Abstract
Osteocytes are the primary mechanosensitive cells in bone. However, their location in mineralized matrix has limited the in vivo study of osteocytic genes induced by mechanical loading. Laser Capture Microdissection (LCM) allows isolation of intracortical bone (Intra-CB), enriched for osteocytes, from bone tissue for gene expression analysis. We used microarray to analyze gene expression from mouse tibial Intra-CB dissected using LCM 4 hours after a single loading bout or after 5 days of loading. Osteocyte enrichment was supported by greater expression of Sost, Dmp1, Dkk1, and Mepe in Intra-CB regions vs. Mixed regions containing periosteum and muscle (fold-change (FC) = 3.4, 2.2, 5.1, 3.0, respectively). Over 150 differentially expressed genes (DEGs) due to loading (loaded vs. contralateral control) in Intra-CB were found on Day 1 and Day 5, but only 10 genes were differentially expressed on both days, including Ngf (Day 1 FC= 13.5, Day 5 FC=11.1) and Wnt1 (Day 1 FC=1.5, Day 5 FC=5.1). The expression of Ngf and Wnt1 within Intra-CB was confirmed by in situ hybridization, and a significant increase in number of Wnt1 mRNA molecules occurred on day 1. We also found changes in extracellular matrix remodeling with Timp1 (FC=3.1) increased on day 1 and MMP13 (FC=0.3) decreased on day 5. Supporting this result, IHC for osteocytic MMP13 demonstrated a marginal decrease due to loading on day 5. Gene Ontology (GO) biological processes for loading DEGs indicated regulation of vasculature, neuronal and immune processes while cell-type specific gene lists suggested regulation of osteoclast, osteoblast, and endothelial related genes. In summary, microarray analysis of microdissected Intra-CB revealed differential regulation of Ngf, Wnt1, and MMP13 due to loading in osteocytes.
Keywords: Osteocytes, Laser Capture Microdissection, Mechanical Loading, Bone Formation, Gene Expression
2. Introduction
Bone mass is regulated by mechanical loading. Bone loss occurs with conditions like bed rest due to decreased loads on the skeleton [1], while increased skeletal loads can increase bone mass [2]–[4]. These load-adaptive changes are controlled at the cellular level by bone forming osteoblasts, bone resorbing osteoclasts, and mechano-sensitive osteocytes. The importance of osteocytes as the primary mechanosensitive cell in bone has been well described [5]–[7]. Osteocytes make up 90–95% of the cells within bone and are well distributed in a network of dendrites and cell bodies [8], making them efficient at sending signals to osteoblasts at the bone surface. Their position within the bone lacunocanalicular (LCN) network allows them to sense and respond to changes in loading. Mechanisms whereby osteocytes sense and respond to loading have been described, including calcium signaling [9] [10] and release of nitric oxide (NO) and prostaglandins (COX-2), which are critical for loading-induced bone formation [11], [12]. However, the molecules produced by osteocytes that may signal to osteoblasts (and other cells) to regulate bone formation following mechanical loading are still largely unknown [13].
qPCR and RNAseq analysis have substantially increased our understanding of genes expressed by cells within bulk bone tissue following loading. These findings have demonstrated early downregulation of Sost [14] and upregulation of Wnt1 [14], [15], supporting an array of evidence for the importance of Wnt signaling in mechanotransduction [16]–[18]. Sclerostin downregulation due to loading has been localized to osteocytes by IHC [19]. Wnt1 is upregulated in MLOY4 osteocytes in response to fluid-flow shear stress [20], but whether osteocytes upregulate Wnt1 expression due to in vivo loading is unclear. Another gene upregulated in bulk bone tissue by loading is Ngf [14], [15]. Tomlinson et al. showed that loading increased Ngf-EGFP expression in osteoblasts, but not osteocytes, and that depletion of Ngf attenuated Wnt signaling in osteocytes [21]. These latter findings demonstrate the complex interplay between different cell types in bone mechanotransduction.
Bulk bone tissue, processed by removing muscle and marrow, is thought to represent a predominately osteocyte enriched tissue, but remaining active osteoblasts on the surfaces of cortical and cancellous bone also contribute to gene expression results [22]. Thus, it is difficult to ascribe changes in gene expression in bulk bone samples to osteocytes versus osteoblasts, or even to other bone-resident cells (e.g., endothelial, nerve). Laser Capture Microdissection (LCM) is a technique used to isolate specific regions from tissue sections for a more homogeneous cell population. Others have used LCM to confirm osteocyte-specific knock down of genes in transgenic mouse models [23], and to analyze gene expression in osteocytes following drug administration in rats [24], [25]. However, no previous work has extensively analyzed gene expression of laser microdissected osteocyte-enriched tissue in mouse bone.
While LCM allows separation of intracortical osteocytes from surface osteoblasts for gene expression analysis, difficulty obtaining high concentration and high quality RNA from mouse tissue has limited its use for whole-transcriptome analysis. In particular, RNAseq methods require greater mass (~ 1 ng) and quality of RNA than may be feasible with LCM of mouse bone. Alternatively, some contemporary microarray methods, developed for use on clinical isolates and whole blood samples that are typically degraded and low in concentration [26]–[28], [29], may require as little as 100 pg of RNA. Mouse microarrays enable quantitative assessment of over 20,000 genes representing the majority of well-known/annotated transcripts in the mouse genome.
We utilized mouse tibial loading followed by LCM to isolate intracortical bone (Intra-CB) regions and microarray analysis to discover genes that are regulated by loading in osteocyte-enriched samples. We contrasted the expression in the Intra-CB region with a region containing Mixed tissues (bone, periosteum, muscle). We identified genes specific to Intra-CB regulated by loading, performed pathway analysis on DEG lists and investigated loading effects in cell-type specific gene lists.
3. Methods
3.1. Mice and Mechanical Loading
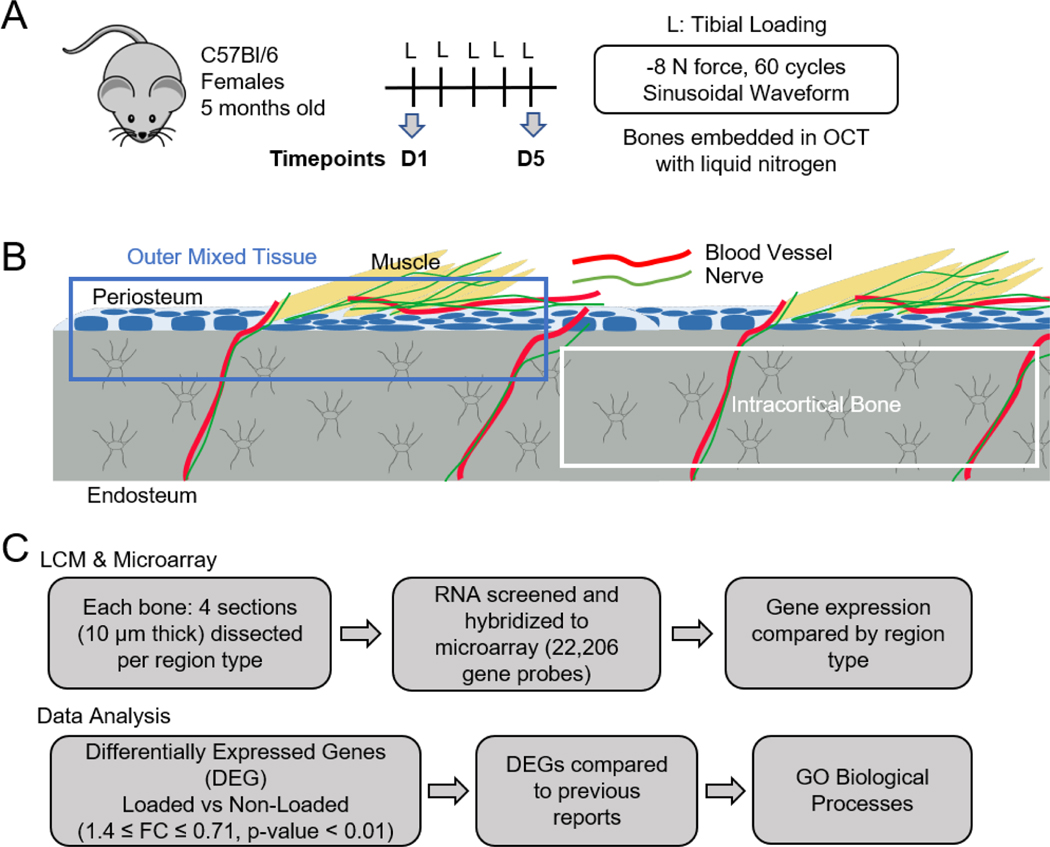
A total of 27 female C57Bl/6NCrl mice were obtained from Charles River and aged to 5 months old. Animals were kept on a 12-hour light/dark cycle and allowed to feed freely on chow containing 20% protein and 5% fat (PicoLab Rodent Diet 20, LabDiet). Mouse strain and sex were chosen for comparison to recently published gene expression studies [15]. Because female C57BL/6 mice are skeletally mature at 5 months [30], they provide a good representation of mechanical loading-induced changes to gene expression in young adults without the confounding effects of growth. Seventeen (17) mice were designated for laser capture microdissection, and 10 for histology.
Axial compression (Electropulse 1000, Instron) was applied to the right hindlimb while mice were under 3% isofluorane. Each loading bout was 60 cycles of a 4 Hz sinusoidal waveform with peak force of −8 N. This force induces a maximal compressive strain magnitude of −2200 με (1200 με peak tensile strain), known to induce lamellar bone formation in young-adult C57Bl/6 female mice [31], [32]. A pre-load of −0.5 N was used for stability of the limb in the device. The contralateral limb was used as a non-loaded control. Following loading, 0.1 mg/kg of buprenorphine was injected subcutaneously for pain management and mice were returned to normal cage activity. Mice were euthanized by CO2 asphyxiation at timepoints described below. All animal experiments were approved by the Washington University IACUC.
3.2. Laser Capture Microdissection
One group (n=9) of mice was sacrificed by CO2 asphyxiation 4 hours after the first loading bout to capture the regulation of genes involved in mechanotransduction shortly after stimulation (Fig. 1A). The second group (n=7, one was excluded due to a lesion on right ankle) was subjected to five daily loading bouts and sacrificed 4 hours after the final bout to represent the genetic regulation occurring during a state of active bone formation [15][16]. Loaded and contralateral control tibias were harvested within 3 min following sacrifice and immediately embedded in Optimal Cutting Temperature compound (Tissue-Tek, Thermo Fisher Scientific) with liquid nitrogen. Bones were kept at −80°C until later use.
Figure 1.
Experimental design and workflow. (A) Flow chart showing experimental parameters for mechanical loading of mice; bones were harvested and embedded 4 h after last loading bout, on either day 1 or day 5. (B) Diagram illustrating Intra-CB (blue outline) and Mixed regions (blue outline) collected for RNA extraction. Yellow=Muscle, Blue=Osteoblasts and Progenitor Cells, Grey=Osteocytes. (C) Workflow from sample collection to microarray data processing and analysis.
For each bone (n=32), eight cryosections 10 μm thick were cut using Kawomoto’s LMD film (Section-Lab Co. Ltd.) and the ends were glued onto a custom slide with the tissue facing down over a window cut into the slide. The sections were kept on dry ice throughout the experiment and LCM was performed on the same day as sectioning to preserve RNA integrity.
A Leica Laser Capture Microdissection microscope (LMD-6000, Leica) was used to dissect two tissue regions, either the intracortical bone (Intra-CB) or outer mixed tissue (Mixed), the latter of which included cortical bone, periosteum, and muscle (Fig. 1B). The Mixed ROI was intended to include the cell types that would contribute to whole-bone samples used for bulk RNA analysis (except marrow), and provided a contrast to the more homogeneous Intra-CB. Because the Mixed ROI includes periosteal osteoblasts, gene expression from these cells was expected to drive many of the changes occurring due to loading in these samples. On the 5x objective, the region of interest (ROI) began approximately 4 mm distal to the tibial plateau (Fig. S1). Laser cuts on the compressive side of the tibia were made on the 10x objective with power set to 45–60, aperture set to 32–35 and frequency set to 1404; the kerf of the laser cut was ~10 μm. The total length of tissue regions dissected was 2–3 mm with a width of approximately 100 μm. The cut-out pieces from each dissected region dropped into a 0.6 ml tube, positioned under the tissue, filled with extraction buffer (Arcturus PicoPure RNA Isolation Kit, Applied Biosystems, Thermo Fisher Scientific). After each section was dissected, the tube was placed on dry ice. Four sections were dissected for each region type. Each section took less than 10 min to dissect and dry ice was used to keep the section cool during the process. Tubes were kept at 80°C until further processing. Residual surface bone tissue was seen on the Intra-CB sections after dissection, which provided a visual check that surface cells were not included in these samples (Fig. S1D). Additional sections were taken from a sample to confirm residual osteoblasts on the slide following dissection of the Intra-CB ROI (Fig. S1F,G). Sections were cut as described, dissected, and Kawomoto’s film was mounted onto glass slides with chitosan adhesive. DAPI staining was performed to localize cells.
Each tube was thawed, vortexed and heated to 42°C for 30 min. Dissected tissue from sections from the same limb and region type were pooled for extraction, resulting in 64 biological samples (from 32 bones; 16 pairs of loaded and control tibias) each eluted in 11 μl of buffer (Arcturus PicoPure RNA Isolation Kit; Applied Biosystems) (Fig. 1C). Agilent RNA 6000 Pico (Bioanalyzer 2100, Agilent Technologies) was used as an initial screening to measure RNA concentration (mean = 126 pg/μl, range: 11 to 644 pg/μl) and RIN (mean = 3, range: 1 to 9). By visual inspection of the bioanalyzer traces, pairs were removed that contained a highly degraded or contaminated sample (Fig. S2). Forty-four (44) samples with adequate Bioanalyzer traces remained for subsequent processing steps (n=6 Intra-CB pairs, n=5 Mixed pairs for each timepoint).
3.3. Microarray
The Genome Technology Access Center (GTAC, Washington University in St. Louis) performed RNA amplification and preparation of RNA, hybridization to microarray chips, and normalization of raw intensity data. GeneChip Whole Transcriptome Pico Kit (Affymetrix, Applied Biosystems, Thermo Fisher Scientific) was used to prepare samples for hybridization. This assay begins with reverse transcription along the length of RNA, making it well suited for samples with low RIN. Total RNA was converted to complementary RNA (cRNA) and amplified for 12 cycles of PCR before amplification with T7 in vitro transcription (IVT) technology. Spectrophotometer (Nanophotometer NP60, Implen) analysis was used to determine if samples needed speed vacuuming (<20 μg, n=5) before continuing. Five samples in total were speed vacuumed. Subsequently, cRNA was converted to ss-cDNA and spectrophotometer analysis confirmed adequate concentration for fragmentation and biotinylation (5.5 μg). Agilent RNA 6000 Nano (Bioanalyzer 2100, Agilent Technologies) traces were visually inspected after ss-cDNA generation for additional quality control before hybridization.
Hybridization of 2.5 μg of ss-cDNA to Clariom S Pico array (Applied Biosystems, Thermo Fisher Scientific) occurred overnight at 45°C for 18 hours in a GeneChip Hybridization Oven 640 (Affymetrix, Thermo Fisher Scientific). The Hybridization, Wash, and Stain Kit for cartridge arrays (Thermo Fisher Scientific) was used to wash and stain the arrays in an Affymetrix Fluidics Station 450 (Thermo Fisher Scientific). Arrays were scanned using an Affymetrix GeneChip 7G 3000 Scanner (Affymetrix, Thermo Fisher Scientific). The Clariom S Pico Assay allowed detection of over 20,000 annotated genes. Probe intensities (.cel files) were normalized and background corrected using Robust Mult-Array Average (RMA) with Partek Genomics Suite (v6.6, Partek, Inc.). Measures of microarray chip quality, including Area Under Curve (AUC) of the Receiver Operator Curve (ROC) and absolute deviation of the residuals from the median (mad residual mean) showed limited RNA degradation and no outliers were identified (Fig. S3).
3.4. Differential Expression Analysis
The normalized intensities (.chp files) were analyzed for differential expression in Transcriptome Analysis Console (TAC) software (Applied Biosystems, Thermo Fisher Scientific). First, Intra-CB gene intensities were compared to Mixed samples. Principal component analysis (PCA) of all the genes and hierarchical clustering analysis of the genes with Log2 intensities greater than 9 (>4,000 genes) were used to confirm primary differences due to region type (Intra-CB vs. Mixed). One sample was not clearly labeled (Day 5 Intra-CB/Mixed) and was removed from analysis, yielding n=4–6 pairs per group/time (Day 1 Intra-CB, n=6; Day 5 Intra-CB, n=5; Day 1 Mixed, n=5; Day 5 Mixed, n=4). Differentially expressed genes (DEGs) due to loading were identified in the Intra-CB and Mixed samples separately at each timepoint. Because differences due to loading were not robust, a relatively permissive DEG criteria was used: fold change (FC, loaded/non-loaded) of FC ≤ 0.71 or ≥ 1.4, p-value < 0.01, using repeated measures for tibias from the same mouse. The eBayes method was used for multiple comparisons and default settings were used for other parameters.
DEG lists and intensity values were exported from TAC and genes shared by region and timepoint were discovered. Lists for each group were compared using the VennDiagram package in R. Loading DEGs were also compared to lists of loading DEGs identified from bulk RNAseq as reported in Chermside-Scabbo et al. [15]. DEGs from Chermside-Scabbo et al. represented bulk tissue gene expression, which included the tibia with muscle removed and marrow spun out; similar mice (female, C57Bl/6, 5 mo), loading parameters (−8 N peak force), and timepoints (4 hr after 1 bout and 5 bouts) were used in their study design [15].
3.5. Gene Ontology and Cell Type Analysis
Differential regulation of biological processes were determined by importing DEG lists into the Gene Ontology (GO) Enrichment Analysis tool (The Gene Ontology Consortium) [33]–[35] and analyzed using Fisher’s Exact test and False Discovery Rate < 0.15. The genes driving each of the top 10 processes (by fold enrichment) were visualized in heatmaps (Prism 8.4.2, GraphPad Software) with their respective loading-induced FC.
Intra-CB regions may include blood vessels that transport immune cells and depend on osteoclastic bone resorption for growth and development [36]. Furthermore, changes in these structures due to disease or drug administration can occur rapidly [36]. To investigate any changes occurring in vasculature due to loading, gene lists for endothelial cells compiled by Coates et al. for RNAseq cell phenotyping [37] were incorporated into our analysis. Additionally, osteocytes have been known to express many genes to remodel their local environment and regulate osteoblasts and osteoclasts [24], [38]–[40]. To determine if osteocyte expression of osteoblast and osteoclast factors are different by timepoint, gene lists associated with osteoblasts and osteoclasts from Coates et al. were also imported into TAC and visualized in heatmaps as previously described. Neuron associated gene lists were also collected from Youlten et al [41] and Paic et al. [42] for analysis.
3.6. In Situ Hybridization and Immunohistochemistry
Loaded and contralateral control tibias were partially trimmed of muscle and fixed in 10% neutral buffered formalin overnight. Bones were washed in PBS and decalcified for 14 days in 14% EDTA (pH=7.4) at 4°C. After full decalcification, bones were paraffin embedded and kept in humidity-controlled containers until sectioning.
Paraffin sections for staining were cut 5 μm thick on Superfrost Plus Microscope Slides (Fisher Scientific) and baked overnight at 60°C. Sections were kept in containers with desiccant until staining. RNA in situ hybridization using RNAscope 2.5HD Duplex Assay (Advanced Cell Diagnostics, Inc.) was performed following manufacturer’s directions with a few modifications. Slides were baked overnight at 37°C before the procedure. Before deparaffinization, slides were baked for 30 min at 60°C. Additional baking steps at 60°C for 10 min were added before and after applying the hydrophobic barrier around the tissue and after target retrieval for 30 min at 60°C. In addition to the baking steps, pepsin treatment was used for target retrieval for 45 min at 40°C in place of the manufacturer’s suggested reagents. Amplification steps 6 and 10 were also increased to 30 min. Ngf (446331) and Wnt1 (401091-C2) probes were selected with channel 1 (C1: HRP-based Green) and channel 2 (C2: AP-based Fast Red), respectively. Positive control probes targeted Ppib (C1) and Polr2a (C2). Negative control probes targeted a bacterial gene (dapB). Both control probe reagents were provided in the Duplex detection kit. Images were acquired at 40x on a Nanozoomer 2.0HT microscope (Hamamatsu) for quantification and on a confocal microscope with a DFC7000 T camera (Leica Microsystems) for closer inspection at 63x. Following initial imaging, the coverslips on a few sections were removed and counterstained with 50% hematoxylin (Hematoxylin Solution, Gill No. 1, Sigma Aldrich) diluted in 0.02% ammonium hydroxide (Sigma Aldrich) after initial imaging, but some loss of staining occurred. NDP.view 2.3 software (Hamamatsu) was used to export 1-mm regions of interest (ROI). The ROI location was approximately 5 mm distal to the tibial plateau (site of peak strain). The precise position was adjusted on some samples to avoid regions with tissue sectioning artifacts, but each tibial pair had matching ROI locations. The number of Ngf (blue) and Wnt1 (pink) positive punctate dots per ROI were counted, as well as the number of lacunae per ROI stained with multiple Ngf or Wnt1 mRNA molecules using BIOQUANT OSTEO software (Bioquant Imaging Analysis Corp.). Samples were blinded.
For immunohistochemistry (IHC), sections were deparaffinized, followed by antigen retrieval incubation with Unitrieve antigen retrieval solution (Innovex Biosciences) in a 60°C water bath for 30 min. Endogenous peroxidases were blocked with 3% H2O2 (Sigma-Aldrich) at room temperature for 10 min and sections were blocked using 20% goat serum (ab7481, Abcam). 1:50 anti-MMP13 rabbit antibody (ab39012, Lot GR3365904–1, Abcam) or normal rabbit IgG (2729S, Lot 8, Cell Signaling Technologies) was applied overnight at 4°C. Sections were incubated in 1:100 goat anti-rabbit HRP secondary antibody (P0448, Lot 20042622, Dako) for 30 min at room temperature. Peroxidase reaction was performed with ImmPACT DAB Substrate Peroxidase kit (SK-4105, Lot ZF0830, Vector Laboratories, Maravai LifeSciences) for 6 min. Sections were counterstained with hematoxylin (Modified Mayer’s, Electron Microscopy Sciences). Images were acquired at 20x on a Nanozoomer 2.0HT microscope. ROIs were obtained as described for RNAscope quantification. Samples were blinded and cells (total and MMP13-positive) within Intra-CB were counted using BIOQUANT OSTEO software.
3.7. Statistics
Statistics for in situ and IHC quantification were conducted using a two-way ANOVA with repeated measures and factors including loading and timepoint. Sidak’s multiple comparisons test was used for post-hoc analysis (Prism 8.4.2, GraphPad Software). Significance was defined as p<0.05, with marginal differences noted at p<0.10.
4. Results
4.1. Regions dissected from Intra-CB are enriched for osteocyte-specific genes
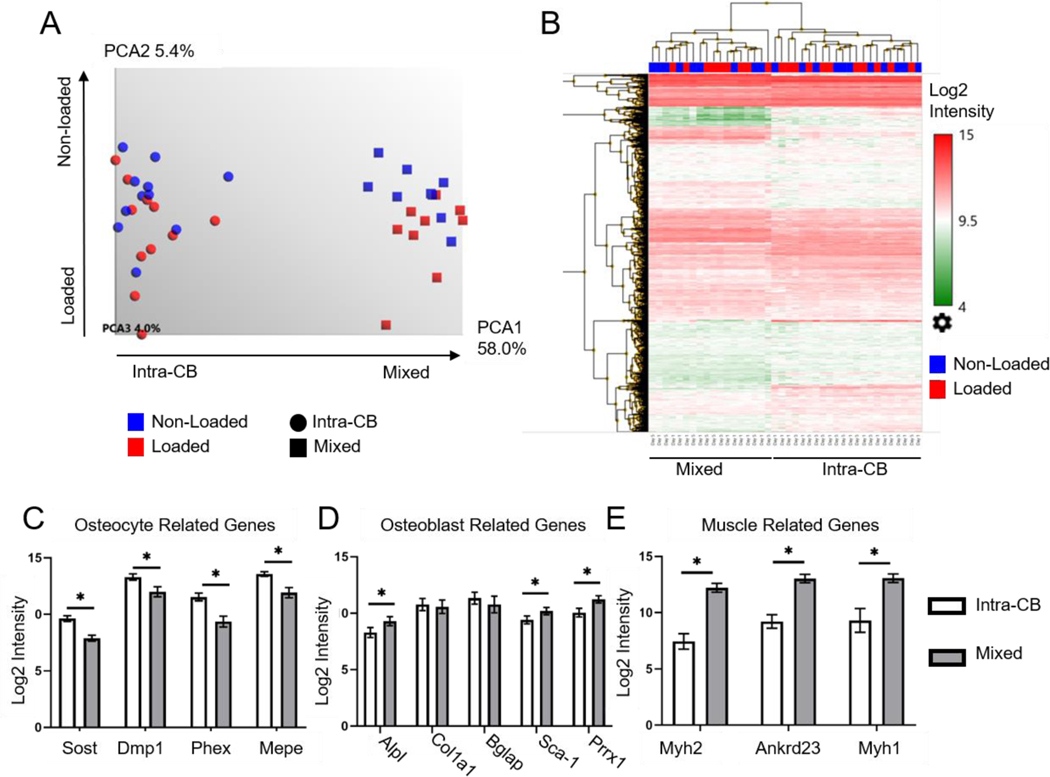
PCA showed that the majority (58%) of variation in gene expression intensities across all samples can be explained by the first principal component (PC1), with samples clearly separated by region type along this axis (Fig. 2A). In contrast, PC2 explained only 5.4% of variation, with non-loaded and loaded samples of the Mixed type marginally separated along this axis. Hierarchical mapping also showed clustering of samples primarily by region type; secondary clustering by loading or timepoint was not apparent (Fig. 2B). Together, PCA and clustering analysis indicate that Intra-CB and Mixed samples have different expression profiles, consistent with Intra-CB samples lacking some of the cells that are contained in Mixed samples. Additional PCA, performed for each tissue type and timepoint separately, demonstrated a strong separation between non-loaded and loaded samples along PC1 (Fig. S4). The greatest amount of sample variation explained by PC1 was seen in the Mixed region type on day 5 (PC1, 55%) compared to other groups (PC1, 30–35%). Greater variance due to loading in the Mixed region on day 5 is consistent with periosteal osteoblasts being more transcriptionally active at this timepoint, consistent with active periosteal bone formation. By contrast, periosteal osteoblasts are not fully active on day 1 [15], nor are the changes associated with active osteoblasts captured within Intra-CB samples.
Figure 2.
LCM allowed dissection of distinct populations of cells from mouse tibias. (A) PCA of gene expression from regions of Intra-CB (n=22) and mixed tissue (n=18) showed separation of region-type along PC1. Loaded and non-loaded samples were not clearly separated. (B) Heat map of Log2 gene expression intensities of genes with average Intra-CB Log2 intensities greater than 9 (4,105 genes). This representation of highest expressing genes clustered primarily by region type. (C-E) Select genes demonstrated (C) higher expression of osteocyte-specific genes in Intra-CB, (D) higher expression of Alpl, Sca-1 and Prrx1 and (E) muscle related genes in mixed regions. *FDR p-value<0.05
Genes known to have higher expression in osteocytes compared to other musculoskeletal cells (Sost, Dmp1, Phex, Mepe) [43] were highly expressed in Intra-CB, each above or near the 90th percentile in terms of signal intensity (Table S1). Moreover, each showed over 2-fold greater expression in Intra-CB compared to Mixed samples (FC=3.4, 2.2, 5.1, 3.0, respectively, p<0.001, Fig. 2C). Dkk1 was also expressed higher in Intra-CB tissue, although Fgf23 and Pdpn were not different (Table S1). Sca-1 and Prrx1, known to be expressed by bone lining cells and osteochondral progenitors in the periosteum [44]–[47], were expressed 1.7 and 2.2 fold higher, respectively, in Mixed regions compared to Intra-CB (p<0.001, Fig. 2D). Alpl which is abundantly expressed by early osteoblasts to provide phosphate for mineralization [48], also showed 2.0-fold higher expression intensity in mixed regions. However, two other genes expressed at high levels during active bone formation, Col1a1 and Bglap, were expressed at similar levels between region type (p=0.58, p=0.096, Fig. 2D). Several other genes typically associated with osteoblasts were either expressed more in Mixed tissue (Col5a1, Tmem119), not different by tissue type (Col1a2, Fkbp10, Gja1, Ibsp, Kdelr3, Maged1, Pard6g, Rcn3, Sparc), or expressed higher in Intra-CB (Runx2, Sp7, Spp1) (Table S1). We found no enrichment for osteoclasts or endothelial cells in either tissue type (Table S1). Three genes expressed highly in skeletal muscle (Myh2, Ankrd23, Myh1) [49], [50] were expressed at over 13-fold higher intensity in Mixed regions compared to Intra-CB (FC=30.7, 15.4, 13.3, respectively, p<0.001, Fig. 2E). The enhanced gene expression intensity of osteocyte specific genes, combined with decreased expression of early osteoblast, osteochondral progenitor, and muscle genes in Intra-CB supports an enrichment for osteocytes within the cortical bone region.
4.2. Loading induces differential expression of >100 genes in intracortical and mixed regions at days 1 and 5
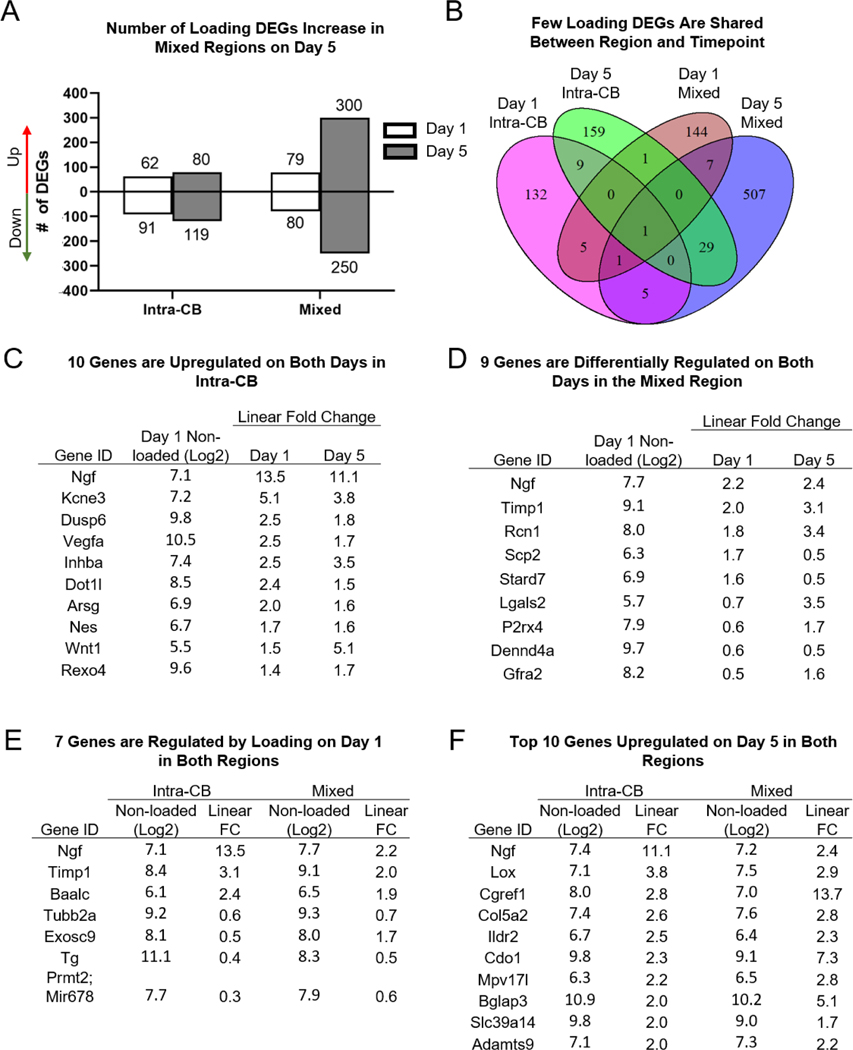
A robust increase in the number of loading DEGs occurred from day 1 (159) to day 5 (550) in the Mixed region, consistent with increased periosteal osteoblast activity during active bone formation at this timepoint (Fig. 3A, Table S2). In the Intra-CB region, the number of loading DEGs was comparable at days 1 and 5, but of the 153 Day 1 DEGs, only 10 remain differentially regulated on day 5, and they are all upregulated due to loading (Fig. 3B, C). Ngf, an important regulator of loading-induced bone formation known to be expressed by osteoblasts [21], had the highest FC on day 1 (13.5) and day 5 (11.1). Kcne3, a gene responsive to Vegfa (also upregulated on both days) and expressed by endothelial tip cells was upregulated over 5-fold on day 1 and over 3-fold on day 5 [51]. The only genes to increase their magnitude by at least 1.0 FC from day 1 to day 5 were Inhba (2.5 to 3.5) and Wnt1 (1.5 to 5.1). Inhba can form subunits for activin or inhibin which both have important roles in modulating TGF-ꞵ and BMP signaling, and have been shown to affect differentiation of osteoblasts and osteoclasts [52]. Wnt1 has been noted previously as a loading-induced gene in bone [14]–[16], [20].
Figure 3.
More than 150 genes are differentially regulated by loading in Intra-CB on each day, with only 10 genes in common at both timepoints. (A) The number of DEGs is similar on both days in Intra-CB samples but an increase in number of DEGs occurs on day 5 in mixed regions. (B) Most genes are not shared between regions or timepoints. (C-D) Differentially regulated genes (C) in Intra-CB and (D) Mixed regions on both days (all 10, 9 shown) (E) in both regions on day1 (all 7 shown) and (F) in both regions on day 5 (top 10 shown). Values are basal gene intensity (Log2) and absolute fold change due to loading (loaded/non-loaded).
Many genes upregulated on day 1 did not remain up on day 5. Four of the most highly upregulated genes in Intra-CB on day 1 (Enkur, Egr1, Tnfrsf12a, Timp1) were not differentially expressed on day 5 (Table 1). Enkur encodes a protein that binds to calmodulin and may be important for signal transduction via calcium channels [53], [54], but has not been identified in bone. Calcium channel signaling is activated early in the mechanotransduction process by osteocytes [10]. Early growth response 1 (Egr1), has been shown to play a role in many factors relating to proliferation and differentiation as well as regulation of Postn, Bglap, and Ctsk [55], [56]. Tnfrsf12a is not well known in the context of bone formation, but this TNF ligand has been reported to be upregulated by loading [14], [15] and to bind to mesenchymal progenitor cells and activate NF-κB [57]. Tissue inhibitor of metallopeptidase inhibitor 1 (Timp1) inhibits matrix metalloproteinases and extracellular remodeling and has been shown to enhance the anabolic response to parathyroid hormone [58]. More recently, TIMP1 was shown to suppress osteoblast and osteoclast differentiation, mediated by Wnt signaling [59], [60].
Table 1.
Intra-CB top 20 upregulated DEGs by fold change
| Day 1 | Day 5 | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Gene ID | Non-Loaded Intensity (Log2) | Linear FC (Loaded/Non-Loaded) | P-val | Gene ID | Non-Loaded Intensity (Log2) | Linear FC (Loaded/Non-Loaded) | P-val |
| Ngf | 7.1 | 13.5 | 1.62E-05 | Ngf | 7.4 | 11.1 | 1.50E-05 |
| Kcne3 | 7.2 | 5.1 | 0.0007 | Anxa8 | 8.4 | 7.1 | 0.0009 |
| Enkur | 5.2 | 3.4 | 0.0052 | Wnt1 | 5.3 | 5.1 | 0.0002 |
| Egr1 | 9.0 | 3.4 | 0.0005 | Pappa | 8.1 | 5.1 | 0.0001 |
| Tnfrsf12a | 9.1 | 3.3 | 0.0003 | Adm | 9.1 | 4.5 | 0.0068 |
| Egr2 | 8.5 | 3.2 | 0.0003 | Kcne3 | 8.1 | 3.8 | 1.79E-05 |
| Timp1 | 8.4 | 3.1 | 0.0007 | Lox | 7.1 | 3.8 | 0.0037 |
| Lmcd1 | 8.8 | 2.8 | 0.0004 | Inhba | 7.4 | 3.5 | 0.0001 |
| Dusp6 | 9.8 | 2.5 | 0.0014 | Apln | 7.7 | 3.1 | 0.0016 |
| Vegfa | 10.5 | 2.5 | 0.0057 | Ptgs2 | 5.3 | 2.9 | 0.0014 |
| Zeb1 | 7.8 | 2.5 | 0.0036 | Cgref1 | 8.0 | 2.8 | 0.0083 |
| Inhba | 7.4 | 2.5 | 0.0038 | Bmp2 | 6.8 | 2.7 | 0.0092 |
| Baalc | 6.1 | 2.4 | 0.0001 | Pik3r3 | 7.0 | 2.7 | 0.0004 |
| Pdgfa | 8.9 | 2.4 | 0.0041 | Col5a2 | 7.4 | 2.6 | 0.0029 |
| Hspb7 | 8.0 | 2.4 | 0.0053 | Prex2 | 5.6 | 2.6 | 0.0012 |
| Dot1l | 8.5 | 2.4 | 0.0029 | Ildr2 | 6.7 | 2.5 | 0.006 |
| Stc1 | 7.0 | 2.4 | 0.0068 | Asic3 | 9.1 | 2.5 | 0.0062 |
| Gem | 6.5 | 2.4 | 0.0092 | Tmie | 8.4 | 2.5 | 0.0049 |
| Mpdz | 5.5 | 2.4 | 0.004 | Rasl11a | 5.4 | 2.3 | 0.0085 |
| Crem | 7.2 | 2.2 | 0.0082 | Tnfrsf11b | 9.7 | 2.3 | 0.0052 |
Note: 90th percentile value of log2 intensity = 9.9
Consistent with an increase in Timp1 on day 1, by day 5 a decrease in Mmp13 expression occurred (0.3-fold, Table 2). Matrix metalloproteinase (MMP)-13 has been identified in osteocyte perilacunar remodeling [61]. Of the genes upregulated by loading on day 5 that were not induced on day 1, Annexin A8 (Anxa8) was the most upregulated (FC=7.1). AnxA8 plays roles in osteoclast differentiation [62], endosome trafficking [63], and Wnt signaling in epithelial cells [64]. Pregnancy-Associated Plasma Protein A, Pappalysin 1 (Pappa) was also upregulated over 5-fold on day 5. This gene is known for its regulation of insulin-like growth factor (IGF) bioavailability [65]. IGF signaling has been shown to increase following loading, mediated by Igf-1 [66]–[69]. Adrenomedullin (Adm) was increased 4.5-fold on day 5 due to loading and has also been shown to play a role in bone formation as an osteoblast expressed protein [70]. Finally, Lox, Apln, and Ptgs2 were also top DEGs upregulated due to loading on day 5. Lox encodes for the protein lysyl oxidase which is important for collagen fiber cross-linking and affects differentiation of osteoblasts and osteoclasts [71]. Interestingly, Apln knockout mice have increased cancellous bone volume and cortical bone perimeter, which was attributed to an increase in osteoblast proliferation mediated by adipose tissue [72]. Cyclooxygenase-2 (Ptgs2 or Cox-2) is well-known for its important role in early mechanotransduction by osteocytes in the context of loading [5], [11], [73]. These genes might play a role in maintaining the response to loading since they were upregulated only on day 5, or their expression could be secondary to earlier changes in transcription.
Table 2.
Intra-CB top 20 downregulated DEGs by fold change
| Day 1 | Day 5 | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Gene ID | Non-Loaded Intensity (Log2) | Linear FC (Loaded/Non-Loaded) | P-val | Gene ID | Non-Loaded Intensity (Log2) | Linear FC (Loaded/Non-Loaded) | P-val |
| Prmt2; Mir678 | 7.7 | 0.3 | 0.0022 | Atp6v0d2 | 8.0 | 0.2 | 5.81E-05 |
| Sep4 | 8.5 | 0.3 | 2.65E-05 | Pax5 | 9.5 | 0.2 | 0.0064 |
| Cd55 | 7.6 | 0.3 | 0.0054 | Hspa4l | 6.3 | 0.2 | 0.0019 |
| Gpn1 | 7.3 | 0.4 | 0.0024 | Jchain | 9.9 | 0.2 | 0.0041 |
| Trim30d | 7.8 | 0.4 | 0.0054 | Lrguk | 6.2 | 0.3 | 0.0075 |
| Aqp9 | 9.1 | 0.4 | 0.0083 | Stk26 | 9.6 | 0.3 | 0.0094 |
| Mapk1 | 7.8 | 0.4 | 0.007 | Acp5 | 11.6 | 0.3 | 0.0005 |
| Xpo1 | 8.8 | 0.4 | 0.0029 | Mmp13 | 10.5 | 0.3 | 0.0003 |
| Tg | 11.1 | 0.4 | 0.0094 | Pqlc3 | 9.3 | 0.3 | 0.0054 |
| Phf20 | 6.3 | 0.4 | 0.002 | Panx1 | 7.8 | 0.4 | 0.0013 |
| Nxt2 | 8.3 | 0.4 | 0.0036 | Pde3b | 7.9 | 0.4 | 0.0009 |
| Drg2 | 8.6 | 0.4 | 0.0067 | Tbc1d13 | 7.4 | 0.4 | 0.0028 |
| Cbr2 | 8.4 | 0.4 | 0.0075 | Tespa1 | 7.7 | 0.4 | 0.0006 |
| Rnf213 | 9.4 | 0.4 | 0.0003 | Lcp2 | 7.9 | 0.4 | 5.19E-05 |
| Ccl8 | 6.8 | 0.5 | 0.0026 | Nckap1l | 9.0 | 0.4 | 0.0062 |
| Cdc6 | 9.0 | 0.5 | 0.0036 | A530088E08Rik | 7.0 | 0.4 | 0.0018 |
| Anpep | 8.8 | 0.5 | 0.0091 | Clec12a | 10.5 | 0.4 | 0.0033 |
| Clasp2 | 8.1 | 0.5 | 0.0062 | Lpar5 | 6.5 | 0.4 | 0.0021 |
| Igtp | 7.6 | 0.5 | 0.0013 | Dcstamp | 7.8 | 0.4 | 0.0055 |
| Lynx1 | 8.5 | 0.5 | 0.0016 | Hyls1 | 8.6 | 0.4 | 0.0063 |
Note: 90th percentile value of log2 intensity = 9.9
Overall, few DEGs were shared between region types on day 1 (7 of 159) or day 5 (30 of 550) (Fig. 3B, Table S3). Of these genes on day 1, the FC was of greater magnitude in Intra-CB compared to Mixed regions, with Ngf and Timp1 having the highest upregulation and Prmt2 having the greatest downregulation (Fig. 3E). Protein Arginine Methyltransferase 2 (Prmt2) is a well-known inhibitor of NF-κB-dependent transcription which is critical for osteoclastogenesis [74], [75]. Of the shared genes by region type on day 5 (Fig. 3F), several genes had greater FC induced by loading in Mixed regions compared to Intra-CB (Cgref1, Col5a2, Cdo1, Mpv17l, Bglap3, Adamts9). These genes are involved in ion binding (Cgref1, Cdo1, Bglap3) [76]–[78], cell signaling (Mpv17l) [79], and extracellular matrix remodeling (Col5a2, Adamts9) [80], [81]. Ngf and Lox were also upregulated in both regions, but with greater FC in Intra-CB. Ngf was the only gene differentially regulated in both regions on both days, although FC was substantially lower when other tissues were present (Day 1, 13.5 vs. 2.2).
To better understand the effect of isolating specific tissue regions on gene expression analysis, we compared Intra-CB and Mixed DEGs to Bulk tissue DEGs from Chermside-Scabbo et al. [15] (Fig. S5). This comparison revealed more similarity between Bulk tissue and Intra-CB on day 1 (37% overlap) compared to day 5 (3% overlap) (Fig. S5A). The genes shared by Bulk tissue and Intra-CB on day 1 included genes also reported by others at early timepoints, including Ngf, Tnfrsf12a, Pdgfa and Wnt1 [14] (Fig. S5B). Most DEGs in Bulk tissue were not detected in Mixed regions, but 37 genes did overlap between day 5 Mixed and Bulk tissue (Fig. S4C). These genes included Cgref1, Cdo1, and Bglap3 (Fig. S4D), which were also more enhanced (by FC) in Mixed regions compared to Intra-CB on day 5.
4.3. Ngf and Wnt1 are expressed by osteocytes
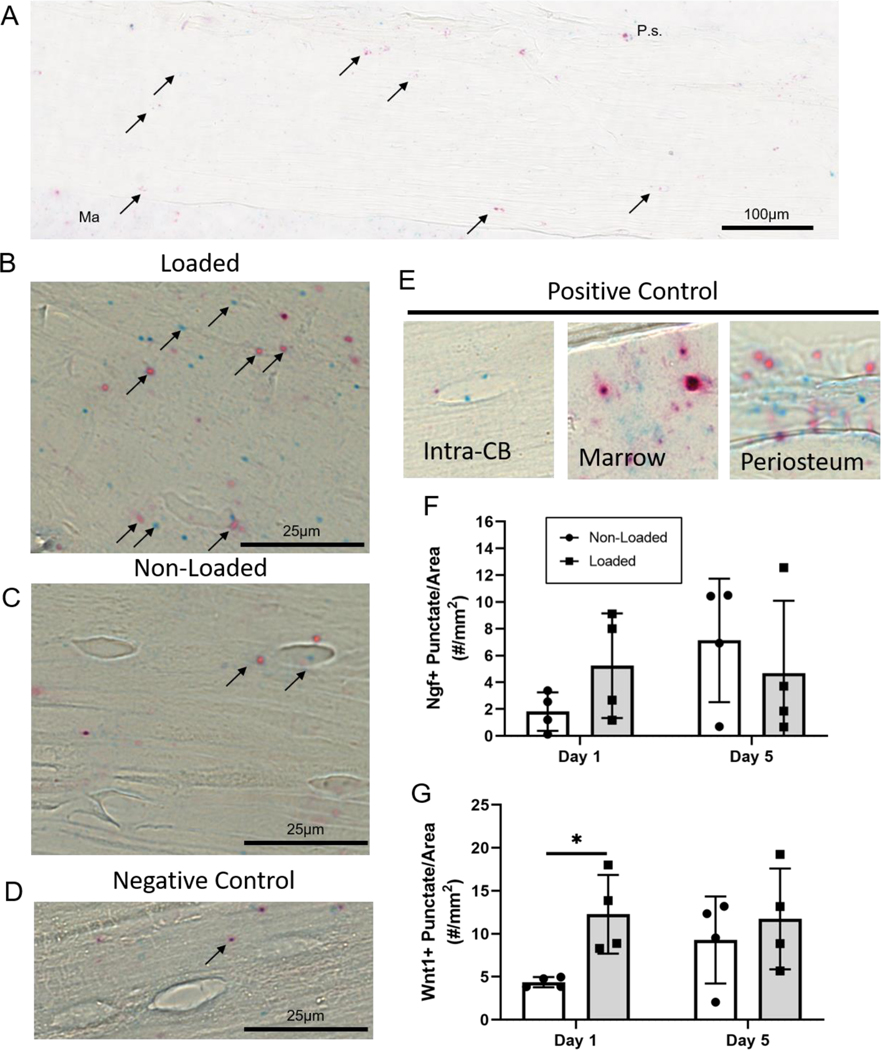
We selected Wnt1 and Ngf for spatial analysis using RNAScope, based on their significant upregulation on days 1 and 5 in Intra-CB. RNA molecules represented by punctate dots, were quantified throughout the 1-mm ROI and normalized by area (Fig. 4A). Wnt1 and Ngf mRNA molecules can be readily seen in the loaded and non-loaded Intra-CB samples (Fig. 4B,C). Negative control probes targeting bacterial genes can be seen to a minimal degree, indicating limited non-specific staining for this assay (Fig. 4D). Positive control probes show more intense staining in osteocytes (although not abundant), marrow, and periosteum (Fig. 4E). Loading did not significantly increase Ngf mRNA molecules within Intra-CB at either timepoint (Fig. 4F), but the number of lacunae within the bone containing multiple Ngf mRNA molecules showed a marginal increase (p=0.09, Fig. S6A). In support of the microarray data, Wnt1 increased over 2-fold with loading on day 1, but unlike the microarray data, no change was detected on day 5 (Fig. 4G). Similarly, Wnt1+ lacunae increased significantly with loading on day 1 but not day 5 (Fig. S6B). We also identified Wnt1 and Ngf mRNA within the periosteum (Fig. S7), but these punctae were not quantified due to limited regions with intact and identifiable periosteum.
Figure 4.
Loading increases Wnt1 expression in Intra-CB. (A) A region approximately 1-mm in length was analyzed for Wnt1 (red) or Ngf (blue) punctate dots representing RNA transcripts targeted by their respective probes (arrows). Representative images show the (B) loaded tibia had more Ngf and Wnt1 expression (in this sample) compared to (C) non-loaded tibias and (D) negative control probes detected minimal RNA transcripts within Intra-CB. (E) Positive control probes show staining in Intra-CB, and more abundantly in marrow, and periosteum. Punctate dots were quantified, showing (F) no significant change in Ngf at either timepoint and (G) a substantial increase in Wnt1 expression on day 1. Ma=Marrow, P.s.=Periosteal Surface.
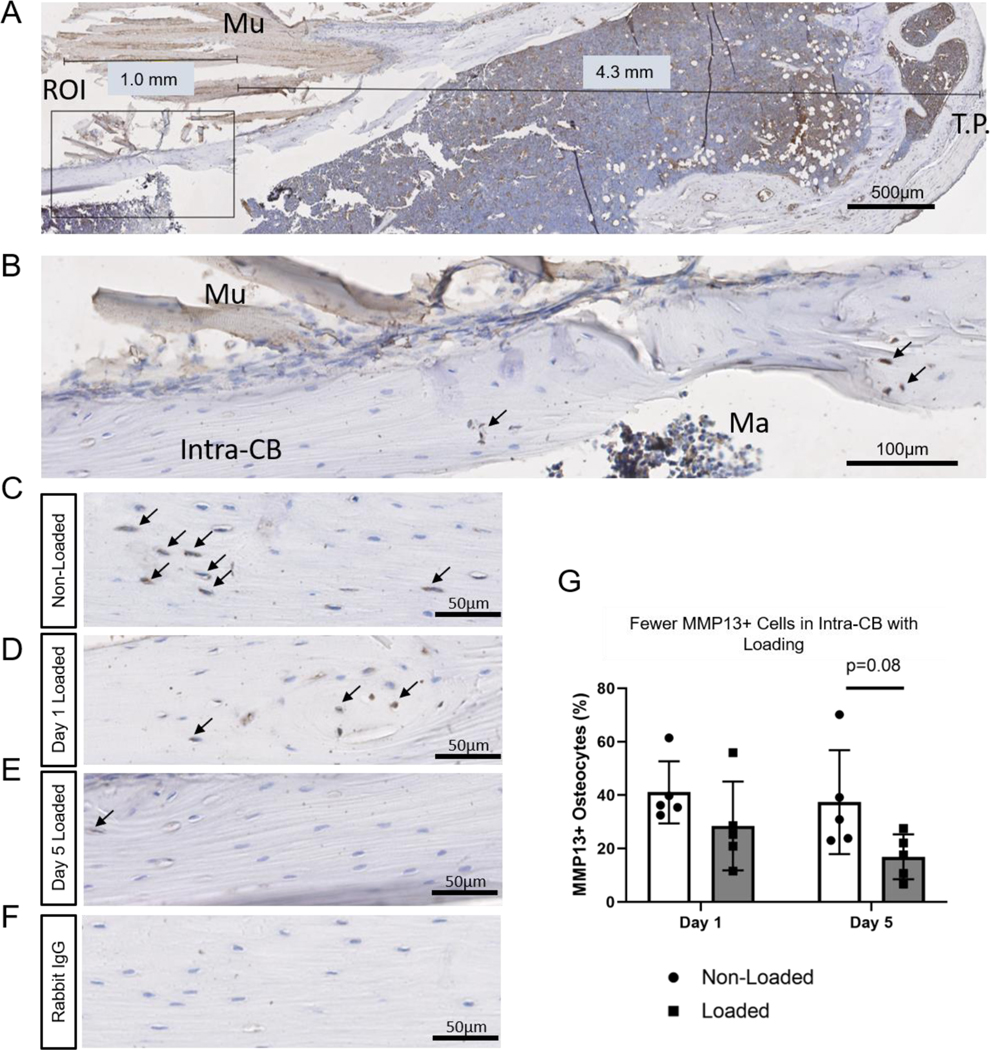
4.4. MMP13 is downregulated in osteocytes from loaded tibias
MMP13 IHC was performed to confirm downregulation shown by microarray in Intra-CB on day 5. MMP13+ osteocytes were quantified within a region close to maximal strain (Fig. 5A-B). Qualitatively, more MMP13+ osteocytes can be seen in the non-loaded compared to loaded tibias (Fig. 5C-E). Rabbit IgG controls indicated negligible non-specific staining (Fig. 5F). Consistent with microarray data, percentage of MMP13+ osteocytes showed a marginal decrease in loaded tibias vs. non-loaded tibias on day 5 (p=0.08, Fig. 5G).
Fig. 5.
MMP13 staining decreases with loading. (A) The region of Interest (ROI) was centered near the region of maximal strain, 5 mm distal to the tibial plateau (T.P.) with adjustments made for tissue artifacts. (B) An approximately 1-mm in length ROI was selected for quantification of MMP13+ osteocytes. (C) MMP13 staining is apparent in non-loaded tibias in the periosteum and Intra-CB. (B-C) MMP13 staining in loaded tibias on day 1 (B) and day 5 (C). (D) Rabbit IgG shows negligible non-specific staining in a non-loaded tibia. (E) Quantification of osteocyte positive cells show a decrease with loading on day 5. N=5/timepoint. Mu=Muscle, Ma=Marrow.
4.5. Vascular development, neuronal, and immune cell biological processes are enriched in Intra-CB bone by loading
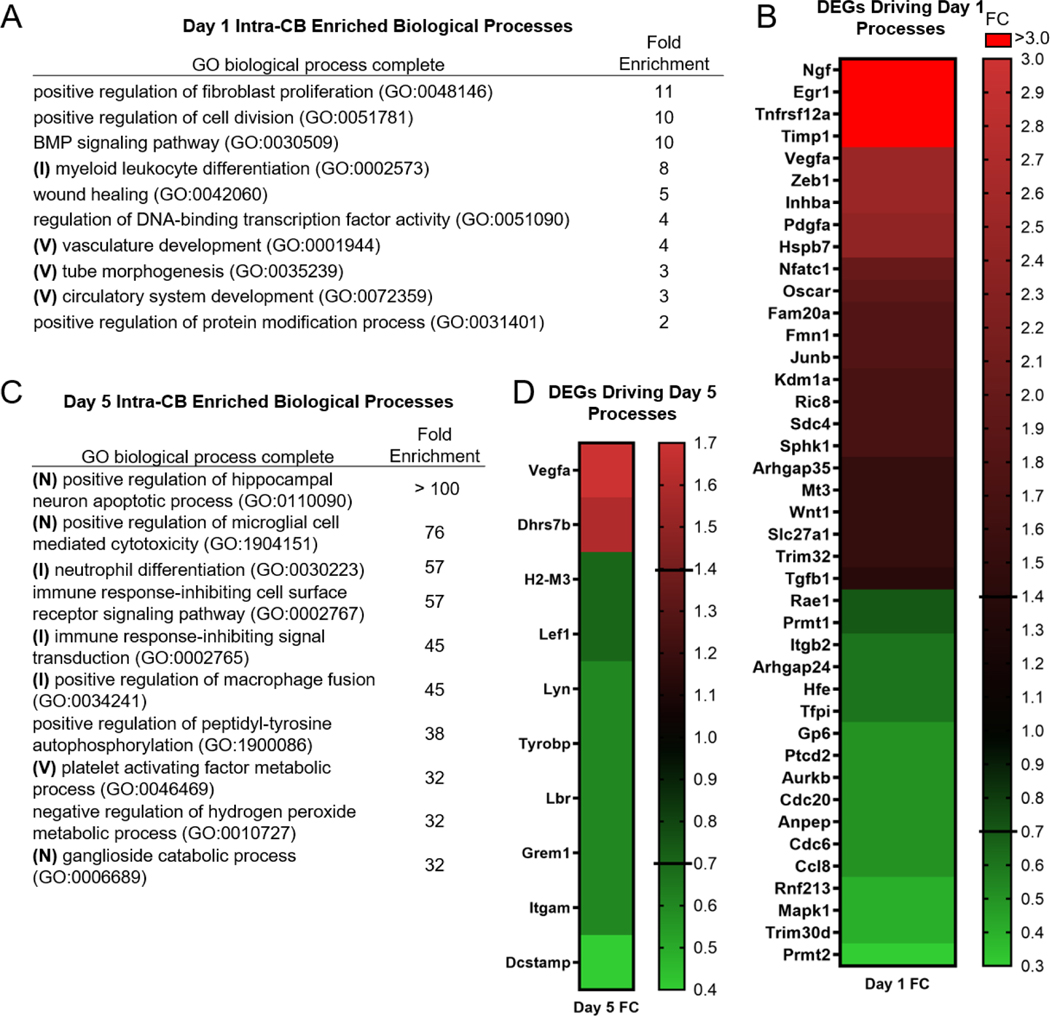
GO biological processes were investigated using DEG lists for Intra-CB on days 1 and 5. Twenty biological processes were enriched on day 1 (Fig. 6A, Table S4), with 41 DEGs driving the top 10 processes by Fold Enrichment (FE) (Fig. 6B, Table S5). Positive regulation of fibroblast proliferation had the greatest FE (11.23) on day 1 and was driven by Pdgfa (FC=2.4), Sphk1 (FC=1.7), Wnt1 (FC=1.5), Tgfb1 (FC=1.4), and Cdc6 (FC=0.5). Myeloid leukocyte differentiation was also highly enriched (FE=7.7) with Vegfa (FC=2.5), Nfatc1 (FC=2.0), Oscar (FC=1.9), Junb (FC=1.8), Kdm1a (FC=1.7), and Tgfb1 (FC=1.4) contributing to its regulation. Three terms within the top 10 were vascular related (vasculature development, tube morphogenesis, circulatory system development) with FE ≥ 3. The genes with the highest FC contributing to these processes included Egr1 (FC=3.4), Tnfrsf12a (FC=3.3), Vegfa and Zeb 1 (FC=2.5), and Pdgfa and Hspb7 (FC=2.4) being the most upregulated, while Mapk1 and Rnf213 (FC=0.4) were the most downregulated DEGs (Table S4). Overall, Ngf, Kcne3, Egr1, Tnfrsf12a, and Timp1 contributed to at least one of the enriched biological processes on day 1 and had the greatest loading-induced FC (FC > 3).
Figure 6.
Vascular (V), neuron (N), and immune (I) regulation are biological processes differentially regulated by loading in Intra-CB. (A) Top 10 (by FC) enriched biological processes on Day 1 are driven by (B) 41 DEGs, while on Day 5 (C) the top 10 enriched biological processes are driven by (D) 10 DEGs.
On day 5, almost 300 biological processes were enriched, but the top 10 biological processes (by FE) (Fig. 6C, Table S4) were attributed to only 10 DEGs (Fig. 6D, Table S5). The genes driving the two most enriched processes (positive regulation of hippocampal neuron apoptotic process and positive regulation of microglial cell mediated cytotoxicity) were Tyrobp and Itgam (FC=0.6). Another neuron related process, Ganglioside catabolic process, was also in the top 10 by FE, attributed to Hexb (FC=0.6) and Gm2a (FC=0.5). Four immune related processes were also highly enriched (FE>45), including neutrophil differentiation and immune response-inhibiting cell surface receptor signaling pathway. These immune processes are driven by Dhrs7b (FC=1.6), Lef1 and H2-M3 (FC=0.7), and Lbr and Lyn (FC=0.6). Consistent with enrichment of vascular processes on Day 1, processes for platelet activating factor metabolic process, primitive hemopoiesis, and leukocyte adhesion to vascular endothelial cell were also within the top 20 enriched processes on day 5 (Table S4). These processes were driven by Lox (FC=3.8), Vegfa (FC=1.7), Pla2g7, Lpcat2 (FC=0.5), and Ptpn1 and Stk4 (FC=0.4). Dcstamp (FC=0.4) was one of the most downregulated DEGs driving day 5 processes and contributed to the positive regulation of macrophage fusion. In summary, immune and vasculature regulation occurred on both days while neuron associated processes were enriched on day 5 only.
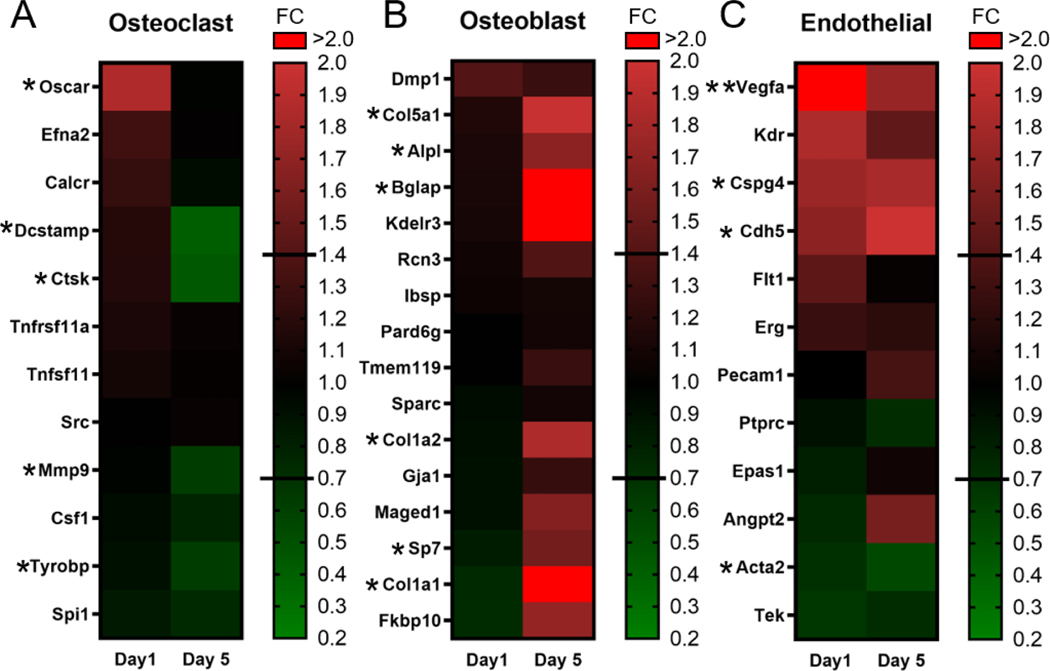
4.6. Osteoclast, Osteoblast, and Endothelial cell-associated genes were differentially expressed due to loading in Intra-CB
Osteoclast, osteoblast, endothelial, immune, and neuronal cell-associated genes were investigated in the Intra-CB specific data set. A priori gene lists for each cell type, except neuronal associated genes (see below), were taken from Coates et. al. [37]. The osteoclast associated gene list revealed upregulation of Oscar (FC=1.9) on day 1, and downregulation of Dcstamp, Ctsk, Mmp9, and Tyrobp on day 5 (FC= 0.4, 0.5, 0.6, 0.6 respectively), indicating a downregulation of osteoclast differentiation factors on day 5 (Fig. 7A, Table S6). The osteoblast associated gene list returned 6 genes (out of 16) that were upregulated on day 5 in Intra-CB (Fig. 7B), including Bglap, Col1a1 (FC=2.1), Col5a1 (FC=2.0), Alpl (FC=1.7), Sp7 (FC=1.6). These results indicate an upregulation of genes classically associated with osteoblast differentiation and bone matrix synthesis on day 5, despite the lack of bone forming surfaces in the Intra-CB region. The endothelial cell associated gene list returned 4 genes (out of 12) on day 1 that were differentially regulated with loading (Fig. 7C). Vegfa (Day 1 FC=2.5, Day 5 FC=1.7) was upregulated on both days while Cspg4 (FC=1.8) and Cdh5 (FC=2.0) were upregulated only on day 5, and Acta2 (FC=0.5) was downregulated on day 5. These data reveal a potential role for endothelial cells in the loading response. Of the immune cell lists, 8 and 4 genes associated with T cells and B cells, respectively, were downregulated with loading while other lists encompassed genes mostly unregulated by loading (Fig. S8.). Finally, a list of neuron related genes was compiled from studies by Paic et. al. [42], and Youlten et. al. [41], but no genes were significantly regulated by loading (Fig. S8A).
Figure 7.
Loading-induced changes in expression of osteoclast, osteoblast, and endothelial cell related genes occur in Intra-CB. (A) Osteoclast genes are downregulated on day 5. (B) Osteoblast genes are upregulated on day 5. (C) Endothelial genes were similarly regulated on both days. Gene Lists from Coates et al. [37] * indicates 1.4 ≤ FC ≤ 0.71 & p<0.05 at 1 timepoint, ** indicates 1.4 ≤ FC ≤ 0.71 & p<0.05 at both timepoints.
5. Discussion
The transcriptional response of osteocytes to in vivo mechanical loading has been difficult to ascertain due to their location within mineralized matrix. Using laser capture microdissection we isolated intracortical bone, without periosteal and endosteal cells, at mid-diaphyseal sites known to experience high mechanical strain with axial tibial loading. We used microarray analysis to evaluate the transcriptional response of this osteocyte-enriched sample to loading, and contrasted this to the response of a mixed tissue sample containing bone as well as periosteum and muscle tissue. We showed enrichment for osteocyte-specific genes in Intra-CB regions compared to Mixed regions, which expressed higher levels of some early osteoblast and osteoprogenitor genes. We identified expression of genes classically associated with other cell types within Intra-CB regions. This pattern has been described by Nioi et al., where Col1a1 and Bglap were expressed higher in osteocytes compared to lining cells and active osteoblasts [24]. With loading, there were hundreds of differentially expressed genes in both regions, although these were mostly non-overlapping. Notably, the set of loading DEGs in Intra-CB samples had little overlap with DEGs reported from bulk tissue samples on day 5, when bone formation is active in this model [15]. Therefore, the analysis of gene expression within Intra-CB samples reveals responses to loading that are not identical to responses detected in more heterogenous tissue samples. Focusing on these Intra-CB DEGs, we find that some previously described responses to loading, namely upregulation of Wnt1 and Ngf, occur in osteocytes. We also find differential regulation of genes related to vasculature and bone remodeling.
Wnt1 is a key regulator of bone mass and skeletal integrity in humans and mice [82], [83]. Wnt1 deletion in Dmp1-Cre mice decreased bone mass while overexpression increased osteoblast number and activity [82]. However, it is unclear if these effects were mediated by osteocytes or osteoblasts, since Dmp1-Cre is known to target both cell types. In support of a role for Wnt1 in osteocytes, our Intra-CB microarray and in situ hybridization results show, for the first time, that a single bout of loading increases Wnt1 expression localized to intracortical bone/osteocytes. Furthermore, our microarray data shows a sustained loading response on day 5 (5.1-fold increase), although in situ hybridization did not detect any differences at this timepoint. Holguin et al. showed sustained Wnt signaling in osteocytes in TOPGAL reporter mice similarly loaded for 1 and 5 days [16]. Others have shown downregulation of SOST in response to loading [14], [16], shown to be critical for increased Wnt signaling and subsequent bone formation [18]. Interestingly, we did not find a significant downregulation in Sost due to loading at either timepoint, although the data showed a marginal change in that direction (Day 1, FC= 0.8, p=0.09; Day 5, FC=0.8, p=0.1). While our results reveal Wnt1 induction in osteocytes with loading, whether this plays a functional role in loading-induced bone formation remains to be shown.
A major finding in this work was the robust loading-induced increase in Ngf expression in Intra-CB on day 1 (13-fold) and day 5 (11-fold). No previous studies have shown Ngf expression within cortical bone. We also found enrichment for positive regulation of hippocampal neuron apoptotic process, which was over 100-fold enriched on day 5 in Intra-CB. Chermside-Scabbo et al. also showed increased Ngf expression in addition to enrichment of pathways for neuron projection [15]. Others have also reported enhanced expression of neuronal phenotypes in osteocyte-enriched gene expression data [41], [42], but genes from these lists were not loading DEGs in our data. Interestingly, the potassium gated channel gene Kcnk13 was identified as more highly expressed in osteocyte cells compared to osteoblasts by Paic et al. [42], and we found a similar gene (Kcne3) to be a top DEG with loading on day 1.
Our results showing Ngf expression in Intra-CB contrasts with work by Tomlinson et al. showing increased expression due to loading in osteoblasts, but not osteocytes, using NGF-EGFP mice and IHC [21]. This lack of expression in osteocytes was later supported in a fracture model [84]. The conflicting results with our work could partially be explained if gene expression is driven mainly by a few high-expressing osteocytes, or due in part to the different assays used. Our RNA extracts included four 10 μm thick sections from each bone which could have captured a few very responsive osteocytes. In situ hybridization using RNAScope was able to confirm presence of Ngf within Intra-CB, but did not confirm the robust increase in expression found in the microarray data, although a marginal increase in Ngf+ lacunae with loading was observed. Additionally, the RNAscope results did not show the abundance of Ngf detected by microarray. At baseline, Ngf was near the median of expression intensities, and after loading expression was above the 90th percentile of expression intensities.
MMP13 plays a role in osteocyte perilacunar remodeling [61], and we showed that loading induced a strong downregulation of MMP13 expression in osteocytes. We also showed a decrease in several osteoclast associated genes within Intra-CB samples due to loading (Dcstamp, Ctsk, Mmp9, Tyrobp). Furthermore, several immune cell differentiation processes were detected in the GO biological process analysis on day 5, with the majority of genes contributing to these processes being downregulated. Osteoclasts derive from the same monocytic lineage as macrophages, and have been shown to regulate immune cell activity [85]. Collectively, our results provide evidence that loading suppresses the expression of factors related to both lacunocanalicular remodeling and osteoclastogenesis, suggesting broad inhibition of bone remodeling.
Interestingly, we found several indications of vasculature changes induced by loading in Intra-CB. Vasculature development was enriched 4-fold as a biological process on day 1, driven primarily by loading-induced increases in Egr1, Tnfrsf12a, Vegfa and Pdgfa. We further showed increases of endothelial cell associated genes (Vegfa, Cspg4, Cdh5). These results may indicate osteocyte regulation of vasculature, induced by loading. Tomlinson et al. demonstrated an increase in vascularity 7 days following loading stimulus in the rat forelimb, although they showed that this increase in blood vessel number was only necessary for woven bone but not lamellar bone formation [86]. Besides driving angiogenesis at sites of new bone, osteocytes may be regulating vascular growth within Intra-CB directly for improved nutrient transport to the bone surface. On the other hand, these vasculature related genes may have been expressed by vascular cells within Intra-CB, not osteocytes. Recent work has described a network of small vessels that can transport nutrients and cells, including immune cells [36]. Intra-CB samples may have contained enough transcortical vessels to reveal these changes.
The tissue samples we analyzed were micro-dissected from the postero-lateral tibial cortex, a region that experiences compressive strain during axial tibial loading [31]. By analyzing this small region, we reduced variability in the mechanical environment within samples; in particular, we did not analyze tissues from the tensile cortex or the neutral axis. Nonetheless, the surface cells in Mixed tissue samples experience fundamentally different mechanical stimuli than cells in the Intra-CB region. Cells on the periosteal surface experience substrate strain as the bone material deforms, whereas intracortical osteocytes experience a combination of substrate strain and effects of fluid movement through the lacunocalicular network [6], [10]. Yet, we emphasize that the transcriptional responses to loading reported here are not attributed solely to direct mechanotransduction events at the individual cell level, but also to the effects of cell-cell communication. In particular, signaling molecules from intracortical osteocytes may largely orchestrate the response of the periosteal osteoblasts [87].
We found several challenges while conducting LCM in the mouse tibia. Due to the small size of the tissue relative to the width of the laser beam, we were not able to compare Intra-CB regions to more localized periosteal regions such as osteoblasts/bone lining cells. These challenges also resulted in poor quality RNA in about half of the samples, resulting in low sample sizes and statistical power. Despite passing our quality control measures, degradation might have biased the results towards more stable transcripts. Furthermore, microarrays have their own disadvantages including a lower dynamic range compared to RNAseq. However, since the concentrations and quality of our samples were not feasible for RNAseq, newer whole-transcriptome microarray technologies which incorporate amplification across the length of RNA and do not require ribosomal depletion or poly-A selection, enabled our investigation into Intra-CB specific gene expression.
In summary, LCM allowed analysis of loading-induced changes of osteocyte-enriched tissue. We demonstrated for the first time an increase in expression of Ngf and Wnt1 by osteocytes due to loading. We also showed an osteocyte-specific decrease in Mmp13 expression due to loading which may implicate a decrease in perilacunar remodeling. Similarly, decreased expression of osteoclast differentiation genes were apparent. Finally, vasculature processes were enriched with loading in Intra-CB.
Supplementary Material
Highlights.
Laser capture microdissection and microarray analysis was used for transcriptomics of osteocyte-enriched intracortical bone in mice following axial tibial loading.
Robust increases in Ngf and Wnt1 occurred with loading in Intra-CB tissue.
Presence of Ngf and Wnt1 in osteocytes was confirmed by in situ hybridization.
Mmp13 downregulation and decreased expression of genes associated with osteoclastogenesis suggests broad inhibition of bone remodeling following loading.
6. Acknowledgments
This work was supported by NIH grants R01AR047867 and T32EB018266 and the Washington University Musculoskeletal Research Center (P30AR074992). We acknowledge the Developmental Biology Microscopy Core at Washington University in St. Louis for use of the laser capture microscope. We thank Crystal Idleburg and Samantha Coleman for assistance with histology. We thank the Genome Technology Access Center (supported by P30CA91842 and UL1TR002345), especially Twyla Juehne and Mala Sharma for their important role in designing and implementing the processing of the RNA for hybridization to microarray probes and Jinsheng Yu for analysis of raw intensity values obtained from microarray chips. We also acknowledge NIH shared instrumentation grant S10 RR0227552 for use of the Nanozoomer 2.0HT microscope. We additionally thank Dr. Farshid Guilak for use of the RNAscope hybridization oven for the in situ assay. For help with protocols for RNAscope we thank Dr. Marc Wein and for MMP13 IHC we thank Dr. Tamara Alliston.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
7 References
- [1].Leblanc AD, Schneider VS, Evans HJ, Engelbretson DA, and Krebs JM, “Bone mineral loss and recovery after 17 weeks of bed rest,” Journal of Bone and Mineral Research, vol. 5, no. 8, pp. 843–850, 1990, doi: 10.1002/jbmr.5650050807. [DOI] [PubMed] [Google Scholar]
- [2].Alfredson H, Nordström P, Pietilä T, and Lorentzon R, “Long-term Loading and Regional Bone Mass of the Arm in Female Volleyball Players,” Calcif Tissue Int, vol. 62, no. 4, pp. 303–308, April. 1998, doi: 10.1007/s002239900436. [DOI] [PubMed] [Google Scholar]
- [3].Ducher G, Daly RM, and Bass SL, “Effects of Repetitive Loading on Bone Mass and Geometry in Young Male Tennis Players: A Quantitative Study Using MRI,” Journal of Bone and Mineral Research, vol. 24, no. 10, pp. 1686–1692, 2009, doi: 10.1359/jbmr.090415. [DOI] [PubMed] [Google Scholar]
- [4].Mosekilde L, “Age-related changes in bone mass, structure, and strength –effects of loading,” Z Rheumatol, vol. 59, no. 1, pp. I1–I9, February. 2000, doi: 10.1007/s003930070031. [DOI] [PubMed] [Google Scholar]
- [5].Ajubi NE, Klein-Nulend J, Nijweide PJ, Vrijheid-Lammers T, Alblas MJ, and Burger EH, “Pulsating fluid flow increases prostaglandin production by cultured chicken osteocytes--a cytoskeleton-dependent process,” Biochem Biophys Res Commun, vol. 225, no. 1, pp. 62–68, August. 1996, doi: 10.1006/bbrc.1996.1131. [DOI] [PubMed] [Google Scholar]
- [6].Burger EH and Klein‐Nulend J, “Mechanotransduction in bone—role of the lacunocanalicular network,” The FASEB Journal, vol. 13, no. 9001, pp. S101–S112, 1999, doi: 10.1096/fasebj.13.9001.s101. [DOI] [PubMed] [Google Scholar]
- [7].Klein‐Nulend J. et al. , “Sensitivity of osteocytes to biomechanical stress in vitro,” The FASEB Journal, vol. 9, no. 5, pp. 441–445, 1995, doi: 10.1096/fasebj.9.5.7896017. [DOI] [PubMed] [Google Scholar]
- [8].Bonewald LF, “The amazing osteocyte,” J. Bone Miner. Res, vol. 26, no. 2, pp. 229–238, February. 2011, doi: 10.1002/jbmr.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hung CT, Pollack SR, Reilly TM, and Brighton CT, “Real-time calcium response of cultured bone cells to fluid flow,” Clin Orthop Relat Res, no. 313, pp. 256–269, April. 1995. [PubMed] [Google Scholar]
- [10].Lewis KJ et al. , “Osteocyte calcium signals encode strain magnitude and loading frequency in vivo,” PNAS, vol. 114, no. 44, pp. 11775–11780, October. 2017, doi: 10.1073/pnas.1707863114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Klein-Nulend J, Semeins CM, Ajubi NE, Nijweide PJ, and Burger EH, “Pulsating fluid flow increases nitric oxide (NO) synthesis by osteocytes but not periosteal fibroblasts--correlation with prostaglandin upregulation,” Biochem Biophys Res Commun, vol. 217, no. 2, pp. 640–648, December. 1995, doi: 10.1006/bbrc.1995.2822. [DOI] [PubMed] [Google Scholar]
- [12].Forwood MR, “Inducible cyclo-oxygenase (COX-2) mediates the induction of bone formation by mechanical loading in vivo,” Journal of Bone and Mineral Research, vol. 11, no. 11, pp. 1688–1693, 1996, doi: 10.1002/jbmr.5650111112. [DOI] [PubMed] [Google Scholar]
- [13].Castillo AB, Chen JC, and Jacobs CR, “Chapter 14 - Cellular and molecular mechanotransduction in bone,” in Marcus and Feldman’s Osteoporosis (Fifth Edition), Dempster DW, Cauley JA, Bouxsein ML, and Cosman F, Eds. Academic Press, 2021, pp. 309–335. [Google Scholar]
- [14].Kelly NH, Schimenti JC, Ross FP, and van der Meulen MCH, “Transcriptional profiling of cortical versus cancellous bone from mechanically-loaded murine tibiae reveals differential gene expression,” Bone, vol. 86, pp. 22–29, May 2016, doi: 10.1016/j.bone.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Chermside-Scabbo CJ et al. , “Old Mice Have Less Transcriptional Activation But Similar Periosteal Cell Proliferation Compared to Young-Adult Mice in Response to in vivo Mechanical Loading,” J Bone Miner Res, vol. 35, no. 9, pp. 1751–1764, September. 2020, doi: 10.1002/jbmr.4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Holguin N, Brodt MD, and Silva MJ, “Activation of Wnt Signaling by Mechanical Loading Is Impaired in the Bone of Old Mice,” J. Bone Miner. Res, vol. 31, no. 12, pp. 2215–2226, December. 2016, doi: 10.1002/jbmr.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Morse A. et al. , “Mechanical Load Increases in Bone Formation via a Sclerostin-Independent Pathway,” Journal of Bone and Mineral Research, vol. 29, no. 11, pp. 2456–2467, 2014, doi: 10.1002/jbmr.2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Tu X. et al. , “Sost downregulation and local Wnt signaling are required for the osteogenic response to mechanical loading,” Bone, vol. 50, no. 1, pp. 209–217, January. 2012, doi: 10.1016/j.bone.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Robling AG et al. , “Mechanical Stimulation of Bone in Vivo Reduces Osteocyte Expression of Sost/Sclerostin *,” Journal of Biological Chemistry, vol. 283, no. 9, pp. 5866–5875, February. 2008, doi: 10.1074/jbc.M705092200. [DOI] [PubMed] [Google Scholar]
- [20].Li X. et al. , “Stimulation of Piezo1 by mechanical signals promotes bone anabolism,” eLife, vol. 8, doi: 10.7554/eLife.49631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Tomlinson RE et al. , “NGF-TrkA signaling in sensory nerves is required for skeletal adaptation to mechanical loads in mice,” Proc Natl Acad Sci U S A, vol. 114, no. 18, pp. E3632–E3641, May 2017, doi: 10.1073/pnas.1701054114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kelly NH, Schimenti JC, Ross FP, and van der Meulen MCH, “A method for isolating high quality RNA from mouse cortical and cancellous bone,” Bone, vol. 68, pp. 1–5, November. 2014, doi: 10.1016/j.bone.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kang KS, Hong JM, and Robling AG, “Postnatal β-catenin deletion from Dmp1-expressing osteocytes/osteoblasts reduces structural adaptation to loading, but not periosteal load-induced bone formation,” Bone, vol. 88, pp. 138–145, July. 2016, doi: 10.1016/j.bone.2016.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Nioi P. et al. , “Transcriptional Profiling of Laser Capture Microdissected Subpopulations of the Osteoblast Lineage Provides Insight Into the Early Response to Sclerostin Antibody in Rats,” J. Bone Miner. Res, vol. 30, no. 8, pp. 1457–1467, August. 2015, doi: 10.1002/jbmr.2482. [DOI] [PubMed] [Google Scholar]
- [25].Taylor S. et al. , “Time-dependent cellular and transcriptional changes in the osteoblast lineage associated with sclerostin antibody treatment in ovariectomized rats,” Bone, vol. 84, pp. 148–159, March. 2016, doi: 10.1016/j.bone.2015.12.013. [DOI] [PubMed] [Google Scholar]
- [26].Nazarov PV et al. , “RNA sequencing and transcriptome arrays analyses show opposing results for alternative splicing in patient derived samples,” BMC Genomics, vol. 18, no. 1, Art. no. 1, December. 2017, doi: 10.1186/s12864-017-3819-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Turnbull AK et al. , “Unlocking the transcriptomic potential of formalin-fixed paraffin embedded clinical tissues: comparison of gene expression profiling approaches,” BMC Bioinformatics, vol. 21, no. 1, p. 30, January. 2020, doi: 10.1186/s12859-020-3365-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ibragimova MK, Tsyganov MM, Pevzner AM, and Litviakov NV, “Transcriptome of Breast Tumors With Different Amplification Status of the Long Arm of Chromosome 8,” Anticancer Research, vol. 41, no. 1, pp. 187–195, January. 2021, doi: 10.21873/anticanres.14764. [DOI] [PubMed] [Google Scholar]
- [29].Schäffler H. et al. , “NOD2- and disease-specific gene expression profiles of peripheral blood mononuclear cells from Crohn’s disease patients,” World J Gastroenterol, vol. 24, no. 11, pp. 1196–1205, March. 2018, doi: 10.3748/wjg.v24.i11.1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Brodt MD, Ellis CB, and Silva MJ, “Growing C57Bl/6 Mice Increase Whole Bone Mechanical Properties by Increasing Geometric and Material Properties,” Journal of Bone and Mineral Research, vol. 14, no. 12, pp. 2159–2166, 1999, doi: 10.1359/jbmr.1999.14.12.2159. [DOI] [PubMed] [Google Scholar]
- [31].Patel TK, Brodt MD, and Silva MJ, “Experimental and finite element analysis of strains induced by axial tibial compression in young-adult and old female C57Bl/6 mice,” J Biomech, vol. 47, no. 2, pp. 451–457, January. 2014, doi: 10.1016/j.jbiomech.2013.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Holguin N, Brodt MD, Sanchez ME, and Silva MJ, “Aging diminishes lamellar and woven bone formation induced by tibial compression in adult C57BL/6,” Bone, vol. 65, pp. 83–91, August. 2014, doi: 10.1016/j.bone.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ashburner M. et al. , “Gene Ontology: tool for the unification of biology,” Nat Genet, vol. 25, no. 1, pp. 25–29, May 2000, doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Mi H, Muruganujan A, Ebert D, Huang X, and Thomas PD, “PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools,” Nucleic Acids Research, vol. 47, no. D1, pp. D419–D426, January. 2019, doi: 10.1093/nar/gky1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].The Gene Ontology Consortium, “The Gene Ontology Resource: 20 years and still GOing strong,” Nucleic Acids Research, vol. 47, no. D1, pp. D330–D338, January. 2019, doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Grüneboom A. et al. , “A network of trans-cortical capillaries as mainstay for blood circulation in long bones,” Nat Metab, vol. 1, no. 2, pp. 236–250, February. 2019, doi: 10.1038/s42255-018-0016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Coates BA et al. , “Transcriptional profiling of intramembranous and endochondral ossification after fracture in mice,” Bone, vol. 127, pp. 577–591, October. 2019, doi: 10.1016/j.bone.2019.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Xiong J, Onal M, Jilka RL, Weinstein RS, Manolagas SC, and O’Brien CA, “Matrix-embedded cells control osteoclast formation,” Nat Med, vol. 17, no. 10, pp. 1235–1241, September. 2011, doi: 10.1038/nm.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Nakashima T. et al. , “Evidence for osteocyte regulation of bone homeostasis through RANKL expression,” Nature Medicine, vol. 17, no. 10, Art. no. 10, October. 2011, doi: 10.1038/nm.2452. [DOI] [PubMed] [Google Scholar]
- [40].Qing H. et al. , “Demonstration of osteocytic perilacunar/canalicular remodeling in mice during lactation,” J Bone Miner Res, vol. 27, no. 5, pp. 1018–1029, May 2012, doi: 10.1002/jbmr.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Youlten SE et al. , “Osteocyte Transcriptome Mapping Identifies a Molecular Landscape Controlling Skeletal Homeostasis and Susceptibility to Skeletal Disease,” bioRxiv, p. 2020.04.20.051409, April. 2020, doi: 10.1101/2020.04.20.051409. [DOI] [PMC free article] [PubMed]
- [42].Paic F. et al. , “Identification of Differentially Expressed Genes Between Osteoblasts and Osteocytes,” Bone, vol. 45, no. 4, pp. 682–692, October. 2009, doi: 10.1016/j.bone.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Dallas SL and Bonewald LF, “Dynamics of the Transition from Osteoblast to Osteocyte,” Ann N Y Acad Sci, vol. 1192, pp. 437–443, March. 2010, doi: 10.1111/j.1749-6632.2009.05246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Cabahug‐Zuckerman P. et al. , “Site‐Specific Load‐Induced Expansion of Sca‐1+Prrx1+ and Sca-1−Prrx1+ Cells in Adult Mouse Long Bone Is Attenuated With Age,” JBMR Plus, vol. 3, no. 9, July. 2019, doi: 10.1002/jbm4.10199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Ouyang Z. et al. , “Prx1 and 3.2kb Col1a1 promoters target distinct bone cell populations in transgenic mice,” Bone, vol. 58, pp. 136–145, January. 2014, doi: 10.1016/j.bone.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Duchamp de Lageneste O. et al. , “Periosteum contains skeletal stem cells with high bone regenerative potential controlled by Periostin,” Nature Communications, vol. 9, no. 1, Art. no. 1, February. 2018, doi: 10.1038/s41467-018-03124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Matic I. et al. , “Quiescent Bone Lining Cells Are a Major Source of Osteoblasts During Adulthood,” Stem Cells, vol. 34, no. 12, pp. 2930–2942, December. 2016, doi: 10.1002/stem.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Blair HC et al. , “Osteoblast Differentiation and Bone Matrix Formation In Vivo and In Vitro,” Tissue Engineering Part B: Reviews, vol. 23, no. 3, pp. 268–280, November. 2016, doi: 10.1089/ten.teb.2016.0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Lindskog C. et al. , “The human cardiac and skeletal muscle proteomes defined by transcriptomics and antibody-based profiling,” BMC Genomics, vol. 16, no. 1, p. 475, June. 2015, doi: 10.1186/s12864-015-1686-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].“The human proteome in skeletal muscle - The Human Protein Atlas.” https://www.proteinatlas.org/humanproteome/tissue/skeletal+muscle (accessed Jan. 24, 2021).
- [51].Deckelbaum RA et al. , “The potassium channel Kcne3 is a VEGFA-inducible gene selectively expressed by vascular endothelial tip cells,” Angiogenesis, vol. 23, no. 2, pp. 179–192, May 2020, doi: 10.1007/s10456-019-09696-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Nicks KM, Perrien DS, Akel NS, Suva LJ, and Gaddy D, “Regulation of osteoblastogenesis and osteoclastogenesis by the other reproductive hormones, Activin and Inhibin,” Mol Cell Endocrinol, vol. 310, no. 1–2, pp. 11–20, October. 2009, doi: 10.1016/j.mce.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kasri NN, Parys JB, Callewaert G, Missiaen L, and De Smedt H, “Calmodulin and calcium-release channels,” Biol Res, vol. 37, no. 4, pp. 577–582, 2004, doi: 10.4067/s0716-97602004000400011. [DOI] [PubMed] [Google Scholar]
- [54].Sutton KA, Jungnickel MK, Wang Y, Cullen K, Lambert S, and Florman HM, “Enkurin is a novel calmodulin and TRPC channel binding protein in sperm,” Dev Biol, vol. 274, no. 2, pp. 426–435, October. 2004, doi: 10.1016/j.ydbio.2004.07.031. [DOI] [PubMed] [Google Scholar]
- [55].Havis E. and Duprez D, “EGR1 Transcription Factor is a Multifaceted Regulator of Matrix Production in Tendons and Other Connective Tissues,” Int J Mol Sci, vol. 21, no. 5, February. 2020, doi: 10.3390/ijms21051664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Li H, Ghazanfari R, Lim HC, Hansson J, Zacharaki D, and Scheding S, “Early Growth Response (EGR)-1 Is a Key Regulator of Human Primary Bone Marrow Stroma Cells with a Dual Role in Proliferation and Hematopoietic Stroma Support Function,” Blood, vol. 130, no. Supplement 1, pp. 3777–3777, December. 2017, doi: 10.1182/blood.V130.Suppl_1.3777.3777. [DOI] [Google Scholar]
- [57].Girgenrath M. et al. , “TWEAK, via its receptor Fn14, is a novel regulator of mesenchymal progenitor cells and skeletal muscle regeneration,” EMBO J, vol. 25, no. 24, pp. 5826–5839, December. 2006, doi: 10.1038/sj.emboj.7601441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Merciris D, Schiltz C, Legoupil N, Marty-Morieux C, de Vernejoul MC, and Geoffroy V, “Over-expression of TIMP-1 in osteoblasts increases the anabolic response to PTH,” Bone, vol. 40, no. 1, pp. 75–83, January. 2007, doi: 10.1016/j.bone.2006.07.013. [DOI] [PubMed] [Google Scholar]
- [59].Xi Y, Huang H, Zhao Z, Ma J, and Chen Y, “Tissue inhibitor of metalloproteinase 1 suppresses growth and differentiation of osteoblasts and differentiation of osteoclasts by targeting the AKT pathway,” Experimental Cell Research, vol. 389, no. 2, p. 111930, April. 2020, doi: 10.1016/j.yexcr.2020.111930. [DOI] [PubMed] [Google Scholar]
- [60].Liang T. et al. , “TIMP-1 inhibits proliferation and osteogenic differentiation of hBMSCs through Wnt/β-catenin signaling,” Bioscience Reports, vol. 39, no. BSR20181290, January. 2019, doi: 10.1042/BSR20181290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Tang S, Herber R-P, Ho S, and Alliston T, “Matrix Metalloproteinase-13 is Required for Osteocytic Perilacunar Remodeling and Maintains Bone Fracture Resistance,” J Bone Miner Res, vol. 27, no. 9, pp. 1936–1950, September. 2012, doi: 10.1002/jbmr.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Crotti TN et al. , “Bone matrix regulates osteoclast differentiation and annexin A8 gene expression,” J Cell Physiol, vol. 226, no. 12, pp. 3413–3421, December. 2011, doi: 10.1002/jcp.22699. [DOI] [PubMed] [Google Scholar]
- [63].Goebeler V, Poeter M, Zeuschner D, Gerke V, and Rescher U, “Annexin A8 Regulates Late Endosome Organization and Function,” MBoC, vol. 19, no. 12, pp. 5267–5278, October. 2008, doi: 10.1091/mbc.e08-04-0383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Lueck K, Carr A-JF, Yu L, Greenwood J, and Moss SE, “Annexin A8 regulates Wnt signaling to maintain the phenotypic plasticity of retinal pigment epithelial cells,” Scientific Reports, vol. 10, no. 1, Art. no. 1, January. 2020, doi: 10.1038/s41598-020-58296-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Conover CA et al. , “Metalloproteinase pregnancy-associated plasma protein A is a critical growth regulatory factor during fetal development,” Development, vol. 131, no. 5, pp. 1187–1194, March. 2004, doi: 10.1242/dev.00997. [DOI] [PubMed] [Google Scholar]
- [66].Lean JM, Jagger CJ, Chambers TJ, and Chow JW, “Increased insulin-like growth factor I mRNA expression in rat osteocytes in response to mechanical stimulation,” Am J Physiol, vol. 268, no. 2 Pt 1, pp. E318–327, February. 1995, doi: 10.1152/ajpendo.1995.268.2.E318. [DOI] [PubMed] [Google Scholar]
- [67].Lau K-HW, Kapur S, Kesavan C, and Baylink DJ, “Up-regulation of the Wnt, estrogen receptor, insulin-like growth factor-I, and bone morphogenetic protein pathways in C57BL/6J osteoblasts as opposed to C3H/HeJ osteoblasts in part contributes to the differential anabolic response to fluid shear,” J Biol Chem, vol. 281, no. 14, pp. 9576–9588, April. 2006, doi: 10.1074/jbc.M509205200. [DOI] [PubMed] [Google Scholar]
- [68].Reijnders CMA, Bravenboer N, Tromp AM, Blankenstein MA, and Lips P, “Effect of mechanical loading on insulin-like growth factor-I gene expression in rat tibia,” J Endocrinol, vol. 192, no. 1, pp. 131–140, January. 2007, doi: 10.1677/joe.1.06880. [DOI] [PubMed] [Google Scholar]
- [69].Sunters A. et al. , “Mechano-transduction in osteoblastic cells involves strain-regulated estrogen receptor alpha-mediated control of insulin-like growth factor (IGF) I receptor sensitivity to Ambient IGF, leading to phosphatidylinositol 3-kinase/AKT-dependent Wnt/LRP5 receptor-independent activation of beta-catenin signaling,” J Biol Chem, vol. 285, no. 12, pp. 8743–8758, March. 2010, doi: 10.1074/jbc.M109.027086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Naot D, Callon KE, Grey A, Cooper GJS, Reid IR, and Cornish J, “A Potential Role for Adrenomedullin as a Local Regulator of Bone Growth*,” Endocrinology, vol. 142, no. 5, pp. 1849–1857, May 2001, doi: 10.1210/endo.142.5.8152. [DOI] [PubMed] [Google Scholar]
- [71].Ida T. et al. , “Extracellular matrix with defective collagen cross-linking affects the differentiation of bone cells,” PLOS ONE, vol. 13, no. 9, p. e0204306, September. 2018, doi: 10.1371/journal.pone.0204306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Wattanachanya L. et al. , “Increased bone mass in mice lacking the adipokine apelin,” Endocrinology, vol. 154, no. 6, pp. 2069–2080, June. 2013, doi: 10.1210/en.2012-2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Bakker AD, Klein-Nulend J, and Burger EH, “Mechanotransduction in bone cells proceeds via activation of COX-2, but not COX-1,” Biochemical and Biophysical Research Communications, vol. 305, no. 3, pp. 677–683, June. 2003, doi: 10.1016/S0006-291X(03)00831-3. [DOI] [PubMed] [Google Scholar]
- [74].Ganesh L. et al. , “Protein methyltransferase 2 inhibits NF-kappaB function and promotes apoptosis,” Mol Cell Biol, vol. 26, no. 10, pp. 3864–3874, May 2006, doi: 10.1128/MCB.26.10.3864-3874.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Boyce BF, Yao Z, and Xing L, “Functions of NF-κB in Bone,” Ann N Y Acad Sci, vol. 1192, pp. 367–375, March. 2010, doi: 10.1111/j.1749-6632.2009.05315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Deng W. et al. , “The novel secretory protein CGREF1 inhibits the activation of AP-1 transcriptional activity and cell proliferation,” The International Journal of Biochemistry & Cell Biology, vol. 65, pp. 32–39, August. 2015, doi: 10.1016/j.biocel.2015.05.019. [DOI] [PubMed] [Google Scholar]
- [77].Stipanuk MH, Ueki I, Dominy JE, Simmons CR, and Hirschberger LL, “Cysteine Dioxygenase: A Robust System for Regulation of Cellular Cysteine Levels,” Amino Acids, vol. 37, no. 1, pp. 55–63, May 2009, doi: 10.1007/s00726-008-0202-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Hauschka PV, “Osteocalcin: The Vitamin K-Dependent Ca2+-Binding Protein of Bone Matrix,” PHT, vol. 16, no. 3–4, pp. 258–272, 1986, doi: 10.1159/000215298. [DOI] [PubMed] [Google Scholar]
- [79].Iida R, Yasuda T, Tsubota E, Matsuki T, and Kishi K, “Cloning, Mapping, Genomic Organization, and Expression of Mouse M-LP, a New Member of the Peroxisomal Membrane Protein Mpv17 Domain Family,” Biochemical and Biophysical Research Communications, vol. 283, no. 2, pp. 292–296, May 2001, doi: 10.1006/bbrc.2001.4769. [DOI] [PubMed] [Google Scholar]
- [80].Somerville RPT et al. , “Characterization of ADAMTS-9 and ADAMTS-20 as a Distinct ADAMTS Subfamily Related to Caenorhabditis elegans GON-1 *,” Journal of Biological Chemistry, vol. 278, no. 11, pp. 9503–9513, March. 2003, doi: 10.1074/jbc.M211009200. [DOI] [PubMed] [Google Scholar]
- [81].Hong D. et al. , “Morphological and proteomic analysis of early stage of osteoblast differentiation in osteoblastic progenitor cells,” Exp Cell Res, vol. 316, no. 14, pp. 2291–2300, August. 2010, doi: 10.1016/j.yexcr.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Joeng KS et al. , “Osteocyte-specific WNT1 regulates osteoblast function during bone homeostasis,” J Clin Invest, vol. 127, no. 7, pp. 2678–2688, June. 2017, doi: 10.1172/JCI92617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Laine CM et al. , “WNT1 Mutations in Early-Onset Osteoporosis and Osteogenesis Imperfecta,” 10.1056/NEJMoa1215458, May 08, 2013. https://www.nejm.org/doi/10.1056/NEJMoa1215458 (accessed Dec. 31, 2020). [DOI] [PMC free article] [PubMed]
- [84].Li Z. et al. , “Fracture repair requires TrkA signaling by skeletal sensory nerves,” J Clin Invest, vol. 129, no. 12, pp. 5137–5150, December. 2019, doi: 10.1172/JCI128428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Madel M-B et al. , “Immune Function and Diversity of Osteoclasts in Normal and Pathological Conditions,” Front. Immunol, vol. 10, 2019, doi: 10.3389/fimmu.2019.01408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Tomlinson RE, Schmieder AH, Quirk JD, Lanza GM, and Silva MJ, “Antagonizing the αvβ3 Integrin Inhibits Angiogenesis and Impairs Woven but Not Lamellar Bone Formation Induced by Mechanical Loading,” J Bone Miner Res, vol. 29, no. 9, pp. 1970–1980, September. 2014, doi: 10.1002/jbmr.2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Schaffler MB, Cheung W-Y, Majeska R, and Kennedy O, “Osteocytes: Master Orchestrators of Bone,” Calcif Tissue Int, vol. 94, no. 1, pp. 5–24, January. 2014, doi: 10.1007/s00223-013-9790-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.