Abstract
The yeast transcriptional activator Gal4p can bind to sites in nucleosomal DNA in vivo which it is unable to access in vitro. One event which could allow proteins to bind to otherwise inaccessible sites in chromatin in living cells is DNA replication. To determine whether replication is required for Gal4p to bind to nucleosomal sites in yeast, we have used previously characterized chromatin reporters in which Gal4p binding sites are incorporated into nucleosomes. We find that Gal4p is able to perturb nucleosome positioning via nucleosomal binding sites in yeast arrested either in G1, with α-factor, or in G2/M, with nocodazole. Similar results were obtained whether Gal4p synthesis was induced from the endogenous promoter by growth in galactose medium or by an artificial, hormone-inducible system. We also examined binding of the Drosophila transcriptional activator Bicoid, which belongs to the homeodomain class of transcription factors. We show that Bicoid, like Gal4p, can bind to nucleosomal sites in SWI+ and swi1Δ yeast and in the absence of replication. Our results indicate that some feature of the intracellular environment other than DNA replication or the SWI-SNF complex permits factor access to nucleosomal sites.
Transcriptional activators work in part by overcoming the repressive influence of chromatin, as suggested by experiments both in vitro and in vivo (27, 55, 78). However, this leaves as an open question the mechanism by which the activators themselves gain access to sites in chromatin. To address this question, we and others have used the yeast Saccharomyces cerevisiae to examine the interactions of transcriptional activators with defined chromatin structures (24, 47, 48, 61, 65, 72, 83), with particular emphasis on the yeast Gal4 protein. Previous work has demonstrated that Gal4p can bind in vitro to reconstituted chromatin containing five Gal4p binding sites, either as an isolated nucleosome or in an array, and form a metastable complex, independent of an activation domain (53, 79). However, Gal4p binding to a single nucleosomal site is strongly inhibited when the binding site is centered 40 or 74 bp from the nucleosome edge; when the site is centered 21 bp from the edge, binding is effective (75). In contrast, studies in yeast have shown that Gal4p can gain access to sites near the center of a positioned nucleosome on a multicopy plasmid when overexpressed (48, 83) and can bind to a site centered 41 bp from the edge of the nucleosome even at endogenous levels (65, 83). In all cases, binding results in perturbation of chromatin (48, 65, 83) and is greatly enhanced by the presence of a functional activation domain (48, 65).
The mechanism by which Gal4p gains access to nucleosomal sites in vivo is unclear. One possibility is that a chromatin remodeling complex, such as SWI-SNF, can recognize sequence or structural elements near the Gal4p binding site and remodel nucleosomes locally to facilitate Gal4p binding (13, 57). However, although binding of Gal4p to a pair of nucleosomal weak binding sites in yeast has been shown to be stronger in SWI+ than swi cells (8), we have found that neither SWI-SNF nor GCN5 is needed for perturbation by Gal4p of a positioned nucleosome containing a strong Gal4p binding site in yeast (61, 67). Another possibility is that during DNA replication, the transient removal of the histones from DNA provides an opportunity for transcription factors such as Gal4p to bind to nucleosomal sites in vivo (7, 70, 77). In vitro studies have shown that in some cases, repression of transcription by chromatin can be relieved by replication in the presence of relevant transcription factors (5, 37). On the other hand, in vivo experiments have shown that transcriptional activators can remodel chromatin in the absence of replication (58, 62, 65, 74, 78, 83, 84). However, several of these examples involve complex promoters in which factors may contribute to chromatin remodeling via nonnucleosomal cis-acting elements (58, 62, 74, 78, 84), and in another case, the activator GAL4-ER-VP16 (see Results for description) was constitutively present, and chromatin remodeling was induced by addition of β-estradiol (65). Only recently has the binding of a single transcriptional activator, Gal4p, to a single nucleosomal site been examined in nonreplicating yeast cells (83). In this case, for technical reasons, cells were first grown to stationary phase and then arrested for 12 h with hydroxyurea, simultaneously with induction of Gal4p synthesis, prior to examination of chromatin perturbation by Gal4p. Thus, although Gal4p perturbation of chromatin was observed, it was not established whether Gal4p binding could occur to a nucleosomal site in a shorter, perhaps more physiologically relevant interval or in cells arrested in log phase. Furthermore, whether Gal4p can bind to a nucleosomal site in yeast arrested in other phases of the cell cycle also remains undetermined. This is not merely a moot point, as for example the ability of a transcriptional activator to overcome repression of a telomeric URA3 gene varies at different points in the cell cycle (1).
In the present work, we test the ability of Gal4p to perturb a positioned nucleosome containing a Gal4p binding site in the absence of replication. We examine two distinct nucleosomes containing Gal4p binding sites, in one case near the nucleosome pseudodyad (i.e., near the center) and in the other case centered 41 bp from the nucleosome’s edge; induce Gal4p synthesis by two distinct routes; and investigate binding and chromatin perturbation at two widely separate points in the cell cycle, G1 and G2/M. We also increase the scope of our conclusions by performing similar experiments with the transcriptional activator Bicoid from Drosophila melanogaster, whose DNA-binding domain is structurally different from that of Gal4p.
MATERIALS AND METHODS
Strains, media, and genetic methods.
We used yeast strains YJ0α (constructed from YJ0 [MATa trp1 ura3-52 leu2-3,112 ade2-101 gal4Δ gal80Δ MEL1], a gift from Stephen Johnston), and FY23bar1Δ (MATa bar1::LEU2 ura3-52 trp1Δ63 leu2Δ1) for experiments involving Gal4p. For experiments with Bicoid protein, we used strains CY296 (MATa gal4Δ::LEU2 lys2-801 leu2-Δ1 his3-Δ200 ura3-Δ99 trp1-Δ99 [8]), CY297b (MATα swi1Δ::LEU2 gal4Δ::LEU2 lys2-801 leu2-Δ1 his3-Δ200 ura3-Δ99 trp1-Δ99 [61]), and YJ0bar1Δ (MATa trp1 ura3-52 leu2-3,112 ade2-101 gal4Δ gal80Δ MEL1 bar1::LEU2). The bar1Δ strains were constructed from FY23 (76) and YJ0 by one-step gene replacement of the bar1 gene, using the BamHI-HindIII fragment from plasmid pZV77 (a gift from David Gross), and the gene replacement was confirmed by Southern blotting. Yeast cells were grown in complete synthetic dropout media (Bio101) containing 2% glucose and transformed by a modification (31) of the method of Ito et al. (33).
For cell cycle arrest experiments, cells were grown in medium containing 2% glucose or 1.5% raffinose and 25 mM phthalic acid (pH 5.5) to an optical density at 600 nm of between 0.2 and 0.5. Cells were then arrested for 3 h, using α-factor at 0.2 to 0.5 μM or nocodazole at 10 μg/ml. Cell cultures were supplemented with fresh α-factor or nocodazole every 3 h; for experiments involving long nocodazole arrest, cells were spun down at 3 and 6 h and resuspended in fresh medium containing 15 μg of nocodazole per ml. Gal4p or Bicoid synthesis was induced 3 h after initiating arrest by addition of 100 or 200 nM β-estradiol, as indicated, or by spinning cells down and resuspending them in 2% galactose medium containing 25 mM phthalic acid (pH 5.5) and α-factor or nocodazole as appropriate. Cell morphology was examined at 3 h and again at intervals following induction of Gal4p or Bicoid. Cells exhibited <5% budded cells and >90% shmoos from 3 to 9 h following addition of α-factor, and cell density (measured with a counting chamber) did not increase after the initial 3-h arrest period. More than 90% of nocodazole-arrested cells exhibited the characteristic dumbbell-shaped (large-budded) morphology for the duration of the arrest, and cell density did not increase. Furthermore, the copy number of the TRP1ARS1-based plasmids did not increase relative to the genome during the arrest, as determined by Southern blotting, consistent with each ARS1-based plasmid replicating once per plasmid molecule and only during S phase (15a).
Plasmids.
To construct pADH/lexA.hER.VP16, the expression vector for LexA-ER-VP16, a 1-kb SalI-NotI fragment containing the coding sequence for ER-VP16 (43) was ligated with pEG202, which contains a full-length lexA gene fused to the ADH1 promoter (26). A 7-bp linker containing a unique BstEII site was introduced at the junction of the lexA and ER sequences. The reporter plasmids containing LexA operators upstream of a lacZ gene (see Fig. 1B) (28) have been described elsewhere (22). Induction via LexA-ER-VP16 was measured 3 h after addition of 100 nM β-estradiol.
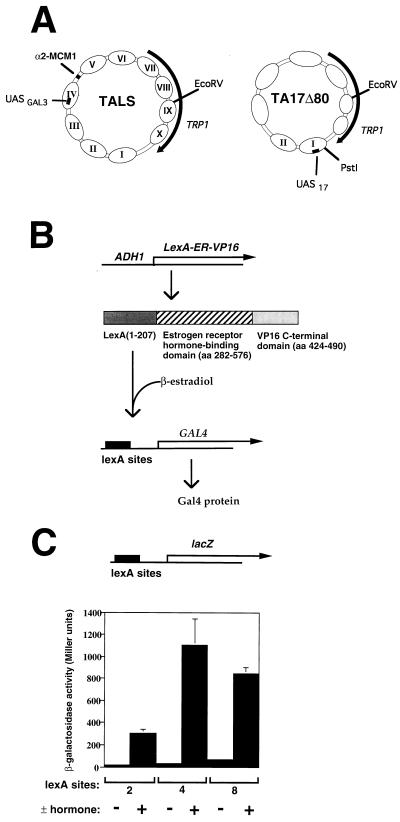
FIG. 1.
Experimental strategy. (A) Schematic depiction of two chromatin reporter plasmids, TALS and TA17Δ80. UASGAL3 is a single Gal4p binding site from the GAL3 promoter (3), and UAS17 is a single near-consensus Gal4p binding site (21) introduced in the TRP1ARS1 derivative TA17Δ80 (48). Only nucleosomes I and II have been shown to be well positioned in TA17Δ80, and so the remaining nucleosomes are not numbered. (B) Scheme for placing Gal4p synthesis under hormonal control. aa, amino acids. See text for details. (C) The chimeric activator LexA-ER-VP16 was tested for activity at a CYC1-lacZ reporter gene having two, four, or eight LexA binding sites upstream. Yeast cells (YJ0α) grown in raffinose medium to mid-log phase were incubated for 3 h after addition of 100 nM β-estradiol before measurements of β-galactosidase activity for plus-hormone samples. The results shown are an average from three independent colonies from each of two independent transformations.
Plasmid pGAL1-10/4 lexA sites/GAL4 was constructed by ligating together (i) a BamHI-HindIII fragment containing a modified GAL1-10 promoter deleted for the four Gal4 binding sites and URS B and C regions and containing four lexA sites (28), (ii) a 40-bp HindIII-SphI oligonucleotide, containing a unique NdeI site at the beginning of the GAL4 open reading frame, (iii) a 2.6-kb SphI-KpnI fragment containing the remainder of the GAL4 coding and termination sequences, and (iv) the shuttle vector pRS426 (11).
The yeast plasmids TALS and TA17Δ80 were excised from bacterial vectors and religated before being transformed into yeast, as described previously (47, 48). To create TABic4Δ80, the sequence 5′-TCCCTATCTAATCCCTATCTAATCCCTATCTAATCCC-3′ was inserted into pRS104 (47) between residues corresponding to 859 and 860 map units of TRP1ARS1 by PCR to create pRS104Bic4; an 80-bp deletion was then made by PCR (63) and verified by DNA sequencing, to create pRS104Bic4Δ80. Yeast sequences were then excised from this plasmid with SacI and HindIII (yielding a fragment carrying the 3′ end of the TRP1 gene) and ligated with the complementary SacI-HindIII fragment from pRS110 (containing the 5′ end of the TRP1 gene [47]) and transformed into yeast to create TABic4Δ80, which was verified by Southern analysis.
The multicopy plasmid pRS426GAL4 contains the GAL4 gene under control of its own promoter (61). The Bicoid expression vector was created by ligating a 3-kb KpnI-XbaI fragment from pDB1 (9) (a gift from D. S. Burz) containing the Bicoid gene under control of the GAL1 promoter into the polylinker of pRS416 (11). Bicoid expression was induced by GAL4-ER-VP16, using 100 nM β-estradiol (Fig. 6), or by GAL4-ER-VP16F442P (66), using 200 nM β-estradiol (Fig. 7).
FIG. 6.
Remodeling of TABic4Δ80 chromatin by Bicoid in unsynchronized SWI+ and swi1Δ yeast cells. (A) MNase cleavage sites were mapped counterclockwise from the EcoRV site in chromatin from yeast cells (CY296) harboring TABic4Δ80 and the Bicoid (Bic) expression system grown in the absence of hormone (lanes 3 and 4) or 4.5 h after addition of 100 nM β-estradiol (lanes 5 and 6). Also shown is chromatin treated with MNase from cells harboring TA17Δ80, grown in glucose (lane 7) or galactose medium (lane 8). DNA samples are TABic4Δ80 (lanes 1 and 2). The locations of nucleosomes I and II are indicated at the top and the right, with the small rectangle in nucleosome I corresponding to binding sites for Bicoid or Gal4p in the various episomes. MNase was used at 5 (lanes 3 and 6) and 20 (lanes 4, 5, 7, and 8) U/ml for chromatin and at 4 (lane 1) and 10 (lane 2) U/ml for naked DNA. (B) MNase cleavage sites were mapped counterclockwise from the EcoRV site in chromatin from swi1Δ yeast cells (CY297b) harboring TABic4Δ80 and the Bicoid expression system grown in the absence of hormone (lanes 3 to 5) or 4.5 h after addition of 100 nM β-estradiol (lanes 6 to 8). MNase was used at 0 (lanes 3 and 8), 2 (lanes 4 and 7), and 5 (lanes 5 and 6) U/ml for chromatin and at 4 (lane 1) and (lane 2) 10 U/ml for naked DNA.
FIG. 7.
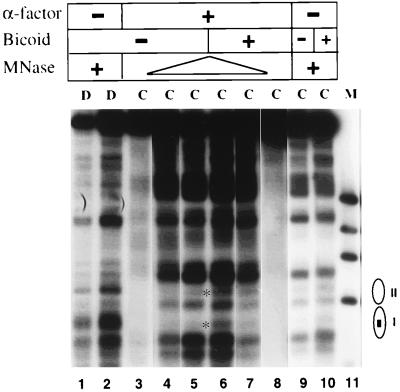
Remodeling of TABic4Δ80 chromatin by Bicoid in arrested cells assayed by indirect end-label analysis of MNase cleavage sites. Yeast cells (YJ0bar1Δ) harboring TABic4Δ80 and the Bicoid expression system were first arrested with α-factor or not, as indicated. Hormone was then added (+Bicoid lanes) or not (−Bicoid lanes), cells were incubated an additional 4.25 h, and chromatin was isolated for MNase digestion. MNase cleavage sites were mapped counterclockwise from the EcoRV site. Lanes C contain chromatin; lanes D contain naked DNA controls. The locations of nucleosomes I and II are indicated schematically at the sides, with the small rectangle in nucleosome I corresponding to the four Bicoid binding sites. The asterisks indicate cleavage sites induced by Bicoid expression. MNase was used at 0 (lanes 3 and 8), 20 (lanes 9 and 10), 50 (lanes 4 and 7), and (lanes 5 and 6) 200 U/ml for chromatin samples and at 4 (lane 1) and 10 (lane 2) U/ml for naked DNA. Lane 11 contains φX DNA digested with HaeIII. A shorter exposure was used for lanes 9 to 11 than for lanes 1 to 8.
Northern analysis.
Total cellular RNA was extracted, electrophoresed on formaldehyde-containing gels, blotted, and hybridized as described previously (12, 85). A 2.9-kb SphI-HindIII fragment from the GAL4 gene was used to probe for the GAL4 transcript. The GAL1 and GAL10 transcripts were visualized by probing with EcoRI-BamHI and SalI-EcoRI restriction fragments, respectively, from pBM48 (a gift from Mark Johnston), which contains the SalI-BamHI fragment (SC4918) from the GAL1-10 locus (69).
Enzyme assays and chromatin characterization.
Assays for α-galactosidase (the MEL1 gene product) and β-galactosidase were performed as described previously (59, 61). For topoisomer analysis, DNA was prepared and electrophoresed on topoisomer-resolving gels, and Gaussian centers of the resulting topoisomer distributions were determined as described previously (46, 49).
For indirect end-label analysis (50, 80), chromatin was prepared by combining two previously described methods (16, 38). Cells (100 to 200 ml) were grown in medium containing 25 mM phthalic acid (pH 5.5) to an optical density at 600 nm of 0.5 to 1.5 and harvested by centrifugation at 4,000 × g for 5 min. The pellet was resuspended in 50 mM Tris (pH 7.4)–0.1% β-mercaptoethanol at a concentration of 5 × 107 cells/ml and incubated with gentle shaking in a water bath for 15 min at 30°C. Cells were centrifuged as before; the pellet was resuspended in medium containing 1 M sorbitol at a concentration of 109 cells/ml and spheroplasted by addition of 1/10 volume of Zymolyase 100T (10 mg/ml; Seikagaku America, Inc., Ijamsville, Md.) and gentle shaking at 30°C for 15 min. The spheroplasted cells were then diluted with 15 volumes of chilled medium containing 1 M sorbitol and collected by centrifugation (2,000 × g for 5 min). Cells were washed with medium containing 1 M sorbitol and spun down in an HB-4 rotor at 3,500 rpm (2,000 × g) for 5 min. The pellet was then taken up at 2 × 109 cells/ml in 1 M sorbitol–50 mM NaCl–10 mM Tris-HCl (pH 7.4)–5 mM MgCl2–1 mM CaCl2–1 mM β-mercaptoethanol (or 10 mM dithiothreitol)–0.5 mM spermidine and transferred in 100- or 150-μl aliquots into 1.5-ml microcentrifuge tubes on ice. Micrococcal nuclease (MNase) or restriction enzyme was added to each tube to an appropriate concentration, and digestion was begun by addition of an equal volume of the same buffer containing 0.15% Nonidet P-40, mixing gently, and immediately placing the tube in a 37°C bath. Digestions were halted (after 5 min for MNase and after 15 and 30 min for PstI) by addition of 55 μl of proteinase K (5 mg/ml) sodium dodecyl sulfate (SDS; 5%). Naked DNA samples were processed as above but immediately digested with proteinase K-SDS, cleaned and precipitated, and digested with MNase in 300 μl of 10 mM HEPES (pH 7.5)–2 mM CaCl2–5 mM MgCl2 for 5 min. Samples were cleaned and precipitated and then analyzed as described below.
After >2 h of incubation with proteinase K-SDS, samples were cleaned with phenol and chloroform and precipitated. Pellets were taken up in 100 μl 10 mM Tris-Cl (pH 8.0)–1 mM EDTA and treated with RNase A. One-third to one-half of each sample was digested with EcoRV, precipitated, and electrophoresed on a 1.2% agarose gel for 5.5 h at 135 V (4.5 V/cm). Blotting and hybridization were as described previously (12, 47). Samples were probed with an EcoRV-HindIII probe from TRP1ARS1 sequences, prepared by PCR. At least two independent analyses were done for each indirect end-label experiment.
RESULTS
Hormone-dependent induction of Gal4p synthesis.
To investigate the ability of Gal4p to bind to nucleosomal sites in the absence of replication, we used two chromatin reporter plasmids, TALS and TA17Δ80, depicted in Fig. 1A. In yeast cells not expressing Gal4p (e.g., cells grown in glucose medium), each of these reporters contains a nucleosomal Gal4p binding site (48, 65). Our strategy was to prevent cells harboring TALS or TA17Δ80 from replicating by arresting them in late G1 (with α-factor) (15) or G2/M (with nocodazole) (1, 25, 34) and then to express Gal4p while maintaining cell cycle arrest and analyze plasmid chromatin structure. Perturbation of chromatin structure accompanying Gal4p expression as is seen in unsynchronized cells (48, 65) would indicate that Gal4p was able to recognize and bind to its site in a positioned nucleosome in the absence of replication.
We first sought to achieve rapid, inducible expression of Gal4p by placing the Gal4p coding sequence under hormone control. This strategy was based on previous work with the chimeric activator GAL4-ER-VP16, which contains the Gal4p DNA-binding domain, the human estrogen receptor hormone-binding domain (ER), and the strong VP16 activation domain (43). Expression of GAL4-ER-VP16 in yeast allows rapid, hormone-dependent expression from promoters containing Gal4p binding sites (43, 65). To allow Gal4p induction by a similar strategy, we replaced the coding sequence for the Gal4p DNA-binding domain of GAL4-ER-VP16 with that for the bacterial LexA protein to create pADH/lexA.hER.VP16 (Fig. 1B). This expression vector was introduced into yeast and was first tested for activity in assays using lacZ reporters containing two, four, or eight lexA sites (Fig. 1C). LexA-ER-VP16 showed low activity in the absence of estradiol, which increased 12- to 33-fold after 3 h induction with β-estradiol.
The GAL4 gene was then placed under control of the same promoter containing two, four, or eight lexA sites in place of the lacZ gene (Fig. 1B). This plasmid and the LexA-ER-VP16 expression vector were introduced into a gal4Δ gal80Δ MEL1+ yeast strain, and the induction of Gal4p upon the addition of hormone was demonstrated by induction of α-galactosidase (the product of the Gal4p-regulated MEL1 gene) (data not shown). The Gal4p expression vector having four LexA binding sites was found to have the best induction characteristics and was used in all subsequent experiments. Proper induction was confirmed in each experiment by monitoring α-galactosidase activity.
Induction of the GAL1 and GAL10 genes in nonreplicating yeast.
Figure 2A shows directly the induction of GAL4 transcripts via LexA-ER-VP16 (lanes 1 and 2) 3 h after addition of 100 nM β-estradiol. Hormone induction of Gal4p synthesis in raffinose medium in the gal4Δ gal80Δ strain YJ0α resulted in strong induction of the GAL1 and GAL10 genes, indicating that the artificially induced Gal4p protein functions normally (Fig. 2A, lane 2). Some expression of GAL1 and GAL10 transcripts was observed even in uninduced cells (Fig. 2A, lanes 1 and 3), corresponding to low levels of Gal4p (weakly detectable at the RNA level and also when assayed for α-galactosidase activity) induced by LexA-ER-VP16 in the absence of β-estradiol. Importantly, this level of Gal4p was not sufficient to cause significant changes in chromatin structure in our reporters (see below).
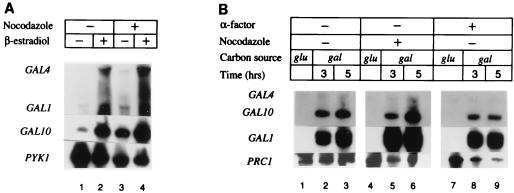
FIG. 2.
Induction of mRNAs by Gal4p in arrested and unsynchronized yeast cells. (A) Induction of GAL4 mRNA under control of LexA-ER-VP16, and consequent induction of GAL1 and GAL10 mRNAs by Gal4p, from unsynchronized and nocodazole-arrested YJ0α yeast cells grown in raffinose medium in the presence (for 3 h) and absence of β-estradiol. The PYK1 message was examined as a control. (B) Induction of GAL4, GAL1, and GAL10 mRNAs under endogenous control in unsynchronized and arrested yeast cells. Cells (FY24 yeast cells containing TALS and pRS426GAL4) were grown in glucose medium and mRNA was harvested (glu lanes), or the cells were spun down and transferred to galactose medium, and mRNA was harvested at indicated times (gal lanes). The PRC1 message was examined as a control.
Figure 2A also shows that the GAL1 and GAL10 genes are induced upon hormone addition in cells arrested in G2/M phase with nocodazole. Similarly, galactose induction of the wild-type GAL4 gene caused approximately equivalent induction of the GAL1 and GAL10 genes in unsynchronized cells and in cells arrested with nocodazole or α-factor (Fig. 2B). These results indicate that GAL4 functions normally in nonreplicating yeast cells, consistent with previous work showing that a lacZ reporter under gal control could be induced in α-factor-arrested cells (32). Our results have additional significance, however, as induction of both the GAL1 and GAL10 genes is normally accompanied by remodeling (albeit subtle [17]) of chromatin structure (2, 42). Thus, our results suggest that Gal4p is capable of remodeling the chromatin structure of the GAL1-10 promoter in the absence of replication.
Remodeling of TALS chromatin by Gal4p.
The results of Fig. 2 demonstrate that Gal4p can induce transcription and therefore, by inference, remodel chromatin at the GAL1-10 locus in nonreplicating yeast. However, since the Gal4p binding sites in the GAL1-10 promoter are nonnucleosomal (41), this experiment yields no information on binding of Gal4p to a nucleosomal site. We therefore next examined Gal4p binding to the TALS chromatin reporter. TALS is a TRP1ARS1-based episome in which nucleosomes are strongly positioned by the α2-MCM1 complex in yeast α cells (60). A single Gal4p binding site, derived from the GAL3 gene (3), is centered 41 bp from the left edge in nucleosome IV, which is immediately adjacent to the α2-MCM1 binding site (Fig. 1A). This region is inaccessible to Escherichia coli Dam methyltransferase expressed in yeast, in the absence of Gal4 protein (39). In the presence of Gal4p, nucleosome IV is perturbed and TALS chromatin is remodeled, as shown by changes in MNase cleavage, SacI accessibility, and plasmid topology (65).
We examined TALS remodeling by monitoring plasmid topology. This assay is based on the fact that packaging of DNA in chromatin causes a change in linking number of −1 per nucleosome, which can be readily visualized and quantified in a closed circular plasmid (20, 49, 64). Perturbation of TALS chromatin by Gal4p expressed in galactose medium results in a loss of nearly one negative supercoil per plasmid (65) (Fig. 3B, lanes 1 and 2). An approximately equivalent change in topology is seen upon hormone induction of Gal4p synthesis via LexA-ER-VP16 (Fig. 3A, lanes 1 and 2; Table 1). Thus, although the LexA-ER-VP16-mediated induction of Gal4p is less stringent than that of the native GAL4 promoter, it closely mimics native induction with respect to TALS remodeling.
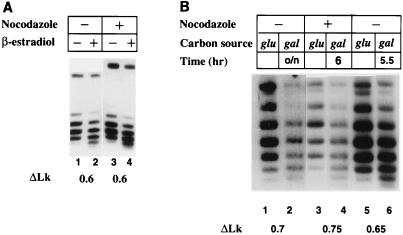
FIG. 3.
Remodeling of TALS chromatin assessed by changes in topology in unsynchronized and arrested yeast cells. (A) Cells (YJ0α) harboring TALS and having GAL4 under control of LexA-ER-VP16 were grown in raffinose medium in the presence or absence of hormone, either unsynchronized or arrested as indicated, and DNA was isolated for analysis of TALS topoisomer distributions. Hormone induction was for 3 h at 100 nM β-estradiol. The band near the top is nicked circular DNA, and the lower bands represent topoisomers differing in linking number from adjacent bands by one; under the conditions used, faster-migrating species are more positively supercoiled. Values shown for ΔLk indicate the differences between the calculated centers of the Gaussian distributions in the lanes indicated. (B) Topoisomer distributions of TALS from cells (FY24) harboring TALS and a multicopy plasmid bearing the GAL4 gene grown in glucose medium in the presence of nocodazole (10 μg/ml) for 3 h or in its absence, as indicated. Cells were spun down and taken up in galactose medium with or without nocodazole and incubated for the additional intervals indicated. The uppermost band corresponds to nicked circular TALS, and faster-migrating topoisomers are more positively supercoiled. The linking number changes between samples grown in glucose and galactose are indicated at the bottom. o/n, overnight.
TABLE 1.
Gal4p-induced changes in TALS topology in unsynchronized and nocodazole-arrested yeast cells
| Cells | ΔLka (mean ± SD)
|
|
|---|---|---|
| Glu/Gal | −E2/+E2 | |
| Unsynchronized | 0.72 ± 0.07 (2) | 0.55 ± 0.34 (4) |
| Nocodazole arrested | 0.8 ± 0.35 (3) | 0.55 ± 0.35 (6) |
Difference in linking number, measured from the Gaussian centers of topoisomer distributions, for TALS topoisomers from yeast cells grown in galactose compared to glucose, or in the presence and absence of 100 nM β-estradiol (E2) in cells in which Gal4p is under hormone-inducible control, as indicated. Values reflect a loss of negative supercoiling in the presence of Gal4p. The number of independent determinations used to obtain each value is shown in parentheses.
When yeast cells harboring TALS were arrested with nocodazole, induction of Gal4p via LexA-ER-VP16 again resulted in loss of negative supercoiling (Fig. 3A, lanes 3 and 4; Table 1). Similar results were observed upon induction of the native GAL4 gene in nocodazole-arrested cells (Fig. 3B, lanes 3 and 4; Table 1). MNase cleavage sites in TALS chromatin induced by Gal4p (65) were also observed when Gal4p was induced by LexA-ER-VP16, in both unsynchronized and nocodazole-arrested cells (4). Thus, a nucleosomal Gal4p binding site could be accessed by Gal4p, with consequent chromatin remodeling, in the absence of replication.
Remodeling of TA17Δ80 chromatin by Gal4p.
Because nucleosome positioning in TALS requires the α2 protein, we could not determine whether Gal4p could bind to a positioned nucleosome in TALS in α-factor-arrested yeast cells. To circumvent this problem and to investigate a different nucleosomal Gal4p binding site, we used a different chromatin reporter, TA17Δ80 (48). This TRP1ARS1-based episome contains a single 17-bp near-consensus Gal4p binding site near the center of a positioned nucleosome (Fig. 1A). In the presence of Gal4p, nucleosome positioning in TA17Δ80 near the Gal4p binding site is perturbed, indicating that Gal4p is able to outcompete histones for occupancy of this site in cycling yeast cells (48). As with TALS, the Gal4p binding site is in a region of the nucleosome which is inaccessible to Gal4p in vitro (75), and so we wished to test whether its accessibility to Gal4p in vivo would occur during replication.
We first attempted to use LexA-ER-VP16 mediated induction of Gal4p as described above. However, for reasons that we do not understand, introduction of TA17Δ80 into yeast harboring the expression vectors for LexA-ER-VP16 and Gal4p resulted in loss of inducible Gal4p activity. We therefore instead turned to galactose induction of Gal4p from the native GAL4 gene on a multicopy plasmid in cells first grown in glucose. This protocol differs from one commonly used for Gal4p-dependent induction from cells grown in raffinose or glycerol-lactate medium. In raffinose or glycerol-lactate medium, the GAL4 gene is active, but Gal4p is repressed by Gal80p (36, 44); addition of galactose to the cells releases Gal4p from Gal80p repression, and genes responsive to Gal4p are rapidly induced (40). In contrast, the GAL4 gene is repressed in glucose medium, and a considerable lag occurs before Gal4p-responsive genes are induced following a shift to galactose medium (23, 35, 51).
Before undertaking experiments with arrested cells, we first determined the time of galactose induction needed to perturb nucleosome positioning in TA17Δ80 in cycling yeast cells. TA17Δ80 shows little if any change in topology upon galactose induction, in contrast to TALS (4). This probably means that nucleosomes are rearranged without net loss upon Gal4p binding to TA17Δ80, whereas Gal4p binding to TALS causes both rearrangement and net loss of nucleosomes. We therefore monitored chromatin remodeling of TA17Δ80 by MNase digestion followed by indirect end labeling (50, 80). Only slight perturbation of nucleosome positioning was seen at 3 and 4.5 h following galactose induction; however, after 6 h, changes in the MNase cleavage pattern reflecting perturbation of nucleosome positioning could clearly be seen (4) (see below). Similarly, between 5 and 6 h of growth in galactose medium sufficed for the maximal change in topology of TALS induced by Gal4p (Fig. 3B). Based on these results, we examined remodeling of TA17Δ80 chromatin by Gal4p in noncycling yeast cells 6 h following galactose induction. FY23bar1Δ cells were grown to mid-log phase and incubated with α-factor or nocodazole for 3 h to ensure arrest. (The bar1 deletion removes a protease which degrades α-factor, allowing α-factor arrest to be maintained for long periods.) Cells were then spun down and resuspended in glucose or galactose medium for an additional 6 h of incubation in the continued presence of α-factor or nocodazole prior to isolation and digestion of chromatin. Cell density was unchanged during the 6-h induction period, and the morphology remained consistent with the arrest (see Materials and Methods).
Figure 4 shows indirect end-label analysis of MNase-digested TA17Δ80 chromatin from cycling cells, and cells arrested with nocodazole or α-factor, grown in glucose and galactose. In cycling cells, enhanced MNase cleavage is seen in the region of nucleosome II following 6 h of growth in galactose medium compared to cells grown in glucose (Fig. 4A, lanes 1 to 6; note the two bands marked by asterisks). The upper of these two new cleavage sites does not correspond to any of the cleavage sites seen in naked DNA (lane 7) and is the most prominent. This cleavage most likely reflects a rearrangement of nucleosome positioning that results from Gal4p binding to its site near the center of nucleosome I, as it depends on growth in galactose and is not seen at endogenous levels of Gal4p (4). Furthermore, a PstI site near the Gal4p binding site which is blocked in cells grown in glucose becomes accessible after 6 h of growth in galactose (Fig. 5; see below), consistent with perturbation of nucleosome positioning by Gal4p. A somewhat different MNase cleavage pattern is seen after longer (overnight) growth in galactose medium, in which enhanced cleavage is seen in both nucleosomes I and II, while the upper cleavage site seen in nucleosome II after 6 h becomes less prominent (4, 48). We do not at present understand the reason for this apparent change in perturbation of TA17Δ80 chromatin occurring between 6 and 24 h. However, since the perturbation seen in the region of nucleosome II was completely reproducible, we used this MNase cleavage pattern to compare with that seen in arrested cells 6 h following galactose induction.
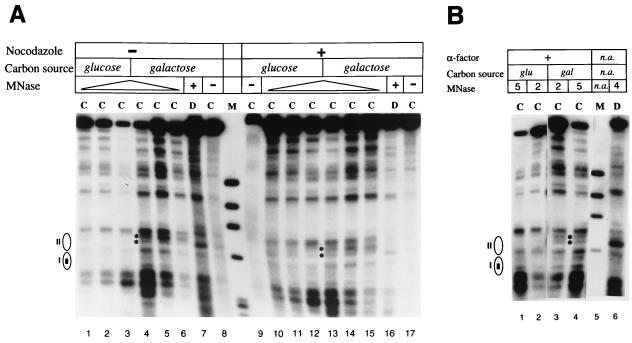
FIG. 4.
Remodeling of TA17Δ80 chromatin by Gal4p in unsynchronized and arrested cells assayed by indirect end-label analysis of MNase cleavage sites. (A) Yeast cells (FY23bar1Δ) harboring TA17Δ80 and pRS426GAL4 were grown in glucose, either unsynchronized or arrested with nocodazole as indicated, or shifted from glucose to galactose medium (still containing nocodazole for arrested cells) and incubated an additional 6 h prior to harvesting of chromatin for MNase digestion. MNase cleavage sites were mapped counterclockwise from the EcoRV site (Fig. 1A). Lanes: C, chromatin; D, naked DNA; M, φX DNA digested with HaeIII. The locations of nucleosomes I and II are indicated at the left, with the rectangle in nucleosome I representing the Gal4p binding site. Cleavage sites induced in galactose medium after 6 h are indicated by asterisks (lanes 4 and 13); the upper site is much more prominent and is not cleaved in naked DNA. MNase was used at 0 (lanes 8, 9, and 17), 2 (lanes 1, 6, 10, and 15), 5 (lanes 2, 5, 11, and 14), and 20 (lanes 3, 4, 12, and 13) U/ml for chromatin and at 4 (lane 7) and 10 (lane 16) U/ml for naked DNA. (B) Cells were grown in glucose and arrested with α-factor, and chromatin was isolated before and after 6 h of additional incubation in galactose medium. n.a., not applicable. MNase concentrations used are given in units per milliliter.
FIG. 5.
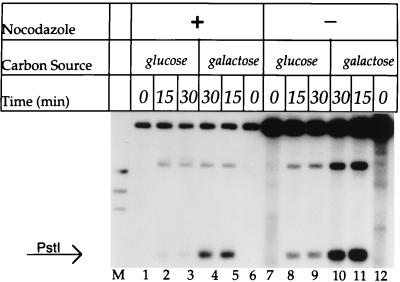
Remodeling of TA17Δ80 chromatin by Gal4p in unsynchronized and arrested cells assayed by restriction enzyme accessibility. Chromatin from cells treated as for Fig. 6 (an aliquot from the same preparation) was incubated in the absence (for 30 min) or presence of PstI at 200 U/ml for 15 min or 30 min, as indicated. Purified DNA was secondarily digested with EcoRV and analyzed by indirect end labeling, probing counterclockwise from the EcoRV site (Fig. 1A). The band at the top is the EcoRV-cut intact plasmid, and the band indicated by the arrow corresponds to cleavage at the PstI site in nucleosome I. The cleavage site at about 1,400 bp corresponds to a second PstI site in TA17Δ80. Lane M contains φX DNA digested with HaeIII.
When cells were arrested with nocodazole or with α-factor and then grown in galactose medium for an additional 6 h, a perturbation in the MNase cleavage pattern similar to that in cycling cells was seen (Fig. 4A, lanes 9 to 15; Fig. 4B). These results suggest that Gal4p can access a nucleosomal binding site even cells arrested in late G1 by α-factor or in G2/M by nocodazole. We also examined accessibility to the restriction enzyme PstI in TA17Δ80 chromatin from cells grown in glucose and galactose. The PstI recognition site is in nucleosome I, 30 bp from the edge of the Gal4p binding site (Fig. 1A). This site is strongly protected against digestion in chromatin from cells grown in glucose and is strongly cleaved after 6 h of growth in galactose (Fig. 5, lanes 7 to 12), corroborating the MNase cleavage data that indicate chromatin remodeling by Gal4p in galactose. Enhanced PstI cleavage is also seen in cells arrested with nocodazole in galactose but not in glucose (Fig. 5, lanes 1 to 6), again in agreement with MNase cleavage results (Fig. 4).
We conclude from these experiments that Gal4p is able to remodel a preexisting positioned nucleosome in TA17Δ80 in nonreplicating yeast cells, in agreement with results obtained with TALS as the chromatin reporter.
The Drosophila transcriptional activator Bicoid can bind to nucleosomal sites in SWI+ and in swi yeast, and in the absence of replication.
Transcriptional activators bind to DNA via domains that exhibit considerable structural diversity (30). The DNA-binding domain of Gal4p is characterized by six cysteine residues that coordinate two Zn2+ ions in a bimetal-thiolate cluster; the two Gal4p molecules comprising the biologically active dimer contact CCG triplets at either end of the 17-bp recognition site through contacts with the major groove (45, 54). Gal4p can bind to nucleosomal sites in yeast, in swi as well as SWI+ cells (61) and in the absence of replication (this work). To determine whether these properties are shared with transcriptional activators having other kinds of DNA-binding domains, we chose to examine interaction of the Drosophila transcriptional activator Bicoid with nucleosomal sites in yeast.
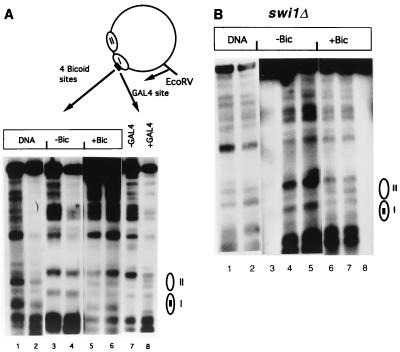
Bicoid is a member of the homeodomain class of transcriptional activators and binds to the consensus site TCTAATCCC (14). However, whereas a binding site for a single Gal4p dimer allows maximal transcriptional activation in yeast (82), maximal activation by Bicoid in yeast requires more than two binding sites (9). To simulate a strong binding site, we therefore replaced the Gal4p binding site of TA17Δ80 with four copies of the Bicoid consensus binding site spaced 11 bp apart (29) to generate TABic4Δ80, which was introduced into yeast. Nucleosomes I and II retained the strong positioning seen with TA17Δ80 in this episome, as seen by MNase digestion followed by indirect end-label analysis (Fig. 6A, lanes 1 to 4).
Bicoid expression was placed under hormone control by introducing into yeast cells a plasmid having the Bicoid coding sequence fused to the GAL1 promoter along with an expression vector for GAL4-ER-VP16 (9, 43). Administration of 100 nM β-estradiol induced activation of a reporter gene containing four Bicoid sites to near maximal levels in 3 to 4 h (4). Induction of Bicoid for 4.5 h in yeast cells harboring TABic4Δ80 resulted in perturbation of nucleosome I, which contains the four Bicoid binding sites, as well as the neighboring nucleosome II (Fig. 6A, lanes 5 and 6), similar to the perturbation of TA17Δ80 caused by Gal4p expression (Fig. 6A, lanes 7 and 8).
Although the SWI-SNF complex can assist binding of transcription factors to nucleosomal sites in vitro (13, 81), Gal4p binds to its site in TA17Δ80 in swi1Δ yeast cells, indicating that interactions with factors other than SWI-SNF may assist its binding in vivo (61). To determine whether Bicoid could also bind to a nucleosomal site in yeast cells lacking a functional SWI-SNF complex, we introduced TABic4Δ80 along with the expression vectors for Bicoid and GAL4-ER-VP16 into the yeast strain CY297b, which is a swi1Δ strain congenic with CY296, the strain used in the experiment of Fig. 6A. Activation of the Bicoid-lacZ reporter gene in these swi1Δ cells was only about half that seen in SWI+ cells (4), but despite this lower activity or expression of Bicoid, perturbation of TABic4Δ80 was still readily seen (Fig. 6B). Thus, Bicoid, like Gal4p, is able to bind to a nucleosomal site in yeast in both SWI+ and swi cells with concomitant perturbation of chromatin.
To determine whether Bicoid could bind to nucleosomal sites in TABic4Δ80 in the absence of replication, we introduced this episome and the Bicoid expression system into YJ0bar1Δ yeast cells. We arrested these cells with α-factor for 3 h and then induced Bicoid expression for 4.25 h. Bicoid induction in α-factor-arrested cells resulted in perturbation of TABic4Δ80 chromatin similar to that seen in cycling cells (Fig. 7). Thus, two transcriptional activators with different kinds of DNA-binding and activation domains are both able to bind to nucleosomal sites in yeast in the absence of replication.
DISCUSSION
The mechanism by which transcriptional activators bind to nucleosomal sites in vivo is presently unknown. One possibility is that an activator binds to sites in chromatin during replication, when histones are transiently removed from the DNA template (18, 70). Several previous studies have shown that chromatin remodeling by transcriptional activators can occur in vivo in the absence of replication (58, 62, 65, 74, 78, 83, 84). These studies, however, have generally examined promoters containing binding sites for multiple factors that could contribute to chromatin remodeling, some of which are likely to interact with nonnucleosomal sites (58, 62, 74, 78, 84). Xu et al. (83) recently examined binding of Gal4p in nonreplicating yeast in a more isolated context by using a derivative of TALS in which a near-consensus Gal4p binding site replaced the UASGAL3 (Fig. 1A) and found no inhibition of Gal4p binding in cells arrested with hydroxyurea for 12 h. However, in this experiment, hydroxyurea arrest was initiated simultaneously with the shift into galactose medium, leaving open the possibility that Gal4p binding initiated before arrest was achieved.
We have considerably extended previous work on the role of replication in allowing binding of activators to nucleosomal sites in vivo by examining binding of Gal4p to nucleosomal sites in two distinct environments (TALS and TA17Δ80), at two stages of the cell cycle, and at shorter intervals following cell cycle arrest. Our results indicate that perturbation of chromatin by Gal4p via nucleosomal binding sites can occur in nonreplicating yeast cells. First, changes in TALS topology (Fig. 3) and MNase cleavage pattern (4) induced by Gal4p are unaltered in cells arrested by nocodazole (α-factor arrest could not be used in these experiments, because strong nucleosome positioning in TALS depends on the yeast α2 protein). Second, changes in the MNase cleavage pattern, as well as PstI accessibility, induced by Gal4p in TA17Δ80 are unchanged in arrested cells (Fig. 4 and 5). The perturbation of chromatin structure in TALS and TA17Δ80 requires both Gal4p and its binding site to be present (48, 65), and so we infer that it is caused by histone displacement or rearrangement which is a consequence of transcription factor binding. This interpretation is consistent with previous studies of transcription factor binding and chromatin remodeling (73, 83). We have also examined binding of the D. melanogaster activator protein Bicoid to a nucleosomal site in a nearly identical context as the Gal4p binding site in TA17Δ80 and find that induction of Bicoid protein elicits similar perturbation in this context as Gal4p does in TA17Δ80, that it can do so without assistance from the SWI-SNF complex, and that it can do so in α-factor-arrested yeast cells.
Nucleosomal sites similar in their location to those used in this study are inaccessible to Gal4p in vitro (75). There are no published reports of Bicoid binding to nucleosomal sites in vitro. However, the full array of contacts made between the closely related homeodomain protein, Antennapedia, and its binding site appears incompatible with nucleosome structure (19), and the MNase cleavages seen in the region of nucleosomes I and II of TABic4Δ80 upon induction of Bicoid indicate nucleosome perturbation. It therefore appears likely that Bicoid, like Gal4p and most other DNA-binding proteins examined to date, cannot access sites near the center of a nucleosome in vitro. Our results imply that some mechanism other than replication must allow Gal4p, and probably Bicoid, to access nucleosomal sites in vivo. One possibility is that activators recruit a chromatin remodeling complex(es) that assists in their binding to nucleosomal sites. We have ruled out a requirement for SWI-SNF (61) or GCN5 (67) in this role, but other candidate activities could be involved (10, 81). Alternatively, binding sites might be in dynamic equilibrium between histone-bound and accessible states (46, 56), thereby allowing activator binding at sufficient concentrations. However, this mechanism would not explain the examples of DNA-binding proteins which do not access nucleosomal sites in vivo (39, 73), nor would it account for a role for activation domains in assisting chromatin perturbation (48, 65, 71).
Interestingly, remodeling of the PHO5 promoter by Pho4p in the absence of replication requires glucose, suggesting an energy-dependent process (62), whereas we observe remodeling of TALS and TA17Δ80 chromatin by Gal4p in arrested cells in both raffinose medium (Fig. 3A) and galactose medium (Fig. 3B and 4). We also observe strong induction of the GAL1 message, which is normally accompanied by chromatin remodeling (2), in arrested cells in both raffinose and galactose (Fig. 2). Perhaps the requirement for glucose in the PHO5 system has to do with changes in protein phosphorylation or some other event specifically needed for activation of PHO5 (72).
Replication has been shown to allow repression of transcription by chromatin to be overcome in vitro (5, 37). An in vivo correlate to these findings has not yet been discovered. In some cases factor binding is inhibited by chromatin, even in replicating cells (24, 73, 83), whereas in other examples in which factor binding does occur in chromatin, replication is not required (references 58, 62, 74, 78, 83, and 84 and this work). The discrepancy between the in vitro and in vivo findings suggests that replication may be important for factor binding to chromatin only under very specific conditions in vivo. Consistent with this notion, the specialized chromatin structure present at yeast telomeres allows transactivation of a URA3 reporter gene in G2/M phase but not in cells arrested in G0 or G1 (1). Further work will be required to determine whether other specific chromatin structures, perhaps involving linker histones (6, 68), resist factor binding in a cell cycle-dependent manner.
ACKNOWLEDGMENTS
We thank Kellie Cummings for initiating construction and characterization of TABic4Δ80; David Gross, Steve Hanes, Dave Burz, Stephen Johnston, Mark Johnston, and Joan Curcio for gifts of yeast strains and plasmids; Michael Kladde and Robert Simpson for helpful discussions; and the Wadsworth Center Molecular Genetics Core for oligonucleotide synthesis and DNA sequencing.
This work was supported by grant GM51993 from the National Institutes of Health.
REFERENCES
- 1.Aparicio O, Gottschling D M. Overcoming telomeric silencing: a trans-activator competes to establish gene expression in a cell cycle-dependent way. Genes Dev. 1994;8:1133–1146. doi: 10.1101/gad.8.10.1133. [DOI] [PubMed] [Google Scholar]
- 2.Axelrod J D, Reagan M S, Majors J. GAL4 disrupts a repressing nucleosome during activation of GAL1 transcription in vivo. Genes Dev. 1993;7:857–869. doi: 10.1101/gad.7.5.857. [DOI] [PubMed] [Google Scholar]
- 3.Bajwa W, Torchia T E, Hopper J E. Yeast regulatory gene GAL3: carbon regulation; UASGAL elements in common with GAL1, GAL2, GAL7, GAL10, GAL80, and MEL1; encoded protein strikingly similar to yeast and Escherichia coli galactokinases. Mol Cell Biol. 1988;8:3439–3447. doi: 10.1128/mcb.8.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balasubramanian, B., and R. H. Morse. Unpublished results.
- 5.Barton M C, Emerson B M. Regulated expression of the β-globin gene locus in synthetic nuclei. Genes Dev. 1994;8:2453–2465. doi: 10.1101/gad.8.20.2453. [DOI] [PubMed] [Google Scholar]
- 6.Bouvet P, Dimitrov S, Wolffe A P. Specific regulation of chromosomal 5S rRNA gene transcription in vivo by histone H1. Genes Dev. 1994;8:1147–1159. doi: 10.1101/gad.8.10.1147. [DOI] [PubMed] [Google Scholar]
- 7.Brown D D. The role of stable complexes that repress and activate eukaryotic genes. Cell. 1984;37:359–365. doi: 10.1016/0092-8674(84)90366-0. [DOI] [PubMed] [Google Scholar]
- 8.Burns L G, Peterson C L. The yeast SWI-SNF complex facilitates binding of a transcriptional activator to nucleosomal sites in vivo. Mol Cell Biol. 1997;17:4811–4819. doi: 10.1128/mcb.17.8.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burz D S, Rivera-Pomar R, Jäckle H, Hanes S D. Cooperative DNA-binding by bicoid provides a mechanism for threshold-dependent gene activation in the Drosophila embryo. EMBO J. 1998;18:5998–6009. doi: 10.1093/emboj/17.20.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cairns B R, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg R D. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 11.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 12.Church G, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Côté J, Quinn J, Workman J L, Peterson C L. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 14.Driever W, Nüsslein-Vollhard C. The bicoid protein is a positive regulator of hunchback transcription in the Drosophila embryo. Nature. 1989;337:138–143. doi: 10.1038/337138a0. [DOI] [PubMed] [Google Scholar]
- 15.Duntze W, MacKay V, Manney T R. Saccharomyces cerevisiae: a diffusible sex factor. Science. 1970;168:1472–1473. doi: 10.1126/science.168.3938.1472. [DOI] [PubMed] [Google Scholar]
- 15a.Fangman W L, Hice R H, Chlebowicz-Sledziewska E. ARS replication during the yeast S phase. Cell. 1983;32:381–838. doi: 10.1016/0092-8674(83)90069-7. [DOI] [PubMed] [Google Scholar]
- 16.Fedor M J, Lue N F, Kornberg R D. Statistical positioning of nucleosomes by specific protein binding to upstream activating sequence in yeast. J Mol Biol. 1988;204:109–127. doi: 10.1016/0022-2836(88)90603-1. [DOI] [PubMed] [Google Scholar]
- 17.Fedor M J, Kornberg R D. Upstream activation sequence-dependent alteration of chromatin structure and transcription activation of the yeast GAL1-GAL10 genes. Mol Cell Biol. 1989;9:1721–1732. doi: 10.1128/mcb.9.4.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gasser R, Koller T, Sogo J M. The stability of nucleosomes at the replication fork. J Mol Biol. 1996;258:224–239. doi: 10.1006/jmbi.1996.0245. [DOI] [PubMed] [Google Scholar]
- 19.Gehring W J, Affolter M, Bürglin T. Homeodomain proteins. Annu Rev Biochem. 1994;63:487–526. doi: 10.1146/annurev.bi.63.070194.002415. [DOI] [PubMed] [Google Scholar]
- 20.Germond J E, Hirt B, Oudet P, Gross-Bellard M, Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci USA. 1975;72:1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giniger E, Varnum S M, Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell. 1985;40:767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- 22.Golemis E A, Khazak V. Alternative yeast two-hybrid systems. The interaction trap and interaction mating. Methods Mol Biol. 1997;63:197–218. doi: 10.1385/0-89603-481-X:197. [DOI] [PubMed] [Google Scholar]
- 23.Griggs D W, Johnston M. Regulated expression of the GAL4 gene in yeast provides a sensitive genetic switch for glucose repression. Proc Natl Acad Sci USA. 1991;88:8597–8601. doi: 10.1073/pnas.88.19.8597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gross D S, Adams C C, Lee S, Stentz B. A critical role for heat shock transcription factor in establishing a nucleosome-free region over the TATA-initiation site of the yeast HSP82 heat shock gene. EMBO J. 1993;12:3931–3945. doi: 10.1002/j.1460-2075.1993.tb06071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guacci V, Hogan E, Koshland D. Chromosome condensation and sister chromatid pairing in budding yeast. J Cell Biol. 1994;125:517–530. doi: 10.1083/jcb.125.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 27.Han M, Grunstein M. Nucleosome loss activates yeast downstream promoters in vivo. Cell. 1988;55:1137–1145. doi: 10.1016/0092-8674(88)90258-9. [DOI] [PubMed] [Google Scholar]
- 28.Hanes, S. D., and R. Brent. Unpublished results.
- 29.Hanes S D, Ridihough G, Ish-Horowicz D, Brent R. Specific DNA recognition and intersite spacing are critical for action of the Bicoid morphogen. Mol Cell Biol. 1994;14:3364–3375. doi: 10.1128/mcb.14.5.3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harrison S C. A structural taxonomy of DNA-binding domains. Nature. 1991;353:715–719. doi: 10.1038/353715a0. [DOI] [PubMed] [Google Scholar]
- 31.Hill J, Ian K A, Donald G, Griffiths D E. DMSO-enhanced whole cell yeast transformation. Nucleic Acids Res. 1991;19:5791. doi: 10.1093/nar/19.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hovland P, Flick J, Johnston M, Sclafani R A. Galactose as a gratuitous inducer of GAL gene expression in yeasts growing on glucose. Gene. 1989;83:57–64. doi: 10.1016/0378-1119(89)90403-4. [DOI] [PubMed] [Google Scholar]
- 33.Ito H, Fukuda Y, Marata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacobs C W, Adams A E, Szaniszlo P J, Pringle J R. Function of microtubules in the Saccharomyces cerevisiae cell cycle. J Cell Biol. 1988;107:1409–1426. doi: 10.1083/jcb.107.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnston M. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol Rev. 1987;51:458–476. doi: 10.1128/mr.51.4.458-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnston S A, Salmeron J M, Jr, Dincher S S. Interaction of positive and negative regulatory proteins in the galactose regulon of yeast. Cell. 1987;50:143–146. doi: 10.1016/0092-8674(87)90671-4. [DOI] [PubMed] [Google Scholar]
- 37.Kamakaka R T, Bulger M, Kadonaga J T. Potentiation of RNA polymerase-II transcription by Gal4-VP16 during but not after DNA replication and chromatin assembly. Genes Dev. 1993;7:1779–1795. doi: 10.1101/gad.7.9.1779. [DOI] [PubMed] [Google Scholar]
- 38.Kent N A, Bird L E, Mellor J. Chromatin analysis in yeast using NP-40 permeabilized spheroplasts. Nucleic Acids Res. 1993;21:4653–4654. doi: 10.1093/nar/21.19.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kladde M P, Simpson R T. Positioned nucleosomes inhibit Dam methylation in vivo. Proc Natl Acad Sci USA. 1994;91:1361–1365. doi: 10.1073/pnas.91.4.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leuther K K, Johnston S A. Nondissociation of GAL4 and GAL80 in vivo after galactose induction. Science. 1992;256:1333–1335. doi: 10.1126/science.1598579. [DOI] [PubMed] [Google Scholar]
- 41.Lohr D. Organization of the GAL1-GAL10 intergenic control region chromatin. Nucleic Acids Res. 1984;12:8457–8474. doi: 10.1093/nar/12.22.8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lohr D, Lopez J. GAL4/GAL80-dependent nucleosome disruption/deposition on the upstream regions of the yeast GAL1-10 and GAL80 genes. J Biol Chem. 1995;270:27671–27678. doi: 10.1074/jbc.270.46.27671. [DOI] [PubMed] [Google Scholar]
- 43.Louvion J-F, Havaux-Copf B, Picard D. Fusion of GAL4-VP16 to a steroid binding domain provides a tool for gratuitous induction of galactose-responsive genes in yeast. Gene. 1993;131:129–134. doi: 10.1016/0378-1119(93)90681-r. [DOI] [PubMed] [Google Scholar]
- 44.Ma J, Ptashne M. The carboxy-terminal 30 amino acids of GAL4 are recognized by GAL80. Cell. 1987;50:137–142. doi: 10.1016/0092-8674(87)90670-2. [DOI] [PubMed] [Google Scholar]
- 45.Marmorstein R, Carey M, Ptashne M, Harrison S C. DNA recognition by GAL4: structure of a protein-DNA complex. Nature. 1992;356:408–416. doi: 10.1038/356408a0. [DOI] [PubMed] [Google Scholar]
- 46.Morse R H. Topoisomer heterogeneity of plasmid chromatin in living cells. J Mol Biol. 1991;222:133–137. doi: 10.1016/0022-2836(91)90198-f. [DOI] [PubMed] [Google Scholar]
- 47.Morse R H, Roth S Y, Simpson R T. A transcriptionally active tRNA gene interferes with nucleosome positioning in vivo. Mol Cell Biol. 1992;12:4015–4025. doi: 10.1128/mcb.12.9.4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morse R H. Nucleosome disruption by transcription factor binding in yeast. Science. 1993;262:1563–1566. doi: 10.1126/science.8248805. [DOI] [PubMed] [Google Scholar]
- 49.Morse, R. H. Analysis of DNA topology in yeast chromatin. Methods Mol. Biol., in press. [DOI] [PubMed]
- 50.Nedospasov S A, Georgiev G P. Non-random cleavage of SV40 DNA in the compact minichromosome and free in solution by micrococcal nuclease. Biochem Biophys Res Commun. 1980;92:532–539. doi: 10.1016/0006-291x(80)90366-6. [DOI] [PubMed] [Google Scholar]
- 51.Nehlin J O, Carlberg M, Ronne H. Control of yeast GAL genes by MIG1 repressor: a transcriptional cascade in the glucose response. EMBO J. 1991;10:3373–3377. doi: 10.1002/j.1460-2075.1991.tb04901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Owen-Hughes T, Workman J L. Experimental analysis of chromatin function in transcriptional control. Crit Rev Eukaryotic Gene Expr. 1994;4:403–441. [PubMed] [Google Scholar]
- 53.Owen-Hughes T, Workman J L. Remodeling the chromatin structure of a nucleosome array by transcription factor-targeted trans-displacement of histones. EMBO J. 1996;15:4702–4712. [PMC free article] [PubMed] [Google Scholar]
- 54.Pan T, Coleman J E. GAL4 transcription factor is not a “zinc finger” but forms a Zn(II)2Cys6 binuclear cluster. Proc Natl Acad Sci USA. 1990;87:2077–2081. doi: 10.1073/pnas.87.6.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paranjape S M, Kamakaka R T, Kadanoga J T. Role of chromatin structure in the regulation of transcription by RNA polymerase II. Annu Rev Biochem. 1994;63:265–297. doi: 10.1146/annurev.bi.63.070194.001405. [DOI] [PubMed] [Google Scholar]
- 56.Polach K J, Widom J. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J Mol Biol. 1995;254:130–149. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- 57.Quinn J, Fyrberg A M, Ganster R W, Schmidt M C, Peterson C L. DNA-binding properties of the yeast SWI/SNF complex. Nature. 1996;379:844–847. doi: 10.1038/379844a0. [DOI] [PubMed] [Google Scholar]
- 58.Reik A, Schütz G, Stewart A F. Glucocorticoids are required for establishment and maintenance of an alteration in chromatin structure: induction leads to a reversible disruption of nucleosomes over an enhancer. EMBO J. 1991;10:2569–2576. doi: 10.1002/j.1460-2075.1991.tb07797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rose M D, Winston F, Hieter P. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 60.Roth S Y, Dean A, Simpson R T. Yeast α2 repressor positions nucleosomes in TRP1/ARS1 chromatin. Mol Cell Biol. 1990;10:2247–2260. doi: 10.1128/mcb.10.5.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ryan M P, Jones R, Morse R H. SWI-SNF complex participation in transcriptional activation at a step subsequent to activator binding. Mol Cell Biol. 1998;18:1774–1782. doi: 10.1128/mcb.18.4.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schmid A, Fascher K D, Hörz W. Nucleosome disruption at the yeast PHO5 promoter upon PHO5 induction occurs in the absence of DNA replication. Cell. 1992;71:853–864. doi: 10.1016/0092-8674(92)90560-y. [DOI] [PubMed] [Google Scholar]
- 63.Simpson R T. Nucleosome positioning can affect the function of a cis-acting element in vivo. Nature. 1990;343:387–389. doi: 10.1038/343387a0. [DOI] [PubMed] [Google Scholar]
- 64.Simpson R T, Thoma F, Brubaker J M. Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: a model system for study of higher order structure. Cell. 1985;42:799–808. doi: 10.1016/0092-8674(85)90276-4. [DOI] [PubMed] [Google Scholar]
- 65.Stafford G A, Morse R H. Chromatin remodeling by transcriptional activation domains in a yeast episome. J Biol Chem. 1997;272:11526–11534. doi: 10.1074/jbc.272.17.11526. [DOI] [PubMed] [Google Scholar]
- 66.Stafford G A, Morse R H. Mutations in the AF-2/hormone binding domain of the chimeric activator GAL4 · ER · VP16 inhibit hormone-dependent transcriptional activation and chromatin remodeling in yeast. J Biol Chem. 1998;273:34240–34246. doi: 10.1074/jbc.273.51.34240. [DOI] [PubMed] [Google Scholar]
- 67.Stafford, G. A., and R. H. Morse. Unpublished results.
- 68.Steinbach O, Wolffe A P, Rupp R. Accumulation of somatic linker histones causes loss of mesodermal competence in Xenopus. Nature. 1997;389:406–412. doi: 10.1038/38755. [DOI] [PubMed] [Google Scholar]
- 69.St. John T P, Davis R W. The organization and transcription of the galactose gene cluster of Saccharomyces. J Mol Biol. 1981;152:285–315. doi: 10.1016/0022-2836(81)90244-8. [DOI] [PubMed] [Google Scholar]
- 70.Svaren J, Chalkley R. The structure and assembly of active chromatin. Trends Genet. 1990;6:52–56. doi: 10.1016/0168-9525(90)90074-g. [DOI] [PubMed] [Google Scholar]
- 71.Svaren J, Schmitz J, Hörz W. The transactivation domain of Pho4 is required for nucleosome disruption at the PHO5 promoter. EMBO J. 1994;13:4856–4862. doi: 10.1002/j.1460-2075.1994.tb06812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Svaren J, Hörz W. Transcription factors vs nucleosomes: regulation of the PHO5 promoter in yeast. Trends Biochem Sci. 1997;22:93–96. doi: 10.1016/s0968-0004(97)01001-3. [DOI] [PubMed] [Google Scholar]
- 73.Venter U, Svaren J, Schmitz J, Schmid A, Hörz W. A nucleosome precludes binding of the transcription factor Pho4 in vivo to a critical target site in the PHO5 promoter. EMBO J. 1994;13:4848–4855. doi: 10.1002/j.1460-2075.1994.tb06811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Verdin E, Paras P, Jr, Van Lint C. Chromatin disruption in the promoter of human immunodeficiency virus type I during transcriptional activation. EMBO J. 1993;12:3249–3259. doi: 10.1002/j.1460-2075.1993.tb05994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vettese-Dady M, Walter P, Chen H, Juan L J, Workman J L. Role of the histone amino termini in facilitated binding of a transcription factor, GAL4-AH, to nucleosome cores. Mol Cell Biol. 1994;14:970–981. doi: 10.1128/mcb.14.2.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Winston F, Dollard C, Ricupero-Hovasse S L. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- 77.Wolffe A P. Implications of DNA replication for eukaryotic gene expression. J Cell Sci. 1991;99:201–206. doi: 10.1242/jcs.99.2.201. [DOI] [PubMed] [Google Scholar]
- 78.Wong J, Shi Y-B, Wolffe A P. A role for nucleosome assembly in both silencing and activation of the Xenopus TRβa gene by the thyroid hormone receptor. Genes Dev. 1995;9:2696–2711. doi: 10.1101/gad.9.21.2696. [DOI] [PubMed] [Google Scholar]
- 79.Workman J L, Kingston R E. Nucleosome core displacement in vitro via a metastable transcription factor-nucleosome complex. Science. 1992;258:1780–1784. doi: 10.1126/science.1465613. [DOI] [PubMed] [Google Scholar]
- 80.Wu C. The 5′ ends of Drosophila heat-shock genes in chromatin are sensitive to DNase I. Nature. 1980;286:854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- 81.Wu C. Chromatin remodeling and the control of gene expression. J Biol Chem. 1997;272:28171–28174. doi: 10.1074/jbc.272.45.28171. [DOI] [PubMed] [Google Scholar]
- 82.Xu E H, Kodadek T, Johnston S A. A single GAL4 dimer can maximally activate transcription under physiological conditions. Proc Natl Acad Sci USA. 1995;92:7677–7680. doi: 10.1073/pnas.92.17.7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xu M, Simpson R T, Kladde M P. Gal4p-mediated chromatin remodeling depends on binding site position in nucleosomes but does not require DNA replication. Mol Cell Biol. 1998;18:1201–1212. doi: 10.1128/mcb.18.3.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zaret K S, Yamamoto K R. Reversible and persistent changes in chromatin structure accompany activation of a glucocorticoid-dependent enhancer element. Cell. 1984;38:29–38. doi: 10.1016/0092-8674(84)90523-3. [DOI] [PubMed] [Google Scholar]
- 85.Zhang M, Rosenblum-Vos L S, Lowry C V, Boayke K A, Zitomer R S. A yeast protein with homology to β-subunits of G proteins is involved in control of heme-regulated and catabolite-repressed genes. Gene. 1991;97:153–161. doi: 10.1016/0378-1119(91)90047-f. [DOI] [PubMed] [Google Scholar]