Abstract
Body composition imaging relies on assessment of tissues composition and distribution. Quantitative data provided by body composition imaging analysis have been linked to pathogenesis, risk, and clinical outcomes of a wide spectrum of diseases, including cardiovascular and oncologic. Manual segmentation of imaging data allows to obtain information on abdominal adipose tissue; however, this procedure can be cumbersome and time-consuming. On the other hand, quantitative imaging analysis based on artificial intelligence (AI) has been proposed as a fast and reliable automatic technique for segmentation of abdominal adipose tissue compartments, possibly improving the current standard of care. AI holds the potential to extract quantitative data from computed tomography (CT) and magnetic resonance (MR) images, which in most of the cases are acquired for other purposes. This information is of great importance for physicians dealing with a wide spectrum of diseases, including cardiovascular and oncologic, for the assessment of risk, pathogenesis, clinical outcomes, response to treatments, and complications. In this review we summarize the available evidence on AI algorithms aimed to the segmentation of visceral and subcutaneous adipose tissue compartments on CT and MR images.
Keywords: Obesity, visceral adiposity, artificial intelligence (AI), deep learning, machine learning
Introduction
Body composition imaging relies on assessment of tissues composition and distribution. Quantitative data of body composition have been linked to pathogenesis and clinical outcomes of a wide spectrum of diseases, including oncological and cardiovascular.
Obesity classification is based on body mass index (BMI) which has the shortcoming not to provide any information on the distribution of adipose tissue and skeletal muscle tissue, nor does it allow to distinguish the two main compartments of abdominal adipose tissue: visceral adipose tissue (VAT) and subcutaneous adipose tissue (SAT). Indeed, patients with the same BMI may have remarkably different amount and distribution of VAT and SAT.
Abdominal obesity is a widespread disorder among the population of all ages, related to various pathological conditions such as cardiovascular disease, metabolic syndrome, diabetes, chronic inflammation, and cancer (1).
It would be of paramount importance for clinicians to stratify patients according to different risk profiles based on obesity phenotypes (e.g., visceral metabolic or non-visceral “benign” obesity).
Computed tomography (CT) and magnetic resonance (MR) imaging (MRI) allow for non-invasive and precise measurement of body composition (2). However, segmentation of the abdominal compartments is not routinely performed since the procedure is cumbersome and time-consuming (3). Different methods for automated and reproducible segmentation of body composition have been proposed. However, due to the complex anatomy of the abdominal compartments, time-consuming manual correction of automatic segmentation is required in most of the cases (3). To address this issue, single slice area segmentation technique on a two-dimensional (2D) abdominal CT image has been proposed, showing good correlation with three-dimensional (3D) data of the entire volume (3). Indeed, it has been shown that segmentation of VAT area on a single slice (located 3 cm above the lower margin of L3 vertebra) correlates with VAT volume and body weight, being a good method for VAT segmentation despite limitations of abdominal anatomy and inter/intra-reader variabilities (4).
Therefore, a fully automated segmentation method for abdominal adipose tissue analysis would be helpful in order to improve accuracy and speed-up the process.
For instance, an early work by Kullberg et al. in 2007 reported on a fully automatic segmentation technique for VAT and SAT on abdominal MR images, excluding intramuscular adipose tissue and bone marrow (5). The method was found to be reproducible, but accuracy was limited in lean subjects (5).
More recently, AMRA profiler (AMRA Medical AB, Linköping, Sweden) has been proposed as a fast and accurate strategy to perform body composition analysis, including adipose tissue, on MR images (6).
Artificial intelligence (AI), including machine learning and deep learning, is a breakthrough technology which has been recently proposed as a solution to obtain automatic and reliable analysis of abdominal adipose tissue (7,8). Machine learning is a class of techniques allowing for computer learning aimed to extrapolate or classify models (7). Machines can also be capable of analyze a series of big data and extract features beyond the human skills (8). Deep learning is a subset of artificial neural network (ANN) algorithms organized with a structure composed by multiple layers to obtain high-level abstractions in the data (9-12) (Figure 1).
Figure 1.
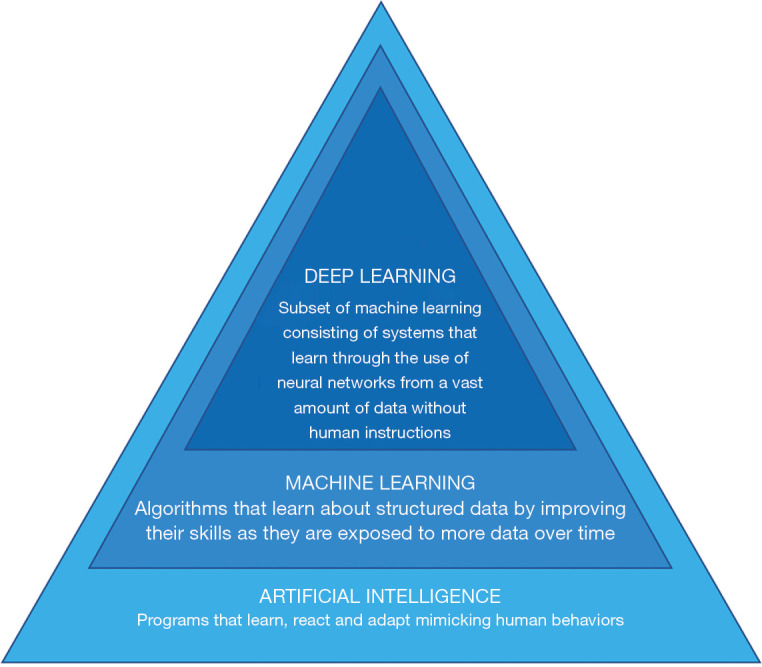
Scheme of AI backbone with subsets: machine learning and deep learning. AI, artificial intelligence.
In this review we summarize the available evidence on the importance of adipose tissue and AI algorithms aimed to segmentation of VAT and SAT compartments on CT and MR images.
Abdominal adipose tissue
Adipose tissue was believed to be a long-term energy depot organ from which free fatty acids can be released to meet the body’s energy needs during fasting. More recently, it has become clear that adipose tissue is to be considered as an endocrine organ that possesses a dynamic and complex activity aimed to regulate whole body homeostasis (13,14).
The adipocytes are cells with metabolic activity expressed by secreting a great number of adipokines active in the regulation of glucose and lipid metabolism, appetite, cardiovascular homeostasis, inflammatory, immune, and reproductive functions, as well as other fundamental biological and physiological functions (15,16).
Abdominal adipose tissue is divided into two main compartments: VAT and SAT. These two compartments with different molecular, biological, and anatomical compositions, have a different meaning and importance. While SAT shows a greater activity for long-term energy storage, VAT has a greater metabolic and hormonal activity through the release of adipokines (17,18). Increased VAT has an impact on health, being connected with a wide spectrum of pathologies.
Patients with a higher amount of VAT show a greater risk of developing insulin resistance (19) and metabolic syndrome (20). Indeed, excessive accumulation of VAT is associated with metabolic disorders such as hyperinsulinemia, hyperglycaemia, hypertriglyceridemia, increased apolipoproteins B-rich lipoproteins, impaired glucose tolerance, constellation of features linked to insulin resistance syndrome (17).
Visceral obesity, hyperinsulinemia and insulin resistance often precede the onset of the metabolic syndrome (21). Elevated blood pressure values in viscerally obese individuals are associated to insulin resistance and compensative hyperinsulinemia (22). High blood pressure can also be induced by visceral obesity through increased activity of the adipose tissue renin-angiotensin-aldosterone system (17).
Increased VAT has also been correlated with cardiovascular disorders. For instance, a correlation was found with hypercoagulability due to increased secretion of plasminogen activator inhibitor 1 (PAI-1), increased intima-media thickness and peripheral arterial disease (23,24). Furthermore, hyperinsulinemia in a clinical picture of visceral obesity is a predictor of coronary artery disease (25). Obese individuals with increased VAT are at risk of developing cardiovascular diseases and type 2 diabetes (26).
Adipokines, growth factors and proinflammatory cytokines secreted by VAT are considered mediating factors linked to the development of obesity-related tumors (27,28). Indeed, specific fingerprints of abdominal adipose tissue amount and distribution have been associated to several disorders (including oncological), in terms of risk, intra-tumoral genetic mutations, prognosis, and post-treatment effects and complications (29-35).
Skeletal muscle
Cachexia is defined as progressive functional deterioration characterized by loss of adipose tissue and skeletal muscle that cannot be fully compensated with nutrition (36).
The loss of skeletal muscle mass is called sarcopenia and can be associated to cancer or chronic diseases including chronic kidney disease (37,38).
Muscle mass quantification is of great clinical importance since sarcopenia correlates with several adverse clinical outcomes (39). This assessment can be performed by means of several imaging technique and using variable methodologies (40-46). For instance, quantitative evaluation of total skeletal muscle area at L3 level or psoas muscle area only are common approaches, being also these two measures correlated one to another (47). Multiple research studies established that psoas muscle area, measured on medical images, is a robust biomarker of sarcopenia showing association with conditions including osteoporotic fractures risk in the elderly, mortality after liver transplantation, mortality after ruptured aneurysm surgery and cortisol levels in patients with hypercortisolism (48-51).
Thus, quantification of adipose tissue distribution as well as of skeletal muscle mass are both relevant targets to be included, separate or combined, in the assessment of body composition.
Artificial intelligence
Machine learning
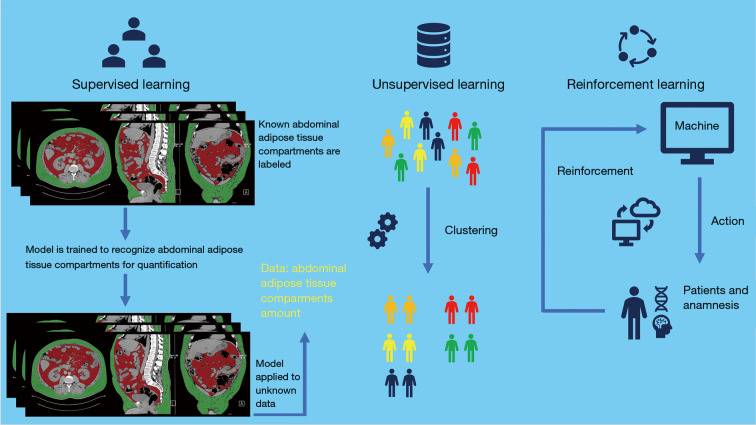
Machine learning is based on algorithms that allow computers to learn and make predictions without programming by explicit rules, possibly improving as a function of experience (7). Depending on task to be accomplished, machine learning is classified into three categories: supervised, unsupervised and reinforcement learning (Figure 2) (7). Supervised machine learning is so called as it consists of training supervision, in which data labels are supplied to the algorithm. In this category of machine learning the expected outputs are usually labeled by human experts. The major function of the algorithm is to associate inputs to outputs.
Figure 2.
Schematic structure of the three sub-categories of machine learning: supervised, unsupervised and reinforcement, according to the task to be accomplished.
In unsupervised machine learning no data labels are supplied to the algorithm. Machine learning aims to find the hidden structure in the data and separate them into clusters or groups.
In reinforcement machine learning, a certain task is performed by the computer program in a dynamic environment in which positive or negative reinforcement feedback is received. In this category, learning is provided by interactions with an environment without explicit instruction (52).
Artificial neural networks
ANNs are subset of automatic learning made up of statistical and mathematical models, inspired to biological nervous system processes information through a number of elements called neurons linked together by multiple connections (i.e., nodes and edges) (7). This type of network is made up of “hidden layers” and an output layer, which in turn consists of neurons connected to all the other neurons of the layers (7). Learning in this type of network can be supervised, partially supervised, or unsupervised (7).
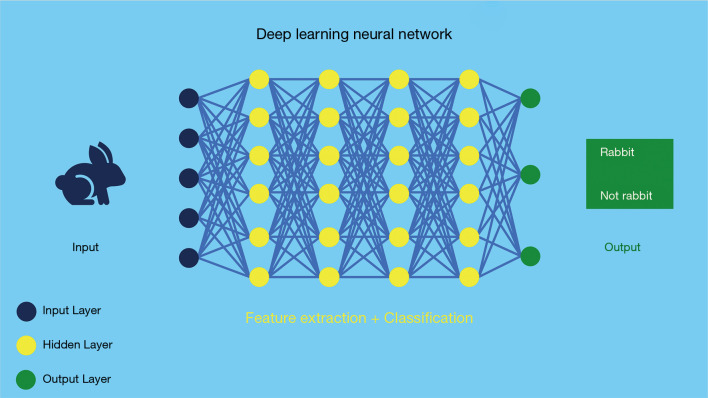
Deep learning
Deep learning is a subset of neural networks algorithms that contains several hidden (hence “deep”) layers. These are algorithms seek to model high-level abstractions in the data. For instance, deep learning is typically applied for recognizing objects in images (Figure 3).
Figure 3.
Deep learning model architecture with neural network and multiple hidden layers. This model includes a complex system of neural networks capable of learning tasks without human training.
There are different deep learning models: there are 1D convolutional neural networks (CNNs) in popular deep learning framework (i.e., pytorch and tensorflow) and 2D or 3D models. These two different models are called fully connected neural network and CNNs respectively, being the latter the most commonly applied learning technique in the medical imaging field.
Methods
The literature research was performed on December 2020 using MEDLINE PubMed Central, taking into account only articles written in English and without limits of time span. Combinations of keywords for articles search were: “adipose tissue artificial intelligence”, “adipose tissue deep learning” or “adipose tissue machine learning”. Relevant articles related to abdominal adipose tissue and AI were selected also from the reference list of each identified article.
AI and abdominal adipose tissue
Mounting evidence in recent years evaluated the automated abdominal adipose tissue segmentation on CT and MRI scans by means of machine learning and deep learning algorithms. On this respect, the Dice Score is a common metric to assess the spatial overlap between the predicted label maps and the ground truth. It provides both size and localization consensus for any type of method, not only AI.
Dice coefficient is the most commonly used tool in order to validate AI-derived algorithms tailored to image segmentation (Figure 4) (53).
Figure 4.
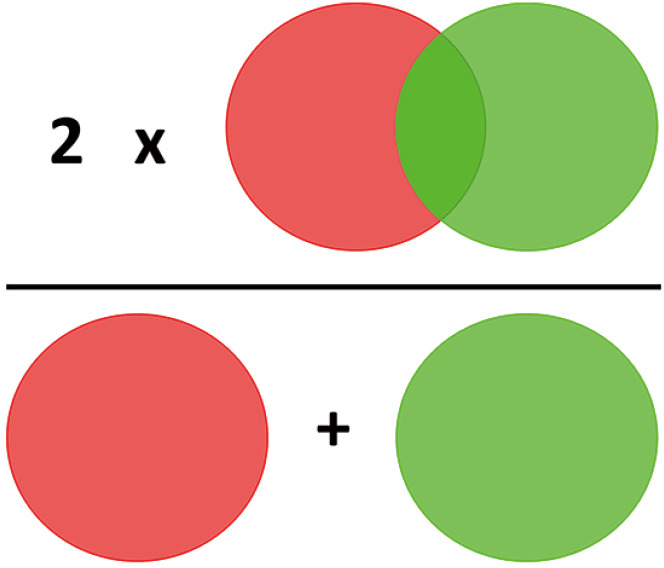
Scheme to explain how Dice coefficient is calculated. The light red and light green circles highlight the concept of 2*overlapped pixels (top row) divided by total number of pixels (bottom row).
Computed tomography
Several papers investigated the impact of AI on the abdominal fat CT-based segmentation (Table 1).
Table 1. Summary of CT comparative studies with proposed networks and results of abdominal VAT and SAT quantification.
| Authors | AI category | Method | Number of patients | Segmentation level | VAT Dice scores (mean ± standard deviation) | SAT Dice scores (mean ± standard deviation) |
|---|---|---|---|---|---|---|
| Weston et al. (2019) (3) | Deep learning (supervised) | U-Net | 2,369 | Transverse processes of the L3 | – | 0.98±0.05 |
| Nowak et al. (2020) (54) | Deep learning (supervised) | Deep19 model | 1,143 | L3/L4 | 0.950±0.053 | 0.974±0.041 |
| Koitka et al. (2021) (55) | Deep learning (supervised) | U-Net 3D | 50 | Whole abdomen | – | 0.9953 |
| Park et al. (2020) (56) | Deep learning (supervised) | FCN-based segmentation system | 946 | Lower somatic limiting of L3 | 0.97 (for internal and external datasets) | 0.97 (for internal and external datasets) |
| Dabiri et al. (2020) (57) | Deep learning (supervised) | U-Net | 2,529 | L3 | Dataset-2 (test set), Dataset-2 (training set): 98.6±4.31 | Dataset-2 (test set), Dataset-2 (training set): 98.94±3.96 |
| Dataset-2 (test set), Dataset-3 (training set): 98.03±4.85 | Dataset-2 (test set), Dataset-3 (training set): 98.16±4.25 | |||||
| Dataset-3 (test set), Dataset-2 (training set) 97.46±4.27 | Dataset-3 (test set), Dataset-2 (training set): 98.85±1.04 | |||||
| Dataset-3 (test set), Dataset-3 (training set): 97.92±3.90 | Dataset-3 (test set), Dataset-3 (training set): 99.16±0.91 | |||||
| Hemke et al. (2020) (58) | Deep learning (supervised) | U-Net | 200 | Immediately cranial to the acetabular roof | – | 500 (augmentation) 0.95 |
| 1,000 (augmentation) 0.97 | ||||||
| 2,000 (augmentation) 0.96 | ||||||
| 3,000a (augmentation) 0.97 | ||||||
| 4,000 (augmentation) 0.97 | ||||||
| Makrogiannis et al. (2013) (59) | Machine learning (supervised) | Fuzzy C-Means; GVF | 168 | L3/L4 | 85.6±5.7 | 93.1±2.9 |
| Wang et al. (2019) (60) | Deep learning-machine learning fusion (supervised) | SVM; DilaLab; DilaLabPlus | 20 | Whole abdomen | DilaLab 0.9278±0.0158 | DilaLabPlus 0.9808±0.0063 |
| Wang et al. (2017) (61) | Deep learning (supervised) | Selection-CNN; Segmentation-CNN | 40 | Whole abdomen | 0.9150±0.0624 | 0.9797±0.0145 |
a, model with highest Dice scores for soft tissue classes used for final experiments. CT, computed tomography; CNN, convolutional neural network; FCN, fully convolutional network; GVF, gradient vector flow; SAT, subcutaneous adipose tissue; SVM, support vector machine; VAT, visceral adipose tissue.
Weston et al. proposed an automatic approach for abdominal segmentation using deep CNN (3). This deep learning model allowed to segment VAT, SAT, muscle, bone and abdominal organs, matching or even outperforming the accuracy reached by manual segmentation (3). Segmentation was performed at the level of L3 vertebra transverse processes (3). When comparing U-Net segmentation and semi-automatic segmentation, no significant differences in performance were found (3). VAT had a larger standard deviation than the other compartments (3). The VAT error was greater, indicating that this algorithm works well and demonstrating that this model is at least as accurate as manual segmentation, regardless patient weight (3). Moreover, model performance exceeded inter-observer agreement on the SAT (3).
Blanc-Durand et al. extracted measurements of VAT, SAT and muscular body mass from CT scan using a deep learning approach, in order to evaluate progression-free survival and overall survival in non-small cell lung cancer patients (62). A five-fold cross-validation was performed in a subset of 35 patients, subsequently the anthropometric data were normalized to the body surface area, to control the different morphologies of the patients (62).
Interestingly, VAT/SAT ratio normalized to the body surface area was shown to be an independent predictor of progression-free survival and overall survival in non-small cell lung cancer patients (62).
Nowak et al. evaluated the accuracy of the body segmentation of VAT, SAT and skeletal muscles using DeepMedic Open-Source CNN, obtaining extremely accurate results in all three body compartments. The analysis was performed on single slice images at the L3/L4 level. Of note, DeepMedic (i.e., deep learning) offers open-source free access to their code or their trained models ready for use in clinical research studies (54).
Koitka et al. developed a deep-learning solution using multi-resolutions U-Net 3D neural networks, a completely automatic and reproducible method for quantifying the volume of body tissue composition. They quantified abdominal cavity, bones, muscle, SAT, and thoracic cavity. Subsequently, the authors subclassified adipose tissue and muscle using Hounsfield unit (HU) thresholds.
One limitation of the study was the differentiation between VAT and adipose tissue within organs. Adipose tissue in the abdominal cavity, having all the same range of HU, was considered entirely as VAT including adipose tissue within organs which should be excluded from the statistics as, by definition, is not part of VAT (55).
Orlov et al. used supervised machine learning and low-level generic image descriptors to evaluate age-related change in adipose tissue, soft tissues (neither bone nor fat) and bone using CT in single slice image at L4 level, in order to assess effects of biological aging and to correlate these changes with metabolic and functional activity. Adipose tissue was shown to be a good aging indicator in both genders. The strongest aging indicator in males was the greater abdominal adipose tissue distribution, while adipose tissue in females was a close second to the fat distribution of the entire torso, presenting a more widespread distribution. Therefore, adipose tissue was found to be a stronger aging indicator in males, despite in this study the adipose tissue compartments, VAT and SAT, were not segmented separately (63).
Park et al. aimed to develop and validate a method of automatic segmentation of adipose and abdominal muscle tissues using a deep learning system, with segmentations at the level of the lower aspect of L3.
The ground truth data consisted of manually segmented maps of abdominal VAT, abdominal SAT, and abdominal muscle tissue. This system, based on fully convolutional network (FCN)-based segmentation, showed good performance and high accuracy in recognizing and quantitatively analyze these tissues (56).
Another deep learning method for the automatic segmentation of VAT, SAT, abdominal muscle and intermuscular adipose tissue was proposed by Dabiri et al. Segmentations were performed on a single slice at L3 level. Three datasets of patients with different types of cancer were used for networks training and validation.
The middle L3 slice was detected by CT scan volume and subsequently sent to the five-class segmentation network (skeletal muscle, SAT, VAT, intermuscular adipose tissue and background). This multi-label segmentation network was created to refine U-Net and FCN to take advantage of both networks and limit the disadvantages. The inputs to this network consist of the L3 axial images, while the targets consist of the corresponding masks of the skeletal muscle, VAT, SAT and intermuscular adipose tissue regions.
The authors suggested that localization and segmentation network showed potential for fully automated VAT, SAT, abdominal muscle and analysis with high accuracy (57).
Hemke et al. developed a deep CNN for the automatic segmentation of body composition (i.e., VAT, SAT, intermuscular adipose tissue, bone and mixed intra-pelvic content) in the pelvic area, precisely at the supra-acetabular level. This model was trained for 50 epochs using the U-Net architecture. The results showed, once again, accurate automated segmentation for pelvic body composition (58).
Makrogiannis et al. used a machine learning method integrating spatial information, intensity and texture for VAT and SAT quantification, allowing for identification and removal of food residues, showing very good accuracy and generalization capacity (59).
Wang et al. provided a novel three-level machine learning technique for the automatic quantification of VAT and SAT. In particular, a Support Vector Machine classifier configured to group abdominal CT images was applied. Consequently, a CNN-based pyramidal dilation network (DilaLab) was used to address the complex internal distribution problems of the non-abdominal internal adipose tissue of the VAT segmentation. Finally, DilaLabPlus network (i.e., the first dilated convolution layer from DilaLab and plus a decoder module) was used for the quantification of the SAT. This method demonstrated improvement in the feasibility and reliability of the automatic quantification of VAT and SAT (60).
Moreover, Wang et al. used a deep learning CNN based computer-aided detection scheme for abdominal VAT and SAT quantification. This scheme was based on a two-step process: abdominal CT slices selection and VAT and SAT areas segmentation. This methodology allowed to process CT volumetric data with high concordance and accuracy as compared to manual segmentation (61).
Finally, Pickhardt et al. used a deep learning approach for quantification of VAT and SAT at L1 level in patients with metabolic syndrome. Fully automated quantitative tissue measures of VAT and SAT, combined with other data (i.e., muscle and liver) obtained from abdominal CT scans, allowed to identify patients at risk for metabolic syndrome (64).
Taken together, available data about abdominal fat segmentation on CT images suggest that AI is a powerful and reliable tool to quickly quantify VAT and SAT.
Magnetic resonance imaging
The impact of AI to improve the process of abdominal fat quantitative analysis has been recently assessed on research studies based on MR images (Table 2).
Table 2. Summary of MR comparative studies with proposed networks and results of abdominal VAT and SAT quantification.
| Authors | AI category | Method | Number of patients | Sequences | Segmentation level | VAT Dice scores (mean ± standard deviation) | SAT Dice scores (mean ± standard deviation) |
|---|---|---|---|---|---|---|---|
| Langner et al. (2019) (65) | Deep learning (supervised) | U-Net; V-Net | Study Tellus: 10; Study BetaJudo: 10 | Study Tellus: mDixon Quant; Study BetaJudo: mDixon Quant | L3–L4 | U-Net: Study Tellus, 0.988±0.007; Study BetaJudo, 0.970±0.010 | U-Net: Study Tellus, 0.992±0.003; Study BetaJudo, 0.987±0.004 |
| V-Net: Study Tellus, 0.982±0.009; Study BetaJudo, 0.916±0.059 | V-Net: Study Tellus, 0.987±0.004; Study BetaJudo, 0.978±0.012 | ||||||
| Küstner et al. (2020) (66) | Deep learning (semisupervised) | DCNet | 300 | Dixon | Whole body | TUEF/DZD (1.5T): 0.92±0.02 | TUEF/DZD (1.5T): 0.98±0.02 |
| TUEF/DZD (3.0T): 0.86±0.06 | TUEF/DZD (3.0T): 0.94±0.03 | ||||||
| NAKO (3.0T): 0.89±0.05 | NAKO (3.0T): 0.95±0.05 | ||||||
| Estrada et al. (2019) (67) | Deep learning (supervised) | FatSegNet | 587 | Dixon | Abdominal region: from inferior somatic margin of twelfth thoracic vertebra (Th12) to the inferior somatic margin of L5 | Axial: 0.826 | Axial: 0.970 |
| Thoracic region: all above the inferior somatic margin of Th12 | Coronal: 0.826 | Coronal: 0.966 | |||||
| Pelvic region: everything below the lower somatic margin of L5 | Sagittal: 0.824 | Sagittal: 0.966 | |||||
| View aggregation: 0.850 | View aggregation: 0.975 | ||||||
| Shen et al. (2019) (68) | Deep learning (supervised and unsupervised) | U-Net | 75 | IDEAL-IQ | Not specified | 0.96; 0.97 based on AFK-MC2 | 0.96 |
| Masoudi et al. (2020) (1) | Deep learning (supervised and unsupervised) | C-GAN; U-Net | 165 | Fat saturated T1-weighted | Whole abdomen | 0.9433 | 0.9746 |
MR, magnetic resonance imaging; DZD, German Center for Diabetes Research database; NAKO, German National Cohort database; AFK-MC2, Markov chain Monte Carlo; SAT, subcutaneous adipose tissue; TUEF, Tuebingen Family Study database; VAT, visceral adipose tissue.
Konigorski et al. used a fully automated approach for the segmentation of the VAT, SAT and for adipose tissue located in the coronary compartment. This system mirrored results of manual segmentation and was reproducible. The quantity of adipose tissue compartments was then correlated with gene expression in SAT and concentration of adipokines. The major result was a correlation between plasma levels of leptin and the amount of adipose tissue, in particular of SAT (69).
Langner et al. used two different neural networks architectures (i.e., U-Net and V-Net) for automatic segmentation of VAT and SAT, in multicenter water-fat scans of the abdomen. U-Net showed a higher performance than V-Net, enabling reliable automatic segmentation of VAT and SAT. Furthermore, with a standardized imaging protocol, this method could be proposed for multi-centric studies including a large number of subjects from different geographic places (65).
Küstner et al. quantified the different compartments of adipose tissue from whole-body MRI, using a 3D densely connected CNN (DCNet) that was compared to a similar 3D U-Net segmentation effectively testing for parameters including precision, sensitivity, specificity and accuracy. The proposed DCNet allowed to perform segmentation of adipose tissue in 3D whole body MRI data sets and was proposed as generalizable for different epidemiological cohort studies (66).
Estrada et al. introduced a new deep learning method that allows for fast and accurate automatic segmentation of VAT and SAT on Dixon MRI scans (FatSegNet). Three phases make up FatSegNet: localization of the abdominal region through two 2D Competitive Dense Fully Convolutional Networks (CDFNet), segmentation of adipose tissue on three views by independent CDFNets, and view aggregation. CDFNet showed greater accuracy and robustness than traditional deep learning networks. FatSegNet was reported as a reliable tool to perform volumetric segmentation of VAT and SAT on 3D Dixon MRI scan (67).
Shen et al. proposed a machine learning-deep learning fusion method for the segmentation of VAT and SAT, based on iterative decomposition of water and fat with echo asymmetry and least square estimation-iron quantification (IDEAL-IQ) technology. This method, which showed high reliability, comprised two main phases: first, a deep network segmented the boundaries of SAT and the contour of the peritoneal cavity; second, the assumption-free method K-means ++ with a Markov chain Monte Carlo (AFK-MC2) clustering was used for VAT quantification (68).
Masoudi et al. developed an innovative algorithm for the automatic quantification of adipose tissue on MR images, based on deep learning techniques, through cross modality adaptation. Specifically, a cycle generative adversarial network (C-GAN) was used to transform the MR scans into their equivalent synthetic CT (s-CT) images. On these images, the segmentation of adipose tissue is easier due to the descriptive nature of HU for CT images (1).
Finally, Rawshani et al. used a machine learning approach to quantify VAT and SAT, on T1 weighted axial images, as potential predictors of cardio-metabolic risk in non-obese male subjects. VAT was found to be a strong predictor of ectopic fat accumulation in the liver and heart (70).
Research studies aimed to quantitatively analyze abdominal fat on MR images, suggest that AI is a promising, robust and reliable innovation, although less explored than applications on CT images up to date.
Impact of AI for body composition analysis in obesity
Several CT and MRI studies of the abdomen are acquired on a daily basis worldwide. These studies, that are mainly carried out for different purposes, contain important “hidden” information that are overlooked and not taken into account in most of the cases.
Work overload is one of the major constraints that hinder a systematic quantitative analysis of body composition. AI holds the potential to solve this issue and possibly enables body composition analysis, including abdominal fat compartments, with quantitative data to be included in all radiological reports. This solution is potentially able to improve the current standard of care with minimal additional cost to the healthcare system and almost no additional workload to physicians.
Various approaches with different AI algorithms have been proposed for the segmentation of abdominal fat compartments. The proposed algorithms, for both CT and MR, allow fast segmentation with accuracy similar or even higher with respect to manual segmentation, making the solutions applicable to clinical practice with a possible positive impact on patient’s health. Moreover, AI is able to provide a rapid whole abdomen assessment which can be more accurate and less variable with respect to single-slice approach for segmentation of VAT and SAT, especially in order to follow-up patients undergoing weight changes (71).
Since individual body components are related to cardio-metabolic risks, the value of assessing VAT and SAT is not the only factor to be taken into account in risk assessment. Indeed, ectopic fat also can have a great impact on metabolism and inflammation. On this respect, AI-derived algorithms to assess ectopic fat in the liver and the pancreas can also be of importance for cardio-metabolic risk assessment.
A meta-analysis evaluating AI-assisted models for analysis of liver fibrosis and steatosis, reported only two studies on AI-assisted diagnosis of non-alcoholic fatty liver disease (NAFLD), with liver biopsy as gold standard (72-74). These two studies used ANN and CNN as AI models, on mixed and obese populations for calculating the amount of fat in the liver through the use of ultrasound, respectively (73,74).
Although these two studies have shown high sensitivity and specificity, the method used for data acquisition does not allow for a correct quantification of VAT and SAT.
To the best of our knowledge, AI-derived algorithms for quantification of ectopic fat in the pancreas through imaging were never explored up to date.
“Functional body composition” is a new concept of body composition, integrating body components with regulatory systems. Adipose tissue and skeletal muscle mass are both determinants of energy consumption at rest. Imbalance between body components and function is linked to metabolic imbalance occurring in response to weight changes and variable diseases (75). Furthermore, the ratio between trunk SAT and VAT and the ratio of VAT (or SAT) and skeletal muscle mass (i.e., the so-called load-capacity model) it should be considered due to cardio-metabolic impact (76).
Future developments
There is no doubt that the use of AI solutions geared to the quantification of body composition will increase progressively over the next few years. In terms of cardio-metabolic risk assessment, it would be important to develop AI-derived algorithms that allow for quantification of fat in the liver and the pancreas together with analysis of functional body composition. In the context of precision medicine, future studied will validate AI algorithms geared to quantitative assessment of body tissues composition on CT and MR images, to obtain impactful information in terms of risk assessment, patient-tailored pre- and post-treatment evaluation, and longitudinal follow-up, for multiple diseases including cardio-vascular and oncological.
Conclusions
AI is quickly emerging as breakthrough innovation for abdominal fat quantitative analysis on CT and MR images, which in most of the cases are acquired for other purposes. Progressively improving algorithms hold the potential to extract quantitative data from existing images, allowing physicians to obtain important information with minimal additional cost to the healthcare system and almost no additional workload to physicians.
This information can have an impact on several diseases, including cardiovascular and oncologic, in terms of risk, pathogenesis, clinical outcomes, response to treatments, and complications, possibly improving the current standard of care.
Acknowledgments
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
Footnotes
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/qims-21-370). The authors have no conflicts of interest to declare.
References
- 1.Masoudi S, Anwar SM, Harmon SA, Choyke PL, Turkbey B, Bagci U. Adipose Tissue Segmentation in Unlabeled Abdomen MRI using Cross Modality Domain Adaptation. Annu Int Conf IEEE Eng Med Biol Soc 2020;2020:1624-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seabolt LA, Welch EB, Silver HJ. Imaging methods for analyzing body composition in human obesity and cardiometabolic disease. Ann N Y Acad Sci 2015;1353:41-59. 10.1111/nyas.12842 [DOI] [PubMed] [Google Scholar]
- 3.Weston AD, Korfiatis P, Kline TL, Philbrick KA, Kostandy P, Sakinis T, Sugimoto M, Takahashi N, Erickson BJ. Automated Abdominal Segmentation of CT Scans for Body Composition Analysis Using Deep Learning. Radiology 2019;290:669-79. 10.1148/radiol.2018181432 [DOI] [PubMed] [Google Scholar]
- 4.Noumura Y, Kamishima T, Sutherland K, Nishimura H. Visceral adipose tissue area measurement at a single level: can it represent visceral adipose tissue volume? Br J Radiol 2017;90:20170253. 10.1259/bjr.20170253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kullberg J, Ahlström H, Johansson L, Frimmel H. Automated and reproducible segmentation of visceral and subcutaneous adipose tissue from abdominal MRI. Int J Obes (Lond) 2007;31:1806-17. 10.1038/sj.ijo.0803671 [DOI] [PubMed] [Google Scholar]
- 6.Borga M, Thomas EL, Romu T, Rosander J, Fitzpatrick J, Leinhard OD, Bell JD. Validation of a fast method for quantification of intra-abdominal and subcutaneous adipose tissue for large-scale human studies. NMR Biomed 2015;28:1747-53. 10.1002/nbm.3432 [DOI] [PubMed] [Google Scholar]
- 7.Choy G, Khalizadeh O, Michalski M, Do S, Samir AE, Pianykh OS, Geis JR, Pandharipande PV, Brink JA, Dreyer KJ. Current Applications and Future Impact of Machine Learning in Radiology. Radiology 2018;288:318-28. 10.1148/radiol.2018171820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang S, Summers RM. Machine learning and radiology. Med Image Anal 2012;16:933-51. 10.1016/j.media.2012.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.LeCun Y, Bengio Y, Hinton G.Deep learning. Nature 2015;521:436-44. 10.1038/nature14539 [DOI] [PubMed] [Google Scholar]
- 10.Quattrocchi CC, Mallio CA, Presti G, Beomonte Zobel B, Cardinale J, Iozzino M, Della Sala SW. The challenge of COVID-19 low disease prevalence for artificial intelligence models: report of 1,610 patients. Quant Imaging Med Surg 2020;10:1891-3. 10.21037/qims-20-782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mallio CA, Quattrocchi CC, Beomonte Zobel B, Parizel PM. Artificial intelligence, chest radiographs, and radiology trainees: a powerful combination to enhance the future of radiologists? Quant Imaging Med Surg 2021;11:2204-7. 10.21037/qims-20-1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mallio CA, Napolitano A, Castiello G, Giordano FM, D’Alessio P, Iozzino M, Sun Y, Angeletti S, Russano M, Santini D, Tonini G, Beomonte Zobel B, Vincenzi B, Quattrocchi CC. Deep Learning Algorithm Trained with COVID-19 Pneumonia Also Identifies Immune Checkpoint Inhibitor Therapy-Related Pneumonitis. Cancers (Basel) 2021;13:652. 10.3390/cancers13040652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galic S, Oakhill JS, Steinberg GR. Adipose tissue as an endocrine organ. Mol Cell Endocrinol 2010;316:129-39. 10.1016/j.mce.2009.08.018 [DOI] [PubMed] [Google Scholar]
- 14.Scherer PE. Adipose tissue: from lipid storage compartment to endocrine organ. Diabetes 2006;55:1537-45. 10.2337/db06-0263 [DOI] [PubMed] [Google Scholar]
- 15.Frühbeck G, Gómez-Ambrosi J. Control of body weight: a physiologic and transgenic perspective. Diabetologia 2003;46:143-72. 10.1007/s00125-003-1053-4 [DOI] [PubMed] [Google Scholar]
- 16.Rodríguez A, Ezquerro S, Méndez-Gimenez L, Becerril S, Frühbeck G.Revisiting the adipocyte: a model for integration of cytokine signaling in the regulation of energy metabolism. Am J Physiol Endocrinol Metab 2015;309:E691-714. 10.1152/ajpendo.00297.2015 [DOI] [PubMed] [Google Scholar]
- 17.Ibrahim MM. Subcutaneous and visceral adipose tissue: structural and functional differences. Obes Rev 2010;11:11-8. 10.1111/j.1467-789X.2009.00623.x [DOI] [PubMed] [Google Scholar]
- 18.Fox CS, Massaro JM, Hoffmann U, Pou KM, Maurovich-Horvat P, Liu CY, Vasan RS, Murabito JM, Meigs JB, Cupples LA, D'Agostino RB, Sr, O'Donnell CJ. Abdominal visceral and subcutaneous adipose tissue compartments: association with metabolic risk factors in the Framingham Heart Study. Circulation 2007;116:39-48. 10.1161/CIRCULATIONAHA.106.675355 [DOI] [PubMed] [Google Scholar]
- 19.Kissebah AH, Vydelingum N, Murray R, Evans DJ, Hartz AJ, Kalkhoff RK, Adams PW. Relation of body fat distribution to metabolic complications of obesity. J Clin Endocrinol Metab 1982;54:254-60. 10.1210/jcem-54-2-254 [DOI] [PubMed] [Google Scholar]
- 20.Després JP, Moorjani S, Lupien PJ, Tremblay A, Nadeau A, Bouchard C. Regional distribution of body fat, plasma lipoproteins, and cardiovascular disease. Arteriosclerosis 1990;10:497-511. 10.1161/01.ATV.10.4.497 [DOI] [PubMed] [Google Scholar]
- 21.Lemieux S, Despres SP. Metabolic complications of visceral obesity: contribution to the etiology of life of type 2 diabetes and implications for prevention and treatment. Diabete Metab 1994;20:375-93. [PubMed] [Google Scholar]
- 22.Sharma AM, Engeli S, Pischon T. New developments in mechansims of obesity-induced hypertension: role of adipose tissue. Curr Hypertens Rep 2001;3:152-6. 10.1007/s11906-001-0030-x [DOI] [PubMed] [Google Scholar]
- 23.Harris MM, Stevens J, Thomas N, Schreiner P, Folsom AR. Association of fat distribution and obesity with hypertension in a bi-ethnic population: the ARIC study. Atherosclerosis Risk in Communities Study. Obes Res 2000;8:516-24. 10.1038/oby.2000.64 [DOI] [PubMed] [Google Scholar]
- 24.Planas A, Clará A, Pou JM, Vidal-Barraquer F, Gasol A, de Moner A, Contreras C, Marrugat J. Relationship of obesity distribution to peripheral arterial occlusive disease in elderly men. Int J Obes Relat Metab Disord 2001;25:1068-70. 10.1038/sj.ijo.0801638 [DOI] [PubMed] [Google Scholar]
- 25.Fontbonne A, Charles MA, Thibult N, Richard JL, Claude JR, Warnet JM, Rosselin GE, Eschwège E. Hyperinsulinaemia as a predictor of coronary heart disease mortality in a healthy population: the Paris Prospective Study, 15-year follow-up. Diabetologia 1991;34:356-61. 10.1007/BF00405009 [DOI] [PubMed] [Google Scholar]
- 26.McFarlane SI, Banerji M, Sowers JR. Insulin resistance and cardiovascular disease. J Clin Endocrinol Metab 2001;86:713-8. [DOI] [PubMed] [Google Scholar]
- 27.Després JP, Lemieux I. Abdominal obesity and metabolic syndrome. Nature 2006;444:881-7. 10.1038/nature05488 [DOI] [PubMed] [Google Scholar]
- 28.Zhang HP, Zou J, Xu ZQ, Ruan J, Yang SD, Yin Y, Mu HJ. Association of leptin, visfatin, apelin, resistin and adiponectin with clear cell renal cell carcinoma. Oncol Lett 2017;13:463-8. 10.3892/ol.2016.5408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seguro LPC, Paupitz JA, Caparbo VF, Bonfa E, Pereira RMR. Increased visceral adipose tissue and altered adiposity fistribution in premenopausal lupus patients: correlation with cardiovascular risk factors. Lupus 2018;27:1001-6. 10.1177/0961203318758504 [DOI] [PubMed] [Google Scholar]
- 30.Greco F, Cirimele V, Mallio CA, Beomonte Zobel B, Grasso RF. Increased visceral adipose tissue in male patients with clear cell renal cell carcinoma. Clin Cancer Investig J 2018;7:132-6. 10.4103/ccij.ccij_62_18 [DOI] [Google Scholar]
- 31.Greco F, Mallio CA, Grippo R, Messina L, Vallese S, Rabitti C, Quarta LG, Grasso RF, Beomonte Zobel B. Increased visceral adipose tissue in male patients with non-clear cell renal cell carcinoma. Radiol Med 2020;125:538-43. 10.1007/s11547-020-01146-6 [DOI] [PubMed] [Google Scholar]
- 32.Greco F, Quarta LG, Grasso RF, Beomonte Zobel B, Mallio CA. Increased visceral adipose tissue in clear cell renal cell carcinoma with and without peritumoral collateral vessels. Br J Radiol 2020;93:20200334. 10.1259/bjr.20200334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greco F, Mallio CA, Cirimele V, Grasso RF, Beomonte Zobel B. Subcutaneous adipose tissue as a biomarker of pancreatic cancer: A pilot study in male patients. Clin Cancer Investig J 2019;8:10-9. 10.4103/ccij.ccij_36_19 [DOI] [Google Scholar]
- 34.Mallio CA, Greco F, Pacella G, Schena E, Beomonte Zobel B. Gender-based differences of abdominal adipose tissue distribution in non-small cell lung cancer patients. Shanghai Chest 2018;2:20. 10.21037/shc.2018.03.03 [DOI] [Google Scholar]
- 35.Greco F, Mallio CA. Relationship Between Visceral Adipose Tissue and Genetic Mutations (VHL and KDM5C) in Clear Cell Renal Cell Carcinoma. Radiol Med 2021;126:645-51. 10.1007/s11547-020-01310-y [DOI] [PubMed] [Google Scholar]
- 36.Tisdale MJ. Mechanisms of cancer cachexia. Physiol Rev 2009;89:381-410. 10.1152/physrev.00016.2008 [DOI] [PubMed] [Google Scholar]
- 37.Martin L, Birdsell L, Macdonald N, Reiman T, Clandinin MT, McCargar LJ, Murphy R, Ghosh S, Sawyer MB, Baracos VE. Cancer cachexia in the age of obesity: skeletal muscle depletion is a powerful prognostic factor, independent of body mass index. J Clin Oncol 2013;31:1539-47. 10.1200/JCO.2012.45.2722 [DOI] [PubMed] [Google Scholar]
- 38.Sabatino A, D'Alessandro C, Regolisti G, di Mario F, Guglielmi G, Bazzocchi A, Fiaccadori E. Muscle mass assessment in renal disease: the role of imaging techniques. Quant Imaging Med Surg 2020;10:1672-86. 10.21037/qims.2020.03.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guglielmi G, Bazzocchi A.Body composition imaging. Quant Imaging Med Surg 2020;10:1576-9. 10.21037/qims-2019-bc-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guerri S, Mercatelli D, Aparisi Gómez MP, Napoli A, Battista G, Guglielmi G, Bazzocchi A. Quantitative imaging techniques for the assessment of osteoporosis and sarcopenia. Quant Imaging Med Surg 2018;8:60-85. 10.21037/qims.2018.01.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giraudo C, Cavaliere A, Lupi A, Guglielmi G, Quaia E. Established paths and new avenues: a review of the main radiological techniques for investigating sarcopenia. Quant Imaging Med Surg 2020;10:1602-13. 10.21037/qims.2019.12.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Messina C, Albano D, Gitto S, Tofanelli L, Bazzocchi A, Ulivieri FM, Guglielmi G, Sconfienza LM. Body composition with dual energy X-ray absorptiometry: from basics to new tools. Quant Imaging Med Surg 2020;10:1687-98. 10.21037/qims.2020.03.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ponti F, De Cinque A, Fazio N, Napoli A, Guglielmi G, Bazzocchi A. Ultrasound imaging, a stethoscope for body composition assessment. Quant Imaging Med Surg 2020;10:1699-722. 10.21037/qims-19-1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cornacchia S, La Tegola L, Maldera A, Pierpaoli E, Tupputi U, Ricatti G, Eusebi L, Salerno S, Guglielmi G. Radiation protection in non-ionizing and ionizing body composition assessment procedures. Quant Imaging Med Surg 2020;10:1723-38. 10.21037/qims-19-1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huber FA, Del Grande F, Rizzo S, Guglielmi G, Guggenberger R. MRI in the assessment of adipose tissues and muscle composition: how to use it. Quant Imaging Med Surg 2020;10:1636-49. 10.21037/qims.2020.02.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ponti F, Soverini V, Plazzi A, Aparisi Gómez MP, Mercatelli D, Guglielmi G, Battista G, Marchesini G, Bazzocchi A. DXA-assessed changes in body composition in obese women following two different weight loss programs. Nutrition 2018;46:13-9. 10.1016/j.nut.2017.07.016 [DOI] [PubMed] [Google Scholar]
- 47.Hamaguchi Y, Kaido T, Okumura S, Kobayashi A, Hammad A, Tamai Y, Inagaki N, Uemoto S. Proposal for new diagnostic criteria for low skeletal muscle mass based on computed tomography imaging in Asian adults. Nutrition 2016;32:1200-5. 10.1016/j.nut.2016.04.003 [DOI] [PubMed] [Google Scholar]
- 48.Sheu Y, Marshall LM, Holton KF, Caserotti P, Boudreau RM, Strotmeyer ES, Cawthon PM, Cauley JA. Abdominal body composition measured by quantitative computed tomography and risk of non-spine fractures: the Osteoporotic Fractures in Men (MrOS) Study. Osteoporos Int 2013;24:2231-41. 10.1007/s00198-013-2322-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Englesbe MJ, Patel SP, He K, Lynch RJ, Schaubel DE, Harbaugh C, Holcombe SA, Wang SC, Segev DL, Sonnenday CJ. Sarcopenia and mortality after liver transplantation. J Am Coll Surg 2010;211:271-8. 10.1016/j.jamcollsurg.2010.03.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee JS, He K, Harbaugh CM, Schaubel DE, Sonnenday CJ, Wang SC, Englesbe MJ, Eliason JL, Michigan Analytic Morphomics group (MAMG). Frailty, core muscle size, and mortality in patients undergoing open abdominal aortic aneurysm repair. J Vasc Surg 2011;53:912-7. 10.1016/j.jvs.2010.10.111 [DOI] [PubMed] [Google Scholar]
- 51.Miller BS, Ignatoski KM, Daignault S, Lindland C, Gauger PG, Doherty GM, Wang SC, University of Michigan Analytical Morphomics group . A quantitative tool to assess degree of sarcopenia objectively in patients with hypercortisolism. Surgery 2011;150:1178-85. 10.1016/j.surg.2011.09.020 [DOI] [PubMed] [Google Scholar]
- 52.Mohri M, Rostamizadeh A, Talwalkar A. Foundations of machine learning. Cambridge, Massachusetts: MIT Press, 2012. [Google Scholar]
- 53.Yao AD, Cheng DL, Pan I, Kitamura F. Deep Learning in Neuroradiology: A Systematic Review of Current Algorithms and Approaches for the New Wave of Imaging Technology. Radiol Artif Intell 2020;2:e190026. 10.1148/ryai.2020190026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nowak S, Faron A, Luetkens JA, Geißler HL, Praktiknjo M, Block W, Thomas D, Sprinkart AM. Fully Automated Segmentation of Connective Tissue Compartments for CT-Based Body Composition Analysis: A Deep Learning Approach. Invest Radiol 2020;55:357-66. 10.1097/RLI.0000000000000647 [DOI] [PubMed] [Google Scholar]
- 55.Koitka S, Kroll L, Malamutmann E, Oezcelik A, Nensa F. Fully automated body composition analysis in routine CT imaging using 3D semantic segmentation convolutional neural networks. Eur Radiol 2021;31:1795-804. 10.1007/s00330-020-07147-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park HJ, Shin Y, Park J, Kim H, Lee IS, Seo DW, Huh J, Lee TY, Park T, Lee J, Kim KW. Development and Validation of a Deep Learning System for Segmentation of Abdominal Muscle and Fat on Computed Tomography. Korean J Radiol 2020;21:88-100. 10.3348/kjr.2019.0470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dabiri S, Popuri K, Ma C, Chow V, Feliciano EMC, Caan BJ, Baracos VE, Beg MF. Deep learning method for localization and segmentation of abdominal CT. Comput Med Imaging Graph 2020;85:101776. 10.1016/j.compmedimag.2020.101776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hemke R, Buckless CG, Tsao A, Wang B, Torriani M. Deep learning for automated segmentation of pelvic muscles, fat, and bone from CT studies for body composition assessment. Skeletal Radiol 2020;49:387-95. 10.1007/s00256-019-03289-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Makrogiannis S, Caturegli G, Davatzikos C, Ferrucci L. Computer-aided assessment of regional abdominal fat with food residue removal in CT. Acad Radiol 2013;20:1413-21. 10.1016/j.acra.2013.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Z, Meng Y, Weng F, Chen Y, Lu F, Liu X, Hou M, Zhang J.An Effective CNN Method for Fully Automated Segmenting Subcutaneous and Visceral Adipose Tissue on CT Scans. Ann Biomed Eng 2020;48:312-28. 10.1007/s10439-019-02349-3 [DOI] [PubMed] [Google Scholar]
- 61.Wang Y, Qiu Y, Thai T, Moore K, Liu H, Zheng B.A two-step convolutional neural network based computer-aided detection scheme for automatically segmenting adipose tissue volume depicting on CT images. Comput Methods Programs Biomed 2017;144:97-104. 10.1016/j.cmpb.2017.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Blanc-Durand P, Campedel L, Mule S, Jegou S, Luciani A, Pigneur F, Itti E. Prognostic value of anthropometric measures extracted from whole-body CT using deep learning in patients with non-small-cell lung cancer. Eur Radiol 2020;30:3528-37. 10.1007/s00330-019-06630-w [DOI] [PubMed] [Google Scholar]
- 63.Orlov NV, Makrogiannis S, Ferrucci L, Goldberg IG. Differential Aging Signals in Abdominal CT Scans. Acad Radiol 2017;24:1535-43. 10.1016/j.acra.2017.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pickhardt PJ, Graffy PM, Zea R, Lee SJ, Liu J, Sandfort V, Summers RM. Utilizing Fully Automated Abdominal CT-Based Biomarkers for Opportunistic Screening for Metabolic Syndrome in Adults Without Symptoms. AJR Am J Roentgenol 2021;216:85-92. 10.2214/AJR.20.23049 [DOI] [PubMed] [Google Scholar]
- 65.Langner T, Hedström A, Mörwald K, Weghuber D, Forslund A, Bergsten P, Ahlström H, Kullberg J. Fully convolutional networks for automated segmentation of abdominal adipose tissue depots in multicenter water-fat MRI. Magn Reson Med 2019;81:2736-45. 10.1002/mrm.27550 [DOI] [PubMed] [Google Scholar]
- 66.Küstner T, Hepp T, Fischer M, Schwartz M, Fritsche A, Häring HU, Nikolaou K, Bamberg F, Yang B, Schick F, Gatidis S, Machann J. Fully Automated and Standardized Segmentation of Adipose Tissue Compartments via Deep Learning in 3D Whole-Body MRI of Epidemiologic Cohort Studies. Radiol Artif Intell 2020;2:e200010. 10.1148/ryai.2020200010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Estrada S, Lu R, Conjeti S, Orozco-Ruiz X, Panos-Willuhn J, Breteler MMB, Reuter M. FatSegNet: A fully automated deep learning pipeline for adipose tissue segmentation on abdominal dixon MRI. Magn Reson Med 2020;83:1471-83. 10.1002/mrm.28022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shen N, Li X, Zheng S, Zhang L, Fu Y, Liu X, Li M, Li J, Guo S, Zhang H. Automated and accurate quantification of subcutaneous and visceral adipose tissue from magnetic resonance imaging based on machine learning. Magn Reson Imaging 2019;64:28-36. 10.1016/j.mri.2019.04.007 [DOI] [PubMed] [Google Scholar]
- 69.Konigorski S, Janke J, Drogan D, Bergmann MM, Hierholzer J, Kaaks R, Boeing H, Pischon T. Prediction of Circulating Adipokine Levels Based on Body Fat Compartments and Adipose Tissue Gene Expression. Obes Facts 2019;12:590-605. 10.1159/000502117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rawshani A, Eliasson B, Rawshani A, Henninger J, Mardinoglu A, Carlsson Å, Sohlin M, Ljungberg M, Hammarstedt A, Rosengren A, Smith U.Adipose tissue morphology, imaging and metabolomics predicting cardiometabolic risk and family history of type 2 diabetes in non-obese men. Sci Rep 2020;10:9973. 10.1038/s41598-020-66199-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schweitzer L, Geisler C, Pourhassan M, Braun W, Glüer CC, Bosy-Westphal A, Müller MJ. What is the best reference site for a single MRI slice to assess whole-body skeletal muscle and adipose tissue volumes in healthy adults? Am J Clin Nutr 2015;102:58-65. 10.3945/ajcn.115.111203 [DOI] [PubMed] [Google Scholar]
- 72.Decharatanachart P, Chaiteerakij R, Tiyarattanachai T, Treeprasertsuk S. Application of artificial intelligence in chronic liver diseases: a systematic review and meta-analysis. BMC Gastroenterol 2021;21:10. 10.1186/s12876-020-01585-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Byra M, Styczynski G, Szmigielski C, Kalinowski P, Michałowski Ł, Paluszkiewicz R, Ziarkiewicz-Wróblewska B, Zieniewicz K, Sobieraj P, Nowicki A.Transfer learning with deep convolutional neural network for liver steatosis assessment in ultrasound images. Int J Comput Assist Radiol Surg 2018;13:1895-903. 10.1007/s11548-018-1843-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kuppili V, Biswas M, Sreekumar A, Suri HS, Saba L, Edla DR, Marinho RT, Sanches JM, Suri JS. Extreme Learning Machine Framework for Risk Stratification of Fatty Liver Disease Using Ultrasound Tissue Characterization. J Med Syst 2017;41:152. 10.1007/s10916-017-0797-1 [DOI] [PubMed] [Google Scholar]
- 75.Müller MJ, Bosy-Westphal A, Later W, Haas V, Heller M. Functional body composition: insights into the regulation of energy metabolism and some clinical applications. Eur J Clin Nutr 2009;63:1045-56. 10.1038/ejcn.2009.55 [DOI] [PubMed] [Google Scholar]
- 76.Siervo M, Prado CM, Mire E, Broyles S, Wells JCK, Heymsfield S, Katzmarzyk PT. Body composition indices of a load-capacity model: gender- and BMI-specific reference curves. Public Health Nutr 2015;18:1245-54. 10.1017/S1368980014001918 [DOI] [PMC free article] [PubMed] [Google Scholar]