Abstract
Background/Aim: Extracellular S100b effects are mediated by the receptor for advanced glycation end products (RAGE), which is the S100b membrane receptor. RAGE belongs to the immunoglobulin superfamily of cell surface molecules and serves as a multiligand receptor and is expressed in high abundance by alveolar type I (AT-I) cells in adult pulmonary tissue. This study aimed to provide an insight into the association between the severity of COVID-19 disease and serum S100b levels during admission to the emergency department (ED). Patients and Methods: A total of 64 patients (34 mild cases; 30 severe cases) were diagnosed with COVID-19 pneumonia and 30 healthy volunteers were admitted to study. Serum S100b levels were measured by using enzymle linked immunoassay method from blood serum samples. Results: Serum S100b levels showed a significantly higher mean value in mild and severe disease cohorts than in healthy controls (p=0.036 and p=0.028 respectively). Receiver operating characteristic (ROC) analysis indicated greater area under the curve (AUC) for serum S100b levels of the COVID-19 patients (AUC=0.663, 95% CI=0.541-0.785; p=0.014). In addition, serum S100b concentration was measured as 151.7 ng/ml at 79.3% sensitivity and 51.7% specificity (p=0.014). Serum S100b protein levels can serve as a valuable clinical marker in establishing diagnosis of patients. Though not useful in identifying different stages of COVID-19 infection, serum S100b concentration along with other known markers can be utilized to reliably predict clinical severity along with other clinical parameters.
Keywords: S100b levels, COVID-19, pneumonia
Transmitted from human to human through droplets, Coronavirus disease-19 (COVID-19) infection can progress asymptomatically. Its clinical manifestations, however, have a wide spectrum in symptomatic patients, ranging from upper respiratory tract infections (URTI) to serious clinical conditions such as pneumonia, acute respiratory disease syndrome (ARDS), sepsis, and septic shock (1,2).
S100b protein is a cytosolic calcium binding biomarker with a molecular mass of 21 kDa. Despite its extra-cranial presence, S100b is mainly expressed in the central nervous system (CNS) (3). As a cytosolic calcium sensor, this protein acts on dynamics of cytoskeletal components, energy metabolism, protein phosphorylation, and calcium homeostasis. When released from damaged cells, extracellular signals could be activated by S100b. It is also structurally released by astrocytes in the brain and can function in an autocrine or paracrine manner (4).
Accumulating data from in vitro experiments have revealed the impact of extracellular S100b on the biology of various cellular types other than the CNS. In vitro trials have provided clinical evidence that S100b, similar to other S100 family of proteins, stimulates inflammatory responses in lymphocytes, vascular smooth muscle cells, endothelial cells, macrophages, and cardiomyocytes (5,6).
Inside and outside the CNS, S100b acts as a damaged molecular model (DAMP) protein, which is released from damaged cells and can activate cells of innate immune response, altering the functions of cells that participate in inflammatory response and induce apoptosis (6). While S100b is expressed at very low levels in normal lungs, it is mainly secreted in intravascular granulocytes and rarely in interstitial dendritic cells, peribronchial nerves, and APUD cells.
This proinflammatory protein is up-regulated in airway dendritic cells and bronchiolar epithelial cells in the course of pulmonary inflammation (7-8). Extracellular S100b effects are mediated by the receptor for advanced glycation end products (RAGE), which is the S100b membrane receptor.
RAGE belongs to the immunoglobulin superfamily of cell surface molecules and serves as a multiligand receptor (9-10). It is expressed in high abundance by alveolar type I (AT-I) cells in adult pulmonary tissue (11). In addition, its activation plays a role in cellular proinflammatory responses and can promote several signal chains that ultimately trigger NFkappaB (NF-kB) activation, thereby inducing the expression of chemokines, cytokines, and cell adhesion molecules. Besides, ligand-mediated RAGE stimulation seems to contribute to the pathogenesis of various human diseases such as inflammatory diseases, including diabetes, Alzheimer’s disease, cancer, and acute lung injury/acute respiratory distress syndrome (ALI/ARDS) (12,13).
Based on the aforementioned clinical data, this report tries to provide an insight into the association between the severity of COVID-19 disease and serum S100b levels during admission to the emergency department (ED).
Patients and Methods
Study population. This is a prospective case-control study, and the required approval was obtained from the Ethics Committee of Pamukkale University. The study population consisted of patients who presented to COVID-19 outpatient clinic of our ED between 26 January -10 March 2021. The inclusion criteria were specified as manifesting symptoms of pneumonia, having PCR-confirmed COVID-19 diagnosis and presenting to ED for further examination and treatment. Those with the following clinical manifestations were excluded from the study:pregnancy, acute pulmonary embolism, chronic inflammatory disease, kidney and liver failure, chronic obstructive pulmonary disease or asthma, presence of any cancer diagnosis, and history of neurologic disease.
The explanatory information about the study was delivered to the patient group and healthy controls. The informed consent forms were collected from all the subjects who agreed to enroll in the study. Our case identification and clinical severity classification was based on the WHO’s guideline on clinical management of COVID-19 disease (14). Accordingly, 64 patients (34 mild cases; 30 severe cases) were diagnosed with COVID-19 infection as a result of clinical evaluation in ED, and their diagnosis was further confirmed by RT-PCR. The patients’ SARS-CoV-2 infection was confirmed by reverse transcriptase-polymerase chain reaction (RT-PCR) assay from nasopharyngeal swab specimen.
On the other hand, 30 healthy volunteers with no symptoms of COVID-19 infection, comorbidities, acute diseases, regular drug regimen, or recent history of infection were included in the study as the control group. Following the case identification and selection of eligible population, they were divided into three cohorts as CT-negative, PCR-positive COVID-19 infection cohort (mild cases), COVID-19 pneumonia cohort (severe cases), and healthy cohort (controls).
Mild disease group. This cohort included the subjects presenting to ED with symptoms of URTI, showing no pneumonia manifestation in imaging, and receiving the diagnosis of “mild cases” according to the WHO guideline (14).
Severe disease group. This cohort was made up of patients admitted to ED with COVID-19 symptoms, having PCR-confirmed COVID-19 diagnosis, showing pneumonia manifestation in imaging, and diagnosed with COVID-19 infection in line with the WHO guideline (14).
Healthy group (Control group). This cohort consisted of the volunteers having no history of acute, subacute or comorbid diseases, not coming down with an infection in the last two weeks, not following a regular drug regimen, presenting to ED with non-infectious complaints, and giving their informed consent to enroll in the study.
Clinical evaluation and data collection. The resulting dataset included patients’ demographic data, medical history, vital signs (fever, blood pressure, sPO2), laboratory findings (complete blood count (CBC), CRP (C-Reactive Protein), D-dimer, Ferritin, and hsTnT parameters), onset time of symptoms, comorbid diseases, hospitalization department (ward or ICU), CURB-65 and Pneumonia Severity index (PSI) scores, and CT severity scores.
S100b level measurement.To measure S100b level, 3 ml blood sample were transferred into a dry tube and centrifuged at 5,000 rpm for 10 min. Its serum section was then isolated for Enzyme-Linked Immunosorbent Assay (ELISA) assay analysis. Serum S100b levels were measured using a commercially available ELISA kits (Human S100B ELISA Kit, Mybiocource, Catalog No: MBS2503148) per the manufacturer’s protocols.
Statistical analysis. Because no similar research exists so as to guide us regarding minimum sample size, a power analysis was conducted. Accordingly, at least 66 people (22 for each cohort) were required to obtain 95% power at 95% confidence interval, assuming that the estimated effect size would be medium (f=0.5).
All statistical analyses were carried out using Statistical Package for the Social Sciences (SPSS) v.25 (IBM Corp., Armonk, NY, USA). The continuous variables were expressed as median (IQR). Normality of the data was checked by the Kolmogrov-Smirnov test. For non-parametric independent group analysis, a Kruskal Wallis or Mann Whitney U-test was performed to compare the differences. Receiver operator curve analysis (ROC) was used to determine spesifity and sensitivity cut-off of the S100B level in the diagnosis of COVID-19 disease. The relationship between the continuous variables was tested by Spearman correlation analysis. Statistical significance was set at p<0.05 in all the analyses.
Ethical approval. This study was approved by the Pamukkale University Ethical Commitee (26.01.2021 dated; 60116787-020-9695 numbered).
Results
The effect size for the difference of serum S100b concentrations between the three groups at 95% power and 95% CI was f=0.3 which can be considered “medium” according to Cohen’s criteria.
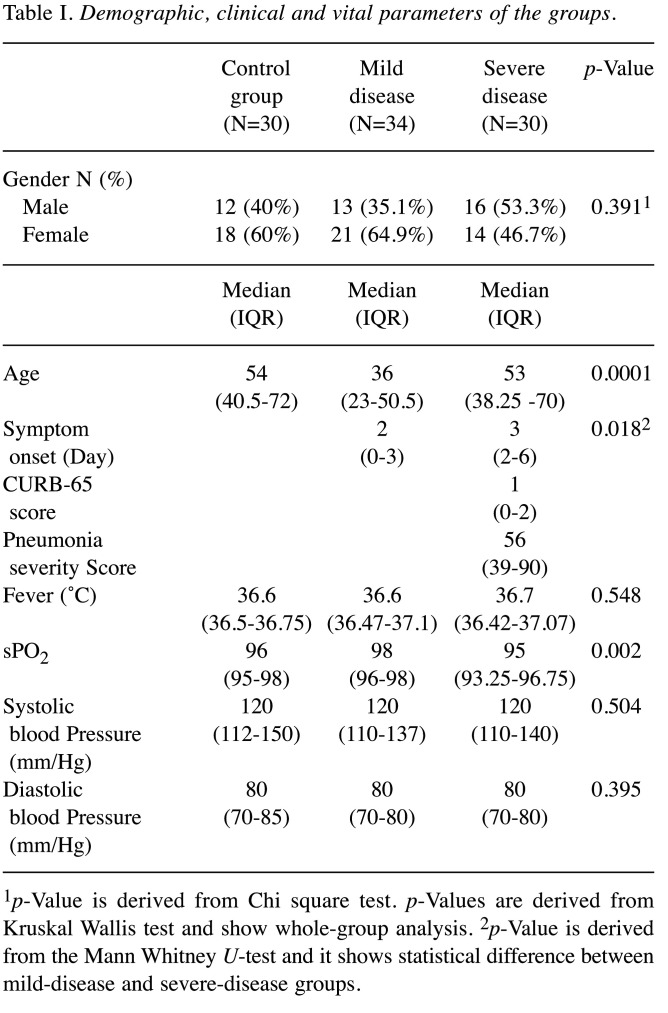
The statistical analyses yielded no significant difference between the three groups with respect to gender (p=0.391). The mean age was lower in the mild disease group than the controls and severe disease group (p=0.0001). Besides, symptom duration in the severe disease cohort turned out to be significantly longer than in the mild disease group (p=0.018). Of vital parameters, only sPO2 levels remained significantly lower in the severe disease cohort than the other two groups (p=0.002) (Table I).
Table I. Demographic, clinical and vital parameters of the groups.
1p-Value is derived from Chi square test. p-Values are derived from Kruskal Wallis test and show whole-group analysis. 2p-Value is derived from the Mann Whitney U-test and it shows statistical difference between mild-disease and severe-disease groups.
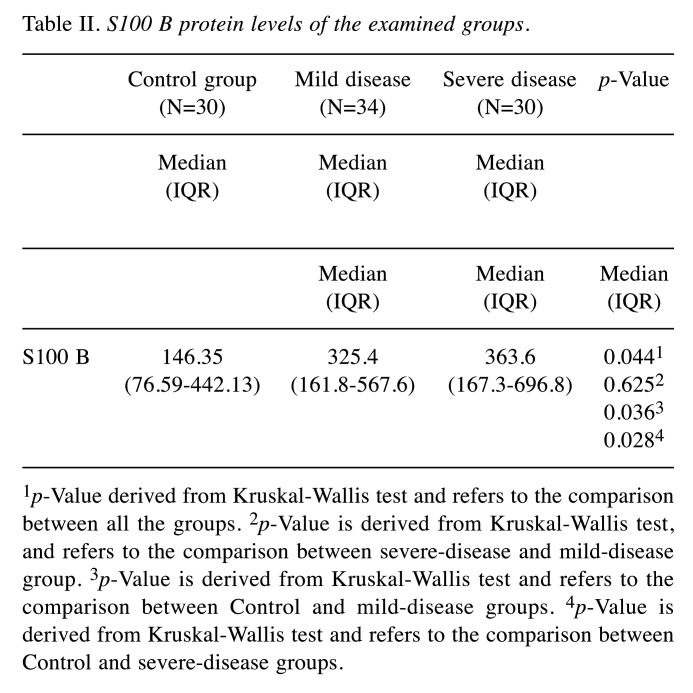
S100b levels were measured as 146.35 ng/ml (76.59-442.13) in the controls, 325.4 ng/ml (161.8-567.6) in the mild disease cohort, and 363.6 ng/ml (167.3-696.8) in the severe disease cohort. This difference between the figures was found to be statistically significant (p=0.044). While serum S100b levels indicated a significantly higher mean value in mild and severe disease cohorts than the healthy controls (p=0.036 and p=0.028 respectively), no significant difference was noted between the patient groups in terms of serum S100b elevation (p=0.625) (Table II).
Table II. S100 B protein levels of the examined groups.
1p-Value derived from Kruskal-Wallis test and refers to the comparison between all the groups. 2p-Value is derived from Kruskal-Wallis test, and refers to the comparison between severe-disease and mild-disease group. 3p-Value is derived from Kruskal-Wallis test and refers to the comparison between Control and mild-disease groups. 4p-Value is derived from Kruskal-Wallis test and refers to the comparison between Control and severe-disease groups.
Our receiver operating characteristic (ROC) analysis indicated greater area under the curve (AUC) for serum S100b levels of the COVID-19 patients (AUC=0.663, 95% CI=0.541-0.785; p=0.014) compared to controls. In addition, serum S100b concentration was measured as 151.7 ng/ml at 79.3% sensitivity and 51.7% specificity (p=0.014) (Figure 1).
Figure 1. Receiver operator curve (ROC) analysis of S100B protein levels.
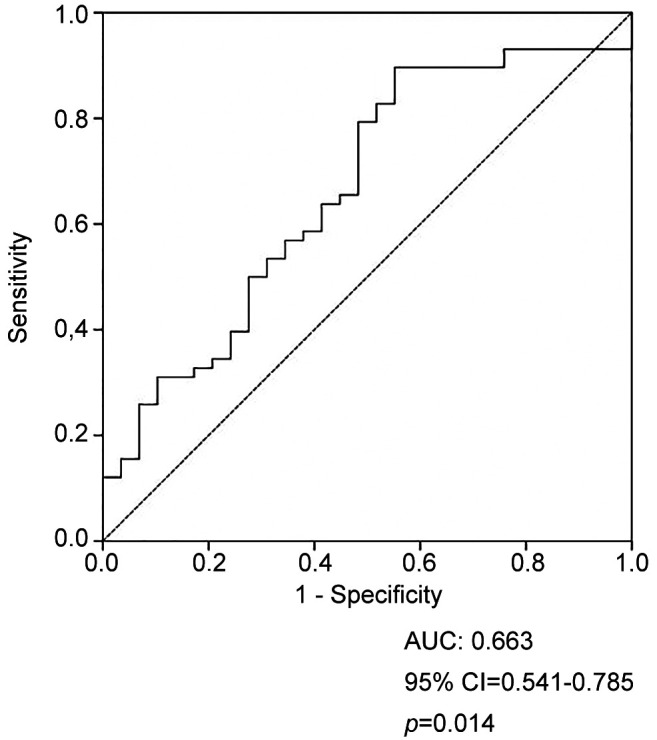
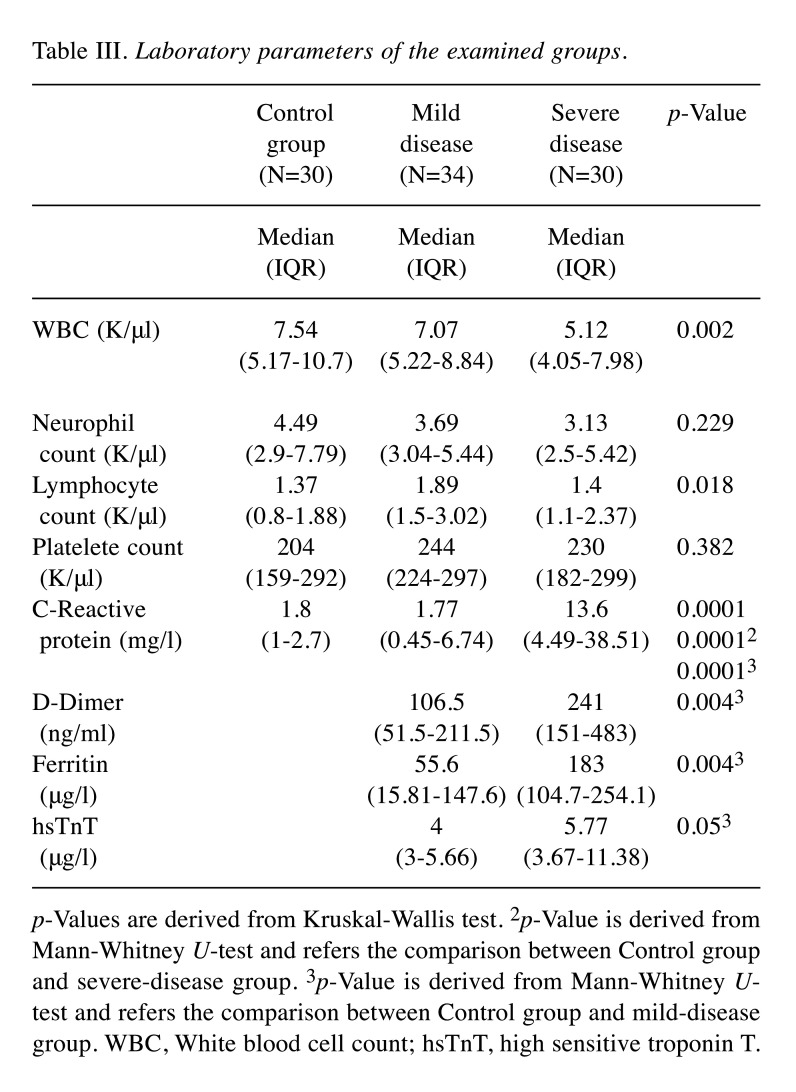
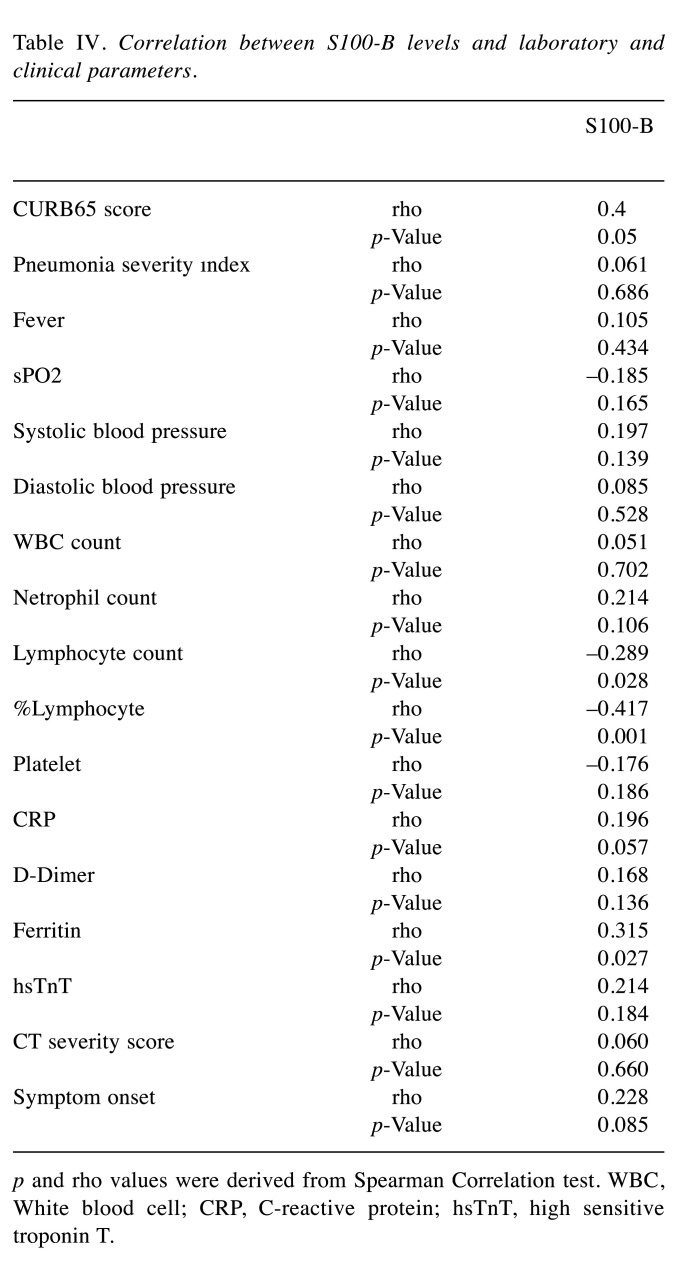
Table III lists the results obtained from the laboratory parameters of each group. In relation to the association between serum S100b concentration and clinical, vital and laboratory parameters in the patient cohorts, a moderate positive correlation was found between CURB65 scores and serum S100b levels of patients with pneumonia in the severe disease cohort (rho=0.4 and p=0.001). A low negative correlation was observed between serum S100b level and lymphocyte count (rho=–0.289 and p=0.028), while a moderate negative correlation was found between serum S100b level and lymphocyte percentage (rho=–0.417 and p=0.001) (Table IV).
Table III. Laboratory parameters of the examined groups.
p-Values are derived from Kruskal-Wallis test. 2p-Value is derived from Mann-Whitney U-test and refers the comparison between Control group and severe-disease group. 3p-Value is derived from Mann-Whitney Utest and refers the comparison between Control group and mild-disease group. WBC, White blood cell count; hsTnT, high sensitive troponin T.
Table IV. Correlation between S100-B levels and laboratory and clinical parameters.
p and rho values were derived from Spearman Correlation test. WBC, White blood cell; CRP, C-reactive protein; hsTnT, high sensitive troponin T.
Discussion
In this report, we tried to address the association between the severity stage of COVID-19 disease and serum S100b level during admission to ED. The most striking finding to emerge from our data comparison is that more elevated serum S100b concentration was evident in the cohorts (severe and mild disease) than the controls. However, our analysis did not confirm any significant differences in S100b levels between the two disease groups. Furthermore, we noted a significant negative correlation between S100b levels and the lymphocyte count and percentage, both of which are confirmed to be crucial parameters in evaluating the clinical severity of COVID-19 infection. Our results also revealed that the amount of serum S100b concentration was negatively correlated with CURB-65 score, an indicator of clinical severity, at a moderate level in the severe disease cohort.
An increasing number of reports are being published on the overexpression of S100b protein in the case of neurological dysfunction (15-18). An independent association is reported between S100b protein and systemic inflammation markers in ischemic stroke (19). The S100b protein also promotes expression of interleukin-1β (IL-1β) enzyme by macrophages, inducing increased retinal inflammation (19). A recent study on the interaction between inflammation and serum S100b protein suggests that the dynamic change of serum S100b protein on the 1st and 3rd days in patients with sepsis is interrelated with sepsis-induced encephalopathy (20). As identified in previous literature reports, S100b concentration might be a reliable predictor of tissue hypoperfusion in critically ill patients without brain damage (21). In the literature, many biological parameters such as P-selectin, endoplasmic reticulum stress markers, ferritin, d-dimer etc. have been examined for their diagnostic values and clinical outcomes (22-24). However, we could not identify relevant research conducted on patients with pneumonia during our literature review.
In a new study on COVID-19 disease, Aceti et al. reported a significant correlation between serum S100b protein and disease severity score and concluded that S100b protein can act as a predictive marker for identifying severity of this disease (25). However, this study scrutinized the S100b levels of hospitalized COVID-19 patients alone and did not investigate the changes in the healthy population. On the other hand, our study revealed that serum S100b protein levels remained higher in the patient cohorts in comparison to the healthy controls. Unlike Aceti et al.’s study, our findings suggest that elevated concentration of serum S100b was also evident in the mild disease cohort made up of unhospitalized COVID-19 patients without pneumonia. We are of the opinion that this situation reveals the clear association between S100b protein and inflammatory processes. Moreover, though serum S100b protein levels did not markedly differ in patients with and without pneumonia, a moderate positive correlation existed between CURB65 score and S100b protein in our severe disease group. As stated by Aceti et al., this association validates the view that S100b protein can be utilized to predict the severity of COVID-19 pneumonia. In addition, serum S100b protein level was measured as 151.7 ng/ml at 79.3% sensitivity in our study, suggesting that this protein might be a reliable marker in distinguishing between patients and healthy individuals in ED admissions.
A substantial body of research analyzing abnormalities in laboratory parameters documents increased ferritin and CRP values as well as low lymphocyte count in COVID-19 disease, and draws attention to the significance of ferritin level in establishing clinical severity. For instance, Yılmaz et al. have reported that lymphocyte count was lower in the patients with COVID-19 pneumonia compared to the CT-negative group, and that CT severity was negatively correlated with lymphocyte count and percentage. Besides, these researchers have also highlighted the importance of serum ferritin levels in assessing clinical and radiological severity (24). A systematic review analyzing the relationship between lymphocyte count and severity of COVID-19 has likewise designated lymphopenia as a useful clinical marker for poor prognosis. In that regard, our findings broadly support the work of other studies in that lymphocyte count was lower while ferritin and CRP values were higher in the cohort with pneumonia manifestation in imaging (severe disease group). Similar to our findings, Aceti et al. have reported a positive correlation between serum ferritin and S100b levels in their study (rho=0.32 and p=0.007) (25). Considering the negative correlation of S100b protein with lymphocyte count and percentage as well as its positive correlation with ferritin levels, it can be concluded that serum S100b level can serve as a valuable biomarker in assessing the clinical severity of COVID-19 disease.
The generalisability of our results is subject to certain limitations. Although the absence of a critical disease group in our study can be considered as a limitation, we aimed to track dynamic S100b changes in the patient group as the primary outcome and investigate its association with clinical severity as the secondary outcome. Another source of weakness which could have affected our results is that serum S100b level was not measured sequentially during the clinical follow-up.
Taken together, our results suggest that serum S100b protein level can serve as a valuable clinical marker in establishing diagnosis of patients. Though not useful in identifying different stages of COVID-19 infection, serum S100b concentration along with other known markers can be utilized to reliably predict clinical severity due to its correlation with other clinical parameters.
Conflicts of Interest
The Authors declare no conflicts of interest.
Authors’ Contributions
Conception: Aylin Koseler, Ergun Mete. Study Design: Ramazan Sabirli, Aylin Koseler. Funding: Aylin Koseler. Materials: Tarik Goren, Ergun Mete and Ibrahim Turkcuer. Data collection and processing: Tarik Goren, Özgür Kurt, Literature Review: Ramazan Sabirli, Ibrahim Turkcuer and Tarik Goren. Composition: Ramazan Sabirli, Ergun Mete and Aylin Koseler. Clinical Review: Ibrahim Turkcuer, Özgür Kurt.
References
- 1.CDC. 2019 Novel Coronavirus, Wuhan, China. CDC. Available at: https://www.cdc.gov/coronavirus/2019-ncov/about/index.html. [Last accessed on January 27, 2020]
- 2.Cascella M, Rajnik M, Cuomo A, Dulebohn SC, Di Napoli R. Features, evaluation, and treatment of Coronavirus (COVID-19) (Updated 2020 Aug 10). In: StatPearls (Internet). Treasure Island (FL): StatPearls Publishing; 2020 Jan. Available at: https://www.ncbi.nlm.nih.gov/books/NBK554776/ [Last accessed on June 27, 2021] [PubMed]
- 3.Donato R. Intracellular and extracellular roles of S100 proteins. Microsc Res Tech. 2003;60(6):540–551. doi: 10.1002/jemt.10296. [DOI] [PubMed] [Google Scholar]
- 4.Donato R, Sorci G, Riuzzi F, Arcuri C, Bianchi R, Brozzi F, Tubaro C, Giambanco I. S100B’s double life: intracellular regulator and extracellular signal. Biochim Biophys Acta. 2009;1793(6):1008–1022. doi: 10.1016/j.bbamcr.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 5.Sorci G, Bianchi R, Riuzzi F, Tubaro C, Arcuri C, Giambanco I, Donato R. S100B protein, a damage-associated molecular pattern protein in the brain and heart, and beyond. Cardiovasc Psychiatry Neurol. 2010;2010:656481. doi: 10.1155/2010/656481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hofmann MA, Drury S, Fu C, Qu W, Taguchi A, Lu Y, Avila C, Kambham N, Bierhaus A, Nawroth P, Neurath MF, Slattery T, Beach D, McClary J, Nagashima M, Morser J, Stern D, Schmidt AM. RAGE mediates a novel proinflammatory axis: a central cell surface receptor for S100/calgranulin polypeptides. Cell. 1999;97(7):889–901. doi: 10.1016/s0092-8674(00)80801-6. [DOI] [PubMed] [Google Scholar]
- 7.Morbini P, Villa C, Campo I, Zorzetto M, Inghilleri S, Luisetti M. The receptor for advanced glycation end products and its ligands: a new inflammatory pathway in lung disease. Mod Pathol. 2006;19(11):1437–1445. doi: 10.1038/modpathol.3800661. [DOI] [PubMed] [Google Scholar]
- 8.Sorci G, Giovannini G, Riuzzi F, Bonifazi P, Zelante T, Zagarella S, Bistoni F, Donato R, Romani L. The danger signal S100B integrates pathogen- and danger-sensing pathways to restrain inflammation. PLoS Pathog. 2011;7(3):e1001315. doi: 10.1371/journal.ppat.1001315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donato R. RAGE: a single receptor for several ligands and different cellular responses: the case of certain S100 proteins. Curr Mol Med. 2007;7(8):711–724. doi: 10.2174/156652407783220688. [DOI] [PubMed] [Google Scholar]
- 10.Leclerc E, Fritz G, Vetter SW, Heizmann CW. Binding of S100 proteins to RAGE: an update. Biochim Biophys Acta. 2009;1793(6):993–1007. doi: 10.1016/j.bbamcr.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 11.Mukherjee TK, Mukhopadhyay S, Hoidal JR. Implication of receptor for advanced glycation end product (RAGE) in pulmonary health and pathophysiology. Respir Physiol Neurobiol. 2008;162(3):210–215. doi: 10.1016/j.resp.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 12.Schmidt AM, Yan SD, Yan SF, Stern DM. The multiligand receptor RAGE as a progression factor amplifying immune and inflammatory responses. J Clin Invest. 2001;108(7):949–955. doi: 10.1172/JCI14002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buckley ST, Ehrhardt C. The receptor for advanced glycation end products (RAGE) and the lung. J Biomed Biotechnol. 2010;2010:917108. doi: 10.1155/2010/917108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clinical management of COVID-19: interim guidance, 27 May 2020 (No. WHO/2019-nCoV/clinical/2020.5). World Health Organization. Available at: https://apps.who.int/iris/handle/10665/332196. [Last accessed on October 22, 2020]
- 15.Yardan T, Erenler AK, Baydin A, Aydin K, Cokluk C. Usefulness of S100B protein in neurological disorders. J Pak Med Assoc. 2011;61(3):276–281. [PubMed] [Google Scholar]
- 16.Rothermundt M, Peters M, Prehn JH, Arolt V. S100B in brain damage and neurodegeneration. Microsc Res Tech. 2003;60(6):614–632. doi: 10.1002/jemt.10303. [DOI] [PubMed] [Google Scholar]
- 17.Cristóvão JS, Gomes CM. S100 Proteins in Alzheimer’s Disease. Front Neurosci. 2019;13:463. doi: 10.3389/fnins.2019.00463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hajduková L, Sobek O, Prchalová D, Bílková Z, Koudelková M, Lukášková J, Matuchová I. Biomarkers of brain damage: S100B and NSE concentrations in cerebrospinal fluid—A normative study. Biomed Res Int. 2015;2015:379071. doi: 10.1155/2015/379071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beer C, Blacker D, Bynevelt M, Hankey GJ, Puddey IB. Systemic markers of inflammation are independently associated with S100B concentration: results of an observational study in subjects with acute ischaemic stroke. J Neuroinflammation. 2010;7:71. doi: 10.1186/1742-2094-7-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu L, Feng Q, Ai ML, Deng SY, Liu ZY, Huang L, Ai YH, Zhang L. The dynamic change of serum S100B levels from day 1 to day 3 is more associated with sepsis-associated encephalopathy. Sci Rep. 2020;10(1):7718. doi: 10.1038/s41598-020-64200-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Routsi C, Stamataki E, Nanas S, Psachoulia C, Stathopoulos A, Koroneos A, Zervou M, Jullien G, Roussos C. Increased levels of serum S100B protein in critically ill patients without brain injury. Shock. 2006;26(1):20–24. doi: 10.1097/01.shk.0000209546.06801.d7. [DOI] [PubMed] [Google Scholar]
- 22.Karsli E, Sabirli R, Altintas E, Canacik O, Sabirli GT, Kaymaz B, Kurt Ö, Koseler A. Soluble P-selectin as a potential diagnostic and prognostic biomarker for COVID-19 disease: A case-control study. Life Sci. 2021;277:119634. doi: 10.1016/j.lfs.2021.119634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Köseler A, Sabirli R, Gören T, Türkçüer I, Kurt Ö. Endoplasmic reticulum stress markers in SARS-COV-2 infection and pneumonia: Case-control study. In Vivo. 2020;34(3 Suppl):1645–1650. doi: 10.21873/invivo.11956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yilmaz A, Sabirli R, Seyit M, Ozen M, Oskay A, Cakmak V, Goren T, Turkcuer I. Association between laboratory parameters and CT severity in patients infected with Covid-19: A retrospective, observational study. Am J Emerg Med. 2021;42:110–114. doi: 10.1016/j.ajem.2021.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aceti A, Margarucci LM, Scaramucci E, Orsini M, Salerno G, Di Sante G, Gianfranceschi G, Di Liddo R, Valeriani F, Ria F, Simmaco M, Parnigotto PP, Vitali M, Romano Spica V, Michetti F. Serum S100B protein as a marker of severity in Covid-19 patients. Sci Rep. 2020;10(1):18665. doi: 10.1038/s41598-020-75618-0. [DOI] [PMC free article] [PubMed] [Google Scholar]