Abstract
There could be two carcinogenetic pathways for lung adenocarcinoma (LADC): the nonsmokers’ pathway and the smokers’ pathway. This review article describes the two pathways with special reference to potential relationships between histological subtypes, malignant grades, and driver mutations. The lung is composed of two different tissue units, the terminal respiratory unit (TRU) and the central airway compartment (CAC). In the nonsmokers’ pathway, LADCs develop from the TRU, and their histological appearances change from lepidic to micropapillary during the progression process. In the smokers’ pathway, LADCs develop from either the TRU or the CAC, and their histological appearances vary among cases in the middle of the progression process, but they are likely converged to acinar/solid at the end. On a molecular genetic level, the nonsmokers’ pathway is mostly driven by EGFR mutations, whereas in the smokers’ pathway, approximately one‐quarter of LADCs have KRAS mutations, but the other three‐quarters have no known driver mutations. p53 mutations are an important factor triggering the progression of both pathways, with unique molecular alterations associated with each, such as MUC21 expression and chromosome 12p13‐21 amplification in the nonsmokers’ pathway, and HNF4α expression and TTF1 mutations in the smokers’ pathway. However, investigation into the relationship between histological progression and genetic alterations is in its infancy. Tight cooperation between traditional histopathological examinations and recent molecular genetics can provide valuable insight to better understand the nature of LADCs.
Keywords: lung adenocarcinoma, micropapillary, solid/acinar, progression, smoking
It has been supposed there could be two carcinogenetic pathways for lung adenocarcinoma (LADC): the nonsmokers’ pathway and the smokers’ pathway. This review article describes the two pathways with special reference to potential relationships between histological subtypes, malignant grades, and driver mutations
1. INTRODUCTION
Lung adenocarcinoma (LADC) shows considerable heterogeneity in patients’ characteristics, clinical courses, histological appearances, and molecular alternations. It is needed to clearly describe their biological nature for precision medicine.
Recent efforts in the field of molecular genetics have uncovered essential driver oncogenes, including EGFR, KRAS, BRAF, ALK, ROS1, RET, and NRG1, that can be targets for newly developed therapeutic agents.1, 2, 3, 4, 5, 6, 7 Subtyping of LADC based on the driver mutations is beneficial in a clinical setting and is becoming increasingly common. However, the driver mutations themselves do not seem to be direct determinants for malignant grades, 7, 8 as subtyping solely based on the driver mutations may not be enough to support a prognosis. On the other hand, traditional histological subtyping is informative in terms of identifying histogenesis and malignant grades.7
It has been supposed there could be two carcinogenetic pathways for LADC: the nonsmokers’ pathway and the smokers’ pathway. This review article describes the two pathways with special reference to potential relationships between histological subtypes, malignant grades, and driver mutations.
2. THE TWO PUTATIVE CARCINOGENETIC PATHWAYS
Current research suggests that there are two distinct pathways involved in the development of LADC in smokers and nonsmokers.7 Recently, Yatabe et al proposed a theory that the lung is composed of two different tissue units, the terminal respiratory unit (TRU) and the central airway compartment (CAC).7, 9, 10 The TRU includes the distal/terminal bronchiole to the alveoli and mainly participates in gas exchange, whereas non‐TRU/CAC includes the trachea, bronchus, and the proximal/lobular bronchioles, which are responsible for directing gas into the alveoli. Most LADCs are developed from the TRU, which constantly expresses thyroid transcription factor 1 (TTF1).7 TRU‐type LADCs predominantly occur in nonsmokers. The predisposition to nonsmokers is particularly interesting, as it is quite different from the other histological types, such as squamous cell, small cell, and large cell carcinomas.7 In contrast, TTF1‐negative, non‐TRU/CAC–type LADCs preferentially occur in smokers.7 Thus, this TRU/non‐TRU concept is a concise theory to better understand the histogenesis of the nonsmokers’ and smokers’ pathways.
3. A PROPOSAL OF FIVE ESSENTIAL GROUPS FOR LADCS
The TRU/non‐TRU concept is a theory based on histogenesis. On the other hand, the nonsmokers’/smokers’ concept is a theory to describe the potential effects of carcinogenetic stimuli on the development/progression of cancers. Actually, TRU‐type LADCs can develop in both smokers and nonsmokers and can differentially progress each other. Thus, we have been considering that the TRU/non‐TRU classification is not enough to better understand the tumor progression. To improve this, we here propose five groups based on histological appearances, which are modified from our previous publications,11, 12 and describe them while linking together the nonsmokers’/smokers’ pathways, the TRU/non‐TRU concept, patients’ baseline characteristics, clinical courses, and driver mutations (Table 1).
TABLE 1.
Essential clinicopathological features in the five groups (A‐E) proposed
| Group A (296) | Group B (18) | Group C (97) | Group D (65) | Group E (230) | |
|---|---|---|---|---|---|
| Histological | Lepidic/papillary | With mucinous cribriform | With micropapillary | Mucinous/enteric | Acinar/solid |
| Cytological | TRU | TRU | TRU | Non‐TRU | TRU/non‐TRU |
| Immunohistochemical | TTF1+/HNF4α‐ | TTF1+/HNF4α‐ | TTF1+/HNF4α‐ | TTF1‐/HNF4α+ | TTF1+‐/HNF4α+‐ |
| Cell growth (Ki‐67 LI, mean ± SD) | Weak (0.08 ± 0.11) | Moderate (0.14 ± 0.09) | Moderate (0.15 ± 0.12) | Moderate (0.23 ± 0.22) | Strong (0.35 ± 0.18) |
| Frequency sW (s ≤ I/s≥II) | 42.0% (54.3%/8.9%) | 2.5% (2.1%/3.6%) | 13.8% (11.5%/20.1%) | 9.3% (9.5%/8.3%) | 32.4% (22.5%/59.5%) |
| Age median (range) | 68 (28‐86) | 61 (29‐79) | 73 (31‐82) | 68 (43‐84) | 69 (36‐87) |
| Gender M/F | 0.7 | 1.3 | 0.42 | 1.8 | 3.5 |
| Smoking S/NS | 0.6 | 0.3 | 0.3 | 1.5 | 7.3 |
| Prognosis (5 y‐DFS sW/s ≤ I) | Favorable (92.6%/94.3%) | Worse (79.8%/100%) | Worst (49.0%/61.3%) | Worse (54.5%/70.0%) | Worst (46.6%/63.2%) |
| Main drivers | EGFR | ALK/ROS1 | EGFR | KRAS/NRG1 | KRAS |
| Potential progressors | p53 loss27; CXCL14 gain;32 E‐cadherin loss, axin1 gain, wnt signal gain;34 MUC21 gain;16 DYRK2 gain;31 c‐met gain33 | TTF1 loss;46, 47 pulmonary surfactant system loss46 | p53 loss;12, 41 DUSP6 loss;43 S100A11 gain;37 FXYF3 loss;42 HDAC9 loss;39 miR31 loss;40 CTSL1 loss;44 TPM1 loss:45 CLIC4 loss38 |
Lung adenocarcinoma patients who underwent surgical operation in Kanagawa prefectural cardiovascular center hospital from 1994 to 2013 (706 cases in total, 514 pathological stage 0/I and 192 II/III) were subjected. Number of cases examined in each of the groups is shown parentheses after the alphabets. Pathological stages were determined according to the AJCC Cancer Staging Manual (8th edition).50 Superscript numbers indicate the reference numbers.
Abbreviations: 5y‐DFS, 5‐y disease‐free survival; F, female; LI, labeling index; M, male; NS, nonsmoker; S, smoker; sW, whole pathological stage; s ≤ I, pathological stage 0 and I; S≥II, pathological stage II and III; TRU, terminal respiratory unit.
Briefly, group A is defined as lepidic or papillary dominant LADC without any mucin‐producing element or micropapillary element (according to the WHO classification system,7 histological elements less than 5% were ignored [judged as none]); group B as LADC with acinar element of mucinous cribriform pattern; group C as LADC with micropapillary elements; group D as mucinous adenocarcinoma (both invasive and in situ) and/or enteric adenocarcinoma; and group E as conventional acinar (not mucinous cribriform pattern) and/or solid dominant LADC. Representative histological appearances are shown with references to the nonsmokers’/smokers’ pathways, essential molecular alterations, and the potential between‐group crosstalk in Figure 1. Specific features in detail for each of the five groups are as follows:
FIGURE 1.
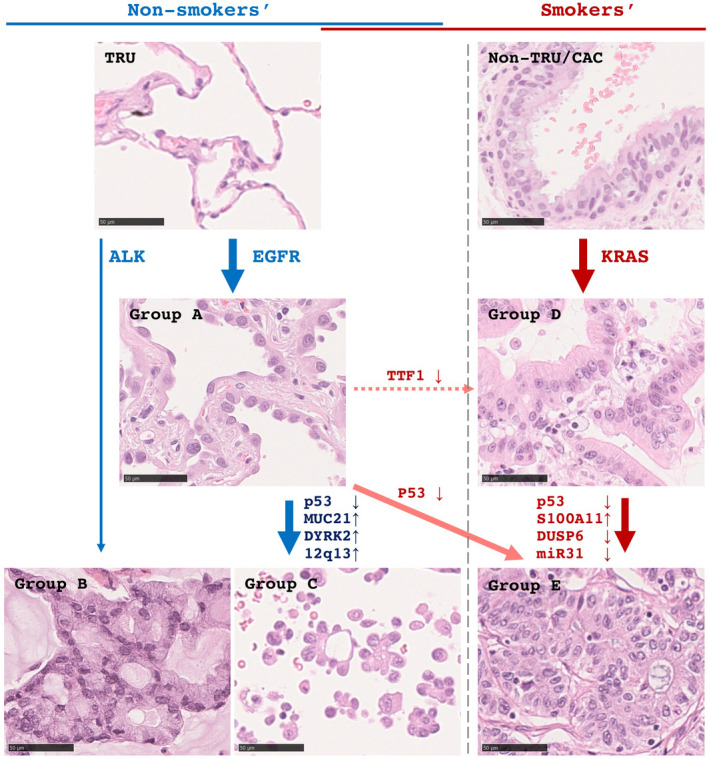
A scheme of histological progression of lung adenocarcinomas (LADCs) in the nonsmokers’ (left panels, blue arrows) and smokers’ (right panels, red arrows) pathway is shown. In the smokers’ pathway, group A (lepidic histology) develops from the terminal respiratory unit (TRU) with EGFR mutations, and some of them can progress to group C (micropapillary) through accumulations of second molecular alterations. Group B (acinar/cribriform with mucin) may directly develop from the TRU with ALK mutations. In the smokers’ pathway, group D (mucinous subtype) develops from the non‐TRU/central airway compartment (CAC), where one major driver in the smoker's pathway is KRAS. Some can progress to group E (conventional acinar/solid) through second alterations. Alternatively, there are putative extra‐bypassing pathways. Under exposing to smoking stimuli, group A can progress to group E (orange arrow). Also, through acquired thyroid transcription factor 1 (TTF1) inactivation, group A can progress to group D (dashed orange arrow). Scale bars, 50 µm
3.1. Group A
Group A includes TRU‐type LADCs that typically affect female nonsmokers (Table 1), mostly show lepidic‐dominant histology including adenocarcinoma in situ and minimally invasive adenocarcinomas, and occasionally show papillary‐dominant histology. LADCs of this group are generally slow growing (Table 1) and not aggressive; hence, they are surgically operable in most cases. Thus, pathologists see this type of LADCs in surgical specimens frequently, where they comprise 42% (Table 1). Surgical removal is enough to completely control this group in most cases, and it ultimately results in favorable outcomes (Figure 2). Although majority of this group of LADCs have EGFR mutations (Table 2 and Figure 3), EGFR tyrosine kinase inhibitors (TKIs) are usually not needed because of the favorable postoperative outcomes.13
FIGURE 2.
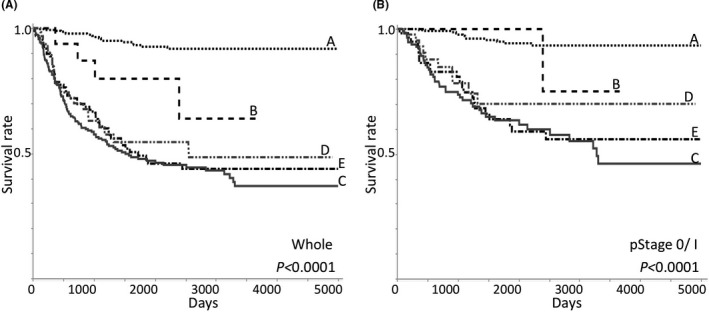
Kaplan‐Meier's disease‐free survival curves from whole (A) and pathological stage 0/I (pStage 0/I) lung adenocarcinomas (LADCs) (B) are shown. A total of 706 cases who underwent surgical resection in Kanagawa Prefectural Cardiovascular and Respiratory Center Hospital from 1994 to 2013 were examined: 514 stage 0/I (279 group A, 11 group B, 59 group C, 49 group D, and 116 group E), 192 stage II/III (17 group A, 7 group B, 38 group C, 16 group D, and 114 group E). Pathological stages were determined according to the AJCC Cancer Staging Manual 8th edition50
TABLE 2.
Frequencies (%) of driver mutations in the five groups (A‐E) proposed
| A (184) | B (15) | C (79) | D (46) | E (166) | |
|---|---|---|---|---|---|
| EGFR | 72.3 | 0.0 | 74.7 | 0.0 | 11.5 |
| ERBB2 | 4.9 | 0.0 | 1.3 | 0.0 | 1.8 |
| MET | 2.7 | 0.0 | 5.1 | 0.0 | 1.2 |
| ALK | 1.1 | 60.0 | 2.5 | 0.0 | 1.2 |
| ROS1 | 1.1 | 20.0 | 0.0 | 0.0 | 0.0 |
| RET | 0.0 | 6.7 | 0.0 | 0.0 | 0.0 |
| BRAF | 0.5 | 0.0 | 1.3 | 0.0 | 3.6 |
| KRAS | 2.2 | 0.0 | 0.0 | 55.4 | 18.1 |
| NRG1 | 0.0 | 0.0 | 0.0 | 10.9 | 0.0 |
| NONE | 13.6 | 13.3 | 16.5 | 34.8 | 63.3 |
Lung adenocarcinomas surgically resected in Kanagawa prefectural cardiovascular center hospital were examined. Number in each of the groups is shown in parentheses after the alphabets.
FIGURE 3.
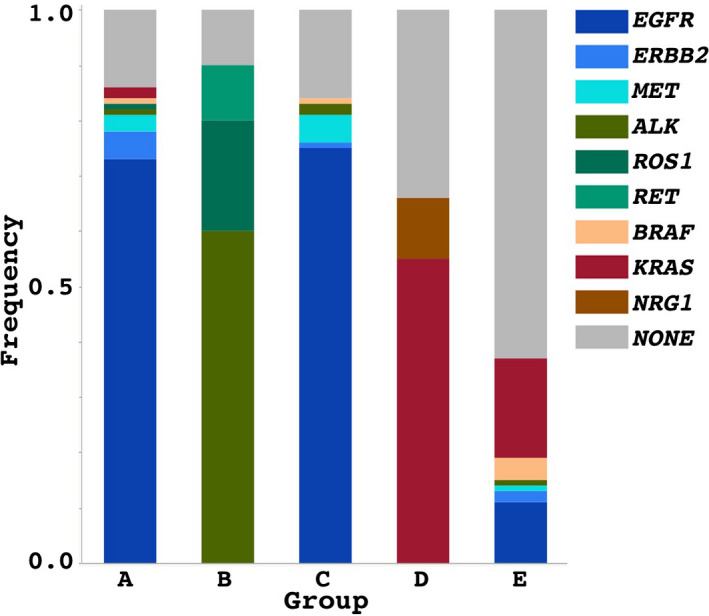
Frequencies of driver mutations are shown in each of the groups. A difference in the driver mutations is remarkable between the groups. EGFR mutations are mostly seen in groups A and C, ALK and ROS1 mutations are seen in group B, and KRAS mutations are more commonly seen in group D, followed by group E
3.2. Group B
This group includes TRU‐type LADCs that entirely or partially show cribriform and solid elements with mucin production. This group affects relatively young nonsmokers (Table 1) and shows relatively favorable prognosis (Figure 2). Gender predisposition seems relatively weak (Table 1). This group is rare and comprises 2.5% of all surgically removed LADCs (Table 1). A particular feature is frequent fusion gene mutations, such as ALK and ROS1 (Table 2 and Figure 3). This type can be described as “solid LADC with mucin production,” “acinar LADC with cribriform structure with mucin production,” or sometimes as “signet ring cell carcinoma,” in pathological diagnosis reports.
3.3. Group C
As group A, group C includes TRU‐type LADCs that predominantly affect female nonsmokers. This type has a micropapillary element in lepidic or papillary backgrounds, where proportions of the micropapillary element vary among cases, but generally are not so large (typically 5 to 25%) in surgically removed LADCs.13, 14 LADCs of this group are usually described as “lepidic or papillary‐dominant LADC with micropapillary element” or simply as “lepidic or papillary‐dominant LADC” or “minimally invasive adenocarcinoma” in pathological diagnosis reports, and they comprise 13.8% of surgically removed LADCs (Table 1). Proliferating activities are generally not as strong (Table 1), but they often show strong lymphatic canal and vascular involvements and aggressive clinical courses 13, 15 (Figure 2). Most inoperable cases from nonsmokers belong to this group.13, 16 Fortunately, this group generally has EGFR mutations (Table 2 and Figure 3) and can respond well to EGFR‐TKI treatments.13, 15, 17, 18
3.4. Group D
This group exclusively comprises specific histological subtypes of mucinous LADC (invasive or in situ) and enteric LADC. They consist of tall columnar neoplastic cells that are usually mucin producing. This group is negative for TTF1 and positive for hepatocyte nuclear factor 4α (HNF4α), an important transcription factor in the upper digestive tract epithelia, implying their gastric/intestinal differentiation; hence they are categorized as non‐TRU–type.19 This group is infrequent and comprises 9.3% of surgically removed LADCs (Table 1). LADCs of this group preferentially develop in male smokers, but gender difference and smoker predisposition seems to be not so significant. This group shows relatively poor clinical courses (Figure 2). LADCs of this group are frequently affected by KRAS mutations but almost never by EGFR mutations (Table 2 and Figure 3).7, 10, 19, 20 Interestingly, NRG1 mutations are specific to group D.21 Another particular feature is a strong relationship to pulmonary fibrosis.20 Most of pulmonary fibrosis patients are smokers.20 Smoking‐related carcinogens, chronic cellular damage, and tissue remodeling might promote this group of LADC in cooperation. LADCs of this group can be described as “mucinous LADC,” “enteric LADC,” or maybe sometimes “acinar LADCs” in pathological diagnosis reports.
3.5. Group E
The fifth group is typically observed in male heavy smokers. This group comprises 32.4% of all surgically removed LADCs (Table 1). LADCs of this group histologically show acinar or solid architecture, where neoplastic cells tend to be larger and show more remarkable nuclear atypism and polymorphism than the other groups.12 Noteworthily, 12.3% of LADCs in this group are negative for both TTF1 and HNF4α (our original data), suggesting their poor degree of differentiation. Proliferating activity is generally high, with a Ki‐67 labeling index of more than 0.3 (Table 1). This group shows significantly worse clinical courses (Figure 2). Genetically, 18.1% of LADCs have KRAS mutations, and also small proportions have EGFR or ALK mutations (Table 2, Figure 3). However, more than 60% of LADCs in this group have no known driver mutations (Table 2, Figure 3). Thus, due to its heterogeneity, group E might be the terminal of the different pathways. LADCs of this group can be described as “acinar dominant LADC,” “solid dominant LADC,” or “poorly differentiated LADC” in pathological diagnosis reports.
4. TUMOR PROGRESSION IN THE NONSMOKERS’ PATHWAY
A micropapillary element is an essential determinant for the malignant activity in TRU‐type LADCs.16, 17, 22, 23 As mentioned previously, group C has EGFR mutations as frequently as group A; both are TRU‐type LADCs and commonly share a lepidic (or papillary) element. Thus, it is suggested that a micropapillary element could generate from lepidic (or papillary) elements. So far, pathologists have investigated morphological features of micropapillary elements and suggested that a micropapillary element is a structure resulting from tumor cells’ focal stacking to form pseudopapillary projections without fibrovascular stalks.17, 24, 25 To form such structures, neoplastic cells are inevitably anchorage independent, detach from basement membranes, can grow, and simultaneously have to retain epithelial polarity and an intercellular adhesion system partially.13, 23, 24, 25, 26 It is essential to question how lepidic (or papillary) elements acquire the micropapillary phenotype in terms of their molecular basis.14, 23, 24, 25, 27 p53 mutations can be an important factor to trigger this process,28, 29, 30 but there also seem to be additional unique events. Our recent study demonstrated differentially expressed genes in micropapillary elements compared with lepidic elements.31 The results showed some alterations in adhesion molecules, basement membrane materials, and kinases.31 In particular, overexpression of oligo‐glycosylated MUC21 protein 22 and amplification of chromosome 12q13‐21 locus covering CDK4, MDM2, and DYRK2 genes 31 were suggested to participate in producing micropapillary elements. Although there are no other studies that comprehensively compare molecular profiles between micropapillary and background lepidic (or papillary) elements in individual tumors, several studies have shown differences in molecular alterations (genetic mutations) between high‐grade and low‐grade components of LADCs (that were not specified as micropapillary).28 They also demonstrated molecular alterations likely to be involved in producing micropapillary morphology, such as gain of CXCL14, MET, AXIN1, and WNT family proteins 18, 26, 32, 33, 34 (Table 1). The mechanisms behind the molecular alterations and how these alterations produce micropapillary elements are still largely unclear. Uncovering these, as well as carcinogens which promote this process, could lead to the establishment of novel therapeutic and prevention strategies.
5. TUMOR PROGRESSION IN THE SMOKERS’ PATHWAY
The smokers’ pathway appears to be more complex. Poorly differentiated LADCs (group E) can arise from other groups mentioned, not only non‐TRU/CAC–type LADCs (group D) but also TRU‐type LADCs (group A), as group E includes both TTF1‐negative (non‐TRU) and positive (TRU) LADCs (Table 1). Interestingly, in group E, even in TTF1‐positive LADCs, EGFR mutations are rather rare (Table 2 and Figure 3). The frequency of KRAS mutations, that are the most common driver mutations in the smokers’ pathway, is also not high (Table 2 and Figure 3),12, 35 suggesting that there could be another alternative tumor‐driving system.7, 28 We propose a hypothesis that there may be no single strong leading mutation in LADCs in the smoker's pathway. Several tiny mutations each could participate in small quantities, the sum of which could be equivalent to one driver mutation to promote tumor development. In fact, smokers’ LADCs have been shown to accumulate more genetic mutations than nonsmokers’.28, 36 It is of great interest to uncover how such small mutations collectively drive the driver mutation–negative smokers’ LADCs. Concentrating efforts in molecular genetics and bioinformatics will solve this molecular puzzle in the near future.
Apart from driver mutations, regarding tumor progression, p53 mutations are an important trigger factor,29, 30 and disruptions of oncogenic KRAS‐induced negative feedback systems, such as downregulation of DUSP6, miR‐31, and upregulation of S100A11, are common events in the smokers’ pathway.37, 38, 39, 40, 41, 42, 43, 44, 45 Moreover, TTF1 mutations and surfactant protein gene mutations are also reported to be unique to the smokers’ pathway.46, 47
6. EXTRA‐BYPASSING PATHWAYS
Alternatively, there could be extra‐bypassing pathways. Actually, LADCs having both TTF1‐positive lepidic and HNF4α‐positive mucinous components are occasionally seen. Interestingly, Matsubara D et al demonstrated that mucinous LADCs with TTF1‐positive lepidic component more often had TTF1 gene mutations and methylations than pure mucinous LADCs.47 The findings suggest that there could be a bypass from A to D (Figure 1). Also, as mentioned afore, group E might be the terminal of the different pathways. Under exposing to smoking stimuli, different groups might progress to group E (Figure 1).
7. ISSUES IN RECENT LUNG CANCER RESEARCH
Large‐scale genetic analyses have provided us with a large volume of information and are helping to uncover a fuller picture of the molecular basis of LADC. However, more care should be taken regarding the processing of analytical results. For example, it is important to take into consideration if the tumor is definitely negative for driver mutations. Analytical results may be false negatives due to extremely small neoplastic cell contents. In general, LADCs, particularly, poorly differentiated ones, have many leukocytes and fibroblasts, and hence this “noise” can easily result in false negatives. Purification of neoplastic cells from tumor tissues is an important process to improve accuracy of mutational analyses. At present, histological (or cytological) appearance is the most reliable biological marker that detects neoplastic cells. However, using artificial intelligence to detect neoplastic cells and the automatic operation of microdissection systems can improve accuracy.
8. FUTURE PERSPECTIVE
Again, the recent advance in molecular genetics has uncovered a lot of molecular targets for therapeutic agents. However, this progress seems to have been slowing down in recent years. Going forward, we should focus on molecular alterations responsible for tumor progression, which would be targets for next‐generation therapies. For such alterations, we here suggest a term “progressor.” We know intratumor subclonal heterogeneity and passenger mutations (nonfunctional noise) are critical obstacles against successful detection of essential molecular alterations to promote tumor progression.48, 49 Thus, simple large‐scale analyses on bulky tumor samples are not sufficient for the success. As is shown in Figure 4, if we comparatively analyze high‐grade components and background low‐grade components from identical tumors, it alleviates the issue of noise. Microdissection systems can be the most dependable way to reach such targets.
FIGURE 4.
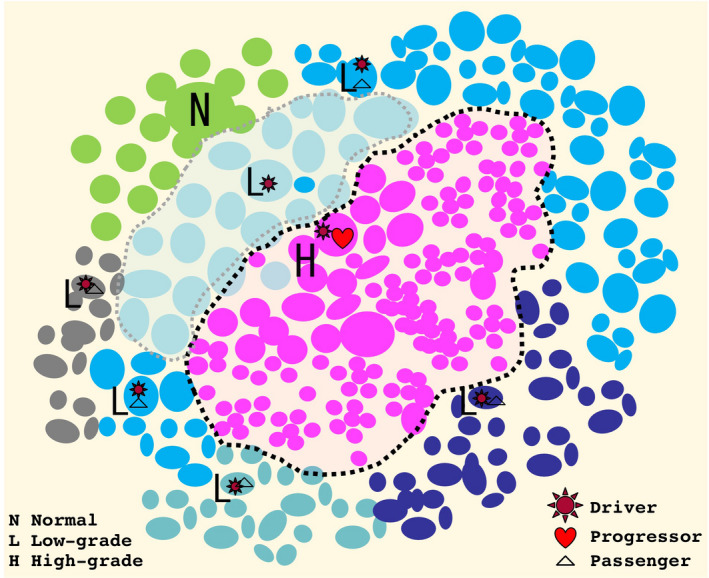
A scheme for progression of lung adenocarcinoma (LADC). During the progression process, one LADC can produce genetically heterogenous subpopulations. At first, driver mutations transform normal counterpart cells (N) to low‐grade neoplastic cells (L). While the neoplastic cells are proliferating, many additional mutations can occur, most of which might be nonfunctional passenger mutations, and some of which can be true progressors to produce high‐grade elements (H)
9. SUMMARY AND CONCLUSION
There are the two major pathways in LADC carcinogenesis. In the nonsmokers’ and smokers’ pathways, the terminals are micropapillary and acinar/solid histology, respectively. LADCs are highly heterogenous even in an identical tumor. Different genetic alterations can participate in each pathway, and identifying essential molecular alterations promoting tumor progression is a critical task. To meet the next goal, focusing on high‐grade elements based on careful histological examinations is necessary.
CONFLICT OF INTEREST STATEMENT
The authors have no conflict of interest.
ACKNOWLEDGEMENTS
We thank Hideaki Mitsui and Takeshisa Suzuki (Department of Pathology, Yokohama City University) and Motoki Sekiya, Misaki Sugiyama, and Emi Honda (Division of Pathology, Kanagawa Prefectural Cardiovascular and Respiratory Center Hospital) for their technical assistance.
Okudela K, Matsumura M, Arai H, Woo T. The nonsmokers’ and smokers’ pathways in lung adenocarcinoma: Histological progression and molecular bases. Cancer Sci. 2021;112:3411–3418. 10.1111/cas.15031
REFERENCES
- 1.Kohno T, Ichikawa H, Totoki Y, et al. KIF5B‐RET fusions in lung adenocarcinoma. Nat Med. 2012;18:375‐377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kosaka T, Yatabe Y, Endoh H, Kuwano H, Takahashi T, Mitsudomi T. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res. 2004;64:8919‐8923. [DOI] [PubMed] [Google Scholar]
- 3.Gainor JF, Shaw AT. Novel targets in non‐small cell lung cancer: ROS1 and RET fusions. Oncologist. 2013;18:865‐875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gainor JF, Varghese AM, Ou SH, et al. ALK rearrangements are mutually exclusive with mutations in EGFR or KRAS: an analysis of 1,683 patients with non‐small cell lung cancer. Clin Cancer Res. 2013;19:4273‐4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lynch TJ. The evolving story of the epidermal growth factor receptor as a target for non‐small‐cell lung cancer. Clin Adv Hematol Oncol. 2004;2:786‐787. [PubMed] [Google Scholar]
- 6.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4‐ALK fusion gene in non‐small‐cell lung cancer. Nature. 2007;448:561‐566. [DOI] [PubMed] [Google Scholar]
- 7.Travis WDNM, Yatabe Y, Brambilla E, Nicholson AG, Aisner SC, Austin JHM. Who classification of tumours of the lun, pleura, thymus and heart, 4th edn. Lyon: IARC Press; 2015. [Google Scholar]
- 8.Brhane Y, Yang P, Christiani DC, et al. Genetic determinants of lung cancer prognosis in never smokers: A Pooled analysis in the international lung cancer consortium. Cancer Epidemiol Biomarkers Prev. 2020;29:1983‐1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yatabe Y. EGFR mutations and the terminal respiratory unit. Cancer Metastasis Rev. 2010;29:23‐36. [DOI] [PubMed] [Google Scholar]
- 10.Yatabe Y, Kosaka T, Takahashi T, Mitsudomi T. EGFR mutation is specific for terminal respiratory unit type adenocarcinoma. Am J Surg Pathol. 2005;29:633‐639. [DOI] [PubMed] [Google Scholar]
- 11.Okudela K, Woo T, Mitsui H, et al. Proposal of an improved histological sub‐typing system for lung adenocarcinoma ‐ significant prognostic values for stage I disease. Int J Clin Exp Pathol. 2010;3:348‐366. [PMC free article] [PubMed] [Google Scholar]
- 12.Okudela K, Woo T, Mitsui H, et al. Morphometric profiling of lung cancers‐its association with clinicopathologic, biologic, and molecular genetic features. Am J Surg Pathol. 2010;34:243‐255. [DOI] [PubMed] [Google Scholar]
- 13.Matsumura M, Okudela K, Kojima Y, et al. A Histopathological Feature of EGFR‐Mutated Lung Adenocarcinomas with Highly Malignant Potential ‐ An Implication of Micropapillary Element. PLoS One. 2016;11:e0166795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monroig‐Bosque PDC, Morales‐Rosado JA, Roden AC, et al. Micropapillary adenocarcinoma of lung: Morphological criteria and diagnostic reproducibility among pulmonary pathologists. Ann Diagn Pathol. 2019;41:43‐50. [DOI] [PubMed] [Google Scholar]
- 15.Li C, Shen Y, Hu F, et al. Micropapillary pattern is associated with the development of brain metastases and the reduction of survival time in EGFR‐mutation lung adenocarcinoma patients with surgery. Lung Cancer. 2020;141:72‐77. [DOI] [PubMed] [Google Scholar]
- 16.Chao L, Yi‐Sheng H, Yu C, et al. Relevance of EGFR mutation with micropapillary pattern according to the novel IASLC/ATS/ERS lung adenocarcinoma classification and correlation with prognosis in Chinese patients. Lung Cancer. 2014;86:164‐169. [DOI] [PubMed] [Google Scholar]
- 17.Saito R, Ninomiya H, Okumura S, Mun M, Sasano H, Ishikawa Y. Novel Histologic Classification of Small Tumor Cell Nests for Lung Adenocarcinoma With Prognostic and Etiological Significance: Small Solid Nests and Pure Micropapillary Nests. Am J Surg Pathol. 2021;45:604‐615. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Wang R, Cai D, et al. A comprehensive investigation of molecular features and prognosis of lung adenocarcinoma with micropapillary component. J Thorac Oncol. 2014;9:1772‐1778. [DOI] [PubMed] [Google Scholar]
- 19.Sugano M, Nagasaka T, Sasaki E, et al. HNF4alpha as a marker for invasive mucinous adenocarcinoma of the lung. Am J Surg Pathol. 2013;37:211‐218. [DOI] [PubMed] [Google Scholar]
- 20.Kojima Y, Okudela K, Matsumura M, et al. The pathological features of idiopathic interstitial pneumonia‐associated pulmonary adenocarcinomas. Histopathology. 2017;70:568‐578. [DOI] [PubMed] [Google Scholar]
- 21.Nakaoku T, Tsuta K, Ichikawa H, et al. Druggable oncogene fusions in invasive mucinous lung adenocarcinoma. Clin Cancer Res. 2014;20:3087‐3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsumura M, Okudela K, Nakashima Y, et al. Specific expression of MUC21 in micropapillary elements of lung adenocarcinomas ‐ Implications for the progression of EGFR‐mutated lung adenocarcinomas. PLoS One. 2019;14:e0215237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Makimoto Y, Nabeshima K, Iwasaki H, et al. Micropapillary pattern: a distinct pathological marker to subclassify tumours with a significantly poor prognosis within small peripheral lung adenocarcinoma (</=20 mm) with mixed bronchioloalveolar and invasive subtypes (Noguchi's type C tumours). Histopathology. 2005;46:677‐684. [DOI] [PubMed] [Google Scholar]
- 24.Fukutomi T, Hayashi Y, Emoto K, Sakamoto M, Kamiya K, Kohno M. Low papillary structures in lepidic lung adenocarcinoma: any relationship with micropapillary lung adenocarcinoma?–reply. Hum Pathol. 2013;44:2867‐2868. [DOI] [PubMed] [Google Scholar]
- 25.Nagano T, Ishii G, Nagai K, et al. Structural and biological properties of a papillary component generating a micropapillary component in lung adenocarcinoma. Lung Cancer. 2010;67:282‐289. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J, Sun J, Zhang Z, et al. Protein overexpression and gene amplification of cellular mesenchymal‐epithelial transition factor is associated with poor prognosis in micropapillary‐predominant subtype pulmonary adenocarcinoma. Hum Pathol. 2018;72:59‐65. [DOI] [PubMed] [Google Scholar]
- 27.Cakir E, Yilmaz A, Demirag F, et al. Prognostic significance of micropapillary pattern in lung adenocarcinoma and expression of apoptosis‐related markers: caspase‐3, bcl‐2, and p53. APMIS. 2011;119:574‐580. [DOI] [PubMed] [Google Scholar]
- 28.Caso R, Sanchez‐Vega F, Tan KS, et al. The Underlying Tumor Genomics of Predominant Histologic Subtypes in Lung Adenocarcinoma. J Thorac Oncol. 2020;15:1844‐1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greathouse KL, White JR, Vargas AJ, et al. Interaction between the microbiome and TP53 in human lung cancer. Genome Biol. 2018;19:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marrogi AJ, Mechanic LE, Welsh JA, et al. TP53 mutation spectrum in lung cancer is not different in women and men. Cancer Epidemiol Biomarkers Prev. 2005;14:1031‐1033. [DOI] [PubMed] [Google Scholar]
- 31.Koike C, Okudela K, Matsumura M, et al. Frequent DYRK2 gene amplification in micropapillary element of lung adenocarcinoma ‐ an implication in progression in EGFR‐mutated lung adenocarcinoma. Histol Histopathol. 2021;36:305–315. [DOI] [PubMed] [Google Scholar]
- 32.Sata Y, Nakajima T, Fukuyo M, et al. High expression of CXCL14 is a biomarker of lung adenocarcinoma with micropapillary pattern. Cancer Sci. 2020;111:2588‐2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao ZR, To KF, Mok TS, Ng CS. Is there significance in identification of non‐predominant micropapillary or solid components in early‐stage lung adenocarcinoma? Interact Cardiovasc Thorac Surg. 2017;24:121‐125. [DOI] [PubMed] [Google Scholar]
- 34.Zhu L, Yang S, Zheng L, Zhang G, Cheng G. WNT/beta‐catenin pathway activation via Wnt1 overexpression and Axin1 downregulation correlates with cadherin‐catenin complex disruption and increased lymph node involvement in micropapillary‐predominant lung adenocarcinoma. J Thorac Dis. 2020;12:5906‐5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woo T, Okudela K, Yazawa T, et al. Prognostic value of KRAS mutations and Ki‐67 expression in stage I lung adenocarcinomas. Lung Cancer. 2009;65:355‐362. [DOI] [PubMed] [Google Scholar]
- 36.Wang X, Ricciuti B, Nguyen T, et al. Association between smoking history and tumor mutation burden in advanced non‐small cell lung cancer. Cancer Res. 2021;81(9):2566‐2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woo T, Okudela K, Mitsui H, et al. Up‐Regulation of S100A11 in Lung Adenocarcinoma ‐ Its Potential Relationship with Cancer Progression. PLoS One. 2015;10:e0142642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okudela K, Katayama A, Woo T, et al. Proteome analysis for downstream targets of oncogenic KRAS–the potential participation of CLIC4 in carcinogenesis in the lung. PLoS One. 2014;9:e87193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okudela K, Mitsui H, Suzuki T, et al. Expression of HDAC9 in lung cancer–potential role in lung carcinogenesis. Int J Clin Exp Pathol. 2014;7:213‐220. [PMC free article] [PubMed] [Google Scholar]
- 40.Okudela K, Tateishi Y, Umeda S, et al. Allelic imbalance in the miR‐31 host gene locus in lung cancer–its potential role in carcinogenesis. PLoS One. 2014;9:e100581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okudela K, Woo T, Kitamura H. KRAS gene mutations in lung cancer: particulars established and issues unresolved. Pathol Int. 2010;60:651‐660. [DOI] [PubMed] [Google Scholar]
- 42.Okudela K, Yazawa T, Ishii J, et al. Down‐regulation of FXYD3 expression in human lung cancers: its mechanism and potential role in carcinogenesis. Am J Pathol. 2009;175:2646‐2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okudela K, Yazawa T, Woo T, et al. Down‐regulation of DUSP6 expression in lung cancer: its mechanism and potential role in carcinogenesis. Am J Pathol. 2009;175:867‐881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okudela K, Mitsui H, Woo T, et al. Alterations in cathepsin L expression in lung cancers. Pathol Int. 2016;66:386‐392. [DOI] [PubMed] [Google Scholar]
- 45.Okudela K, Mitsui H, Woo T, et al. Expression of tropomyosins in lung cancer ‐ a potential role in carcinogenesis and its utility in a histopathological diagnosis. Histol Histopathol. 2016;31:857‐866. [DOI] [PubMed] [Google Scholar]
- 46.Honda T, Sakashita H, Masai K, et al. Deleterious Pulmonary Surfactant System Gene Mutations in Lung Adenocarcinomas Associated With Usual Interstitial Pneumonia. JCO Precision. Oncology. 2018;2:1‐24. [DOI] [PubMed] [Google Scholar]
- 47.Matsubara D, Soda M, Yoshimoto T, et al. Inactivating mutations and hypermethylation of the NKX2‐1/TTF‐1 gene in non‐terminal respiratory unit‐type lung adenocarcinomas. Cancer Sci. 2017;108:1888‐1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jamal‐Hanjani M, Wilson GA, McGranahan N, et al. Tracking the Evolution of Non‐Small‐Cell Lung Cancer. N Engl J Med. 2017;376:2109‐2121. [DOI] [PubMed] [Google Scholar]
- 49.Kumar S, Warrell J, Li S, et al. Passenger Mutations in More Than 2,500 Cancer Genomes: Overall Molecular Functional Impact and Consequences. Cell. 2020;180(5):915‐927.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Amin MB, Gress DM, Vega LM, et al. AJCC Cancer staging manual, Eighth Edition, 8th edn. New York, NY: Springer International; 2018. [Google Scholar]


