Abstract
In recent years, plant biotechnology has witnessed unprecedented technological change. Advances in high-throughput sequencing technologies have provided insight into the location and structure of functional elements within plant DNA. At the same time, improvements in genome engineering tools have enabled unprecedented control over genetic material. These technologies, combined with a growing understanding of plant systems biology, will irrevocably alter the way we create new crop varieties. As the first wave of genome-edited products emerge, we are just getting a glimpse of the immense opportunities the technology provides. We are also seeing its challenges and limitations. It is clear that genome editing will play an increased role in crop improvement and will help us to achieve food security in the coming decades; however, certain challenges and limitations must be overcome to realize the technology’s full potential.
1. INTRODUCTION
1.1. The Role of Genome Engineering in Agriculture
As world population climbs from the current 7.3 billion to a projected 9.7 billion by 2050, there will be an increasing demand to efficiently produce and distribute food. It is predicted that food demand will increase 59%–98% by 2050,1 which will likely necessitate rethinking current agricultural practices. This challenge—along with higher temperatures, drought, flooding, pests, and diseases—places food security at the top of the international political agenda. Alongside challenges in production, there is an increasing awareness and interest in functional foods—those that have healthier characteristics beyond basic nutrition.2 Whereas a solution to these challenges is unlikely to come from a single technological advance, it is important to critically evaluate new technologies to determine their role in a solution.
One potential solution to improve food security and enhance food quality relates to the use of genome engineering to create new crop varieties. Genome engineering (or genome editing) can generally be defined as the targeted modification of DNA within living organisms. Due to the wide-ranging utility of modifying an organism’s genome, the breadth of applications that fall under the genome engineering umbrella is enormous. Examples of such applications for agricultural purposes can range from basic biology (e.g., understanding gene function) to applied biology (e.g., altering plant structure or characteristics to produce a useful product). In general, the common ground for most genome engineering projects is their reliance on tools that are capable of recognizing and altering a user-selected DNA sequence. This user-selected DNA sequence can include coding regions within genes to noncoding intergenic sequences; the modifications can range from single-nucleotide substitutions to large deletions or insertions. Being able to introduce a wide range of targeted DNA changes, in turn, results in a wide range of potential products, including those that could help address concerns related to food security or quality.
To apply genome engineering to produce useful agricultural products, three major questions need to be addressed: (i) what new, useful traits are to be introduced; (ii) what DNA modifications are required to generate the traits; (iii) how are these modifications physically introduced into a desired crop’s genome? Unfortunately, answering these questions can be challenging, particularly for questions (ii) and (iii). For example, limited knowledge of the biology underlying certain complex plant traits (e.g., drought tolerance), and the inability to transform certain crop varieties can create significant bottlenecks when trying to generate new products. Nonetheless, significant progress has been made in applying genome engineering in agriculture. Within the last 5 years, numerous products have emerged from genome-editing platforms, including those with higher yield, drought tolerance, and improved oil characteristics. Here, we review the different types of genome modifications that can be introduced in plants and their potential cellular consequences (Section 2), the successes of applying genome editing in agriculture (Sections 3 and 4), and the current limitations and challenges within this field (Section 5).
2. GENOME EDITING IN PLANTS: POTENTIAL DNA MODIFICATIONS
2.1. Single-Nucleotide Polymorphisms
Perhaps the most subtle targeted genome edit is one that results in a single-nucleotide polymorphism. Here, the total size and organization of the crop genome remains unchanged; however, one nucleotide (out of the millions or billions within a plant’s genome) is changed to a different nucleotide. Surprisingly, this subtle change can have profound impacts on cellular function. For example, if positioned appropriately within a gene, a single-nucleotide polymorphism can result in the complete inactivation of gene activity or protein function. This so-called gene knockout can occur when a single-nucleotide polymorphism transforms a codon that normally codes for an amino acid into an early stop codon (Fig. 1B). Having an early stop codon within a gene’s coding sequence results in the premature termination of protein synthesis, thereby producing a truncated protein with potentially reduced or no activity.3–5 Notably, if there are introns downstream of an early stop codon, the mRNA can be subjected to nonsense-mediated decay—a cellular response to aberrantly processed mRNA, which results in degradation of the message.6 Alternatively, a single-nucleotide polymorphism can destroy protein function if a nonsynonymous substitution is introduced within a codon that normally encodes an amino acid required for protein function.3,7,8 This substitution, for example, can be within an active site where a substrate molecule binds.
Fig. 1.
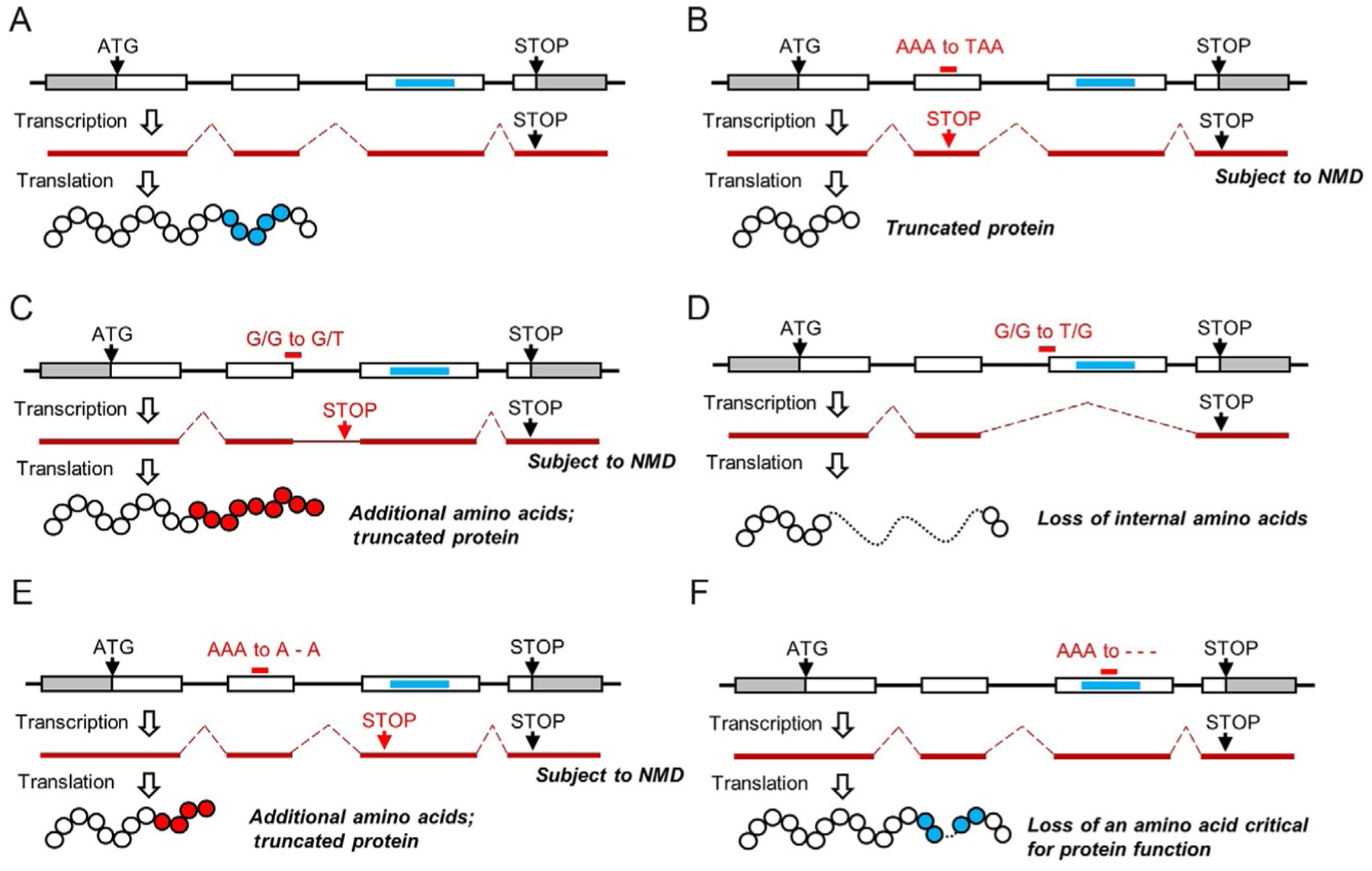
Examples of genome edits that can alter gene or protein activity. (A) Illustration of transcription and translation of a “normal” gene. (B) Illustration of a single-nucleotide polymorphism leading to an early stop codon. (C) Illustration of a single-nucleotide polymorphism within the conserved 5′ splice site that can result in missplicing and inclusion of an intron. Within this example, the intron is shown to comprise an early stop codon. (D) Illustration of a single-nucleotide polymorphism within the conserved 3′ splice site that can result in missplicing and exclusion of a downstream exon. (E) Illustration of a single-nucleotide deletion that can result in a frameshift and early stop codon. (F) Illustration an inframe deletion within a site that encodes an amino acid critical for protein function. Gray boxes, 5′ and 3′ untranslated regions; white boxes, exons; blue rectangle, DNA region encoding amino acids critical for protein function; red lines, mRNA; white circles, amino acids; blue circles, amino acids critical for protein function; red circles, amino acids not normally encoded by the wild-type gene; NMD, nonsense-mediated decay; forward slash, intron–exon or exon–intron junction; dash, deleted nucleotide.
Additionally, a single-nucleotide polymorphism can destroy gene function if it occurs within conserved intron splice/donor sequences. Plant genes contain highly conserved intron splice/donor sequences, which include the 5′ splice site (AG/GTAAG) and 3′ splice site (TGCAG/G).9–11 Single-nucleotide polymorphisms within these splice sites may significantly affect mRNA processing, and potentially result in production of a nonfunctional protein.3,12 For instance, mutations in the 5′ splice site can abolish splicing, resulting in inclusion of intron sequence within the mRNA transcript.13,14 If stop codons are present within the intron, a truncated protein will be produced (Fig. 1C). Alternatively, mutations in the 3′ splice sites can result in exon skipping, wherein a single splicing event removes an exon and flanking intron sequences (Fig. 1D). Also, mutations in splice sites can activate cryptic splice sites, thereby affecting the processing of neighboring exons.15 An example of where a single-nucleotide polymorphism affected intron splicing and destroyed protein function can be seen with the mutant FAD3C gene in canola.13 FAD3C is a fatty acid desaturase partially responsible for the conversion of linoleic to linolenic acid. A G-to-A base substitution was identified at the 5′ splice site within intron 6 of FAD3C. This mutation resulted in impaired splicing, wherein intron 6 was retained in the mature transcript. Stop codons present within intron 6 were predicted to result in early termination of translation and production of a truncated and inactive FAD3C polypeptide. Consistent with this prediction, seed oil from plants with the FAD3C mutation had significantly lower linolenic acid content than wild type—a phenotype expected from having a nonfunctional FAD3C protein.
In addition to destroying protein function, single-nucleotide polymorphisms can attenuate or alter protein function. For example, it is possible to dampen or weaken protein activity if a polymorphism results in a missense mutation within a codon that code for an amino acid not entirely critical for protein function.16,17 Whereas the protein still retains function, the overall activity level has been reduced. On the other hand, a polymorphism can change a protein’s properties if a structural change occurs that enables or prevents a certain activity from taking place. For example, a single-nucleotide polymorphism in a gene’s coding sequence can result in an amino acid change that prevents molecules from interacting with a certain protein domain. This is seen in the case of the EPSPS (5-enolpyruvylshikimate-3-phosphate synthase) gene, which produces a protein involved in branched-chain amino acid synthesis. One or two amino acid substitutions can change the structure of one of the protein’s substrate-binding sites, such that the natural substrate continues to bind; however, a chemical analog, glyphosate, is prevented from binding or binding is attenuated.18
2.2. Indels
Another class of subtle genome edits are small deletions or insertions (indels), which are typically less than 100 nt. To provide perspective, a deletion of 1–100 nucleotides in a crop plant like maize represents 0.00000004%–0.000004% of the total genome (maize has a 2.5 Gbp genome). However, like substitutions, small indels can have profound impacts on cellular function. For example, gene knockouts can be attained if an indel is introduced into a gene’s coding sequence, where the deletion or insertion has a size of −3N+1 or 3N−1 (where N is a whole number), respectively. This type of mutation, also known as a frameshift mutation, frequently leads to early termination of translation and a truncated protein, wherein the truncated protein has attenuated or no activity (Fig. 1E).19–22 On the other hand, gene knockouts can be attained if the deletion is inframe (−3N) and removes codons encoding amino acids critical for protein function (Fig. 1F).19
In another application, small indels can be introduced to prevent DNA-binding proteins (e.g., transcription factors) from recognizing their DNA target. One possible use for this approach is to alter expression of a target gene. An example of this application was shown with the Os11N3 gene in rice. Os11N3 is a gene that encodes a member of the SWEET sucrose-efflux transporter family. Activation of Os11N3 (or so-called OsSWEET14) gene expression results in export of sucrose from plant cells, which can help support pathogen growth. Gene expression of Os11N3 is induced by pathogenic transcription factors, which are injected into rice cells by bacteria. One of these pathogen transcription factors binds to a site within the promoter region of Os11N3. When small deletions were introduced within the binding site, the transcription factor no longer induced Os11N3 gene expression, thereby attenuating pathogenicity.23
2.3. Large Deletions
Genome editing can also generate large deletions (~100 nt to >1 MB). Introducing large deletions can enable valuable basic plant biology research, as removal of gene clusters, cis-regulatory modules, or noncoding RNA genes can provide information on the function of these elements as a group.24 Alternatively, large deletions can be valuable for applied research, including crop breeding, by providing a means to remove unwanted loci that may be detrimental to fitness or food quality. Notably, introducing large deletions may be one of the most “risky” gene edits, as there is a greater chance to remove genes or functional elements that are necessary for normal plant growth and function.
2.4. Insertions
One of the more practical types of gene edits involves the targeted insertion of DNA sequences into predetermined locations within a plant genome. This is in direct comparison to random insertion using biolistics or Agrobacterium-mediated transformation methods, where DNA sequences are randomly integrated into a host’s genome. Random integration presents numerous challenges, including (i) the need to screen through numerous events to find those with stable gene expression, (ii) the possibility that the insertion site causes negative effects on agronomic performance, and (iii) the breeding difficulties that arise when there are two or more unlinked transgenes. Targeted insertion overcomes most of these challenges. A DNA sequence of interest, potentially a gene-coding sequence or promoter, is inserted into the host genome, with or without the subsequent removal of host DNA.25–31 The ability to choose where a gene integrates allows users to select a “safe harbor,” where gene expression is reliable and agronomic performance is not affected, or to select a region that already contains desired functional elements for the purpose of trait stacking.
2.5. The Tools
Numerous tools are available to achieve the gene edits described earlier, including sequence-specific nucleases. Sequence-specific nucleases introduce targeted DNA double-strand breaks (DSBs). Normally, DSBs are toxic to cells if unrepaired; therefore, cells have evolved pathways to repair such breaks. In general, plant cells have two major DNA-repair pathways, non-homologous end joining (NHEJ) and homologous recombination (HR). Both pathways can be exploited to introduce sequence changes into a genome. For example, NHEJ is naturally error-prone, frequently resulting in small indels at the site of repair. Therefore, introducing a DSB at a locus of interest can result in targeted mutagenesis. On the other hand, HR copies information to the break site using a donor DNA template. The donor template is usually a sister chromatid or homologous chromosome. Any sequence differences in the donor will be copied to the broken chromosome. It is possible to “trick” the cell into repairing the break with a user-supplied donor template, thereby allowing user-specified DNA sequence changes to be incorporated into the genome.
Currently, most types of genome edits rely on introduction of DSBs. To introduce a DSB, users have the choice between multiple technologies, including meganucleases, zinc-finger nucleases, TALENs, and CRISPR/Cas systems, with the most popular technologies being CRISPR/Cas and TALENs. Readers are directed elsewhere for reviews about these technologies.32,33 In addition to nucleases, users also have the option to use Cre-lox and other site-specific recombination systems for gene edits,34 or CRISPR/Cas deaminases, which include Cas9:AID and Cas9:APOBEC1.35 However, both technologies are limited in the types of genome edits that can be introduced. Deaminases can be used to create targeted SNPs (targeted G→A or C→T changes) at frequencies that are potentially higher than performing nuclease-mediated HR. Cre-lox systems are limited to targeted insertion or precise deletion at a location, where necessary recombination sequences are already present. Together, these tools provide users with the ability to introduce diverse types of DNA modifications.
3. AGRICULTURAL DEMANDS AND GENOME EDITING SUCCESSES
3.1. Increasing Food Production
One means to address rising food demands from a growing population is to increase crop yield. However, crop yield is a complex trait governed by many different factors, both environmental and genetic. With regard to the environment, crop yield can be significantly influenced by agricultural practices, which includes nutrient and water supply, and management of weeds and pests. With regard to genetics, crop productivity can be influenced not only by selecting for crops with higher yield but also by selecting for crops that have increased resistance to drought, bacteria, fungi, and viruses, or other factors, such as optimal flowering time. Although the use of genome editing to improve crop productivity is still in its infancy, there have been numerous successes already reported.
3.2. Increasing Crop Productivity
Perhaps the most direct way to increase yield is to increase the productivity of the crop, which includes increasing grain size and weight. Grain weight is mainly attributed to the size of the grain (volume) and the degree of grain filling (plumpness).36,37 The genetics underlying grain size and weight have been correlated with hundreds of quantitative trait loci (QTLs). Several of the major QTLs have been molecularly characterized, unveiling the underlying functional elements. In rice, these functional elements include G-protein gamma subunit (GS3), serine carboxypeptidase (GS5), protein phosphatase (qGL3/qGL3.1), RING-type E3 ligase (GW2), polyubiquitin-interacting protein (GW5/qSW5), IAA-glucose hydrolase (TGW6), and Snakin/GASA protein (GASR7).38 In effort to increase yield in rice, CRISPR/Cas9 was used to simultaneously knockout three major genes that negatively regulate grain size: GW2, GW5, and TGW6.39 The resulting plants displayed an increased grain size and weight between 20% and 30% of wild type. In a different study, CRISPR/Cas was employed in wheat to knockout the three homoeoalleles of GASR7.40 The resulting wheat plants had significantly elevated thousand kernel weight compared to wild-type plants.
Alternatively, yield can be influenced by manipulating flowering time. This was demonstrated in tomato by modifying flower repressor genes. Tomato is classified as a day-neutral plant, although many cultivars flower during long-day conditions.41,42 Tomato is a sympodial plant with indeterminate plant architecture, wherein the plant continually grows and produces fruit until it dies. Using CRISPR/Cas9, the flowering repressor SELF-PRUNING 5G (SP5G) gene was knocked out, resulting in a day-length insensitive tomato.43 The modified plants exhibited early yield due to rapid flowering and compact determinate growth in long-day conditions. In addition to the SPG5G gene, mutations were introduced into SELF PRUNING (SP), known to switch plant architecture from indeterminate to “bushy” determinate.44 The resulting double mutants displayed rapid flowering bursts and earlier fruit ripening when compared to control lines. Whereas overall yield was lower, harvest index—defined as total yield divided by plant weight—was increased. These results provide information useful to engineer crops for cultivation in different geographical areas, to shorten the cycle of crops to minimize impacts of drought, and to open the possibility of domesticating weed species with agronomical interest.
3.3. Increasing Resistance to Plant Pathogens
Crop losses caused by pathogens, animals, and weeds are estimated to reduce global agricultural productivity between 20% and 40%.45,46 Efforts to reduce crop losses can indirectly result in an increased yield. Genome editing has been successfully deployed to engineer crops with increased resistance to bacteria and fungi. For example, genome editing was used to improve plant disease resistance in rice to thwart bacterial blight. Bacterial blight, caused by Xanthomonas oryzae, is a devastating disease that affects millions of hectares throughout the world. Yield losses in Southeast Asian countries are estimated to range from 10%–20% in moderate conditions to up to 50% in conducive conditions.47 To facilitate pathogenesis, X. oryzae injects effector proteins via a Type III secretion system into the cytoplasm of plant cells. Some of the effector proteins are TAL effectors, which specialize in activating expression of certain genes that help provide a suitable environment for the pathogen to persist. One of these genes, Os11N3 (also called OsSWEET14) is activated when TAL effectors bind to a DNA sequence within the gene’s promoter. To disrupt the binding of the X. oryzae-derived TAL effectors, TALENs were designed to mutate disease susceptibility elements in the Os11N3 promoter.23 Rice plants with mutations in the TAL effector binding elements showed increased resistance to bacterial blight.
Another example of using genome editing to confer disease resistance involved modifying the wheat genome to confer resistance to powdery mildew. Powdery mildew is a fungal disease caused by Blumeria graminis f.sp. tritici (Bgt).48 Yield losses range from 10% to 15%, but can reach as high as 40% under heavy epidemics.49 It was known in barley, Arabidopsis and tomato that the mildew-resistance gene (MLO) was associated with increased resistance to Bgt.50–52 To introduce Bgt resistance in wheat, the three MLO homoeoalleles were targeted using TALENs. Wheat plants containing homozygous knockout mutations in all three homoeoalleles displayed heritable, broad-spectrum resistance to powdery mildew.22
3.4. Weed Management
Effective weed management is critical to achieve maximum crop growth and productivity. One management method is to spray herbicides that kill or stunt the growth of weeds, but have limited impact on crop plants. Herbicides generally work by binding to and inactivating proteins necessary for normal cellular function. For example, the most widely used herbicide, glyphosate, functions by inactivating the enzyme 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS), thereby shutting down the shikimate pathway. Specifically, glyphosate binds to a pocket within the EPSPS protein that normally binds phosphoenol pyruvate. The binding of glyphosate to the phosphoenol pyruvate site mimics an intermediate state of the ternary enzyme–substrate complex.53 By preventing the conversion of the substrate shikimate 3-phosphate to 5-enolpyruvylshikimate-3-phosphate, the plant is unable to produce aromatic amino acids required for survival.
Genome editing has been employed to generate plants that are tolerant to herbicides, including glyphosate. One general mechanism for introducing herbicide tolerance involves protein-structure changes, such that the herbicide no longer binds, but the protein remains functional. For example, certain mutations within the phosphoenol pyruvate binding site within EPSPS can prevent glyphosate binding, but still permit phosphoenol pyruvate binding. This was demonstrated in Linum usitatissimum (flax) using CRISPR/Cas and single-stranded oligo repair templates to introduce the mutations.54 Two SNPs were introduced within the EPSPS gene, resulting in a T178I and P182A (TIPA) substitution within the phosphoenol pyruvate binding site. When sprayed with glyphosate, edited flax lines showed significantly higher levels of tolerance as compared to wild-type plants. Mutations were also introduced into the rice EPSPS gene using CRISPR/Cas and an intronreplacement strategy.55 Whereas plants heterozygous for the gene edit had significantly higher levels of glyphosate tolerance, plants containing homozygous gene edits were not viable, suggesting that modification of the endogenous EPSPS gene in rice resulted in insufficient EPSPS protein activity to maintain plant viability.
In addition to EPSPS and glyphosate tolerance, protein-structure changes can be introduced into acetolactate synthase (ALS) gene(s) for increasing tolerance to ALS-inhibiting herbicides. ALS, also called acetohydroxyacid synthase (AHAS), is a conserved protein required for biosynthesis of the branched-chain amino acids, namely, valine, leucine, and isoleucine. ALS is the primary target for five structurally distinct classes of herbicides, including sulfonylureas (e.g., chlorsulfuron), imidazolinones, triazolopyrimidines, sulfonyl-aminocarbonyl triazolinones, and pyrimidinyl thiobenzoates. Common mutations in ALS that confer resistance to these herbicides occur at positions A122, P197, 205, W574, and S653, in reference to the Arabidopsis ALS protein. There have been several examples of the use of ZFNs and CRISPR/Cas to introduce mutations in ALS, including in tobacco, soybean, maize, and rice.56–58 Results from these studies demonstrate the power of genome editing to rapidly introduce herbicide tolerance into plants.
3.5. Increasing Resistance to Abiotic Stress
Other factors influencing yield are abiotic stresses, including drought. Unfortunately, the biological basis of drought tolerance is complex and not well understood, thereby slowing progress toward developing drought-tolerant crops. Nonetheless, one potential method to introduce drought tolerance includes modulating levels of phytohormones, chemicals that control plant growth and development. Of the plant phytohormones, ethylene has been demonstrated to play a role in a wide variety of physiological processes, including stresses due to drought and temperature change.59 Reducing the sensitivity of the ethylene response in maize resulted in enhanced drought tolerance. Reduced sensitivity was initially achieved in maize by overexpressing ARGOS genes—negative regulators of the ethylene response.60,61 Recently, a genome-editing approach was used to generate drought-tolerant maize.31 Here, the native maize GOS2 promoter was inserted upstream of the ARGOS8-coding sequence, thereby increasing the expression of ARGOS8. Modified plants showed an increase in grain yield by five bushels per acre under flowering stress conditions, with no yield loss under optimal conditions. This work demonstrates not only that genome editing can be used to enhance drought tolerance, but also that a single endogenous gene can be modified to positively affect a complex trait.
4. CREATING HEALTHIER AND MORE NUTRITIOUS FOOD
Whereas increasing food production is paramount, consumers are also looking for improvements in food nutrition and quality. In developing countries, as household incomes rise, consumers demand not only a greater quantity of food but also food of higher quality. Globally, consumers are showing unprecedented interest in how food is produced, and processed, with increased awareness of claims such as “gluten-free,” “reduced cholesterol,” “excellent source of vitamins and minerals,” or “100% whole grain.” Later we provide examples of genome editing successes related to food quality.
4.1. Improving Oil Composition
Oils are ubiquitous ingredients in most processed foods and play an important role in food quality and nutrition. Oils high in monounsaturated fats are considered heart healthy and can help reduce bad cholesterol levels. Such oils typically have a longer shelf life and enhanced oxidative stability.62 In addition, higher intakes of specific dietary fats, including polyunsaturated fatty acids, have been associated with lower risk of heart disease.63 Unfortunately, high levels of polyunsaturated fats are associated with low oxidative and frying stability. This problem was conventionally solved by partial hydrogenation—a process that reduces levels of polyunsaturated fatty acids but also produces trans fats, which have been linked with coronary heart disease and the buildup of plaque in arteries.64 In June 2015, the Food and Drug Administration announced that partially hydrogenated oils are not “generally recognized as safe” for use in human food. A problem with certain plant oils, including soybean oil, is that they have high levels of polyunsaturated fatty acids and frequently undergo partial hydrogenation to improve stability. Improvements in fatty acid composition within plants may avoid the need for partial hydrogenation, which, in-turn, can result in healthier oils.
In an effort to improve oil composition in soybean, the fatty acid synthesis pathway was modified using TALENs. Soybean oil is primarily composed of palmitic acid (unsaturated), stearic acid (unsaturated), oleic acid (monounsaturated), linoleic acid (polyunsaturated), and linolenic acid (polyunsaturated), wherein these five fatty acids are present in soybean oil at approximately 13%, 4%, 20%, 55%, and 8%, respectively. Two gene families, FAD2 and FAD3, are responsible for the production of most of the polyunsaturated fatty acids found in seed oil: FAD2 is responsible for converting oleic acid into linoleic acid, and FAD3 is responsible for converting linoleic into linolenic acid. Using TALENs, two FAD2 genes, FAD2–1A and FAD2–1B, were simultaneously knocked out, resulting in soybean oil with significantly higher levels of oleic acid (~80%, compared to ~20% in wild type), and significantly lower levels of both linoleic (~5%, compared to ~55% in wild type) and linolenic acid (~5%, compared to ~8% in wild type).19 To further improve oil composition, TALENs were used to create knockout mutations in the FAD3 gene, FAD3A, on top of the FAD2–1A and FAD2–1B mutations.20 Triple homozygous mutant soybean plants exhibited increased oleic acid levels (82.2%) and reduced linoleic (2.7%) and linolenic acid (2.5%). In addition to soybean, the fatty acid composition of the oil seed plant Camelina sativa was modified using genome editing.65 Here, CRISPR/Cas was employed to knockout the FAD2 gene, resulting in plants with increased oleic acid (50%, compared to 16% in wild type), decreased linoleic acid (<4%, compared to 16% in wild type), and decreased linolenic (<10%, compared to 35% in wild type). The methods and materials within these studies provide solutions for the demand for oil with increased oxidative stability.
4.2. Reduced-Acrylamide Potatoes
Acrylamide—a suspected human carcinogen—is produced from carbohydrate-rich foods during high-temperature cooking, with a large percentage of intake coming from French fries and potato chips. The accumulation of acrylamide in potato is directly related to storage and processing practices. To ensure a continuous supply of potatoes for direct consumption and processing, potatoes are stored in cold temperatures. During cold storage, potatoes accumulate the reducing sugars glucose and fructose, which are generated from the cleavage of sucrose by the vacuolar invertase protein (VInv).66 However, subsequent cooking of the potatoes results in a Maillard reaction between the reducing sugars and free amino acids, thereby producing acrylamide. In effort to reduce the accumulation of acrylamide in potatoes, TALENs were used to knockout the VInv gene. Nontransgenic potato plants comprising mutations in all four VInv alleles were identified. Cold-stored tubers from modified potato plants exhibited lower levels of fructose and glucose, as compared to wild-type plants. Further, after frying, the resulting chips exhibited less browning and a 73% reduction in acrylamide content.21 Results from this study suggest that genome editing is likely to be an effective approach for minimizing acrylamide formation in processed potatoes.
4.3. Fruits With Increased Antioxidants
One way to improve nutritional characteristics in crops is to increase and stabilize the levels of certain secondary metabolites, including anthocyanins. Anthocyanins are pigments found in leaves, stems, roots, flowers, and fruit tissues, and are known to act as effective antioxidants. Increasing intake of anthocyanins in animal disease models results in increased cardioprotection, life extension in cancer-prone mice, and inhibition of adipocyte development.67 Anthocyanin production within plant cells is controlled primarily by the activity of transcription factors, particularly the Myb transcription factors. In an effort to increase anthocyanin production in tomatoes, TALENs and CRISPR/Cas were employed to increase the expression of the Myb transcription factor gene, ANT1. Here, a strong promoter (CaMV 35S) was inserted upstream of the ANT1 gene in the tomato variety, Micro Tom.68 The resulting plants displayed increased levels of anthocyanins as determined by the deep purple pigmentation found in many tissues, including stems, leaves, and fruits.
5. CHALLENGES AND FUTURE OUTLOOK
5.1. Challenges in Delivering Genome-Editing Reagents
One of the most important steps in practicing genome editing in crops, and also one of the most challenging steps, is the delivery of genome-editing reagents and subsequent generation of modified plants. Unfortunately, transformation and regeneration methods for most crops are low efficiency, time consuming and labor intensive, or they are simply unavailable. These shortcomings have created significant bottlenecks in implementing genome editing in crops. Furthermore, each transformation method has limitations and undesirable features, which can reduce the likelihood of obtaining a properly modified plant. The three most popular delivery techniques are Agrobacterium, biolistics, and protoplast transformation. Understanding the strengths and limitations of each system is necessary for implementing genome editing in crops.
5.2. Agrobacterium
Agrobacterium-mediated transformation has been extensively used for delivering genome-editing reagents. The transformation process begins when plant cells are contacted with Agrobacterium. Agrobacterium then injects T-DNA/protein complexes into the cytoplasm of the plant cell, and the T-DNA/protein complexes then traffic into the nucleus. Once inside the nucleus, the T-DNA/protein complex can randomly integrate into the host’s genome, or it can remain extrachromosomal. If the T-DNA/protein complex remains extrachromosomal, then transient gene expression can be achieved. For most genome engineering applications, T-DNA is stably integrated into the plant genome. Examples of genome editing by stable integration of T-DNA can be found in corn, soybean, barley, Brassica oleracea, potato, and rice.69–77
Advantages of using Agrobacterium to deliver genome-editing reagents include the high probability of achieving single-copy insertions, and protocols have been developed and optimized for transforming many different crops. Disadvantages include: (i) the need for long tissue culture periods to recover transgenic plants, (ii) low frequency of stably transformed plants, (iii) the narrow range of genotypes within a crop species that can be transformed, and (iv) the host-range limitations of certain Agrobacterium species.78 Further, transformation efficiency and regeneration capacity of target cells are usually negatively correlated.79 Methods to expand the number of genotypes that can be transformed, as well as reducing the need for tissue culture, may help increase the utility of Agrobacterium for delivering genome editing reagents to crops.
5.3. Biolistics
Biolistics, or particle bombardment, is a common method for nuclear plant transformation, and for transformations that require DNA to be delivered to chloroplasts and mitochondria. Biolistics is based on the direct delivery of DNA into plant cells using gold or tungsten particles.80 The gold or tungsten particles are coated with DNA which are shot at plant tissue at high velocity and can become lodged inside plant cells. Once inside the cell, the DNA elutes off the particles and becomes transiently expressed or stably integrates into the host genome. The physical delivery of DNA to cells circumvents host-range limitations, as is sometimes encountered with Agrobacterium. Biolistics has been employed to successfully deliver genome-editing reagents in multiple crop plants, including corn, cotton, soybean, and wheat.22,25,28,58,81
Advantages of biolistics include: (i) many different tissues and cell types can be transformed, (ii) no binary vector is required, (iii) it is possible to deliver multiple plasmids with high frequencies of cotransformation, (iv) the transformation protocol is simple, (v) large DNA fragments can be delivered, and (vi) the system can be adapted to deliver mRNA or protein.40,82 Disadvantages include: (i) messy integration patterns, (ii) relatively low throughput, (iii) high input cost, and (iv) the cellular target cannot be controlled (i.e., cytoplasm, nucleus, mitochondria, or plastid).
5.4. Protoplast Transformation
Protoplast transformation involves the direct delivery of DNA to individual plant cells using polyethylene glycol or electroporation. Once inside the nucleus, the DNA can be transiently expressed or stably integrated into the genome. Of the three methods discussed here, protoplast transformation has arguably the highest efficacy, where it is possible to achieve over 70% transformation frequency. Protoplast transformation has been used to successfully deliver genome-editing reagents in multiple crop plants, including potato, wheat, rice, and flax.21,54,83–85
Advantages of protoplast transformation include: (i) delivery of multiple plasmids with high levels of cotransformation, (ii) no binary vector required, (iii) high frequency transformation, and (iv) most plant species are amenable to protoplast isolation and transformation. Disadvantages include limited plant species that are amenable to regeneration from protoplasts, and among those that are amenable, the technique is time consuming and labor intensive.
5.5. Overcoming Delivery Limitations
Most transformation methods are limited to one or a few genotypes per crop because only one or a few genotypes are amenable to regeneration. Unfortunately, amenability to transformation and regeneration is usually not correlated with acceptable agronomic performance. In an effort to expand the frequency of transformation and range of genotypes, Lowe et al. describe a new method which uses the Baby boom (Bbm) and Wuschel2 (Wus2) transcription factors.86 Agrobacterium- and biolistic-mediated transformation were employed to deliver Bbm and Wus2 to tissues from maize, rice, sorghum, and sugarcane varieties. Frequencies of transformation increased in rice, sugarcane, and sorghum from 0%–3% to 15%–885%. Whereas this method may not have achieved a complete and universal genotype-independent transformation system for all crops, this is arguably one of the biggest steps in recent years toward this goal.
An alternative means to edit crops that are recalcitrant to transformation is to use virus-based in planta delivery systems. Here, a crop plant is infected with an engineered virus that harbors genome-editing reagents. As the virus spreads from cell to cell and throughout the plant, the genome-editing reagents are expressed and introduce genome modifications. Preferably, the virus will spread to meristematic tissue where it can facilitate the introduction of heritable genome modifications. As proof of concept, the geminivirus vector, Cabbage leaf curl virus, was used to deliver gRNAs for Cas9 in Nicotiana benthamiana.87 Although high frequencies of mutations were observed in somatic tissues, the use of geminiviruses as delivery vectors precluded production of heritable mutations, as geminiviruses are specifically excluded from meristematic cells. In addition to geminiviruses, an RNA virus, Tobacco rattle virus (TRV), was modified to deliver genome-editing reagents to N. benthamiana and petunia plants.88–90 Unlike geminiviruses, TRV has been shown to enter meristematic cells.91 In agreement with this property, mutations (albeit at low frequencies) were found in seed from plants infected with engineered TRV, demonstrating that TRV is capable of entering meristem cells and delivering genome-editing reagents to create heritable DNA modifications. Improvements in virus-based delivery systems may expand the number of crops that can be edited and improve the frequency that heritable mutations are created.
5.6. Nontransgenic Genome Engineering
When choosing a delivery method, it is also important to consider governmental regulations that may dictate which genome-edited plants can be deployed in the field. In many jurisdictions, introduction of foreign DNA into a cell is a trigger for regulation.92 Further, the process of delivery may result in undesired off-target modifications due to random insertion of nuclease-encoding DNA. Editing plant genomes by transiently delivering DNA to cells, or by delivering mRNA or protein, may reduce regulatory and off-target concerns related to genome-edited plants. Several approaches have been employed to address this concern, including delivery of sequence-specific nucleases as mRNA,40,93 protein,94 mRNA/protein Cas9-gRNA complexes,57,95 or transient delivery of plasmid DNA or mRNA.21,40 Results from these studies provide novel approaches to reduce the likelihood of unwanted, random DNA integration.
5.7. Trait Gene/Locus Identification
Another significant challenge for genome editing in agriculture is determining types of genetic modification(s) needed to create a trait of interest. Identifying a target gene or DNA region requires both an understanding of the crop’s genome and gene function. However, many of the traits and targets mentioned herein are based on gene-function studies that were performed in model plants. In an effort to provide more information about the genetic makeup of crop plants, many genomes are being, or have been sequenced, including bread wheat, rice, soybean, corn, and potato.96–100 However, genome sequences are mostly limited to one variety within each crop. Due to genomic differences between varieties, there is a need to resequence target genes or DNA regions, as SNPs, indels or gross chromosomal rearrangements could be present. This resequencing is necessary to carry out accurate genome editing, as using genome editing tools relies on having the precise sequence of the target region in the variety being modified. Further, due to many crops being highly recalcitrant to transformation and regeneration, molecular characterization of gene function has been limited. To help address these concerns, our understanding of gene function and systems biology in crop plants needs to be advanced.
5.8. Frequency of Introducing a Desired Gene Edit
An additional challenge in implementing genome editing is the efficiency that certain edits are introduced. The simplest gene edit, with respect to nuclease technology, is an indel. Indels are created by delivering nucleases to plant cells, wherein the nucleases create targeted DNA DSBs followed by imprecise repair by the NHEJ pathway. Frequency of this type of edit can be extremely high, reaching over 70%.101 However, indels are only one type of the many edits that can be made. Other edits include large deletions, single-nucleotide polymorphisms, gene insertions, gene replacements, and inversions. Frequencies of creating these types of edits are generally much lower than frequencies of creating indels.
Perhaps the most frequent gene edit outside of indels are large deletions, where two DSBs are introduced within the same chromosome, and intervening sequence is deleted. As proof of concept in plants, Zhou et al. created two DSBs using CRISPR/Cas in the rice genome to delete large stretches of DNA, approximately 245 kb, at a high frequency (~4/24 transgenic calli).24 It was later demonstrated in Arabidopsis that the frequency of the deletion is inversely correlated with its size (i.e., the larger the deletion, the lower the efficiency). Whereas plants comprising small deletions of <100 bp were identified at fairly high frequencies (~10%), the frequency of plants with phenotypically selected deletions of 5–120 kb was below 1%.102
Low frequencies of gene edits also plague the introduction of single-nucleotide polymorphisms, gene replacements, and gene insertions, as these edits rely on the use of the HR machinery. To achieve HR, both nucleases and donor molecules are provided to plant cells; the resulting DSBs are repaired by the HR machinery using the information within the donor molecule as a template. If the donor molecule has sequence differences from the chromosomal target, the differences will be stably incorporated into the host genome. In general, the efficiency of HR is about two orders of magnitude lower than NHEJ,103 with reports of frequencies ranging from ~10% to 0.08% of transformed cells.54,104 Wide ranges of gene targeting frequencies are most likely due to numerous confounding variables, including nuclease efficiency, nuclease type, DSB overhang, plant species, cell type, cell-cycle stage, donor homology-arm length, donor structure (e.g., single stranded vs double stranded vs circular vs linear), composition of nucleotides within the homology arms, the length of sequence being modified, the way the donor was delivered (T-DNA vs plasmid vs virus), and the epigenetic status of the target locus. One solution to address the low frequencies of gene targeting is to establish an efficient transformation pipeline and screen hundreds of transgenic plants—a brute-force approach. However, this is not practical for most research labs, as high-throughput platforms are costly, resource intensive and laborious. An alternative solution is to increase the frequency that gene editing events occur. Qi et al. demonstrated in Arabidopsis that knockout of genes involved in classical NHEJ, ku70, or lig4, resulted in 3- to 16-fold enhancement in HR-based gene targeting frequencies.105 Further, knockout of a gene involved in HR-based DSB repair using sister chromatids, smc6b, resulted in a three- to fourfold enhancement in HR frequencies. It was later shown in rice that concurrent knockout DNA ligase 4 alongside the delivery of Cas9 and donor molecules, resulted in increased frequencies of HR.56 Several other strategies to increase HR frequencies have been demonstrated, including overexpression of RecA and RAD54,106–108 overexpression of the DNA-resecting proteins OsRecQI4 and OsExo1,109 and delivery of donor molecules on replicons.68,110–112
5.9. Breeding Challenges With Genome-Edited Crops
Because only a limited number of genotypes of a given species are amenable to transformation, the majority of gene edits are introduced into varieties with poor agronomic performance. This usually requires genome-edited crops to be placed into breeding programs to transfer the mutations into elite varieties. However, breeding suffers from several drawbacks, including an investment of several years for backcrossing, laborious crossing protocols for certain crops, cost of continual genotyping, and the potential cost and labor of relocating the plants to a different environment (e.g., winter and summer nurseries). Whereas breeding can be a viable option for transferring one or a few mutations, challenges and costs greatly increase as the number of unlinked gene edits increase. For example, when selfing a plant that is heterozygous for a single gene edit, the frequency of progeny comprising homozygous edits is high (1 out of every 4 plants); however, if the plant is heterozygous for four gene edits, the frequency decreases to 1 out of every 256 plants, a frequency that is cumbersome to handle for many breeding platforms. In addition to low frequency challenges, increasing the number of gene edits simultaneously increases the chance that an edit occurs near a locus that negatively effects the overall performance of the plant, thereby creating a linkage drag when crossed with elite varieties.113 Lastly, for traits such as yield, one needs to carefully compare the value of a product generated by backcrossing to a product that could be generated through a traditional breeding and selection programs, and the annual yield increases associated with such program.
6. CONCLUDING THOUGHTS
Genome editing is poised to change agriculture in the 21st century and beyond. We are already seeing the first wave of genome-edited products emerge, ranging from soybean with improved oil characteristics to drought-tolerant maize. With the continued application of genome editing in crops, we will better understand the constraints and limitations of the technology. These constraints must be addressed before realizing the technology’s full potential: two key constraints perhaps being our understanding of plant systems biology and our ability to transform and regenerate elite crop varieties. Reducing or eliminating these constraints gets us closer to the ultimate goal of genome editing in agriculture—having the ability to truly “design” crops from the ground up.
REFERENCES
- 1.Valin H, Sands RD, van der Mensbrugghe D, et al. The future of food demand: understanding differences in global economic models. Agric Econ. 2014;45(1):51–67. [Google Scholar]
- 2.Siro I, Kapolna E, Kapolna B, Lugasi A. Functional food. Product development, marketing and consumer acceptance—a review. Appetite. 2008;51(3):456–467. [DOI] [PubMed] [Google Scholar]
- 3.Slade AJ, McGuire C, Loeffler D, et al. Development of high amylose wheat through TILLING. BMC Plant Biol. 2012;12:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gady ALF, Vriezen WH, Van de Wal MHBJ, et al. Induced point mutations in the phytoene synthase 1 gene cause differences in carotenoid content during tomato fruit ripening. Mol Breed. 2012;29(3):801–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boden SA, Cavanagh C, Cullis BR, et al. Ppd-1 is a key regulator of inflorescence architecture and paired spikelet development in wheat. Nat Plants. 2015;1(2):14016. [DOI] [PubMed] [Google Scholar]
- 6.Isshiki M, Yamamoto Y, Satoh H, Shimamoto K. Nonsense-mediated decay of mutant waxy mRNA in rice. Plant Physiol. 2001;125:1388–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dierking EC, Bilyeu KD. New sources of soybean seed meal and oil composition traits identified through TILLING. BMC Plant Biol. 2009;9:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Acevedo-Garcia J, Spencer D, Thieron H, et al. Mlo-based powdery mildew resistance in hexaploid bread wheat generated by a non-transgenic TILLING approach. Plant Biotechnol J. 2017;15(3):367–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown JWS. Arabidopsis intron mutations and pre-mRNA splicing. Plant J. 1996;10(5):771–780. [DOI] [PubMed] [Google Scholar]
- 10.Lorković ZJ, Kirk DAW, Lambermon MHL, Filipowicz W. Pre-mRNA splicing in higher plants. Trends Plant Sci. 2000;5(4):160–167. [DOI] [PubMed] [Google Scholar]
- 11.Bilyeu K, Palavalli L, Sleper D, Beuselinck P. Mutations in soybean microsomal omega-3 fatty acid desaturase genes reduce linolenic acid concentration in soybean seeds. Crop Sci. 2005;45(5):1830–1836. [Google Scholar]
- 12.Sestili F, Palombieri S, Botticella E, Mantovani P, Bovina R, Lafiandra D. TILLING mutants of durum wheat result in a high amylose phenotype and provide information on alternative splicing mechanisms. Plant Sci. 2015;233:127–133. [DOI] [PubMed] [Google Scholar]
- 13.Hu X, Sullivan-Gilbert M, Gupta M, Thompson SA. G-to-A mutation at a 5′ splice site of fad3c caused impaired splicing in a low linolenic mutant of canola (Brassica napus L.). Plant Biotechnol. 2007;24(4):397–400. [Google Scholar]
- 14.Bradley JM, Whitelam GC, Harberd NP. Impaired splicing of phytochrome B pre-mRNA in a novel phyB mutant of Arabidopsis. Plant Mol Biol. 1995;27: 1133–1142. [DOI] [PubMed] [Google Scholar]
- 15.McCullough AJ, Baynton CE, Schuler MA. Interactions across exons can influence splice site recognition in plant nuclei. Plant Cell. 1996;8(12):2295–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Je BI, Gruel J, Lee YK, et al. Signaling from maize organ primordial via FASCIATED EAR3 regulates stem cell proliferation and yield traits. Nat Genet. 2016;48(7):785–791. [DOI] [PubMed] [Google Scholar]
- 17.Krieger U, Lippman ZB, Zamir D. The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nat Genet. 2010;42(5):459–463. [DOI] [PubMed] [Google Scholar]
- 18.Sammons RD, Gaines TA. Glyphosate resistance: state of knowledge. Pest Manag Sci. 2014;70(9):1367–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haun W, Coffman A, Clasen BM, et al. Improved soybean oil quality by targeted mutagenesis of the fatty acid desaturase 2 gene family. Plant Biotechnol J. 2014;12(7):934–940. [DOI] [PubMed] [Google Scholar]
- 20.Demorest ZL, Coffman A, Baltes NJ, et al. Direct stacking of sequence-specific nuclease-induced mutations to produce high oleic and low linolenic soybean oil. BMC Plant Biol. 2016;16(1):225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clasen BM, Stoddard TJ, Luo S, et al. Improving cold storage and processing traits in potato through targeted gene knockout. Plant Biotechnol J. 2016;14:169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Cheng X, Shan Q, et al. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol. 2014;32(9):947–951. [DOI] [PubMed] [Google Scholar]
- 23.Li T, Liu B, Spalding MH, Weeks DP, Yang B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat Biotechnol. 2012;30(5):390–392. [DOI] [PubMed] [Google Scholar]
- 24.Zhou H, Liu B, Weeks DP, Spalding MH, Yang B. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 2014;42(17):10903–10914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ainley WM, Sastry-Dent L, Welter ME, et al. Trait stacking via targeted genome editing. Plant Biotechnol J. 2013;11:1126–1134. [DOI] [PubMed] [Google Scholar]
- 26.Cai CQ, Doyon Y, Ainley WM, et al. Targeted transgene integration in plant cells using designed zinc finger nucleases. Plant Mol Biol. 2009;69(6):699–709. [DOI] [PubMed] [Google Scholar]
- 27.Ayar A, Wehrkamp-Richter S, Laffaire JB, et al. Gene targeting in maize by somatic ectopic recombination. Plant Biotechnol J. 2012;11:305–314. [DOI] [PubMed] [Google Scholar]
- 28.D’Halluin K, Vanderstraeten C, Van Hulle J, et al. Targeted molecular trait stacking in cotton through targeted double-strand break induction. Plant Biotechnol J. 2013;11(8):933–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shukla VK, Doyon Y, Miller JC, et al. Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature. 2009;459(7245):437–441. [DOI] [PubMed] [Google Scholar]
- 30.Zhao Y, Zhang C, Liu W, et al. An alternative strategy for targeted gene replacement in plants using a dual-sgRNA/Cas9 design. Sci Rep. 2016;6:23890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi J, Gao H, Wang H, et al. ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol J. 2017;15(2):207–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaj T, Gersbach CA, Barbas CF. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31(7):397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baltes NJ, Voytas DF. Enabling plant synthetic biology through genome engineering. Trends Biotechnol. 2015;33(2):120–131. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Yau YY, Perkins-Balding D, Thomson JG. Recombinase technology: applications and possibilities. Plant Cell Rep. 2011;30(3):267–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J, Sun Y, Du J, Zhao Y, Xia L. Generation of targeted point mutations in rice by a modified CRISPR/Cas9 system. Mol Plant. 2017;10(3):526–529. [DOI] [PubMed] [Google Scholar]
- 36.Xing Y, Zhang Q. Genetic and molecular bases of rice yield. Annu Rev Plant Biol. 2010;61:421–442. [DOI] [PubMed] [Google Scholar]
- 37.Sakamoto T, Matsuoka M. Identifying and exploiting grain yield genes in rice. Curr Opin Plant Biol. 2008;11(2):209–214. [DOI] [PubMed] [Google Scholar]
- 38.Zuo J, Li J. Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annu Rev Genet. 2014;48:99–118. [DOI] [PubMed] [Google Scholar]
- 39.Xu R, Yang Y, Qin R, et al. Rapid improvement of grain weight via highly efficient CRISPR/Cas9-mediated multiplex genome editing in rice. J Genet Genomics. 2016;43(8):529–532. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y, Liang Z, Zong Y, et al. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat Commun. 2016;7:12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Binchy A, Morgan JV. Influence of light intensity and photoperiod on inflorescence initiation in tomatoes. Isr J Agric Res. 1970;9:261–269. [Google Scholar]
- 42.Hurd RG. Long-day effects on growth and flower initiation of tomato plants in low light. Ann Appl Biol. 1973;73(2):221–228. [Google Scholar]
- 43.Soyk S, Müller NA, Park SJ, et al. Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato. Nat Genet. 2017;49(1):162–168. [DOI] [PubMed] [Google Scholar]
- 44.Carmel-Goren L, Liu YS, Lifschitz E, Zamir D, Zamir D. The SELF-PRUNING gene family in tomato. Plant Mol Biol. 2003;52:1215–1222. [DOI] [PubMed] [Google Scholar]
- 45.Savary S, Ficke A, Aubertot JN, Hollier C. Crop losses due to diseases and their implications for global food production losses and food security. Food Secur. 2012;4(4):519–537. [Google Scholar]
- 46.Oerke E-C. Crop losses to pests. J Agric Sci. 2006;144:31–43. [Google Scholar]
- 47.Mew TW, Alvarez AM, Leach JE, Swings J. Focus on bacterial blight of rice. Plant Dis. 1993;77:5–12. [Google Scholar]
- 48.Glawe DA. The powdery mildews: a review of the world’s most familiar (yet poorly known) plant pathogens. Annu Rev Phytopathol. 2008;46:27–51. [DOI] [PubMed] [Google Scholar]
- 49.Singh RP, Singh PK, Rutkoski J, et al. Disease impact on wheat yield potential and prospects of genetic control. Annu Rev Phytopathol. 2016;54:13.1–13.20. [DOI] [PubMed] [Google Scholar]
- 50.Consonni C, Humphry ME, Hartmann HA, et al. Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat Genet. 2006;38(6):716–720. [DOI] [PubMed] [Google Scholar]
- 51.Bai Y, Pavan S, Zheng Z, et al. Naturally occurring broad-spectrum powdery mildew resistance in a central American tomato accession is caused by loss of mlo function. Mol Plant Microbe Interact. 2008;21(1):30–39. [DOI] [PubMed] [Google Scholar]
- 52.Piffanelli P, Ramsay L, Waugh R, et al. A barley cultivation-associated polymorphism conveys resistance to powdery mildew. Nature. 2004;430(7002):887–891. [DOI] [PubMed] [Google Scholar]
- 53.Schönbrunn E, Eschenburg S, Shuttleworth WA, et al. Interaction of the herbicide glyphosate with its target enzyme 5-enolpyruvylshikimate 3-phosphate synthase in atomic detail. Proc Natl Acad Sci USA. 2001;98(4):1376–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sauer NJ, Narváez-Vásquez J, Mozoruk J, et al. Oligonucleotide-mediated genome editing provides precision and function to engineered nucleases and antibiotics in plants. Plant Physiol. 2016;170:1917–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li J, Meng X, Zong Y, et al. Gene replacements and insertions in rice by intron targeting using CRISPR-Cas9. Nat Plants. 2016;2(10):16139. [DOI] [PubMed] [Google Scholar]
- 56.Endo M, Mikami M, Toki S. Biallelic gene targeting in rice. Plant Physiol. 2016;170(2):667–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Svitashev S, Young JK, Schwartz C, Gao H, Falco SC, Cigan AM. Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol. 2015;169(2):931–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Z, Liu Z-B, Xing A, et al. Cas9-guide RNA directed genome editing in soybean. Plant Physiol. 2015;169:960–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arraes FBM, Beneventi MA, Lisei de Sa ME, et al. Implications of ethylene biosynthesis and signaling in soybean drought stress tolerance. BMC Plant Biol. 2015;15(1):213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shi J, Habben JE, Archibald RL, et al. Overexpression of ARGOS genes modifies plant sensitivity to ethylene, leading to improved drought tolerance in both Arabidopsis and maize. Plant Physiol. 2015;169(1):266–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guo M, Rupe MA, Wei J, et al. Maize ARGOS1 (ZAR1) transgenic alleles increase hybrid maize yield. J Exp Bot. 2014;65(1):249–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Clemente TE, Cahoon EB. Soybean oil: genetic approaches for modification of functionality and total content. Plant Physiol. 2009;151(3):1030–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mozaffarian D, Willett WC. Trans fatty acids and cardiovascular risk: a unique cardiometabolic imprint? Curr Atheroscler Rep. 2007;9(6):486–493. [DOI] [PubMed] [Google Scholar]
- 64.Ascherio A, Katan MB, Zock PL, Stampfer MJ, Willett WC. Trans fatty acids and coronary heart disease. N Engl J Med. 1999;340:1994–1998. [DOI] [PubMed] [Google Scholar]
- 65.Jiang WZ, Henry IM, Lynagh PG, Comai L, Cahoon EB, Weeks DP. Significant enhancement of fatty acid composition in seeds of the allohexaploid, Camelina sativa, using CRISPR/Cas9 gene editing. Plant Biotechnol J. 2017;. 10.1111/pbi.12663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sturm A Primary structures, functions, and roles in plant development and sucrose partitioning. Plant Physiol. 1999;121:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Glover BJ, Martin C. Anthocyanins. Curr Biol. 2012;22(5):147–150. [DOI] [PubMed] [Google Scholar]
- 68.Čermák T, Baltes NJ, Čegan R, Zhang Y, Voytas DF. High-frequency, precise modification of the tomato genome. Genome Biol. 2015;16:232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Char SN, Unger-Wallace E, Frame B, et al. Heritable site-specific mutagenesis using TALENs in maize. Plant Biotechnol J. 2015;13(7):1002–1010. [DOI] [PubMed] [Google Scholar]
- 70.Curtin SJ, Zhang F, Sander JD, et al. Targeted mutagenesis of duplicated genes in soybean with zinc-finger nucleases. Plant Physiol. 2011;156(2):466–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Butler NM, Atkins PA, Voytas DF, Douches DS. Generation and inheritance of targeted mutations in potato (Solanum tuberosum L.) using the CRISPR/Cas system. PLoS One. 2015;10(12):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Miao J, Guo D, Zhang J, et al. Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 2013;23(10):1233–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lawrenson T, Shorinola O, Stacey N, et al. Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guided Cas9 nuclease. Genome Biol. 2015;16(1):258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gao H, Smith J, Yang M, et al. Heritable targeted mutagenesis in maize using a designed endonuclease. Plant J. 2010;61:176–187. [DOI] [PubMed] [Google Scholar]
- 75.Djukanovic V, Smith J, Lowe K, et al. Male-sterile maize plants produced by targeted mutagenesis of the cytochrome P450-like gene (MS26) using a re-designed I-CreI homing endonuclease. Plant J. 2013;76:888–899. [DOI] [PubMed] [Google Scholar]
- 76.Wendt T, Holm PB, Starker CG, et al. TAL effector nucleases induce mutations at a pre-selected location in the genome of primary barley transformants. Plant Mol Biol. 2013;83(3):279–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gurushidze M, Hensel G, Hiekel S, Schedel S, Valkov V, Kumlehn J. True-breeding targeted gene knock-out in barley using designer TALE-nuclease in haploid cells. PLoS One. 2014;9:e92046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nam J, Matthysse AG, Gelvin SB. Differences in susceptibility of Arabidopsis ecotypes to crown gall disease may result from a deficiency in T-DNA integration. Plant Cell. 1997;9(3):317–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Altpeter F, Springer NM, Bartley LE, et al. Advancing crop transformation in the era of genome editing. Plant Cell. 2016;28(7):1510–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sanford JC. The biolistic process. Trends Biotechnol. 1988;6(12):299–302. [Google Scholar]
- 81.Jacobs TB, LaFayette PR, Schmitz RJ, Parrott WA. Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol. 2015;15:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Svitashev S, Schwartz C, Lenderts B, Young JK, Mark Cigan A. Genome editing in maize directed by CRISPR-Cas9 ribonucleoprotein complexes. Nat Commun. 2016;7:13274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shan Q, Wang Y, Li J, et al. Targeted genome modification of crop plants using the CRISPR-Cas system. Nat Biotechnol. 2013;31(8):686–688. [DOI] [PubMed] [Google Scholar]
- 84.Shan Q, Wang Y, Chen K, et al. Rapid and efficient gene modification in rice and Brachypodium using TALENs. Mol Plant. 2013;6(4):1365–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liang Z, Zhang K, Chen K, Gao C. Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J Genet Genomics. 2014;41:63–68. [DOI] [PubMed] [Google Scholar]
- 86.Lowe K, Wu E, Wang N, et al. Morphogenic regulators baby boom and Wuschel improve monocot transformation. Plant Cell. 2016;28:1998–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yin K, Han T, Liu G, et al. A geminivirus-based guide RNA delivery system for CRISPR/Cas9 mediated plant genome editing. Sci Rep. 2015;5:14926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ali Z, Abul-Faraj A, Piatek M, Mahfouz MM. Activity and specificity of TRV-mediated gene editing in plants. Plant Signal Behav. 2015;10(10):e1044191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ali Z, Abul-Faraj A, Li L, et al. Efficient virus-mediated genome editing in plants using the CRISPR/Cas9 system. Mol Plant. 2015;8(8):1288–1291. [DOI] [PubMed] [Google Scholar]
- 90.Marton I, Zuker A, Shklarman E, et al. Nontransgenic genome modification in plant cells. Plant Physiol. 2010;154(3):1079–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Martín-Hernández AM, Baulcombe DC. Tobacco rattle virus 16-kilodalton protein encodes a suppressor of RNA silencing that allows transient viral entry in meristems. J Virol. 2008;82(8):4064–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Voytas DF, Gao C. Precision genome engineering and agriculture: opportunities and regulatory challenges. PLoS Biol. 2014;12(6):e1001877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stoddard TJ, Clasen BM, Baltes NJ, et al. Targeted mutagenesis in plant cells through transformation of sequence-specific nuclease mRNA. PLoS One. 2016;11(5):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Luo S, Li J, Stoddard TJ, et al. Non-transgenic plant genome editing using purified sequence-specific nucleases. Mol Plant. 2015;8(9):1425–1427. [DOI] [PubMed] [Google Scholar]
- 95.Woo JW, Kim J, Kwon SI, et al. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat Biotechnol. 2015;33(11):1162–1164. [DOI] [PubMed] [Google Scholar]
- 96.Schmutz J, Cannon SB, Schlueter J, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–183. [DOI] [PubMed] [Google Scholar]
- 97.Schnable PS, Ware D, Fulton RS, et al. The B73 maize genome: complexity, diversity, and dynamics. Nature. 2005;326(5956):1112–1115. [DOI] [PubMed] [Google Scholar]
- 98.Xu X, Pan S, Cheng S, et al. Genome sequence and analysis of the tuber crop potato. Nature. 2011;475(7355):189–195. [DOI] [PubMed] [Google Scholar]
- 99.Mayer KFX, Rogers J, Dole el J, et al. A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science. 2014;345(6194):1251788. [DOI] [PubMed] [Google Scholar]
- 100.Matsumoto T, Wu JZ, Kanamori H, et al. The map-based sequence of the rice genome. Nature. 2005;436:793–800. [DOI] [PubMed] [Google Scholar]
- 101.Li J, Stoddard TJ, Demorest ZL, et al. Multiplexed, targeted gene editing in Nicotiana benthamiana for glyco-engineering and monoclonal antibody production. Plant Biotechnol J. 2016;14(2):533–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ordon J, Gantner J, Kemna J, et al. Generation of chromosomal deletions in dicotyledonous plants employing a user-friendly genome editing toolkit. Plant J. 2017;89(1):155–168. [DOI] [PubMed] [Google Scholar]
- 103.Steinert J, Schiml S, Puchta H. Homology-based double-strand break-induced genome engineering in plants. Plant Cell Rep. 2016;35(7):1429–1438. [DOI] [PubMed] [Google Scholar]
- 104.Wright DA, Townsend JA, Winfrey RJ, et al. High-frequency homologous recombination in plants mediated by zinc-finger nucleases. Plant J. 2005;44(4):693–705. [DOI] [PubMed] [Google Scholar]
- 105.Qi Y, Zhang Y, Zhang F, et al. Increasing frequencies of site-specific mutagenesis and gene targeting in Arabidopsis by manipulating DNA repair pathways. Genome Res. 2013;23(3):547–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Reiss B, Klemm M, Kosak H, Schell J. RecA protein stimulates homologous recombination in plants. Proc Natl Acad Sci USA. 1996;93:3094–3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shaked H, Melamed-Bessudo C, Levy AA. High-frequency gene targeting in Arabidopsis plants expressing the yeast RAD54 gene. Proc Natl Acad Sci USA. 2005;102(34):12265–12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Even-Faitelson L, Samach A, Melamed-Bessudo C, Avivi-Ragolsky N, Levy AA. Localized egg-cell expression of effector proteins for targeted modification of the Arabidopsis genome. Plant J. 2011;68:929–937. [DOI] [PubMed] [Google Scholar]
- 109.Kwon Y-I, Abe K, Osakabe K, et al. Overexpression of OsRecQl4 and/or OsExo1 enhances DSB-induced homologous recombination in rice. Plant Cell Physiol. 2012;53(12):2142–2152. [DOI] [PubMed] [Google Scholar]
- 110.Baltes NJ, Gil-Humanes J, Cermak T, Atkins PA, Voytas DF. DNA replicons for plant genome engineering. Plant Cell. 2014;26(1):151–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Butler NM, Baltes NJ, Voytas DF, Douches DS. Geminivirus-mediated genome editing in potato (Solanum tuberosum L.) using sequence-specific nucleases. Front Plant Sci. 2016;7:1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gil-Humanes J, Wang Y, Liang Z, et al. High efficiency gene targeting in hexaploid wheat using DNA replicons and CRISPR/Cas9. Plant J. 2017;89(6):1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Peleman JD, van der Voort JR. The challenges in marker assisted breeding. Cgn. 2003;125:125–130. [Google Scholar]


