Abstract
The Mediator complex of Saccharomyces cerevisiae is required for both general and regulated transcription of RNA polymerase II (PolII) and is composed of two stable subcomplexes (Srb4 and Rgr1 subcomplexes). To decipher the function of each Mediator subcomplex and to delineate the functional relationship between the subcomplexes, we characterized the compositions and biochemical activities of PolII-Mediator complexes (holoenzymes) prepared from several Mediator mutant strains of S. cerevisiae. We found that holoenzymes devoid of a functional Gal11 module were defective for activated but not basal transcription in a reconstituted in vitro system. This activation-specific defect was correlated with a crippled physical interaction to transcriptional activator proteins, which could be bypassed by artificial recruitment of a mutant holoenzyme to a promoter. Consistent with this observation, a direct interaction between Gal11 and gene-specific transcriptional activator proteins was detected by far-Western analyses and column binding assays. In contrast, the srb5 deletion mutant holoenzyme was defective for both basal and activated transcription, despite its capacity for activator binding that is comparable to that of the wild-type holoenzyme. These results demonstrate that the Gal11 module of the Rgr1 subcomplex is required for the efficient recruitment of PolII holoenzyme to a promoter via activator-specific interactions, while the Srb4 subcomplex functions in the modulation of general polymerase activity.
The activator-squelching assay, in which the addition of excess amounts of one activator interferes with transcriptional stimulation by another activator, suggests the existence of a common target for the two activators (20). The fact that a crude yeast fraction devoid of all the basal transcription factors could relieve the squelching effect demonstrated that a distinct intermediary molecule is involved in the mediation of signal transfer between transcriptional activator proteins and basal transcription machinery (7, 20). Monitoring of this intermediary activity throughout the biochemical fractionation of a yeast whole-cell extract led to the purification of a multiprotein complex called Mediator (21). The Mediator complex is tightly associated with RNA polymerase II (PolII) and enables PolII to respond to transcriptional activators in an in vitro system reconstituted with pure general transcription factors. In addition, Mediator stimulates basal transcription, as well as transcription factor IIH (TFIIH)-dependent, in vitro phosphorylation of the carboxy-terminal domain (CTD) of the largest subunit of PolII, Rpb1 (21).
An independent genetic approach for identifying CTD-interacting proteins led to the discovery of the SRB gene family (31) and the identification of a PolII complex that contains all of the SRB gene products (16, 27, 40). Srb- and Mediator-containing PolII complexes contain Srb proteins (Srb2, Srb4, Srb5, Srb6, and Srb7) as well as the products of previously described transcriptional regulatory genes (GAL11, SIN4, RGR1, ROX3, and HRS1/PGD1/MED3) (13, 21, 26, 30). In addition, several novel subunits (Med1, Med2, Med4, Med6, Med7, Med8, Med9, Med10, and Med11) were identified as components of both PolII complexes (12, 15, 25, 30). However, some components differ between the two PolII complexes; specifically, certain Srb proteins (Srb8, Srb9, Srb10, and Srb11) and the Swi-Snf complex are absent from the Mediator-PolII complex (holoenzyme) (25, 30, 46).
Genetic studies revealed that some of the genes that encode Mediator subunits are required for the transcriptional regulation of specific genes, whereas others are necessary for general transcription in vivo (reviewed in reference 3). Differential dissociation of the Mediator components by high-urea treatment revealed that functionally related Mediator subunits physically associate to form stable Srb4 and Rgr1 subcomplexes (24). The Gal11 module proteins (Gal11, Sin4, and Hrs1) are found in the Rgr1 subcomplex together with several Med proteins (Med1, Med4, Med7, Med9, and Med10) and Srb7. We showed recently that newly identified Med proteins (Med9 and Med10) are required for the regulation of a group of genes that are different from those regulated by the Gal11 module (15). These results strongly suggest that other Mediator proteins in the Rgr1 subcomplex may also be involved in the transcriptional regulation of distinct subsets of genes.
While the Rgr1 subcomplex is composed of Mediator subunits required for the transcriptional regulation of distinct subsets of genes, the Srb4 subcomplex is composed of Mediator subunits required for general transcription events (Srb2, Srb4, Srb5, Srb6, and Med6). The Srb4 subcomplex was successfully reconstituted with recombinant proteins in vitro (22), and the functional interactions between the components of this subcomplex were deciphered genetically by determining the suppressor relationships among SRB4, SRB6, and MED6 (23, 24). The general requirement for Srb4 in PolII transcription suggests that Srb4 and its associated proteins function in the modulation of the basic activity of PolII, rather than in the reception of gene-specific activator signals. However, weak binding affinity between Srb4 and gene-specific transcriptional activators was detected in an in vitro biochemical assay (22).
Although Srb4 was suggested as an activator binding target on the basis of genetic and physical interactions (22), the general requirement of Srb4 for the expression of most PolII-transcribed genes makes it difficult to explain how activator specificity is achieved. Therefore, in order to identify the activator binding targets of Mediator and to elucidate the mechanism by which Mediator interacts with specific transcriptional activator proteins, we sought to determine the distinct functions of the two Mediator subcomplexes, the Rgr1 and Srb4 subcomplexes. As a means of identifying the specific function of each Mediator module, we purified and analyzed Mediator-PolII complexes (holoenzymes) from yeast strains that contained a mutation in one of the Mediator components. Our data reveal that activated transcription via the Gal11 module occurs through a specific interaction of the Gal11 module with a gene-specific transcriptional activator in vitro. On the basis of these observations, we propose a transcriptional activation mechanism that involves (i) the recruitment of a holoenzyme by an activator through an interaction with a distinct Mediator module and (ii) the modulation of PolII activity by the Srb-containing Mediator subcomplex upon activator-Mediator interaction.
MATERIALS AND METHODS
Protein purification.
The Mediator-PolII complex (holoenzyme) was purified from Saccharomyces cerevisiae wild-type cells (YCL10; MATα ade2 ura3 lys2 trp1 his3 leu2 med6Δ::LEU2 [MED6 on pRS313, HIS3]), rgr1Δ2 mutant cells (DY2010; MATa rgr1-Δ2::TRP1 can1 leu2 trp1 ura3), srb5Δ mutant cells (CTY153; MATa ura3 his3 leu2 lys2 srb5Δ::URA3 hisG), gal11Δ mutant cells (HS301; MATa ura3 trp1 leu2 prb1 pep4 prc1 gal2 gal11Δ::LEU2), and hrs1Δ mutant cells (SSAB-2CF; MATα ade2 ura3 his3 leu2 hrs1Δ::LEU2). The cultured cells were harvested, washed with cold water, and dissolved in 0.5 ml of 3× lysis buffer (21) per g of wet cells. All subsequent steps were carried out at 4°C. After freezing-thawing, the cells were disrupted by 20 cycles of bead beating (one cycle: a 30-s burst followed by 90 s of chilling at 4°C) in a stainless steel chamber with an equal volume of 0.5-mm-diameter glass beads, and cell debris was removed from the lysate by centrifugation at 12,000 × g for 20 min. To the supernatant, 0.1 volume of 4 M potassium acetate (pH 7.6) and 0.01 volume of 10% (wt/vol) polyethyleneimine (pH 8.0) were added slowly, and the mixture was stirred gently for 30 min. The resulting lysate was clarified by centrifugation in a Beckman Ti45 rotor at 42,000 rpm for 90 min and then subjected to four consecutive chromatographic steps that included BioRex 70 (Bio-Rad), DEAE-Sepharose FF (Pharmacia), Biogel-HTP hydroxyapatite (Bio-Rad), and MonoQ HR 5/5 (Pharmacia) as described previously (21). Holoenzyme activity was monitored with a specific transcription assay and immunoblot analysis. MonoQ column fractions that contained peak holoenzyme activity (1 M potassium acetate eluate) were pooled and used in the in vitro transcription and CTD phosphorylation assays. The amount of each type of holoenzyme used in the assays was normalized on the basis of its nonspecific transcription activity as described previously (21). Recombinant Gal4VP16, glutathione S-transferase (GST)–VP16, GCN4, and GST-Gal11 were isolated from bacterial expression strains described previously (5, 28, 32, 36).
In order to tag the Gal4VP16 fusion protein with a site for phosphorylation, an adapter DNA molecule coding for a phosphorylation motif sequence (Arg-Arg-Ala-Ser-Val) was prepared by annealing oligonucleotides AdapC (5′-CTAGTCGTCGTGCATCTGTTGGATCCCA-3′) and AdapD (5′-TATGGGATCCAACAGATGCACGACGA-3′); the 28-bp adapter DNA fragment was flanked by SpeI and NdeI sites. This adapter DNA fragment and the NdeI-BamHI fragment of the Gal4VP16 gene obtained from pET-Gal4VP16 were cloned together into the SpeI-BamHI sites of pEHB1 (N-terminal six-histidine fusion system constructed by the modification of pET-3a) to create pEh-KGVP. The recombinant Gal4VP16 doubly tagged with a hexahistidine stretch and a phosphorylation motif at its N terminus was purified from Escherichia coli BL21(DE3) carrying pEh-KGVP as described previously (5).
In vitro transcription and CTD phosphorylation.
The reconstituted in vitro transcription assay was performed as described elsewhere (21). In order to examine the CTD phosphorylation event during transcription initiation complex formation, transcription buffer (25) supplemented with cold ATP (8 μM) and [γ-32P]ATP (3 μCi) was added to the DNA template JJ470, TFIIH, and other protein components. The mixture was incubated for 10 or 40 min at 25°C. In order to examine CTD phosphorylation during the transcription reaction, CTP and UTP (0.8 mM each) were also added to the above reaction mixture, which was incubated for 10 min at 25°C. Except for the ribonucleotides, all the other components were as in the reconstituted transcription assay. Reactions were stopped by the addition of 4× sodium dodecyl sulfate (SDS) gel loading buffer (8 μl; 200 mM Tris-Cl [pH 6.8], 400 mM dithiothreitol, 8% SDS, 40% glycerol, 0.4% bromophenol blue), and half of the reaction products were analyzed on an SDS–7.5% polyacrylamide gel. RNA transcripts and 32P-labeled Rpb1 were quantitated with the use of a PhosphorImager (Molecular Dynamics).
Immunoprecipitation and immunoblotting.
Crude anti-Rgr1 antiserum (200 μl) (24) was conjugated with protein G-agarose beads (200 μl) (GIBCO BRL) as described previously (26), and each aliquot of antibody-beads (20 μl) was incubated for 6 to 12 h at 4°C with the holoenzyme fraction (MonoQ column). The beads were washed three times with IP buffer-100 (400 μl; 20 mM Tris-HCl [pH 7.8], 0.1 mM EDTA, 0.2 mM Nonidet P-40, 1 mM dithiothreitol, 10% [vol/vol] glycerol, 100 mM potassium acetate), and the bound proteins were eluted twice with 100 mM glycine (pH 2.5) (25 μl). The eluates were treated with 10% trichloroacetate, and the precipitated proteins were resolved on an SDS-polyacrylamide gel and analyzed by silver staining or immunoblotting.
Immunoblot analysis was performed with monoclonal antibody 8WG16 (for Rpb1), rabbit antiserum directed against the Gal4 DNA binding region (for Gal4VP16; Santa Cruz Biotechnology), and antisera directed against various Mediator components. Anti-Sin4 antiserum was generated in rats with a recombinant six-histidine-tagged Sin4 protein fragment (N-terminal 430 amino acids) as an antigen.
SRP.
Surface plasmon resonance (SPR) measurements for the detection of protein-protein interactions were taken with a BIAcore Biosensor (Biosensor). All measurements were taken at 25°C in running buffer HBP-150 (40 mM HEPES-KOH [pH 7.6], 7.5 mM MgCl2, 150 mM potassium acetate, 0.005% Surfactant P-20) at a flow rate of 5 μl/min. Purified proteins were immobilized on Sensor Chip CM-5 with the use of an amine coupling kit (Biosensor) in accordance with the manufacturer’s instructions at a flow rate of 5 μl/min. Gal4VP16 (100 ng/μl, 50 μl) and GST-VP16 (50 ng/μl, 70 μl) were each coupled in 100 mM sodium formate (pH 3.0). GCN4 protein (100 ng/μl, 50 μl) was coupled in 100 mM sodium formate (pH 4.0), and TATA binding protein (TBP) (100 ng/μl, 50 μl) was immobilized in running buffer. Analyte proteins were diluted in running buffer to the appropriate concentrations and were dialyzed against running buffer for 3 h. All injected proteins were centrifuged for 5 min just prior to injection. After injection, bound proteins were removed by injection of 30 μl of 100 mM glycine (pH 3.0 or 4.0). The data were analyzed with the use of BIAevaluation software (version 2.1; Biosensor).
Far-Western analysis and GST column binding assay.
For far-Western blotting, ∼2 μg of either wild-type or gal11Δ mutant holoenzyme (immunopurified from the active MonoQ fraction by use of anti-Rgr1 antibody–beads) was electrophoresed through an SDS–10% polyacrylamide gel and transferred to a nitrocellulose membrane. The proteins bound to the filter were denatured and renatured as described previously (43) and were incubated for 8 h at 4°C in binding buffer (5 ml) (17) containing 32P-labeled Gal4VP16 protein (5 × 103 to 1 × 104 cpm/ng; total, 200 ng). The blot was washed three times with binding buffer and twice with phosphate-buffered saline containing 0.2% Triton X-100 and was subjected to autoradiography. Phosphorylation of Gal4VP16 was performed with 1 U of the catalytic subunit of bovine heart kinase (Sigma) per μl and 1 μCi of [γ-32P]ATP per μl in 50 μl of HMK buffer as described previously (19).
For the GST column binding assay, purified recombinant GST-Gal11 protein or GST protein was bound to glutathione-agarose beads (Sigma) at a concentration of 1 μg of protein per 5 μl of beads. In vitro-translated (35S-labeled) Gal4VP16, Gal4VP16Δ456FP442 (2), or GCN4 (32) (10 μl from each in vitro translation reaction) was incubated overnight at 4°C with 25 μl of GST- or GST-Gal11-conjugated agarose beads in IP buffer-100. After binding, the beads were washed three times with binding buffer (250 μl) and boiled in SDS sample buffer, and the bound proteins were analyzed by SDS–12.5% polyacrylamide gel electrophoresis (PAGE) and autoradiography.
Artificial recruitment assay.
To generate a gal11Δ::TRP1 strain (designated JMP1), the GAL11 targeting plasmid pRS304-GAL11KO was constructed by inserting a GAL11 gene fragment into the SpeI and EcoRI sites of pRS304. The GAL11 gene fragment was generated by PCR (the two oligonucleotides used in the PCR were gal11-3spe [5′-TAACTAGTTGGAATAATTGGACAAGTGCTACTTGAACATTTGAAGTTAAC-3′] and gal11-5ri [5′-TGGAATTCAGGAGCAGCAGACATAGCAGATTTAAAAGAAATAGCGTTAAC-3′]) and digestion with SpeI and EcoRI. pRS304-GAL11KO was linearized by digestion with HincII and transformed into S. cerevisiae YCL4 (MATα ade2 ura3 lys2 trp1 his3 leu2 med6Δ::LEU2 [MED6 on pRS316, URA3]). The URA3-based MED6 plasmid (pRS316-MED6) in YCL4 and JMP1 was replaced with pRS313-MED6 (strains YCL10 and JMP2) and pRS313-LexA-MED6 (strains JMP3 and JMP4), respectively, by the plasmid shuffling method as described previously (25).
To construct strains JMP5, JMP6, and JMP7, wild-type cells (YPH499; MATa ade2 ura3 lys2 trp1 his3 leu2 GAL+), gal11Δ mutant cells (HS16; MATα ade2 his3 ura3 trp1 leu2 can1 gal11Δ::LEU2), and srb5Δ mutant cells (CTY153) were transformed with pRS313-LexA-MED6, respectively. The srb5 null strain CTY153 was transformed with pRS313-MED6 to make JMP8. In order to construct pRS313-LexA-MED6, the 1.6-kb ScaI-EcoRI fragment from pEG202 (14) and the 1.3-kb EcoRI-NaeI fragment from pGBT-MED6 were inserted sequentially between the EcoRV-EcoRI and EcoRI-SmaI sites in pRS313.
An episomal lacZ reporter plasmid (pLGSD5 [11] for control experiments, pSH18-34T [6] for strains JMP2 to JMP6, or pSH18-34TLeu for strains JMP7 and JMP8) with an appropriate selective marker was also introduced into the resulting strains. In order to construct pSH18-34TLeu, the SmaI fragment containing the lacZ gene was inserted into the SmaI site of pRS425. Transformant cells were grown in selective synthetic complete medium containing 2% glucose until the mid-log phase. At an optical density at 600 nm of 1.0, the cells were harvested by centrifugation, resuspended in yeast extract-peptone medium containing 2% glucose, and further shaken for 3 h. β-Galactosidase activity was measured from permeabilized cells as described previously (10).
RESULTS
Subunit compositions of mutant holoenzymes.
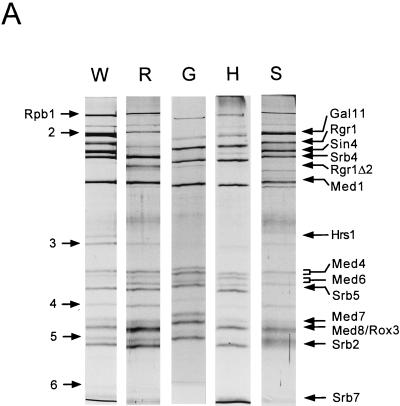
In order to identify a distinct function for each Mediator subunit, we purified holoenzymes from wild-type and mutant yeast strains carrying either an Rgr1 C-terminal truncation (rgr1Δ2), a gal11 deletion (gal11Δ), or an srb5 deletion (srb5Δ). Despite the expected differences in subunit composition, the chromatographic properties of the mutant holoenzymes were identical to those of the wild-type holoenzyme. In order to determine more precisely the composition of each variation of holoenzyme, the holoenzyme fractions (MonoQ) were immunoprecipitated with anti-Rgr1 antibodies. Silver staining and immunoblot analyses of the immunoprecipitated holoenzymes revealed mutant-specific deficiencies in Mediator subunits. For example, the rgr1Δ2 holoenzyme was deficient in the Sin4, Gal11, and Hrs1 polypeptides (Fig. 1A and B, lanes R) and had substoichiometric amounts of Med7 to Med11, while the immunopurified gal11Δ holoenzyme (lanes G) was completely deficient in Gal11 and devoid of most of Hrs1 but retained all of the other Mediator components in stoichiometric amounts (lanes W). Although a reduced amount of Sin4 was associated with the immunoprecipitated gal11Δ holoenzyme, immunoblot analysis of the gal11Δ holoenzyme fraction (MonoQ) revealed that a wild-type amount of Sin4 copurified with other Mediator components (Fig. 1B, lane G). This result indicates that Sin4 does associate with the gal11Δ holoenzyme in vivo but that their interaction is weakened when Gal11 and Hrs1 are absent; this weakening may cause a partial dissociation of Sin4 during immunoprecipitation.
FIG. 1.
Subunit compositions of mutant holoenzymes. (A) Immunoprecipitation analysis. Holoenzymes (MonoQ fractions) were prepared from wild-type (YCL10; W), rgr1Δ2 (DY2010; R), gal11Δ (HS301; G), hrs1Δ (SSAB-2CF; H), and srb5Δ (CTY153; S) strains and immunoprecipitated with anti-Rgr1 antibody–beads as described in Materials and Methods. Proteins were resolved on an SDS–10% polyacrylamide gel and visualized by silver staining. The positions of core polymerase subunits (Rpb) and Mediator components are indicated at the left and right, respectively. (B) Immunoblot analysis. Wild-type (W) and mutant (R, rgr1Δ2; S, srb5Δ; G, gal11Δ; H, hrs1Δ) holoenzymes (MonoQ fractions) were subjected to immunoblot analysis with antisera specific for the PolII and Mediator components indicated between the panels.
Although HRS1 was isolated originally as an extragenic suppressor of the hyperdeletion phenotype of hpr1Δ cells (37), it has been reported that hrs1Δ mutant strains have transcriptional defects similar to those of gal11 and sin4 null mutant strains (33). Therefore, the loss of Hrs1 from the gal11Δ holoenzyme prompted us to examine whether the hrs1Δ holoenzyme has similar defects. To this end, we purified the hrs1Δ holoenzyme and examined its subunit composition (Fig. 1A and B, lanes H). Interestingly, Gal11 is the only other Mediator component deficient in the hrs1Δ holoenzyme besides Hrs1 (Fig. 1B, lanes G and H) (immunoblot analysis also revealed that Sin4 was associated with the hrs1Δ holoenzyme in vivo). These results strongly suggest that the similar mutant phenotypes exhibited by rgr1Δ2, sin4Δ, gal11Δ, and hrs1Δ mutant strains resulted mainly from the common loss of Gal11 and Hrs1 in these strains.
The subunit composition of the srb5Δ holoenzyme was quite different from that of the rgr1Δ2 holoenzyme (Fig. 1A and B, lanes S). The srb5Δ holoenzyme was completely devoid of Srb2 and Srb5 but retained all of the other Mediator subunits, although certain subunits were present in substoichiometric amounts (Hrs1, Med7 to Med9, and Med11). In addition, the loss of the Srb2 and Srb5 proteins from the holoenzyme appeared to have a secondary effect on the overall integrity of the holoenzyme. The absolute amount of each Mediator subunit was one-fifth the wild-type level, as judged from immunoblot analyses of both purified holoenzymes (MonoQ fraction; Fig. 1B) and crude chromatographic fractions (BioRex70 and DEAE-Sepharose fractions; data not shown). Therefore, the phenotype of the srb5Δ mutant strain may result from the combined effects of the loss of Srb2- and Srb5-specific functions, as well as the reduced amount of the holoenzyme itself.
Transcriptional activities of mutant holoenzymes in vitro.
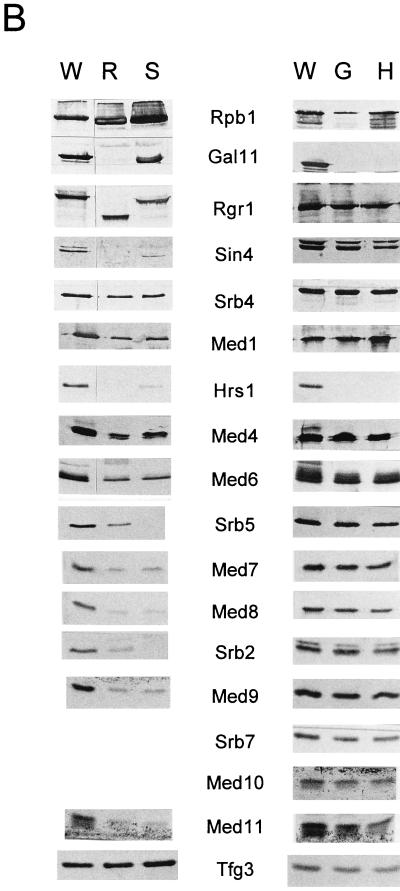
In order to determine which of the compositional defects of the mutant holoenzymes are directly responsible for each of the transcriptional defects, we examined the transcriptional activities of the mutant holoenzymes by using a reconstituted in vitro system. When equivalent amounts of each holoenzyme (based on nonspecific transcription activity) were used to assess basal transcription, the rgr1Δ2 holoenzyme displayed basal transcription activity comparable to that of the wild-type holoenzyme (Fig. 2A, lanes 1 and 2). The gal11Δ and hrs1Δ holoenzymes also displayed basal transcription levels similar to those of the wild-type holoenzymes (Fig. 2B, lanes 1, 4, and 7). However, the srb5Δ holoenzyme consistently showed basal transcription activity three- to fourfold weaker than those of the other enzymes (Fig. 2A, lane 3). These results indicate that Srb5 and Srb2 are required for basal transcription, whereas the Gal11 module proteins are dispensable for basal transcription in vitro.
FIG. 2.
In vitro transcription of mutant holoenzymes. Wild-type and mutant holoenzymes (700 ng each) containing the same levels of nonspecific transcriptional elongation activities were analyzed for their promoter-specific transcriptional activities in the presence of the indicated activators in an in vitro transcription system reconstituted with pure general transcription factors and other supplements as described previously (21). RNA transcripts from templates containing either the Gal4 binding site (GAL4:G−) or the GCN4 binding site (GCN4:G−) are indicated. (A) Transcriptional defects of rgr1Δ2 and srb5Δ holoenzymes. The specifically initiated transcripts from reactions that contained wild-type (W), rgr1Δ2 (R), and srb5Δ (S) holoenzymes in the absence (none) or presence of activator protein (Gal4VP16 or GCN4; 30 ng each) are shown. (B) Transcriptional defects of gal11Δ and hrs1Δ holoenzymes. The specifically initiated transcripts from reactions that contained wild-type, gal11Δ, and hrs1Δ holoenzymes in the absence (−) or presence of activator protein (Gal4VP16 [V] or GCN4 [N]; 30 ng each) are shown.
When the transcriptional activator protein Gal4VP16 was added to basal transcription reaction mixtures containing the wild-type holoenzyme under permissive conditions, transcription from the activator-specific template was increased more than 20-fold (Fig. 2A, lane 4, and 2B, lane 2). However, all of the other mutant holoenzymes that we tested were completely defective for transcriptional activation by Gal4VP16, even under permissive conditions (Fig. 2A, lanes 5 and 6, and 2B, lanes 5 and 8) (1.1- to 2-fold activation). Even when five times more srb5Δ holoenzyme fraction was used to make the amounts of other Mediator proteins in that fraction equivalent to those in the other holoenzyme fractions, the srb5Δ holoenzyme was not able to mediate transcriptional activation (data not shown). Only when recombinant Srb2 and Srb5 proteins were added to the transcription reaction mixture was the srb5 mutant holoenzyme able to respond to the activator (data not shown). These results show that Rgr1, Sin4, Gal11, and Hrs1 are required mainly for activated transcription, while Srb5 and Srb2 are required for both activated transcription and basal transcription in vitro.
Transcriptional activation by GCN4 was affected similarly in that the rgr1Δ2 and srb5Δ holoenzymes were nearly incapable of responding to the activator (Fig. 2A, lanes 8 and 9) (one- and twofold activation, respectively). However, the gal11Δ and hrs1Δ holoenzymes retained some level of activation by GCN4 compared to the rgr1Δ2 and srb5Δ holoenzymes (Fig. 2B, lanes 5 to 9) (three- and fivefold activation for the gal11Δ and hrs1Δ holoenzymes, respectively). Whether this small but activator-specific difference in transcriptional activation between the mutant holoenzymes reflects an activator-specific requirement for certain Mediator subunits is currently under investigation (see Discussion).
CTD phosphorylation of mutant holoenzymes.
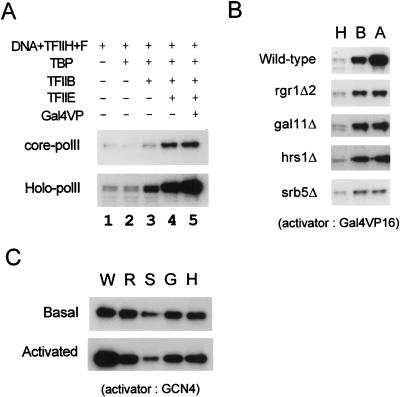
The unphosphorylated form of PolII is recruited to the transcriptional preinitiation complex (PIC) and then phosphorylated at the CTD by TFIIH during the transition from transcriptional initiation to elongation (reviewed in reference 35). TFIIH-dependent CTD phosphorylation of both the core polymerase and the holoenzyme was stimulated by conditions that promote PIC formation (Fig. 3A, lanes 1 to 4). However, due to the presence of Mediator activity, phosphorylation of the holoenzyme was more than 15-fold higher than that of the core polymerase under each of the conditions tested (Fig. 3A, lanes 1 to 4). Furthermore, the addition of Gal4VP16 increased the efficiency of phosphorylation of the holoenzyme threefold but had no effect on the phosphorylation of the core polymerase (Fig. 3A, lanes 4 and 5). The various conditions that prevent activated transcription (such as the omission of a general transcription factor or DNA template) all yielded a complete loss of activator-dependent stimulation of CTD phosphorylation (data not shown).
FIG. 3.
TFIIH-dependent CTD phosphorylation of mutant holoenzymes. (A) Core polymerase (core-polII, 0.3 μg) and holoenzyme (Holo-polII, 0.9 μg) were incubated with [γ-32P]ATP and without (−) or with (+) the indicated supplements (Gal4VP16 [Gal4VP, 30 ng], TBP [50 ng], TFIIB [50 ng], TFIIE [60 ng], TFIIH [60 ng], TFIIF [F, 20 ng], and DNA [pJJ470, 200 ng]) in transcription buffer for 10 min (holoenzyme) or 40 min (core polymerase) at 25°C. Phosphorylated Rpb1 was analyzed by SDS-PAGE (7.5% gel) and visualized by autoradiography. (B) CTD phosphorylation of mutant holoenzymes during Gal4VP16-mediated transcriptional activation. The degrees of CTD phosphorylation of the indicated holoenzymes (900 ng) under various transcription reaction conditions are shown. CTD phosphorylation by TFIIH (60 ng) only in the presence of the DNA template (pJJ470, 200 ng; lane H) and under basal (lane B) or activated (Gal4VP16 mediated, lane A) transcription conditions is shown. (C) CTD phosphorylation of mutant holoenzymes during GCN4-mediated transcriptional activation. CTD of wild-type (W), rgr1Δ2 (R), srb5Δ (S), gal11Δ (G), and hrs1Δ (H) holoenzymes was phosphorylated by TFIIH under basal or activated (GCN4-mediated) transcription conditions.
In order to investigate whether the mutant holoenzymes were defective in any aspect of CTD phosphorylation, we examined their phosphorylation efficiencies under conditions that support basal and activated transcription. Under basal transcription conditions, the levels of CTD phosphorylation displayed by the rgr1Δ, gal11Δ, and hrs1Δ holoenzymes were equal to that of the wild-type holoenzyme; however, the srb5Δ holoenzyme consistently exhibited only one-third the level of CTD phosphorylation exhibited by the wild-type holoenzyme (Fig. 3B, lane B, and 3C, lane S). Under conditions that support activated transcription, all of the mutant holoenzymes lost their ability to respond to Gal4VP16, in that no activator-dependent stimulation of CTD phosphorylation was observed (Fig. 3B, lane A). These results reveal a positive correlation between the rate of in vitro transcription and the level of TFIIH-dependent CTD phosphorylation and confirm the specific requirements for each Mediator subunit in transcriptional activation that were suggested by the in vitro transcription assays (Fig. 2).
We also tested whether this simple correlation between transcriptional activation and CTD phosphorylation efficiency holds true for GCN4-mediated reactions. The mutant holoenzymes were also defective for the stimulation of CTD phosphorylation in response to GCN4 (Fig. 3C). Although we detected a small increase in the CTD phosphorylation efficiencies of the gal11Δ and hrs1Δ holoenzymes (Fig. 3C, lanes G and H) (1.3- to 1.5-fold stimulation) in response to GCN4-mediated transcriptional activation (Fig. 2B) (3- to 5-fold activation), the effect was not significant. Therefore, for unknown reasons there appears to be no simple correlation between activation and CTD phosphorylation under the conditions that we used.
Direct interactions between transcriptional activators and holoenzymes.
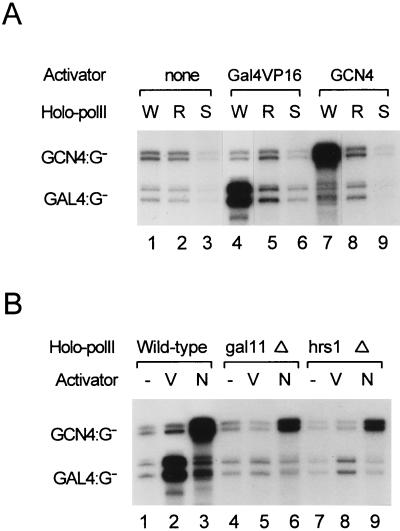
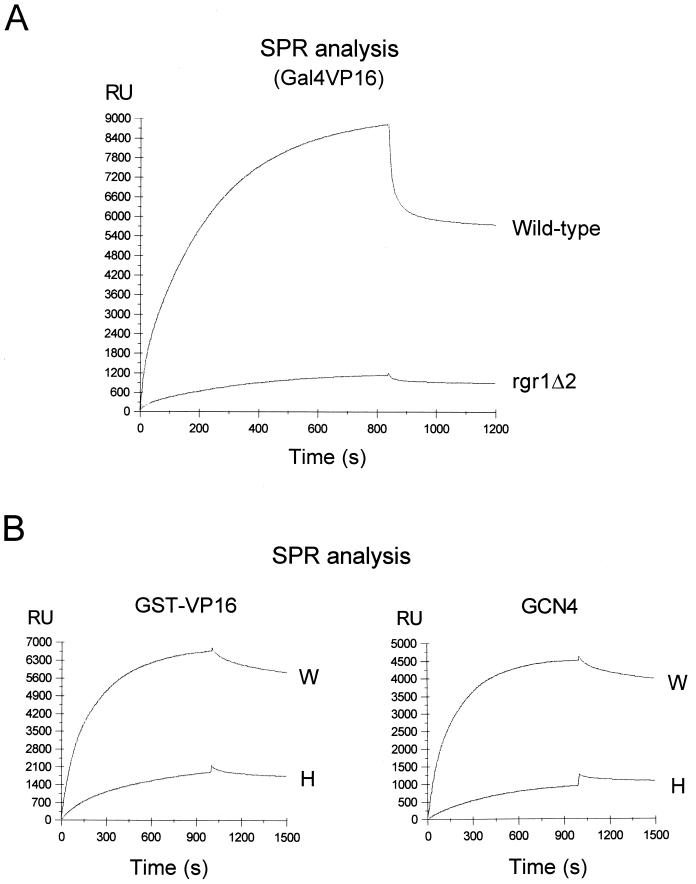
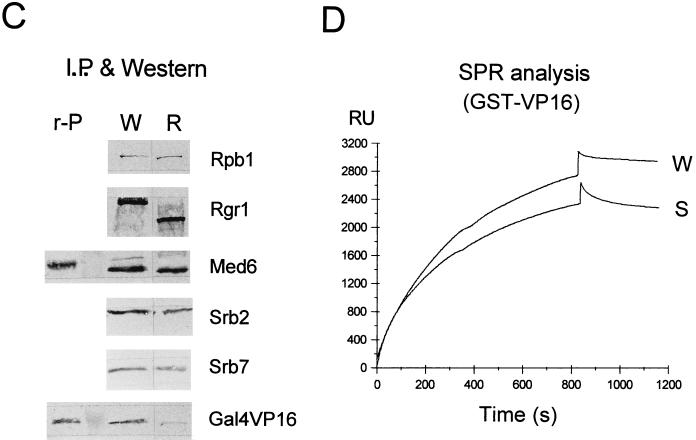
Although the VP16 activation domain has been shown to interact physically with a number of general transcription factors (18, 34, 39, 47), this type of interaction (that is, in the absence of the Mediator complex) does not elicit an activator response in a reconstituted in vitro transcription system. In order to explore the possibility of an activator-Mediator interaction and its relevance in transcriptional activation, we examined the activator binding strengths of wild-type and mutant holoenzymes by using a conventional immunoprecipitation analysis (Fig. 4C). The wild-type holoenzyme was coimmunoprecipitated with a stoichiometric amount of Gal4VP16 (Fig. 4C, lane W), while the amount of the activator that was coimmunoprecipitated with the rgr1Δ2 holoenzyme was less than 15% that immunoprecipitated with the wild-type holoenzyme (lane R). This result showed that the activator binds to at least one of the Mediator subunits associated with the C-terminal region of Rgr1. In order to quantitate the activator binding strengths of wild-type and mutant holoenzymes in equilibrium, we used the SPR assay (performed with a BIAcore instrument; see Materials and Methods). When a saturating amount of the wild-type holoenzyme was passed over a surface-immobilized Gal4VP16 chip (application was followed by extensive washing), the refractory index was increased more than 5,500 ΔRU (change in the refractory index units), indicating a very strong interaction between the activator and the holoenzyme (Fig. 4A). This interaction was specific for a functional activation domain, as the holoenzyme bound efficiently to the VP16 activation domain alone (GST-VP16) but not to a mutated, nonfunctional activation domain (GST-VP16Δ456FP442) (Fig. 4B and data not shown). The holoenzyme also bound efficiently to GCN4 (4,000 ΔRU), indicating the activator-holoenzyme interaction to be a general phenomenon (Fig. 4B).
FIG. 4.
Interactions of transcriptional activators with wild-type and mutant holoenzymes. (A, B, and D) Plotted is the change in the refractory index units (RU) versus time after injection of the indicated holoenzymes onto an activator-immobilized Biosensor chip. (A) SPR analysis of the interactions between Gal4VP16 and wild-type and rgr1Δ2 holoenzymes (70 μl of a MonoQ fraction at a concentration of 200 μg/ml). (B) SPR analysis of the interactions between wild-type (W) and hrs1Δ (H) holoenzymes (50 μl of a MonoQ fraction at a concentration of 250 μg/ml) and GST-VP16 or GCN4. (C) Coimmunoprecipitation (I.P.) of Gal4VP16 with wild-type (W) and mutant (R, rgr1Δ2) holoenzymes followed by Western blotting. Each holoenzyme (MonoQ fraction, 10 μg) was mixed with Gal4VP16 (300 ng) and immunoprecipitated with anti-Rgr1 antibody–beads (see Materials and Methods). In order to measure the amounts of precipitated Gal4VP16 and holoenzymes, we included equimolar amounts of recombinant Gal4VP16 and Med6 proteins (r-P; Med6, 80 ng; Gal4VP16, 60 ng) as quantitative standards. Immunoblot analyses with antisera specific for the proteins indicated to the right of the panel are shown. (D) SPR analysis of the interactions between wild-type (W, 150 μg/ml) and srb5Δ (S, 450 μg/ml) holoenzymes and an activator (GST-VP16). Due to the low specific concentration of the srb5Δ holoenzyme in the MonoQ fraction (one-fifth the wild-type level), a threefold-larger amount of srb5Δ MonoQ fraction was injected to supply an amount of the srb5Δ holoenzyme comparable to that of the wild-type enzyme.
In contrast to the strong interaction between the activator and the wild-type holoenzyme, the rgr1Δ2 holoenzyme caused only a minor increase in the refractory index under identical conditions (ΔRU, 880; corresponding to ∼15% of the ΔRU observed with the wild-type holoenzyme), indicating that the mutant holoenzyme bound poorly to Gal4VP16. The gal11Δ and hrs1Δ holoenzymes were also defective in activator interaction, according to the SPR analysis (Fig. 4B and data not shown). The fact that these three mutant holoenzymes all lacked Gal11 and Hrs1 suggests that Gal11 and Hrs1 are required for activator binding and may serve as the major binding targets for transcriptional activators.
In order to test whether the transcriptional defect of the srb5Δ holoenzyme resulted from a defect in activator binding, we measured the interaction between the activator (Gal4VP16) and the srb5Δ holoenzyme by using SPR analysis. As shown in Fig. 4D, the srb5Δ holoenzyme displayed activator binding activity comparable to that of the wild-type holoenzyme. This result implies that the crippled transcriptional activation observed with the srb5Δ holoenzyme did not result from a defective activator interaction. Rather, this result and the CTD phosphorylation data (Fig. 3B) suggest that Srb2 and Srb5 function at a later step to modulate PolII activity through an interaction with the CTD.
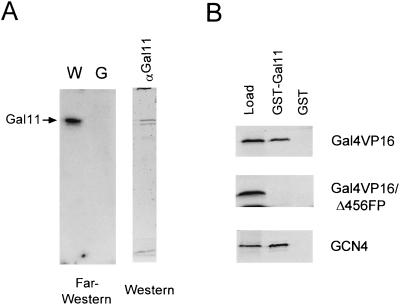
Identification of Gal11 as a target for activator binding.
In order to determine which Mediator subunit(s) interacts directly with the activator, we probed an immunoaffinity-purified holoenzyme with radioactive Gal4VP16 by using far-Western analysis (see Materials and Methods). Two polypeptides with an apparent molecular size equivalent to that of Gal11 interacted strongly with the Gal4VP16 probe (Fig. 5A, lane W). These polypeptides cross-reacted with an anti-Gal11 antibody and were absent in the gal11Δ holoenzyme (Fig. 5A, lane G). These results demonstrate that two polypeptides are encoded by GAL11 (the faster-migrating band might be a breakdown product of Gal11) and that Gal11 alone is sufficient for activator binding. In order to examine whether this interaction requires a functional activator, we analyzed the interactions between GST-Gal11 and 35S-labeled Gal4VP16 mutant derivatives (Fig. 5B). A substantial amount of wild-type Gal4VP16 (10% of the amount in the loaded column fraction) bound specifically to the GST-Gal11 column but not to the GST column. However, nonfunctional mutant Gal4VP16Δ456FP442 did not bind at all to the GST-Gal11 column. These results clearly show a specific and direct interaction between Gal11 and a functional VP16 activation domain. Furthermore, we obtained similar results when GCN4 was used as the activator in the GST-Gal11 column binding assay. Therefore, Gal11 appears to constitute a general binding site for acidic transcriptional activation domains.
FIG. 5.
Direct interaction of Gal11 with acidic activators. (A) Far-Western analysis of holoenzymes with Gal4VP16. Wild-type (W) and gal11Δ (G) holoenzymes (immunopurified from a MonoQ fraction with an anti-Rgr1 antibody column, ∼2 μg each) were resolved on an SDS–10% polyacrylamide gel and transferred to a nitrocellulose membrane. The proteins were renatured and allowed to bind to 32P-labeled Gal4VP16 as described in Materials and Methods. The position of the Gal11 protein was revealed by Western analysis of the same blot with the use of anti-Gal11 antiserum (αGal11). (B) Column binding assay for interactions between Gal11 and acidic activators. Agarose beads conjugated with purified GST or GST-Gal11 (5 μg each) were incubated overnight at 4°C with 10 μl of in vitro-translated, 35S-labeled Gal4VP16, Gal4VP16Δ456FP442, or GCN4. After extensive washing, the beads were boiled in SDS sample buffer, and the proteins were resolved by SDS–12.5% PAGE and subjected to autoradiography. Load represents 10% of the amount of in vitro-translated products loaded on the beads.
Artificial recruitment of the gal11 mutant holoenzyme.
The fact that the Rgr1 subcomplex, in particular, the Gal11 module interacts directly with acidic transcriptional activators suggests that the Gal11 module functions in the recruitment of the holoenzyme to the promoter via a specific interaction with activator proteins. Therefore, it is conceivable that transcriptional defects in gal11 mutant cells would originate mainly from the inefficient recruitment of PolII to the promoter. In order to test this hypothesis, we examined whether artificial recruitment of the gal11 mutant holoenzyme to the promoter could bypass the requirement for the Gal11 module in transcriptional activation.
Transformation of a LexA-Med6 fusion construct on a CEN-ARS plasmid into a wild-type strain as an extragenic copy caused 300-fold transcriptional activation of the reporter gene containing LexA binding sites at the promoter. The addition of the LexA-Med6 fusion construct to the gal11 null strain also induced more than 90-fold transcriptional activation of the reporter gene (Table 1). Interestingly, the substitution of wild-type Med6 with a LexA-Med6 fusion protein in the above strains clearly demonstrated that the artificial recruitment of the holoenzyme could bypass the requirement for the Gal11 module in transcriptional activation. The artificial recruitment of the holoenzyme via LexA-Med6 to the LexA binding sites of the reporter gene caused almost 2,000-fold transcriptional activation in both the wild-type and the gal11 null mutant strains (Table 1). When similar experiments were performed with the srb5 null mutant strain, artificial recruitment of the srb5 holoenzyme via LexA-Med6 to LexA binding sites did not rescue the transcriptional defect of the srb5Δ holoenzyme (Table 1). These results suggest that Gal11 functions mainly in the holoenzyme recruitment step and may be dispensable for subsequent steps in the transcription process once the holoenzyme is brought to a promoter through specific interactions with an enhancer-bound activator. In contrast, Srb5 appears to be required for the postrecruitment steps of transcriptional activation.
TABLE 1.
Artificial recruitment of the RNA PolII holoenzyme restores the transcriptional defects in a gal11 null strain but not in an srb5 null strain
| Strain | Genotype | LexA-Med6a | β-Gal activityb |
|---|---|---|---|
| YCL10 | GAL11+ SRB5+ MED6+ | − | <1 |
| JMP5 | GAL11+ SRB5+ MED6+ | + | 299 |
| JMP3 | GAL11+ SRB5+ med6Δ | + | 1,843 |
| JMP2 | gal11Δ SRB5+ MED6+ | − | <1 |
| JMP6 | gal11Δ SRB5+ MED6+ | + | 90 |
| JMP4 | gal11Δ SRB5+ med6Δ | + | 2,012 |
| JMP8 | GAL11+ srb5Δ MED6+ | − | 3 |
| JMP7 | GAL11+ srb5Δ MED6+ | + | 12 |
+, present; −, absent.
β-Galactosidase (β-Gal) activity from a lacZ reporter gene under the control of a GAL1 core promoter with eight LexA binding sites (LexAop-GAL1C-lacZ) was measured. Yeast cells were grown in appropriate selective media containing 2% glucose and transferred to yeast extract-peptone broth containing 2% glucose prior to harvest. β-Galactosidase assays were performed with permeabilized cells in triplicate, and standard errors were less than 20%.
Activator-mediated interactions between holoenzymes and TBP.
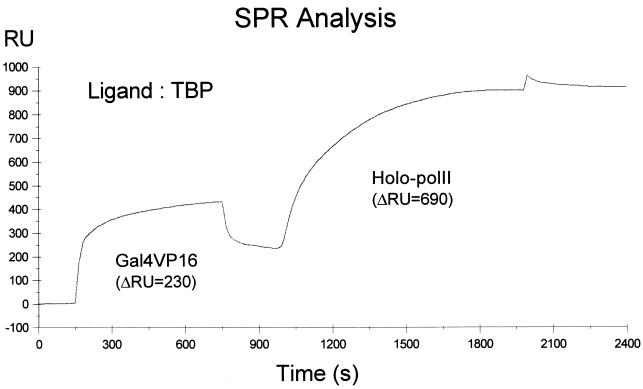
The physical interaction between the activator and Gal11, which can be functionally replaced by the artificial recruitment of the mutant holoenzyme to the promoter, suggests that transcriptional activators facilitate PIC formation chiefly by recruiting a holoenzyme to the promoter. TBP binding to the TATA promoter element is also a rate-limiting step of PIC formation, and it has been suggested that transcriptional activators promote TBP binding by interacting with basal transcriptional factors. Therefore, the concurrent recruitment of the holoenzyme and TBP to a promoter should greatly enhance transcriptional efficiency. In order to test whether an activator could bind to Mediator and TBP simultaneously, the interactions between TBP, Mediator, and a transcriptional activator were examined. When the holoenzyme was passed over a TBP-immobilized Biosensor chip for SPR analysis, we did not observe any specific interactions (data not shown), whereas Gal4VP16 bound efficiently to immobilized TBP (Fig. 6). However, when the holoenzyme was injected after the activator was allowed to bind to immobilized TBP, a significant increase in the refractory index was observed (Fig. 6). Three independent experiments performed with various concentrations of protein gave rise to similar results (data not shown). These results suggest that there is no direct physical interaction between TBP and the holoenzyme; however, the two could associate with each other indirectly through their simultaneous interactions with a transcriptional activator.
FIG. 6.
Simultaneous interactions of an activator with a holoenzyme and TBP. The ΔRU during the time course of the SPR analysis with a TBP-immobilized Biosensor chip is shown. Gal4VP16 (30 μg/ml, 30 μl) and wild-type holoenzyme (Holo-polII; 300 μg/ml, 50 μl) were injected sequentially after washing of the chip with binding buffer. The ΔRU for activator binding to TBP and that for holoenzyme binding to activator bound to TBP are shown.
DISCUSSION
Extensive biochemical studies have demonstrated the essential role of TBP-associated factors in transcriptional activation and selective activator interactions (8, 9, 42). However, in vivo analyses with yeasts have revealed that TBP-associated factors are dispensable for general transcription (29, 44) but are required for the transcription of a subset of essential genes (1, 38, 45). On the other hand, Mediator has been shown to be required for the general regulation of PolII transcription in vivo (41), but the mechanism by which Mediator enables the basal transcription machinery to respond to gene-specific transcriptional regulatory proteins is not known. From the biochemical analyses of various mutant holoenzymes, we have identified the Gal11 module as a specific and direct target for activator binding and have characterized the functional relevance of this interaction with a reconstituted in vitro transcription system. Our results clearly show that Mediator is involved in transcriptional regulation through its direct and specific interactions with activators.
The major activator binding module of Mediator.
Analyses of physical interactions within the Mediator complex have revealed that about two-thirds of the Mediator subunits are tightly bound to Rgr1 (the so-called Rgr1 subcomplex [24]). Within the Rgr1 subcomplex, a group of genetically characterized transcriptional regulators (Sin4, Gal11, and Hrs1) appears to form a separate functional module (Gal11 module). The direct binding of activator proteins to Gal11 supports the idea that the Gal11 module may participate in the regulation of transcription by its interaction with activator and repressor (or corepressor) proteins. The severe reduction in the activator binding efficiency of the gal11Δ holoenzyme and its accompanying inability to respond to the activator in a reconstituted transcription system argue strongly for the physiological significance of the activator-Gal11 interaction. Furthermore, the fact that the artificial tethering of the gal11 mutant holoenzyme to promoters can bypass the requirement for Gal11 in transcriptional activation also supports that notion. Once enhancer-bound activators contact a holoenzyme, they may hold the enzyme in the appropriate conformation for stable PIC formation. Although free activator domains that do not associate with an enhancer may still interact with Gal11, the low specific concentration of activator proteins probably limits this interaction to only the relevant promoters in vivo.
Activator-specific transcriptional activation of Mediator.
Although the gal11Δ and hrs1Δ holoenzymes were defective in their interaction with GCN4 and Gal4VP16, both of the mutant holoenzymes supported some transcriptional activation by GCN4 but not by Gal4VP16 (Table 2). This result suggests that GCN4 has additional targets either in the holoenzyme complex or among the other general transcription factors, whereas Gal4VP16 utilizes the Gal11 module as its major target during the transcription initiation step (4). We recently obtained a result that supports this notion. During the analysis of novel Mediator subunits, we found that a newly identified Mediator component, Med10, is absolutely required for GCN4-mediated transcriptional activation in vivo, whereas the loss of the Gal11 module has a relatively minor effect on GCN4-mediated transcriptional activation (15). Therefore, the Med10-mediated GCN4 response may be more crucial to the activation process than activator binding by the Gal11 module.
TABLE 2.
Quantitation of functional activities of wild-type and mutant holoenzymes in vitroa
| Holoenzyme | Transcription (fold activation) by:
|
CTD phosphorylation (fold stimulation) by:
|
||
|---|---|---|---|---|
| Gal4VP16 | GCN4 | Gal4VP16 | GCN4 | |
| Wild type | 19–21 | 15–17 | 4.3 | 3.6 |
| rgr1Δ2 | 1.3 | 1.0 | 1.1 | 1.2 |
| srb5Δ | 1.6 | 2.3 | 1.0 | 1.0 |
| gal11Δ | 1.1 | 3.0 | 1.0 | 1.3 |
| hrs1Δ | 2.3 | 5.1 | 1.1 | 1.5 |
Although the mechanism by which Med10 mediates GCN4-induced transcriptional activation is not yet known, the presence of stoichiometric amounts of the Med10 subunit in the gal11Δ and hrs1Δ holoenzymes may enable the mutant holoenzymes to respond to GCN4 to some degree. This idea is further supported by the fact that the rgr1Δ2 holoenzyme contains substoichiometric amounts of Rgr1-associated Mediator subunits, including Med10 (Fig. 1B and data not shown). Therefore, the loss of the major activator binding module (Gal11 module) and GCN4-specific Mediator subunit Med10 in the rgr1Δ2 holoenzyme may result in an additive (or synergistic) defect in GCN4-mediated transcriptional activation.
Although our immunoblot analyses of the gal11Δ and hrs1Δ holoenzymes revealed no difference in their subunit compositions, the hrs1Δ holoenzyme showed a higher response to the activator than did the gal11Δ holoenzyme (Table 2). One possible explanation for this observation is that small amounts of Hrs1 and Gal11 may remain associated with the gal11Δ and hrs1Δ holoenzymes, respectively. A residual amount of Gal11 in the hrs1Δ holoenzyme may be responsible for the slightly higher activator response observed with the hrs1Δ holoenzyme.
Mediator subunits involved in the postrecruitment process of transcriptional activation.
Our previous studies revealed the absolute and specific requirement for Med6 in transcriptional activation (25). However, the Mediator-activator interaction was not affected by a deficiency in Med6, indicating that Med6 has no apparent binding affinity for activators in vitro (data not shown). It was also shown that Med6 associates with Srb proteins rather than with the components of the Rgr1 subcomplex (24). In addition, while analyses of the srb5Δ holoenzyme revealed a requirement for Srb2 and Srb5 in both basal and activated transcription in vitro, mutant holoenzymes that lacked these subunits showed a capacity for activator binding that was comparable to that of the wild-type holoenzyme (Fig. 4D). Taken together with the results of an artificial recruitment experiment with the srb5Δ holoenzyme (Table 1), these findings suggest that facilitated recruitment of a holoenzyme to a promoter is necessary but not sufficient for transcriptional activation and that an unknown biochemical activity other than physical recruitment to the promoter is required for transcriptional activation. As mentioned previously, the genetic interaction between the SRB genes and the CTD of PolII indicates that SRB gene products (the Srb4 subcomplex) may function in the modulation or isomerization of holoenzyme activity, possibly subsequent to activator binding. The identification of the mechanism of SRB function is essential for the elucidation of the transcriptional activation mechanism.
Despite the requirement for an activator-Gal11 interaction in transcriptional activation in a defined system, all of the components of the Gal11 module (Sin4, Gal11, and Hrs1) are dispensable for cell viability (30 and references therein). Both SPR and coimmunoprecipitation analyses of mutant holoenzymes that lack Gal11 have shown that these holoenzymes retain a weak ability to interact with activators. Thus, the Gal11 module may constitute the major activator binding site, but secondary activator interaction sites may provide enough binding to sustain cell viability in vivo. Srb4, which was shown to have a low binding affinity for transcriptional activators (22), and other members of the Rgr1 subcomplex may be good candidates for secondary Mediator subunits that interact with activators.
TBP-activator-Mediator interactions.
It is interesting that an activator can bind to TBP and Mediator simultaneously. This result indicates that the activator accelerates and stabilizes PIC formation by interacting with both TBP and a holoenzyme at the same time. Although the TFIIB-PolII interaction helps to stabilize the PIC (reviewed in reference 35), the affinity of the TFIIB-PolII interaction alone may not be high enough to sustain stable PIC formation. The strong connection between TBP and PolII established by the interactions of an activator and Mediator may provide an important contribution to stable PIC formation during transcriptional activation. Therefore, recruitment of TBP and a holoenzyme to the promoter, the stabilization of PIC formation, and isomerization (modulation) of holoenzyme activity appear to constitute the major mechanisms of transcriptional activation by the holoenzyme in vitro.
ACKNOWLEDGMENTS
We thank Jeong Kon Seo and Juri Kim for technical help, Kelly LaMarco for careful reading of the manuscript, and other members of Young-Joon Kim’s laboratory for helpful comments. We also thank Roger Kornberg, Richard Young, Toshio Fukasawa, David Stillman, and Andres Aguilera for providing Mediator mutant strains, antibodies, and related plasmids. We are grateful to Michael Green for GST-VP16 fusion plasmids and Steve Hanes for reporter plasmids.
This work was supported by grants from SBRI (B-96-004) and the Ministry of Health and Welfare, Republic of Korea (HMP-97-B-3-0030 of the 1997 Good Health R&D Project), to Y.-J.K.
REFERENCES
- 1.Apone L M, Virbasius C M, Reese J C, Green M R. Yeast TAF(II)90 is required for cell-cycle progression through G2/M but not for general transcription activation. Genes Dev. 1996;10:2368–2380. doi: 10.1101/gad.10.18.2368. [DOI] [PubMed] [Google Scholar]
- 2.Berger S L, Cress W D, Cress A, Triezenberg S J, Guarente L. Selective inhibition of activated but not basal transcription by the acidic activation domain of VP16: evidence for transcriptional adaptors. Cell. 1990;61:1199–1208. doi: 10.1016/0092-8674(90)90684-7. [DOI] [PubMed] [Google Scholar]
- 3.Bjorklund S, Kim Y J. Mediator of transcriptional regulation. Trends Biochem Sci. 1996;21:335–337. doi: 10.1016/s0968-0004(96)10051-7. [DOI] [PubMed] [Google Scholar]
- 4.Brown S A, Weirich C S, Newton E M, Kingston R E. Transcriptional activation domains stimulate initiation and elongation at different times and via different residues. EMBO J. 1998;17:3146–3154. doi: 10.1093/emboj/17.11.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chasman D I, Leatherwood J, Carey M, Ptashne M, Kornberg R D. Activation of yeast polymerase II transcription by herpesvirus VP16 and GAL4 derivatives in vitro. Mol Cell Biol. 1989;9:4746–4749. doi: 10.1128/mcb.9.11.4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Estojak J, Brent R, Golemis E A. Correlation of two-hybrid affinity data with in vitro measurements. Mol Cell Biol. 1995;15:5820–5829. doi: 10.1128/mcb.15.10.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flanagan P M, Kelleher III R J, Sayre M H, Tschochner H, Kornberg R D. A mediator required for activation of RNA polymerase II transcription in vitro. Nature. 1991;350:436–438. doi: 10.1038/350436a0. [DOI] [PubMed] [Google Scholar]
- 8.Goodrich J A, Cutler G, Tjian R. Contacts in context: promoter specificity and macromolecular interactions in transcription. Cell. 1996;84:825–830. doi: 10.1016/s0092-8674(00)81061-2. [DOI] [PubMed] [Google Scholar]
- 9.Goodrich J A, Tjian R. TBP-TAF complexes: selectivity factors for eukaryotic transcription. Curr Opin Cell Biol. 1994;6:403–409. doi: 10.1016/0955-0674(94)90033-7. [DOI] [PubMed] [Google Scholar]
- 10.Guarente L. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 1983;101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- 11.Guarente L, Yocum R R, Gifford P. A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc Natl Acad Sci USA. 1982;79:7410–7414. doi: 10.1073/pnas.79.23.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gustafsson C M, Myers L C, Beve J, Spahr H, Lui M, Erdjument-Bromage H, Tempst P, Kornberg R D. Identification of new mediator subunits in the RNA polymerase II holoenzyme from Saccharomyces cerevisiae. J Biol Chem. 1998;273:30851–30854. doi: 10.1074/jbc.273.47.30851. [DOI] [PubMed] [Google Scholar]
- 13.Gustafsson C M, Myers L C, Li Y, Redd M J, Lui M, Erdjument-Bromage H, Tempst P, Kornberg R D. Identification of Rox3 as a component of mediator and RNA polymerase II holoenzyme. J Biol Chem. 1997;272:48–50. doi: 10.1074/jbc.272.1.48. [DOI] [PubMed] [Google Scholar]
- 14.Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 15.Han S J, Lee Y C, Gim B S, Ryu G-H, Park S J, Lane W S, Kim Y-J. Activator-specific requirement of yeast Mediator proteins for RNA polymerase II transcriptional activation. Mol Cell Biol. 1999;19:979–988. doi: 10.1128/mcb.19.2.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hengartner C J, Thompson C M, Zhang J, Chao D M, Liao S M, Koleske A J, Okamura S, Young R A. Association of an activator with an RNA polymerase II holoenzyme. Genes Dev. 1995;9:897–910. doi: 10.1101/gad.9.8.897. [DOI] [PubMed] [Google Scholar]
- 17.Henry N L, Campbell A M, Feaver W J, Poon D, Weil P A, Kornberg R D. TFIIF-TAF-RNA polymerase II connection. Genes Dev. 1994;8:2868–2878. doi: 10.1101/gad.8.23.2868. [DOI] [PubMed] [Google Scholar]
- 18.Ingles C J, Shales M, Cress W D, Triezenberg S J, Greenblatt J. Reduced binding of TFIID to transcriptionally compromised mutants of VP16. Nature. 1991;351:588–590. doi: 10.1038/351588a0. [DOI] [PubMed] [Google Scholar]
- 19.Kaelin W G, Jr, Krek W, Sellers W R, DeCaprio J A, Ajchenbaum F, Fuchs C S, Chittenden T, Li Y, Farnham P J, Blanar M A, Livingston D M, Flemington E K. Expression cloning of a cDNA encoding a retinoblastoma-binding protein with E2F-like properties. Cell. 1992;70:351–364. doi: 10.1016/0092-8674(92)90108-o. [DOI] [PubMed] [Google Scholar]
- 20.Kelleher R J, III, Flanagan P M, Kornberg R D. A novel mediator between activator proteins and the RNA polymerase II transcription apparatus. Cell. 1990;61:1209–1215. doi: 10.1016/0092-8674(90)90685-8. [DOI] [PubMed] [Google Scholar]
- 21.Kim Y J, Bjorklund S, Li Y, Sayre M H, Kornberg R D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 22.Koh S S, Ansari A Z, Ptashne M, Young R A. An activator target in the RNA polymerase II holoenzyme. Mol Cell. 1998;1:895–904. doi: 10.1016/s1097-2765(00)80088-x. [DOI] [PubMed] [Google Scholar]
- 23.Lee T I, Wyrick J J, Koh S S, Jennings E G, Gadbois E L, Young R A. Interplay of positive and negative regulators in transcription initiation by RNA polymerase II holoenzyme. Mol Cell Biol. 1998;18:4455–4462. doi: 10.1128/mcb.18.8.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee Y C, Kim Y-J. Requirement for a functional interaction between mediator components Med6 and Srb4 in RNA polymerase II transcription. Mol Cell Biol. 1998;18:5364–5370. doi: 10.1128/mcb.18.9.5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee Y C, Min S, Gim B S, Kim Y J. A transcriptional mediator protein that is required for activation of many RNA polymerase II promoters and is conserved from yeast to humans. Mol Cell Biol. 1997;17:4622–4632. doi: 10.1128/mcb.17.8.4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, Bjorklund S, Jiang Y W, Kim Y J, Lane W S, Stillman D J, Kornberg R D. Yeast global transcriptional regulators Sin4 and Rgr1 are components of mediator complex/RNA polymerase II holoenzyme. Proc Natl Acad Sci USA. 1995;92:10864–10868. doi: 10.1073/pnas.92.24.10864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao S M, Zhang J, Jeffery D A, Koleske A J, Thompson C M, Chao D M, Viljoen M, van Vuuren H J, Young R A. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature. 1995;374:193–196. doi: 10.1038/374193a0. [DOI] [PubMed] [Google Scholar]
- 28.Lin Y S, Green M R. Mechanism of action of an acidic transcriptional activator in vitro. Cell. 1991;64:971–981. doi: 10.1016/0092-8674(91)90321-o. [DOI] [PubMed] [Google Scholar]
- 29.Moqtaderi Z, Bai Y, Poon D, Weil P A, Struhl K. TBP-associated factors are not generally required for transcriptional activation in yeast. Nature. 1996;383:188–191. doi: 10.1038/383188a0. [DOI] [PubMed] [Google Scholar]
- 30.Myers L C, Gustafsson C M, Bushnell D A, Lui M, Erdjument-Bromage H, Tempst P, Kornberg R D. The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev. 1998;12:45–54. doi: 10.1101/gad.12.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nonet M L, Young R A. Intragenic and extragenic suppressors of mutations in the heptapeptide repeat domain of Saccharomyces cerevisiae RNA polymerase II. Genetics. 1989;123:715–724. doi: 10.1093/genetics/123.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Neil K, Shuman J D, Ampe C, DeGrado W F. DNA-induced increase in the alpha-helical content of C/EBP and GCN4. Biochemistry. 1991;30:9030–9034. doi: 10.1021/bi00101a017. [DOI] [PubMed] [Google Scholar]
- 33.Piruat J I, Chavez S, Aguilera A. The yeast HRS1 gene is involved in positive and negative regulation of transcription and shows genetic characteristics similar to SIN4 and GAL11. Genetics. 1997;147:1585–1594. doi: 10.1093/genetics/147.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roberts S G, Ha I, Maldonado E, Reinberg D, Green M R. Interaction between an acidic activator and transcription factor TFIIB is required for transcriptional activation. Nature. 1993;363:741–744. doi: 10.1038/363741a0. [DOI] [PubMed] [Google Scholar]
- 35.Roeder R G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 36.Sakurai H, Hiraoka Y, Fukasawa T. Yeast GAL11 protein is a distinctive type transcription factor that enhances basal transcription in vitro. Proc Natl Acad Sci USA. 1993;90:8382–8386. doi: 10.1073/pnas.90.18.8382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Santos-Rosa H, Clever B, Heyer W D, Aguilera A. The yeast HRS1 gene encodes a polyglutamine-rich nuclear protein required for spontaneous and hpr1-induced deletions between direct repeats. Genetics. 1996;142:705–716. doi: 10.1093/genetics/142.3.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen W C, Green M R. Yeast TAF(II)145 functions as a core promoter selectivity factor, not a general coactivator. Cell. 1997;90:615–624. doi: 10.1016/s0092-8674(00)80523-1. [DOI] [PubMed] [Google Scholar]
- 39.Stringer K F, Ingles C J, Greenblatt J. Direct and selective binding of an acidic transcriptional activation domain to the TATA-box factor TFIID. Nature. 1990;345:783–786. doi: 10.1038/345783a0. [DOI] [PubMed] [Google Scholar]
- 40.Thompson C M, Koleske A J, Chao D M, Young R A. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell. 1993;73:1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- 41.Thompson C M, Young R A. General requirement for RNA polymerase II holoenzymes in vivo. Proc Natl Acad Sci USA. 1995;92:4587–4590. doi: 10.1073/pnas.92.10.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verrijzer C P, Tjian R. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem Sci. 1996;21:338–342. [PubMed] [Google Scholar]
- 43.Vinson C R, LaMarco K L, Johnson P F, Landschulz W H, McKnight S L. In situ detection of sequence-specific DNA binding activity specified by a recombinant bacteriophage. Genes Dev. 1988;2:801–806. doi: 10.1101/gad.2.7.801. [DOI] [PubMed] [Google Scholar]
- 44.Walker S S, Reese J C, Apone L M, Green M R. Transcription activation in cells lacking TAF(II)s. Nature. 1996;383:185–188. doi: 10.1038/383185a0. [DOI] [PubMed] [Google Scholar]
- 45.Walker S S, Shen W C, Reese J C, Apone L M, Green M R. Yeast TAF(II)145 required for transcription of G1/S cyclin genes and regulated by the cellular growth state. Cell. 1997;90:607–614. doi: 10.1016/s0092-8674(00)80522-x. [DOI] [PubMed] [Google Scholar]
- 46.Wilson C J, Chao D M, Imbalzano A N, Schnitzler G R, Kingston R E, Young R A. RNA polymerase II holoenzyme contains SWI/SNF regulators involved in chromatin remodeling. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- 47.Xiao H, Pearson A, Coulombe B, Truant R, Zhang S, Regier J L, Triezenberg S J, Reinberg D, Flores O, Ingles C J, Greenblatt J. Binding of basal transcription factor TFIIH to the acidic activation domains of VP16 and p53. Mol Cell Biol. 1994;14:7013–7024. doi: 10.1128/mcb.14.10.7013. [DOI] [PMC free article] [PubMed] [Google Scholar]