Abstract
The rBC2LCN lectin, known as a stem cell marker probe that binds to an H type 3 fucosylated trisaccharide motif, was recently revealed to also bind to pancreatic ductal adenocarcinoma (PDAC) cells. A lectin‐drug conjugate was generated by fusing rBC2LCN with a cytocidal toxin, and it showed a strong anticancer effect in in vitro and in vivo PDAC models. However, it is unclear which molecules are carrier proteins of rBC2LCN on PDAC cells. In this study, we identified a rBC2LCN‐positive glycoprotein expressed in PDAC. Tumor lysates of PDAC patient‐derived xenografts (PDXs) were coprecipitated with rBC2LCN lectin and analyzed by liquid chromatography–mass spectrometry. A total of 343 proteins were initially identified. We used a web‐based database to select five glycoproteins and independently evaluated their expression in PDAC by immunohistochemistry (IHC). Among them, we focused on carcinoembryonic antigen 5 (CEA) as the most cancer‐specific carrier protein in PDAC, as it showed the most prominent difference in expression rate between PDAC cells (74%) and normal pancreatic duct cells (0%, P > .0001). rBC2LCN lectin and CEA colocalization in PDAC samples was confirmed by double‐staining analysis. Furthermore, rBC2LCN‐precipitated fractions were blotted with an anti‐CEA polyclonal antibody (pAb), and CEA pAb–precipitated fractions were blotted with rBC2LCN lectin. The results demonstrate that CEA is in fact a ligand of rBC2LCN lectin.
Keywords: CEA, glycoprotein, lectin, pancreatic ductal adenocarcinoma, rBC2LCN
This study reports that carcinoembryonic antigen 5 (CEA) is a cancer‐specific rBC2LCN‐positive glycoprotein expressed in pancreatic ductal adenocarcinoma (PDAC). The rBC2LCN lectin, reported as a stem cell marker probe, has been detected in some cancers, and a glycan epitope of rBC2LCN, H type 3 fucosylated motif (Fucα1‐2Galβ1‐3GalNAc), could be a therapeutic target for cancers. This study showed for the first time that CEA is modified with α1‐2 fucose.

Abbreviations
- ABC
avidin‐biotin complex
- CEA
carcinoembryonic antigen 5
- GLUT1
glucose transporter type 1
- hPSCs
human pluripotent stem cells
- HRP
horseradish peroxidase
- IHC
immunohistochemistry
- iPSCs
induced pluripotent stem cells
- ITGB1
integrin beta 1
- LDC
lectin‐drug conjugate
- mAb
monoclonal antibody
- MUC16
mucin 16
- MW
molecular weight
- pAb
polyclonal antibody
- PBS
phosphate buffer saline
- PDAC
pancreatic ductal adenocarcinoma
- PDX
patient‐derived xenograft
- PSCA
prostate stem cell antigen
1. INTRODUCTION
Despite numerous attempts to disclose the weaknesses of pancreatic ductal adenocarcinoma (PDAC), useful and definitive targets have not been elucidated to date.1 In the fight against PDAC, we have focused on cell surface glycans because all cells are covered with a glycan layer referred to as the glycocalyx.2 In addition, glycans are known to be altered in various states of cancer development and progression, and glycosylation is a hallmark of malignant transformation.3
We have previously revealed that rBC2LCN lectin strongly binds to PDAC cells 4 and its fucosylated trisaccharide epitope, H type 3 (Fucα1‐2Galβ1‐3GalNAc), is expressed on PDAC cells.5 We do not yet know which molecules are the carrier proteins of this glycan epitope, but it could be a therapeutic target for PDAC. In fact, a lectin‐drug conjugate (LDC) generated by fusing rBC2LCN with the 38‐kDa domain of Pseudomonas aeruginosa exotoxin A showed a strong anticancer effect in in vitro and in vivo PDAC models.
rBC2LCN lectin was initially identified as a stem cell marker for induced pluripotent stem cells (iPSCs) that binds to the H type 3 glycan motif; it is predominantly expressed on cells possessing pluripotent capacities.6, 7 When this lectin was conjugated with cytocidal toxin and added to cocultures of undifferentiated pluripotent cells and differentiated somatic cells, only undifferentiated human pluripotent stem cells (hPSCs) were eliminated.8 In the context of cancer cell research, rBC2LCN lectin–reacting cells have been detected in some cancers, and rBC2LCN is suggested to be associated with cancer stem–like features, such as slow proliferation, increased cell motility, and drug resistance.4, 9 In fact, the number of reports of rBC2LCN lectin in cancers, such as PDAC and colon, breast, and prostate cancers, has recently increased.9, 10, 11 However, the molecular mechanism of rBC2LCN lectin in cancer cells and more specifically the molecules that act as carrier proteins decorated with the H type 3 glycan motif in cancers have not been elucidated. In addition, in terms of cancer progression, rBC2LCN lectin–positive cells may retain cancer stemness features and thus could be therapeutic targets for cancers.
In this study, we aimed to identify glycoprotein ligands of rBC2LCN expressed in PDAC. We used the cell lysates of cell lines, patient‐derived xenografts (PDXs), and patient‐derived tissues of PDAC to identify rBC2LCN‐positive glycoproteins, and we successfully identified carcinoembryonic antigen 5 (CEA) as one of the major ligands of rBC2LCN lectin.
2. MATERIALS AND METHODS
2.1. Clinical samples and PDX samples in mice
Tumor and normal tissues from patients were obtained from Tsukuba University Hospital. PDAC PDX models were produced as described previously.4 All tumor specimens from PDAC patients were minced into 2‐mm fragments, and three to four pieces were implanted into subcutaneous pockets on the bellies of CB17/Icr‐scid/SCID mice (female, 6‐8 weeks old, CLEA Japan). All animal experiments were conducted in accordance with ethical regulations approved by the Animal Care Committee University Health Network, University of Tsukuba, Tsukuba, Ibaraki, Japan, and the mice were maintained according to the animal use guidelines of the University of Tsukuba. For the use of clinical samples for this research, written informed consent was obtained from all patients, and approval was obtained from the Institutional Review Board of the University of Tsukuba Hospital (IRV code: H28‐90).
2.2. Cell lines and cell culture
The human pancreatic cancer cell line Capan‐1 was purchased from the American Type Tissue Culture Collection (ATCC), and SUIT‐2 cells were obtained from the Japanese Collection of Research Bioresources Cell Bank. The culture media and conditions were used as previously described.4 Capan‐1 cells were cultured in Iscove's modified Dulbecco's medium (FUJIFILM Wako Pure Chemical Co) supplemented with 20% fetal bovine serum (Thermo Fisher Scientific) and 1% penicillin‐streptomycin (FUJIFILM Wako Pure Chemical Co). SUIT‐2 cells were cultured in Dulbecco's modified Eagle's medium (FUJIFILM Wako Pure Chemical Co) supplemented with 10% FBS and 1% penicillin‐streptomycin (FUJIFILM Wako Pure Chemical Co)
2.3. Flow cytometry
The cells were incubated with rBC2LCN‐635 (FUJIFILM Wako Pure Chemical Co) and FITC‐labeled anti‐CEA monoclonal antibody (mAb; Cat No. ab106739, Abcam) for 30 minutes at room temperature. After washing twice, the cells were analyzed with a FACSJazz cytometer (BD) and FlowJo v. 7.6.1 (FlowJo LLC).
2.4. Immunofluorescence staining
Cells were seeded and cultured on 35‐mm glass‐bottom dishes. After the cells reached 50%‐70% confluency, they were washed with phosphate buffer saline (PBS) and fixed for 10 minutes with paraformaldehyde (FUJIFILM Wako Pure Chemical Co). After blocking with bovine serum albumin at room temperature for 60 minutes, the cells were incubated overnight at 4°C with anti‐CEA mAb (GTX17254, GeneTex). Following incubation with biotinylated secondary polyclonal antibody (pAb) for 10 minutes, streptavidin‐Alexa 594 (s32256, Invitrogen) was added for 30 minutes. After washing with PBS, FITC‐labeled rBC2LCN was added for 60 minutes at room temperature. Nuclear staining was performed using 4′,6‐diamidino‐2‐phenylindole (Dojindo Molecular Technologies Inc). Images of stained cells were acquired by a BZX‐710 microscope (Keyence Corporation).
2.5. Immunohistochemistry (IHC) and double‐staining analysis
Formalin‐fixed, paraffin‐embedded tissues were sliced into 3‐μm slide sections and deparaffinized. Antigen retrieval was performed by autoclaving sections immersed in 10 mmol/L citrate‐Na buffer. The tissue sections were then treated with 3% hydrogen peroxidase in methanol for 15 minutes to inactivate endogenous peroxidase activity. Following blocking with normal serum, the tissues were incubated with the following primary antibodies for 1 hour at room temperature: anti‐CEA mAb (1:50, GTX17254, GeneTex), anti‐ITGB1 mAb (1:50, Cat No. 9699, Cell Signaling Technology), anti‐MUC16 pAb (1:500, Cat No. 20077‐1‐AP, Proteintech), anti‐PSCA mAb (4 μg/mL, Cat No. MAB7636, R&D Systems), and anti‐GLUT1 pAb (1:50, Cat No. ab14683, Abcam). IHC was conducted using the avidin‐biotin complex (ABC) technique with a SAB‐PO kit (Nichirei Bioscience) according to the manufacturers' specifications. Tissue sections were visualized by 3,3′‐diaminobenzidine (Nichirei Bioscience) and counterstained with hematoxylin. Images were captured with a BZ‐X710 microscope (Keyence Co).
Tissue staining was then evaluated by two investigators independently in a blinded manner.12, 13 Tissue staining was scored according to the proportion of positive cells as follows: 0, 0%; 1, <25%; 2, 25%‐50%; 3, 51%‐75%; and 4, 76%‐100%. The staining intensity was scored as follows: 0, none; 1, weak; 2, moderate; and 3, strong. The total staining score was calculated by multiplying the above two subscores. Samples with scores of 0‐3 were considered to have low expression, and those with scores ≥4 were considered to have high expression. Normal pancreatic tissues were divided into low‐ or high‐expression groups based on the intensity of staining in the pancreatic duct.
For double‐staining with rBC2LCN and primary antibody, the sections were visualized by streptavidin‐594 (Invitrogen) and FITC‐labeled rBC2LCN. Fluorescence images were obtained using a BZ‐X710 microscope (Keyence Co).
2.6. Liquid chromatography–mass spectrometry (LC‐MS/MS) analysis
Two different PDX lines (PC3 and PC42) were analyzed by LC/MS/MS analysis. Proteins were extracted from tumor tissues by incubation with 1% Triton X‐100 on ice for 1 hour. The lysates (320 mg) were incubated with biotinylated rBC2LCN immobilized on Dynabeads M‐280 Streptavidin (VERITAS Corporation). Proteins precipitated with rBC2LCN were separated by 12.5% SDS‐PAGE (DRC) and silver stained. Whole protein bands were cut out from the SDS‐PAGE gel following silver staining. The gel fragments were incubated with trypsin at 37°C for 16 hours. After the peptides were extracted from the gel, the peptide solution was dried and subjected to LC‐MS/MS. LC‐MS/MS analysis was performed using a Paradigm MS4 liquid chromatograph (Michrom Bioresources) and an LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific). The sequence data set for the search was constructed using protein entries from the human (Homo sapiens) output from Swiss‐Prot version 2017_08 (20 192 total cases; Swiss Institute of Bioinformatics; European Bioinformatics Institute) and porcine (Sus scrofa) trypsin amino acid sequences. The sequence data were queried using Mascot (ver. 2.5; Matrix Science), and the search results were entered into the Scaffold browsing software (Proteome Software).
The localization of each candidate protein in cells was confirmed with the UniProtKB database. N‐ and O‐glycosylation sites were predicted using NetNGlyc 1.0 (http://www.cbs.dtu.dk/services/NetNGlyc/) and NetOGlyc 4.0 (http://www.cbs.dtu.dk/services/NetOGlyc/). The tissue expression of the proteins was analyzed with The Human Protein Atlas (https://www.proteinatlas.org/).
2.7. Precipitation with rBC2LCN or anti‐CEA pAb
The hydrophobic fractions were prepared using the CelLytic MEM Protein Extraction Kit (Sigma‐Aldrich) in accordance with the manufacturer's procedures. Protein concentrations were measured with the Micro BCA Protein Assay Kit (Thermo Fisher Scientific). Biotinylated rBC2LCN or biotinylated anti‐CEA pAb (Cat No. PA5‐16665, Thermo Fisher Scientific) immobilized on Dynabeads M‐280 Streptavidin (10 µL) was incubated with the hydrophilic fractions (10 µg) at 4°C overnight with agitation. After the beads were washed four times with PBS containing 1% Triton X‐100, the bound proteins were eluted with 20 µL of tris buffer saline containing 0.2% SDS at 95°C for 5 minutes.
2.8. Western and lectin blot analysis
One microgram of the extracted proteins or 4 µL of the samples precipitated with rBC2LCN or anti‐CEA pAb described above were loaded for electrophoretic separation on a 5%‐20% polyacrylamide gel (DRC). The separated proteins were transferred to a PVDF membrane. For Western blot analysis, the membranes were blocked with Block Ace (Funakoshi). The membranes were incubated with horseradish peroxidase (HRP)‐conjugated rBC2LCN (0.1 mg/mL) or anti‐CEA pAb (1:200, Cat No. PA5‐16665, Thermo Fisher Scientific) followed by HRP‐conjugated goat anti‐rabbit IgG (1:5000, Cat No. 111‐035‐003, Jackson ImmunoResearch). Silver staining of the gel was also performed using a silver staining MS kit (Wako Pure Chemical Co) according to the manufacturer's instructions. HRP labeling was performed using a peroxidase labeling kit (Dojindo Molecular Technologies Inc).
2.9. Statistical analysis
The data were analyzed using GraphPad Prism 6.0. The positive ratio of PDAC relative to normal tissues in IHC was analyzed using Fisher's exact test. The P‐values are expressed as follows: *P < .05 and ***P < .0001.
3. RESULTS
3.1. Identification of rBC2LCN‐positive glycoproteins expressed in PDAC by LC‐MS/MS analysis
To identify rBC2LCN‐positive glycoproteins expressed in PDAC, the lysates of tumor tissues from two PDX mouse models were incubated with rBC2LCN‐immobilized beads, and the bound proteins were eluted and analyzed by LC‐MS/MS. rBC2LCN‐positive glycoproteins expressed in PDAC were selected via the following strategy (Figure S1). Among the 343 proteins identified, membrane and glycoproteins were selected using the UniProtKb database, and 12 membrane glycoproteins with N‐ and/or O‐glycosylation sites were identified with the NetNGlyc 1.0 server and NetOGlyc 4.0 server (Table 1). We assessed the expression levels of the proteins in the normal pancreas and PDAC by employing the Human Protein Atlas. Based on these findings, five membrane glycoproteins that are expressed in PDAC tissues but minimally expressed in normal pancreatic tissues were selected: CEA, integrin beta 1 (ITGB1), mucin 16 (MUC16), prostate stem cell antigen (PSCA), and glucose transporter type 1 (GLUT1).
TABLE 1.
rBC2LCN‐precipitated membrane glycoproteins in pancreatic ductal adenocarcinoma (PDAC) identified by LC‐MS/MS analysis
| No. | Protein name | Gene | Unique peptides | N‐glycosylation sitesa | O‐glycosylation sitesb |
|---|---|---|---|---|---|
| 1 | Integrin beta 1 | ITGB1 | 7 | 12 | 13 |
| 2 | Lysosome‐associated membrane glycoprotein 1 | LAMP1 | 3 | 18 | 7 |
| 3 | Prostate stem cell antigen | PSCA | 3 | 3 | 0 |
| 4 | 4F2 cell surface antigen heavy chain | SLC3A2 | 3 | 4 | 1 |
| 5 | Spectrin beta chain, nonerythrocytic 1 | SPTBN1 | 3 | 6 | 134 |
| 6 | CD44 antigen | CD44 | 2 | 11 | 145 |
| 7 | CD63 antigen | CD63 | 2 | 3 | 3 |
| 8 | Carcinoembryonic antigen‐related cell adhesion molecule 5 | CEA/CEACAM5 | 2 | 29 | 2 |
| 9 | Carcinoembryonic antigen‐related cell adhesion molecule 6 | CEACAM6 | 2 | 12 | 2 |
| 10 | Glucose transporter type 1 | GLUT1 | 2 | 2 | 1 |
| 11 | Transferrin receptor protein 1 | TFRC | 2 | 5 | 11 |
| 12 | Mucin 16 | MUC16 | 2 | 117 | 4876 |
The number of N‐glycosylation sites was predicted by the NetNGlyc 1.0 server (http://www.cbs.dtu.dk/services/NetNGlyc/).
The number of O‐glycosylation sites was predicted by the NetOGlyc 4.0 server (http://www.cbs.dtu.dk/services/NetOGlyc/).
3.2. Immunohistochemical analysis of five membrane glycoproteins in PDAC
We then performed IHC for CEA, ITGB1, MUC16, PSCA, and GLUT15 in PDAC tissues (n = 19) and normal pancreatic tissues (n = 22; Figure 1). Antibodies against the five glycoproteins showed stronger staining in PDAC tissues than in normal pancreatic duct tissues. The percentage of samples with high expression of CEA, MUC16, and GLUT1 was significantly higher among PDAC samples than among normal pancreatic samples (CEA [tumor vs normal, 74% vs 0%, P < .0001], MUC16 [53% vs 22%, P = .0262], GLUT1 [63% vs 0%, P < .0001]), whereas no significant difference was observed for ITGB1 (48% vs 23%, P = .1147) or PSCA (37% vs 9%, P = .0570). Among the proteins assessed, CEA was highly expressed in the largest proportion of PDAC samples, so we focused on CEA for further analysis in this study. CEA consists of 702 amino acids and has a theoretical molecular weight (MW) of 76.8 kDa.14, 15 As CEA is a heavily glycosylated protein containing 29 N‐ and 2 O‐glycosylation sites, its apparent MW is approximately 180 kDa.16, 17
FIGURE 1.
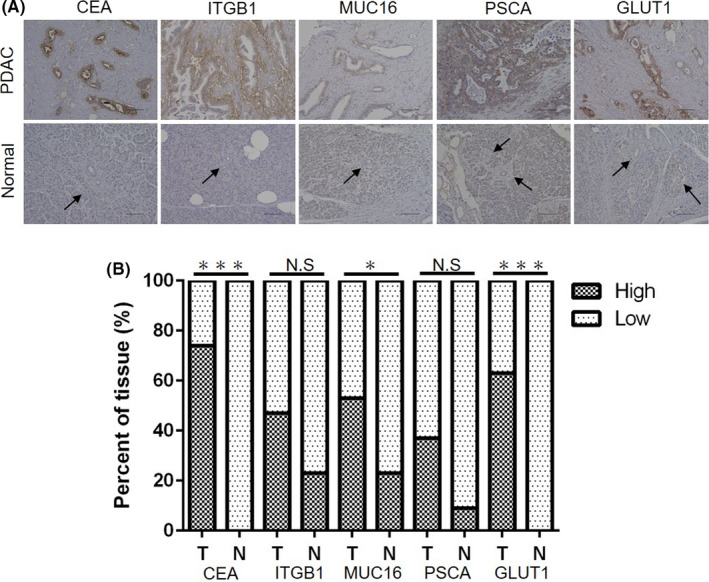
Immunohistochemistry analysis of five membrane glycoproteins (carcinoembryonic antigen 5 [CEA], integrin beta 1 [ITGB1], mucin 16 [MUC16], prostate stem cell antigen [PSCA], glucose transporter type 1 [GLUT1]) identified by LC‐MS/MS analysis of rBC2LCN‐precipitated fractions in pancreatic ductal adenocarcinoma (PDAC) tissues (n = 19) and normal pancreatic tissues (n = 22). A, Representative images of immunohistochemical staining of five membrane glycoproteins (CEA, ITGB1, MUC16, PSCA, GLUT1) in PDAC tissues and normal pancreatic tissues. Black arrow: pancreatic duct. Scale bars, 100 μm. B, The percentage of cells with high expression of each membrane glycoprotein was quantified in PDAC tissues (T; n = 19) and normal pancreatic tissues (N; n = 22). NS: not significant, *P < .05, ***P < .0001
3.3. Carcinoembryonic antigen 5 is a glycoprotein ligand of rBC2LCN in a pancreatic cancer cell line
We first evaluated whether CEA is a glycoprotein ligand of rBC2LCN in pancreatic cancer cell lines. rBC2LCN has been reported to bind to Capan‐1 cells but not to SUIT‐2 cells.4 Consistently, rBC2LCN and anti‐CEA mAb were visualized by fluorescence staining in Capan‐1 (Figure 2A). rBC2LCN bound the cell surface of Capan‐1 cells, whereas anti‐CEA mAb stained the cell surface and the cytoplasm. rBC2LCN ligands were partially colocalized with CEA. In contrast, SUIT‐2 cells were negative for both rBC2LCN and anti‐CEA mAb staining. In the flow cytometry analysis, rBC2LCN and anti‐CEA mAb were positively correlated in Capan‐1 cells (Figure 2B). Again, SUIT‐2 cells were not positive for rBC2LCN nor anti‐CEA mAb. In terms of lectin blotting, rBC2LCN showed smeared bands ranging from 37 to 250 kDa in Capan‐1 cells, but no signal was observed for SUIT‐2 cells. The anti‐CEA pAb showed protein bands at 75 and 160 kDa in Capan‐1 cells but not in SUIT‐2 cells (Figure 2C). After precipitation with rBC2LCN, 75‐ and 160‐kDa protein bands were visualized with anti‐CEA pAb in Capan‐1 cells but not in SUIT‐2 cells (Figure 2D). As the apparent MW of CEA is known to be ~180 kDa,16, 17 the 160‐kDa protein band might correspond to glycosylated CEA. Indeed, the anti‐CEA mAb showed a specific band at 160 kDa in tumor tissue lysates from the PDAC PDX models (Figure S2), indicating that the 160‐kDa protein band corresponds to glycosylated CEA. The 75‐kDa protein band might be caused by the cross‐reactivity of the anti‐CEA pAb with other CEA isoforms.18 These results demonstrate that CEA is a glycoprotein ligand of rBC2LCN in Capan‐1 cells.
FIGURE 2.
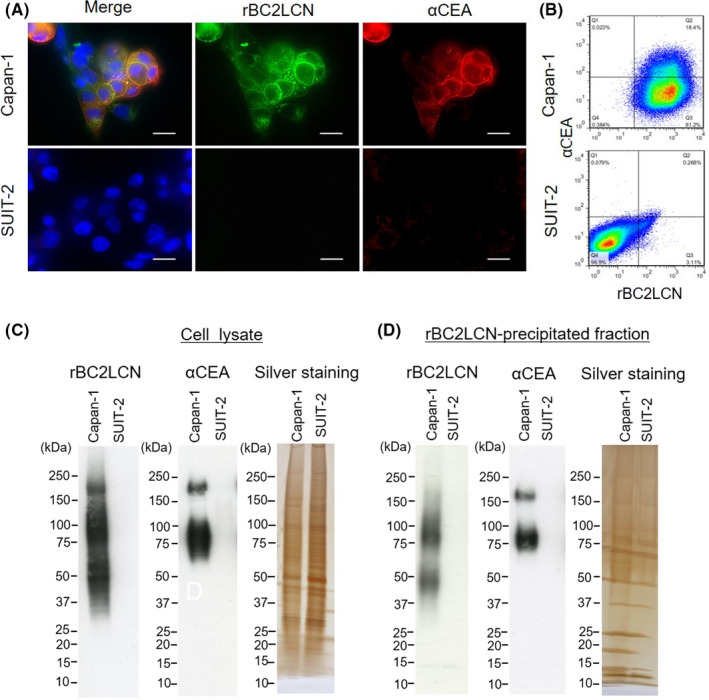
Carcinoembryonic antigen 5 (CEA) is a glycoprotein ligand of rBC2LCN in a pancreatic cancer cell line. A, Immunofluorescence staining of rBC2LCN (green), CEA (red), and nuclei (blue) in Capan‐1 and SUIT‐2 cells. Merge: yellow. Original magnification: ×1000. Scale bars, 50 μm. B, Flow cytometry analysis of CEA and rBC2LCN ligands in Capan‐1 and SUIT‐2 cells. C, Lectin and Western blotting of cell lysates for rBC2LCN and CEA and silver staining of cell lysate. D, rBC2LCN‐precipitated fractions were blotted with rBC2LCN or anti‐CEA polyclonal antibody (pAb) (αCEA) and silver stained
3.4. Carcinoembryonic antigen 5 is a glycoprotein ligand of rBC2LCN in tumor tissues from the PDAC PDX model mice
We then evaluated whether CEA is a glycoprotein ligand of rBC2LCN in tumors from the PDAC PDX model mice. The tumor tissues of three different PDX model mice were stained with rBC2LCN and anti‐CEA mAb (Figure 3A and Table S1). Both rBC2LCN and anti‐CEA mAb were positively stained in tumor tissues derived from all PDX model mice, in agreement with previous reports.4, 19 Interestingly, the fluorescence intensity of rBC2LCN correlated with that of the anti‐CEA mAb: Both rBC2LCN and the anti‐CEA mAb showed stronger staining in PDX2 and PDX3 than in PDX1. rBC2LCN ligands were partially colocalized with CEA in epithelial cells of duct structure. In the lectin blotting experiment, rBC2LCN showed strong smeared bands in the tumor lysates of all PDX model mice, indicating that many rBC2LCN‐positive glycoproteins are expressed in tumors (Figure 3B, left panel). The anti‐CEA pAb showed reactivity to the 160‐kDa protein band as well as the 50‐100–kDa protein bands (Figure 3B, right panel). After lectin precipitation with rBC2LCN, rBC2LCN showed smeared bands at 50‐250 kDa (Figure 3C, left panel), whereas the anti‐CEA pAb showed reactivity to the 160‐kDa protein band as well as the 50‐150–kDa protein bands (Figure 3C, middle panel). After immunoprecipitation with the anti‐CEA pAb, both rBC2LCN and the anti‐CEA pAb showed reactivity to the 160‐kDa protein band corresponding to CEA as well as the 50‐150–kDa protein bands. The MW of rBC2LCN‐positive CEA (~160 kDa) was slightly different among the different PDX model mice. This might be due to the different degrees of glycosylation among the distinct PDX model mice. These results demonstrate that CEA is a glycoprotein ligand of rBC2LCN in PDAC PDX model mice.
FIGURE 3.
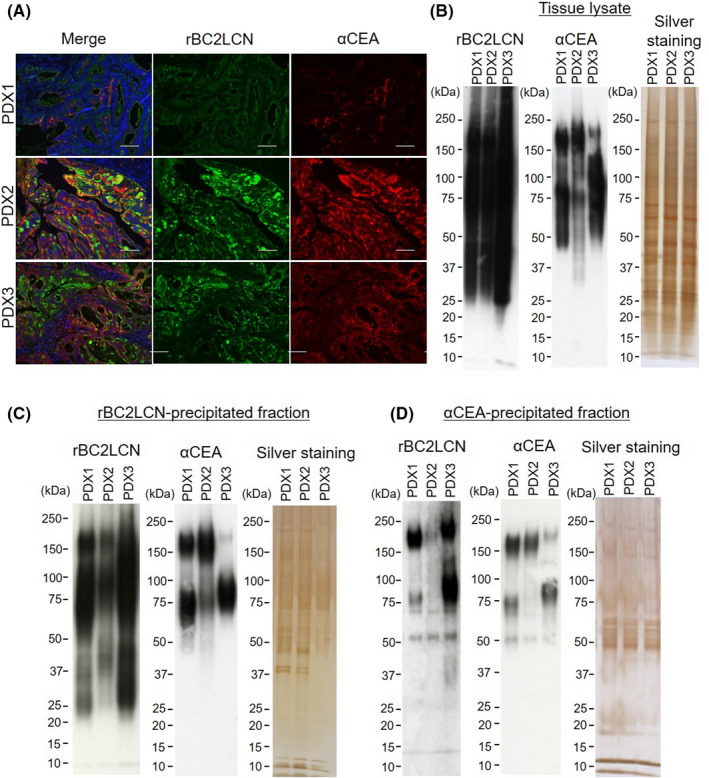
Carcinoembryonic antigen 5 (CEA) is a glycoprotein ligand of rBC2LCN in pancreatic ductal adenocarcinoma (PDAC) patient‐derived xenograft (PDX) models. A, PDAC PDX tissues were stained with rBC2LCN (green) and anti‐CEA monoclonal antibody (mAb) (red). Nuclei: blue. Merge: yellow. Scale bars, 100 μm. B, Lectin and Western blotting of tumor lysates of PDAC PDX model mice with rBC2LCN and anti‐CEA polyclonal antibody (pAb) (n = 3) and silver staining. C, rBC2LCN‐precipitated fractions were blotted with rBC2LCN or anti‐CEA pAb and silver stained. D, Anti‐CEA‐precipitated fractions were blotted with rBC2LCN and anti‐CEA pAb and silver stained
3.5. Carcinoembryonic antigen 5 is a glycoprotein ligand of rBC2LCN in clinical PDAC tissues
Finally, we evaluated whether CEA is a glycoprotein ligand of rBC2LCN in clinical PDAC tissues. Significant colocalization of rBC2LCN and the anti‐CEA mAb was observed in ductal epithelial tissues (Figure 4A). Figure S4 shows the results of staining for all cases. Further validation was performed via lectin and Western blotting with six PDAC tissues and three normal pancreatic tissues. rBC2LCN and anti‐CEA pAb showed stronger intensity in PDAC tissue lysates than in normal pancreatic tissue lysates. A protein band at 160 kDa was detected in PDAC by both rBC2LCN and the anti‐CEA pAb (Figure 4B). rBC2LCN‐precipitated proteins were then blotted with the anti‐CEA pAb (Figure 4C). An ~160‐kDa protein band corresponding to CEA was detected in PDAC tissues but not in normal pancreatic tissues.
FIGURE 4.
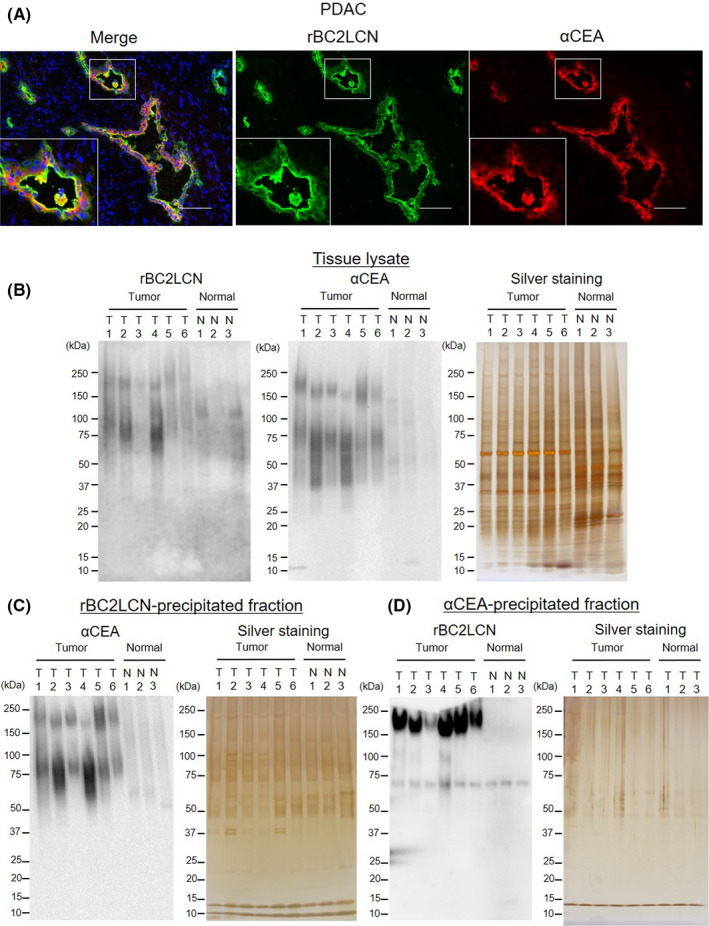
Carcinoembryonic antigen 5 (CEA) is a glycoprotein ligand of rBC2LCN in clinical pancreatic ductal adenocarcinoma (PDAC) samples. A, Representative immunofluorescence staining of rBC2LCN (green) and CEA (red) in clinical PDAC tissues. Nuclei: blue. Merge: yellow. Scale bars, 100 μm. B, Lectin blot and Western blot analysis of rBC2LCN and CEA in PDAC tissues (n = 6) and normal pancreatic tissues (n = 3) and silver staining. C, rBC2LCN‐precipitated fractions were blotted with anti‐CEA polyclonal antibody (pAb) and silver stained. D, αCEA‐precipitated fractions were blotted with rBC2LCN and silver stained
In addition, lectin blotting of the αCEA‐precipitated samples also showed an ~160‐kDa protein band (Figure 4D). These results demonstrate that CEA is a glycoprotein ligand of rBC2LCN in PDAC. rBC2LCN‐positive CEA is specifically expressed in PDAC tumors but not normal tissues.
To validate other normal tissues, we performed lectin blotting for the αCEA‐precipitated fractions of normal colon tissues (Figure S5). According to the findings of previous studies, CEA is expressed in normal colon tissues. However, the reactivity of the CEA‐precipitated fraction was lower than that of pancreatic cancer fraction. These results suggest that targeting rBC2LCN‐positive CEA may improve tumor specificity in other organs as well.
4. DISCUSSION
As expected, PDAC showed strong affinity to the rBC2LCN lectin, which was previously revealed to bind iPSCs with pluripotent differentiation capacities. Although podocalyxin was identified as one of the main ligands of rBC2LCN on the iPSC membrane,7 its ligands on PDAC cells have not been identified. We therefore aimed to identify the cancer‐specific glycoprotein ligands of rBC2LCN. Using rBC2LCN‐precipitated cell lysates from PDX model mice as a starting source, our initial LC‐MS/MS screen identified 343 proteins. We utilized a web‐based database to narrow the list of candidates down to 12 glycoproteins. We finally focused on CEA as one of the major ligands of the rBC2LCN lectin on PDAC cells.
It should be discussed whether our selected glycans, including CEA, which was identified as the most prominent glycoprotein, are meaningful glycoproteins expressed on the PDAC cell surface that could become therapeutic targets and/or diagnostic markers. The cell lysates of PDXs included various noise proteins other than our desired rBC2LCN ligands. Some proteins were considered to originate from noncancerous mouse cells, including fibroblasts, endothelial cells and/or immune cells, while the others were considered to be cytoplasmic proteins of human PDAC cells. Using a web‐based database, we eliminated meaningless proteins, and 12 glycoproteins were identified as rBC2LCN‐positive glycoproteins on PDAC. These 12 proteins should all be glycoproteins on PDAC cells that are capable of binding to the rBC2LCN lectin because they include N‐ or O‐glycosylated sites (Table 1). The 12 selected glycoproteins included cell surface receptors (TFRC, CD44), transporters (SLC3A2, GLUT1), LAMP family proteins (LAMP1, CD63), CEA family proteins (CEACAM5 and CEACAM6), mucin family proteins (MUC16) and others (ITGB1, PSCA, SPTNB1). Our analysis assumed that rBC2LCN lectin binding ligands (a) would be strongly expressed on PDAC cells and (b) should not bind to noncancerous cells because we would like to use this ligand as a therapeutic target and/or diagnostic marker. Based on The Human Protein Atlas database, we further narrowed our 12 protein candidates down to five glycoproteins (CEA, ITGB1, MUC16, PSCA, GLUT1) with good signal/noise ratios; these glycoproteins showed strong expression in PDAC tissues and minimal expression in normal pancreatic tissues. These features were independently further confirmed by IHC analysis, which also showed that the positive rates for the five membrane glycoproteins were higher in PDAC tissues than in normal tissues (Figure 1). Among them, we focused on CEA for further validation because CEA showed the highest positive rate for PDAC tissues and no positivity for normal pancreatic tissues (Figure 1).
Carcinoembryonic antigen 5 is one of the most popular cancer markers and is used in the assessment of many solid cancers, including PDAC.20 In gastric and colorectal cancers, glycans on CEA have been reported to change depending on the degree of malignancy.21, 22 However, little is known regarding glycans on CEA in PDAC. IHC staining of PDAC samples demonstrated that CEA colocalized with rBC2LCN ligands, and lectin precipitation and Western blotting demonstrated that CEA was bound by rBC2LCN (Figures 2, 3, 4). Therefore, we concluded that CEA is a glycoprotein ligand of rBC2LCN in PDAC. In addition, the lectin and Western blotting results for clinical samples showed that protein bands at 75‐100 kDa and 160 kDa were observed in PDAC with rBC2LCN, while no 160‐kDa bands were observed in normal tissues (Figure 4B). As a protein band was detected at 160 kDa for CEA in PDAC, these results also suggest that CEA is a cancer‐specific ligand of rBC2LCN.
In terms of the result for rBC2LCN lectin, which was the focus of this study, lectin staining with rBC2LCN was positive in a pancreatic cancer cell line (Capan‐1), PDAC PDX lysates, and clinical samples, as previously reported.4 Lectin blotting with rBC2LCN also showed a strong affinity for all PDAC samples. Clinical specimens showed stronger reactivity for PDAC samples than normal pancreatic tissue samples (Figure 4B). These results reaffirmed that the glycan epitopes of rBC2LCN have potential as diagnostic and therapeutic targets for PDAC.
We previously reported that a glycan epitope of rBC2LCN, H type 3 (Fucα1‐2Galβ1‐3GalNAc), is detected exclusively in O‐glycans but not N‐glycans on podocalyxin in iPSCs.7 CEA is a glycoprotein consisting of ~60% carbohydrate and has 29 N‐glycosylated sites and two O‐glycosylated sites according to the predictions of the NetNGlyc 1.0 server and NetOGlyc 4.0 server (Table 1). Recently, we used HPLC and matrix‐assisted laser desorption/ionization time‐of‐flight (MALDI‐TOF) MS to demonstrate that a glycan epitope of rBC2LCN, H type 3, is expressed in the tumors of PDAC PDX model mice.5 Therefore, CEA expressed in PDAC might be modified with the H type 3 O‐glycan because CEA has two O‐glycosylation sites.
The glycan epitope of rBC2LCN, Fucα1‐2Galβ1‐3GalNAc, is expressed on PDAC. The α1‐2 fucose is transferred by fucosyltransferases 1 and 2.6, 23 There are various reports about the functions of α1‐2 fucosylation in cancer progression.24 In melanoma, oral/head and neck, gastric, and hepatocellular carcinomas, α1‐2 fucosylation is reported to exert tumor‐suppressive effects.25, 26, 27, 28 In contrast, in breast, ovarian, and prostate tumors, α1‐2 fucosylation is reported to exert tumor‐promoting effects; for example, it can promote TGFβ signaling and cellular proliferation.29, 30, 31, 32 In breast cancer, FUT1 and FUT2 are suggested to play important roles in the regulation of cancer stem cell properties, such as proliferation, tumorigenesis, and metastasis.33 In addition, FUT1 and FUT2 are overexpressed in iPSCs.5 These results suggest that α1‐2 fucose might be related to stemness. However, there are few reports about α1‐2 fucose in PDAC. It has been demonstrated that hypoxia suppresses cell surface α1‐2 fucosylation, which promotes cancer cell motility and migration.34 This study showed for the first time that CEA is modified with α1‐2 fucose. In Figures 4, S4, S6, and TableS 2, we examined the staining pattern, lectin blotting results, and clinicopathological features of clinical samples to verify the relationship with malignancy. Colocalization was observed in all PDAC cases, although the degree of colocalization varied. It is unclear whether the expression level of rBC2LCN‐positive CEA varies in each malignancy classification of PDAC and serum CEA level. The verification of this relevance will be undertaken in future studies. In addition, the functions of α1‐2 fucosylation of CEA in cancer progression remain unknown and should be analyzed in future studies.
Antibody‐drug conjugates (ADCs) targeting CEA are in clinical trials for the treatment of colorectal cancer.35, 36, 37 However, CEA is also expressed in normal tissues such as those of the esophagus, stomach, and colon.38 In view of the results of Figure S5, the response of rBC2LCN‐positive CEA is reduced in some cases. In this respect, rBC2LCN‐positive CEA could be a more specific target for tumors. Recently, we successfully generated mAbs that bind to rBC2LCN‐positive podocalyxin but not to rBC2LCN‐negative podocalyxin.39 Such mAbs that can discriminate between glycans of glycoproteins could be generated against rBC2LCN‐positive CEA for the highly specific targeting of PDAC in future studies.
In conclusion, we identified that CEA is an rBC2LCN‐positive glycoprotein expressed in PDAC. rBC2LCN‐positive CEA could be a novel drug target for PDAC. Our findings may lay a foundation for understanding the molecular basis of the cancer stemness of the notorious disease PDAC and may provide insight for the development of a new diagnostic and/or therapeutic tool for PDAC.
CONFLICT OF INTEREST
All the authors declare no conflict of interest.
Supporting information
Supplementary Material
Supplementary Material
ACKNOWLEDGMENTS
This research was supported in part by the Japan Agency for Medical Research and Development (AMED) under grant number 20ae0101027h0005, and in part by Japan Society for the Promotion of Science KAKENHI Grant Number JP18H04057.
Furuta T, Oda T, Kiyoi K, et al. Carcinoembryonic antigen as a specific glycoprotein ligand of rBC2LCN lectin on pancreatic ductal adenocarcinoma cells. Cancer Sci. 2021;112:3722–3731. 10.1111/cas.15023
Contributor Information
Tatsuya Oda, Email: tatoda@md.tsukuba.ac.jp.
Hiroaki Tateno, Email: h-tateno@aist.go.jp.
REFERENCES
- 1.Yu Y, Yang G, Huang H, et al. Preclinical models of pancreatic ductal adenocarcinoma: challenges and opportunities in the era of precision medicine. J Exp Clin Cancer Res. 2021;40:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tarbell JM, Cancel LM. The glycocalyx and its significance in human medicine. J Intern Med. 2016;280:97‐113. [DOI] [PubMed] [Google Scholar]
- 3.Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer. 2015;15:540‐555. [DOI] [PubMed] [Google Scholar]
- 4.Shimomura O, Oda T, Tateno H, et al. A novel therapeutic strategy for pancreatic cancer: targeting cell surface glycan using rBC2LC‐N lectin‐drug conjugate (LDC). Mol Cancer Ther. 2018;17:183‐195. [DOI] [PubMed] [Google Scholar]
- 5.Hasehira K, Furuta T, Shimomura O, Asada M, Oda T, Tateno H. Quantitative structural analysis of glycans expressed within tumors derived from pancreatic cancer patient‐derived xenograft mouse models. Biochem Biophys Res Commun. 2021;534:310‐316. [DOI] [PubMed] [Google Scholar]
- 6.Tateno H, Toyota M, Saito S, et al. Glycome diagnosis of human induced pluripotent stem cells using lectin microarray. J Biol Chem. 2011;286:20345‐20353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tateno H, Matsushima A, Hiemori K, et al. Podocalyxin is a glycoprotein ligand of the human pluripotent stem cell‐specific probe rBC2LCN. Stem Cells Transl Med. 2013;2:265‐273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tateno H, Saito S. Engineering of a potent recombinant lectin‐toxin fusion protein to eliminate human pluripotent stem cells. Molecules. 2017;22:1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mawaribuchi S, Onuma Y, Aiki Y, et al. The rBC2LCN‐positive subpopulation of PC‐3cells exhibits cancer stem‐like properties. Biochem Biophys Res Commun. 2019;515:176‐182. [DOI] [PubMed] [Google Scholar]
- 10.Kitaguchi D, Oda T, Enomoto T, et al. Lectin drug conjugate therapy for colorectal cancer. Cancer Sci. 2020;111:4548‐4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mawaribuchi S, Haramoto Y, Tateno H, Onuma Y, Aiki Y, Ito Y. rBC2LCN lectin as a potential probe of early‐stage HER2‐positive breast carcinoma. FEBS Open Bio. 2020;10:1056‐1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu JY, Jiang L, Liu JJ, et al. AEBP1 promotes epithelial‐mesenchymal transition of gastric cancer cells by activating the NF‐kappaB pathway and predicts poor outcome of the patients. Sci Rep. 2018;8:11955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu L, Liu X, Dong Z, et al. N6‐methyladenosine‐related genomic targets are altered in breast cancer tissue and associated with poor survival. J Cancer. 2019;10:5447‐5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beauchemin N, Benchimol S, Cournoyer D, Fuks A, Stanners CP. Isolation and characterization of full‐length functional cDNA clones for human carcinoembryonic antigen. Mol Cell Biol. 1987;9:3221‐3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammarstrom S, Engvall E, Johansson BG, et al. Nature of the tumor associated determinant(s) of carcinoembryonic antigen. Proc Natl Acad Sci USA. 1975;72:1528‐1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamashita K, Totani K, Kuroki M, et al. Structural studies of the carbohydrate moieties of carcinoembryonic antigens. Cancer Res. 1987;13:3451‐3459. [PubMed] [Google Scholar]
- 17.Chandrasekaran EV, Davila M, Nixon DW, et al. Isolation and structures of the oligosaccharide units of carcinoembryonic antigen. J Biol Chem. 1983;11:7213‐7222. [PubMed] [Google Scholar]
- 18.Hatakeyama K, Wakabayashi‐Nakao K, Ohsima K, et al. Novel protein isoforms of carcinoembryonic antigen are secreted from pancreatic, gastric and colorectal cancer cells. BMC Res Notes. 2013;6:381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiroshima Y, Maawy A, Zhang Y, et al. Photoimmunotherapy inhibits tumor recurrence after surgical resection on a pancreatic cancer patient‐derived orthotopic xenograft (PDOX) nude mouse model. Ann Surg Oncol. 2015;22(Suppl 3):S1469‐S1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gold P, Shuster J, Freedman SO. Carcinoembryonic antigen (CEA) in clinical medicine: historical perspectives, pitfalls and projections. Cancer. 1978;42:1399‐1405. [DOI] [PubMed] [Google Scholar]
- 21.Gomes C, Almeida A, Barreira A, et al. Carcinoembryonic antigen carrying SLe(X) as a new biomarker of more aggressive gastric carcinomas. Theranostics. 2019;9:7431‐7446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saeland E, Belo AI, Mongera S, van Die I , Meijer GA, van Kooyk Y . Differential glycosylation of MUC1 and CEACAM5 between normal mucosa and tumour tissue of colon cancer patients. Int J Cancer. 2012;131:117‐128. [DOI] [PubMed] [Google Scholar]
- 23.Nolfi D, Capone A, Rosati F, Della GC. The alpha‐1,2 fucosylated tubule system of DU145 prostate cancer cells is derived from a partially fragmented Golgi complex and its formation is actin‐dependent. Exp Cell Res. 2020;396:112324. [DOI] [PubMed] [Google Scholar]
- 24.Keeley TS, Yang S, Lau E. The diverse contributions of fucose linkages in cancer. Cancers. 2019;11:1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keeley T, Lin S, Lester DK, Lau EK, Yang S. The fucose salvage pathway inhibits invadopodia formation and extracellular matrix degradation in melanoma cells. PLoS One. 2018;13:e0199128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hotta H, Hamamura K, Yamashita K, et al. Lewis y antigen is expressed in oral squamous cell carcinoma cell lines and tissues, but disappears in the invasive regions leading to the enhanced malignant properties irrespective of sialyl‐Lewis x. Glycoconj J. 2013;30:585‐597. [DOI] [PubMed] [Google Scholar]
- 27.Chandrasekaran EV, Xue J, Piskorz C, et al. Potential tumor markers for human gastric cancer: an elevation of glycan:sulfotransferases and a concomitant loss of alpha1,2‐fucosyltransferase activities. J Cancer Res Clin Oncol. 2007;133:599‐611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huthinson WL, Du MQ, Johnson PJ, Wiliams R. Fucosyltransferases: differential plasma and tissue alterations in hepatocellular carcinoma and cirrhosis. Hepatology. 1991;13:683‐688. [PubMed] [Google Scholar]
- 29.Tan KP, Ho MY, Cho HC, Yu J, Hung JT, Yu AL. Fucosylation of LAMP‐1 and LAMP‐2 by FUT1 correlates with lysosomal positioning and autophagic flux of breast cancer cells. Cell Death Dis. 2016;7:e2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hao Y, Zhu L, Yan L, et al. c‐Fos mediates alpha1, 2‐fucosyltransferase 1 and Lewis y expression in response to TGF‐beta1 in ovarian cancer. Oncol Rep. 2017;38:3355‐3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li FF, Liu JJ, Liu DW, et al. Lewis Y regulates signaling molecules of the transforming growth factor beta pathway in ovarian carcinoma‐derived RMG‐I cells. Int J Oncol. 2012;40:1196‐1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kosanovic MM, Jankovic MM. Sialylation and fucosylation of cancer‐associated prostate specific antigen. J BUON. 2005;10:247‐250. [PubMed] [Google Scholar]
- 33.Lai TY, Chen IJ, Lin RJ, et al. Fucosyltransferase 1 and 2 play pivotal roles in breast cancer cells. Cell Death Discov. 2019;5:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aubert M, Panicot L, Crotte C, et al. Restoration of alpha (1,2) fucosyltransferase activity decreases adhesive and metastatic properties of human pancreatic cancer cells. Cancer Res. 2000;60:1449‐1456. [PubMed] [Google Scholar]
- 35.Sousa AR, Oliveira MJ, Sarmento B. Impact of CEA‐targeting nanoparticles for drug delivery in colorectal cancer. J Pharmacol Exp Ther. 2019;370:657‐670. [DOI] [PubMed] [Google Scholar]
- 36.Dotan E, Cohen SJ, Statodub AN, et al. Phase I/II trial of labetuzumab govitecan (Anti‐CEACAM5/SN‐38 antibody‐drug conjugate) in patients with refractory or relapsing metastatic colorectal cancer. J Clin Oncol. 2017;35:3338‐3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang C, Wang Z, Yang Z, et al. Phase I escalating‐dose trial of CAR‐T therapy targeting CEA (+) metastatic colorectal cancers. Mol Ther. 2017;25:1248‐1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hammarstrom S. The carcinoembryonic antigen (CEA) family: structures, suggested functions and expression in normal and malignant tissues. Semin Cancer Biol. 1999;9:67‐81. [DOI] [PubMed] [Google Scholar]
- 39.Watanabe T, Kakuta J, Saito S, et al. Monoclonal antibodies specific for podocalyxin expressed on human induced pluripotent stem cells. Biochem Biophys Res Commun. 2020;532:647‐654. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Supplementary Material


