Abstract
Homeobox genes generally encode transcription factors involved in regulating developmental processes. In the pineal gland, a brain structure devoted to nocturnal melatonin synthesis, a number of homeobox genes are also expressed postnatally; among these is the LIM homeobox 4 gene (Lhx4). We here report that Lhx4 is specifically expressed in the postnatal pineal gland of rats and humans. Circadian analyses revealed a four-fold rhythm in Lhx4 expression in the rat pineal gland, with rhythmic expression detectable from postnatal day 10. Pineal Lhx4 expression was confirmed to be positively driven by adrenergic signaling, as evidenced by in vivo modulation of Lhx4 expression by pharmacological (isoprenaline injection) and surgical (superior cervical ganglionectomy) interventions. In cultured pinealocytes, Lhx4 expression was upregulated by cyclic AMP, a second messenger of norepinephrine. By use of RNAscope technology, Lhx4 transcripts were found to be exclusively localized in melatonin-synthesizing pinealocytes. This prompted us to investigate the possible role of Lhx4 in regulation of melatonin-producing enzymes. By use of siRNA technology, we knocked down Lhx4 by 95% in cultured pinealocytes; this caused a reduction in transcripts encoding the melatonin-producing enzyme arylalkylamine N-acetyl transferase (Aanat). Screening the transcriptome of siRNA-treated pinealocytes by RNAseq revealed a significant impact of Lhx4 on the phototransduction pathway and on transcripts involved in development of the nervous system and photoreceptors. These data suggest that rhythmic expression of Lhx4 in the pineal gland is controlled via an adrenergic-cyclic AMP mechanism and that Lhx4 acts to promote nocturnal melatonin synthesis.
Keywords: homeobox, melatonin, pineal gland, RNAscope, siRNA, RNA sequencing
Introduction
Members of the homeobox gene family encode transcription factors that regulate developmental processes, cellular differentiation and morphogenesis in all metazoans(Slack, Holland, & Graham, 1993). A specific set of homeobox genes are known to regulate development of the pineal gland, a neuroendocrine organ producing the nighttime hormone melatonin (Rath, Rohde, Klein, & Møller, 2013). Among these, we have previously shown that the LIM homeobox 9 gene (Lhx9) is essential for pineal gland morphogenesis (Yamazaki et al., 2015). However, a number of homeobox genes are expressed continuously or even restricted to postnatal stages (Rath et al., 2013). Large-scale analyses of the rat pineal gland transcriptome have identified another LIM homeobox gene, Lhx4, as being expressed in the adult gland (Bailey et al., 2009; Mays et al., 2018). In the mouse embryo, Lhx4 is expressed in ventral areas along the neural tube, where it directs cell-type specification in the pituitary (Sheng et al., 1997), but expression in this location ceases before birth (Li et al., 1994).
Recent analyses of Lhx4 in the pineal gland of adult rats show a daily rhythm with increased transcript levels during nighttime (Hartley et al., 2015; Yamazaki et al., 2015). The pineal gland receives neural input from the endogenous circadian clock located in the suprachiasmatic nucleus of the hypothalamus via a neural pathway that includes the superior cervical ganglia (SCG), which innervate the pineal gland (Klein et al., 2010; Møller & Baeres, 2002). During nighttime, the projections from the SCG release norepinephrine (NE); this causes an increase in intracellular cyclic AMP, which in turn induces transcription and posttranslational activation of the penultimate enzyme in melatonin synthesis arylalkylamine N-acetyltransferase (Aanat)(Klein, 2007). Large scale screening-analyses of the pineal transcriptome suggest that this mechanism guides pineal Lhx4 expression7, but the role of Lhx4 in the mature pineal gland is unknown. The current study was undertaken to clarify the mechanisms driving rhythmic Lhx4 expression in the pineal gland and the possible regulatory impact of Lhx4 on gene transcripts involved in melatonin synthesis and other biological processes.
Materials and methods
Animals
Sprague-Dawley rats were obtained from Charles River (Sulzfeld, Germany) or Janvier (Saint Berthevin Cedex, France) as either adult males (150–250g) or timed-pregnant females. Animals were housed under a 12L:12D light:dark (LD) schedule and sacrificed by CO2anesthesia followed by decapitation. For circadian experiments, rats were kept for two days in constant darkness (DD). Decentralization of the superior cervical ganglia (SCGdcn) and removal of the superior cervical ganglia (SCGx) were performed as previously described (Møller, Phansuwan-Pujito, Morgan, & Badiu, 1997; Savastano et al., 2010); animals were sacrificed 10 days post-surgery. For in vivo adrenergic stimulation experiments, rats were injected (ip) with isoprenaline (10mg/kg; Tocris Bioscience, Bristol, UK) or PBS at ZT5 and sacrificed at ZT8. For developmental experiments, heads (embryonic day 15 (E15) to postnatal day 2 (P2)) or brains (P10-P30) were removed and immersion-fixed in cold 4% paraformaldehyde, cryoprotected in 25% sucrose and frozen on crushed dry ice. E0 is defined as the plug date and P0 as the day of birth. For analyses of adult animals, whole brains or dissected tissues were removed and frozen on dry ice. For each preparation of primary pinealocyte cell cultures, 12 to 22 pineal glands were collected at Zeitgeber time (ZT) 4. All experiments were performed in accordance with the guidelines of EU Directive 2010/63/EU. Specific procedures were approved by the Danish Council for Animal Experiments (2017-15-0201-01190) and the University of Copenhagen (P17–311).
Human tissue
Sections of two human male pineal glands (no. 233: age 54, time of death 4:25AM [cause of death: heart failure], and no. 249: age 22, time of death 11:10 PM [cause of death: suicide]) were obtained from the Human Brain Tissue Bank of Semmelweis University (Budapest, Hungary). The human brain microdissection procedures were approved by the Regional and Institutional Committee of Science and Research Ethics of Scientific Council of Health (34/2002/TUKEB-13716/2016/EHR, Budapest, Hungary) and the Code of Ethics of the World Medical Association (Declaration of Helsinki).
Radiochemical in situ hybridization
Cryostat brain sections, 12 μm (adult rats) or 14 μm (rat developmental series) or 18 μm (human), were hybridized with a [35S]dATP-labelled DNA antisense probe corresponding to position 668-633 on rat Lhx4 mRNA (NM_001108348.1), 5’-CTTTGGGGAGTTCTTGTATGCATTCTTTAGTGTCTC-3’ or position 221-186 on human LHX4 mRNA (NM_033343.4), 5’-TCTCTCGCTCTAGCTCGATTCAGAAATTTCAAAATA-3’ as previously described(Klitten et al., 2008). X-ray images were digitalized and signals quantified using Scion Image Beta 4.0.2 (Scion, Frederick, MD, USA), comparing the optical density with a C14 standard.
RNAscope in situ hybridization
Cryostat sections of the brain and retina (12 μm) were pretreated according to the manufacturer’s protocol for fresh frozen sections (ACDBio, Newark, CA; #320513). Sections were subjected to RNAscope (Wang et al., 2012) following the manufacturer’s protocol (ACDBio; #320293). The RNAscope Fluorescent Multiplex Reagent Kit (ACDBio; #320850) was used with probe sets detecting Lhx4 (positions 23–1176 on NM_001108348.1) and Aanat (positions 161–1506 on NM_012818.2), respectively.
Pinealocyte cell culture and siRNA transfection
Pinealocyte primary cultures were prepared as described (Rohde, Rovsing, Ho, Møller, & Rath, 2014) with 105cells per sample. Pinealocytes were stimulated with isoprenaline (10 μM; Tocris Bioscience) or dibutyryl cyclic AMP (DBcAMP) (500μM; Sigma-Aldrich, Steinheim, Germany) for the indicated duration of time prior to being harvested. Pretreatment with 30μg/ml Actinomycin D (Sigma Aldrich) was done for one hour. For knocking down Lhx4, Stealth siRNA targeting position 1221–1245 on Lhx4 mRNA (siLhx4) (Life Technologies, Nærum, Denmark; #RSS323070) was transfected into pinealocytes with a final siRNA concentration of 40 nM using Lipofectamine RNAiMAX (Life Technologies); non-targeting siRNA (siNT) (#12935300) was used as negative control (Rohde, Hertz, & Rath, 2019). The medium was changed 24h after initiation of transfection. On the next day, pinealocytes were treated with DBcAMP or isoprenaline and/or actinomycin D prior to being harvested.
RT-qPCR
RNA was extracted using TriZOL (Life Technologies). 250 ng RNA from cultures or 500 ng RNA from tissue samples were DNAseI-treated and used for cDNA synthesis with Superscript III (Life Technologies). Real-time PCR was performed using a Lightcycler 96 (Roche, Hvidovre, Denmark) with 10 μl reactions containing 0.5 μM primers (Table 1), Faststart Essential DNA Green Master (Roche) and 0.2 μl cDNA. The following program was used: 95°C for 10 min; 40 cycles of 95°C for 10s, 63°C for 10s, 72°C for 15s. Standard curves were generated by 10-fold serial dilutions of pUC57 plasmids (Genscript, Piscataway, NJ) containing the PCR target sequence. Transcripts of interest were normalized to expression of Gapdh and Actb.
Table 1.
RT-qPCR primer sequences
| Transcript | NCBI reference sequence | Position | Forward primer (5’-3’) | Reverse primer (5’-3’) |
|---|---|---|---|---|
| Gapdh | NM_017008.4 | 77–386 | TGGTGAAGGTCGGTGTGAACGGAT | TCCATGGTGGTGAAGACGCCAGTA |
| Actb | NM_031144.3 | 26–184 | ACAACCTTCTTGCAGCTCCTC | CCACGATGGAGGGGAAGAC |
| Lhx4 | NM_001108348.1 | 52–223 | TGATACTTGGCGTGCCCCCAC | CAACCGGCGCACTGGGGAAT |
| Aanat | NM_012818.2 | 525–682 | TGCTGTGGCGATACCTTCACCA | CAGCTCAGTGAAGGTGAGAGAT |
| Asmt | NM_144759.2 | 696–860 | GCAAGACCCAGTGTGAGGTT | CAGTAGTGCACCACCTGGC |
| Tph1 | NM_001100634.2 | 702–814 | CAGAAACCTTCCTCTGCTCTCA | CAGGACGGATGGAAAACCCT |
Statistical analyses of radiochemical in situ hybridization and qPCR data
Statistical analyses were performed in Prism 7.00 (GraphPad, La Jolla, CA) by one- or two-way ANOVA followed by Bonferroni-corrected Fisher’s LSD multiple-comparison post hoc tests or by unpaired Welch’s t-test. A p-value of 0.05 was considered significant unless stated otherwise. Number of replicates (n) are given in figure legends.
RNA sequencing
RNA was extracted using TriZOL (Life Technologies) and purified with a Qiagen RNeasy® Micro Kit including DNAse I treatment (Qiagen, Frederick, MD, USA). RNA quality was determined on a Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA) with RIN-values ranging from 7.7–8.9. cDNA library preparation with ribosomal RNA depletion was performed with the SMARTer® Stranded Total RNA-Seq Pico Input Mammalian Kit (Takara Bio, Mountain View, CA, USA) using 3.4 ng total RNA. Nine barcoded libraries were pooled and sequenced on one FlowCell of a HiSeq2500 (Illumina, San Diego, CA, USA) in Rapid Run Mode yielding a total of 410 million 2 ∙ 100-bp read-pairs (average reads per sample: 45.6 ∙ 106± 3.3 ∙ 106 SEM).
The RNAseq data was aligned with RNA STAR (2.5.4a)(Dobin et al., 2013) against RefSeq rn6. The mean alignment rate was 94% with 66% uniquely assigned to the transcriptome. Read quantitation was performed using FeatureCounts of the Subread package (1.5.2)(Liao, Smyth, & Shi, 2014) with Refseq rn6 gene definitions as the template. Of the total 28,341 genes in the database, 26,553 had at least 1 read mapped to them. Differential expression analysis was performed using DESeq2 (v.1.16.1)(Love, Huber, & Anders, 2014) running Wald tests (Anders & Huber, 2010). To identify genes with a greatly altered expression for a DAVID analysis, the following parameters were used: a cutoff on Benjamini-Hochberg adjusted p-value of ≤0.001, percent change of expression of ≥+/−20% and mean read-pair count of ≥100. These genes were run through DAVID software (Huang da, Sherman, & Lempicki, 2009a, 2009b). From the output, the gene ontology readout BP_5 was examined along with the KEGG_PATHWAY readout with a significance cutoff of p<0.01 (Bonferroni-corrected) applied.
Results
Lhx4 is expressed in melatonin-producing pinealocytes of the postnatal pineal gland
To investigate tissue distribution of Lhx4 expression, transcript levels were analyzed by RT-qPCR in 14 tissues at ZT6 and ZT18 (Fig. 1A). Of the 14 investigated tissues, Lhx4 was found to be specifically expressed at high levels in the pineal gland and at lower levels in the retina (<1:100). Day-night differences in expression were detected in both the pineal gland (p<0.01) and the retina (p<0.05). Specific expression of Lhx4 in the pineal gland was confirmed by radiochemical in situ hybridization of coronal and sagittal brain sections (Fig. 1B). LHX4 expression signal was also detectable in the parenchyma of the human pineal gland (Fig. 1C).
Figure 1. Lhx4 is expressed in pinealocytes of the adult rat and human pineal gland.
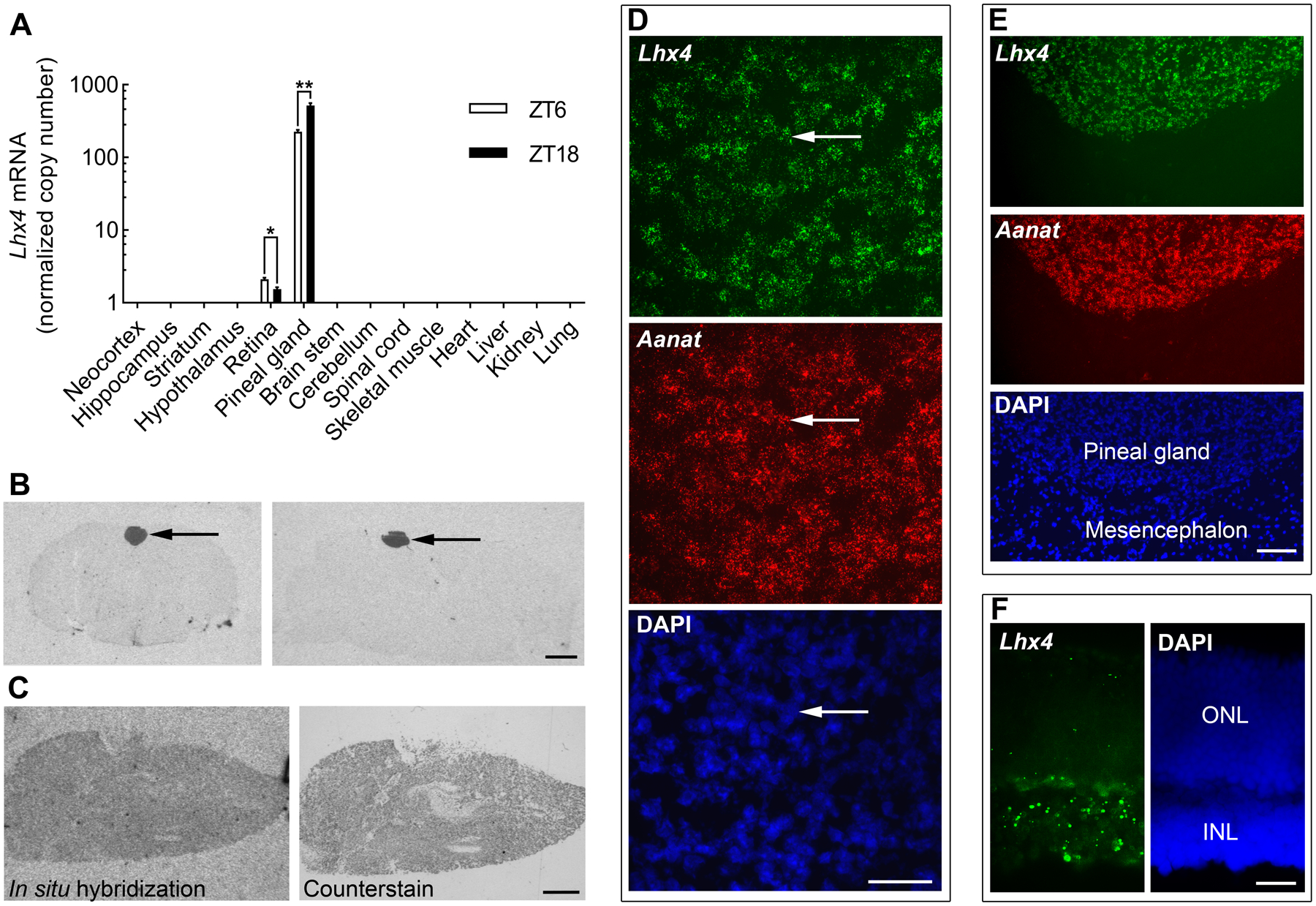
(A)Lhx4 mRNA levels in rat tissues obtained at midday (ZT6) or midnight (ZT18) as determined by RT-qPCR analysis. Lhx4 transcript quantity was normalized to a normalization factor based on the geometric mean of Gapdh and Actb expression. Gapdh and Actb were detected in all samples. Bars represent the mean ± SEM. n = 3–6. Data were analyzed by Bonferroni-corrected unpaired two-tailed Welch’s t-tests. *, p<0.05; **, p<0.01. (B) Representative autoradiographs generated by radiochemical in situ hybridization for detection of Lhx4 mRNA in coronal (left) and sagittal (right) brain sections from adult rats sacrificed at ZT18. Arrows point to the pineal gland. Scale bar, 2 mm. (C) Autoradiograph of radiochemical in situ hybridization for detection of LHX4 in the human pineal gland (left). Following exposure, the hybridized section was counterstained in cresyl violet (right). Sections shown are from no. 233, Scale bar, 1 mm. (D) RNAscope in situ hybridization for detection of Lhx4 mRNA and Aanat mRNA in coronal sections of the pineal gland from rats sacrificed at ZT18. Lower panel shows a DAPI stain for comparison. High magnification fluorescence micrograph image showing expression in individual pinealocytes. Arrows indicate the same cell photographed with different filter settings. Scale bar, 50 μm. (E) RNAscope in situ hybridization for detection of Lhx4 mRNA and Aanat mRNA in coronal sections of the pineal gland from rats sacrificed at ZT18. Low magnification image with the pineal gland and adjacent parts of the mesencephalic tectum. Scale bar, 100 μm. (F) RNAscope in situ hybridization for detection of Lhx4 mRNA in the retina of rats sacrificed at ZT18. A DAPI stain is shown for identification of retinal layers. ONL, outer nuclear layer; INL, inner nuclear layer. Scale bar, 20 μm.
To obtain cellular resolution of Lhx4 transcript distribution, RNAscope in situ hybridization was performed on rat brain sections (Fig. 1D,E). This analysis revealed specific expression of Lhx4 in cells of the pineal gland; Lhx4 transcripts exhibited full cellular colocalization with Aanat transcripts, thus showing that Lhx4 is specifically expressed in melatonin-producing pinealocytes of the pineal gland. RNAscope in situ hybridization on sections of the retina revealed expression of Lhx4 in the inner nuclear layer and in the outer nuclear layer, whereas the transcript was not detectable in other retinal layers (Fig. 1F).
Prompted by the developmental role of Lhx4 in other neuroendocrine structures (Sheng et al., 1997), we investigated the ontogenetic expression pattern of Lhx4 in the rat pineal gland using radiochemical in situ hybridization (Fig. 2). From E15 to E19, Lhx4 transcripts were not detectable in the developing pineal gland or any other adjacent brain areas, but from E21 expression was seen specifically in the pineal gland. Rhythmic expression of Lhx4 in the pineal was detectable from P10 and onwards (Fig. 3). These data support previous findings (Yamazaki et al., 2015) and suggest that pineal Lhx4 exerts its function at postnatal stages only.
Figure 2. Expression of Lhx4 in the developing rat pineal gland.
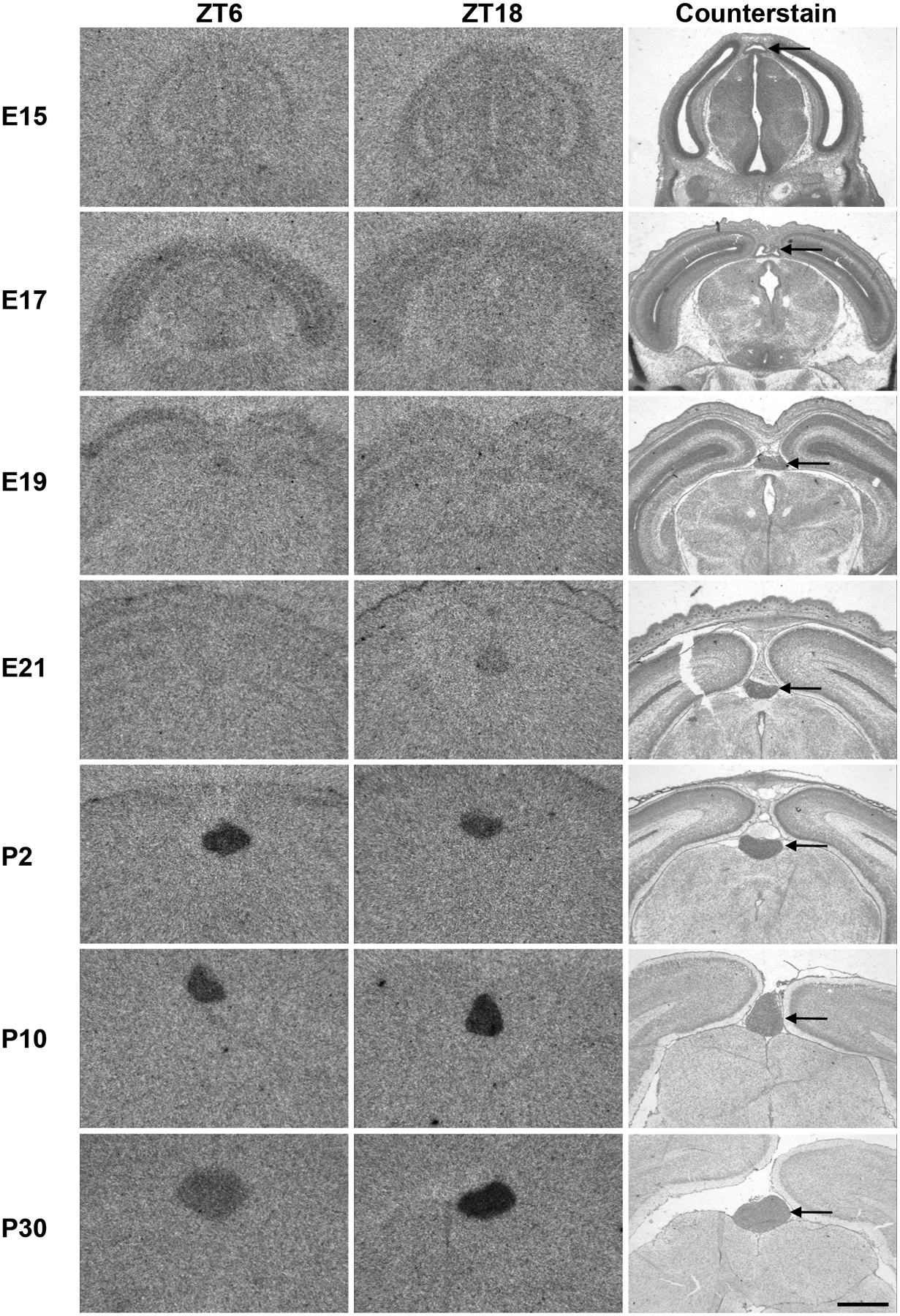
Radiochemical in situ hybridization for detection of Lhx4 mRNA in coronal sections of the brain from rats sacrificed at ZT6 (left) and ZT18 (middle) at the indicated developmental stages (one per row) ranging from E15 to P30. ZT18 sections were counterstained in cresyl violet for comparison (right). Arrow points to the pineal gland. Scale bar, 1 mm; E, embryonal day; P, postnatal day.
Figure 3. Quantification of Lhx4 expression in the developing rat pineal gland.
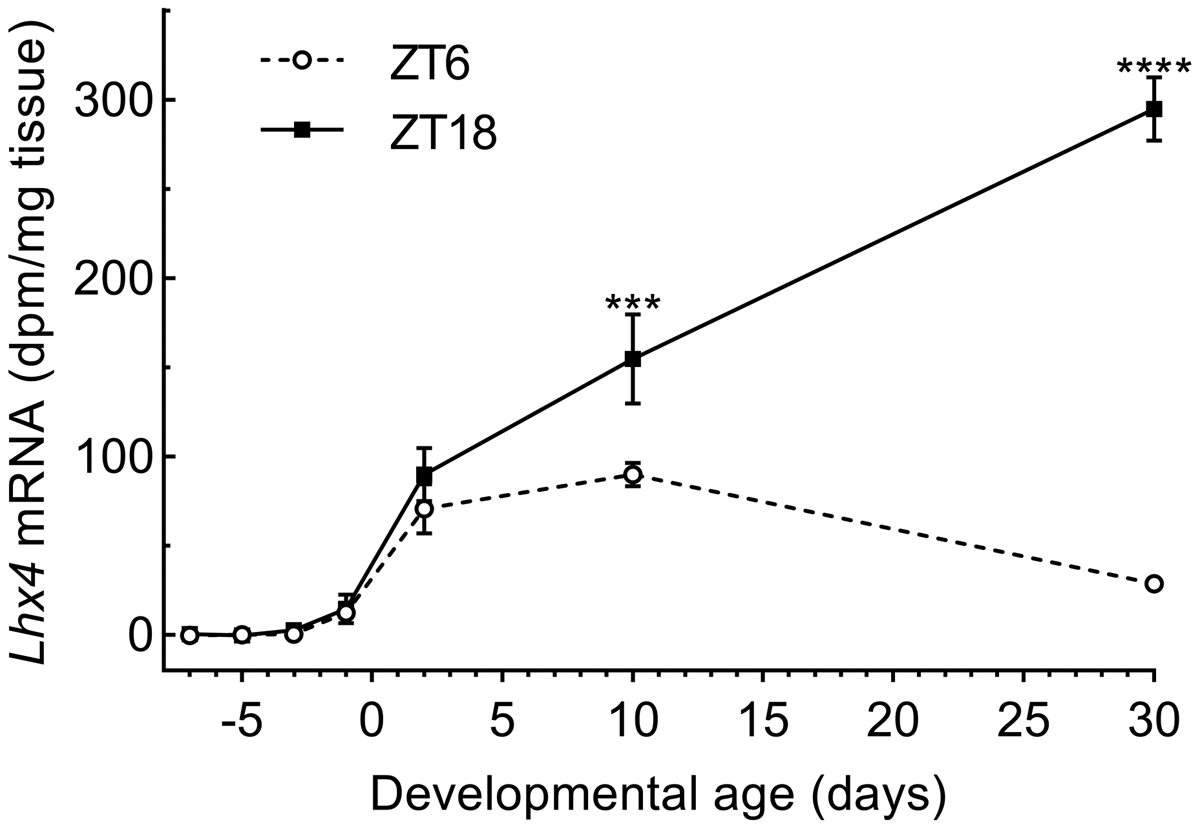
Densitometric quantification of the Lhx4 transcript hybridization signals shown in Figure 2. White circles indicate ZT6 and squares indicate ZT18. Data represent the mean ± SEM. n = 4. E22 is designated day 0 on the x-axis and the data points span from E15 to P30. Two-way ANOVA showed significant changes in expression both during development (effect of age) (p<0.0001) and between ZT6 and ZT18 (effect of time of day)(p<0.0001). Bonferroni-corrected Fisher’s LSD multiple comparisons post hoc tests showed a significant daily rhythm starting from P10: ***, p<0.001; ****, p<0.0001.
Circadian Lhx4 expression in the adult pineal gland is controlled by adrenergic signaling via the superior cervical ganglia
To map daily expression of Lhx4 in the adult pineal gland, Lhx4 transcript levels were analyzed by use of radiochemical in situ hybridization in rats kept in LD 12h:12h (Fig. 4A) and DD (Fig. 4B). Lhx4 exhibited a four-fold diurnal rhythm (p<0.0001) with high levels at midnight (ZT18) (Fig. 4C) in accord with previous observations (Hartley et al., 2015; Yamazaki et al., 2015); this rhythmic pattern was unaltered in constant darkness (p<0.01) suggesting that pineal expression of Lhx4 is regulated by the internal clock of the brain.
Figure 4. Lhx4 expression exhibits a circadian rhythm in the adult rat pineal gland.
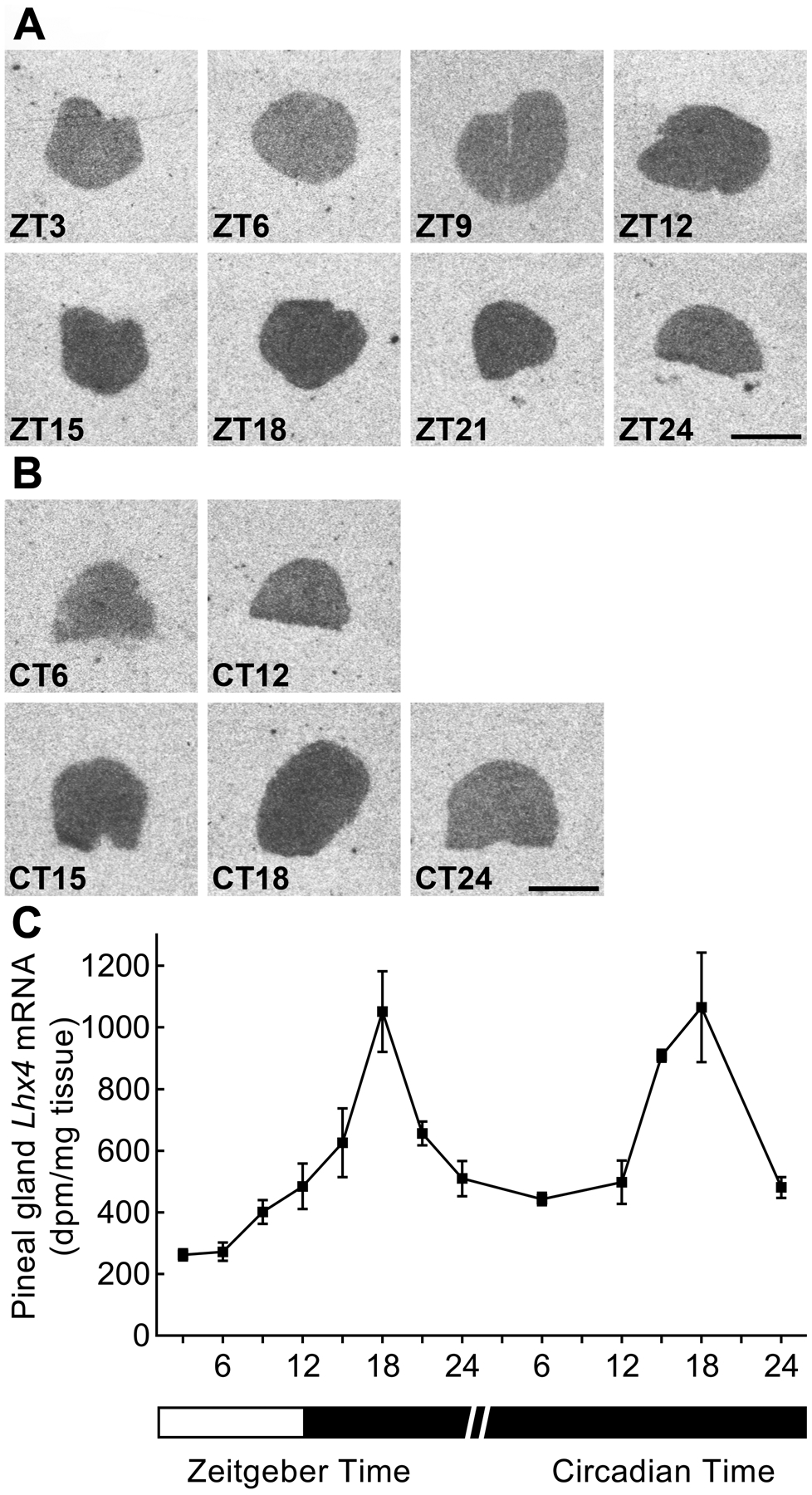
(A) Radiochemical in situ hybridization for detection of Lhx4 mRNA in coronal sections of the pineal gland from rats kept under an 12h:12h LD schedule. Scale bar, 1 mm. (B) Radiochemical in situ hybridization for detection of Lhx4 mRNA in coronal sections of the pineal gland from rats kept in DD. Scale bar, 1 mm; CT, circadian time. (C) Densitometric quantification of the Lhx4 transcript hybridization signals. The white area of the horizontal bar indicates light conditions, whereas black areas indicate dark conditions. Values represent the mean ± SEM. n = 3–4. One-way ANOVA showed a diurnal rhythm in LD (p<0.0001) and a circadian rhythm in constant darkness (p<0.01).
To examine if neural fibers from the SCG control rhythmic Lhx4 expression in the pineal gland, brain sections from rats having undergone ganglionectomy (SCGx) or decentralization (SCGdcn) were subjected to radiochemical in situ hybridization (Fig. 5A). Quantitative analyses of pineal transcript levels (Fig. 5B) revealed a significant difference in Lhx4 mRNA expression in control rats between ZT6 and ZT18 (p<0.0001) with highest Lhx4 mRNA levels at ZT18. The Lhx4 transcript level did not vary between the groups (p-values >0.05) except between the ZT18 control rats and the SCGdcn/SCGx ZT18 rats (p-values <0.0001), thus showing that SCGx and SCGdcn eliminate the rhythm in pineal Lhx4 expression and that the input to and from the SCG is required for the daily rhythm in expression. To investigate if pineal expression of Lhx4 is controlled by adrenergic signaling, rats were injected with the β-adrenergic agonist isoprenaline during daytime (Fig. 5C). Quantitative analyses of radiochemical in situ hybridization results revealed a significant increase in Lhx4 expression in the pineal gland following isoprenaline injection (Fig. 5D; p<0.05), suggesting that nocturnal pineal Lhx4 expression is induced by adrenergic signaling.
Figure 5. Rhythmic expression of Lhx4 is controlled by sympathetic input to the pineal gland.
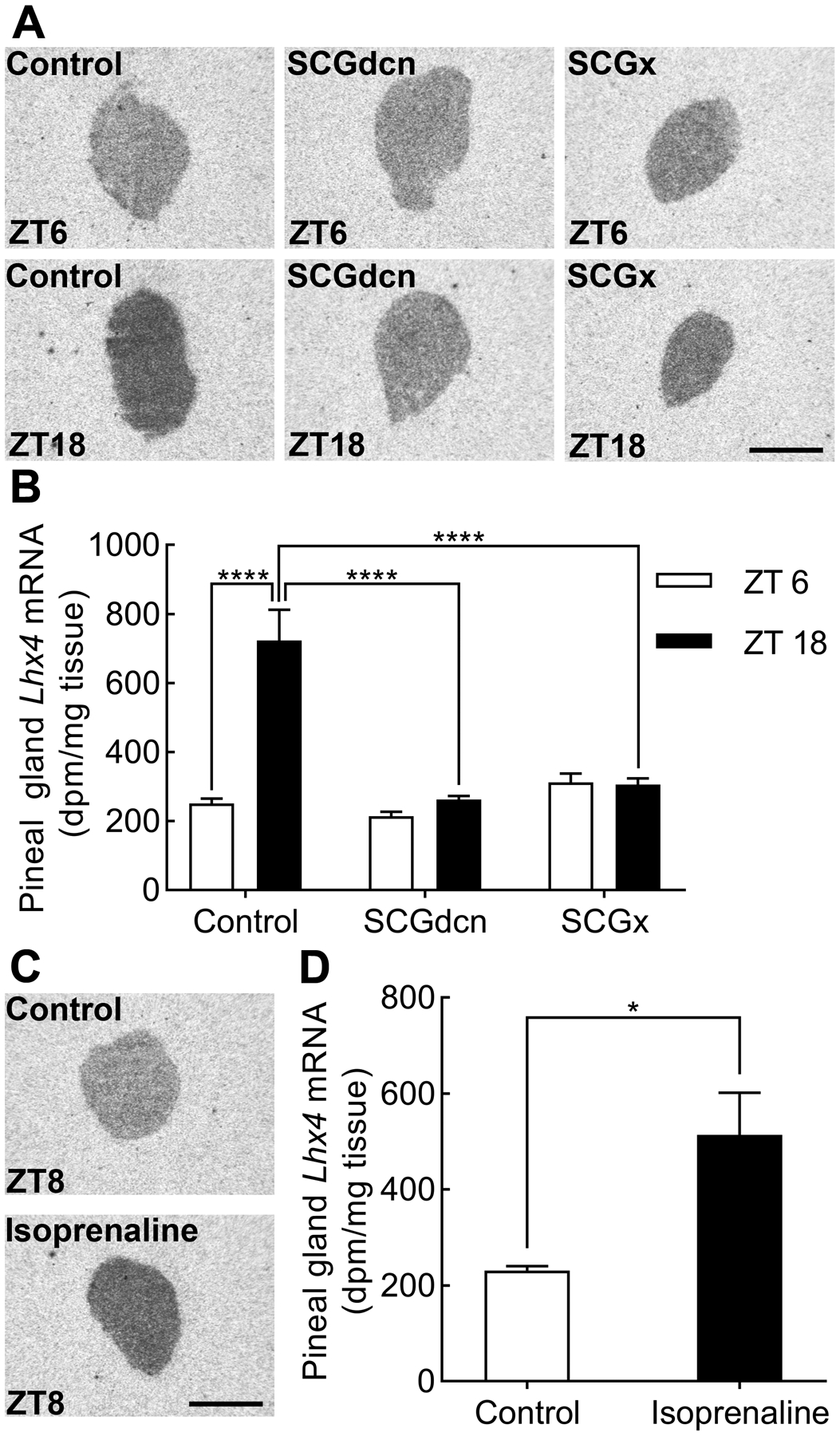
(A) Radiochemical in situ hybridization for detection of Lhx4 in sagittal sections through the pineal gland of SCGdcn, SCGx or control rats sacrificed at ZT6 and ZT18. Scale bar, 1 mm. (B) Densitometric quantification of Lhx4 expression in the pineal gland of SCGdcn, SCGx and control rats. Bars represent the mean ± SEM. n = 3–4. Two-way ANOVA showed significant effects of both surgical procedures (p<0.0001) and time of day (p<0.0001). Bonferroni-corrected Fisher’s LSD multiple-comparison tests showed a significant rhythm in the control rats, no rhythms in SCGdcn and SCGx rats (p-values>>0.05), and reduced Lhx4 expression levels in the surgically modified rats: ****, p<0.0001. (C) Representative autoradiographs from sagittal brain sections through the pineal gland of rats injected with PBS (control) or isoprenaline at ZT5 and sacrificed at ZT8. Scale bar, 1 mm. (D) Densitometric quantification of Lhx4 mRNA in the pineal glands of isoprenaline-injected rats and controls. Bars represent the mean ± SEM. n=3–7. Two-tailed unpaired student’s t-test revealed an effect of isoprenaline on pineal Lhx4 expression: *, p<0.05.
Isoprenaline and the second messenger cyclic AMP induce Lhx4 transcription in pinealocyte cell cultures
In transcriptional activation of Aanat (Klein, 2007; Roseboom et al., 1996) and other pineal transcripts (Bailey et al., 2009; Coon et al., 2012), adrenergic signaling acts via the second messenger cyclic AMP. To test if Lhx4 is also regulated via this pathway, and to verify the in vivo regulation with isoprenaline, cultured pinealocytes were treated with isoprenaline or DBcAMP (Fig. 6A). Both DBcAMP and isoprenaline stimulation significantly increased Lhx4 transcript levels (p-values <0.01 and <0.001). Blocking transcription by pretreating the pinealocytes with actinomycin D prevented the DBcAMP-induced increase in Lhx4 expression (Fig. 6B), thus suggesting that the induced adrenergic-cAMP elevation of Lhx4 expression in the pineal gland requires de novo mRNA synthesis.
Figure 6. DBcAMP stimulation increases Lhx4 mRNA levels through transcription.
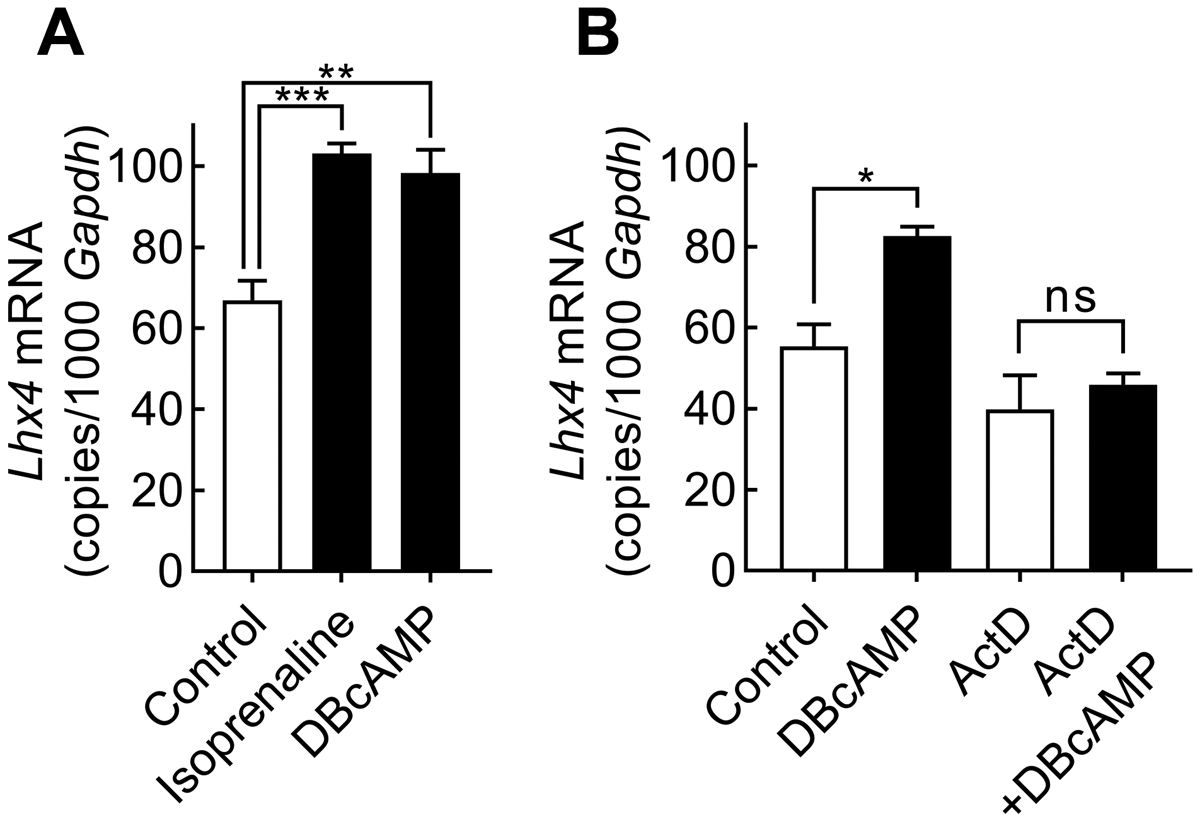
RT-qPCR analysis of Lhx4 expression in cultured pinealocytes. Bars represent the mean ± SEM. (A) Pinealocytes were stimulated with isoprenaline or DBcAMP for 6h. One-way ANOVA followed by Bonferroni-corrected Fisher’s LSD multiple comparisons post hoc tests showed a positive effect of isoprenaline and DBcAMP on Lhx4 expression, n = 4. (B) The effect of actinomycin D (ActD) on Lhx4 expression was tested by 1 h treatment prior to addition of DBcAMP for another 6 h. Two-way ANOVA with Bonferroni-corrected Fisher’s LSD multiple comparisons post hoc tests showed that DBcAMP increased Lhx4 expression (p<0.05) but had no effect in the presence of actinomycin D (p>0.05), n = 3: ns, not significant, p>0.05; *, p<0.05; **, p<0.01; ***, p<0.001.
siRNA-mediated knockdown of Lhx4 reduces expression of Aanat and Tph1 in cyclic AMP-stimulated pinealocytes
To investigate if Lhx4 controls expression of the transcripts encoding melatonin-producing enzymes, Lhx4 was knocked down in cultured pinealocytes using RNAi technology (Fig. 7). siRNA targeting Lhx4 (siLhx4) significantly reduced Lhx4 transcript levels by more than 95% compared to samples treated with a non-targeting siRNA control (siNT) in both DBcAMP-stimulated and unstimulated pinealocytes as revealed by RT-qPCR (p-values <0.001; Fig. 6A).
Figure 7. siRNA-mediated knockdown of Lhx4 expression affects transcripts encoding melatonin-producing enzymes.
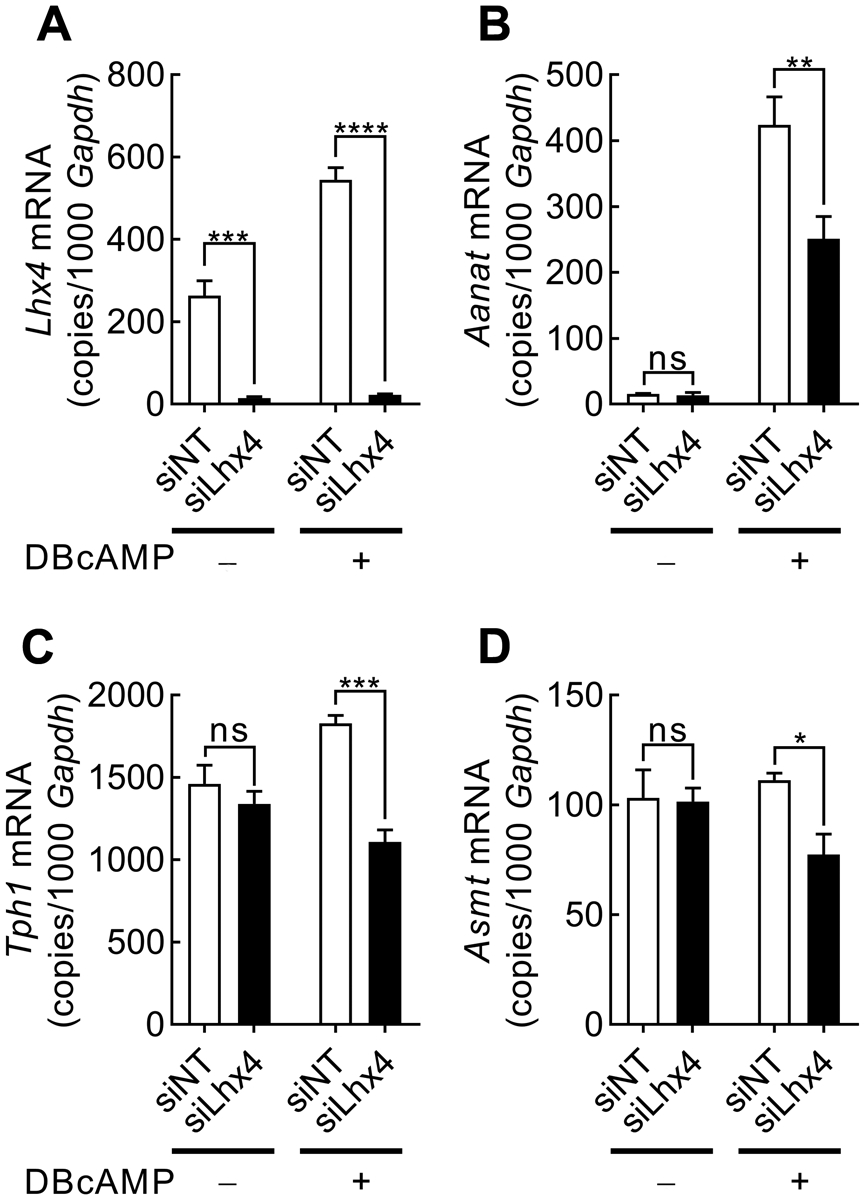
RT-qPCR quantification of expression of Lhx4 (A), Tph1 (B), Aanat (C) and Asmt (D) normalized to Gapdh transcript levels in rat pinealocyte cultures treated with non-targeting siRNA (siNT) or Lhx4-targeting siRNA (siLhx4) and stimulated with DBcAMP for 6 h. Values represent the mean ± SEM. n=3. Data were analyzed by two-way ANOVA with Bonferroni-corrected Fisher’s LSD multiple comparisons post hoc tests: ns, not significant; *, p<0.05; **, p<0.01; ***, p<0.001; ****, p<0.0001.
The addition of siLhx4 significantly reduced the expression of Aanat (p<0.01), tryptophan hydroxylase 1 (Tph1) (p<0.001), and acetylserotonin O-methyltransferase (Asmt) (p<0.05) following 6 h of DBcAMP stimulation (Fig. 7B–D), thus suggesting that Lhx4 contributes to the production of melatonin in pinealocytes by positively regulating the nocturnal expression of melatonin-producing enzymes.
RNA sequencing data indicate Lhx4 knockdown affects transcripts involved in the phototransduction pathway and neuronal and photoreceptor development
To evaluate genome-wide effects of Lhx4 knockdown, RNAseq was performed on DBcAMP-stimulated pinealocyte cultures treated with either siLhx4 or siNT. The two experimental groups were found to represent separate clusters in a PCA plot (Supporting Information, Fig. S1). RNAseq data confirmed the knockdown of Lhx4 and showed a reduction in the expression of the transcripts encoding melatonin-producing enzymes, i.e. Tph1, Aanat and Asmt (Fig. 8A). In preparation for a DAVID analysis, the RNAseq data was filtered to identify 963 highly differentially expressed genes between the experimental groups (Supporting Information, Table S1). Among these genes, 532 were downregulated and 431 were upregulated in the siLhx4-treated samples compared to controls. This list of filtered genes was analyzed using DAVID software (Supporting Information, Table S2) yielding annotations for 925 of the filtered genes. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway output from DAVID revealed two pathways to be significantly (p<0.01) enriched, the phototransduction pathway (p<0.001, Bonferroni-corrected) and the aldosterone synthesis and secretion pathway (p<0.01, Bonferroni-corrected). Details regarding the change in expression of the 10 filtered genes from the phototransduction pathway were extracted from the RNAseq data and revealed most genes to be down regulated after siLhx4 treatment (Fig. 8B). The biological processes (BP5) output from DAVID revealed seven highly significantly (p<0.0001, Bonferroni-corrected) enriched GOTERMS, mostly related to neuronal development, including eye development (Fig. 8C). Thus, in addition to melatonin synthesis, these data suggest that Lhx4 is involved in regulating phototransduction and developmental processes.
Figure 8. Analysis of RNA sequencing data comparing cultured rat pinealocytes treated with siNT and siLhx4.
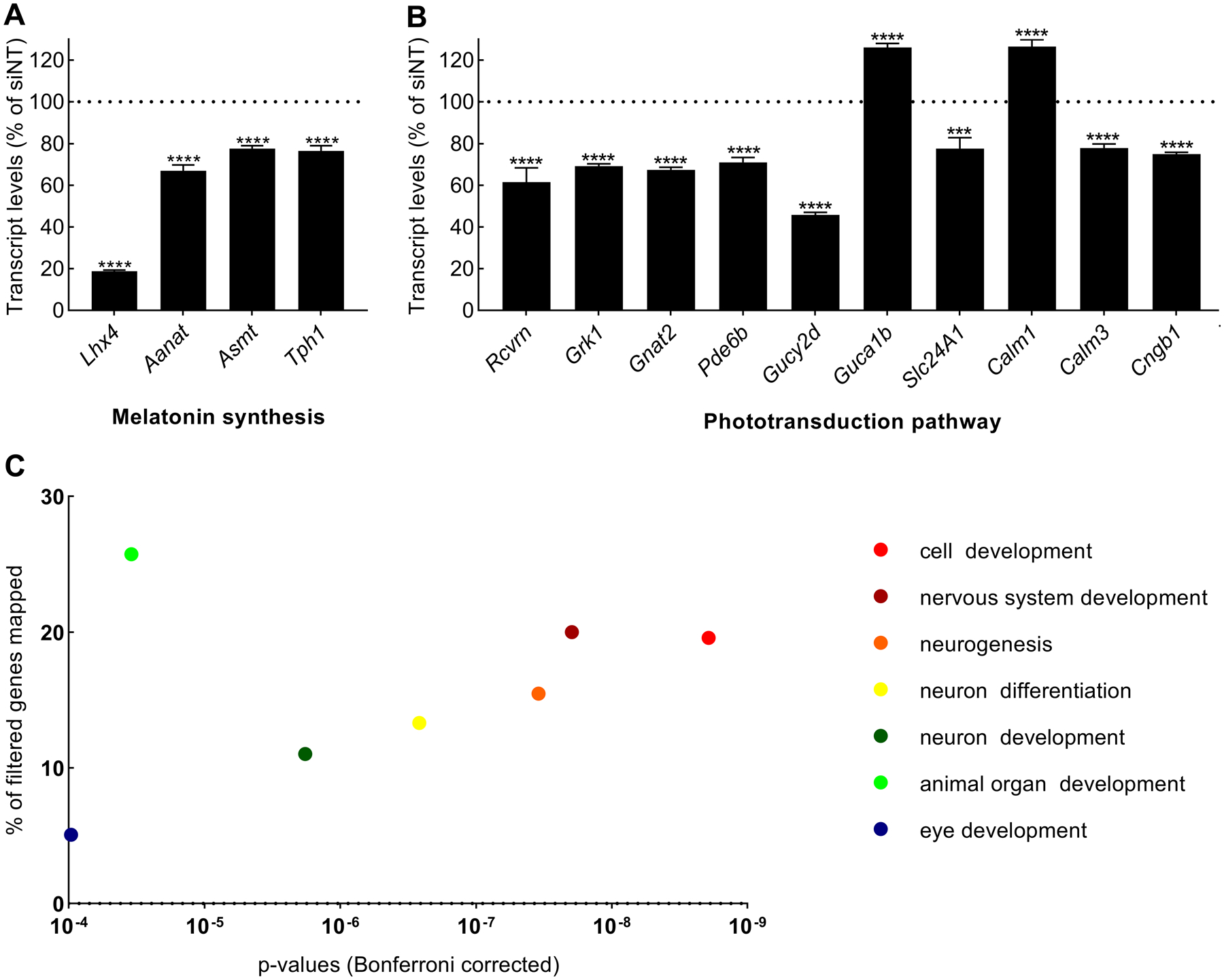
(A)Lhx4 and melatonin synthesis. Lhx4, Tph1, Aanat and Asmt transcript levels presented as normalized counts as percent in siLhx4-treated cultures relative to siNT. Displayed significance levels are based on the Bonferroni-Dunn method: ****, p<0.0001. (B) Phototransduction pathway identified in KEGG from the DAVID output. Expression levels of phototransduction pathway filtered genes presented as normalized counts as percent in siLhx4-treated cultures relative to siNT. Displayed significance levels are based on the Bonferroni-Dunn method: ****, p<0.0001. (C) Biological processes. GOTERM_BP_5 terms with a p<0.0001 (Bonferroni-corrected) from the DAVID output are displayed with the p-value of each term and the percentage of the filtered genes assigned to them. The order of terms in the accompanying legend reflects ascending p-values (from right to left on the graph). Rcvrn, Recoverin; Grk1, G protein-coupled receptor kinase 1; Gnat2, G protein subunit alpha transducing 2; Pde6b, Phosphodiesterase 6b; Gucy2d, Guanylate cyclase 2d; Guca1b, Guanylate cyclase activator 1b; Slc24A1, Solute carrier family 24 member 1; Cngb1, Cyclic nucleotide gated channel beta 1; Calm1, Calmodulin 1; Calm3, Calmodulin 3.
Discussion
The current study establishes that the homeobox gene Lhx4 regulates expression of the genes that encode melatonin-synthesizing enzymes in the pineal gland. The data further support the concept that the sympathetic nervous input to the pineal gland is the signaling mechanism that drives the circadian rhythm of Lhx4 expression in the gland.
The Lhx4 homeobox gene is known to control the development of the pituitary (Sheng et al., 1997), but the current report is the first to establish a role of Lhx4 in postnatal neuroendocrinology. The developmental expression pattern of Lhx4 in the pineal gland, as reported here, suggests that pineal Lhx4 function is restricted to postnatal stages. Developmental appearance of Lhx4 transcripts in the pineal gland coincides with those of Tph1 (Besancon et al., 1995), Aanat (Pfeffer & Stehle, 1998) and Asmt (Ribelayga, Gauer, Pevet, & Simonneaux, 1998). Our detection of rhythmic Lhx4 expression starting between P2 and P10 is also in accord with the appearance of rhythmic Aanat expression (Ellison, Weller, & Klein, 1972; Pfeffer & Stehle, 1998) and the presence of sympathetic nerve fibers (Håkanson, Lombard des Gouttes, & Owman, 1967) in the rat pineal gland during the first postnatal week. The Lhx4 developmental profile corresponds to that of the paired box 4 (Pax4) homeobox gene (Rath, Bailey, Kim, Ho, et al., 2009). In contrast to Lhx4, the related Lhx9 gene is expressed exclusively in the prenatal pineal and acts to promote development of the gland (Yamazaki et al., 2015), as does the Pax6 gene (Estivill-Torrus, Vitalis, Fernandez-Llebrez, & Price, 2001; Rath, Bailey, Kim, Ho, et al., 2009). The orthodenticle 2 (Otx2) and cone-rod (Crx) homeobox genes are expressed both in the developing and the mature pineal gland (Rath et al., 2006), and at least Otx2 plays dual roles in biology of the pineal gland, being essential for both gland development (Nishida et al., 2003) and melatonin synthesis (Rohde et al., 2019). In contrast, the dual roles of Lhx4 are separated both temporally and spatially in different neuroendocrine tissues with a developmental role in the pituitary (Sheng et al., 1997) and a physiological regulatory role in the pineal gland, where its neuroendocrine function in melatonin synthesis acts to maintain cell identity of the pinealocyte. The classical developmental role of Lhx4 seems to be reflected in the Lhx4-affected biological processes identified in our RNAseq results. Notably, the fact that the DAVID software used here revealed a number of developmental biological processes does not mean that Lhx4 controls development of the pineal gland; it merely reflects differential expression of a number of traceable transcripts, which are otherwise involved in development of neural structures.
The involvement of Lhx4 in regulation of melatonin synthesis, as reported here, adds Lhx4 to a growing number of homeobox genes which exhibit circadian rhythms in the adult pineal gland and further control pineal melatonin synthesis (Rath et al., 2013). The retinal and anterior neural fold homeobox gene (Rax) has an expression peak late in the day (Rohde, Klein, Møller, & Rath, 2011) and is essential for high expression of Aanat in the pineal gland (Rohde, Bering, Furukawa, & Rath, 2017). The Otx2 and Crx homeobox genes peak sequentially early in the night (Rohde et al., 2014), and act both individually and in concert to control expression of Tph1, Aanat and Asmt (Rohde et al., 2019). It is clear from the data presented here, both RT-qPCR and RNAseq analyses, that Lhx4 also controls pineal gene expression; however, further studies are needed to clarify the individual roles of LHX4 and other pineal homeobox gene-encoded transcription factors in regulation of melatonin synthesis. Notably, the siRNA-based method applied in the current study does not completely inactivate Lhx4 and therefore does not allow quantitative statements on the relative contribution of individual transcription factors; however, as opposed to global and conditional knockout mouse, we can here analyze the tissue-specific role of homeobox genes in a melatonin-proficient system.
It would seem appropriate to propose the rhythm in Lhx4 expression as a steering factor in rhythmic melatonin synthesis; acting in concert, rhythmic homeobox genes may shape the daily melatonin profile, but the driving force still seems to be nocturnal adrenergic signaling acting directly on AANAT (Klein, 2007) with Lhx4 contributing to melatonin production. On the other hand, homeobox gene expression in the pinealocytes is indeed essential for expression of the melatonin-synthesizing enzymes, thus pointing towards an important role in conferring tissue specificity of melatonin synthesis.
With regard to the cellular distribution of Lhx4 expression, we here used RNAscope in situ hybridization to show that Lhx4 and Aanat transcripts fully colocalize in the pinealocytes. This finding suggests that Lhx4 exerts its regulatory function directly in the melatonin-producing pinealocytes in vivo. It was previously shown by immunohistochemistry that OTX2 and CRX are also present in melatonin-producing pinealocytes, as identified by ASMT-immunoreactivity (Rohde et al., 2019), but from a methodological point of view, the RNAscope method used here overcomes the lack of reliable antisera for cellular detection of AANAT. This report represents the first use of RNAscope technology on the pineal gland and demonstrates its importance in optimizing gene product detection in the gland. Notably, specific expression of Crx and Lhx4 in pinealocytes was also confirmed in recent large-scale single cell RNAseq analyses (Mays et al., 2018). Further, our findings strengthen the concept that homeobox gene-encoded transcription factors are expressed in melatonin-producing pinealocytes to promote hormone synthesis. This report also represents the first demonstration of homeobox gene expression in the mature human pineal gland and thus points towards a conserved role of Lhx4 in pineal biology.
The combined data from in vivo and in vitro experiments presented here further establish that regulation of rhythmic Lhx4 expression in the rat pineal gland is regulated by a positive input from the SCG which acts via adrenergic signaling and cyclic AMP to induce nocturnal gene transcription. These findings are in line with the regulatory mechanism controlling rhythmic expression of Aanat and other transcripts in the rat pineal gland (Bailey et al., 2009; Klein et al., 2010) and adds to previous largescale screening efforts on regulation of gene expression in the rat pineal gland (Bailey et al., 2009; Hartley et al., 2015).
In the current study, the Lhx4 transcript was also found in the retina. In contrast to our findings in the pineal gland, retinal expression levels of Lhx4 were higher during daytime as compared to nighttime, probably reflecting that different regulatory mechanisms for circadian regulation exists in the retina (Iuvone et al., 2005) and that it serves another biological function, as discussed below. The relatively low expression of Lhx4 in the retina as compared to the pineal gland can be explained by the higher degree of cellular heterogeneity in the retina; Lhx4 is expressed in a subset of retinal cell types only, namely in the outer part of the neural retina, whereas it is present in the majority of cells in the pineal gland. Notably, recent data suggest that Lhx4 is specifically expressed in early developing cone photoreceptors of the mouse retina (Buenaventura, Corseri, & Emerson, 2019). The finding of Lhx4 transcripts in the retina seems to reflect a general molecular similarity between the pinealocyte and the retinal photoreceptors, both of which express a common set of genes devoted to phototransduction and melatonin synthesis (Klein, 2004, 2006; Rath et al., 2013). The pineal homeobox genes Crx, Otx2, Rax, Pax4 and Pax6 are also expressed in the retina (Rath, Bailey, Kim, Coon, et al., 2009; Rath, Morin, Shi, Klein, & Møller, 2007; Rohde et al., 2011). Within the retina, melatonin plays an important role in adaptation of the photoreceptor to darkness (Iuvone et al., 2005). The retinal photoreceptor and the pinealocyte appear to have evolved from a common ancestral photodetector cell (Klein, 2004) with the expression of phototransduction genes in the mammalian pineal gland presumably resembling a rudimentary molecular trait or representing acquired roles in pineal regulation.
DAVID analysis of the RNAseq results identified the phototransduction pathway as being most heavily influenced by Lhx4 knockdown in the cultured pinealocytes; the expression of all genes directly involved in the phototransduction cascade was affected (Vinberg, Chen, & Kefalov, 2018). Since there is no phototransduction in the mammalian pineal gland, this finding does not imply that Lhx4 plays an important role in regulating phototransduction in the pineal gland per se. However, our findings do point to the possibility that the persistent molecular similarities between the mammalian retinal photoreceptor and the pinealocyte have the advantage of allowing us to use a pinealocyte-based model system to investigate the regulation of genes involved in retinal biology. Further studies, beyond the scope of the current report, will be needed to confirm the possible role of Lhx4 in controlling melatonin synthesis and especially phototransduction in the mature retina.
Supplementary Material
Acknowledgment
Funding was provided by the Independent Research Fund Denmark (grant number 8020-00037B to MFR), the Novo Nordisk Foundation (grant numbers NNF15OC0015988 and NNF17OC0026938 to MFR), the Lundbeck Foundation (grant number R249-2017-931 to MBC and MFR) and the Carlsberg Foundation (grant numbers CF15-0515 and CF17-0070 to MFR). We wish to thank Rikke Lundorf (University of Copenhagen) for expert histological assistance, the Molecular Genomics Core at NICHD (NIH) for technical support on RNA sequencing analyses, Dr. Alejandro Anaya Rocha and Dr. Matt Kelley (NIH) for advice on RNAscope procedures. We thank Prof. Éva Dobolvine Renner and Prof. Miklós Palkovits, the Human Brain Tissue Bank at Semmelweis University for providing sections of the human pineal gland supported by NAP 2017-1.2.1-NKP-2017-00002.
Abbreviations
- Aanat
aralkylamine N-acetyltransferase
- Actb
beta-actin
- ActD
actinomycin D
- Asmt
acetylserotonin O-methyltransferase
- Crx
cone rod homeobox
- CT
circadian time
- DBcAMP
dibutyryl cyclic AMP
- DD
constant darkness
- E
embryonic day
- Gapdh
glyceraldehyde 3-phosphate dehydrogenase
- LD
12h light:12h dark
- Lhx
LIM homeobox
- NE
norepinephrine
- Otx
orthodenticle homeobox
- P
postnatal day
- Pax
paired box
- Rax
retinal and anterior neural fold homeobox
- RNAseq
RNA sequencing
- SCGdcn
decentralization of the superior cervical ganglia
- SCGx
superior cervical ganglionectomy
- siNT
non-targeting siRNA
- siRNA
short-interference RNA
- Tph1
tryptophan hydroxylase 1
- ZT
Zeitgeber time
Footnotes
Disclosure statement: The authors declare no conflicts of interests.
References
- Anders S, & Huber W (2010). Differential expression analysis for sequence count data. Genome Biol, 11(10), R106. doi: 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey MJ, Coon SL, Carter DA, Humphries A, Kim JS, Shi Q, Gaildrat P, Morin F, Ganguly S, Hogenesch JB, Weller JL, Rath MF, Møller M, Baler R, Sugden D, Rangel ZG, Munson PJ, & Klein DC (2009). Night/day changes in pineal expression of >600 genes: central role of adrenergic/cAMP signaling. J Biol Chem, 284(12), 7606–7622. doi: 10.1074/jbc.M808394200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besancon R, Chouaf L, Jouvet A, Sliwinski S, Belin MF, & Fevre-Montange M (1995). Developmental expression of tryptophan hydroxylase mRNAs in the rat pineal gland: an in situ hybridization study. Brain Res Mol Brain Res, 29(2), 253–262. [DOI] [PubMed] [Google Scholar]
- Buenaventura DF, Corseri A, & Emerson MM (2019). Identification of genes with enriched expression in early developing mouse cone photoreceptors. Invest Ophthalmol Vis Sci, 60(8), 2787–2799. doi: 10.1167/iovs.19-26951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coon SL, Munson PJ, Cherukuri PF, Sugden D, Rath MF, Møller M, Clokie SJ, Fu C, Olanich ME, Rangel Z, Werner T, Mullikin JC, & Klein DC (2012). Circadian changes in long noncoding RNAs in the pineal gland. Proc Natl Acad Sci U S A, 109(33), 13319–13324. doi: 10.1073/pnas.1207748109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, & Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics, 29(1), 15–21. doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison N, Weller JL, & Klein DC (1972). Development of a circadian rhythm in the activity of pineal serotonin N-acetyltransferase. J Neurochem, 19(5), 1335–1341. [DOI] [PubMed] [Google Scholar]
- Estivill-Torrus G, Vitalis T, Fernandez-Llebrez P, & Price DJ (2001). The transcription factor Pax6 is required for development of the diencephalic dorsal midline secretory radial glia that form the subcommissural organ. Mech Dev, 109(2), 215–224. [DOI] [PubMed] [Google Scholar]
- Håkanson R, Lombard des Gouttes MN, & Owman C (1967). Activities of tryptophan hydroxylase, dopa decarboxylase, and monoamine oxidase as correlated with the appearance of monoamines in developing rat pineal gland. Life Sci, 6(24), 2577–2585. [DOI] [PubMed] [Google Scholar]
- Hartley SW, Coon SL, Savastano LE, Mullikin JC, Program NCS, Fu C, & Klein DC (2015). Neurotranscriptomics: The Effects of Neonatal Stimulus Deprivation on the Rat Pineal Transcriptome. PLoS One, 10(9), e0137548. doi: 10.1371/journal.pone.0137548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, & Lempicki RA (2009a). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res, 37(1), 1–13. doi: 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, & Lempicki RA (2009b). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc, 4(1), 44–57. doi: 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Iuvone PM, Tosini G, Pozdeyev N, Haque R, Klein DC, & Chaurasia SS (2005). Circadian clocks, clock networks, arylalkylamine N-acetyltransferase, and melatonin in the retina. Prog Retin Eye Res, 24(4), 433–456. doi: 10.1016/j.preteyeres.2005.01.003 [DOI] [PubMed] [Google Scholar]
- Klein DC (2004). The 2004 Aschoff/Pittendrigh lecture: Theory of the origin of the pineal gland--a tale of conflict and resolution. J Biol Rhythms, 19(4), 264–279. doi: 10.1177/0748730404267340 [DOI] [PubMed] [Google Scholar]
- Klein DC (2006). Evolution of the vertebrate pineal gland: the AANAT hypothesis. Chronobiol Int, 23(1–2), 5–20. doi: 10.1080/07420520500545839 [DOI] [PubMed] [Google Scholar]
- Klein DC (2007). Arylalkylamine N-acetyltransferase: “the Timezyme”. J Biol Chem, 282(7), 4233–4237. doi: 10.1074/jbc.R600036200 [DOI] [PubMed] [Google Scholar]
- Klein DC, Bailey MJ, Carter DA, Kim JS, Shi Q, Ho AK, Chik CL, Gaildrat P, Morin F, Ganguly S, Rath MF, Møller M, Sugden D, Rangel ZG, Munson PJ, Weller JL, & Coon SL (2010). Pineal function: impact of microarray analysis. Mol Cell Endocrinol, 314(2), 170–183. doi: 10.1016/j.mce.2009.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klitten LL, Rath MF, Coon SL, Kim JS, Klein DC, & Møller M (2008). Localization and regulation of dopamine receptor D4 expression in the adult and developing rat retina. Exp Eye Res, 87(5), 471–477. doi: 10.1016/j.exer.2008.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Witte DP, Branford WW, Aronow BJ, Weinstein M, Kaur S, Wert S, Singh G, Schreiner CM, Whitsett JA, & et al. (1994). Gsh-4 encodes a LIM-type homeodomain, is expressed in the developing central nervous system and is required for early postnatal survival. EMBO J, 13(12), 2876–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, & Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics, 30(7), 923–930. doi: 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, & Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol, 15(12), 550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mays JC, Kelly MC, Coon SL, Holtzclaw L, Rath MF, Kelley MW, & Klein DC (2018). Single-cell RNA sequencing of the mammalian pineal gland identifies two pinealocyte subtypes and cell type-specific daily patterns of gene expression. PLoS One, 13(10), e0205883. doi: 10.1371/journal.pone.0205883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller M, & Baeres FM (2002). The anatomy and innervation of the mammalian pineal gland. Cell Tissue Res, 309(1), 139–150. doi: 10.1007/s00441-002-0580-5 [DOI] [PubMed] [Google Scholar]
- Møller M, Phansuwan-Pujito P, Morgan KC, & Badiu C (1997). Localization and diurnal expression of mRNA encoding the beta1-adrenoceptor in the rat pineal gland: an in situ hybridization study. Cell Tissue Res, 288(2), 279–284. [DOI] [PubMed] [Google Scholar]
- Nishida A, Furukawa A, Koike C, Tano Y, Aizawa S, Matsuo I, & Furukawa T (2003). Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat Neurosci, 6(12), 1255–1263. doi: 10.1038/nn1155 [DOI] [PubMed] [Google Scholar]
- Pfeffer M, & Stehle JH (1998). Ontogeny of a diurnal rhythm in arylalkylamine-N-acetyltransferase mRNA in rat pineal gland. Neurosci Lett, 248(3), 163–166. [DOI] [PubMed] [Google Scholar]
- Rath MF, Bailey MJ, Kim JS, Coon SL, Klein DC, & Møller M (2009). Developmental and daily expression of the Pax4 and Pax6 homeobox genes in the rat retina: localization of Pax4 in photoreceptor cells. J Neurochem, 108(1), 285–294. doi: 10.1111/j.1471-4159.2008.05765.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath MF, Bailey MJ, Kim JS, Ho AK, Gaildrat P, Coon SL, Møller M, & Klein DC (2009). Developmental and diurnal dynamics of Pax4 expression in the mammalian pineal gland: nocturnal down-regulation is mediated by adrenergic-cyclic adenosine 3’,5’-monophosphate signaling. Endocrinology, 150(2), 803–811. doi: 10.1210/en.2008-0882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath MF, Morin F, Shi Q, Klein DC, & Møller M (2007). Ontogenetic expression of the Otx2 and Crx homeobox genes in the retina of the rat. Exp Eye Res, 85(1), 65–73. doi: 10.1016/j.exer.2007.02.016 [DOI] [PubMed] [Google Scholar]
- Rath MF, Munoz E, Ganguly S, Morin F, Shi Q, Klein DC, & Møller M (2006). Expression of the Otx2 homeobox gene in the developing mammalian brain: embryonic and adult expression in the pineal gland. J Neurochem, 97(2), 556–566. doi: 10.1111/j.1471-4159.2006.03773.x [DOI] [PubMed] [Google Scholar]
- Rath MF, Rohde K, Klein DC, & Møller M (2013). Homeobox genes in the rodent pineal gland: roles in development and phenotype maintenance. Neurochem Res, 38(6), 1100–1112. doi: 10.1007/s11064-012-0906-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribelayga C, Gauer F, Pevet P, & Simonneaux V (1998). Ontogenesis of hydroxyindole-O-methyltransferase gene expression and activity in the rat pineal gland. Brain Res Dev Brain Res, 110(2), 235–239. [DOI] [PubMed] [Google Scholar]
- Rohde K, Bering T, Furukawa T, & Rath MF (2017). A modulatory role of the Rax homeobox gene in mature pineal gland function: Investigating the photoneuroendocrine circadian system of a Rax conditional knockout mouse. J Neurochem, 143(1), 100–111. doi: 10.1111/jnc.14120 [DOI] [PubMed] [Google Scholar]
- Rohde K, Hertz H, & Rath MF (2019). Homeobox genes in melatonin-producing pinealocytes: Otx2 and Crx act to promote hormone synthesis in the mature rat pineal gland. J Pineal Res, 66(4), e12567. doi: 10.1111/jpi.12567 [DOI] [PubMed] [Google Scholar]
- Rohde K, Klein DC, Møller M, & Rath MF (2011). Rax: developmental and daily expression patterns in the rat pineal gland and retina. J Neurochem, 118(6), 999–1007. doi: 10.1111/j.1471-4159.2011.07385.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde K, Rovsing L, Ho AK, Møller M, & Rath MF (2014). Circadian dynamics of the cone-rod homeobox (CRX) transcription factor in the rat pineal gland and its role in regulation of arylalkylamine N-acetyltransferase (AANAT). Endocrinology, 155(8), 2966–2975. doi: 10.1210/en.2014-1232 [DOI] [PubMed] [Google Scholar]
- Roseboom PH, Coon SL, Baler R, McCune SK, Weller JL, & Klein DC (1996). Melatonin synthesis: analysis of the more than 150-fold nocturnal increase in serotonin N-acetyltransferase messenger ribonucleic acid in the rat pineal gland. Endocrinology, 137(7), 3033–3045. [DOI] [PubMed] [Google Scholar]
- Savastano LE, Castro AE, Fitt MR, Rath MF, Romeo HE, & Munoz EM (2010). A standardized surgical technique for rat superior cervical ganglionectomy. J Neurosci Methods, 192(1), 22–33. doi: 10.1016/j.jneumeth.2010.07.007 [DOI] [PubMed] [Google Scholar]
- Sheng HZ, Moriyama K, Yamashita T, Li H, Potter SS, Mahon KA, & Westphal H (1997). Multistep control of pituitary organogenesis. Science, 278(5344), 1809–1812. [DOI] [PubMed] [Google Scholar]
- Slack JM, Holland PW, & Graham CF (1993). The zootype and the phylotypic stage. Nature, 361(6412), 490–492. doi: 10.1038/361490a0 [DOI] [PubMed] [Google Scholar]
- Vinberg F, Chen J, & Kefalov VJ (2018). Regulation of calcium homeostasis in the outer segments of rod and cone photoreceptors. Prog Retin Eye Res, 67, 87–101. doi: 10.1016/j.preteyeres.2018.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, Wu X, Vo HT, Ma XJ, & Luo Y (2012). RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn, 14(1), 22–29. doi: 10.1016/j.jmoldx.2011.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki F, Møller M, Fu C, Clokie SJ, Zykovich A, Coon SL, Klein DC, & Rath MF (2015). The Lhx9 homeobox gene controls pineal gland development and prevents postnatal hydrocephalus. Brain Struct Funct, 220(3), 1497–1509. doi: 10.1007/s00429-014-0740-x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


