the ntg1 ntg2 apn1 triple mutant are due to the TLS pathway.
Growth phenotypes of mutants defective in multiple pathways.
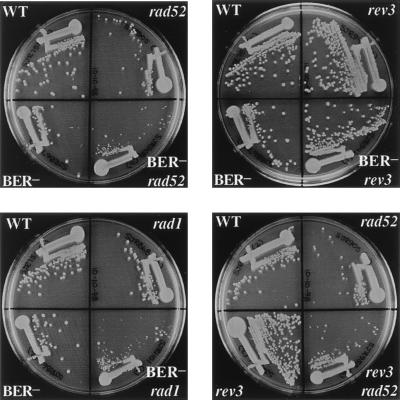
The relative growth rates of strains defective in one or more of the pathways implicated in survival after spontaneous DNA damage were assessed by examining the sizes of colonies formed on rich medium. Strains defective in the BER pathway (ntg1 ntg2 apn1 triple mutant), the NER pathway (rad1 mutant), or the TLS pathway (rev3 mutant) grew at rates comparable to that of the wild-type parental strain, whereas a recombination-defective strain (rad52 mutant) exhibited a slow-growth phenotype (Fig. 2). The simultaneous elimination of recombination and either the BER or TLS pathway (ntg1 ntg2 apn1 rad52 quadruple mutant or rad52 rev3 double mutant, respectively) and the simultaneous loss of BER and NER (ntg1 ntg2 apn1 rad1 quadruple mutant) each resulted in a synergistic decrease in growth rate, with such mutants growing much slower than the single or triple mutants (Fig. 2). In contrast, simultaneous elimination of the BER and TLS pathways (ntg1 ntg2 apn1 rev3 quadruple mutant) did not have an obvious impact on growth rate (Fig. 2). The mutant growth rates resembled the responses of the various mutants to oxidizing agents (Fig. 1), where synergism was observed in the ntg1 ntg2 apn1 rad1 quadruple mutant, the rad52 rev3 double mutant, and the ntg1 ntg2 apn1 rad52 quadruple mutant but not in the ntg1 ntg2 apn1 rev3 quadruple mutant. It should be noted that the ntg1 ntg2 apn1 rad1 quadruple mutant (but not the other mutants) consistently produced large and small colonies when purified on rich medium. Upon repurification, the small colonies again produced a mixture of large and small colonies, while the large colonies produced only large colonies. The basis of this phenomenon is unknown but is being investigated further.
FIG. 2.
Growth rates of mutant strains. Yeast strains were streaked onto YEPD media and incubated at 30°C for 2 days. BER, BER-rad52, BER-rev3, and BER-rad1 represent the ntg1 ntg2 apn1, ntg1 ntg2 apn1 rad52, ntg1 ntg2 apn1 rev3, and ntg1 ntg2 apn1 rad1 mutants, respectively. WT, wild type.
DISCUSSION
The yeast proteins Ntg1 and Ntg2 are N-glycosylase-associated AP lyases that recognize a wide spectrum of oxidized pyrimidines in vitro and participate in the repair of oxidative lesions via the BER pathway. In order to examine the in vivo role of these proteins, relevant genes were disrupted, and the resulting mutants were examined for increased sensitivity to DNA damage and for spontaneous hyper-rec and mutator phenotypes. Although Eide et al. previously reported a slight sensitivity of an ntg1 mutant to menadione (11), we were unable to reproduce this phenotype in our strain background. Neither ntg1 nor ntg2 single mutants had any detectable mutant phenotype, suggesting a functional redundancy between the proteins. Such redundancy has been previously reported to occur in E. coli, where sensitivity to DNA-damaging agents is not observed until multiple BER proteins are eliminated (7, 8). Even with an ntg1 ntg2 double mutant, however, we were not able to discern any alteration in phenotype relative to the wild-type parental strain. We reasoned that a more global disruption of the yeast BER pathway might be required in order to discern the in vivo roles of Ntg1p and Ntg2p, and so Apn1p was eliminated alone and in combination with Ntg1p and Ntg2p. In contrast to a previous report, no increased sensitivity to oxidizing agents was observed in an apn1Δ mutant derived from our strain background (SJR864) (28). Even using the strain background (DBY747) used in the previous study, we have not been able to reproduce the H2O2 sensitivity (data not shown). The reason for this discrepancy is unclear.
Although no increase in sensitivity to oxidizing agents was observed in the ntg1 ntg2 apn1 triple mutant, this strain was found to undergo homologous recombination at a greatly increased rate, as well as to exhibit a mutator phenotype. The hyper-rec phenotype was dependent on the simultaneous disruption of NTG1, NTG2, and APN1; no other combination of single or double mutants showed an increase in recombination or mutation rates. The fact that these mutant phenotypes are observed only in the triple mutant suggests competition for a common intermediate, where any of these three proteins can compensate for a loss of the other two. Based on the known biochemical activities of these proteins, we suggest that this common intermediate is an AP site and that the AP lyase activity of Ntg1p or Ntg2p is sufficient to allow cells to process AP sites that accumulate with the loss of Apn1p. If the increased recombination and mutation rates in the ntg1 ntg2 apn1 triple mutant were due to unrepaired base damage caused by the loss of Ntg1 and Ntg2 proteins, then the ntg1 ntg2 double mutant should also have exhibited increased recombination and mutation rates, but this was not the case. These data thus provide the first evidence for the biological relevance of AP lyase activity in the removal of AP sites in yeast. We further propose that the accumulation of endogenous AP sites in the ntg1 ntg2 apn1 triple mutant (hereafter referred to as the BER-deficient mutant) leads directly to an increase in recombination and mutation rates. Consistent with this interpretation, recent studies have shown that overexpression of 3-methyladenine DNA glycosylase (encoded by MAG1) in S. cerevisiae leads to a strong mutator phenotype (15). This effect was attributed to the increased level of AP sites generated by an imbalance between the DNA glycosylase activity of Mag1p and AP site processing activities. We predict that such cells also should exhibit a strong hyper-rec phenotype. Recently, a second AP endonuclease (encoded by APN2) was discovered in S. cerevisiae (21). While loss of APN2 alone did not cause cells to become more sensitive to damaging agents, loss of both AP endonucleases (apn1Δ apn2Δ double mutant) caused cells to become highly sensitive to the alkylating agent methyl methanesulfonate (21). Based on our data, we suggest that a loss of APN2 in the mutant backgrounds used in this study might further increase recombination and mutation rates, as well as increase the sensitivity of such strains to oxidizing agents.
The increase in recombination and mutation rates of the BER-deficient cells suggests that at least two other pathways, recombination and TLS, are involved in the processing of spontaneous DNA damage and that all three pathways may compete for a common structure. This was further substantiated by the simultaneous elimination of two of these three pathways. In these experiments, the recombination or TLS pathway was eliminated by the disruption of the RAD52 or REV3 gene, respectively. Simultaneous elimination of the BER and recombination pathways (ntg1 ntg2 apn1 rad52 quadruple mutant) resulted in a much stronger mutator phenotype than was observed when only one pathway was eliminated. The effect was synergistic rather than additive, suggesting competition between the two pathways for a common intermediate. Similarly, simultaneous elimination of the BER and TLS pathways (ntg1 ntg2 apn1 rev3 quadruple mutant) resulted in a synergistic increase in the recombination rate. Finally, the elimination of the TLS pathway in BER- or recombination-defective cells lowered the mutation rate observed in these strains, thereby demonstrating that the majority of mutations probably result from translesion synthesis by DNA Pol ζ. This finding is further supported by the work of Johnson et al. (21), demonstrating that AP sites are readily bypassed by DNA Pol ζ in vivo.
In addition to the BER, recombination, and TLS pathways, the NER pathway also processes spontaneous DNA damage in yeast. Therefore, we examined interactions of the NER pathway with the other three repair pathways. NER was eliminated by the disruption of the RAD1 gene. Although loss of NER did not significantly increase recombination or mutation rates, a synergistic effect was observed when BER and NER were simultaneously eliminated. Such synergism again indicates competition between BER and NER for the repair of a common intermediate, which we suggest is an AP site. This implicates NER in the removal of AP sites that would be expected to accumulate in an AP endonuclease- and AP lyase-deficient background, and this conclusion is supported by evidence that prokaryotic and eukaryotic NER proteins recognize and process AP sites in vitro (29, 34). However, Rad1p has also been implicated in homologous recombination where the Rad1p-Rad10p complex cleaves 3′ single-stranded ends of exposed DNA (10, 12). The involvement of Rad1p in recombination may also contribute to the sensitivity of cells to oxidizing agents and the increased occurrence of mutator phenotypes that we have observed in our mutant strains.
The repair of oxidative DNA damage in S. cerevisiae has been assumed to proceed mainly through the BER pathway (13), although alternative pathways for the repair of this type of damage have also been proposed (24, 29). The clear interactions between the BER, recombination, TLS, and NER pathways with regard to the spontaneous mutation and recombination phenotypes suggested that the insensitivity of BER-deficient yeast strains to oxidative DNA damage might be due to the action of one or more repair and tolerance pathways. If competition for a common lesion is occurring, then a synergistic increase in damage sensitivity would be expected when the relevant pathways are eliminated simultaneously. This is exactly the type of response that was observed when cells were treated with either menadione or H2O2 (Fig. 1). Cells defective in any one of the BER, recombination, TLS, and NER pathways were no more sensitive to these oxidizing agents than were wild-type cells. The simultaneous elimination of recombination and either BER or TLS resulted in significant sensitivity to both agents. In contrast, a strain defective in both BER and TLS was no more sensitive to either menadione or H2O2 than was the wild type. This suggests that the oxidizing agents used here may cause two classes of lesions, one of which is processed predominantly via the BER or recombination pathway and the other of which is most often bypassed by the TLS or recombination pathway. Alternatively, the sensitivity patterns to these damaging agents may be due to changes in the cells’ abilities to sense DNA damage, resulting in a loss of control of the cell cycle. Interestingly, these damage sensitivity patterns mirror the relative growth rates of strains in the absence of exogenous DNA-damaging agents. For example, the simultaneous elimination of the BER and recombination pathways results in both synergistic menadione sensitivity and an extremely low growth rate.
We have examined the ability of yeast to process both spontaneous and oxidative DNA damage via the BER, NER, TLS, and recombination pathways. As illustrated in Fig. 3, genetic analyses indicate that all four pathways have overlapping specificities and that all compete for the repair of base damage. Given that simultaneous loss of Ntg1p, Ntg2p, and Apn1p is required to cause an increase in recombination and mutation rates, we propose that an AP site is the common recognized lesion and that AP sites can be processed by the NER, TLS, or recombination pathway. If the damaged bases themselves, rather than the subsequent abasic sites, were the target of these alternative pathways, then mutant phenotypes should have been observed with the ntg1 ntg2 double mutant. No mutant phenotype was observed for an ntg1 ntg2 double mutant, and none was observed when these mutations were combined with mutations of other pathways (i.e., ntg1 ntg2 rad1 triple mutant). We also note that the overlapping pathway specificities reported here for oxidative and spontaneous DNA damages resemble those involved in the repair of UV damage in yeast.
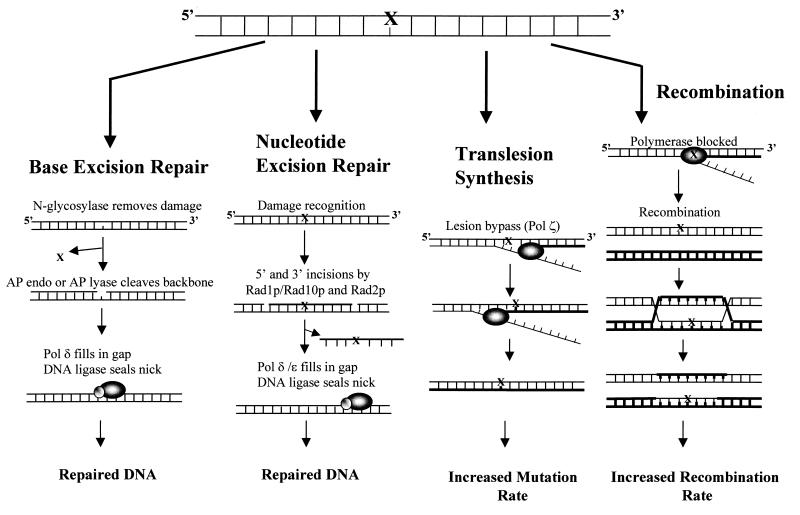
FIG. 3.
Processing of oxidative and spontaneous DNA damage in S. cerevisiae. “X” represents either a base damage which can be recognized and removed by the BER or NER pathway, can be bypassed by TLS, or can block replication causing recombination to occur or an AP site which feeds into any one of the four pathways illustrated. The amount of damage that is processed by each pathway varies depending on the repair background genotype.
Although multiple pathways clearly can process or tolerate certain DNA lesions, the relative contribution of each pathway is likely to vary under differing circumstances. For the removal of a particular damage, pathway selection may depend on the stage of the cell cycle or the context of the lesion in relation to other DNA transactions at a given time, such as during replication or transcription. Recombination, for example, might be preferentially utilized to bypass a lesion that stalls the replication machinery, or it might be used to reestablish collapsed replication forks when DNA nicks are encountered (3). DNA lesions that arrest transcription elongation have been shown to elicit transcription-coupled repair (6, 35). This includes both UV-induced lesions subject to NER and thymine glycol subject to BER, so the removal of oxidative damage by either of these excision repair pathways may be the predominant pathway for transcribed regions. Finally, other environmental stresses may require tolerance to DNA damage, leading to preferential deployment of TLS. Whether the overlapping nature of the pathways involved in the repair and tolerance of oxidative or spontaneous DNA damage in yeast exists in higher eukaryotes remains to be determined. However, we note that many of the yeast proteins investigated in this study have human homologs (e.g., Ntg2p, Apn1p, Rad52p, and Rev3p). The existence of multiple repair and bypass pathways underscores both the prevalence of DNA damage and the importance of dealing effectively with that damage via multiple pathways.
ACKNOWLEDGMENTS
We thank Philip C. Hanawalt for critically reading the manuscript.
This work was supported by NIH grant GM38464 (S.J.R.) and NCI grants CA73041 (P.W.D.) and CA78622 (P.W.D.). R.L.S. was supported by the NIH Predoctoral Training Program in Biochemistry, Cell and Molecular Biology (grant T32 GM08367).
R.L.S. and N.J.M. contributed equally to this work.
REFERENCES
- 1.Augeri L, Lee Y M, Barton A B, Doetsch P W. Purification, characterization, gene cloning and expression of Saccharomyces cerevisiae redoxyendonuclease, a homolog of Escherichia coli endonuclease III. Biochemistry. 1997;36:721–729. doi: 10.1021/bi9625511. [DOI] [PubMed] [Google Scholar]
- 2.Barton A B, Kaback D B. Molecular cloning of chromosome I DNA from Saccharomyces cerevisiae: analysis of the genes in the FUN38-MAK16-SPO7 region. J Bacteriol. 1994;176:1872–1880. doi: 10.1128/jb.176.7.1872-1880.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bierne H, Michel B. When replication forks stop. Mol Microbiol. 1994;13:17–23. doi: 10.1111/j.1365-2958.1994.tb00398.x. [DOI] [PubMed] [Google Scholar]
- 4.Boeke J D, Trueheart J, Natsoulis G, Fink G R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 5.Brennan R J, Swoboda B E, Schiestl R H. Oxidative mutagens induce intrachromosomal recombination in yeast. Mutat Res. 1994;308:159–167. doi: 10.1016/0027-5107(94)90151-1. [DOI] [PubMed] [Google Scholar]
- 6.Cooper P K, Nouspikel T, Clarkson S G, Leadon S A. Defective transcription-coupled repair of oxidative base damage in Cockayne syndrome patients from XP group G. Science. 1997;275:990–993. doi: 10.1126/science.275.5302.990. [DOI] [PubMed] [Google Scholar]
- 7.Cunningham R P, Saporito S M, Spitzer S G, Weiss B. Endonuclease IV (nfo) mutants of Escherichia coli. J Bacteriol. 1986;168:1120–1127. doi: 10.1128/jb.168.3.1120-1127.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cunningham R P, Weiss B. Endonuclease III (nth) mutants of Escherichia coli. Proc Natl Acad Sci USA. 1985;82:474–478. doi: 10.1073/pnas.82.2.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Datta A, Jinks-Robertson S. Association of increased spontaneous mutation rates with high levels of transcription in yeast. Science. 1995;268:1616–1619. doi: 10.1126/science.7777859. [DOI] [PubMed] [Google Scholar]
- 10.Davies A A, Friedberg E C, Tomkinson A E, Wood R D, West S C. Role of Rad1 and Rad10 proteins in nucleotide excision repair and recombination. J Biol Chem. 1995;270:24638–24641. doi: 10.1074/jbc.270.42.24638. [DOI] [PubMed] [Google Scholar]
- 11.Eide L, Bjoras M, Pirovano M, Alseth I, Berdal K G, Seeberg E. Base excision of oxidative purine and pyrimidine DNA damage in Saccharomyces cerevisiae by a DNA glycosylase with sequence similarity to endonuclease III from Escherichia coli. Proc Natl Acad Sci USA. 1996;93:10735–10740. doi: 10.1073/pnas.93.20.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fishman-Lobell J, Haber J E. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science. 1992;258:480–484. doi: 10.1126/science.1411547. [DOI] [PubMed] [Google Scholar]
- 13.Friedberg E C, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C: ASM Press; 1995. [Google Scholar]
- 14.Gietz D, Jean A S, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glassner B J, Rasmussen L J, Najarian M T, Posnick L M, Samson L D. Generation of a strong mutator phenotype in yeast by imbalanced base excision repair. Proc Natl Acad Sci USA. 1998;95:9997–10002. doi: 10.1073/pnas.95.17.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greene C N, Jinks-Robertson S. Frameshift intermediates in homopolymer runs are removed efficiently by yeast mismatch repair proteins. Mol Cell Biol. 1997;17:2844–2850. doi: 10.1128/mcb.17.5.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Habraken Y, Sung P, Prakash L, Prakash S. Yeast excision repair gene RAD2 encodes a single strand endonuclease. Nature. 1993;366:365–368. doi: 10.1038/366365a0. [DOI] [PubMed] [Google Scholar]
- 18.Hatahet Z, Kow Y W, Purmal A A, Cunningham R P, Wallace S S. New substrates for old enzymes: 5-hydroxy-2′-deoxycytidine and 5-hydroxy-2′-deoxyuridine are substrates for Escherichia coli endonuclease III and formamidopyrimidine DNA N-glycosylase, while 5-hydroxy-2′-deoxyuridine is a substrate for uracil DNA N-glycosylase. J Biol Chem. 1994;269:18814–18820. [PubMed] [Google Scholar]
- 19.Huang J C, Hsu D S, Kazantsev A, Sancar A. Substrate spectrum of human excinuclease: repair of abasic sites, methylated bases, mismatches, and bulky adducts. Proc Natl Acad Sci USA. 1994;91:12213–12217. doi: 10.1073/pnas.91.25.12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jinks-Robertson S, Petes T D. Experimental determination of rates of concerted evolution. Methods Enzymol. 1993;224:631–646. doi: 10.1016/0076-6879(93)24047-x. [DOI] [PubMed] [Google Scholar]
- 21.Johnson R E, Torres-Ramos C A, Izumi T, Mitra S, Prakash S, Prakash L. Identification of APN2, the Saccharomyces cerevisiae homolog of the major AP endonuclease HAP1, and its role in the repair of abasic sites. Genes Dev. 1998;12:3137–3143. doi: 10.1101/gad.12.19.3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaytor M D, Livingston D M. Saccharomyces cerevisiae RAD52 alleles temperature-sensitive for the repair of DNA double-strand breaks. Genetics. 1994;137:933–944. doi: 10.1093/genetics/137.4.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lea D E, Coulson C A. The distribution of the numbers of mutants in bacterial populations. J Genet. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 24.Moller P, Wallin H. Adduct formation, mutagenesis and nucleotide excision repair of DNA damage produced by reactive oxygen species and lipid peroxidation. Mutat Res. 1998;410:271–290. doi: 10.1016/s1383-5742(97)00041-0. [DOI] [PubMed] [Google Scholar]
- 25.Nelson J R, Lawrence C W, Hinkle D C. Thymine-thymine dimer bypass by yeast DNA polymerase ζ. Science. 1996;272:1646–1649. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- 26.Nutter L M, Ngo E O, Fisher G R, Gutierrez P L. DNA strand scission and free radical production in menadione-treated cells. J Biol Chem. 1992;267:2474–2479. [PubMed] [Google Scholar]
- 27.Petes T D, Malone R E, Symington L S. Recombination in yeast. In: Broach J R, Pringle J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces, vol. I. Genome dynamics, protein synthesis, and energetics. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1991. pp. 407–521. [Google Scholar]
- 28.Ramotar D, Popoff S C, Gralla E B, Demple B. Cellular role of yeast Apn1 apurinic endonuclease/3′-diesterase: repair of oxidative and alkylation DNA damage and control of spontaneous mutation. Mol Cell Biol. 1991;11:4537–4544. doi: 10.1128/mcb.11.9.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reardon J T, Bessho T, Kung H C, Bolton P H, Sancar A. In vitro repair of oxidative DNA damage by human nucleotide excision repair system: possible explanation for neurodegeneration in xeroderma pigmentosum patients. Proc Natl Acad Sci USA. 1997;94:9463–9468. doi: 10.1073/pnas.94.17.9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- 31.Saparbaev M, Prakash L, Prakash S. Requirement of mismatch repair genes MSH2 and MSH3 in the RAD1-RAD10 pathway of mitotic recombination in Saccharomyces cerevisiae. Genetics. 1996;142:727–736. doi: 10.1093/genetics/142.3.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Senturker S, van der Kemp P A, You H J, Doetsch P W, Dizdaroglu M, Boiteux S. Substrate specificity of the Ntg1 and Ntg2 proteins of Saccharomyces cerevisiae for modified bases in oxidatively damaged DNA. Nucleic Acids Res. 1998;26:5270–5276. doi: 10.1093/nar/26.23.5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–20. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 34.Snowden A, Kow Y W, Houten B V. Damage repertoire of Escherichia coli UvrABC nuclease complex includes abasic sites, base-damage analogues, and lesions containing adjacent 5′ or 3′ nicks. Biochemistry. 1990;29:7254–7259. doi: 10.1021/bi00483a013. [DOI] [PubMed] [Google Scholar]
- 35.Sweder K S, Hanawalt P C. Preferential repair of cyclobutane pyrimidine dimers in the transcribed strand of a gene in yeast chromosomes and plasmids is dependent on transcription. Proc Natl Acad Sci USA. 1992;89:10696–106700. doi: 10.1073/pnas.89.22.10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tishkoff D X, Filosi N, Gaida G M, Kolodner R D. A novel mutation avoidance mechanism dependent on Saccharomyces cerevisiae RAD27 is distinct from DNA mismatch repair. Cell. 1997;88:253–263. doi: 10.1016/s0092-8674(00)81846-2. [DOI] [PubMed] [Google Scholar]
- 37.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 38.Xiao W, Chow B L. Synergism between yeast nucleotide and base excision repair pathways in the protection against DNA methylation damage. Curr Genet. 1998;33:92–99. doi: 10.1007/s002940050313. [DOI] [PubMed] [Google Scholar]
- 39.Yap W Y, Kreuzer K N. Recombination hotspots in bacteriophage T4 are dependent on replication origins. Proc Natl Acad Sci USA. 1991;88:6043–6047. doi: 10.1073/pnas.88.14.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.You H J, Swanson R L, Doetsch P W. Saccharomyces cerevisiae possesses two functional homologs of Escherichia coli endonuclease III. Biochemistry. 1998;37:6033–6040. doi: 10.1021/bi973042h. [DOI] [PubMed] [Google Scholar]