Abstract
We studied a child with severe viral, bacterial, fungal, and parasitic diseases, who was homozygous for a loss-of-function mutation of REL, encoding c-Rel, which is selectively expressed in lymphoid and myeloid cells. The patient had low frequencies of NK, effector memory cells reexpressing CD45RA (Temra) CD8+ T cells, memory CD4+ T cells, including Th1 and Th1*, Tregs, and memory B cells, whereas the counts and proportions of other leukocyte subsets were normal. Functional deficits of myeloid cells included the abolition of IL-12 and IL-23 production by conventional DC1s (cDC1s) and monocytes, but not cDC2s. c-Rel was also required for induction of CD86 expression on, and thus antigen-presenting cell function of, cDCs. Functional deficits of lymphoid cells included reduced IL-2 production by naive T cells, correlating with low proliferation and survival rates and poor production of Th1, Th2, and Th17 cytokines by memory CD4+ T cells. In naive CD4+ T cells, c-Rel is dispensable for early IL2 induction but contributes to later phases of IL2 expression. The patient’s naive B cells displayed impaired MYC and BCL2L1 induction, compromising B cell survival and proliferation and preventing their differentiation into Ig-secreting plasmablasts. Inherited c-Rel deficiency disrupts the development and function of multiple myeloid and lymphoid cells, compromising innate and adaptive immunity to multiple infectious agents.
Keywords: Genetics, Immunology
Keywords: Genetic diseases
Introduction
In humans, severe combined immunodeficiency (SCID) results from inborn errors of immunity causing a cell-intrinsic lack of autologous αβ and γδ T cells (1). Combined immunodeficiency (CID) results from inborn errors of αβ T cells, which are present, but in abnormally small numbers, or functionally compromised, at least in terms of their ability to proliferate in response to antigens in vitro. Patients also display a B cell defect in vivo that is either intrinsic or secondary to the T cell deficit (2). The T cell defect in patients with CID may be intrinsic or secondary to a deficiency of antigen-presenting cell (APC) function, such as inherited HLA class II deficiency (3, 4). Antigen-dependent T cell proliferation in vitro is normal for patients with other inborn errors of T cells not classified as CID (5). In the 2019 International Union of Immunological Societies (IUIS) classification of inborn errors of immunity, there were 57 CIDs with (syndromic CID) and 35 CIDs without (isolated CID) extrahematopoietic manifestations (5, 6). Patients with isolated or syndromic CID experience a broad range of infectious, autoimmune, autoinflammatory, allergic, and/or malignant phenotypes. Clinical manifestations vary among cases and are dependent on various host and environmental factors. These manifestations largely reflect the T and B cell phenotypes, which are dependent on the patient’s genome, the gene mutated, and the mutant genotype itself. The severe infections observed in CID include a broad range of viral, bacterial, fungal, and parasitic diseases, in addition to pyogenic bacterial infections typically caused by the B cell defect (7). Age at onset and the range and severity of infections also depend on T and B cell phenotypes and the extent of abnormalities affecting other cell types, particularly myeloid cells. It is difficult to obtain a complete understanding of the pathogenesis of most of these infections from studies performed in mouse models alone (8).
The molecular and cellular bases of some of the infections associated with T cell defects observed in patients with SCID or CID have been clarified by the identification of inborn errors of immunity underlying specific infections in otherwise healthy individuals (9, 10). For example, severe disease caused by weakly virulent mycobacteria, including Bacillus Calmette-Guérin (BCG) vaccines, in otherwise healthy patients is known as Mendelian susceptibility to mycobacterial disease (MSMD) (11, 12). Patients with MSMD carry inborn errors of IL-12–, IL-23–, or ISG15-dependent IFN-γ–mediated immunity (12). Patients with SCID are therefore probably vulnerable to BCG because of a lack of T cell–dependent IFN-γ production. Another example is provided by chronic mucocutaneous candidiasis (CMC), a fungal infection seen in patients with SCID and various CIDs. It is also seen in otherwise healthy patients with selective defects of IL-17F–, IL-17RA–, and IL-17RC–dependent immunity, implying that impaired IL-17A and IL-17F production by αβ T cells is probably at least partly responsible for this disease in patients with SCID or CID (13–18). The parasitic infection cryptosporidiosis was recently described in several patients with inherited IL-21R deficiency, potentially implicating the lymphokine IL-21 in other patients with CID, such as those with MHC class II or X-linked (XR) CD40 ligand (CD40L) deficiency, who are also susceptible to this parasite (5, 19). The molecular and cellular basis of the other infections seen in patients with CID, such as CMV disease and recurrent shingles, remains largely elusive, apart from the observation that multiple effector functions of T cells are defective (20). Genetic studies of CID patients infected with multiple pathogens or of patients with isolated infections therefore constitute 2 powerful and complementary approaches to determining the molecular basis of T cell immunity to infection (8). In this context, we studied a child with BCG disease, CMC, cryptosporidiosis, CMV disease, and shingles, whose overt T and B cell deficiencies suggested that she had a rare form of CID.
Results
A private homozygous essential splice site mutation of REL.
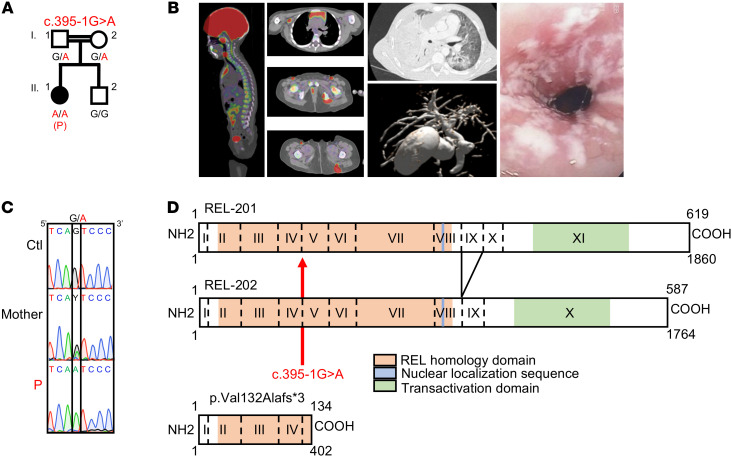
We performed whole-exome sequencing on a patient (P; II.1) born in 2011 to first-cousin parents originating from and living in Morocco (Figure 1A). P had various infections from early childhood onwards: CMC, including recurrent esophagitis; chronic CMV viremia with recurrent fever; recurrent oral herpes simplex virus 1 (HSV-1) lesions; chronic replication of adenovirus and enterovirus in the gastrointestinal tract; 1 episode of thoracic shingles; disseminated BCG disease following vaccination (BCG-osis), with bone, lung, and lymph node involvement; and chronic cholangitis due to Cryptosporidium parvum (Figure 1B). P had no overt extrahematopoietic clinical manifestations, such as neurological anomalies, growth retardation, or anhidrotic ectodermal dysplasia (21). PBMCs from P did not proliferate, or only weakly proliferated, in vitro in response to any of the physiological stimuli tested: tuberculin (PPD), candidin, tetanus toxoid, and CMV antigens (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI150143DS1). The patient’s infectious phenotypes were cured by hematopoietic stem cell transplantation (HSCT) performed in 2017. The rate of homozygosity in her exome was high (3.26%), consistent with parental consanguinity (22). On the basis of this finding, we selected homozygous nonsynonymous and essential splice site variants with a minor allele frequency (MAF) of less than 0.01 and a combined annotation-dependent depletion (CADD) score above the mutation significance cutoff (MSC) (Supplemental Figure 1A and refs. 23, 24). We also excluded variants that were common (MAF >0.01) in our in-house database of more than 9000 exomes, but absent from public databases (1kG, ExAC), corresponding to the “black list” (25). We identified variants of 22 genes, including a candidate nucleotide substitution of REL (c.395-1G>A), which encodes c-Rel, 1 of the 5 canonical and noncanonical NF-κB proteins (26). Rel–/– mice have no overt spontaneous infectious phenotype, but rather display a profound impairment of T and B cell proliferation in vitro and impaired Ab responses in vivo (27). None of the other 21 variants was a plausible candidate, given the expression and function of the corresponding genes (Supplemental Table 2). Sanger sequencing confirmed the homozygous nature of P’s REL variant (c.395-1G>A), located in the essential acceptor splice site of exon 5 common to the 2 known REL isoforms, which differ solely by the alternative splicing of exon 9 (Figure 1, C and D). The skipping of exon 5 is predicted to generate a frameshift and a premature stop codon at position 134 in the mRNA (Figure 1D). The parents were heterozygous (Figure 1C). The variant was not found in the gnomAD, BRAVO, or Greater Middle East (GME) databases, or in our in-house database of exomes from more than 9000 patients with various infectious diseases. The CADD score of 26.7 for this allele (Supplemental Figure 1B) is well above the MSC of 2.31 for REL (24). Moreover, no predicted loss-of-function (pLOF) REL alleles (nonsense, frameshift deletions/insertions, or essential splice variants) in the homozygous state were reported in gnomAD. The consensus negative score (CoNeS) (28) for this gene is –0.9725, indicating that REL is under strong negative selection, consistent with an autosomal-recessive (AR) inborn error of immunity (Supplemental Figure 1C). Together, these findings suggested that P had AR c-Rel deficiency.
Figure 1. A private homozygous essential splice site mutation of REL.
(A) Familial segregation. (B) From left to right: Disseminated BCG-osis with bone and lung lesions, cholangitis due to C. parvum, esophageal candidiasis. (C) REL genomic DNA electropherograms for a control (Ctl), P, and her mother. (D) Schematic representation of REL isoforms. The mutation is in the acceptor splice site of exon 5. The impact of the mutation is indicated.
An aberrant transcript underlies a loss of c-Rel production and function.
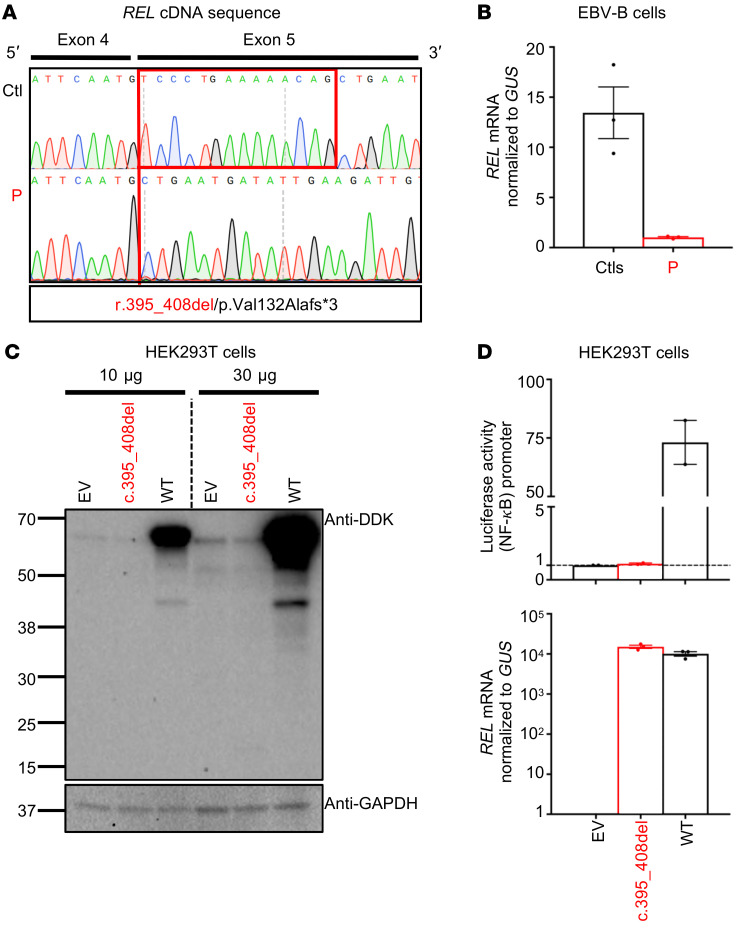
We studied the functional consequences of the variant for REL RNA splicing by performing reverse transcription PCR (RT-PCR) on mRNA extracted from the patient’s EBV-transformed B cells (EBV-B cells). The full-length REL cDNA was of similar MW in P, her heterozygous mother, and in healthy controls (Supplemental Figure 2A). As predicted in silico, the cloning and Sanger sequencing of these PCR products showed that the c.395-1G>A REL variant was associated with a deletion of 14 nucleotides (r.395_408del) in exon 5 and the presence of a premature termination codon in the region encoding the REL homology domain of the protein (p.Val132Alafs*3; Figure 1D and Figure 2A). We performed TA cloning of the cDNAs and found that 100% of the clones analyzed lacked the 14 nucleotides. The impact of the nucleotide substitution was confirmed by exon trapping, which also showed a lack of leakiness (Supplemental Figure 2, B–D). Real-time quantitative PCR with a probe spanning exons 5 and 6 showed that EBV-B cells from P had about a tenth as much REL mRNA as healthy control cells, suggesting that the mutant transcript undergoes nonsense-mediated mRNA decay (Figure 2B). We then transfected 293T human embryonic kidney (HEK) cells with plasmids containing a cDNA corresponding to either the WT or the mutant (c.395_408del) REL allele (Mut), with an N-terminal DDK tag, and analyzed the protein products of the plasmid by Western blotting (Figure 2C; see complete unedited blots in the supplemental material). We detected no protein product for the mutant allele and no product of lower MW suggestive of a truncated protein. We also assessed the ability of WT and Mut c-Rel to induce a luciferase transgene under the control of the NF-κB promoter. WT c-Rel induced high levels of luciferase activity, unlike Mut c-Rel, which did not induce luciferase activity, despite the production of similar amounts of the mutant and WT mRNAs (Figure 2D). Overall, these findings indicate that the private c.395-1G>A variant disrupted REL mRNA splicing, with no leakiness, resulting in a loss of expression (LOE) and a LOF of c-Rel due to the presence of a premature termination codon in the aberrant transcript that was the sole product of the mutant allele.
Figure 2. An aberrant transcript underlies a loss of c-Rel production and function.
(A) PCR of full-length REL cDNA from EBV-B cells, showing a deletion of 14 nucleotides (r.395_408del) in exon 5 in P and a predicted termination codon (p.Val132Alafs*3). (B) RT-qPCR on total mRNA extracted from EBV-B cells from 3 controls (Ctls) and P. Data are displayed as 2-ΔCt after normalization to GUS expression. Data indicate the mean ± SEM. n = 3. (C) Western blot of extracts from HEK293T cells transfected with a pcDNA3.1 plasmid encoding an empty N-terminal DDK tag (empty vector [EV]), or encoding the WT or mutant (c.395_408del) REL allele. Two Abs were used: an Ab against the DDK tag and an Ab against GAPDH. Results are representative of 3 independent experiments. (D) Top panel: Luciferase activity of HEK293T cells cotransfected with an NF-κB reporter plasmid plus the pcDNA3.1 EV and a plasmid encoding the WT or c.395_408del REL allele. Results show the fold induction of activity relative to EV-transfected cells. Data indicate the mean ± SEM. n = 2. Each dot represents the mean of 9 technical replicates. Bottom panel: RT-qPCR results. Data are displayed as 2-ΔCt after normalization to GUS expression. Data indicate the mean ± SEM. n = 1. Each dot represents the mean of 3 technical replicates.
c-Rel deficiency alters the development of some lymphoid subsets.
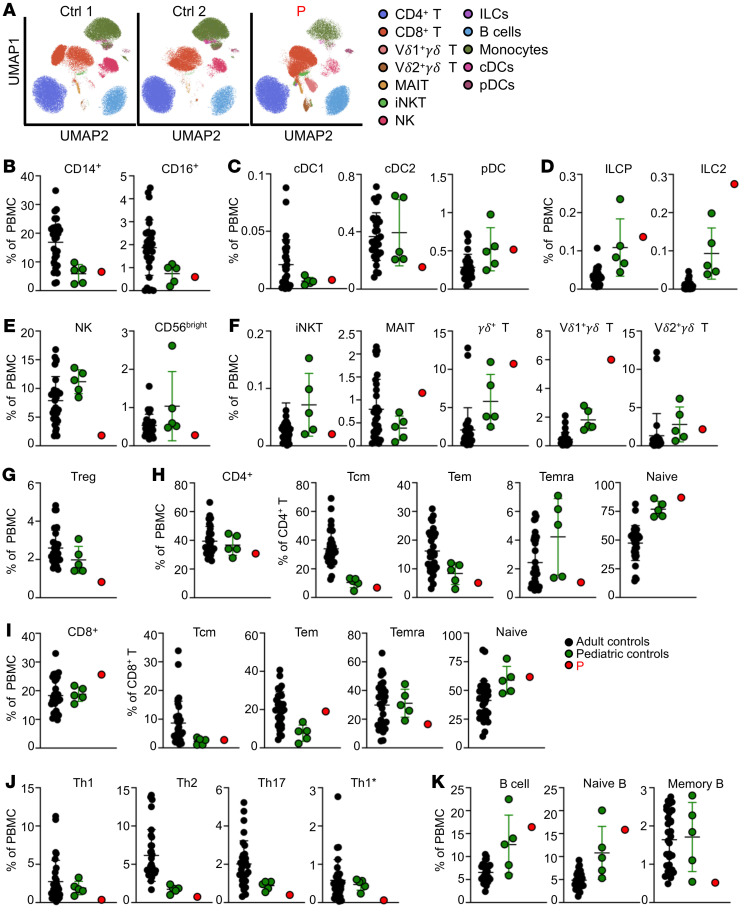
REL mRNA is expressed in various human tissues, but its levels are 3 to 10 times higher in nucleated hematopoietic cells relative to levels in other cell types (BioGPS, The Human Protein Atlas). We assessed the impact of the homozygous c.395-1G>A mutation on endogenous c-Rel expression. No c-Rel protein was detected in EBV-B cells from P, by Western blotting or flow cytometry (Supplemental Figure 3, A and B; see complete unedited blots in the supplemental material), or in her leukocytes by flow cytometry (Supplemental Figure 3C). By contrast, c-Rel was strongly expressed by control EBV-B cells and was detected in all leukocyte subsets from healthy donors, the highest levels being recorded for γδ T cells, naive CD19+ B cells, CD16+ monocytes, and CD141+ conventional DCs (cDC1s) (Supplemental Figure 3D). We performed flow cytometry on whole blood (Supplemental Tables 3 and 4) and PBMCs (Figures 3, A–K) from P to evaluate the impact of c-Rel deficiency on the distribution of the various leukocyte subsets relative to age-matched controls. P had normal counts of granulocytes, monocytes, and DCs, with frequencies of CD14+ and CD16+ monocytes, cDC1s, cDC2s and plasmacytoid DCs (pDCs) within the range of pediatric controls (Supplemental Table 3 and Figure 3, A–C). The total helper innate lymphoid cell (ILC) frequency was high, with normal frequencies of ILC precursors (ILCP) and IFN-γ+ ILC type 1 cells, and a high frequency of IL-13+ ILC type 2 cells (Figure 3D and Supplemental Figure 4A). NK cell counts and proportions were low (Supplemental Table 4 and Figure 3E), with a normal distribution of the CD56dim NK and CD56bright subsets on evaluation of a separate batch (Supplemental Figure 4B). Analysis of other innate-like lymphocyte subsets revealed levels of iNKT and Vδ2+γδ T cells similar to those in pediatric controls, whereas the levels of MAIT and Vδ1+γδ T cells were higher (Figure 3F). Tregs, defined as CD4+CD25+FOXP3+ cells, were present at levels below the lower limit of the range for the adult and pediatric controls (Figure 3G). P had normal counts of total CD4+ and CD8+ T cells (Supplemental Table 4), with a diversified T cell receptor (TCR) Vβ repertoire (Supplemental Table 5). Despite chronic infections with BCG, fungal, viral, and parasitic pathogens, P had relatively low frequencies of memory CD4+ T cells, in the lower range of those of pediatric controls (Supplemental Table 4). By contrast, P had a high frequency of effector memory (Tem) cells and a low frequency of terminally differentiated effector memory CD45RA+ (Temra) CD8+ T cells (Supplemental Table 4). In-depth immunophenotyping of PBMCs from P revealed naive and memory subsets of CD4+ T cells within the range for pediatric controls (Figure 3H). Analysis of CD8+ T cells in P showed that the frequencies of naive and central memory T (Tcm) cells were similar to those in the pediatric controls, whereas Tem cell frequencies were higher and Temra cell frequencies were lower than those in the pediatric controls (Figure 3I). For memory CD4+ T cells, P had low frequencies of Th1 and Th1* cells, while Th2 and Th17 cells were in the lower part of the range for pediatric controls (Figure 3J). Total B cell counts were high, with very high frequencies of CD24++CD38++ transitional B cells and lower frequencies of CD24–CD38++ plasmablasts and CD27+ memory B cells than those in healthy pediatric controls (Supplemental Table 4 and Figure 3K). Overall, these findings suggest that c-Rel is essential for the generation and/or maintenance of NK cells, CD8+ Temra cells, and memory CD4+ T cells, including Th1* and Th1* cells, Tregs, and memory B cells.
Figure 3. c-Rel deficiency alters the development of some lymphoid subsets.
Immunophenotyping of PBMCs from 31 adult controls, 5 pediatric controls, and P. (A) UMAP (uniform manifold approximation and projection) plots. The legends indicate the different leukocyte subsets, as defined by surface markers (provided in Methods). (B) Frequencies of monocyte subsets in total PBMCs. (C) Frequencies of cDCs and pDCs in total PBMCs. (D) ILC phenotyping showing the frequencies of ILC progenitors (ILCP) and ILC type 2 in total PBMCs. (E) NK cell phenotyping showing total NK cells and the frequency of CD56bright cells in total PBMCs. (F) Frequencies of iNKT cells, MAIT cells, γδ T cells, and Vδ1 and Vδ2 subsets in total PBMCs. (G) Frequency of Tregs in total PBMCs. (H and I) Frequencies of total CD4+ (H) and CD8+ (I) T cells in total PBMCs, and of naive, Tcm, Tem, and Temra cells within the CD4+ and CD8+ T cell compartments. (J) Frequencies of Th cell subsets in total PBMCs. (K) Frequencies of total B cells, naive B cells, and memory B cells in total PBMCs. Error bars represent the mean and SD.
Normal baseline transcriptome in c-Rel–deficient leukocytes at the single-cell level.
We further investigated the development and phenotype of leukocyte subsets in P, by performing single-cell RNA-Seq (scRNA-Seq) on cryopreserved PBMCs from P and her mother, together with cryopreserved PBMCs from 6 other healthy controls. Integrated sample clustering of the various immune subsets identified 13 distinct major subsets: CD14+ and CD16+ monocytes, pDCs, mature (CD56dim) and immature (CD56bright) NK cells, γδ T cells, Tregs, naive and memory CD4+ T cells, naive and memory CD8+ T cells, and naive and memory B cells expressing either the κ or λ light chain (Figure 4A). We then compared the frequencies of cells attributed to the different clusters for P and the healthy controls. Consistent with the results obtained by immunophenotyping, P had low frequencies of Tregs and memory B cells (Figure 4B and Supplemental Figure 5A). She also had relatively low frequencies of memory CD4+ T cells, contrasting with her higher frequency of memory CD8+ T cells (Figure 4B and Supplemental Figure 5A). The frequencies of the other immune subsets were similar between P and the controls (Supplemental Figure 5A). We then evaluated the effect of c-Rel deficiency on the baseline transcriptional program of PBMCs by performing a differential expression analysis for each leukocyte cluster. Single-cell gene expression levels were extracted and aggregated for each individual and for each cell subset. We then focused on genes with expression levels of more than 10 copies per million (cpm) and compared normalized gene expression between P and the controls. No significant differences in transcription were observed between the cells of P and the control cells for any of the subset clusters (Supplemental Figure 5B). The sc-RNA-Seq data provided additional evidence that c-Rel deficiency results in poor development of memory CD4+ T, Tregs, and memory B cells. They also suggested that c-Rel deficiency had no effect on the overall transcriptomic profile in the basal state.
Figure 4. Normal baseline transcriptome in c-Rel–deficient leukocytes at the single-cell level.
(A) UMAP clustering integrating scRNA-Seq data for PBMCs from P and 7 controls. Major cell clusters are labeled on the basis of enrichment in cell markers. B cells were extracted and clustered with UMAP to identify subsets on the basis of heavy- and light-chain usage and the naive versus memory state. Mo, monocytes; progenitors, hematopoietic stem cells. (B) Overlay of P’s cells captured by scRNA-Seq over the cells from controls (same UMAP as in A).
c-Rel deficiency affects the production of IL-12 by some, but not all, myeloid cells.
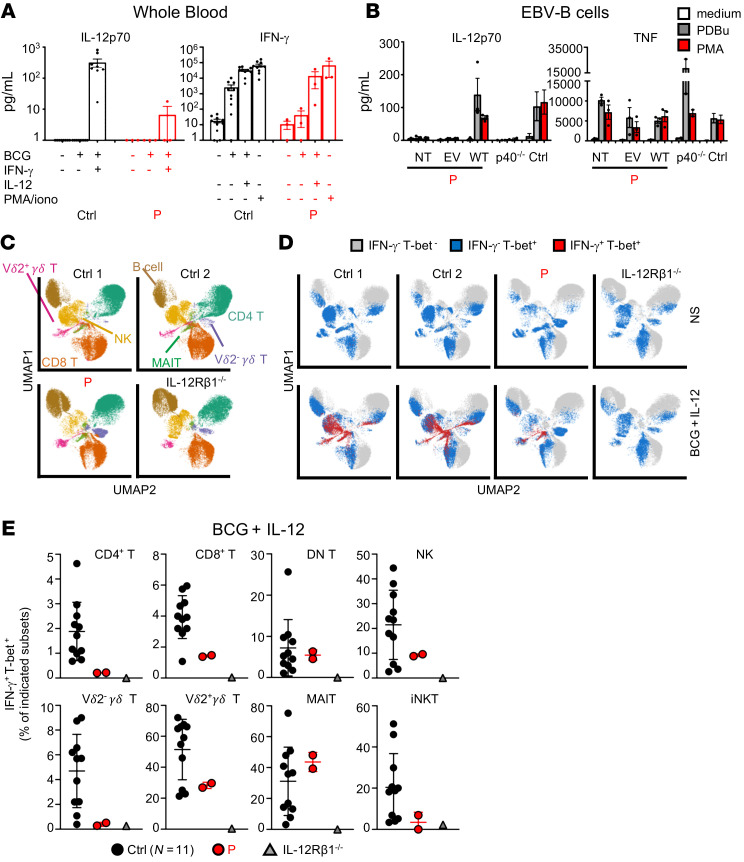
We then investigated the cellular mechanisms underlying severe BCG disease in P. Mouse c-Rel is essential for the production of IL-12 and IL-23 by DCs ex vivo, or by in vitro–derived macrophages and DCs, following stimulation with IFN-γ, TLR agonists (LPS, CpG, poly-IC), anti-CD40 Ab, or CD40L (29–35). We therefore investigated the impact of human c-Rel deficiency on the production of these cytokines. We first measured the production of IL-12p40 and IL-12p70 (comprising IL-12p40 and IL-12p35) in whole blood from P in response to BCG alone or together with IFN-γ. BCG plus IFN-γ substantially increased the secretion of IL-12p40 and IL-12p70 in whole blood from healthy controls (Figure 5A and Supplemental Figure 6A). By contrast, IL-12p40 production was strongly impaired, and that of IL-12p70 was barely detectable in whole blood from P. CD1c+ (cDC2s), derived in vitro from CD34+ cells isolated from P, produced normal amounts of IL-12p40, IL-12p70, IL-23, and CXCL10 in response to BCG plus IFN-γ, whereas all these cytokines, except CXCL10, were barely detectable in the supernatants of cDC1s derived in vitro from CD34+ cells isolated from P (Supplemental Figure 6B). Similarly, monocytes derived from control cells stimulated with LPS produced large amounts of IL-12p40, IL-23, and IL-1β (but no IL-12p70, data not shown), whereas the monocytes of P produced IL-1β but not the other cytokines tested (Supplemental Figure 6C). This impaired production of IL-12p40 and IL-12p70 we observed in cells from P was c-Rel dependent, as EBV-B cells from P transduced with an empty vector displayed normal TNF induction, but no induction of IL-12p40 or IL-12p70 in response to phorbol esters (PMA or PDBu), in contrast to the EBV-B cells from controls, and this defect was rescued by transduction with WT REL cDNA (Figure 5B and Supplemental Figure 6D). We then evaluated IFN-γ production by whole blood. Stimulation with BCG alone strongly induced IFN-γ production in whole blood from heathy individuals, whereas whole blood from P failed to respond to BCG (Figure 5A). The addition of IL-12 to BCG further increased IFN-γ secretion by whole-blood cells from controls and rescued the secretion of this cytokine by whole-blood cells from P. PMA plus ionomycin also induced a robust and comparable production of IFN-γ in P and controls (Figure 5A). Overall, these results suggest that the endogenous IL-12 production defect of the myeloid cells of P accounts, at least partly, for the impaired production of IFN-γ by whole blood in response to BCG.
Figure 5. c-Rel deficiency affects the production of IL-12 and IFN-γ.
(A) ELISA results for IL-12p70 and IFN-γ production in whole blood from controls (n = 10) and P, after a 48-hour incubation with different ligands. Data indicate the mean ± SEM. n = 3. iono, ionomycin. (B) ELISA results showing IL-12p70 and TNF production in P’s EBV-B cells that were not transfected (NT), or transfected with an empty retroviral plasmid (EV) or a plasmid encoding the WT c-Rel (WT); in cells from patients with complete AR IL-12p40 deficiency (p40–/–, n = 2); and in cells from controls (n = 3), after a 24-hour incubation with PDBu (10–7 M) or PMA (400 ng/mL). Data indicate the mean ± SEM. n = 3. (C–E) IFN-γ expression after 24 hours of stimulation of leukocytes from 11 controls, 1 patient with AR IL-12Rβ1 deficiency, and P. (C) UMAP plots are presented with legends indicating the different leukocyte subsets, as defined by surface markers (indicated in Methods). (D) UMAP plots show Tbet and IFN-γ expression in leukocyte subsets from 2 representative controls, 1 patient with AR IL-12Rβ1 deficiency, and P. (E) Dot plot graphs are shown with the values obtained in D. Error bars represent the mean and SD. Technical replicates are shown for P.
c-Rel deficiency affects the production of IFN-γ by some, but not all, lymphoid cells in response to BCG plus IL-12.
We then assessed the production of IFN-γ by the innate lymphoid cells, innate-like lymphoid cells, and T cells of P. We first evaluated the Mycobacterium-specific IFN-γ response of Tbet+ lymphoid cells. We previously showed, in healthy donors, that NK, MAIT, Vγ2+γδ T, CD4+, and CD8+ αβ T cells were the dominant producers of IFN-γ upon BCG stimulation, whereas invariant NKT (iNKT), Vγ1+γδ T, and B cells produced much smaller amounts (36). We detected IFN-γ production by Vγ2+γδ T, Vγ2-γδ T, mucosal-associated invariant T (MAIT), iNKT, NK, and CD4+ and CD8+ T cells from healthy donors in response to BCG plus IL-12, whereas cells from an IL-12Rβ1–deficient patient did not respond to such stimulation (Figure 5, C–E). By contrast, the production of IFN-γ by Tbet+ lymphoid cells from P was subnormal to normal relative to that in controls, except for the patient’s CD4+ T cells, which produced IFN-γ in quantities below the lower limit of the control range (Figure 5, D and E). Stimulation with PMA and ionomycin also resulted in poor IFN-γ production by CD4+ and CD8+ T cells and, to a lesser extent, by NK and Vγ2+γδ T cells (Supplemental Figure 6E). These results suggest that the weakened IFN-γ production in response to BCG observed in whole blood from P resulted from the impairment of IL-12 production observed in whole blood and in in vitro–derived cDC1s and monocytes. Furthermore, the addition of exogenous IL-12 rescued to some extent the impaired IFN-γ production in response to BCG in all lymphoid cells except CD4+ T cells (Figure 5E). In contrast to what was observed for CD4+ T cells, c-Rel deficiency did not substantially affect the intrinsic abilities of innate and innate-like lymphoid cells to produce IFN-γ.
c-Rel deficiency impairs the upregulation of costimulatory molecules on peripheral cDCs and contributes to poor T cell responses to antigen.
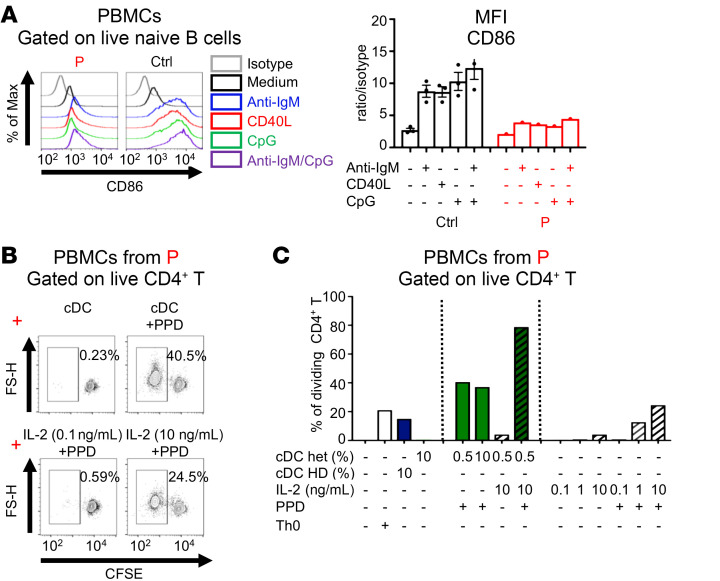
Monocytes and DCs display high levels of c-Rel expression (Supplemental Figure 3, C and D). We therefore investigated the effects of c-Rel deficiency on DC function. We evaluated the basal expression of HLA-DR, CD80, CD83, and CD86 on sorted pDCs, cDC2s, and cDC1s derived in vitro from CD34+ cells isolated from P and healthy controls and found that the expression levels were similar between P and the controls (Supplemental Figure 7, A and B). Given the rarity of these cells ex vivo, it was not possible to assess the induction of these costimulatory molecules on primary DCs. Instead, we used primary naive B cells, which are also potent APCs, to evaluate the induction of these molecules following stimulation with anti-IgM Ab, CD40L, CpG, or CpG plus anti-IgM Ab (Figure 6A and Supplemental Figure 8, A and B). The basal levels of CD40, CD80, and CD86 were similar between P and the controls. Upon stimulation, the control B cells exhibited a strong induction of CD86 under all conditions tested, whereas CD40 was upregulated principally by CpG (alone or in combination with anti-IgM Ab), and CD80 was induced only slightly by CpG plus anti-IgM. The induction of CD86 and, to a lesser extent, the upregulation of CD40, was strongly impaired in cells from P, whereas the weak induction of CD80 in these cells was similar to that observed in controls. The expression and upregulation of costimulatory molecules at the surface of APCs and T cells are important for antigen-specific T cell responses (37). We therefore measured the proliferation of CD4+ T cells from P by stimulating her PBMCs with tuberculin in the presence of syngeneic HLA-similar sorted CD11c+ cDCs from her mother, at a physiological ratio of 1:200, in the presence or absence of IL-2 (Figure 6, B and C, and Supplemental Figure 8C). The mother’s cDCs were not alloreactive, whereas the control HLA–mismatched cells were. After 7 days of culturing, CD4+ T cells from P displayed a robust response to tuberculin (PPD) in the presence of her mother’s cDCs, whereas PPD in the presence of high doses of exogenous IL-2 induced a response that was half as strong (Figure 6C). However, the addition of both IL-2 and syngenic cDCs to P’s PBMCs induced a strong response to PPD. These data show that the impaired upregulation of CD40 and CD86 by c-Rel–deficient APCs partially account for the defective antigen-specific CD4+ T cell responses observed in P (Supplemental Table 1).
Figure 6. c-Rel deficiency impairs the upregulation of costimulatory molecules on peripheral cDCs and contributes to poor T cell responses to antigen.
(A) PBMCs from controls (n = 3) and P were incubated with the indicated ligands for 12 hours. Expression of CD86 was assessed on CD19+CD27– naive B cells. Max, maximum. (B and C) CFSE-stained PBMCs from P were cultured for 7 days under Th0 conditions, or with sorted Lin–HLA-DR+CD11c+ cDCs from a control donor (cDC HD) or P’s mother (cDC het) in 2 different proportions (0.5% or 10%), with or without various doses of IL-2 (ng/mL) or tuberculin (PPD; 5 μg /mL). (B) Flow cytometric plots showing CFSE dilution for viable CD4+ T cells and (C) analysis of the percentage of dividing cells. n = 1.
c-Rel deficiency impairs CD4+ T cell proliferation, IL-2 production, and effector function.
We then investigated the impact of c-Rel deficiency on T-lymphocyte function. The polyclonal stimulation of fresh PBMCs from P with beads coated with mAbs against CD3 and CD28, or CD2, CD3, and CD28 (Th0), resulted in levels of proliferation of CD4+ and CD8+ T cells from P that were apparently similar to those for control cells, as shown by CFSE dilution (Figure 7A and Supplemental Figure 9A, left panels). However, the response of P’s CD4+ and CD8+ T cells to phytohemagglutinin (PHA) was markedly weaker than that of the control cells (Figure 7A and Supplemental Figure 9A, right panels). The addition of exogenous IL-2 to PHA-stimulated PBMCs from P restored both CD4+ and CD8+ T cell proliferation and the expression of CD25 on CD4+ T cells (Figure 7A and Supplemental Figure 9, A and B). Despite the apparently normal proliferation under polyclonal stimulation, the number of live CD4+ T cells was much lower for P, in all conditions, as shown by labeling with tracking dyes (Supplemental Figure 9C). IL-2 is required for T cell survival and proliferation. We thus assessed IL-2 expression in fresh PBMCs from P, in terms of both frequency and MFI in CD4+ T cells, following stimulation with anti-CD3/anti-CD28 mAbs or PMA and ionomycin (P/I). A smaller proportion of CD4+ T cells from P expressed IL-2, with a much lower MFI than that seen in control cells (Figure 7B). We assessed TNF expression levels in P’s cells and found that they were slightly lower, but within the range of expression levels in control cells, whereas IFN-γ levels were lower following stimulation with anti-CD3/anti-CD28 mAbs (Supplemental Figure 9D). We then analyzed the function of sorted naive CD4+ T cells from P. Under Th0 conditions, we observed markedly lower proliferation of naive CD4+ T cells from P (Figure 7C) and less IL-2 production compared with naive CD4+ T cells from healthy controls, as shown by flow cytometry and ELISA (Figure 7D and Supplemental Figure 9E). Exogenous IL-2 increased the proliferation of naive CD4+ T cells from P, but not to the levels observed in healthy controls (Supplemental Figure 9F). We then assessed the ability of P’s naive CD4+ T cells to differentiate into the Th1 and Th17 lineages in vitro after 5 days in Th-polarizing conditions. Naive CD4+ T cells from P secreted only very small amounts of IFN-γ, IL-17A, and IL-17F, despite normal induction of the TBX21 and RORC genes, which encode canonical transcription factors for Th1 (Tbet) and Th17 (RORγt) cells, respectively (Figure 7E, Supplemental Figure 9G, and ref. 38). We then measured the production of effector cytokines and IL-2 by sorted memory CD4+ T cells from P and healthy controls that had been cultured under Th0 conditions for 5 days. We found that cells from P secreted much smaller amounts of Th1, Th2, and Th17 cytokines (Figure 7F) and of IL-2 (Figure 7G). This likely resulted from poor expansion of memory Th cells, as the frequencies of the cells expressing most cytokines were within the control range, with the exception of IFN-γ–expressing cells (Supplemental Figure 9H), consistent with the lower frequencies of Th1 and Th1* cells, as determined by surface phenotype (Figure 3J). Overall, these results show that c-Rel–deficient naive and memory CD4+ T cells have intrinsically defective IL-2 production and proliferation, contributing to the broader defect of cytokine production by memory CD4+ T cells ex vivo and the compromised in vitro differentiation of naive CD4+ T cells into Th1 and Th17 cells.
Figure 7. c-Rel deficiency impairs CD4+ T cell proliferation, IL-2 production, and effector function.
(A) Fresh PBMCs from controls (results for 1 control are shown) and P were incubated for 4 days with beads coated with anti-CD3/anti-CD28 mAbs, anti-CD2/anti-CD3/anti-CD28 mAbs (Th0), or PHA (1 μg/mL) with or without IL-2. Histograms show CFSE dilution for CD4+ T cells. Representative results from 3 independent experiments are shown. (B) Fresh PBMCs from 4 controls and P were cultured for 6 hours with anti-CD3/anti-CD28 mAbs or PMA (20 ng/mL) and ionomycin (10–7 M) (P/I). The percentage of IL-2+ cells and MFI of IL-2 in CD4+ T cells are shown. (C) Sorted naive CD4+ T cells from controls (n = 4) and P were incubated for 4 days under Th0 conditions. Flow cytometric analyses of CFSE dilution were performed, and the percentage of cells in each division was analyzed. Data indicate the mean ± SEM. Representative results from 2 independent experiments are shown. (D) Percentage of IL-2+ cells, analyzed by flow cytometry, in sorted naive CD4+ T cells from controls (n = 11) and P, after 4 days culturing under Th0 conditions. Data indicate the mean ± SEM. n = 3. (E) Analysis of cytokine production by sorted naive CD4+ T cells cultured under Th0 or Th1- or Th17-polarizing conditions. Cells were obtained from 40 (Th1 culture) and 30 (Th17 culture) controls and P, and cytokine production was measured after 5 days. Data indicate the mean ± SEM. Representative results from 2 independent experiments are shown. (F and G) Cytokine production by sorted memory CD4+ T cells from controls (n = 32) and P was measured after 5 days of culturing under Th0 conditions. Data indicate the mean ± SEM. Representative results from 2 independent experiments are shown.
c-Rel regulates the expression of a restricted set of genes at early stages of CD4+ T cell activation.
In mouse and human T cells, treatment with anti-CD28 mAb or PMA leads to nuclear translocation of c-Rel, where it binds the IL2 gene promoter (39–42). In these conditions, mouse c-Rel–deficient T cells display impaired IL-2 production and lower levels of proliferation after 72 hours (27). We characterized human c-Rel function at the early stages of T cell activation by performing RNA-Seq on purified naive CD4+ T cells from P, 2 hours after activation with PMA, anti-CD3 mAb alone or together with anti-CD2 or anti-CD28 mAbs. Dimension reduction analysis showed that the type of stimulation accounted for most of the differences observed between samples (principal component analysis [PCA], PC1: 75.1% and PC2: 6.3%), whereas the genetic status for the REL gene accounted for 2.3% of the difference (PC3) (Supplemental Figure 10A). As a result, no overt major differences were observed between P and healthy controls in terms of the genes that were significantly induced or repressed under each set of stimulation conditions (Supplemental Figure 10B). PMA is a potent activator of NF-κB. In naive CD4+ T cells from healthy donors, 479 genes were upregulated, while 394 genes were downregulated, with a log2 fold change (FC) threshold of at least 2 (4-fold change; Supplemental Table 6A, GSE166873) upon stimulation with PMA. The list of upregulated transcripts includes those encoding for the high-affinity IL-2Rα receptor chain (rank no. 10), and IL-2 itself (rank no. 36); NF-κB proteins and their cognate negative regulators (REL, rank no. 46; NFKB1, no. 128; NFKBIZ, no. 146; NFKB2, no. 260; TNFAIP3, no. 284); members of the AP-1 complex (FOSL1, no. 12; FOSL2, no. 39; FOS, no. 74; JUNB, no. 117; FOSB, no. 197; JUND, no. 301; JUN, no. 305); surface costimulatory or inhibitory receptors, such as PDCD1 (no. 30), and CTLA4 (no. 90); and BHLHE40 (no. 185), a transcription factor involved in IFN-γ production and in the proinflammatory activity of CD4+ T cells (43). Among these top-ranking genes, we identified those for which induction differed significantly between P and the controls. We compiled a list of 23 genes with lower levels of expression in P’s cells and 1 gene with higher levels of expression in P’s cells than in control cells (Figure 8A). Under these conditions, IL2 was induced at normal levels in P’s cells after 2 hours of stimulation (Supplemental Table 6A, GSE166873). However, induction was significantly impaired for REL itself, TNFAIP3, and NFKBIZ (two NF-κB negative regulators); FOSL2, FOS, FOSB, and JUN (AP-1 proteins); and BHLHE40. All these genes harbor NF-κB consensus binding sequences in their promoter regions. We then performed ChIP-Seq on purified naive CD4+ T cells from 3 donors after 2 hours of PMA stimulation (Supplemental Table 7, A and B, GSE167185). We detected strong enrichment in c-Rel binding sites upon stimulation, with 1309 peaks displaying enrichment of at least 5-fold relative to the medium (Supplemental Figure 10C). NF-κB and AP-1 binding motifs ranked number 2 and number 3 among the motifs with the greatest enrichment (Figure 8B). Statistical analysis (see Supplemental Methods) revealed a strong correlation between these peaks and genes with lower levels of expression in P’s cells (P = 9.4 × 10–9; Figure 8, C and D). RT-qPCR on control purified naive CD4+ T cells stimulated with anti-CD3 mAb alone or with anti-CD28 mAb showed that the expression of REL and all AP-1 genes peaked after 2 hours, whereas the expression of IL2 and BHLHE40 peaked after 8 hours. We further confirmed the lower levels of expression of REL and AP-1 genes in activated naive CD4+ T cells from P, together with lower levels of IL2 expression and the abolition of BHLHE40 upregulation at 8 hours under the same conditions of activation (Figure 8E and Supplemental Figure 10D). Thus, c-Rel controls its own upregulation and that of a restricted set of genes, including AP-1 genes and BHLHE40, at early stages of CD4+ T cell activation, but it is not directly involved in early (2 hours) IL2 induction.
Figure 8. c-Rel regulates the expression of a restricted set of genes at early stages of CD4+ T cell activation.
(A) Heatmap showing log2 FC of expression in PMA-stimulated naive CD4+ T cell at 2 hours. Only genes both differentially expressed in response to stimulation in controls (adjusted P < 0.05 and | log2 FC | >1) and differentially expressed in P (adjusted P < 0.05 and | log2 FC | >2) are shown. For selected genes, the log2 FC between controls and P is indicated. (B) DNA binding motifs at PMA-induced c-Rel binding peaks. The de novo motif discovery P value and family of transcription factors are shown. (C) Bubble histogram showing the association of P’s dysregulated genes with c-Rel genomic binding sites as defined by ChIP-Seq in PMA-stimulated naive CD4+ T cells and located within 10 kb of their transcription start site. The association was computed for all c-Rel peaks and for a subgroup of peaks for which c-Rel binding increased by more than 5-fold following stimulation (1309 peaks). The circle size represents the significance of the association (–log10 Fisher’s exact test P value). The color gradient shows the ratio of the number of peaks proximal (within 10 kb) to these genes relative to randomly selected gene sets. (D) Genomic snapshots of selected genes with lower levels of expression in P. The top 3 tracks show the control input DNA and ChIP-Seq for c-Rel in nonstimulated (NS) and PMA-treated naive CD4+ T cells from 3 controls; blue boxes indicate significant c-Rel binding peaks. Below the gene structure are shown the normalized RNA-Seq profiles for naive CD4+ T cells from controls (overlaid) and P, at steady state or after stimulation. (E) RT-qPCR after 2 hours of activation of naive CD4+ T cells from 4 controls and P with anti-CD3 mAb with or without anti-CD28 mAb. Data are displayed as 2-ΔCt after normalization to GUS expression. Error bars represent the SD. n = 1.
The B cell response is intrinsically and extrinsically impaired in c-Rel deficiency.
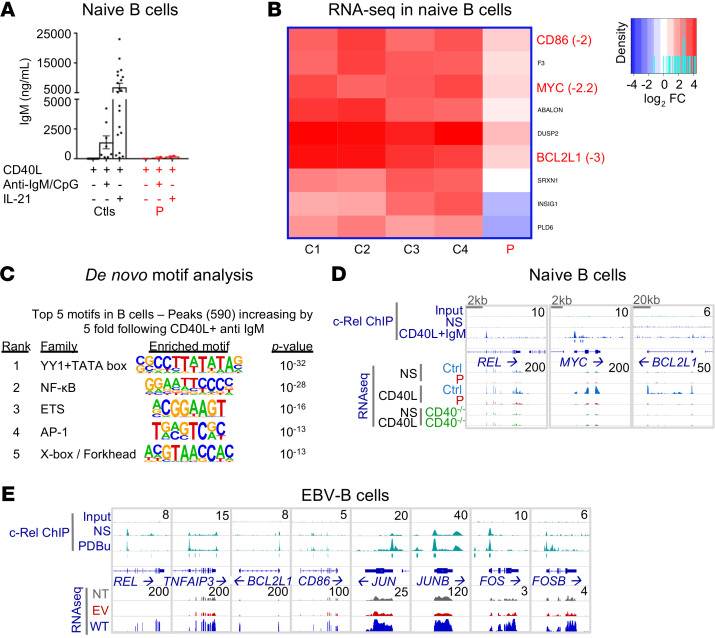
Despite having normal serum IgG concentrations and normal numbers of circulating CD19+ B cells, P had no detectable specific Ab responses following vaccination with protein and polysaccharide vaccines and had abnormally low allohemagglutinin titers (Supplemental Table 8). P also had extremely low levels of circulating memory B cells (Supplemental Table 4). We assessed the ability of P’s naive B cells to differentiate in vitro into Ig-secreting plasmablasts. After 7 days of stimulation with CD40L together with CpG and B cell receptor (BCR) agonists, naive B cells from healthy donors produced substantial amounts of IgM, but no IgG or IgA (Figure 9A and Supplemental Figure 11A). However, addition of IL-21 to cultures of CD40L-stimulated naive B cells from healthy donors induced the production of large amounts of IgM, as well as switched Ig isotypes (Figure 9A and Supplemental Figure 11A). In marked contrast to cells from healthy donors, c-Rel–deficient naive B cells produced very low, if at all detectable, levels of IgM (Figure 9A) and failed to undergo class switching to produce IgG or IgA upon in vitro stimulation with CD40L/CpG/BCR agonists or CD40L/IL-21 (Supplemental Figure 11A). Similar results were obtained when we assessed the ability of transitional B cells from P to differentiate into Ig-secreting cells in vitro (data not shown). We were unable to investigate the functional defects of memory B cells because of the scarcity of these cells in P. As class switching and the differentiation of B cells into Ig-secreting cells increase with cell division (44), we assessed the proliferative capacity of naive B cells from P. Her naive B cells underwent cell division in response to diverse stimuli, but the extent of proliferation observed was always lower for these cells compared with naive B cells from healthy donors (Supplemental Figure 11B, upper panels). Coupled with this proliferation defect, the survival of B cells from P following CD40L stimulation was severely compromised, as shown by flow cytometric determinations of the frequency of dead cells (Supplemental Figure 11B, lower panels). The survival of c-Rel–deficient B cells was partly rescued by additional stimuli (i.e., CpG, BCR agonist, IL-21), but remained markedly lower than that of naive B cells from healthy controls. Thus, the overall survival and proliferation of naive B cells from P were substantially compromised. Together, these defects prevented effective B cell differentiation, resulting in poor plasma cell generation and low levels of Ig secretion in vitro. These defects are also consistent with the extremely low counts of circulating memory B cells in this patient in vivo. We then investigated the possible contribution of B cell–extrinsic defects to the impairment of B cell responses in vivo. T follicular helper cells, which express CD40L, ICOS, and IL-21, are the key CD4+ T cell subset responsible for T-dependent B cell responses, class switching, and long-lived humoral memory B cells (45–47). We therefore measured the induction of the expression of these molecules on naive CD4+ T cells. ICOS upregulation in response to Th0 conditions was unaffected by c-Rel deficiency (Supplemental Figure 11, C and D, upper panels). By contrast, CD40L expression on activated naive CD4+ T cells from P was reduced. This reduction was even more noticeable when CD40L expression was measured on ICOS-expressing CD4+ T cells (Supplemental Figure 11, C and D, lower panels). Furthermore, the acquisition of expression of IL-21, a potent inducer of human B cell proliferation and differentiation (45–48), by naive CD4+ T cells was markedly impaired by c-Rel deficiency under Th1-polarizing conditions (Supplemental Figure 11E). Thus, the Ab deficiency seen in P probably results from both T cell– and B cell–intrinsic defects.
Figure 9. The B cell response is impaired in c-Rel deficiency.
(A) Naive B cells from controls (n = 20) and P, cultured with CD40L, alone or in combination with anti-IgM mAb and CpG or IL-21 for 7 days. IgM secretion was assessed by IgH chain–specific ELISA. n = 2. Data indicate the mean ± SEM. (B) Heatmap representing the log2 FC of expression in naive B cells stimulated with CD40L at 2 hours. Only the genes both differentially expressed in response to stimulation in controls (adjusted P < 0.05 and | log2 FC | >1) and differentially expressed in P (adjusted P < 0.05 and | log2 FC | >2) are shown. For selected genes, the log2 FC difference between controls and P is indicated. (C) DNA binding motifs enriched at CD40L plus anti-IgM–induced c-Rel binding peaks. The de novo motif discovery P value and family of transcription factors are shown. (D) Genomic snapshots of selected genes with altered responses to CD40L in P. Shown, from top to bottom, are the normalized sequence read profiles for control input DNA and ChIP-Seq for c-Rel in nonstimulated and CD40L plus anti-IgM–treated naive B cells from controls (blue boxes indicate significant c-Rel binding peaks), together with RNA-Seq profiles for nonstimulated and CD40L-treated controls (overlaid), P, and a CD40-deficient patient. (E) Genomic snapshots of selected genes for which expression in P’s EBV-B cells was rescued by retroviral transduction with a plasmid encoding the WT c-Rel. Shown, from top to bottom, are normalized profiles for control input DNA and ChIP-Seq for c-Rel in nonstimulated and PDBu-treated EBV-B cells from controls (blue boxes indicate significant c-Rel binding peaks), together with RNA-Seq profiles for nonstimulated EBV-B cells from P that were not transfected, or that were transfected with an empty retroviral plasmid or a plasmid encoding the WT c-Rel.
c-Rel deficiency compromises the expression of CD40-dependent target genes involved in naive B cell maturation.
In mice, c-Rel is crucial for B cell proliferation, which precedes differentiation in Ab-secreting cells and efficient humoral immunity (27). Our data on P’s naive B cells support the existence of both proliferation and survival defects. We further studied transcriptomic changes in naive B cells isolated from P, after incubation for 2 hours with CD40L, anti-IgM mAb, or PMA, all of which trigger NF-κB activation. Cells from a patient with AR CD40 deficiency were used as a negative control for CD40L simulation. We observed no overt major difference between the P’s cells and control cells in any of these conditions (Supplemental Figure 12A). Upon CD40L stimulation of control cells, 216 genes were significantly upregulated, with at least a 4-fold increase in expression levels (Supplemental Table 6B, GSE166873). The upregulated genes included those encoding transcription factors and cytokine receptors involved in B cell maturation, such as MYC, BCL2L1, IRF4, IL21R, and IL2RA. Nine of these genes were significantly less strongly induced in P’s cells (by a factor of at least 4), and these genes included MYC, BCL2L1, and CD86 (Figure 9B). In marked contrast, we observed no significant difference between cells from P and those from controls following BCR or PMA stimulation (Supplemental Table 6B, GSE166873). MYC expression was upregulated in all conditions, whereas BCL2L1 expression was induced only upon CD40 stimulation in the controls’ cells. REL was also upregulated in all conditions tested in cells from controls. Although a 4-fold difference relative to controls was not achieved, the levels of induction of REL in cells from P were lower in response to stimulation with CD40L (P = 0.04; log2 FC: –0.9) and PMA (P = 0.003; log2 FC: –1.3) (Supplemental Table 6B, GSE166873). These data suggest that c-Rel may control the expression of a particular set of genes downstream from CD40L. ChIP-Seq on control naive B cells following a 2-hour stimulation with CD40L/anti-IgM mAb or PMA revealed strong enrichment at c-Rel binding sites, with 1509 and 304 peaks, respectively, and an enrichment of at least 5-fold relative to the medium (Supplemental Table 7, C and D, GSE167185; Supplemental Figure 12B). The NF-κB site was the second most enriched motif (P = 10–28; Figure 9C). Notably, c-Rel bound to the promoters of MYC, BCL2L1, and REL, and the levels of expression of these genes were lower in cells from P and a CD40-deficient patient (Figure 9D). Furthermore, an isogenic comparison of RNA-Seq results for EBV-B cells from P, which were stably transduced with an empty vector or WT REL cDNA (Supplemental Figure 12C), demonstrated a statistically significant c-Rel–dependent rescue of basal expression levels for REL (P = 2.7 × 10–65), BCL2L1 (2.2 × 10–22), and CD86 (5.5 × 10–9) (Supplemental Table 6C, GSE166873; Figure 9E), and for expression levels of TNFAIP3, JUNB, JUN, FOS, and FOSB, which were found to be impaired in the RNA-Seq experiments performed on purified naive CD4+ T cells from P after 2 hours of stimulation with PMA (Supplemental Table 6A, GSE166873; Figure 9E). These findings are consistent with the significant enrichment in c-Rel binding sites in the promoters of these genes, as shown by ChiP-Seq on control EBV-B cells (Supplemental Table 7, E and F, GSE167185; Figure 9E). Overall, the combined RNA-Seq results for P’s naive B cells and the ChIP-Seq results for control naive B cells after short periods of stimulation support the hypothesis that CD40L/CD40 signaling relies on c-Rel to regulate the expression of a small set of transcription factors involved in B cell maturation and survival, such as MYC, BCL2L1, and REL itself.
Discussion
We describe AR complete c-Rel deficiency as a genetic etiology of CID in a single child. Our study fulfills the prerequisites of the 2014 criteria for single-patient genetic studies (49), thereby establishing a causal relationship between the patient’s REL genotype and phenotype. Since 2014, a total of 29 new inborn errors of immunity have been reported in individual patients, highlighting similarities to and differences from the corresponding mouse mutant, when such mutants are available, as recently exemplified by a study in a patient with inherited Tbet deficiency (36). Moreover, another c-Rel–deficient patient was recently reported with an infectious phenotype consistent with that of our patient (50) (Supplemental Table 9). We show that c-Rel was essential for the development and/or maintenance of NK cells, Temra CD8+ T cells, memory CD4+ T cells, including Th1 and Th1*, Tregs, and memory B cells. In myeloid cells, c-Rel was required for the production of IL-12 and IL-23 and for antigen-specific T cell responses, at least through the regulation of CD86 expression and, therefore, APC function. In lymphoid cells, c-Rel regulates a later phase of IL-2 expression by CD4+ T cells, which probably accounts for the lower frequencies of Tregs (51), memory CD4+ T cells, and Temra CD8+ T cells (52, 53). The low levels of IL-12, IL-23, and IL-2 production together contribute to the poor development of Th1 and Th1* cells in vivo (54–56). The IL-2 production defect also affects the patient’s T cell proliferation, accounting for the broader defect in cytokine production by memory CD4+ T cells ex vivo and the defective differentiation of naive CD4+ T cells into Th1 and Th17 cells in vitro. Finally, c-Rel deficiency affects the CD40-mediated induction of antiapoptotic MYC and BCL2L1 in naive B cells, compromising the survival of these cells and thereby affecting their proliferation and differentiation into Ab-secreting cells (57). This deficiency also affects the expression of ICOS, CD40L, and IL-21 by activated CD4+ T cells, suggesting that defective Tfh cells also contribute to abnormal mature B cell responses.
Our results confirm the salient observations made in Rel–/– mice (Supplemental Table 9). Mouse c-Rel binds to the promoter of the IL12b gene (29), and its absence leads to lower IL-12 production by Rel–/– macrophages and DCs (31–35). Rel–/– mice have low frequencies of Tregs (58), and Rel–/– T cells display impaired IL-2 production following stimulation with anti-CD3/anti-CD28 mAbs or PMA, and their T cell proliferation capacity is low (27). Rel–/– mice also have an impaired Th1 response to challenge with Leishmania (59) or Toxoplasma (60), and this impaired response is also seen in models of experimental autoimmune encephalomyelitis (30). Finally, Rel–/– mice display poor Ab responses (27), memory B cell development (61), and Tfh cell function (62, 63). Our study also sheds new light on functions of human c-Rel that have not, to our knowledge, been documented in mice. We describe the impact of c-Rel deficiency on the development of leukocyte subsets not studied (ILCs, iNKT cells, MAIT cells, γδ T cells), or not characterized (Th1* cells; refs. 54, 55) in mice. We also studied myeloid subsets derived in vitro. Our results indicate that, in humans, c-Rel is essential for the production of IL-12 and IL-23 in monocytes and cDC1s, but not in cDC2s. This is consistent with the normal levels of IL-12 in whole blood of individuals who selectively lack cDC2s because of AD IRF8 or AR SPPL2A deficiency (64, 65). Consistently, exogenous IL-12 compensated for the impaired IFN-γ production by c-Rel–deficient innate and innate-like lymphoid cells in response to BCG, but not for that of CD4+ T cells, highlighting the distinct c-Rel requirements of lymphoid cells for intrinsic IFN-γ production. We also focused on IL2, the transcription of which is mediated by the nuclear factor of activated T cells (NFAT), AP-1, and NF-κB proteins (66). By coupling ChIP-Seq and RNA-Seq, we showed that c-Rel acted indirectly as a rheostat at early stages and promoted the optimal induction of IL2 at later stages by inducing both AP-1 and REL. However, exogenous IL-2 only partially rescued antigen-specific T cell proliferation, due to the previously unsuspected role of c-Rel in the control of CD86 and, to a lesser extent, CD40 upregulation in DCs.
Our findings provide a cellular explanation for the patient’s susceptibility to BCG. Complete AR deficiencies of IL-12p40 or IL-12Rβ1 underlie MSMD, due to the impairment of IL-12–and IL-23–dependent IFN-γ production by T cells (11, 12, 55). In this context, mycobacterial disease in patients with complete AR c-Rel deficiency probably results from impaired production of IL-12 and IL-23 by cDC1s and monocytes as well as from impaired IFN-γ production by lymphoid cells. As in IL-12Rβ1–deficient patients, we found that the in vivo development of Th1 and Th1* cells was impaired, with Th1* cells being enriched in Mycobacterium-responsive clones (54, 55). Furthermore, c-Rel deficiency altered the in vitro development of Th1 cells from naive CD4+ T cells, probably reflecting poor expansion in culture. We also assessed the genotype-dependent effect on the generation of cytokine-producing CD4+ T cells by culturing purified memory (CD45RA–) CD4+ T cells under polyclonal nonpolarizing conditions (Th0). The patient’s cells produced only very small amounts of all cytokines, reflecting the poor expansion in vitro, as the overall frequencies of cytokine-expressing cells were similar to those of controls, except for IFN-γ, for which the frequency was lower in P, probably because of the low frequencies of Th1 and Th1* cells in the patient. We further studied in vivo Mycobacterium-specific IFN-γ production. In controls, upon BCG infection, all IFN-γ –producing cells produce large amounts of Tbet (36). These cells consist predominantly of CD4+ and CD8+ T cells and NK, MAIT, and Vγ2+γδ T cells (36). IFN-γ production in response to BCG plus IL-12 was much more strongly impaired in CD4+ T cells than in other lymphoid cells. The role of c-Rel in IFN-γ expression in CD4+ T cells has yet to be determined. ChIP-Seq did not detect c-Rel binding to the IFNG promoter in activated CD4+ T cells, and we observed no significant induction of IFNG mRNA after 2 hours of activation in controls. We found, however, that BHLHE40 TF was one of the most strongly induced genes in activated CD4+ T cells, that its expression was significantly lower in the patient’s cells than in control cells, and that c-Rel bound to its promoter in CD4+ T cells and in other cell types (activated naive B cells or EBV-B cells; Supplemental Table 6, A–C, GSE166873). Interestingly, mice lacking BHLHE40 are highly susceptible to Mycobacterium tuberculosis because of poor IFN-γ production (67, 68). BHLHE40 is also predominantly expressed by Th1 and Th1* cells in humans, and the association of its expression with NF-κB activation defines a core signature associated with inflammatory properties of human CD4+ T cells (43).
Similarly, the CMC in this c-Rel–deficient patient probably resulted from impaired IL-17A and IL-17F production by Th17 cells. CMC was more severe in P than in patients with IL-12p40 or IL-12Rβ1 deficiency, who have low frequencies of Th17 cells (18, 69, 70). This suggests that the CMC observed in this patient reflects poor expansion of Th17 cells and low production of IL-17A and IL-17F and/or that the production of IL-17 cytokines by other lymphocytes is profoundly affected. The patient had no pyogenic infections. However, the patient’s c-Rel deficiency would be predicted to predispose her to pyogenic infections through its effects on Ab responses and Tfh cell function. The occurrence of such infections was probably prevented by early IgG replacement therapy. The pathogenesis of biliary cryptosporidiosis is less clear. It may not be coincidental that the few inherited genetic defects conferring a predisposition to Cryptosporidium parvum infection identified to date are XR CD40L, AR CD40, HLA class II, NIK, and IL-21R deficiencies (3, 19, 71–73), although the cellular mechanism controlling C. parvum infection remains elusive. Susceptibility to cryptosporidiosis in this patient may be secondary to the impairment of CD40L and IL-21 production by activated CD4+ T cells. In addition, or alternatively, CD40-CD40L interaction may trigger c-Rel expression in myeloid cells in a NF-κB–inducing kinase–dependent (NIK-dependent) or NIK-independent manner, potentially controlling IL-21 production by T cells and immunity to the parasite. Finally, the infectious diseases of the other recently reported c-Rel–deficient patient include tuberculosis, chronic cryptosporidiosis, and CMV disease, consistent with our patient’s infectious phenotype (50). Overall, this experiment of nature demonstrates the essential role of human c-Rel in diverse effector functions across the 3 key leukocyte subsets governing adaptive immunity: DCs, T cells, and B cells. It is also essential for certain functions of innate immunity. The simultaneous correction of the immunological and infectious phenotypes by HSCT confirmed that the patient’s immunodeficiency was caused by severe intrinsic defects of leukocytes. In the absence of human c-Rel, innate and adaptive immunity to various infectious agents is severely compromised by a broad and profound defect of myeloid and lymphoid leukocytes.
Methods
RNA-Seq and ChIP-Seq.
ChIP and RNA-Seq experiments are detailed in the Supplemental Methods. The ChIP-Seq data are available in the NCBI’s Gene Expression Omnibus (GEO) database (GEO GSE167185), and the RNA-Seq data are available in the NCBI’s SuperSeries (GEO GSE166873).
Study approval.
The experiments were conducted in accordance with local, national, and international regulations and were approved by the local ethics committee (Comités de Protection des Personnes [CPP] Ile-de-France 2, ID-RCB: 2010-A00636-33), the ANSM (French National Agency for Medicines and Health Products Safety, B100712-40), and by the French Ministry of Research (IE-2010-547). Informed consent was obtained from the patient’s family, as the patient is a minor, in accordance with World Medical Association rules, the Declaration of Helsinki, and European Union directives.
Author contributions
Designing research RL, JPDS, CSM, NM, KL, PG, FG, SGT, JLC, and AP. Conducting experiments RL, VB, DL, GR, TL, YJZ, KM, DTA, JM, AIL, MO, RY, SP, MM, CD, and KP. Analyzing data RL, FR, AG, TH, ME, DL, MB, AB, and LA. Reagents CB, FA, LRS, EAM, BB, SB, RMB, JR, SB, IB, APl, BN, JB, and AAB. Writing RL, JLC, and AP.
Supplementary Material
Acknowledgments
We thank the patient and her family, the health care professionals from the Immunology-Hematology and Rheumatology unit at the Necker Hospital for Sick Children, and the members of our laboratory. This work was supported by the French National Research Agency (ANR-14-CE15-0006-01; ANR-14-RARE-0005-02; ANR-18-CE93-0008-01; ANR-16-CE17-0005-01); the National Institute of Allergy and Infectious Diseases (NIAID), NIH (R01AI127564; P01AI061093; 5R21OD023291); “Investissement d’avenir” (ANR-10-IAHU-01); the Integrative Biology of Emerging Infectious Diseases Laboratoire d’Excellence (ANR-10-LABX-62-IBEID); the Jeffrey Modell Centers Network; the St. Giles Foundation; the Sidra Genomics Core; the National Health and Medical Research Council of Australia; the New South Wales Ministry of Health, Canada Institute of Health Research (no. 168959), FRQ-S, SIDRA Genomics core, and SIDRA Medicine. RL was supported by the INSERM (Poste d’accueil), a Fulbright commission grant, the Philipp Foundation, and the Bettencourt Foundation.
Version 1. 09/01/2021
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(17):e150143.https://doi.org/10.1172/JCI150143.
Contributor Information
Romain Lévy, Email: romain.levy@inserm.fr.
David Langlais, Email: david.langlais@mcgill.ca.
Vivien Béziat, Email: vivien.beziat@inserm.fr.
Franck Rapaport, Email: frapaport@mail.rockefeller.edu.
Geetha Rao, Email: g.rao@garvan.org.au.
Tomi Lazarov, Email: lazarovt@mskcc.org.
Mathieu Bourgey, Email: mathieu.bourgey@mcgill.ca.
Coralie Briand, Email: coralie.briand@aphp.fr.
Kunihiko Moriya, Email: kunihikomoriya@gmail.com.
Fatima Ailal, Email: drailalfatima@gmail.com.
Janet Markle, Email: janet.markle@vumc.org.
Ai Ing Lim, Email: aiing.lim@nih.gov.
Masato Ogishi, Email: mogishi@rockefeller.edu.
Rui Yang, Email: ryang@mail.rockefeller.edu.
Mehdi Emam, Email: seyedmehdi.emam@mcgill.ca.
Caroline Deswarte, Email: caroline.deswarte@inserm.fr.
Tanwir Habib, Email: tanwir.habib@icloud.com.
Luis R. Saraiva, Email: lsaraiva@sidra.org.
Andrea Guennoun, Email: aguennoun@sidra.org.
Bertrand Boisson, Email: bebo283@mail.rockefeller.edu.
Serkan Belkaya, Email: sbelkaya@bilkent.edu.tr.
Ruben Martinez-Barricarte, Email: ruben.m.barricarte@vumc.org.
Jérémie Rosain, Email: jeremie.rosain@gmail.com.
Aziz Belkadi, Email: abdelaziz.belkadi@gmail.com.
Sylvain Breton, Email: sylvain.breton@aphp.fr.
Ibtihal Benhsaien, Email: ibtihalbenhsaien@gmail.com.
Alessandro Plebani, Email: plebani@med.unibs.it.
Vassilios Lougaris, Email: vlougarisbs@yahoo.com.
James P. Di Santo, Email: james.di-santo@pasteur.fr.
Bénédicte Neven, Email: benedicte.neven@aphp.fr.
Laurent Abel, Email: laurent.abel@inserm.fr.
Cindy S. Ma, Email: c.ma@garvan.org.au.
Ahmed Aziz Bousfiha, Email: profbousfiha@gmail.com.
Nico Marr, Email: nmarr@sidra.org.
Jacinta Bustamante, Email: jacinta.bustamante@inserm.fr.
Kang Liu, Email: kangbelsky@gmail.com.
Philippe Gros, Email: philippe.gros@mcgill.ca.
Frédéric Geissmann, Email: geissmaf@mskcc.org.
Jean-Laurent Casanova, Email: Jean-Laurent.Casanova@rockefeller.edu.
Anne Puel, Email: anne.puel@inserm.fr.
References
- 1.Fischer A, et al. Notarangelo LD, et al. Human inborn errors of immunity: an expanding universe. Science Immunology. 2020;5(49):eabb1662. doi: 10.1126/sciimmunol.abb1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Notarangelo LD. Combined immunodeficiencies with nonfunctional T lymphocytes. Adv Immunol. 2014;121:121–190. doi: 10.1016/B978-0-12-800100-4.00004-0. [DOI] [PubMed] [Google Scholar]
- 3.Ouederni M, et al. Major histocompatibility complex class II expression deficiency caused by a RFXANK founder mutation: a survey of 35 patients. Blood. 2011;118(19):5108–5118. doi: 10.1182/blood-2011-05-352716. [DOI] [PubMed] [Google Scholar]
- 4.Al-Herz W, et al. Major histocompatibility complex class II deficiency in Kuwait: clinical manifestations, immunological findings and molecular profile. J Clin Immunol. 2013;33(3):513–519. doi: 10.1007/s10875-012-9831-8. [DOI] [PubMed] [Google Scholar]
- 5.Tangye SG, et al. Human inborn errors of immunity: 2019 update on the classification from the International Union of Immunological Societies Expert Committee. J Clin Immunol. 2020;40(1):24–64. doi: 10.1007/s10875-019-00737-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bousfiha A, et al. Human inborn errors of immunity: 2019 update of the IUIS phenotypical classification. J Clin Immunol. 2020;40(1):66–81. doi: 10.1007/s10875-020-00758-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Notarangelo LD. Functional T cell immunodeficiencies (with T cells present) Annu Rev Immunol. 2013;31:195–225. doi: 10.1146/annurev-immunol-032712-095927. [DOI] [PubMed] [Google Scholar]
- 8.Casanova JL, et al. Immunology taught by human genetics. Cold Spring Harb Symp Quant Biol. 2013;78:157–172. doi: 10.1101/sqb.2013.78.019968. [DOI] [PubMed] [Google Scholar]
- 9.Casanova JL. Severe infectious diseases of childhood as monogenic inborn errors of immunity. Proc Natl Acad Sci U S A. 2015;112(51):E7128–E7137. doi: 10.1073/pnas.1521651112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casanova JL. Human genetic basis of interindividual variability in the course of infection. Proc Natl Acad Sci U S A. 2015;112(51):E7118–E7127. doi: 10.1073/pnas.1521644112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosain J, et al. Mendelian susceptibility to mycobacterial disease: 2014-2018 update. Immunol Cell Biol. 2019;97(4):360–367. doi: 10.1111/imcb.12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bustamante J. Mendelian susceptibility to mycobacterial disease: recent discoveries. Hum Genet. 2020;139(6–7):993–1000. doi: 10.1007/s00439-020-02120-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puel A, et al. Chronic mucocutaneous candidiasis in humans with inborn errors of interleukin-17 immunity. Science. 2011;332(6025):65–68. doi: 10.1126/science.1200439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lévy R, et al. Genetic, immunological, and clinical features of patients with bacterial and fungal infections due to inherited IL-17RA deficiency. Proc Natl Acad Sci U S A. 2016;113(51):E8277–E8285. doi: 10.1073/pnas.1618300114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ling Y, et al. Inherited IL-17RC deficiency in patients with chronic mucocutaneous candidiasis. J Exp Med. 2015;212(5):619–631. doi: 10.1084/jem.20141065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu L, et al. Gain-of-function human STAT1 mutations impair IL-17 immunity and underlie chronic mucocutaneous candidiasis. J Exp Med. 2011;208(8):1635–1648. doi: 10.1084/jem.20110958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boisson B, et al. An ACT1 mutation selectively abolishes interleukin-17 responses in humans with chronic mucocutaneous candidiasis. Immunity. 2013;39(4):676–686. doi: 10.1016/j.immuni.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Puel A. Human inborn errors of immunity underlying superficial or invasive candidiasis. Hum Genet. 2020;139(6–7):1011–1022. doi: 10.1007/s00439-020-02141-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kotlarz D, et al. Loss-of-function mutations in the IL-21 receptor gene cause a primary immunodeficiency syndrome. J Exp Med. 2013;210(3):433–443. doi: 10.1084/jem.20111229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jouanguy E, et al. Human inborn errors of immunity to herpes viruses. Curr Opin Immunol. 2020;62:106–122. doi: 10.1016/j.coi.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Döffinger R, et al. X-linked anhidrotic ectodermal dysplasia with immunodeficiency is caused by impaired NF-kappaB signaling. Nat Genet. 2001;27(3):277–285. doi: 10.1038/85837. [DOI] [PubMed] [Google Scholar]
- 22.Belkadi A, et al. Whole-exome sequencing to analyze population structure, parental inbreeding, and familial linkage. Proc Natl Acad Sci U S A. 2016;113(24):6713–6718. doi: 10.1073/pnas.1606460113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Itan Y, et al. The mutation significance cutoff: gene-level thresholds for variant predictions. Nat Methods. 2016;13(2):109–110. doi: 10.1038/nmeth.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maffucci P, et al. Blacklisting variants common in private cohorts but not in public databases optimizes human exome analysis. Proc Natl Acad Sci U S A. 2019;116(3):950–959. doi: 10.1073/pnas.1808403116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Q, et al. 30 Years of NF-κB: a blossoming of relevance to human pathobiology. Cell. 2017;168(1–2):37–57. doi: 10.1016/j.cell.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Köntgen F, et al. Mice lacking the c-rel proto-oncogene exhibit defects in lymphocyte proliferation, humoral immunity, and interleukin-2 expression. Genes Dev. 1995;9(16):1965–1977. doi: 10.1101/gad.9.16.1965. [DOI] [PubMed] [Google Scholar]
- 28.Rapaport F, et al. Negative selection on human genes underlying inborn errors depends on disease outcome and both the mode and mechanism of inheritance. Proc Natl Acad Sci U S A. 2021;118(3):e2001248118. doi: 10.1073/pnas.2001248118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanjabi S, et al. Selective requirement for c-Rel during IL-12 P40 gene induction in macrophages. Proc Natl Acad Sci U S A. 2000;97(23):12705–12710. doi: 10.1073/pnas.230436397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hilliard BA, et al. Critical roles of c-Rel in autoimmune inflammation and helper T cell differentiation. J Clin Invest. 2002;110(6):843–850. doi: 10.1172/JCI15254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mason N, et al. Cutting edge: identification of c-Rel-dependent and -independent pathways of IL-12 production during infectious and inflammatory stimuli. J Immunol. 2002;168(6):2590–2594. doi: 10.4049/jimmunol.168.6.2590. [DOI] [PubMed] [Google Scholar]
- 32.Grumont R, et al. c-Rel regulates interleukin 12 p70 expression in CD8(+) dendritic cells by specifically inducing p35 gene transcription. J Exp Med. 2001;194(8):1021–1032. doi: 10.1084/jem.194.8.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carmody RJ, et al. Essential roles of c-Rel in TLR-induced IL-23 p19 gene expression in dendritic cells. J Immunol. 2007;178(1):186–191. doi: 10.4049/jimmunol.178.1.186. [DOI] [PubMed] [Google Scholar]
- 34.Mise-Omata S, et al. A proximal kappaB site in the IL-23 p19 promoter is responsible for RelA- and c-Rel-dependent transcription. J Immunol. 2007;179(10):6596–6603. doi: 10.4049/jimmunol.179.10.6596. [DOI] [PubMed] [Google Scholar]
- 35.Visekruna A, et al. Transcription factor c-Rel plays a crucial role in driving anti-CD40-mediated innate colitis. Mucosal Immunol. 2015;8(2):307–315. doi: 10.1038/mi.2014.68. [DOI] [PubMed] [Google Scholar]
- 36.Yang R, et al. Human T-bet governs innate and innate-like adaptive IFN-γ immunity against mycobacteria. Cell. 2020;183(7):1826–1847. doi: 10.1016/j.cell.2020.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen L, Flies DB. Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat Rev Immunol. 2013;13(4):227–242. doi: 10.1038/nri3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okada S, et al. Immunodeficiencies. Impairment of immunity to Candida and Mycobacterium in humans with bi-allelic RORC mutations. Science. 2015;349(6248):606–613. doi: 10.1126/science.aaa4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghosh P, et al. The interleukin 2 CD28-responsive complex contains at least three members of the NF kappa B family: c-Rel, p50, and p65. Proc Natl Acad Sci U S A. 1993;90(5):1696–1700. doi: 10.1073/pnas.90.5.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou XY, et al. Molecular mechanisms underlying differential contribution of CD28 versus non-CD28 costimulatory molecules to IL-2 promoter activation. J Immunol. 2002;168(8):3847–3854. doi: 10.4049/jimmunol.168.8.3847. [DOI] [PubMed] [Google Scholar]
- 41.Maggirwar SB, et al. Regulation of the interleukin-2 CD28-responsive element by NF-ATp and various NF-kappaB/Rel transcription factors. Mol Cell Biol. 1997;17(5):2605–2614. doi: 10.1128/MCB.17.5.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bryan RG, et al. Effect of CD28 signal transduction on c-Rel in human peripheral blood T cells. Mol Cell Biol. 1994;14(12):7933–7942. doi: 10.1128/mcb.14.12.7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emming S, et al. A molecular network regulating the proinflammatory phenotype of human memory T lymphocytes. Nat Immunol. 2020;21(4):388–399. doi: 10.1038/s41590-020-0622-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bryant VL, et al. Cytokine-mediated regulation of human B cell differentiation into Ig-secreting cells: predominant role of IL-21 produced by CXCR5+ T follicular helper cells. J Immunol. 2007;179(12):8180–8190. doi: 10.4049/jimmunol.179.12.8180. [DOI] [PubMed] [Google Scholar]
- 45.Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity. 2014;41(4):529–542. doi: 10.1016/j.immuni.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tangye SG, et al. The good, the bad and the ugly - TFH cells in human health and disease. Nat Rev Immunol. 2013;13(6):412–426. doi: 10.1038/nri3447. [DOI] [PubMed] [Google Scholar]
- 47.Ma CS, et al. The origins, function, and regulation of T follicular helper cells. J Exp Med. 2012;209(7):1241–1253. doi: 10.1084/jem.20120994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leonard WJ, et al. The γc Family of cytokines: basic biology to therapeutic ramifications. Immunity. 2019;50(4):832–850. doi: 10.1016/j.immuni.2019.03.028. [DOI] [PubMed] [Google Scholar]
- 49.Casanova JL, et al. Guidelines for genetic studies in single patients: lessons from primary immunodeficiencies. J Exp Med. 2014;211(11):2137–2149. doi: 10.1084/jem.20140520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beaussant-Cohen S, et al. Combined immunodeficiency in a patient with c-Rel deficiency. J Allergy Clin Immunol. 2019;144(2):606–608. doi: 10.1016/j.jaci.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Malek TR, Castro I. Interleukin-2 receptor signaling: at the interface between tolerance and immunity. Immunity. 2010;33(2):153–165. doi: 10.1016/j.immuni.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kalia V, et al. Prolonged interleukin-2Ralpha expression on virus-specific CD8+ T cells favors terminal-effector differentiation in vivo. Immunity. 2010;32(1):91–103. doi: 10.1016/j.immuni.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 53.Farber DL, et al. Human memory T cells: generation, compartmentalization and homeostasis. Nat Rev Immunol. 2014;14(1):24–35. doi: 10.1038/nri3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Acosta-Rodriguez EV, et al. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat Immunol. 2007;8(6):639–646. doi: 10.1038/ni1467. [DOI] [PubMed] [Google Scholar]
- 55.Martinez-Barricarte R, et al. Human IFN-γ immunity to mycobacteria is governed by both IL-12 and IL-23. Sci Immunol. 2018;3(30):eaau6759. doi: 10.1126/sciimmunol.aau6759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liao W, et al. Modulation of cytokine receptors by IL-2 broadly regulates differentiation into helper T cell lineages. Nat Immunol. 2011;12(6):551–559. doi: 10.1038/ni.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roy K, et al. A regulatory circuit controlling the dynamics of NFκB cRel transitions B cells from proliferation to plasma cell differentiation. Immunity. 2019;50(3):616–628. doi: 10.1016/j.immuni.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Isomura I, et al. c-Rel is required for the development of thymic Foxp3+ CD4 regulatory T cells. J Exp Med. 2009;206(13):3001–3014. doi: 10.1084/jem.20091411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reinhard K, et al. c-Rel promotes type 1 and type 17 immune responses during Leishmania major infection. Eur J Immunol. 2011;41(5):1388–1398. doi: 10.1002/eji.201041056. [DOI] [PubMed] [Google Scholar]
- 60.Mason NJ, et al. T cell-intrinsic expression of c-Rel regulates Th1 cell responses essential for resistance to Toxoplasma gondii. J Immunol. 2004;172(6):3704–3711. doi: 10.4049/jimmunol.172.6.3704. [DOI] [PubMed] [Google Scholar]
- 61.Milanovic M, et al. Differential requirements for the canonical NF-κB transcription factors c-REL and RELA during the generation and activation of mature B cells. Immunol Cell Biol. 2017;95(3):261–271. doi: 10.1038/icb.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu WH, et al. A miR-155-Peli1-c-Rel pathway controls the generation and function of T follicular helper cells. J Exp Med. 2016;213(9):1901–1919. doi: 10.1084/jem.20160204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen G, et al. Regulation of the IL-21 gene by the NF-κB transcription factor c-Rel. J Immunol. 2010;185(4):2350–2359. doi: 10.4049/jimmunol.1000317. [DOI] [PubMed] [Google Scholar]
- 64.Kong XF, et al. Disruption of an antimycobacterial circuit between dendritic and helper T cells in human SPPL2a deficiency. Nat Immunol. 2018;19(9):973–985. doi: 10.1038/s41590-018-0178-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hambleton S, et al. IRF8 mutations and human dendritic-cell immunodeficiency. N Engl J Med. 2011;365(2):127–138. doi: 10.1056/NEJMoa1100066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liao W, et al. Interleukin-2 at the crossroads of effector responses, tolerance, and immunotherapy. Immunity. 2013;38(1):13–25. doi: 10.1016/j.immuni.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu F, et al. The transcription factor Bhlhe40 is a switch of inflammatory versus antiinflammatory Th1 cell fate determination. J Exp Med. 2018;215(7):1813–1821. doi: 10.1084/jem.20170155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huynh JP, et al. Bhlhe40 is an essential repressor of IL-10 during Mycobacterium tuberculosis infection. J Exp Med. 2018;215(7):1823–1838. doi: 10.1084/jem.20171704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Puel A, et al. Inborn errors of human IL-17 immunity underlie chronic mucocutaneous candidiasis. Curr Opin Allergy Clin Immunol. 2012;12(6):616–622. doi: 10.1097/ACI.0b013e328358cc0b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.de Beaucoudrey L, et al. Mutations in STAT3 and IL12RB1 impair the development of human IL-17-producing T cells. J Exp Med. 2008;205(7):1543–1550. doi: 10.1084/jem.20080321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Al-Saud BK, et al. Clinical, immunological, and molecular characterization of hyper-IgM syndrome due to CD40 deficiency in eleven patients. J Clin Immunol. 2013;33(8):1325–1335. doi: 10.1007/s10875-013-9951-9. [DOI] [PubMed] [Google Scholar]
- 72.DiSanto JP, et al. CD40 ligand mutations in X-linked immunodeficiency with hyper-IgM. Nature. 1993;361(6412):541–543. doi: 10.1038/361541a0. [DOI] [PubMed] [Google Scholar]
- 73.Willmann KL, et al. Biallelic loss-of-function mutation in NIK causes a primary immunodeficiency with multifaceted aberrant lymphoid immunity. Nat Commun. 2014;5:5360. doi: 10.1038/ncomms6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.