Abstract
Background
The detection of somatic mutations in cell-free DNA (cfDNA) from liquid biopsy has emerged as a noninvasive tool to monitor the follow-up of cancer patients. However, the significance of cfDNA clinical utility remains uncertain in patients with brain tumors, primarily because of the limited sensitivity cfDNA has to detect real tumor-specific somatic mutations. This unresolved challenge has prevented accurate follow-up of glioma patients with noninvasive approaches.
Methods
Genome-wide DNA methylation profiling of tumor tissue and serum cfDNA of glioma patients.
Results
Here, we developed a noninvasive approach to profile the DNA methylation status in the serum of patients with gliomas and identified a cfDNA-derived methylation signature that is associated with the presence of gliomas and related immune features. By testing the signature in an independent discovery and validation cohorts, we developed and verified a score metric (the “glioma-epigenetic liquid biopsy score” or GeLB) that optimally distinguished patients with or without glioma (sensitivity: 100%, specificity: 97.78%). Furthermore, we found that changes in GeLB score reflected clinicopathological changes during surveillance (eg, progression, pseudoprogression, and response to standard or experimental treatment).
Conclusions
Our results suggest that the GeLB score can be used as a complementary approach to diagnose and follow up patients with glioma.
Keywords: cell-free DNA, epigenetics, glioma, pseudoprogression, serum
Key Points.
Serum is a noninvasive source of glioma DNA.
Serum DNA methylation can be used to detect glioma and associated immune signatures.
Machine learning using serum-based epigenetic markers can be applied to classify a patient’s serum methylome for glioma or not.
Importance of the Study.
Current glioma diagnostic and classification criteria that guide clinical management relies on the tissue profiling obtained by invasive surgical approaches (tissue biopsy or excision). We investigated whether glioma could be identified in a noninvasive manner by profiling epigenome-wide methylation of cell-free DNA in serum. We defined highly specific and sensitive noninvasive epigenetic markers that distinguish gliomas from non-gliomas. This study provides the framework to develop a blood-based epigenetic panel marker to complement the standard of care to diagnose, prognosticate, and monitor disease recurrence or progression as well as to stratify therapy which ultimately may improve the quality of life of these patients.
Gliomas are a heterogeneous group of intracranial tumors that constantly evolve, generally recur, and frequently progress to more malignant subtypes. Specific genomic and epigenomic alterations define subtypes of glioma that have distinct prognostic outcomes1–3 (eg, isocitrate dehydrogenase [IDH] mutation, 1p19q chromosomal deletion, and the glioma-CpG island methylator phenotype [G-CIMP]). Currently, the detection of such molecular abnormalities relies on profiling tissue obtained invasively by surgical approach (tissue biopsy or excision). However, this surgical approach does not allow for serial tissue evaluation to study the dynamic molecular evolution of these tumors; might not be feasible to obtain sufficient tumor material for molecular analysis in deep or surgically inaccessible tumors thereby delaying disease diagnosis. Currently, magnetic resonance imaging (MRI) is the preferred noninvasive method to diagnose and follow up the evolution of gliomas; however, besides being costly and cumbersome, MRI has limited ability to distinguish (1) gliomas from other tumors (eg, primary central nervous system [CNS] lymphoma), (2) progression from pseudoprogression resulting from therapy-induced necrosis, or (3) minimal or remnant tumoral burden.4,5
A liquid biopsy (LB) constitutes an attractive minimally or noninvasive method that may overcome some of the challenges and limitations faced by the other approaches. LB allows the detection of materials shed by tumors, including circulating tumor cells and genomic specimens6 in biofluids (eg, blood and cerebrospinal fluid [CSF]), enabling the identification of biomarkers that reflect tumor burden and dynamic evolution earlier or in real-time. In the past decade, many researchers have investigated the diagnostic, prognostic, and predictive applications of LB for many tumor types throughout a patient’s disease course.7–11 For instance, in CNS neoplasms, including gliomas, CSF has shown to be a relevant source of molecular markers6,12–21 and was used to track the evolution of gliomas.22 However, the procedure to obtain CSF is invasive and, to some extent, complex and risky in patients harboring CNS tumors, which limits its use for serial assessment of the disease. In contrast, LB of blood is minimally invasive, quick, and thus can be easily performed longitudinally.
One of the main limitations of blood-derived LB is the dismal and often low yield of molecular material released into the blood by CNS tumors, which may hinder the detection of targeted molecular features.6,21,23 A more comprehensive “omics” approach, such as the whole-genome DNA methylation profile, was proposed to overcome that limitation6,24 and was implemented for certain cancer types.23,25,26 Not only is DNA methylation a stable and tissue-specific marker, but it is also clinically relevant to gliomas.2,27 The detection of tumor-specific DNA methylation alterations in LB-derived specimens has already been described in non-CNS neoplasms.25,28 We hypothesized that the profiling of the genome-wide methylome of circulating cell-free DNA (cfDNA) via microarray allows the identification of specific markers associated with glioma.2,29
In this study, we identified a set of epigenetic signatures in the serum that reflects the tissue methylome landscape of glioma as well as immune signatures specifically associated with the detection of glioma. Using these signatures, we developed a model through a supervised machine learning (ML) that predicted the diagnosis of glioma in an independent validation dataset and reflected tumor dynamic changes throughout tumor burden surveillance with high accuracy. Our results lay the groundwork to apply serum-derived methylation signatures to diagnose and follow up patients with gliomas.
Materials and Methods
Patients
This study included 208 serum samples collected at primary visit only (n = 15), primary and recurrence or follow-up visits (n = 33 patients with 71 serum samples), or recurrence visit only (n = 14 patients with 28 serum samples) from 149 patients who underwent surgery to resect gliomas, as well as serum from patients with brain metastatic tumor (n = 4), colloid cyst (n = 1), pituitary adenoma (n = 14), primary meningioma (n = 42), recurrent meningioma (n = 21), CNS lymphoma (n = 4), and non-tumor brain diseases (n = 8) stored at the Department of Neurosurgery, Hermelin Brain Tumor Center (HBTC) and at the Henry Ford Health System (HFHS, Detroit, MI) from 2011 to 2019. In addition, publicly available data from serum cfDNA from two patients with colorectal cancer30,31 were included. Longitudinal clinical annotation for 71 time points (primary, recurrent, and/or off-treatment follow-up) across 33 HBTC patients with glioma was rereviewed by two neuro-oncologists (J.S. and T.W.), one neuro-surgeon (I.L.), and two neuro-pathologists (A.M. and D.A.C.). Detailed demographic information about the study cohort is given in Table 1 and Supplementary Table S1. The project was approved by the HFHS Institutional Review Board (#12490), and patients consented to allow their specimens to be used for research purposes. Deidentified IDs were used to protect patient privacy.
Table 1.
Summary of the Clinical Features
| Characteristics | Total Primary (n = 48) | IDH Mutant (n = 19) | IDH Wild Type (n = 7) | IDH Unknown (n = 22) |
|---|---|---|---|---|
| Clinical | ||||
| Anaplastic astrocytoma | 6 (12.5%) | 3 (15.8%) | 2 (28.6%) | 1 (4.5%) |
| Anaplastic oligodendroglioma | 5 (10.4%) | 3 (15.8%) | 2 (28.6%) | 0 (0%) |
| Glioblastoma | 27 (56.3%) | 3 (15.8%) | 3 (42.8%) | 21 (95.5%) |
| Astrocytoma | 5 (10.4%) | 5 (26.3%) | 0 (0%) | 0 (0%) |
| Oligodendroglioma | 5 (10.4%) | 5 (26.3%) | 0 (0%) | 0 (0%) |
| Age | ||||
| Mean (SD) | 54.5 ± 15.7 | 47.8 ± 17.2 | 59.7 ± 20.4 | 58.6 ± 10.6 |
| Range | 21-82 | 28-78 | 21-79 | 44-82 |
| Race | ||||
| Black or African American | 8 (16.7%) | 3 (15.8%) | 3 (42.9%) | 2 (9.1%) |
| Caucasian | 38 (79.1%) | 14 (73.6%) | 4 (57.1%) | 20 (90.9%) |
| Hispanic | 1 (2.1%) | 1 (5.3%) | 0 (0%) | 0 (0%) |
| Indian | 1 (2.1%) | 1 (5.3%) | 0 (0%) | 0 (0%) |
| Survival | ||||
| Mean (LQ-UQ) (months) | 38 (20.1-55.7) | 50.4 (40.9-65.6) | 27.1 (8.9-33.7) | 30.8 (18.1-37.4) |
| Gender | ||||
| Female | 17 (35.4%) | 9 (47.4%) | 5 (71.4%) | 3 (13.6%) |
| Male | 31 (64.6%) | 10 (52.6%) | 2 (28.6%) | 19 (86.4%) |
| 1p19q Codel status | ||||
| Intact | 24 (50.0%) | 10 (52.6%) | 7 (100%) | 7 (31.8%) |
| Co-deleted | 9 (18.8%) | 7 (36.9%) | 0 (0%) | 2 (9.1%) |
| Unknown | 15 (31.2%) | 2 (10.5%) | 0 (0%) | 13 (59.1%) |
| MGMT promoter status | ||||
| Methylated | 27 (56.2%) | 15 (78.9%) | 4 (57.1%) | 8 (36.4%) |
| Unmethylated | 21 (43.8%) | 4 (21.1%) | 3 (42.9%) | 14 (63.6%) |
Abbreviations: LQ-UQ, lower quartiles-upper quartiles; MGMT, O-6-methylguanine-DNA methyltransferase.
Serum Collection and Preprocessing
Peripheral blood (15 mL) was drawn from each subject at the time of surgery (both for primary or recurrent samples), before opening the dura mater. The peripheral blood of six patients was also drawn at follow-up (off-treatment, 7-42 months from diagnosis). Serum was separated within 1 h of collection by centrifugation at 1300 × g for 10 min at 20°C, aliquoted into up to five 2-mL cryovials, and stored at −80°C until cfDNA isolation and processing.
Molecular (DNA/RNA) Isolation and Data Generation and Processing
All patient cfDNA was extracted from 1.2- to 9.3-mL aliquots of serum (Supplementary Table S1) by using the Quick-cfDNA Serum & Plasma Kit according to the manufacturer’s protocol (Zymo Research, catalog # D4076). DNA concentration and integrity were measured with a Qubit (Thermo Fisher Scientific) or a 4200 TapeStation (Agilent Technologies). For 22 patients with available tumor tissue, DNA and RNA were isolated from the tissue, and DNA methylation profiling (Illumina Human 450K array—HM450K), whole-genome sequencing, and RNA sequencing were performed as we reported previously1,2,32,33 (Supplementary Table S1). From the entire cohort, serum cfDNA (2-100 ng) was extracted, bisulfite-converted (Zymo EZ DNA methylation Kit; Zymo Research), and profiled using an Illumina Human EPIC array (EPIC). Prior to profiling, the isolated DNA was restored using a restoration kit provided by Illumina. This allowed us to restore fragmented DNA and concentrate the low yield. This is the same kit and approach described by Moss et al.30 They show reproducible results using low cfDNA (up to 50 ng). The raw cfDNA methylation intensity data files (IDAT) have been deposited to Mendeley Data (ID: cgrz6zztfg). Serum cfDNA methylation data from patients without glioma were obtained from previously published reports.30,31 IDAT files (derived from HBTC and published data) were processed using the minfi package34 in R using a protocol described previously.2
Deconvolution
We used a published method30 to deconvolute the relative contribution of different cell types to a given sample based on the top 100 of the most specific hypermethylated and hypomethylated CpG probes for each cell type. Using those 100 probe signatures, we applied a nonnegative least squares method to deconvolute the cell types of our serum and tissue cohorts by using the standalone program provided by Moss and colleagues.30 We then normalized the percentages generated by the standalone program for each cell type from 0 to 100 by serum or tissue separately.
Development of the Glioma-Epigenetic-Liquid Biopsy (GeLB) Score
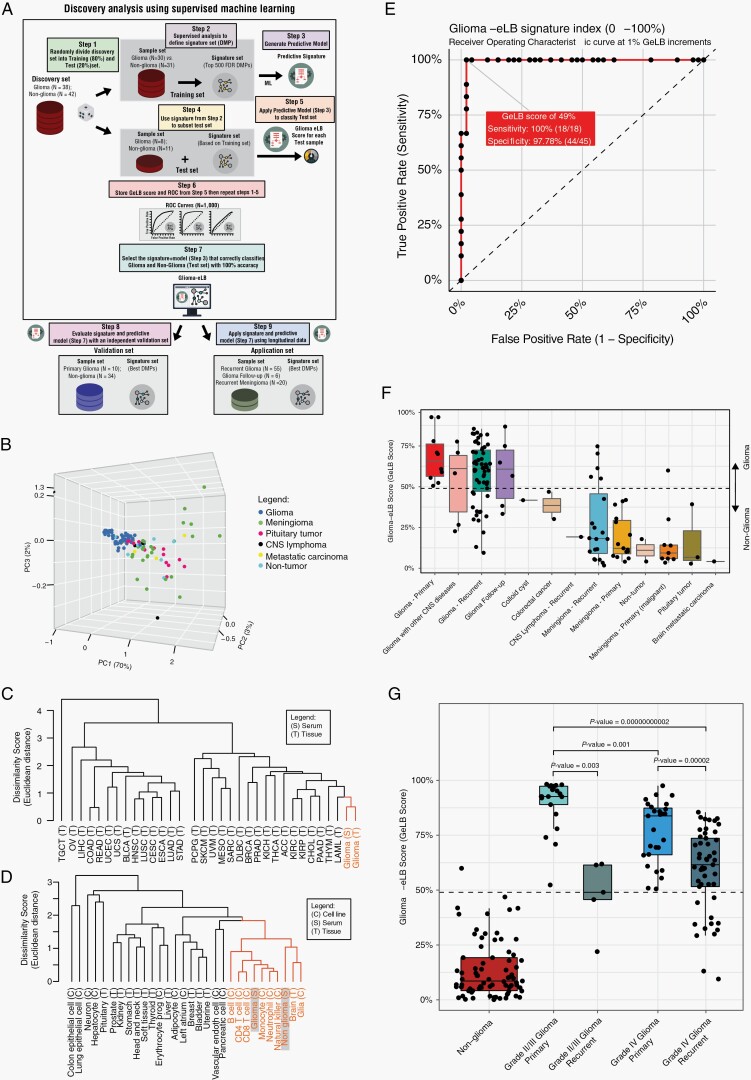
We performed a supervised epigenome-wide differential analysis to identify specific, serum-based, epigenetic markers associated with glioma and then applied the signatures to a ML-based model to predict the presence of glioma. In order to capture glioma-specific DNA methylation-based signatures, we compared the methylome of matched serum and tissue from glioma patients and selected CpGs in which the methylation levels were similar (<5% average DNA methylation difference). In step 1, we randomly selected 38 samples from patients with primary glioma and 42 from those without glioma and classified them as our discovery set (n = 80). The remaining serum cfDNA methylation data from patients with primary or recurrent gliomas or without glioma were included in the validation and application sets (n = 125). In step 2, we randomized our discovery set into training (80%) and test (20%) sets. In step 3, using the training set and a supervised method, we selected the top 500 differentially methylated probes sorted on the basis of false discovery rate (FDR)-corrected Wilcoxon rank-sum test P value. We confirmed the glioma origin of the eLB signature by evaluating the measured DNA methylation for each CpG in the glioma tissue profiled as part of The Cancer Genome Atlas (TCGA) project. We named these signatures “glioma-eLB” or “GeLB.” In step 4, with this training and signature (GeLB) dataset, we created a predictive ML model using a random forest (RF) method. In steps 5 and 6, we applied the model to the test set (using the same selected GeLB data as the training set [step 5]) to classify the samples as glioma or non-glioma. This analysis provided us with a GeLB score, which estimates the probability that a serum cfDNA sample originates from a patient with glioma (which depicts a high GeLB score, close to 100%) or not (low GeLB score, close to 0%). A score was generated for each patient, and a receiver operating characteristic (ROC) curve was analyzed to identify the optimal cutoff score for maximum sensitivity and specificity. We repeated steps 2 through 5 a thousand times, each time reshuffling the discovery set into training and test sets (step 2) and storing the RF scores for each testing sample per iteration (Supplementary Figure S2A). The iteration (or GeLB selection) that correctly identified gliomas (n = 8) from non-gliomas (n = 11) in the test set, along with the greatest difference in score (minimum score of the gliomas minus the maximum score of the non-gliomas) was then identified as the optimal GeLB signature. We selected the top 500 most significantly different CpGs (FDR < 0.01) based on DNA methylation levels in the serum. We identified 476 CpG sites (Supplementary Figure S2B), now called as GeLB signature, which was used to discriminate the serum of glioma from non-glioma patients (Figure 2B). Using the GeLB signature, we determined that a GeLB score cutoff of 49% accurately distinguished serum from a patient with glioma from another with non-glioma condition in our test set (step 7).
Fig. 2.
Identification of glioma-specific epigenetic liquid biopsy (GeLB) as a diagnostic marker for gliomas. (A) Schematic diagram of the method (supervised machine learning) and approach (discovery and independent validation) to identify the glioma-eLB CpG signatures used to generate the GeLB score (see Materials and Methods for details). (B) Principal component analysis. (C, D) Hierarchical clustering for (C) primary tumor tissue and (D) non-tumorous cell and tissue types. (E) Receiver operating characteristic curve derived from the independent validation set. (F) GeLB score calculated for each tumor tissue and available cfDNA methylation serum. (G) GeLB score distribution by glioma grade (II/III and IV). Abbreviation: cfDNA, cell-free DNA.
Statistical Analysis
All processing and statistical analyses were done using R (3.6.3). The Wilcoxon rank-sum test and adjustments for multiple testing (eg, FDR) were used to identify differentially methylated sites. To identify tissue-specific methylated probes, we calculated the mean DNA methylation difference for each CpG probe between serum and the tissue and set the cutoff difference at less than 5%. We used RF analysis as the ML method to classify and predict glioma.
Results
Serum cfDNA Methylome Profile Derived From Primary Glioma
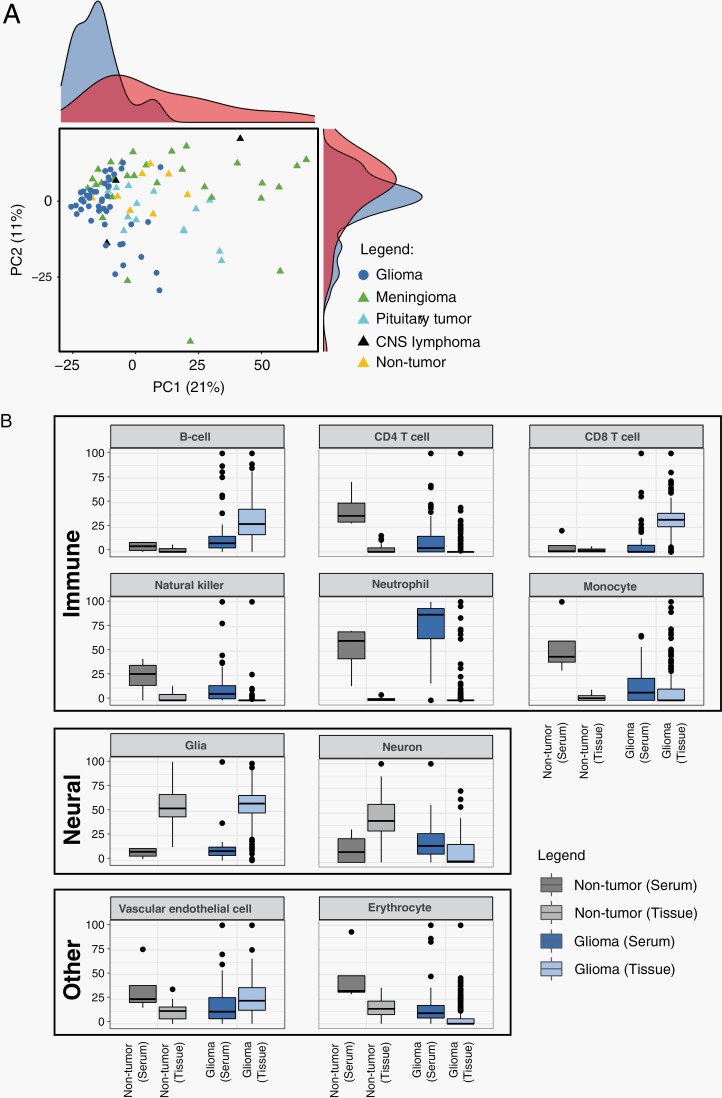
The total cfDNA quantity extracted from serum, normalized by the genome size (genomic equivalents [GE]/ml), showed that patients with glioma had significantly higher serum cfDNA levels than patients with other tumors or conditions (mean ± SE: 15 416.48 ± 2221.5 vs 2694 ± 638.9 GE/ml, Student’s t-test, P = .000001). The mean DNA input used to profile methylation with the Illumina microarray (EPIC) was 117.7 ng. Principal component analysis (PCA) of the genome-wide cfDNA methylation level showed a distinct separation between primary gliomas (n = 48) and non-tumorous or other neoplasms specimens (Figure 1A, Supplementary Table S1).
Fig. 1.
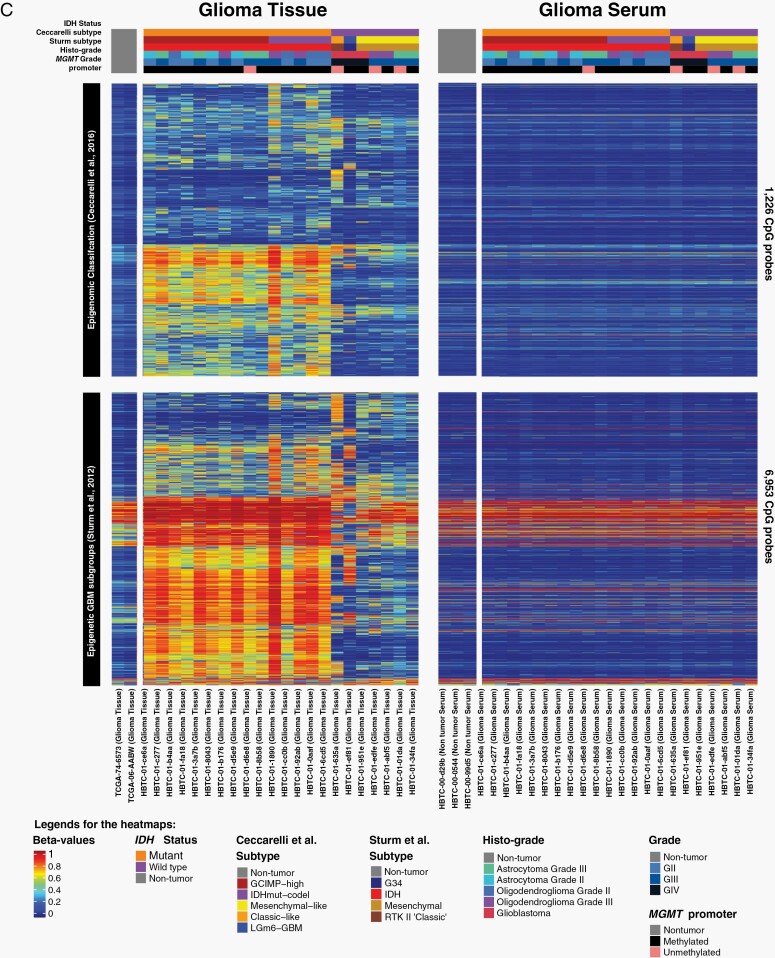
Genome-wide DNA methylation profile of serum cell-free DNA (cfDNA) from patients with glioma. (A) Principal component analysis of serum-based cfDNA methylation. (B) Cell-type-specific CpG methylation-based deconvolution. (C) Published tissue-derived epigenetic signatures are shown as a heatmap.
The composition of immune cell-specific methylation-based signatures in the serum from patients with glioma was distinct from the non-tumor serum and the glioma tissue counterparts. For instance, B-cell-related signatures were higher on average in patients with glioma than in patients without tumor, both in serum (average 1.45-fold higher, P < .05) and tissue samples (20- and 18.88-fold higher, P < .05) (Figure 1B, Supplementary Table S2). Conversely, CD4 T cells, monocytes, and erythrocyte-related signatures were depleted in serum from patients with tumors compared with those without tumors (0.8-, 0.34-, and 0.17-fold lower, P < .05). In tissue, signatures from B cells, CD8 T cells, monocytes, vascular cells, natural killer cells, and neutrophils were >1-fold higher (P < .05), and signatures from neurons were lower (P < .05) in the tumorous than in non-tumor tissues. The serum from glioma patients contained high and variable amounts of neutrophil- and CD8 T-cell-related signatures.
GeLB Score to Predict Patients With Glioma
The majority of the published glioma-epigenetic signatures derived from tissue were undetectable in the serum methylome (Figure 1C, Supplementary Figure S1). In order to explore the glioma specificity of the GeLB signature, we used methylomes from independent cohorts of primary glioma tissue and other non-glioma tumors and showed that the glioma serum methylome clustered with the primary glioma tissue counterpart (Figure 2C, Supplementary Figure S2C, D), thereby corroborating our initial observation that the eLB measurements reflected the glioma tissue methylome. To further investigate the content of the GeLB signature, we analyzed the similarity matrix across available non-cancer cell-type data based on methylation (brain-, neural-, and immune-associated cell types) and found that the serum cfDNA methylome from patients with glioma was similar to cell signatures related to neutrophils, monocytes, and normal glia- and neuronal-cell methylation than to other non-neoplastic cell types originating from different cell lineages (Figure 2D). The non-tumor brain serum segregated with brain and glial cell types, suggesting that the GeLB signature reflected tissue-of-origin signatures.
Next, we annotated the genomic location of the GeLB signatures (N = 476). We found that 147 CpG probes (31%) overlapped with known CpG Island; 107/147 were hypermethylated and enriched in CpG Island (OR = 0.45, 95% CI: [0.30, 0.68], chi-square P < .0001) and 40/147 were hypomethylated and enriched in CpG Island (OR = 1.94, 95% CI: [1.31-2.86], chi-square P < .001) above the expected distribution for the EPIC platform (data not shown). The lack of gene expression data matched to normal brain tissue DNA methylation limited our ability to investigate the biological context of these GeLB signatures.
Independent Validation of the GeLB Score
Given the tissue specificity of the detectable serum GeLB signature, we applied a supervised ML model to determine the robustness of the GeLB signature to detect the presence of glioma in an independent validation set (Figure 2E, F). The validation set consisted of serum cfDNA methylation data from primary (n = 18) and recurrent (n = 55) gliomas, and LB from patients with non-tumorous diseases of the brain (epilepsy and colloid cyst) or either intracranial (pituitary adenoma, meningioma, brain metastasis, and CNS lymphoma) and extracranial tumors (colorectal cancer). Using the GeLB signature previously obtained by RF analysis on the discovery set, we generated a GeLB score, which ranged from 0% to 100%, for each sample in the validation set (Figure 2F, Supplementary Table S1). Interestingly, from the discovery set, which had an equal distribution between grade II/III and grade IV glioma, the GeLB score was significantly higher in low-grade (II/III) than high-grade (IV) glioma at diagnosis, a condition in which the impact of the hypermethylated epigenome (G-CIMP)1 maximally differentiates the two groups of tumors (Figure 2G). We evaluated the GeLB score as an independent validation at increments of 1% and determined that, at our defined optimal cutoff of 49%, the signature could accurately (accuracy = 98%) distinguish a patient with primary glioma from one without (sensitivity = 100% [18/18]; specificity = 97.78% [44/45], Figure 2E). The only false-positive was a primary aggressive and atypical meningioma misclassified as glioma. Notably, the four CNS lymphomas included in our non-glioma cohort were correctly classified as non-glioma.
Identification of IDH Glioma Subtypes by Noninvasive eLB
Somatic mutation in one of the IDH genes (IDH1, IDH2) is a prognostic marker for adult glioma (World Health Organization [WHO] grade II-IV) and is traditionally identified from excised brain tissue. IDH status was available for 26 patients through targeted IDH sequencing or immuno-histochemistry (IDH mutation [IDHmut], n = 19 and wild-type IDH [IDHwt], n = 7) (Table 1, Supplementary Table S1) while 22 patients had unknown IDH status. There was a significant difference in overall survival between patients with IDHmut and IDHwt tumors (median [95% CI]: 50.4 months [41.4-59.4] vs 27.1 months [4.7-49.5], respectively, Supplementary Figure S1A). We combined IDHmut 1p19q codeletion (n = 5, Codel) and IDHmut 1p19q intact (n = 10) into one class, IDHmut (n = 15).
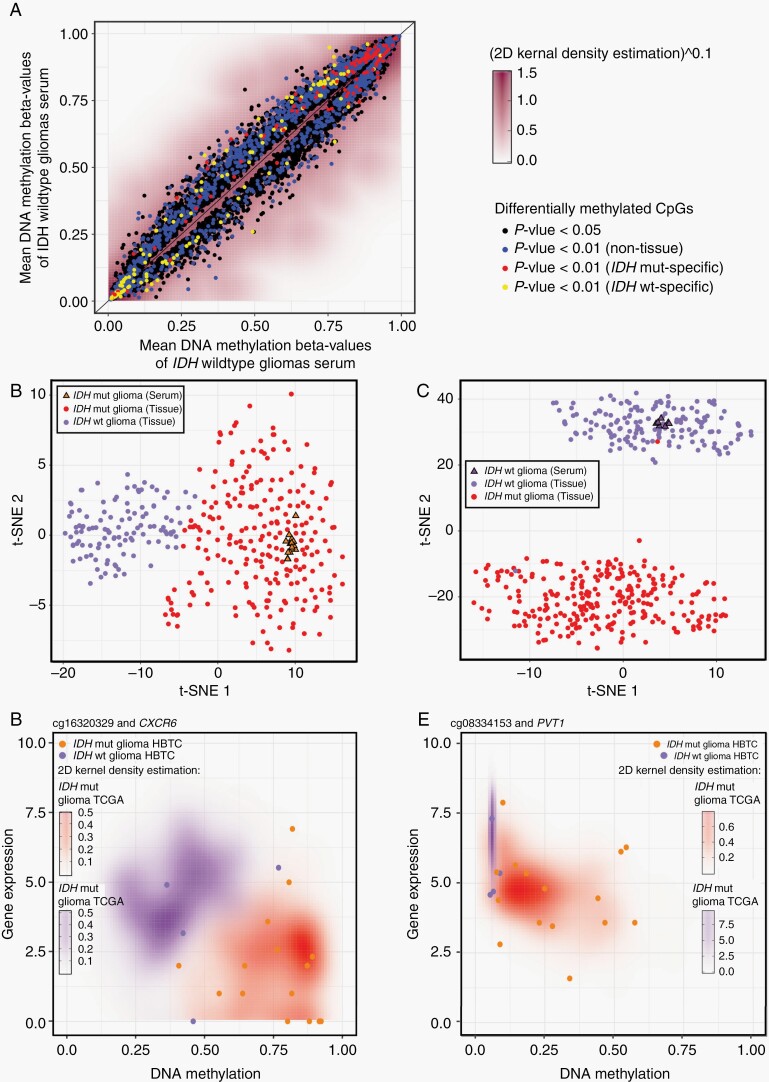
Applying a supervised method and restricting our analysis to CpGs within autosomal chromosomes (chrom 1-22), we identified 2647 IDH-eLB signatures (unadjusted P < .01) that distinguished IDHmut from IDHwt gliomas (Figure 3A). To improve our specificity, we refined our analysis further by selecting serum samples with a similar methylation pattern in the matching tumor tissue (CpG overlap HM450K = 1525/2647) which generated specific IDHmut-eLB and IDHwt-eLB signatures (n = 114/1525 [7.5%] and n = 124/1525 [8%], respectively; Figure 3A, Supplementary Figure S3A). Harnessing the matching tissue methylome as well as pan-glioma methylome data from adult patients (Supplementary Figure S2E), we found that the IDH-specific eLB distinguished the two IDH subtypes at the tissue level and the IDH-serum methylome (IDHwt and IDHmut) clustered with the respective IDH tissue subtype (Figure 3B, C, Supplementary Figure S3B, D), corroborating the specificity of the identified IDH-eLB.
Fig. 3.
IDH-specific eLB prognostic markers. (A) Mean genome-wide DNA methylation beta values in patient-derived serum from 15 with IDHmut (x-axis) and 7 with IDHwt (y-axis). t-Distributed stochastic neighbor embedding (t-SNE) using (B) IDHmut and (C) IDHwt tissue-specific eLB signatures. (D and E) Scatter plots of DNA methylation (x-axis) vs gene expression (y-axis) for all pan-glioma primary tumor tissues for promoter CpGs-associated with CXCR6 (D) and PVT1 (E). Abbreviations: eLB, epigenetic liquid biopsy; IDH, isocitrate dehydrogenase.
We defined IDH mutation score by analyzing the cfDNA methylation data of serum from patients with IDH wild-type or IDH-mutant tumors. We used the same approach to define IDH-eLB as described for GeLB (ie, supervised epigenome-wide differential analysis). We then investigated the potential functional or biological role of the IDH-eLB signature by analyzing the methylome and transcriptome (RNA-seq) from the matching glioma tissue. Consistent with others who have shown enrichment in islands and shores and depletion in open seas,25 we observed that the overlapping CpG islands, shores, and open seas of IDHwt-specific eLB were significantly enriched or depleted compared to the expected distribution set used by the methylation platform (chi-square test P value 3.7E-04 enriched [CpG islands], 7.8E-05 enriched [shores], 8.9E-05 depleted [open seas], respectively, Supplementary Figure S3E). This observation is consistent with others who have shown enrichment in islands and shores and not in open seas.25 We identified 28 IDH-eLB-specific signatures linked to a gene promoter (Supplementary Table S3), of which 14 transcripts were differentially expressed in IDHmut vs IDHwt tissues. Ten out of 14 transcripts were inversely expressed in relation to the promoter methylation state (ie, hypermethylated promoter and down-regulated expression or vice versa). For instance, IDH-eLB signatures include the promoter hypomethylation of CXCR6, a gene that was reported to be associated with poor prognosis when upregulated and with longer survival when knocked out in an animal model.35,36 Congruent with those reports, the hypomethylated promoter of CXCR6 was associated with its overexpression in IDHwt glioma tissues from both TCGA and HBTC cohorts (Figure 3D, Supplementary Figure S3F).
Another example detected in our IDH-eLB signatures was the promoter hypomethylation of the PVT1 long noncoding RNA (lncRNA) that when highly expressed was associated with progression and poor prognosis in a pan-cancer cohort from TCGA and poor response to chemotherapy in patients with gliomas or squamous cell carcinoma of the head and neck.37 In line with these findings, hypomethylation of the PVT1 promoter was detected in our IDH-eLB in association with overexpression of the corresponding gene in the subtype with the worst prognosis (IDHwt) in samples from both the TCGA and the current cohort (Figure 3E, Supplementary Figure S3G).
The GeLB Score Changes Associated With Treatment and Progression—Case Studies
The evolutionary history of glioma patients is typically characterized by tumor recurrence, an event that can also be associated with progression toward higher grade glioma. Therefore, we investigated whether the GeLB score was able to predict recurrence and/or progression. To evaluate the application of the GeLB score as a potential tool to monitor glioma progression, we assessed the GeLB score in 33 HBTC patients with glioma from whom we had at least two serum samples from different time points (primary and either recurrence or off-treatment follow-up), comprising a total of 71 samples. The serum and clinical history were acquired between 2011 and 2019 (eg, before and after treatment, pathology report, and at signs of progression on MRI scans) (Supplementary Table S1). Patients’ treatment followed standard therapy paradigms. The majority of patients received radiation with concurrent chemotherapy followed by adjuvant chemotherapy with temozolomide38; however, three patients participated in clinical trials prior to a second or third surgery.
Compared to the baseline GeLB score at primary diagnosis (n = 33, median GeLB: 78.41%), we observed a continuous decrease in the GeLB score of these patients during longitudinal evaluation of recurrence (n = 27, median GeLB: 61.1 at first recurrence and 56.1% at second or third recurrence, P = .0001) (Figure 4A, Supplementary Figure S4A). With the exception of eight cases (four of which are described below), longitudinal samples represent persistent or recurrent gliomas and these conditions are consistently captured by the value invariably above 49%. However, the shift in score from primary to recurrence suggests that the methylation profile of recurrent samples differs from the primary samples, a change that is likely induced by treatment intervention.
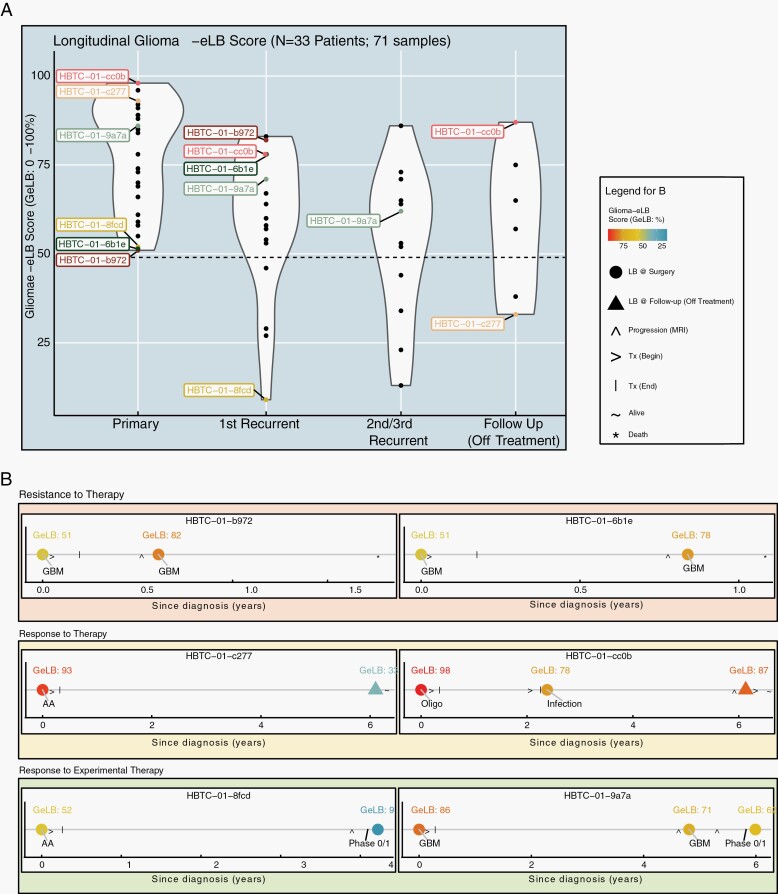
Fig. 4.
Use of GeLB as a surveillance tool during follow-up treatment and off-treatment. (A) Violin plot represents the distributions of GeLB scores (y-axis) for surgical (primary, first recurrent, second/third recurrence) and follow-up (off-treatment) time points from 33 patients with glioma (n = 71 samples). (B) The longitudinal profiles for 6 (of 33) cases are separated into three groups defined by resistance or response to therapy. Abbreviation: AA, anaplastic astrocytoma; GeLB, glioma-epigenetic liquid biopsy.
Several HBTC patients were enrolled in different clinical trials during the time of serum collection. Similar to patient HBTC-01-c2777 who responded favorably to treatment and the GeLB score dropped, we observed negative changes in GeLB score (high to low) in all cases successfully treated with various experimental agents consistent with the absence of the tumor in MRI for more than 6 months. Patient HBTC-01-8fcd (Figure 4B) had initially been diagnosed with anaplastic astrocytoma (AA), a baseline GeLB score of 52%, and treated with standard chemoradiation and surgery. Four years after surgery the tumor recurred and was successfully treated with an experimental drug. After 6 months, reflecting a progression-free state of this patient the GeLB score, remarkably dropped to 9%. In another case, patient HBTC-01-9a7a’s GeLB score decreased during the first recurrence (86%-71%) (Figure 4B). Following the inclusion of this patient in an experimental study that led to disease stabilization as ascertained by MRI, the GeLB score decreased from 71% to 62%.
For all patients who underwent recurrence, the GeLB score remained high or even increased further, and this observation was associated with progression of the tumor shown by MRI (Figure 4B, patient HBTC-01-b972). For example, in the case of patient HBTC-01-6b1e (Figure 4B), the increase in GeLB score coincides with glioma progression. The patient’s initial tumor was diagnosed as glioblastoma (GBM) and was treated with standard of care (chemo-radiotherapy) for 6 weeks. Approximately, 8 months later, the first sign of progression by MRI resulted in the second line of treatment (surgery). The patient’s baseline GeLB score corresponding to the primary diagnosis (GBM) was low but still indicative of a glioma (51%) which increased to 78% at progression. This patient died 7 months later. The opposite scenario was observed in patient HBTC-01-c277, initially diagnosed with AA and presenting a baseline GeLB score of 93% (Figure 4B). Following surgery and chemo-radiotherapy, after 6 years, the patient experienced long-term remission, without signs of recurrence on MRI and a concurrent serum GeLB score of 33%, considered a non-glioma based on our defined cutoff (<49%). Patient HBTC-01-cc0b (Figure 4B) who had been diagnosed with a grade II oligodendroglioma had a baseline GeLB score of 98% at diagnosis, indicative of the presence of a glioma. Two years after the initial diagnosis, the patient developed a cranial infection at the surgical site without evidence of tumor recurrence and with a GeLB score of 78% (decrease of 20% from baseline). After 4 years, this patient showed clear signs of recurrence confirmed by MRI when and the patient initiated chemotherapy for tumor progression. Cases HBTC-01-6b1e and HBTC-01-b972 experienced recurrence as documented by MRI imaging, GeLB score increased to 53% and 61% from baseline, respectively.
The GeLB Score may Discriminate Pseudoprogression From True Progression of Glioma
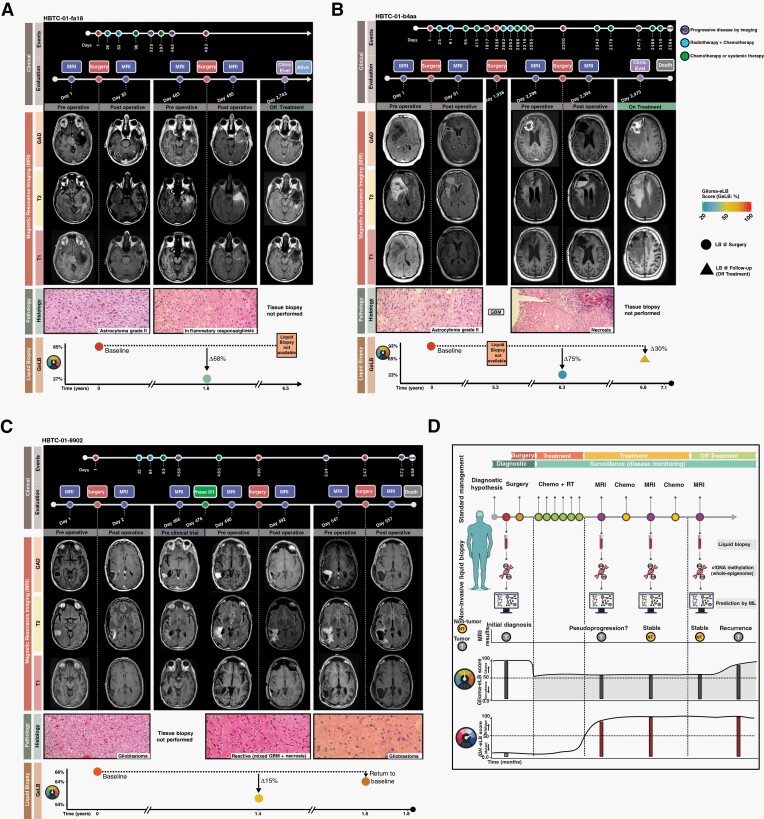
One of the most significant unmet challenges in the clinical follow-up of patients with brain tumors is the ability to distinguish pseudoprogression from true progression with nonsurgical procedures. Currently, there are no reliable diagnostic noninvasive approaches that can discriminate pseudoprogression from true progression. We tested whether the GeLB score could distinguish both states. Our cohort included three patients who presented bonafide pseudoprogression as confirmed by MRI imaging and histology. Patient HBTC-01-fa18 (Figure 5A) had initially been diagnosed with astrocytoma grade II and treated with standard chemoradiation. At diagnosis, the patients presented a baseline GeLB score of 85%, consistent with the diagnosis of glioma. Following the standard of care treatment and monitoring with MRI assessment, the patient showed signs of progression (MRI: T1, T2, and Gadolinium) that required a second surgical resection. Following surgery, pathology reports for the resected tumor indicated necrosis or reactive tissue (ie, inflammatory response mixed with gliosis). Interestingly, the concurrent GeLB score at that time point dropped to a score consistent with a non-glioma specimen (27%). Congruent with the diagnosis of pseudoprogression, this patient is alive for more than 6 years with no signs of progression. The baseline GeLB score for patient HBTC-01-b4aa (Figure 5B) was 93% and initially diagnosed as astrocytoma grade II. Following initial standard treatment, after 5 years, the tumor progressed and the patient underwent surgery confirming GBM recurrence. One year later, a follow-up MRI (T1, T2, and Gadolinium) indicated tumor recurrence, but the pathology report of the new surgical specimen revealed only the presence of extensive necrosis and the patient’s associated GeLB score at that time point was 23% (75% drop from baseline) (Figure 5B) indicative of a non-glioma state. However, a routine MRI performed 5 months after the second surgery showed a massive tumor regrowth and the corresponding GeLB increased to 65%. The patient died 4 months later. Patient HBTC-01-8902 presented a baseline GeLB score of 66% at diagnosis (GBM, Figure 5C). After 12 months, the patient showed signs of tumor recurrence treated with an experimental agent (Phase 0/1), followed by a second surgery that revealed predominant reactive gliosis and necrosis with minimal tumor. At the time of surgery, GeLB score was 54%, a 15% drop from baseline. After 1 month, GeLB increased to 64% consistent with the signs of progression in the MRI. Based on the imaging results, the patient underwent a third surgery and the histology analysis diagnosed a recurrent GBM.
Fig. 5.
GeLB score discriminates pseudoprogression from true progression of glioma. Clinical findings, magnetic resonance imaging, pathology, and liquid biopsy (GeLB score) features available for three patients with evidence of necrosis at multiple time points post-primary diagnosis. (A) HBTC-01-fa18, (B) HBTC-01-b4aa, and (C) HBTC-01-8902. (D) Our proposed approach to use noninvasive serum cell-free DNA methylome as a surveillance tool to complement current management for diagnosis and follow-up. Abbreviation: GeLB, glioma-epigenetic liquid biopsy.
Discussion
It is established that tissue-derived methylation markers have a role in stratifying patients with glioma. Additionally, widespread and specific DNA methylation patterns reflect the cell of origin both in tissue- and blood-derived specimens.1,2,25,27,39 Here, we tested whether, by profiling the methylome of circulating cfDNA in the serum from a cohort of patients with gliomas and other tumors and non-neoplastic conditions, using a methylation microarray, we would identify methylation markers that recapitulated the epigenetic features of glioma tissue. We defined a set of serum signatures (GeLB) that distinguished patients with glioma from their counterparts with high sensitivity and specificity, suggesting its potential diagnostic role of GeLB.40 Applied to publicly available cohorts of tumor tissue,2 these signatures were more similar to the methylation of the matching glioma tissue than the cfDNA methylome of other neoplasias or non-neoplastic conditions (Figures 1 and 2, Supplementary Figures S1 and S2), suggesting that GeLB is glioma-specific (Figures 1 and 2, Supplementary Figures S1 and S2). Thus, using GeLB signatures as input, we applied a ML approach and developed the GeLB score that accurately predicted whether the serum cfDNA methylome originated from a patient with glioma (GeLB score >50%) or a non-glioma condition (Figure 2). Considering the high accuracy and specificity of the GeLB score we observed, these signatures could potentially be complementary to imaging in the pre-surgical diagnosis of challenging cases faced in neuro-oncology such as the differentiation of glioma and other CNS conditions with similar features on imaging (eg, primary CNS lymphoma; demyelinating disease, or metastatic disease) and the diagnosis of tumors not amenable to resection due to comorbidities or neuroanatomic location (eg, deep or eloquent regions in the brain).
We also investigated the application of a GeLB score in a subset of patients with gliomas from which serum was withdrawn during surveillance (Figure 5D). The absolute level of the GeLB score did not seem to be significantly associated with cfDNA amount, tumor size, glioma subtype, or grading (eg, GBM, oligodendroglioma, AA, low-grade astrocytoma, etc.) (Figures 2G and 4B, Supplementary Figure 4). At the pre-surgical time point (primary), all the patients were categorized as having gliomas by the GeLB score (Figure 4B). During follow-up, the score decreased from the baseline in the majority of the cases (22/33; 67%), 64% of which kept GeLB scores within the glioma class and the remainder (36%) fell into the non-glioma score (Supplementary Figure 4). In some cases, the variation of the GeLB score levels was associated with specific features of the disease course such as progression, pseudoprogression, and response to treatment (standard or experimental) as shown in Figures 4 and 5. For instance, the GeLB score steadily increased as the tumor evolved (patient HBTC-01-b972 and HBTC-01-6b1e); decreased during an infectious process (patient HBTC-01-cc0b) followed by a later increase in association with tumor progression as shown by imaging assessment and decreased in association with treatment with an experimental drug (patient HBTC-01-9a7a). In three cases of pseudoprogression confirmed by the presence of necrotic tissue by pathology (eg, patient HBTC-01-fa18, HBTC-01-b4aa, and HBTC-01-8902), pre-surgical imaging suggested tumor progression whilst the concurrent GeLB score decreased in relation to baseline, two of them reaching a non-glioma score (eg, patient HBTC-01-fa18 and HBTC-01-b4aa). As for diagnosis, these promising results underscore the potential use of the GeLB score as a personalized surveillance tool, in which variation in the score’s value from the patient’s baseline could be useful to monitor disease evolution to differentiate between true disease progression and pseudoprogression secondary to therapy (radiation- or chemotherapy-induced necrosis or immunotherapy agents) as a complementary approach to imaging methods.
We also defined an IDH-status methylation signature in the serum from a small cohort of patients with IDH status assessed by standard methods that distinguished patients harboring IDH-mutant or wild-type glioma (Figure 3B, C, Supplementary Figure S3B, D). Interestingly, by analyzing the methylome and transcriptome (RNA-seq) from the matching glioma tissue, these methylation signatures were negatively associated with the expression of genes reported to have prognostic role in glioma and other tumors from the TCGA cohort (eg, CXCR6 and PVT1) (Figure 3D, Supplementary Figure S3F). Further validation is required to determine if this noninvasive approach can resolve an unmet need to detect the IDH tumor switch (IDHmut to IDHwt) upon recurrence, a phenotype that has important clinical implication as shown by our group and others.41
The use of serum or plasma for the molecular characterization of cfDNA is a point of discussion due to genomic contamination and dilution of the circulating nucleic acids in serum-derived specimens. Although these factors may hinder the detection of somatic mutations derived from the tumors, the choice of serum may not interfere with the survey of specific methylation marks, as cfDNA preserves information from tissue of origin as the fragmentation of the DNA released by healthy or tumorous cells is not a random event42,43; in addition, low input of DNA (eg, <250 ng) does not prevent the detection of methylation alterations using microarrays.30 Therefore, we investigated whether using serum would allow for the detection of epigenetic alterations directly or indirectly related to the glioma. Interestingly, the plasma GeLB score was lower in plasma than in serum (GeLB score mean values ~25% vs >50%, respectively), possibly reflecting the presence of immune cell-related signatures in the serum but not in plasma. By deconvoluting the cell composition of the serum using cell-type-specific methylation signatures, we found that the serum from patients with glioma contained more neuronal, glial, as well as a distinct immune cell landscape, compared to the serum of patients with other intra- or extracranial tumors or non-neoplastic conditions, suggesting that the serum methylome captured tumor-related markers (glial and neuronal cells) as well as local and/or systemic immune response to the tumor (immune cells).44–46 In addition, we found that applying the GeLB score to plasma specimens was highly correlated to the results using serum specimens to classify samples as glioma or non-gliomas (average Pearson’s r = 0.965, Supplementary Figure S4B) in a small cohort of paired (from 6 patients with glioma off-treatment) or unpaired (colorectal cancer or non-neoplastic brain conditions) serum and plasma samples. Despite promising, these results warrant validation in a larger cohort and by concurrently deconvoluting the serum or plasma cell composition of patients with gliomas using standard methods to compare with the methylation-based deconvolution methods. These encouraging results provide a framework for prospective studies with larger cohorts involving a blood-based epigenetic panel marker to assess tumor burden and monitor the progression and response to treatment of an ever-evolving disease such as malignant gliomas using a minimally invasive approach.
Supplementary Material
Acknowledgments
The authors are grateful to the patients who participated in this study. We thank Nancy Takacs for administrative support; Daniel Weisenberger and the team at USC Epigenome Center for assistance with DNA methylation profiling; Susan MacPhee and Kate Lawrenson for critical reading of this manuscript; and Bartow Thomas and Kelly Tundo for assisting with serum collection at follow-up off-treatment.
Funding
This work was supported by the Henry Ford Health System, Department of Neurosurgery at the Hermelin Brain Tumor Center, and Head for the Cure Foundation. Additionally, H.N., T.S.S., T.M.M., L.M.P., and A.C.D. are supported by the Department of Defense (CA170278), H.N., A.C.D., M.W., A.M., and L.M.P. are supported by the National Institutes of Health (R01CA222146), and A.C.D., L.M.P., and J.R. are supported by the Department of Defense (CA180174).
Conflict of interest statement. The authors have no competing interests.
Author contributions. The contributions of the authors are as follows: Serum and tissue collection and storing: K.N. and A.C.D.; surgical procedure and pathology review: J.S., A.M., D.A.C., A.R., M.L.R., T.M., J.R., I.L., T.W., and S.K.; cfDNA extraction and methylome generation: T.M.M., K.N., and A.C.D.; glioma-eLB and IDH-eLB signatures: H.N., T.S.S., T.M.M., and A.V.C.; methodology: H.N., T.S.S., T.M.M., and A.V.C.; statistical analysis: H.N. and T.S.S. with input from L.M.P. and A.S.; pituitary, meningioma tissue, and serum curation: H.N., T.M.M., M.W., M.S.M., and A.V.C.; clinical analysis and interpretation: H.N., T.S.S., T.M.M., L.M.P., J.S., K.P.A., T.M., I.L., T.W., A.I., S.K., and A.V.C.; data interpretation: H.N., T.S.S., T.M.M., J.S., A.I., and A.V.C.; visualization: T.S.S. and T.M.M. with input from J.S., A.I., A.V.C., and H.N.; writing—original draft: H.N. and A.V.C.; and overall concept and coordination: H.N. All authors read and approved the final manuscript.
References
- 1. Noushmehr H, Weisenberger DJ, Diefes K, et al. ; Cancer Genome Atlas Research Network . Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ceccarelli M, Barthel FP, Malta TM, et al. ; TCGA Research Network . Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse Glioma. Cell. 2016;164(3):550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sturm D, Witt H, Hovestadt V, et al. . Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22(4):425–437. [DOI] [PubMed] [Google Scholar]
- 4. Verma N, Cowperthwaite MC, Burnett MG, Markey MK. Differentiating tumor recurrence from treatment necrosis: a review of neuro-oncologic imaging strategies. Neuro Oncol. 2013;15(5):515–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Parvez K, Parvez A, Zadeh G. The diagnosis and treatment of pseudoprogression, radiation necrosis and brain tumor recurrence. Int J Mol Sci. 2014;15(7):11832–11846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fontanilles M, Duran-Peña A, Idbaih A. Liquid biopsy in primary brain tumors: looking for stardust! Curr Neurol Neurosci Rep. 2018;18(3):13. [DOI] [PubMed] [Google Scholar]
- 7. Dor Y, Cedar H. Principles of DNA methylation and their implications for biology and medicine. Lancet. 2018;392(10149):777–786. [DOI] [PubMed] [Google Scholar]
- 8. Bettegowda C, Sausen M, Leary RJ, et al. . Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li J, Han X, Yu X, et al. . Clinical applications of liquid biopsy as prognostic and predictive biomarkers in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. J Exp Clin Cancer Res. 2018;37(1):213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fernandez-Cuesta L, Perdomo S, Avogbe PH, et al. . Identification of circulating tumor DNA for the early detection of small-cell lung cancer. EBioMedicine. 2016;10:117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vatandoost N, Ghanbari J, Mojaver M, et al. . Early detection of colorectal cancer: from conventional methods to novel biomarkers. J Cancer Res Clin Oncol. 2016;142(2):341–351. [DOI] [PubMed] [Google Scholar]
- 12. Martínez-Ricarte F, Mayor R, Martínez-Sáez E, et al. . Molecular diagnosis of diffuse gliomas through sequencing of cell-free circulating tumor DNA from cerebrospinal fluid. Clin Cancer Res. 2018;24(12):2812–2819. [DOI] [PubMed] [Google Scholar]
- 13. Mouliere F, Mair R, Chandrananda D, et al. . Detection of cell-free DNA fragmentation and copy number alterations in cerebrospinal fluid from glioma patients. EMBO Mol Med. 2018;10(12):e9323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Panditharatna E, Kilburn LB, Aboian MS, et al. . Clinically relevant and minimally invasive tumor surveillance of pediatric diffuse midline gliomas using patient-derived liquid biopsy. Clin Cancer Res. 2018;24(23):5850–5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pan W, Gu W, Nagpal S, Gephart MH, Quake SR. Brain tumor mutations detected in cerebral spinal fluid. Clin Chem. 2015;61(3):514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. De Mattos-Arruda L, Mayor R, Ng CKY, et al. . Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat Commun. 2015;6:8839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang J, Bettegowda C. Applications of DNA-based liquid biopsy for central nervous system neoplasms. J Mol Diagn. 2017;19(1):24–34. [DOI] [PubMed] [Google Scholar]
- 18. Best MG, Sol N, Zijl S, Reijneveld JC, Wesseling P, Wurdinger T. Liquid biopsies in patients with diffuse glioma. Acta Neuropathol. 2015;129(6):849–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shankar GM, Balaj L, Stott SL, Nahed B, Carter BS. Liquid biopsy for brain tumors. Expert Rev Mol Diagn. 2017;17(10):943–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Adamczyk LA, Williams H, Frankow A, et al. . Current understanding of circulating tumor cells - potential value in malignancies of the central nervous system. Front Neurol. 2015;6:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Heitzer E, Haque IS, Roberts CES, Speicher MR. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat Rev Genet. 2019;20(2):71–88. [DOI] [PubMed] [Google Scholar]
- 22. Miller AM, Shah RH, Pentsova EI, et al. . Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid. Nature. 2019;565(7741):654–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zill OA, Banks KC, Fairclough SR, et al. . The landscape of actionable genomic alterations in cell-free circulating tumor DNA from 21,807 advanced cancer patients. Clin Cancer Res. 2018;24(15):3528–3538. [DOI] [PubMed] [Google Scholar]
- 24. Wan JCM, Massie C, Garcia-Corbacho J, et al. . Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17(4):223–238. [DOI] [PubMed] [Google Scholar]
- 25. Shen SY, Singhania R, Fehringer G, et al. . Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature. 2018;563(7732):579–583. [DOI] [PubMed] [Google Scholar]
- 26. Kis O, Kaedbey R, Chow S, et al. . Circulating tumour DNA sequence analysis as an alternative to multiple myeloma bone marrow aspirates. Nat Commun. 2017;8:15086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Malta TM, de Souza CF, Sabedot TS, et al. . Glioma CpG island methylator phenotype (G-CIMP): biological and clinical implications. Neuro Oncol. 2018;20(5):608–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Uehiro N, Sato F, Pu F, et al. . Circulating cell-free DNA-based epigenetic assay can detect early breast cancer. Breast Cancer Res. 2016;18(1):129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jaunmuktane Z, Capper D, Jones DTW, et al. . Methylation array profiling of adult brain tumours: diagnostic outcomes in a large, single centre. Acta Neuropathol Commun. 2019;7(1):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moss J, Magenheim J, Neiman D, et al. . Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat Commun. 2018;9(1):5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gallardo-Gómez M, Moran S, Páez de la Cadena M, et al. . A new approach to epigenome-wide discovery of non-invasive methylation biomarkers for colorectal cancer screening in circulating cell-free DNA using pooled samples. Clin Epigenetics. 2018;10:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brennan CW, Verhaak RG, McKenna A, et al. ; TCGA Research Network . The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brat DJ, Verhaak RGW, et al. ; Cancer Genome Atlas Research Network . Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372(26):2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fortin JP, Triche TJ Jr, Hansen KD. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics. 2017;33(4):558–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lepore F, D’Alessandro G, Antonangeli F, et al. . CXCL16/CXCR6 axis drives microglia/macrophages phenotype in physiological conditions and plays a crucial role in glioma. Front Immunol. 2018;9:2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gao Q, Zhao YJ, Wang XY, et al. . CXCR6 upregulation contributes to a proinflammatory tumor microenvironment that drives metastasis and poor patient outcomes in hepatocellular carcinoma. Cancer Res. 2012;72(14):3546–3556. [DOI] [PubMed] [Google Scholar]
- 37. Zhang Y, Yang G, Luo Y. Long non-coding RNA PVT1 promotes glioma cell proliferation and invasion by targeting miR-200a. Exp Ther Med. 2019;17(2):1337–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stupp R, Mason WP, van den Bent MJ, et al. ; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group . Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 39. de Souza CF, Sabedot TS, Malta TM, et al. . A distinct DNA methylation shift in a subset of glioma CpG island methylator phenotypes during tumor recurrence. Cell Rep. 2018;23(2):637–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Capper D, Jones DTW, Sill M, et al. . DNA methylation-based classification of central nervous system tumours. Nature. 2018;555(7697):469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mazor T, Chesnelong C, Pankov A, et al. . Clonal expansion and epigenetic reprogramming following deletion or amplification of mutant IDH1. Proc Natl Acad Sci U S A. 2017;114(40):10743–10748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sun J, Zhou W, Mao K, et al. . Examination of plasma cell-free DNA of glioma patients by whole exome sequencing. World Neurosurg. 2019;125:e424–e428. [DOI] [PubMed] [Google Scholar]
- 43. Sun K, Jiang P, Cheng SH, et al. . Orientation-aware plasma cell-free DNA fragmentation analysis in open chromatin regions informs tissue of origin. Genome Res. 2019;29(3):418–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Amankulor NM, Kim Y, Arora S, et al. . Mutant IDH1 regulates the tumor-associated immune system in gliomas. Genes Dev. 2017;31(8):774–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wiencke JK, Koestler DC, Salas LA, et al. . Immunomethylomic approach to explore the blood neutrophil lymphocyte ratio (NLR) in glioma survival. Clin Epigenetics. 2017;9:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Thorsson V, Gibbs DL, Brown SD, et al. ; Cancer Genome Atlas Research Network . The immune landscape of cancer. Immunity. 2018;48(4):812–830.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.