ABSTRACT
Background
The Mediterranean diet is a well-recognized healthy diet that has shown to induce positive changes in gut microbiota. Lifestyle changes such as diet along with physical activity could aid in weight loss and improve cardiovascular risk factors.
Objectives
To investigate the effect of an intensive lifestyle weight loss intervention on gut microbiota.
Methods
This is a substudy of the PREDIMED-Plus (Prevención con Dieta Mediterránea-Plus), a randomized controlled trial conducted in overweight/obese men and women (aged 55–75 y) with metabolic syndrome. The intervention group (IG) underwent an intensive weight loss lifestyle intervention based on an energy-restricted Mediterranean diet (MedDiet) and physical activity promotion, and the control group (CG) underwent a non-energy-restricted MedDiet for 1 y. Anthropometric, biochemical, and gut microbial 16S rRNA sequencing data were analyzed at baseline (n = 362) and 1-y follow-up (n = 343).
Results
IG participants had a weight loss of 4.2 (IQR, –6.8, –2.5) kg compared with 0.2 (IQR, –2.1, 1.4) kg in the CG (P < 0.001). Reductions in BMI, fasting glucose, glycated hemoglobin, and triglycerides and an increase in HDL cholesterol were greater in IG than in CG participants (P < 0.05). We observed a decrease in Butyricicoccus, Haemophilus, Ruminiclostridium 5, and Eubacterium hallii in the IG compared with the CG. Many genera shifted in the same direction within both intervention groups, indicating an overall effect of the MedDiet. Decreases in Haemophilus, Coprococcus 3, and few other genera were associated with a decrease in adiposity parameters in both intervention groups. Changes in Lachnospiraceae NK4A136 were positively associated with changes in MedDiet adherence.
Conclusions
Weight loss induced by an energy-restricted MedDiet and physical activity induce changes in gut microbiota. The role of MedDiet-induced changes on the host might be via short-chain fatty acid producing bacteria, whereas with energy restriction, these changes might be modulated with other mechanisms, which need to be explored in future studies. This trial was registered at http://www.isrctn.com/ISRCTN89898870 as ISRCT 89898870.
Keywords: weight loss, gut microbiota, Mediterranean diet, energy restriction, obesity
Introduction
Microbiota colonizes the human gut during or shortly after birth and continues to grow and develop until it establishes a stable environment in adults. During adulthood, the variability and complexity of the human gut microbiome are influenced by several lifestyle choices, including dietary and nondietary factors such as physical activity, stress, or smoking habits (1). Also, environmental factors, aging, medications, and diseases shift the composition and functionality of our microbes. Individuals with conditions such as diabetes, metabolic syndrome (MetS), and cardiovascular risks have shown to have a dysbiotic gut with opportunistic pathogens (2). Obesity has been associated with lower diversity and richness of the microbiota, as well as a decreased Bacteroidetes-to-Firmicutes ratio (B/F) (3), but this remains inconclusive as some studies have failed to show this association (4, 5). Different studies support gut microbiota as an environmental factor related to the progress of obesity and metabolic disturbances (2, 6), even though the causal nature of this has not been completely understood.
Weight loss is an effective strategy for obese and overweight individuals to reduce the risk of developing metabolic disorders and cardiovascular diseases (CVDs). Lifestyle changes using different dietary strategies and increasing physical activity promotion have been recommended to lose weight (7). Diet is an important factor in modulating not only weight but also gut microbiota composition and function. Several studies have shown a change in the gut microbiota associated with specific dietary factors or patterns (8–10). A recent study conducted in the NU-AGE (New dietary strategies addressing the specific needs of elderly population for an healthy ageing in Europe) trial demonstrated that higher adherence to a Mediterranean diet (MedDiet) pattern for 1 y was associated with specific gut microbiome changes that were associated with improved health status and reduced frailty (11). Another recent study, conducted among overweight and obese participants adhering to the MedDiet or an isocaloric control diet for 8 wk, showed significant improvements in a decrease in circulating total cholesterol, insulin resistance, and fecal bile acids related to changes in gut microbiota (12). Combining the beneficial effects of an energy-restricted MedDiet and physical activity in a weight loss perspective could aid in the betterment of cardiometabolic risk factors through changing gut microbiota profile. In this substudy conducted in the framework of the PREDIMED-Plus (Prevención con Dieta Mediterránea-Plus) randomized trial, as the primary objective, we evaluated the 1-y effect of an energy-reduced MedDiet weight loss lifestyle intervention program compared with non-energy-restricted MedDiet intake on gut microbiota composition in overweight/obese adults with MetS. As a secondary objective, we explored the associations of the gut microbiota composition with respect to the components of the intervention.
Methods
Study design and participants
The present study was conducted in the frame of the PREDIMED-Plus study, with further details in Supplemental Method 1. The primary outcome of the parent study, PREDIMED-Plus, is weight loss and a composite of CVD incidence. Evaluation of gut microbiota composition is an intermediate outcome of the PREDIMED-Plus study. Eligible participants were community-dwelling men and women aged 55–75 y and 60–75 y, respectively, without a documented history of CVD at baseline of overweight/obesity [BMI (in kg/m2) ≥27 and ≤40] and with at least 3 components of MetS according to the American Heart Association and National Heart, Lung, and Blood Institute. Details of the trial have been described elsewhere (13). Further details on the study can be found at https://www.predimedplus.com/. This trial was registered at http://www.isrctn.com/ISRCTN89898870 as ISRCT 89898870. Participants were not involved in the design, conduct, or reporting of the study; further information can be found in Supplemental Method 1.
In this substudy, a total of 400 participants matched for age, sex, and BMI were randomly selected from the intervention group (IG, n = 200) and control group (CG, n = 200) from 2 PREDIMED-Plus study centers (Reus and Malaga). Briefly, participants randomly allocated to the IG were instructed to adhere to an energy-reduced MedDiet, accompanied by physical activity promotion, to accomplish specific weight loss objectives. Trained dietitians conducted an individual motivational interview, a group session, and a phone call each month during the intervention follow-up (1 y). The IG received an intensive intervention consisting of individualized behavioral support, and participants in the CG received information on maintaining ad libitum unrestricted caloric MedDiet with no advice on weight loss strategies such as to increase physical activity. In the case of the CG, participants received only 1 individual session and 1 group session every 6 mo to motivate and adhere to the intervention. Trained dietitians and nurses conducted the intervention and collected baseline and 1-y measurements and biological samples.
Evaluation of food consumption and anthropometric and biochemical measurements
At baseline and 12-mo follow-up visits, nurses measured waist circumference (midway between the lowest rib and the iliac crest, using an anthropometric tape), weight (using electronic calibrated scales), and height (using a wall-mounted stadiometer) twice. Dietary consumption was estimated by the dietitians using a validated FFQ, and energy and nutrient consumption were calculated using the Spanish food composition tables. Mediterranean diet adherence score (MedScore) was calculated from a modified version of a previously validated questionnaire (14) (17-point validated tool), and information on physical activity was collected using a validated questionnaire (15). Serum and plasma samples were collected at baseline and 1 y following the intervention after an overnight fast and then aliquoted and stored at –80°C. Standard enzymatic methods were conducted to evaluate serum total cholesterol, HDL cholesterol, and triglyceride concentrations. LDL cholesterol was calculated by the Friedewald formula whenever triglycerides were <300 mg/dL.
Fecal sample collection and processing
Supplemental Method 2 describes fecal sample collection. Fecal DNA extraction was conducted using the QIAamp PowerFecal DNA Kit (Qiagen) according to the manufacturer's protocol, and an additional bead-beating step of 5 min using the FastPrep-24 5G Homogenizer (MP Biomedicals) was added to the first lysing step. The quantity of DNA was evaluated using Qubit 2.0 Fluorometer-dsDNA (High Sensitivity Kit; Invitrogen). After extraction, the DNA was stored at –20°C until further processing.
16S rRNA sequencing and processing
Supplemental Method 3 provides detail on 16S rRNA gene sequencing. Briefly, we used the Ion Ribosomal 16S Kit (Thermo Fisher Scientific) that includes 2 primer sets selectively amplifying the corresponding hypervariable regions of the 16S region in bacteria: primer set V2–4–8 and primer set V3–6, 7–9. After sequencing, the individual sequence reads were filtered using Ion Reporter Software V4.0 (Thermo Fisher Scientific)to remove low-quality and polyclonal sequences. Data were processed and separated into 6 hypervariable regions using an adapted script available from Mas-Lloret et al (16). Only variable region V4 was used for further analyses. These files were imported to QIIME2, and the DADA2 pipeline was followed (see Supplemental Method 3). Taxonomy was assigned to the clustered sequences with SILVA 132 as the 16S classifier database. Mitochondrial features and features unidentified at the phylum level were removed in the preprocessing step in R (v 3.6) (17). The MetagenomeSeq package was used to normalize the samples using the cumulative sum scaling and log transformation method.
Bioinformatics and statistical analysis
Baseline characteristics of study participants were described as mean and SD or median with 25% and 75% IQR (based on distribution) for quantitative variables and as percentages for categorical variables. Differences in baseline characteristics were evaluated with χ2 tests for categorical variables, t tests (for normally distributed variables), and Wilcoxon tests (for nonnormally distributed variables). Effects of intervention on changes in different variables were evaluated using Wilcoxon tests and are shown appropriately according to their distribution. Abovementioned Wilcoxon and t tests were evaluated using package MatrixTests in R (v 3.6.2) (18), and significance was determined at P < 0.05.
For the microbiome analysis, normalized data from the MetagenomeSeq package were used (19). The α diversity (chao1, Shannon index), B/F, log of Prevotella-to-Bacteroides ratio (P/B) [adapted from Roager et al. (20)], and phylogenetic distance were evaluated using packages microbiome and picante (21, 22). Effect of intervention (Time × Treatment) adjusted by baseline weight, sex, and study center was used to estimate the changes in α diversity, B/F, P/B, and phylogenetic distance by a linear mixed model. In addition, for the B/F and P/B, we also adjusted by baseline ratio values. Principal coordinate analysis (PCoA)–based β diversity (weighted UniFrac distance, unweighted UniFrac, Bray–Curtis dissimilarity) was evaluated with the vegan package in R (v 3.6.2), and PERMANOVA (permutational multivariate analysis of variance) was conducted with the adonis function (999 permutations) using participants as strata and also adjusting for baseline weight, sex, and study center (23). The condition for homogeneity was verified using the betadisper function.
To investigate the changes in microbial genera between the intervention groups, the fitZig function from the MetagenomeSeq package that implements a zero-inflated Gaussian model was used. We accounted for the repeated measures with a mixed model, and the analysis was carried out at the genus level. According to the authors’ recommendation (24), we calculated effective sample sizes and retained only the genera that had an effective sample size more than the median of all samples. To reduce the type I error rate in multiple testing, we used the false discovery rate (FDR) approach to correct P values. An FDR of 10% was set for the between-group analysis. For the within-group analysis, we used the fitfeature function that uses a zero-inflated lognormal model. Log-fold changes in fitfeature were calculated from the coefficients of the zero-inflated lognormal model. In addition, we calculated effective sample sizes and report the only genus that passes the threshold. For this analysis, an FDR of 5% was set.
We also used a second approach using sparse partial least squares discriminant analysis (sPLS-DA) to compare the results from those obtained in MetagenomeSeq. This supervised method from the mixOmics package selects features that can best discriminate the 2 intervention groups at the end of the intervention (25). The samples were center log-ratio transformed (using package Hotelling) and indexed with respect to their baseline samples, which accounts for within-participant variations [adapted from Lee et al. (26)]. The number of components and features per component were calculated using the tune.splsda function, based on minimum balanced error rate. Each feature selected has an associated loading representing the relative importance of that feature on the component for discriminating the groups. This is represented as variable importance in projection (VIP), and a feature with a VIP of >1 is regarded as important for discrimination. Features having VIP >1 were chosen to be compared with the results of MetagenomeSeq.
The associations between changes in measured biochemical variables and changes in microbial genera that significantly changed in the IG or the CG (and VIP >1) were analyzed using a NBZIMM package in R, which uses a negative binomial mixed model and allows to adjust for covariates (27). Coefficients obtained from this along with adjusted P values were visualized in R software using ggplots2 (28). To detect the associations in the overall population, we adjusted for group of intervention, study center, sex, and baseline weight. P values were corrected by FDR for multiple testing.
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States analysis
Predicted metagenome functions were performed using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States plugin (29) within QIIME2 with the q2-picrust2 plugin. MetaCyc pathways (30) were normalized within QIIME2 and analyzed using the open-source software STAMP with Welch's t test option (31). Those pathways with a P < 0.05 were posteriorly analyzed in QIIME2 with the longitudinal plugin for paired sampled comparisons. For this analysis, an FDR of 10% was set.
Results
Characteristics of the study population
A flowchart of selected participants is represented in Supplemental Figure 1. A total of 400 participants matched by age, sex, and BMI were randomly allocated to this study (200 per intervention group). After preprocessing steps (as mentioned in Supplemental Figure 1 and Supplemental Method 3), data at baseline were available for 183 participants in the IG and 179 participants in the CG, corresponding to 171 participants in the IG and 172 participants in the CG after 1 y. There were no significant differences in the measured baseline variables between groups (Table 1), except for higher body weight in the IG (P = 0.03). Diet, food groups (Supplemental Table 1), and physical activity changes (Table 2) were in the expected direction, with significant improvements in the IG compared with the CG.
TABLE 1.
Baseline characteristics of study participants1
| Characteristic | IG | CG |
|---|---|---|
| No. | 183 | 179 |
| Age, y | 64.3 (5.1) | 65.1 (4.9) |
| Sex, M/F, n | 97/86 | 77/102 |
| Weight, kg | 89.7 (13.6) | 86.7 (11.56)2 |
| BMI, kg/m2 | 33.4 (30.8, 36.0) | 32.9 (30.5, 35.6) |
| Waist circumference, cm | 110.7 (9.8) | 108.9 (9.55) |
| Diabetes (yes), % (n) | 26.2 (48/135) | 20.6 (37/142) |
| Hypercholesteremia, (no/yes), % (n) | 94.5 (10/173) | 93.8 (11/168) |
| Total cholesterol, mg/dL | 203.0 (177.0, 224.5) | 197.0 (172.5, 226.5) |
| LDL cholesterol, mg/dL | 116.0 (94.5, 140.8) | 115.0 (97.0, 139.5) |
| HDL cholesterol, mg/dL | 46.0 (40.0, 57.0) | 47.0 (42.0, 54.0) |
| Triglycerides, mg/dL | 151.0 (55.3, 246.8) | 152.0 (68.5, 235.5) |
| Glucose, mg/dL | 104.0 (92.5, 118.0) | 103.0 (94.0, 116.0) |
| Glycated hemoglobin, % | 5.8 (5.6, 6.3) | 5.8 (5.5, 6.3) |
| Physical activity, METs-min/wk | 1627 (682, 3650) | 1767 (839, 3308) |
| Energy intake, kcal/d | 2546.5 (543.7) | 2416.6 (514.7) |
| 17-point Mediterranean adherence score | 7.7 (2.1) | 8 (2.4) |
| Smoking, n | ||
| Current smoker | 32 | 24 |
| Former smoker | 65 | 68 |
| Never | 85 | 87 |
| No data | 1 | |
| Study center (Malaga/Reus), n | 66/117 | 73/106 |
1Values expressed as Mean (SD) for normally distributed variables and Median (25%, 75% IQR) for non-normal distributions unless otherwise indicated. Chi-square, Wilcoxon, and t tests were conducted for categorical, nonnormal, and normally distributed variables, respectively. CG, control group; IG, intervention group; METs, Metabolic equivalent of task.
2Significant difference < 0.05.
TABLE 2.
Effects of intervention on anthropometric and biochemical variables measured1
| Characteristic | Changes in IG (n = 171) | Changes in CG (n = 172) | P value |
|---|---|---|---|
| Weight, kg | –4.2 (–6.8, –2.5) | –0.2 (–2.1, 1.4) | <0.001 |
| BMI, kg/m2 | –1.6 (–2.5, –0.9) | –0.05 (–0.8, 0.6) | <0.001 |
| Waist circumference, cm | –5 (–9.0, –1.8) | 0.0 (–2.5, 2.0) | <0.001 |
| Total cholesterol, mg/dL | –1.0 (–17.5, 14.0) | –2.0 (–22.0, 14.0) | 0.767 |
| LDL cholesterol, mg/dL | 1.0 (–14.5, 14.5) | –2.0 (–19.0, 13.0) | 0.577 |
| HDL cholesterol, mg/dL | 3.0 (–0.5, 6.0) | 2.0 (–2.3, 6.0) | 0.012 |
| Triglycerides, mg/dL | –19.0 (–52.5, 9.5) | –3.5 (–41.5, 28.0) | 0.028 |
| Glucose, mg/dL | –5.0 (–14.0, 2.0) | 0.5 (–7.3, 8.0) | <0.001 |
| Glycated hemoglobin, % | –0.1 (–0.3, 0.1) | 0.0 (–0.1, 0.2) | 0.002 |
| Physical activity, METs-min/wk | 1154 (0, 2633) | 0 (–787, 743) | <0.001 |
| 17-point Mediterranean adherence score | 6.0 (4.0, 8.5) | 2.0 (1.0, 5.0) | <0.001 |
| Energy intake, kcal/d | –318.2 (–655.6, 3.2) | 44.3 (–329.3, 391.7) | <0.001 |
1Values expressed as Median (25%, 75% IQR). Wilcoxon test was conducted for evaluating the differences between 2 groups of intervention. CG, control group; IG, intervention group; METs, Metabolic equivalent of task.
After 1 y (Table 2), IG participants lost an average of 4.2 (IQR, –6.8, –2.5) kg compared with 0.2 (IQR, –2.1, 1.4) kg in the CG (P < 0.001). Reductions in BMI, waist circumference, and concentrations of triglycerides, glucose, and glycated hemoglobin were greater in IG than in CG participants (all, P < 0.05), whereas a significantly higher increase in HDL cholesterol was observed in the IG compared with the CG (P < 0.05) (Table 2). Even though participants belonged to a Mediterranean region, the baseline MedDiet score was equal to or below the median MedDiet adherence score (low, ≤7; medium, 8–10; and high, 11–17) in both arms of intervention (32). This adherence increased with 1 y of intervention in both groups.
Changes in α and β diversity
No significant differences in α diversity indices (Chao1, Shannon) adjusted for body weight at baseline between the 2 intervention groups or within groups were observed (Table 3). Time and treatment interaction did not vary significantly for weighted UniFrac, unweighted UniFrac, or Bray–Curtis dissimilarity (Table 3, Figure 1). Likewise, no differences were noted at baseline and the 1-y time point (Supplemental Table 2). B/F increased significantly in the IG compared with the CG (P < 0.05), but no changes in P/B were observed (Supplemental Table 3, Supplemental Figure 2A,B). No differences in baseline α and β diversity, B/F, and P/B were observed between the groups.
TABLE 3.
Effects intervention on changes in α and β diversity metrics1
| Diversity measures (n = 343) | Treatment * Time (P value) |
|---|---|
| Chao12 | 0.16 |
| Shannon diversity2 | 0.15 |
| Phylogenetic distance2 | 0.21 |
| Weighted UniFrac3 | 0.72 |
| Unweighted UniFrac3 | 0.23 |
| Bray Curtis dissimilarity3 | 0.33 |
Effect of intervention (Treatment * Time) evaluated by linear mixed model adjusted for sex, study center, and baseline weight for chao1, Shannon diversity, and phylogenetic distance. Weighted UniFrac, unweighted UniFrac, and Bray–Curtis dissimilarity were evaluated by PERMANOVA (permutational multivariate analysis of variance) adjusted for sex, study center and baseline weight, and participants as strata.
α diversity indexes.
β diversity indexes.
FIGURE 1.
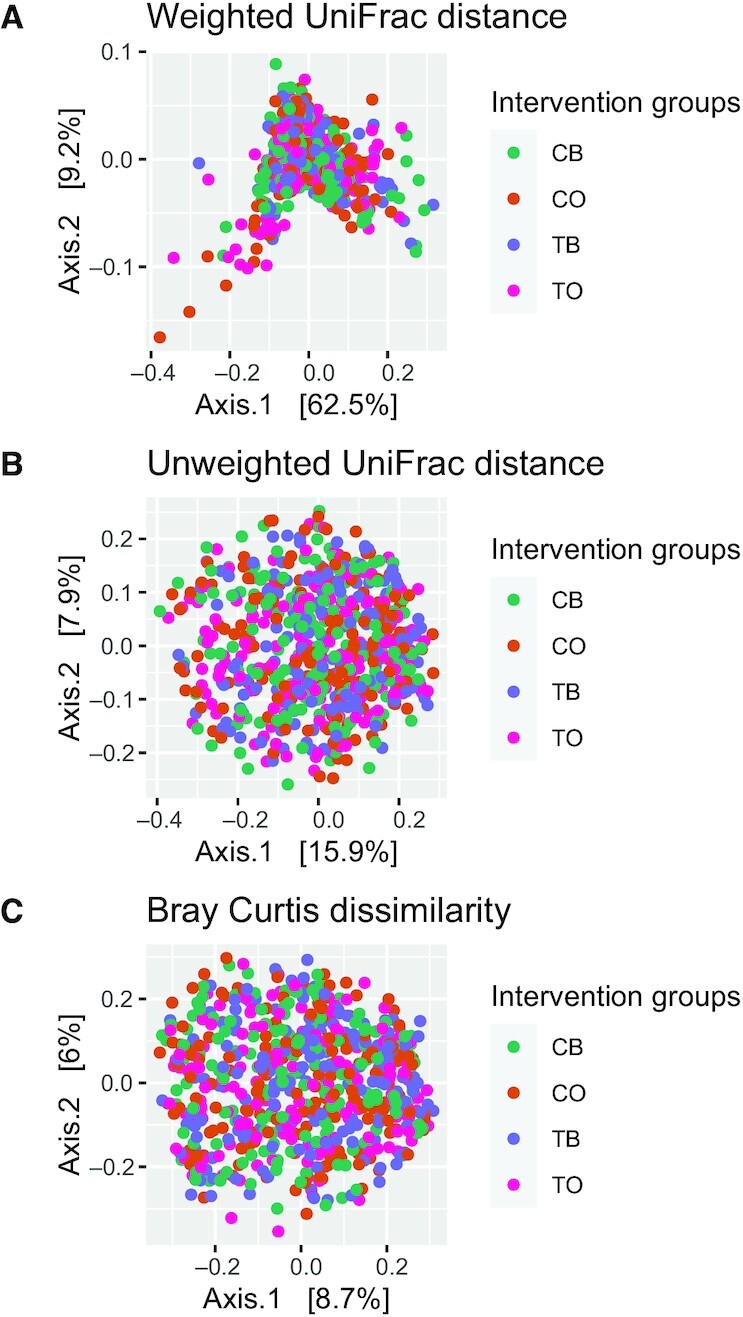
(A) Principal coordinate analysis (PCoA) of weighted UniFrac distance showing 2 groups of intervention at 2 time points. (B) PCoA of unweighted UniFrac distance. (C) PCoA of Bray–Curtis distance (n = 343). CB, control group at baseline; CO, control group at year 1; TB, intervention group at baseline; TO, intervention group at year 1.
Effect of intervention on changes in gut microbiota
Differential abundance analysis between the 2 groups of intervention conducted at the genus level showed Haemophillus, Butyricicoccus, Eubacterium hallii, and Ruminiclostridium 5 were reduced and Coprobacter and uncultured bacterium (from Rhodospirillales order) increased in the IG compared with the CG (all FDR P < 0.1) (Table 4, Supplemental Figure 3A–H) while adjusting for sex, study center, and baseline weight. LogFC represents the coefficient of change in the MetagenomeSeq model evaluated comparing the IG with the CG. Some of the genera (Haemophillus, E. halii, Ruminococcus NK4A214) that were found to vary significantly between the groups in the MetagenomeSeq model also contributed to characterizing the IG and the CG in the sPLS-DA model (Supplemental Figure 4).
TABLE 4.
Differentially abundant genus between groups of intervention1
| Genus | logFC (ΔIG–ΔCG) | P value | Adjusted P value |
|---|---|---|---|
| Haemophilus | –7.6 | <0.001 | <0.001 |
| Butyricicoccus | –4.2 | <0.001 | <0.001 |
| Ruminiclostridium 5 | –2.2 | 0.003 | 0.09 |
| Eubacterium hallii | –2.2 | 0.006 | 0.08 |
| O_Rhodospirillales_F_uncultured_uncultured bacterium | 4.3 | 0.006 | 0.05 |
| Ruminococcaceae NK4A214 | 2.6 | 0.007 | 0.08 |
| Coprobacter | 2.3 | 0.030 | 0.08 |
Model adjusted for baseline weight, sex, and study center. logFC is the β estimate of the adjusted model. P value adjusted by false discovery rate for multiple testing. CG, control group; IG, intervention group.
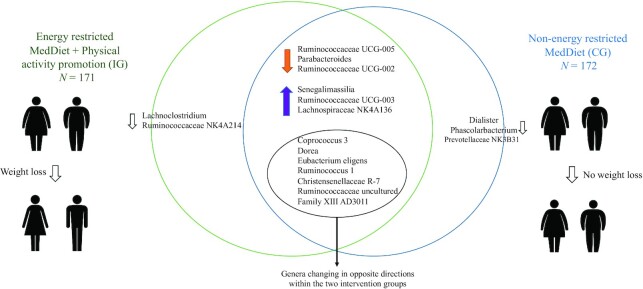
Figure 2 shows the Venn diagram of genera that shifted within both groups. Fifteen genera in the IG (Figure 2) and 16 genera in the CG (Figure 2) were significantly different from baseline to 1 y within each intervention group and had a VIP >1 from the sPLS-DA model. Within IG analysis, 7 among 15 genera reducing in relative abundance belonged to the family Lachnospiraceae, whereas some of these such as Roseburia and Dorea increased in the CG (Supplemental Tables 4–5). An increase in some short-chain fatty acid (SCFA) producers such as Lachnospira and Lachnospiraceae NK4A136 group was observed in both intervention groups (Supplemental Tables 4–5). Overall predominant changes in both groups belonged to genera from Lachnospiraceae and Ruminococcaceae families.
FIGURE 2.
Venn diagram representing IG and CG by genera varying within groups evaluated with MetagenomeSeq and having a variable importance in projection >1 from the sparse partial least squares discriminant analysis model. Genera shifting in the same direction as well in opposite directions within each intervention groups are shown. CG, control group; IG, intervention group.
Associations between changes in gut microbiota and measured variables
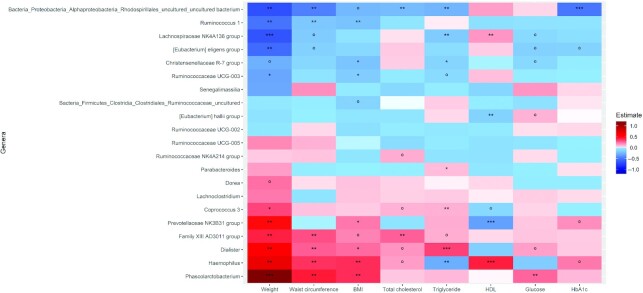
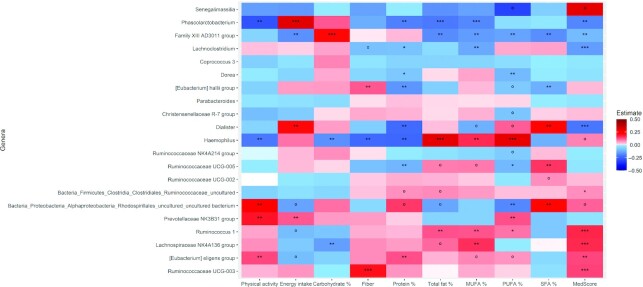
In the overall population, as well individually within groups, changes in Eubacterium eligens were negatively associated with changes in weight (FDR P < 0.05), waist circumference (insignificant FDR), glucose (insignificant FDR), and HbA1c (insignificant FDR) (Figure 3, Supplemental Figures 5A, 6A). Haemophilus, which varied significantly between the groups of intervention, was positively associated with weight changes in the overall population (Figure 3). Parabacteroides was positively associated with triglyceride concentrations in the overall population, as well as in the IG and the CG (Figure 3, Supplemental Figures 5A, 6A). Interestingly, Phascolarbacterium, which was positively associated with energy intake, also followed the same direction for weight, BMI, waist circumference, and glucose but was negatively associated with physical activity. Changes in fiber intake were negatively associated with changes in Haemophilus but positively associated with changes in E. hallii and Ruminococcaceae UCG-003 (Figure 4). Lachnospiraceae NK4A136 group was also positively associated with MedScore (Figure 4). Few other associations within the IG and the CG were observed (Supplemental Figures 5A,B, 6A,B).
FIGURE 3.
Heat plot showing associations in overall study population (n = 343) between changes in microbial genera and clinical variables. Model evaluated by negative binomial mixed model, adjusting for covariates sex, study center, and baseline body weight; adjusted P value denoted by ***P < 0.001, **P < 0.05, *P < 0.1, and ºP < 0.2.
FIGURE 4.
Heat plot showing associations in overall study population (n = 343) between changes in microbial genera and energy intake variables. Model evaluated by negative binomial mixed model, adjusting for covariates sex, study center, and baseline body weight; adjusted P value denoted by ***P < 0.001, **P < 0.05, *P < 0.1, and ºP < 0.2.
Changes in bacterial predicted metagenomics functions during intervention
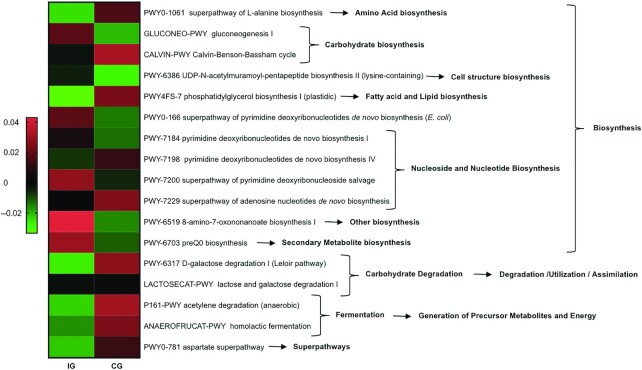
Metabolic pathways belonging to the biosynthesis of nucleotides, nucleosides, and amino acids and carbohydrates changed significantly between the 2 intervention groups (Figure 5). Compared with the CG, fermentation pathways leading to the generation of energy were reduced in the IG.
FIGURE 5.
Heat map representing median values of significantly increasing or decreasing predicted metagenome pathways in the IG (n = 171) and CG (n = 172). CG, control group; IG, intervention group.
Discussion
We report for the first time, to our knowledge, the effect of a large long-term lifestyle-based weight loss intervention with energy-reduced MedDiet and increased physical activity on gut microbiota. Several changes in the relative abundance of genera have been observed within and between the intervention groups that can be attributed to weight loss, diet, and physical activity. Changes observed in the gut microbiota profile were also associated with changes in some CVD risk factors.
We observed a significant change in the relative abundance of members belonging to the Firmicutes phylum (decreasing: Butyricoccus, Ruminiclostridium 5, and E. hallii; increasing: Ruminococcaceae NK4A214, Coprobacter) and a significant increase in the B/F in the IG compared with the CG, which could partly be explained by higher weight loss in the IG compared with the CG. Even though widely debated, it has been reported that during weight loss, the B/F increases, suggesting that it may respond to energy restriction (3, 33, 34). An increase in B/F has also been reported with higher adherence to MedDiet as well as low animal protein intake (35).
Other results from the above MedDiet adherence study (35), indicating an increase in the relative abundance of Dorea, Roseburia, and Coprococcus (all reported as SCFA producers of the Lachnospiraceae family), also were in line with our results only in the non-energy-restricted MedDiet group (CG). However, in the IG, we observed these taxa to reduce in 1 y of intervention. Correspondingly, we also observed a decrease in the predicted fermentation pathways in the IG compared with the CG. Although the reduction in these carbohydrate/fiber-using SCFA producers could indicate contradictory findings, some studies have observed an increase in SCFA gut production in obese compared with normal-weight individuals (36, 37). Whether this increase in SCFA producers may be the cause or the consequence of obesity remains to be elucidated. The high-energy deriving capacities of carbohydrate/polysaccharide-using bacteria could create a net energy excess for the host, contributing to obesity. However, SCFAs, especially butyrate and their producers, have been well associated with several beneficial health effects (38); hence, a careful evaluation of their composition as well quantity is required to infer further.
Even though there were reductions in certain SCFA producers in the IG, we observed within the same group a selective increase in other SCFA producers (39) such as the Lachnospiraceae NK4A136 group and Ruminococcaceae (UCG-003, UCG-002), which also were associated positively with MedScore. We also observed that some SCFA-producing genera (Lachnospira, Lachnospiraceae NK4A136 group, and Alistipes) shifted in the same direction within both intervention groups, reflecting overall the effect of MedDiet on gut microbiota. Increases of proteins, polyphenols, and unsaturated fats have shown inhibitory activities to certain bacterial genera (40–42). In parallel, in the IG, participants consumed higher protein, polyphenols, and unsaturated fats compared with those in the CG, possibly leading to selective enrichment in certain SCFA producers compared with others that might be inhibited by a synergy of the abovementioned components. It has been demonstrated in a mice study that calorie restriction could limit butyrogenic enzymes and promote propiogenic enzymes, which could lead to competition and selective growth of SCFA producers (43, 44).
Changes in Coprococcus 3 were positively associated with changes in weight, total cholesterol, and triglycerides and negatively with HDL cholesterol in the overall population. In line with our results, enrichment of the Coprococcus genus has been associated with a high lifetime CVD risk profile in Bogalusa Heart study participants, as well as with the obese phenotype (45).
Not only Coprococcus but also other genera majorly belonging to the Lachnospiraceae family (Blautia, Dorea, Roseburia, Coprococcus 3) and Ruminococcus 1 were observed to be changing in opposite directions in the IG and the CG. We observed a positive association for changes in the relative abundance of Coprococcus 3 and Dorea with changes in weight significantly in the overall population and nonsignificantly in both intervention groups, consistent with a Swedish study (46). This study also reported a positive association of these genera with plasma branched-chain amino acids (BCAAs), usually increased in type 2 diabetes (T2D) and MetS (47, 48). Similar observations were made in the METSIM (METabolic Syndrome In Men) cohort, in which Blautia was associated with higher BMI and also higher circulating BCAAs, whereas Chistensenellaceae R-7 group abundance was negatively associated with BCAAs (49). Consistently, we found a negative association in the Chistensenellaceae R-7 group with changes in weight, BMI, triglycerides, and plasma glucose. It has been demonstrated that following the MedDiet enriched with extra virgin olive oil reduced circulating concentrations of BCAAs and was associated with a lower risk of T2D (50). Taking these findings into consideration, we suspect BCAAs as one of the pathways for glucose regulation in the IG via MedDiet-associated weight loss and corresponding changes in gut microbiota (51, 52). These results could also indicate that even with following the same dietary pattern, factors such as energy restriction and physical activity could play an additional beneficial role in overweight/obese individuals by altering glucose regulation via BCAAs (53).
In the IG, we also observed changes in some previously bile acid–associated bacteria, such as Lachnoclostridium (containing members of 7α-dehydroxylating capacity) and Bilophila (deconjugator of taurine–bile acid), that have shown to control lipid and glucose metabolism in mice studies (54, 55). Consistently, we observed a positive (nonsignificant) association between Lachnoclostridium and glucose. The observations we make above are specific to the IG, indicating that calorie restriction along with an increase in physical activity could modulate bile-related bacteria (56). Compared with dietary interventions, very few studies have been conducted studying the effect of physical activity on gut microbiota, with contradictory results. Haemophilus and Phascolarctobacterium, which overall had shown a positive association in this study with risk factors assessed, were also negatively associated with changes in physical activity. We suspect the associations we observe here are not solely dependent on physical activity but rather a synergy between energy homeostasis and nutrient intake.
Predicted metagenomics functions have been shown to differ between adults with different body weight and health status. In our study, we observed that predicted functions of the bacterial community in the gut of the IG were trying to adapt to energy restriction by increasing biosynthesis pathways, especially carbohydrate and nucleotide biosynthesis. However, as protein and fat intake increased in the IG, we also observed a decrease in amino acids and lipid biosynthesis, indicating an adaptation to diet. Many of the observations made in this study should also be interpreted in terms of calorie restriction as it has been reported that calorie restriction could alter gut microbiota and their functionality independent of a dietary regimen (57, 58).
With the exception of a few landmark studies (11, 59), our study explores the effect of a healthy lifestyle intervention on gut microbiota in a comparatively large sample population and follow-up (35, 44, 60). The randomized controlled trial design of our study allows us to establish causality when assessing the effect of the interventions, being one of the most important strengths, but this does not apply when we assess associations as secondary analyses. Another strength of the present study is that we have observed significant differences between groups in all components of the intervention (weight loss, adherence to MedDiet, and physical activity) in the expected direction, allowing us to test for potential effects of the intervention on gut microbiota. The nature of the intervention comprising dietary intervention, behavioral therapy, and physical activity promotion indicates the multilevel intervention strategy that promotes participants to follow the intervention and obtain clinical benefits.
As much as this multifaceted intervention strategy is beneficial, it implies a limitation on the inference of results that cannot be attributed solely to a single component of the intervention. Along with this, some limitations of this study also deserve to be mentioned. First, our findings are limited to adults with high BMI who also met the criteria for MetS and were living in a Mediterranean country. Therefore, they cannot be generalized to other populations or all individuals with MetS. Second, the lack of data on fecal metabolites and species-level taxonomy does not allow us to infer further the pathways associated with the associations we have observed. Third, the dietary records were collected from a self-reported questionnaire, which might over/underestimate the intake of certain food groups. Future studies with a comprehensive set of metabolomics, metagenomics, and intermediate time points would allow us to better understand the transition of gut microbiota during the weight loss period.
Overall, in this 1-y lifestyle-based intervention, we observed that an energy-restricted Mediterranean diet with physical activity and behavioral support induced weight loss and improved CVD-associated risk factors. A decrease in several members of Firmicutes, especially belonging to the Lachnospiraceae, and a selective increase in some SCFA producers were observed in the IG. This work identifies that even with similar healthy dietary patterns, the addition of an intervention program enhancing calorie restriction and physical activity could have a significant benefit on the CVD risk factors potentially modulated via the gut microbiota.
Supplementary Material
Acknowledgments
We thank all the volunteers for their participation and personnel for their contribution in the PREDIMED-Plus trial. We also thank all the investigators of the PREDIMED-Plus study and the PREDIMED-Plus Biobank Network, as a part of the National Biobank Platform of the Instituto de Salud Carlos III, for storing and managing the PREDIMED-Plus biological samples.
The authors’ contributions were as follows—JS-S, MB, MAM-G, FJT, DC, JVL, JV, and MF: designed the study; JM, SG, AA, IM-I, JCF-G, LT-C, RF-C, RO, AMG-P, OC and MRB-L: provided sample collection and processing; JM, IMI, SG, and AA: conducted the statistical analysis; MB, FT, and JS-S: provided supervision; JM, IMI, JS-S, MB, and FJT: wrote the manuscript; and all authors: read and approved the final manuscript.
Author disclosures: JS-S declares that he is a member of Danone S.A.’s Advisory Board and a member of the Danone Institute, has received payments from Danone S.A. for the purposes of scientific and technical consulting, has served on the board of and received grant support through his institution from the International Nut and Dried Fruit Council and Eroski Foundation, and has received consulting fees or travel expenses from Nuts for Life and Australian Nut Industry Council. All other authors report no conflicts of interest.
Notes
Supported by the official Spanish institutions for funding scientific biomedical research, CIBER Fisiopatología de la Obesidad y Nutrición (CIBEROBN) and Instituto de Salud Carlos III (ISCIII), through the Fondo de Investigación para la Salud (FIS), which is cofunded by the European Regional Development Fund (3 coordinated FIS projects led by JS-S, including the following projects: PI13/00462, PI16/00501, and PI19/00576, 2 led by JV, including PI17/01441 and PI14/01206); the Especial Action Project, entitled “Implementación y evaluación de una intervención intensiva sobre la actividad física Cohorte,” PREDIMED-Plus grant (OBN16PE01) to JS-S; and the Recercaixa (number 2013ACUP00194) grant to JS-S. DC obtained a grant from the Generalitat Valenciana (PROMETEO 2017/17) and a grant from the Ministry of Science and Innovation/Instituto de Salud Carlos III (reference: PI19/00781). Eat2BeNICE project (European Union’s Horizon 2020 research and innovation program under the grant agreement No 728018) was obtained by Alejandro Arias Vazquez (PI) (Radboud University Medical Center) and Third Party [PI: JS-S (Institut d’Investigacions Sanitàries Pere I Virgili)]. This study also was supported by the Fondo de Investigacion Sanitaria (PI16/00516), cofounded by the Fondo Europeo de Desarrollo Regional (FEDER) and Fundacion Danone. MRB-L was supported by “Miguel Servet Type I” program (CP15/00028) from the ISCIII-Madrid (Spain), cofinanced by the Fondo Europeo de Desarrollo Regional-FEDER. SG received support from Agaur (Comissió Executiva d'Ajuts de Recerca de l'Agència de Gestió d'Ajuts Universitaris i de Recerca de la Generalitat de Catalunya, number 2018FI_B_00444). JM and AA were supported by the European Union's Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No. 713679, cofunded from the Universitat Rovira i Virgili (URV) and Fundació Catalunya La Pedrera. JS-S, senior author of this article, was partially supported by ICREA under the ICREA Academia program.
Food companies Hojiblanca (Lucena, Spain) and Patrimonio Comunal Olivarero (Madrid, Spain) donated extra virgin olive oil, and the Almond Board of California (Modesto, CA), American Pistachio Growers (Fresno, CA), and Paramount Farms (Wonderful Company, LLC, Los Angeles, CA) donated nuts for the PREDIMED-Pilot study.
None of the funding sources took part in the design, collection, analysis, interpretation of the data, writing of the report, or the decision to submit the manuscript for publication.
JS-S and FJT are senior authors.
The data sets generated and analyzed during the current study are not expected to be made available outside the core research group, as neither participants’ consent forms nor ethics approval included permission for open access. However, the researchers will follow a controlled data-sharing collaboration model, as in the informed consent participants agreed with a controlled collaboration with other investigators for research related to the project's aims. Therefore, investigators who are interested in this study can contact the PREDIMED Steering Committee by sending a request letter to predimed_scommittee@googlegroups.com. A data-sharing agreement indicating the characteristics of the collaboration and data management will be completed for the proposals that are approved by the Steering Committee.
Supplemental Tables 1–5, Supplemental Figures 1–6, and Supplemental Methods 1–3 are available from the “Supplementary data” link in the online posting of the article at https://academic.oup.com/ajcn/.
Abbreviations used: BCAA, branched-chain amino acid; B/F, Bacteroidetes-to-Firmicutes ratio; CG, control group; CVD, cardiovascular disease; FDR, false discovery rate; IG, intervention group; MedDiet, Mediterranean diet; MetS, metabolic syndrome; MedScore, Mediterranean diet adherence score; P/B, Prevotella-to-Bacteroides ratio; PCoA, principal coordinate analysis; SCFA, short-chain fatty acid; sPLS-DA, sparse partial least squares discriminant analysis; T2D, type 2 diabetes; VIP, variable importance in projection.
Contributor Information
Jananee Muralidharan, Universitat Rovira i Virgili, Department of Biochemistry and Biotechnology, Hospital Universitari de Sant Joan de Reus, Institut d’Investigacions Sanitàries Pere i Virgili, Human Nutrition Unit, Reus, Spain; CIBER de Fisiopatología de la Obesidad y la Nutrición (CIBEROBN), Instituto de Salud Carlos III, Madrid, Spain.
Isabel Moreno-Indias, CIBER de Fisiopatología de la Obesidad y la Nutrición (CIBEROBN), Instituto de Salud Carlos III, Madrid, Spain; Unidad de Gestion Clínica de Endocrinología y Nutrición, Laboratorio del Instituto de Investigación Biomédica de Málaga (IBIMA), Hospital Universitario de Málaga (Virgen de la Victoria), Universidad de Málaga, Málaga, Spain.
Mónica Bulló, Universitat Rovira i Virgili, Department of Biochemistry and Biotechnology, Hospital Universitari de Sant Joan de Reus, Institut d’Investigacions Sanitàries Pere i Virgili, Human Nutrition Unit, Reus, Spain; CIBER de Fisiopatología de la Obesidad y la Nutrición (CIBEROBN), Instituto de Salud Carlos III, Madrid, Spain.
Jesús Vioque Lopez, Instituto de Investigación Sanitaria y Biomédica de Alicante, ISABIAL-UMH, Alicante, Spain; CIBER Epidemiología y Salud Pública (CIBERESP), Instituto de Salud Carlos III (ISCIII), Madrid, Spain.
Dolores Corella, CIBER de Fisiopatología de la Obesidad y la Nutrición (CIBEROBN), Instituto de Salud Carlos III, Madrid, Spain; Department of Preventive Medicine, University of Valencia, Valencia, Spain.
Olga Castañer, CIBER de Fisiopatología de la Obesidad y la Nutrición (CIBEROBN), Instituto de Salud Carlos III, Madrid, Spain; Cardiovascular Risk and Nutrition (Regicor Study Group), Hospital del Mar Research Institute (IMIM), Barcelona, Spain.
Josep Vidal, Endocrinology and Nutrition Department, Hospital Clinic Universitari, Barcelona, Spain; August Pi i Sunyer Biomedical Research Institute (IDIBAPS), Barcelona, Spain.
Alessandro Atzeni, Universitat Rovira i Virgili, Department of Biochemistry and Biotechnology, Hospital Universitari de Sant Joan de Reus, Institut d’Investigacions Sanitàries Pere i Virgili, Human Nutrition Unit, Reus, Spain; CIBER de Fisiopatología de la Obesidad y la Nutrición (CIBEROBN), Instituto de Salud Carlos III, Madrid, Spain.
Jose Carlos Fernandez-García, CIBER de Fisiopatología de la Obesidad y la Nutrición (CIBEROBN), Instituto de Salud Carlos III, Madrid, Spain; Unidad de Gestion Clínica de Endocrinología y Nutrición, Laboratorio del Instituto de Investigación Biomédica de Málaga (IBIMA), Hospital Universitario de Málaga (Virgen de la Victoria), Universidad de Málaga, Málaga, Spain.
Laura Torres-Collado, Instituto de Investigación Sanitaria y Biomédica de Alicante, ISABIAL-UMH, Alicante, Spain; CIBER Epidemiología y Salud Pública (CIBERESP), Instituto de Salud Carlos III (ISCIII), Madrid, Spain.
Rebeca Fernández-Carrión, CIBER de Fisiopatología de la Obesidad y la Nutrición (CIBEROBN), Instituto de Salud Carlos III, Madrid, Spain; Department of Preventive Medicine, University of Valencia, Valencia, Spain.
Monsterrat Fito, CIBER de Fisiopatología de la Obesidad y la Nutrición (CIBEROBN), Instituto de Salud Carlos III, Madrid, Spain; Cardiovascular Risk and Nutrition (Regicor Study Group), Hospital del Mar Research Institute (IMIM), Barcelona, Spain.
Romina Olbeyra, August Pi i Sunyer Biomedical Research Institute (IDIBAPS), Barcelona, Spain.
Ana Maria Gomez-Perez, CIBER de Fisiopatología de la Obesidad y la Nutrición (CIBEROBN), Instituto de Salud Carlos III, Madrid, Spain; Unidad de Gestion Clínica de Endocrinología y Nutrición, Laboratorio del Instituto de Investigación Biomédica de Málaga (IBIMA), Hospital Universitario de Málaga (Virgen de la Victoria), Universidad de Málaga, Málaga, Spain.
Serena Galiè, Universitat Rovira i Virgili, Department of Biochemistry and Biotechnology, Hospital Universitari de Sant Joan de Reus, Institut d’Investigacions Sanitàries Pere i Virgili, Human Nutrition Unit, Reus, Spain; CIBER de Fisiopatología de la Obesidad y la Nutrición (CIBEROBN), Instituto de Salud Carlos III, Madrid, Spain.
Maria Rosa Bernal-López, CIBER de Fisiopatología de la Obesidad y la Nutrición (CIBEROBN), Instituto de Salud Carlos III, Madrid, Spain; UGC Medicina Interna, Hospital Regional Universitario de Málaga, Malaga, Spain.
Miguel Angel Martinez-Gonzalez, CIBER de Fisiopatología de la Obesidad y la Nutrición (CIBEROBN), Instituto de Salud Carlos III, Madrid, Spain; University of Navarra, Department of Preventive Medicine and Public Health, IdiSNA, Pamplona, Spain; Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, MA, USA.
Jordi Salas-Salvadó, Universitat Rovira i Virgili, Department of Biochemistry and Biotechnology, Hospital Universitari de Sant Joan de Reus, Institut d’Investigacions Sanitàries Pere i Virgili, Human Nutrition Unit, Reus, Spain; CIBER de Fisiopatología de la Obesidad y la Nutrición (CIBEROBN), Instituto de Salud Carlos III, Madrid, Spain.
Francisco Jose Tinahones, CIBER de Fisiopatología de la Obesidad y la Nutrición (CIBEROBN), Instituto de Salud Carlos III, Madrid, Spain; Unidad de Gestion Clínica de Endocrinología y Nutrición, Laboratorio del Instituto de Investigación Biomédica de Málaga (IBIMA), Hospital Universitario de Málaga (Virgen de la Victoria), Universidad de Málaga, Málaga, Spain.
References
- 1.Rodriguez-Castaño GP, Caro-Quintero A, Reyes A, Lizcano F. Advances in gut microbiome research, opening new strategies to cope with a Western lifestyle. Front Genet. 2017;7:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lippert K, Kedenko L, Antonielli L, Kedenko I, Gemeier C, Leitner M, Kautzky-Willer A, Paulweber B, Hackl E. Gut microbiota dysbiosis associated with glucose metabolism disorders and the metabolic syndrome in older adults. Benef Microbes. 2017;8:545–56. [DOI] [PubMed] [Google Scholar]
- 3.Crovesy L, Masterson D, Rosado EL. Profile of the gut microbiota of adults with obesity: a systematic review. Eur J Clin Nutr. 2020;74:1251–62. [DOI] [PubMed] [Google Scholar]
- 4.Hu H-J, Park S-G, Jang HB, Choi M-G, Park K-H, Kang JH, Park SI, Lee H-J, Cho S-H. Obesity alters the microbial community profile in Korean adolescents. PLoS One. 2015;10:e0134333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duncan SH, Lobley GE, Holtrop G, Ince J, Johnstone AM, Louis P, Flint HJ. Human colonic microbiota associated with diet, obesity and weight loss. Int J Obes. 2008;32:1720–4. [DOI] [PubMed] [Google Scholar]
- 6.Baothman OA, Zamzami MA, Taher I, Abubaker J, Abu-Farha M. The role of gut microbiota in the development of obesity and Diabetes. Lipids Health Dis. 2016;15:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weinsier RL, Hunter GR, Heini AF, Goran MI, Sell SM. The etiology of obesity: relative contribution of metabolic factors, diet, and physical activity. Am J Med. 1998;105:145–50. [DOI] [PubMed] [Google Scholar]
- 8.Wong JMW. Gut microbiota and cardiometabolic outcomes: influence of dietary patterns and their associated components. Am J Clin Nutr. 2014;100:369S–77S. [DOI] [PubMed] [Google Scholar]
- 9.Matijašić BB, Obermajer T, Lipoglavšek L, Grabnar I, Avguštin G, Rogelj I. Association of dietary type with fecal microbiota in vegetarians and omnivores in Slovenia. Eur J Nutr. 2014;53:1051–64. [DOI] [PubMed] [Google Scholar]
- 10.Haro C, Montes-Borrego M, Rangel-Zúñiga OA, Alcalã-Diaz JF, Gamez-Delgado F, Pérez-Martinez P, Delgado-Lista J, Quintana-Navarro GM, Tinahones FJ, Landa BBet al. . Two healthy diets modulate gut microbial community improving insulin sensitivity in a human obese population. J Clin Endocrinol Metab. 2015;101:jc20153351. [DOI] [PubMed] [Google Scholar]
- 11.Ghosh TS, Rampelli S, Jeffery IB, Santoro A, Neto M, Capri M, Giampieri E, Jennings A, Candela M, Turroni Set al. . Mediterranean diet intervention alters the gut microbiome in older people reducing frailty and improving health status: the NU-AGE 1-year dietary intervention across five European countries. Gut. 2020;69:1218–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meslier V, Laiola M, Roager HM, De Filippis F, Roume H, Quinquis B, Giacco R, Mennella I, Ferracane R, Pons Net al. . Mediterranean diet intervention in overweight and obese subjects lowers plasma cholesterol and causes changes in the gut microbiome and metabolome independently of energy intake. Gut. 2020;69:1258–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martínez-González MA, Buil-Cosiales P, Corella D, Bulló M, Fitó M, Vioque J, Romaguera D, Martínez JA, Wärnberg J, López-Miranda Jet al. . Cohort profile: design and methods of the PREDIMED-Plus randomized trial. Int J Epidemiol. 2019;48:387–388. [DOI] [PubMed] [Google Scholar]
- 14.Schröder H, Fitó M, Estruch R, Martínez-González MA, Corella D, Salas-Salvadó J, Lamuela-Raventós R, Ros E, Salaverría I, Fiol Met al. . A short screener is valid for assessing Mediterranean diet adherence among older Spanish men and women. J Nutr. 2011;141:1140–5. [DOI] [PubMed] [Google Scholar]
- 15.Molina L, Sarmiento M, Peñafiel J, Donaire D, Garcia-Aymerich J, Gomez M, Ble M, Ruiz S, Frances A, Schröder Het al. . Validation of the Regicor Short Physical Activity Questionnaire for the Adult Population. PLoS One. 2017;12:e0168148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mas-Lloret J, Obón-Santacana M, Ibáñez-Sanz G, Guinó E, Pato ML, Rodriguez-Moranta F, Mata A, García-Rodríguez A, Moreno V, Pimenoff VN. Gut microbiome diversity detected by high-coverage 16S and shotgun sequencing of matched stool and colon biopsy samples. Sci Data. 2020;7:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koncevicius K. matrixTests: fast statistical hypothesis tests on rows and columns of matrices. [Internet]. 2020; ; [cited 2020 Aug 3]. Available from: https://cran.r-project.org/package=matrixTests. [Google Scholar]
- 19.Paulson JN, Stine OC, Bravo HC, Pop M. Differential abundance analysis for microbial marker-gene surveys. Nat Methods. 2013;10:1200–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roager HM, Licht TR, Poulsen SK, Larsen TM, Bahl MI. Microbial enterotypes, inferred by the Prevotella-to-Bacteroides ratio. Appl Environ Microbiol. 2014;80:1142 LP–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lahti L, Shetty S. microbiome R package. [Internet] [cited 2020 Jul 15]. Available from: http://microbiome.github.io. [Google Scholar]
- 22.Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, Blomberg SP, Webb CO. Picante: r tools for integrating phylogenies and ecology. Bioinformatics. 2010;26:1463–4. [DOI] [PubMed] [Google Scholar]
- 23.Oksanen J, Kindt R, Legendre P, O'Hara R, Stevens MHH, Oksanen MJ, Suggests M. The vegan package. Community Ecol Packag. 2007:10:719. [Google Scholar]
- 24.Paulson JN, Stine OC, Bravo HC, Pop M. Differential abundance analysis for microbial marker-gene surveys. Nat Methods. 2013;10:1200–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rohart F, Gautier B, Singh A, Lê Cao K-A. mixOmics: an R package for ‘omics feature selection and multiple data integration. PLoS Comput Biol. 2017;13:e1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee AH, Shannon CP, Amenyogbe N, Bennike TB, Diray-Arce J, Idoko OT, Gill EE, Ben-Othman R, Pomat WS, van Haren SDet al. . Dynamic molecular changes during the first week of human life follow a robust developmental trajectory. Nat Commun. 2019;10:1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X, Yi N. NBZIMM: negative binomial and zero-inflated mixed models, with application to microbiome/metagenomics data analysis. BMC Bioinformatics. 2020;21:488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wickham H. ggplot2: elegant graphics for data analysis. New York: Springer-Verlag; 2016. [Google Scholar]
- 29.Douglas GM, Maffei VJ, Zaneveld JR, Yurgel SN, Brown JR, Taylor CM, Huttenhower C, Langille MGI. PICRUSt2 for prediction of metagenome functions. Nat Biotechnol. 2020;38:685–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caspi R, Billington R, Keseler IM, Kothari A, Krummenacker M, Midford PE, Ong WK, Paley S, Subhraveti P, Karp PD. The MetaCyc database of metabolic pathways and enzymes—a 2019 update. Nucleic Acids Res. 2020;48:D445–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30:3123–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Álvarez-Álvarez I, Martínez-González MÁ, Sánchez-Tainta A, Corella D, Díaz-López A, Fitó M, Vioque J, Romaguera D, Martínez JA, Wärnberg J. Adherence to an energy-restricted Mediterranean diet score and prevalence of cardiovascular risk factors in the PREDIMED-plus: a cross-sectional study. Rev Española Cardiol. 2019;72:925–34. [DOI] [PubMed] [Google Scholar]
- 33.Remely M, Tesar I, Hippe B, Gnauer S, Rust P, Haslberger AG. Gut microbiota composition correlates with changes in body fat content due to weight loss. Benef Microbes. 2015;6:431–9. [DOI] [PubMed] [Google Scholar]
- 34.Fragiadakis GK, Wastyk HC, Robinson JL, Sonnenburg ED, Sonnenburg JL, Gardner CD. Long-term dietary intervention reveals resilience of the gut microbiota despite changes in diet and weight. Am J Clin Nutr. 2020;111:1127–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia-Mantrana I, Selma-Royo M, Alcantara C, C MC. Shifts on gut microbiota associated to Mediterranean diet adherence and specific dietary intakes on general adult population. Front Microbiol. 2018;9:890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Teixeira TFS, Grześkowiak Ł, Franceschini SCC, Bressan J, Ferreira C, Peluzio MCG. Higher level of faecal SCFA in women correlates with metabolic syndrome risk factors. Br J Nutr. 2013;109:914–9. [DOI] [PubMed] [Google Scholar]
- 37.Rahat-Rozenbloom S, Fernandes J, Gloor GB, Wolever TMS. Evidence for greater production of colonic short-chain fatty acids in overweight than lean humans. Int J Obes. 2014;38:1525–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanna S, van Zuydam NR, Mahajan A, Kurilshikov A, Vila AV, Võsa U, Mujagic Z, Masclee AAM, Jonkers D, Oosting M. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat Genet. 2019;51:600–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Louis P, Flint HJ. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol. 2017;19:29–41. [DOI] [PubMed] [Google Scholar]
- 40.Zhao J, Zhang X, Liu H, Brown MA, Qiao S. Dietary protein and gut microbiota composition and function. Curr Protein Pept Sci. 2019;20:145–54. [DOI] [PubMed] [Google Scholar]
- 41.Duda-Chodak A. The inhibitory effect of polyphenols on human gut microbiota. J Physiol Pharmacol. 2012;63:497–503. [PubMed] [Google Scholar]
- 42.Wolters M, Ahrens J, Romaní-Pérez M, Watkins C, Sanz Y, Benítez-Páez A, Stanton C, Günther K. Dietary fat, the gut microbiota, and metabolic health—a systematic review conducted within the MyNewGut project. Clin Nutr. 2018;38(6):2504–20. [DOI] [PubMed] [Google Scholar]
- 43.Tanca A, Abbondio M, Palomba A, Fraumene C, Marongiu F, Serra M, Pagnozzi D, Laconi E, Uzzau S. Caloric restriction promotes functional changes involving short-chain fatty acid biosynthesis in the rat gut microbiota. Sci Rep. 2018;8:14778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Filippis F, Pellegrini N, Vannini L, Jeffery IB, La Storia A, Laghi L, Serrazanetti DI, Di Cagno R, Ferrocino I, Lazzi Cet al. . High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut. 2016;65:1812 LP–1821. [DOI] [PubMed] [Google Scholar]
- 45.Castaner O, Goday A, Park Y-M, Lee S-H, Magkos F, Shiow S-ATE, Schröder H. The gut microbiome profile in obesity: a systematic review. Int J Endocrinol. 2018;2018:4095789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ottosson F, Brunkwall L, Ericson U, Nilsson PM, Almgren P, Fernandez C, Melander O, Orho-Melander M. Connection between BMI-related plasma metabolite profile and gut microbiota. J Clin Endocrinol Metab. 2018;103:1491–501. [DOI] [PubMed] [Google Scholar]
- 47.Floegel A, Stefan N, Yu Z, Mühlenbruch K, Drogan D, Joost H-G, Fritsche A, Häring H-U, Hrabě de Angelis M, Peters Aet al. . Identification of serum metabolites associated with risk of type 2 diabetes using a targeted metabolomic approach. Diabetes. 2013;62:639–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Würtz P, Tiainen M, Mäkinen V-P, Kangas AJ, Soininen P, Saltevo J, Keinänen-Kiukaanniemi S, Mäntyselkä P, Lehtimäki T, Laakso Met al. . Circulating metabolite predictors of glycemia in middle-aged men and women. Diabetes Care. 2012;35:1749–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Org E, Blum Y, Kasela S, Mehrabian M, Kuusisto J, Kangas AJ, Soininen P, Wang Z, Ala-Korpela M, Hazen SLet al. . Relationships between gut microbiota, plasma metabolites, and metabolic syndrome traits in the METSIM cohort. Genome Biol. 2017;18:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ruiz-Canela M, Guasch-Ferré M, Toledo E, Clish CB, Razquin C, Liang L, Wang DD, Corella D, Estruch R, Á Het al. . Plasma branched chain/aromatic amino acids, enriched Mediterranean diet and risk of type 2 diabetes: case-cohort study within the PREDIMED Trial. Diabetologia. 2018;61:1560–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, Haqq AM, Shah SH, Arlotto M, Slentz CAet al. . A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9:311–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karusheva Y, Koessler T, Strassburger K, Markgraf D, Mastrototaro L, Jelenik T, Simon M-C, Pesta D, Zaharia O-P, Bódis Ket al. . Short-term dietary reduction of branched-chain amino acids reduces meal-induced insulin secretion and modifies microbiome composition in type 2 diabetes: a randomized controlled crossover trial. Am J Clin Nutr. 2019;110:1098–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pedersen HK, Gudmundsdottir V, Nielsen HB, Hyotylainen T, Nielsen T, Jensen BAH, Forslund K, Hildebrand F, Prifti E, Falony Get al. . Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016;535:376–81. [DOI] [PubMed] [Google Scholar]
- 54.Li F, Jiang C, Krausz KW, Li Y, Albert I, Hao H, Fabre KM, Mitchell JB, Patterson AD, Gonzalez FJ. Microbiome remodelling leads to inhibition of intestinal farnesoid X receptor signalling and decreased obesity. Nat Commun. 2013;4:2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parséus A, Sommer N, Sommer F, Caesar R, Molinaro A, Ståhlman M, Greiner TU, Perkins R, Bäckhed F. Microbiota-induced obesity requires farnesoid X receptor. Gut. 2017;66:429–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cerdá B, Pérez M, Pérez-Santiago JD, Tornero-Aguilera JF, González-Soltero R, Larrosa M. Gut microbiota modification: another piece in the puzzle of the benefits of physical exercise in health?. Front Physiol. 2016;7:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fabbiano S, Suárez-Zamorano N, Chevalier C, Lazarević V, Kieser S, Rigo D, Leo S, Veyrat-Durebex C, Gaïa N, Maresca Met al. . Functional gut microbiota remodeling contributes to the caloric restriction-induced metabolic improvements. Cell Metab. 2018;28:907–921.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu J, Yang L, Li S, Huang P, Liu Y, Wang Y, Tang H. Metabolomics insights into the modulatory effects of long-term low calorie intake in mice. J Proteome Res. 2016;15:2299–308. [DOI] [PubMed] [Google Scholar]
- 59.Heianza Y, Sun D, Smith SR, Bray GA, Sacks FM, Qi L. Changes in gut microbiota-related metabolites and long-term successful weight loss in response to weight-loss diets: the POUNDS Lost Trial. Diabetes Care. 2018;41:413–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haro C, Montes-Borrego M, Rangel-Zuniga OA, Alcala-Diaz JF, Gomez-Delgado F, Perez-Martinez P, Delgado-Lista J, Quintana-Navarro GM, Tinahones FJ, Landa BBet al. . Two healthy diets modulate gut microbial community improving insulin sensitivity in a human obese population. J Clin Endocrinol Metab. 2016;101:233–42. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.