Abstract
Background
The volvocine algae, which include the single-celled species Chlamydomonas reinhardtii and the colonial species Volvox carteri, serve as a model in which to study the evolution of multicellularity and cellular differentiation. Studies reconstructing the history of this group have by and large relied on datasets of one to a few genes for phylogenetic inference and ancestral character state reconstruction. As a result, volvocine phylogenies lack concordance depending on the number and/or type of genes (i.e., chloroplast vs nuclear) chosen for phylogenetic inference. While multiple studies suggest that multicellularity evolved only once in the volvocine algae, that each of its three colonial families is monophyletic, and that there have been at least three independent origins of cellular differentiation in the group, other studies call into question one or more of these conclusions. An accurate assessment of the evolutionary history of the volvocine algae requires inference of a more robust phylogeny.
Results
We performed RNA sequencing (RNA-seq) on 55 strains representing 47 volvocine algal species and obtained similar data from curated databases on 13 additional strains. We then compiled a dataset consisting of transcripts for 40 single-copy, protein-coding, nuclear genes and subjected the predicted amino acid sequences of these genes to maximum likelihood, Bayesian inference, and coalescent-based analyses. These analyses show that multicellularity independently evolved at least twice in the volvocine algae and that the colonial family Goniaceae is not monophyletic. Our data further indicate that cellular differentiation arose independently at least four, and possibly as many as six times, within the volvocine algae.
Conclusions
Altogether, our results demonstrate that multicellularity and cellular differentiation are evolutionarily labile in the volvocine algae, affirming the importance of this group as a model system for the study of major transitions in the history of life.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12915-021-01087-0.
Keywords: Phylotranscriptomics, Volvocine algae, Green algae, Phylogeny, Multicellularity
Background
The evolution of multicellularity is widely considered a major transition in the history of life [1–4]. Multicellularity not only gave rise to most of the visible life forms on the planet, but also opened the door to cellular differentiation, including that between somatic and reproductive cells, a hallmark feature of sexual reproduction in eukaryotes that exhibit morphological complexity [3, 5, 6]. Questions regarding the evolution of multicellularity and cellular differentiation have been approached using the fossil record [7–9], laboratory evolution [10–13], and comparative approaches that include superimposing cell biology upon molecular phylogeny [14–16]. The last of these approaches is predicated on the assumption that the cell biology and molecular phylogeny are mutually informative, an assumption that requires the phylogeny itself to be accurate.
The volvocine green algae have proved especially useful for investigating the major transition leading to multicellularity. The group consists of ~ 50 extant species, which exhibit a range of body plans, cell numbers, sizes, and forms of sexual reproduction. The smallest of these are single-celled (e.g., Chlamydomonas reinhardtii); the largest, at up to 3 mm in diameter and up to 50,000 cells, are spheroidal, swimming colonies in the genus Volvox. Since the initial “very pleasant sight” of swimming Volvox colonies described by Van Leeuwenhoek more than 300 years ago [17], the volvocine algae have come to be accepted as a useful model system in which to address questions related to the origins of multicellularity and cellular differentiation [18, 19]. Multiple species have now had their genomes sequenced [20–24], and those of unicellular C. reinhardtii and multicellular V. carteri forma nagariensis are well-annotated [23, 24]. However, the volvocine algae encompass more than two organisms representing alternative forms of life in terms of size and development. Vegetative forms range in characteristic cell number from 1 to ~ 50,000 and exhibit intermediate degrees of complexity likely similar to extinct ancestors. Further, multicellularity and cellular differentiation arose within the volvocine algae much more recently than those traits arose in animals: ~ 220 million years ago [25] versus ~ 600 million years ago [26], respectively.
Evolution of the volvocine algae has sometimes been viewed as a linear progression in size and complexity [27, 28]. Unicellular taxa such as Chlamydomonas occupy one end of this continuum, while fully differentiated, multicellular taxa such as Volvox occupy the other. This concept, the “volvocine lineage hypothesis”, used a streamlined phylogeny of the volvocine algae to help explain how a multicellular species with complete germ-soma differentiation such as Volvox might evolve from a unicellular, Chlamydomonas-like ancestor. However, morphological and molecular phylogenetic studies suggest that the history of the volvocine algae may be more complicated, as cellular differentiation, different modes of sexual reproduction, and varying body plans appear to have evolved multiple times within the group [29, 30].
Current understanding of the major evolutionary relationships within this group has often been based on the analysis of five chloroplast gene sequences [14, 25, 31–35]. Chloroplast gene-based phylogenies have also been used to carry out ancestral-state reconstructions [14, 29, 30, 36], opening a window on how multicellularity and cellular differentiation evolved within the volvocine algae. Overall, the branching order of most chloroplast gene-based phylogenies is defined by two related groups: (i) a set of unicellular species (e.g., Chlamydomonas reinhardtii) that are paraphyletic with respect to (ii) a clade that encompasses the three major families of colonial volvocine algae: Tetrabaenaceae (Tetrabaena and Basichlamys), Goniaceae (Gonium and Astrephomene), and Volvocaceae (Colemanosphaera, Eudorina, Pandorina, Platydorina, Pleodorina, Volvox, Volvulina, and Yamagishiella) (Fig. 1a, d). In this scheme, the Tetrabaenaceae is a sister group to the clade formed by the Goniaceae and Volvocaceae. Although this framework only takes into account family-level relationships, several conclusions can be drawn. First, the colonial species form a clade. Second, each of the three families is monophyletic. Third, monophyly among the colonial species implies that multicellularity evolved only once within that group with no reversion to unicellularity.
Fig. 1.
Phylogenies of the volvocine algae are not concordant: Four volvocine green algae phylogenies based on different types of data, displayed in chronological order of their appearance in the literature. Species highlighted in shades of gray exhibit somatic cell differentiation. The varying colors to the right of each phylogeny have been arbitrarily assigned to particular genera and are intended to be used as a visual aid to highlight differences among the phylogenies
Two recent studies have called into question the monophyly of the colonial volvocine algae (Fig. 1c). Pröschold et al. [37] based their inferences on two datasets: one consisting of SSU rDNA sequences plus internal transcribed spacer (ITS) sequences 1 and 2, the other consisting of ITS sequences alone. Nakada et al. [38] used a single-gene 18S rRNA dataset. Both studies inferred that the colonial species are paraphyletic with respect to certain unicells in the genera Chlamydomonas and Vitreochlamys.
The taxonomic status of the Goniaceae has also been called into question by studies (Fig. 1b) that indicate the group is either not monophyletic [39] or that there is low support for a sister relationship between Astrephomene and Gonium [33, 37, 38]. Moreover, a number of recent volvocine algal phylogenies leave uncertainty as to how many times cellular differentiation evolved within the group. Chloroplast sequence data suggest at least 3 independent origins of cellular differentiation: in Astrephomene, in Volvox section Volvox (sometimes referred to as Euvolvox), and in the Eudorina, Volvox, Pleodorina (EVP) clade (Fig. 1b–d). Within the EVP clade it is unclear whether cellular differentiation in Pleodorina thompsonii, Volvox gigas and V. powersii, and Pleodorina starrii and P. indica arose independently from that in V. carteri (Fig. 1b–d).
The foregoing uncertainties highlight the need for a new and more robust molecular phylogeny of the volvocine algae. These uncertainties may arise from incomplete taxonomic sampling, limited genetic sampling, or both. While five volvocine algal species have had their genomes sequenced, most taxonomically comprehensive phylogenetic inferences about this evolutionarily important group have been constructed using relatively small datasets. Most consist of the sequence of five chloroplast genes [14, 31, 34, 40] representing an aggregate of ~ 6000 nucleotide positions. Others consist of small (≤ 6) multi-gene datasets consisting of chloroplast gene(s), ribosomal molecular markers, or both [37, 38]. Moreover, the use of chloroplast genes in phylogenetic reconstruction can be problematic because they are effectively a single linkage group, they vary little among recently diverged species [41], and they are at increased risk of incomplete lineage sorting due to the retention of ancestral polymorphisms [42, 43].
Of special concern is the observation that volvocine phylogenies inferred using chloroplast genes (Fig. 1d) conflict with those constructed using nuclear genes (Fig. 1b, c) [37–39]. While conflicts between chloroplast and nuclear phylogenies are not unusual [44–46], they do foster ambiguity.
Here, we seek to resolve volvocine relationships using taxonomically dense sampling of multiple, unlinked loci. We have adopted a phylotranscriptomic approach that uses a concatenated amino acid alignment of 40 nuclear protein-coding, single-copy genes. We sequenced whole transcriptomes of 55 strains encompassing 47 nominal species and used previously published RNA-Seq data for 9 strains and amino acid alignments for 4 strains that were shared with our group by the De Clerck laboratory. Our goal was to derive a robust phylogeny of the volvocine algae that would enable inferences about the evolution of multicellularity, cellular differentiation, sexual dimorphism, and other traits in this group. Our results represent the most taxonomically comprehensive phylogeny yet produced of the volvocine algae using a nuclear dataset, including all described genera and multiple representatives of all genera that are not monotypic. Our results show that the colonial species do not form a clade, that the Goniaceae are not monophyletic, and that multicellularity has independently evolved at least twice and cellular differentiation at least four times within the volvocine algae.
Results and discussion
De novo transcriptome data makes possible 40 single-gene alignments
We sampled 68 taxa representing all presumed major lineages of the colonial volvocine algae and 9 of their nearest unicellular relatives. Because the phylogenetic position of Chlamydomonas reinhardtii has recently been called into question [37, 38], we used a member of the Trebouxiophyceae, Chlorella variabilis, as an outgroup (Table 1). All described volvocine genera were included, with multiple species represented for every genus that is not monotypic. Truly comprehensive taxon sampling was not possible, since several described species, especially in the genus Volvox, are no longer available in culture collections. While our main focus was to resolve relationships within the colonial volvocine algae, our study included several closely related unicellular taxa from the genera Chlamydomonas and Vitreochlamys in order to provide better phylogenetic resolution of the volvocine algae as a whole.
Table 1.
List of taxa used in this study and summary of sequencing and assembly. Under Strain or Pubmed ID, “CC” refers to Chlamydomonas Culture Collection at the University of Minnesota (CC, USA), “F” refers to Culture Collection of Freshwater Algae at the Institute of Hydrobiology, Chinese Academy of Sciences (FACHB, China), “N” refers to National Institute for Environmental Studies (NIES, Japan), “S” refers to Culture Collection of Algae at the University of Göttingen (SAG, Germany), and “U” refers to The Culture Collection of Algae at the University of Texas at Austin (UTEX, USA). “QRPMK” and “TR” under RNA Extraction Method refer to QIAGEN RNeasy Plant Mini Kit and TRizol RNeasy, respectively. Strains assigned an asterisk represent data from previously published studies, with accession numbers shown in Supplementary Materials: Table S1 [21, 47]
| Taxa | Strain or Pubmed ID | RNA extraction method | Total # of raw reads | Total # of clean PE reads | Total # of Contigs | N50 | G/C % |
|---|---|---|---|---|---|---|---|
| Astrephomene gubernaculifera | N-418 | QRPMK | 61460551 | 44350080 | 78105 | 3894 | 0.61 |
| A. perforata | N-564 | QRPMK | 63350620 | 46811342 | 72913 | 3613 | 0.61 |
| Basichlamys sacculifera | N-566 | QRPMK | 66205749 | 48327288 | 134880 | 1945 | 0.69 |
| Colemanosphaera angeleri | F 2363* | 28119810 | 27000176 | 120763 | 3348 | 0.62 | |
| C. charkowiensis | F 2326* | 27967726 | 26876865 | 103286 | 3382 | 0.62 | |
| N-3383 | TR | 58777161 | 40550696 | 119848 | 2523 | 0.62 | |
| Chlamydomonas debaryana | S 11-55a | QRPMK | 87455695 | 64552440 | 105096 | 2776 | 0.67 |
| S 70.81 | QRPMK | 50406749 | 42095108 | 103962 | 2858 | 0.64 | |
| C. globosa | S 81.72 | TR | 73820020 | 49419928 | 120062 | 2079 | 0.65 |
| C. moewusii | S 11-16f | QRPMK | 54728573 | 40436359 | 118833 | 2498 | 0.6 |
| C. reinhardtii | CC-503* | ||||||
| C. schloesseri | S 2486 | QRPMK | 70163393 | 49255021 | 64233 | 3094 | 0.66 |
| Chlorella variabilis NC64a | 20852019* | ||||||
| Eudorina cylindrica | F 2322* | 26442110 | 25262148 | 103026 | 2846 | 0.64 | |
| N-722 | QRPMK | 44479173 | 36297801 | 105029 | 2927 | 0.64 | |
| E. elegans | F 2321* | ||||||
| N-456 | TR | 84549961 | 56756776 | 122975 | 2108 | 0.63 | |
| N-458 | QRPMK | 67451343 | 46725012 | 131335 | 2944 | 0.65 | |
| N-568 | QRPMK | 83433797 | 69539684 | 120487 | 3614 | 0.64 | |
| N-717 | QRPMK | 47625429 | 37616922 | 89118 | 2094 | 0.64 | |
| N-719 | QRPMK | 57157324 | 43565312 | 98180 | 2976 | 0.63 | |
| N-720 | QRPMK | 38134498 | 31388413 | 95662 | 3699 | 0.61 | |
| E. illinoisensis | N-460 | QRPMK | 62485922 | 49396583 | 97014 | 2615 | 0.64 |
| E. minodii | N-856 | QRPMK | 83574258 | 66413244 | 128802 | 4127 | 0.61 |
| E. peripheralis | N-725 | QRPMK | 46607143 | 36528930 | 100934 | 2322 | 0.62 |
| E. unicocca | S 24-1c | TR | 54482446 | 37679863 | 110391 | 2267 | 0.62 |
| Gonium multicoccum | N-737 | QRPMK | 65984190 | 44301578 | 122279 | 3275 | 0.64 |
| G. pectorale | N-2863* | ||||||
| G. octonarium | N-851 | TR | 60029472 | 39826211 | 104857 | 2302 | 0.65 |
| G. quadratum | N-653 | QRPMK | 62431743 | 48230702 | 100468 | 3789 | 0.64 |
| G. viridistellatum | N-654 | QRPMK | 43286980 | 31852410 | 97991 | 2837 | 0.65 |
| Pandorina colemaniae | F 2361* | 25181003 | 23823370 | 80843 | 3292 | 0.62 | |
| N-572 | TR | 49862534 | 37589048 | 84053 | 2237 | 0.62 | |
| P. morum | F 2362* | 29837807 | 28442854 | 147990 | 2674 | 0.61 | |
| N-890 | QRPMK | 39337532 | 24490235 | 133742 | 3123 | 0.61 | |
| Platydorina caudata | N-728 | QRPMK | 48702213 | 36896556 | 125206 | 3503 | 0.61 |
| Pleodorina indica | N-736 | QRPMK | 57564627 | 44082516 | 192570 | 2006 | 0.63 |
| P. japonica | U 2523 | QRPMK | 64897803 | 49495315 | 83662 | 3499 | 0.61 |
| P. starrii | N-1362 | TR | 68790016 | 45935282 | 87641 | 2438 | 0.64 |
| N-1363 | TR | 59466876 | 44726861 | 128732 | 1695 | 0.63 | |
| P. thompsonii | N-4126 | QRPMK | 81077847 | 61986735 | 118940 | 4005 | 0.62 |
| Tetrabaena socialis | N-571* | 42302450 | 41232209 | 62014 | 1012 | 0.65 | |
| Vitreochlamys aulata | N-878 | QRPMK | 80710519 | 54651118 | 132115 | 2673 | 0.65 |
| S 80.81 | QRPMK | 41008694 | 32585611 | 84834 | 2100 | 0.65 | |
| V. nekrassovii | S 11-10 | QRPMK | 65658295 | 48341108 | 109360 | 3304 | 0.62 |
| V. ordinata | N-882 | TR | 60631467 | 42981449 | 75434 | 2778 | 0.69 |
| Volvox africanus | N-863 | QRPMK | 70739371 | 52378949 | 110066 | 4586 | 0.55 |
| V. aureus | N-541 | QRPMK | 80821130 | 64315625 | 106245 | 4065 | 0.55 |
| V. barberi | N-730 | QRPMK | 49805303 | 37197105 | 152910 | 2217 | 0.58 |
| V. carteri f. kawasakiensis | N-732 | TR | 53994112 | 34239609 | 86004 | 1982 | 0.58 |
| V. carteri f. nagariensis | 20616280* | ||||||
| N-865 | QRPMK | 65624222 | 50641434 | 117314 | 3947 | 0.57 | |
| V. carteri f. weismannia | N-866 | QRPMK | 25913610 | 20987137 | 78230 | 2636 | 0.57 |
| V. dissipatrix | N-4128 | QRPMK | 77137081 | 51045569 | 112190 | 5146 | 0.55 |
| V. ferrisii | N-3986 | QRPMK | 49810357 | 37522413 | 91045 | 3281 | 0.57 |
| V. gigas | N-867 | QRPMK | 50554912 | 37592146 | 151302 | 2499 | 0.63 |
| V. globator | S 199.80 | QRPMK | 67413919 | 52861449 | 100861 | 4005 | 0.56 |
| V. kirkiorum | N-543 | QRPMK | 44941633 | 34650729 | 89533 | 3081 | 0.56 |
| V. obversus | N-868 | QRPMK | 67394604 | 50823546 | 99171 | 3881 | 0.58 |
| V. ovalis | N-2569 | QRPMK | 52242003 | 35319700 | 107999 | 3643 | 0.53 |
| V. powersii | N-4127 | QRPMK | 55232784 | 42410688 | 140237 | 2908 | 0.62 |
| V. tertius | N-544 | QRPMK | 86222840 | 62563240 | 108551 | 5454 | 0.54 |
| Volvulina boldii | N-893 | QRPMK | 25665262 | 20161297 | 158265 | 1593 | 0.62 |
| V. compacta | F 2337* | 27543035 | 26374210 | 90261 | 3550 | 0.61 | |
| N-582 | QRPMK | 53045384 | 38991781 | 112583 | 3380 | 0.63 | |
| V. pringsheimii | N-895 | TR | 82233959 | 55849890 | 128304 | 3122 | 0.62 |
| V. steinii | S 90-1 | QRPMK | 46133638 | 37699635 | 89515 | 3635 | 0.62 |
| Yamagishiella unicocca | F 2364* | 30922957 | 29574420 | 64415 | 3609 | 0.63 |
The total number of raw reads generated from RNA sequencing for each species ranged from 25,665,262 to 87,455,695 reads with an average of 60,194,849 reads per species. After quality trimming of the raw reads (see “Methods”), the total number of clean paired-end reads ranged from 20,161,297 to 69,539,684 with an average of 44,416,935 reads per species (Table 1). From the RNA-seq data, we assembled a total of 40 single-gene alignments that were later concatenated to a single alignment representing an aggregate of 12,650 amino acids, equivalent to 37,950 nucleotide positions, with a total of 5972 parsimony-informative sites. Numbers of informative positions in the single-gene alignments ranged from 40 to 446. Trees inferred using maximum likelihood (ML), Bayesian inference (BI), and coalescence-based (CB) analyses were generally well-supported with some topological differences between the ML and BI analyses relative to the CB analysis, as described below.
Our results conflict with prior volvocine algal phylogenies in four respects
First, we find that the colonial volvocine algae are paraphyletic with respect to some unicellular species. Second, monophyly of the family Goniaceae is not supported. Third, section Volvox is inferred to be sister to the remaining Volvocaceae. Fourth, cellular differentiation independently arose at least four and perhaps as many as six times within the volvocine algae.
Colonial volvocine algae are not monophyletic
All three of our phylogenetic analyses indicate that the colonial volvocines are not monophyletic (Figs. 2 and 3); further, an approximately unbiased (AU) test strongly rejected monophyly for this group (p = 2.82e− 38) (Additional file 1: Fig. S1a). These findings represent a major departure from earlier chloroplast gene-based volvocine phylogenies [14, 25, 31–34, 40, 48], phylogenies based on morphological characters [49, 50], phylogenies inferred using ITS 1 and 2 sequences [39], as well as less taxonomically comprehensive phylogenies inferred using nuclear data [51], all of which suggest that the colonial volvocine algae are monophyletic.
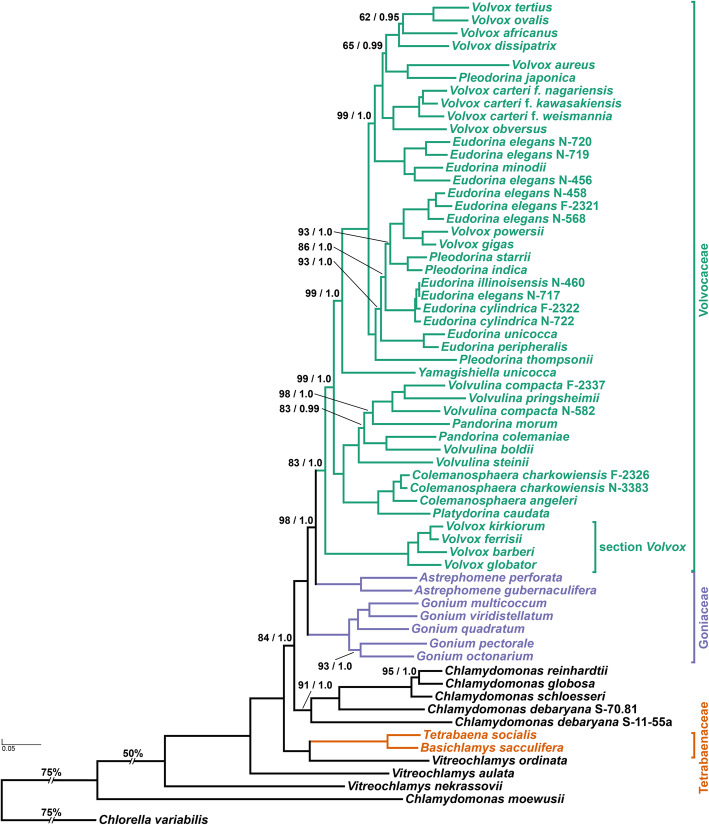
Fig. 2.
Molecular phylogeny of the colonial volvocine algae (Tetrabaenaceae, Goniaceae, and Volvocaceae) and closely related unicellular taxa represented by Chlamydomonas and Vitreochlamys with Chlorella variabilis as the outgroup. The phylogenetic tree shown is based on a multi-gene dataset of single-copy, protein-coding nuclear genes (12,650 aligned amino acid positions of 68 taxa) inferred using the maximum likelihood method, the branching order of which is identical to that inferred in the Bayesian Inference using MrBayes. Numbers on branches represent bootstrap values and Bayesian posterior probabilities, respectively (all support values not shown are MLBS = 100, BPP = 1.0). Branch lengths correspond to genetic divergence, as indicated by the scale bar. Members of the Tetrabaenaceae, Goniaceae, and Volvocaceae are denoted in orange, purple, and green, respectively; unicellular species are denoted in black
Fig. 3.
Phylogeny of the volvocine algae inferred using a coalescent-based analysis of 40 single-gene phylogenies. Numbers on branches represent posterior probabilities (support values not shown are CPP = 1.0). Members of the Tetrabaenaceae, Goniaceae, and Volvocaceae are denoted in orange, purple, and green, respectively; unicellular species are denoted in black
Consistent with Pröschold et al. [37], our results support the view that multicellularity evolved independently in the Tetrabaenaceae and in the Goniaceae + Volvocaceae. In each analytical framework, the Tetrabaenaceae was found to be sister to Vitreochlamys ordinata rather than to the Goniaceae + Volvocaceae (Maximum Likelihood Bootstrap [MLBS] = 100, Bayesian Posterior Probabilities [BPP] = 1.0, Coalescent Posterior Probabilities [CPP] = 1.0). A sister relationship between the Tetrabaenaceae and V. ordinata was inferred in 17/39 of our single-gene phylogenies and in 27/39 of our 4-taxa, unrooted, single-gene phylogenies (Fig. 4). These results imply one independent origin of multicellularity in the Tetrabaenaceae and another origin in the Goniaceae + Volvocaceae.
Fig. 4.
Phylogenetic relationships between Gonium and Astrephomene and between the Tetrabaenaceae and Vitreochlamys ordinata. Four-taxon, unrooted trees were generated by collapsing our single-gene phylogenies. The percentage of single-gene phylogenies representing a specific four-taxon, unrooted tree is represented by the purple, orange, and green bars for trees containing Gonium and Astrphomene, and red, orange, and blue for trees containing the Tetrabaenaceae. A percentage of single-gene phylogenies that show Gonium not sister to Astrephomene represented by the black bar. B percentage of four-taxon, unrooted trees representing specific relationships between Gonium and Astrephomene. C percentage of single-gene phylogenies that show Tetrabaenaceae sister to V. ordinata represented by the black bar. D percentage of four-taxon, unrooted trees representing specific relationships between the Tetrabaenaceae and V ordinata in four-taxon, unrooted trees. For all relationships involving V. ordinata, 39 out of 40 single-gene phylogenies were used due to V. ordinata not appearing in one of the inferences. All single-gene phylogenies were inferred using maximum likelihood under the appropriate evolutionary model as estimated by ProtTest
Our results differ in key respects from a recent volvocine algal phylogeny inferred by Zhang et al. [51], which like ours is based on single-copy nuclear genes. Zhang et al. [51] sought to understand the evolutionary relationships between two psychrophilic algae: Chlamydomonas sp. ICE-L and Tetrabaena socialis N-691. To do so, they constructed a phylogeny consisting of ICE-L, N-691, three colonial Volvox strains, and eight unicellular species, including C. reinhardtii. Among their conclusions was that T. socialis N-691 is sister to the Volvocaceae, which is at odds with results shown in Figs. 2 and 3. These results indicate that the Tetrabaenaceae is sister to V. ordinata, and together they are sister to C. reinhardtii + Goniaceae + Volvocaceae.
We hypothesized that the lack of concordance between our findings and those of Zhang et al. [51] could be attributed to limited taxon sampling. To test this hypothesis, we first confirmed that T. socialis N-691 and T. socialis N-571 are conspecific (Additional file 2: Confirming the conspecificity of Tetrabaena socialis N-571 and N-691) [52, 53]. Once we confirmed that N-691 and N-571 were conspecific, we were able to replicate the branching order produced by Zhang et al. [51] using our concatenated 40-gene dataset (Additional file 1: Fig. S2a) [51]. For our initial tree, we sampled our strains of Chlamydomonas reinhardtii, C. moewusii, T. socialis, Volvox aureus, V. carteri f. nagariensis, and V. globator to match taxa that were used in that study. For an outgroup species, we sampled Chlorella variabilis. Multiple studies have shown that the accuracy of phylogenetic reconstruction can be improved by increasing the number of taxa sampled [54–56]. When we added more taxa and performed ML analysis on the new dataset, the three colonial volvocine families were no longer monophyletic. The Tetrabaenaceae were sister to Vitreochlamys ordinata, and this clade appeared sister to C. reinhardtii + Goniaceae + Volvocaceae (Additional file 1: Fig. S2b) [51]. These analyses confirm that the placement of T. socialis N-691 as sister to the Volvocaceae is an artifact of limited taxon sampling. From this, we draw three conclusions: First, the colonial volvocine algae are not monophyletic; second, at least two independent origins of multicellularity occurred within the volvocine algae; third, once multicellularity evolved no extant lineage reverted to the ancestral unicellular state (see Figs. 2 and 3).
The family Goniaceae is not monophyletic
Multiple volvocine phylogenies have concluded that the Goniaceae is monophyletic [14, 25, 29, 31, 33, 37, 38, 49, 50, 57, 58]. Our analyses suggest otherwise (Figs. 2 and 3): we find that Astrephomene is sister to the Volvocaceae (MLBS = 98, BPP = 1.0, CPP = 0.81) rather than to Gonium. This inference is strengthened by observations that 37/40 of our single-gene phylogenies show that Gonium and Astrephomene are not sister taxa, as do 20/40 of our four-taxon, unrooted phylogenies (Fig. 4). All three of our analyses indicate that Astrephomene is monophyletic and sister to the Volvocaceae clade (MLBS = 98, BPP = 1.0, CPP = 0.81), with Gonium sister to Astrephomene + Volvocaceae (MLBS = 100, BPP = 1.0, CPP = 0.86). Furthermore, we performed an AU test where the monophyly of the Goniaceae was tested against our finding of paraphyly for the Goniaceae. The null hypothesis, monophyly of the Goniaceae, was rejected (p = 0.0446) (Additional file 1: Fig. S1b). The inferred sister relationship between Astrephomene and the Volvocaceae is also consistent with the apparent synapomorphy of zygote germination producing a single gone cell, which is unique to these two taxa [50]
Prior studies have produced mixed results regarding monophyly of the Goniaceae, sometimes with low support values for the relevant relationships. Nozaki and colleagues [59] published four phylogenies inferred using a single chloroplast gene and different inference methods; all four trees either showed low support for monophyly of the Goniaceae or suggested a topology where Astrephomene is sister to Gonium + Volvocaceae. Coleman [39] inferred a volvocine phylogeny based on ITS-1 and ITS-2 sequences that showed Astrephomene sister to Tetrabaenaceae + Gonium + Volvocaceae; however, the bootstrap support for this suggested relationship was between 50 and 75%, indicating weak support for the branching order. Other phylogenies suggesting monophyly in the Goniaceae do so with weak or contradictory support [33, 37, 38].
Our inference that the Goniaceae are not monophyletic is consistent with some — but not all — of the analyses recently reported by Pröschold et al. [37] and Nakada et al. [38]. However, we should not disregard past morphological and ultrastructural studies suggesting a close relationship between Astrephomene and Gonium [50, 60, 61]. These taxa differ from the Volvocaceae in that each cell, rather than the entire colony, is surrounded by a tripartite boundary [62]. This feature distinguishes their mode of colony formation from all other colonial algae within the Volvocaceae; our results suggest that it is ancestral to the Goniaceae + Volvocaceae and lost in the Volvocaceae.
Volvox section Volvox is sister to the remaining Volvocaceae
Our data indicate that Volvox section Volvox is not a subclade within either the Pandorina + Volvulina + Colemanosphaera (PVC) or Eudorina + Volvox + Pleodorina (EVP) subclades. Older studies based on the rbcL chloroplast gene [49], ITS-1 and ITS-2 sequences [39], and morphology [50] suggest that section Volvox belongs to a clade that encompasses Eudorina, Pleodorina, and other Volvox species. More recent studies of the volvocine algae based on 5 chloroplast genes, or based on multiple datasets that include 1 chloroplast gene [37], suggest that section Volvox belongs to a clade that includes Pandorina, Volvulina, and Platydorina [14, 31], and (in the studies where it was included) Colemanosphaera [34, 40]. By contrast, all of our analyses indicate that section Volvox is monophyletic and sister to the remaining Volvocaceae (MLBS = 83, BPP = 1.0, CPP = 0.73). AU tests rejected the monophyly of section Volvox + Colemanosphaera + Platydorina (p-AU = 4.64e− 88) and the monophyly of section Volvox + the PVC clade (p-AU = 0.0332) (Additional file 1: Fig. S1c). These results bolster our finding that section Volvox is sister to the remaining Volvocaceae (Figs. 2 and 3).
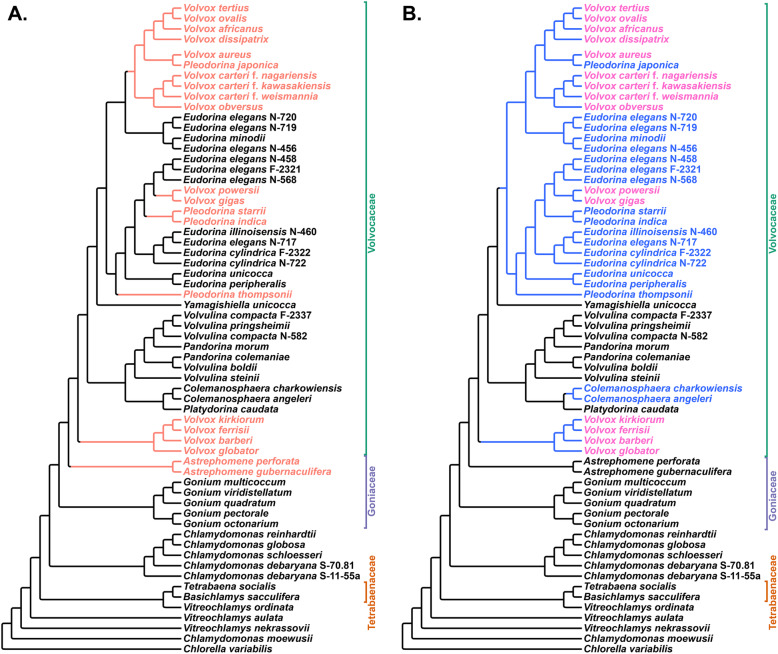
Cellular differentiation independently arose at least four times in the volvocine algae
The last major difference between our results and earlier phylogenies concerns the number of independent origins of cellular differentiation. Prior literature suggests that cellular differentiation independently evolved at least three times: once in Astrephomene, once in section Volvox, and at least once in the EVP clade [14, 36]. By contrast, our results show a minimum of four independent origins of cellular differentiation: one in Astrephomene, one in section Volvox, and at least two in the EVP clade (Fig. 5a). We cannot exclude the possibility of two additional independent origins in the branches leading to Pleodorina starrii and Volvox gigas (Fig. 5a). In Astrephomene, section Volvox, Pleodorina, and Volvox dissipatrix, differentiated cells carry out the function of motility, whereas undifferentiated cells participate in both motility and reproduction [15]. The remaining Volvox species within the EVP clade have all evolved specialized germ cells for reproduction and somatic cells for motility [25, 30].
Fig. 5.
A Phylogeny of the volvocine algae highlighting the lineages in which soma differentiation has evolved (peach). This tree indicates a minimum of four and maximum of six independent origins of cellular differentiation. B Phylogeny of the volvocine algae highlighting the lineages that are isogamous (black), anisogamous (blue), and oogamous (names in pink only). Both phylogenies were inferred using maximum likelihood
Isogamy is the ancestral mode of sexual reproduction
Consistent with past studies, our results suggest that isogamy, the production of similar sized, motile gametes, is the ancestral mode of sexual reproduction among the volvocine algae (Fig. 5b and Additional file 1: Table S2). Isogamy is present in the unicellular genera Chlamydomonas and Vitreochlamys and is retained within the multicellular genera Astrephomene, Basichlamys, Gonium, Pandorina, Platydorina, Tetrabaena, Volvulina, and Yamagishiella. Colemanosphaera, Eudorina, Pleodorina, and Volvox have all evolved either anisogamy or oogamy [34, 63–65]. Anisogamy appears to have independently evolved at least three times from an isogamous ancestor: in section Volvox and in both Colemanosphaera and EVP. Conventional anisogamy, which consists of two motile gamete types of unequal size, appears in Colemanosphaera, Eudorina, and Pleodorina. This finding differs from those of Hanschen et al. [29], who reported that anisogamy independently evolved twice among the volvocine algae from isogamous ancestors. Oogamy, a specialized form of anisogamy where the female gamete is immotile and significantly larger than the motile, male gamete, is inferred to have independently evolved at least three times in lineages leading to section Volvox, V. gigas + V. powersii, and in the clade containing V. africanus, V. aureus, V. carteri, V. dissipatrix, V. obversus, V. ovalis, and V. tertius [34, 63, 64]. This last finding confirms results from Hanschen et al. [29], who also reported at least three independent origins of oogamy among the volvocine algae.
Platydorina caudata is sister to Colemanosphaera, and Pandorina is paraphyletic with respect to Volvulina
Within the PVC clade, our results add further support to the view that Pandorina is paraphyletic with respect to Volvulina (Figs. 2 and 3) [14, 25, 29, 33, 34, 39, 66]. Also, consistent with other multi-gene analyses Colemanosphaera appears to be monophyletic with high support (MLBS = 100, BPP = 1.0, CPP = 1.0) and sister to Platydorina (MLBS = 100, BPP = 1.0, CPP = 1.0) (Figs. 2 and 3) [31, 34, 35].
The genera Eudorina, Volvox, and Pleodorina are polyphyletic
Yamagishiella unicocca is sister to the Eudorina+Volvox+Pleodorina (EVP) clade, which encompasses two large subclades (MLBS=99, BPP=1.0, CPP=0.88) (Figs. 2 and 3). Our results support prior work suggesting that the genera Volvox, Eudorina and Pleodorina are not monophyletic [14, 25, 29–31, 33–36, 39, 67, 68]. The genus Volvox appears to be polyphyletic, with members represented across the two EVP subclades and the section Volvox clade. Members of both the Pleodorina and Eudorina genera are inferred to be polyphyletic across the two EVP subclades.
Historically, the genus Volvox has been divided into 4 sections – Copelandosphaera, Janetosphaera, Merrillosphaera, and Volvox – based on morphological [69] and molecular data [67]. A recent section-level revision of the genus Volvox [35] resulted in the creation and deletion of sections Besseyosphaera and Copelandosphaera, respectively. Hereafter, we will only refer to the revised taxonomic sections proposed by Nozaki et al. [35], with which our maximum likelihood, Bayesian inference, and coalescent-based results are in agreement (Additional file 1: Fig. S3) [35]. Our coalescentbased analysis suggests that each of the four sections is monophyletic, and that none encompass novel taxa not listed by Nozaki et al. [35] (Fig. 3). The branching order of our ML and BI analyses, however, suggests that section Merrillosphaera is not monophyletic (Additional file 1: Fig. S3) [35]. Our ML and BI analyses indicate that V. africanus, V. dissipatrix, V. ovalis, and V. tertius form a clade with V. aureus and P. japonica that is separate from the other Merrillosphaera taxa (MLBS=65, BPP=0.99) (Additional file 1: Fig. S3) [35]. In contrast, our CB analysis provides strong support (CPP=0.99) for the inference that the Merrillosphaera species are monophyletic (Fig. 3). Heeding our support values rather than only the branching order, we propose that the taxonomic system of the genus Volvox as outlined by Nozaki and colleagues [35] be retained.
Unicellular taxa are nested within the clade containing the colonial volvocine algae
Of the unicellular taxa, Chlamydomonas debaryana, C. globosa, C. reinhardtii, C. schloesseri, and Vitreochlamys ordinata are nested within the clade containing the colonial volvocine algae. Our results confirm prior studies showing the genus Vitreochlamys to be polyphyletic [38, 48]. The closest unicellular relative to the clade that contains the colonial algae + C. reinhardtii is suggested to be V. aulata (Figs. 2 and 3). This suggests that at least some members of Vitreochlamys are very closely related to the colonial volvocine algae. This relationship had been previously suggested by other studies [38, 70] including Nakazawa et al. [48], whose ultrastructural studies uncovered striking similarities in how these taxa formed pyrenoids and eyespot apparati (stigma), and established their tripartite cell walls.
Chlamydomonas is a polyphyletic genus [20, 38, 71, 72] composed of at least 500 species [72]. Although we sampled only a handful of Chlamydomonas species, our data support this view and broadly agree with the Chlamydomonas relationships inferred by Pröschold et al. [37], who used a combination of molecular phylogenetic analyses, sporangium wall lysis tests, and ultrastructural analyses. Our data strongly support C. schloesseri being sister to C. reinhardtii + C. globosa (MLBS = 100, BPP = 1.0, CPP = 1.0) and designating C. schloesseri as a “true” Chlamydomonas species, as suggested by Pröschold et al. [37]. Our study is also in agreement with a recent study by Craig et al. [20] that shows C. schloesseri being sister to C. reinhardtii + C. globosa. Also, like Pröschold et al. [37], our analyses indicate that C. debaryana SAG 70.81 is sister to Chlamydomonas schloesseri and its relatives (MLBS = 100, BPP = 1.0, CPP = 1.0). However, unlike the Pröschold et al. [37] study, which proposed that strain C. debaryana/Edaphochlamys debaryana (SAG 11-55a) is sister to the Tetrabaenaceae, our analyses support the view that C. debaryana/Edaphochlamys debaryana is more closely related to C. reinhardtii (MLBS = 91, BPP = 1.0, CPP = 0.81) than to the colonial algae. Our finding is further supported by Craig et al. [20] who inferred that C. debaryana/Edaphochlamys debaryana + Chlamydomonas sphaeroides is sister to the clade containing C. schloesseri + C. reinhardtii + C. globosa. Our placement of C. debaryana (SAG 11-55a) could be a result of limited (N = 6) sampling within the Chlamydomonas genus, which was more extensively sampled by Pröschold et al. [37] (N > 30). Consistent with a prior study, C. moewusii appears to be more distantly related to the colonial volvocines than is Vitreochlamys nekrassovii [14].
Conclusions
Using a 40-protein dataset, we have shown that the Tetrabaenaceae and the Goniaceae + Volvocaceae likely represent two independent origins of multicellularity and that cellular differentiation has independently evolved at least four, and possibly six times within the volvocine algae. The separate origin of multicellularity within the Tetrabaenaceae highlights the need for certain volvocine genomes, such as Vitreochlamys ordinata, to be sequenced, assembled and annotated. Because Vitreochlamys ordinata is the unicellular sister taxon to the multicellular Tetrabaenaceae, detailed analysis of its genome could give future researchers insight into how the simple form of multicellularity observed among the Tetrabaenaceae might have evolved.
Our results suggest that both multicellularity and cellular differentiation are evolutionarily labile traits within the volvocine algae. We have established a robust phylogeny of this group, which we hope will assist future efforts aimed at re-evaluating ancestral character states and understanding the origins of multicellularity and cellular differentiation in the volvocine green algae. The fruit of such efforts could then be used to carry out ancestral-state reconstruction of traits related to cellularity, differentiation, and gamete size as well as to discern the evolutionary history of gene families across the volvocine algae as a whole and within its major clades.
Methods
Strains and culture conditions
Algal strains used in this study were obtained from the National Institute for Environmental Studies (NIES, Japan), the Culture Collection of Algae at the University of Göttingen (SAG, Germany), and the Culture Collection of Algae at the University of Texas at Austin (UTEX, USA). Strain provenance and culture collection ID numbers are shown in Table 1, with previously published data designated with an asterisk. All cultures were grown at 20–26 °C under cool-white LED lamps (4300K) with an intensity of 2500–2700 lux under a 14-h light/10-h dark cycle. A detailed description of each strain’s morphology, degree of cellular differentiation, and gamete size, as well as the medium used to culture each strain is provided in Additional file 1: Tables S2 and S3 [73–77], respectively.
RNA extraction procedures
Two protocols were used to isolate total RNA: a modified version of the TRizol RNeasy method described by Matt and Umen [78] and a slightly modified QIAGEN RNeasy Plant Mini Kit protocol. For a detailed description of each, please see Additional file 2: RNA extraction procedures. Information on the protocol used for each strain is provided in Table 1.
Library preparation and sequencing
Before generating a sequencing library, RNA quality and quantity were assessed by Nanodrop and Qubit (Thermo Fisher Scientific, Waltham, MA 02451 USA). RNA integrity was evaluated using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA 95051, USA). mRNA was isolated using poly T beads, whereafter Illumina libraries were prepared using the NEBNext Ultra II Directional RNA Library Prep Kit. Library concentrations were determined fluorometrically; sequencing was carried out on the Illumina NovaSeq 6000 platform (Illumina, Inc., San Diego, CA 92122 USA) to generate 151 bp paired-end reads.
Quality control of reads
Raw read quality was assessed through FastQC v.0.11.8 with an additional FastQC assessment post-trimming. Quality control of the raw reads was completed with Trimmomatic v.0.39 [79] where the bases at the 5′ and 3′ end of each read are trimmed if found to be below a quality score of 3. A 4-base sliding window approach was used to trim the rest of the read once average quality fell below a score of 15; reads that were below a minimum length of 36 bases were discarded (LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36). If adapter content was detected by FastQC the additional ILLUMINACLIP step was used with the “TruSeq3-PE-2.fa” file provided by the Trimmomatic developers. If performed, the following ILLUMINACLIP parameters were used: 2:30:10 at the beginning of each command line. This allows for 2 “seed” mismatches where the seed is a short segment of the adapter that is being aligned in every section of the read. If more than 2 mismatches occurred, no trimming of the read occurred. Additionally, there had to be at least 30 matched bases in the paired-end palindrome read alignment and at least 10 matched bases between an adapter sequence and read.
De novo assembly
SOAPdenovo-Trans v1.0.4 [80] was used to assemble de novo transcriptomes from the quality filtered, paired-end reads using a k-mer size of 25 (SOAPdenovo-Trans-31mer all -s <config input file> -o <outfile> -K 25). GapCloser from the SOAPdenovo package was utilized to close gaps in each transcriptome using the same configuration file, which contains read-specific information and file paths, from the previous step (-b <config file> -a <.scafSeq file output by SOAPdenovo-Trans> -o <outfile> -l <max read length, int value> -t <thread number>). Default parameters were used for CD-HIT v4.8.1 [81] to reduce redundant transcripts from our de novo transcriptomes.
Orthologous gene identification for phylotranscriptomic analysis
The evolutionary history of the volvocine algae dates back at least 200 million years [25]. Over this timescale nucleotide sequences become saturated with substitutions, diminishing their phylogenetic utility [82]. Amino acid sequences were therefore chosen for our alignments, as they are known to be more reliable for ascertaining distant evolutionary relationships [83]. De Clerck and colleagues identified 58 nuclear protein-coding, single-copy genes that were members of highly conserved gene families across the green algae (Chlorophyceae, Prasinophytes, and Trebouxiophyceae) and land plants (Streptophyta) [84]. Their amino acid alignment of the 58 nuclear protein-coding genes that includes Chlamydomonas reinhardtii CC-503, Chlorella variabilis NC64A, Gonium pectorale NIES-2863, and Volvox carteri HK10 was kindly shared with our research team. Out of the 58 genes shared, we used 40 for our gene alignments. In order to identify those specific genes in the de novo transcriptomes of our taxa, a Basic Local Alignment Search Tool (BLAST) server was established in our lab, and a unique BLAST database for each taxon was created following the instructions in the BLAST manual. A BLASTP search using the C. reinhardtii CC-503, G. pectorale NIES-2863, and V. carteri HK10 genes from De Clerck et al. [84] as our query sequences enabled us to identify the orthologous genes for each of our taxa.
Gene sequence alignments and phylotranscriptomic analysis
The BLASTP results were used to identify the scaffold and open read frame where each gene was located in a strain’s transcriptome. Using a custom Python script (Additional file 3), each scaffold was extracted from its transcriptome and translated in the appropriate reading frame; then, the translated scaffold was added to an alignment file. For consistency, we generated de novo transcriptomes since we lacked a reference genome for most of our sequenced strains. At times, a gene was found to be incomplete for a given taxon due to assembler or sequencing error after manual examination. When this was determined to be the case, the gene was manually stitched together. This was done in a highly conservative manner: if we could not ascertain whether or not a gene was incomplete due to assembler or sequencing error, then it was excluded from the alignment for the given species. We treated the data from previously published studies in the same fashion as data generated in our lab by filtering the raw reads through quality trimming, then assembling de novo transcriptomes using the same programs and parameters (Table 1).
Amino acid sequences were aligned using MUSCLE v3.8.31 [85]. Alignments were also subjected to manual alignment in Aliview v1.26 [86]; extraneous data were trimmed, leaving only the aligned genes. Ambiguously aligned regions were eliminated from each alignment leaving only conserved and reliably aligned regions for phylogenetic analysis using the following parameters in Gblocks v0.91b [87]: -t=p -b3=8 -b4=2 -b5=h -b6=y. Phyutility v2.7.1 [88] was used to concatenate all gene alignment files.
Single-gene alignments were subjected to ML and BI analyses in order to infer single-gene phylogenies. Single-gene phylogenies were then further analyzed using a coalescent-based approach. The concatenated multi-gene alignment was partitioned so that the appropriate model of protein substitution was applied to each gene for the supermatrix phylogenetic approach under ML and BI.
The ML and BI analyses of the concatenated dataset used a partitioning strategy where the best evolutionary model for each gene was predicted by ProtTest v3.4.2 under the Akaike Information Criterion (AIC). For information regarding each predicted evolutionary model, please refer to Additional file 1: Table S4 [84, 89–91]. The ML analysis was conducted using IQtree v1.6.12 [92] under partition models [93]. Support values reported for the IQtree ML analysis were estimated through the bootstrap technique where 1000 ultrafast bootstrap replicates were generated [94]. The BI analysis was performed with MrBayes 3.2.7a [95] with 3 heated and 1 cold Markov chains, where trees were sampled every 1000 generations for a total of 1,000,000 generations with 1000 trees discarded at the beginning of each chain (ngen = 100000000, samplefreq = 1000, burnin = 1000, nruns = 4, nchains = 4, starttree = random).
ASTRAL [96] was used to perform the coalescent-based analysis where all 40 single-gene phylogenies produced by IQtree were used as the input after collapsing branches with low bootstrap support (< 10) using Newick Utilities v1.6 [97]. Posterior probabilities were assessed for the Bayesian and coalescent-based analyses in MrBayes and ASTRAL, respectively. Lastly, approximately unbiased (AU) tests with 100,000 RELL re-samplings were conducted to test certain key topologies and hypotheses using IQtree (-zw 100000 -au) (Additional file 1: Fig. S1).
Supplementary Information
Additional file 1 Table S1. Accession numbers of previously published RNA-seq data. Table S2. Information on sampled genera regarding cellularity, typical cell number, differentiation, and gamete size. Table S3. Medium used to culture each sequenced strain. Table S4. 40 genes with best predicted evolutionary model under the Akaike information criterion (AIC). Table S5. National Center for Biotechnology Institute (NCBI) accession numbers for BioProject PRJNA701495. Fig. S1. Approximately Unbiased (AU) tests comparing key hypotheses for our 40-protein concatenated dataset. Fig. S2. (A) Phylogeny that represents a replication of the Zhang et al. [51] results as they relate to the volvocine algae. (B) Phylogeny of the volvocine algae that represents a change in the branching order once more volvocine taxa are sampled for phylogenetic inference. Fig. S3. Phylogeny of the volvocine algae which highlights the four sections of genus Volvox as recognized by Nozaki et al. [35].
Additional file 2 Additional methods. Provides additional detail for methods used during RNA extractions, and of how we confirmed that Tetrabaena socialis N-571 and N-691 are conspecific.
Additional file 3. Python script. Python script that was used to mine individual de novo transcriptomes for single-copy genes located through local BLAST databases. This script reads BLAST input files in XML format and extracts and translates a scaffold in all six reading frames based on the scaffold ID of the first BLAST hit. Once the scaffold is translated in all six reading frames, the ‘hit sequence’ is located and written to a file with the ‘query sequence’ in FASTA format.
Acknowledgements
The authors gratefully acknowledge the core facilities at the Parker H. Petit Institute for Bioengineering and Bioscience at the Georgia Institute of Technology for the use of their shared equipment, services, and expertise. We are also grateful for useful discussion with and comments on the manuscript provided by Andy Chea, Emily Cook, Sa Geng, Armin Hallmann, Joseph Lachance, Bradley Olson, and James Umen. We thank Olivier De Clerck for kindly providing amino acid alignments. This material is based upon work while Herron was serving at the National Science Foundation.
Authors’ contributions
Performed the experiments (CRL), carried out the analyses (CRL, MDH), and wrote the manuscript (CRL, MDH, RFR). The authors have read and approved the final manuscript.
Funding
This work used the Hive cluster, which is supported by the National Science Foundation under grant number OAC-1828187. This research was supported in part through research cyberinfrastructure resources and services provided by the Partnership for an Advanced Computing Environment (PACE) at the Georgia Institute of Technology, Atlanta, Georgia, USA. This project was funded by NASA Astrobiology Grant# NNA17BB05A to Rosenzweig (PI) Herron (co-I); NSF DEB-1723293 to Herron (PI) Rosenzweig (co-PI); Rosenzweig NASA Exobiology Grant # 80NSSC20K0621 to Rosenzweig (PI); Rosenzweig Georgia Tech start-up account DE00000308.
Availability of data and materials
All raw data generated and used for this study have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under BioProject PRJNA701495 [98]. Accession numbers for our raw RNA-Seq reads range from SRR13719233 to SRR13719287, and accession numbers for our assembled contigs have been provided in Additional file 1: Table S4. For detailed information regarding accession number assignment to a specific taxon, please refer to Additional file 1: Table S5. Files containing our single-gene phylogenies and amino acid alignments, our IQtree partition file, and MrBayes configuration file have been uploaded to Dryad [99]. Previously published data used in this study from Hu et al. [47] and Featherston et al. [21] can be found under BioProject number PRJNA532307 [100] and PRJNA393411 [101], respectively.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Grosberg RK, Strathmann RR. The evolution of multicellularity: a minor major transition? Annu Rev Ecol Evol Syst. 2007;38(1):621–654. doi: 10.1146/annurev.ecolsys.36.102403.114735. [DOI] [Google Scholar]
- 2.Maynard Smith J, Szathmáry E. The major transitions in evolution. Oxford University Press; 1995. [Google Scholar]
- 3.Szathmáry E, Maynard Smith J. The major evolutionary transitions. Nature. 1995;374(6519):227–232. doi: 10.1038/374227a0. [DOI] [PubMed] [Google Scholar]
- 4.West SA, Fisher RM, Gardner A, Kiers ET. Major evolutionary transitions in individuality. Proc Natl Acad Sci. 2015;112(33):10112–10119. doi: 10.1073/pnas.1421402112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hallmann A. Evolution of reproductive development in the volvocine algae. Sex Plant Reprod. 2011;24(2):97–112. doi: 10.1007/s00497-010-0158-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wahl ME, Murray AW. Multicellularity makes somatic differentiation evolutionarily stable. Proc Natl Acad Sci. 2016;113(30):8362–8367. doi: 10.1073/pnas.1608278113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen L, Xiao S, Pang K, Zhou C, Yuan X. Cell differentiation and germ–soma separation in Ediacaran animal embryo-like fossils. Nature. 2014;516(7530):238–241. doi: 10.1038/nature13766. [DOI] [PubMed] [Google Scholar]
- 8.Han T, Runnegar B. Megascopic eukaryotic algae from the 2.1-billion-year-old negaunee iron-formation, Michigan. Science. 1992;257(5067):232–235. doi: 10.1126/science.1631544. [DOI] [PubMed] [Google Scholar]
- 9.Schirrmeister BE, de Vos JM, Antonelli A, Bagheri HC. Evolution of multicellularity coincided with increased diversification of cyanobacteria and the Great Oxidation Event. Proc Natl Acad Sci. 2013;110(5):1791–1796. doi: 10.1073/pnas.1209927110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herron MD, Borin JM, Boswell JC, Walker J, Chen I-CK, Knox CA, Boyd M, Rosenzweig F, Ratcliff WC. De novo origins of multicellularity in response to predation. Sci Rep. 2019;9(1):2328. doi: 10.1038/s41598-019-39558-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quintero-Galvis JF, Paleo-López R, Solano-Iguaran JJ, Poupin MJ, Ledger T, Gaitan-Espitia JD, Antoł A, Travisano M, Nespolo RF. Exploring the evolution of multicellularity in Saccharomyces cerevisiae under bacteria environment: an experimental phylogenetics approach. Ecol Evol. 2018;8(9):4619–4630. doi: 10.1002/ece3.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ratcliff WC, Denison RF, Borrello M, Travisano M. Experimental evolution of multicellularity. Proc Natl Acad Sci. 2012;109(5):1595–1600. doi: 10.1073/pnas.1115323109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ratcliff WC, Herron MD, Howell K, Pentz JT, Rosenzweig F, Travisano M. Experimental evolution of an alternating uni- and multicellular life cycle in Chlamydomonas reinhardtii. Nat Commun. 2013;4:1–7. doi: 10.1038/ncomms3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herron MD, Michod RE. Evolution of complexity in the volvocine algae: transitions in individuality through Darwin’s eye. Evolution. 2008;62(2):436–451. doi: 10.1111/j.1558-5646.2007.00304.x. [DOI] [PubMed] [Google Scholar]
- 15.Kirk DL. A twelve-step program for evolving multicellularity and a division of labor. BioEssays. 2005;27(3):299–310. doi: 10.1002/bies.20197. [DOI] [PubMed] [Google Scholar]
- 16.Koyanagi KO. Inferring cell differentiation processes based on phylogenetic analysis of genome-wide epigenetic information: hematopoiesis as a model case. Genome Biol Evol. 2015;7(3):699–705. doi: 10.1093/gbe/evv024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Leeuwenhoek A. Part of a letter from Mr Antony van Leeuwenhoek, concerning the worms in sheeps livers, gnats, and animalcula in the excrements of frogs. Philos Trans R Soc Lond. 1700;22:509–518. [Google Scholar]
- 18.Kirk DL. Germ–soma differentiation in Volvox. Dev Biol. 2001;238(2):213–223. doi: 10.1006/dbio.2001.0402. [DOI] [PubMed] [Google Scholar]
- 19.Schmitt R. Differentiation of germinal and somatic cells in Volvox carteri. Curr Opin Microbiol. 2003;6(6):608–613. doi: 10.1016/j.mib.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 20.Craig RJ, Hasan AR, Ness RW, Keightley PD. Comparative genomics of Chlamydomonas. Plant Cell. 2021;33(4):1016–1041. doi: 10.1093/plcell/koab026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Featherston J, Arakaki Y, Hanschen ER, Ferris PJ, Michod RE, Olson BJSC, Nozaki H, Durand PM. The 4-celled Tetrabaena socialis nuclear genome reveals the essential components for genetic control of cell number at the origin of multicellularity in the volvocine lineage. Mol Biol Evol. 2018;35(4):855–870. doi: 10.1093/molbev/msx332. [DOI] [PubMed] [Google Scholar]
- 22.Hanschen ER, Marriage TN, Ferris PJ, et al. The Gonium pectorale genome demonstrates co-option of cell cycle regulation during the evolution of multicellularity. Nat Commun. 2016;7:1–10. doi: 10.1038/ncomms11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Marechal-Drouard L, Marshall WF, Qu LH, Nelson DR, Sanderfoot AA, Spalding MH, Kapitonov VV, Ren Q, Ferris P, Lindquist E, Shapiro H, Lucas SM, Grimwood J, Schmutz J, Cardol P, Cerutti H, Chanfreau G, Chen CL, Cognat V, Croft MT, Dent R, Dutcher S, Fernandez E, Fukuzawa H, Gonzalez-Ballester D, Gonzalez-Halphen D, Hallmann A, Hanikenne M, Hippler M, Inwood W, Jabbari K, Kalanon M, Kuras R, Lefebvre PA, Lemaire SD, Lobanov AV, Lohr M, Manuell A, Meier I, Mets L, Mittag M, Mittelmeier T, Moroney JV, Moseley J, Napoli C, Nedelcu AM, Niyogi K, Novoselov SV, Paulsen IT, Pazour G, Purton S, Ral JP, Riano-Pachon DM, Riekhof W, Rymarquis L, Schroda M, Stern D, Umen J, Willows R, Wilson N, Zimmer SL, Allmer J, Balk J, Bisova K, Chen CJ, Elias M, Gendler K, Hauser C, Lamb MR, Ledford H, Long JC, Minagawa J, Page MD, Pan J, Pootakham W, Roje S, Rose A, Stahlberg E, Terauchi AM, Yang P, Ball S, Bowler C, Dieckmann CL, Gladyshev VN, Green P, Jorgensen R, Mayfield S, Mueller-Roeber B, Rajamani S, Sayre RT, Brokstein P, Dubchak I, Goodstein D, Hornick L, Huang YW, Jhaveri J, Luo Y, Martinez D, Ngau WCA, Otillar B, Poliakov A, Porter A, Szajkowski L, Werner G, Zhou K, Grigoriev IV, Rokhsar DS, Grossman AR. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318(5848):245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prochnik SE, Umen J, Nedelcu AM, Hallmann A, Miller SM, Nishii I, Ferris P, Kuo A, Mitros T, Fritz-Laylin LK, Hellsten U, Chapman J, Simakov O, Rensing SA, Terry A, Pangilinan J, Kapitonov V, Jurka J, Salamov A, Shapiro H, Schmutz J, Grimwood J, Lindquist E, Lucas S, Grigoriev IV, Schmitt R, Kirk D, Rokhsar DS. Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science. 2010;329(5988):223–226. doi: 10.1126/science.1188800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herron MD, Hackett JD, Aylward FO, Michod RE. Triassic origin and early radiation of multicellular volvocine algae. Proc Natl Acad Sci. 2009;106(9):3254–3258. doi: 10.1073/pnas.0811205106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brunet T, King N. The origin of animal multicellularity and cell differentiation. Dev Cell. 2017;43(2):124–140. doi: 10.1016/j.devcel.2017.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lang NJ. Electron microscopy of the Volvocaceae and Astrephomenaceae. Am J Bot. 1963;50(3):280–300. doi: 10.1002/j.1537-2197.1963.tb12235.x. [DOI] [Google Scholar]
- 28.Larson A, Kirk MM, Kirk DL. Molecular phylogeny of the volvocine flagellates. Mol Biol Evol. 1992;9(1):85–105. doi: 10.1093/oxfordjournals.molbev.a040710. [DOI] [PubMed] [Google Scholar]
- 29.Hanschen ER, Herron MD, Wiens JJ, Nozaki H, Michod RE. Multicellularity drives the evolution of sexual traits. Am Nat. 2018;192(3):E93–E105. doi: 10.1086/698301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herron MD, Desnitskiy AG, Michod RE. Evolution of developmental programs in Volvox (Chlorophyta) J Phycol. 2010;46(2):316–324. doi: 10.1111/j.1529-8817.2009.00803.x. [DOI] [Google Scholar]
- 31.Hanschen ER, Herron MD, Wiens JJ, Nozaki H, Michod RE. Repeated evolution and reversibility of self-fertilization in the volvocine green algae*. Evolution. 2018;72(2):386–398. doi: 10.1111/evo.13394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakada T, Tomita M, Nozaki H. Volvulina compacta (Volvocaceae, Chlorophyceae), new to Japan, and its phylogenetic position. J Jpn Bot. 2010;85:364–369. [Google Scholar]
- 33.Nozaki H, Misawa K, Kajita T, Kato M, Nohara S, Watanabe MM. Origin and evolution of the colonial Volvocales (Chlorophyceae) as inferred from multiple, chloroplast gene sequences. Mol Phylogenet Evol. 2000;17(2):256–268. doi: 10.1006/mpev.2000.0831. [DOI] [PubMed] [Google Scholar]
- 34.Nozaki H, Yamada TK, Takahashi F, Matsuzaki R, Nakada T. New “missing link” genus of the colonial volvocine green algae gives insights into the evolution of oogamy. BMC Evol Biol. 2014;14:1–11. doi: 10.1186/1471-2148-14-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nozaki H, Matsuzaki R, Yamamoto K, Kawachi M, Takahashi F. Delineating a new heterothallic species of Volvox (Volvocaceae, Chlorophyceae) using new strains of “Volvox africanus.”. PLoS ONE. 2015;10:e0142632. doi: 10.1371/journal.pone.0142632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grochau-Wright ZI, Hanschen ER, Ferris PJ, Hamaji T, Nozaki H, Olson BJSC, Michod RE. Genetic basis for soma is present in undifferentiated volvocine green algae. J Evol Biol. 2017;30(6):1205–1218. doi: 10.1111/jeb.13100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pröschold T, Darienko T, Krienitz L, Coleman AW. Chlamydomonas schloesseri sp. nov. (Chlamydophyceae, Chlorophyta) revealed by morphology, autolysin cross experiments, and multiple gene analyses. Phytotaxa. 2018;362(1):21–38. doi: 10.11646/phytotaxa.362.1.2. [DOI] [Google Scholar]
- 38.Nakada T, Tsuchida Y, Tomita M. Improved taxon sampling and multigene phylogeny of unicellular chlamydomonads closely related to the colonial volvocalean lineage Tetrabaenaceae-Goniaceae-Volvocaceae (Volvocales, Chlorophyceae) Mol Phylogenet Evol. 2019;130:1–8. doi: 10.1016/j.ympev.2018.09.013. [DOI] [PubMed] [Google Scholar]
- 39.Coleman AW. Phylogenetic analysis of “Volvocacae” for comparative genetic studies. Proc Natl Acad Sci. 1999;96(24):13892–13897. doi: 10.1073/pnas.96.24.13892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu Y, Xing W, Song H, Zhu H, Liu G, Hu Z. Evolutionary analysis of unicellular species in chlamydomonadales through chloroplast genome comparison with the colonial volvocine algae. Front Microbiol. 2019;10:1351. doi: 10.3389/fmicb.2019.01351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong W, Liu J, Yu J, Wang L, Zhou S. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS ONE. 2012;7(4):e35071. doi: 10.1371/journal.pone.0035071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jakob SS, Blattner FR. A chloroplast genealogy of Hordeum (Poaceae): long-term persisting haplotypes, incomplete lineage sorting, regional extinction, and the consequences for phylogenetic inference. Mol Biol Evol. 2006;23(8):1602–1612. doi: 10.1093/molbev/msl018. [DOI] [PubMed] [Google Scholar]
- 43.Xu B, Wu N, Gao X-F, Zhang L-B. Analysis of DNA sequences of six chloroplast and nuclear genes suggests incongruence, introgression, and incomplete lineage sorting in the evolution of Lespedeza (Fabaceae) Mol Phylogenet Evol. 2012;62(1):346–358. doi: 10.1016/j.ympev.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 44.Rose J, Toledo CA, Lemmon EM, Lemmon AR, Sytsma K. Out of sight, out of mind: widespread nuclear and plastid-nuclear discordance in the flowering plant genus Polemonium (Polemoniaceae) suggests widespread historical gene flow despite limited nuclear signal. Syst Biol. 2020;70:162–180. doi: 10.1093/sysbio/syaa049. [DOI] [PubMed] [Google Scholar]
- 45.Soltis DE, Kuzoff RK. Discordance between nuclear and chloroplast phylogenies in the Heuchera group (Saxifragaceae) Evolution. 1995;49(4):727–742. doi: 10.1111/j.1558-5646.1995.tb02309.x. [DOI] [PubMed] [Google Scholar]
- 46.Yu W-B, Huang P-H, Li D-Z, Wang H. Incongruence between nuclear and chloroplast DNA phylogenies in Pedicularis section Cyathophora (Orobanchaceae) PLoS ONE. 2013;8(9):e74828. doi: 10.1371/journal.pone.0074828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu Y, Xing W, Song H, Hu Z, Liu G. Comparison of colonial volvocine algae based on phylotranscriptomic analysis of gene family evolution and natural selection. Eur J Phycol. 2020;55(1):100–112. doi: 10.1080/09670262.2019.1663269. [DOI] [Google Scholar]
- 48.Nakazawa A, Krienitz L, Nozaki H. Taxonomy of the unicellular green algal genus Vitreochlamys (Volvocales), based on comparative morphology of cultured material. Eur J Phycol. 2001;36(2):113–128. doi: 10.1080/09670260110001735268. [DOI] [Google Scholar]
- 49.Nozaki H. Morphology and evolution of sexual reproduction in the Volvocaceae (Chlorophyta) J Plant Res. 1996;109(3):353–361. doi: 10.1007/BF02344484. [DOI] [Google Scholar]
- 50.Nozaki H, Itoh M. Phylogenetic relationships within the colonial Volvocales (Chlorophyta) inferred from cladistic analysis based on morphological data. J Phycol. 1994;30(2):353–365. doi: 10.1111/j.0022-3646.1994.00353.x. [DOI] [Google Scholar]
- 51.Zhang Z, Qu C, Yao R, Nie Y, Xu C, Miao J, Zhong B. The parallel molecular adaptations to the Antarctic cold environment in two psychrophilic green algae. Genome Biol Evol. 2019;11(7):1897–1908. doi: 10.1093/gbe/evz104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nozaki H, Ohtani S. Gonium sociale (Volvocales, Chlorophyta) from Antarctica. Jpn J Phycol. 1992;40:267–271. [Google Scholar]
- 53.Mai JC, Coleman AW. The internal transcribed spacer 2 exhibits a common secondary structure in green algae and flowering plants. J Mol Evol. 1997;44(3):258–271. doi: 10.1007/PL00006143. [DOI] [PubMed] [Google Scholar]
- 54.Hedtke SM, Townsend TM, Hillis DM. Resolution of phylogenetic conflict in large data sets by increased taxon sampling. Syst Biol. 2006;55(3):522–529. doi: 10.1080/10635150600697358. [DOI] [PubMed] [Google Scholar]
- 55.Pollock DD, Zwickl DJ, McGuire JA, Hillis DM. Increased taxon sampling is advantageous for phylogenetic inference. Syst Biol. 2002;51(4):664–671. doi: 10.1080/10635150290102357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zwickl DJ, Hillis DM. Increased taxon sampling greatly reduces phylogenetic error. Syst Biol. 2002;51(4):588–598. doi: 10.1080/10635150290102339. [DOI] [PubMed] [Google Scholar]
- 57.Nozaki H, Itoh M, Watanabe MM, Kuroiwa T. Ultrastructure of the vegetative colonies and systematic position of Basichlamys (Volvocales, Chlorophyta) Eur J Phycol. 1996;31(1):67–72. doi: 10.1080/09670269600651211a. [DOI] [Google Scholar]
- 58.Nozaki H, Ito M, Uchida H, Watanabe MM, Takahashi H, Kuroiwa T. Phylogenetic analysis of Yamagishlella and Platydorina (volvocaceae, chlorophyta) based on rbcl gene sequences. J Phycol. 1997;33(2):272–278. doi: 10.1111/j.0022-3646.1997.00272.x. [DOI] [Google Scholar]
- 59.Nozaki H, Itoh M, Sano R, Uchida H, Watanabe MM, Kuroiwa T. Phylogenetic relationships within the colonial volvocales (chlorophyta) inferred from rbcl gene sequence data. J Phycol. 1995;31(6):970–979. doi: 10.1111/j.0022-3646.1995.00970.x. [DOI] [Google Scholar]
- 60.Nozaki H. Ultrastructure of the extracellular matrix of Gonium (Volvocales, Chlorophyta) Phycologia. 1990;29(1):1–8. doi: 10.2216/i0031-8884-29-1-1.1. [DOI] [Google Scholar]
- 61.Nozaki H, Kuroiwa T. Ultrastructure of the extracellular matrix and taxonomy of Eudorina, Pleodorina and Yamagishiella gen. nov.(Volvocaceae, Chlorophyta) Phycologia. 1992;31(6):529–541. doi: 10.2216/i0031-8884-31-6-529.1. [DOI] [Google Scholar]
- 62.Kirk DL, Birchem R, King N. The extracellular matrix of Volvox: a comparative study and proposed system of nomenclature. J Cell Sci. 1986;80(1):207–231. doi: 10.1242/jcs.80.1.207. [DOI] [PubMed] [Google Scholar]
- 63.Herron MD. Origins of multicellular complexity: Volvox and the volvocine algae. Mol Ecol. 2016;25(6):1213–1223. doi: 10.1111/mec.13551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.da Silva J, Drysdale VL. Isogamy in large and complex volvocine algae is consistent with the gamete competition theory of the evolution of anisogamy. Proc R Soc B Biol Sci. 2018;285(1890):20181954. doi: 10.1098/rspb.2018.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Umen JG. Evolution of sex and mating loci: an expanded view from Volvocine algae. Curr Opin Microbiol. 2011;14(6):634–641. doi: 10.1016/j.mib.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Coleman AW. Biogeography and speciation in the Pandorina/Volvulina (chlorophyta) superclade. J Phycol. 2001;37(5):836–851. doi: 10.1046/j.1529-8817.2001.01043.x. [DOI] [Google Scholar]
- 67.Nozaki H. Origin and evolution if the genera Pleodorina and Volvox (Volvocales) Biologia. 2003;58:425–431. [Google Scholar]
- 68.Nozaki H, Ott FD, Coleman AW. Morphology, molecular phylogeny and taxonomy of two new species of Pleodorina (Volvoceae, Chlorophyceae) J Phycol. 2006;42(5):1072–1080. doi: 10.1111/j.1529-8817.2006.00255.x. [DOI] [Google Scholar]
- 69.Smith GM. A comparative study of the species of Volvox. Trans Am Microsc Soc. 1944;63(4):265–310. doi: 10.2307/3223302. [DOI] [Google Scholar]
- 70.Nozaki H, Ohta N, Takano H, Watanabe MM. Reexamination of phylogenetic relationships within the colonial Volvocales (Chlorophyta): an analysis of atpB and rbcL gene sequences. J Phycol. 1999;35(1):104–112. doi: 10.1046/j.1529-8817.1999.3510104.x. [DOI] [Google Scholar]
- 71.Nakada T, Misawa K, Nozaki H. Molecular systematics of Volvocales (Chlorophyceae, Chlorophyta) based on exhaustive 18S rRNA phylogenetic analyses. Mol Phylogenet Evol. 2008;48(1):281–291. doi: 10.1016/j.ympev.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 72.Pröschold T, Marin B, Schlösser UG, Melkonian M. Molecular phylogeny and taxonomic revision of Chlamydomonas (Chlorophyta). I. Emendation of Chlamydomonas Ehrenberg and Chloromonas Gobi, and description of Oogamochlamys gen. nov. and Lobochlamys gen. nov. Protist. 2001;152:36. doi: 10.1078/1434-4610-00068. [DOI] [PubMed] [Google Scholar]
- 73.Kato S. Laboratory culture and morphology of Colacium vesiculosum Ehrb. (Euglenophyceae) Jpn J Phycol. 1982;30:63–67. [Google Scholar]
- 74.Ichimura T, Watanabe M. The Closterium calosporum complex from the Ryukyu Islands - variation and taxonomical problems. Mem Natn Sci Mus Tokyo. 1974. p. 7.
- 75.Ichimura T. The life cycle and its control in some species of Closterium, with special reference to the biological species problems. 1973. [Google Scholar]
- 76.Starr RC. In: Special methods-dry soil samples. Stein JR, editor. Cambridge: Cambridge University Press; 1973. [Google Scholar]
- 77.Sueoka N. Mitotic replication of deoxyribonucleic acid in Chlamydomonas reinhardtii. Natl Acad Sci U S Am. 1960;46(1):83–91. doi: 10.1073/pnas.46.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Matt GY, Umen JG. Cell-type transcriptomes of the multicellular green alga Volvox carteri yield insights into the evolutionary origins of germ and somatic differentiation programs. G3amp58 GenesGenomesGenetics. 2018;8:531–550. doi: 10.1534/g3.117.300253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xie Y, Wu G, Tang J, Luo R, Patterson J, Liu S, Huang W, He G, Gu S, Li S, Zhou X, Lam TW, Li Y, Xu X, Wong GKS, Wang J. SOAPdenovo-Trans: de novo transcriptome assembly with short RNA-Seq reads. Bioinformatics. 2014;30(12):1660–1666. doi: 10.1093/bioinformatics/btu077. [DOI] [PubMed] [Google Scholar]
- 81.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28(23):3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hasegawa M, Hashimoto T. Ribosomal RNA trees misleading? Nature. 1993;361(6407):23. doi: 10.1038/361023b0. [DOI] [PubMed] [Google Scholar]
- 83.Loomis WF, Smith DW. Molecular phylogeny of Dictyostelium discoideum by protein sequence comparison. Proc Natl Acad Sci. 1990;87(23):9093–9097. doi: 10.1073/pnas.87.23.9093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.De Clerck O, Kao S-M, Bogaert KA, et al. Insights into the evolution of multicellularity from the sea lettuce genome. Curr Biol. 2018;28:2921–2933.e5. doi: 10.1016/j.cub.2018.08.015. [DOI] [PubMed] [Google Scholar]
- 85.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Larsson A. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics. 2014;30(22):3276–3278. doi: 10.1093/bioinformatics/btu531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17(4):540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 88.Smith SA, Dunn CW. Phyutility: a phyloinformatics tool for trees, alignments and molecular data. Bioinformatics. 2008;24(5):715–716. doi: 10.1093/bioinformatics/btm619. [DOI] [PubMed] [Google Scholar]
- 89.Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008;25(7):1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 90.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Bioinformatics. 1992;8(3):275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 91.Dayhoff MO, Schwartz RM, Orcutt BC. A model of evolutionary change in proteins. Atlas Protein Seq Struct. 1978;5:345–352. [Google Scholar]
- 92.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32(1):268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chernomor O, von Haeseler A, Minh BQ. Terrace aware data structure for phylogenomic inference from supermatrices. Syst Biol. 2016;65(6):997–1008. doi: 10.1093/sysbio/syw037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 2018;35(2):518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang C, Rabiee M, Sayyari E, Mirarab S. ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinform. 2018;19(S6):153. doi: 10.1186/s12859-018-2129-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Junier T, Zdobnov EM. The Newick utilities: high-throughput phylogenetic tree processing in the UNIX shell. Bioinformatics. 2010;26(13):1669–1670. doi: 10.1093/bioinformatics/btq243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lindsey C, Rosenzweig F, Herron MD. Phylotranscriptomics points to multiple independent origins of multicellularity and cellular differentiation in the volvocine algae. NCBI BioProjects. https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA701495. [DOI] [PMC free article] [PubMed]
- 99.Lindsey C, Rosenzweig F, Herron MD. Phylotranscriptomics points to multiple independent origins of multicellularity and cellular differentiation in the volvocine algae. Dryad. 10.5061/dryad.v9s4mw6w5. [DOI] [PMC free article] [PubMed]
- 100.Hu Y, Xing W, Song H, Hu Z, Liu G. Comparison of colonial volvocine algae based on phylotranscriptomic analysis of gene family evolution and natural selection. NCBI BioProjects. https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA532307.
- 101.Featherston J, Arakaki Y, Hanschen ER, Ferris PJ, Michod RE, Olson BJSC, et al. The 4-celled Tetrabaena socialis nuclear genome reveals the essential components for genetic control of cell number at the origin of multicellularity in the volvocine lineage. NCBI BioProjects. https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA393411. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1 Table S1. Accession numbers of previously published RNA-seq data. Table S2. Information on sampled genera regarding cellularity, typical cell number, differentiation, and gamete size. Table S3. Medium used to culture each sequenced strain. Table S4. 40 genes with best predicted evolutionary model under the Akaike information criterion (AIC). Table S5. National Center for Biotechnology Institute (NCBI) accession numbers for BioProject PRJNA701495. Fig. S1. Approximately Unbiased (AU) tests comparing key hypotheses for our 40-protein concatenated dataset. Fig. S2. (A) Phylogeny that represents a replication of the Zhang et al. [51] results as they relate to the volvocine algae. (B) Phylogeny of the volvocine algae that represents a change in the branching order once more volvocine taxa are sampled for phylogenetic inference. Fig. S3. Phylogeny of the volvocine algae which highlights the four sections of genus Volvox as recognized by Nozaki et al. [35].
Additional file 2 Additional methods. Provides additional detail for methods used during RNA extractions, and of how we confirmed that Tetrabaena socialis N-571 and N-691 are conspecific.
Additional file 3. Python script. Python script that was used to mine individual de novo transcriptomes for single-copy genes located through local BLAST databases. This script reads BLAST input files in XML format and extracts and translates a scaffold in all six reading frames based on the scaffold ID of the first BLAST hit. Once the scaffold is translated in all six reading frames, the ‘hit sequence’ is located and written to a file with the ‘query sequence’ in FASTA format.
Data Availability Statement
All raw data generated and used for this study have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under BioProject PRJNA701495 [98]. Accession numbers for our raw RNA-Seq reads range from SRR13719233 to SRR13719287, and accession numbers for our assembled contigs have been provided in Additional file 1: Table S4. For detailed information regarding accession number assignment to a specific taxon, please refer to Additional file 1: Table S5. Files containing our single-gene phylogenies and amino acid alignments, our IQtree partition file, and MrBayes configuration file have been uploaded to Dryad [99]. Previously published data used in this study from Hu et al. [47] and Featherston et al. [21] can be found under BioProject number PRJNA532307 [100] and PRJNA393411 [101], respectively.