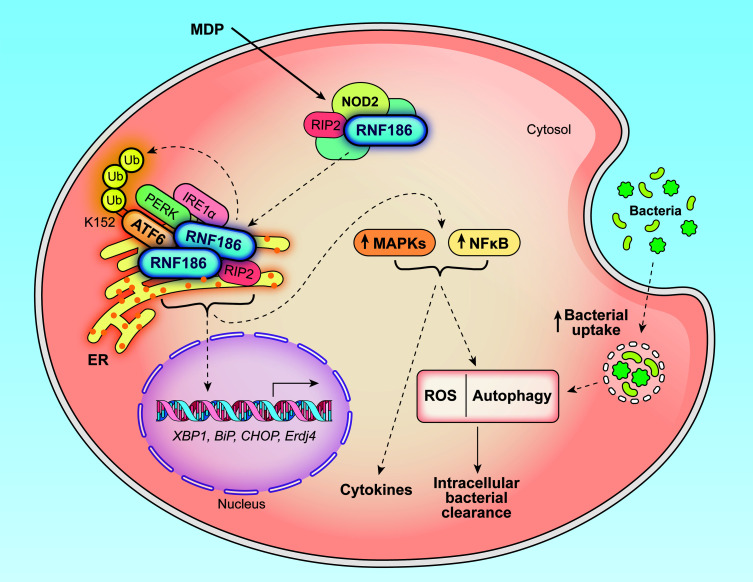
Abstract
Properly balancing microbial responses by the innate immune system through pattern recognition receptors (PRRs) is critical for intestinal immune homeostasis. Ring finger protein 186 (RNF186) genetic variants are associated with inflammatory bowel disease (IBD). However, functions for the E3 ubiquitin ligase RNF186 are incompletely defined. We found that upon stimulation of the PRR nucleotide-binding oligomerization domain containing 2 (NOD2) in human macrophages, RNF186 localized to the ER, formed a complex with ER stress sensors, ubiquitinated the ER stress sensor activating transcription factor 6 (ATF6), and promoted the unfolded protein response (UPR). These events, in turn, led to downstream signaling, cytokine secretion, and antimicrobial pathway induction. Importantly, RNF186-mediated ubiquitination of K152 on ATF6 was required for these outcomes, highlighting a key role for ATF6 ubiquitination in PRR-initiated functions. Human macrophages transfected with the rare RNF186-A64T IBD risk variant and macrophages from common rs6426833 RNF186 IBD risk carriers demonstrated reduced NOD2-induced outcomes, which were restored by rescuing UPR signaling. Mice deficient in RNF186 or ATF6 demonstrated a reduced UPR in colonic tissues, increased weight loss, and less effective clearance of bacteria with dextran sodium sulfate–induced injury and upon oral challenge with Salmonella Typhimurium. Therefore, we identified that RNF186 was required for PRR-induced, UPR-associated signaling leading to key macrophage functions; defined that RNF186-mediated ubiquitination of ATF6 was essential for these functions; and elucidated how RNF186 IBD risk variants modulated these outcomes.
Keywords: Immunology
Keywords: Innate immunity, Macrophages
Introduction
Balancing microbially induced cytokines and microbial clearance is critical at mucosal sites, such as the intestine. Pattern recognition receptors (PRRs) are key in mediating the recognition and responses to microbes (1). Consistent with the importance of balancing these PRR responses, both loss of function and gain of function in PRR-initiated outcomes are associated with inflammatory bowel disease (IBD) (1, 2). We and others have identified IBD risk loci that lead to loss of innate immune-associated outcomes across a broad range of PRRs (3–9), thereby leading to a reduced ability to clear microbes. Despite the success in identification of IBD risk loci, altered functions for most of these loci are not known. Defining these functions can provide important insight into mechanisms mediating homeostasis at mucosal surfaces.
The rs6426833 A variant in the ring finger protein 186 (RNF186) region confers an increased risk for ulcerative colitis (UC) and is observed at a 0.395–0.548 frequency in healthy individuals of European ancestry (per Single Nucleotide Polymorphism Database [dbSNP] as of July 2019) (9). A rare RNF186 A64T variant confers a 1.49-fold increased risk for developing UC (10). The combination of rare coding mutations and common noncoding variants in the RNF186 region conferring risk for IBD highlights contributions for this gene in IBD pathogenesis. RNF186 is an E3 ubiquitin ligase. Reports have focused on RNF186 functions mainly in epithelial cells (11–16). RNF186-deficient mice and mice with a knockin of the rare RNF186 A64T variant demonstrate more severe dextran sodium sulfate–induced (DSS-induced) colitis associated with intestinal epithelial dysregulation (12, 14). Yet another study identified a role for RNF186 in regulating nutrient sensing in epithelial cells (16). RNF186 localizes to the ER and enhances ER stress–associated apoptotic signaling in HeLa cells (13) and promotes ER stress leading to impaired insulin signaling in mouse hepatocytes (15). Therefore, while some functions for RNF186 have been identified in epithelial cells, ER-associated roles for RNF186 in mediating outcomes in innate immunity, including downstream of PRRs, have not been well defined; ER-dependent pathways can lead to very distinct outcomes in myeloid-derived cells when compared with epithelial cells. Our protein sequence-based analysis through PSORTII predicting that a fraction of RNF186 localizes to the ER, combined with 2 earlier studies reporting a role for RNF186 regulation of ER stress–associated signaling in epithelial cells (13, 15), led us to hypothesize that RNF186 might localize to the ER in human macrophages and in turn regulate innate immune outcomes in these cells through ER-associated mechanisms.
Activation of unfolded protein response (UPR) signaling is a canonical response of the ER to resolve stress emanating from accumulation of misfolded proteins (17). UPR signaling is coordinated by 3 ER-resident sensors: inositol-requiring protein 1 (IRE1α), PKR-like ER kinase (PERK), and activating transcription factor 6 (ATF6) (18, 19). Dysregulated UPR signaling can contribute to various diseases, including IBD (18). However, much of the focus on how dysregulated UPR signaling contributes to IBD pathogenesis has been on its role in epithelial cells (20, 21). In particular, deletion of IRE1 (22, 23) or X-box binding protein 1 (XBP1) (24) in mouse intestinal epithelial cells promotes intestinal inflammation. In addition, loss-of-function rare variants in XBP1 are associated with IBD (24). In contrast to its well-described role in epithelial cells, the role of UPR signaling in innate immunity and cell subsets critical for mediating innate immune defenses is less well understood. The UPR has been implicated in PRR-initiated responses in macrophages; however, mechanisms regulating the UPR in macrophages are incompletely defined. IRE1α has generally been reported to be activated and contribute to PRR-induced cytokines in mouse macrophages (25, 26). In contrast, ATF6 has been found to contribute to this outcome in some studies (27) but not other studies (25, 26). PERK signaling has generally not been implicated downstream of PRRs in mouse macrophages (25, 26). These studies predominantly focused on mouse macrophages; mouse and human macrophages can demonstrate significant differences in immune-mediated pathways (28). We recently reported that each of the 3 ER stress sensors is required for optimal NLR-induced cytokines in human macrophages (29). We therefore hypothesized that RNF186 would regulate innate immune responses to microbes in human macrophages through regulation of ER-associated functions, in particular through modulating UPR signaling.
In this study using primary human monocyte-derived macrophages (MDMs), we defined a critical role for RNF186 in mediating PRR-induced, UPR-dependent inflammatory and antimicrobial outcomes in human macrophages; established that RNF186-dependent ATF6 ubiquitination was required for these outcomes; defined structural regions in RNF186 mediating the defined outcomes; identified that the RNF186 rare and common UC risk genetic variants modulated these responses; and established that both RNF186 and ATF6 deficiency in mice resulted in impaired clearance of resident intestinal microbes and enteric pathogens.
Results
RNF186 localizes to the ER and promotes the nucleotide-binding oligomerization domain containing 2–induced UPR in human macrophages.
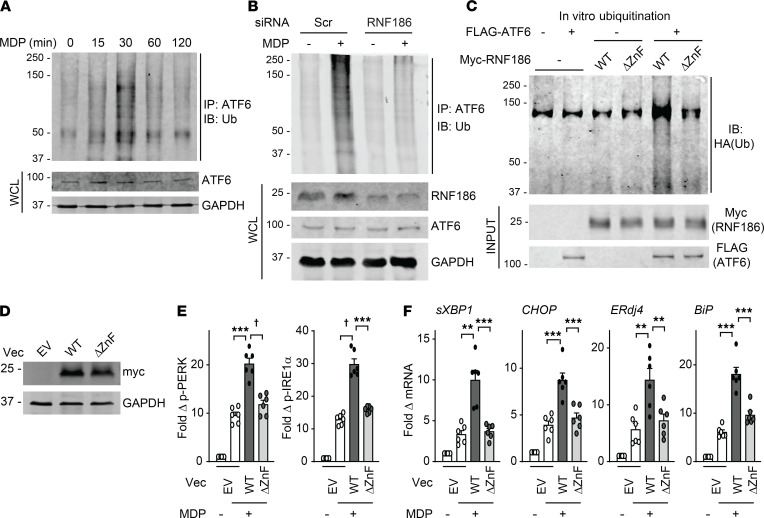
We first assessed whether RNF186 could localize to the ER in human macrophages and whether this was regulated upon PRR stimulation. We examined the PRR nucleotide-binding oligomerization domain containing 2 (NOD2) as intestinal tissues are exposed in an ongoing manner to the NOD2 ligand peptidoglycan (30, 31), with muramyl dipeptide (MDP) being the minimal component of peptidoglycan, which activates NOD2. Through a cellular fractionation approach, we observed that whereas the majority of RNF186 was in the cytosol, a small portion of RNF186 localized to the ER at baseline in human monocyte-derived macrophages (MDMs) (Figure 1A and Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI145472DS1). With NOD2 stimulation, the fraction of RNF186 localized to the ER increased transiently, peaked at 30 minutes, and then returned to the levels observed at baseline by 60–120 minutes (Figure 1A and Supplemental Figure 1A). Cytosolic RNF186 was regulated in a reciprocal pattern over this time period, suggesting that the cytosolic fraction was transiently localizing to the ER (Figure 1A and Supplemental Figure 1A).
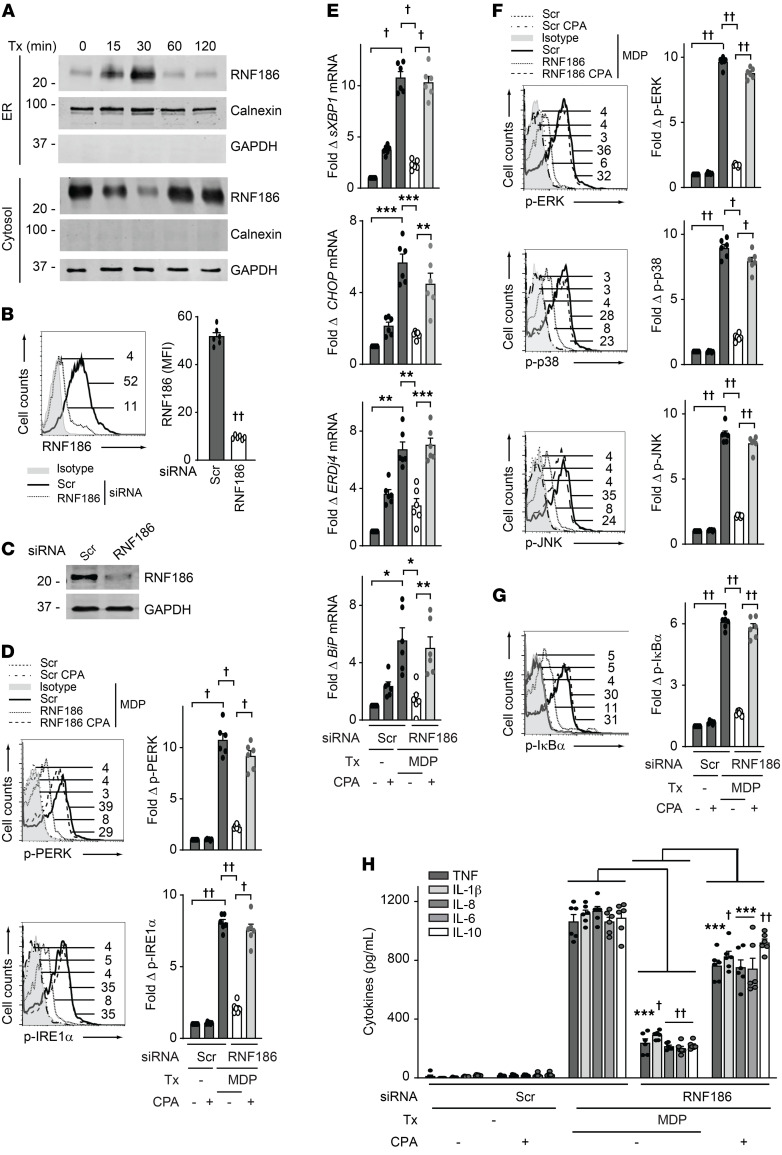
Figure 1. RNF186 localizes to the ER and is required for NOD2-induced UPR pathway activation.
(A) MDMs were treated with 100 μg/mL MDP for the indicated times. ER and cytosolic fractions were assessed for RNF186 expression by Western blot. Markers for ER (calnexin) and cytosolic (GAPDH) fractions are shown. (B and C) MDMs were transfected with scrambled or RNF186 siRNA. RNF186 protein expression by (B) flow cytometry with representative histogram with MFI values shown with horizontal lines and summary graph (n = 6 donors; similar results were observed in an additional n = 6) and (C) Western blot. (D–H) MDMs were transfected with scrambled or RNF186 siRNA and then treated with 100 μg/mL MDP ± 10 μM cyclopiazonic acid (CPA). (D) Fold phospho-PERK and phospho-IRE1α induction at 30 minutes (n = 6; similar results in an additional n = 6). (E) Fold change mRNA expression at 6 hours (n = 6; similar results in an additional n = 6). (F and G) Fold phospho-protein induction at 15 minutes (n = 6; similar results in an additional n = 6). (H) Cytokines at 12 hours (n = 6; similar results in an additional n = 6). Mean + SEM. Marker positions are shown (kDa) for Western blots. *P < 0.05; **P < 0.01; ***P < 0.001; †P < 1 × 10−4; ††P < 1 × 10−5 determined by 2-tailed Student’s t test with a Bonferroni-Holm correction for multiple comparisons. Scr, scrambled; Tx, treatment.
Given the PRR-dependent regulation of RNF186 ER localization, we next sought to determine ER-dependent mechanisms through which RNF186 might be modulating PRR-initiated outcomes in MDMs. In particular, we asked whether RNF186 regulates PRR-initiated activation of the UPR pathway given the ability of UPR signaling to contribute to PRR-initiated signaling (26, 29). We used siRNA to knock down RNF186 expression in human macrophages and confirmed effective reduction in expression of both RNF186 mRNA (Supplemental Figure 1B) and protein expression as assessed by flow cytometry and Western blot (Figure 1, B and C). Cells remained viable under these knockdown conditions (Supplemental Figure 1C) and remained functional in response to Dectin-1 stimulation (Supplemental Figure 1D). We then examined activation of the key UPR proteins PERK and IRE1α. NOD2 stimulation led to PERK and IRE1α activation in MDMs, and this was significantly reduced with RNF186 knockdown (Figure 1D). Moreover, NOD2 stimulation induced a UPR transcriptional program (32), including spliced XBP1, occurring downstream of IRE1α pathway activation, and CHOP, ERdj4, and BiP, and this was significantly reduced with RNF186 knockdown (Figure 1E).
We next assessed whether the RNF186-dependent UPR occurring upon NOD2 stimulation was required for key outcomes downstream of NOD2. We therefore examined MAPK and NF-κB signaling pathways and subsequent secretion of cytokines (33, 34); each of these outcomes was reduced in RNF186-deficient MDMs (Figure 1, F–H). To clearly establish that RNF186 induction of the UPR was contributing to these outcomes, we sought to complement the impaired UPR in RNF186-deficient MDMs. Cyclopiazonic acid (CPA), a pharmacological inducer of the UPR (35), was able to restore NOD2-induced PERK and IRE1α activation (Figure 1D) and the downstream UPR transcriptional program (Figure 1E) in RNF186-deficient MDMs. We ensured that CPA treatment did not reduce viability of MDMs (Supplemental Figure 1E). Importantly, complementation of the UPR pathway through CPA treatment in RNF186-deficient MDMs significantly restored NOD2-induced signaling (Figure 1, F and G) and cytokine secretion (Figure 1H). We observed similar outcomes using tunicamycin as an alternative pharmacological UPR inducer (Supplemental Figure 1, F–J). We ensured cell viability was intact with tunicamycin treatment (Supplemental Figure 1E).
To extend the role for the RNF186-dependent UPR to additional PRRs, we examined the PRR NOD1 through treatment with TriDAP (L-Ala-γ-D-Glu-mDAP). NOD1 stimulation led to induction of the UPR, and this was reduced with RNF186 knockdown (Supplemental Figure 2, A and B). Moreover, NOD1-induced MAPK and NF-κB activation and cytokine secretion were reduced with RNF186 knockdown (Supplemental Figure 2, C–E), and complementation of the UPR with CPA treatment in RNF186-deficient MDMs (Supplemental Figure 2, A and B) restored these NOD1-induced outcomes (Supplemental Figure 2, C–E). Taken together, PRR stimulation resulted in a transient localization of RNF186 to the ER with subsequent induction of RNF186-dependent UPR activation, which was required for PRR-induced signaling and cytokine secretion in human MDMs.
RNF186 interacts with UPR sensors and promotes assembly of NOD2 signaling complex components with UPR sensors.
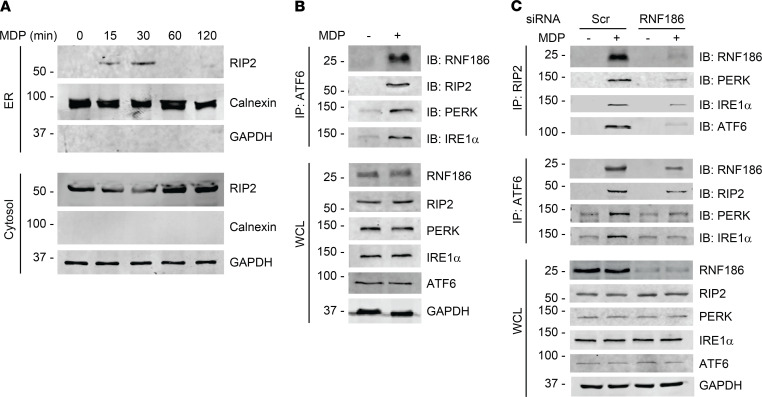
NOD2 stimulation induces formation of a signaling complex composed of the adaptor protein receptor-interacting protein 2 (RIP2) (36). An earlier study reported that NOD2 stimulation leads to RIP2 translocation to ER in epithelial cells (37), and we found that NOD2 stimulation induces RIP2 translocation to the ER and establishment of a complex with UPR sensors in human macrophages (29). We therefore asked whether RNF186 associates with and regulates the NOD2-induced RIP2 and UPR sensor complex in human MDMs. First, we confirmed that NOD2 stimulation leads to localization of RIP2 to the ER; peak RIP2 ER translocation occurred 30 minutes after NOD2 stimulation of human macrophages (Figure 2A). We next assessed whether RNF186 associates with UPR sensors along with RIP2. At 30 minutes after NOD2 stimulation, the time of peak RNF186 (Figure 1A) and RIP2 translocation to the ER, RNF186 associated with a complex composed of ATF6, PERK, IRE1α, and RIP2 (Figure 2B). Importantly, RNF186 expression was required for optimal formation of the NOD2-induced RIP2 complex with UPR sensors (Figure 2C and Supplemental Figure 3A); we observed a similar requirement for RNF186 expression using a distinct immunoprecipitation approach (Figure 2C and Supplemental Figure 3B). In contrast, and consistent with the lack of RNF186 regulation of Dectin-1–induced cytokines (Supplemental Figure 1D), RNF186 did not associate with the Dectin-1–induced signaling complex (Supplemental Figure 3C). Taken together, these results indicated that RNF186 was associated with and required for optimal formation of the NOD2-induced UPR signaling complex in human MDMs.
Figure 2. NOD2 stimulation induces an RNF186 complex with UPR sensors.
(A) MDMs were treated with 100 μg/mL MDP for the indicated times. ER and cytosolic fractions were assessed for RIP2 by Western blot. Markers for ER (calnexin) and cytosolic (GAPDH) fractions are shown. Representative of 2 independent experiments. (B) MDMs were treated with 100 μg/mL MDP for 30 minutes. ATF6 was immunoprecipitated followed by immunoblotting (IB) of the indicated proteins. Representative of 2 independent experiments. (C) MDMs were transfected with scrambled or RNF186 siRNA and then treated with 100 μg/mL MDP for 30 minutes. RIP2 or ATF6 was immunoprecipitated, and recruitment of the indicated proteins was assessed by IB. Representative of 2–3 independent experiments. Expression of the respective proteins and GAPDH in whole-cell lysates (WCLs) served as loading controls. Marker positions are shown (kDa). Scr, scrambled.
RNF186 localization to the ER is required for NOD2-induced UPR pathway activation and downstream outcomes.
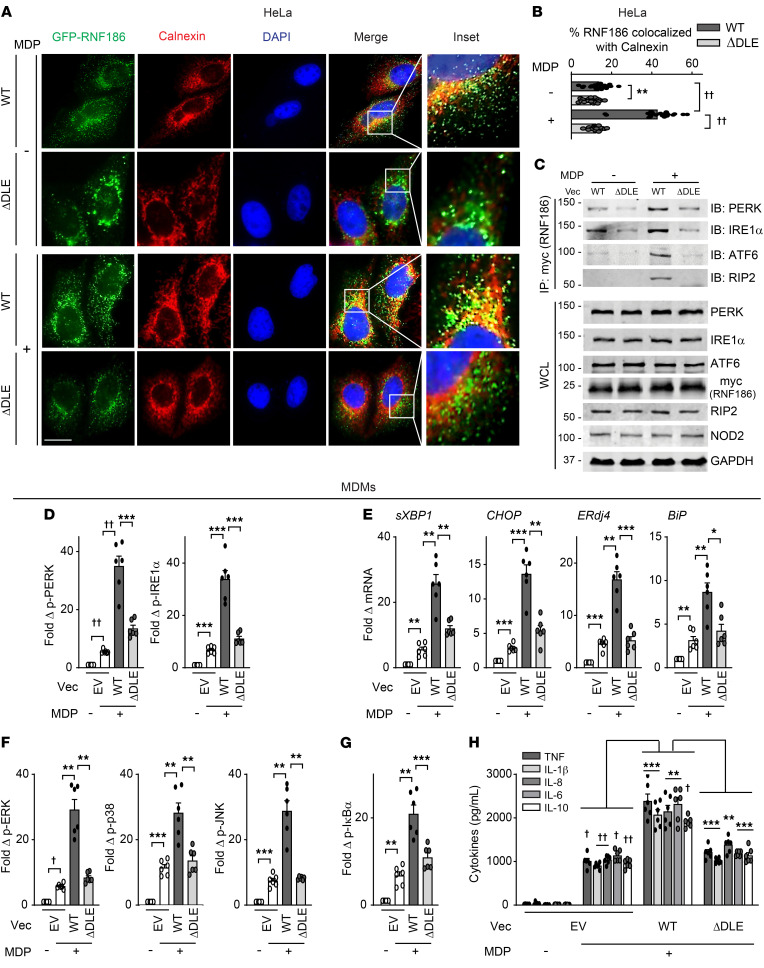
We next sought to clearly establish that RNF186 localization to ER is required for NOD2-induced UPR activation and downstream outcomes. Our sequence-based analysis predicted an ER retention motif, DLE, at the N-terminus of RNF186. We substituted these 3 amino acids with alanines in RNF186 (RNF186-ΔDLE) (Supplemental Figure 4A). We conducted a time course analysis of NOD2-induced RNF186 localization to the ER in HeLa cells to ensure a similar pattern of RNF186 ER localization in epithelial cells to that observed in MDMs. RNF186 localization to the ER in HeLa cells peaked 30 minutes after NOD2 stimulation (Supplemental Figure 4, B and C), similar to what was observed in MDMs. We next transfected both the WT and RNF186-ΔDLE constructs into HeLa cells and ensured equal expression levels of the RNF186 constructs as assessed by Western blot (Figure 3C). In untreated cells, the already low level of WT RNF186 localization to the ER was reduced with RNF186-ΔDLE (Figure 3, A and B). Furthermore, upon NOD2 stimulation, RNF186-ΔDLE did not localize to the ER as effectively as did WT RNF186 (Figure 3, A and B). Consistent with the reduced ER localization, RNF186-ΔDLE did not as effectively associate with the UPR sensors PERK, IRE1α, and ATF6 after NOD2 stimulation when compared with WT RNF186 (Figure 3C and Supplemental Figure 4D). RNF186-ΔDLE association with RIP2 was similarly reduced under these conditions (Figure 3C and Supplemental Figure 4D). These studies showed that in addition to macrophages, PRR stimulation regulated RNF186 interactions with UPR sensors in epithelial cells.
Figure 3. RNF186 localization to the ER is required for NOD2-induced UPR signaling and downstream outcomes.
(A and B) HeLa cells were transfected with GFP-tagged RNF186-WT or RNF186-ΔDLE and NOD2 and then treated with 100 μg/mL MDP for 30 minutes. Cells were immunostained for ER (calnexin, red) and nucleus (DAPI, blue). (A) Representative micrographs; scale bar: 10 μm. Inset represents 3× enlarged images from the merged panel. (B) Summary graph of percentage of RNF186 colocalized with calnexin (ER) (25 cells quantified). Representative of 2 independent experiments. (C) HeLa cells were transfected with myc-RNF186 WT or ΔDLE and NOD2 followed by treatment with 100 μg/mL MDP for 30 minutes. Myc (RNF186) was immunoprecipitated, and the recruitment of the indicated proteins was assessed by immunoblotting (IB). Whole-cell lysates (WCLs) of the indicated proteins served as loading controls. Marker positions are shown (kDa). Representative of 3 independent experiments. (D–H) MDMs (rs6426833 AA low RNF186-expressing carriers) (n = 6) were transfected with empty vector (EV) or myc-tagged RNF186 WT or ΔDLE and then treated with 100 μg/mL MDP. (D) Fold phospho-PERK and phospho-IRE1α induction at 30 minutes. (E) Fold change mRNA expression at 4 hours. (F and G) Fold phospho-protein induction at 15 minutes. (H) Cytokines at 24 hours. Mean + SEM. *P < 0.05; **P < 0.01; ***P < 0.001; †P < 1 × 10−4; ††P < 1 × 10−5 determined by 2-tailed Student’s t test with a Bonferroni-Holm correction for multiple comparisons. Vec, vector.
We next sought to address whether RNF186 localization to the ER is required for NOD2-induced UPR activation and downstream signaling and cytokines in human MDMs. To determine this, we first confirmed RNF186 expression regulation with NOD2 stimulation and the IBD risk rs6426833 genotype (noncoding; ~31 kb from RNF186) regulation of RNF186 expression in human MDMs. With NOD2 stimulation, RNF186 mRNA peaked at 4 hours (Supplemental Figure 5A), and RNF186 protein expression peaked at 12–24 hours (Supplemental Figure 5B). MDMs from rs6426833 AA disease risk carriers demonstrated lower baseline and particularly NOD2-induced RNF186 mRNA (Supplemental Figure 5C) and protein (Supplemental Figure 5D) expression compared with GG carriers. Therefore, we confirmed that RNF186 expression increased with PRR stimulation and was modulated by the IBD risk rs6426833 genotype. We next transfected RNF186-WT or RNF186-ΔDLE (ER localization mutant) into MDMs expressing low levels of RNF186 (rs6426833 AA carriers) so as to minimize endogenous RNF186 expression. We ensured equal expression levels as assessed by flow cytometry (Supplemental Figure 5E) and Western blot (Supplemental Figure 5F). We further ensured reduced NOD2-mediated RNF186-ΔDLE localization to the ER in MDMs compared with WT RNF186 (Supplemental Figure 5G) and consistently reduced NOD2-induced RNF186-ΔDLE association with the UPR sensors compared with WT RNF186 in MDMs (Supplemental Figure 5H). RNF186-WT–transfected MDMs demonstrated increased NOD2-induced UPR activation compared with empty vector–transfected MDMs (Figure 3, D and E), thereby providing a complementary approach to the RNF186 knockdown studies above. Importantly, NOD2-induced UPR activation was reduced in RNF186-ΔDLE compared with RNF186-WT–transfected MDMs (Figure 3, D and E). In turn, NOD2-induced MAPK and NF-κB activation (Figure 3, F and G) and cytokine secretion (Figure 3H) were reduced in RNF186-ΔDLE–transfected compared with RNF186-WT–transfected MDMs. We further identified a putative ER localization motif at position K219 of RNF186 and substituted K219 with an alanine (Supplemental Figure 6A) to confirm the importance of RNF186 localization to the ER for NOD2-induced outcomes through a second approach. RNF186-K219A demonstrated reduced ER localization in HeLa cells upon NOD2 stimulation (Supplemental Figure 6, B and C) and in turn reduced NOD2-induced UPR signaling, MAPK and NF-κB activation, and cytokine secretion in MDMs (Supplemental Figure 7, A–E). These data indicated that RNF186 localization to the ER was required for NOD2-induced UPR pathway activation and subsequent signaling and cytokines in human macrophages.
RNF186 E3 ubiquitin ligase activity is required for optimal NOD2-induced UPR-associated outcomes.
Posttranslational modifications (e.g., phosphorylation, ADP-ribosylation, glycosylation, and acetylation) play an integral role in UPR signaling (38). To the best of our knowledge, only a few studies have identified ubiquitin-mediated regulation of UPR sensors, and in some cases, this has been correlated with subsequent degradation of the targeted sensor (39–41). Ubiquitin modifications of UPR sensors during innate immune responses and with PRR stimulation are incompletely defined. Further, how RNF186-mediated ubiquitination of UPR sensors might regulate outcomes downstream of PRRs is not clear. NOD2 stimulation led to ubiquitination of the ATF6 complex in human MDMs, with peak ubiquitination occurring 30 minutes after stimulation (Figure 4A). Notably, RNF186 expression was required for optimal NOD2-induced ubiquitination of the ATF6 complex (Figure 4B). To establish that ATF6 could serve as a direct substrate for RNF186-mediated ubiquitination, we conducted in vitro ubiquitination assays with purified RNF186 and ATF6 (Supplemental Figure 8B for purified proteins). Importantly, RNF186 directly ubiquitinated ATF6 (Figure 4C).
Figure 4. RNF186 promotes NOD2-induced ubiquitination of the ATF6 complex, and RNF186 E3 ubiquitin ligase activity is required for the NOD2-induced UPR.
(A) MDMs were treated with 100 μg/mL MDP for the indicated times. ATF6 was immunoprecipitated, and ubiquitinated proteins (Ubs) were assessed by Western blot (representative of 3 independent experiments). (B) MDMs were transfected with scrambled or RNF186 siRNA and then treated with 100 μg/mL MDP for 30 minutes. ATF6 was immunoprecipitated, and Ubs were assessed by Western blot (representative of 2 independent experiments). Whole-cell lysates (WCLs) of the indicated proteins served as loading controls. (C) In vitro ubiquitination was assessed as per Methods with purified HA-ubiquitin with (+) or without (-) purified FLAG-ATF6 with/without purified myc-RNF186 WT or ΔZnF. Ubiquitin protein (α-HA) was detected by Western blot (representative of 2 independent experiments). Marker positions are shown (kDa). (D–F) MDMs were transfected with empty vector (EV) or myc-tagged RNF186 WT or ΔZnF. (D) RNF186 protein expression (myc) by Western blot. (E and F) Transfected MDMs (n = 6) were treated with 100 μg/mL MDP. (E) Fold phospho-PERK and phospho-IRE1α induction at 30 minutes. (F) Fold change mRNA expression at 4 hours. Mean + SEM. **P < 0.01; ***P < 0.001; †P < 1 × 10−4 determined by 2-tailed Student’s t test with a Bonferroni-Holm correction for multiple comparisons. Scr, scrambled; Vec, vector; ZnF, zinc finger.
The N-terminal RING domain of RNF186 has a conserved C3HC4-type ZnF motif that can catalyze ubiquitination of downstream substrates in epithelial cells (13, 14). To assess whether the ZnF motif in the RING domain of RNF186 is required for ubiquitination of ATF6 and NOD2-induced, UPR-dependent outcomes, we generated a ZnF mutant of RNF186 (RNF186-ΔZnF) (see Supplemental Figure 8A for structural region and Supplemental Figure 8B for purified protein). RNF186-ΔZnF demonstrated impaired ubiquitination of ATF6 in vitro (Figure 4C). We then transfected either RNF186 WT or RNF186-ΔZnF into MDMs and ensured equal levels of RNF186 expression (Figure 4D and Supplemental Figure 8C). Upon NOD2 stimulation, MDMs transfected with RNF186-ΔZnF demonstrated less effective PERK and IRE1α activation (Figure 4E) and UPR-associated transcript induction (Figure 4F) and in turn less effective MAPK and NF-κB activation (Supplemental Figure 8, D and E) and cytokine secretion (Supplemental Figure 8F) compared with RNF186 WT-transfected MDMs. Therefore, the E3 ligase activity of RNF186 was critical for NOD2-induced activation of the UPR and UPR-dependent downstream signaling and cytokines.
RNF186 ubiquitinates ATF6 at K152, which is required for PRR-induced, UPR-associated outcomes.
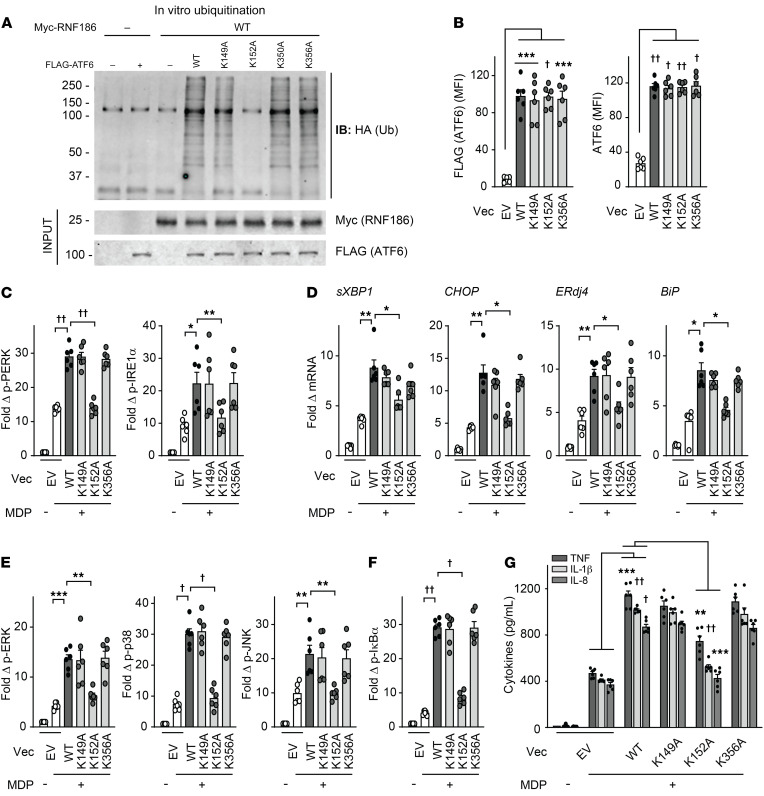
As we had found that RNF186 directly ubiquitinates ATF6, we sought to determine the site(s) on ATF6 at which this ubiquitination occurs to establish whether ATF6 ubiquitination is required for the PRR-induced, RNF186-dependent outcomes we had defined. To our knowledge, a clear requirement for ATF6 ubiquitination in functional outcomes has not been previously established and would have important implications for a broad range of cellular and biological responses. Our in silico analysis predicted various lysine residues in human ATF6 that might serve as ubiquitin conjugating sites, including K149, K152, K350, and K356 (Supplemental Figure 9A). We therefore constructed mutants for each of these residues where the lysine was substituted with an alanine (as per Supplemental Figure 9A). We transfected the FLAG-tagged WT or each of the lysine mutants (K149A, K152A, K350A, or K356A) of ATF6 into HEK293 cells and ensured equivalent expression (Supplemental Figure 9B). We then purified each of the ATF6 mutants (Supplemental Figure 9C) and examined in vitro ubiquitination of FLAG-tagged WT or ATF6 mutants by RNF186. ATF6-K152A showed reduced RNF186-mediated ubiquitination compared with ATF6-WT (Figure 5A), whereas the remaining ATF6 lysine mutants (K149A, K350A, and K356A) were not impaired in this ubiquitination (Figure 5A). Of note is that K152 is highly conserved in ATF6 across species (through zebrafish) (Supplemental Figure 9D); overall protein identity between human and zebrafish ATF6 is 34% (Supplemental Figure 9E).
Figure 5. Lysine 152 in ATF6 is critical for RNF186-dependent ubiquitination of ATF6 and PRR-induced, RNF186-dependent downstream outcomes.
(A) In vitro ubiquitination was assessed as per Methods with purified HA-ubiquitin with/without purified myc-RNF186 WT with/without purified FLAG-tagged WT or lysine mutants (K149A, K152A, K350A, K356A) of ATF6. Ubiquitin protein (α-HA) was detected by Western blot (representative of 3 independent experiments). Marker positions are shown (kDa). (B–G) MDMs were transfected with empty vector (EV) or FLAG-tagged WT or mutants of ATF6. (B) ATF6 protein expression as detected by flow cytometry (n = 6). (C–G) Transfected MDMs (n = 6) were treated with 100 μg/mL MDP. (C) Fold phospho-PERK and phospho-IRE1α induction at 30 minutes. (D) Fold change mRNA expression at 4 hours. (E and F) Fold phospho-protein induction at 15 minutes. (G) Cytokines at 24 hours. Similar results in an additional n = 6 for B–G. Mean + SEM. *P < 0.05; **P < 0.01; ***P < 0.001; †P < 1 × 10−4; ††P < 1 × 10−5 determined by 2-tailed Student’s t test with a Bonferroni-Holm correction for multiple comparisons. Vec, vector.
To assess whether the K152A residue in the ATF6 protein is required for PRR-induced, RNF186-dependent outcomes, we expressed either WT or ATF6-K152A in MDMs. We also examined ATF6-K149A and ATF6-K356A as negative controls. These constructs expressed at similar levels in MDMs (Figure 5B). Upon NOD2 stimulation, WT ATF6 transfection increased PERK and IRE1α activation (Figure 5C) and downstream UPR-associated transcript induction (Figure 5D) and, in turn, MAPK and NF-κB activation (Figure 5, E and F) and cytokine secretion (Figure 5G and Supplemental Figure 9F). However, each of these measures was less effectively induced in ATF6-K152A–transfected MDMs (Figure 5, C–G, and Supplemental Figure 9F). Importantly, ATF6 expression was unchanged after NOD2 stimulation, and this was not altered with ATF6-K152A (Supplemental Figure 9G). Taken together, the ATF6-K152 residue was critical for RNF186-mediated ubiquitination and was required for NOD2-induced activation of the UPR and downstream signaling and cytokines; these findings have important implications for posttranslational ubiquitin modification of ATF6 in contributing to ATF6-dependent outcomes more broadly.
The rare RNF186-A64T disease risk variant shows impaired NOD2-induced ubiquitination of ATF6 and UPR-dependent outcomes.
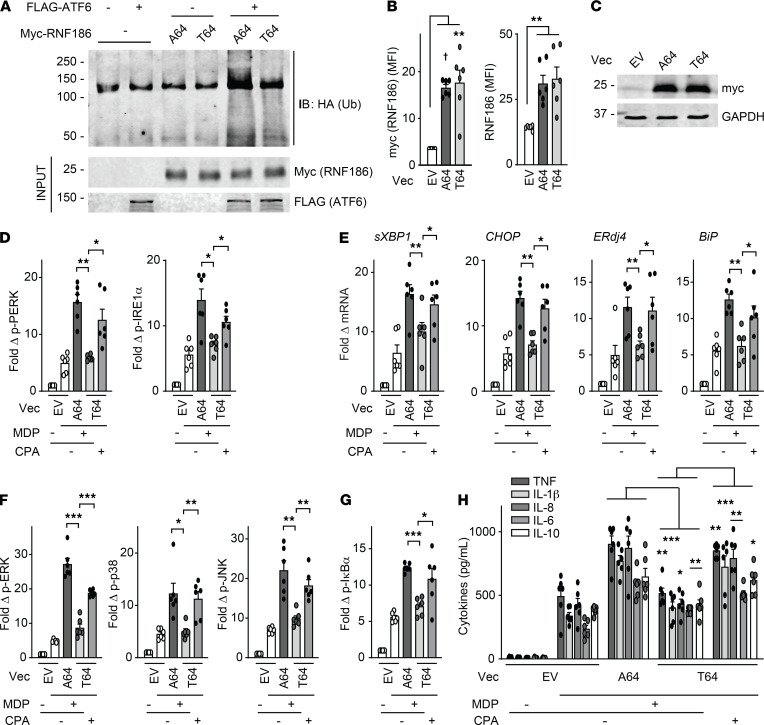
We next investigated the consequences of the rare RNF186 A64T IBD risk variant on the RNF186-dependent mechanisms we had identified. The RNF186 A64T mutation is located in the RING domain (required for E3 ubiquitin ligase activity) of RNF186 (Supplemental Figure 10A). Because this is a rare coding variant (0.8% allele frequency per dbSNP as of July 2019), we utilized a transfection approach to assess this disease risk coding mutation. We expressed GFP-RNF186-A64 (WT) or GFP-RNF186-T64 (risk variant) with NOD2 and ensured that these respective constructs expressed at similar levels in HeLa cells (Supplemental Figure 10B). RNF186-A64 and RNF186-T64 demonstrated similar frequencies of localization to the ER at baseline and upon NOD2 stimulation (Supplemental Figure 10, C and D). However, the RNF186-T64 risk variant demonstrated an impaired ability to directly ubiquitinate ATF6 in vitro (Figure 6A and Supplemental Figure 10E). To assess the consequences of the rare RNF186-T64 variant on NOD2-induced, RNF186- and UPR-dependent outcomes, we transfected either A64 or T64 RNF186 into MDMs. We ensured equal expression of RNF186-A64 and RNF186-T64 (Figure 6, B and C). RNF186-T64 demonstrated an impaired ability to enhance NOD2-induced UPR activation (Figure 6, D and E), along with downstream signaling (Figure 6, F and G) and cytokine secretion (Figure 6H) compared with RNF186-A64 (WT). Importantly, complementation of the UPR through CPA treatment in RNF186-T64–transfected MDMs (Figure 6, D and E) restored downstream signaling and cytokine secretion (Figure 6, F–H). Taken together, the rare disease risk variant RNF186-A64T in the RING domain of RNF186 demonstrated a reduced ability to ubiquitinate ATF6 and activate the UPR and, in turn, mediate UPR-dependent downstream signaling and cytokines upon PRR stimulation.
Figure 6. The rare RNF186 A64T IBD risk variant demonstrates lower levels of NOD2-induced, UPR-dependent outcomes.
(A) In vitro ubiquitination with purified HA-ubiquitin with/without purified FLAG-ATF6 with/without purified myc-RNF186-A64 or myc-RNF186-T64. Ubiquitin protein (α-HA) was detected by Western blot. Representative of 2 independent experiments. Marker positions are shown (kDa). (B–H) MDMs (rs6426833 AA low RNF186-expressing carriers) were transfected with empty vector (EV) or myc-tagged RNF186-A64 (WT) or RNF186-T64 (risk variant). RNF186 protein expression as detected by (B) flow cytometry (n = 6) or (C) Western blot. (D–H) Transfected MDMs were treated with 100 μg/mL MDP (n = 6) ± 10 μM CPA. (D) Fold phospho-PERK and phospho-IRE1α induction at 30 minutes. (E) Fold change mRNA expression at 6 hours. (F and G) Fold phospho-protein induction at 15 minutes. (H) Cytokines at 12 hours. Mean + SEM. *P < 0.05; **P < 0.01; ***P < 0.001; †P < 1 × 10−4 determined by 2-tailed Student’s t test with a Bonferroni-Holm correction for multiple comparisons. Vec, vector.
RNF186-dependent UPR signaling is required for NOD2-induced antimicrobial pathways.
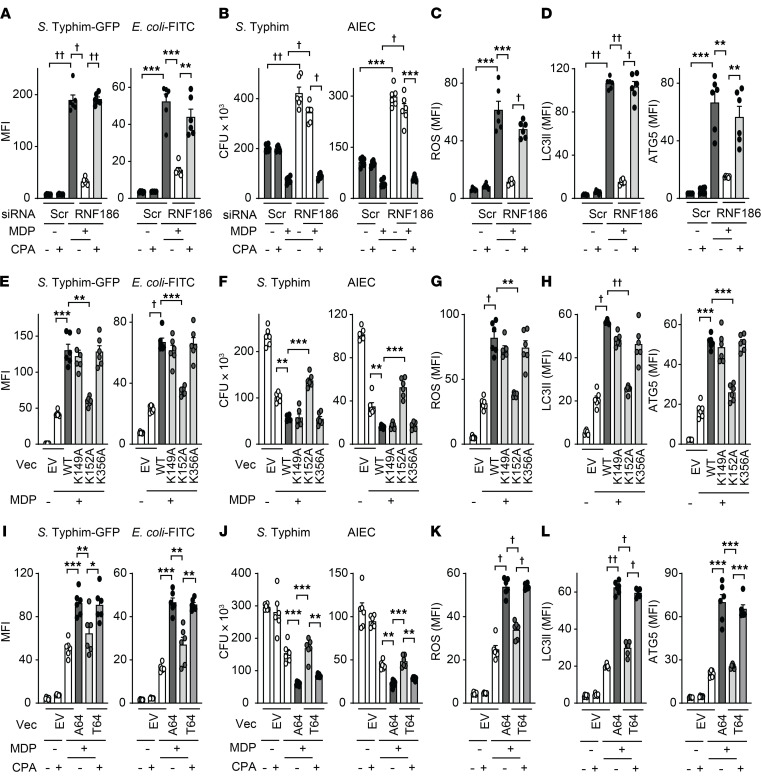
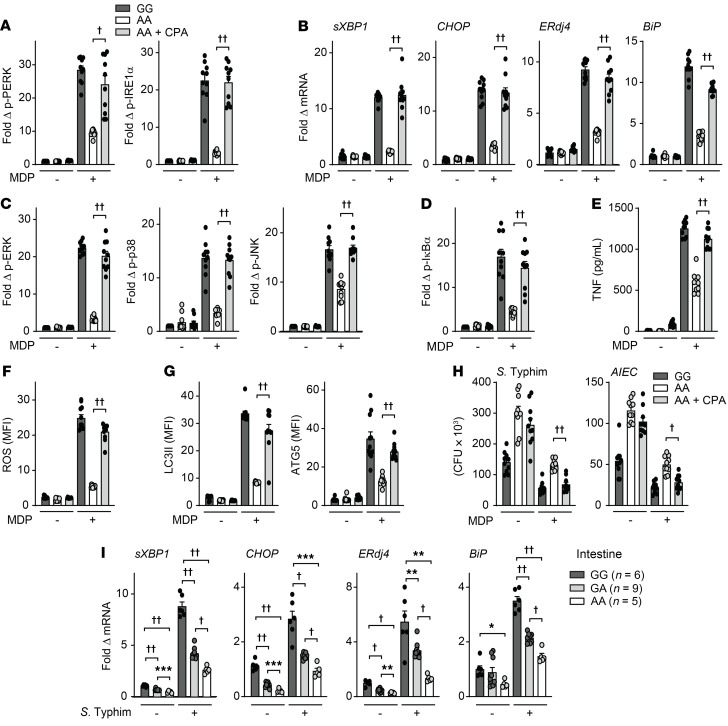
Impaired bacterial clearance can contribute to IBD pathogenesis (6, 8, 42) and PRR-initiated signaling promotes antimicrobial pathways (1). Given that the rare loss-of-function RNF186 A64T IBD risk variant resulted in reduced UPR activation and in turn reduced PRR-induced signaling and cytokines, and that MDMs from common RNF186 IBD risk variant carriers showed reduced RNF186 expression (Supplemental Figure 5, C and D), we sought to assess whether RNF186-dependent UPR signaling is required for optimal induction of antimicrobial pathways and bacterial clearance. Chronic PRR stimulation of macrophages enhances microbial clearance and simulates the ongoing exposure to PRR ligands by macrophages in intestinal tissue (1, 43, 44). We therefore examined antimicrobial pathways at baseline and after chronic NOD2 stimulation of human macrophages. Bacterial uptake is an initial step for clearing bacteria by macrophages. With prolonged (48 hours) NOD2 stimulation of human MDMs, the uptake of Salmonella Typhimurium–GFP (S. Typhimurium–GFP) was increased (Figure 7A and Supplemental Figure 11A). However, this enhanced uptake was impaired in RNF186-deficient MDMs (Figure 7A and Supplemental Figure 11A). Importantly, complementing UPR signaling through CPA in chronic NOD2-stimulated, RNF186-deficient MDMs restored bacterial uptake (Figure 7A and Supplemental Figure 11A). We observed similar outcomes with fluorophore-labeled E. coli bioparticles (Figure 7A and Supplemental Figure 11A). We next assessed the ability of RNF186-deficient MDMs to clear the bacteria that had entered into the cells. As per our prior studies (43), intracellular levels of S. Typhimurium were reduced in MDMs after chronic NOD2 stimulation (Figure 7B), consistent with more effective bacterial clearance. However, despite having taken up less bacteria, chronic NOD2-stimulated, RNF186-deficient MDMs were less effective at clearing the existing intracellular S. Typhimurium compared with WT MDMs (Figure 7B). Similar impaired bacterial clearance in RNF186-deficient MDMs was observed with adherent-invasive E. coli (Figure 7B), which colonizes the ilea of patients with Crohn’s disease with increased frequency (45). Importantly, complementing UPR signaling in RNF186-deficient MDMs with CPA was able to restore bacterial clearance (Figure 7B), thereby clearly establishing the role of UPR signaling in these RNF186-dependent outcomes.
Figure 7. RNF186-dependent UPR signaling is required for NOD2-induced bacterial clearance and antimicrobial pathways.
(A–D) MDMs (n = 6) were transfected with scrambled or RNF186 siRNA, then treated with 100 μg/mL MDP for 48 hours ± 10 μM CPA. (E–H) MDMs (n = 6; similar results in an additional n = 6) were transfected with empty vector (EV) or FLAG-tagged WT or lysine mutants (K149A, K152A, or K356A) of ATF6 and then treated with 100 μg/mL MDP for 48 hours. (I–L) MDMs (n = 6) (rs6426833 AA low RNF186-expressing carriers) were transfected with EV or myc-tagged RNF186-A64 (WT) or RNF186-T64 (risk variant) and then treated with 100 μg/mL MDP for 48 hours ± 10 μM CPA. (A, E, and I) Bacterial uptake. (B, F, and J) Intracellular bacterial clearance. (C, G, and K) ROS production. (D, H, and L) LC3II and ATG5 expression. Mean + SEM. *P < 0.05; **P < 0.01; ***P < 0.001; †P < 1 × 10−4; ††P < 1 × 10−5 determined by 2-tailed Student’s t test with a Bonferroni-Holm correction for multiple comparisons. Scr, scrambled; Vec, vector.
We next investigated mechanisms through which RNF186-dependent UPR signaling contributes to NOD2-induced intracellular bacterial clearance. ROS contributes to bacterial clearance and to maintenance of homeostasis in the intestine (46). Furthermore, chronic PRR stimulation induces intracellular ROS (3, 4). Upon knockdown of RNF186 in MDMs, chronic NOD2-induced ROS was impaired, and complementation of UPR signaling through CPA largely restored NOD2-induced ROS (Figure 7C and Supplemental Figure 11B).
Induction of autophagy is an additional critical antimicrobial pathway regulated through PRR stimulation (1), and loss-of-function variants in autophagy-associated genes confer risk for IBD (9). Chronic NOD2 stimulation of MDMs induced autophagy as assessed by the autophagy marker LC3II and the autophagy-associated molecule ATG5, and this induction was impaired in RNF186-deficient MDMs (Figure 7D and Supplemental Figure 11C). Restoring UPR signaling through CPA in RNF186-deficient MDMs rescued the autophagy induction observed with chronic NOD2 stimulation (Figure 7D and Supplemental Figure 11C). Taken together, these data indicated that RNF186 promoted antimicrobial mechanisms in human macrophages in a UPR-dependent manner.
Given that the ATF6-K152 residue is critical for RNF186-mediated ubiquitination of ATF6 and is required for PRR induced UPR signaling, we next examined whether ATF6-K152 is required for the NOD2-induced, RNF186-dependent antimicrobial pathways we had defined. We transfected MDMs with either ATF6-WT or ATF6-lysine mutants (K152A, or K149A or K356A as negative controls). ATF6-WT–transfected MDMs demonstrated enhanced NOD2-mediated bacterial uptake (Figure 7E), intracellular bacterial clearance (Figure 7F and Supplemental Figure 9H [for baseline clearance]), and antimicrobial pathways (Figure 7, G and H) compared with empty vector–transfected cells. In contrast, each of these outcomes was impaired in ATF6-K152A–transfected MDMs (Figure 7, E–H). These data indicated that ATF6-K152 was required for PRR-induced, RNF186-dependent antimicrobial pathways in human MDMs.
Given that the rare RNF186 A64T IBD risk variant demonstrated impaired NOD2-induced UPR activation and downstream signaling and cytokines, we next assessed whether this risk variant also led to reduced NOD2-mediated antimicrobial pathways in a UPR-dependent manner. We transfected MDMs (rs6426833 AA low RNF186-expressing carriers to minimize endogenous RNF186 expression) with either RNF186-WT or RNF186-T64 (IBD risk variant). Compared with RNF186-WT–transfected MDMs, NOD2-mediated induction of bacterial uptake (Figure 7I), intracellular bacterial clearance (Figure 7J), and antimicrobial pathways (Figure 7, K and L) were impaired in RNF186-T64–transfected MDMs but could be restored with complementation of UPR signaling through CPA treatment (Figure 7, I–L). Taken together, these data indicated that RNF186-dependent UPR signaling was required for NOD2-induced antimicrobial pathways in human MDMs, this required ubiquitination of ATF6 at K152, and impaired antimicrobial pathways with the RNF186-T64 IBD risk variant could be restored with complementation of the UPR.
MDMs from IBD risk rs6426833 A carriers show reduced NOD2-induced UPR-dependent outcomes, and these can be restored with complementation of the UPR.
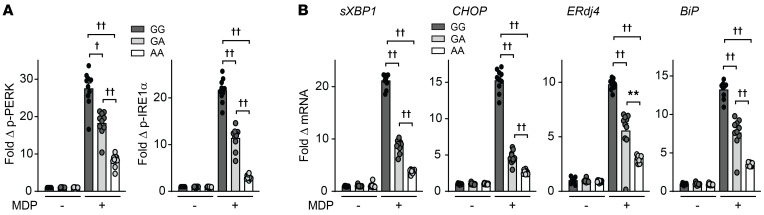
Given that the rs64268331 A risk variant leads to reduced RNF186 expression (Supplemental Figure 5, C and D), we examined whether the NOD2-induced UPR was regulated in an rs64268331 genotype–dependent manner. MDMs from rs64268331 A risk carriers demonstrated reduced NOD2-induced activation of UPR signaling relative to G carrier cells (Figure 8, A and B). We next sought to determine whether restoring UPR signaling in these lower RNF186-expressing rs64268331 A risk carrier MDMs could restore key PRR-initiated outcomes. We therefore used CPA to restore UPR signaling in rs64268331 A risk carrier MDMs to the levels observed in GG carrier MDMs (Figure 9, A and B). While MDMs from rs64268331 A risk carriers demonstrated reduced NOD2-induced MAPK (Supplemental Figure 12A) and NF-κB (Supplemental Figure 12B) activation, cytokine secretion (Supplemental Figure 12C), antimicrobial pathways (Supplemental Figure 12, D and E), and intracellular bacterial clearance (Supplemental Figure 12F) compared with G carriers, complementation of the UPR with CPA (Figure 9, A and B) increased each of these outcomes in AA carrier MDMs to the levels observed in GG carrier MDMs (Figure 9, C–H, and Supplemental Figure 12G). We further assessed whether the rs6426833 IBD risk genotype modulates the UPR in intestinal myeloid cells. Intestinal myeloid cells from rs6426833 AA risk carriers demonstrated reduced RNF186 expression (Supplemental Figure 12H) and reduced UPR transcripts (Figure 9I) at baseline and showed a less effective induction of these pathways upon coculture with S. Typhimurium. Finally, we analyzed colonic biopsies from UC patients using an existing data set (NCBI’s Gene Expression Omnibus [GEO] GSE59071). At this chronic phase of disease, colonic RNF186 mRNA expression was not different between the control group and UC group, whereas expression of the downstream UPR pathway transcripts XBP1, ERdj4, and BiP was increased in UC colon compared with control colon (Supplemental Figure 13, A and B). Expression of these UPR transcripts decreased during inactive UC (Supplemental Figure 13B). We confirmed similar upregulation of XBP1, ERdj4, and BiP transcripts in UC colon in 4 additional cohorts, 2 of which were pediatric cohorts (GEO GSE117993, GSE109142), and one of which also showed an increase in CHOP mRNA in UC colon compared with control colon (Supplemental Figure 13, C–F). Taken together, MDMs from rs6426833 AA risk carriers in the RNF186 region demonstrated less effective PRR-induced UPR signaling, which in turn led to a reduction in multiple key PRR-induced outcomes; these outcomes could be restored with complementation of the deficient UPR.
Figure 8. IBD risk rs6426833 AA disease risk carriers demonstrate reduced PRR-induced UPR signaling.
MDMs from rs6426833 GG, GA, or AA carriers (n = 10 donors/genotype) were left untreated or treated with 100 μg/mL MDP. (A) Fold phospho-PERK and phospho-IRE1α induction at 30 minutes. (B) Fold change mRNA expression at 4 hours. Mean + SEM. **P < 0.01; †P < 1 × 10−4; ††P < 1 × 10−5 determined by 2-tailed Student’s t test with a Bonferroni-Holm correction for multiple comparisons.
Figure 9. Complementation of the UPR in MDMs from IBD risk rs6426833 AA carriers increases NOD2-induced, RNF186-dependent outcomes to levels observed in GG carrier MDMs.
(A–H) MDMs from rs6426833 GG and AA carriers (n = 10 donors/genotype) were left untreated or treated with 100 μg/mL MDP ± 10 μM CPA. (A) Fold phospho-PERK and phospho-IRE1α induction at 30 minutes. (B) Fold change mRNA expression at 4 hours. (C and D) Fold phospho-protein induction at 15 minutes. (E) Cytokines at 12 hours. (F) ROS at 48 hours. (G) LC3II and ATG5 expression at 48 hours. (H) After 48 hours, intracellular bacterial clearance was assessed. (I) Intestinal myeloid cells were cocultured with S. Typhimurium. Fold change mRNA at 2 hours stratified on rs6426833 genotype. Mean + SEM. *P < 0.05; **P < 0.01; ***P < 0.001; †P < 1 × 10−4; ††P < 1 × 10−5. Significance is between AA carriers with/without CPA in A–H determined by 2-tailed Student’s t test and with a Bonferroni-Holm correction for multiple comparisons in I.
RNF186 promotes the UPR, cytokine secretion, and intracellular bacterial clearance in intestinal lamina propria macrophages from mice.
To assess whether RNF186 regulates intestinal macrophage outcomes in a manner similar to that observed in peripheral macrophages, we utilized mouse models. We first ensured that similar to human MDMs, RNF186 promoted PRR-induced UPR signaling and UPR-associated transcripts, downstream signaling, cytokine secretion, and intracellular bacterial clearance in mouse bone marrow–derived macrophages as assessed through an RNF186 knockdown approach (Supplemental Figure 14, A–E). RNF186 was also required for live S. Typhimurium–induced UPR transcripts (Supplemental Figure 14F). ATF6 similarly promoted PRR-induced UPR signaling and downstream outcomes and live bacterial–induced UPR transcripts in mouse bone marrow–derived macrophages (Supplemental Figure 15). Whereas intestinal macrophages respond poorly to PRR stimulation (47), they do respond to live bacteria (48). Colonic lamina propria macrophages (Supplemental Figure 16A) demonstrated increased Rnf186 expression and Atf6 expression as well as increased UPR transcripts upon coculture with live S. Typhimurium (Supplemental Figure 16, B and C). Upon in vivo delivery of RNF186 siRNA to mice, RNF186 expression was effectively reduced in colonic tissues (Supplemental Figure 17A) and colonic macrophages (Supplemental Figure 17B) compared with mice administered scrambled siRNA. Ex vivo isolated RNF186-deficient intestinal macrophages cocultured with S. Typhimurium demonstrated reduced UPR transcripts (Supplemental Figure 17C) and decreased cytokine secretion (Supplemental Figure 17D). They further demonstrated a reduced ability to take up bacteria, and they were unable to as effectively clear these lower levels of intracellular bacteria (Supplemental Figure 17E). Therefore, RNF186 was required for the bacterially induced UPR and bacterial clearance in intestinal macrophages.
RNF186 promotes the UPR in intestinal macrophages during acute intestinal injury.
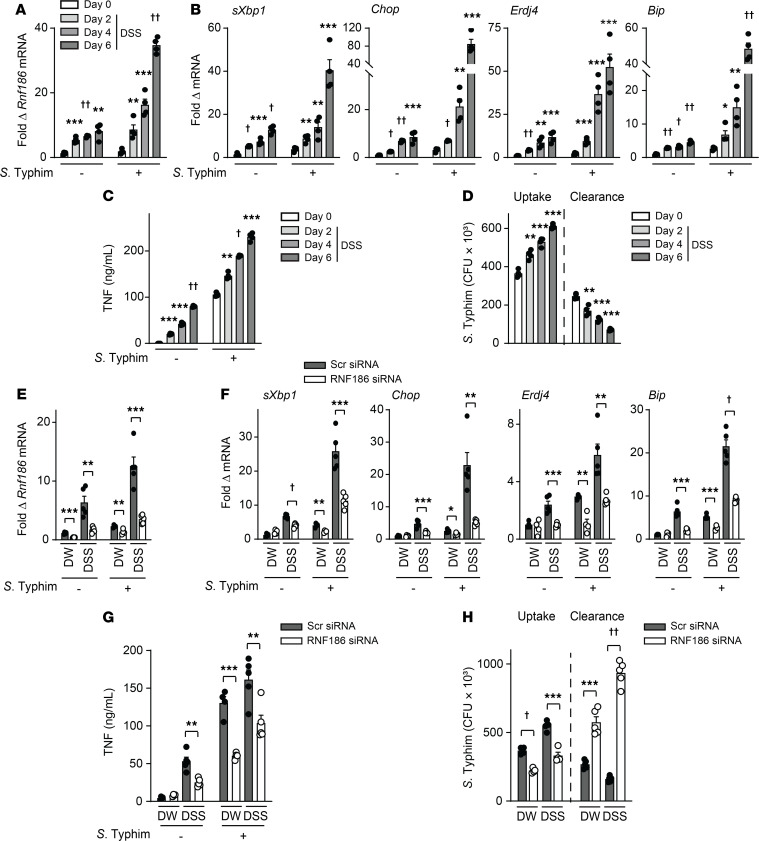
To assess how RNF186 regulates intestinal macrophage outcomes during intestinal injury in vivo, we utilized the DSS model of acute injury; this model allows for assessing handling of high levels of intestinal resident microbes, which we hypothesize will be impaired with RNF186 deficiency. Of note is that 2 prior studies identified increased DSS-induced disease severity with RNF186 deficiency (12, 14), but the focus of these studies was on epithelial responses; they did not examine myeloid cell regulation and did not evaluate bacterial handling, which is an important determinant of intestinal injury. We first assessed the regulation of pathways in ex vivo isolated intestinal macrophages in a DSS time course; the isolated macrophages were further cocultured ex vivo with S. Typhimurium so as to examine the inducible responses in these cells. Over the 6-day time period examined, Rnf186 expression progressively increased, as did UPR transcripts, cytokine secretion, bacterial uptake, and bacterial clearance in colonic macrophages (Figure 10, A–D, and Supplemental Figure 18A), consistent with the progressive influx and increased exposure to PRR ligands. We confirmed similar regulation of Rnf186 expression and UPR transcripts in colonic tissues over this time period (Supplemental Figure 18, B and C). To assess how RNF186 regulates these intestinal macrophage outcomes during acute injury, we examined DSS-induced injury on day 4 as an intermediate time point in RNF186-deficient mice (through in vivo delivery of RNF186 siRNA; we will refer to these mice as RNF186-deficient mice for the ensuing studies). Intestinal macrophages from RNF186-deficient mice (Figure 10E) given DSS demonstrated reduced UPR transcripts (Figure 10F) and reduced cytokine secretion (Figure 10G and Supplemental Figure 19A) when compared with WT mice (in vivo delivery of scrambled siRNA), and these outcomes were more pronounced upon coculture with S. Typhimurium (Figure 10, F and G; and Supplemental Figure 19A). The cells further demonstrated reduced uptake and clearance of bacteria compared with intestinal macrophages from WT mice (Figure 10H); this would have implications for clearing the dramatic translocation of resident intestinal microbes occurring with intestinal injury. A reduction in Rnf186 expression (Supplemental Figure 19B) and in UPR transcripts (Supplemental Figure 19C) was similarly observed in colonic tissues of these RNF186-deficient mice. Given the parallel regulation of UPR transcripts in colonic tissues and colonic macrophages upon DSS-induced intestinal injury, we will examine colonic tissues in the studies that follow. Taken together, RNF186 regulates the UPR and efficacy of bacterial uptake and bacterial clearance in intestinal macrophages during acute intestinal injury.
Figure 10. RNF186 promotes bacterially induced UPR pathways and cytokines and bacterial clearance in mouse intestinal macrophages during intestinal injury.
(A–D) WT mice were given 2.5% DSS in drinking water (DW) for the indicated times (n = 4/time point). (E–H) Mice given scrambled or RNF186 siRNA (i.p.) were administered 2.5% DSS in DW for 4 days (n = 5). (A–H) Colonic macrophages were isolated, cocultured with S. Typhimurium, and assessed for (A, B, E, and F) fold change mRNA expression at 4 hours; (C and G) TNF secretion at 24 hours; (D and H) bacterial uptake and intracellular bacterial clearance. Mean + SEM. Scr, scrambled. *P < 0.05; **P < 0.01; ***P < 0.001; †P < 1 × 10−4; ††P < 1 × 10−5 determined by 2-tailed Student’s t test.
RNF186 reduces DSS-induced intestinal injury and promotes clearance of resident intestinal microbes in mice.
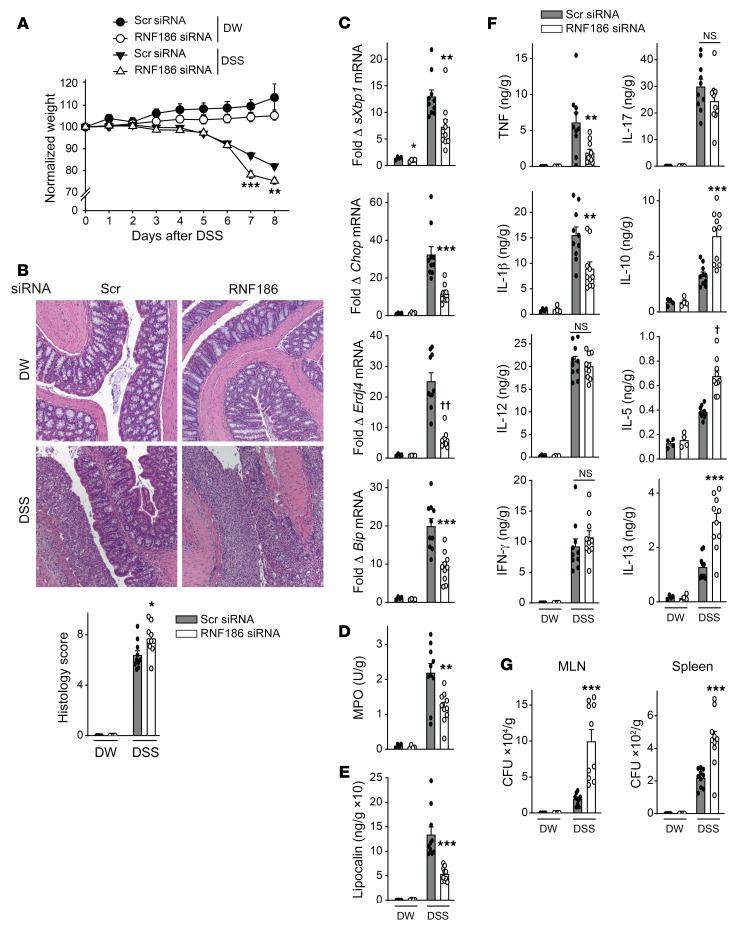
We next assessed how RNF186 regulates disease outcomes with acute intestinal injury. Upon DSS administration, RNF186-deficient mice (Supplemental Figure 20A) demonstrated increased weight loss and increased histologic injury (Figure 11, A and B) compared with WT mice. They further demonstrated reduced colonic UPR transcripts (Figure 11C), reduced colonic myeloperoxidase (MPO) activity (myeloid cell measure) (Figure 11D), and reduced fecal lipocalin (myeloid cell measure) (Figure 11E) compared with WT mice. Consistently, the RNF186-deficient mice demonstrated reduced colonic TNF and IL-1β protein expression (Figure 11F). In contrast, colonic Th1 (IL-12, IFN-γ) and Th17 (IL-17) cytokines were equivalently expressed in WT and RNF186-deficient mice (Figure 11F). Inflammatory cytokines can demonstrate a reciprocal pattern of regulation when compared with antiinflammatory cytokines and Th2 cytokines. The antiinflammatory cytokine IL-10 and the Th2 cytokines IL-5 and IL-13 were increased in the colon of RNF186-deficient compared to WT mice given DSS (Figure 11F). We confirmed the regulation of a subset of these cytokines in serum (Supplemental Figure 20B). RNF186-deficient mice given DSS demonstrated more severe disease despite decreased inflammatory mediators, and we had found that both peripheral (Figure 7, A and B, and Supplemental Figure 14E) and intestinal (Figure 10H) RNF186-deficient macrophages were impaired in bacterial uptake and intracellular bacterial clearance. We therefore assessed bacterial burden and found that RNF186-deficient mice demonstrated an increased bacterial burden in the mesenteric lymph nodes (MLNs) and spleen during DSS-induced injury compared with WT mice (Figure 11G). Therefore, during intestinal injury, RNF186 promoted the UPR and bacterial clearance mechanisms; optimal bacterial clearance is critical for limiting intestinal injury.
Figure 11. RNF186 promotes clearance of resident intestinal microbiota during DSS-induced injury.
Mice given scrambled or RNF186 siRNA (i.p.) were administered 2.5% DSS in drinking water (DW) (n = 4–10). (A) Body weights. (B) Representative H&E of colon sections and histology scores; original magnification, 100×. (C) Colon mRNA expression. (D) Colon myeloperoxidase. (E) Fecal lipocalin. (F) Colon cytokines. (G) Bacteria in MLN and spleen. (A–G) DSS for 6 days and then water for an additional 2 days; independent experiment for A–E, and G with DSS for 7 days. Mean + SEM. NS, not significant. *P < 0.05; **P < 0.01; ***P < 0.001; †P < 1 × 10–4; ††P < 1 × 10−5 determined by 2-tailed Student’s t test.
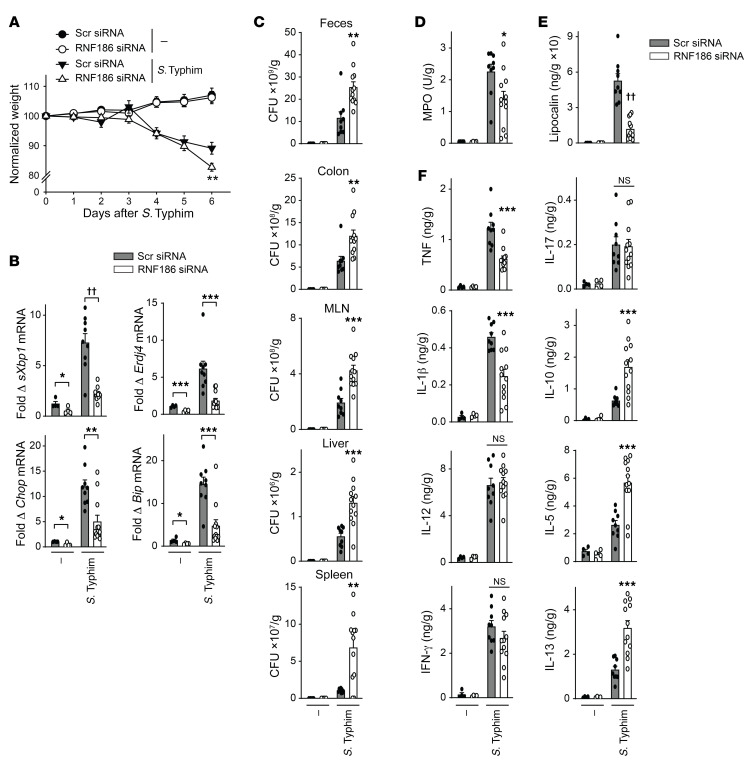
RNF186 is required for clearance of enteric pathogens in vivo.
To clearly establish the role for RNF186 in bacterial challenges in vivo, we assessed RNF186-deficient mice upon oral challenge with S. Typhimurium. RNF186-deficient mice (Supplemental Figure 21A) demonstrated greater weight loss (Figure 12A), reduced colonic UPR transcripts (Figure 12B), and a greater burden of S. Typhimurium in the feces, colon, and MLN, as well as systemically in the liver and spleen (Figure 12C) compared with WT mice. They further demonstrated reduced colonic MPO activity (Figure 12D), reduced fecal lipocalin (Figure 12E), and reduced colonic TNF and IL-1β (Figure 12F), consistent with reduced myeloid cell responses. In contrast, colonic Th1 (IL-12, IFN-γ) and Th17 (IL-17) cytokines did not differ, and the antiinflammatory cytokine IL-10 and Th2 cytokines IL-5 and IL-13 were increased in the colon of RNF186-deficient compared with WT mice with S. Typhimurium infection (Figure 12F), similar to the pattern observed with DSS-induced injury. A similar cytokine pattern was observed in serum (Supplemental Figure 21B). Therefore, RNF186 was required for regulating oral challenge with invasive enteric bacteria in mice in vivo.
Figure 12. RNF186 is required for optimal clearance of S. Typhimurium upon oral infection in mice.
Mice given scrambled or RNF186 siRNA (i.p.) were orally inoculated with S. Typhimurium (n = 4–12). Six days later mice were assessed. (A) Body weights. (B) Colon mRNA expression. (C) S. Typhimurium CFU in the indicated tissues. (D) Colon myeloperoxidase. (E) Fecal lipocalin. (F) Colon cytokines. Mean + SEM. NS, not significant. *P < 0.05; **P < 0.01; ***P < 0.001; ††P < 1 × 10–5 determined by 2-tailed Student’s t test.
ATF6 is required for clearance of resident luminal microbes and enteric pathogens in vivo.
As we had found that RNF186-dependent ATF6 ubiquitination promotes downstream UPR signaling, cytokines, and bacterial clearance, we wished to assess whether ATF6 regulates intestinal injury in a manner similar to RNF186. We first reduced ATF6 expression in mice through siRNA delivery in vivo and confirmed effective ATF6 deletion in colonic tissues (Supplemental Figure 22A) and colonic macrophages (Supplemental Figure 22B) (we will refer to these as ATF6-deficient mice in the studies that follow) compared with mice administered scrambled siRNA. ATF6-deficient intestinal macrophages demonstrated reduced UPR signaling, cytokine secretion, and bacterial uptake and clearance upon coculture ex vivo with S. Typhimurium (Supplemental Figure 22, C–E).
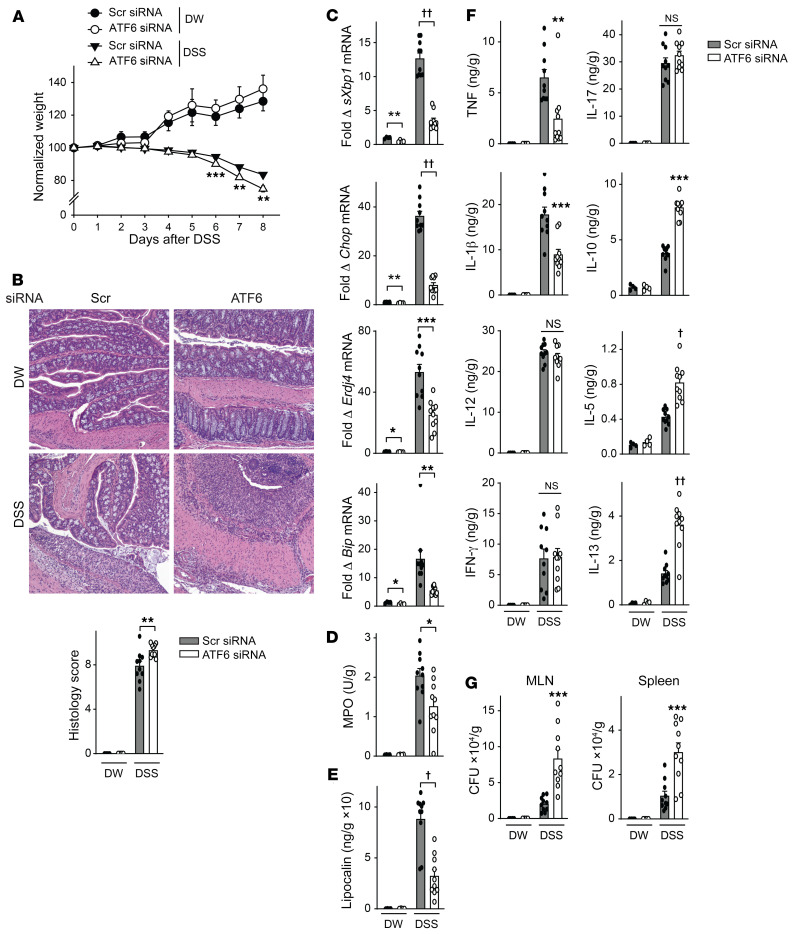
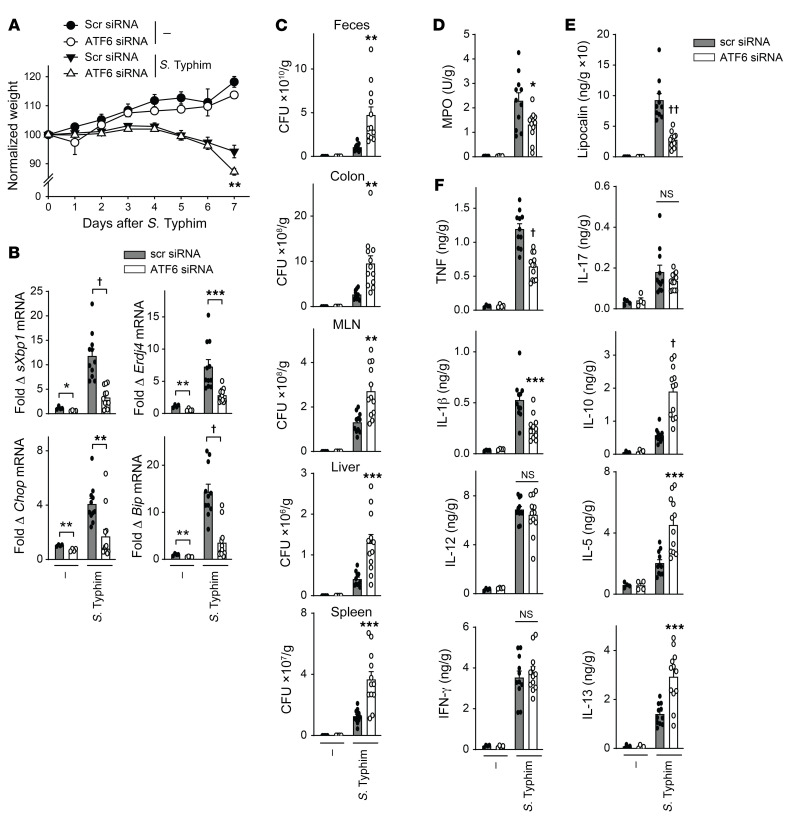
Importantly, similar to RNF186-deficient mice, ATF6-deficient mice (Supplemental Figure 23A) demonstrated more severe weight loss and intestinal injury with DSS-induced injury (Figure 13, A and B). This was in the context of a reduced colonic UPR, reduced myeloid cell responses, reduced colonic inflammatory cytokines, and increased antiinflammatory and Th2 cytokines and, in turn, an increased burden of bacteria in the MLN and spleen (Figure 13, C–G). A similar cytokine pattern was observed in serum (Supplemental Figure 23B). Moreover, ATF6-deficient mice demonstrated more severe disease, reduced UPR measures, reduced myeloid cell responses, and an impaired ability to clear the enteric pathogen S. Typhimurium (Figure 14 and Supplemental Figure 24). Therefore, ATF6 was required for regulating bacteria with acute intestinal injury and upon oral challenge with invasive enteric bacteria in mice in vivo.
Figure 13. ATF6 promotes clearance of resident intestinal microbiota during DSS-induced injury.
Mice given scrambled or ATF6 siRNA (i.p.) were administered 2.5% DSS in drinking water (DW) (n = 4–10). (A) Body weights. (B) Representative H&E of colon sections and histology scores; original magnification, 100×. (C) Colon mRNA expression. (D) Colon myeloperoxidase. (E) Fecal lipocalin. (F) Colon cytokines. (G) Bacteria in MLN and spleen. (A–G) DSS for 6 days and then water for an additional 2 days; independent experiment for A–E and G with DSS for 8 days. Mean + SEM. NS, not significant. *P < 0.05; **P < 0.01; ***P < 0.001; †P < 1 × 10–4; ††P < 1 × 10–5 determined by 2-tailed Student’s t test.
Figure 14. ATF6 is required for optimal clearance of S. Typhimurium upon oral infection in mice.
Mice given scrambled or ATF6 siRNA (i.p.) were orally inoculated with S. Typhimurium (n = 4–12). Seven days later mice were assessed. (A) Body weights. (B) Colon mRNA expression. (C) S. Typhimurium CFU in the indicated tissues. (D) Colon myeloperoxidase. (E) Fecal lipocalin. (F) Colon cytokines. Mean + SEM. NS, not significant. *P < 0.05; **P < 0.01; ***P < 0.001; †P < 1 × 10–4, ††P < 1 × 10–5 determined by 2-tailed Student’s t test.
Discussion
In this study in primary human MDMs, we found that with NOD2 stimulation, there was a transient increase in RNF186 localization to the ER where it associated with ER stress sensors (IRE1α, PERK, ATF6) and mediated ubiquitination and assembly of the UPR signaling complex along with induction of UPR signaling. RNF186-dependent UPR pathway induction, in turn, was required for PRR-induced MAPK and NF-κB signaling, cytokine secretion, and a range of antimicrobial pathways, including bacterial uptake, ROS production, and autophagy pathway induction; the MAPK and NF-κB signaling pathways are required for these implicated antimicrobial pathways (3, 4, 49, 50). Both RNF186 localization to the ER and RNF186 E3 ubiquitin ligase activity are required for these outcomes. Importantly, we established that RNF186 directly ubiquitinated the ER stress sensor ATF6 at position K152 and that this was critical for the PRR-induced, RNF186-dependent outcomes defined. Both the RNF186 A64T IBD risk rare variant and the RNF186 common IBD risk rs6426833 A variant demonstrated a loss of function in RNF186, with a reduction in PRR-induced UPR signaling and the UPR-dependent outcomes identified. Importantly, RNF186 deficiency in mice, as well as deficiency of the ER stress sensor ATF6 ubiquitinated by RNF186, resulted in lower induction of the UPR in intestinal myeloid cells and colonic tissues, as well as more severe disease with less effective clearance of intestinal microbes with acute intestinal injury and upon challenge with oral enteric pathogens. These findings are consistent with a loss of function in innate pathways leading to risk for infection and inflammation (3–9). Taken together, we defined a critical role for RNF186 in orchestrating the innate immune response by promoting PRR-induced, UPR-associated outcomes in macrophages; established that RNF186-dependent ATF6 ubiquitination was required for these outcomes; identified that the RNF186 rare and common UC risk genetic variants modulated these responses; and found that deficiency of RNF186 and ATF6 in vivo led to impaired clearance of intestinal bacteria (Supplemental Figure 25).
A dysregulated UPR has been increasingly implicated in a variety of diseases, including IBD. However, the major focus of the dysregulated ER stress response in IBD has been in the context of epithelial cells (20, 21). Further, a recent study found that unresolved ER stress in intestinal epithelial cells activates the ATF6 pathway, which promotes intestinal inflammation (51). The focus of the limited reports examining functions for RNF186 has also been predominantly in epithelial cells (11–16). Within epithelial cell lineages, a couple of reports have described a role for RNF186 in ER stress. In HeLa cells, RNF186 localizes to the ER and promotes ER stress–associated apoptosis signaling (13). In hepatocytes, RNF186 promotes ER stress and in turn impairs insulin sensitivity (15). However, UPR signaling in macrophages can lead to outcomes that are very distinct from those observed in epithelial cells, consistent with the different function of these cell types. We found that RNF186 promoted a transient, physiological increase in the UPR in human macrophages in response to microbial stimuli, which was required for key macrophage functions, including antimicrobial pathways. We identified that RNF186 needed to specifically localize to the ER to mediate these outcomes. Only a few studies have identified ubiquitin-mediated modifications of UPR sensor proteins (39, 41, 52, 53). The E3 ligase TRAF6 ubiquitinates IRE1α in mouse macrophages (41), and CHIP ubiquitinates IRE1α in HEK293 cells (53). To our knowledge, ubiquitin modifications of ATF6 in innate immunity have not been described. Outside of innate immunity, the E3 ubiquitin ligase HRD1 promotes ATF6 ubiquitination in pancreatic β cells, and this correlates with proteasome-mediated degradation (52). ATF6 ubiquitination in mouse fibroblasts also correlates with proteasome-mediated degradation (40). In contrast, upon PRR stimulation of human macrophages, we did not observe reduced ATF6 expression over the time of the various functional measures assessed. Importantly, to our knowledge, prior studies did not define the specific sites(s) in ATF6 undergoing ubiquitin modification and required for downstream functional outcomes. Clearly implicating the functional consequences of ATF6 ubiquitination requires preventing ubiquitination at the implicated site within ATF6; reducing ubiquitination in general leads to effects on many molecules and pathways that may be contributing to outcomes independent of ATF6. We found that ATF6 was ubiquitinated at K152 by RNF186, and this was required for PRR-induced outcomes in human macrophages. Whether K152 serves as a site for ubiquitination by additional E3 ubiquitin ligases and is required for ATF6-dependent outcomes in additional cell types and processes will need to be determined. Therefore, our identification of ATF6 K152 as the site for RNF186-mediated ubiquitination in human macrophages, with this site being highly conserved across species, highlights potential important implications for ATF6 ubiquitination in various cell subsets and in a broad range of ATF6-dependent functions.
We found that PRR-induced, RNF186-dependent UPR signaling and downstream outcomes in human macrophages required intact RNF186 E3 ubiquitin ligase activity in the RING domain. The rare and common IBD-associated genetic variants in RNF186 led to a loss of function through 2 distinct mechanisms. The RNF186 A64T rare variant is in the RING domain and leads to a loss of E3 ligase activity in RNF186 and in turn reduced PRR-induced ubiquitination of ATF6 along with reduced downstream outcomes. On the other hand, primary MDMs from rs6426833 A carriers in the RNF186 region showed reduced RNF186 expression and in turn lower levels of PRR-induced UPR signaling and downstream outcomes; in both cases, complementing UPR signaling could restore downstream outcomes.
Dysregulation of the UPR is observed in multiple immune-mediated diseases, and defining disease-associated genes regulating the UPR may provide insight into mechanisms contributing to these diseases. However, few genetic disease variants directly regulating UPR pathways have been described to date, with XBP1 variants (24) and LACC1 variants (29) being notable exceptions. We identified that RNF186 is an important regulator of the physiological and transient UPR observed with PRR stimulation in human macrophages. Whereas an increased UPR is observed in many immune-mediated diseases, a failure to properly induce pathways required for the UPR can also increase risk for immune-mediated diseases, as is observed with IRE1β and XBP1 deficiencies (22, 24). We observed similar outcomes with ATF6 deficiency; ATF6 deficiency impaired clearance of intestinal bacteria and increased intestinal injury. Future studies will be needed to define the roles of RNF186 and ATF6 in distinct cell subsets in vivo in disease through conditional expression regulation in animal models. Taken together, we identified that loss-of-function, immune-mediated disease risk variants in RNF186 resulted in a reduced PRR-induced UPR in human macrophages, which then resulted in reduction of multiple key downstream outcomes, including microbial clearance. Our study indicates that optimally balancing the UPR in macrophages might be an effective therapeutic strategy in infection and immune-mediated diseases.
Methods
Human myeloid cell isolation and cell culture.
Genotyping was conducted by TaqMan (Life Technologies, Thermo Fisher Scientific). Monocytes were purified from human PBMCs by adhesion and cultured with 10 ng/mL M-CSF (Shenandoah Biotechnology) in 10% FBS containing RPMI media for 7 days to generate MDMs. Intestinal lamina propria cells were isolated from colonic resection specimens from uninvolved intestine in patients without IBD undergoing surgery for diverticular disease or colon cancer as in Hedl et al. (5).
MDM stimulation.
MDMs were treated with MDP (Bachem). In some cases, 10 μM CPA (MP Biomedicals) was also added. Supernatants were assayed for TNF (clones MAb1 and MAb11), IL-8 (clones G265-5 and G265-8), IL-10 (clones JES3-9D7 and JES3-12G8) (all from BD Biosciences), IL-1β (clones CRM56 and CRM57), and IL-6 (clone 8.6) (both eBioscience, Thermo Fisher Scientific) by ELISA.
mRNA expression.
RNA was isolated using TRIzol (Thermo Fisher Scientific). Quantitative PCR was performed using All-In-One qPCR Mix (Genecopoeia) on the ABI Prism 7000 (Applied Biosystems, Thermo Fisher Scientific). Samples were normalized to GAPDH. Primer sequences are in Supplemental Table 1.
Transfection of siRNAs and DNA vectors.
First, 100 nM scrambled or ON-TARGETplus SMARTpool siRNA against RNF186 (Dharmacon) (4 pooled siRNAs for each gene) was transfected into MDMs using Amaxa nucleofector technology (Lonza). Forty-eight hours later, cells were used in experimental studies. WT RNF186 was amplified from HEK293 cells (ATCC) and subcloned into pEGFP-C1 to generate pEGFP-WT-RNF186. Then, pEGFP-RNF186-ΔDLE (Asp37Ala, Leu38Ala, Glu39Ala) localization mutant, pcDNA.3-WT-RNF186-myc-tagged (C-terminus), pcDNA3-RNF186-ΔZnF mutant (Cys58Ala, His60Trp, Cys63Ala, Cys66Ala), pcDNA3-RNF186-ΔDLE (Asp37Ala, Leu38Ala, Glu39Ala), and pcDNA3-RNF186-Ala64Thr myc-tagged mutants were generated through site-directed mutagenesis (Herculase II Fusion enzyme, Agilent Technologies) of the RNF186 parent vector and confirmed by sequencing. p3xFLAG-hATF6 (FLAG, N-terminus) was deposited to Addgene (plasmid 11975) by Ron Prywes (54). p3xFLAG-hATF6 lysine mutants (K149A, K152A, K350A, and K356A) were generated through site-directed mutagenesis (GenScript) of the ATF6 parent vector and confirmed by sequencing, and 1.5 μg myc-RNF186 (WT or mutants) and 1 μg of NOD2 were transfected into HeLa cells by Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific) for 48 hours. RNF186 constructs (1.5 μg) and ATF6 constructs (1 μg) were transfected into MDMs by Amaxa nucleofector technology (Lonza) for 48 hours.
Immunoprecipitation and immunoblotting assays.
Immunoblotting was performed with antibodies against RNF186 (Aviva Systems Biology and Novus Biologicals) or GAPDH (6C5, MilliporeSigma). Immunoprecipitation was carried out with anti-RIP2 (BD Biosciences), anti-myc (Cell Signaling Technology), and anti-ATF6 (ProteinTech) conjugated to protein A or protein G–agarose. Associated proteins were examined with antibodies to RIP2 (25/RIG-G, BD Biosciences), ATF6 (ProteinTech), IRE1α (14C10), PERK (D11A8), ubiquitin, HA (C29F4), FLAG (9A3), Dectin-1 (E1X3Z), or Syk (4D10) (Cell Signaling Technology). In some cases, ER fractionation was carried out using a 1.3 M sucrose gradient and ultracentrifugation as per Williamson et al. (55). Calnexin (AF18, Santa Cruz Biotechnology) and GAPDH (6C5, MilliporeSigma) served as fractionation controls.
Flow cytometry staining.
Permeabilized MDMs were assessed for protein expression by flow cytometry using antibodies to RNF186 (Aviva Systems Biology), ATG5 (C-1), and MAP LC3β (G-9) (both from Santa Cruz Biotechnology), and phospho antibodies for p44/42 (Erk1/2) (197G2), p38 (28B10), SAPK/JNK (G9), IκBα (14D4) (all from Cell Signaling Technology), IRE1α (EPR5253) (Abcam), and PERK (Thermo Fisher Scientific).
Recombinant protein purification.
HEK293 cells were transfected with 5 μg of myc-RNF186 constructs (WT or mutants) or FLAG-ATF6 (WT or mutants) using Lipofectamine 2000. After 72 hours, cells were lysed and myc-RNF186 or FLAG-ATF6 were purified with anti-myc or anti-FLAG antibodies (Cell Signaling Technology) conjugated with protein G–agarose beads. The proteins were eluted with myc or FLAG peptides (both 200 ng/mL, APExBIO).
In vitro ubiquitination assays.
Purified recombinant myc-RNF186 (750 ng) was incubated with FLAG-ATF6 (500 ng) in a 50 μL reaction mixture containing HA-ubiquitin (5 μg), UBE1 (100 nM), UBE2N (Ubc13/Uev1a) (500 nM), 2 mM MgCl2, 2 mM ATP, and 1× reaction buffer (all from Boston Biochem). Reaction mixtures were incubated at 30°C for 1 hour and terminated by the addition of 1× loading dye followed by heating to 95°C for 5 minutes. The samples were probed by Western blot with antibodies to HA tag.
Intracellular ROS measurement.
Intracellular ROS was measured by incubating MDMs with 10 μM 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA, Life Technologies, Thermo Fisher Scientific) for 30 minutes and assessed by flow cytometry.
Intracellular bacterial clearance.
Human MDMs were infected in triplicate for 1 hour with adherent-invasive E. coli strain LF82 (a gift from Emiko Mizoguchi, Kurume University, Kurume, Fukuoka, Japan) or Salmonella enterica serovar Typhimurium (S. Typhimurium) at 10:1 MOI, washed with PBS, and incubated in HBSS medium containing 50 μg/mL gentamicin for an additional hour. Mouse cells were cocultured with bacteria for 20 minutes, washed with PBS, and incubated in HBSS medium containing 50 μg/mL gentamicin for a total time of 4 hours after initial bacteria coculture. Cells were washed, lysed with 1% Triton X-100 (MilliporeSigma), and plated on Luria-broth or MacConkey agar. CFUs were quantified.
Bacterial entry/phagocytosis.
FITC-labeled E. coli bioparticles (1.5 × 106) (Invitrogen, Thermo Fisher Scientific) or 5:1 MOI S. Typhimurium–GFP (provided by Jorge E. Galan, Yale University, New Haven, Connecticut, USA) were cocultured with MDMs for 20 minutes, cell surface fluorescence was quenched with 0.25 mg/mL trypan blue, and cells were then analyzed by flow cytometry.
Microscopy.
HeLa cells (ATCC) were transfected with 250 ng of GFP-tagged WT and mutants (ΔDLE or T64) of RNF186 for 48 hours followed by NOD2 stimulation. Cells were fixed in 4% paraformaldehyde and then permeabilized with 0.1% Triton X-100 and incubated with anti-calnexin (AF18) (Santa Cruz Biotechnology) or DAPI (Acros Organics) and detected by secondary antibodies labeled with Cy5 (Jackson ImmunoResearch). Fluorescence microscopy was conducted with the Zeiss Axio Observer microscope and pixel density analysis was conducted with ZEN lite (Carl Zeiss Microscopy). Quantification of colocalization was performed using ImageJ (NIH).
Mouse lamina propria cell isolation.
C57BL/6N mice were maintained in a specific pathogen–free facility. First, 15 μg/mouse ON-TARGETplus RNF186 (sequence per J-050361-12), ATF6 (sequence as per J-044894-08) siRNA, or control (sequence per D-001810-02) with UU overhang/in vivo design (Dharmacon) was i.p. administered in AteloGene Systemic Use (Cosmo Bio USA), and then studies were conducted 48 hours later. For in vivo studies extending beyond 4 days, a second dose of in vivo siRNA was i.p. administered on day 4 of the disease model. Colonic lamina propria cells were isolated as previously described (56) and CD11b+ cells were then purified (Miltenyi Biotec). Cytokines were detected by ELISA: TNF-α (clone 6B8, MP6-XT22), IL-1β (B122), IFN-γ (AN-18, XMG1.2), IL-5 (TRFK5, TRFK4), IL-12 (C15.6, C17.8), IL-10 (JES5-16E3, JES5-2A5) (BioLegend); IL-13 (eBio13A, eBio1316H), IL-17 (eBio17CK15A5, eBio17B7) (Thermo Fisher Scientific).
DSS colitis.
DSS (MW 36,000–50,000; MPBio) 2.5% (w/v) was added to the drinking water of mice with ad libitum access. Tissues were plated on Luria-broth agar for bacterial colony counts. H&E-stained colon sections were examined by a pathologist blinded to treatment and scored for epithelial injury (scale 0–4), inflammation (scale 0–4), and disease extent (scale 0–5). Mouse colon tissue was homogenized and cytokines (ELISA) were normalized to tissue weight.
S. Typhimurium in vivo infection.
Mice were orally inoculated with streptomycin (20 mg/mouse) (VWR International) and 24 hours later orally inoculated with 1 × 106 CFU of S. Typhimurium (strain SL1344). Mice were euthanized and organs collected in PBS.
Myeloperoxidase assay.
Colonic tissue was homogenized in hexadecyltrimethylammonium bromide (MilliporeSigma) buffer. MPO was assayed using o-dianisidine dihydrochloride (MilliporeSigma) and H2O2 (change in optical density at 450 nm).
Lipocalin.
Frozen fecal samples were reconstituted in PBS/0.1% Tween 20. Supernatants were assessed for lipocalin by ELISA (BioLegend).
Statistics.
Significance was assessed using a 2-tailed Student’s t test. A Bonferroni-Holm correction was applied for multiple corrections where appropriate. A P value less than 0.05 was considered significant.
Study approval.
Human cell studies were performed as approved by the IRB at Yale University. Mouse studies were performed in accordance with Yale University IACUC and NIH guidelines.
Author contributions
KR, MH, and CA designed research studies, conducted experiments, and analyzed data; SS conducted experiments and analyzed data; XZ analyzed data; and KR and CA wrote the manuscript. CA supervised the study and obtained funding.
Supplementary Material
Acknowledgments
This work was supported by the NIH (R01DK099097; P30DK034989) and a gift from the S. Sydney DeYoung Foundation.
Version 1. 09/01/2021
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(17):e145472.https://doi.org/10.1172/JCI145472.
Contributor Information
Kishu Ranjan, Email: kishu.ranjan@yale.edu.
Matija Hedl, Email: matija.hedl@yale.edu.
Saloni Sinha, Email: saloni.sinha@yale.edu.
Xuchen Zhang, Email: xuchen.zhang@yale.edu.
Clara Abraham, Email: clara.abraham@yale.edu.
References
- 1.Abraham C, Medzhitov R. Interactions between the host innate immune system and microbes in inflammatory bowel disease. Gastroenterology. 2011;140(6):1729–1737. doi: 10.1053/j.gastro.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med. 2009;361(21):2066–2078. doi: 10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lahiri A, et al. Human LACC1 increases innate receptor-induced responses and a LACC1 disease-risk variant modulates these outcomes. Nat Commun. 2017;8:15614. doi: 10.1038/ncomms15614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan J, et al. An inflammatory bowel disease-risk variant in INAVA decreases pattern recognition receptor-induced outcomes. J Clin Invest. 2017;127(6):2192–2205. doi: 10.1172/JCI86282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hedl M, et al. Pattern recognition receptor signaling in human dendritic cells is enhanced by ICOS ligand and modulated by the Crohn’s disease ICOSLG risk allele. Immunity. 2014;40(5):734–746. doi: 10.1016/j.immuni.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Homer CR, et al. ATG16L1 and NOD2 interact in an autophagy-dependent antibacterial pathway implicated in Crohn’s disease pathogenesis. Gastroenterology. 2010;139(5):1630–1641. doi: 10.1053/j.gastro.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Travassos LH, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11(1):55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 8.Dhillon SS, et al. Variants in nicotinamide adenine dinucleotide phosphate oxidase complex components determine susceptibility to very early onset inflammatory bowel disease. Gastroenterology. 2014;147(3):680–689. doi: 10.1053/j.gastro.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 9.Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491(7422):119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beaudoin M, et al. Deep resequencing of GWAS loci identifies rare variants in CARD9, IL23R and RNF186 that are associated with ulcerative colitis. PLoS Genet. 2013;9(9):e1003723. doi: 10.1371/journal.pgen.1003723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ji Y, et al. The role and mechanism of action of RNF186 in colorectal cancer through negative regulation of NF-κB. Cell Signal. 2020;75:109764. doi: 10.1016/j.cellsig.2020.109764. [DOI] [PubMed] [Google Scholar]
- 12.Zhang H, et al. RNF186 regulates EFNB1 (ephrin B1)-EPHB2-induced autophagy in the colonic epithelial cells for the maintenance of intestinal homeostasis. Autophagy. doi: 10.1080/15548627.2020.1851496. [published online December 17, 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang P, et al. A novel RING finger E3 ligase RNF186 regulate ER stress-mediated apoptosis through interaction with BNip1. Cell Signal. 2013;25(11):2320–2333. doi: 10.1016/j.cellsig.2013.07.016. [DOI] [PubMed] [Google Scholar]
- 14.Fujimoto K, et al. Regulation of intestinal homeostasis by the ulcerative colitis-associated gene RNF186. Mucosal Immunol. 2017;10(2):446–459. doi: 10.1038/mi.2016.58. [DOI] [PubMed] [Google Scholar]
- 15.Tong X, et al. RNF186 impairs insulin sensitivity by inducing ER stress in mouse primary hepatocytes. Cell Signal. 2018;52:155–162. doi: 10.1016/j.cellsig.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 16.Lear TB, et al. The RING-type E3 ligase RNF186 ubiquitinates Sestrin-2 and thereby controls nutrient sensing. J Biol Chem. 2019;294(45):16527–16534. doi: 10.1074/jbc.AC119.010671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8(7):519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 18.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13(2):89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 19.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334(6059):1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 20.Bettigole SE, Glimcher LH. Endoplasmic reticulum stress in immunity. Annu Rev Immunol. 2015;33:107–138. doi: 10.1146/annurev-immunol-032414-112116. [DOI] [PubMed] [Google Scholar]
- 21.Kaser A, et al. Endoplasmic reticulum stress: implications for inflammatory bowel disease pathogenesis. Curr Opin Gastroenterol. 2010;26(4):318–326. doi: 10.1097/MOG.0b013e32833a9ff1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bertolotti A, et al. Increased sensitivity to dextran sodium sulfate colitis in IRE1beta-deficient mice. J Clin Invest. 2001;107(5):585–593. doi: 10.1172/JCI11476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang HS, et al. The endoplasmic reticulum stress sensor IRE1α in intestinal epithelial cells is essential for protecting against colitis. J Biol Chem. 2015;290(24):15327–15336. doi: 10.1074/jbc.M114.633560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaser A, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134(5):743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bronner DN, et al. Endoplasmic reticulum stress activates the inflammasome via NLRP3- and caspase-2-driven mitochondrial damage. Immunity. 2015;43(3):451–462. doi: 10.1016/j.immuni.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinon F, et al. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat Immunol. 2010;11(5):411–418. doi: 10.1038/ni.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rao J, et al. ATF6 mediates a pro-inflammatory synergy between ER stress and TLR activation in the pathogenesis of liver ischemia-reperfusion injury. Am J Transplant. 2014;14(7):1552–1561. doi: 10.1111/ajt.12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seok J, et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 2013;110(9):3507–3512. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang C, et al. LACC1 required for NOD2-induced, ER stress-mediated innate immune outcomes in human macrophages and LACC1 risk variants modulate these outcomes. Cell Rep. 2019;29(13):4525–4539. doi: 10.1016/j.celrep.2019.11.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng BS, et al. Mast cells play a crucial role in Staphylococcus aureus peptidoglycan-induced diarrhea. Am J Pathol. 2007;171(2):537–547. doi: 10.2353/ajpath.2007.061274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng S, Abraham C. NF-κB1 inhibits NOD2-induced cytokine secretion through ATF3-dependent mechanisms. Mol Cell Biol. 2013;33(24):4857–4871. doi: 10.1128/MCB.00797-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grootjans J, et al. The unfolded protein response in immunity and inflammation. Nat Rev Immunol. 2016;16(8):469–484. doi: 10.1038/nri.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hedl M, Abraham C. A TNFSF15 disease-risk polymorphism increases pattern-recognition receptor-induced signaling through caspase-8-induced IL-1. Proc Natl Acad Sci U S A. 2014;111(37):13451–13456. doi: 10.1073/pnas.1404178111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hedl M, Abraham C. Nod2-induced autocrine interleukin-1 alters signaling by ERK and p38 to differentially regulate secretion of inflammatory cytokines. Gastroenterology. 2012;143(6):1530–1543. doi: 10.1053/j.gastro.2012.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fumagalli F, et al. Translocon component Sec62 acts in endoplasmic reticulum turnover during stress recovery. Nat Cell Biol. 2016;18(11):1173–1184. doi: 10.1038/ncb3423. [DOI] [PubMed] [Google Scholar]
- 36.Ogura Y, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411(6837):603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 37.Bist P, et al. E3 Ubiquitin ligase ZNRF4 negatively regulates NOD2 signalling and induces tolerance to MDP. Nat Commun. 2017;8:15865. doi: 10.1038/ncomms15865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hetz C, Papa FR. The unfolded protein response and cell fate control. Mol Cell. 2018;69(2):169–181. doi: 10.1016/j.molcel.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 39.Gao B, et al. Synoviolin promotes IRE1 ubiquitination and degradation in synovial fibroblasts from mice with collagen-induced arthritis. EMBO Rep. 2008;9(5):480–485. doi: 10.1038/embor.2008.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hong M, et al. Endoplasmic reticulum stress triggers an acute proteasome-dependent degradation of ATF6. J Cell Biochem. 2004;92(4):723–732. doi: 10.1002/jcb.20118. [DOI] [PubMed] [Google Scholar]
- 41.Qiu Q, et al. Toll-like receptor-mediated IRE1α activation as a therapeutic target for inflammatory arthritis. EMBO J. 2013;32(18):2477–2490. doi: 10.1038/emboj.2013.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holland SM. Chronic granulomatous disease. Clin Rev Allergy Immunol. 2010;38(1):3–10. doi: 10.1007/s12016-009-8136-z. [DOI] [PubMed] [Google Scholar]
- 43.Lahiri A, Abraham C. Activation of pattern recognition receptors up-regulates metallothioneins, thereby increasing intracellular accumulation of zinc, autophagy, and bacterial clearance by macrophages. Gastroenterology. 2014;147(4):835–846. doi: 10.1053/j.gastro.2014.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Foster SL, et al. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447(7147):972–978. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- 45.Darfeuille-Michaud A, et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology. 2004;127(2):412–421. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- 46.Shiloh MU, et al. Phenotype of mice and macrophages deficient in both phagocyte oxidase and inducible nitric oxide synthase. Immunity. 1999;10(1):29–38. doi: 10.1016/S1074-7613(00)80004-7. [DOI] [PubMed] [Google Scholar]
- 47.Smythies LE, et al. Human intestinal macrophages display profound inflammatory anergy despite avid phagocytic and bacteriocidal activity. J Clin Invest. 2005;115(1):66–75. doi: 10.1172/JCI19229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Franchi L, et al. NLRC4-driven production of IL-1β discriminates between pathogenic and commensal bacteria and promotes host intestinal defense. Nat Immunol. 2012;13(5):449–456. doi: 10.1038/ni.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kang JW, et al. Myeloid cell expression of LACC1 is required for bacterial clearance and control of intestinal inflammation. Gastroenterology. 2020;159(3):1051–1067. doi: 10.1053/j.gastro.2020.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun R, et al. TNFSF15 promotes antimicrobial pathways in human macrophages and these are modulated by TNFSF15 disease-risk variants. Cell Mol Gastroenterol Hepatol. 2021;11(1):249–272. doi: 10.1016/j.jcmgh.2020.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stengel ST, et al. Activating transcription factor 6 mediates inflammatory signals in intestinal epithelial cells upon endoplasmic reticulum stress. Gastroenterology. 2020;159(4):1357–1374. doi: 10.1053/j.gastro.2020.06.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fonseca SG, et al. Wolfram syndrome 1 gene negatively regulates ER stress signaling in rodent and human cells. J Clin Invest. 2010;120(3):744–755. doi: 10.1172/JCI39678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu X, et al. Ubiquitination of inositol-requiring enzyme 1 (IRE1) by the E3 ligase CHIP mediates the IRE1/TRAF2/JNK pathway. J Biol Chem. 2014;289(44):30567–30577. doi: 10.1074/jbc.M114.562868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen X, et al. The luminal domain of ATF6 senses endoplasmic reticulum (ER) stress and causes translocation of ATF6 from the ER to the Golgi. J Biol Chem. 2002;277(15):13045–13052. doi: 10.1074/jbc.M110636200. [DOI] [PubMed] [Google Scholar]
- 55.Williamson CD, et al. Isolation of endoplasmic reticulum, mitochondria, and mitochondria-associated membrane and detergent resistant membrane fractions from transfected cells and from human cytomegalovirus-infected primary fibroblasts. Curr Protoc Cell Biol. 2015;68(68):3.27.1–3.27.33. doi: 10.1002/0471143030.cb0327s68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu X, et al. NOD2 regulates CXCR3-dependent CD8+ T cell accumulation in intestinal tissues with acute injury. J Immunol. 2014;192(7):3409–3418. doi: 10.4049/jimmunol.1302436. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.