Abstract
A rapid detection test for SARS-CoV-2 is urgently required to monitor virus spread and containment. Here, we describe a test that uses nanoprobes, which are gold nanoparticles functionalized with an aptamer specific to the spike membrane protein of SARS-CoV-2. An enzyme-linked immunosorbent assay confirms aptamer binding with the spike protein on gold surfaces. Protein recognition occurs by adding a coagulant, where nanoprobes with no bound protein agglomerate while those with sufficient bound protein do not. Using plasmon absorbance spectra, the nanoprobes detect 16 nM and higher concentrations of spike protein in phosphate-buffered saline. The time-varying light absorbance is examined at 540 nm to determine the critical coagulant concentration required to agglomerates the nanoprobes, which depends on the protein concentration. This approach detects 3540 genome copies/μl of inactivated SARS-CoV-2.
Keywords: SARS-CoV-2, Spike protein, Aptamer, Biosensing, Surface plasmon resonance, Gold nanoparticle
Graphical abstract
1. Introduction
Control over a viral pandemic requires widespread testing to arrest infection spread. The most widely used method is based on nucleic acid amplification, where viral ribonucleic acid (RNA) extracted from a patient sample, such as a nasal swab or saliva, is used to make complementary deoxyribonucleic acid (DNA) strands through reverse transcriptase. Specific regions of the target nucleic acid are then amplified using polymerase chain reaction (PCR) [1,2].
Although molecular diagnostic point of care devices are available [3,4], these tests are typically verified in labs through expensive and complex methods that take hours to complete. The relative lack of testing centers in many jurisdictions, requirement for trained personnel and specialized equipment, and a global reagent shortage have hindered widespread testing, allowing the SARS-CoV-2 infection to persist [5]. As the virus continues to spread and infect, it naturally mutates over time, creating more potent variants of concern that fuel secondary outbreaks, e.g., those first identified in the UK (B.1.1.7), South Africa (B.1.351), Brazil (P.1), and most recently India (B.1.617). Hence, there is an immediate need for a scalable, instrument-free, inexpensive, and accessible rapid test for this viral infection [6].
Immunoassay identifies SARS-CoV-2 infection by targeting virus-specific antigens or antibodies (Abs). For instance, immunoglobulin G (IgG) Abs generated by the immune system and present in the blood or nasal swab samples can be detected using enzyme-linked immunosorbent assay (ELISA) [7], lateral flow assays [8], luminescent immunoassays [9], either in a lab or through a point of care (POC) test. Direct instrument-free antigen detection of SARS-CoV-2 fragments from patient samples is possible [10,11], but such a test is less sensitive than one based on nucleic acid amplification.
We use aptamers to improve the detection sensitivity. Aptamers are more stable than Abs and offer better functionality for protein detection [12]. These single-stranded DNA or RNA molecules bind to protein targets by folding into a three-dimensional (3D) conformation. They are developed using the systematic evolution of ligands by exponential enrichment (SELEX). A large random library of single-stranded nucleic acids (RNA or DNA), typically 35–50 nucleotides in length [13], is screened against a target of interest, and the ones that bind are amplified and screened repeatedly [14]. Aptamers are used for biosensing with electrochemical impedance spectroscopy, colorimetric and fluorescent methods [15]. and to detect the nucleocapsid [16] and spike [17] proteins of the SARS-CoV-2 virus.
Metal nanoparticles have size-dependent optical properties. When a small spherical metallic nanoparticle is irradiated with light, the corresponding oscillating electric field causes conduction electrons to also oscillate coherently, thus displacing the conduction electron charge cloud relative to the nanoparticle nuclei. When the frequency of the oscillating electric field matches the oscillations of these electrons, resonant energy transfer occurs, which is referred to as dipole particle plasmon resonance [18]. Pristine ∼20 nm AuNPs suspended in a colloid have a plasmon resonance peak at ∼520 nm wavelength. As the nanoparticle size increases, this peak shifts to higher wavelengths since scattering and absorption increase [19].
Since gold nanoparticles (AuNPs) exhibit superior chemical stability, they are used as a biosensing material for colorimetric assays [20]. Nanoparticles conjugated with aptamers have been used as protein labels to image and track endocytosis [21], as immunosensors [22], and to detect the SARS-CoV-2, influenza A [23], hantaan, H5N1 and HIV [24] viruses, thrombin [25], cancer cells [26], and IL-6 [27].
Nanoparticle agglomeration assays are used to detect protein-protein binding [28], nucleic acids [[29], [30], [31], [32], [33]], divalent heavy metal ions, such as Pb2+, Cd2+, Hg2+, and small organic molecules [34]. Many AuNP agglomeration tests have no specific affinity to the target but there are examples of functional AuNPs. Nanoparticle surfaces capped with different surfactants can sense differential protein adsorption [35]. Pathogens can also be detected based on their affinity to different shaped AuNPs [36]. An agglomeration test based on molecular recognition uses an aptamer-linked AuNP assay that detects thrombin [37] and ATP [38].
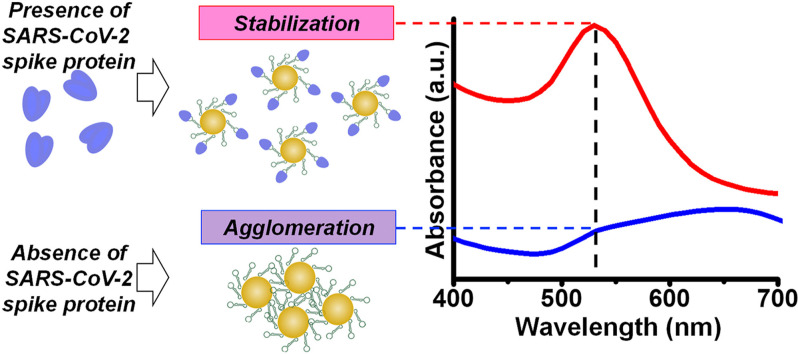
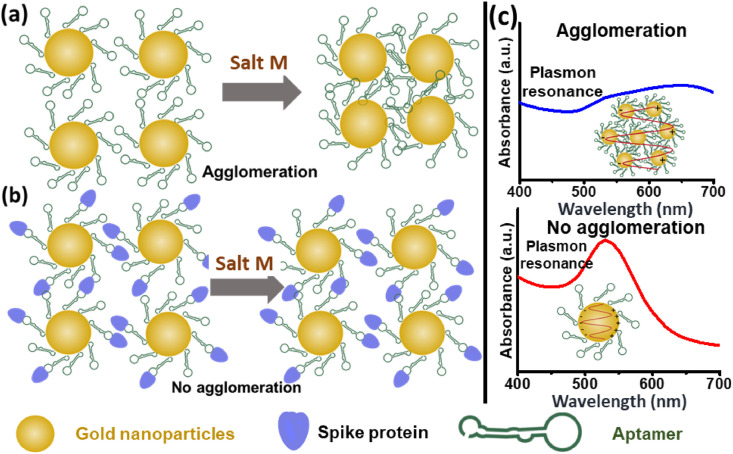
Here, we use molecular recognition by functionalizing AuNPs with molecules with specific affinity to the target analyte. We use a colloidal aqueous suspension of AuNPs functionalized with specific aptamers to detect the spike protein on the SARS-CoV-2 membrane by binding with it. The sensing signal is enhanced by adding a coagulant, MgCl2 salt solution (Salt M), to induce nanoparticle agglomeration, which depends on the amount of aptamer-protein binding. The extent of agglomeration influences the optical properties of the nanoparticle-containing suspension. For control samples with no spike protein, the nanoprobes aggregate as illustrated in Fig. 1 (a). We hypothesize that the counterions from the coagulant Salt M form complexes with the surface charges on the AuNPs [39], reducing the net charge of a nanoprobe, which promotes agglomeration and shifts surface plasmon absorbance towards higher wavelengths [40].
Fig. 1.
Schematic illustrating the principle of the SARS-CoV-2 test. (a) Nanoprobes are AuNPs functionalized with aptamers in an aqueous suspension. When the SARS-CoV-2 spike protein is absent from the colloid, addition of the coagulant Salt M neutralizes surface charges on the nanoprobes, inducing their agglomeration. (b) Nanoprobes with spike protein bind with aptamers and resist agglomeration, which depends on the extent of this binding. Protein binding provides additional charge to the nanoparticle, enhancing steric stabilization. (c) Plasmon absorbance spectra for the nanoprobes show how agglomeration in a colloidal suspension broadens the absorbance spectrum and shifts peak absorbance to higher wavelengths. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In contrast, when the nanoprobe suspension is mixed with a sample containing spike protein, the aptamers on the surfaces of nanoprobes bind specifically with the spike protein, adding electrostatic charge [41] and enhancing steric stabilization [42], as illustrated in Fig. 1(b). The net effect of the binding is to diminish nanoprobe agglomeration, decreasing the wavelength shift in surface plasmon absorbance. The absorbance intensity spectra with and without agglomeration are schematically illustrated in Fig. 1(c). Changes in the absorbance intensity, a surrogate for nanoprobe agglomeration, are measured with a spectrophotometer. Depending on the amount of spike protein bound to the aptamers on the nanoprobe surfaces, a critical coagulant concentration (C c) induces complete nanoparticle agglomeration for all cases. Hence, the Salt M concentration is held below the value of C c for a specific sample.
2. Materials and methods
2.1. Aptamers
Aptamer-4C(5′-ATCCAGAGTGACGCAGCATTTCATCGGGTCCAAAAGGGG-CTGCTCGGGATTGCGGATATGGACACGT-3′) [43] was obtained from Aptagen LLC, Jacobus, USA. The manufacturer synthesized the aptamers with thiols on their 5′ ends, using high performance liquid chromatography (HPLC) for purification and verification. The secondary structure is provided in Fig. S1 of the supplementary information. The thiolated-end group allowed facile immobilization of these aptamers on gold surfaces. The aptamer dilution buffer was phosphate buffer saline (PBS, Thermofisher, Canada) with 0.55 mM of MgCl2 (Sigma Aldrich). Before aptamer immobilization, the disulfide bonds between the thiols at the end of these aptamers were reduced by incubating with Tris(2-carboxyethyl)phosphine hydrochloride (TCEP, Sigma Aldrich, Canada) using 10 times the molar concentration of the aptamer, as suggested by the manufacturer. Hence, 100 μM of aptamer solution was incubated with 1 mM TCEP for an hour in darkness. After reduction, these aptamers were used without further purification or dialysis since TCEP does not interfere with thiol-gold binding. These reduced aptamers were refolded and returned to active conformation in a water bath at 87 °C for 5 min and then cooled over an ice bath.
2.2. ELISA on the gold-coated glass slide
Glass slides, each with a 100 nm gold film and 15 nm Cr adhesion layer, were purchased from Angstrom Engineering, Kitchener, Canada. 18-well sticky-Slides were purchased from Ibidi, Germany. 0.55 mM MgCl2 was added to PBS and used as a buffer. The receptor-binding domain of the spike protein that has a fusion Fc protein used in this study was synthesized by the Structural Genomics Center at the University of Toronto. Secondary rabbit anti-human horseradish peroxidase (HRP) antibody was procured from Jackson Immunoresearch, US. Casein was used as the ELISA blocking agent and TMB was purchased from Sigma Aldrich, Canada.
100 μM of the reduced and refolded aptamer in the buffer was diluted to 50 nM. An 18-well sticky-slide was attached to the as-purchased pristine gold-coated glass slides. 200 μl of the 50 nM aptamer solution was incubated in each well for 1 h at room temperature in the dark. This solution was pipetted out and the wells rinsed with 0.05% Tween-20 (Sigma Aldrich) in PBS (PBST). 200 μl of casein blocking agent in deionized water was added to each well and incubated on a hot plate (covered, to avoid evaporation) at 43 °C for 15 min. The wells were then pipetted out and rinsed with PBST. Spike protein with an Fc tag was prepared in PBS, added to each well, and incubated at 43 °C for 15 min. The wells were again pipetted out and rinsed with PBST. 1/20,000 dilution of secondary antibody in the buffer was added and incubated for 10 min at 43 °C on a hot plate in darkness. The liquid in the wells was aspirated out and the wells were rinsed with PBST. 100 μl of TMB was added to each well and incubated for 4 min in darkness and at room temperature after which 50 μl of 0.18 M sulfuric acid was added to arrest the color change. 100 μl of fluid from each well was then transferred into a 96-well plate and the absorbance was read at 450 nm with a plate reader (Tecan M200).
2.3. Validation of binding between nanoprobe and aptamer
The nanoprobes were synthesized and supplied by Genemis Laboratories Inc. and used without further modification. MgCl2 dissolved in deionized water was used as the coagulant (Salt M). The S1 subunit of spike protein that contains the receptor-binding domain was purchased from Acrobiosystems, USA. 100 μl of the nanoprobe colloid was mixed with 100 μl of varying concentration of the S1 subunit spike protein in the buffer and incubated for 30 min at room temperature in a 96-well plate. 75 μl of 100 mM of coagulant Salt M was added and incubated for 10 more minutes. The absorbance spectra were measured between 400 nm and 700 nm with the Tecan M200 plate reader for 10 min.
2.4. Nanoprobe characterization
TEM imaging was performed on as-supplied nanoprobes using a Talos 200X scanning transmission electron microscope from Thermo Scientific. The TEM images were analyzed with ImageJ image processing software. Particle sizes were determined for ten particles from a representative TEM image and the mean and standard deviations were determined. High-resolution transmission electron micrographs were obtained with a JEOL 2010F field emission microscope and EDX elemental analysis was performed using EDX through mapping analysis.
DLS measurements were performed on an aliquot of the nanoprobe suspension to characterize the particle size of the non-agglomerated nanoprobes. For the study of particle aggregation dynamics, mixtures of 20 μl of the nanoprobe suspension were added to a quartz cuvette with 20 μl of 67 nM of S1 subunit spike protein mixtures either in PBS or PBS buffer only and incubated at room temperature in darkness for 30 min. 15 μl of 100 mM Salt M was added to a clean quartz cuvette. DLS measurements were performed every 15 s for 900 s. A DelsaMax Pro BCI-3217-DMP analyzer was used for DLS measurements.
The as-supplied nanoprobes were washed by ultracentrifugation at 50,000×g and resuspended in deionized water. The aptamer was also suspended in deionized water. About 5 μl from each suspension was drop cast on a clean gold surface and air-dried for FTIR characterization. A Bruker HYPERION 3000 FT-IR imaging instrument with a 15× objective was used with a mercury cadmium telluride detector, and 256 scans were collected for both the background and samples. The background scans were performed on a pristine gold surface and sample scans were performed for the dried aptamer and nanoprobe samples. 50 μl of the washed nanoprobe sample was dried on a diced glass surface and XRD spectra were obtained using a Bruker D8 DISCOVER instrument containing a DAVINCI diffractometer and Co-Kα radiation.
2.5. Critical coagulation concentration: an index of binding
100 μl of 1, 10, and 100 nM of S1 subunit spike protein concentrations in buffer were mixed with a 100 μl nanoprobe suspension in a 96-well plate and separately incubated for 15 min. 75 μl of 0, 20, 40, 60, and 100 mM of Salt M diluted with PBS were added to separate samples and the absorbance measured at 540 nm for 15 min.
2.6. Detection of inactivated virus samples
The 2019-nCoV/USA-WA1/2020 strain of SARS-CoV-2 virus cultured in Vero E6 cells and subsequently heat-inactivated was obtained from ATCC, USA (ARCC VR-1986-HK). The concentration of the SARS-CoV-2 virus in the sample was represented by genome copies/μL based on the digital droplet PCR values provided by the manufacturer. The virus lysis buffer containing Tween-20, Casein, BSA, and other components was purchased from Acrobiosystems, USA (LY-13). This as-purchased lysis buffer was diluted to 30% v/v in PBS and used as a lysis buffer to prepare dilutions of the virus and incubated at room temperature for 30 min for lysis. In a 96-well plate, 100 μl of lysed virus samples containing 8850, 3540, 1770, and 177 genome copies/μL of the virus and PBS were mixed with 100 μl of the nanoprobe and incubated at room temperature for a further 30 min. Salt M was added and the absorbance measured at 540 nm over 15 min.
3. Results and discussion
3.1. ELISA for aptamer selection
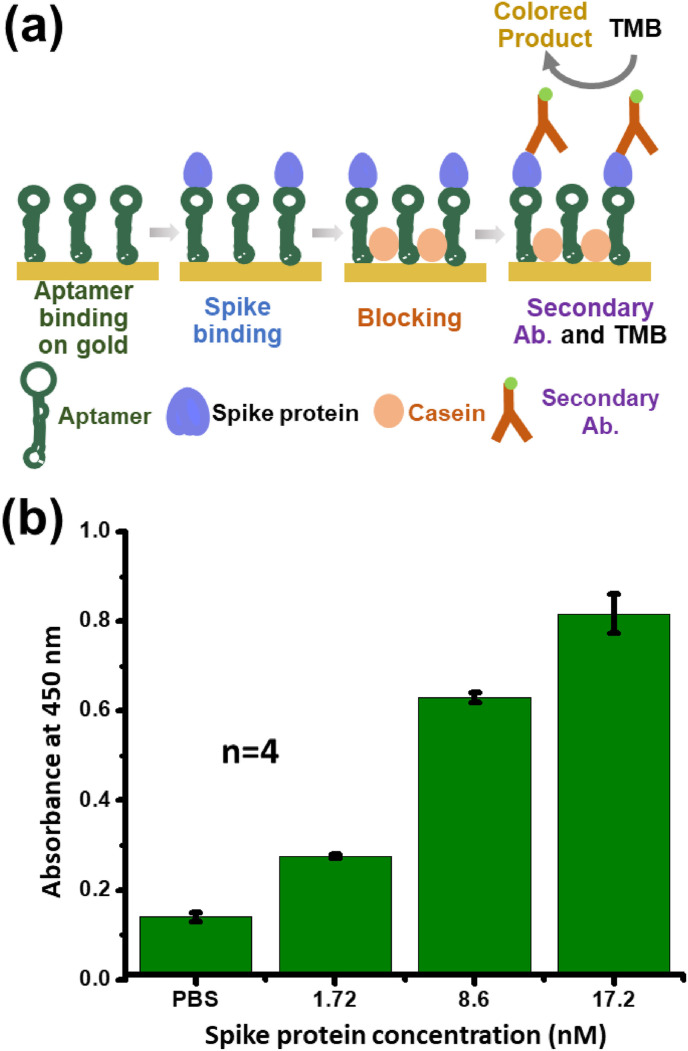
We consider two aptamers, developed previously with SELEX, that target the receptor-binding domain (RBD) of the SARS-CoV-2 spike protein [43] which interacts with the host cell at the outset of infection. Of these, the CoV-2-RBD-4C aptamer with higher fluorescence [43] is selected and synthesized with thiols at its 5′ end. The disulfide bonds formed during storage are reduced with TCEP. Through a refolding process [44], these aptamers are returned to their 3D confirmation with a specific binding affinity to the spike protein. Subsequently, the aptamers are immobilized on a flat gold-coated glass slide. The surface of the slide is blocked with casein to prevent non-specific binding of sample molecules to the bare surface between the aptamers. Samples containing spike protein in phosphate-buffered saline (PBS) with a monomeric fragment crystallizable region (Fc) tag are then deposited on the surface.
The binding of the spike protein with the aptamer is confirmed as follows. An Fc binding secondary antibody with HRP reporter is bound to the aptamer-spike protein complex, following which 3,3′,5,5′-Tetramethylbenzidine (TMB) is added to generate a color signal proportional to the amount of HRP present. The addition of 0.18 M sulfuric acid solution stops the reaction and produces a yellow color signal. The signal is quantified by measuring the absorbance at 450 nm which corresponds to the absorbance maximum of the TMB reaction product.
This process of aptamer immobilization that results in the generation of the TMB signal forms an ELISA that is illustrated in Fig. 2 (a). The measured absorbance at 450 nm is presented in Fig. 2(b) for different concentrations of the spike protein. The absorbance increases monotonically with increasing spike protein concentration. A 1.72 nM concentration is clearly distinguishable from the PBS background and there is a monotonic increase in absorbance as the concentration is increased further to 8.6 nM and 17.2 nM.
Fig. 2.
Results of ELISA for aptamer and spike protein binding. (a) Schematic of the assay steps that indicate aptamer binding to the flat gold surface, spike protein binding to the aptamer, blocking with casein, secondary Ab binding to the spike protein with the monomeric Fc tag, and addition of the HRP chromogenic substrate (TMB). (b) Response of the colorimetric assay with spike protein diluted in PBS buffer, where a 1.72 nM spike protein concentration in PBS is clearly distinguishable from the background. Bars represent the standard error of the mean of four replicates. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. Spike protein detection with nanoprobe
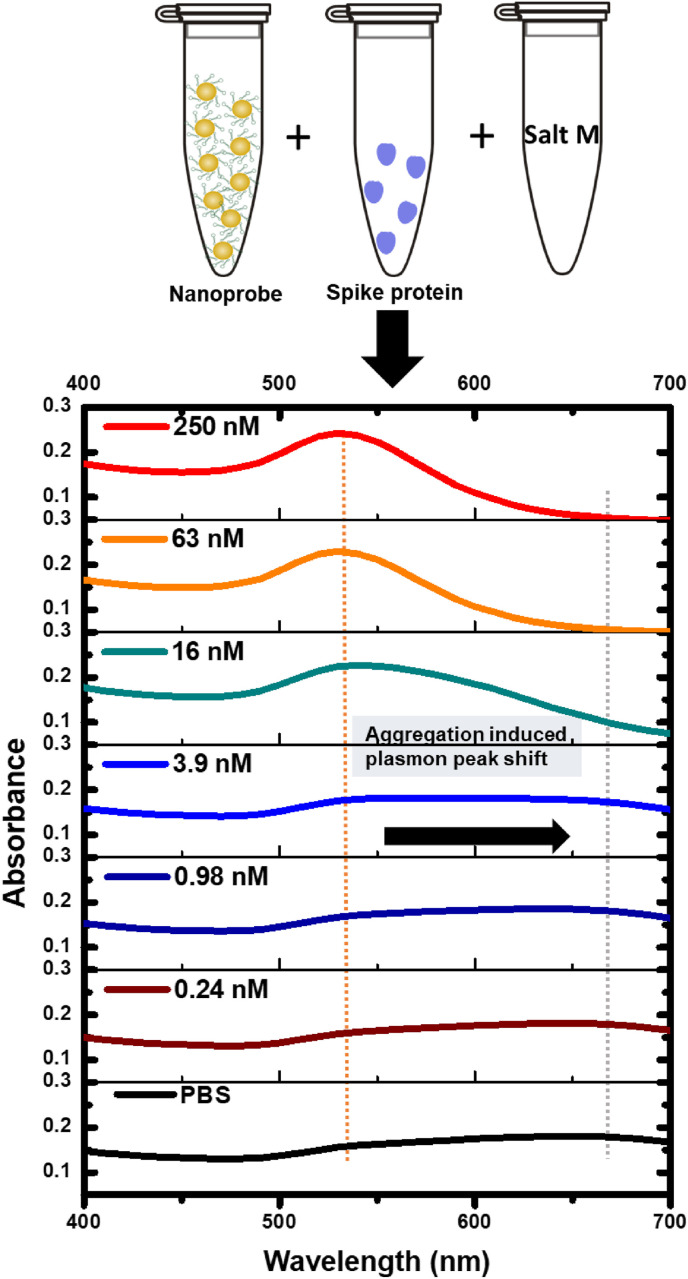
A colloidal suspension of AuNPs functionalized with the SARS-CoV-2-RBD-4C aptamer, or nanoprobes, is used. The absorbance spectrum for the suspension is sensitive to the extent of nanoprobe agglomeration, which introduces absorption differences based on the characteristic agglomerate sizes. These spectra are measured between 400 and 700 nm. The resistance to agglomeration correlates with the spike protein concentration in PBS and is characterized by comparing the plasmon absorbance peaks that depend on characteristic nanoparticle size. The absorbance peak for fully suspended nanoprobes, i.e., when there is no agglomeration, occurs around ∼540 nm. The onset of agglomeration broadens this peak and shifts it towards higher wavelengths. An agglomeration assay is performed where a fixed concentration of coagulant Salt M is added to the nanoprobe suspensions containing varying spike protein concentrations. The schematic of the assay and resulting spectra for different cases when 100 mM of Salt M is introduced to initiate agglomeration are shown in Fig. 3 .
Fig. 3.
Absorbance spectra for various concentrations of spike protein in PBS. The nanoprobe and sample mixtures are incubated for 30 min after which 100 mM of the coagulant Salt M is added. The mixture is then incubated for an additional 10 min. The absorbance spectrum broadens considerably for spike protein concentrations in PBS lower than 16 nM. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
For spike protein concentrations in PBS up to 3.9 nM, the absorbance spectra broaden significantly, and the absorbance peak shifts toward wavelengths greater than ∼540 nm, which is indicative of agglomeration. This spectral broadening is less significant for a 16 nM spike protein concentration, indicating resistance to agglomeration. There is even far less broadening for the 63 and 250 nM samples when the absorbance peak is anchored at ∼540 nm, confirming the colloidal stability of the nanoprobes and their resistance to agglomeration in the presence of sufficient spike protein. Hence, increasing the spike protein concentrations stabilizes the nanoprobe suspension.
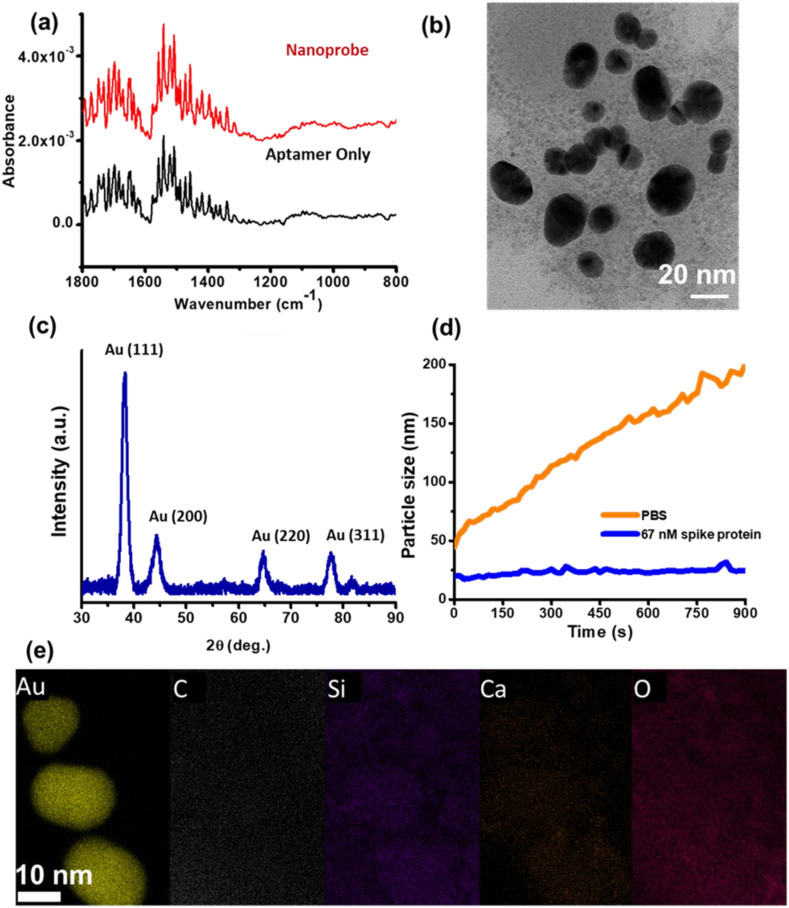
3.3. FTIR, TEM, XRD and DLS characterization
Fourier transform infrared spectroscopy (FTIR) spectra were measured on gold surfaces with dried nanoprobe and aptamer. Fig. 4 (a) shows that, within the fingerprint region [45], the spectral signature of the aptamer lies between 800 cm−1 and 1800 cm−1, which is identical to that for the nanoprobes, thus confirming that aptamers are present on the nanoprobes. The characteristic size of the nanoprobes is obtained from transmission electron microscopy (TEM). An aliquot of as-supplied nanoprobes is dried on a copper grid and imaged. Fig. 4(b) shows that the characteristic size of the nanoparticles is 18 ± 6 nm. This characteristic size is also measured with dynamic light scattering (DLS) and found to be 18 ± 2 nm, in agreement with the TEM micrograph. The crystallinity of dried nanoprobes is confirmed with powder X-ray diffraction (XRD) for which results are presented in Fig. 4(c). The diffraction peaks at 38.1°, 44.3°, 64.5°, and 77.7° respectively correspond to the standard Bragg reflections for the (111), (200), (220), and (311) crystalline planes of a gold nanoparticle. The highest intensity peak is for the (111) plane, which indicates the preferred growth direction.
Fig. 4.
FTIR, TEM, XRD, and DLS characterization of the nanoprobes. (a) IR spectra measured using FTIR in the fingerprint region for aptamers. The spectral signature of aptamers and nanoprobes is identical, indicating that aptamers are present on the nanoprobes. (b) Representative TEM image of the nanoprobe, where the nanoparticle characteristic size, quantified by measuring the longest dimension of 10 non-overlapping particles is 18 ± 6 nm. (c) XRD measurements reveal four distinct peaks corresponding to standard Bragg reflections for a crystalline gold nanoparticle with the highest intensity peak at Au (111). (d) The nanoprobe suspensions are mixed with samples, one containing only PBS and the other with 67 nm of spike protein added to PBS. The two mixtures are incubated for 30 min after which 100 mM of Salt M is added and DLS measurements performed at 15 s intervals over 900 s. Nanoprobes bound to spike protein do not agglomerate while those in PBS alone steadily agglomerate, reaching a 190 nm size at 900 s. (e) EDX mapping of elemental gold (Au), carbon (C), silicon (Si), calcium (Ca), and oxygen (O) on the nanoprobes. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The agglomeration kinetics of the nanoprobes are investigated by placing 20 μl of the nanoprobe suspension in a quartz cuvette with either 67 nM spike protein in PBS or PBS alone, which is then incubated for 30 min. Subsequently, 15 μl of 100 mM Salt M solution is added and DLS measurements obtained at 15 s intervals over 900 s, as shown in Fig. 4(d). We note that the addition of 100 mM Salt M to PBS increases the mean particle diameter from 40 nm to 190 nm, providing evidence of nanoprobe agglomeration in the absence of spike protein. On the other hand, with 67 nM spike protein added to PBS, the mean particle diameter remains constant at 25 nm even after 100 mM of Salt M is added, indicating aptamer-mediated spike protein binding that impedes nanoprobe agglomeration in suspension. Elemental analysis using EDX of high-resolution transmission electron micrographs of gold nanoprobes is presented in Fig. 4(e), which shows the presence of elemental Au, C, Ca, Si and O. Although, the nanoprobes are washed in an ultracentrifuge, the presence of these elements is attributed to the synthesis method and the reagents used to prepare and stabilize the nanoprobes.
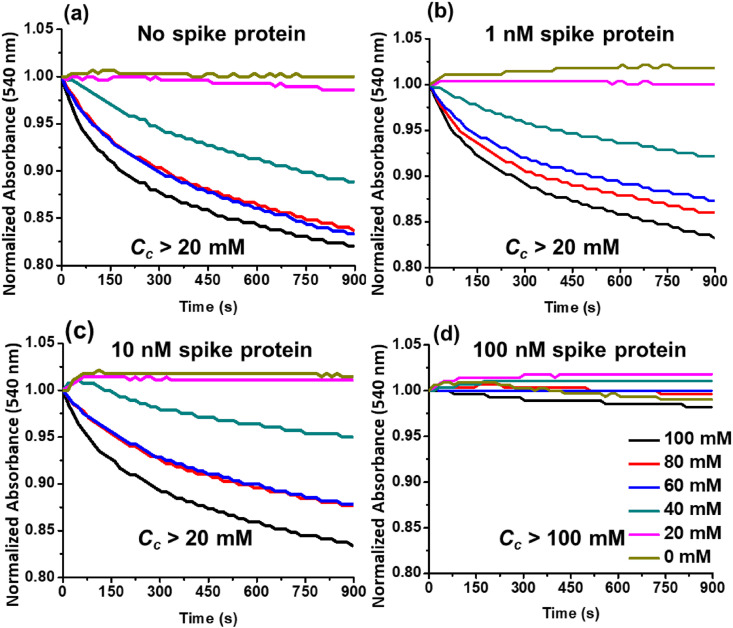
3.4. Critical coagulation concentration
The nanoparticle size increases with the binding of spike protein as shown in Fig. S2 of the supplementary material. The temporal variations in particle size as nanoprobe agglomeration proceeds are shown in Fig. 4(d). For the PBS only sample, the characteristic particle size increases steadily from 40 nm to 190 nm, where the initial 40 nm size is attributed to the small but finite delay between addition of Salt M to the nanoprobe-PBS mixture and the initiation of the DLS measurement. No nanoprobe agglomeration is observed for the sample with 67 nM spike protein where the characteristic nanoparticle size is virtually unchanged over time. Hence, we infer that the spike protein in the sample binds with the nanoprobes, stabilizing them against the agglomeration induced by the addition of the coagulant Salt M. Since spike protein binding with the aptamer moderates nanoprobe agglomeration, the minimum concentration of Salt M required to completely aggregate all nanoprobes in a mixture, C c is an indicator of the presence of spike protein.
To determine the extent of agglomeration, the temporal absorbance is measured at 540 nm. The nanoprobes are mixed with 1 nM, 10 nM, and 100 nM of spike protein concentrations in PBS and incubated for 15 min at room temperature. Different concentrations of Salt M are then added to these mixtures and the absorbance is measured over 15 min. These temporal absorbance measurements are normalized with their initial values at t = 0 s. Since the absorbance is steady for a stable colloidal suspension, a reduction in the normalized absorbance implies nanoparticle agglomeration.
Experimental measurements for a negative control containing PBS alone, i.e., with no spike protein, are shown in Fig. 5 (a). Nanoprobe agglomeration occurs for Salt M concentrations of 40 mM and higher. When 1 and 10 nM of spike protein are added to the nanoprobe-containing PBS suspension and incubated for 15 min, the absorbance is similar, as shown in Fig. 5(b and c), indicating that binding of these small amounts of spike protein to the nanoprobes does not significantly alter C c. Fig. 5(d) shows that C c > 100 mM for nanoprobes bound to spike protein in a 100 nM sample when there is very little agglomeration.
Fig. 5.
Nanoprobe agglomeration kinetics for different spike protein concentrations diluted in PBS. (a) 0 nM, (b) 1 nM, (c) 10 nM, and (d) 100 nM mixtures of spike protein in PBS are mixed with nanoprobe suspensions and incubated for 15 min, following which different concentrations of coagulant Salt M are introduced into the mixtures and their absorbance measured at 540 nm for 15 min. Increasing the spike concentration from 10 to 100 nM in PBS also increases Cc from a value greater than 20 mM to one greater than 100 mM. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.5. Virus detection
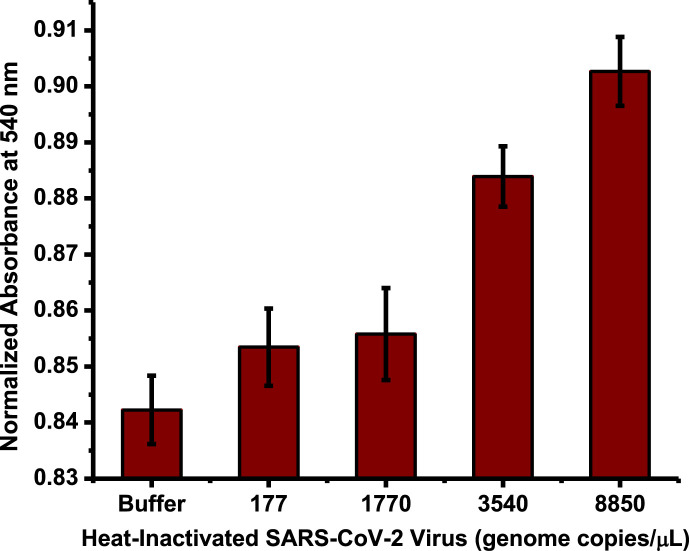
Different concentrations (genome copies/μL) of heat-inactivated SARS-CoV-2 are mixed with the lysing buffer and incubated for 30 min at room temperature. This lysed heat-inactivated SARS-CoV-2 is mixed with the nanoprobes following another incubation for 30 min. Subsequently, Salt M is added and the absorbance of the suspension is measured at 540 nm over 15 min. The presence of lysing buffer and ions within does not affect the aggregation kinetics of the nanoprobes as shown in Fig. S3 of the supplementary information.
Fig. 6 shows that the absorbances of the buffer and samples containing inactivated SARS-CoV-2 virus are different, where the absorbance increases with increasing virus concentration. It is not possible to distinguish between the buffer and 177 and 1770 genome copies/μL of the virus, but 3540 and 8850 genome copies/μL of the virus are readily distinguished from the buffer. Moreover, 8850, 3540, and 1770 genome copies/μL can be differentiated from each other.
Fig. 6.
The response of nanoprobe agglomeration to heat-inactivated SARS-CoV-2 in the buffer at t = 900 s after addition of coagulant Salt M. As the number of heat-inactivated SARS-CoV-2 genome copies increases, the agglomeration decreases. Although it is not possible to distinguish between PBS, 177, and 1770 genome copies/μL of the virus, higher concentrations of 3540 and 8850 genome copies/μL are distinguishable from the buffer. The error bars are the standard errors of the mean for six samples. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
With all reagents and consumables supplied with the test kit, the assay can be readily adopted in a POC setting. The test kit would include a collection tube, sample dilution buffer, nanoprobes, and Salt M, where the assay could be validated with SARS-CoV-2 infected samples in the future. A commercial absorbance reader would detect the color change due to nanoprobe agglomeration in the presence of the spike protein. The user would be instructed to collect the sample in a collection tube and add a specific volume of the diluting buffer, such as PBS together with the lysing buffer, to the collected saliva. Following preparation, this mixture would be added to a nanoprobe container and left undisturbed for 30 min. This vessel containing the mixture would be inserted into an absorbance reader for an initial measurement, followed by the addition of Salt M and continuing with measurements over another 15 min. Excluding the time taken for sample collection, the total test time would likely be 75 min.
4. Conclusions
The CoV-2-RBD-4C aptamer binds specifically to the SARS-CoV-2 spike protein, which is a surrogate for the coronavirus during the test. Gold nanoparticles functionalized with this aptamer form nanoprobes that are suspended in a colloid. An agglomeration assay using the nanoprobes detects 16 nM and higher concentrations of spike protein in PBS by measuring the absorbance spectra of the samples. The critical coagulant salt concentration, C c, that induces agglomeration is a useful metric to determine spike protein binding with the aptamer. Its value increases with increasing spike protein concentration in the sample. The nanoprobes detect 3540 genome copies/μL and higher concentrations of inactivated SARS-CoV-2 virus by examining the absorbance at 540 nm. With further validation using real world samples, this test could form the basis for a test that can be performed by a user with an inexpensive spectrophotometer.
Credit author contribution statement
Srivatsa Aithal: Conceptualization, Methodology, Investigation, Writing - Original Draft, Writing - Review & Editing Sarah Mishriki: Methodology, Investigation Rohit Gupta: Methodology, Investigation, Writing - Review & Editing Rakesh P. Sahu: Conceptualization, Methodology, Writing - Original Draft, Writing - Review & Editing George Botos: Conceptualization, Methodology Shazia Tanvir: Conceptualization, Methodology Russell W. Hanson: Resources Ishwar K. Puri: Conceptualization, Methodology, Writing - Review & Editing.
Funding sources
This work was supported by the MITACS Accelerate Fund (IT18550) and Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant (RGPIN-2019-06571).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Dr. Jason McLellan for providing us with the spike-receptor binding domain expression construct through the Toronto Open Access Covid-19 Protein Manufacturing Center (comprising BioZone and the Structural Genomics Consortium (SGC)) under an Open Science Trust Agreement, https://www.biozone.utoronto.ca/sgc-open-science-trust-agreement/. The Center received funding from the Toronto COVID-19 Action Fund. We thank Dr. Carmen Andrei of the Canadian Centre for Electron Microscopy for the TEM imaging and EDX analysis.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.talanta.2021.122841.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Chau C.H., Strope J.D., Figg W.D. COVID‐19 clinical diagnostics and testing technology, pharmacother. J. Hum. Pharmacol. Drug Ther. 2020;40:857–868. doi: 10.1002/phar.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rabiee N., Bagherzadeh M., Ghasemi A., Zare H., Ahmadi S., Fatahi Y., Dinarvand R., Rabiee M., Ramakrishna S., Shokouhimehr M., Varma R.S. Point-of-Use rapid detection of SARS-CoV-2: nanotechnology-enabled solutions for the COVID-19 pandemic. Int. J. Mol. Sci. 2020;21:5126. doi: 10.3390/ijms21145126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kilic T., Weissleder R., Lee H. Molecular and immunological diagnostic tests of COVID-19: current status and challenges. IScience. 2020;23:101406. doi: 10.1016/j.isci.2020.101406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Afzal A. Molecular diagnostic technologies for COVID-19: limitations and challenges. J. Adv. Res. 2020;26:149–159. doi: 10.1016/j.jare.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esbin M.N., Whitney O.N., Chong S., Maurer A., Darzacq X., Tjian R. Overcoming the bottleneck to widespread testing: a rapid review of nucleic acid testing approaches for COVID-19 detection. RNA. 2020;26:771–783. doi: 10.1261/rna.076232.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacKay M.J., Hooker A.C., Afshinnekoo E., Salit M., Kelly J., Feldstein J.V., Haft N., Schenkel D., Nambi S., Cai Y., Zhang F., Church G., Dai J., Wang C.L., Levy S., Huber J., Ji H.P., Kriegel A., Wyllie A.L., Mason C.E. The COVID-19 XPRIZE and the need for scalable, fast, and widespread testing. Nat. Biotechnol. 2020;38:1021–1024. doi: 10.1038/s41587-020-0655-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas S.N., Altawallbeh G., Zaun C.P., Pape K.A., Peters J.M., Titcombe P.J., Dileepan T., Rapp M.J., Bold T.D., Schacker T.W., Arbefeville S., Ferrieri P., Thyagarajan B., Jenkins M.K., Karger A.B. Initial determination of COVID-19 seroprevalence among outpatients and healthcare workers in Minnesota using a novel SARS-CoV-2 total antibody ELISA. Clin. Biochem. 2021;90:15–22. doi: 10.1016/j.clinbiochem.2021.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu J.-L., Tseng W.-P., Lin C.-H., Lee T.-F., Chung M.-Y., Huang C.-H., Chen S.-Y., Hsueh P.-R., Chen S.-C. Four point-of-care lateral flow immunoassays for diagnosis of COVID-19 and for assessing dynamics of antibody responses to SARS-CoV-2. J. Infect. 2020;81:435–442. doi: 10.1016/j.jinf.2020.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai X., Chen J., li Hu J., Long Q., Deng H., Liu P., Fan K., Liao P., Liu B., Wu G., Chen Y., Li Z., Wang K., Zhang X., Tian W., Xiang J., Du H., Wang J., Hu Y., Tang N., Lin Y., Ren J., Huang L., Wei J., Gan C., Chen Y., Gao Q., Chen A., He C., Wang D.-X., Hu P., Zhou F., Huang A., Wang D. A peptide-based magnetic chemiluminescence enzyme immunoassay for serological diagnosis of coronavirus disease 2019. J. Infect. Dis. 2020;222:189–193. doi: 10.1093/infdis/jiaa243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diao B., Wen K., Zhang J., Chen J., Han C., Chen Y., Wang S., Deng G., Zhou H., Wu Y. Accuracy of a nucleocapsid protein antigen rapid test in the diagnosis of SARS-CoV-2 infection. Clin. Microbiol. Infect. 2021;27:289. doi: 10.1016/j.cmi.2020.09.057. e1-289.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee J.-H., Choi M., Jung Y., Lee S.K., Lee C.-S., Kim J., Kim J., Kim N.H., Kim B.-T., Kim H.G. A novel rapid detection for SARS-CoV-2 spike 1 antigens using human angiotensin converting enzyme 2 (ACE2) Biosens. Bioelectron. 2021;171:112715. doi: 10.1016/j.bios.2020.112715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McConnell E.M., Nguyen J., Li Y. Aptamer-based biosensors for environmental monitoring. Front. Chem. 2020;8:434. doi: 10.3389/fchem.2020.00434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maio G.E., Enweronye O., Zumrut H.E., Batool S., Van N.A., Mallikaratchy P.R. Systematic optimization and modification of a DNA aptamer with 2’-O-methyl RNA analogues. Chemistry. 2017;2:2335–2340. doi: 10.1002/slct.201700359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shembekar N. In: Receptors in Immunodiagnostics: Antibodies, Antibody Fragments, Single Domain Antibodies and Aptamers BT - Immunodiagnostic Technologies from Laboratory to Point-Of-Care Testing. Suman P., Chandra P., editors. Springer Singapore; Singapore: 2021. pp. 223–245. [DOI] [Google Scholar]
- 15.Jo H., Ban C. Aptamer–nanoparticle complexes as powerful diagnostic and therapeutic tools. Exp. Mol. Med. 2016;48 doi: 10.1038/emm.2016.44. e230–e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Z., Wu Q., Chen J., Ni X., Dai J. A DNA aptamer based method for detection of SARS-CoV-2 nucleocapsid protein. Virol. Sin. 2020;35:351–354. doi: 10.1007/s12250-020-00236-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zakashansky J.A., Imamura A.H., Salgado D.F., Romero Mercieca H.C., Aguas R.F.L., Lao A.M., Pariser J., Arroyo-Currás N., Khine M. Detection of the SARS-CoV-2 spike protein in saliva with Shrinky-Dink© electrodes. Anal. Methods. 2021;13:874–883. doi: 10.1039/D1AY00041A. [DOI] [PubMed] [Google Scholar]
- 18.Kelly K.L., Coronado E., Zhao L.L., Schatz G.C. The optical properties of metal nanoparticles: the influence of size, shape, and dielectric environment. J. Phys. Chem. B. 2003;107:668–677. doi: 10.1021/jp026731y. [DOI] [Google Scholar]
- 19.Jain P.K., Lee K.S., El-Sayed I.H., El-Sayed M.A. Calculated absorption and scattering properties of gold nanoparticles of different size, shape, and composition: applications in biological imaging and biomedicine. J. Phys. Chem. B. 2006;110:7238–7248. doi: 10.1021/jp057170o. [DOI] [PubMed] [Google Scholar]
- 20.Doria G., Conde J., Veigas B., Giestas L., Almeida C., Assunção M., Rosa J., Baptista P.V. Noble metal nanoparticles for biosensing applications. Sensors. 2012;12:1657–1687. doi: 10.3390/s120201657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen L.Q., Xiao S.J., Hu P.P., Peng L., Ma J., Luo L.F., Li Y.F., Huang C.Z. Aptamer-mediated nanoparticle-based protein labeling platform for intracellular imaging and tracking endocytosis dynamics. Anal. Chem. 2012;84:3099–3110. doi: 10.1021/ac202810b. [DOI] [PubMed] [Google Scholar]
- 22.Iarossi M., Schiattarella C., Rea I., De Stefano L., Fittipaldi R., Vecchione A., Velotta R., Della Ventura B. Colorimetric immunosensor by aggregation of photochemically functionalized gold nanoparticles. ACS Omega. 2018;3:3805–3812. doi: 10.1021/acsomega.8b00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y., Zhang L., Wei W., Zhao H., Zhou Z., Zhang Y., Liu S. Colorimetric detection of influenza A virus using antibody-functionalized gold nanoparticles. Analyst. 2015;140:3989–3995. doi: 10.1039/C5AN00407A. [DOI] [PubMed] [Google Scholar]
- 24.Draz M.S., Shafiee H. Applications of gold nanoparticles in virus detection. Theranostics. 2018;8 doi: 10.7150/thno.23856. 1985–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho H., Baker B.R., Wachsmann-Hogiu S., Pagba C.V., Laurence T.A., Lane S.M., Lee L.P., Tok J.B.H. Aptamer-based SERRS sensor for thrombin detection. Nano Lett. 2008;8:4386–4390. doi: 10.1021/nl802245w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Medley C.D., Bamrungsap S., Tan W., Smith J.E. Aptamer-conjugated nanoparticles for cancer cell detection. Anal. Chem. 2011;83:727–734. doi: 10.1021/ac102263v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giorgi-Coll S., Marín M.J., Sule O., Hutchinson P.J., Carpenter K.L.H. Aptamer-modified gold nanoparticles for rapid aggregation-based detection of inflammation: an optical assay for interleukin-6. Microchim. Acta. 2020;187:13. doi: 10.1007/s00604-019-3975-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsai C.-S., Yu T.-B., Chen C.-T. Gold nanoparticle-based competitive colorimetric assay for detection of protein–protein interactions. Chem. Commun. 2005:4273. doi: 10.1039/b507237a. [DOI] [PubMed] [Google Scholar]
- 29.Jung Y.L., Jung C., Parab H., Li T., Park H.G. Direct colorimetric diagnosis of pathogen infections by utilizing thiol-labeled PCR primers and unmodified gold nanoparticles. Biosens. Bioelectron. 2010;25:1941–1946. doi: 10.1016/j.bios.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 30.Liu M., Yuan M., Lou X., Mao H., Zheng D., Zou R., Zou N., Tang X., Zhao J. Label-free optical detection of single-base mismatches by the combination of nuclease and gold nanoparticles. Biosens. Bioelectron. 2011;26:4294–4300. doi: 10.1016/j.bios.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 31.Moitra P., Alafeef M., Alafeef M., Alafeef M., Dighe K., Frieman M.B., Pan D., Pan D., Pan D. Selective naked-eye detection of SARS-CoV-2 mediated by N gene targeted antisense oligonucleotide capped plasmonic nanoparticles. ACS Nano. 2020;14 doi: 10.1021/acsnano.0c03822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karami A., Hasani M., Azizi Jalilian F., Ezati R. Conventional PCR assisted single-component assembly of spherical nucleic acids for simple colorimetric detection of SARS-CoV-2. Sensor. Actuator. B Chem. 2021;328 doi: 10.1016/j.snb.2020.128971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karami A., Hasani M., Azizi Jalilian F., Ezati R. Hairpin-spherical nucleic acids for diagnosing COVID-19: a simple method to generalize the conventional PCR for molecular assays. Anal. Chem. 2021;93:9250–9257. doi: 10.1021/acs.analchem.1c01515. [DOI] [PubMed] [Google Scholar]
- 34.Saha K., Agasti S.S., Kim C., Li X., Rotello V.M. Gold nanoparticles in chemical and biological sensing. Chem. Rev. 2012;112:2739–2779. doi: 10.1021/cr2001178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rogowski J.L., Verma M.S., Gu F.X. Discrimination of proteins using an array of surfactant-stabilized gold nanoparticles. Langmuir. 2016;32:7621–7629. doi: 10.1021/acs.langmuir.6b01339. [DOI] [PubMed] [Google Scholar]
- 36.Verma M.S., Chen P.Z., Jones L., Gu F.X. “Chemical nose” for the visual identification of emerging ocular pathogens using gold nanostars. Biosens. Bioelectron. 2014;61:386–390. doi: 10.1016/j.bios.2014.05.045. [DOI] [PubMed] [Google Scholar]
- 37.Peng Y., Li L., Mu X., Guo L. Aptamer-gold nanoparticle-based colorimetric assay for the sensitive detection of thrombin. Sensor. Actuator. B Chem. 2013;177:818–825. doi: 10.1016/j.snb.2012.12.004. [DOI] [Google Scholar]
- 38.Wang J., Wang L., Liu X., Liang Z., Song S., Li W., Li G., Fan C. A gold nanoparticle-based aptamer target binding readout for ATP assay. Adv. Mater. 2007;19:3943–3946. doi: 10.1002/adma.200602256. [DOI] [Google Scholar]
- 39.Dominguez-Medina S., Blankenburg J., Olson J., Landes C.F., Link S. Adsorption of a protein monolayer via hydrophobic interactions prevents nanoparticle aggregation under harsh environmental conditions. ACS Sustain. Chem. Eng. 2013;1:833–842. doi: 10.1021/sc400042h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zook J.M., Rastogi V., MacCuspie R.I., Keene A.M., Fagan J. Measuring agglomerate size distribution and dependence of localized surface plasmon resonance absorbance on gold nanoparticle agglomerate size using analytical ultracentrifugation. ACS Nano. 2011;5:8070–8079. doi: 10.1021/nn202645b. [DOI] [PubMed] [Google Scholar]
- 41.Hassanzadeh K., Perez Pena H., Dragotto J., Buccarello L., Iorio F., Pieraccini S., Sancini G., Feligioni M. Considerations around the SARS-CoV-2 spike protein with particular attention to COVID-19 brain infection and neurological symptoms. ACS Chem. Neurosci. 2020;11:2361–2369. doi: 10.1021/acschemneuro.0c00373. [DOI] [PubMed] [Google Scholar]
- 42.Tadros T. Encycl. Colloid Interface Sci. Springer Berlin Heidelberg; Berlin, Heidelberg: 2013. Steric stabilization; pp. 1048–1049. [DOI] [Google Scholar]
- 43.Song Y., Song J., Wei X., Huang M., Sun M., Zhu L., Lin B., Shen H., Zhu Z., Yang C. Discovery of aptamers targeting the receptor-binding domain of the SARS-CoV-2 spike glycoprotein. Anal. Chem. 2020;92:9895–9900. doi: 10.1021/acs.analchem.0c01394. [DOI] [PubMed] [Google Scholar]
- 44.Lee K., Nojoomi A., Jeon J., Lee C.Y., Yum K. Near-infrared fluorescence modulation of refolded DNA aptamer-functionalized single-walled carbon nanotubes for optical sensing. ACS Appl. Nano Mater. 2018;1:5327–5336. doi: 10.1021/acsanm.8b01377. [DOI] [Google Scholar]
- 45.Mello M.L.S., Vidal B.C. Changes in the infrared microspectroscopic characteristics of DNA caused by cationic elements, different base richness and single-stranded form. PloS One. 2012;7 doi: 10.1371/journal.pone.0043169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.