Abstract
V(D)J recombination is initiated by introduction of site-specific double-stranded DNA breaks by the RAG-1 and RAG-2 proteins. The broken DNA ends are then joined by the cellular double-strand break repair machinery. Previous work has shown that truncated (core) versions of the RAG proteins can catalyze V(D)J recombination, although less efficiently than their full-length counterparts. It is not known whether truncating RAG-1 and/or RAG-2 affects the cleavage step or the joining step of recombination. Here we examine the effects of truncated RAG proteins on recombination intermediates and products. We found that while truncated RAG proteins generate lower levels of recombination products than their full-length counterparts, they consistently generate 10-fold higher levels of one class of recombination intermediates, termed signal ends. Our results suggest that this increase in signal ends does not result from increased cleavage, since levels of the corresponding intermediates, coding ends, are not elevated. Thus, removal of the “dispensable” regions of the RAG proteins impairs proper processing of recombination intermediates. Furthermore, we found that removal of portions of the dispensable regions of RAG-1 and RAG-2 affects the efficiency of product formation without altering the levels of recombination intermediates. Thus, these evolutionarily conserved sequences play multiple, important roles in V(D)J recombination.
Immunoglobulin and T-cell receptor gene segments are rearranged by V(D)J recombination to generate a diverse repertoire of antigen binding domains. The recombinase binds recombination signal sequences (hereafter termed signals) which flank the gene segments and introduces a double-stranded break (DSB) precisely between each signal and gene segment. This cleavage event produces two types of DNA termini, signal ends that terminate in signals and coding ends that contain the gene segment. Signal ends join to form a signal joint, whereas coding ends join to form a coding joint encoding the antigen binding domain (Fig. 1A) (24, 27, 28, 33, 38, 40).
FIG. 1.
(A) Schematic diagram of V(D)J recombination intermediates (coding ends and signal ends) and products (coding joints and signal joints) generated from the plasmid substrate pJH290. Signals are represented by triangles, and coding segments are represented by rectangles. (B) Conservation of RAG-1 and RAG-2. Each protein is shown as a rectangle, with individual amino acids represented as uniformly sized dark or light bands. Dark bands represent amino acids that are absolutely conserved in human, rabbit, mouse, chicken, xenopus, and trout proteins (2, 5, 6, 8, 10, 22, 32). Full-length RAG-1 (FL1) contains 1,040 amino acids; full-length RAG-2 (FL2) contains 527 amino acids. Truncated RAG-1 (TR1) consists of amino acids 384 to 1040, and truncated RAG-2 (TR2) consists of amino acids 1 to 387.
The V(D)J recombinase minimally consists of the highly conserved, lymphoid-cell-specific proteins RAG-1 and RAG-2 (22, 32). Transfection of the genes encoding RAG-1 and RAG-2 into cultured fibroblasts renders these cells competent to rearrange extrachromosomal recombination substrates, indicating that the RAG proteins are the only lymphoid-cell-specific factors necessary for recombination (22). DSBs with the same characteristics as in vivo intermediates (27, 28, 33, 35, 40) are generated in cell-free reactions containing purified, truncated RAG-1 and RAG-2 and the appropriate divalent metal ion (20). After cleavage, the RAG proteins remain associated with the broken DNA ends. Stable complexes have been isolated that contain the RAG proteins and a pair of cleaved signal ends (1). More recently, complexes containing the RAG proteins and all four DNA ends (two signal ends and two coding ends) have been isolated (9). We and others have suggested that disassembly or remodeling of these postcleavage complexes may be necessary to allow the joining machinery to complete formation of coding or signal joints (1, 39).
Mutational analyses revealed that RAG-1 and RAG-2 proteins truncated by 30 and 25%, respectively, are still capable of recombining plasmid substrates in fibroblasts, although generally with lower efficiency than full-length RAG proteins (3, 12, 21, 26, 30, 31, 34). These truncated proteins, which contain residues 384 to 1008 of 1,040 amino acids (RAG-1) and 1 to 387 of 527 amino acids (RAG-2) (Fig. 1B), are more soluble than their full-length counterparts and are, therefore, the forms of the RAG proteins used in cell-free systems (4, 13, 20, 23, 37, 38).
Sequence analysis of the portions of RAG-1 and RAG-2 that have been considered dispensable for recombination (amino acids 1 to 383 and 1009 to 1040 of RAG-1 and 388 to 527 of RAG-2) reveals many amino acid residues that are conserved across diverse species (Fig. 1B), suggesting that they may play an important role(s) in recombination. This proposal is supported by recent work demonstrating that regions within the N terminus of RAG-1 enhance signal joint formation (21, 26). However, these studies did not address which step of recombination, cleavage or joining, is affected by truncation of RAG-1 and RAG-2. Specifically, the effects of the truncations on recombination intermediates were not examined.
Here, we analyze the levels of both recombination intermediates (signal ends and coding ends) and products (signal joints and coding joints). In agreement with previous work (3, 12, 21, 26, 31, 34), we found that the levels of V(D)J recombination products, coding and signal joints, are reduced when truncated RAG proteins are used. We expected that the levels of recombination intermediates would be similarly reduced, since the characterized functions of the RAG proteins affect the cleavage step. However, the truncated RAG proteins consistently produced levels of signal ends that were 10-fold-higher than those produced by the full-length RAG proteins. In contrast, levels of coding ends were generally not affected. These observations suggest that the truncated RAG proteins do not increase cleavage but, rather, stabilize the signal end intermediates. Analysis of additional RAG deletion mutants allowed us to separate the effects on intermediates and products, showing that these effects map to distinct regions of the RAG proteins. Thus, these studies have uncovered several unexpected functions of the “dispensable” regions of the RAG proteins, suggesting that they have roles both in the processing of postcleavage complexes and in product formation.
MATERIALS AND METHODS
Plasmids.
Plasmid recombination substrates used in this study are pJH290, which produces coding joints on the plasmid backbone, and pJH289 (18), which produces signal joints on the plasmid backbone. The expression vector encoding full-length RAG-1 is pJH548 (30). Full-length RAG-2 is encoded by pJH549 (30), which is not epitope tagged, and pMS201 (31), which is tagged with three copies of c-myc epitope. The expression vector pMS127b (30) encodes truncated core RAG-1, and pMS216 (31) encodes truncated RAG-2. Vectors containing smaller deletions within RAG-1 are as follows: pMS119C (30) encodes amino acids 1 to 1008; pMS126 (30) encodes 332 to 1008; pSK151 (12) encodes 331 to 1040. Vectors containing smaller deletions within RAG-2 are as follows: pMS211 (31) encodes amino acids 1 to 517; pR2CC11 (3) encodes 1 to 481; pR2CC15 (3) encodes 1 to 425; pR2CC10 (3) encodes 1 to 418; pMS215 (31) encodes del388 to 413.
Transient-transfection assay.
Transient transfections were performed as previously described (35, 36). Briefly, 2 μg of recombination substrate, 2.1 μg of full-length RAG-1 expression vector (or the molar equivalent of smaller versions of RAG-1), and 2.5 μg of full-length RAG-2 expression vector (or the molar equivalent of smaller versions of RAG-2) were transfected into Chinese hamster ovary cells (RMP41, CHOK1, or xrs-6) by using either the calcium phosphate method (CellPhect kit; Pharmacia) or FuGene6 (Boehringer Mannheim). DNA was harvested after 40 to 48 h. DNA was harvested according to the method of Hirt as described previously (35, 36). All substrates and RAG vectors were analyzed in a minimum of three independent transfections.
PCR assays.
Ligation-mediated PCR (LMPCR) was performed as previously described (29, 35, 36). Briefly, ligations were incubated overnight at 14°C with 2 U of T4 DNA ligase (Gibco BRL), 200 pmol of annealed oligonucleotides DR19/DR20 (35), and 1/50 of the DNA harvested from a single transfection. Ligated samples were amplified for 24 cycles with primers specific to the 12-signal (DR20 and DR55) or 23-signal (DR20 and ML68) (36). Primers ML68 and DR55 were used to amplify signal joints (24 cycles) generated on plasmid pJH290. Blots were probed with the 32P end-labeled oligonucleotide probe DR55, ML68, or the junction-specific probe DR69 (35).
Southern blot analysis.
Two- to three-fifths of each transfection sample was subjected to PvuII digestion for 4 h. Samples were subjected to Southern blot analysis, and blots were probed with a 32P random-primed 693-nucleotide (nt) PvuII fragment of pJH290 (36). PhosphorImager quantitation was performed with a Storm860 (Molecular Dynamics).
Western blot analysis.
Lysates from RMP41 cells transfected with recombination substrate and RAG expression vectors were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and probed with anti-human c-myc antibody (PharMingen). Detection involved use of horseradish peroxidase-conjugated anti-mouse immunoglobulin G (IgG) (PharMingen) as the secondary antibody and enhanced chemiluminescence Western blotting detection reagents (Amersham).
RESULTS
Truncated RAG proteins generate high levels of signal ends.
Previously we developed an assay to measure levels of recombination intermediates and products derived from extrachromosomal substrates (35). A plasmid recombination substrate and expression vectors encoding RAG-1 and RAG-2 are transiently transfected into RMP41 fibroblast cells; after 40 to 48 h, products and intermediates are analyzed either by PCR amplification or directly by Southern blot analysis, as described previously (7, 35, 36).
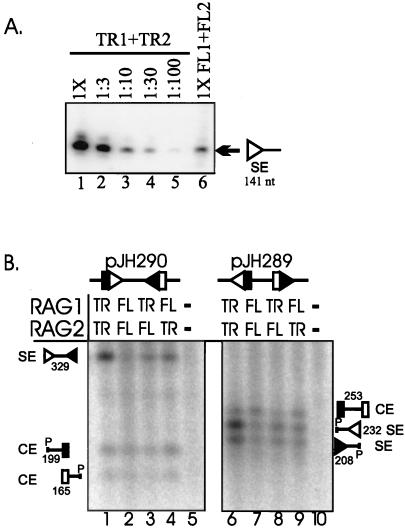
Initial analysis by LMPCR revealed that truncated RAG proteins generated approximately 10-fold-higher levels of signal ends than full-length RAG proteins (Fig. 2A, compare lanes 3 and 6). To confirm and extend these findings, samples from transfections of truncated or full-length RAG proteins were analyzed directly by Southern blotting. Cleavage at both signals of the substrate pJH290 generates a 329-nt excised fragment terminating in signal ends (diagrammed in Fig. 1A), whereas cleavage of the pJH289 substrate generates a pair of coding ends on an excised fragment and a pair of signal ends on the plasmid backbone. As shown in Fig. 2B, levels of the excised signal end fragment derived from cleavage of pJH290 were substantially increased in transfections containing truncated RAG proteins compared to full-length RAG proteins (329-nt species labeled SE; compare lanes 1 and 2). A similar effect was seen with the pJH289 substrate, in which levels of the signal ends on the plasmid backbone were increased in the presence of truncated RAG proteins (232- and 208-nt species; compare lanes 6 and 7). Quantitation of data from several transfections revealed that signal ends produced by truncated RAG proteins were consistently about 10-fold more abundant than signal ends derived from full-length RAG proteins (Table 1). Similar results were obtained in another wild-type CHO cell line, CHOK1, and in the mutant CHO cell line, xrs-6, which fails to carry out signal joint formation due to lack of functional Ku86 (7) (data not shown).
FIG. 2.
Signal end levels are 10-fold higher in transfections with truncated RAG proteins compared with full-length proteins. (A) LMPCR analysis of a signal end derived from pJH290. DNA from a transfection containing truncated RAG proteins (TR1+TR2) was diluted, as indicated above each lane, prior to ligation. DNA from a transfection containing full-length RAG proteins (FL1+FL2) was undiluted (1×). The blot was hybridized with radiolabeled oligonucleotide DR69 (35). The expected position of the LMPCR product (141 nt) is marked. The signal analyzed in this blot contains a 12-nt spacer; LMPCR analysis of a signal with a 23-nt spacer gave comparable results (data not shown). (B) Southern blot analysis of the two recombination substrates pJH290 and pJH289 (diagrams shown above blot). Samples were cut with restriction enzyme PvuII. Molecular species generated by cleavage and/or restriction enzyme digestion are identified to the side of each blot (restriction site represented by P). Lanes 5 and 10 contain transfections of recombination substrate in the absence of RAG constructs. Each lane contains three-fifths of the total DNA harvested from a transfection sample; all samples in panel B were transfected in parallel. The blot was hybridized with a radiolabeled PvuII fragment of pJH290 containing the unrearranged signal pair (36). Open triangle, signal with 12-nt spacer (12-signal); closed triangle, signal with 23-nt spacer (23-signal); SE, fragments containing a signal end(s); CE, fragments containing a coding end(s). This blot is representative of more than four experiments that all gave similar results. Although the 12-signal end fragment in lane 6 appears to be present at slightly higher levels than the 23-signal end fragment, three independent experiments (data not shown) show no difference between the levels of the 12- and 23-signal end fragments. In this and some subsequent gel photographs, all samples were run on the same gel, but some lanes were removed for clarity.
TABLE 1.
Quantitation of levels of signal end intermediates in several transfections containing truncated and/or full-length RAG proteins
| RAG vectors | Signal end quantitationa | No. of independent trans-fections quantitated |
|---|---|---|
| FL1 + FL2 | 1.0b | 4 |
| TR1 + TR2 | 9.8 ± 3.7 | 4 |
| TR1 + FL2 | 3.0 ± 1.2 | 3 |
| FL1 + TR2 | 2.2 ± 0.5 | 3 |
| None | 0.01 ± 0.03 | 4 |
Levels of pJH290 signal end species (329 nt) on Southern blots were quantitated by PhosphorImager analysis. Means and standard deviations are reported.
The level of the signal end species in transfections containing full-length RAG proteins was set to 1; signal end levels in other transfections are normalized to the full-length RAG transfection value.
To determine whether truncated RAG-1 and RAG-2 are jointly responsible for the difference in signal end levels, we transfected combinations of truncated and full-length RAG proteins (TR1 plus FL2 or FL1 plus TR2). Both combinations gave moderately (two- to threefold) increased levels of signal ends on the excised fragment (Fig. 2B, lanes 3 and 4) and on the plasmid backbone (lanes 8 and 9), as summarized in Table 1. These results indicate that both RAG-1 and RAG-2 contain elements that affect signal end levels.
One potential explanation for increased levels of signal end intermediates is that truncated RAG proteins may simply perform DNA cleavage more efficiently than their full-length counterparts. The transient transfection system allows detection of coding ends, which have also been observed previously in lymphoid cells expressing high levels of full-length RAG-1 and RAG-2 (24). Unlike signal ends, levels of coding ends on neither the plasmid backbone (Fig. 2B, 199- and 165-nt species labeled CE in lanes 1 and 2) nor the excised fragment (253-nt species in lanes 6 and 7) were affected by the use of truncated RAG proteins. This experiment was repeated more than 10 times, with similar results. These data suggest that the truncated RAG proteins do not generate increased levels of signal ends simply by performing cleavage more efficiently. An alternative possibility is that the high levels of signal ends reflect altered processing, such as increased stability of these recombination intermediates or increased protection from degradation, as discussed below.
Truncated RAG proteins generate low levels of signal and coding joints.
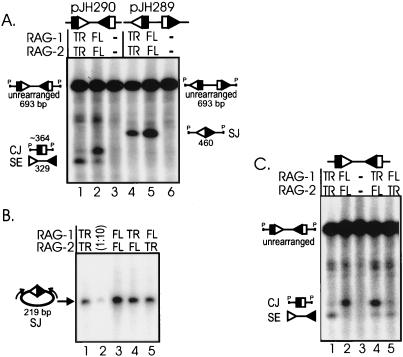
Since truncated RAG proteins unexpectedly generated increased levels of signal ends, we measured the abundance of coding and signal joints by Southern blotting. Coding joints were reduced in transfections of truncated RAG proteins (Fig. 3A; ∼364-nt species labeled CJ; compare lanes 1 and 2). Quantitation of five independent experiments revealed an average sevenfold decrease in levels of coding joints generated by truncated RAG proteins in comparison to full-length RAG proteins. Signal joints were also reduced three- to fivefold in the presence of truncated RAG proteins (460-nt species labeled SJ; compare lanes 4 and 5). Thus, although we observed increased signal ends in transfections containing truncated RAG proteins, levels of recombination products were actually decreased.
FIG. 3.
Coding joint and signal joint levels are reduced in transfections containing truncated RAG proteins. (A) Southern blot analysis of the two recombination substrates (pJH290 and pJH289). Molecular species are identified on the sides of the blot. Unlabeled species present between the coding joint and unrearranged fragments correspond to singly cleaved species (36). Techniques were performed as described for the experiment shown in Fig. 2B. (B) PCR analysis of signal joints generated by truncated and full-length RAG proteins. Lane 2, a 10-fold dilution of the transfection in lane 1. PCR samples were subjected to gel electrophoresis and Southern blot analysis as previously described (36). The blot was hybridized with the radiolabeled oligonucleotide DR55. These same transfection samples were analyzed in panel C. (C) Southern blot analysis of coding joints generated by truncated and full-length RAG proteins. Techniques were performed as in the experiment shown in Fig. 2B. SE, signal end fragment; CJ, coding joint fragment; SJ, signal joint fragment; P, PvuII restriction site.
We next asked whether truncated RAG-1, RAG-2, or both are responsible for the effects on coding and signal joints. The PCR analysis shown in Fig. 3B indicates that transfections of both combinations of truncated plus full-length RAG proteins moderately reduced levels of signal joints (lanes 4 and 5). Therefore, both truncated RAG-1 and RAG-2 are responsible for the reduced signal joint levels. These data are summarized in Table 2.
TABLE 2.
Effects of truncated RAG proteins on recombination intermediates and products
| Product | Effect |
|---|---|
| Signal ends | 10-fold increasea |
| Coding ends | None |
| Signal joints | 3- to 5-fold decrease |
| Coding joints | 7-fold decrease |
Expressed relative to values obtained with full-length RAG-1 and RAG-2.
Southern blot analysis of coding joints is shown in Fig. 3C. Transfection of truncated RAG-1 plus full-length RAG-2 (lane 4) generated levels of coding joints as high as those seen with full-length RAG proteins (lane 2). However, transfection of full-length RAG-1 and truncated RAG-2 (Fig. 3C, lane 5) gave low levels of coding joints, as seen with truncated RAG proteins (lane 1). These results establish that truncated RAG-2, but not RAG-1, is responsible for decreased coding joint formation and provide the first evidence specifically implicating RAG-2 in coding joint formation.
Different regions of the RAG proteins are responsible for increased signal ends and decreased joints.
Since our data show that removal of the dispensable regions of the RAG proteins affects levels of both ends and joints, one might expect to find a correlation between the two; that is, decreased joining might result in increased ends. Such a correlation is not always observed in vivo. Signal ends are not increased in mice (39) or in cultured cells (7) bearing mutations in the DNA repair machinery that virtually eliminate signal joint formation. Nevertheless, we considered the possibility that decreased formation of signal joints seen with truncated RAG proteins might be directly related to the increased levels of signal ends. To explore the relationships between the effects of the truncations on signal ends, signal joints, and coding joints, we used a series of RAG constructs containing smaller deletions within the “dispensable” portions of these proteins. In this manner, we mapped the regions responsible for each effect. In these experiments, the various truncations are tested paired with the standard truncated form of RAG-1 or RAG-2. The same results were obtained when the deletion mutants were paired with full-length RAG proteins (data not shown).
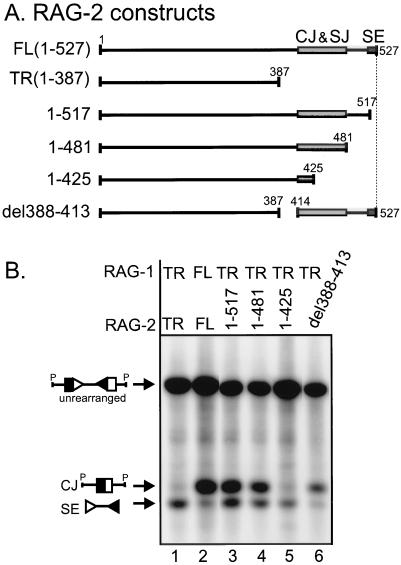
To determine whether the same regions of RAG-2 are responsible for decreased signal and coding joints and increased signal ends, we used the deletion mutants diagrammed in Fig. 4A. An expression vector encoding amino acids 1 to 517 of RAG-2 is sufficient to increase signal ends to levels seen with truncated RAG proteins (Fig. 4B, compare lanes 1 and 3). Therefore, the region responsible for the increased signal ends generated by truncated RAG-2 is localized to the 10 C-terminal amino acids. However, unlike truncated RAG-2, this construct (amino acids 1 to 517) does not cause decreased coding joint formation (compare lanes 1 and 3) or signal joint formation (data not shown), indicating that the reduced levels of joints are not necessarily associated with increased levels of signal ends.
FIG. 4.
Deletion analysis of the regions of RAG-2 responsible for reduced coding joint levels and increased signal end levels. (A) Schematic diagram of the RAG-2 constructs analyzed. Proteins are represented by lines, with spaces indicating regions deleted from the construct. Numbers above each line identify the terminal amino acids. The gray box between amino acids 414 and 481 illustrates the area that affects coding joint and signal joint formation, and the dark box between amino acid 517 to 527 illustrates the area that affects signal end levels, as demonstrated in panel B. Drawings are not to scale. (B) Representative Southern blotting data that map the regions responsible for the decreased levels of coding joint products and increased levels of signal end intermediates produced by truncated RAG proteins.
Additional deletion mutants were used to further map the regions of RAG-2 responsible for decreased coding and signal joints. Coding joints generated by a RAG-2 construct retaining amino acids 1 to 481 are not decreased to the low levels seen with truncated RAG-2 (Fig. 4B, compare lanes 1 and 4). However, removal of an additional 56 amino acids (1 to 425) reduces coding joint levels to those seen with truncated RAG-2 (compare lanes 1 and 5). Similar results are generated by a construct retaining amino acids 1 to 418 (data not shown). Since a mutant with an internal deletion (del388-413) did not decrease coding joint levels to the extent seen with truncated RAG-2 (compare lanes 1 and 6), we conclude that a region within amino acids 414 to 481 is responsible for the lower levels of coding joints observed with truncated RAG-2. This region, which contains many conserved residues, is also responsible for the decreased signal joint levels seen with truncated RAG-2 (data not shown). Additional evidence supports our conclusion that this region of RAG-2 is important for joint formation, as alteration of amino acids 436 to 441 causes a greater-than-15-fold reduction in both signal and coding joints (31). Therefore, nonoverlapping regions of RAG-2 are responsible for the decreased coding and signal joints (residues 414 to 481) and the increased signal ends (residues 517 to 527) caused by truncated RAG-2.
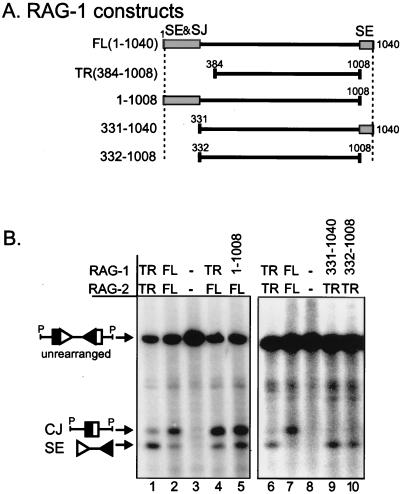
To map the regions of RAG-1 responsible for effects on recombination intermediates (signal ends), we employed the series of RAG-1 deletion mutants diagrammed in Fig. 5A. A construct containing residues 1 to 1008 generated signal ends at levels similar to truncated RAG-1 (Fig. 5B, compare lanes 1 and 5), indicating that removal of the C-terminal “dispensable” region of RAG-1 is sufficient to produce this effect. However, removal of the N-terminal “dispensable” region of RAG-1 (amino acids 331 to 1040) is also sufficient to cause the high levels of signal ends seen with truncated RAG-1 (Fig. 5B, compare lanes 6 and 9). As expected, signal ends were increased when a construct with deletions at both ends (332 to 1008) was used (Fig. 5B, lane 10). Therefore, removal of either the N-terminal 330 amino acids or the C-terminal 32 amino acids of RAG-1 is sufficient to reproduce the effect of truncated RAG-1 on levels of signal ends.
FIG. 5.
Deletion analysis of regions of RAG-1 responsible for increased levels of signal ends. (A) Schematic diagram of the RAG-1 constructs analyzed. Shaded boxes indicate regions that affect signal end (SE) or signal joint (SJ) levels. (B) Representative Southern blotting data that map the portions of RAG-1 responsible for increased signal ends. Techniques were performed as in the experiment shown in Fig. 2B. Signal end levels in the del1-1008 construct are unusually high on this blot compared to those in other independent experiments (data not shown).
We also analyzed the effects of the RAG-1 deletion mutants on recombination products. Since coding joint levels were not affected by the use of truncated RAG-1, as shown above, only analysis of signal joint levels is discussed here. Removal of the N terminus but not the C terminus decreased signal joints to levels seen with truncated RAG-1 (data not shown). In agreement with our findings, others have also mapped the region responsible for reduced signal joints to the N terminus of RAG-1 (21, 26). Therefore, the regions of RAG-1 responsible for the high levels of signal ends seen with truncated RAG-1 do not strictly correlate with the regions responsible for low levels of signal joints.
Truncation increases levels of RAG proteins.
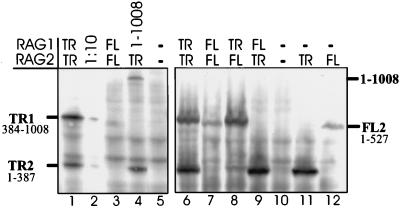
Previous work has shown that levels of truncated RAG-1 protein are about 10-fold higher than levels of a RAG-1 construct missing only the C-terminal 32 amino acids (21). However, in that study, levels of RAG-2 were not measured. We measured levels of the RAG proteins by Western blot analysis with antibodies to c-myc epitope tags present on both truncated proteins, full-length RAG-2, and a RAG-1 construct missing only the C-terminal 32 amino acids (del1-1008) (30, 31). As shown in Fig. 6, truncated RAG-1 and RAG-2 were expressed at approximately 10-fold-higher levels than full-length RAG-2 protein and RAG-1 (del1-1008) (compare lane 1 to lanes 3 and 4; lanes 6 and 7). Therefore, like RAG-1, RAG-2 protein is present at higher levels when truncated. When transfections contained full-length RAG-1 and truncated RAG-2 or vice versa (lanes 8 and 9), levels of truncated proteins remained high and levels of full-length RAG-2 protein remained low. Thus, the intermediate effects of the combinations of one truncated plus one full-length RAG protein on signal ends are not simply caused by intermediate protein levels. These experiments, which have been repeated three times with the same results, also show that the truncated proteins do not influence the levels of their full-length partners.
FIG. 6.
Truncated RAG proteins are present at 10-fold-higher levels than full-length RAG proteins. Western blot analysis of transfections containing truncated or full-length RAG proteins. Truncated RAG-1 (TR1), truncated RAG-2 (TR2), full-length RAG-2 (FL2), and a RAG-1 construct containing amino acids 1 to 1008 each have three copies of the c-myc epitope on their carboxy termini and are identified to the left and right of the blots. Full-length RAG-1 cannot be detected, since it has no c-myc epitope tag. Two independent experiments are shown. Blots were hybridized with anti-human c-myc primary antibody. Lane 2, 10-fold dilution of the sample in lane 1; lanes 5 and 10, no RAG proteins in transfections.
Protein levels are not responsible for the effects of truncated RAG proteins on signal ends or joints.
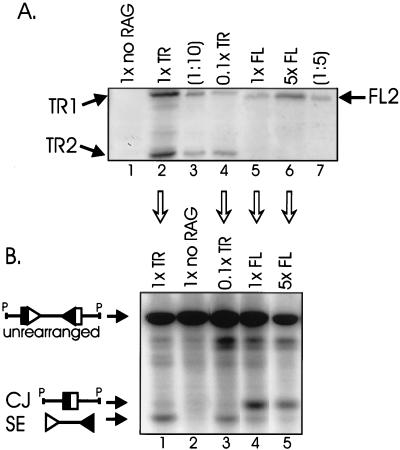
The results described above suggest that the effects of truncated RAG proteins on signal ends and recombination products may be secondary to the effects of these truncations on protein levels. This hypothesis was tested in two ways: by decreasing the levels of truncated RAG proteins and by increasing the levels of full-length RAG proteins. As shown in Fig. 7A, transfections of 10-fold-less truncated RAG expression vector (0.1×) produced 10-fold-lower protein levels than a standard (1×) transfection (compare lanes 2 and 4). These reduced levels of truncated proteins were similar to standard (1×) levels of full-length RAG proteins (compare lanes 4 and 5). Similarly, transfecting fivefold-more (5×) full-length RAG expression vector produced fivefold-higher protein levels (compare lanes 5 and 6). The effects of altered RAG protein levels on V(D)J recombination were assessed by Southern blotting (Fig. 7B). Levels of signal ends and coding joints were comparable in transfections containing the standard (1×) amounts of truncated RAG proteins and 10-fold-smaller (0.1×) amounts (Fig. 7B, compare lanes 1 and 3). Transfections containing either the standard amounts of full-length RAG proteins or fivefold-larger (5×) amounts also generated similar levels of signal ends and coding joints (compare lanes 4 and 5). Therefore, altered protein levels are not responsible for the differences in levels of signal ends and recombination products.
FIG. 7.
Higher levels of truncated RAG proteins are not responsible for the effects on signal end and coding joint levels. (A) Western blot analysis showing that the amount of transfected expression vector (indicated above each lane) closely correlates with levels of protein. Different amounts of RAG expression vectors were transfected (although the total amount of transfected DNA was held constant): 1× denotes the standard amounts of RAG-1 and RAG-2 (see Materials and Methods); 0.1× denotes transfections containing 10-fold-smaller amounts of RAG constructs. Lane 1, recombination substrate but no RAG constructs; lane 3, the same transfection sample as lane 2 but 10-fold-less sample was analyzed by electrophoresis; lane 7, the same transfection sample as lane 6 but 10-fold-less sample was analyzed by electrophoresis. Western blotting was performed as in the experiment shown in Fig. 6. (B) Southern blot analysis of transfections identical to those shown in panel A (performed simultaneously).
Additional evidence supporting this conclusion is provided by the RAG-1 construct 1-1008. This protein was expressed at levels much lower than truncated RAG-1 (Fig. 6, lane 4), comparable to levels of full-length RAG-2 (Fig. 6, lane 3). However, levels of signal ends were similar to levels produced by truncated RAG-1 (Fig. 5B, compare lanes 1 and 5). Thus, increased levels of RAG proteins are not responsible for the increased levels of signal ends seen with truncated RAG proteins.
DISCUSSION
Here we report the first analysis of the effects of truncated and full-length RAG proteins on formation of both the intermediates and the products of V(D)J recombination. The ability to examine the effects of these proteins on recombination intermediates allows us to more precisely identify steps in the reaction pathway that are affected by truncation of RAG-1 and RAG-2. Since the characterized functions of the RAG proteins affect the cleavage step, we expected that the decreased levels of coding and signal joints associated with truncated RAG proteins would be a result of decreased cleavage. However, we did not observe decreased levels of either signal or coding ends, indicating that the truncations affect a step (or steps) subsequent to cleavage. Instead, levels of signal ends are substantially increased in cells transfected with truncated RAG proteins, while coding end levels are not affected. It is surprising that these intermediates are so differently affected by RAG truncation (discussed below). Regions of RAG-2 responsible for decreased levels of joints and increased levels of signal ends can be mapped to nonoverlapping regions within the C terminus, indicating that these effects are not linked. These data indicate that, in addition to performing DNA cleavage, the RAG proteins play previously unsuspected roles in the processing and joining of V(D)J recombination intermediates. Interestingly, the C-terminal one-fourth of RAG-2 also appears to play a role in the regulation of endogenous immunoglobulin gene rearrangement (11).
Possible mechanisms for the effects of truncated RAG proteins on signal ends.
Several lines of evidence suggest that coding and signal ends are processed along different pathways. For example, coding joints undergo loss and/or addition of nucleotides much more frequently than signal joints (16) and form with faster kinetics in vivo than signal joints (24, 25). Furthermore, in cell-free systems, signal joint formation is enhanced by removal of the RAG proteins through deproteinization or heating before the addition of joining factors, while coding joint formation is actually enhanced by the continued presence of RAG proteins during the joining phase of the reaction (13, 23). In addition, the RAG proteins are more stably associated with signal ends than coding ends in cell-free reactions (1, 9). These observations support a model in which signal ends remain in a stable two-ended RAG-DNA complex that persists after coding ends are joined. For joining to occur, the RAG proteins must be removed from the signal ends, a reaction which may be mediated by specific disassembly or remodeling factors (1, 39).
Here we show, for the first time, that truncation of the RAG proteins affects processing of coding and signal ends in different ways. We suggest that, in the absence of their “dispensable” termini, truncated RAG-1 and RAG-2 stabilize the signal end intermediates such that postcleavage complexes containing signal ends are poor substrates for disassembly, resulting in their accumulation. Several mechanisms could be responsible for the effect of truncated RAG proteins on signal ends. Since truncation increases levels of RAG proteins, one possibility is that the 10-fold excess of truncated RAG proteins obstructs joining or disassembly simply by mass action. However, we have shown that decreasing truncated RAG protein levels 10-fold or increasing full-length RAG protein levels fivefold does not affect signal end levels. In addition, regions of the RAG proteins responsible for increased protein levels differ from those responsible for increased signal ends (for example, deletion of the C-terminal 32 amino acids of RAG-1 reproduced the increased signal ends but not the increased protein levels seen with truncated RAG-1). Thus, increased levels of RAG proteins are not responsible for increased levels of signal ends.
Since signal ends might also be generated by recleavage of signal joints, we considered the possibility that more efficient recleavage might account for the increased levels of signal ends observed with truncated RAG proteins. This model predicts that truncated RAG proteins should not raise the levels of signal ends in cells that are incapable of forming signal joints. However, we found that truncated RAG proteins increased signal end levels in xrs-6 cells, which are virtually completely defective for signal joint formation. These data argue strongly against a model involving recleavage of signal joints.
Another possibility is that the termini of the RAG proteins could facilitate disassembly of the postcleavage complex. For example, the “dispensable” regions of RAG-1 and RAG-2 may contain protein degradation signals or chaperone recognition sites that aid in removal of the RAG proteins from signal ends. A phosphorylation site that targets cell cycle-specific degradation (T490) has been characterized in the C terminus of RAG-2 (17, 19); however, this site is not necessary for the effects reported here (Fig. 4 and data not shown). A precedent for a chaperone recognition site is provided by the bacteriophage Mu transposition system. The MuA transposase is recognized by the chaperone ClpX to allow removal of the transposase from a stable transposition intermediate, the strand transfer complex (14). A 10-amino-acid, positively charged sequence at the C terminus of MuA (LEQNRRKKAI) is sufficient to direct ClpX activity to MuA, indicating that the ClpX chaperone recognizes a discrete signal (15). We have mapped the region of RAG-2 responsible for increased signal end levels to the 10 C-terminal amino acids: KKSFLRRLFD. Seven of these ten residues are absolutely conserved among all six species examined (underlined). Like the ClpX recognition site, which requires four basic amino acids for proper function (15), four of the conserved residues are positively charged. Perhaps these 10 C-terminal amino acids of RAG-2 serve as a recognition site for a disassembly factor.
A role for RAG proteins in joining.
We have shown that RAG-1 and RAG-2 contain regions important for efficient coding and signal joint formation. According to our mapping data, a region within amino acids 1 to 330 of RAG-1 is responsible for the decreased levels of signal joints seen with truncated RAG-1, and a region within amino acids 414 to 481 of RAG-2 is responsible for the decreased levels of signal joints and coding joints seen with truncated RAG-2.
It may seem surprising that these regions that affect joining do not also cause the increased levels of signal ends generated by truncated RAG proteins. Similarly, the modest decrease in coding joint formation is not accompanied by a discernible increase in levels of coding ends. However, in this complex in vivo system, levels of ends reflect input from several different factors, including the efficiency of cleavage and the use of alternative pathways such as DNA degradation, in addition to joining. A precedent for this situation is seen in Ku86-deficient mice, in which the abundance of signal ends does not differ from that in wild-type mice, despite severely impaired formation of signal joints (39).
According to recent in vitro data, coding joint formation is stimulated up to 50-fold in the presence of truncated RAG proteins (23), and the RAG proteins are capable of opening hairpin coding ends (1a). Here we show that the “dispensable” regions of the RAG proteins also affect joining. It is unclear what roles RAG-1 and RAG-2 may play in coding and signal joint formation. Interestingly, removal of the “dispensable” regions of RAG-1 only affects signal joint formation, while removal of the “dispensable” region of RAG-2 affects levels of both signal and coding joints, suggesting that these regions have different functions. Biochemical studies of the full-length RAG proteins should further illuminate the roles of these proteins in the joining reaction.
ACKNOWLEDGMENTS
We thank Mary Lowe for assistance in manuscript preparation and Mary Purugganan and Tania Baker for helpful comments on the manuscript.
This work was supported in part by NIH grant AI-36420. S.S. was supported in part by NIH Predoctoral Fellowship T32-AI07495. D.B.R. is a Charles E. Culpeper Medical Research Scholar.
REFERENCES
- 1.Agrawal A, Schatz D G. RAG1 and RAG2 form a stable postcleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell. 1997;89:43–53. doi: 10.1016/s0092-8674(00)80181-6. [DOI] [PubMed] [Google Scholar]
- 1a.Besmer E, Mansilla-Soto J, Cassard S, Sawchuk D J, Brown G, Sadofsky M, Lewis S M, Nussenzweig M C, Cortes P. Hairpin coding end opening is mediated by RAG1 and RAG2 proteins. Mol Cell. 1998;2:817–828. doi: 10.1016/s1097-2765(00)80296-8. [DOI] [PubMed] [Google Scholar]
- 2.Carlson L M, Oettinger M A, Schatz D G, Masteller E L, Hurley E A, McCormack W T, Baltimore D, Thompson C B. Selective expression of RAG-2 in chicken B cells undergoing immunoglobulin gene conversion. Cell. 1991;64:201–208. doi: 10.1016/0092-8674(91)90221-j. [DOI] [PubMed] [Google Scholar]
- 3.Cuomo C A, Oettinger M A. Analysis of regions of RAG-2 important for V(D)J recombination. Nucleic Acids Res. 1994;22:1810–1814. doi: 10.1093/nar/22.10.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eastman Q M, Leu T M J, Schatz D G. Initiation of V(D)J recombination in vitro obeying the 12/23 rule. Nature. 1996;380:85–88. doi: 10.1038/380085a0. [DOI] [PubMed] [Google Scholar]
- 5.Fuschiotti P, Harindranath N, Mage R G, McCormack W T, Dhanarajan P, Roux K H. Recombination activating genes-1 and -2 of the rabbit: cloning and characterization of germline and expressed genes. Mol Immunol. 1998;30:1021–1035. doi: 10.1016/0161-5890(93)90127-w. [DOI] [PubMed] [Google Scholar]
- 6.Greenhalgh P, Olesen C E M, Steiner L A. Characterization and expression of recombination activating genes (RAG-1 and RAG-2) in Xenopus laevis. J Immunol. 1998;151:3100–3110. [PubMed] [Google Scholar]
- 7.Han J-O, Steen S B, Roth D B. Ku86 is not required for protection of signal ends or for formation of nonstandard V(D)J recombination products. Mol Cell Biol. 1997;17:2226–2234. doi: 10.1128/mcb.17.4.2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hansen J D, Kaattari S L. The recombination activating gene 1 (RAG1) of rainbow trout (Oncorhynchus mykiss): cloning, expression, and phylogenetic analysis. Immunogenetics. 1998;42:188–195. doi: 10.1007/BF00191224. [DOI] [PubMed] [Google Scholar]
- 9.Hiom K, Gellert M. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol Cell. 1998;1:1011–1019. doi: 10.1016/s1097-2765(00)80101-x. [DOI] [PubMed] [Google Scholar]
- 10.Ichihara Y, Hirai M, Kurosawa Y. Sequence and chromosome assignment to 11p13-p12 of human RAG genes. Immunol Lett. 1992;33:277–284. doi: 10.1016/0165-2478(92)90073-w. [DOI] [PubMed] [Google Scholar]
- 11.Kirch S A, Rathbun G, Oettinger M A. Dual role of RAG2 in V(D)J recombination: catalysis and regulation of ordered Ig gene assembly. EMBO J. 1998;17:4881–4886. doi: 10.1093/emboj/17.16.4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirch S A, Sudarsanam P, Oettinger M A. Regions of RAG1 protein critical for V(D)J recombination. Eur J Immunol. 1996;26:886–891. doi: 10.1002/eji.1830260425. [DOI] [PubMed] [Google Scholar]
- 13.Leu T M, Eastman Q M, Schatz D G. Coding joint formation in a cell-free V(D)J recombination system. Immunity. 1997;7:303–314. doi: 10.1016/s1074-7613(00)80532-4. [DOI] [PubMed] [Google Scholar]
- 14.Levchenko I, Luo L, Baker T A. Disassembly of the Mu transposase tetramer by the ClpX chaperone. Genes Dev. 1995;9:2399–2408. doi: 10.1101/gad.9.19.2399. [DOI] [PubMed] [Google Scholar]
- 15.Levchenko I, Yamauchi M, Baker T A. ClpX and MuB interact with overlapping regions of Mu transposase: implications for control of the transposition pathway. Genes Dev. 1997;11:1561–1572. doi: 10.1101/gad.11.12.1561. [DOI] [PubMed] [Google Scholar]
- 16.Lewis S M. The mechanism of V(D)J joining: lessons from molecular, immunological and comparative analyses. Adv Immunol. 1994;56:27–150. doi: 10.1016/s0065-2776(08)60450-2. [DOI] [PubMed] [Google Scholar]
- 17.Li Z, Dordai D I, Lee J, Desiderio S. A conserved degradation signal regulates RAG-2 accumulation during cell division and links V(D)J recombination to the cell cycle. Immunity. 1996;5:575–589. doi: 10.1016/s1074-7613(00)80272-1. [DOI] [PubMed] [Google Scholar]
- 18.Lieber M R, Hesse J E, Lewis S, Bosma G C, Rosenberg N, Mizuuchi K, Bosma M J, Gellert M. The defect in murine severe combined immune deficiency: joining of signal sequences but not coding segments in V(D)J recombination. Cell. 1988;55:7–16. doi: 10.1016/0092-8674(88)90004-9. [DOI] [PubMed] [Google Scholar]
- 19.Lin W-C, Desiderio S. Cell cycle regulation of V(D)J recombination-activating protein RAG-2. Proc Natl Acad Sci USA. 1994;91:2733–2737. doi: 10.1073/pnas.91.7.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McBlane J F, van Gent D C, Ramsden D A, Romeo C, Cuomo C A, Gellert M, Oettinger M A. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 21.McMahan C J, Difilippantonio J, Rao N, Spanopoulou E, Schatz D G. A basic motif in the N-terminal region of RAG1 enhances V(D)J recombination activity. Mol Cell Biol. 1997;17:4544–4552. doi: 10.1128/mcb.17.8.4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oettinger M A, Schatz D G, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 23.Ramsden D A, Paull T T, Gellert M. Cell-free V(D)J recombination. Nature. 1997;388:488–491. doi: 10.1038/41351. [DOI] [PubMed] [Google Scholar]
- 24.Ramsden D A, Gellert M. Formation and resolution of double-strand break intermediates in V(D)J rearrangement. Genes Dev. 1995;9:2409–2420. doi: 10.1101/gad.9.19.2409. [DOI] [PubMed] [Google Scholar]
- 25.Roman C A J, Baltimore D. Genetic evidence that the RAG1 protein directly participates in V(D)J recombination through substrate recognition. Proc Natl Acad Sci USA. 1996;93:2333–2338. doi: 10.1073/pnas.93.6.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roman C A J, Cherry S R, Baltimore D. Complementation of V(D)J recombination deficiency in RAG-1−/− B cells reveals a requirement for novel elements in the N-terminus of RAG-1. Immunity. 1997;7:13–24. doi: 10.1016/s1074-7613(00)80506-3. [DOI] [PubMed] [Google Scholar]
- 27.Roth D B, Menetski J P, Nakajima P B, Bosma M J, Gellert M. V(D)J recombination: broken DNA molecules with covalently sealed (hairpin) coding ends in scid mouse thymocytes. Cell. 1992;70:983–991. doi: 10.1016/0092-8674(92)90248-b. [DOI] [PubMed] [Google Scholar]
- 28.Roth D B, Nakajima P B, Menetski J P, Bosma M J, Gellert M. V(D)J recombination in mouse thymocytes: double-strand breaks near T cell receptor δ rearrangement signals. Cell. 1992;69:41–53. doi: 10.1016/0092-8674(92)90117-u. [DOI] [PubMed] [Google Scholar]
- 29.Roth D B, Zhu C, Gellert M. Characterization of broken DNA molecules associated with V(D)J recombination. Proc Natl Acad Sci USA. 1993;90:10788–10792. doi: 10.1073/pnas.90.22.10788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sadofsky M J, Hesse J E, McBlane J F, Gellert M. Expression and V(D)J recombination activity of mutated RAG-1 proteins. Nucleic Acids Res. 1993;21:5644–5650. doi: 10.1093/nar/21.24.5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sadofsky M J, Hesse J E, Gellert M. Definition of a core region of RAG-2 that is functional in V(D)J recombination. Nucleic Acids Res. 1994;22:1805–1809. doi: 10.1093/nar/22.10.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schatz D G, Oettinger M A, Baltimore D. The V(D)J recombination activating gene, RAG-1. Cell. 1989;59:1035–1048. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- 33.Schlissel M, Constantinescu A, Morrow T, Baxter M, Peng A. Double-strand signal sequence breaks in V(D)J recombination are blunt, 5′-phosphorylated, RAG-dependent, and cell cycle regulated. Genes Dev. 1993;7:2520–2532. doi: 10.1101/gad.7.12b.2520. [DOI] [PubMed] [Google Scholar]
- 34.Silver D P, Spanopoulou E, Mulligan R C, Baltimore D. Dispensable sequence motifs in the RAG-1 and RAG-2 genes for plasmid V(D)J recombination. Proc Natl Acad Sci USA. 1993;90:6100–6104. doi: 10.1073/pnas.90.13.6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steen S B, Gomelsky L, Roth D B. The 12/23 rule is enforced at the cleavage step of V(D)J recombination in vivo. Genes Cells. 1996;1:543–553. doi: 10.1046/j.1365-2443.1996.d01-259.x. [DOI] [PubMed] [Google Scholar]
- 36.Steen S B, Gomelsky L, Speidel S L, Roth D B. Initiation of V(D)J recombination in vivo: role of recombination signal sequences in formation of single and paired double-strand breaks. EMBO J. 1997;16:2656–2664. doi: 10.1093/emboj/16.10.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Gent D C, Ramsden D A, Gellert M. The RAG1 and RAG2 proteins establish the 12/23 rule in V(D)J recombination. Cell. 1996;85:107–113. doi: 10.1016/s0092-8674(00)81086-7. [DOI] [PubMed] [Google Scholar]
- 38.van Gent D C, McBlane J F, Ramsden D A, Sadofsky M J, Hesse J E, Gellert M. Initiation of V(D)J recombination in a cell-free system. Cell. 1995;81:925–934. doi: 10.1016/0092-8674(95)90012-8. [DOI] [PubMed] [Google Scholar]
- 39.Zhu C, Bogue M A, Lim D-S, Hasty P, Roth D B. Ku86-deficient mice exhibit severe combined immunodeficiency and defective processing of V(D)J recombination intermediates. Cell. 1996;86:379–389. doi: 10.1016/s0092-8674(00)80111-7. [DOI] [PubMed] [Google Scholar]
- 40.Zhu C, Roth D B. Characterization of coding ends in thymocytes of scid mice: implications for the mechanism of V(D)J recombination. Immunity. 1995;2:101–112. doi: 10.1016/1074-7613(95)90082-9. [DOI] [PubMed] [Google Scholar]