Abstract
A major sex difference in Alzheimer’s disease (AD) is that men with the disease die earlier than do women. In aging and preclinical AD, men also show more cognitive deficits. Here, we show that the X chromosome affects AD-related vulnerability in mice expressing the human amyloid precursor protein (hAPP), a model of AD. XY-hAPP mice genetically modified to develop testicles or ovaries showed worse mortality and deficits than did XX-hAPP mice with either gonad, indicating a sex chromosome effect. To dissect whether the absence of a second X chromosome or the presence of a Y chromosome conferred a disadvantage on male mice, we varied sex chromosome dosage. With or without a Y chromosome, hAPP mice with one X chromosome showed worse mortality and deficits than did those with two X chromosomes. Thus, adding a second X chromosome conferred resilience to XY males and XO females. In addition, the Y chromosome, its sex-determining region Y gene (Sry), or testicular development modified mortality in hAPP mice with one X chromosome such that XY males with testicles survived longer than did XY or XO females with ovaries. Furthermore, a second X chromosome conferred resilience potentially through the candidate gene Kdm6a, which does not undergo X-linked inactivation. In humans, genetic variation in KDM6A was linked to higher brain expression and associated with less cognitive decline in aging and preclinical AD, suggesting its relevance to human brain health. Our study suggests a potential role for sex chromosomes in modulating disease vulnerability related to AD.
INTRODUCTION
The expansion of translational neuroscience to investigate sex differences and their mechanistic underpinnings is of major consequence to human health (1). Understanding what makes one sex more vulnerable (or resilient) to aging and disease unravels new pathways to target with treatments that could benefit both sexes.
Alzheimer’s disease (AD) is the most common neurodegenerative condition and a global health threat. In the absence of effective medical treatments, more than 50 million men and women worldwide will suffer from this devastating condition by 2050 (2). The burdens of the disease combined with failed clinical trials (3) warrant a deeper understanding of the heterogeneous nature of AD, with the goal of developing better therapies.
Being male or female, defined here as harboring a different sex chromosome complement (XY versus XX), is an understudied biologic variable that contributes heterogeneity to AD. Sex differences in AD reveal differing vulnerabilities in men and women (4, 5). Many more women have AD, largely due to their longevity (6) as they live to advanced ages, when AD risk and incidence is highest. In contrast, men with the disease die earlier in populations worldwide, indicating a male disadvantage with early-onset (7-9) and late-onset (10, 11) subtypes of AD. Furthermore, in aging and preclinical AD before the age of 85 years, men show worse cognition (12), more cognitive decline (13-15), and increased measures of neurodegeneration (16), despite similar deposition of amyloid and tau (15, 17), the pathological hallmarks of AD. This could underlie higher prevalence (18) and earlier onset of mild cognitive impairment (MCI) in men compared to women in some populations (19, 20). Here, we assess sex-biased mortality in AD by meta-analysis, investigate whether sex chromosomes affect vulnerability in a mouse model of AD, and test whether an X chromosome gene influences cognition in this mouse model.
RESULTS
Male sex and increased mortality in AD and the hAPP mouse model
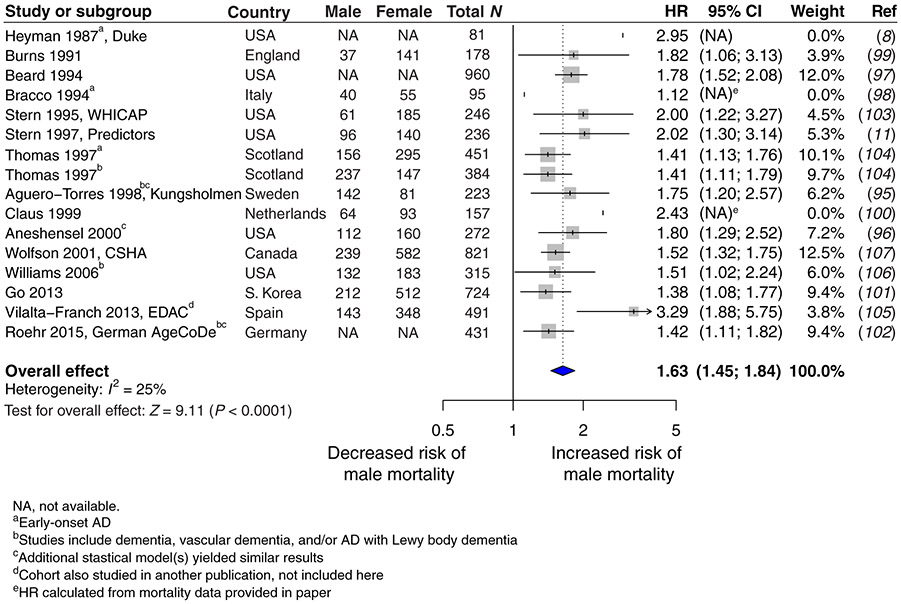
We conducted a meta-analysis of data collected on mortality in human populations worldwide. Only longitudinal studies that defined the time variable as age of disease onset or duration of disease after onset were included; cross-sectional studies were excluded. Our meta-analysis showed that male sex increased risk for death in AD by 62% compared to female sex [male hazard ratio (HR) 1.63, CI 1.45 to 1.84, P < 0.0001; Fig. 1]. We then examined mortality in transgenic mice that expressed mutated forms of the human amyloid precursor protein (hAPP) (line J20) (21) and exhibited premature death, cognitive impairments, and pathological markers of the disease. Male hAPP mice died significantly earlier than did female hAPP mice on two genetic backgrounds, C57BL6/J (P < 0.001; Fig. 2A) and a mixed F1 generation of C57BL6/J crossed with FVB/N (P < 0.05; fig. S1).
Fig. 1. A meta-analysis of hazard ratios for male and female mortality in AD populations worldwide.
Hazard ratios (HRs) and 95% CIs are shown in a forest plot for studies (8, 11, 95-107) reporting male risk, compared to female risk, for death in longitudinal (and not cross-sectional) analysis of individuals with AD. Overall HR with 95% CI shown in bold indicates increased risk of male mortality (male, HR 1.63, CI 1.45 to 1.84; P < 0.0001). WHICAP, Washington Heights-Inwood Columbia Aging Project; CSHA, Canadian Study of Health and Aging; EDAC, Evolution of Dementia of the Alzheimer-type and Caregiver burden; AgeCoDe, Aging, Cognition, and Dementia in Primary Care Patients.
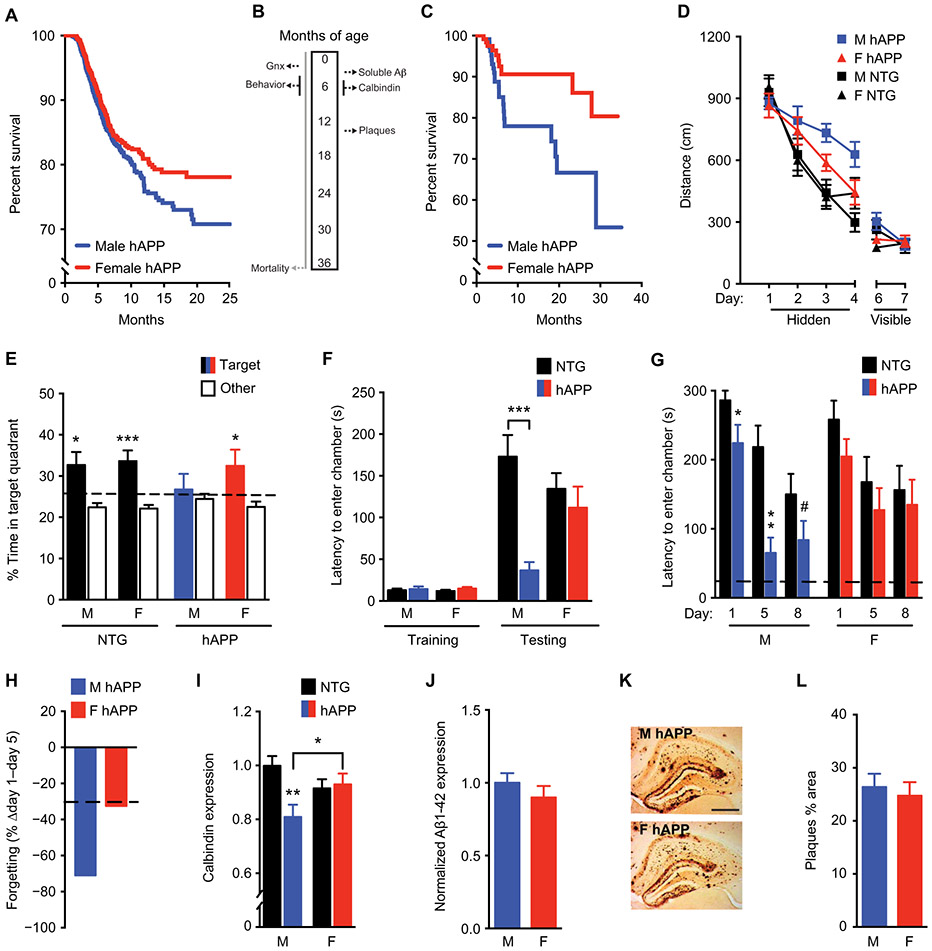
Fig. 2. Male sex increases mortality, cognitive deficits, and synaptic protein abnormalities in hAPP mice.
(A) Shown are Kaplan-Meier survival curves of male hAPP mice (n = 1572, blue) compared with female hAPP mice (n = 1589, red); all mice had intact gonads (log-rank test, P < 0.001). (B) All mice except those in (A) underwent gonadectomy (Gnx) at about 2.5 months of age; this was followed by behavioral testing conducted from 4 to 7 months of age and survival analysis conducted until 3 years of age. (C) Shown are Kaplan-Meier survival curves of male (n = 116) compared to female (n = 123) hAPP mice after gonadectomy (log-rank test, P < 0.05). (D) Shown are spatial learning curves of mice (age 4 to 7 months; n = 10 to 15 per group) tested in the Morris water maze during hidden platform training and when the platform was visible. Data points are daily average of total distance traveled to reach the platform over four trials. Mixed-model ANOVA for hidden training: female hAPP versus male hAPP mice, P < 0.05. (E) A probe trial was conducted after hidden platform learning and removal of the escape platform. Percentage of time mice spent in the target quadrant of the maze, indicating memory for platform location, versus the average time spent in the other three quadrants is shown; *P < 0.05; ***P < 0.001. The dashed line represents chance performance (25%). (F) Shown is passive avoidance, fear memory of mice (age 3 to 3.5 months; n = 7 to 10 per group) reflected by latency to enter the dark chamber during training and testing 1 day after an electric shock to the foot. Two-way ANOVA: hAPP effect, P < 0.01; hAPP by sex interaction, P < 0.05. (G) Forgetting of passive avoidance memory in a separate cohort of mice (age 5 to 6 months; n = 10 to 12 per group), reflected by latency to enter a dark chamber 1, 5, and 8 days after a foot shock, was measured. The dashed line represents latency to enter the dark chamber during training, which did not differ among groups. (H) Percentage loss of fear memory from days 1 to 5 is shown. The dashed line represents the average for nontransgenic (NTG) animals. (I) Shown is quantitation of calbindin immunoreactivity in mouse dentate gyrus (age 5 to 7 months; n = 11 to 14 mice per group). Two-way ANOVA: hAPP effect, P < 0.05; hAPP by sex interaction, P < 0.05. Means are relative to NTG male control mice, arbitrarily defined as 1. (J) Soluble Aβ1-42 amounts in the mouse hippocampus determined by enzyme-linked immunosorbent assay (ELISA) are shown (age 3 months; n = 8 to 11 mice per group). (K) Representative immunostaining of hippocampal Aβ deposits in coronal brain sections from a male (top, M) and female (bottom, F) hAPP mouse (age 14.5 to 15 months). Scale bar, 200 μm; magnification, ×4. (L) Quantitation of percentage area covered by Aβ deposits in hAPP mice (age 14.5 to 15 months; n = 11 per group). Behavioral studies in male and female NTG and hAPP mice were performed across seven independent cohorts including in fig. S4. #P = 0.06; *P < 0.05; **P < 0.01; ***P < 0.001 [Bonferroni-Holm for (F), (G), and (I)]. Data are presented as means ± SEM.
Men and women undergo depletion of circulating gonadal hormones with aging (22-24), but mice do not (fig. S2) (25, 26). Because AD is a disease of aging, we simulated human reproductive aging in male and female nontransgenic and hAPP mice by gonadectomy to deplete circulating hormones (Fig. 2B) and assessed survival in gonadectomized male and female hAPP mice. Male hAPP mice still died significantly faster than did female mice (P < 0.05; Fig. 2C). We explored whether hAPP mice showed a sex difference in cognitive functions independent of gonadal hormones. To reduce confounders, equalize hormones between sexes, and model reproductive aging of humans, we gonadectomized all nontransgenic and hAPP mice.
Male sex increases cognitive and molecular deficits in hAPP mice
We tested spatial learning and memory of gonadectomized mice in the Morris water maze and found that hAPP mice were impaired (P < 0.05; Fig. 2D). However, male hAPP mice traveled significantly longer distances to find the hidden platform than did females, indicating poorer learning capacity (P < 0.05; Fig. 2D). In a probe trial, male hAPP mice lacked memory retention, in contrast to all other groups (Fig. 2E). All mice located the target platform equally well when visible (Fig. 2D), and male and female mice within each group swam at equal speeds, although hAPP mice overall swam marginally slower (P < 0.001; fig. S3A).
In passive avoidance testing, which measures hippocampus- and amygdala-dependent fear memory, male hAPP mice, but not females, quickly reentered the dark chamber where they received a shock during training (P < 0.05; Fig. 2F). Male hAPP mice, but not females, lost the fear memory (P < 0.05; Fig. 2, G and H). Male vulnerability to deficits was significant with gonadectomy at young, middle, or old life stage (P < 0.05 to P < 0.001; fig. S4), across a range of cognitive and behavioral tasks (P < 0.05 to P < 0.001; fig. S4), and in an independent transgenic line of hAPP mice, hAPP-J9, which showed milder deficits (P < 0.05; fig. S5) (21, 27, 28).
Male hAPP mice showed significantly decreased expression of the neuronal activity–related protein calbindin (P < 0.05; Fig. 2I) in the hippocampus. Male and female hAPP mice did not differ in soluble β-amyloid (Aβ) (Fig. 2J) or protein expression of hAPP, total tau, and phospho-tau in the hippocampus (figs. S6, A to D, and S7) when cognitive and behavioral deficits had emerged (3 to 4 months). They also did not differ in amyloid plaque deposition (Fig. 2, K and L, and fig. S8) during middle age (14.5 to 15 months); however, females tended to show more plaques at a very old age (24 to 27 months) (fig. S9) as previously observed (29), despite decreased behavioral deficits compared to males.
We examined hAPP mRNA expression in the presence and absence of gonads and found that hAPP mRNA expression was equivalent across the experimental groups (fig. S10). Therefore, any unintentional gonadal hormone influences at the promoter of hAPP-J20 mice were not observed, a critical measure when directly comparing sexes in transgenic disease models.
Sex chromosomes mediate increased male vulnerability in hAPP mice
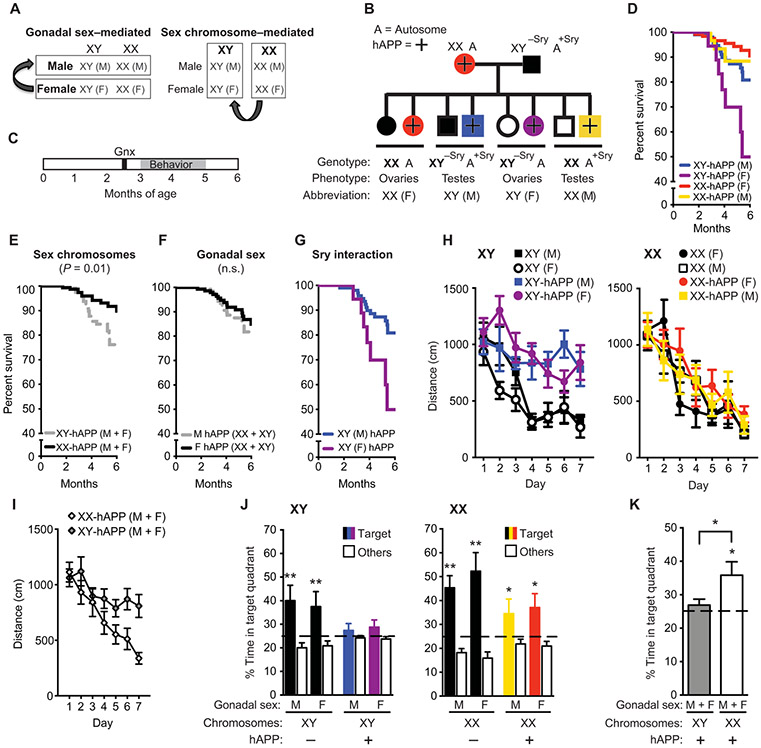
To dissect the etiology of male disadvantage related to AD after gonadectomy, we examined Four Core Genotype (FCG) (30, 31) mice. In normal mice and humans, the Sry gene on the Y chromosome encodes a protein that initiates development of testes followed by perinatal masculinization of the body and brain (32). In the FCG mouse model, Sry is transposed onto an autosome from the Y chromosome. This genetic manipulation enables generation of XX and XY mice, each with either female ovarian (F, −Sry) or male testicular (M, +Sry) development: XX(F) ovaries, XX(M) testes, XY(F) ovaries, and XY(M) testes. A sex difference that varies by gonads is gonadal sex–mediated; one that varies by chromosome complement is sex chromosome–mediated (Fig. 3A).
Fig. 3. Sex chromosomes mediate increased male vulnerability to mortality and cognitive impairments in hAPP mice.
(A) Strategy to identify the cause of sexual dimorphism using the FCG mouse model. (B) Diagram of the cross between hAPP and FCG transgenic mice is presented. FCG mice harbor a transposition of the Sry gene from the Y chromosome onto an autosome (A, autosome). Progeny include XX and XY mice, each with either ovarian (F) or testicular (M) development and with or without hAPP expression (hAPP, +). (C) Experimental strategy: All mice underwent gonadectomy at about 2.5 months of age, followed by behavioral testing and survival studies at 3 to 6 months of age. (D to G) In the Kaplan-Meier survival curves, (D) all groups of hAPP mice showed (E) a main effect of sex chromosomes on mortality (XY, HR 2.49, CI 1.21 to 5.14, P < 0.01) and (F) no main effect of gonadal sex on mortality (P = 0.45). (G) An interaction between sex chromosomes and gonadal sex indicated lower mortality in XY (male, M) compared to XY (female, F) mice (XY-M, HR 0.18, CI 0.03 to 0.92, P < 0.05). Analyses were by Cox proportional hazards for all groups: (XY-M: n = 101; XX-F: n = 122; XY-F: n = 18; XX-M: n = 31). (H and I) Spatial learning curves from the eight genotypes of mice tested altogether in the Morris water maze (age 3 to 5 months; n = 5 to 6 per group) show that (H) XY-hAPP mice (M or F) traveled longer distances to find the target platform, enabling escape from the water maze, than did XX-hAPP mice (M or F). This is highlighted in (I), where all XY-hAPP (M + F) mice were compared with all XX-hAPP (M + F) mice. XX or XY mice without hAPP (M or F) learned similarly well. Data points are daily averages of total distance traveled to reach the platform over four trials. Mixed-model ANOVA: XX-hAPP versus XY-hAPP, P < 0.01. (J and K) A probe trial, during which the escape platform in the target quadrant was removed, tested for memory of the platform location in the eight genotypes of mice. Percentage of time spent in the target quadrant, indicating memory of the platform location, versus the average time spent in the other three quadrants showed that (J) XY-hAPP (M or F) mice did not favor the target quadrant, whereas XX-hAPP (M or F) mice did. The greater impairment of learning and memory in XY-hAPP mice is highlighted in (K) where all XY-hAPP (M + F) mice are compared with all XX-hAPP (M + F) mice. The dashed line represents chance performance. These findings were replicated in an independent cohort (fig. S11). *P < 0.05; **P < 0.01 versus chance performance of 25% (one-sample t tests) or as indicated by bracket (t test). Data are presented as means ± SEM. n.s., not significant.
We crossed FCG mice with hAPP mice to produce eight genotypes that included the four sex genotypes with or without hAPP (Fig. 3B). After sexual differentiation and reproductive maturity, we gonadectomized mice and assessed survival, cognition, and biochemical markers (Fig. 3C). XY-hAPP mice sexually differentiated as either male (M, testicular phenotype, +Sry) or female (F, ovarian phenotype, −Sry) died faster than did XX-hAPP mice of either gonadal phenotype (Fig. 3, D to F). In addition to the main effect of sex chromosomes, sex chromosomes interacted with gonadal phenotype in XY-hAPP mice. That is, XY-hAPP males (+Sry) survived longer than XY-hAPP females (−Sry) (P < 0.05; Fig. 3G), an effect not observed in XX-hAPP mice.
To determine whether sex chromosomes mediate male vulnerability to Aβ-related cognitive deficits, we tested mice in the Morris water maze. In finding the hidden platform, male or female XY-hAPP mice showed significantly worse learning than did male or female XX-hAPP mice (P < 0.01; Fig. 3, H and I, and fig. S11A). In contrast, all nontransgenic mice without hAPP learned similarly (Fig. 3, H and I, and fig. S11A). In a probe trial, XY-hAPP mice lacked memory retention (Fig. 3, J and K, and fig. S11B), whereas all XX (nontransgenic and hAPP) mice remembered, regardless of being male or female (P < 0.05; Fig. 3J and fig. S11B). All mice swam at equal speeds and located a visible target platform equally (fig. S3, B and C). In passive avoidance testing, male or female XY-hAPP mice showed significantly worse fear memory than did male or female XX-hAPP and nontransgenic mice (P < 0.001 and P < 0.01, respectively; fig. S12). As in non-FCG hAPP mice, male or female XX and XY mice did not differ in the amount of soluble Aβ in the hippocampus (fig. S6E) at the age of cognitive and behavioral testing.
A second X chromosome confers resilience to AD-related vulnerability in XY (male) and XO (female) hAPP mice
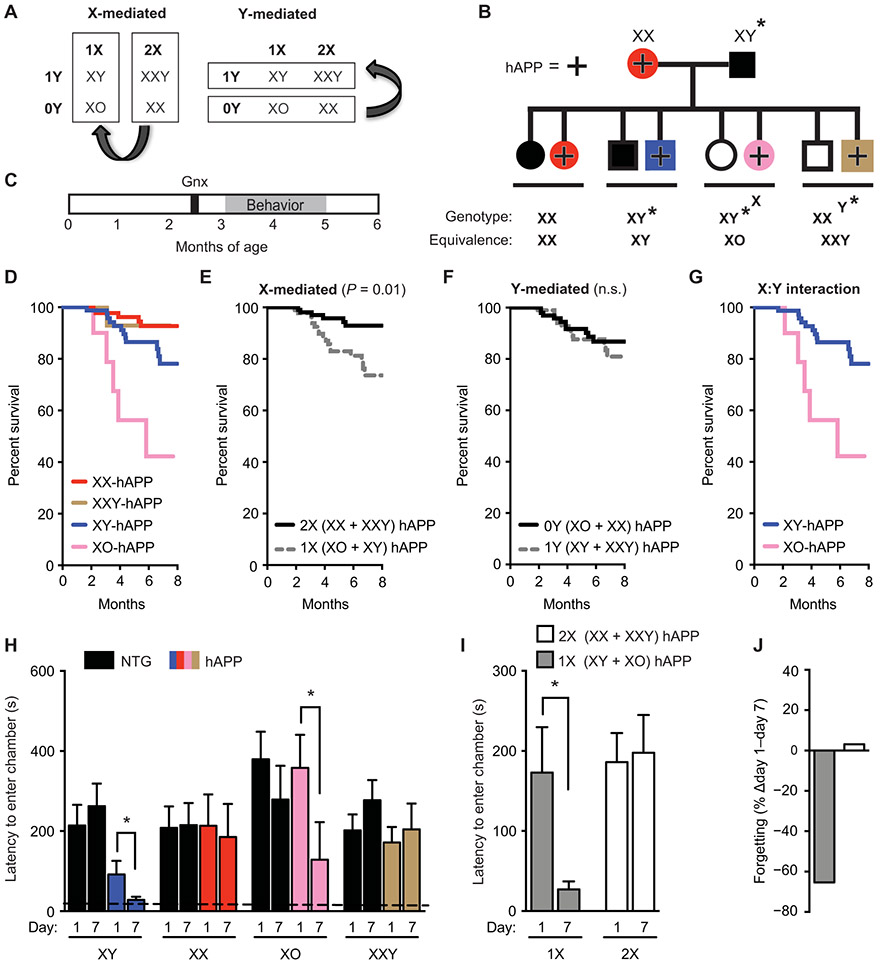
To further dissect causes of the sex chromosomal effects, we determined whether the presence of a Y or the lack of a second X chromosome conferred male disadvantage in hAPP mice. We investigated the XY* model (33, 34) of sex chromosomal biology in mice with and without hAPP. The Y* chromosome in XY* males contains an altered pseudoautosomal region that recombines abnormally with the X chromosome during meiosis. Progeny of XY* males crossed with XX females include four sex genotypes roughly equivalent to the following: XX and XO mice with ovaries and XY and XXY mice with testes. A sexual dimorphism that varies by the presence or absence of a Y is Y chromosome–mediated; one that varies by the presence of one versus two X’s is X chromosome–mediated (Fig. 4A).
Fig. 4. A second X chromosome confers resilience against AD-related cognitive impairments in XY (male) and XO (female) hAPP mice.
(A) Strategy to identify whether the sex chromosome effect depends on the X or Y chromosome. (B) Diagram of mouse cross used in this experiment. hAPP females (XX, hAPP) were crossed with XY* males that harbored an altered pseudoautosomal region on the Y chromosome, allowing abnormal crossover with the X chromosome during meiosis (33, 34). The cross resulted in offspring of eight genotypes, each of the sex chromosome genotypes, with or without hAPP. The equivalent number of X and Y chromosomes for each genotype is shown. (C) Experimental strategy: All mice underwent gonadectomy at 2.5 months of age followed by behavioral testing and survival studies between 3 and 6 months of age. (D to G) In the Kaplan-Meier survival curves in (D), all hAPP mice show (E) a main effect of X chromosome dose on mortality (2X, HR 0.2, P < 0.01, CI 0.12 to 0.75) and (F) no main effect of a Y chromosome on mortality (P = 0.53). (G) An interaction between X and Y chromosomes showed lower mortality in the presence of Y (or male gonadal type) when X dose = 1 (XY versus XO, HR 0.23, P < 0.01, CI 0.08 to 0.64). Analyses were by Cox proportional hazards for all groups (XY: n = 79, XX: n = 88; XO: n = 10; XXY: n = 15 mice). (H to J) Shown is testing of mice in the passive avoidance task, measured by latency to enter the dark chamber 1 and 7 days after a foot shock (age 3 to 5 months; n = 4 to 16 per group). (H) Abnormal loss of fear memory in hAPP mice of XY and XO genotypes is shown. Two-way repeated measures ANOVA: X dose effect, P < 0.05. The dashed line represents latency to enter the dark chamber during training, which did not differ among the groups. (I) Greater loss of fear memory in hAPP mice with 1X compared to 2X chromosomes is presented. (J) Percent loss of fear memory in hAPP mice with 1X compared to 2X chromosomes is shown. *P < 0.05 as indicated by bracket (Bonferroni-Holm). Data are presented as means ± SEM.
We crossed XY* males with hAPP females to produce eight genotypes of mice exhibiting varying dosages of X and Y chromosomes, with or without hAPP (Fig. 4B). We gonadectomized mice and then assessed survival, cognition, and biochemistry (Fig. 4C). Mice with one X chromosome (XY-hAPP and XO-hAPP) died significantly faster than did those with two X chromosomes (XX-hAPP and XXY-hAPP) (P < 0.01; Fig. 4, D to F). Therefore, the addition of an X chromosome to XY-hAPP mice prevented male vulnerability, extending survival to that observed in XX-hAPP females. In addition to the main effect of X dose (P < 0.01; Fig. 4E), but not of Y (Fig. 4F), the Y interacted with the X; that is, XY-hAPP mice survived longer than XO-hAPP mice (P < 0.01; Fig. 4G).
We then tested whether the addition of an X to XY-hAPP mice reduced male vulnerability to cognitive deficits in the passive avoidance task (Fig. 4, H to J). Both male and female hAPP mice with one X chromosome (XY-hAPP and XO-hAPP) showed significant forgetting of fear memory (P < 0.05; Fig. 4, H to J), whereas those with two X chromosomes (XX-hAPP and XXY-hAPP) did not forget (Fig. 4, H to J). In contrast, all mice without hAPP had comparable and robust fear memory. As in FCG-hAPP mice, XY*-hAPP mice with 1X or 2X chromosomes did not differ in the amount of soluble Aβ in the hippocampus (fig. S6F). Thus, although hAPP mice with 1X or 2X chromosomes had comparable amounts of Aβ, hAPP mice with 2X chromosomes were less impaired.
A second X chromosome elevates Kdm6a expression independent of gonadal phenotype or the Y chromosome
We sought to understand how a second X chromosome could confer resilience, because XY and XX mice express only one active X due to X-chromosome inactivation in females. Whereas X-chromosome inactivation silences one X chromosome in mammalian XX cells, a small subset of X-linked genes escape X-chromosome inactivation and show transcription from both alleles, leading to higher expression in females (35-38). Of those, we focused on the gene lysine-specific demethylase 6a (Kdm6a; also known as Utx) encoding an H3K27 demethylase that consistently escapes X-chromosome inactivation in both mice and humans (39, 40). Loss-of-function mutations in KDM6A cause cognitive deficits in humans (41-46), and Kdm6a plays a post-developmental role in mouse synaptic plasticity and cognition (47).
We therefore examined Kdm6a expression in mouse brains. We first confirmed that Kdm6a escaped X-chromosome inactivation in the XX mouse brain through RNA fluorescence in situ hybridization (RNA FISH) (48) in mouse primary cortical neurons. Isolated XX neuronal nuclei with Xist RNA coating the inactive X chromosome, indicating X-chromosome inactivation, showed Kdm6a labeling at two sites, marking its transcription from both the active and inactive X chromosomes (Fig. 5A). In contrast, XY neurons showed only one site for transcription (Fig. 5A). Immunolabeling of Kdm6a protein in the adult hippocampus of XX and XY mice with a well-characterized antibody (49) showed a largely neuronal cytoplasmic staining pattern that was diffuse in both XX and XY mouse brains (Fig. 5B).
Fig. 5. A second X chromosome elevates Kdm6a expression independent of gonads or the Y chromosome in mice.
(A) Representative fluorescence in situ hybridization images for Kdm6a and Xist (RNA FISH) expression in XX (top) and XY (bottom) primary mouse neuronal nuclei. Kdm6a is shown in red, Xist is shown in green, and 4’,6-diamidino-2-phenylindole (DAPI) nuclear stain is shown in blue. Nascent Kdm6a transcripts appear as red fluorescent puncta at the site of transcription (indicated by white arrows). Xist RNA remains associated with the inactive X chromosome and is detected only in XX cells. Inset numbers indicate the percentage of nuclei with two sites of nascent Kdm6a accumulation in XX cells and one site in XY cells (n = 100 cells). Scale bar, 2 μm. (B) Representative confocal images of Kdm6a staining (left), Kdm6a with DAPI staining (middle), and Kdm6a with Neuronal nuclei (NeuN) staining (right) in the hippocampal dentate gyrus region of a gonadectomized nontransgenic (NTG) female XX mouse (top row) and a gonadectomized NTG male XY mouse (bottom row). Kdm6a is shown in red, DAPI nuclear stain is shown in blue, and NeuN is shown in green. Scale bar, 50 μm; magnification, ×100. (C and D) Western blot representative image (C) and subsequent quantification (D) of Kdm6a protein expression in the hippocampus of gonadectomized NTG XX female and XY male mice. Bands represent individual mouse samples. (C) Representative images show samples bound by the GeneTex antibody, and (D) quantification is given for both GeneTex and Abcam rabbit anti-Kdm6a antibodies; Kdm6a was normalized using glyceraldehyde phosphate dehydrogenase (GAPDH) as a loading control. Means are relative to NTG XY male control mice, arbitrarily defined as 1 (age 3.4 to 3.6 months; n = 3 mice per group). Gonadectomized NTG XX female mice show higher Kdm6a protein expression. Two-tailed t test, *P < 0.05. (E and F) Hippocampal Kdm6a mRNA expression in (E) FCG mice (age 3.5 to 5.5 months; n = 6 to 26 mice per group) and (F) XY* mice (age 5.5 to 7.5 months; n = 4 to 17 mice per group) with and without hAPP, shown relative to XY male mice without hAPP. Two-way ANOVA: sex chromosome effect, ***P < 0.001 and X dose effect, ***P < 0.001. Data are presented as means ± SEM in (D) to (F). *P < 0.05; ***P < 0.001 (Bonferroni-Holm).
We assessed whether two X chromosomes increased expression of Kdm6a protein and mRNA in mouse hippocampus. Kdm6a protein expression was significantly higher in XX mice than in XY mice as measured by two antibodies (P < 0.05; Fig. 5, C and D). To determine whether the second X chromosome primarily governed higher expression, we assessed Kdm6a mRNA in FCG and XY* mice. As anticipated (50, 51), hippocampal Kdm6a was significantly elevated in XX mice with testes and ovaries (P < 0.001; Fig. 5E). The presence of neither hAPP nor the Y chromosome altered this primary X-chromosome effect (Fig. 5F).
KDM6A expression is elevated in the brains of women, and KDM6A genetic variation in humans associates with cognitive resilience
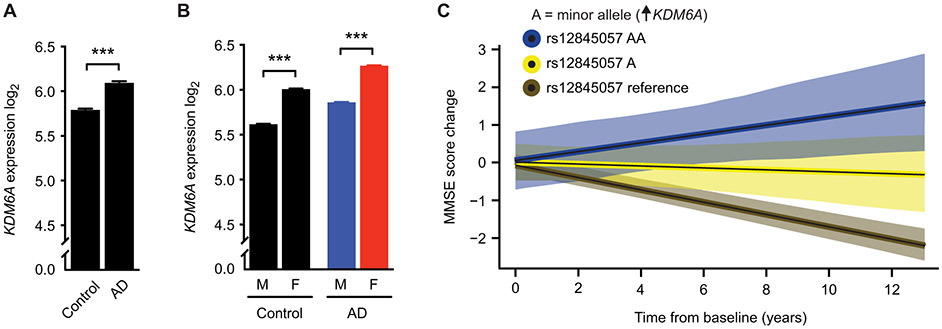
We explored whether KDM6A mRNA expression was altered by sex in the brains of individuals with and without AD. We queried gene expression from a public dataset (GSE 15222; tables S1 and S2) accounting for age, postmortem interval, and sex. KDM6A expression was significantly higher in pathologically confirmed AD cases relative to controls in the temporal cortex, an area affected in early AD (P = 3.64 × 10−4; Fig. 6A). This increase was independently confirmed in two other public datasets of human postmortem gene expression in the temporal cortex, parahippocampal gyrus, and superior temporal gyrus (tables S1 and S3). In contrast, regions typically affected later or spared in AD such as the cerebellum showed no changes (tables S1 to S3). We then assessed KDM6A expression in brains of individuals identified as male or female in the GSE 15222 dataset. KDM6A expression was higher in females with (P = 4.83 × 10−4; Fig. 6B) and without AD (P = 9.79 × 10−4; Fig. 6B).
Fig. 6. KDM6A genetic variation associates with cognitive resilience in humans.
(A) Shown is human KDM6A RNA expression via RNA sequencing and microarray in the temporal and parahippocampal cortex of individuals without (control, n = 135) and with AD (n = 86) (***P = 3.64 × 10−4). (B) Shown is human KDM6A RNA expression via RNA sequencing and microarray in individuals identified as male (M) or female (F) without (M, n = 75; F, n = 60; ***P = 9.79 × 10−4) and with AD (M, n = 37; F, n = 49; ***P = 4.83 × 10−4). Expression data were analyzed by linear models accounting for effects of postmortem interval and age at death. (C) Shown is cognitive change with 95% CIs in 778 individuals of the ADNI cohort (cognitively normal, 268; MCI, 465; AD, 45), who carried two alleles (AA, blue, n = 8 all female), one allele (A, yellow, n = 78), or no allele (noncarriers, reference, brown, n = 692) for the rs12845057 variant of the KDM6A gene associated with increased KDM6A RNA expression in brain (table S4). Cognition was measured by the MMSE score. Increasing dose of the minor allele was associated with slower rates of cognitive decline over time (β = 0.141, SE 0.035, P = 0.00005). Cognitive data were analyzed by linear models accounting for effects of baseline age, sex, education, and APOEε4 dose. Data are presented as means ± SEM in (A) and (B). ***P < 0.001 (Bonferroni-Holm).
We then queried whether KDM6A expression, by proxy of a genetic variation, was associated with cognitive change over time. Using the Genotype-Tissue Expression project (GTEx) online portal of gene expression across tissues of nearly 1000 individuals (52), we searched for common variants associated with altered expression of KDM6A. The minor allele of one genetic variant, rs12845057, was associated with increased expression of KDM6A in the brain (P = 7.0 × 10−6). Frequency of the minor allele (A) is about 14% globally and 7% in Europeans (53). To test associations between the KDM6A variant and cognitive change, we queried the Alzheimer’s Disease Neuroimaging Initiative (ADNI) dataset derived from a multisite study of individuals with both whole-genome sequencing and serial neuropsychological examinations (n = 778) that is enriched for individuals with MCI, a transition phase to AD. The minor allele was distributed equally among categories of cognitively normal (n = 268), MCI (n = 465), and AD (n = 45) individuals, indicating that it did not associate with disease risk (cohort demographics; table S4). Next, we used linear mixed-effects regression models to test for an association between the minor allele A of the KDM6A variant and cognitive change, accounting for baseline age, sex, education, and APOEε4 dose. Increasing dose of the minor allele of the KDM6A variant was significantly associated with less cognitive decline over time using the Mini-Mental State Examination (MMSE) (β = 0.141, SE 0.035, P = 0.00005; Fig. 6C). This finding was consistent in another cognitive measure using the Alzheimer’s Disease Assessment Scale (ADAS-cog) in overall function using the clinical dementia rating sum of boxes score (CDR), when assessing women only in all measures (fig. S13), and when assessing cognition in cognitively normal and in MCI individuals as subgroups (table S5).
Kdm6a knockdown in XX mouse neurons worsens, whereas Kdm6a overexpression in XY neurons attenuates Aβ toxicity in vitro
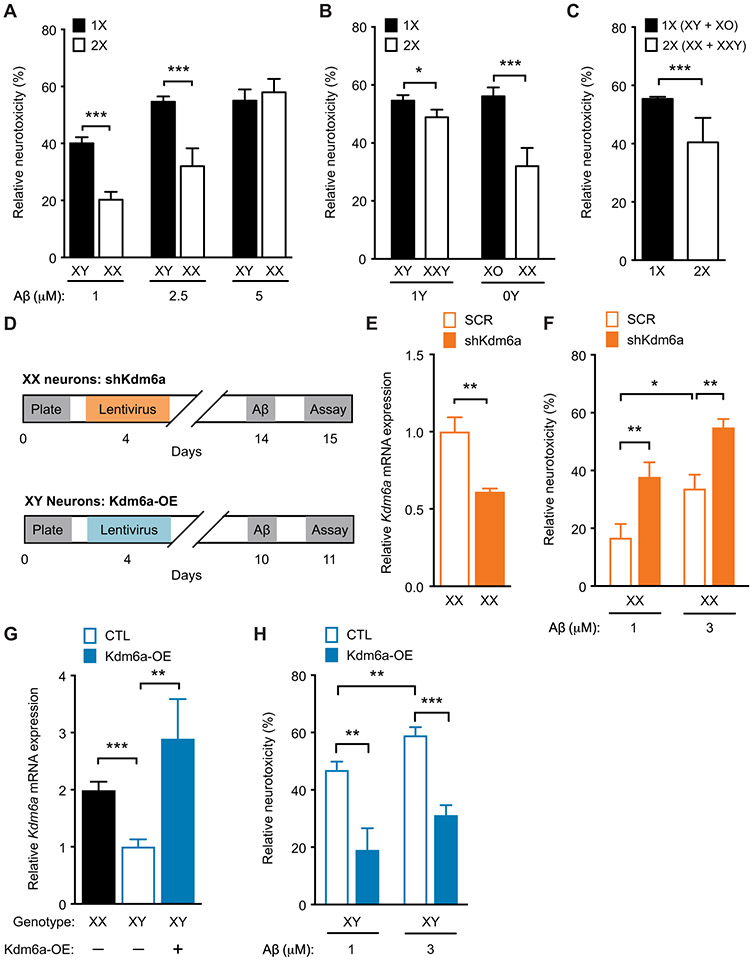
We next turned to experiments with primary wild-type mouse neurons exposed to recombinant Aβ1-42. The Aβ preparation was enriched for oligomers during the experimental time frame, based on our previous characterization (54). XY mouse neurons were more vulnerable to Aβ-induced toxicity, in a dose-dependent manner, compared to XX neurons, using both the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (P < 0.001; Fig. 7A) and the lactate dehydrogenase (LDH) assay (P < 0.01; fig. S14). In parallel with in vivo findings, neurons derived from XY* mice with one X chromosome (XY and XO) were significantly more vulnerable to Aβ toxicity than those with two X chromosomes (XX and XXY) (P < 0.001; Fig. 7, B and C). The protective effect of two X chromosomes was decreased by the Y chromosome (P < 0.05; Fig. 7B), indicating an X:Y interaction.
Fig. 7. Kdm6a knockdown in XX mouse neurons worsens, whereas Kdm6a overexpression in XY neurons attenuates Aβ toxicity in vitro.
(A to C) Vulnerability of mouse primary neurons was tested by the MTT assay. For each genotype, cell toxicity was calculated as a percentage of the corresponding vehicle-treated group, 24 hours after treatment with increasing doses of Aβ. (A) Mouse primary cortical XY neurons showed greater vulnerability than did XX neurons after exposure to vehicle or increasing doses of Aβ (n = 8 to 40 wells per experimental group from 8 to 10 pups per genotype, from four independent litters). Two-way ANOVA: sex chromosome effect, P < 0.01; Aβ dose effect, P < 0.001; interaction, P < 0.05. (B) Toxicity of Aβ in neurons of varying X and Y chromosome dosage derived from littermate pups of XY* males crossed with nontransgenic (NTG) females, with genotypes roughly equivalent to XO, XX, XY, and XX, exposed to vehicle or Aβ (2.5 μM) (n = 15 to 45 wells per experimental group from 7 to 10 pups per genotype, from four independent litters). Two-way ANOVA: X effect, P < 0.0001; Y effect, not significant; X by Y interaction, P < 0.05. (C) Main effect of X chromosome dose shows increased Aβ toxicity in neurons with 1X (XO and XY combined) compared to those with 2X chromosomes (XX and XXY combined). (D) Experimental strategy of lentivirus-mediated knockdown of Kdm6a in XX mouse primary cortical neurons (top) and Kdm6a overexpression in XY mouse primary cortical neurons (bottom). (E) Shown is Kdm6a mRNA expression in neurons transfected with lentivirus expressing scrambled (SCR) or short hairpin (sh) Kdm6a for knockdown expressed relative to XX SCR (n = 5 to 6 wells per experimental group from eight XX pups, from two litters). Two-tailed t test, **P < 0.01. (F) Shown is Aβ toxicity in XX neurons treated with SCR or shKdm6a and exposed to vehicle or Aβ (1 and 3 μM); knockdown of Kdm6a worsened Aβ toxicity (n = 24 to 25 wells per experimental group from 14 XX pups, from three independent litters). Two-way ANOVA: Kdm6a effect, P < 0.001; Aβ effect, P < 0.001; Kdm6a by Aβ interaction, P = 0.99. (G) Kdm6a mRNA expression in neurons transfected with lentivirus expressing control (CTL) or overexpressing Kdm6a (Kdm6a-OE), shown relative to control XY neurons (n = 3 to 8 wells per experimental group from 12 XY pups, from two independent litters). One-way ANOVA, P < 0.001. (H) Shown is Aβ toxicity in XY neurons transfected with lentivirus expressing control or overexpressing Kdm6a (Kdm6a OE) and exposed to vehicle or Aβ (1 and 3 μM); overexpression of Kdm6a attenuated Aβ toxicity (n = 12 to 13 wells per experimental group from 26 XY pups, from three independent litters). Two-way ANOVA: Kdm6a effect, P < 0.001; Aβ effect, P = 0.01; Kdm6a by Aβ interaction, P = 0.99. *P < 0.05; **P < 0.01; ***P < 0.001 (Bonferroni-Holm). Data are presented as means ± SEM.
Given that Kdm6a escapes X-chromosome inactivation in XX mouse neurons and is increased in XX compared to XY mouse brains, we tested directly whether Kdm6a modulates neuronal susceptibility to Aβ toxicity in vitro. In XX mouse neurons, we decreased Kdm6a expression (P < 0.01; Fig. 7, D and E) to that found in XY neurons via lentivirus-mediated knockdown. Knockdown of Kdm6a in XX mouse neurons significantly worsened dose-dependent Aβ toxicity (P < 0.01; Fig. 7F) to a range observed in XY neurons. In XY mouse neurons, we increased Kdm6a expression to that found in XX neurons or higher (P < 0.01; Fig. 7, D and G) via lentivirus-mediated overexpression. Overexpression of Kdm6a in XY mouse neurons significantly attenuated dose-dependent Aβ toxicity (P < 0.001; Fig. 7H) to a range observed in XX neurons.
Kdm6a attenuates male vulnerability to cognitive impairments in XY-hAPP mice
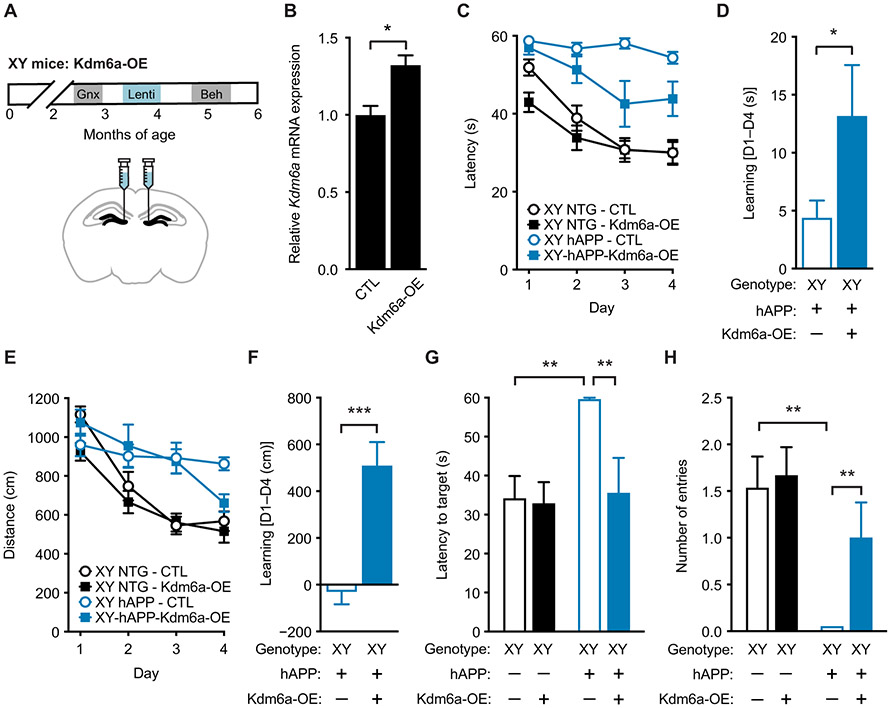
We next determined whether increasing expression of Kdm6a attenuated male vulnerability to cognitive deficits in XY-hAPP mice. We gonadectomized XY nontransgenic and hAPP mice, injected lentivirus with (Kdm6A-OE) or without (control) the Kdm6a transgene bilaterally into the dentate gyrus, a region that affects spatial learning and memory, and analyzed mice behaviorally 1 month later (Fig. 8A). Lentiviral-mediated overexpression of Kdm6a in XY males increased Kdm6a mRNA expression in the dentate gyrus (P < 0.05; Fig. 8B) to that expected in XX females. In finding the hidden platform of the Morris water maze, XY-hAPP-Kdm6a-OE mice showed significantly better performance than XY-hAPP control mice measured by latency (P < 0.001; Fig. 8C) and learning (P < 0.05; Fig. 8D), quantified by comparing the last day of training to the first. Similarly, XY-hAPP-Kdm6a-OE mice showed significantly better learning than did XY-hAPP control mice measured by distance (P < 0.001; Fig. 8, E and F), although distance curves did not statistically differ. In a probe trial, XY-hAPP-Kdm6A-OE mice showed robust spatial memory retention, compared to XY-hAPP control mice (P < 0.01; Fig. 8, G and H) performing similarly to unimpaired nontransgenic mice. With the visible platform, hAPP mice swam marginally faster with longer distance than did nontransgenic mice; however, overexpression of Kdm6a did not alter either measure in either genotype (fig. S3, D and E). Further, increasing Kdm6a expression in XY mice did not alter hAPP-induced hyperactivity in the open field task and increased time spent in open arms in the elevated plus maze in hAPP mice (fig. S15).
Fig. 8. Kdm6a overexpression in hippocampus attenuates male vulnerability to cognitive impairments in XY-hAPP mice.
(A) Experimental strategy: XY mice were gonadectomized and injected with lentivirus expressing control or overexpressing Kdm6a (Kdm6a OE) into the dentate gyrus of the hippocampus; animals were then tested on behavioral tasks. (B) Shown is Kdm6a mRNA expression measured in dentate gyrus of mice injected with lentivirus expressing control or overexpressing Kdm6a (Kdm6a OE) (n = 3 mice per experimental group), relative to XY control; t test, *P < 0.05. (C to F) Spatial learning task results for the four experimental groups of XY mice tested in the Morris water maze (age 5 to 5.5 months; n = 7 to 15 per group). XY-hAPP-Kdm6a-OE mice exhibited (C) decreased latency to find the target escape platform (mixed-model ANOVA: XY-hAPP-CTL versus XY-hAPP-Kdm6a-OE, P < 0.001) and (D) a better learning index of latency during hidden platform training, measured by the difference in performance of each mouse at day 4 from average group performance on day 1 (D1 to D4). (E) XY-hAPP-Kdm6a-OE mice did not travel a statistically decreased distance to find the target platform but (F) showed better learning in the distance traveled during hidden platform training. (G and H) Probe trial results 24 hours after completion of hidden platform learning, indicating spatial memory of the escape platform location, showed that XY-hAPP-Kdm6a-OE mice had attenuated spatial deficits including decreased (G) latency to target platform and (H) increased number of entries into the target zone, compared to XY-hAPP-CTL mice. *P < 0.05; **P < 0.01; ***P < 0.001 [Bonferroni-Holm for (G) and (H)]. Data are presented as means ± SEM.
DISCUSSION
Our data suggest a role for sex chromosomes in mice in countering deficits and toxicity related to AD in both sexes. A second X chromosome decreased mortality and brain dysfunction in gonadectomized male and female hAPP mice, without altering soluble Aβ or co-pathogenic proteins. A second X chromosome conferred resilience, in part, through the candidate gene Kdm6a, a histone demethylase gene that escapes X-chromosome inactivation, causing higher expression in cells with two X’s compared to one X. Genetic variation of KDM6A linked to its increased brain expression was associated with slower cognitive decline in an aging population of individuals, including those with MCI.
Dissection of sex differences and their mechanistic underpinnings with powerful genetic tools provides opportunities to understand disease and unravel new sex-based pathways (55). Male sex is a major, underappreciated risk factor for rapid progression to death in AD (7-11), as confirmed by our meta-analysis (Fig. 1), and in other neurodegenerative conditions (56-59). These findings do not contradict the fact that more women have AD due to their longevity (6) and their increased risk or incidence after age 85 (4, 5, 12, 60), which together contribute to a higher lifetime risk of AD in women compared to men (61). When men get AD, they die faster (7-11). The male brain may be biologically older and more vulnerable, an idea supported by epigenetic (62) and metabolic studies (63) of humans.
In aging and preclinical AD, male sex may increase the likelihood of abnormalities favoring transition to clinical dementia. Men show worse memory function (12) and cognitive decline than do women (13-15), implying less compensation for similar subclinical brain pathology measured by positron emission tomography imaging of amyloid (12, 15). In studies of AD biomarkers (64), men show increased neurodegeneration (16, 17), a precursor for dementia. These findings could underlie earlier onset and increased incidence or prevalence of MCI observed in men from many (19, 20, 65-67), although not all (68-70), populations.
Recent studies of aging and AD [reviewed in (4)] indicate similar amyloid amounts in the brain (12, 15, 17, 71, 72) and cerebrospinal fluid (CSF) (71) of men and women, similar overall tau burden (73), but increased CSF and regional tau in women with high amyloid (71, 73). Likewise, AD pathology is similar between the sexes, up until older ages (72), when both pathology and risk of AD increases in women. Each sex may respond differently to comparable amounts of pathogenic proteins, a possibility observed in mice (74), which may explain why with similar tau loads, men show less neuro-structural preservation (75) and more cognitive impairment (76).
Congruent with human observations, soluble Aβ and amyloid deposition were similar between the sexes in our mice until very old age and did not explain male vulnerability at the neuronal or cognitive level. Other AD mouse models show very high amounts of Aβ with increased mortality in female mice (77-80) and are thus incongruent with our mouse findings. Given that no single model of AD fully recapitulates human AD, a disease with a wide clinical spectrum, we conducted cellular viability, cognitive, behavioral, synaptic, and mortality studies that collectively showed worse outcomes in primary neurons and gonadectomized male mice, a sex bias that persisted in our hAPP mice regardless of age at hormone depletion, mouse strain, or genetic background. Our mouse studies focused on hAPP/Aβ-dependent abnormalities, representing a specific component of AD, a complex disease comprising multiple pathogenic proteins and risk factors.
We used gonadectomy to equate gonadal hormones between the sexes and simulate human reproductive aging, an approach distinctly different from previous studies of sex in AD-related models [reviewed in (81)]. Gonadectomy enables direct comparison of the sexes without confounding due to activational (short-acting) effects of ovarian and testicular hormones. This is of value, because ovarian hormones modulate Aβ, network dysfunction, and cognitive deficits in female hAPP mice (82, 83). Our experiments did not test the activational effects of hormonal treatments [reviewed in (82, 83)].
Sex chromosomes largely governed sex differences in vulnerability to mortality, cognitive dysfunction, molecular impairments, and cellular dysfunction in the FCG-hAPP mouse model. The XY genotype in hAPP mice that developed with ovaries or testes worsened measures, compared to the XX genotype that developed with ovaries or testes. Similarly, we recently found that sex chromosomes influenced mortality in mice during normal aging (84), suggesting action on fundamental pathways converging in aging and disease. The lack of a second X chromosome, rather than the presence of a Y chromosome, caused male disadvantage in animal and cellular models of AD in the XY* model. The presence of only one X chromosome (in XO females and XY males) consistently worsened hAPP/Aβ-related mortality, cognitive deficits, and cellular viability in both males and females, compared to two X chromosomes (in XX females and XXY males). The Y chromosome, the Y chromosome gene Sry, testes, or some combination of these decreased mortality in hAPP mice with one but not two X chromosomes. XY-hAPP males (+Sry) survived longer than did XY-hAPP females (−Sry) or XO-hAPP females (−Sry), indicating a potential protective role of the Sry protein or of testicular development itself in the XY, but not XX, genotype. Given that the X and Y chromosomes share homologous genes in pseudoautosomal regions, select Y genes may partially compensate for the lack of a second X chromosome.
Many factors influencing neural function reside on the X chromosome (85). Two X chromosomes could confer neural advantage through increased X dose arising from baseline escape of the inactive X chromosome. Whereas XY and XX organisms express one active X due to X-chromosome inactivation in females, select factors like the Kdm6a gene escape inactivation. Kdm6a is a histone demethylase that robustly and consistently escapes X-chromosome inactivation in female mice and humans (39, 40) and is enriched in the brain (51, 86, 87). The second X chromosome increased Kdm6a expression, independent of gonads or the Y chromosome, in our mice. This is important, because the Y paralog of Kdm6a, UTY (88), has high homology to Kdm6a (89) but a nearly inactive histone demethylation domain (90, 91). The presence of UTY in XY neurons and mice did not modify Kdm6a-mediated attenuation of AD-related toxicity in vitro or in vivo.
KDM6A expression in human brain was higher in females compared to males and in those with AD compared to controls. Because KDM6A loss-of-function mutations cause intellectual disability in humans (42-46) and Kdm6a elevation caused neural and cognitive resilience in our mouse studies, it is interesting to speculate that increased KDM6A in AD could be a protective, compensatory response.
A common genetic variant in an intergenic region near KDM6A, rs12845057, was associated with greater expression in human brain. The minor allele frequency varies across populations, and about 13% of females and 6.5% of males carry it globally (53). In the current study of the ADNI cohort, increasing the minor allele dose was associated with cognitive resilience in individuals undergoing longitudinal testing over a decade, a finding consistent across clinical measures and when we assessed females only. Our analysis in males, who carry half the frequency, was likely limited by statistical power. In our subgroup analyses by clinical diagnosis, individuals with MCI showed the most resilience associated with the KDM6A minor allele, suggesting that increased KDM6A could modify clinical trajectory during the transitional period from MCI to AD. Whereas the ADNI cohort includes longitudinal data and multisite investigations, its limitations include a study of predominantly non-Hispanic, Caucasian populations within the United States. How broadly our findings extend to other populations remains to be determined.
In the current study, modestly increasing Kdm6a expression in XY mouse primary neurons and hippocampus of XY-hAPP mice attenuated hAPP/Aβ neurotoxicity and cognitive impairment. These findings suggest that minor elevation in Kdm6a transcription was sufficient to functionally increase neural resilience and partially reverse deficits in the XY-hAPP mice. Whether this requires histone demethylase activity is currently unknown. Kdm6a may act differently across cell types and biological systems. Whereas Kdm6a deletion in hippocampus impairs synaptic plasticity and cognition in mice (47), its deletion in immune CD4+ T cells ameliorates the neuroimmune response in a mouse model of autoimmune encephalomyelitis (92). Thus, downstream actions of Kdm6a may be cell type specific.
Our study has several caveats and limitations. Our experiments do not exclude other potential contributions of X- or Y-based biological functions. A second X chromosome could contribute resilience through other baseline X escapee genes, epigenetic diversity derived from parent-of-X origin, or reactivation of the silent X chromosome. Furthermore, we did not study how the Y chromosome, its Sry gene, or testicular development contributed to a decreased mortality in hAPP mice with one X chromosome. Last, there are limitations to modeling AD in mice, including in each mouse model we used. Thus, we investigated several models and approaches, including mouse primary neurons, hAPP mice, human brain tissue expression data, and human cognitive data, and included several AD-related measures to increase the potential relevance of our findings. Collectively, these results imply that a second X chromosome, or genes that an X chromosome harbors, could contribute to counteracting AD vulnerability in both sexes.
MATERIALS AND METHODS
Study design
The objectives of our study were to probe the association of sex-based mortality risk in AD using meta-analysis; investigate whether sex chromosomes modify vulnerability related to AD in mice using molecular, cellular, neurogenetic, and behavioral approaches; and test in mice whether an X chromosome factor decreased male vulnerability related to AD. We used experimental models of AD (mice and their primary neurons) and human databases of both brain tissue expression and of clinical cognitive performance. All animal studies were approved by the Institutional Animal Care and Use Committee of the University of California, San Francisco and conducted in compliance with the National Institutes of Health guidelines. For animal experiments, all studies were conducted in a blinded manner and included male and female mice across the lifespan in multiple cohorts at the ages and background strains indicated. Mouse studies used littermate controls along with randomization of mice, and experimentalists were blinded to the genotypes of the mice. In mouse studies, exclusion criteria (greater than 2 SDs above or below the mean) were defined a priori to ensure unbiased exclusion of outliers. We used transgenic mouse models of sex biology crossed with hAPP mice and also used mouse primary neurons exposed to varying doses of Aβ. We assessed several outcome measures including mortality, cognition, cell death, pathology, RNA and protein measures, and biochemistry. Cell culture treatments were carried out with vehicle or synthetic Aβ1-42 peptide previously characterized by atomic force microscopy, and relative neurotoxicity was assessed with MTT and LDH assays.
Our findings showing a statistical effect of the second X chromosome in contributing resilience across measures in mice and mouse primary neurons led us to study Kdm6a, an X-linked gene that escapes inactivation in mice and humans. We established that Kdm6a escapes X-chromosome inactivation in mouse primary neurons using RNA FISH. We then queried KDM6A expression in humans using established databases of brain tissues including the Mayo Clinic Brain Bank and Mount Sinai School of Medicine Brain Bank (RNA sequencing), Gene Expression Omnibus (RNA microarray), and GTEx. We examined clinical and cognitive trajectories using the ADNI database to assess the relevance of our findings to the human condition. Last, we tested whether elevating the expression of Kdm6a causally contributed resilience to AD-related deficits in mouse primary neurons and hAPP mice using lentiviral gene delivery methods.
Statistical analyses
Statistical analyses were carried out with GraphPad Prism (version 5.0) for t tests and log-rank tests for survival analyses. For FCG-hAPP mouse and XY*-hAPP mouse survival statistical analysis, Cox proportional hazards models were applied to determine main effects, and a multivariate Cox model was used to test interactions of main variables on survival. R (nmle package) was used for analyses of variance (ANOVAs), post hoc tests, and meta-analysis. Differences between two means were assessed by two-tailed t tests for all experiments unless indicated otherwise in a replication cohort. Differences among multiple means were assessed by two-way ANOVA. A mixed-model ANOVA was used for analyses of Morris water maze data and included effects of repeated measures. Only significant P values were stated for two-way ANOVA results. Unless indicated otherwise, multiple comparisons of post hoc t tests were corrected for with the Bonferroni-Holm (stepwise Bonferroni) procedure to control for a family-wise error rate of α = 0.05. Linear mixed-effects models were fit in R (93) using the standard lme4 (94) package. In mouse studies, exclusion criteria (greater than 2 SDs above or below the mean) were defined a priori to ensure unbiased exclusion of outliers. Error bars represent ±SEM. Null hypotheses were rejected at or below a P value of 0.05. All analyses for KDM6A human studies were performed using R version 3.5.2 unless otherwise stated. We used linear mixed-effects modeling with random intercepts to test whether the genetic variant identified via GTEx as a modifier of KDM6A expression in brain also affected cognitive and clinical changes in the ADNI cohort. We covaried for baseline age, sex, education, and APOEε4 dose.
Supplementary Material
Fig. S1. Male and female hAPP mouse mortality on a mixed genetic background.
Fig. S2. Human and mouse gonadal hormones in aging.
Fig. S3. Swim speeds and visible platform performance of mice in a water maze test.
Fig. S4. Male sex and cognitive deficits under several conditions.
Fig. S5. Male sex worsens cognition in another transgenic hAPP line.
Fig. S6. Aβ and related proteins in XX- and XY-hAPP mice crossed with FCG and XY* models.
Fig. S7. Western blot image of hAPP, t-Tau, p-Tau, and loading controls.
Fig. S8. 3D6 immunostaining specificity for Aβ plaques.
Fig. S9. Aβ plaque area in very old female hAPP mice.
Fig. S10. hAPP mRNA expression with and without gonadectomy.
Fig. S11. Independent, replicate water maze cohort of FCG mice crossed with hAPP mice.
Fig. S12. Sex chromosomes and fear memory impairment in male hAPP mice.
Fig. S13. KDM6A variant, other clinical exam scores, and assessment of women only.
Fig. S14. Cell death induced by Aβ toxicity and assessed by LDH release assay.
Fig. S15. Kdm6a overexpression and other behavioral tasks in male XY-hAPP mice.
Table S1. KDM6A in AD and control brain tissues.
Table S2. GSE 15222 cohort characteristics.
Table S3. Mayo and Mount Sinai Brain Bank sample characteristics.
Table S4. ADNI cohort characteristics.
Table S5. Longitudinal and subgroup analyses in the ADNI cohort.
Data file S1. Primary data for figures.
Acknowledgments:
We thank S. Cheung for quantification of plaque data, C. Chen for cryostat sectioning of brains, and S. Gupta for assistance with R programming. Data used in this article were obtained from the ADNI database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in the analysis or writing of this report. A complete listing of ADNI investigators is found at http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
Funding:
Primary support for mouse studies and human analyses was provided by NSF grant 1650113 (E.J.D.); NIH grants NS092918 and AG034531 (D.B.D.), AG049152 and AG062588 (J.S.Y.), GM128431 (B.P.), and CTSI UL1 TR000004 (I.L.); the Larry L. Hillblom Foundation (L.W.B. and J.S.Y.); the Coulter-Weeks Foundation (D.B.D.); the Bakar Family Foundation (D.B.D.); the American Federation for Aging Research (E.J.D. and D.B.D.); and the Glenn Foundation for Medical Research (D.B.D.). Additional support was provided by NIH grants AG023501 (B.L.M.); NS043196 and HD076125 (A.P.A.); AG011385 and NS065780 (L.M.); P30AG10161, R01AG15819, and R01AG17917 (D.A.B.); U01AG46152 and U01AG61356 (P.L.D.J. and D.A.B.); R01AG36836 (P.L.D.J.); and AG062234 and AG062629 (J.J.P.). Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (NIH grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging and the National Institute of Biomedical Imaging and Bioengineering and through generous contributions from the following: AbbVie, Alzheimer’s Association, Alzheimer’s Drug Discovery Foundation, Araclon Biotech, BioClinica Inc., Biogen, Bristol-Myers Squibb Company, CereSpir Inc., Cogstate, Eisai Inc., Elan Pharmaceuticals Inc., Eli Lilly and Company, EuroImmun, F. Hoffmann-La Roche Ltd. and its affiliated company Genentech Inc., Fujirebio, GE Healthcare, IXICO Ltd., Janssen Alzheimer Immunotherapy Research & Development LLC., Johnson & Johnson Pharmaceutical Research & Development LLC, Lumosity, Lundbeck, Merck & Co. Inc., Meso Scale Diagnostics LLC, NeuroRx Research, Neurotrack Technologies, Novartis Pharmaceuticals Corporation, Pfizer Inc., Piramal Imaging, Servier, Takeda Pharmaceutical Company, and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the NIH (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Footnotes
SUPPLEMENTARY MATERIALS
Competing interests: D.A.B. has consulted for AbbVie Inc., Data Monitoring Committee, ABBV-8E12 Study and serves on the Scientific Advisory Board for Vigorous Minds Inc.; L.M. has consulted for Abingworth, Eisai, and Sangamo Therapeutics and serves on the Scientific Advisory Board for Alkahest, Arvinas Operations, Biogen, and Dolby Family Ventures; D.B.D. has consulted for Unity Biotechnology. The other authors declare that they have no competing interests.
Data and materials availability: All data associated with this study are present in the paper or the Supplementary Materials. hAPP-J20 mice can be obtained from the Jackson Laboratory.
REFERENCES AND NOTES
- 1.McCarthy MM, Woolley CS, Arnold AP, Incorporating sex as a biological variable in neuroscience: What do we gain? Nat. Rev. Neurosci 18, 707–708 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Prince M, Wimo A, Guerchet M, Ali G-C, Wu Y-T, Prina M, World Alzheimer Report 2015: The Global Impact of Dementia (Alzheimer’s Disease International (ADI), 2015). [Google Scholar]
- 3.Elmaleh DR, Farlow MR, Conti PS, Tompkins RG, Kundakovic L, Tanzi RE, Developing effective Alzheimer's disease therapies: Clinical experience and future directions. J. Alzheimers Dis 71, 715–732 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dubal DB, Sex Differences in Alzheimer’s disease: An updated, balanced, and emerging perspective on differing vulnerabilities, in Sex Differences in Neurology and Psychiatry, Lanzberger R, Kranz GS, Savic-Berglund I, Eds., Handbook of Clinical Neurology, Vol. 175 (3rd series) (Elsevier, 2020); 10.1016/B978-0-444-64123-6.00018-7. [DOI] [PubMed] [Google Scholar]
- 5.Mielke MM, Vemuri P, Rocca WA, Clinical epidemiology of Alzheimer's disease: Assessing sex and gender differences. Clin. Epidemiol 6, 37–48 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hebert LE, Scherr PA, McCann JJ, Beckett LA, Evans DA, Is the risk of developing Alzheimer's disease greater for women than for men? Am. J. Epidemiol 153, 132–136 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Claus JJ, van Gool WA, Teunisse S, Walstra GJM, Kwa VIH, Hijdra A, Verbeeten B Jr., Koelman JHTM, Bour LJ, Ongerboer De Visser BW, Predicting survival in patients with early Alzheimer's disease. Dement. Geriatr. Cogn. Disord 9, 284–293 (1998). [DOI] [PubMed] [Google Scholar]
- 8.Heyman A, Wilkinson WE, Hurwitz BJ, Helms MJ, Haynes CS, Utley CM, Gwyther LP, Early-onset Alzheimer's disease: Clinical predictors of institutionalization and death. Neurology 37, 980–984 (1987). [DOI] [PubMed] [Google Scholar]
- 9.Ueki A, Shinjo H, Shimode H, Nakajima T, Morita Y, Factors associated with mortality in patients with early-onset Alzheimer's disease: A five-year longitudinal study. Int. J. Geriatr. Psychiatry 16, 810–815 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Lapane KL, Gambassi G, Landi F, Sgadari A, Mor V, Bernabei R, Gender differences in predictors of mortality in nursing home residents with AD. Neurology 56, 650–654 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Stern Y, Tang M-X, Albert MS, Brandt J, Jacobs DM, Bell K, Marder K, Sano M, Devanand D, Albert SM, Bylsma F, Tsai W-Y, Predicting time to nursing home care and death in individuals with Alzheimer disease. JAMA 277, 806–812 (1997). [PubMed] [Google Scholar]
- 12.Buckley RF, Mormino EC, Amariglio RE, Properzi MJ, Rabin JS, Lim YY, Papp KV, Jacobs HIL, Burnham S, Hanseeuw BJ, Doré V, Dobson A, Masters CL, Waller M, Rowe CC, Maruff P, Donohue MC, Rentz DM, Kirn D, Hedden T, Chhatwal J, Schultz AP, Johnson KA, Villemagne VL, Sperling RA; Alzheimer's Disease Neuroimaging Initiative; Australian Imaging, Biomarker and Lifestyle study of ageing; Harvard Aging Brain Study, Sex, amyloid, and APOE ε4 and risk of cognitive decline in preclinical Alzheimer's disease: Findings from three well-characterized cohorts. Alzheimers Dement. 14, 1193–1203 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Casaletto KB, Elahi FM, Staffaroni AM, Walters S, Contreras WR, Wolf A, Dubal D, Miller B, Yaffe K, Kramer JH, Cognitive aging is not created equally: Differentiating unique cognitive phenotypes in "normal" adults. Neurobiol. Aging 77, 13–19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cerhan JR, Folsom AR, Mortimer JA, Shahar E, Knopman DS, McGovern PG, Hays MA, Crum LD, Heiss G, Correlates of cognitive function in middle-aged adults. Atherosclerosis Risk in Communities (ARIC) Study Investigators. Gerontology 44, 95–105 (1998). [DOI] [PubMed] [Google Scholar]
- 15.Jack CR Jr., Wiste HJ, Weigand SD, Knopman DS, Vemuri P, Mielke MM, Lowe V, Senjem ML, Gunter JL, Machulda MM, Gregg BE, Pankratz VS, Rocca WA, Petersen RC, Age, sex, and APOE ε4 effects on memory, brain structure, and β-amyloid across the adult life span. JAMA Neurol. 72, 511–519 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jack CR Jr., Therneau TM, Wiste HJ, Weigand SD, Knopman DS, Lowe VJ, Mielke MM, Vemuri P, Roberts RO, Machulda MM, Senjem ML, Gunter JL, Rocca WA, Petersen RC, Transition rates between amyloid and neurodegeneration biomarker states and to dementia: A population-based, longitudinal cohort study. Lancet Neurol. 15, 56–64 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jack CR Jr., Wiste HJ, Weigand SD, Therneau TM, Knopman DS, Lowe V, Vemuri P, Mielke MM, Roberts RO, Machulda MM, Senjem ML, Gunter JL, Rocca WA, Petersen RC, Age-specific and sex-specific prevalence of cerebral β-amyloidosis, tauopathy, and neurodegeneration in cognitively unimpaired individuals aged 50-95 years: A cross-sectional study. Lancet Neurol. 16, 435–444 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jack CR Jr., Therneau TM, Weigand SD, Wiste HJ, Knopman DS, Vemuri P, Lowe VJ, Mielke MM, Roberts RO, Machulda MM, Graff-Radford J, Jones DT, Schwarz CG, Gunter JL, Senjem ML, Rocca WA, Petersen RC, Prevalence of biologically vs clinically defined Alzheimer spectrum entities using the National Institute on Aging-Alzheimer's Association Research Framework. JAMA Neurol. 76, 1174–1183 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petersen RC, Roberts RO, Knopman DS, Geda YE, Cha RH, Pankratz VS, Boeve BF, Tangalos EG, Ivnik RJ, Rocca WA, Prevalence of mild cognitive impairment is higher in men. The Mayo Clinic Study of Aging. Neurology 75, 889–897 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roberts RO, Geda YE, Knopman DS, Cha RH, Pankratz VS, Boeve BF, Tangalos EG, Ivnik RJ, Rocca WA, Petersen RC, The incidence of MCI differs by subtype and is higher in men: The Mayo Clinic Study of Aging. Neurology 78, 342–351 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mucke L, Masliah E, Yu G-Q, Mallory M, Rockenstein EM, Tatsuno G, Hu K, Kholodenko D, Johnson-Wood K, McConlogue L, High-level neuronal expression of Aβ1–42 in wild-type human amyloid protein precursor transgenic mice: Synaptotoxicity without plaque formation. J. Neurosci 20, 4050–4058 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferrini RL, Barrett-Connor E, Sex hormones and age: A cross-sectional study of testosterone and estradiol and their bioavailable fractions in community-dwelling men. Am. J. Epidemiol 147, 750–754 (1998). [DOI] [PubMed] [Google Scholar]
- 23.Morley JE, Kaiser F, Raum WJ, Perry III HM, Flood JF, Jensen J, Silver AJ, Roberts E, Potentially predictive and manipulable blood serum correlates of aging in the healthy human male: Progressive decreases in bioavailable testosterone, dehydroepiandrosterone sulfate, and the ratio of insulin-like growth factor 1 to growth hormone. Proc. Natl. Acad. Sci. U.S.A 94, 7537–7542 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Veldhuis JD, Aging and hormones of the hypothalamo-pituitary axis: Gonadotropic axis in men and somatotropic axes in men and women. Ageing Res. Rev 7, 189–208 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson JF, Felicio LS, Osterburg HH, Finch CE, Differential contributions of ovarian and extraovarian factors to age-related reductions in plasma estradiol and progesterone during the estrous cycle of C57BL/6J mice. Endocrinology 130, 805–810 (1992). [DOI] [PubMed] [Google Scholar]
- 26.Nelson JF, Latham KR, Finch CE, Plasma testosterone levels in C57BL/6J male mice: Effects of age and disease. Acta Endocrinol. 80, 744–752 (1975). [DOI] [PubMed] [Google Scholar]
- 27.Chin J, Palop JJ, Puoliväli J, Massaro C, Bien-Ly N, Gerstein H, Scearce-Levie K, Masliah E, Mucke L, Fyn kinase induces synaptic and cognitive impairments in a transgenic mouse model of Alzheimer's disease. J. Neurosci 25, 9694–9703 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberson ED, Halabisky B, Yoo JW, Yao J, Chin J, Yan F, Wu T, Hamto P, Devidze N, Yu G-Q, Palop JJ, Noebels JL, Mucke L, Amyloid-β/Fyn–induced synaptic, network, and cognitive impairments depend on tau levels in multiple mouse models of Alzheimer's disease. J. Neurosci 31, 700–711 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Callahan MJ, Lipinski WJ, Bian F, Durham RA, Pack A, Walker LC, Augmented senile plaque load in aged female β-amyloid precursor protein-transgenic mice. Am. J. Pathol 158, 1173–1177 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arnold AP, Chen X, What does the “four core genotypes” mouse model tell us about sex differences in the brain and other tissues? Front. Neuroendocrinol 30, 1–9 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mahadevaiah SK, Odorisio T, Elliott DJ, Rattigan Á, Szot M, Laval SH, Washburn LL, McCarrey JR, Cattanach BM, Lovell-Badge R, Burgoyne PS, Mouse homologues of the human AZF candidate gene RBM are expressed in spermatogonia and spermatids, and map to a Y chromosome deletion interval associated with a high incidence of sperm abnormalities. Hum. Mol. Genet 7, 715–727 (1998). [DOI] [PubMed] [Google Scholar]
- 32.McCarthy MM, Arnold AP, Reframing sexual differentiation of the brain. Nat. Neurosci 14, 677–683 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arnold AP, Mouse models for evaluating sex chromosome effects that cause sex differences in non-gonadal tissues. J. Neuroendocrinol 21, 377–386 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eicher EM, Hale DW, Hunt PA, Lee BK, Tucker PK, King TR, Eppig JT, Washburn LL, The mouse Y* chromosome involves a complex rearrangement, including interstitial positioning of the pseudoautosomal region. Cytogenet. Cell Genet 57, 221–230 (1991). [DOI] [PubMed] [Google Scholar]
- 35.Berletch JB, Yang F, Disteche CM, Escape from X inactivation in mice and humans. Genome Biol. 11, 213 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carrel L, Willard HF, X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434, 400–404 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Tukiainen T, Villani A-C, Yen A, Rivas MA, Marshall JL, Satija R, Aguirre M, Gauthier L, Fleharty M, Kirby A, Cummings BB, Castel SE, Karczewski KJ, Aguet F, Byrnes A; GTEx Consortium; Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group; Statistical Methods groups—Analysis Working Group; Enhancing GTEx (eGTEx) groups; NIH Common Fund; NIH/NCI; NIH/NHGRI; NIH/NIMH; NIH/NIDA; Biospecimen Collection Source Site—NDRI; Biospecimen Collection Source Site—RPCI; Biospecimen Core Resource—VARI; Brain Bank Repository—University of Miami Brain Endowment Bank; Leidos Biomedical—Project Management; ELSI Study; Genome Browser Data Integration &Visualization—EBI; Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz, Lappalainen T, Regev A, Ardlie KG, Hacohen N, MacArthur DG, Landscape of X chromosome inactivation across human tissues. Nature 550, 244–248 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang F, Babak T, Shendure J, Disteche CM, Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res. 20, 614–622 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berletch JB, Ma W, Yang F, Shendure J, Noble WS, Disteche CM, Deng X, Escape from X inactivation varies in mouse tissues. PLOS Genet. 11, e1005079 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Greenfield A, Carrel L, Pennisi D, Philippe C, Quaderi N, Siggers P, Steiner K, Tam PPL, Monaco AP, Willard HF, Koopman P, The UTX gene escapes X inactivation in mice and humans. Hum. Mol. Genet 7, 737–742 (1998). [DOI] [PubMed] [Google Scholar]
- 41.Bögershausen N, Gatinois V, Riehmer V, Kayserili H, Becker J, Thoenes M, Simsek-Kiper PÖ, Barat-Houari M, Elcioglu NH, Wieczorek D, Tinschert S, Sarrabay G, Strom TM, Fabre A, Baynam G, Sanchez E, Nürnberg G, Altunoglu U, Capri Y, Isidor B, Lacombe D, Corsini C, Cormier-Daire V, Sanlaville D, Giuliano F, Le Quan Sang K-H, Kayirangwa H, Nürnberg P, Meitinger T, Boduroglu K, Zoll B, Lyonnet S, Tzschach A, Verloes A, Di Donato N, Touitou I, Netzer C, Li Y, Geneviève D, Yigit G, Wollnik B, Mutation update for Kabuki syndrome genes KMT2D and KDM6A and further delineation of X-linked Kabuki syndrome subtype 2. Hum. Mutat 37, 847–864 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Lederer D, Grisart B, Digilio MC, Benoit V, Crespin M, Ghariani SC, Maystadt I, Dallapiccola B, Verellen-Dumoulin C, Deletion of KDM6A, a histone demethylase interacting with MLL2, in three patients with Kabuki syndrome. Am. J. Hum. Genet 90, 119–124 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miyake N, Koshimizu E, Okamoto N, Mizuno S, Ogata T, Nagai T, Kosho T, Ohashi H, Kato M, Sasaki G, Mabe H, Watanabe Y, Yoshino M, Matsuishi T, Takanashi J.-i., Shotelersuk V, Tekin M, Ochi N, Kubota M, Ito N, Ihara K, Hara T, Tonoki H, Ohta T, Saito K, Matsuo M, Urano M, Enokizono T, Sato A, Tanaka H, Ogawa A, Fujita T, Hiraki Y, Kitanaka S, Matsubara Y, Makita T, Taguri M, Nakashima M, Tsurusaki Y, Saitsu H, Yoshiura K.-i., Matsumoto N, Niikawa N, MLL2 and KDM6A mutations in patients with Kabuki syndrome. Am. J. Med. Genet. A 161A, 2234–2243 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Miyake N, Mizuno S, Okamoto N, Ohashi H, Shiina M, Ogata K, Tsurusaki Y, Nakashima M, Saitsu H, Niikawa N, Matsumoto N, KDM6A point mutations cause Kabuki syndrome. Hum. Mutat 34, 108–110 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Van Laarhoven PM, Neitzel LR, Quintana AM, Geiger EA, Zackai EH, Clouthier DE, Artinger KB, Ming JE, Shaikh TH, Kabuki syndrome genes KMT2D and KDM6A: Functional analyses demonstrate critical roles in craniofacial, heart and brain development. Hum. Mol. Genet 24, 4443–4453 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang P, Tan H, Xia Y, Yu Q, Wei X, Guo R, Peng Y, Chen C, Li H, Mei L, Huang Y, Liang D, Wu L, De novo exonic deletion of KDM6A in a Chinese girl with Kabuki syndrome: A case report and brief literature review. Am. J. Med. Genet. A 170, 1613–1621 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Tang G-B, Zeng Y-Q, Liu P-P, Mi T-W, Zhang S-F, Dai S-K, Tang Q-Y, Yang L, Xu Y-J, Yan H-L, Du H-Z, Teng Z-Q, Zhou F-Q, Liu C-M, The histone H3K27 demethylase UTX regulates synaptic plasticity and cognitive behaviors in mice. Front. Mol. Neurosci 10, 267 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Panning B, X inactivation in mouse ES cells: Histone modifications and FISH. Methods Enzymol. 376, 419–428 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Wiedemuth R, Thieme S, Navratiel K, Dorschner B, Brenner S, UTX—Moonlighting in the cytoplasm? Int. J. Biochem. Cell Biol 97, 78–82 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Berletch JB, Deng X, Nguyen DK, Disteche CM, Female bias in Rhox6 and 9 regulation by the histone demethylase KDM6A. PLOS Genet. 9, e1003489 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lei X, Jiao J, UTX affects neural stem cell proliferation and differentiation through PTEN signaling. Stem Cell Rep. 10, 1193–1207 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.GTEx Consortium; Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group; Statistical Methods groups—Analysis Working Group; Enhancing GTEx (eGTEx) groups; NIH Common Fund; NIH/NCI; NIH/NHGRI; NIH/NIMH; NIH/NIDA; Biospecimen Collection Source Site—NDRI; Biospecimen Collection Source Site—RPCI; Biospecimen Core Resource—VARI; Brain Bank Repository—University of Miami Brain Endowment Bank; Leidos Biomedical—Project Management; ELSI Study; Genome Browser Data Integration &Visualization—EBI; Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz; Lead analysts; Laboratory, Data Analysis &Coordinating Center (LDACC); NIH program management; Biospecimen collection; Pathology; eQTL Manuscript Working Group, Battle A, Brown CD, Engelhardt BE, Montgomery SB, Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017).29022597 [Google Scholar]
- 53.Sherry ST, Ward M-H, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K, dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheng JS, Dubal DB, Kim DH, Legleiter J, Cheng IH, Yu G-Q, Tesseur I, Wyss-Coray T, Bonaldo P, Mucke L, Collagen VI protects neurons against Aβ toxicity. Nat. Neurosci 12, 119–121 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sampathkumar NK, Bravo JI, Chen Y, Danthi PS, Donahue EK, Lai RW, Lu R, Randall LT, Vinson N, Benayoun BA, Widespread sex dimorphism in aging and age-related diseases. Hum. Genet 139, 333–356 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buter TC, van den Hout A, Matthews FE, Larsen JP, Brayne C, Aarsland D, Dementia and survival in Parkinson disease: A 12-year population study. Neurology 70, 1017–1022 (2008). [DOI] [PubMed] [Google Scholar]
- 57.Forsaa EB, Larsen JP, Wentzel-Larsen T, Alves G, What predicts mortality in Parkinson disease?: A prospective population-based long-term study. Neurology 75, 1270–1276 (2010). [DOI] [PubMed] [Google Scholar]
- 58.Kihira T, Yoshida S, Okamoto K, Kazimoto Y, Ookawa M, Hama K, Miwa H, Kondo T, Survival rate of patients with amyotrophic lateral sclerosis in Wakayama Prefecture, Japan, 1966 to 2005. J. Neurol. Sci 268, 95–101 (2008). [DOI] [PubMed] [Google Scholar]
- 59.Sejvar JJ, Holman RC, Bresee JS, Kochanek KD, Schonberger LB, Amyotrophic lateral sclerosis mortality in the United States, 1979-2001. Neuroepidemiology 25, 144–152 (2005). [DOI] [PubMed] [Google Scholar]
- 60.Jorm AF, Jolley D, The incidence of dementia: A meta-analysis. Neurology 51, 728–733 (1998). [DOI] [PubMed] [Google Scholar]
- 61.Brookmeyer R, Abdalla N, Estimation of lifetime risks of Alzheimer's disease dementia using biomarkers for preclinical disease. Alzheimers Dement. 14, 981–988 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Horvath S, Gurven M, Levine ME, Trumble BC, Kaplan H, Allayee H, Ritz BR, Chen B, Lu AT, Rickabaugh TM, Jamieson BD, Sun D, Li S, Chen W, Quintana-Murci L, Fagny M, Kobor MS, Tsao PS, Reiner AP, Edlefsen KL, Absher D, Assimes TL, An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome Biol. 17, 171 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goyal MS, Blazey TM, Su Y, Couture LE, Durbin TJ, Bateman RJ, Benzinger TL-S, Morris JC, Raichle ME, Vlassenko AG, Persistent metabolic youth in the aging female brain. Proc. Natl. Acad. Sci. U.S.A 116, 3251–3255 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jack CR Jr., Bennett DA, Blennow K, Carrillo MC, Feldman HH, Frisoni GB, Hampel H, Jagust WJ, Johnson KA, Knopman DS, Petersen RC, Scheltens P, Sperling RA, Dubois B, A/T/N: An unbiased descriptive classification scheme for Alzheimer disease biomarkers. Neurology 87, 539–547 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Caracciolo B, Palmer K, Monastero R, Winblad B, Bäckman L, Fratiglioni L, Occurrence of cognitive impairment and dementia in the community: A 9-year-long prospective study. Neurology 70, 1778–1785 (2008). [DOI] [PubMed] [Google Scholar]
- 66.Ganguli M, Dodge HH, Shen C, DeKosky ST, Mild cognitive impairment, amnestic type: An epidemiologic study. Neurology 63, 115–121 (2004). [DOI] [PubMed] [Google Scholar]
- 67.Koivisto K, Reinikainen KJ, Hanninen T, Vanhanen M, Helkala E-L, Mykkanen L, Laakso M, Pyorala K, Riekkinen PJ Sr., Prevalence of age-associated memory impairment in a randomly selected population from eastern Finland. Neurology 45, 741–747 (1995). [DOI] [PubMed] [Google Scholar]
- 68.Di Carlo A, Lamassa M, Baldereschi M, Inzitari M, Scafato E, Farchi G, Inzitari D, CIND and MCI in the Italian elderly: Frequency, vascular risk factors, progression to dementia. Neurology 68, 1909–1916 (2007). [DOI] [PubMed] [Google Scholar]
- 69.Kivipelto M, Helkala E-L, Hänninen T, Laakso MP, Hallikainen M, Alhainen K, Soininen H, Tuomilehto J, Nissinen A, Midlife vascular risk factors and late-life mild cognitive impairment: A population-based study. Neurology 56, 1683–1689 (2001). [DOI] [PubMed] [Google Scholar]
- 70.Solfrizzi V, Panza F, Colacicco AM, D'Introno A, Capurso C, Torres F, Grigoletto F, Maggi S, Del Parigi A, Reiman EM, Caselli RJ, Scafato E, Farchi G, Capurso A; Italian Longitudinal Study on Aging Working Group, Vascular risk factors, incidence of MCI, and rates of progression to dementia. Neurology 63, 1882–1891 (2004). [DOI] [PubMed] [Google Scholar]
- 71.Hohman TJ, Dumitrescu L, Barnes LL, Thambisetty M, Beecham G, Kunkle B, Gifford KA, Bush WS, Chibnik LB, Mukherjee S, De Jager PL, Kukull W, Crane PK, Resnick SM, Keene CD, Montine TJ, Schellenberg GD, Haines JL, Zetterberg H, Blennow K, Larson EB, Johnson SC, Albert M, Bennett DA, Schneider JA, Jefferson AL; Alzheimer’s Disease Genetics Consortium and the Alzheimer’s Disease Neuroimaging Initiative, Sex-specific association of apolipoprotein E with cerebrospinal fluid levels of tau. JAMA Neurol. 75, 989–998 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jansen WJ, Ossenkoppele R, Knol DL, Tijms BM, Scheltens P, Verhey FRJ, Visser PJ; Amyloid Biomarker Study Group, Aalten P, Aarsland D, Alcolea D, Alexander M, Almdahl IS, Arnold SE, Baldeiras I, Barthel H, van Berckel BNM, Bibeau K, Blennow K, Brooks DJ, van Buchem MA, Camus V, Cavedo E, Chen K, Chetelat G, Cohen AD, Drzezga A, Engelborghs S, Fagan AM, Fladby T, Fleisher AS, van der Flier WM, Ford L, Förster S, Fortea J, Foskett N, Frederiksen KS, Freund-Levi Y, Frisoni GB, Froelich L, Gabryelewicz T, Gill KD, Gkatzima O, Gómez-Tortosa E, Gordon MF, Grimmer T, Hampel H, Hausner L, Hellwig S, Herukka S-K, Hildebrandt H, Ishihara L, Ivanoiu A, Jagust WJ, Johannsen P, Kandimalla R, Kapaki E, Klimkowicz-Mrowiec A, Klunk WE, Köhler S, Koglin N, Kornhuber J, Kramberger MG, Van Laere K, Landau SM, Lee DY, de Leon M, Lisetti V, Lleó A, Madsen K, Maier W, Marcusson J, Mattsson N, de Mendonça A, Meulenbroek O, Meyer PT, Mintun MA, Mok V, Molinuevo JL, Møllergård HM, Morris JC, Mroczko B, Van der Mussele S, Na DL, Newberg A, Nordberg A, Nordlund A, Novak GP, Paraskevas GP, Parnetti L, Perera G, Peters O, Popp J, Prabhakar S, Rabinovici GD, Ramakers IHGB, Rami L, Resende de Oliveira C, Rinne JO, Rodrigue KM, Rodríguez-Rodríguez E, Roe CM, Rot U, Rowe CC, Rüther E, Sabri O, Sanchez-Juan P, Santana I, Sarazin M, Schröder J, Schütte C, Seo SW, Soetewey F, Soininen H, Spiru L, Struyfs H, Teunissen CE, Tsolaki M, Vandenberghe R, Verbeek MM, Villemagne VL, Vos SJB, van Waalwijk van Doorn LJC, Waldemar G, Wallin A, Wallin AK, Wiltfang J, Wolk DA, Zboch M, Zetterberg H, Prevalence of cerebral amyloid pathology in persons without dementia: A meta-analysis. JAMA 313, 1924–1938 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Buckley RF, Mormino EC, Rabin JS, Hohman TJ, Landau S, Hanseeuw BJ, Jacobs HIL, Papp KV, Amariglio RE, Properzi MJ, Schultz AP, Kirn D, Scott MR, Hedden T, Farrell M, Price J, Chhatwal J, Rentz DM, Villemagne VL, Johnson KA, Sperling RA, Sex differences in the association of global amyloid and regional tau deposition measured by positron emission tomography in clinically normal older adults. JAMA Neurol. 76, 542–551 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kodama L, Guzman E, Etchegaray JI, Li Y, Sayed FA, Zhou L, Zhou Y, Zhan L, Le D, Udeochu JC, Clelland CD, Cheng Z, Yu G, Li Q, Kosik KS, Gan L, Microglial microRNAs mediate sex-specific responses to tau pathology. Nat. Neurosci 23, 167–171 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ossenkoppele R, Lyoo CH, Jester-Broms J, Sudre CH, Cho H, Ryu YH, Choi JY, Smith R, Strandberg O, Palmqvist S, Kramer J, Boxer AL, Gorno-Tempini ML, Miller BL, La Joie R, Rabinovici GD, Hansson O, Assessment of demographic, genetic, and imaging variables associated with brain resilience and cognitive resilience to pathological tau in patients with Alzheimer disease. JAMA Neurol. 77, 632–642 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Digma LA, Madsen JR, Rissman RA, Jacobs DM, Brewer JB, Banks SJ; Alzheimer’s Disease Neuroimaging Initiative, Women can bear a bigger burden: Ante- and postmortem evidence for reserve in the face of tau. Brain Commun. 2, fcaa025 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Carroll JC, Rosario ER, Kreimer S, Villamagna A, Gentzschein E, Stanczyk FZ, Pike CJ, Sex differences in β-amyloid accumulation in 3xTg-AD mice: Role of neonatal sex steroid hormone exposure. Brain Res. 1366, 233–245 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Halford RW, Russell DW, Reduction of cholesterol synthesis in the mouse brain does not affect amyloid formation in Alzheimer's disease, but does extend lifespan. Proc. Natl. Acad. Sci. U.S.A. 106, 3502–3506 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hirata-Fukae C, Li H-F, Hoe H-S, Gray AJ, Minami SS, Hamada K, Niikura T, Hua F, Tsukagoshi-Nagai H, Horikoshi-Sakuraba Y, Mughal M, Rebeck GW, LaFerla FM, Mattson MP, Iwata N, Saido TC, Klein WL, Duff KE, Aisen PS, Matsuoka Y, Females exhibit more extensive amyloid, but not tau, pathology in an Alzheimer transgenic model. Brain Res. 1216, 92–103 (2008). [DOI] [PubMed] [Google Scholar]
- 80.Wang J, Tanila H, Puolivali J, Kadish I, van Groen T, Gender differences in the amount and deposition of amyloidβ in APPswe and PS1 double transgenic mice. Neurobiol. Dis 14, 318–327 (2003). [DOI] [PubMed] [Google Scholar]
- 81.Dubal DB, Broestl L, Worden K, Sex and gonadal hormones in mouse models of Alzheimer's disease: What is relevant to the human condition? Biol. Sex Differ 3, 24 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Broestl L, Worden K, Moreno AJ, Davis EJ, Wang D, Garay B, Singh T, Verret L, Palop JJ, Dubal DB, Ovarian cycle stages modulate Alzheimer-related cognitive and brain network alterations in female mice. eNeuro 5, ENEURO.0132-17.2018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pike CJ, Sex and the development of Alzheimer's disease. J. Neurosci. Res 95, 671–680 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Davis EJ, Lobach I, Dubal DB, Female XX sex chromosomes increase survival and extend lifespan in aging mice. Aging Cell 18, e12871 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Skuse DH, X-linked genes and mental functioning. Hum. Mol. Genet 14Spec No 1, R27–R32 (2005). [DOI] [PubMed] [Google Scholar]
- 86.Vawter MP, Evans S, Choudary P, Tomita H, Meador-Woodruff J, Molnar M, Li J, Lopez JF, Myers R, Cox D, Watson SJ, Akil H, Jones EG, Bunney WE, Gender-specific gene expression in post-mortem human brain: Localization to sex chromosomes. Neuropsychopharmacology 29, 373–384 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xu J, Deng X, Watkins R, Disteche CM, Sex-specific differences in expression of histone demethylases Utx and Uty in mouse brain and neurons. J. Neurosci 28, 4521–4527 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Greenfield A, Scott D, Pennisi D, Ehrmann I, Ellis P, Cooper L, Simpson E, Koopman P, An H-YDb epitope is encoded by a novel mouse Y chromosome gene. Nat. Genet 14, 474–478 (1996). [DOI] [PubMed] [Google Scholar]
- 89.Gažová I, Lengeling A, Summers KM, Lysine demethylases KDM6A and UTY: The X and Y of histone demethylation. Mol. Genet. Metab 127, 31–44 (2019). [DOI] [PubMed] [Google Scholar]
- 90.Shpargel KB, Sengoku T, Yokoyama S, Magnuson T, UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLOS Genet. 8, e1002964 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Walport LJ, Hopkinson RJ, Vollmar M, Madden SK, Gileadi C, Oppermann U, Schofield CJ, Johansson C, Human UTY(KDM6C) is a male-specific NЄ-methyl lysyl demethylase. J. Biol. Chem 289, 18302–18313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Itoh Y, Golden LC, Itoh N, Matsukawa MA, Ren E, Tse V, Arnold AP, Voskuhl RR, The X-linked histone demethylase Kdm6a in CD4+ T lymphocytes modulates autoimmunity. J. Clin. Invest 129, 3852–3863 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.R. C. Team, R: A Language and Environment for Statistical Computing version 3.6.1, R Foundation for Statistical Computing, Vienna, Austria: (2019). [Google Scholar]
- 94.Bates D, Mächler M, Bolker B, Walker S, lme4: Linear mixed-effects models using Eigen and S4, Journal of Statistical Software, 67 (2014). [Google Scholar]
- 95.Agüero-Torres H, Fratiglioni L, Guo Z, Viitanen M, Winblad B, Prognostic factors in very old demented adults: A seven-year follow-up from a population-based survey in Stockholm. J. Am. Geriatr. Soc 46, 444–452 (1998). [DOI] [PubMed] [Google Scholar]
- 96.Aneshensel CS, Pearlin LI, Levy-Storms L, Schuler RH, The transition from home to nursing home mortality among people with dementia. J. Gerontol. B Psychol. Sci. Soc. Sci 55, S152–S162 (2000). [DOI] [PubMed] [Google Scholar]
- 97.Beard CM, Kokmen E, O’Brien PC, Kurland LT, Are patients with Alzheimer's disease surviving longer in recent years? Neurology 44, 1869–1871 (1994). [DOI] [PubMed] [Google Scholar]
- 98.Bracco L, Gallato R, Grigoletto F, Lippi A, Lepore V, Bino G, Lazzaro MP, Carella F, Piccolo T, Pozzilli C, Giometto B, Amaducci L, Factors affecting course and survival in Alzheimer's disease. A 9-year longitudinal study. Arch. Neurol 51, 1213–1219 (1994). [DOI] [PubMed] [Google Scholar]
- 99.Burns A, Lewis G, Jacoby R, Levy R, Factors affecting survival in Alzheimer's disease. Psychol. Med 21, 363–370 (1991). [DOI] [PubMed] [Google Scholar]
- 100.Claus JJ, Walstra GJM, Bossuyt PM, Teunisse S, Van Gool WA, A simple test of copying ability and sex define survival in patients with early Alzheimer's disease. Psychol. Med 29, 485–489 (1999). [DOI] [PubMed] [Google Scholar]
- 101.Go SM, Lee KS, Seo SW, Chin J, Kang SJ, Moon SY, Na DL, Cheong H-K, Survival of Alzheimer's disease patients in Korea. Dement. Geriatr. Cogn. Disord 35, 219–228 (2013). [DOI] [PubMed] [Google Scholar]
- 102.Roehr S, Luck T, Bickel H, Brettschneider C, Ernst A, Fuchs A, Heser K, König H-H, Jessen F, Lange C, Mösch E, Pentzek M, Steinmann S, Weyerer S, Werle J, Wiese B, Scherer M, Maier W, Riedel-Heller SG; AgeCoDe Study Group, Mortality in incident dementia—Results from the German Study on Aging, Cognition, and Dementia in Primary Care Patients. Acta Psychiatr. Scand 132, 257–269 (2015). [DOI] [PubMed] [Google Scholar]
- 103.Stern Y, Tang MX, Denaro J, Mayeux R, Increased risk of mortality in Alzheimer's disease patients with more advanced educational and occupational attainment. Ann. Neurol 37, 590–595 (1995). [DOI] [PubMed] [Google Scholar]
- 104.Thomas BM, McGonigal G, McQuade CA, Starr JM, Whalley LJ, Survival in early onset dementia: Effects of urbanization and socio-economic deprivation. Neuroepidemiology 16, 134–140 (1997). [DOI] [PubMed] [Google Scholar]
- 105.Vilalta-Franch J, López-Pousa S, Calvó-Perxas L, Garre-Olmo J, Psychosis of Alzheimer disease: Prevalence, incidence, persistence, risk factors, and mortality. Am. J. Geriatr. Psychiatry 21, 1135–1143 (2013). [DOI] [PubMed] [Google Scholar]
- 106.Williams MM, Xiong C, Morris JC, Galvin JE, Survival and mortality differences between dementia with Lewy bodies vs Alzheimer disease. Neurology 67, 1935–1941 (2006). [DOI] [PubMed] [Google Scholar]
- 107.Wolfson C, Wolfson DB, Asgharian M, M'Lan CE, Østbye T, Rockwood K, Hogan DB; Clinical Progression of Dementia Study Group, A reevaluation of the duration of survival after the onset of dementia. N. Engl. J. Med 344, 1111–1116 (2001). [DOI] [PubMed] [Google Scholar]
- 108.Viechtbauer W, Conducting meta-analyses in R with the metafor package. J. Stat. Softw 36, 1–48 (2010). [Google Scholar]
- 109.Hartung J, Knapp G, A refined method for the meta-analysis of controlled clinical trials with binary outcome. Stat. Med 20, 3875–3889 (2001). [DOI] [PubMed] [Google Scholar]
- 110.Hartung J, Knapp G, On tests of the overall treatment effect in meta-analysis with normally distributed responses. Stat. Med 20, 1771–1782 (2001). [DOI] [PubMed] [Google Scholar]
- 111.Hedges LV, Vevea JL, Fixed- and random-effects models in meta-analysis. Psychol. Methods 3, 486–504 (1998). [Google Scholar]
- 112.Sidik K, Jonkman JN, A simple confidence interval for meta-analysis. Stat. Med 21, 3153–3159 (2002). [DOI] [PubMed] [Google Scholar]
- 113.Rockenstein EM, McConlogue L, Tan H, Power M, Masliah E, Mucke L, Levels and alternative splicing of amyloid β protein precursor (APP) transcripts in brains of APP transgenic mice and humans with Alzheimer's disease. J. Biol. Chem 270, 28257–28267 (1995). [DOI] [PubMed] [Google Scholar]
- 114.Dubal DB, Yokoyama JS, Zhu L, Broestl L, Worden K, Wang D, Sturm VE, Kim D, Klein E, Yu G-Q, Ho K, Eilertson KE, Yu L, Kuro-o M, De Jager PL, Coppola G, Small GW, Bennett DA, Kramer JH, Abraham CR, Miller BL, Mucke L, Life extension factor klotho enhances cognition. Cell Rep. 7, 1065–1076 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dubal DB, Zhu L, Sanchez PE, Worden K, Broestl L, Johnson E, Ho K, Yu G-Q, Kim D, Betourne A, Kuro-o M, Masliah E, Abraham CR, Mucke L, Life extension factor klotho prevents mortality and enhances cognition in hAPP transgenic mice. J. Neurosci 35, 2358–2371 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cissé M, Halabisky B, Harris J, Devidze N, Dubal DB, Sun B, Orr A, Lotz G, Kim DH, Hamto P, Ho K, Yu G-Q, Mucke L, Reversing EphB2 depletion rescues cognitive functions in Alzheimer model. Nature 469, 47–52 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Palop JJ, Jones B, Kekonius L, Chin J, Yu G-Q, Raber J, Masliah E, Mucke L, Neuronal depletion of calcium-dependent proteins in the dentate gyrus is tightly linked to Alzheimer's disease-related cognitive deficits. Proc. Natl. Acad. Sci. U.S.A 100, 9572–9577 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Klein WL, Aβ toxicity in Alzheimer's disease: Globular oligomers (ADDLs) as new vaccine and drug targets. Neurochem. Int 41, 345–352 (2002). [DOI] [PubMed] [Google Scholar]
- 119.Aras MA, Hartnett KA, Aizenman E, Assessment of cell viability in primary neuronal cultures. Curr. Protoc. Neurosci Chapter 7, Unit 7.18 (2008). [DOI] [PubMed] [Google Scholar]
- 120.Allen M, Carrasquillo MM, Funk C, Heavner BD, Zou F, Younkin CS, Burgess JD, Chai H-S, Crook J, Eddy JA, Li H, Logsdon B, Peters MA, Dang KK, Wang X, Serie D, Wang C, Nguyen T, Lincoln S, Malphrus K, Bisceglio G, Li M, Golde TE, Mangravite LM, Asmann Y, Price ND, Petersen RC, Graff-Radford NR, Dickson DW, Younkin SG, Ertekin-Taner N, Human whole genome genotype and transcriptome data for Alzheimer's and other neurodegenerative diseases. Sci. Data 3, 160089 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang M, Beckmann ND, Roussos P, Wang E, Zhou X, Wang Q, Ming C, Neff R, Ma W, Fullard JF, Hauberg ME, Bendl J, Peters MA, Logsdon B, Wang P, Mahajan M, Mangravite LM, Dammer EB, Duong DM, Lah JJ, Seyfried NT, Levey AI, Buxbaum JD, Ehrlich M, Gandy S, Katsel P, Haroutunian V, Schadt E, Zhang B, The Mount Sinai cohort of large-scale genomic, transcriptomic and proteomic data in Alzheimer's disease. Sci Data 5, 180185 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Webster JA, Gibbs JR, Clarke J, Ray M, Zhang W, Holmans P, Rohrer K, Zhao A, Marlowe L, Kaleem M, McCorquodale DS III, Cuello C, Leung D, Bryden L, Nath P, Zismann VL, Joshipura K, Huentelman MJ, Hu-Lince D, Coon KD, Craig DW, Pearson JV; NACC-Neuropathology Group, Heward CB, Reiman EM, Stephan D, Hardy J, Myers AJ, Genetic control of human brain transcript expression in Alzheimer disease. Am. J. Hum. Genet 84, 445–458 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Carithers LJ, Ardlie K, Barcus M, Branton PA, Britton A, Buia SA, Compton CC, DeLuca DS, Peter-Demchok J, Gelfand ET, Guan P, Korzeniewski GE, Lockhart NC, Rabiner CA, Rao AK, Robinson KL, Roche NV, Sawyer SJ, Segrè AV, Shive CE, Smith AM, Sobin LH, Undale AH, Valentino KM, Vaught J, Young TR, Moore HM; GTEx Consortium, A novel approach to high-quality postmortem tissue procurement: The GTEx Project. Biopreserv. Biobank 13, 311–319 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Petersen RC, Aisen PS, Beckett LA, Donohue MC, Gamst AC, Harvey DJ, Jack CR Jr., Jagust WJ, Shaw LM, Toga AW, Trojanowski JQ, Weiner MW, Alzheimer's Disease Neuroimaging Initiative (ADNI): Clinical characterization. Neurology 74, 201–209 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Saykin AJ, Shen L, Foroud TM, Potkin SG, Swaminathan S, Kim S, Risacher SL, Nho K, Huentelman MJ, Craig DW, Thompson PM, Stein JL, Moore JH, Farrer LA, Green RC, Bertram L, Jack CR Jr., Weiner MW; Alzheimer's Disease Neuroimaging Initiative, Alzheimer's Disease Neuroimaging Initiative biomarkers as quantitative phenotypes: Genetics core aims, progress, and plans. Alzheimers Dement. 6, 265–273 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, Green RC, Harvey D, Jack CR Jr., Jagust W, Liu E, Morris JC, Petersen RC, Saykin AJ, Schmidt ME, Shaw L, Siuciak JA, Soares H, Toga AW, Trojanowski JQ; Alzheimer’s Disease Neuroimaging Initiative, The Alzheimer's Disease Neuroimaging Initiative: A review of papers published since its inception. Alzheimers Dement. 8, S1–S68 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bonham LW, Desikan RS, Yokoyama JS; Alzheimer’s Disease Neuroimaging Initiative, The relationship between complement factor C3, APOE ε4, amyloid and tau in Alzheimer's disease. Acta Neuropathol. Commun 4, 65 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Folstein MF, Folstein SE, McHugh PR, “Mini-mental state”: A practical method for grading the cognitive state of patients for the clinician. J. Psychiatr. Res 12, 189–198 (1975). [DOI] [PubMed] [Google Scholar]
- 129.Saykin AJ, Shen L, Yao X, Kim S, Nho K, Risacher SL, Ramanan VK, Foroud TM, Faber KM, Sarwar N, Munsie LM, Hu X, Soares HD, Potkin SG, Thompson PM, Kauwe JSK, Kaddurah-Daouk R, Green RC, Toga AW, Weiner MW; Alzheimer's Disease Neuroimaging Initiative, Genetic studies of quantitative MCI and AD phenotypes in ADNI: Progress, opportunities, and plans. Alzheimers Dement. 11, 792–814 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ongen H, Buil A, Brown AA, Dermitzakis ET, Delaneau O, Fast and efficient QTL mapper for thousands of molecular phenotypes. Bioinformatics 32, 1479–1485 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.GTEx Consortium, Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 348, 648–660 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Robinson MD, Oshlack A, A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Stegle O, Parts L, Durbin R, Winn J, A Bayesian framework to account for complex non-genetic factors in gene expression levels greatly increases power in eQTL studies. PLOS Comput. Biol 6, e1000770 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wright FA, Sullivan PF, Brooks AI, Zou F, Sun W, Xia K, Madar V, Jansen R, Chung W, Zhou Y-H, Abdellaoui A, Batista S, Butler C, Chen G, Chen T-H, D'Ambrosio D, Gallins P, Ha MJ, Hottenga JJ, Huang S, Kattenberg M, Kochar J, Middeldorp CM, Qu A, Shabalin A, Tischfield J, Todd L, Tzeng J-Y, van Grootheest G, Vink JM, Wang Q, Wang W, Wang W, Willemsen G, Smit JH, de Geus EJ, Yin Z, Penninx BWJH, Boomsma DI, Heritability and genomics of gene expression in peripheral blood. Nat. Genet 46, 430–437 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Male and female hAPP mouse mortality on a mixed genetic background.
Fig. S2. Human and mouse gonadal hormones in aging.
Fig. S3. Swim speeds and visible platform performance of mice in a water maze test.
Fig. S4. Male sex and cognitive deficits under several conditions.
Fig. S5. Male sex worsens cognition in another transgenic hAPP line.
Fig. S6. Aβ and related proteins in XX- and XY-hAPP mice crossed with FCG and XY* models.
Fig. S7. Western blot image of hAPP, t-Tau, p-Tau, and loading controls.
Fig. S8. 3D6 immunostaining specificity for Aβ plaques.
Fig. S9. Aβ plaque area in very old female hAPP mice.
Fig. S10. hAPP mRNA expression with and without gonadectomy.
Fig. S11. Independent, replicate water maze cohort of FCG mice crossed with hAPP mice.
Fig. S12. Sex chromosomes and fear memory impairment in male hAPP mice.
Fig. S13. KDM6A variant, other clinical exam scores, and assessment of women only.
Fig. S14. Cell death induced by Aβ toxicity and assessed by LDH release assay.
Fig. S15. Kdm6a overexpression and other behavioral tasks in male XY-hAPP mice.
Table S1. KDM6A in AD and control brain tissues.
Table S2. GSE 15222 cohort characteristics.
Table S3. Mayo and Mount Sinai Brain Bank sample characteristics.
Table S4. ADNI cohort characteristics.
Table S5. Longitudinal and subgroup analyses in the ADNI cohort.
Data file S1. Primary data for figures.