Abstract
Background
Stereotactic radiosurgery (SRS), a validated treatment for brain tumors, requires accurate tumor contouring. This manual segmentation process is time-consuming and prone to substantial inter-practitioner variability. Artificial intelligence (AI) with deep neural networks have increasingly been proposed for use in lesion detection and segmentation but have seldom been validated in a clinical setting.
Methods
We conducted a randomized, cross-modal, multi-reader, multispecialty, multi-case study to evaluate the impact of AI assistance on brain tumor SRS. A state-of-the-art auto-contouring algorithm built on multi-modality imaging and ensemble neural networks was integrated into the clinical workflow. Nine medical professionals contoured the same case series in two reader modes (assisted or unassisted) with a memory washout period of 6 weeks between each section. The case series consisted of 10 algorithm-unseen cases, including five cases of brain metastases, three of meningiomas, and two of acoustic neuromas. Among the nine readers, three experienced experts determined the ground truths of tumor contours.
Results
With the AI assistance, the inter-reader agreement significantly increased (Dice similarity coefficient [DSC] from 0.86 to 0.90, P < 0.001). Algorithm-assisted physicians demonstrated a higher sensitivity for lesion detection than unassisted physicians (91.3% vs 82.6%, P = .030). AI assistance improved contouring accuracy, with an average increase in DSC of 0.028, especially for physicians with less SRS experience (average DSC from 0.847 to 0.865, P = .002). In addition, AI assistance improved efficiency with a median of 30.8% time-saving. Less-experienced clinicians gained prominent improvement on contouring accuracy but less benefit in reduction of working hours. By contrast, SRS specialists had a relatively minor advantage in DSC, but greater time-saving with the aid of AI.
Conclusions
Deep learning neural networks can be optimally utilized to improve accuracy and efficiency for the clinical workflow in brain tumor SRS.
Keywords: artificial intelligence, brain tumor, deep learning, randomization, stereotactic radiosurgery
Keys Points.
This randomized, cross-modal reader study shows AI can facilitate brain tumor SRS.
AI assistance improves tumor contouring accuracy, especially for less-experienced clinicians.
AI assistance improves efficiency and reduces variability among physicians.
Importance of the Study.
Stereotactic radiosurgery (SRS), a validated treatment for brain metastases and brain tumors, requires accurate tumor contouring for treatment planning. This manual segmentation process is time-consuming and prone to substantial inter-practitioner variability even amongst experts, leading to large variation in care quality.
Artificial intelligence (AI) has been proposed for use in lesion detection and segmentation. However, the potential impact of integrating such technology into SRS workflows remains largely unexplored. In this paper, we deploy an expert-level AI-based assistive tool into the SRS workflows and conduct a randomized, cross-modal, multi-reader, multispecialty, multi-tumor-type reader study on AI-assisted tumor contouring. The study demonstrates AI assistance increases inter-clinician agreement, improves tumor detection by 10.5%, and shortens mean contouring time by 30.8%. This multi-reader evaluation provides comprehensive comparisons of the impacts of AI on different experience levels of clinicians. The improvement of accuracy is more apparent among less-experienced physicians.
Each year, around 3 million people are diagnosed with primary or metastatic brain tumors worldwide.1–3 Stereotactic radiosurgery (SRS), which delivers high doses of ionizing radiation in a single or few fractions to small targets, has become one of the validated treatments for brain tumors.4,5 To effectively eradicate tumors while minimizing damage to surrounding tissues, SRS requires delicate manual delineation of tumors. This tumor contouring process is extremely time-consuming and is prone to high inter- and intra-practitioner variability.6,7 Moreover, some challenging cases, such as tiny brain metastases, are hard to identify even for experienced physicians, leading to suboptimal treatment outcomes.
Driven by the ever-increasing capability of artificial intelligence (AI), a variety of computer-aided detection techniques have been proposed over the past decade for potential applications to clinical practice. Specifically, deep learning based on convolutional neural networks to automatically learn representative features from raw medical images has been used in different specialties,8 including radiology,9,10 dermatology,11 ophthalmology,12 pathology,13,14 and oncology.15,16 As neural networks can perform very well on segmentation tasks, they have been proposed to facilitate radiotherapy, especially for contouring of organs at risk17–21 and tumors.22–25 In particular, convolutional U-Net and DeepMedic architecture have been utilized for brain metastasis segmentation.26–32
While deep learning has shown promise in automated contouring of radiotherapy treatment planning, most previous studies focused on standalone performance testing of systems, reporting performance metrics using retrospective data. However, an accurate system alone will not necessarily assist physicians in radiotherapy treatment planning. A prospective study assessing the efficiency, accuracy, and reproducibility of such AI tools within the radiation therapy clinical workflow is warranted.33 To fully understand the potential clinical capability of AI, it is essential to integrate the system into clinical workflow and investigate its impact on practitioners. In this research, we present a thorough reader study where multiple medical professionals with different levels of experience and specialties perform target contouring on different types of brain tumors, using a state-of-the-art algorithm, automated brain tumor segmentation (ABS). We evaluate inter-reader variability, accuracy, and speed of physicians performing assisted contouring for brain tumor SRS.
Materials and Methods
Study Design
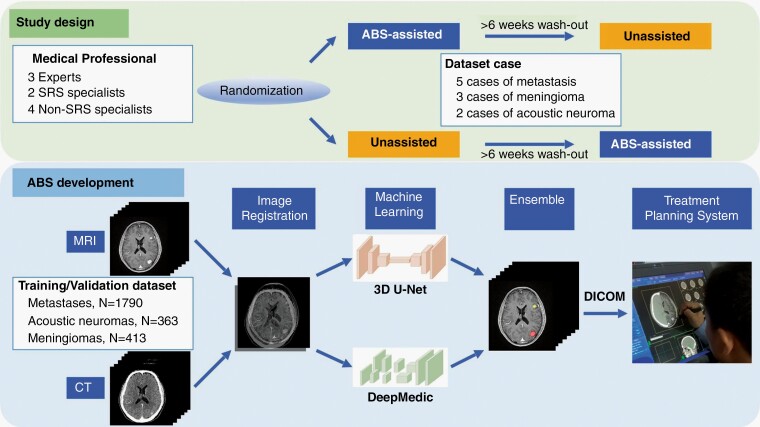
The study was designed as a cross-modal, multi-reader study as illustrated in Figure 1. All medical professionals contoured the selected dataset cases with or without auto-segmentation assistance. To reduce possible bias from the memory of previous contouring, the participants were randomized into one of the two segmentation modes: assisted-first or unassisted-first. The two contour sessions were separated by a washout period of at least 6 weeks along with ongoing full-time clinical practice. During the second session, the physicians were blinded to their first sets of segmentations and those undertaken by other participants. The readers were allowed to access patients’ histories and previous imaging studies, but no further information or radiology report about the simulation magnetic resonance (MR)/computed tomography (CT) images for SRS was provided. To simulate a contouring pace similar to normal clinical workflow, readers were not instructed to complete all cases in a single contouring session. Instead, they were asked to conclude all cases within 3 days. The segmentation time was recorded for each of the selected cases. The protocol of the prospective reader study was approved by the Institutional Research Ethics Committee.
Fig. 1.
Schematic diagrams of the reader study design and the machine learning model for automated brain tumor segmentation (ABS).
Machine Learning Models
The ABS system, extended from our previous collaboration work,26 in this study is a deep learning-based segmentor using multimodal imaging from MR and CT, and ensemble neural networks as illustrated in Figure 1. Specifically, we combined DeepMedic34 and three-dimensional (3D) U-Net35 architecture. The two neural networks were optimized with different objectives independently, such that the DeepMedic model focused on small lesions with a high sensitivity, whereas the 3D U-Net addressed overall tumor segmentation with a high specificity. By adopting the ensemble technique, the overall performance in both lesion segmentation and detection can be further improved and reach a balance between high sensitivity and high specificity.26,36 We preprocessed the images for training and validation and trained the neural networks in accordance with a previous work,26 but in a multitasking fashion where brain metastases, meningiomas, and acoustic neuromas were handled simultaneously. For each case, after performing rigid image registration between CT and MR images using mutual information optimization (mean registration error is on the order of submillimeter),37,38 we resampled the images to an isotropic resolution of 1 × 1 × 1 mm3. We extracted only the brain region, removed background, and then standardized the images with zero-mean and unit-variance normalization. Brain window (width: 80, level: 40) is applied to the CT images. The image preprocessing techniques ensure model performance and generalizability by standardizing the data acquired from different scanners and scanning protocols. The development dataset consisted of a cohort of 1288 patients (570 with brain metastases, 353 with acoustic neuromas, and 365 with meningiomas) with 2566 tumors (1790 brain metastases, 363 acoustic neuromas, and 413 meningiomas). The median tumor volume was 0.7 ml (range: 0.01-83.4 ml). The ground-truth tumor masks for ABS model development were the gross tumor volumes (GTVs) for clinical treatment, delineated by attending radiation oncologists or neurosurgeons on associated contrast-enhanced CT- and T1-weighted MR scans.
System Integration
A key factor in clinical adoption of machine learning is seamless integration of the model with existing hospital information systems. In this study, we integrated ABS with CyberKnife MultiPlan, routing corresponding MR and CT images, and GTVs with Digital Imaging and Communications in Medicine (DICOM) standards. The end-to-end inference time of ABS, including image preprocessing, neural network inference, and data post-processing, was approximately 90 seconds. Clinicians were then able to validate and modify tumor contours using the existing treatment planning system.
Study Dataset
A separate dataset consisting of five metastases cases (ranging from one to eight tumors per patient), three meningioma cases, and two acoustic neuroma cases were randomly selected for the prospective reader study. The composition of the dataset was compatible with that of the clinical practice at the institutional CyberKnife center, and the 10 selected cases were all independent of the development dataset for model training. The characteristics of the 10 cases in the reader study, as outlined in Table 1, included 16 metastases, 2 acoustic neuromas, and 5 meningiomas. The median tumor volume was 0.89 ml (range: 0.006-3.745 ml) for metastases, 9.62 ml (range: 1.23-18.02 ml) for acoustic neuromas, and 2.19 ml for meningiomas (range: 0.15-2.96 ml).
Table 1.
Dataset Case Characteristics
| Metastasis | Acoustic Neuroma | Meningioma | |
|---|---|---|---|
| Median age, years (range) | 67 (55-81) | 37 (32-42) | 58 (45-74) |
| Number of cases | 5 | 2 | 3 |
| Median tumor volume, ml | 0.89 | 9.62 | 2.19 |
| Number of tumors | 16 (8, 4, 2, 1, 1 tumor in each case) | 2 | 5 (3, 1, 1 tumor in each case) |
| Small tumor (volume <0.7 ml) | 7 | 0 | 2 |
| Large tumor (volume ≥0.7 ml) | 9 | 2 | 3 |
| Primary malignancy, number of cases | Lung 3 Breast 1 NPC 1 |
NA | NA |
Abbreviations: NA, not applicable; NPC, nasopharyngeal carcinoma.
Medical Professionals
A total of nine medical doctors, including one diagnostic neuro-radiologist, five radiation oncologists, and two neurosurgeons participated in the reader study. Among them, three physicians with over 14 years’ professional experience in neuroimaging or intracranial SRS were designated as the expert group. The expert group consisted of one diagnostic neuro-radiologist, one radiation oncologist, and one neurosurgeon. Among the other six physicians (nonexpert), the years of experience ranged from 2 to 20, with two chief resident physicians. Only two trained radiation oncologists had a specialization in intracranial SRS, with an average of 15 SRS cases per year (SRS specialists). The other four physicians did not often practice SRS (less than six SRS cases per year, designated as non-SRS specialists).
Expert Consensus for Ground-Truth Establishment
Two months after the completion of all contouring sessions, the ground truth of segmentation was established with consensus from the three experts. They had access to the patients’ histories and further follow-up imaging studies. The ground truth of contouring was determined slice-by-slice on the simulation MR and CT images. The GTVs for clinical treatment were not disclosed to the ABS system, the expert group, or other participants. Since the experts participated in the ground-truth determination, only the contours from nonexpert readers were compared with the ground truth for further analyses.
Statistical Analysis
Lesion-wise sensitivity was calculated for both contouring modes (detected tumors divided by all tumors contained in the ground truth). A metastasis was considered detected when the contours of the tumor overlapped with the corresponding ground-truth segmentation. The false-positive rate was also reported by the average number of false-positive lesions per patient. The degree of overlap with ground truth was then measured by the Dice similarity coefficient (DSC), defined as , where X is the predicted volume and Y is the corresponding ground-truth volume.
We used Wilcoxon paired-signed tests to compare the DSC, sensitivity of lesion detection, and time consumed between two contouring modes. The statistical significance was set at two-tailed P < 0.05. All analyses were performed using MedCalc Statistical Software version 19.1.3 (MedCalc Software, Ostend, Belgium).
Results
Ground-Truth Determination and AI Standalone Performance
For the 10 AI-unseen cases, the ABS system alone had a high overall sensitivity of 95.7% for lesion detection, with a sensitivity of 88.9% for small tumors (GTV < 0.7 ml, corresponding to 1.1 cm in diameter). The average false-positive rate was 0.5 lesions per patient. The median DSC was 0.836 (95% confidence interval [CI]: 0.783, 0.855), and the pixel-wise area under the receiver operating characteristic curve was 0.995.
Impact of Computer Assistance for the Experts
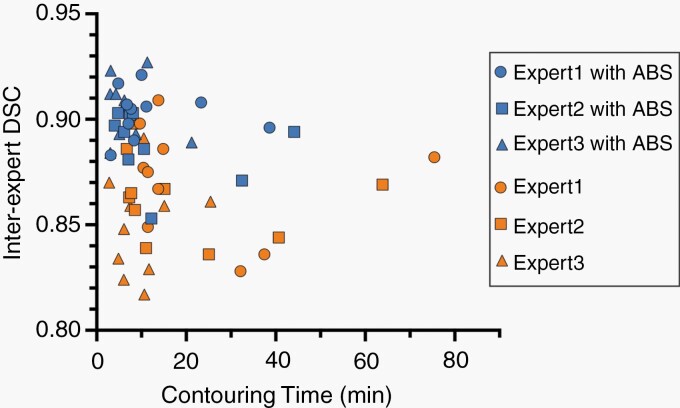
The inter-expert DSC was calculated to evaluate inter-reader agreement according to the degree of contours overlapping with the other two experts. With the aid of the ABS system, the agreement of tumor segmentation was significantly increased among the experts. The median inter-reader DSC increased from 0.862 (95% CI: 0.848, 0.870) to 0.900 (95% CI: 0.893, 0.906, P < 0.001) among the expert-level physicians. Supplementary Table 1 shows a reduction in inter-observer variability for all three experts. The contouring time per case was significantly shorter with assistance than without assistance (median time: 7.3 vs 11.2 minutes; P < 0.001). The median relative time-saving was 32.6% of working hours (95% CI: 17.7%, 46.6%). This time-saving benefit was consistent for every case contoured, with more pronounced effects found for metastases (6.1 vs 9.6 minutes; P < 0.001) and meningioma (7.1 vs 14.9 minutes; P = .004) than for acoustic neuroma (10.9 vs 11.5 minutes; P = .03). There was no difference in the median contouring time per case between the first and second sessions (8.6 vs 10.0 minutes; P = .52). As shown in Figure 2, the ABS-assisted group demonstrated a global trend of improvement on both inter-reader agreement and contouring time, indicating that the system could increase inter-expert agreement as well as efficiency of SRS treatment planning.
Fig. 2.
Scatterplot of inter-expert Dice similarity coefficient (DSC) against contouring time of individual dataset cases among experts. The inter-expert DSC was calculated using the degree of contours overlapping with other experts. The blue symbols depict the performance using automated brain tumor segmentation (ABS) assistance, whereas the orange symbols represent the performance of experts alone.
Impact of Computer Assistance for the Nonexperts
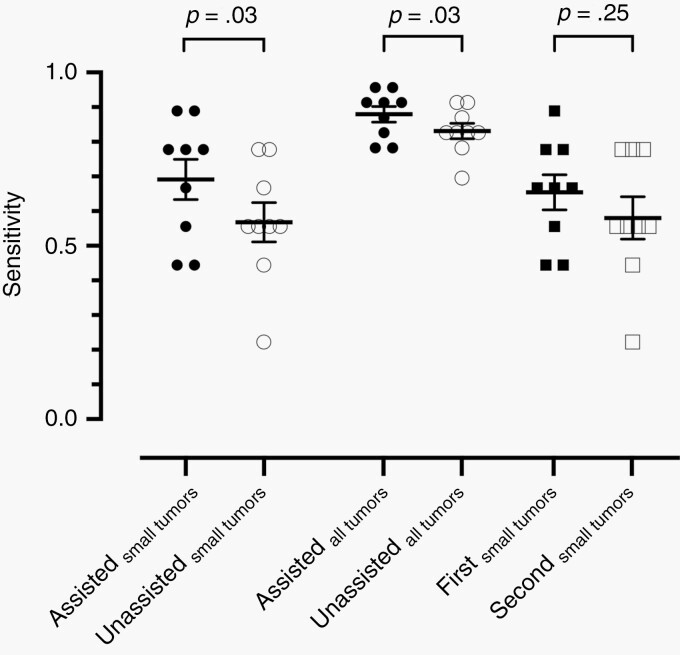
The contours from six nonexpert readers (two SRS specialists and four non-SRS specialists) were compared with the ground truth for further analyses. The overall sensitivity was significantly improved with AI assistance, with a median sensitivity of 91.3% (95% CI: 78.8%, 95.1%) for assisted contouring and 82.6% (95% CI: 78.9%, 90.7%) for unassisted reads (P = .03; Figure 3). Note that AI-assisted physicians exhibited a better lesion-detection rate for small tumors (69% vs 56.8%; P = .03). Nonetheless, there was no significant difference in detection sensitivity between the first and second sessions of contouring (66.7% vs 55.6%; P = 0.11).
Fig. 3.
Sensitivity of lesion detection with or without algorithm assistance among nonexperts for small tumors, all tumors, and order of contouring session. Error bars indicate standard error of means. While algorithm assistance significantly improved detection sensitivity, the order of contouring sessions did not affect the performance.
While physicians generally exhibit precision and a low false-positive rate during tumor identification, the algorithm would provide a higher lesion-wise sensitivity with the cost of a higher false-positive rate (0.5 lesions per case). Despite this, the false-positive lesions generated by ABS could be easily identified and deleted by the physicians, leading to a low average false-positive rate in both assisted and unassisted modes (median: 0 vs 0.1; P = 0.875). The false-positive lesions all came from the metastatic cases analyzed by less-experienced physicians.
In addition, algorithm assistance improved contouring accuracy. The average increase in DSC was 0.028, with a median DSC of 0.866 for unassisted mode, to 0.873 for assisted mode (P = 0.002).
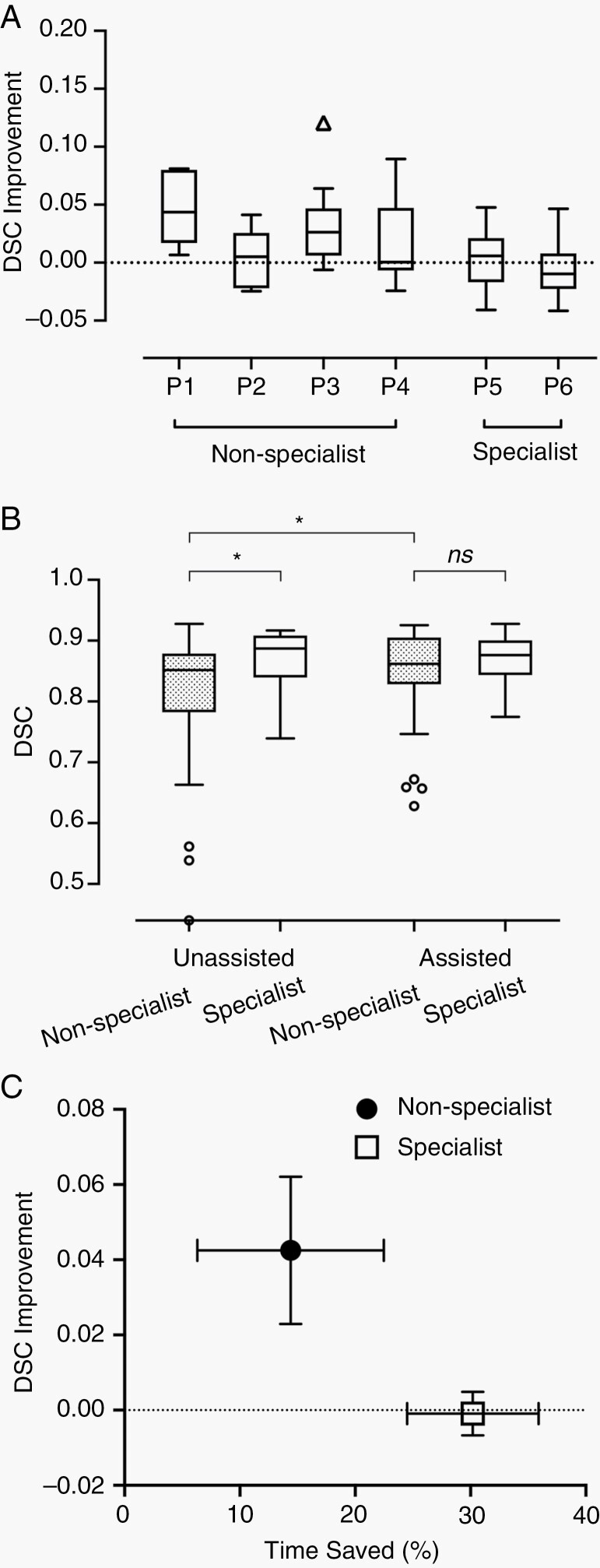
In particular, the improvement of DSC was more apparent among the four non-SRS specialists (median DSC of 0.847-0.865; P < 0.001). Figure 4A demonstrates the improvement in DSC with AI assistance for each of the six physicians. Non-SRS specialists (designated as P1-P4) gained greater refinement of contouring accuracy as compared to SRS specialists (P5 and P6) who regularly performed cranial SRS (Mann-Whitney test; P = 0.006). When unassisted, the SRS specialists had a significantly higher contouring accuracy than non-SRS specialists (median DSC 0.889 vs 0.847; P = 0.03), as shown in Figure 4B. Using ABS, both groups performed comparably well (0.876 vs 0.865; P = 0.54). The order of contouring mode (assisted-first vs unassisted-first) had no significant impact on contouring accuracy (median 0.868 vs 0.866; P = 0.36).
Fig. 4.
Improvement of Dice similarity coefficient (DSC) by experience level and algorithm assistance. (A) Tukey box plots of DSC improvement with algorithm assistance among six physicians. Compared with experienced stereotactic radiosurgery (SRS) specialists (P5 and P6), physicians with less SRS practice (P1-P4, non-SRS specialists) gained more DSC improvement upon assistance. (B) Tukey box plots of DSC between SRS specialists and non-SRS specialists with or without algorithm assistance. Less-experienced physicians demonstrated comparable contouring accuracy to those with more SRS experience upon algorithm assistance. Dotted boxes represent nonspecialists. (C) The mean DSC improvement and average time-saving using the algorithm among physicians of different experience levels. Nonspecialists gained more accuracy refinement but saved less time, while SRS specialists had less DSC improvement, but higher efficiency improvements. Error bars indicate standard error of means. * indicates a significant difference.
In accordance with the improved efficiency found for experts, the median contouring time per case was significantly shorter with ABS assistance than without assistance (6.7 minutes vs 7.6 minutes; P < 0.001). The median time-saving was 30.5% (95% CI: 17.8%, 39.9%). There was no significant difference in contouring time between the first and the second session (7.1 vs 7.3 minutes; P = 0.08). While the benefit of time-saving was universal for all the experts across all cases, the nonexpert group exhibited a more inhomogeneous effect on efficiency improvement (Supplementary Figure S1). Figure 4C shows a different pattern of DSC improvement and time-saving between SRS specialists and less-experienced physicians. The four non-SRS specialists (P1-P4) gained more improvement on contouring accuracy but less benefit in reduction of working hours. By contrast, the two SRS specialists (P5 and P6) had little gain in DSC, but high time-saving with the aid of the algorithm.
Discussion
Although recent studies have described the advancement of learning-based algorithms for brain tumor detection and segmentation, the potential impacts of the computer assistance tool in clinical practice have not yet been examined. In this cross-modal, multi-reader study, we demonstrated that ABS-assisted segmentation during brain tumor contouring resulted in a significant reduction of inter-reader variability and working hours for expert-level physicians. With the assistance of ABS, lesion-detection sensitivity and contouring accuracy were significantly improved for nonexpert-level clinicians. To our knowledge, the present work is the first to address the real benefits of AI assistance with multiple physicians at different levels of experience and in different specialties. Our work outlines a scheme for clinical adoption of a learning-based algorithm in brain tumor SRS practice.
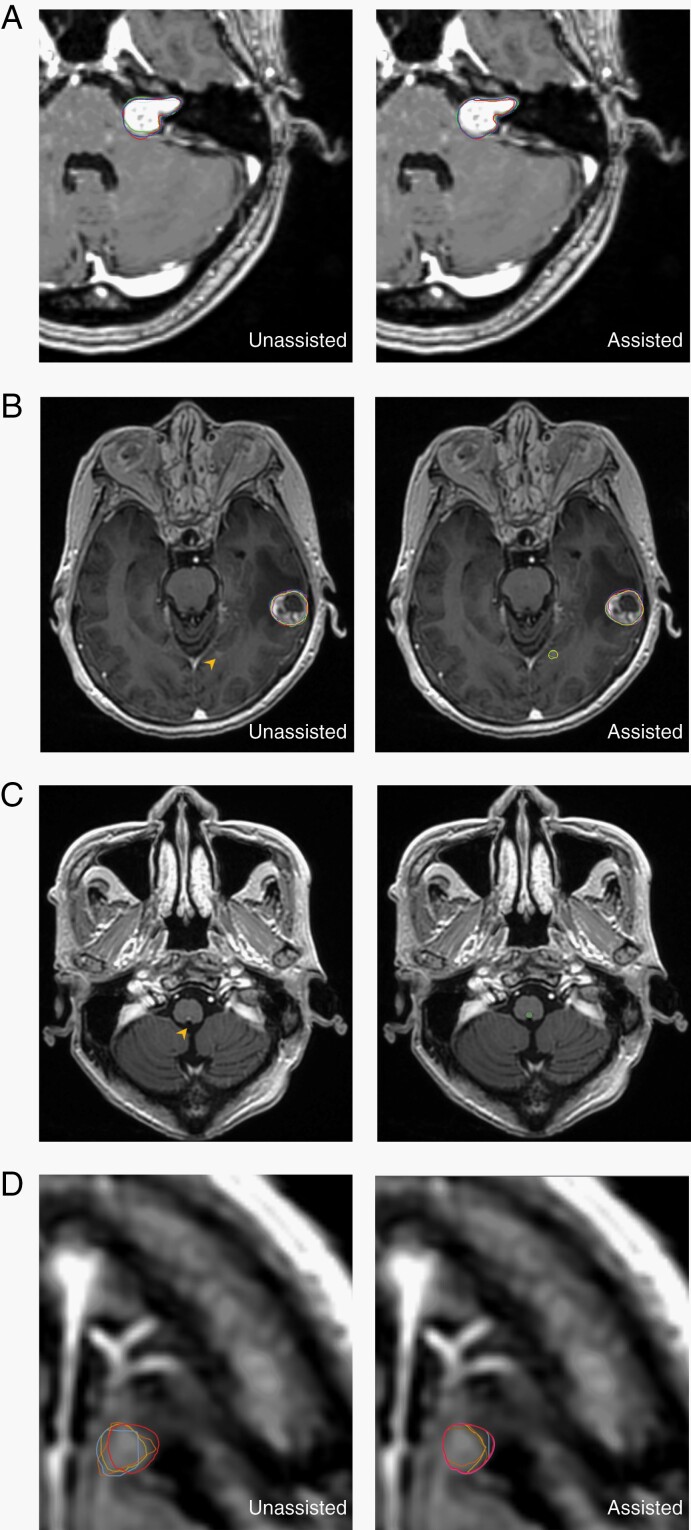
Variability of target delineation has been identified as the most important issue for the quality assurance of brain tumor SRS,6,39 but good strategies to address the issue are lacking. While previous studies found that inter-rater variability differs among different types of tumors,6,7 our results showed that ABS assistance significantly increased inter-reader agreement across brain metastases, acoustic neuromas, and meningiomas (Supplementary Table 1). One representative case of acoustic neuroma is shown in Figure 5A. In addition, the gain on inter-observer agreement was consistent for all experts. The present study demonstrates that collaborative intelligence could be a key advancement in high-quality brain SRS.
Fig. 5.
Representative case of increased inter-reader agreement, enhanced lesion detection, and improved contouring accuracy with algorithm assistance. (A) Stereotactic radiosurgery (SRS) experts showed higher inter-reader agreement for a left acoustic neuroma upon collaboration with the algorithm. Colored lines indicate each contour from three experts. (B) The small metastatic tumor (4 mm in diameter, arrowhead) was missed by all six nonexpert physicians when unassisted. With algorithm assistance, three of them contoured the tumor, while less-experienced physicians (non-SRS specialists) deleted the algorithm-generated segmentation. (C) False negative of a tiny metastasis. Left, the 2.2 mm tumor at the posterior aspect of brainstem was missed by the algorithm (arrowhead). Right, the ground truth is illustrated by green contours. All nonexpert physicians did not detect the lesion, either in assisted or unassisted mode. (D) Algorithm assistance improved the contouring accuracy of a metastatic nodule at left frontal lobe. The red line indicates ground truth, and the magenta one is the algorithm-generated segmentation in assisted mode. Other colored lines indicate each contour from non-SRS specialists.
Regarding lesion detection, the ABS assistance improved the sensitivity from 82.6% to 91.3%, with all improvement coming from the higher detection rate for small tumors. A major advantage of ABS is that the algorithm can provide steady detection, even when multiple metastases are present. In contrast, physicians are prone to neglecting tiny lesions when contouring a much more apparent tumor nearby. Figure 5B demonstrates a representative example of how clinicians would most benefit from incorporating ABS. During unassisted contouring, the attention of the six clinicians was drawn to the large tumor, and they all missed the small tumor (diameter 4 mm; Figure 5B, arrowhead). While three clinicians detected the lesion with the assistance of ABS, the other three less-experienced readers stuck to their opinions and deleted the ABS-generated segmentation of the lesion. This representative scenario contributed to the finding that the sensitivity of lesion detection of the algorithm alone (95%) was higher than that of the assisted mode (91.3%). Generally speaking, tiny tumors are more challenging for both ABS and physicians. For example, one brain metastasis at the posterior border of medulla (2.2 mm in diameter; Figure 5C) was not detected by the algorithm or physicians, both with and without computer assistance. The lesion was one of the three tiny lesions that required adjudications during ground-truth establishment.
The improvement of sensitivity with ABS assistance all derived from identifying additional small lesions, mostly metastases. The benefit in detection sensitivity improvement might be limited, if neuro-radiology report was available when performing contouring tasks. However, SRS usually demands fast segmentation for a rapid clinical workflow.27 With the increasing usage of SRS for multiple brain metastases, neurosurgeons or radiation oncologists might have to contour many brain lesions alone once simulation was done to facilitate the further treatment planning process. Meanwhile, the worldwide shortage of radiologist is a pressing issue, and the community of radiology also urges collaborative works trying to leverage AI to compensate for the lack of qualified radiologists.40
In the present study, ABS had a higher average false-positive rate than human physicians. Fortunately, false positives generally are not a major issue for physicians in the current clinical workflow, nor would ABS mislead physicians to create more false positives, regardless of experience level. With growing data supporting SRS alone, rather than whole-brain radiotherapy for multiple brain metastases,41 the shifting paradigm indicates rising demands for SRS to target numerous metastases. The present study demonstrates a promising utility of collaborative intelligence for future SRS workflow.
Algorithm assistance improved the contouring accuracy in terms of DSC. ABS-assisted clinicians demonstrated a higher delineation accuracy than ABS or the clinicians alone, especially for the less-experienced clinicians (Figure 5D). This implies that ABS and clinicians would complement each other and the collaboration between AI and medical professionals should lead to better and more reliable radiotherapy. The highly conformal radiation dose gradient surrounding the target implies that a small improvement of delineation may result in a greater potential clinical impact in terms of increased tumor control and reduced normal tissue toxicity. The present study demonstrates that the integration of a state-of-the-art algorithm into a clinical workflow for collaborative intelligence has the potential to improve patient care.
The most significant benefit of ABS assistance in the present study was related to efficiency, with a relative time-saving of 32.6% among experts and 30.5% for the nonexpert physicians. In the nonexpert group, the physicians who were experienced in SRS (P5 and P6) derived a similar efficiency improvement to the expert group. While experienced readers may feel confident refining the contours already generated by ABS, physicians with less experience may spend more time verifying the lesion and editing the segmentations. Notably, there was no difference in time spent between the first or the second session, either among the expert or nonexpert physicians.
There are several limitations of the present study. The ABS was developed using a database of upfront SRS for brain metastases, acoustic neuromas, and meningiomas. Other brain tumors or vascular lesions suitable for intracranial SRS, such as pituitary tumors, gliomas, and arteriovenous malformation, were not evaluated in the current study. Further work should explore the use of ABS to contour other brain tumor types. In addition, the present study was conducted in a single tertiary center. Besides, the patient volume of SRS specialists in the present study was relatively low. Although the physician volume is important for the performance in target contouring, there is no consensus or criteria on case load or frequency for defining a SRS specialist. Investigations with even more wide ranges of experiences in SRS shall be conducted in the future studies of AI-assisted segmentation. Given the improvements on tumor delineation reported here, understanding the actual impact on tumor control or radiation necrosis is another important consideration. Ultimately, further studies demonstrating the clinical value of collaborative intelligence in SRS are warranted for prospective clinical evaluation.
Conclusion
This work provides a cross-modal, multi-reader, multispecialty, multi-experience, multi-tumor type reader study on AI-assisted tumor contouring. The impact of assisted contouring is comprehensive, including improvements in accuracy and efficiency as well as reductions in contouring variability among clinicians. This study paves the first step toward understanding how deep learning neural networks can be best designed and utilized to facilitate treatment planning of radiotherapy, and how clinicians can best interact with such assistive tools to deliver better patient care. It is expected that physicians will be able to shift their focus from manual contouring to quality control of AI outputs, enabling them to concentrate more on high-quality patient care and counseling.
Supplementary Material
Acknowledgments
The authors acknowledge the contributions of Dr Haun-Chih Wang, MD, Dr I-Han Lee, MD, and Dr How-Chung Cheng, MD for their participation and advice to enable this work. Gratitude also goes to Dr Chia-Chun Wang, MD, PhD for data statistics recommendations. We also appreciate the assistance and support from Department of Medical Research, National Taiwan University Hospital, Taipei, Taiwan.
Conflict of interest statement. F.-R.X. and F.-M.H. received research grants from Vysioneer Inc., Cambridge, MA, USA.
Author contributions. Conceptualization: S.-L.L., F.-R.X., J.C.-H.C., W.-C.Y., J.-T.L., Y.-F.C., and F.-M.H. Data curation: S.-L.L., F.-R.X., Y.-H. Cheng, Y.-C.C., J.-Y.L., C.-H.L., and J.-T.L. Investigation: S.-L.L., F.-R.X., J.C.-H.C., W.-C.Y., J.-T.L., Y.-F.C., and F.-M.H. Methodology: S.-L.L., W.-C.Y., Y.-F.C., and F.-M.H. Formal analysis: S.-L.L., J.C.-H.C., Y.-H.C., Y.-C.C., J.-Y.L., C.-H.L., and J.-T.L. Project administration: S.-L.L., J.-T.L., Y.-F.C., and F.-M.H. Visualization: S.-L.L., J.C.-H.C., Y.-H.C., Y.-C.C., J.-Y.L., C.-H.L., J.-T.L., and F.-M.H. Resources: F.-R.X., J.C.-H.C., W.-C.Y., J.-T.L., Y.-F.C., and F.-M.H. Software: F.-R.X., Y.-H.C., Y.-C.C., J.-Y.L., C.-H.L., and J.-T.L. Writing—original draft: S.-L.L., W.-C.Y., and J.-T.L. Writing—review and editing: F.-R.X., J.C.-H.C., Y.-F.C., and F.-M.H.
References
- 1.Patel AP, Fisher JL, Nichols E, et al. . Global, regional, and national burden of brain and other CNS cancer, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019;18(4):376–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.American Cancer Society. Global Cancer Facts & Figures.2020. https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/global-cancer-facts-and-figures/global-cancer-facts-and-figures-4th-edition.pdf. Accessed September 25, 2020.
- 3.Achrol AS, Rennert RC, Anders C, et al. . Brain metastases. Nat Rev Dis Primers. 2019;5(1):1–26. [DOI] [PubMed] [Google Scholar]
- 4.National Comprehensive Cancer Network. Central Nervous System Cancers (Version 3.2020).https://www.nccn.org/professionals/physician_gls/pdf/CNS.pdf. Accessed September 25, 2020.
- 5.Lin X, DeAngelis LM. Treatment of brain metastases. J Clin Oncol. 2015;33(30):3475–3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Growcott S, Dembrey T, Patel R, Eaton D, Cameron A. Inter-observer variability in target volume delineations of benign and metastatic brain tumours for stereotactic radiosurgery: results of a national quality assurance programme. Clin Oncol (R Coll Radiol). 2020;32(1):13–25. [DOI] [PubMed] [Google Scholar]
- 7.Sandström H, Jokura H, Chung C, Toma-Dasu I. Multi-institutional study of the variability in target delineation for six targets commonly treated with radiosurgery. Acta Oncol. 2018;57(11):1515–1520. [DOI] [PubMed] [Google Scholar]
- 8.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–444. [DOI] [PubMed] [Google Scholar]
- 9.Pasa F, Golkov V, Pfeiffer F, Cremers D, Pfeiffer D. Efficient deep network architectures for fast chest X-ray tuberculosis screening and visualization. Sci Rep. 2019;9(1):6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng JZ, Ni D, Chou YH, et al. . Computer-aided diagnosis with deep learning architecture: applications to breast lesions in US images and pulmonary nodules in CT scans. Sci Rep. 2016;6:24454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esteva A, Kuprel B, Novoa RA, et al. . Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gulshan V, Peng L, Coram M, et al. . Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316(22):2402–2410. [DOI] [PubMed] [Google Scholar]
- 13.Nagpal K, Foote D, Liu Y, et al. . Development and validation of a deep learning algorithm for improving Gleason scoring of prostate cancer. NPJ Digit Med. 2019;2:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steiner DF, MacDonald R, Liu Y, et al. . Impact of deep learning assistance on the histopathologic review of lymph nodes for metastatic breast cancer. Am J Surg Pathol. 2018;42(12):1636–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu SH, Kim MS, Chung HS, et al. . Early experience with Watson for Oncology: a clinical decision-support system for prostate cancer treatment recommendations. World J Urol. 2021;39(2):407–413. [DOI] [PubMed] [Google Scholar]
- 16.McKinney SM, Sieniek M, Godbole V, et al. . International evaluation of an AI system for breast cancer screening. Nature. 2020;577(7788):89–94. [DOI] [PubMed] [Google Scholar]
- 17.Nikolov S, Blackwell S, Mendes R, et al. . Deep learning to achieve clinically applicable segmentation of head and neck anatomy for radiotherapy. arXiv preprint arXiv:1809.04430. 2018. [DOI] [PMC free article] [PubMed]
- 18.Fritscher K, Raudaschl P, Zaffino P, Spadea MF, Sharp GC, Schubert R.. Deep Neural Networks for Fast Segmentation of 3D Medical Images. Cham: Springer; 2016. doi: 10.1007/978-3-319-46723-8_19 [DOI] [Google Scholar]
- 19.Liu Z, Liu X, Xiao B, et al. . Segmentation of organs-at-risk in cervical cancer CT images with a convolutional neural network. Phys Med. 2020;69:184–191. [DOI] [PubMed] [Google Scholar]
- 20.Lustberg T, van Soest J, Gooding M, et al. . Clinical evaluation of atlas and deep learning based automatic contouring for lung cancer. Radiother Oncol. 2018;126(2):312–317. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Zhao L, Wang M, Song Z. Organ at risk segmentation in head and neck ct images using a two-stage segmentation framework based on 3D U-Net. IEEE Access. 2019;7:144591–144602. [Google Scholar]
- 22.Lin L, Dou Q, Jin YM, et al. . Deep learning for automated contouring of primary tumor volumes by MRI for nasopharyngeal Carcinoma. Radiology. 2019;291(3):677–686. [DOI] [PubMed] [Google Scholar]
- 23.Men K, Dai J, Li Y. Automatic segmentation of the clinical target volume and organs at risk in the planning CT for rectal cancer using deep dilated convolutional neural networks. Med Phys. 2017;44(12):6377–6389. [DOI] [PubMed] [Google Scholar]
- 24.Liang Y, Schott D, Zhang Y, et al. . Auto-segmentation of pancreatic tumor in multi-parametric MRI using deep convolutional neural networks. Radiother Oncol. 2020;145:193–200. [DOI] [PubMed] [Google Scholar]
- 25.Guo Z, Guo N, Gong K, Zhong S, Li Q. Gross tumor volume segmentation for head and neck cancer radiotherapy using deep dense multi-modality network. Phys Med Biol. 2019;64(20):205015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu S-Y, Weng W-H, Lu S-L, et al. . Multimodal volume-aware detection and segmentation for brain metastases radiosurgery. Paper presented at: Workshop on Artificial Intelligence in Radiation Therapy; October 17, 2019; Shenzhen, China.
- 27.Liu Y, Stojadinovic S, Hrycushko B, et al. . A deep convolutional neural network-based automatic delineation strategy for multiple brain metastases stereotactic radiosurgery. PLoS One. 2017;12(10):e0185844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grøvik E, Yi D, Iv M, Tong E, Rubin D, Zaharchuk G. Deep learning enables automatic detection and segmentation of brain metastases on multisequence MRI. J Magn Reson Imaging. 2020;51(1):175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xue J, Wang B, Ming Y, et al. . Deep learning-based detection and segmentation-assisted management of brain metastases. Neuro Oncol. 2020;22(4):505–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bousabarah K, Ruge M, Brand JS, et al. . Deep convolutional neural networks for automated segmentation of brain metastases trained on clinical data. Radiat Oncol. 2020;15(1):87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang G, Li W, Ourselin S, Vercauteren T. Automatic brain tumor segmentation based on cascaded convolutional neural networks with uncertainty estimation. Front Comput Neurosci. 2019;13:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Charron O, Lallement A, Jarnet D, Noblet V, Clavier JB, Meyer P. Automatic detection and segmentation of brain metastases on multimodal MR images with a deep convolutional neural network. Comput Biol Med. 2018;95:43–54. [DOI] [PubMed] [Google Scholar]
- 33.Huynh E, Hosny A, Guthier C, et al. . Artificial intelligence in radiation oncology. Nat Rev Clin Oncol. 2020;17(12):771–781. [DOI] [PubMed] [Google Scholar]
- 34.Kamnitsas K, Ledig C, Newcombe VFJ, et al. . Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med Image Anal. 2017;36:61–78. [DOI] [PubMed] [Google Scholar]
- 35.Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O. 3D U-Net: learning dense volumetric segmentation from sparse annotation. Paper presented at: International Conference on Medical Image Computing and Computer-Assisted Intervention; October 17–21, 2016; Athens, Greece.
- 36.Dietterich TG. Ensemble methods in machine learning. Paper presented at: International Workshop on Multiple Classifier Systems; June 21–23, 2000; Cagliari, Italy.
- 37.Marstal K, Berendsen F, Staring M, Klein S. SimpleElastix: a user-friendly, multi-lingual library for medical image registration. Paper presented at: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops; June 26–July 1, 2016; Las Vegas, NV, USA.
- 38.Wang X, Li L, Hu C, Qiu J, Xu Z, Feng Y. A comparative study of three CT and MRI registration algorithms in nasopharyngeal carcinoma. J Appl Clin Med Phys. 2009;10(2):3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stanley J, Dunscombe P, Lau H, et al. . The effect of contouring variability on dosimetric parameters for brain metastases treated with stereotactic radiosurgery. Int J Radiat Oncol Biol Phys. 2013;87(5):924–931. [DOI] [PubMed] [Google Scholar]
- 40.Allyn J. International Radiology Societies Tackle Radiologist Shortage.2020. https://www.rsna.org/news/2020/february/international-radiology-societies-and-shortage. Accessed February 14, 2021.
- 41.Kann BH, Park HS, Johnson SB, Chiang VL, Yu JB. Radiosurgery for brain metastases: changing practice patterns and disparities in the United States. J Natl Compr Canc Netw. 2017;15(12):1494–1502. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.