Abstract
Inteins, or intervening proteins, are mobile genetic elements translated within host polypeptides and removed through protein splicing. This self-catalyzed process breaks two peptide bonds and rejoins the flanking sequences, called N- and C-exteins, with the intein scarlessly escaping the host protein. As these elements have traditionally been viewed as purely selfish genetic elements, recent work has demonstrated that the conditional protein splicing (CPS) of several naturally occurring inteins can be regulated by a variety of environmental cues relevant to the survival of the host organism or crucial to the invading protein function. The RadA recombinase from the archaeon Pyrococcus horikoshii represents an intriguing example of CPS, whereby protein splicing is inhibited by interactions between the intein and host protein C-extein. Single-stranded DNA (ssDNA), a natural substrate of RadA as well as signal that recombinase activity is needed by the cell, dramatically improves the splicing rate and accuracy. Here, we investigate the mechanism by which ssDNA exhibits this influence and find that ssDNA strongly promotes a specific step of the splicing reaction, cyclization of the terminal asparagine of the intein. Interestingly, inhibitory interactions between the host protein and intein that block splicing localize to this asparagine, suggesting that ssDNA binding alleviates this inhibition to promote splicing. We also find that ssDNA directly influences the position of catalytic nucleophiles required for protein splicing, implying that ssDNA promotes assembly of the intein active site. This work advances our understanding of how ssDNA accelerates RadA splicing, providing important insights into this intriguing example of CPS.
Inteins are intervening proteins that are removed from host proteins through protein splicing following translation. During this process, the intein precisely excises itself from the surrounding sequences (called N- and C-exteins), leaving a regular peptide bond.1,2 As self-contained enzymes, with the capacity to seamlessly break and re-form peptide bonds, inteins have been utilized in numerous biotechnological applications, including bioseparations, bioconjugation, biosensing, and protein cyclization.3,4 Thus, the primary motivation for investigation of inteins has been applied. Recently, however, there has been a push to better understand the biology of these enigmatic sequences.
Some inteins are mobile genetic elements, by virtue of resident endonuclease domains, and thousands have been identified across the genomes of archaea, bacteria, unicellular eukaryotes, phages, and viruses.5,6 Inteins are often within proteins essential for host organism fitness and viability, with >70% found in ATP binding proteins and >60% localized in proteins involved in DNA replication, recombination, and repair.5 Remarkably, inteins have been independently acquired by non-orthologous bacterial and archaeal homologues, such as the replicative helicases DnaB and MCM.5 This intein localization invites many questions about the possible roles of these unique elements in nature. Interestingly, many inteins have lost the ability to be mobile, through loss or degeneration of the homing endonuclease domain, but are nonetheless retained in crucial genes.
As splicing must occur prior to the host protein achieving full activity, protein splicing has tremendous regulatory potential. However, inteins have long been considered mere parasites.7 While protein splicing is self-catalyzed, the rate and accuracy of the process are highly variable between inteins and can be modulated by the environment (refs 8–15; reviewed in refs 16 and 17). Recent work has demonstrated several examples of conditional protein splicing (CPS) in vitro and in the native organism.18 These include splicing inhibition imposed by disulfide redox traps, reactive oxygen/nitrogen species and metals, and splicing activation by salt, temperature, and substrate of the intein-containing host protein.8–15 We contend that not all inteins are molecular parasites but rather that some have adapted to regulate their host proteins in response to the environment, providing greater control over host protein activity. This represents a potentially widespread, and understudied, form of posttranslational regulation.
Our group has discovered a unique example of CPS with the RadA recombinase intein from the deep-sea hyperthermophilic archaeon Pyrococcus horikoshii (Pho) (UniprotKB 058001).10,14,19 The Pho RadA intein first garnered interest following work that measured protein splicing in three different foreign extein reporters, where it ranked among the highest of 20 inteins tested for splicing efficiency.20 In a manner consistent with these findings, our group determined that splicing of the RadA intein proceeds rapidly and accurately in yet another foreign extein reporter, but curiously, splicing was blocked in the native extein context.10 To induce splicing in native exteins, a high temperature (>75 °C) nearing environmental relevance for Pho or solution conditions that disrupt protein–protein interactions were required.10 On the basis of structural modeling and mutagenesis, we determined that splicing inhibition was due to C-extein–intein interactions that must be broken to allow splicing.10
RadA is homologous to Rad51 in eukaryotes and RecA in bacteria, ancient and highly conserved proteins crucial to the repair and maintenance of DNA.21–23 RadA recognizes single-stranded DNA (ssDNA), a critical signal of genome stress, and forms a nucleoprotein filament, which activates the ATPase of RadA.21–23 Notably, we discovered that this biologically relevant RadA substrate, ssDNA, specifically accelerated Pho RadA intein splicing by ~50-fold compared to the acceleration caused by only a high temperature as well as improved splicing accuracy.14,24 Interestingly, splicing activates recombinase ATPase activity, as the Pho RadA intein is located in the ATP binding P-loop.10 Here, we further probe the mechanism of ssDNA-induced splicing. We find that ssDNA strongly promotes activity of the intein terminal asparagine, a catalytic residue in which inhibitory interactions that block splicing localize. In addition, we report that ssDNA binding decreases the distance between the two catalytic nucleophiles required for splicing, suggesting ssDNA promotes assembly of the intein active site.
ssDNA Only Modestly Accelerates Pho RadA Linear Thioester Formation.
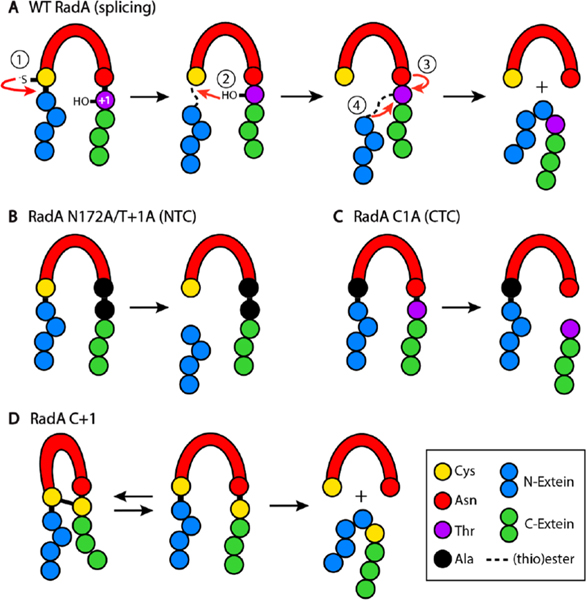
The Pho RadA intein employs a class 1 splicing mechanism (Figure 1A).25
Figure 1.
(A) Mechanism of class 1 intein splicing, with steps described in the text. Pho RadA variants used in this study and products from splicing, cleavage, and disulfide bonding reactions. Exteins, intein, and specific amino acid features of each protein are listed in the key.
Specifically, the first residue of the intein, a cysteine (C1; residue numbering is intein-centric), performs a nucleophilic attack on the preceding peptide bond, resulting in a thioester linkage (step 1). Next, the first residue of the C-extein, a threonine (T+1), initiates a second nucleophilic attack on the aforementioned thioester, resulting in a branched intermediate (step 2). Next, the last residue of the intein, an asparagine, cyclizes to free the intein (step 3). Finally, the ester connecting the N- and C-exteins rearranges to a peptide bond (step 4).1
As ssDNA can greatly accelerate Pho RadA splicing, we sought to determine which steps of the splicing reaction were influenced.14 For these studies, we used an N-terminal truncation that maintains the ATPase domain and ssDNA binding loops and responds to ssDNA like the wild type (WT) does.14 To test whether ssDNA promotes the first step of splicing, we substituted the last residue of the intein, N172, and the first residue of the C-extein, T+1, with alanine (RadA N172A/T+1A). While this variant cannot complete the splicing reaction, it can initiate the first nucleophilic attack by C1 on the preceding amide bond, leading to formation of a linear thioester.1 This thioester, which would normally undergo a nucleophilic attack by T+1, can undergo N-terminal cleavage (NTC). NTC results in the free N-extein [4.4 kDa; not resolved via sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE)] and the intein–C-extein complex (42.7 kDa) (Figure 1B). An external nucleophile, in this case dithiothreitol (DTT), is present to induce cleavage of the thioester.26
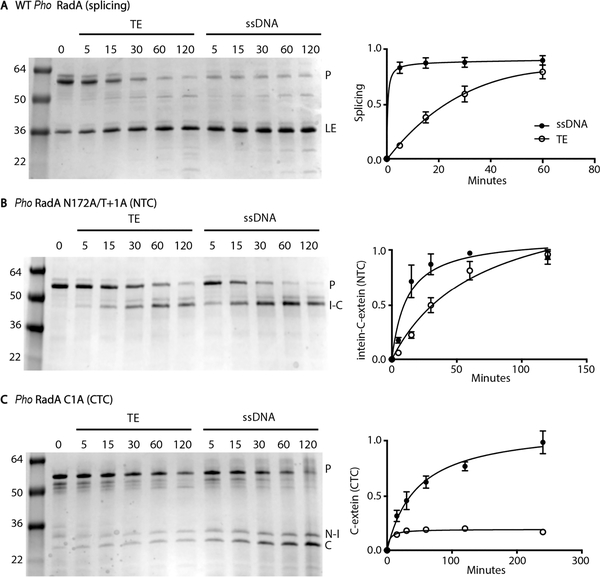
We measured both splicing of WT RadA and cleavage of RadA N172A/T+1A over time at 63 °C with or without ssDNA (Figure 2A,B).
Figure 2.
Pho RadA splicing stimulated by ssDNA: (A) splicing of WT RadA, (B) N-terminal cleavage of RadA N172A/T+1A, and (C) C-terminal cleavage of RadA C1A with or without ssDNA. For each panel, a representative gel is on the left and a graph of reaction kinetics on the right. For each graph, data are based on three independent experiments, with the error being the standard deviation. Reaction mixtures were incubated at 63 °C for the indicated times, separated by nonreducing SDS–PAGE, and stained with Coomassie. Levels of precursor (P), ligated extein (LE), intein–C-extein (I–C), N-extein–intein (N–I), and C-extein (C) proteins were measured by densitometry. Tris-EDTA (TE) was used as a buffer control for samples without ssDNA.
While ssDNA accelerates the rate of WT RadA splicing by ~50-fold (Figure 2A),14 ssDNA has a more modest impact on the rate of RadA N172A/T+1A cleavage, accelerating it by ~4- fold. While clearly influenced by ssDNA, this acceleration in rate is insufficient to explain the effect of ssDNA on splicing.
Terminal Asparagine Cyclization of Pho RadA Requires ssDNA.
Dissection of the second step of splicing is hampered because for class 1 inteins, the (thio)ester formed at the junction between the N-extein and intein in step 1 must be present for the nucleophilic attack in step 2 (Figure 1A).1 The third step of splicing, however, whereby the terminal asparagine of the intein cyclizes to cleave itself from the branched intermediate, can be uncoupled from step 2.1 This results in C-terminal cleavage (CTC), producing the N-extein-intein compound (23.7 kDa) and free C-extein (23.0 kDa) (Figure 1C).
To measure whether ssDNA accelerates uncoupled asparagine cyclization, we substituted the cysteine at position 1 of the intein with alanine (RadA C1A), which blocks the first two steps of splicing. We incubated Pho RadA C1A at 63 °C with or without ssDNA and measured CTC at various times. While the level of the precursor diminishes over time both with and without ssDNA, ssDNA is required for significant cleavage product accumulation, implying that Pho RadA C1A degrades nonspecifically without ssDNA (Figure 2C). The rate of uncoupled Asn cyclization for RadA C1A is slower than the overall rate of WT RadA splicing (Figure 2). Conformational changes in the first two steps of splicing can greatly accelerate Asn cyclization,27 suggesting ssDNA binding may induce similar changes. In contrast to NTC, which can proceed without ssDNA (Figure 2B), asparagine cyclization is largely unable to occur in the absence of ssDNA (Figure 2C), suggesting that this step is significantly influenced by ssDNA and central to explaining its role.
ssDNA Promotes the Proximity of Splicing Nucleophiles.
We sought an additional measure of how ssDNA directs RadA splicing. An absolute requirement for splicing is the proper positioning of catalytic residues proximal to one another within the intein active site. Previously, a disulfide bond trap was discovered in the Pyrococcus abyssi PolIII intein, where a bond forms between native C1 and C+1 residues.15 Thus, we reasoned that if we substituted RadA T+1 for a cysteine (C+1), a disulfide may form between C1 and C+1, as C1 is the only native cysteine in the protein. This disulfide could then be used to measure the relative proximity of the two catalytic nucleophiles. Specifically, an increase in the level of intramolecular disulfide bond formation would indicate that C1 and C+1 are closer in space and/or within bonding distance for a longer period of time (Figure 1D).
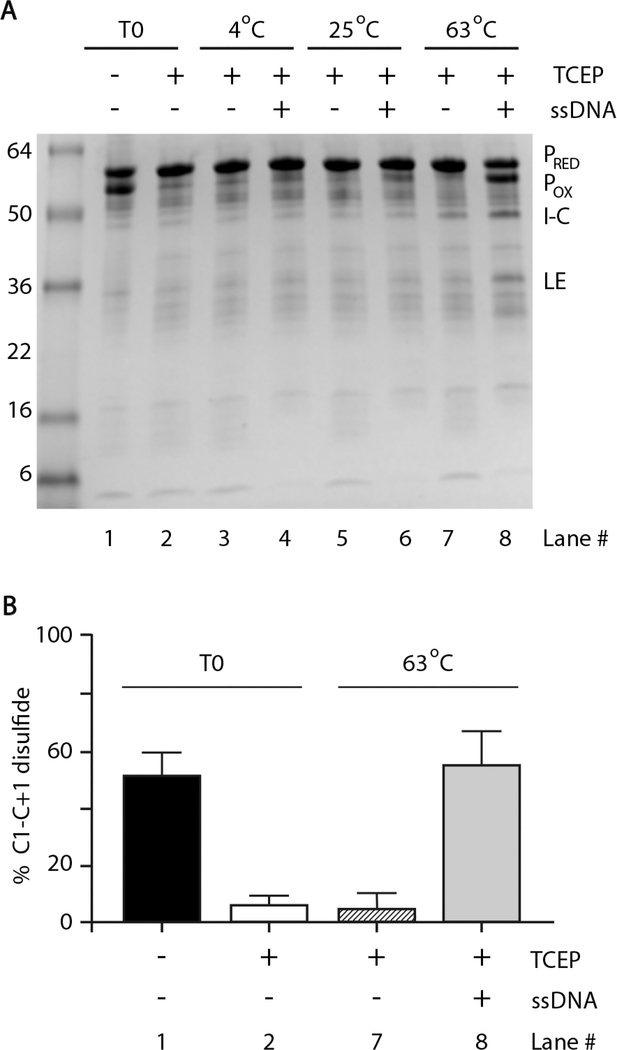
Indeed, following overexpression and purification of RadA C +1, there was a band migrating slightly faster than the precursor on SDS–PAGE under oxidizing conditions (Figure 3A). This band disappeared in the presence of the reducing agent tris(2-carboxyethyl)phosphine (TCEP) and the level of catalytically active, reduced precursor concomitantly increased (Figure 3A; lane 2), demonstrating that the faster migrating band is intramolecular disulfide-bonded RadA precursor. TCEP was used as a reducing agent, rather than DTT, as it is will not induce cleavage reactions.26
Figure 3.
ssDNA promotes disulfide between catalytic nucleophiles. (A) Gel demonstrating the internal disulfide bond between C1 and C +1 and influence of ssDNA. Pho RadA C+1 was purified under nonreducing conditions (–TCEP). Reaction mixtures were incubated at the indicated temperatures for 5 min, separated by SDS–PAGE, and stained with Coomassie. Levels of the oxidized precursor (POX) and reduced precursor (PRED) were measured by densitometry. Tris-EDTA (TE) was used as a buffer control in the absence of ssDNA. (B) Quantification of C1–C+1 disulfide bond formation with or without ssDNA and TCEP prior to incubation (T0) or after 5 min at 63 °C. Error bars represent the standard deviation from three independent experiments.
Disulfide bonding is reversible depending on redox conditions, while protein splicing is not (Figure 1A,D). Therefore, we monitored disulfide bond formation 5 min after the addition of TCEP to prevent a significant decrease in the level of the precursor through splicing or cleavage. At time zero, approximately half of the RadA C+1 precursor was intramolecularly disulfide bonded in the absence of TCEP (Figure 3A,B, lane 1), while the precursor was fully reduced in the presence of TCEP (Figure 3A,B, lane 2). With ssDNA and TCEP present, disulfide re-formed between C1 and C+1 to a level similar to that prior to addition of TCEP at 63 °C(Figure 3A,B, lane 8). Under the same conditions without ssDNA, no disulfide re-formed (Figure 3A,B, lane 7). Significant disulfide re-formation required both a high temperature (63 °C) and ssDNA, although ssDNA could promote some disulfide at 25 °C(Figure 3A, lane 6), while incubation at 63 °C alone could not (Figure 3A, lane 7). We conclude that ssDNA promotes the relative proximity of catalytic nucleophiles required for splicing, thereby facilitating the assembly of the intein active site.
Model of ssDNA-Induced Pho RadA Intein Splicing.
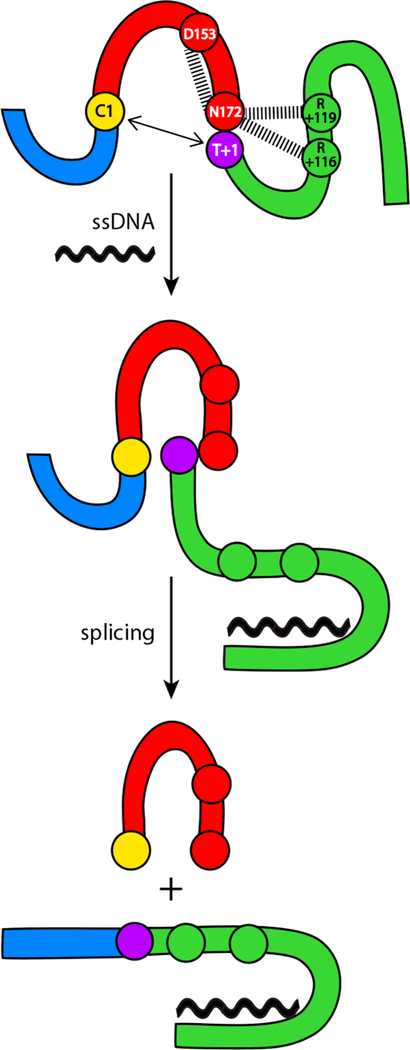
On the basis of previous structure modeling and biochemical analysis,10 interactions between the intein and C-extein “mask” the intein–C-extein junction to block splicing of Pho RadA. Interestingly, the strongest predicted interactions occur between the side chains of two C-extein arginine residues, R +116 and R+119, and the side chain of the terminal asparagine of the intein, N172 (Figure 4).10
Figure 4.
Model of ssDNA-based acceleration of Pho RadA protein splicing. We propose that inhibitory interactions (dashed lines) between intein residue N172 and intein (D153) and extein (R+116 and R+119) residues and the distance between nucleophiles (bidirectional arrows) block splicing. Upon binding to ssDNA, inhibitory interactions are broken, allowing Asn cyclization and splicing to proceed (Figure 2). Additionally, we propose that ssDNA binding compresses the intein active site, bringing C1 and T+1 into the proximity for catalysis (Figure 3). The color scheme is the same as that in Figure 1.
Mutation of R+116 and R+119 strongly promoted splicing at lower temperatures, consistent with the alleviation of inhibitory interactions between the C-extein and intein.10 In addition, in the RadA intein structure, a highly conserved aspartate, D153, forms a hydrogen bond with N172, keeping the side chain of N172 away from the intein–C-extein amide bond and preventing premature asparagine cyclization.25 Together, these data point to the catalytic N172 as a hub for interactions that block splicing.
While inactive as an ATPase prior to splicing, RadA retains the ability to bind to ssDNA via recognition loops in the C-extein.14 We propose that binding of ssDNA to the C-extein disrupts the interactions of R+116 and R+119 with N172 (Figure 4). As ssDNA induces asparagine cyclization (Figure 2C), it is likely to also disrupt the D153–N172 interaction (Figure 4), which is predicted to block cyclization in the absence of conformational changes.22 We thus argue that accelerating cyclization (Figure 2B) and promoting the proximity of catalytic nucleophiles (Figure 3) combine to explain the role of ssDNA in RadA splicing acceleration (Figure 4).
In summary, our results provide mechanistic insights into this important example of CPS, whereby the natural substrate of the host protein and cellular signal of DNA damage promote faster, more accurate splicing of the very protein needed for the DNA damage response. We are hopeful that our findings may inform yet to be discovered examples of conditional protein splicing and the development of ssDNA- or DNA damage-based biosensors.
Supplementary Material
Acknowledgments
Funding
This work was supported by National Institutes of Health Grants GM39422 and GM44844 to M.B. and F32GM121000 to C.W.L.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.bio-chem.9b00506.
Experimental details (PDF)
Accession Codes
P. horikoshii RadA, UniprotKB 058001.
REFERENCES
- (1).Mills KV, Johnson MA, and Perler FB (2014) Protein splicing: how inteins escape from precursor proteins. J. Biol. Chem. 289, 14498–14505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Paulus H (2001) Inteins as enzymes. Bioorg. Chem. 29, 119–129. [DOI] [PubMed] [Google Scholar]
- (3).Shah NH, and Muir TW (2014) Inteins: nature’s gift to protein chemists. Chem. Sci. 5, 446–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Wood DW, and Camarero JA (2014) Intein applications: from protein purification and labeling to metabolic control methods. J. Biol. Chem. 289, 14512–14519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Novikova O, Jayachandran P, Kelley DS, Morton Z, Merwin S, Topilina NI, and Belfort M (2016) Intein clustering suggests functional importance in different domains of life. Mol. Biol. Evol. 33, 783–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Kelley DS, Lennon CW, Belfort M, and Novikova O (2016) Mycobacteriophages as incubators for intein dissemination and evolution. mBio 7, e01537–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Naor A, Altman-Price N, Soucy SM, Green AG, Mitiagin Y, Turgeman-Grott I, Davidovich N, Gogarten JP, and Gophna U (2016) Impact of a homing intein on recombination frequency and organismal fitness. Proc. Natl. Acad. Sci. U. S. A. 113, E4654–E4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Mills KV, and Paulus H (2001) Reversible inhibition of protein splicing by Zinc ion. J. Biol. Chem. 276, 10832–10838. [DOI] [PubMed] [Google Scholar]
- (9).Callahan BP, Topilina NI, Stanger MJ, Van Roey P, and Belfort M (2011) Structure of catalytically competent intein caught in a redox trap with functional and evolutionary implications. Nat. Struct. Mol. Biol. 18, 630–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Topilina NI, Novikova O, Stanger M, Banavali NK, and Belfort M (2015) Post- translational environmental switch of RadA activity by extein-intein interactions in protein splicing. Nucleic Acids Res. 43, 6631–6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Topilina NI, Green CM, Jayachandran P, Kelley DS, Stanger MJ, Piazza CL, Nayak S, and Belfort M (2015) SufB intein of Mycobacterium tuberculosis as a sensor for oxidative and nitrosative stresses. Proc. Natl. Acad. Sci. U. S. A. 112, 10348–10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Reitter JN, Cousin CE, Nicastri MC, Jaramillo MV, and Mills KV (2016) Salt-dependent conditional protein splicing of an intein from Halobacterium salinarum. Biochemistry 55, 1279–1282. [DOI] [PubMed] [Google Scholar]
- (13).Ciragan A, Aranko AS, Tascon I, and Iwaï H (2016) Salt-inducible protein splicing in cis and trans by inteins from extremely halophilic archaea as a novel protein-engineering tool. J. Mol. Biol. 428, 4573–4588. [DOI] [PubMed] [Google Scholar]
- (14).Lennon CW, Stanger M, and Belfort M (2016) Protein splicing of a recombinase intein induced by ssDNA and DNA damage. Genes Dev. 30, 2663–2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Chen W, Li L, Du Z, Liu J, Reitter JN, Mills KV, Linhardt RJ, and Wang C (2012) Intermolecular disulfide bond between catalytic cysteines in an intein precursor. J. Am. Chem. Soc. 134, 2500–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Lennon CW, and Belfort M (2017) Inteins. Curr. Biol. 27, R204–R206. [DOI] [PubMed] [Google Scholar]
- (17).Belfort M (2017) Mobile self-splicing introns and inteins as environmental sensors. Curr. Opin. Microbiol. 38, 51–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Kelley DS, Lennon CW, Li Z, Miller MR, Banavali NK, Li H, and Belfort M (2018) Mycobacterial DnaB helicase intein as oxidative stress sensor. Nat. Commun. 9, 4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).González JM, Masuchi Y, Robb FT, Ammerman JW, Maeder DL, Yanagibayashi M, Tamaoka J, and Kato C (1998) Pyrococcus horikoshii sp. nov., a hyperthermophilic archaeon isolated from a hydrothermal vent at the Okinawa Trough. Extremophiles 2, 123–130. [DOI] [PubMed] [Google Scholar]
- (20).Ellilä S, Jurvansuu JM, and Iwaï H(2011) Evaluation and comparison of protein splicing by exogenous inteins with foreign exteins in Escherichia coli. FEBS Lett. 585, 3471–3477. [DOI] [PubMed] [Google Scholar]
- (21).Seitz EM, Brockman JP, Sandler SJ, Clark AJ, and Kowalczykowski SC (1998) RadA protein is an archaeal RecA protein homolog that catalyzes DNA strand exchange. Genes Dev. 12, 1248–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Cox MM (2007) Regulation of bacterial RecA protein function. Crit. Rev. Biochem. Mol. Biol. 42, 41–63. [DOI] [PubMed] [Google Scholar]
- (23).Bell JC, and Kowalczykowski SC (2016) RecA: Regulation and mechanism of a molecular search engine. Trends Biochem. Sci. 41, 491–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Lennon CW, Stanger M, Banavali NK, and Belfort M (2018) Conditional protein splicing switch in hyperthermophiles through intein-extein partnership. mBio 9, e02304–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Oeemig JS, Zhou D, Kajander T, Wlodawer A, and Iwaï H (2012) NMR and crystal structures of the Pyrococcus horikoshii RadA intein guide a strategy for engineering a highly efficient and promiscuous intein. J. Mol. Biol. 421, 85–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Amitai G, Callahan BP, Stanger MJ, Belfort G, and Belfort M (2009) Modulation of intein activity by its neighboring extein substrates. Proc. Natl. Acad. Sci. U. S. A. 106, 11005–11010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Frutos S, Goger M, Giovani B, Cowburn D, and Muir TW (2010) Branched intermediate formation stimulates peptide bond cleavage in protein splicing. Nat. Chem. Biol. 6, 527–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.