Abstract
Somatic mutations in the PRKACA gene encoding the catalytic α subunit of protein kinase A (PKA-C) are responsible for cortisol-producing adrenocortical adenomas. These benign neoplasms contribute to the development of Cushing’s syndrome. The majority of these mutations occur at the interface between the two lobes of PKA-C and interfere with the enzyme’s ability to recognize substrates and regulatory (R) subunits, leading to aberrant phosphorylation patterns and activation. Rarely, patients with similar phenotypes carry an allosteric mutation, E31V, located at the C-terminal end of the αA-helix’s and adjacent to the αC-helix, but structurally distinct from the PKA-C/R subunit interface mutations. Using a combination of solution NMR, thermodynamics, kinetic assays, and molecular dynamics simulations, we show that the E31V allosteric mutation disrupts central communication nodes between the N- and C- lobes of the enzyme as well as nucleotide-substrate binding cooperativity, a hallmark for kinases’ substrate fidelity and regulation. For both orthosteric (L205R and W196R) and allosteric (E31V) Cushing’s syndrome mutants, the loss of binding cooperativity is proportional to the density of the intramolecular allosteric network. This structure-activity relationship suggests a possible common mechanism for Cushing’s syndrome driving mutations in which decreased nucleotide/substrate binding cooperativity is linked to loss in substrate fidelity and dysfunctional regulation.
Keywords: cAMP-dependent protein kinase A, Cushing’s syndrome, allostery, binding cooperativity
Graphical Abstract
INTRODUCTION
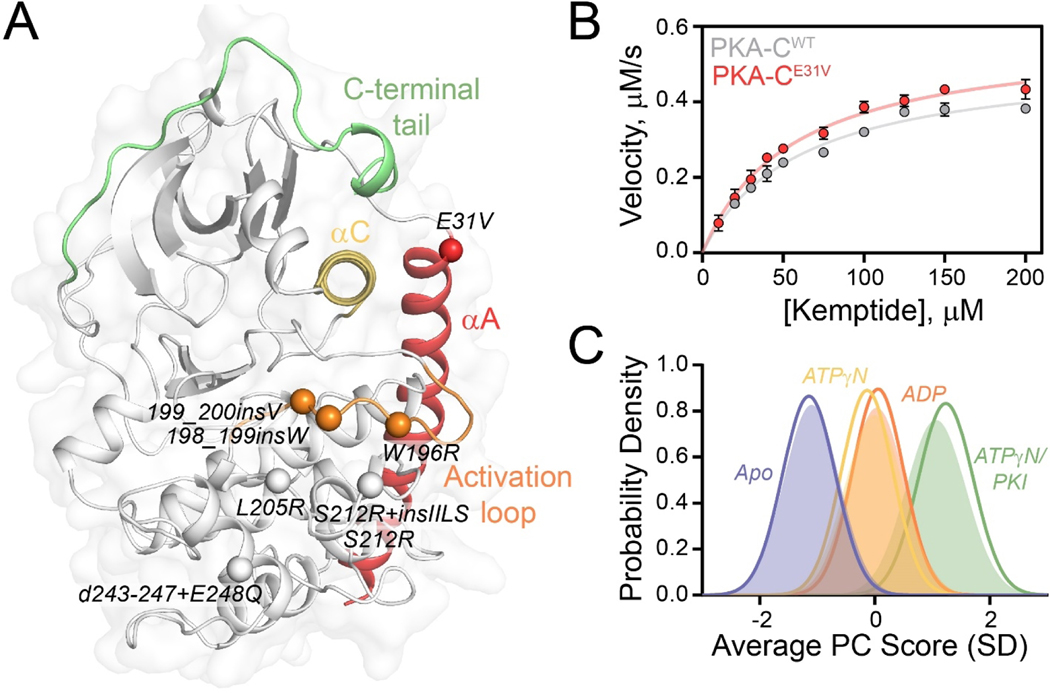
Cushing’s syndrome is defined by a collection of symptoms that result from prolonged exposure to high cortisol levels, leading to cardiovascular and metabolic complications [1]. Playing a fundamental role in regulating metabolism and cell proliferation in endocrine tissues, the cAMP signaling pathway and its aberrant activation are linked to several endocrine diseases [2–4]. The role of one component of this signaling cascade, cAMP-dependent protein kinase A (PKA), in Cushing’s syndrome was not appreciated until recently when somatic mutations were identified in the PRKACA gene encoding the catalytic α subunit of PKA (PKA-C) [5–12]. To date, a total of eight mutations have been discovered, which give the same disease phenotypes [13]. Except for E31V, all mutations are located near the substrate-binding cleft adjacent to the catalytic/regulatory (R) subunit interface (Figure 1A).
Figure 1. Structural and kinetic characterization of PKA-CE31V.
(A) X-ray structure of PKA-C bound to endogenous inhibitor, PKI (PDB: 1ATP) highlighting important structural elements and locations of Cushing’s syndrome mutations in relation to E31V. (B) Steady state phosphorylation kinetics of PKA-CE31V with Kemptide. (C) CONCISE analysis on the apo, ATPγN-, ADP- and ATPγN/PKI-bound forms of PKA-CWT (opaque gaussian) and PKA-CE31V (outlined gaussian). Note that the probability distribution of PKA-CWT when bound to ATPγN (yellow) and ADP (orange) are overlapping.
PKA is the principal intracellular effector of the second messenger, cAMP. Inactive PKA exists as a holoenzyme (R2:C2) containing an R-subunit dimer bound to two catalytic (C) subunits [14]. Each R-subunit contains an inhibitory sequence that occupies the active site of the enzyme. Following stimulation of adenylate cyclase, two cAMP molecules bind to each R-subunit, initiating a conformational change and releasing active PKA-C. While R-subunits are the primary intracellular regulator of PKA-C, the endogenous inhibitor, protein kinase inhibitor (PKI), restricts PKA-C activity within the nucleus and controls nuclear exportation [15]. Spatiotemporal regulation is controlled by various ancillary proteins such as A-kinase anchoring proteins (AKAPs) that, via interactions with R-subunits, localize PKA-C near substrates [16].
PKA-C toggles between three major conformational states: open (apo), intermediate (nucleotide-bound), and closed (nucleotide/substrate-bound) [17]. This bean-shaped enzyme consists of a conserved catalytic core comprised of two lobes. The N-lobe of the kinase is smaller and contains mostly β-sheets and the αC-helix and harboring the ATP binding site, while the C-lobe comprises mostly α-helices and contains the substrate-binding cleft [18]. In contrast to other Ser/Thr kinases, PKA-C contains an αA-helix at its N-terminus, which anchors the N-lobe to the C-lobe and contributes to the tethering/positioning of the αC-helix. This important structural motif is recognized for its role in the activation and inactivation of protein kinases [17]. E31V is located at the C-terminus of the αA-helix and adjacent to the C-terminus of the αC-helix. While other Cushing’s syndrome mutations have been shown to disrupt R-subunit/PKA-C interactions, alter the enzyme’s catalytic efficiency, and/or change its substrate specificity, E31V does not prevent R-subunit binding and its activity against Kemptide is identical to the wild-type kinase [13]. Therefore, the mechanism of dysfunction for PKA-CE31V has remained elusive [13, 19, 20].
Recently, we discovered that the most common Cushing’s syndrome mutation, PKA-CL205R, abrogates the nucleotide/pseudosubstrate binding cooperativity by reducing the intramolecular allostery between the small and large lobe [20]. Based on these findings, we suggested that this dysfunctional binding cooperativity and altered allostery disrupt substrate recognition and interactions with R-subunits, thereby perturbing canonical cAMP signaling. Despite E31V and L205R being spatially distant, our previous NMR analysis suggested these residues are allosterically coupled [20]. Therefore, we surmised that E31V may affect the kinase’s function in a manner similar to L205R, i.e., the non-conservative mutation may disrupt the allosteric network and the binding cooperativity.
To dissect this allosteric mutation’s effects, we carried out solution NMR spectroscopy along with isothermal titration calorimetry (ITC), kinetic assays, and molecular dynamics (MD) simulations. We found that the E31V mutation ablates the canonical positive cooperativity, typically seen for PKA-C while maintaining the kinase’s catalytic efficiency. Specifically, the E31V mutation directly affects the allosteric node that connects the αA-, αC-helix, and activation loop, thereby disrupting nucleotide-substrate binding cooperativity. Finally, by comparing PKA-CWT with three drivers for Cushing’s syndrome, PKA-CE31V, PKA-CL205R, and PKA-CW196R, we found a direct relationship between the loss in binding cooperativity and the reduction of allosteric communication within the enzyme. Altogether, our results suggest the existence of a common dysfunctional mechanism for PKA-C Cushing’s mutations discovered thus far.
RESULTS
E31V mutation ablates nucleotide-substrate binding cooperativity in PKA-C.
To evaluate the effects of E31V on the thermodynamics of nucleotide (ATPγN) and pseudosubstrate (PKI) binding, we used isothermal titration calorimetry (ITC) [21]. Values of ΔG, ΔH, -TΔS, Kd, and cooperativity coefficients (σ) obtained for PKA-CE31V are summarized in Table S1 and S2 [20]. We found PKA-CWT and PKA-CE31V have similar binding affinities for ATPγN (Kd = 83 ± 8 and 91 ± 9 μM, respectively). To evaluate the effects of E31V on the binding cooperativity between nucleotide and pseudosubstrate, PKI5–24, we compared the binding affinity of PKI5–24 in the absence and presence of ATPγN and calculated σ (defined mathematically in Materials and Methods). A 7-fold higher binding affinity is observed for PKA-CE31V compared to PKA-CWT when binding PKI5–24 to their apo forms (Kd = 2.5 ± 0.5 and 17 ± 2 μM, respectively). In contrast to PKA-CWT, upon saturation with ATPγN, PKA-CE31V displays a 12-fold reduction in binding affinity (Kd = 0.16 ± 0.02 and 2 ± 1 μM, respectively).
As previously determined, the binding of PKI5–24 to PKA-CWT is highly cooperative (σ = 106 ± 18); in contrast, PKA-CE31V displays no cooperativity with σ = 1.3 ± 0.7. To evaluate the effects of E31V on the kinase’s catalytic efficiency, we carried out steady-state coupled enzyme assays using the standard substrate, Kemptide. Despite the dramatic effects on binding cooperativity, PKA-CE31V displayed only a slight increase in Vmax and a slight decrease in KM, resulting in similar catalytic efficiencies (kcat/KM = 0.41 ± 0.05 and 0.46 ± 0.04 for PKA-CWT and PKA-CE31V, respectively; Figure 1B and Table S3). Interestingly, mutagenesis of residues adjacent to the E31 site has shown similar kinetic behavior [22].
NMR mapping of nucleotide/PKI binding response.
To analyze the binding response of PKA-CE31V to nucleotide and pseudosubstrate, we mapped the amide backbone fingerprint of the enzyme using [1H, 15N]-TROSY-HSQC experiments [23]. The amide fingerprints of the kinase in different ligated forms are displayed in Figure S1. The global response of the two kinases to ligand binding was determined using CONCISE (COordiNated ChemIcal Shifts bEhavior) [24], which performs a statistical analysis on linear chemical shift trajectories of amide resonances to identify the position of each state along the conformational equilibrium, shows that nucleotide and pseudosubstrate shift the overall populations from an open state to an intermediate and fully closed state. Upon binding both ADP and ATPγN, the probability density of the amide resonances from the apo shifts toward an intermediate state, and the subsequent saturation with PKI peptide further shifts toward the fully closed state (Figure 1C). Globally wild-type and mutant behave similarly; however, upon binding ATPγN, PKA-CE31V adopts a more open conformation compared to PKA-CWT and subsequent binding of PKI shifts the probability distribution toward a more closed state.
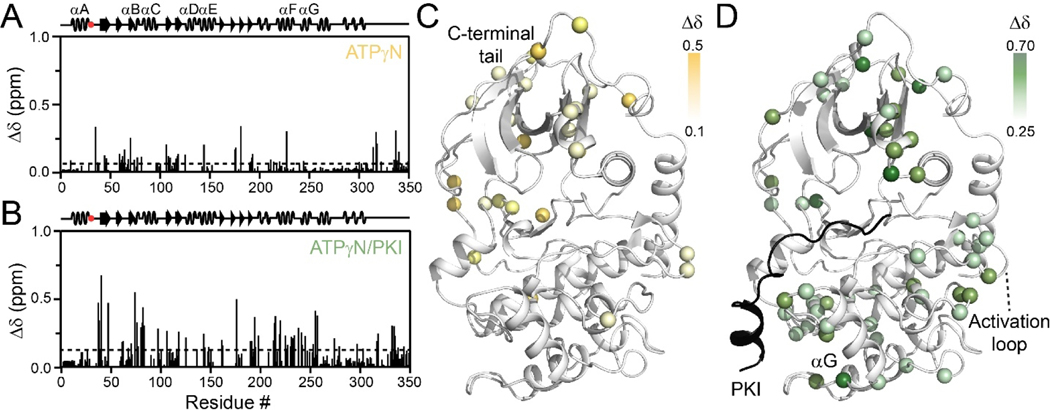
To further confirm the changes in the global response of PKA-C induced by E31V, we mapped the chemical shift perturbations (CSP, Δδ) of PKA-CE31V. Upon binding ATPγN, PKA-CE31V exhibits similar CSP patterns as wild-type (Figure 2A,C) with larger CSPs occurring throughout the N-lobe and in the c-terminal tail, though to a lesser extent than PKA-CWT. Further analysis of the ΔCSP (ΔδWT-ΔδE31V) shows regions of positive ΔCSP confirming that upon binding ATPγN, PKA-CE31V does not adopt as closed of a conformation as the wild-type kinase (Figure S2A). Subsequent binding of PKI to ATPγN-saturated PKA-CE31V also exhibits similar CSPs compared to wild-type (Figure 2B,D), though to a larger extent as reflected in the negative ΔCSP values (Figure S2B). Indeed, these results are consistent with the CONCISE analysis indicating that PKI binding shifts the kinase to a more closed conformation for PKA-CE31V.
Figure 2. Structural response of PKA-CE31V binding to nucleotide and protein kinase inhibitor.
Chemical shift perturbation (CSP) of amide fingerprint of PKA-CE31V upon binding (A) ATPγN and subsequent binding of (B) PKI. The average CSP is shown as a dashed line. CSPs of PKA-CE31V amide resonances mapped onto the structure (PDB: 1ATP).
Rearrangement of the allosteric network of PKA-CE31V is linked to a decrease in nucleotide-substrate binding cooperativity.
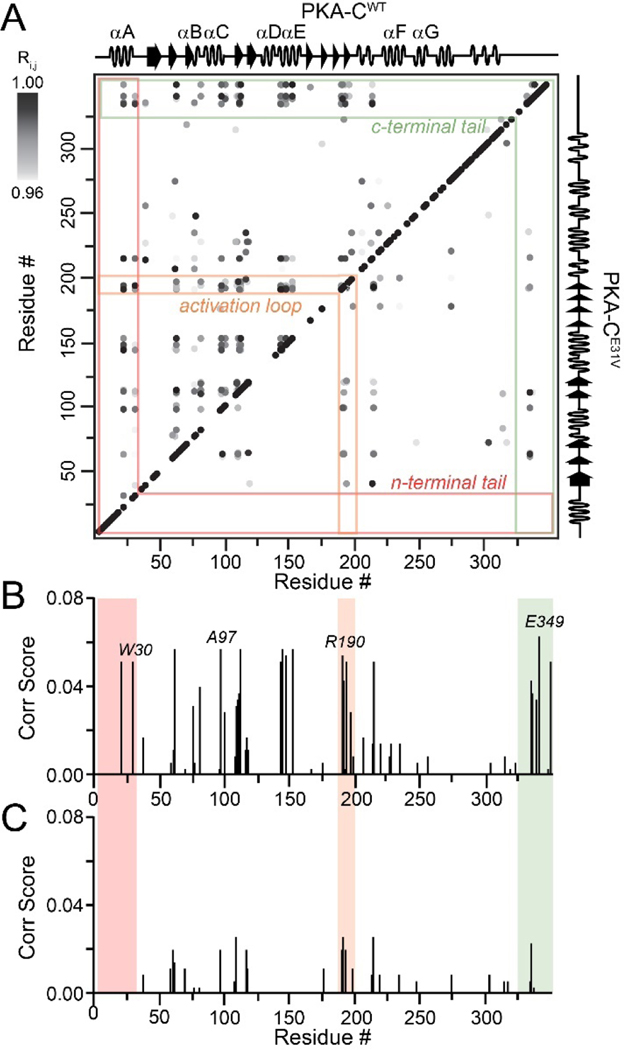
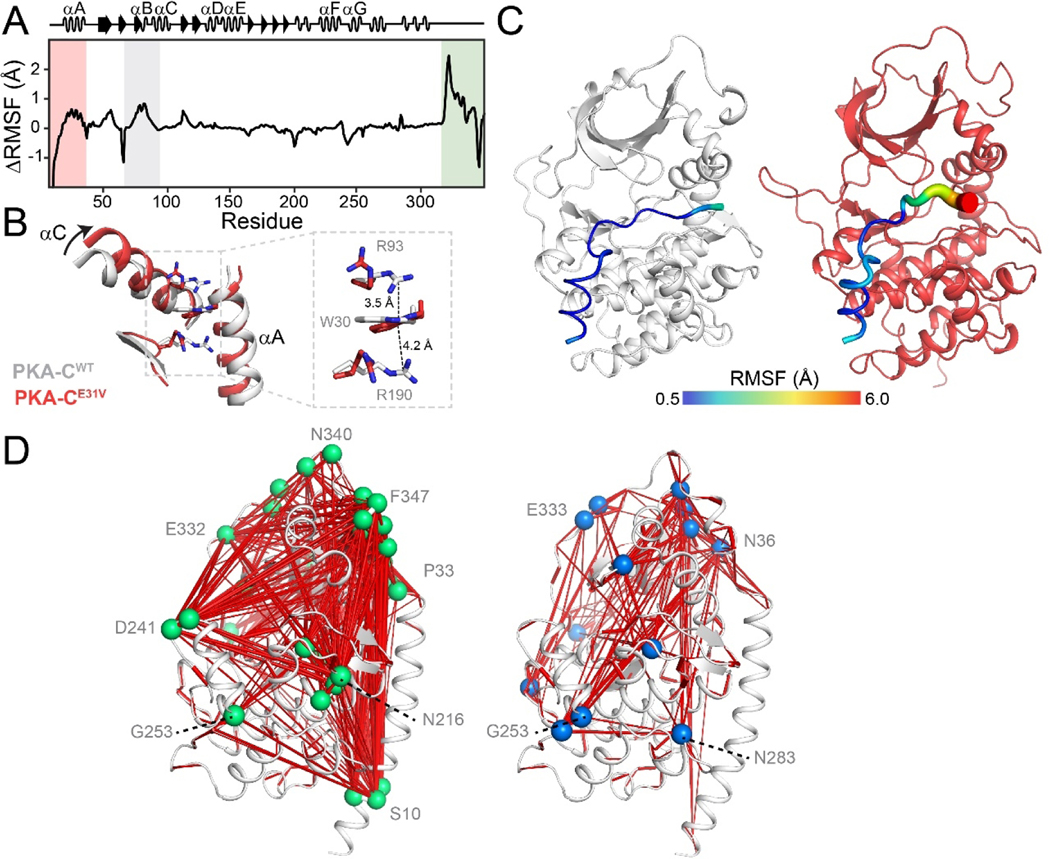
Since cooperativity is often manifested as structural rearrangements upon ligand binding, we used CHEmical Shift Covariance Analysis (CHESCA) to map the allosteric network of PKA-CE31V and compared our results to the wild-type kinase. Briefly, this statistical method identifies covariant residues networks involved in a concerted response upon ligand binding and help tracing allosteric pathways [25–27]. The [1H, 15N]-TROSY-HSQC spectra of four forms of wild-type and PKA-CE31V (apo, ATPγN-bound, ADP-bound, and ATPγN/PKI-bound) were used for CHESCA. Previously, we analyzed the chemical shift changes of PKA-CWT and identified a well-organized communication network in which spatially distinct clusters of residues responding to ATP and PKI binding in a coordinated manner [20, 28]. In contrast, we observed a dramatic reduction in the intramolecular allosteric network of PKA-CE31V similar to PKA-CL205R, a Cushing’s syndrome mutation with significantly higher occurrence [19, 20, 29]. In particular, highly correlated groups of residues in the N-lobe of PKA-CE31V, including the αA-helix (K28, W30) and αC-helix (A97), display a dramatic reduction in the number of correlations for distal regions of the kinase, including the activation loop (R190, G193, L198), αF-helix (K217, V219, G225), and C-terminal tail (E334, N340, E349) (Figure 3). Notably, the loss of correlations occurs in structural elements surrounding the αC-helix.
Figure 3. Intramolecular allosteric network as mapped by correlated chemical shift changes.
(A) CHESCA correlation matrix of PKA-CWT (top diagonal) and PKA-CE31V (bottom diagonal) upon binding PKI highlighting the notable reductions in correlations within the n-terminal tail (red), activation loop (orange), and c-terminal tail (green). Only correlations with Rij> 0.98 are shown. Plot of correlation score vs. residue for (B) PKA-CWT and (C) PKA-CE31V emphasizing the residues that show the largest reductions in correlation score that make contacts with the αC-helix. See material and methods for the calculation of correlation scores.
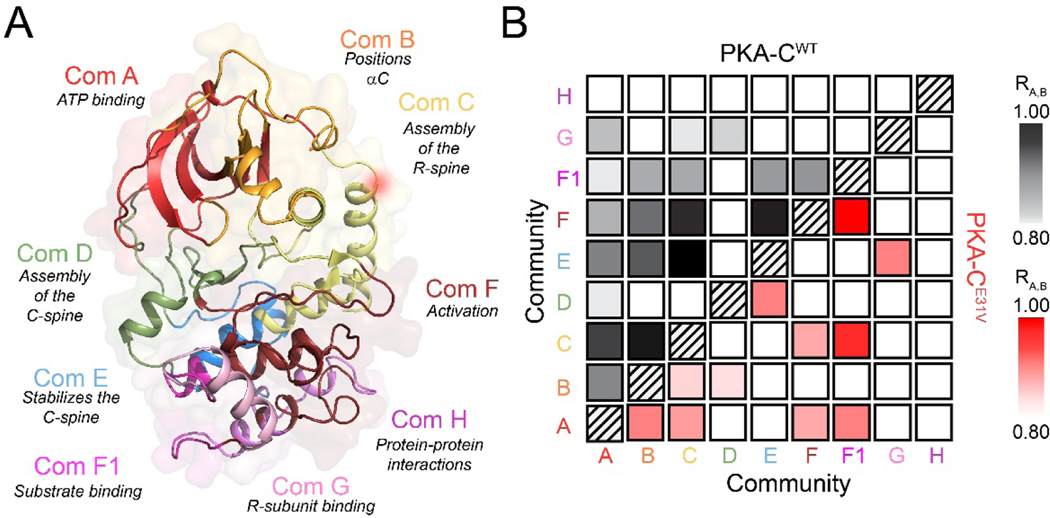
The typical CHESCA analysis gives pairwise correlations along the primary protein sequences. Therefore, we adopted the definition of structural ‘communities’ introduced by McClendon et al. [30] to obtain a three-dimensional view of the correlated structural changes. Using this analysis, we found strong correlations among the major communities in response to nucleotide and pseudosubstrate binding (Figure 4) [30]. In particular, ComA, ComB, and ComC show average correlation coefficients higher than 0.95, indicating that these communities respond to ligand binding in a concerted manner. Notably, there are long-range correlations between ComA, ComB, and ComC with ComE, ComF and ComF1. ComC, which encompasses the αA- and αC-helix including E31, acts as a central hub, connecting six other communities as it is centered around a critical allosteric mediator the αC-helix, which bridges both lobes of the kinase. The density of these correlations underscores the concerted response of the N- and C-lobe to ligand binding. While the E31V mutation exhibits some of the local and long-range correlations, the values of the correlation coefficients are lower. ComC and parts of the activation loop of the mutant exhibit the most noticeable reduction in correlation to both local (ComA and ComB) and distal (ComE and ComF) communities. Indeed ComC acts as a hub in the wild-type community map, having connections to six other communities and centered around a critical allosteric mediator of the kinase regulation, the αC-helix.
Figure 4. Allosteric network among the different functional communities of the kinase for both PKA-CWT and PKA-CE31V.
(A) Community map of PKA-C highlighting functional/regulatory role of each community as defined by [30]. (B) Community CHESCA analysis of PKA-CWT (top diagonal, black) and PKA-CE31V (bottom diagonal, red). Only correlations with RA,B > 0.8 are shown.
MD simulations corroborate the reduction of intramolecular correlations in the allosteric network of PKA-CE31V. To determine the effects of the E31V mutation on the conformational energy landscape of nucleotide-bound PKA-C, we carried out parallel MD simulations in explicit water. We set up the simulations starting from the X-ray coordinates of PKA-CWT (PDB: 4WB5 [31]) mutating E31 into a valine and removing PKI [18]. After initial equilibration, we produced an MD trajectory and analyzed the conformational dynamics of PKA-CE31V. Relative to PKA-CWT, we observed large changes of root mean squared fluctuations (RMSF) of the backbone coordinates with effects that propagate to distal motifs [32], including the αA-helix, αC-helix, as well as the C-terminal tail (Figure 5A). The most noticeable effect of the E31V mutation is the increase in the αA-helix and αC-helix motions, presumably due to V31 moving towards the kinase’s hydrophobic interior and disrupting the cation-π interactions between W30, R93 and R190. These ‘sandwiched’ cation-π interactions are partially responsible for positioning the indole ring of W30 in a conserved pocket that can be exploited to regulate kinases activity [22, 33]. As the αA-helix is displaced, the indole ring of W30 undergoes a 180° flip, maintaining only one cation-π interaction with R190 (Figure 5B). The alteration of local structure is accompanied by the reduction of the global allostery in PKA-CE31V with respect to PKA-CWT, as is characterized by the dihedral mutual information of all residue pairs. The most prominent changes are the loss of correlation between N-terminal αA-helix and other motifs (Figure S3A). The mutual information matrices are further mapped onto the kinase structure via graph analysis, highlighting the attenuated network and vanish of key hubs throughout the kinase, particularly for S10 at the tip of αA-helix, N216 at αF-helix, D241 at αG-helix, and N340, F347 at the C-terminal tail (Figure S3B), in good accordance with the CHESCA analysis. How does the attenuated allosteric network between N-lobe and C-lobe affect the binding of pseudosubstrate PKI5–24 and cooperativity? We first compared the dynamics of PKI in the ternary complexes. Whereas PKI5–24 is rather rigid with an average RMSF of about 1 Å for PKA-CWT, the short peptide is much more dynamic for PKA-CE31V, especially in its C-terminal end (Figure 5C). We next analyzed interaction profile of PKI5–24 by computing the contact probability of all residue pairs between the short peptide and kinase. The stable interaction at the high affinity region (HAR) and the recognition sequence (P-3, P-2 and P+1 sites) is largely preserved, yet the contact probability between the C-terminal end of PKI and the peptide-positioning loop (residues 198–208) decreases (Figure S4), indicative of dynamic C-terminal flipping out of the hydrophobic pocket. Loss of anchoring of C-terminal end echoes the over 100-fold reduction in binding affinity measured by ITC.
Figure 5. Altered conformational dynamics of PKA-CE31V revealed by MD simulations.
(A) ΔRMSF of PKA-CE31V in the binary form over 1.0 μs of simulation. (B) Overlay of PKA-CWT (PDB: 1ATP; gray) and PKA-CE31V (from MD simulations; red) highlighting the structural rearrangements of the αA- and αC-helices caused by the E31V mutation. The inset shows the cation-π stacking interactions altered in response to the mutation. (C) Distinct dynamics of the pseudosubstrate, PKI5–24, in the ternary complex. Putty representation of RMSF for PKI5–24 for a typical MD snapshot of PKA-CWT (gray) and PKA-CE31V (red). (D)Comparison of allosteric mapping from mutual information matrices (MIcutoff = 0.3), with the key hub residues labeled for PKA-CWT (right) and PKA-CE31V (left).
To link the MD simulations to the binding thermodynamics derived from the ITC experiments, we computed the difference in free energy of binding (ΔΔGbinding) for PKA-CWT and PKA-CE31V using the free energy perturbation (FEP) method [34], as detailed in the thermodynamic cycle shown in Figure S5. The ratio of the cooperativity coefficients can be expressed in terms of free energy:
Where represents the ratio of the cooperativity coefficients for wild-type and E31V, and the free energy change of a mutation in different states ΔG1 to ΔG4 is illustrated in Figure S5A. Using this expression for the binding of PKI to apo PKA-CE31V, the FEP method calculates a free energy difference between the free and bound state of −1.1 ± 0.3 kcal/mol, corresponding to a ~7-fold reduction in the binding affinity of PKA-CE31V for PKI. This value is in excellent agreement with the experimental results (Table S4). On the other hand, the binding of PKI to the nucleotide-bound E31V mutant resulted in a free energy perturbation of 0.3 ± 0.2 kcal/mol, indicating a reduction in binding affinity. From the differences of these two values, it is possible to estimate approximately a 11-fold reduction in the nucleotide/PKI binding cooperativity (Figure S5B), which is in qualitative agreement with ITC experiments. We also calculated the change in cooperativity for binding PKI first and then the nucleotide (Figure S5C). These calculations confirmed the reduction in cooperativity with a value that is approximately 24-fold lower for the ATP binding by PKA-CE31V, further supporting the experimentally observed loss in cooperativity for PKA-CE31V (Table S4).
Nucleotide-substrate binding cooperativity and extent of allosteric communication are directly correlated.
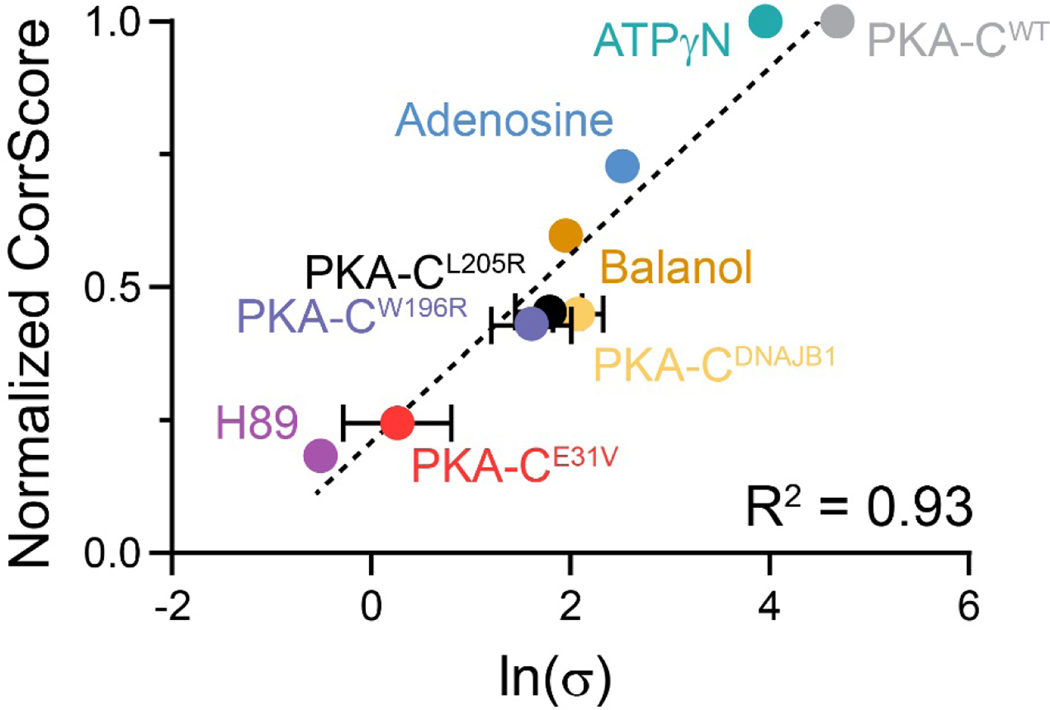
Both thermodynamic and NMR data show that Cushing’s syndrome mutants, PKA-CL205R and PKA-CE31V, exhibit reduced binding cooperativity and intramolecular allosteric communication[20]. Therefore, we hypothesized that the coordinated structural changes might be correlated to the nucleotide-substrate cooperative binding response. Hence, a disruption of the allosteric network would directly affect the nucleotide-substrate binding cooperativity. To test this, we analyzed the thermodynamics and structural response of PKA-CW196R, another mutant that has been found in 3% of Cushing’s patients [13]. This mutation is in the activation loop and is adjacent to the T197 phosphorylation site. We repeated both ITC and NMR analyses for PKA-CW196R, and similarly to PKA-CE31V and PKA-CL205R we found a significant attenuation in both binding cooperativity and extent of intramolecular allosteric communication (Table S5, Figure S6). From the CHESCA matrices of these three mutants and PKA-CWT, we extracted the relative correlation score (see Material and Methods), which can be used to estimate the density of the intramolecular allosteric networks. Included in the analysis were the thermodynamic and allosteric values obtained for the binding of inhibitors, H89 and Balanol, and nucleotides, ATPγN and adenosine, to PKA-CWT as well as those for a chimeric fusion of the PRKACA and DNAJB1 genes (PKA-CDNAJB1)[28], which has been reported to be a driver of fibrolamellar hepatocellular carcinoma[35]. Plotting the relative correlation scores versus ln(σ), we found that they are linearly correlated (R2 = 0.93, Figure 6). This relationship suggests that the extent of the nucleotide-substrate binding cooperativity depends on the coordinated structural changes of the two lobes of the enzyme upon nucleotide and substrate binding. Importantly, the Cushing’s mutants analyzed suggest that the loss of allosteric cooperativity between nucleotide and substrate binding may be a common feature.
Figure 6. Relationship between coordinated structural changes identified by NMR chemical shifts and the nucleotide-substrate binding cooperativity (σ).
Values used for plotting σ of PKA-CL205R and PKA-CDNAJB1 have been published in [20] and [28], respectively. Data points without error bars indicate that error is contained within the point itself.
DISCUSSION
The cAMP/PKA pathway plays a central role in the regulation of adrenocortical growth and steroidogenesis [3, 4]. Although multiple components of the cAMP/PKA pathway have been implicated in various endocrine disease states, it was not until recently that PKA-C was shown to be mutated in disease states [36]. To date, eight mutations have been discovered in PRKACA in cortisol-producing ACA’s responsible for Cushing’s syndrome [5–12]. Except for E31V, all mutations are positioned in the substrate binding cleft or at the R/C interface, providing a justification for the loss in substrate fidelity and regulation of the kinase [13, 20]. However, the few studies performed on the E31V mutant have found that the enzyme’s activity is similar to the wild-type and still able to bind the R-subunits and respond to cAMP activation, making it difficult to rationalize why this mutant results in the same phenotype of the other orthosteric mutations.
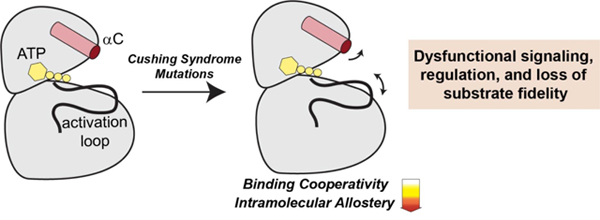
The present study shows that small local conformational changes induced by E31V alter key allosteric interactions that link the terminal regions of the C-terminal tail, αA- and αC-helix to elicit dysfunction. MD simulations revealed that PKA-CE31V experiences increased conformational dynamics within the αA-helix, causing it to dislodge from the kinase core and thereby disrupting canonical cation-π interactions between W30 and R93 and R190 [37, 38]. These structural alterations cause the N-lobe of the kinase to swing outward adopting a more open conformation and thereby disrupting a critical allosteric node responsible for conferring inter-lobe allosteric cooperativity [37].
With allosteric cooperativity playing fundamental role, specifically in regards to macromolecular assembly and signal amplification, it is likely that the loss in nucleotide/pseudosubstrate binding cooperativity has implication on the kinases interactions with its regulatory partners and substrates [39–41]. Indeed, the significant crosstalk amongst signaling pathways alone suggests that dysfunction of one component, in this case PKA-CE31V, can elicit multiple disease consequences. Despite this, the implications of dysfunctional allosteric cooperativity on regulation are currently not well established. Nevertheless, this study lays the foundation for future work addressing the fundamental questions about the linkage between dysfunctional allostery and regulation of PKA. Specifically, how do changes in allosteric cooperativity manifest as alterations in the regulation of PKA? Or, do changes in the allosteric network of the kinase induced by mutation drive non-canonical protein-protein interactions?
In conclusion, our study highlights the intricate regulation of allosteric information transfer and demonstrates that small perturbations in the form of mutations can disrupt enzymatic function through altercations of allosteric pathways and/or conformational ensembles [42, 43]. Indeed, both orthosteric and allosteric mutations to Cushing’s syndrome display reduced binding cooperativity coupled to a loss in intramolecular allosteric communication. Nonetheless, the effects of reduced allosteric cooperativity are likely multifaceted including a loss in substrate fidelity, alterations in regulation, and modulations in catalytic activity. Altogether, these studies provide a foundation for future studies on the allosteric mechanisms of proteins and their disease-associated variations with a particular emphasis on how mutations exert control over allosteric cooperativity to manifest (dys)function.
MATERIALS AND METHODS
Sample Preparation.
Recombinant human Cα subunit of cAMP-dependent protein kinase A cDNA (PKA-CWT and PKA-CE31V) was cloned into a pET-28a vector. A tobacco etch virus (TEV) cleavage site was incorporated via mutagenesis into the vector between the cDNA coding for the kinase and a thrombin cleavage site. The kinase was expressed in Escherichia coli BL21 (DE3) according to procedures previously published [20]. PKI (full-length) was expressed and purified according to procedures previously published [44]. Peptides (Kemptide/PKI5–24) were synthesized using standard Fmoc chemistry on a CEM Liberty Blue microwave synthesizer, cleaved with Reagent K (82.5% TFA, 5% phenol, 5% thioanisole, 2.5% ethanedithiol, and 5% water) for 3 h and purified using a semipreparative Supelco C18 reverse-phase HPLC column at 3 mL/min. Molecular weight and the quantity of the peptides were verified by LC-MS and/or amino acid analysis (Texas Tech Protein Chemistry Laboratory).
ITC Measurements.
ITC measurements were performed with a low-volume NanoITC (TA Instruments). PKA-CWT and PKA-CE31V were dialyzed into 20 mM MOPS, 90 mM KCl, 10 mM DTT, 10 mM MgCl2, and 1 mM NaN3 (pH 6.5). PKA-C concentrations for ITC measurements were between 100–110 μM as confirmed by A280 = 53860 M−1cm-1. All measurements with ATPγN saturated PKA-CWT and PKA-CE31V were performed at 2 mM ATPγN. ITC measurements were performed at 300K in triplicates. Approximately 300 μL of PKA-C was used for each experiment, and 50 μL of 2 mM ATPγN and 0.6–0.8 mM PKI in the titrant syringe. The heat of dilution of the ligand into the buffer was taken into account for all experiments and subtracted. Curves were analyzed with the NanoAnalyze software (TA Instruments) using the Wiseman Isotherm [21]:
| (1) |
where d[MX] is the change in total complex with respect to change in total protein concentration, d[Xtot] is dependent on r, the ratio of Kd with respect to the total protein concentration, and RM, the ratio between total ligand and total protein concentration. The free energy of binding was determined using the following:
where R is the universal gas constant and T is the temperature at measurement (300K). The entropic contribution to binding was calculated using the following:
Calculations for the cooperativity constant (σ) were calculated as follows:
where Kd Apo is the Kd of PKI5–24 binding to the apoenzyme and Kd Nucleotide is the Kd of PKI5–24 binding to the nucleotide-bound enzyme.
Enzyme Assays.
Steady-state activity assays with Kemptide were performed under saturating ATP concentrations and spectrophotometrically at 298K as described by Cook et al [45]. The values of Vmax and KM were obtained from a nonlinear fit of the initial velocities to the Michaelis-Menten equation.
NMR Spectroscopy.
Uniformly 15N-labeled PKA-CWT and PKA-CE31V were overexpressed and purified as described above. NMR experiments were performed in 90 mM KCl, 20 mM KH2PO4, 10 mM dithiothreitol (DTT), 10 mM MgCl2, and 1 mM NaN3 at pH 6.5. Standard [1H-15N]-TROSY-HSQC experiments were carried out for PKA-CE31V and PKA-CWT on a 900-MHz Bruker Advance III spectrometer equipped with a TCI cryoprobe. Concentrations for samples were 0.2–0.3 mM as determined by A280 measurements, 12 mM ATPγN was added for the nucleotide-bound form, and 0.2–1.2 mM PKI for the ternary complex. Spectra were collected at 300K, processed using NMRPipe [46], and visualized using Sparky [47].
All [1H-15N]-TROSY-HSQC experiments were acquired with 2048 (proton) and 256 (nitrogen) complex points. Combined chemical shift perturbations were calculated using 1H and 15N chemical shifts according to the following:
Chemical Shift Analyses.
COordiNated ChemIcal Shift bEhavior (CONCISE).
CONCISE was used to monitor chemical shift trajectories and measure the change in equilibrium position using each PKA-C construct (apo, ATPγN, ADP, ATPγN/PKI). This method uses principal component analysis to identify sets of residues whose chemical shifts respond linearly to a conformational transition (i.e. open, intermediate, and closed). Each residue provides a measure of the equilibrium position for each PKA-C construct in the form of scores along the first principal component (PC1). To identify the residues whose chemical shifts follow a linear trajectory, a threshold of 3.0 for the ratio of the standard deviations of PC1 over PC2 was used, and residues not exhibiting a significant chemical shift were excluded based on the linewidth.
CHEmical Shift Covariance Analysis (CHESCA).
CHESCA was used to identify and functionally characterize allosteric networks of residues eliciting concerted responses to, in this case, nucleotide and pseudosubstrate. A total of four states were used to identify inter-residue correlations: apo, ADP-bound, ATPγN-bound, and ATPγN/PKI-bound. Identification of inter-residue correlations by CHESCA relies on agglomerative clustering (AC) and singular value decomposition (SVD). Pairwise correlations between chemical shift variations experienced by different residues are analyzed to identify networks of coupled residues and when plotted on a correlation matrix, allows for the identification of regions that are correlated to one another. A correlation coefficient (Rij) cutoff of 0.96 was used to filter non-linear residues. Residues not exhibiting a significant chemical shift (small shifts in ppm) were excluded based on linewidth. For each residue the max change in chemical shift was calculated in both the 1H (x) and 15N (y) dimension (Δδx,y). Residues were included in CHESCA analysis only if they satisfied the following: Δδx,y > ½ ΔνxA,yA+½ Δν xB,yB, where A and B correspond to two different forms analyzed (note there is no dependence on which two forms satisfied this statement) and Δv denotes the linewidth. Correlation scores were used to quantify the CHESCA correlation of a single residue or a group of residues with another group. Correlation scores were evaluated for both (a) a single residue and (b) the full protein. The generalized mathematical expression for evaluating either case is given as follows:
where R is the CHESCA correlation matrix and i and j denote (a) a single residue and all other assigned residues in the protein, respectively, or (b) both represent all the assigned residues in the protein. Community CHESCA analysis is a chemical shift based correlation map between various functional communities within the kinase. Each community is a group of residues (McClendon et al.[30]) associated with a function or regulatory mechanism. Mathematically, this community-based CHESCA analysis is a selective interpretation of CHESCA, where we evaluate a correlation score between residues in various communities as shown below. In order to represent community-based CHESCA analysis we lowered the correlation cutoff such that Rcutoff > 0.8. Suppose community A and community B has nA and nB number of assigned residues respectively, the correlation score between A and B is defined as,
Where Rij is the CHESCA correlation coefficient between residue i (belongs to community A) and residue j (belongs to community B). Rcutoff is the correlation value cutoff. RA,B can take values from 0 (no correlation between residues in A and B) to 1 (all residues in A has correlation > cutoff with all residues in B).
MD Simulations.
We used the crystal structure of PKA-CWT (PDB ID: 4WB5[31]) as the template. We further aligned the current structure with the full length PKA-CWT and added the missing residues 1–12 at the N terminus. The protonation state of histidine residues followed our previous settings [32]. The protein was solvated in a rhombic dodecahedron solvent box with TIP3P water molecule layer extended approximately 10 Å away from the surface of the proteins. Counter ions (K+ and Cl-) were added to ensure electrostatic neutrality corresponding to an ionic concentration of ~150 mM. All covalent bonds involving a hydrogen atom of the protein were constrained with the LINCS[48]algorithm. and long-range electrostatic interactions were treated with the particle-mesh Ewald [49] method with a real-space cutoff of 10 Å. Parallel simulations on the apo form, the binary form with one Mg2+ ion and one ATP, and the ternary form with two Mg2+ ions, one ATP and one PKI5–24 were performed simultaneously using GROMACS 5.1.4 [50] with the CHARMM36a1 force field [51]. Each system was minimized using the steepest decent algorithm to remove bad contacts, and then gradually heated to 300K at a constant volume over 1 ns, using harmonic restraints with a force constant 1000 kJ/(mol*Å2) on heavy atoms of both proteins and nucleotides. Over the following 12 ns of simulations at constant pressure (1 atm) and temperature (300K), the restraints were gradually released. The systems were equilibrated for an additional 20 ns without positional restraints. The Parrinello-Rahman[52] barostat was used to keep the pressure constant, while a V-rescale thermostat with a time step of 2 fs was used to keep the temperature constant. Each system was simulated for 1.0 μs, with snapshots recorded every 50 ps. To characterize the allosteric network, the dihedral mutual information was computed using MDEntropy [53] for every residue pair, and the normalized mutual information matrix was mapped onto the structure for graph analysis using Xpyder [54] with MIcutoff = 0.3.
Relative change of cooperativity from free energy perturbation calculations.
The cooperativity can be defined for both nucleotide and pseudosubstrate PKI, respectively. For nucleotide, the change of cooperativity upon mutation can be rewritten as the difference in ΔΔG between the apo and the PKI-bound state, as shown in the following equation and illustrated in Figure S5:
The free energies ΔG due to amino acid mutations were determined following a protocol based on the Bennett acceptance ratio (BAR) implemented in the GROMACS and PMX [34]. To avoid the artifacts by introducing a charged mutation, the double-system/single-box setup was used. The procedure employs dual protein topologies that include both residues of the wild-type (λ = 0) and the mutant protein (λ = 1) coupled by the progressing variable λ. Of course, both the complex and unbound structures were used to obtain the change in binding free energies using standard thermodynamic cycle approach. Single-site mutations were performed based on the well-equilibrated structure of PKA-CWT from simulations. The computational details are identical to those detailed above, except that after 40 ns of equilibration of both initial and final states for each mutation, 200 additional trajectories, each lasting 100 ps, were initiated from the last 20 ns simulations both in the forward and in the backward transformations to accumulate statistical averages and fluctuations.
Supplementary Material
Research Highlights.
A Cushing’s disease mutation of protein kinase A disrupts nucleotide-substrate binding cooperativity
The E31V mutation attenuates the intramolecular allosteric network mapped by NMR
The reduction of intramolecular allostery is log-linearly correlated with the loss of binding cooperativity
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health, GM100310 (G.V.), S10 OD021536 (G.V.) and GM046736 (J.G), the American Heart Association, 20PRE35120253 (C.W), and the National Natural Science Foundation of China No. 22007069 (Y.W.). NMR experiments were carried out at the Minnesota NMR Center and MD calculations at the Minnesota Supercomputing Institute.
Footnotes
Catlin Walker: investigation, formal analysis, data curation, writing original draft, Yingjie Wang: investigation, formal analysis, data curation writing original draft, Cristina Olivieri: formal analysis, data curation. Manu V.S: formal analysis, data curation. Jiali Gao: conceptualization, review draft and editing. David Bernlohr: conceptualization, review, and editing. Davide Calebiro: review and editing. Susan S. Taylor: conceptualization, review & editing. Gianluigi Veglia: conceptualization, review, funding acquisition, writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no competing financial interests or personal relationships that could have appeared to influence the work reported in this paper
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Lacroix A, Feelders RA, Stratakis CA, Nieman LK. Cushing’s syndrome. Lancet (London, England). 2015;386:913–27. [DOI] [PubMed] [Google Scholar]
- [2].Lodish M, Stratakis CA. A genetic and molecular update on adrenocortical causes of Cushing syndrome. Nature reviews Endocrinology. 2016;12:255–62. [DOI] [PubMed] [Google Scholar]
- [3].Calebiro D, Di Dalmazi G, Bathon K, Ronchi CL, Beuschlein F. cAMP signaling in cortisol-producing adrenal adenoma. European Journal of Endocrinology. 2015;173:M99–M106. [DOI] [PubMed] [Google Scholar]
- [4].Calebiro D, Bathon K, Weigand I. Mechanisms of Aberrant PKA Activation by Cα Subunit Mutations. Horm Metab Res. 2017;49:307–14. [DOI] [PubMed] [Google Scholar]
- [5].Goh G, Scholl UI, Healy JM, Choi M, Prasad ML, Nelson-Williams C, et al. Recurrent activating mutation in PRKACA in cortisol-producing adrenal tumors. Nature genetics. 2014;46:613–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Cao Y, He M, Gao Z, Peng Y, Li Y, Li L, et al. Activating hotspot L205R mutation in PRKACA and adrenal Cushing’s syndrome. Science. 2014;344:913–7. [DOI] [PubMed] [Google Scholar]
- [7].Di Dalmazi G, Kisker C, Calebiro D, Mannelli M, Canu L, Arnaldi G, et al. Novel somatic mutations in the catalytic subunit of the protein kinase A as a cause of adrenal Cushing’s syndrome: a European multicentric study. The Journal of clinical endocrinology and metabolism. 2014;99:E2093–100. [DOI] [PubMed] [Google Scholar]
- [8].Beuschlein F, Fassnacht M, Assié G, Calebiro D, Stratakis CA, Osswald A, et al. Constitutive Activation of PKA Catalytic Subunit in Adrenal Cushing’s Syndrome. New England Journal of Medicine. 2014;370:1019–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ronchi CL, Di Dalmazi G, Faillot S, Sbiera S, Assié G, Weigand I, et al. Genetic Landscape of Sporadic Unilateral Adrenocortical Adenomas Without PRKACA p.Leu206Arg Mutation. The Journal of Clinical Endocrinology & Metabolism. 2016;101:3526–38. [DOI] [PubMed] [Google Scholar]
- [10].Sato Y, Maekawa S, Ishii R, Sanada M, Morikawa T, Shiraishi Y, et al. Recurrent somatic mutations underlie corticotropin-independent Cushing’s syndrome. Science (New York, NY). 2014;344:917–20. [DOI] [PubMed] [Google Scholar]
- [11].Thiel A, Reis AC, Haase M, Goh G, Schott M, Willenberg HS, et al. PRKACA mutations in cortisol-producing adenomas and adrenal hyperplasia: a single-center study of 60 cases. European journal of endocrinology. 2015;172:677–85. [DOI] [PubMed] [Google Scholar]
- [12].Nakajima Y, Okamura T, Gohko T, Satoh T, Hashimoto K, Shibusawa N, et al. Somatic mutations of the catalytic subunit of cyclic AMP-dependent protein kinase (PRKACA) gene in Japanese patients with several adrenal adenomas secreting cortisol [Rapid Communication]. Endocrine journal. 2014;61:825–32. [DOI] [PubMed] [Google Scholar]
- [13].Bathon K, Weigand I, Vanselow JT, Ronchi CL, Sbiera S, Schlosser A, et al. Alterations in Protein Kinase A Substrate Specificity as a Potential Cause of Cushing Syndrome. Endocrinology. 2019;160:447–59. [DOI] [PubMed] [Google Scholar]
- [14].Taylor SS, Ilouz R, Zhang P, Kornev AP. Assembly of allosteric macromolecular switches: lessons from PKA. Nature Reviews Molecular Cell Biology. 2012;13:646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Taylor SS, Ilouz R, Zhang P, Kornev AP. Assembly of allosteric macromolecular switches: lessons from PKA. Nature reviews Molecular cell biology. 2012;13:646–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Langeberg LK, Scott JD. Signalling scaffolds and local organization of cellular behaviour. Nature Reviews Molecular Cell Biology. 2015;16:232–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Johnson DA, Akamine P, Radzio-Andzelm E, Taylor SS. Dynamics of cAMP-Dependent Protein Kinase. Chemical Reviews. 2001;101:2243–70. [DOI] [PubMed] [Google Scholar]
- [18].Knighton DR, Zheng JH, Ten Eyck LF, Ashford VA, Xuong NH, Taylor SS, et al. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science (New York, NY). 1991;253:407–14. [DOI] [PubMed] [Google Scholar]
- [19].Calebiro D, Hannawacker A, Lyga S, Bathon K, Zabel U, Ronchi C, et al. PKA catalytic subunit mutations in adrenocortical Cushing’s adenoma impair association with the regulatory subunit. Nature Communications. 2014;5:5680. [DOI] [PubMed] [Google Scholar]
- [20].Walker C, Wang Y, Olivieri C, Karamafrooz A, Casby J, Bathon K, et al. Cushing’s syndrome driver mutation disrupts protein kinase A allosteric network, altering both regulation and substrate specificity. Science Advances. 2019;5:eaaw9298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wiseman T, Williston S, Brandts JF, Lin L-N. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Analytical Biochemistry. 1989;179:131–7. [DOI] [PubMed] [Google Scholar]
- [22].Herberg FW, Zimmermann B, Mcglone M, Taylor SS. Importance of the A-helix of the catalytic subunit of cAMP-dependent protein kinase for stability and for orienting subdomains at the cleft interface. Protein Science. 1997;6:569–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Pervushin K, Riek R, Wider G, Wüthrich K. Attenuated T2 relaxation by mutual cancellation of dipole–dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proceedings of the National Academy of Sciences. 1997;94:12366–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Cembran A, Kim J, Gao J, Veglia G. NMR mapping of protein conformational landscapes using coordinated behavior of chemical shifts upon ligand binding. Physical Chemistry Chemical Physics. 2014;16:6508–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Selvaratnam R, Chowdhury S, VanSchouwen B, Melacini G. Mapping allostery through the covariance analysis of NMR chemical shifts. Proceedings of the National Academy of Sciences. 2011;108:6133–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Boulton S, Akimoto M, Selvaratnam R, Bashiri A, Melacini G. A Tool Set to Map Allosteric Networks through the NMR Chemical Shift Covariance Analysis. Scientific Reports. 2014;4:7306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Byun JA, Melacini G. NMR methods to dissect the molecular mechanisms of disease-related mutations (DRMs): Understanding how DRMs remodel functional free energy landscapes. Methods (San Diego, Calif). 2018;148:19–27. [DOI] [PubMed] [Google Scholar]
- [28].Olivieri C, Walker C, Karamafrooz A, Wang Y, Manu VS, Porcelli F, et al. Defective internal allosteric network imparts dysfunctional ATP/substrate-binding cooperativity in oncogenic chimera of protein kinase A. Communications Biology. 2021;4:321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Di Dalmazi G, Kisker C, Calebiro D, Mannelli M, Canu L, Arnaldi G, et al. Novel somatic mutations in the catalytic subunit of the protein kinase A as a cause of adrenal Cushing’s syndrome: a European multicentric study. The Journal of clinical endocrinology and metabolism. 2014;99:E2093–100. [DOI] [PubMed] [Google Scholar]
- [30].McClendon CL, Kornev AP, Gilson MK, Taylor SS. Dynamic architecture of a protein kinase. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E4623–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Cheung J, Ginter C, Cassidy M, Franklin MC, Rudolph MJ, Robine N, et al. Structural insights into mis-regulation of protein kinase A in human tumors. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:1374–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Masterson LR, Shi L, Metcalfe E, Gao J, Taylor SS, Veglia G. Dynamically committed, uncommitted, and quenched states encoded in protein kinase A revealed by NMR spectroscopy. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:6969–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Thompson EE, Kornev AP, Kannan N, Kim C, Ten Eyck LF, Taylor SS. Comparative surface geometry of the protein kinase family. Protein science : a publication of the Protein Society. 2009;18:2016–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Gapsys V, Michielssens S, Seeliger D, de Groot BL. pmx: Automated protein structure and topology generation for alchemical perturbations. Journal of Computational Chemistry. 2015;36:348–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Honeyman JN, Simon EP, Robine N, Chiaroni-Clarke R, Darcy DG, Lim IIP, et al. Detection of a recurrent DNAJB1-PRKACA chimeric transcript in fibrolamellar hepatocellular carcinoma. Science (New York, NY). 2014;343:1010–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Stratakis CA . Cyclic AMP-dependent protein kinase catalytic subunit A (PRKACA): the expected, the unexpected, and what might be next. J Pathol. 2018;244:257–9. [DOI] [PubMed] [Google Scholar]
- [37].Taylor SS, Kornev AP. Protein kinases: evolution of dynamic regulatory proteins. Trends in Biochemical Sciences. 2011;36:65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Cembran A, Masterson LR, McClendon CL, Taylor SS, Gao J, Veglia G. Conformational equilibrium of N-myristoylated cAMP-dependent protein kinase A by molecular dynamics simulations. Biochemistry. 2012;51:10186–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Williamson JR. Cooperativity in macromolecular assembly. Nature Chemical Biology. 2008;4:458–65. [DOI] [PubMed] [Google Scholar]
- [40].Wu H. Higher-order assemblies in a new paradigm of signal transduction. Cell. 2013;153:287–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Tsai CJ, Del Sol A, Nussinov R. Protein allostery, signal transmission and dynamics: a classification scheme of allosteric mechanisms. Mol Biosyst. 2009;5:207–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Masterson LR, Mascioni A, Traaseth NJ, Taylor SS, Veglia G. Allosteric cooperativity in protein kinase A. Proc Natl Acad Sci U S A. 2008;105:506–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Foda ZH, Shan Y, Kim ET, Shaw DE, Seeliger MA. A dynamically coupled allosteric network underlies binding cooperativity in Src kinase. Nature communications. 2015;6:5939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Olivieri C, Wang Y, Li GC, V S M, Kim J, Stultz BR, et al. Multi-state recognition pathway of the intrinsically disordered protein kinase inhibitor by protein kinase A. eLife. 2020;9:e55607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Cook PF, Neville ME, Vrana KE, Hartl FT, Roskoski R. Adenosine cyclic 3’,5’monophosphate dependent protein kinase: kinetic mechanism for the bovine skeletal muscle catalytic subunit. Biochemistry. 1982;21:5794–9. [DOI] [PubMed] [Google Scholar]
- [46].Delaglio F, Grzesiek S, Vuister G, Zhu G, Pfeifer J, Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. Journal of Biomolecular NMR. 1995;6. [DOI] [PubMed] [Google Scholar]
- [47].Lee W, Tonelli M, Markley JL. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics. 2015;31:1325–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: A linear constraint solver for molecular simulations. Journal of Computational Chemistry. 1997;18:1463–72. [Google Scholar]
- [49].Darden T, York D, Pedersen L. Particle mesh Ewald: An N.log(N) method for Ewald sums in large systems. The Journal of Chemical Physics. 1993;98:10089. [Google Scholar]
- [50].Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. Journal of Chemical Theory and Computation. 2008;4:435–47. [DOI] [PubMed] [Google Scholar]
- [51].Best RB, Zhu X, Shim J, Lopes PEM, Mittal J, Feig M, et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. Journal of chemical theory and computation. 2012;8:3257–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Parrinello M, Rahman A. Crystal Structure and Pair Potentials: A Molecular-Dynamics Study. Physical Review Letters. 1980;45:1196–9. [Google Scholar]
- [53].Hernández CX, Pande VS. MDEntropy: Information-Theoretic Analyses for Molecular Dynamics. The Journal of Open Source Software. 2017;2:427. [Google Scholar]
- [54].Pasi M, Tiberti M, Arrigoni A, Papaleo E. xPyder: a PyMOL plugin to analyze coupled residues and their networks in protein structures. Journal of chemical information and modeling. 2012;52:1865–74. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.