Abstract
Background
We identified host single-nucleotide variants (SNVs) associated with neurocognitive impairment (NCI) in perinatally HIV-infected (PHIV) children.
Methods
Whole-exome sequencing (WES) was performed on 217 PHIV with cognitive score for age (CSA) < 70 and 247 CSA ≥ 70 (discovery cohort [DC]). SNVs identified in DC were evaluated in 2 validation cohorts (VC). Logistic regression was used to estimate adjusted odds ratios (ORs) for NCI. A human microglia NLRP3 inflammasome assay characterized the role of identified genes.
Results
Twenty-nine SNVs in 24 genes reaching P ≤ .002 and OR ≥ 1.5 comparing CSA < 70 to CSA ≥ 70 were identified in the DC, of which 3 SNVs were identified in VCs for further study. Combining the 3 cohorts, SNV in CCRL2 (rs3204849) was associated with decreased odds of NCI (P < .0001); RETREG1/FAM134B (rs61733811) and YWHAH (rs73884247) were associated with increased risk of NCI (P < .0001 and P < .001, respectively). Knockdown of CCRL2 led to decreased microglial release of IL-1β following exposure to ssRNA40 while knockdown of RETREG1 and YWHAH resulted in increased IL-1β release.
Conclusions
Using WES and 2 VCs, and gene silencing of microglia we identified 3 genetic variants associated with NCI and inflammation in HIV-infected children.
Keywords: HIV, neurocognitive impairment, HIV perinatal infection, genome-wide exome sequencing, CCRL2, RETREG1, YWHAH, 14-3-3η protein
CCRL2, RETREG1, and YWHAH genetic variants (rs3204849, rs61733811, rs73884247) are associated with neurocognitive impairment in HIV-infected children. Consistent with the role of inflammation in cognitive impairment, knockdown of RETREG1 and YWHAH increase NLRP3 activation, while CCRL2 knockdown decreases NLRP3 activation.
The pathogenesis of human immunodeficiency virus (HIV)-associated neurocognitive disorder (HAND) appears to be multifactorial [1–3]. Although there is an association in both adults and children with high quantities of virus in plasma and cerebrospinal fluid with the development of neurological disorders, only a subset of infected persons develop such deficits. Additionally, no consistent HIV variant has been identified that would explain why only a subset of persons develop cognitive impairment. Thus, it has been hypothesized that host genetic variation plays a central role in driving the host-virus interactions resulting in central nervous system (CNS) disease.
Multiple genome-wide association studies (GWAS) have been performed in adults designed to identify genetic factors associated with HAND [4–6]. In a GWAS performed by the CHARTER Group on antiretroviral therapy (ART)-era participants, using the Global Deficit Score to define neurocognitive impairment, no single-nucleotide variant (SNV) was identified that met the predetermined criterion for significance [5]. However, multiple SNVs were identified that had previously been found to be associated with T-cell function and neurological diseases.
In the research presented here, a discovery cohort of HIV-infected children evaluated prior to initiation of ART had whole-exome sequencing to identify potential genetic variants associated with cognitive impairment. The genetic variants of interest were progressively applied to 2 validation cohorts that yielded 3 potential gene candidates. To determine the biological link of these polymorphisms to neuroinflammation, knockdown of these genes was evaluated in a microglial cell model of inflammation with each of the 3 genes found to affect the activation of the NLRP3 inflammasome and release of inflammatory cytokines.
METHODS
Study Populations
Three cohorts were assessed, 1 discovery cohort and 2 validation cohorts.
Discovery Cohort
Participants in the discovery cohort were from the Pediatric AIDS Clinical Trials Group (PACTG) P152 and P300 studies. These patients formed a subset of children with symptomatic HIV infection who participated in randomized, double-blinded, multicenter protocols P152 (n = 831) or P300 (n = 471) (for details see [7, 8]).
Validation Cohort 1
Pediatric HIV/AIDS Cohort Study (PHACS) Adolescent Master Protocol (AMP) is a contemporary cohort of perinatally HIV-infected (PHIV) and perinatally HIV exposed uninfected (PHEU) children who have been followed longitudinally since their enrollment in 2007–2009 to better understand the effects of HIV and ART on children perinatally infected with HIV.
Validation Cohort 2
PACTG P338 and P377 were multicenter, randomized clinical trials that enrolled HIV-infected children younger than 18 years, had Centers for Disease Control and Prevention (CDC) immune category 1 or 2 disease, experienced no new CDC clinical category C diagnosis in the previous 12 months, had not previously received protease inhibitors, and had received continuous nucleoside reverse transcriptase ART for at least 16 weeks before study entry (for details see [9, 10]). This study was approved by the Human Research Protection Program of the University of California, San Diego, and written informed consent was obtained from all patients, their parents, or legal guardians (Supplementary Methods).
Neuropsychological Measures
Neuropsychological testing was conducted by trained psychologists for all studies using a battery of tests appropriate for age (for details see [7–11]). For each age-standardized score (mean = 100, SD = 15), neurocognitive impairment was defined as cognitive score for age (CSA) < 70.
Genome-Wide Exome Sequencing
Exome Library Preparation, Sequencing, and Variant Calling
Sequencing libraries were prepared and captured using SureSelect Human All Exon 50 Mb kit (246 samples) or v4 kit (248 samples) (Agilent Technologies) following the manufacturer’s instructions. Sequencing was performed using the Illumina HiSeq 2000 system, generating 100-bp paired-end reads. The SNVs and indels were called using GATK v2.1-9-gb90951c UnifiedGenotyper with parameters -stand_call_conf 20.0, -stand_emit_conf 10.0, and -dcov 800. University of California Santa Cruz assembly hg19 and dbSNP137 were used as references (Supplementary Methods).
Cell Culture
Peripheral blood mononuclear cells were isolated from whole blood of HIV-seronegative donors and human microglial cells were differentiated from primary human monocytes using methods as described previously [12] (Supplementary Methods).
Small Interfering RNA Transfections
Predesigned Dicer-substrate small interfering RNA (DsiRNA; Integrated DNA Technologies) were used for silencing C-C motif chemokine receptor like 2 (CCRL2), reticulophagy regulator 1 (RETREG1, FAM134B), and tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein eta (YWHAH, 14-3-3 protein eta) genes in primary human microglia cells. The DsiRNA transfections were performed using Lipofectamine RNAiMAX transfection reagent (Thermo Fisher Scientific) following the manufacturer’s protocol.
RNA Isolation and Real-Time PCR
Total RNA was isolated using RNeasyMini kit (Qiagen) according to the manufacturer’s protocol and 200–500 ng RNA was used in 20–40 µL of reverse transcription reaction using high-capacity cDNA Reverse Transcription kit (Applied Biosystems). TaqMan Gene Expression Assay for CCRL2, RETREG1, YWHAH, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) genes were used for quantitative polymerase chain reaction (qPCR) analysis (CCRL2-Hs00243702_s1, RETREG1-Hs00375273_m1, YWHAH-Hs00607046_m1, and GAPDH-Hs02786624_g1; Applied Biosystems) [13].
Inflammation Activation Assay
For NLRP3 inflammasome activation, microglial cells transfected with DsiRNA targeting CCRL2, RETREG1, YWHAH, or negative control siRNA were incubated for 24 hours with GU-rich single-stranded RNA within the HIV long terminal repeat (ssRNA40) or vehicle (LyoVec) (Invivogen). Both cell culture supernatants for interleukin-1β (IL-1β) enzyme-linked immunosorbent assay (ELISA; R&D Systems) and cell lysates for western blot analysis of proIL-1β were collected as previously described [14, 15].
Statistical Analysis
Data analysis began from a variant call format (VCF) file of 918 090 variants across 464 samples from study ART-naive participants with neurocognitive data. The phenotype of interest was standardized global CSA < 70. The discovery cohort consisted of 217 cases and 247 controls that were successfully exome sequenced. The 217 cases were P152/P300 study participants that had a CSA < 70 and had peripheral blood mononuclear cells (PBMC) available for testing. The 247 controls (CSA ≥ 70) were matched to cases with regards to age ≤ 2 years or >2 years, sex, and self-reported race. Each variant was tested for association with CSA < 70. Fisher exact test was applied using PLINK/SEQ (http://atgu.mgh.harvard.edu/plinkseq).
Counts, means, and percentages were used to summarize analyses. Logistic regression and a dominance analysis model were used to estimate adjusted odds ratios. To account for multiple assessments over time in validation cohort 1, generalized estimating equations with a logistic link was used. The combined, across-study odds ratio (OR) estimate was computed using inverse variance weights. Type I error rates were minimized by using multiple validation cohorts. Analyses were conducted using SAS 9.4 (SAS Institute) and R 3.2.2 (R Core Team, 2015).
For microglial assays, statistical significance was assessed using the 2-tailed Student t test, ANOVA, and Wilcoxon rank test as appropriate.
RESULTS
Characteristics of Study Populations
Discovery Cohort
Of the 464 evaluable participants, 51% were female, 62% were identified as black, and 27% were Hispanic (Table 1). The median age was 1.5 years, with a median CD4+ count of 859/mm3 and median log10 HIV RNA load of 5.4 copies/mL.
Table 1.
Baseline Characteristics of Discovery and Validation Cohorts
| Characteristic | Discovery Cohort (PHIV) | Validation Cohort 1 (PHIV) | Validation Cohort 1 (PHEU) | Validation Cohort 2 (PHIV) |
|---|---|---|---|---|
| Number | 464 | 394 | 214 | 357 |
| Female, No. (%) | 236 (50.9) | 216 (53.4) | 103 (48) | 192 (53.8) |
| Race/ethnicity, No. (%)a | ||||
| Hispanic | 123 (26.5) | 100 (25.4) | 76 (36.0) | 120 (33.6) |
| Black | 286 (61.6) | 279 (70.8) | 132 (61.7) | 190 (53.2) |
| White | 51 (11) | 89 (22.6) | 73 (34.1) | 44 (12.3) |
| Unknown/other | 4 (0.9) | 26 (6.6) | 9 (4.2) | 3 (.80.8) |
| CD4+, count/mm3, median (Q1, Q3) | 859 (416, 1490) | 730 (522, 953) | NA | 668 (466, 924) |
| Log HIV RNA, median (Q1, Q3) | 5.4 (4.7, 6.0) | 2.5 (1.7, 3.1) | NA | 4.2 (3.6, 4.7) |
| Age, y, median (Q1, Q3) | 1.5 (0.7, 3.6) | 12 (9, 14) | 10 (8, 11) | 6 (5, 9) |
| CSA, median (Q1, Q3) | 71.5 (58, 89) | 86 (75, 95) | 87 (78, 99) | 83 (74, 93) |
| CSA < 70, No. (%) | 217 (46.8) | 54 (13.7) | 24 (11.2) | 54 (15.1) |
| CSA ≥ 70, No. (%) | 247 (53.2) | 340 (86.3) | 190 (88.8) | 303 (84.9) |
Abbreviations: CSA, cognitive score for age; NA, not available; PHEU, perinatally HIV exposed uninfected; PHIV, perinatally HIV infected.
aRace and ethnicity were reported from a single question for the discovery cohort and validation cohort 2. Hispanic ethnicity and race were reported as 2 separate questions for validation cohort 1.
Validation cohort 1 consisted of 394 PHIV children with a median age of 12 years and 214 PHEU children with a median age of 10 years that participated in the PHACS AMP protocol (Table 1). In the PHIV group, 53% of participants were female, 25% Hispanic, and 71% black, which did not differ significantly from the uninfected controls. The median log10 HIV RNA was 2.5 copies/mL with a median CD4+ lymphocyte count of 730/mm3. Of 394 PHIV, the median CSA was 86, with 54 (14%) having a CSA < 70 while 24 (11%) of PHEU had a CSA < 70.
Validation cohort 2 consisted of 357 PHIV children from 2 PACTG studies that enrolled children who were clinically stable for ≥6 months into 3-drug combination therapies. The median age for the combined cohort was 6 years with 54% female, 53% black, and 34% Hispanic. The median log10 HIV RNA was 4.2 copies/mL with a median CD4+ lymphocyte count of 668/mm3. The median CSA for the cohort was 83 with 54 (15.1%) having a CSA < 70 (Table 1).
Genome-Wide Exome Sequencing
A total of 485 samples including 7 duplicates were processed. Of the 478 samples from unique individuals, 14 were excluded either because of extreme missing rates, extreme heterogeneity, or sex discrepancy using PLINK. Thus, as described above, 464 samples from unique study participants were available for analysis.
To identify the potential genes of greatest interest and decrease the risk of type 1 error, the variants most associated with cognitive impairment, reaching P values lower than .002, odds ratios greater than 1.5 or lower than 0.6, and present in at least 5 participants were selected for validation, yielding 29 variants in 24 genes (Table 2). Of these 29 SNVs, 2 pairs were in linkage disequilibrium, TMPRSS11B (rs12331141 and rs2319797), and PPL (rs61734749) and UBN1 (rs35575708).
Table 2.
Specific Single-Nucleotide Variants and Genes With Percent Of Homozygous Major Allele Identified in Discovery Cohort Reaching P ≤ .002 and Odds Ratios ≥ 1.5 or <0.6 With ≥5 Cases With Cognitive Score for Age < 70
| Chromosome Number | Gene | rs Number | Major Allele | Minor Allele | Homozygous Major Allele, % | P Value | Odds Ratio (95% CI) |
|---|---|---|---|---|---|---|---|
| 1 | CR1L | rs3085 | A | G | 91 | 1.60 × 10−3 | 3.02 (1.50–6.08) |
| 1 | IGSF3 | rs201676764 | C | T | 67 | 3.11 × 10−5 | 3.61 (1.93–6.76) |
| 1 | IGSF3 | rs61786588 | A | G | 63 | 1.95 × 10−6 | 3.84 (2.17–6.79) |
| 1 | IGSF3 | rs61786589 | C | G | 88 | 3.33 × 10−5 | 11.59 (2.61–51.47) |
| 2 | GRHL1 | rs2303920 | G | A | 53 | 5.00 × 10−3 | 0.58 (.39–.83) |
| 2 | LINC01237 | rs4973668 | C | G | 39 | 4.83 × 10−4 | 2.19 (1.40–3.41) |
| 2 | STEAP3 | rs113158407 | G | A | 96 | 2.70 × 10−4 | 8.98 (2.03–39.77) |
| 3 | CCRL2 | rs3204849 | T | A | 64 | 8.07 × 10−4 | 0.52 (.35–.76) |
| 4 | TMPRSS11B | rs12331141 | C | T | 36 | 1.81 × 10−4 | 0.48 (.32–.70) |
| 4 | TMPRSS11B | rs2319797 | A | T | 36 | 1.67 × 10−4 | 0.48 (.32–.70) |
| 5 | FAM134B | rs61733811 | C | G | 85 | 3.13 × 10−4 | 2.62 (1.53–4.50) |
| 6 | PREP | rs1051484 | T | C | 30 | 3.72 × 10−4 | 0.48 (.32–.72) |
| 7 | ESYT2 | rs2305473 | T | C | 52 | 5.73 × 10−4 | 1.92 (1.32–2.80) |
| 7 | ESYT2 | rs2305477 | A | C | 54 | 3.26 × 10−4 | 1.98 (1.36–2.88) |
| 7 | KMT2C | rs111493987 | C | A | 59 | 1.95 × 10−6 | 3.77 (2.15–6.60) |
| 7 | VWDE | rs73294382 | T | C | 81 | 1.80 × 10−3 | 0.46 (.29–.75) |
| 8 | PABPC1 | rs76261471 | A | C | 81 | 4.11 × 10−5 | 0.35 (.21–.59) |
| 10 | DCLRE1A | rs17235066 | T | C | 83 | 3.82 × 10−4 | 2.50 (1.40–4.19) |
| 10 | FAM21A | rs199520696 | C | T | 57 | 2.99 × 10−4 | 2.68 (1.56–4.62) |
| 11 | MUC5B | rs202127660 | A | G | 83 | 2.46 × 10−5 | 3.93 (2.00–7.75) |
| 14 | IGHV7-81 | rs201928713 | C | T | 72 | 1.38 × 10−4 | 0.44 (.29–.68) |
| 16 | PPL | rs61734749 | C | T | 96 | 2.46 × 10−4 | 9.10 (2.06–40.27) |
| 16 | UBN1 | rs35575708 | C | T | 96 | 2.46 × 10−4 | 9.10 (2.06–40.27) |
| 17 | CDC27 | rs201613328 | C | A | 82 | 4.04 × 10−5 | 2.79 (1.68–4.62) |
| 17 | CDC27 | rs78525224 | A | T | 84 | 8.21 × 10−6 | 3.97 (2.08–7.58) |
| 17 | ICT1 | rs34496172 | C | T | 76 | 1.25 × 10−4 | 2.36 (1.51–3.68) |
| 19 | ZNF358 | rs11555037 | A | G | 95 | 2.16 × 10−4 | 7.40 (2.15–25.49) |
| 21 | LTN1 | rs61735768 | T | C | 85 | 1.80 × 10−3 | 2.16 1.34–3.50) |
| 22 | YWHAH | rs73884247 | A | G | 93 | 5.29 × 10−4 | 4.09 (1.73–9.73) |
Abbreviation: CI, confidence interval.
Validation Cohorts
The 29 variants identified in the discovery cohort were evaluated in validation cohort 1. Of these 29, 20 variants had sufficient distribution of frequencies to provide odds ratios, of which 3 variants were identified of interest; rs3204849 in the gene encoding for CCRL2, rs61733811 encoding for RETREG1 (FAM134B), and rs73884247 encoding for YWHAH (14-3-3η) (Table 3). The strongest association was identified for rs3204849 (CCRL2) whose minor allele had an odds ratio of 0.22. For rs61733811 and rs73884247, the odds ratios for cognitive impairment were 1.82 and 1.19, respectively. Although not statistically stable, the odds ratios were in the same direction as the discovery cohort and considered possible risk alleles. Additionally, in the discovery cohort, each of these 3 SNVs remained statistically associated with cognitive impairment when study participants with a CSA of <70 were compared to those with a score of ≥70, ≥85, or ≥100 (Table 4). To provide further support for the inclusion of the 3 SNVs of interest, we examined the effect of the SNVs on CSA by HIV status. The interaction test P values between HIV status and the SNV were: rs3204849 (CCRL2) P = .054, rs61733811 (RETREG1/FAM134B) P = .01, and rs73884247 (YWHAH/14-3-3η) P = .89. Among the PHEU group, for each SNV, OR = 0.69 (95% confidence interval [CI], .31–1.54; P = .37) for rs3204849 (CCRL2), OR = 0.24 (95% CI, .05–1.10; P = .066) for rs61733811 (RETREG1/FAM134B), and OR = 1.87 (95% CI, .46–7.57; P = .38) for rs73884247 (YWHAH/14-3-3η).
Table 3.
Specific Single Nucleotide Variants and Genes Identified in the Discovery Cohort That Could Be Examined Using Percent Homozygous Major Allele in Validation Cohort 1
| Chromosome Number | Gene | rs Number | Major Allele | Minor Allele | Homozygous Major Allele, % | Odds Ratio (95% CI) | P Value |
|---|---|---|---|---|---|---|---|
| 1 | CR1L | rs3085 | A | G | 93 | 0.95 (.26–3.48) | .94 |
| 1 | IGSF3 | rs201676764 | C | T | 93 | 1.52 (.54–4.33) | .42 |
| 2 | GRHL1 | rs2303920 | G | A | 41 | 0.96 (.54–1.70) | .89 |
| 2 | LINC01237 | rs4973668 | C | G | 83 | 1.32 (.72–2.43) | .37 |
| 2 | STEAP3 | rs113158407 | G | A | 94 | 0.21 (.03–1.52) | .12 |
| 3 | CCRL2 | rs3204849 | T | A | 66 | 0.22 (.09–.55) | .001 |
| 4 | TMPRSS11B | rs12331141 | C | T | 35 | 1.18 (.63–2.21) | .61 |
| 4 | TMPRSS11B | rs2319797 | A | T | 35 | 1.07 (.58–1.99) | .82 |
| 5 | FAM134B | rs61733811 | C | G | 82 | 1.82 (.94–3.53) | .08 |
| 6 | PREP | rs1051484 | T | C | 32 | 1.28 (.69–2.37) | .43 |
| 7 | ESYT2 | rs2305473 | T | C | 52 | 1.12 (.63–1.99) | .69 |
| 7 | ESYT2 | rs2305477 | A | C | 54 | 1.15 (.65–2.02) | .63 |
| 7 | VWDE | rs73294382 | T | C | 95 | 0.55 (.12–2.47) | .43 |
| 10 | DCLRE1A | rs17235066 | T | C | 85 | 0.48 (.19–1.22) | .12 |
| 10 | FAM21A | rs199520696 | C | T | 28 | 1.09 (.57–2.06) | .80 |
| 11 | MUC5B | rs202127660 | A | G | 90 | 0.50 (.15–1.68) | .26 |
| 16 | PPL | rs61734749 | C | T | 96 | 0.34 (.04–2.89) | .32 |
| 17 | ICT1 | rs34496172 | C | T | 78 | 0.94 (.48–1.84) | .86 |
| 19 | ZNF358 | rs11555037 | A | G | 68 | 0.99 (.53–1.84) | .98 |
| 22 | YWHAH | rs73884247 | A | G | 96 | 1.19 (.32–4.39) | .79 |
Abbreviation: CI, confidence interval.
Table 4.
Odds Ratios and P Values for the 3 SNVs of Interest Comparing CSAs < 70 vs ≥70, <70 vs ≥85, and <70 vs ≥100 in the Discovery Cohort
| Gene SNV | CSA | ||
|---|---|---|---|
| <70 vs ≥70 | <70 vs ≥85 | <70 vs ≥100 | |
| CCRL2 rs3204849 | |||
| Odds ratio | 0.52 | 0.49 | 0.33 |
| 95% CI | .35–.76 | .31–.73 | .17–.64 |
| P value | 8.1 × 10−4 | 1.2 × 10−3 | 9.1 × 10−4 |
| RETREG1 (FAM134B) rs61733811 | |||
| Odds ratio | 2.62 | 2.84 | 3.95 |
| 95% CI | 1.53–4.5 | 1.47–5.47 | 1.17–13.31 |
| P value | 3.1 × 10−4 | 9.0 × 10−4 | 9.1 × 10−3 |
| YWHAH (14-3-3η) rs73884247 | |||
| Odds ratio | 4.09 | 5.84 | 5.48 |
| 95% CI | 1.72–9.73 | 1.72–19.83 | .72–41.61 |
| P value | 5.3 × 10−4 | 6.2 × 10−4 | 3.4 × 10−2 |
Abbreviations: CI, confidence interval; CSA, cognitive score for age; SNV, single nucleotide variant.
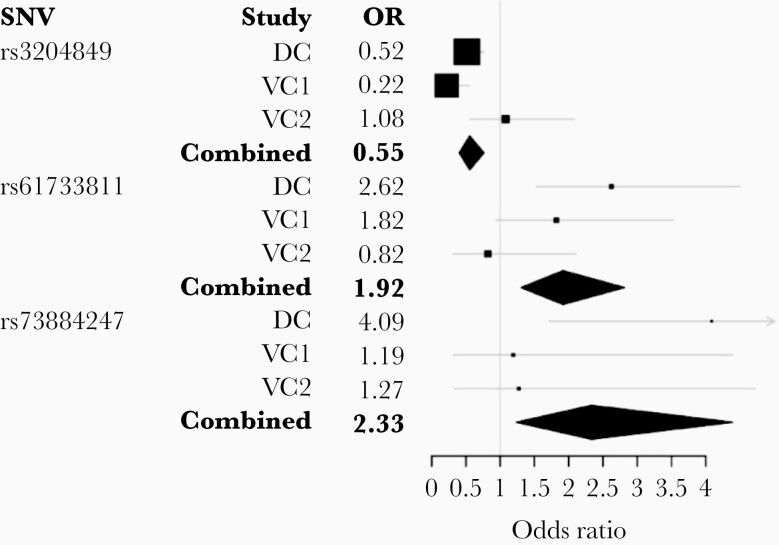
To further validate the SNVs identified in validation cohort 1, the 3 SNVs of interest were assessed in the second validation cohort. Of the 3 SNVs, only YWHAH (rs73884247) was in the same direction as observed in the first 2 cohorts of HIV-infected children. However, combining the 3 cohorts, the 3 SNVs remained significantly associated with the risk of having a cognitive score < 70 (Figure 1). For the combined cohorts the overall odds ratio for rs3204849 genetic variant was 0.55, indicating that the A allele was associated with a decreased risk for cognitive impairment. In contrast, the odds ratios for rs61733811 (FAM134B) G allele and rs73884247 (YWHAH) G allele were 1.92 and 2.33, respectively indicating an increased risk associated with the genetic variant.
Figure 1.
Combined and cohort-specific ORs by SNVs associated with neurocognitive impairment of children. Gray lines indicate 95% confidence intervals and larger box size indicates more precision. Abbreviations: DC, discovery cohort; OR, odds ratio; SNV, single nucleotide variant; VC, validation cohort.
Genotype-Function Correlation
To determine the biological plausibility that the specific genetic variants identified have a biological role that can affect HIV pathogenesis in the brain, we examined the potential that each of the 3 identified SNVs might have an impact on chronic inflammation associated with HIV and neurocognitive impairment. These experiments are based on our laboratory’s findings and that of others indicating that the NLRP3 inflammasome plays an important role in HAND [15, 16]. We assumed that the genetic variants observed to be associated with severe cognitive impairment of children resulted in a loss of function. Thus, genetic variants associated with higher cognitive function would exhibit less inflammation when microglial cells were silenced for the gene of interest, while genetic variants associated with CSA < 70 would exhibit greater inflammation when the gene was knocked down.
Knockdown of CCRL2, RETREG1 (FAM134B), and YWHAH (14-3-3η) in human microglial cells alters the inflammatory response to HIV ssRNA and autophagic flux.
Recently, we identified that exposure of primary human microglial cells to ssRNA40 from the long terminal repeat region of HIV induces the NLRP3 inflammasome with release of inflammatory cytokines including IL-1β [15]. Here, we hypothesized in a dominant model of gene expression that knockdown of genes leading to an increase in release of IL-1β from microglia would be associated with an increased risk for development of neurocognitive impairment (ie, an increase in inflammation), while a decrease in release of IL-1β (ie, a decrease in inflammation) would be associated with a decreased risk for neurocognitive impairment [15].
Having identified that SNVs in CCRL2 (rs3204849), FAM134B (rs61733811), and YWHAH (rs73884247) are associated with neurocognitive impairment in HIV-infected children, we knocked down expression of each of these proteins in microglial cells using RNAi and confirmed that there was no cytotoxicity (Supplementary Figure 1A). Transfected cells were analyzed for knockdown efficiency by qPCR at 48 hours posttransfection, following which cells were incubated for 24 hours with vehicle (LyoVec) or ssRNA40. IL-1β was quantified in culture supernatants by ELISA, and cell lysates were prepared and analyzed for intracellular expression of IL-1β (proIL-1β) by immunoblotting (Supplementary Figure 1B).
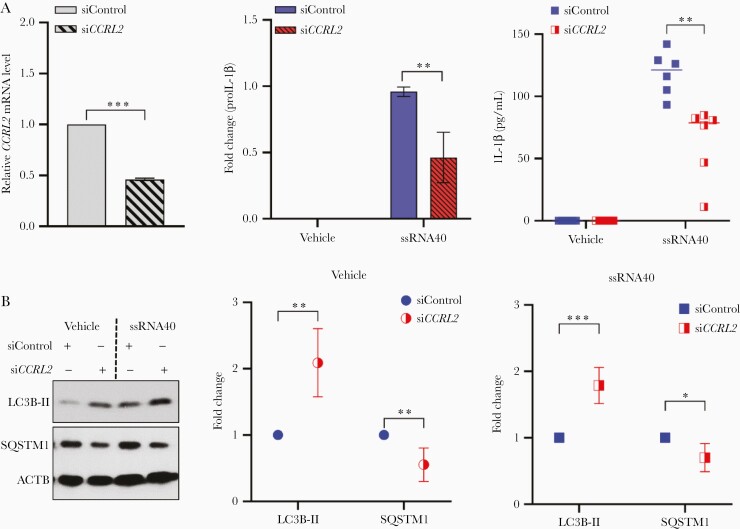
CCRL2 is a 7-transmembrane protein that is expressed in macrophages, dendritic cells, microglia, and neutrophils and its expression is upregulated by many proinflammatory stimuli such as lipopolysaccharide [17–22]. Following knockdown of CCRL2 by approximately 50% in microglial cells (Figure 2A), exposure of cells to HIV ssRNA40 led to a 2-fold reduction in proIL-1β expression levels (P = .002; Figure 2A) and 2.9-fold lower release of IL-1β levels in culture supernatants (P = .002; Figure 2A).
Figure 2.
Knockdown of CCRL2 in human microglial cells results in reduced inflammatory response to HIV ssRNA40 and increased autophagy levels. A, Left, Nonspecific control siRNA (a nontargeting DsiRNA that does not interact with any sequence in human genome; siControl) and siCCRL2-transfected human microglia were assessed for CCRL2 mRNA expression at 48 hours by qPCR. Both control and CCRL2-depleted microglial cultures were incubated with LyoVec (vehicle) or ssRNA40 (5 µg/mL) for an additional 24 hours. Culture supernatants were collected for ELISA, and cells were collected and proteins extracted for immunoblotting. Center, densitometry quantification values were collected for proIL-1β immunoblots after ACTB normalization (see Supplementary Figure1B for representative immunoblots) and plotted based on 4 different donors as means ± SEM. Right, quantitative measurement of IL-1β in the culture supernatants by ELISA (n = 6); median is plotted as horizontal line. B, Left, representative immunoblots for LC3B lipidation (LC3B-II) and SQSTM1. Center and right, densitometry quantification values were collected after ACTB normalization (see Supplementary Methods for details) and plotted based on 6 different donors as means ± SEM. *P < .05, **P < .005, ***P < .001. Abbreviations: ACTB, β-actin; DsiRNA, Dicer-substrate small interfering RNA; ELISA, enzyme-linked immunosorbent assay; HIV, human immunodeficiency virus; IL-1β, interleukin-1β; LC3B, light chain 3B; qPCR, quantitative polymerase chain reaction; SEM, standard error of the mean; SQSTM1, sequestosome 1.
Previous studies from our group and others have identified dysfunctional autophagy as a mechanism that drives chronic inflammation [15, 23–25]. Therefore, we also analyzed if inhibition of the NLRP3 inflammasome as indicated by a decrease in IL-1β release in CCRL2-depleted cells is associated with alterations in autophagy, as indicated by light chain 3 (LC3) lipidation and SQSTM1/p62 degradation. Following CCRL2 knockdown, LC3B lipidation was increased 2-fold and 1.8-fold in vehicle- or ssRNA40-treated cells, respectively (Figure 2B). Similarly, there was a 50% and 30% increase in SQSTM1 degradation in vehicle- or ssRNA40-treated cells, respectively (Figure 2B). These findings are consistent with enhanced autophagy following the knockdown of CCRL2 associated with decreased inflammation on exposure to HIV ssRNA40.
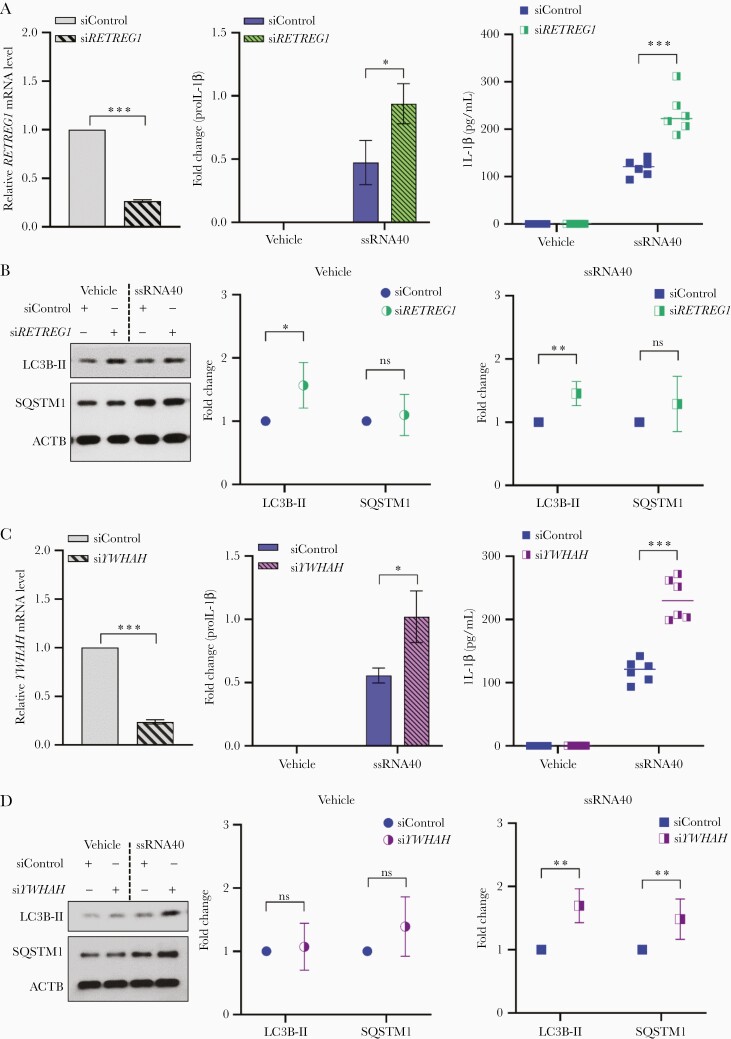
RETREG1 (FAM134B) gene belongs to a family of 3 genes (FAM134A, FAM134B, and FAM134C) that encodes cis-Golgi transmembrane endoplasmic reticulum (ER) receptor protein. These proteins have a conserved (DDFELL) microtubule-associated protein 1A/1B-LC3 and γ-aminobutyric acid receptor-associated protein (GABARAP)-interacting region that facilitate their interaction with LC3 and GABARAP proteins and regulate the turnover of ER [26–28]. FAM134B inhibits Ebola, Dengue, Zika, and West Nile virus replication [29, 30]. Following the methods described above, microglia were depleted of RETREG1 using RNAi. After confirming RETREG1 knockdown (70% knockdown, P < .001; Figure 3A), control (siControl) and RETREG1-depleted cells (siRETREG1) were exposed to ssRNA40. Loss of RETREG1 expression enhanced intracellular IL-1β expression and secretion in supernatants of ssRNA40-treated cells (Supplementary Figure 1C; 2-fold, P = .008; 1.9-fold, P < .001, respectively; Figure 3A). Associated with the increased release of IL-1β in RETREG1-depleted microglia, there was a concomitant increase in LC3B lipidation and inhibition of SQSTM1 degradation, indicating the inhibition of autophagic flux in both vehicle (P < .05, P = .4, respectively Figure 3B) and ssRNA40-treated cells (P < .003, P = .13, respectively Figure 3B). Consistent with the effects of RETREG1 variant (rs61733811) on HIV cognitive impairment (OR = 1.92), the gene silencing data confirm that loss of function for RETREG1 increases inflammation in HIV ssRNA40-exposed microglia and this effect is associated with the inhibition of autophagy in these cells (Figure 3A and 3B).
Figure 3.
Knockdown of RETREG1 (FAM134B) and YWHAH (14-3-3η) in human microglial cells results in enhanced inflammatory response to HIV ssRNA40 and inhibition of autophagic flux. A, Left, nonspecific control siRNA (a nontargeting DsiRNA that does not interact with any sequence in human genome; siControl) and siRETREG1 transfected human microglia were assessed for RETREG1 mRNA expression at 48 hours by qPCR. Both control and RETREG1-depleted microglial cells were incubated in culture with LyoVec (vehicle) or ssRNA40 (5 µg/mL) for an additional 24 hours. Culture supernatants were collected, microglia were harvested, proteins extracted and assessed by immunoblotting. Center, densitometry quantification values were collected for proIL-1β immunoblots after ACTB normalization (see Supplementary Figure 1C for representative immunoblots and Supplementary Methods for details) and plotted based on 4 different donors as means ± SEM. Right, quantitative measurement of IL-1β in the culture supernatants by ELISA (n = 6); median is plotted as horizontal line. B, Left, representative immunoblots for LC3B lipidation (LC3B-II) and SQSTM1 degradation assessment. Center and right, densitometry quantification values were collected after ACTB normalization (see Supplementary Methods for details) and plotted based on 6 different donors as means ± SEM. C, Left, nonspecific control siRNA (a nontargeting DsiRNA that does not interact with any sequence in human genome; siControl) and YWHAH siRNA (siYWHAH) transfected human microglia were collected at 48 hours for YWHAH mRNA expression analysis by qPCR. Both control and YWHAH-depleted cells were incubated with LyoVec (vehicle) or ssRNA40 (5 µg/mL) for an additional 24 hours. Culture supernatants were collected for ELISA, cells were collected, lysed, and lysates were used for immunoblotting. Center, densitometry quantification values were collected for proIL-1β immunoblots after ACTB normalization (see Supplementary Figure 1D for representative immunoblots and Supplementary Methods for details) and plotted based on 4 different donors as means ± SEM. Right, quantitative measurement of IL-1β in the culture supernatants by ELISA (n = 6); median is plotted as horizontal line. D, Left, representative immunoblots for LC3B lipidation (LC3B-II) and SQSTM1 degradation assessment. Center and right, densitometric quantification values were collected after ACTB normalization (see Supplementary Methods for details) and plotted based on 6 different donors as means ± SEM. *P < .05, **P < .005, ***P < .001. Abbreviations: ACTB, β-actin; DsiRNA, Dicer-substrate small interfering RNA; ELISA, enzyme-linked immunosorbent assay; HIV, human immunodeficiency virus; IL-1β, interleukin-1β; LC3B, light chain 3 B; qPCR, quantitative polymerase chain reaction; SEM, standard error of the mean; SQSTM1, sequestosome 1.
YWHAH (14-3-3η) belongs to the 14-3-3 protein family, which consist of 7 isoforms (β, ε, γ, η, τ, σ, and ζ) in humans encoded by 7 genes named YWHAx (x being either B, E, G, H, Q, S, or Z) [31]. These proteins are highly abundant in brain and bind to several serine/threonine phosphorylated ligands from cell cycle and other signal transduction pathways to modulate their functions by altering their localization and interactions with other proteins [32, 33]. Studies have identified the involvement of 14-3-3 proteins in several neurological disorders including Parkinson disease, amyotrophic lateral sclerosis, and epilepsy [34–37]. Following the approach described above for CCRL2 and RETREG1, we examined the induction of IL-1β in microglial cells by HIV ssRNA40 following knockdown of YWHAH (Figure 3C). YWHAH-depleted microglia exhibited increased proIL-1β intracellular expression and IL-1β release in the culture supernatants (Supplementary Figure 1D; 1.8-fold, P < .05; 2.1-fold, P < .001, respectively Figure 3C) following ssRNA40 treatment when compared to control cells. In addition to their role in inflammation, 14-3-3 proteins are also known to regulate the autophagy activity and expression of TFEB target genes by binding to the phosphorylated TFEB (pS211) and controlling its cytoplasmic localization [38]. Here, the knockdown of YWHAH in microglial cells altered autophagy and was associated with increased LC3B lipidation and inhibition of SQSTM1 degradation (1.7-fold, P = .001; >1.5-fold, P < .005, respectively Figure 3D) following ssRNA40 treatment. These findings are consistent with our hypothesis that the minor allele encoded by YWHAH rs73884247 is associated with increased inflammation and impaired autophagy resulting in cognitive impairment (Figure 3C and 3D).
DISCUSSION
Numerous studies have examined host and viral genetics in an attempt to identify virus with specific neurotropism and host genetic variants that lead to increased susceptibility to CNS disease [39–44]. Foremost among the genes identified in adults has been genetic variants that affect inflammation and immune regulation. However, to our knowledge, none of the genome-wide studies performed in adults has attempted to identify a genotype-function association in a biological system to support the findings, and no GWAS of neurocognitive disease has been performed in HIV-infected children. Thus, to our knowledge, this is the first GWAS of cognitive outcomes of HIV-infected children. Additionally, we have performed genotype-function studies that suggest a biological explanation for how each of the SNVs identified could affect neurocognitive outcome.
In the current study, we identified SNVs within 3 genes, CCRL2, RETREG1, and YWHAH, which are associated with severe neurocognitive impairment of children with perinatal HIV infection. These associations are largely driven by the discovery cohort with modest support provided by the validation cohorts. Several factors, however, suggest that the findings of the discovery and validation cohorts when combined have identified genes of importance in the development of cognitive impairment in HIV-infected children. The discovery cohort is the most robust of the 3 cohorts studied although more statistical tests were conducted in the discovery cohort. The discovery cohort were younger (median age 1.5 years) and occurred at a time when children had received no ART and could have progressive disease at the time of enrollment. Additionally, all children with CSA < 70 with PBMCs available (n = 213) were evaluated with controls matched as closely as possible for age, sex, and self-reported race. Although the 2 validation cohorts were the best available groups to study within the United States, each has limitations. Both cohorts had proportionately a lower number of children with CSA < 70 (validation cohort 1, 13.7% and validation cohort 2, 15.1%) and were older than the discovery cohort. Although the PHACS AMP cohort is the most comprehensively studied group of HIV-infected children in the United States, when it enrolled in 2007 to 2009, children at entry were required to be between 7 and 15 years of age. Thus, most of the children were already on effective ART initiated shortly after birth, which likely had a significant effect on the number of children with CSA < 70. For validation cohort 2, entry into the studies required that children had stable infection and CDC category 1 or 2 only. Thus, children with recognized more advanced or progressing disease were not enrolled into the study. Cohort differences, therefore, might partially explain variability in the cohort-specific point estimates, although the 95% CIs generally overlapped, indicating that sampling variability is an additional reason why the estimates differed by cohort. However, given the above caveats, when validation cohorts 1 and 2 were combined with the discovery cohort, the 3 genes of interest remained associated with children having a CSA < 70.
Having identified SNVs within 3 genes associated with severe cognitive impairment of children, the identification of a potential biological mechanism for each of these genes was important to establish a gene-function relationship that can explain these findings. Because inflammation is thought to be an important mechanism that drives HAND, we sought to determine if the genes identified were potentially involved with inflammation within the CNS. Consistent with the hypothesis that increased inflammation would lead to an increased risk for neurocognitive impairment and decreased inflammation would lead to a decreased risk, knockdown of RETREG1 and YWHAH were associated with increased NLRP3 inflammasome activation, while knockdown of CCRL2 was associated with decreased NLRP3 activation. Additionally, we found that knockdown of RETREG1, YWHAH, and CCRL2 alter autophagy, which is an important regulator of NLRP3 inflammasome activity [23, 45].
In summary, 3 SNVs in a dominant genetic model have been identified within CCRL2, RETREG1, and YWHAH that are associated with neurocognitive impairment of children perinatally infected with HIV. Whereas a variant of CCRL2 is associated with a decreased risk of cognitive impairment and decreased inflammation, variants of RETREG1 and YWHAH are associated with an increased risk of cognitive impairment and increased inflammation. These findings suggest that therapeutic approaches that decrease inflammation may be of benefit to HIV-infected children with impaired cognitive function.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author contributions. S. A. S., S. S. B., P. R., P. L. W., G. R. S., and R. B. V. designed the study. K. A. F. and O. H. performed genome-wide exome sequencing. P. R. and R. N. T. performed laboratory assays. S. A. S., S. S. B., K. K. S., P. R., J. K., and S. N. performed analysis of data and data interpretation. All authors contributed to writing and editing of manuscript, and approved submission of the manuscript.
Disclaimer. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Financial support. This work was supported by the National Institute of Neurological Disorders and Stroke, National Institutes of Health (grant numbers R01 NS084912 and R01 NS104015 to S. A. S.). The Pediatric HIV/AIDS Cohort Study was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development; with cofunding from the National Institute on Drug Abuse, National Institute of Allergy and Infectious Diseases, Office of AIDS Research, National Institute of Mental Health, National Institute of Neurological Disorders and Stroke, National Institute on Deafness and Other Communication Disorders, National Heart, Lung, and Blood Institute, National Institute of Dental and Craniofacial Research, and National Institute on Alcohol Abuse and Alcoholism, through cooperative agreements with the Harvard T.H. Chan School of Public Health (grant number HD052102) and the Tulane University School of Medicine (grant number HD052104) and the International Maternal Pediatric Adolescent AIDS Clinical Trials Network. The International Maternal Pediatric Adolescent AIDS Clinical Trials Network was supported by the National Institute of Allergy and Infectious Diseases (grant numbers UM1AI068632 [IMPAACT LOC], UM1AI068616 [IMPAACT SDMC], and UM1AI106716 [IMPAACT LC]); National Institute of Child Health and Human Development, and National Institute of Mental Health.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Presented in part: Conference on Retroviruses and Opportunistic Infections, 4–7 March 2019, Seattle, WA.
References
- 1.Saylor D, Dickens AM, Sacktor N, et al. . HIV-associated neurocognitive disorder–pathogenesis and prospects for treatment. Nat Rev Neurol 2016; 12:234–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.González-Scarano F, Martín-García J. The neuropathogenesis of AIDS. Nat Rev Immunol 2005; 5:69–81. [DOI] [PubMed] [Google Scholar]
- 3.Kaul M, Garden GA, Lipton SA. Pathways to neuronal injury and apoptosis in HIV-associated dementia. Nature 2001; 410:988–94. [DOI] [PubMed] [Google Scholar]
- 4.Levine AJ, Panos SE, Horvath S. Genetic, transcriptomic, and epigenetic studies of HIV-associated neurocognitive disorder. J Acquir Immune Defic Syndr 2014; 65:481–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jia P, Zhao Z, Hulgan T, et al. ; CHARTER Study Group . Genome-wide association study of HIV-associated neurocognitive disorder (HAND): a CHARTER group study. Am J Med Genet B Neuropsychiatr Genet 2017; 174:413–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levine AJ, Service S, Miller EN, et al. . Genome-wide association study of neurocognitive impairment and dementia in HIV-infected adults. Am J Med Genet B Neuropsychiatr Genet 2012; 159B:669–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Englund JA, Baker CJ, Raskino C, et al. . Zidovudine, didanosine, or both as the initial treatment for symptomatic HIV-infected children. AIDS Clinical Trials Group (ACTG) Study 152 Team. N Engl J Med 1997; 336:1704–12. [DOI] [PubMed] [Google Scholar]
- 8.McKinney RE Jr, Johnson GM, Stanley K, et al. . A randomized study of combined zidovudine-lamivudine versus didanosine monotherapy in children with symptomatic therapy-naive HIV-1 infection. The Pediatric AIDS Clinical Trials Group Protocol 300 Study Team. J Pediatr 1998; 133:500–8. [DOI] [PubMed] [Google Scholar]
- 9.Nachman SA, Stanley K, Yogev R, et al. . Nucleoside analogs plus ritonavir in stable antiretroviral therapy-experienced HIV-infected children: a randomized controlled trial. Pediatric AIDS Clinical Trials Group 338 Study Team. JAMA 2000; 283:492–8. [DOI] [PubMed] [Google Scholar]
- 10.Wiznia A, Stanley K, Krogstad P, et al. . Combination nucleoside analog reverse transcriptase inhibitor(s) plus nevirapine, nelfinavir, or ritonavir in stable antiretroviral therapy-experienced HIV-infected children: week 24 results of a randomized controlled trial–PACTG 377. Pediatric AIDS Clinical Trials Group 377 Study Team. AIDS Res Hum Retroviruses 2000; 16:1113–21. [DOI] [PubMed] [Google Scholar]
- 11.Crowell CS, Huo Y, Tassiopoulos K, et al. ; PACTG 219C Study Team and the Pediatric HIVAIDS Cohort Study (PHACS) . Early viral suppression improves neurocognitive outcomes in HIV-infected children. AIDS 2015; 29:295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rawat P, Spector SA. Development and characterization of a human microglia cell model of HIV-1 infection. J Neurovirol 2017; 23:33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001; 25:402–8. [DOI] [PubMed] [Google Scholar]
- 14.Campbell GR, Bruckman RS, Chu YL, Trout RN, Spector SA. SMAC mimetics induce autophagy-dependent apoptosis of HIV-1-infected resting memory CD4+ T cells. Cell Host Microbe 2018; 24:689–702.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rawat P, Teodorof-Diedrich C, Spector SA. Human immunodeficiency virus type-1 single-stranded RNA activates the NLRP3 inflammasome and impairs autophagic clearance of damaged mitochondria in human microglia. Glia 2019; 67:802–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bandera A, Masetti M, Fabbiani M, et al. . The NLRP3 inflammasome is upregulated in HIV-infected antiretroviral therapy-treated individuals with defective immune recovery. Front Immunol 2018; 9:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brouwer N, Zuurman MW, Wei T, Ransohoff RM, Boddeke HW, Biber K. Induction of glial L-CCR mRNA expression in spinal cord and brain in experimental autoimmune encephalomyelitis. Glia 2004; 46:84–94. [DOI] [PubMed] [Google Scholar]
- 18.Galligan CL, Matsuyama W, Matsukawa A, et al. . Up-regulated expression and activation of the orphan chemokine receptor, CCRL2, in rheumatoid arthritis. Arthritis Rheum 2004; 50:1806–14. [DOI] [PubMed] [Google Scholar]
- 19.Yin F, Xu Z, Wang Z, et al. . Elevated chemokine CC-motif receptor-like 2 (CCRL2) promotes cell migration and invasion in glioblastoma. Biochem Biophys Res Commun 2012; 429:168–72. [DOI] [PubMed] [Google Scholar]
- 20.Del Prete A, Bonecchi R, Vecchi A, Mantovani A, Sozzani S. CCRL2, a fringe member of the atypical chemoattractant receptor family. Eur J Immunol 2013; 43:1418–22. [DOI] [PubMed] [Google Scholar]
- 21.Bondue B, Wittamer V, Parmentier M. Chemerin and its receptors in leukocyte trafficking, inflammation and metabolism. Cytokine Growth Factor Rev 2011; 22:331–8. [DOI] [PubMed] [Google Scholar]
- 22.Yoshimura T, Oppenheim JJ. Chemokine-like receptor 1 (CMKLR1) and chemokine (C-C motif) receptor-like 2 (CCRL2); two multifunctional receptors with unusual properties. Exp Cell Res 2011; 317:674–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi CS, Shenderov K, Huang NN, et al. . Activation of autophagy by inflammatory signals limits IL-1β production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol 2012; 13:255–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki T, Franchi L, Toma C, et al. . Differential regulation of caspase-1 activation, pyroptosis, and autophagy via Ipaf and ASC in Shigella-infected macrophages. PLoS Pathog 2007; 3:e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu J, Nagasu H, Murakami T, et al. . Inflammasome activation leads to caspase-1-dependent mitochondrial damage and block of mitophagy. Proc Natl Acad Sci U S A 2014; 111:15514–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khaminets A, Heinrich T, Mari M, et al. . Regulation of endoplasmic reticulum turnover by selective autophagy. Nature 2015; 522:354–8. [DOI] [PubMed] [Google Scholar]
- 27.Rubinsztein DC. Cell biology: receptors for selective recycling. Nature 2015; 522:291–2. [DOI] [PubMed] [Google Scholar]
- 28.Islam F, Gopalan V, Lam AK. RETREG1 (FAM134B): a new player in human diseases: 15 years after the discovery in cancer. J Cell Physiol 2018; 233:4479–89. [DOI] [PubMed] [Google Scholar]
- 29.Chiramel AI, Dougherty JD, Nair V, Robertson SJ, Best SM. FAM134B, the selective autophagy receptor for endoplasmic reticulum turnover, inhibits replication of Ebola virus strains Makona and Mayinga. J Infect Dis 2016; 214:319–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lennemann NJ, Coyne CB. Dengue and Zika viruses subvert reticulophagy by NS2B3-mediated cleavage of FAM134B. Autophagy 2017; 13:322–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ichimura T, Isobe T, Okuyama T, et al. . Molecular cloning of cDNA coding for brain-specific 14-3-3 protein, a protein kinase-dependent activator of tyrosine and tryptophan hydroxylases. Proc Natl Acad Sci U S A 1988; 85:7084–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muslin AJ, Tanner JW, Allen PM, Shaw AS. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 1996; 84:889–97. [DOI] [PubMed] [Google Scholar]
- 33.Yaffe MB, Rittinger K, Volinia S, et al. . The structural basis for 14-3-3:phosphopeptide binding specificity. Cell 1997; 91:961–71. [DOI] [PubMed] [Google Scholar]
- 34.Foote M, Zhou Y. 14-3-3 proteins in neurological disorders. Int J Biochem Mol Biol 2012; 3:152–64. [PMC free article] [PubMed] [Google Scholar]
- 35.Maksymowych WP, Naides SJ, Bykerk V, et al. . Serum 14-3-3η is a novel marker that complements current serological measurements to enhance detection of patients with rheumatoid arthritis. J Rheumatol 2014; 41:2104–13. [DOI] [PubMed] [Google Scholar]
- 36.Wakabayashi H, Yano M, Tachikawa N, Oka S, Maeda M, Kido H. Increased concentrations of 14-3-3 epsilon, gamma and zeta isoforms in cerebrospinal fluid of AIDS patients with neuronal destruction. Clin Chim Acta 2001; 312:97–105. [DOI] [PubMed] [Google Scholar]
- 37.Morales D, Hechavarria R, Wojna V, Acevedo SF. YWHAE/14-3-3ε: a potential novel genetic risk factor and CSF biomarker for HIV neurocognitive impairment. J Neurovirol 2013; 19:471–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu Y, Ren J, He X, Chen H, Wei T, Feng W. YWHA/14-3-3 proteins recognize phosphorylated TFEB by a noncanonical mode for controlling TFEB cytoplasmic localization. Autophagy 2019; 15:1017–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alvarez S, Jiménez JL, Serramía MJ, González M, Cantó-Nogués C, Muñoz-Fernández MA. Lack of association of HIV-1 biological or molecular properties with neurotropism for brain cells. J Mol Neurosci 2006; 29:131–44. [DOI] [PubMed] [Google Scholar]
- 40.Dunfee R, Thomas ER, Gorry PR, Wang J, Ancuta P, Gabuzda D. Mechanisms of HIV-1 neurotropism. Curr HIV Res 2006; 4:267–78. [DOI] [PubMed] [Google Scholar]
- 41.González-Scarano F, Strizki JM, Albright A, Shieh J. Use of primary CNS cultures to investigate HIV neurotropism. J Neurovirol 1997; 3 (Suppl 1):S11–3. [PubMed] [Google Scholar]
- 42.Power C, McArthur JC, Johnson RT, et al. . Distinct HIV-1 env sequences are associated with neurotropism and neurovirulence. Curr Top Microbiol Immunol 1995; 202:89–104. [DOI] [PubMed] [Google Scholar]
- 43.O’Brien WA. Genetic and biologic basis of HIV-1 neurotropism. Res Publ Assoc Res Nerv Ment Dis 1994; 72:47–70. [PubMed] [Google Scholar]
- 44.Olivier IS, Cacabelos R, Naidoo V. Risk factors and pathogenesis of HIV-associated neurocognitive disorder: the role of host genetics. Int J Mol Sci 2018; 19:3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakahira K, Haspel JA, Rathinam VA, et al. . Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol 2011; 12:222–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.