Abstract
Patients with mental disorders often suffer from comorbidity. Transdiagnostic understandings of mental disorders are expected to provide more accurate and detailed descriptions of psychopathology and be helpful in developing efficient treatments. Although conventional clustering techniques, such as latent profile analysis, are useful for the taxonomy of psychopathology, they provide little implications for targeting specific symptoms in each cluster. To overcome these limitations, we introduced Gaussian graphical mixture model (GGMM)-based clustering, a method developed in mathematical statistics to integrate clustering and network statistical approaches. To illustrate the technical details and clinical utility of the analysis, we applied GGMM-based clustering to a Japanese sample of 1,521 patients (Mage = 42.42 years), who had diagnostic labels of major depressive disorder (MDD; n = 406), panic disorder (PD; n = 198), social anxiety disorder (SAD; n = 116), obsessive-compulsive disorder (OCD; n = 66), comorbid MDD and any anxiety disorder (n = 636), or comorbid anxiety disorders (n = 99). As a result, we identified the following four transdiagnostic clusters characterized by i) strong OCD and PD symptoms, and moderate MDD and SAD symptoms; ii) moderate MDD, PD, and SAD symptoms, and weak OCD symptoms; iii) weak symptoms of all four disorders; and iv) strong symptoms of all four disorders. Simultaneously, a covariance symptom network within each cluster was visualized. The discussion highlighted that the GGMM-based clusters help us generate clinical hypotheses for transdiagnostic clusters by enabling further investigations of each symptom network, such as the calculation of centrality indexes.
Introduction
Approximately forty-five percent of individuals with mental disorders suffer from comorbidity, or multiple mental disorders [1]. Comorbidity predicts poorer prognosis and health-related quality of life [2, 3]. There is a substantial clinical need to establish treatment guidelines targeting comorbid cases (e.g., [4–6]). As frequently discussed in psychopathology studies (e.g., [7–9]), such high proportion of comorbidity result from traditional diagnostic criteria―such as the Diagnostic and Statistical Manual of Mental Disorders (DSM) [10] and the International Classification of Diseases (ICD) [11]―that do not fit the empirical data surrounding mental disorder symptoms. The lack of fit between traditional diagnostic criteria and empirical data results in low symptom specificity, marked diagnostic heterogeneity, and poor reliability, as well as high comorbidity [12–15]. Recent studies have begun to use data-driven approaches to understand psychopathology from a transdiagnostic (i.e., across-diagnostic; see [16] for detailed definitions of the term transdiagnostic) perspective (e.g., [7, 12, 17, 18]).
Studies that seek a transdiagnostic understanding of psychopathology often apply clustering techniques, which classify patients with mental disorders into several clusters based on quantitative symptom data. For example, latent class analysis (LCA) [19], which identifies several latent classes behind discrete symptom variables, has been used to derive subgroups within major depressive disorder [20] and postpartum depression [21]. Moreover, latent profile analysis (LPA) [22], which is an extension of LCA that uses continuous variables as indicators, has been used to identify latent classes among comorbid cases of psychopathology [23, 24]. These studies have frequently achieved a quantitative classification of mental disorders that is inconsistent with conventional diagnostic systems. For example, Kircanski et al. [24] identified latent classes characterized by i) mixed symptoms of anxiety and depression and by ii) intense symptoms of irritability, anxiety, depression, and attention deficit hyperactivity disorder (ADHD) using clinical data from children and adolescents.
Although conventional clustering techniques have provided a useful taxonomy of psychopathology, these techniques only identify each cluster’s profile (e.g., Cluster 1 is characterized by intense symptoms of irritability, anxiety, depression, and ADHD). More importantly, these techniques do not provide direct treatment implications for each cluster (e.g., clinicians cannot judge treatment target priorities across symptoms of irritability, anxiety, depression, and ADHD). A technique that identifies not only symptom item mean scores but also rich information regarding symptom interplays in latent class estimations would help clinicians better understand cluster-specific symptom structures and identify key target symptoms in each cluster, both of which are indispensable for clinical reasoning and case formulation. Such a novel technique would become a gateway that connect clustering techniques and network approaches of psychopathology, which are rapidly developing to identify complex symptom interplays and calculate centrality indexes for each symptom [25–28].
To the best of our knowledge, no previous psychopathological (or psychological) studies have achieved the mixture of network estimation and cluster identification. Although Brusco et al. [29] have used a stepwise approach to analyze binary symptom data of depression and anxiety, in which they first performed p-median clustering [30, 31] and subsequently used so-called eLasso procedure [32] to explore the network structure within each cluster, they admit that such stepwise approaches can potentially bias the resulted network structures. The main aim of Brusco et al. [29] was to demonstrate the limitations of using network modeling without accounting for unobserved heterogeneity of participants; they noted that mixture modeling approaches should be developed in future to achieve more accurate estimations of networks within participant clusters.
In the field of computational statistics, on the other hand, a hybrid of LPA and cross-sectional network analysis has recently been developed [33]. As detailed later in this section, this hybrid analysis became possible after the recent development of model-based clustering—a popular framework in mathematical statistics for clustering multivariate data [34, 35]—and use of the Gaussian graphical mixture model (GGMM) [33, 36]. In this paper, we introduce the GGMM-based clustering technique with reference to Fop et al. [33], apply the technique to an existing dataset of patients with depressive and anxiety disorders, and discuss the clinical utility of the techniques and future directions for integrating clustering and network approaches.
GGMM-based clustering technique
Clustering techniques are statistical analyses that identify several groups within a multivariate dataset, and earlier techniques largely depended on heuristic procedures, including the Ward method [37] and k-means clustering [38]. As detailed in Fraley and Raftery [34] and McNicholas [35], model-based clustering is a counter movement of heuristic clustering. It assumes that multivariate data are generated by a finite mixture of a certain number of distributions and determines the number of clusters solely from quantitative criteria, such as the Bayesian Information Criterion (BIC) [39, 40]. The framework of model-based clustering includes many conventional clustering techniques: LCA [19], for example, can be understood as a latent class model-based clustering that assumes multivariate data arise from a mixture of discrete distributions. To cluster multivariate continuous data, the finite Gaussian mixture model (GMM) [34, 35, 41, 42], which assumes that data are generated by finite mixtures of multivariate normal (Gaussian) distributions, are widely used. As described in Fop et al. [33], the finite GMM estimates a mean vector and a covariance matrix for each identified cluster k (for 1 ≤ k ≤ K). The density of each data point in the finite GMM is given by:
| (1) |
where xi is the vector of observed variables, K is the fixed number of clusters, τk are mixing proportions for cluster k, Φ(·) is the multivariate Gaussian density function, μk is the mean vector for cluster k, Σk is the covariance matrix for cluster k, and Θ is the vector of model parameters. The EM algorithm [43] is typically used to estimate the model with a fixed number of clusters K, and the most appropriate number for K is determined through model comparisons based on fit indexes, such as BIC [39, 40].
Finite GMM-based clustering can easily be over-parameterized when large numbers of variables are included in the model. Several remedies have been developed to deal with potential over-parametrization, such as eigenvalue decomposition [44, 45] and factorizing the covariance matrix [46]. However, these remedies do not fully consider the covariance matrix of original variables in cluster specification. Worse still, finite GMM-based clustering does not visualize interplays between variables within each identified cluster. To overcome such limitations, Fop et al. [33] developed GGMM-based clustering, which combines GMM-based clustering with the Gaussian graphical model (GGM) [47, 48]. In GGM, interplays between variables are visualized as a graph (i.e., network), G = (V, E), where V is the set of nodes (i.e., variables), and E is the set of edges (i.e., associations between paired variables). The graph estimation is achieved through covariance matrices of variables with penalty terms, such as the graphical least absolute shrinkage and selection operator (GLASSO) procedure [49], in which all edge parameters are first estimated, then the small ones are shrunk to zero. In GGMM-based clustering, graph structures estimated in GGM are considered in identifying variable clusters. This is expressed in the following definition of each data point’s density in the GGMM:
| (2) |
in which the graph for cluster k, Gk, the collection of graphs, , and the cone of positive definite matrices for the graphs, C+ (Gk) are simply added to formula (1) in the GMM.
GGMM-based clustering can be executed with the mixggm package [33] for R software. In this package, the structural EM algorithm [50, 51], which incorporates the structure learning step into the EM algorithm [43], is used to estimate the model with a fixed number of clusters K, and models with different numbers of clusters are compared using BIC [39, 40]. The penalty function used in the graph calculations is specified by the penalized likelihood approach [52, 53], and graphs with edges that represent covariances between nodes are calculated as a result.
The present study
To demonstrate the clinical and statistical implications of GGMM-based clustering, we re-analyzed an existing dataset of 1,521 Japanese patients with depressive and anxiety disorders (see the Methods section for details). Depressive and anxiety disorders are the most comorbid clinical conditions among mental disorders [54–56]. Therefore, we expected that this dataset, which included comorbid participants with depressive and anxiety disorders, was suitable for demonstrating the discrepancies (or similarities) between latent classes obtained by GGMM-based clustering and conventional diagnostic labels, and for simultaneously visualizing complex symptom interplays in comorbid disorders.
Materials and methods
Dataset
We used an existing dataset collected for a large research project launched in Japan that aimed to validate several measures assessing depressive and anxiety symptoms and related constructs, such as emotion regulation skills [57–60]. The minimal dataset needed to replicate our results is freely available via the Open Science Framework (OSF; https://osf.io/6jnf4/). Collection of the data used in this study was approved by the National Center of Neurology and Psychiatry Institutional Review Board (Approval number: A2013-022; Title of the project: An online survey using clinical and non-clinical samples to validate the Overall Anxiety/Depression and Impairment Scales [OASIS/ODSIS]). In this project, an online survey was conducted in January 2014, using an internet marketing research company’s panelist pool in Japan (Macromill Inc; https://group.macromill.com/). When the survey was conducted, 1,095,443 panelists were registered in this pool, and 389,265 of them were labeled as “disease panelists” based on their self-reported clinical status in February 2013. A total of 2,459 Japanese anonymous disease panelists 18 years or older participated in the survey, and their labels were as follows: major depressive disorder (MDD; n = 619), panic disorder (PD; n = 619), social anxiety disorder (SAD; n = 576), obsessive-compulsive disorder (OCD; n = 645). In addition, data from “non-disease panelists” (n = 371) were collected as counterparts of those from disease panelists.
Since these diagnostic labels were based on self-descriptions made about one year before the study, a series of items designed to check panelists’ current diagnostic status were used during the survey (e.g., “Are you currently diagnosed as having major depressive disorder and being treated for the problem in a medical setting?”). According to the responses to these items, a total of 2,830 participants, including both disease and non-disease panelists, were divided into the following categories: MDD (n = 406), PD (n = 198), SAD (n = 116), OCD (n = 66), comorbid MDD and any anxiety disorder (n = 636), comorbid anxiety disorders (n = 99), other mental disorders (n = 146), and non-clinical (n = 1,163). The analyses used only the data from the 1,521 participants (775 female, 746 male; Mage = 42.42, SD = 9.50) with one or more depressive or anxiety disorder diagnoses. In other words, we excluded the data from participants categorized as other mental disorders (n = 146) or non-clinical (n = 1,163) from the analyses. As the online survey required the participants to respond to all the items, the resulting dataset included no missing values.
Measures
We selectively used the MDD, PD, SAD, and OCD symptom data for the present study. For detailed information on measures included in the dataset, see [59, 60].
MDD symptoms
MDD symptoms were assessed using the Japanese version [61] of the Patient Health Questionnaire [62]. This 9-item measure asks participants about the frequency of depressive symptoms over the past two weeks, using a 4-point Likert scale from 1 (Not at all) to 4 (Nearly every day). Doi et al. [57] found that this Japanese measure has the following two-factor structure: i) cognitive/affective symptoms (6 items; e.g., “feeling down, depressed, or hopeless”) and ii) somatic symptoms (3 items; e.g., “poor appetite or overeating”).
PD symptoms
PD symptoms were assessed using the Japanese version [60] of the Anxiety Sensitivity Index-3 [63]. This 18-item measure assesses participants’ anxiety sensitivity, which is defined as fear of arousal-related physical and psychological sensations [64, 65], using a 5-point Likert scale from 1 (very little) to 5 (very much). Ebesutani et al. [66] found that these 18 items reflect a general factor of anxiety sensitivity plus the following three subfactors: i) physical concerns (6 items; e.g., “It scares me when my heart beats rapidly”), ii) cognitive concerns (6 items; e.g., “When I cannot keep my mind on a task, I worry that I might be going crazy”), and iii) social concerns (6 items; e.g., “When I tremble I fear what people might think of me”).
SAD symptoms
SAD symptoms were assessed using the Japanese version [67] of the Fear of Negative Evaluation Scale-Short Form [68]. This 12-item measure assesses participants’ tendency to feel threatened by the prospect of negative evaluation from others, using a 5-point Likert scale from 1 (not at all) to 5 (very likely). Sasagawa et al. [67] showed that these 12 items exhibit a high internal consistency and have a one-factor structure. Sample items included “I worry about what other people will think of me even when I know it doesn’t make any difference.”
OCD symptoms
OCD symptoms were assessed using the Japanese version [69] of the Obsessive-Compulsive Inventory-Short Form [70]. This 18-item measure assesses participants’ obsessive and compulsive symptoms using a 5-point Likert scale from 1 (not at all) to 5 (very much). Koike et al. [71] showed that the Japanese version has the following six-factor structure: i) hoarding (3 items; e.g., “I have saved up so many things that they get in the way”), ii) checking (3 items; e.g., “I check things more often than necessary”), iii) ordering (3 items; e.g., “I get upset if objects are not arranged properly”), iv) neutralizing (3 items; e.g., “I feel I have to repeat certain numbers”), v) washing (3 items; e.g., “I sometimes have to wash or clean myself simply because I feel contaminated”), and vi) obsessing (3 items; e.g., “I find it difficult to control my own thoughts”).
Data analysis
The data analysis plan was preregistered on the OSF (https://osf.io/6jnf4/), and R script used in the analyses are freely available there. First, we conducted a series of confirmatory factor analyses to calculate factor scores that reflected the factor structure for each symptom measure (see the Measures subsection for details). Second, using the mixggm package [33] for R software, we conducted GGMM-based clustering with the default setting (i.e., tuning parameter β was set to 0) to i) determine the number of clusters that can be obtained from MDD, PD, SAD, and OCD symptom data; ii) summarize a profile of symptom factor scores for each cluster; iii) evaluate the concordance between the cluster allocation of patients and their diagnostic labels; and iv) visualize interplays of individual symptoms as a covariance network for each cluster. Third, using the qgraph [72] and bootnet [27] packages for R, we estimated a partial correlation network of symptoms for each cluster and computed centrality indexes, including bridge ones, for each symptom. To estimate the partial correlation networks, we used the GLASSO procedure [49] with Extended BIC (EBIC) [73]. Finally, using the networktools package [74] for R software, we computed bridge centrality indexes in each cluster to specify symptoms that connect different disorders.
As explained on OSF (https://osf.io/6jnf4/), the data analysis procedures noted above were slightly changed from those planned a priori. First, when conducting GGMM-based clustering, we entered factor scores instead of individual symptom items. This change was made after we found that GGMM-based clustering could not achieve a convergence when using items with ordinal scales and that continuous variables need to be entered. Second, we estimated partial correlation networks and calculated centrality indexes in addition to estimating covariance networks computed in GGMM-based clustering. We expected that partial correlation networks and centrality indexes (i.e., strength, closeness, and betweenness) would be more informative and potentially reveal richer clinical implications than covariance networks alone. Although closeness (i.e., the inverse of the sum of geodesic distances from one node to the other nodes) and betweenness (i.e., number of times one node lies in the shortest paths between other nodes) centrality were widely used in previous investigations of psychopathology networks (e.g., [75–77]), Bringmann et al. [78] recently argued that these two indexes are unstable in psychopathology networks and do not fit common assumptions underlying psychological research. We, therefore, reported closeness and betweenness centrality as supplementary information and interpret only strength centrality (i.e., the sum of absolute values of edge weights connected to each node).
We also have some notes regarding the availability of the R package we used. Although we have conducted GGMM-based clustering on 12 February 2021 using the mixggm package, that package has been removed from the CRAN repository (https://cran.rstudio.com/web/packages/mixggm/index.html) on 21 April 2021 by the developers. To increase the transparency of our analyses, we have uploaded the users’ manual of the mixggm package, which has been initially provided by the developers via the CRAN repository, on OSF (https://osf.io/6jnf4/).
Results
GGMM-based clustering
The results of a series of confirmatory factor analyses on symptom measures were detailed in the R Markdown file on OSF (https://osf.io/6jnf4/), and the factor loadings obtained were used to calculate factor scores of MDD, PD, SAD, and OCD symptoms. Correlations between factor scores included in GGMM-based clustering are displayed in Fig 1. The mixGGM function built in the mixggm package [33] revealed that GGMM-based clustering with a five-cluster solution could not be properly calculated since the variance and covariance matrices became not positive definite within one or more clusters. Furthermore, it automatically compared BIC [39, 40] across GGMMs with one- to four-cluster solutions and then returned the results on the four-cluster solution with the lowest BIC. The participants’ cluster membership is presented in Table 1, and symptom score factor profile for each cluster is displayed in the left panel of Fig 2. As shown, Cluster 4 (n = 791; 52.0% of the participants), the largest cluster, was characterized by strong symptoms of all four disorders. By contrast, Cluster 3 (n = 138; 9.1% of the participants) was characterized by weak symptoms of all four disorders. Cluster 1 (n = 235; 15.5% of participants) was characterized by strong OCD and PD symptoms and moderate MDD and SAD symptoms, and Cluster 2 (n = 357; 23.5% of participants) was characterized by moderate MDD, PD, and SAD symptoms and weak OCD symptoms. Of note, GGMM cluster allocation was inconsistent with participants’ diagnostic labels. As shown in Table 1, all clusters included a certain number of participants with each diagnostic label.
Fig 1. Correlations between factor scores of depressive and anxiety symptoms.
MDD = major depressive disorder, PD = panic disorder, SAD = social anxiety disorder, OCD = obsessive-compulsive disorder. MDD1 = cognitive/affective symptoms, MDD2 = somatic symptoms, PD1 = physical concerns, PD2 = cognitive concerns, PD3 = social concerns, OCD1 = hoarding, OCD2 = checking, OCD3 = ordering, OCD4 = neutralizing, OCD5 = washing, OCD6 = obsessing.
Table 1. Participants’ allocations to the Gaussian graphical mixture model (GGMM)-based clusters (N = 1,521).
| Cluster 1 | Cluster 2 | Cluster 3 | Cluster 4 | Total | |
|---|---|---|---|---|---|
| Major depressive disorder (MDD) | 61 | 125 | 51 | 169 | 406 |
| Panic disorder (PD) | 20 | 63 | 35 | 80 | 198 |
| Social anxiety disorder (SAD) | 15 | 45 | 9 | 47 | 116 |
| Obsessive-compulsive disorder (OCD) | 2 | 6 | 6 | 52 | 66 |
| Comorbid MDD and any anxiety disorder | 120 | 101 | 33 | 382 | 636 |
| Comorbid anxiety disorders | 17 | 17 | 4 | 61 | 99 |
| Total | 235 | 357 | 138 | 791 | 1,521 |
Fig 2. The symptom-profile of patient clusters identified by GGMM (left panel) and the covariance symptom network estimated for each cluster (right panel).
The thickness of edges in the symptom networks reflects the values of covariances. MDD = major depressive disorder, PD = panic disorder, SAD = social anxiety disorder, OCD = obsessive-compulsive disorder. MDD1 = cognitive/affective symptoms, MDD2 = somatic symptoms, PD1 = physical concerns, PD2 = cognitive concerns, PD3 = social concerns, OCD1 = hoarding, OCD2 = checking, OCD3 = ordering, OCD4 = neutralizing, OCD5 = washing, OCD6 = obsessing.
The covariance symptom networks, which were estimated simultaneously with the cluster allocation, are displayed in the right panel of Fig 2. The four symptom network shapes differed markedly from each other, and several features could be identified from visual inspection across the networks. For example, OCD symptoms were strongly interconnected in each cluster. SAD symptoms were characteristically located near the center of each network, except the Cluster 4 network, which had strong symptoms for all four disorders. Moreover, MDD symptoms were relatively isolated (i.e., located far apart from anxiety-related symptoms and directly connected with a small number of symptoms) in every symptom network except Cluster 3, which had low levels of symptomatology.
Partial correlation network estimations via GLASSO regulation with EBIC
The partial correlation network for each cluster identified in GGMM-based clustering is displayed in Fig 3. Several marked features of Cluster 2 and 4 networks can be identified from visual inspections. These networks consisted of both positive and negative edges (i.e., partial correlations), and there were negative edges even within the community of OCD symptoms. By contrast, the Cluster 1 and 3 networks consisted only of positive edges. It is also noteworthy that larger numbers of nonzero edges were obtained for Clusters 2 and 4, compared to Clusters 1 and 3. To summarize, network structures were much more complex in Clusters 2 and 4 than in Clusters 1 and 3.
Fig 3. Partial correlation network estimated for each patient cluster.
Nodes represent symptoms, and edges represent partial correlations between them. The thickness of an edge reflects the absolute value of the regularized partial correlation. Blue and red edges represent positive and negative regularized partial correlations, respectively.
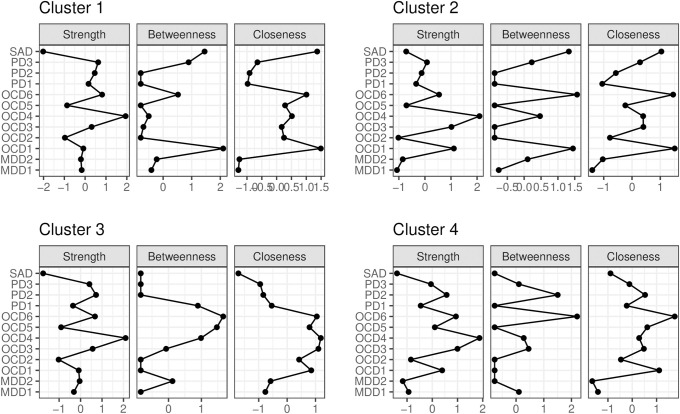
Centrality indexes for each symptom in each cluster are summarized in Fig 4. Interestingly, the OCD neutralizing symptom (OCD4) had the highest strength in each cluster. It is also noteworthy that MAD and SAD symptoms had relatively low strength in each cluster.
Fig 4. Centrality indexes (standardized z-scores) calculated in each partial correlation network.
MDD = major depressive disorder, PD = panic disorder, SAD = social anxiety disorder, OCD = obsessive-compulsive disorder. MDD1 = cognitive/affective symptoms, MDD2 = somatic symptoms, PD1 = physical concerns, PD2 = cognitive concerns, PD3 = social concerns, OCD1 = hoarding, OCD2 = checking, OCD3 = ordering, OCD4 = neutralizing, OCD5 = washing, OCD6 = obsessing.
Bridge centrality indexes calculations
Bridge centrality (i.e., a variant of centrality that takes communities of nodes into account), including expected influence (i.e., the sum of raw values of edge weights connected to each node), for each symptom in each cluster is summarized in Fig 5. As shown, SAD symptoms had the nearly highest bridge strength and expected influence in every network. Of interest, the physical concerns of PD (PD1), as well as SAD symptoms, had the nearly highest bridge strength and expected influence in Cluster 4, with strong symptoms of all four disorders.
Fig 5. Bridge centrality indexes (standardized z-scores) calculated in each partial correlation network.
MDD = major depressive disorder, PD = panic disorder, SAD = social anxiety disorder, OCD = obsessive-compulsive disorder. MDD1 = cognitive/affective symptoms, MDD2 = somatic symptoms, PD1 = physical concerns, PD2 = cognitive concerns, PD3 = social concerns, OCD1 = hoarding, OCD2 = checking, OCD3 = ordering, OCD4 = neutralizing, OCD5 = washing, OCD6 = obsessing.
Discussion
Clinical utility of GGMM-based clustering
In this study, we introduced GGMM-based clustering—developed by Fop et al. [33] in the mathematical statistics field—in the context of psychopathology research. Using an existing dataset of patients with depressive and anxiety disorders, our application demonstrated several lines of clinical utility of GGMM-based clustering. First, GGMM-based clustering can identify several transdiagnostic clusters (e.g., Cluster 1 with strong OCD and PD symptoms and moderate MDD and SAD symptoms; see the left panel of Fig 2). Second, GGMM-based clustering can visualize the symptom network in each cluster simultaneously with cluster identification. We believe that such a mixture of clustering and network approaches has the potential to advance transdiagnostic understanding of psychopathology. In fact, from the right panel of Fig 2, we can identify several network structure similarities and differences across the identified clusters (e.g., symptoms of OCD strongly interconnected in each cluster; MDD symptoms were relatively isolated in each cluster, except Cluster 3 with low levels of symptomatology). By capturing such network structure information, one can move beyond conventional clustering techniques that show only each cluster’s profile to detailed network descriptions for each cluster.
Third, and most importantly, GGMM-based clustering serves as a primer to further investigate cluster characteristics from a network perspective. Since GGMM-based clustering takes graph structures of variables into account in identifying clusters, one can safely conduct intensive network-based analyses that can potentially guide clinical hypotheses, including estimation of partial correlation networks and calculation of centrality indexes. In fact, we can elicit several clinical implications from such intensive analyses conducted in this study: we can i) assume that we need to carefully identify target symptoms when intervening patients categorized in Cluster 2 and 4 cases, which include both positive and negative edges (see Fig 3); ii) judge the neutralizing symptom of OCD (OCD4) with the highest strength as the target symptom in every identified cluster (see Fig 4); and iii) hypothesize that SAD symptoms, with nearly the highest bridge strength and expected influence, sustain the comorbid networks in every cluster (Fig 5). These clinical implications are useful for developing new transdiagnostic treatment for patients with each cluster. We believe that such intensive investigations cannot be adequately achieved with a simple combination of conventional clustering techniques and intensive network analyses, because it is theoretically inconsistent to impose network structures on the clusters that are identified without accounting for the interplays among items.
Statistical implications of GGMM-based clustering
The clinical utility of GGMM-based clustering was achieved by a statistical integration of clustering and network approaches. Such integration is relatively new in the network sciences, and most of the previous integration has been achieved to identify clusters of nodes within an estimated network [79, 80]. By contrast, the techniques to describe a network within each of the identified clusters are scarce in previous studies of psychopathology networks (for a review, see [81]), and the necessity of introducing mixture modeling with accounting for unobserved heterogeneity has been discussed [29]. The GGMM-based clustering introduced here fills such a blank area in network sciences and may lead to further development of statistical models and tools to investigate networks within clusters.
More broadly, GGMM-based clustering can be regarded as a successful integration of latent variable and network models, because the cluster is an exemplar of latent variables. Although the network approach is frequently discussed in contrast to the latent variables approach (e.g., [75, 82–84]), these approaches are mathematically equivalent (i.e., a latent variable model can be converted to a network model with the same free parameters [85–88]). These models, therefore, are open to integration. Some integrated models and analyses―including the residual network model [89] and the exploratory graph analysis used for estimating the dimensionality of psychological constructs [90]―have recently been developed. We hope that the GGMM-based clustering will accelerate such innovative integrations of latent variable and network models.
Limitations and future directions
GGMM-based clustering and the mixggm package [33] have several limitations that should be noted when developing future research directions. First, the current mixggm package visualizes only the structure of a network within each cluster and provides neither partial correlations between nodes nor centrality indexes, both of which have been widely investigated in cross-sectional analyses of psychopathology networks. To increase the clinical utility of GGMM-based clustering and promote clinical hypothesizing, future studies should either develop new packages of GGMM-based clustering or revise the analysis itself to enable partial correlation network and centrality index estimation. Developing such statistical tools would help users determine whether each edge is positive or negative and identify target and bridge symptoms. Second, in our study, GGMM-based clustering could be conducted not with individual symptom items but with symptom factor scores. We hope that future studies will identify why and how often estimation errors in GGMM-based clustering occur with single items and provide remedies for such estimation errors. Such studies will lead to further development of techniques to investigate networks within clusters and increase options for users.
There were also limitations regarding the application study design. First, because our data were collected through an online panel survey, our results might be affected by biases endemic in internet-based clinical research, such as inflation in reported symptoms [91], and participants’ carelessness or deliberate fraud in item response [92]. Second, the diagnostic labels included in the dataset were based on participants’ self-report on single items (e.g., “Are you currently diagnosed as having major depressive disorder and being treated for the problem in a medical setting?”) and, therefore, may be less accurate than those based on structured clinical interviews or well-validated screening tools. For a more accurate transdiagnostic understanding of depressive and anxiety disorders, future studies should use rigorous inclusion criteria to replicate the present application study. Third, our dataset included limited numbers of diagnostic labels (i.e., MDD, PD, SAD, OCD); therefore, our findings cannot be generalized to the entire network of depressive and anxiety disorders. To depict a full picture of comorbidity in depressive and anxiety disorders, future research should include all diagnostic labels for these two disorders, or curate empirical studies of network approaches that include some of the diagnostic labels from these two disorders. The latter approach may become possible in the near future, considering the increasing number of empirical studies investigating networks of: i) MDD and generalized anxiety disorder [93–98], ii) MDD and OCD [99], and iii) MDD and posttraumatic stress disorder [100].
Despite these limitations, our application study successfully demonstrated both the clinical utility and statistical implications of GGMM-based clustering. We hope that GGMM-based clustering will promote a better understanding of comorbid mental disorder structures and that further integrations of clustering and network approaches will be achieved in the future.
Acknowledgments
We would like to thank Editage by Cactus Communications Inc. (www.editage.com) for English language editing. Portions of the present study were presented as a poster at the 84th Annual Convention of the Japanese Psychological Association held online (September–November 2020).
Data Availability
The dataset is available via the Open Science Framework (OSF; https://osf.io/6jnf4/), along with the preregistered data analysis plan and the R script.
Funding Statement
JK, YT, YK, and MI were supported by Japan Society for the Promotion of Science KAKENHI (Grant numbers: 20K14171, 19K14419, 20K20870, and 17H04788, respectively,). MI was supported also by Japan Agency for Medical Research and Development Grant (Reference number: JP20dk0307084) and by National Center of Neurology and Psychiatry (NCNP) Intramural Research Grant (Reference number: 30-2). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kessler RC, Chiu WT, Demler O, Walters EE. Prevalence, severity, and comorbidity of 12-month DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiat. 2005; 62(6):617–27. doi: 10.1001/archpsyc.62.6.617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albert U, Rosso G, Maina G, Bogetto F. Impact of anxiety disorder comorbidity on quality of life in euthymic bipolar disorder patients: Differences between bipolar I and II subtypes. J Affect Disorders. 2008; 105(1–3):297–303. doi: 10.1016/j.jad.2007.05.020 [DOI] [PubMed] [Google Scholar]
- 3.Schoevers RA, Deeg DJH, van Tilburg W, Beekman ATF. Depression and generalized anxiety disorder—Co-occurrence and longitudinal patterns in elderly patients. Am J Geriat Psychiat. 2005; 13(1):31–9. doi: 10.1176/appi.ajgp.13.1.31 [DOI] [PubMed] [Google Scholar]
- 4.Bond DJ, Hadjipavlou G, Lam RW, McIntyre RS, Beaulieu S, Schaffer A, et al. The Canadian Network for Mood and Anxiety Treatments (CANMAT) task force recommendations for the management of patients with mood disorders and comorbid attention-deficit/hyperactivity disorder. Ann Clin Psychiatry. 2012; 24(1):23–37. [PubMed] [Google Scholar]
- 5.Ramasubbu R, Taylor VH, Saaman Z, Sockalingham S, Li M, Patten S, et al. The Canadian Network for Mood and Anxiety Treatments (CANMAT) task force recommendations for the management of patients with mood disorders and select comorbid medical conditions. Ann Clin Psychiatry. 2012; 24(1):91–109. [PubMed] [Google Scholar]
- 6.Schaffer A, McIntosh D, Goldstein BI, Rector NA, McIntyre RS, Beaulieu S, et al. The Canadian Network for Mood and Anxiety Treatments (CANMAT) task force recommendations for the management of patients with mood disorders and comorbid anxiety disorders. Ann Clin Psychiatry. 2012; 24(1):6–22. [PubMed] [Google Scholar]
- 7.Conway CC, Forbes MK, Forbush KT, Fried EI, Hallquist MN, Kotov R, et al. A hierarchical taxonomy of psychopathology can transform mental health research. Perspect Psychol Sci. 2019; 14(3):419–36. doi: 10.1177/1745691618810696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cramer AOJ, Waldorp LJ, van der Maas HLJ, Borsboom D. Comorbidity: A network perspective. Behav Brain Sci. 2010; 33(2–3):137–93. doi: 10.1017/S0140525X09991567 [DOI] [PubMed] [Google Scholar]
- 9.Gordon JA, Redish AD. On the cusp. Current challenges and promises in psychiatry. In: Redish AD, Gordon JA, editors. Computational psychiatry: New perspectives on mental illness. Cambridge, MA: MIT Press; 2016. pp. 3–14. [Google Scholar]
- 10.American Psychiatric Association. Diagnostic and statistical manual of mental disorders. 5th ed. Washington, DC: American Psychiatric Association; 2013. [Google Scholar]
- 11.World Health Organization. International classification of diseases for mortality and morbidity statistics. 11th Revision. World Health Organization. 2018. https://icd.who.int/browse11/l-m/en. [Google Scholar]
- 12.Clark LA, Cuthbert B, Lewis-Fernandez R, Narrow WE, Reed GM. Three approaches to understanding and classifying mental disorder: ICD-11, DSM-5, and the National Institute of Mental Health’s Research Domain Criteria (RDoC). Psychol Sci Public Interest. 2017; 18(2):72–145. doi: 10.1177/1529100617727266 [DOI] [PubMed] [Google Scholar]
- 13.Fried EI, Nesse RM. Depression is not a consistent syndrome: An investigation of unique symptom patterns in the STAR*D study. J Affect Disorders. 2015; 172:96–102. doi: 10.1016/j.jad.2014.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Markon KE, Chmielewski M, Miller CJ. The reliability and validity of discrete and continuous measures of psychopathology: A quantitative review. Psychol Bull. 2011; 137(5):856–79. doi: 10.1037/a0023678 [DOI] [PubMed] [Google Scholar]
- 15.Regier DA, Narrow WE, Clarke DE, Kraemer HC, Kuramoto SJ, Kuhl EA, et al. DSM-5 field trials in the United States and Canada, part II: Test-retest reliability of selected categorical diagnoses. Am J Psychiat. 2013; 170(1):59–70. doi: 10.1176/appi.ajp.2012.12070999 [DOI] [PubMed] [Google Scholar]
- 16.Fusar-Poli P, Solmi M, Brondino N, Davies C, Chae C, Politi P, et al. Transdiagnostic psychiatry: A systematic review. World Psychiatry. 2019; 18(2):192–207. doi: 10.1002/wps.20631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barlow DH, Farchione TJ, Bullis JR, Gallagher MW, Murray-Latin H, Sauer-Zavala S, et al. The unified protocol for transdiagnostic treatment of emotional disorders compared with diagnosis-specific protocols for anxiety disorders: A randomized clinical trial. JAMA Psychiatry. 2017; 74(9):875–84. doi: 10.1001/jamapsychiatry.2017.2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krueger RF, Kotov R, Watson D, Forbes MK, Eaton NR, Ruggero CJ, et al. Progress in achieving quantitative classification of psychopathology. World Psychiatry. 2018; 17(3):282–93. doi: 10.1002/wps.20566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lazarsfeld PF, Henry NW. Latent structure analysis. Boston, MA: Houghton Mifflin; 1968. [Google Scholar]
- 20.Lee CT, Leoutsakos JM, Lyketsos CG, Steffens DC, Breitner JCS, Norton MC. Latent class-derived subgroups of depressive symptoms in a community sample of older adults: The Cache County Study. Int J Geriatr Psych. 2012; 27(10):1061–9. doi: 10.1002/gps.2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Putnam K, Robertson-Blackmore E, Sharkey K, Payne J, Bergink V, Munk-Olsen T, et al. Heterogeneity of postpartum depression: A latent class analysis. Lancet Psychiatry. 2015; 2(1):59–67. doi: 10.1016/S2215-0366(14)00055-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robins RW, John OP, Caspi A. The typological approach to studying personality. In: Cairns RB, Bergman L, Kagan J, editors. Methods and models for studying the individual. Thousand Oaks, CA: Sage; 1998. pp. 135–160. [Google Scholar]
- 23.Contractor AA, Roley-Roberts ME, Lagdon S, Armour C. Heterogeneity in patterns of DSM-5 posttraumatic stress disorder and depression symptoms: Latent profile analyses. J Affect Disorders. 2017; 212:17–24. doi: 10.1016/j.jad.2017.01.029 [DOI] [PubMed] [Google Scholar]
- 24.Kircanski K, Zhang S, Stringaris A, Wiggins JL, Towbin KE, Pine DS, et al. Empirically derived patterns of psychiatric symptoms in youth: A latent profile analysis. J Affect Disorders. 2017; 216:109–116. doi: 10.1016/j.jad.2016.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borsboom D, Cramer AOJ. Network analysis: an integrative approach to the structure of psychopathology. Annu Rev Clin Psychol. 2013; 9(9):91–121. doi: 10.1146/annurev-clinpsy-050212-185608 [DOI] [PubMed] [Google Scholar]
- 26.Costantini G, Epskamp S, Borsboom D, Perugini M, Mottus R, Waldorp LJ, et al. State of the aRt personality research: A tutorial on network analysis of personality data in R. J Res Pers. 2015; 54:13–29. doi: 10.1016/j.jrp.2014.07.003 [DOI] [Google Scholar]
- 27.Epskamp S, Borsboom D, Fried EI. Estimating psychological networks and their accuracy: A tutorial paper. Behav Res Methods. 2018; 50(1):195–212. doi: 10.3758/s13428-017-0862-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Opsahl T, Agneessens F, Skvoretz J. Node centrality in weighted networks: Generalizing degree and shortest paths. Soc Networks. 2010; 32(3):245–51. doi: 10.1016/j.socnet.2010.03.006 [DOI] [Google Scholar]
- 29.Brusco MJ, Steinley D, Hoffman M, Davis-Stober C, Wasserman S. On Ising models and algorithms for the construction of symptom networks in psychopathological research. Psychol Methods. 2019; 24(6):735–53. doi: 10.1037/met0000207 [DOI] [PubMed] [Google Scholar]
- 30.Brusco MJ, Shireman E, Steinley D. A comparison of latent class, K-means, and K-median methods for clustering dichotomous data. Psychol Methods. 2017; 22(3):563–80. doi: 10.1037/met0000095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Köhn HF, Steinley D, Brusco MJ. The p-median model as a tool for clustering psychological data. Psychol Methods. 2010; 15(1):87–95. doi: 10.1037/a0018535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Borkulo CD, Borsboom D, Epskamp S, Blanken TF, Boschloo L, Schoevers RA, et al. A new method for constructing networks from binary data. Sci Rep. 2014; 4:5918. doi: 10.1038/srep05918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fop M, Murphy TB, Scrucca L. Model-based clustering with sparse covariance matrices. Stat Comput. 2019; 29(4):791–819. doi: 10.1007/s11222-018-9838-y [DOI] [Google Scholar]
- 34.Fraley C, Raftery AE. Model-based clustering, discriminant analysis, and density estimation. J Am Stat Assoc. 2002; 97(458):611–31. doi: 10.1198/016214502760047131 [DOI] [Google Scholar]
- 35.McNicholas PD. Model-based clustering. J Classif. 2016; 33(3):331–73. doi: 10.1007/s00357-016-9211-9 [DOI] [Google Scholar]
- 36.Lotsi A, Wit E. Sparse Gaussian graphical mixture model. Afrika Statistika. 2016; 11(2):1041–59. [Google Scholar]
- 37.Ward JH. Hierarchical grouping to optimize an objective function. J Am Stat Assoc. 1963; 58(301):236–44. doi: 10.2307/2282967 [DOI] [Google Scholar]
- 38.MacQueen J. Some methods for classification and analysis of multivariate observations. In: Cam LML, Neyman J, editors. Proceedings of the 5th Berkeley symposium on mathematical statistics and probability. Vol. 1. Berkeley, CA: University of California Press; 1967. pp. 281–297.
- 39.Koller D, Friedman N. Probabilistic graphical models: Principles and techniques. Cambridge, MA: MIT Press; 2009. [Google Scholar]
- 40.Schwartz G. Estimating the dimension of a model. Ann Stat. 1978; 6(2):461–4. doi: 10.1214/aos/1176344136 [DOI] [Google Scholar]
- 41.McLachlan G, Peel D. Finite Mixture Models. Wiley; 2000. [Google Scholar]
- 42.Zivkovic Z. Improved adaptive Gaussian mixture model for background subtraction. Paper presented at the 17th International Conference on Pattern Recognition (ICPR), British Machine Vis Assoc, Cambridge, England. 2004. https://ieeexplore.ieee.org/stamp/stamp.jsp?arnumber=1333992&casa_token=dabc20bZx98AAAAA:2trYXKCBtGAwDhBBsnVMKixYnjF-T35Z9tD-ML6IiFfUqOxZWBUX_oZEsTt-mpBflott8eyXK3TvXQ.
- 43.Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm. J R Stat Soc Series B. 1977; 39(1):1–38. doi: 10.1111/j.2517-6161.1977.tb01600.x [DOI] [Google Scholar]
- 44.Banfield JD, Raftery AE. Model-based Gaussian and non-Gaussian clustering. Biometrics. 1993; 49(3):803–21. doi: 10.2307/2532201 [DOI] [Google Scholar]
- 45.Celeux G, Govaert G. Gaussian parsimonious clustering models. Pattern Recognit. 1995; 28(5):781–93. doi: 10.1016/0031-3203(94)00125-6 [DOI] [Google Scholar]
- 46.McNicholas PD, Murphy TB. Parsimonious Gaussian mixture models. Stat Comput. 2008; 18(3):285–96. doi: 10.1007/s11222-008-9056-0 [DOI] [Google Scholar]
- 47.Epskamp S, Waldorp LJ, Mõttus R, Borsboom D. The Gaussian graphical model in cross-sectional and time-series data. Multivar Behav Res. 2018; 53(4):453–80. doi: 10.1080/00273171.2018.1454823 [DOI] [PubMed] [Google Scholar]
- 48.Lauritzen SL. Graphical Models. Oxford, UK: Clarendon Press; 1996. [Google Scholar]
- 49.Friedman J, Hastie T, Tibshirani R. Sparse inverse covariance estimation with the graphical lasso. Biostatistics. 2008; 9(3):432–41. doi: 10.1093/biostatistics/kxm045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Friedman N. Learning belief networks in the presence of missing values and hidden variables. In: Fisher D, editor. Proceedings of the Fourteenth International Conference on Machine Learning. Burlington, MA: Morgan Kaufmann; 1997. pp. 125–133.
- 51.Friedman N. The Bayesian structural EM algorithm. In: Cooper GF, Moral S, editors. Proceedings of the Fourteenth Conference on Uncertainty in Artificial Intelligence. Burlington, MA: Morgan Kaufmann; 1998. pp. 129–138.
- 52.Green PJ. On use of the EM for penalized likelihood estimation. J R Stat Soc Series B. 1990; 52(3):443–52. doi: 10.1111/j.2517-6161.1990.tb01798.x [DOI] [Google Scholar]
- 53.Mohan K, London P, Fazei M, Witten D, Lee SI. Node-based learning of multiple Gaussian graphical models. J Mach Learn Res. 2014; 15:445–88. [PMC free article] [PubMed] [Google Scholar]
- 54.Andrade L, Caraveo-Anduaga JJ, Berglund P, Bijl RV, De Graaf R, Vollebergh W, et al. The epidemiology of major depressive episodes: Results from the International Consortium of Psychiatric Epidemiology (ICPE) Surveys. Int J Methods Psychiatr Res. 2003; 12(1):3–21. doi: 10.1002/mpr.138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaufman J, Charney D. Comorbidity of mood and anxiety disorders. Depress Anxiety. 2000; 12:69–76. [DOI] [PubMed] [Google Scholar]
- 56.Lamers F, van Oppen P, Comijs HC, Smit JH, Spinhoven P, van Balkom A, et al. Comorbidity patterns of anxiety and depressive disorders in a large cohort study: The Netherlands Study of Depression and Anxiety (NESDA). J Clin Psychiatry. 2011; 72(3):341–8. doi: 10.4088/JCP.10m06176blu [DOI] [PubMed] [Google Scholar]
- 57.Doi S, Ito M, Takebayashi Y, Muramatsu K, Horikoshi M. Factorial validity and invariance of the 7-item Generalized Anxiety Disorder Scale (GAD-7) among populations with and without self-reported psychiatric diagnostic status. Front Psychol. 2018; 9:1741. doi: 10.3389/fpsyg.2018.01741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fujisato H, Ito M, Takebayashi Y, Hosogoshi H, Kato N, Nakajima S, et al. Reliability and validity of the Japanese version of the Emotion Regulation Skills Questionnaire. J Affect Disorders. 2017; 208:145–52. doi: 10.1016/j.jad.2016.08.064 [DOI] [PubMed] [Google Scholar]
- 59.Ito M, Bentley KH, Oe Y, Nakajima S, Fujisato H, Kato N, et al. Assessing depression related severity and functional impairment: The Overall Depression Severity and Impairment Scale (ODSIS). PLOS ONE. 2015; 10(4):e0122969. doi: 10.1371/journal.pone.0122969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ito M, Oe Y, Kato N, Nakajima S, Fujisato H, Miyamae M, et al. Validity and clinical interpretability of Overall Anxiety Severity and Impairment Scale (OASIS). J Affect Disorders. 2015; 170:217–24. doi: 10.1016/j.jad.2014.08.045 [DOI] [PubMed] [Google Scholar]
- 61.Muramatsu K, Miyaoka H, Kamijima K, Muramatsu Y, Yoshida M, Otsubo T, et al. The Patient Health Questionnaire, Japanese version: Validity according to the mini-international neuropsychiatric interview-plus. Psychol Rep. 2007; 101(3):952–60. doi: 10.2466/pr0.101.3.952-960 [DOI] [PubMed] [Google Scholar]
- 62.Kroenke K, Spitzer RL. The PHQ-9: A new depression diagnostic and severity measure. Psychiatric Annals. 2002; 32(9):509–15. doi: 10.3928/0048-5713-20020901-06 [DOI] [Google Scholar]
- 63.Taylor S, Zvolensky MJ, Cox BJ, Deacon B, Heimberg RG, Ledley DR, et al. Robust dimensions of anxiety sensitivity: Development and initial validation of the Anxiety Sensitivity Index-3. Psychol Assess. 2007; 19(2):176–88. doi: 10.1037/1040-3590.19.2.176 [DOI] [PubMed] [Google Scholar]
- 64.McNally RJ. Anxiety sensitivity and panic disorder. Biol Psychiatry. 2002; 52(10):938–46. doi: 10.1016/s0006-3223(02)01475-0 [DOI] [PubMed] [Google Scholar]
- 65.Reiss S, McNally R. (1985). Expectancy model of fear. In Reiss S. & Bootzin R. R. (Eds.), Theoretical issues in behavior therapy (pp. 107–122). New York: Academic. [Google Scholar]
- 66.Ebesutani C, McLeish AC, Luberto CM, Young J, Maack DJ. A bifactor model of anxiety sensitivity: Analysis of the Anxiety Sensitivity Index-3. J Psychopathol Behav Assess. 2014; 36(3):452–64. doi: 10.1007/s10862-013-9400-3 [DOI] [Google Scholar]
- 67.Sasagawa S, Kanai Y, Muranaka Y, Suzuki S, Shimada H, Sakano Y. [Development of a Short Fear of Negative Evaluation Scale for Japanese using item response theory]. Japanese Journal of Behavior Therapy. 2004; 30(2):87–98. Japanese. doi: 10.24468/jjbt.30.2_87 (in Japanese with English abstract). [DOI] [Google Scholar]
- 68.Leary MR. A brief version of the Fear of Negative Evaluation Scale. Pers Soc Psychol Bull. 1983; 9(3):371–5. doi: 10.1177/0146167283093007 [DOI] [Google Scholar]
- 69.Sugiura Y, Sugiura T, Tanno Y. [Reliability and validity of the Japanese version of the Frost Indecisiveness Scale]. Studies in Humanities and Human Sciences. 2007; 41:79–89. Japanese. [Google Scholar]
- 70.Foa EB, Huppert JD, Leiberg S, Langner R, Kichic R, Hajcak G, et al. The Obsessive-Compulsive Inventory: Development and validation of a short version. Psychol Assess. 2002; 14(4):485–96. doi: 10.1037//1040-3590.14.4.485 [DOI] [PubMed] [Google Scholar]
- 71.Koike H, Tsuchiyagaito A, Hirano Y, Oshima F, Asano K, Sugiura Y, et al. Reliability and validity of the Japanese version of the Obsessive-Compulsive Inventory-Revised (OCI-R). Curr Psychol. 2020; 39:89–95. doi: 10.1007/s12144-017-9741-2 [DOI] [Google Scholar]
- 72.Epskamp S, Cramer AOJ, Waldorp LJ, Schmittmann VD, Borsboom D. qgraph: Network visualizations of relationships in psychometric data. J Stat Softw. 2012: 48:1–18. [Google Scholar]
- 73.Chen JH, Chen ZH. Extended Bayesian information criteria for model selection with large model spaces. Biometrika. 2008; 95(3):759–71. doi: 10.1093/biomet/asn034 [DOI] [Google Scholar]
- 74.Jones P. networktools: Tools for identifying important nodes in networks. R package version 1.2.1. The Comprehensive R Archive Network. 2019. https://CRAN.R-project.org/package=networktools.
- 75.Borsboom D, Cramer AOJ, Schmittmann VD, Epskamp S, Waldorp LJ. The small world of psychopathology. PLOS ONE. 2011; 6(11):e27407. doi: 10.1371/journal.pone.0027407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McNally RJ, Robinaugh DJ, Wu GWY, Wang L, Deserno MK, Borsboom D. Mental disorders as causal systems: A network approach to posttraumatic stress disorder. Clin Psychol Sci. 2015; 3(6):836–49. doi: 10.1177/2167702614553230 [DOI] [Google Scholar]
- 77.van Borkulo C, Boschloo L, Borsboom D, Penninx B, Waldorp LJ, Schoevers RA. Association of symptom network structure with the course of longitudinal depression. JAMA Psychiatry. 2015; 72(12):1219–26. doi: 10.1001/jamapsychiatry.2015.2079 [DOI] [PubMed] [Google Scholar]
- 78.Bringmann LF, Elmer T, Epskamp S, Krause RW, Schoch D, Wichers M, et al. What do centrality measures measure in psychological networks? J Abnorm Psychol. 2019; 128(8):892–903. doi: 10.1037/abn0000446 [DOI] [PubMed] [Google Scholar]
- 79.Krivitsky P, Handcock M. latentnet: Latent position and cluster models for statistical networks. R package version 2.9.0. The Comprehensive R Archive Network. 2018. https://CRAN.R-project.org/package=latentnet>.
- 80.Krivitsky PN, Handcock MS, Raftery AE, Hoff PD. Representing degree distributions, clustering, and homophily in social networks with latent cluster random effects models. Soc Networks. 2009; 31(3):204–13. doi: 10.1016/j.socnet.2009.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Robinaugh DJ, Hoekstra RHA, Toner ER, Borsboom D. The network approach to psychopathology: A review of the literature 2008–2018 and an agenda for future research. Psychol Med. 2019; 50(3):353–66. doi: 10.1017/S0033291719003404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Borsboom D. Psychometric perspectives on diagnostic systems. J Clin Psychol. 2008; 64(9):1089–108. doi: 10.1002/jclp.20503 [DOI] [PubMed] [Google Scholar]
- 83.Cramer AOJ, Borsboom D, Aggen SH, Kendler KS. The pathoplasticity of dysphoric episodes: Differential impact of stressful life events on the pattern of depressive symptom inter-correlations. Psychol Med. 2012; 42(5):957–65. doi: 10.1017/S003329171100211X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jones PJ, Heeren A, McNally RJ. Commentary: A network theory of mental disorders. Front Psychol. 2017; 8:1305. doi: 10.3389/fpsyg.2017.01305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bringmann LF, Eronen MI. Don’t blame the model: Reconsidering the network approach to psychopathology. Psychol Rev. 2018; 125(4):606–15. doi: 10.1037/rev0000108 [DOI] [PubMed] [Google Scholar]
- 86.Epskamp S, Fried EI, Borkulo C, Robinaugh DJ, Marsman M, Dalege J, et al. Investigating the utility of fixed-margin sampling in network psychometrics. Multivar Behav Res. 2018; Advance online publication. doi: 10.1080/00273171.2018.1489771 [DOI] [PubMed] [Google Scholar]
- 87.Kruis J, Maris G. Three representations of the Ising model. Sci Rep. 2016; 6:34175. doi: 10.1038/srep34175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.van der Maas HLJ, Dolan CV, Grasman R, Wicherts JM, Huizenga HM, Raijmakers MEJ. A dynamical model of general intelligence: The positive manifold of intelligence by mutualism. Psychol Rev. 2006; 113(4):842–61. doi: 10.1037/0033-295X.113.4.842 [DOI] [PubMed] [Google Scholar]
- 89.Epskamp S, Rhemtulla M, Borsboom D. Generalized network psychometrics: combining network and latent variable models. Psychometrika. 2017; 82(4):904–27. doi: 10.1007/s11336-017-9557-x [DOI] [PubMed] [Google Scholar]
- 90.Golino HF, Epskamp S. Exploratory graph analysis: A new approach for estimating the number of dimensions in psychological research. PLOS ONE. 2017; 12(6):e0174035. doi: 10.1371/journal.pone.0174035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ophir Y, Sisso I, Asterhan CSC, Tikochinski R, Reichart R. The Turker blues: Hidden factors behind increased depression rates among Amazon’s Mechanical Turkers. Clin Psychol Sci. 2020; 8(1):65–83. doi: 10.1177/2167702619865973 [DOI] [Google Scholar]
- 92.Chandler J, Sisso I, Shapiro D. Participant carelessness and fraud: Consequences for clinical research and potential solutions. J Abnorm Psychol. 2020; 129(1):49–55. doi: 10.1037/abn0000479 [DOI] [PubMed] [Google Scholar]
- 93.Beard C, Millner AJ, Forgeard MJC, Fried EI, Hsu KJ, Treadway MT, et al. Network analysis of depression and anxiety symptom relationships in a psychiatric sample. Psychol Med. 2016; 46(16):3359–69. doi: 10.1017/S0033291716002300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bekhuis E, Schoevers RA, van Borkulo CD, Rosmalen JGM, Boschloo L. The network structure of major depressive disorder, generalized anxiety disorder and somatic symptomatology. Psychol Med. 2016; 46(14):2989–98. doi: 10.1017/S0033291716001550 [DOI] [PubMed] [Google Scholar]
- 95.Curtiss J, Klemanski DH. Taxonicity and network structure of generalized anxiety disorder and major depressive disorder: An admixture analysis and complex network analysis. J Affect Disorders. 2016; 199:99–105. doi: 10.1016/j.jad.2016.04.007 [DOI] [PubMed] [Google Scholar]
- 96.Everaert J, Joormann J. Emotion regulation difficulties related to depression and anxiety: A network approach to model relations among symptoms, positive reappraisal, and repetitive negative thinking. Clin Psychol Sci. 2019; 7(6):1304–18. doi: 10.1177/2167702619859342 [DOI] [Google Scholar]
- 97.Fisher AJ, Reeves JW, Lawyer G, Medaglia JD, Rubel JA. Exploring the idiographic dynamics of mood and anxiety via network analysis. J Abnorm Psychol. 2017; 126(8):1044–56. doi: 10.1037/abn0000311 [DOI] [PubMed] [Google Scholar]
- 98.Garabiles MR, Lao CK, Xiong YX, Hall BJ. Exploring comorbidity between anxiety and depression among migrant Filipino domestic workers: A network approach. J Affect Disorders. 2019; 250:85–93. doi: 10.1016/j.jad.2019.02.062 [DOI] [PubMed] [Google Scholar]
- 99.Afzali MH, Sunderland M, Teesson M, Carragher N, Mills K, Slade T. A network approach to the comorbidity between posttraumatic stress disorder and major depressive disorder: The role of overlapping symptoms. J Affect Disorders. 2017; 208:490–96. doi: 10.1016/j.jad.2016.10.037 [DOI] [PubMed] [Google Scholar]
- 100.McNally RJ, Mair P, Mugno BL, Riemann BC. Co-morbid obsessive-compulsive disorder and depression: A Bayesian network approach. Psychol Med. 2017; 47(7):1204–14. [DOI] [PubMed] [Google Scholar]