Abstract
Using PCR-coupled subtractive screening-representational difference analysis, we have cloned a novel gene from AML1-ETO knockin mice. This gene is highly expressed in the yolk sac and fetal liver of the knockin mice. Nucleotide sequence analysis indicates that its cDNA contains an 1,107-bp open reading frame encoding a 368-amino-acid polypeptide. Further protein sequence and protein translation analysis shows that it belongs to a family of ubiquitin-specific proteases (UBP), and its molecular mass is 43 kDa. Therefore, we have named this gene UBP43. Like other ubiquitin proteases, the UBP43 protein has deubiquitinating enzyme activity. Protein ubiquitination has been implicated in many important cellular events. In wild-type adult mice, UBP43 is highly expressed in the thymus and in peritoneal macrophages. Among nine different murine hematopoietic cell lines analyzed, UBP43 expression is detectable only in cell lines related to the monocytic lineage. Furthermore, its expression is regulated during cytokine-induced monocytic cell differentiation. We have investigated its function in the hematopoietic myeloid cell line M1. UBP43 was introduced into M1 cells by retroviral gene transfer, and several high-expressing UBP43 clones were obtained for further study. Morphologic and cell surface marker examination of UBP43/M1 cells reveals that overexpression of UBP43 blocks cytokine-induced terminal differentiation of monocytic cells. These data suggest that UBP43 plays an important role in hematopoiesis by modulating either the ubiquitin-dependent proteolytic pathway or the ubiquitination state of another regulatory factor(s) during myeloid cell differentiation.
t(8;21) is associated with 15% of de novo acute myelogenous leukemia cases and creates the AML1-ETO fusion protein. AML1-ETO knockin mice have been generated to study the effect of the leukemia-associated fusion protein AML1-ETO on hematopoiesis and the pathogenesis of leukemia (34, 53). AML1-ETO/+ embryos died between 11.5 and 13.5 days postcoitus (d.p.c.) and exhibited severely disrupted hematopoiesis and hemorrhages associated with the central nervous system. This phenotype is very similar to that resulting from homozygous disruption of the AML1 gene (35, 49), suggesting that AML1-ETO dominantly blocks normal AML1 function during embryonic development. In contrast to AML1 knockout mice, which do not have any definitive hematopoiesis in the yolk sac, cells isolated from the yolk sacs of AML1-ETO/+ 11.5-d.p.c. embryos did give rise to macrophage-like colonies in vitro (53), and cells isolated from the liver of AML1-ETO/+ 12.5- to 13.5-d.p.c. embryos showed dysplastic hematopoiesis (34). These results suggest that AML1-ETO expression may have other effects on hematopoiesis besides blocking wild-type AML1.
In this study, we employed representational difference analysis (RDA) to identify genes differentially expressed in AML1-ETO knockin mice and have isolated a novel gene, which belongs to a family of ubiquitin-specific proteases (UBPs).
Ubiquitin is a 76-amino-acid (aa) polypeptide with a molecular mass of 8.5 kDa that was first isolated in 1975 from bovine thymus (13). Ubiquitin is present in all eukaryotic cells and is one of the most highly conserved proteins from humans to insects. Ubiquitin occurs in cells either in the free form or covalently coupled via its carboxyl-terminal glycine residue to the ɛ-amino groups of lysine residues in a wide variety of intracellular proteins or of Lys48 of another ubiquitin (1, 17). Ubiquitination is an important protein modification. Two major groups of enzymes, ubiquitin-conjugating enzymes and ubiquitin-specific proteases (deubiquitinating enzymes), regulate the balance of protein ubiquitination (20). Protein ubiquitination is important in a variety of cellular events, one of which is ubiquitin-dependent proteolysis. Proteolysis regulated by the ubiquitin pathway has been implicated in control of the cell cycle (25), transcription activation (48), and antigen presentation (41), as well as in cell fate and growth (22, 54). In ubiquitin-associated proteolysis, ubiquitin targets many critical proteins, including p53 (5, 43), cyclins (12, 52), and the yeast MATα2 repressor (2), for proteolytic degradation by the proteasome. In addition to its important role in proteasome-mediated proteolysis, protein ubiquitination has other regulatory functions. It has been shown that histone ubiquitination is increased in actively transcribing chromatin and transformed cells (7, 29, 47). Ubiquitination also plays a critical role in endocytosis of cell surface receptors (19) and in activation of IκBα kinase (3).
There are two distinct families of deubiquitinating enzymes—ubiquitin carboxyl-terminal hydrolases (UCHs) and ubiquitin-processing proteases (UBPs). The enzymes of the UCH family are small proteins that cleave ubiquitin from its relatively short C-terminal extensions but that are virtually inactive with larger protein substrates (27). The UBP enzymes are a group of larger proteins that can cleave ubiquitin from a wide range of protein substrates. While this family of enzymes shows little amino acid sequence similarity, there are short consensus sequences surrounding cysteine residues (Cys domain) and histidine residues (His domain) that are highly conserved. These conserved domains are important in generating the active site of the enzyme (6, 20). The high degree of divergence in this large family of proteins implies that different UBP family members may have unique biochemical properties, substrate specificities, and cellular localizations. So far, little is known about the function of protein ubiquitination in hematopoiesis. Here we report the cloning of UBP43, a novel member of the ubiquitin-specific proteases, from AML1-ETO knockin mice. UBP43 encodes a functional deubiquitinating enzyme. Its expression is upregulated by AML1-ETO. The expression pattern of UBP43 in normal adult mice and in hematopoietic cell lines suggests that UBP43 may be involved in hematopoiesis. Furthermore, the block of monocytic cell differentiation by constitutive expression of UBP43 suggests an important role for this gene in hematopoiesis.
MATERIALS AND METHODS
RDA of cDNA and clone of UBP43 cDNA.
RDA of cDNA was performed based on a protocol by Hubank and Schatz (23). Generation of AML1-ETO knockin chimeric mice and production of the germ line transmitted AML1-ETO knockin embryos have been reported previously (53). Total RNA was isolated from yolk sacs of both AML1-ETO knockin embryos and their wild-type littermates at 11.5 d.p.c. by guanidine isothiocyanate extraction and CsCl gradient purification (4). Poly(A)+ RNA was purified from total RNA with oligo(dT) cellulose columns (New England Biolabs, Beverly, Mass.). cDNA was synthesized from 5 to 10 μg of poly(A)+ RNA by using a cDNA synthesis kit according to the manufacturer’s instructions (GIBCO-BRL, Grand Island, N.Y.). cDNA prepared from yolk sacs of AML1-ETO knockin embryos was used as a tester, and cDNA prepared from control yolk sacs was used as a driver in RDA. A partial cDNA fragment isolated by RDA was used as a probe to obtain full-length UBP43 cDNA from a mouse thymus cDNA library. UBP43 cDNA was sequenced on both strands by the dideoxy DNA sequencing method (U.S. Biochemicals, Cleveland, Ohio). The cDNA sequence was confirmed by sequencing another UBP43 clone isolated from a murine macrophage library.
Assay for ubiquitin-specific protease activity.
The assay for a ubiquitin-specific protease to deubiquitinate a ubiquitin–β-galactosidase fusion protein has been previously described (38, 55). A plasmid expressing the glutathione S-transferase (GST)–UBP43 fusion protein was constructed by in-frame linkage of a murine UBP43 cDNA to plasmid pGEX-4T-3 (pBR322 Ampr replicon). Plasmid pAC-M-β-gal expresses the Ub-Met-β-gal fusion protein substrate in a pACYC184 Cmr replicon. Escherichia coli BL21 (DE3) bacteria harboring pGEX-4T-3-UBP43 were transformed with pAC-M-β-gal, Ampr Cmr colonies were grown and induced with IPTG (isopropyl-β-d-thiogalactopyranoside), and total protein extracts were analyzed by Western blotting with anti-β-galactosidase rabbit polyclonal antibody (Cappel, Aurora, Ohio) and by the enhanced chemiluminescence system (Amersham, Little Chalfont, Buckinghamshire, England).
Northern blot analysis.
Total RNA was prepared from different cell lines and mouse tissues by the guanidinium isothiocyanate extraction method followed by cesium chloride gradient purification. RNA (10 μg/lane) was denatured in formamide-formaldehyde, followed by electrophoresis in 1% agarose-formaldehyde gels. The RNA was then transferred to a positively charged nylon membrane (ICN Biomedicals, Inc., Costa Mesa, Calif.). The cDNA inserts, purified from low-melting-point agarose gels, were radiolabeled by the random priming method and hybridized with membranes in Church-Gilbert hybridization buffer (7% sodium dodecyl sulfate [SDS] and 1% bovine serum albumin in 0.5 M NaPO4, pH 7.2) for at least 18 h at 65°C. The hybridized membranes were washed in 1× SSC (0.15 M sodium chloride–0.015 M sodium citrate, pH 7.0) and 0.2% SDS at room temperature and then in 0.2× SSC and 0.1% SDS at 65°C. Autoradiography was performed with Kodak XAR-5 film at −80°C.
In situ hybridization.
Embryos were dissected from the peritoneum at 11.5 d.p.c., immediately placed in freshly prepared ice-cold 4% paraformaldehyde in phosphate-buffered saline and fixed overnight at 4°C. Then, the embryos were dehydrated through ethanol into xylene, embedded in paraffin with a Tissue-Tek V.I.P. automatic processor, and sectioned. The sections were dewaxed, rehydrated, and treated with proteinase K to enhance probe accessibility and with acetic anhydride to reduce nonspecific background. Single-stranded 33P-labeled antisense RNA probes were prepared by standard techniques (30) with specific activities of 5 × 108 dpm/μg and hydrolyzed by alkaline treatment to approximately 200 bp. The sense probe was synthesized to the same specific activity as the antisense probe and served as a control for nonspecific background. The sections were hybridized with 33P-labeled UBP43 antisense and sense riboprobes, as described previously (37). Multiple embryos and exposures of different lengths were examined for each sense and antisense probe. Autoradiography with NTB-2 emulsion (Eastman Kodak, Rochester, N.Y.) was performed for 7 to 28 days. Slides were developed in D19 (Eastman Kodak), and the tissues were counterstained with toluidine blue.
Cell culture and differentiation.
Murine monocytic cell lines (RAW264.7 and M1), T-cell lines (BW5147 and EL4), B-cell lines (Ba/F3 and 18.8), erythroid cell lines (MEL and 416B), and a granulocytic cell line (32Dc13) were routinely grown in RPMI 1640 medium (BioWhittaker, Inc., Walkersville, Md.) containing 10% fetal bovine serum (GIBCO-BRL) and 2 mM l-glutamine (GIBCO-BRL). In addition, 10% WEHI3 conditioned medium (source of interleukin 3 [IL-3]) was added to the culture medium for Ba/F3 and 32Dc13 cells. NIH 3T3 cells and 293T cells were grown in Dulbecco’s modified Eagle medium (BioWhittaker, Inc.) containing 10% fetal bovine serum and 2 mM l-glutamine. All cells were cultured at 37°C under 5% CO2. To induce macrophage differentiation, M1 cells were cultured in the presence of recombinant human IL-6 (Amgen, Inc., Thousand Oaks, Calif.) at 100 ng/ml or leukemia inhibitory factor (LIF) (GIBCO-BRL) at 50 ng/ml. To induce granulocyte differentiation, 32Dc13 cells were cultured in medium containing 500 units of granulocyte colony-stimulating factor (G-CSF) (Amgen, Inc.) in the absence of IL-3. To check cell morphology, 104 cells were cytocentrifuged onto each slide, stained with Wright-Giemsa solution, and examined under a microscope.
Retrovirus production and infection.
The 1.4-kb EcoRI-DraI murine UBP43 cDNA fragment was inserted into the MSCVneo retroviral vector at EcoRI/HpaI sites to form UBP43/MSCVneo (16). This vector contains the Moloney murine leukemia virus long terminal repeat to control expression of the inserted cDNA and the simian virus 40 promoter to control expression of the selectable marker neomycin phosphotransferase (neo) (32). Viruses were generated from the recombinant retroviral vector (MSCVneo) or from the UBP43 expression construct UBP43/MSCVneo, as described previously (32). Briefly, 10 μg of UBP43/MSCVneo or MSCVneo plasmid and 2 μg of MSCVEcoPac packaging construct were cotransfected by the Ca3(PO4)2 precipitation method into 293T cells. Forty-eight hours after transfection, the cell culture medium containing the produced viruses was collected and centrifuged at 800 × g for 10 min. The supernatant containing retroviruses was passed through a 0.2-μm filter (Millipore, Bedford, Mass.), aliquoted, and stored at −80°C. To infect cells, 107 CFU of neo retrovirus per ml were incubated with 107 M1 cells in the presence of polybrene (8 μg/ml) (Sigma, St. Louis, Mo.) at 37°C for 12 h. Fresh medium was then applied and the cells allowed recovering. Two days later, G418 (GIBCO-BRL) was added to the medium at 0.2 mg/ml for selection of positively infected cells.
Nucleotide sequence accession number.
The DNA sequence of murine UBP43 has been submitted to the GenBank database under accession no. AF069502.
RESULTS
UBP43, a novel member of the ubiquitin-specific protease family, was cloned from the yolk sac of AML1-ETO knockin mice.
As reported previously, AML1-ETO knockin mice were generated to evaluate the role of AML1-ETO in hematopoiesis (53). To identify genes whose expression is either directly or indirectly affected by AML1-ETO expression, we studied the change of gene expression in the yolk sac of knockin embryos by using PCR-coupled subtractive screening-RDA (23). Total RNA was prepared from the yolk sacs of both AML1-ETO knockin embryos and their wild-type littermates. Reverse transcription reactions were then performed to generate cDNAs. To identify genes whose expression is upregulated by AML1-ETO expression, cDNA from knockin mice was used as the tester and cDNA from wild-type mice was used as the driver. After three rounds of subtractive hybridization between tester cDNA and an excess amount of driver cDNA, several cDNA fragments were obtained. Based on sequence analysis, one of the cDNA fragments was from an unknown gene. Subsequently, a 1,748-bp cDNA was isolated from a cDNA library by using this fragment as a DNA probe. This cDNA contained an 1,107-bp open reading frame encoding a polypeptide of 368 aa (Fig. 1). Comparison of the nucleotide sequence of this gene to entries in the GenBank database revealed that it had no significant similarity to any previously sequenced genes. However, the predicted amino acid sequence revealed a 20 to 25% similarity to members of the ubiquitin-specific protease (UBP) family, including the yeast proteins Ubp8, Ubp15 (20), and Doa4 (38); human Tre-2 (36); and the mouse proteins Dub-1 (54) and Unp (15). The sequence similarity was largely restricted to six conserved domains recently identified in all UBP family members (51), including the Cys box, a QQDAQEF motif, a region with the consensus sequence LPQILVIHLKRF, and three blocks from the His box region (Fig. 2). The sequence alignments of the Cys box and the His box between UBP43 and other indicated family members gave the highest similarity. The putative active-site nucleophile is a cysteine residue in the Cys box (38), found in all known family members as well as in UBP43 (Cys61). Beyond the six conserved regions, UBP43 has little homology to any known UBP43 family members or non-UBP proteins. Since the molecular mass calculated from the predicted 368 aa is approximately 43 kDa, as is the mass of its in vitro-translated product (data not shown), we have named this novel member of the UBP family UBP43.
FIG. 1.
Sequence of murine UBP43. The nucleotide sequence was obtained from both strands by the dideoxy method. The polyadenylation signal AATAAA is underlined. The deduced amino acid sequence is shown below the nucleotide sequence.
FIG. 2.
Sequence alignments of conserved domains between UBP43 and selected members of the UBP family. Sequences conserved among these proteins are shaded. The GenBank accession numbers of these proteins are as follows: Dub-1, U41636; Unp, L00681; Tre-2, X63547; Faf, P55824; Ubp8, P50102; Ubp15, P50101.
UBP43 encodes a functional ubiquitin-specific protease.
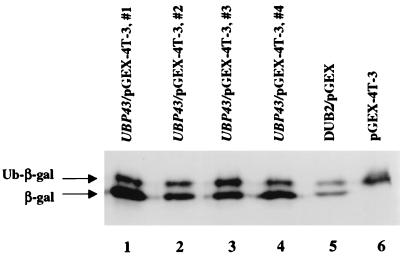
In order to determine whether UBP43 has ubiquitin-specific protease enzymatic activity, we expressed UBP43 as a GST fusion protein in the expression vector pGEX-4T-3. pGEX-4T-3-UBP43 was cotransformed into E. coli BL21 (DE3) with a plasmid expressing the protein Ub-Met-β-gal, in which ubiquitin is fused to the N terminus of β-galactosidase. As shown by immunoblot analysis (Fig. 3), four independent clones expressing the GST-UBP43 fusion protein demonstrated efficient cleavage of Ub-Met-β-gal (lanes 1, 2, 3, and 4), comparable to that observed with DUB-2, a known murine deubiquitinating enzyme (55) (lane 5). As expected, cells with the pGEX-4T-3 vector (lane 6) failed to cleave Ub-Met-β-gal. Therefore, the new member of deubiquitinating enzyme UBP family, UBP43, has deubiquitinating enzyme activity.
FIG. 3.
UBP43 encodes a functional deubiquitinating enzyme. A ubiquitin–β-galactosidase (Ub-Met-β-gal) fusion protein expressed in bacteria was deubiquitinated. Shown is a Western blot with anti-β-galactosidase antiserum. Coexpressed plasmids were pGEX-4T-3-UBP43 (lanes 1 to 4), the positive control pGEX-DUB-2 (lane 5), and the negative control pGEX-4T-3 (vector) (lane 6).
UBP43 is highly expressed in the fetal hematopoietic organs of AML1-ETO knockin embryos but not in AML1 knockout embryos.
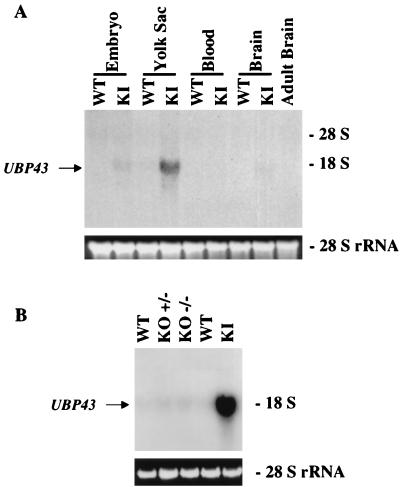
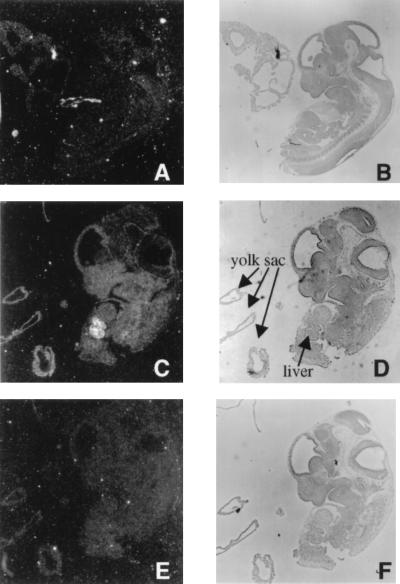
UBP43 was initially cloned by subtractive hybridization screening of RNA from the yolk sacs of wild-type and AML1-ETO knockin embryos. To further study the biological function of UBP43, the pattern of its expression was compared between knockin embryos and wild-type embryos. Total RNA was isolated from 11.5-d.p.c. whole embryo, yolk sac, blood, and brain of knockin and wild-type mice and analyzed by Northern blotting. UBP43 was highly expressed in the yolk sacs of AML1-ETO knockin embryos compared to those of wild-type embryos (Fig. 4A). A trace amount of UBP43 expression was also detected in wild-type yolk sacs. Furthermore, UBP43 expression was also detected in whole embryos and brain RNA of AML1-ETO knockin mice, although the level was much lower than the expression level seen in the yolk sacs of knockin mice. No UBP43 mRNA was detected in the blood of either knockin or wild-type mice or in whole embryo proper and brain tissue of wild-type mice. These results demonstrate that UBP43 is abnormally upregulated in AML1-ETO-expressing mice, particularly in the yolk sac. Since the major phenotypes of AML1-ETO knockin and AML1 knockout mice are similar, UBP43 expression was also studied with RNA from yolk sacs of 11.5-d.p.c. AML1 knockout embryos. No upregulation of UBP43 was detected (Fig. 4B). To further study UBP43 expression in AML1-ETO knockin mice, in situ hybridization analysis was performed to examine the spatial pattern of UBP43 expression (Fig. 5). Both antisense strand and sense strand riboprobes were used in the analysis. UBP43 was highly expressed in the liver and yolk sac of 11.5-d.p.c. AML1-ETO knockin embryos, with highest expression in the fetal liver. There was also a lesser degree of UBP43 transcript accumulation in much of the embryo proper, with differential accumulation in the central nervous system. A more extensive temporal and spatial analysis revealed no detectable UBP43 mRNA expression in 7.5-, 8.5-, 11.5-, and 12.5-d.p.c. wild-type embryos (Fig. 5 and data not shown). Taken together, these results demonstrate that UBP43 expression is upregulated in midgestation AML1-ETO knockin mice, particularly in the two fetal hematopoietic organs, yolk sac and liver.
FIG. 4.
UBP43 is highly expressed in the yolk sac of AML1-ETO knockin mice. (A) Total RNA prepared from various tissues of both wild-type (WT) and AML1-ETO knockin (KI) embryos at 11.5 d.p.c.; (B) total RNA prepared from the yolk sac of WT, heterozygous (+/−), and homozygous (−/−) AML1 knockout (KO) and AML1-ETO KI embryos at 11.5 d.p.c. RNA samples (10 μg/lane) were subjected to Northern blot analysis with 32P-labeled UBP43 cDNA. Ethidium bromide-stained 28S rRNA is shown to demonstrate equivalent RNA loading.
FIG. 5.
In situ hybridization analysis demonstrates high-level UBP43 expression in two fetal hematopoietic organs, liver and yolk sac, of AML1-ETO knockin mice. Both AML1-ETO knockin and wild-type embryos at 11.5 d.p.c. were subjected to in situ hybridization analysis with 33P-labeled UBP43 antisense and sense riboprobes. Shown are dark-field (left) and light-field (right) views of wild-type embryo hybridized to UBP43 antisense probe (A and B), AML1-ETO knockin embryo hybridized to UBP43 antisense probe (C and D), and AML1-ETO knockin embryo hybridized to UBP43 sense probe (E and F).
Expression of UBP43 is restricted to monocytic cell lines and several tissues.
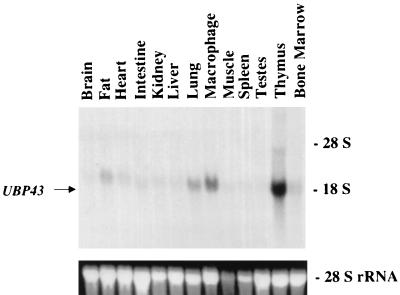
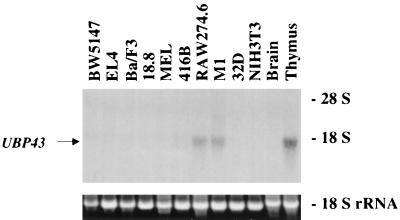
As shown in Fig. 4 and 5, UBP43 was expressed at extremely low levels during normal embryogenesis. However, UBP43 was highly expressed in the two fetal hematopoietic organs of AML1-ETO knockin mice that lack normal hematopoiesis. To further characterize the potential function of UBP43, we first analyzed its expression in various tissues of wild-type adult mice. When total cellular RNA was used in Northern blot hybridization, the highest level of UBP43 expression was detected in the thymus (Fig. 6). UBP43 was also highly expressed in peritoneal macrophages collected from intraperitoneal thioglycolate-treated mice. A low, but clearly detectable, level of UBP43 expression was also seen in the bone marrow, as well as in adipose tissue and lung. We also examined UBP43 expression in a variety of hematopoietic cell lines of different lineages. These included T-cell lines (BW5147 and EL4), B-cell lines (Ba/F3 and 18.8), an erythroid cell line (MEL), an early myeloid cell line (416B), a granulocytic cell line (32Dc13), a monocyte/macrophage cell line (RAW 264.7), and a myeloblastic cell line that can be differentiated to macrophage-like cells (M1). UBP43 expression was detected only in the two monocyte-related cell lines, RAW 264.7 and M1 (Fig. 7), suggesting that UBP43 may play an important role in the monocytic lineage.
FIG. 6.
Northern blot analysis of UBP43 expression in different tissues of adult mice. Total RNA was prepared from different tissues of wild-type adult mice as indicated at the top of the figure. Samples (10 μg/lane) were subjected to Northern blot analysis with a 32P-labeled UBP43 cDNA probe. Ethidium bromide-stained 28S rRNA is shown to demonstrate equivalent RNA loading.
FIG. 7.
UBP43 is highly expressed in monocytic cell lines. Total RNAs from nine different murine hematopoietic cell lines and one fibroblast-like cell line were used for Northern blot analysis. BW5147 and EL4 are T-cell lines, Ba/F3 and 18.8 are B-cell lines, MEL and 416B are erythroid cell lines, RAW 264.7 and M1 are monocytic cell lines, 32D is a granulocytic cell line, and NIH 3T3 is a fibroblast-like cell line. RNAs from brain and thymus were used as negative and positive controls, respectively. Each lane was loaded with 10 μg of total RNA. The blot was hybridized with a 32P-labeled UBP43 cDNA probe. Ethidium bromide-stained 18S rRNA is shown to demonstrate equivalent RNA loading.
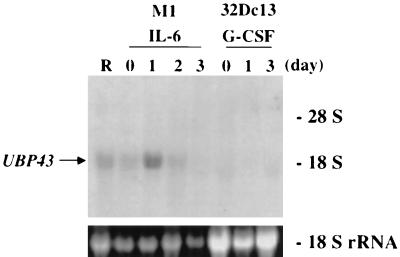
Expression of UBP43 is regulated during cytokine-induced macrophage differentiation of M1 cells.
The M1 myeloblastic leukemia cell line was derived in vitro from a spontaneous myeloblastic leukemia arising in SL strain mice (24). This cell line proliferates autonomously and can be induced with IL-6 or LIF to terminally differentiate along the macrophage lineage (21). Having determined that UBP43 is expressed in M1 cells (Fig. 7), we next studied whether there was any change in UBP43 expression during M1 cell macrophage differentiation. M1 cells were cultured in the presence or absence of IL-6, and total RNA was extracted at various time points and analyzed by Northern blotting. Expression of UBP43 peaked by day 1, gradually declined, and was eventually lost upon the induction of terminal differentiation at day 3 (Fig. 8). Similar results were seen when cells were treated with LIF (data not shown). The murine IL-3-dependent cell line 32Dc13 can be induced towards granulocytic differentiation by G-CSF (11, 46). Although UBP43 expression was not detected in 32Dc13 cells (Fig. 7), we tested whether UBP43 expression could be induced during differentiation of 32Dc13 cell to granulocytes by G-CSF. As shown in Fig. 8, RNA hybridization analysis did not give any detectable UBP43 expression throughout the entire differentiation of 32Dc13 cells to granulocytes by G-CSF. These results indicate that UBP43 expression is lineage restricted and is regulated during monocytic cell differentiation.
FIG. 8.
Expression of UBP43 is regulated during M1 cell differentiation toward macrophages. Total RNAs were isolated from RAW264.7 cells (R); M1 cells cultured in the presence of 100 ng of IL-6 per ml for 0, 1, 2, and 3 days for macrophage differentiation; and 32Dc13 cells cultured in the presence of 500 units of G-CSF per ml for granulocytic cell differentiation. Each lane was loaded with 10 μg of total RNA. The blot was hybridized with a 32P-labeled UBP43 cDNA probe. Ethidium bromide-stained 18S rRNA is shown to demonstrate equivalent RNA loading.
Constitutive expression of UBP43 blocks terminal differentiation of M1 cells.
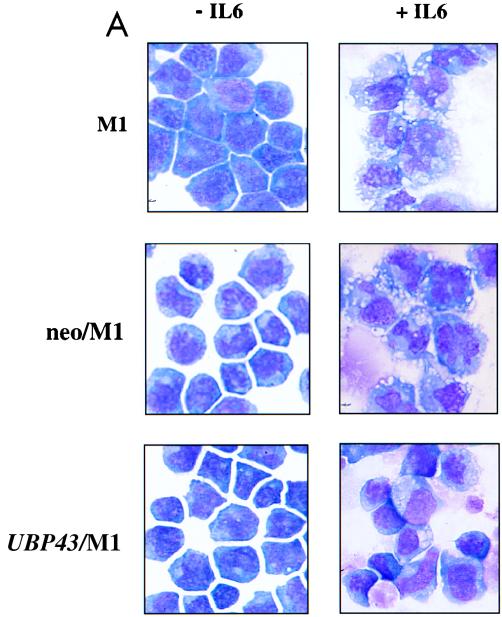
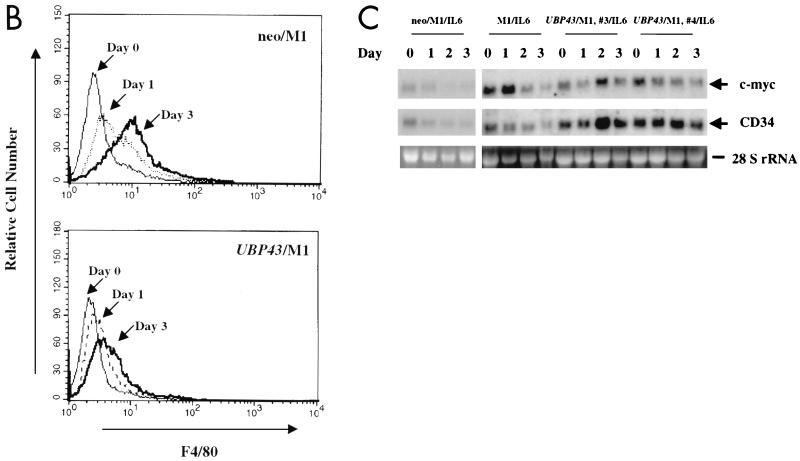
To ascertain whether the observed alteration of UBP43 expression during IL-6- or LIF-dependent M1 cell differentiation was related to the process of differentiation, stable cell lines constitutively expressing UBP43 were established by retroviral infection and G418 selection. Polyclonal populations of M1 cells expressing UBP43 and control populations containing the retroviral vector alone were confirmed by both Southern and Northern blot analysis (data not shown). G418-resistant pools of infected cells were evaluated for morphological evidence of macrophage differentiation and for the expression of F4/80, a macrophage-specific marker, after 3 days in either IL-6 or LIF. Upon IL-6 treatment, parental M1 cells and control M1 cells infected with the vector alone (neo/M1) displayed the morphological features of mature macrophages: abundant, heavily vacuolated cytoplasms; a reduced nuclear to cytoplasmic ratio; distinct heterochromatin; and the lack of prominent nucleoli (Fig. 9A). In contrast, cells infected with the UBP43 retrovirus (UBP43/M1) either remained undifferentiated or displayed an intermediate level of maturation (i.e., promonoblast) characterized by minimal vacuolation, a high nuclear to cytoplasmic ratio, and prominent nucleoli (Fig. 9A), as described previously for the block of M1 cell differentiation (8, 21). Similar morphological changes were seen with treatment of UBP43/M1, neo/M1, and parental cells with LIF (data not shown). Furthermore, UBP43/M1 cells failed to terminally differentiate even after 5 days in culture with IL-6 or LIF. Enumeration of the proportion of cell types within each population upon IL-6 or LIF treatment is given in Table 1. Since UBP43/M1 cells were less morphologically mature than parental M1 cells or control neo/M1 cells, they were evaluated for expression of the cell surface marker F4/80, which is specific for macrophages (14). As shown by flow cytometry analysis (Fig. 9B), control M1 cells (neo/M1) had a significant increase in F4/80 expression upon IL-6-induced differentiation. UBP43/M1 cells displayed only 20% F4/80 expression compared to control neo/M1 cells after 3 days of incubation with IL-6 (Fig. 9B). We also analyzed expression of c-myc and CD34 during IL-6-induced cell differentiation. As reported by other groups (8, 21), CD34 was normally expressed in M1 cells and its expression was decreased during M1 cell differentiation, while c-myc expression was upregulated in day 1 and then downregulated (see M1/IL6, 0 to 3 days, in Fig. 9C). However, M1 cells with constitutive UBP43 expression showed disrupted gene regulation (Fig. 9C). There was continuous expression of CD34 and delayed upregulation of c-myc. These data are consistent with the results from morphological analysis and indicate that constitutive expression of UBP43 blocks the terminal differentiation program of M1 cells, perhaps by disturbing the normal variation of UBP43 expression during terminal macrophage differentiation of M1 cells.
FIG. 9.
Constitutive expression of UBP43 in M1 cells blocks terminal macrophage differentiation. (A) Morphological analysis. After 3 days of culture in the absence or presence of IL-6, cells were evaluated for morphological evidence of terminal differentiation. As detected by Wright-Giemsa stain, the majority of parental M1 cells or control M1 cells infected with retroviral vector alone (neo/M1) showed the morphological features of mature macrophages: heavily vacuolated cytoplasm, a reduced nuclei to cytoplasmic ratio, distinct heterochromatin, and the lack of prominent nucleoli. In contrast, cells overexpressing UBP43 (UBP43/M1) showed evidence of arrested maturation, with most cells having the morphology of promonoblasts or undifferentiated blasts. UBP43-overexpressing cells that were not treated with IL-6 remained entirely undifferentiated, similar to parental and control cells. (B) Flow cytometry analysis of F4/80 expression on IL-6 treated control M1 cells (neo/M1) and UBP43-overexpressing M1 cells (UBP43/M1) by using a phycoerythrin-conjugated monoclonal antibody specific for F4/80. Results from representative clones are shown. (C) Northern blot analysis of CD34 and c-myc expression. Total RNA from M1, neo/M1, and UBP43/M1 clone 3 as well as UBP43/M1 clone 4 cells cultured for the times indicated in the presence of 100 ng of IL-6 per ml were prepared for analysis by Northern blotting of the expression of c-myc and CD34. Each lane was loaded with 10 μg of total RNA. The blot was hybridized with a 32P-labeled murine c-myc or murine CD34 cDNA probe. Ethidium bromide-stained 28S rRNA is shown to demonstrate equivalent RNA loading.
TABLE 1.
Induction of terminal differentiation in control M1 cells and UBP43-overexpressing M1 cells
| Cells used (treatment)a | Cell type (%)b
|
||
|---|---|---|---|
| Blast | Intermediate | Mature | |
| neo/M1 (IL-6) | 14.7 ± 0.76 | 32.5 ± 2.4 | 52.8 ± 2.2 |
| UBP43/M1 (IL-6) | 65.3 ± 1.7 | 29.2 ± 0.9 | 5.5 ± 2.5 |
| neo/M1 (LIF) | 10.7 ± 1.75 | 39.3 ± 2.7 | 50 ± 1.1 |
| UBP43/M1 (LIF) | 67.4 ± 2.7 | 27.9 ± 3.1 | 4.7 ± 2.5 |
M1 cells infected with a retroviral vector containing UBP43 cDNA (UBP43/M1) were compared to control M1 cells infected with a retroviral vector alone (neo/M1) after culture in the presence of IL-6 (100 ng/ml) or LIF (50 ng/ml) for 3 days.
Morphologies of different cell populations were enumerated by Wright-Giemsa staining of cytospin preparations. Cells were categorized as either immature blasts, intermediate monocytes, or mature macrophages; the relative proportions of each category are shown. Data are means ± standard deviations of three independent clones of each virus construct and are from a representative experiment that was repeated three times.
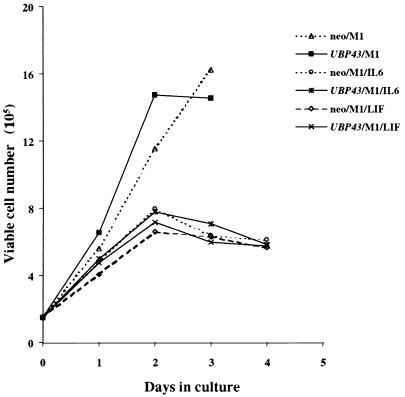
When cells are induced to terminal differentiation, they become growth arrested. As expected, control M1 cells (neo/M1) became growth arrested after 2 days of incubation with IL-6 or LIF (Fig. 10). Interestingly, M1 cells with UBP43 overexpression (UBP43/M1) also showed growth arrest after 2 days incubation with IL-6 or LIF (Fig. 10), although their terminal differentiation was partially blocked. There was no significant difference between the proliferation rates of neo/M1 cells and UBP43/M1 cells. This indicates that constitutive expression of UBP43 does not affect the proliferation program of M1 cells.
FIG. 10.
Overexpression of UBP43 does not affect cell proliferation. Cells (1.5 × 105) were seeded with or without the appropriate treatment, and numbers of viable cells were determined at the indicated times by hemocytometer count. Each time point is the average of three independent experiments of each of the three populations of M1 cells either carrying retrovirus vector alone (neo/M1) or containing the UBP43-expressing retrovirus construct (UBP43/M1).
DISCUSSION
The AML1-ETO chimeric protein is produced by t(8;21)(q22;q22), which is one of the most frequent chromosomal abnormalities detected in acute myelogenous leukemia (42). The AML1-ETO fusion protein has been shown to function primarily as a transcriptional repressor of AML1 target genes and to block AML1 function in vitro and in vivo (10, 31, 34, 53). However, the fusion protein can also activate the BCL-2 promoter and cooperate with AML1 to activate the macrophage CSF (M-CSF) receptor promoter (26, 40). To characterize the effect of AML1-ETO on hematopoiesis, we used RDA to identify its target genes in AML1-ETO-expressing mice. A novel gene, UBP43, has been isolated. It encodes a functional enzyme that belongs to the ubiquitin-specific protease family. UBP43 is highly expressed in the fetal liver and yolk sac of AML1-ETO knockin mice but not in wild-type mice. In wild-type adult mice, UBP43 expression is associated with hematopoietic tissues—the thymus and peritoneal macrophages—further suggesting a biological function of UBP43 in blood cells. The appropriate cell-type- and stage-specific expression of UBP43 may play a critical role in hematopoietic differentiation. Furthermore, we have shown that dysregulated overexpression of UBP43 in a myeloblastic cell line blocks its terminal differentiation. Taken together, our results suggest an important role of UBP43 in hematopoiesis.
UBP43 could be a target gene of AML1-ETO or an upregulated gene related to AML1-ETO expression. Recent experiments have also suggested a direct link between UBP43 gene expression and AML1-ETO expression. We have generated transgenic mice that have inducible AML1-ETO expression in the bone marrow and macrophages under the control of tetracycline. Compared to littermates that did not express AML1-ETO, AML1-ETO-positive transgenic mice showed a higher level of UBP43 expression (18). There are several possible pathways through which AML1-ETO may regulate UBP43 expression: (i) the AML1-ETO fusion protein may directly upregulate UBP43 expression; (ii) the AML1-ETO fusion protein may block AML1 regulation and thereby result in the upregulation of its target gene, UBP43, if AML1 is a repressor for UBP43; (iii) the AML1-ETO fusion protein may mediate other transcriptional activities, which in turn affect UBP43 gene expression. Further studies will be necessary to better define the mechanism by which AML1-ETO upregulates UBP43 expression.
It is apparent that the mechanisms of, and the balance between, ubiquitination and deubiquitination are an important determinant in ubiquitin-dependent cellular events. Just as protein ubiquitination is mediated by ubiquitin-conjugating enzymes, protein deubiquitination is mediated by deubiquitinating enzymes or ubiquitin-specific proteases (UBPs). There are a large number of UBP family members present in yeasts, animals, and plants (20). All UBP family members have several highly homologous domains, but there is little similarity among these members outside these domains. It has been postulated that these variable regions are responsible for their substrate specificity and their cellular localization. To fully understand the function of UBP43, it will be very important to identify its biological substrates. Coexpression of UBP fusion proteins and an artificial UBP substrate, Ub-Met-β-gal, has been used to identify UBP enzyme activity. Most UBPs, such as Dub-1 (54), Ubp1 (45), Doa4 (38), and UBP43 exhibit enzyme activity in this assay. The presence of such a large family of UBP enzymes is indicative of functional diversity. Recent evidence confirms that UBPs do in fact play distinct roles in regulating biological processes including cell growth and differentiation. For instance, the Drosophila melanogaster UBP, Faf (fat facets), is involved in cell fate determination (9); the mammalian Tre-2 oncoprotein, a member of the UBP superfamily (38), acts normally as a growth suppressor within the cell; and yeast Ubp3 is linked to a chromatin regulatory process of transcriptional silencing (33). Here, we provide evidence to suggest an additional biological role for UBPs in hematopoietic cell differentiation. Expression of a leukemogenesis-related fusion protein, AML1-ETO, directly or indirectly increased the level of UBP43 enzyme inappropriately. It is possible that such an increase in enzyme activity blocks the normal ubiquitination of UBP43-specific substrates. These substrates must be normally degraded through the ubiquitination proteasome pathway for differentiation to occur. This model, that UBP43 negatively regulates the turnover of specific substrates of the ubiquitin pathway, adds credence to the hypothesis explaining the reasons for the diversity and abundance of UBPs. This is consistent with genetic interactions observed between the fat facets mutations and proteasome mutations in Drosophila (9).
The upregulation of UBP43 in two hematopoiesis-related tissues in AML1-ETO-expressing embryos (Fig. 4 and 5), the dysplastic hematopoiesis in the yolk sac and liver of AML1-ETO-expressing embryos (34, 52), and the enriched expression of UBP43 in the thymus and thioglycolate-stimulated macrophages of wild-type adult mice (Fig. 6) all suggest a role for UBP43 in hematopoiesis. To investigate whether UBP43 is important in hematopoiesis, we have studied its expression in hematopoietic cell lines of various lineages. Among the nine murine hematopoietic cell lines tested, UBP43 was expressed only in the monocyte-related cell lines M1 and RAW264.7. Furthermore, expression of UBP43 was regulated in IL-6- or LIF-induced macrophage differentiation in M1 cells. UBP43 expression was significantly increased during the first day of IL-6 induction and decreased to below the basal level of untreated M1 cells upon macrophage maturation. These data suggest that regulated expression of UBP43 might be required for terminal macrophage differentiation. To test this hypothesis, we constitutively expressed UBP43 in M1 cells and found that constitutive expression of UBP43 blocked terminal macrophage differentiation in M1 cells. UBP43/M1 cells did not acquire a mature macrophage phenotype, as judged by morphology; failed to express a macrophage surface marker, F4/80; and lacked the ability to suppress c-myc and CD34 expression upon IL-6 induction. Thus, constitutive expression of UBP43 can contribute to a block of terminal myeloid differentiation, one of the characteristics of acute leukemia. Since UBP43 overexpression did not change cell proliferation and cells become unhealthy after 3 days treatment with IL-6 or LIF, it is possible that this “block of terminal differentiation” is a delay of cell differentiation. The relationship between leukemogenesis and UBP43 expression requires further investigation.
It has been postulated that, in general, terminal differentiation is accompanied by cell growth arrest (39). M1 cells, when induced to terminal differentiation by IL-6 or LIF, exit the cell cycle and become growth arrested (data not shown) (21). Interestingly, in spite of a lack of terminal differentiation, M1 cells overexpressing UBP43 still undergo growth arrest similar to control cells, suggesting that UBP43 primarily interferes with a differentiation process independent of cell cycle regulation. Distinct regulation of differentiation and proliferation in the maturation process has been suggested by others (44). Constitutive expression of WT1 in U937 cells abrogates cell differentiation but is still accompanied by cell accumulation in the G1/G0 phase of the cell cycle. Overall, these data support the notion that AML1-ETO expression disrupts expression of a number of genes, some of which may play important roles in cell differentiation and others of which may be important for cell proliferation. Abnormal expression of these genes may cooperate to cause cell transformation.
Since UBP43 expression was initially increased upon IL-6 treatment, we also analyzed multiple cell lines from various tissue origins to examine whether this increase in UBP43 was due to IL-6 stimulation. Northern blot analysis with RNA prepared from these cells showed that UBP43 was not an IL-6-responsive gene (data not shown). This indicates that the up- and downregulation of UBP43 in M1 cells is related to cell differentiation. Currently, we do not know which cells in the thymus are positive for UBP43 expression. However, the high level of UBP43 expression in the thymus suggest a possible link between UBP43 and immature T-cell apoptosis. Therefore, we also analyzed UBP43 expression in a mouse model with regulated thymocyte apoptosis (28). There were no observed changes in UBP43 expression in thymic RNA derived from either untreated mice or mice treated with anti-CD3ɛ monoclonal antibody to induce thymocyte apoptosis (data not shown).
In summary, we have cloned a novel member of the UBP family, which has a highly restricted tissue distribution in normal adult mice and is highly expressed in the fetal hematopoietic organs of AML1-ETO knockin mice. Most importantly, the experimental data suggest a potential role of UBP43 in hematopoiesis. The cloning of the human UBP43 gene and the study of its expression in normal tissues and in leukemia cells and other cancer cells will provide important information related to leukemogenesis and cancer biology in general. In addition, the discovery of this new member of the UBP protein family will facilitate our studies of protein ubiquitination in hematopoiesis and cell differentiation.
ACKNOWLEDGMENTS
We thank Daniel Tenen and Stuart Orkin for many helpful discussions and critical reading of the manuscript; Linda Clayton for providing the murine thymus cDNA library; Christopher Hetherington, Donald Yergeau, Kathy Maltby, and Pu Zhang for technical help; Alan D’Andrea for providing the substrate Ub-Met-β-gal; Nancy Speck for providing AML1 knockout mice; and Marie-Thérèse Little for editing the manuscript.
This work was supported by National Institutes of Health grants CA/AI59589 (D.E.Z.), CA/72009 (D.E.Z.) and HL 59484-01 (J.P.); American Cancer Society grant DHP-166 (D.E.Z.); and the Japan Research Foundation for Clinical Pharmacology and a long-term fellowship from the Human Frontier Science Program (A.I.). D.E.Z. is a Leukemia Society of America Scholar.
REFERENCES
- 1.Chau V, Tobias J W, Bachmair A, Marriott D, Ecker D J, Gonda D K, Varshavsky A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 2.Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MAT alpha 2 repressor. Cell. 1993;74:357–369. doi: 10.1016/0092-8674(93)90426-q. [DOI] [PubMed] [Google Scholar]
- 3.Chen Z J, Parent L, Maniatis T. Site-specific phosphorylation of IkappaBalpha by a novel ubiquitination-dependent protein kinase activity. Cell. 1996;84:853–862. doi: 10.1016/s0092-8674(00)81064-8. [DOI] [PubMed] [Google Scholar]
- 4.Chirgwin J M, Przybyla A E, MacDonald R J, Rutter W J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 5.Chowdary D R, Dermody J J, Jha K K, Ozer H L. Accumulation of p53 in a mutant cell line defective in the ubiquitin pathway. Mol Cell Biol. 1994;14:1997–2003. doi: 10.1128/mcb.14.3.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D’Andrea A, Pellman D. Deubiquitinating enzymes: a new class of biological regulators. Crit Rev Biochem Mol Biol. 1998;33:337–352. doi: 10.1080/10409239891204251. [DOI] [PubMed] [Google Scholar]
- 7.Davie J R, Murphy L C. Level of ubiquitinated histone H2B in chromatin is coupled to ongoing transcription. Biochemistry. 1990;29:4752–4757. doi: 10.1021/bi00472a002. [DOI] [PubMed] [Google Scholar]
- 8.Fackler M J, Krause D S, Smith O M, Civin C I, May W S. Full-length but not truncated CD34 inhibits hematopoietic cell differentiation of M1 cells. Blood. 1995;85:3040–3047. [PubMed] [Google Scholar]
- 9.Fischer-Vize J A, Rubin G M, Lehmann R. The fat facets gene is required for Drosophila eye and embryo development. Development. 1992;116:985–1000. doi: 10.1242/dev.116.4.985. [DOI] [PubMed] [Google Scholar]
- 10.Frank R, Zhang J, Uchida H, Meyers S, Hiebert S W, Nimer S D. The AML1/ETO fusion protein blocks transactivation of the GM-CSF promoter by AML1B. Oncogene. 1995;11:2667–2674. [PubMed] [Google Scholar]
- 11.Friedman A D, Krieder B L, Venturelli D, Rovera G. Transcriptional regulation of two myeloid-specific genes, myeloperoxidase and lactoferrin, during differentiation of the murine cell line 32D C13. Blood. 1991;78:2426–2432. [PubMed] [Google Scholar]
- 12.Glotzer M, Murray A W, Kirschner M W. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- 13.Goldstein G, Scheid M, Hammerling U, Schlesinger D H, Niall H D, Boyse E A. Isolation of a polypeptide that has lymphocyte-differentiating properties and is probably represented universally in living cells. Proc Natl Acad Sci USA. 1975;72:11–15. doi: 10.1073/pnas.72.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gordon S, Keshav S, Chung L P. Mononuclear phagocytes: tissue distribution and functional heterogeneity. Curr Opin Immunol. 1988;1:26–35. doi: 10.1016/0952-7915(88)90047-7. [DOI] [PubMed] [Google Scholar]
- 15.Gupta K, Chevrette M, Gray D A. The Unp proto-oncogene encodes a nuclear protein. Oncogene. 1994;9:1729–1731. [PubMed] [Google Scholar]
- 16.Hawley R G, Lieu F H, Fong A Z, Hawley T S. Versatile retroviral vectors for potential use in gene therapy. Gene Ther. 1994;1:136–138. [PubMed] [Google Scholar]
- 17.Hershko A, Ciechanover A. The ubiquitin system for protein degradation. Annu Rev Biochem. 1992;61:761–807. doi: 10.1146/annurev.bi.61.070192.003553. [DOI] [PubMed] [Google Scholar]
- 18.Hetherington, C. J., N. Harakawa, D. A. Yergeau, L.-Q. Liu, D. G. Tenen, and D. E. Zhang. Inducible AML1-ETO expression in transgenic mice. Submitted for publication.
- 19.Hicke L, Riezman H. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell. 1996;84:277–287. doi: 10.1016/s0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- 20.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 21.Hoffman-Liebermann B, Liebermann D A. Interleukin-6- and leukemia inhibitory factor-induced terminal differentiation of myeloid leukemia cells is blocked at an intermediate stage by constitutive c-myc. Mol Cell Biol. 1991;11:2375–2381. doi: 10.1128/mcb.11.5.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang Y, Baker R T, Fischer-Vize J A. Control of cell fate by a deubiquitinating enzyme encoded by the fat facets gene. Science. 1995;270:1828–1831. doi: 10.1126/science.270.5243.1828. [DOI] [PubMed] [Google Scholar]
- 23.Hubank M, Schatz D G. Identifying differences in mRNA expression by representational difference analysis of cDNA. Nucleic Acids Res. 1994;22:5640–5648. doi: 10.1093/nar/22.25.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ichikawa Y. Further studies on the differentiation of a cell line of myeloid leukemia. J Cell Physiol. 1970;76:175–184. doi: 10.1002/jcp.1040760207. [DOI] [PubMed] [Google Scholar]
- 25.King R W, Deshaies R J, Peters J M, Kirschner M W. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 26.Klampfer L, Zhang J, Zelenetz A O, Uchida H, Nimer S D. The AML1/ETO fusion protein activates transcription of BCL-2. Proc Natl Acad Sci USA. 1996;93:14059–14064. doi: 10.1073/pnas.93.24.14059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larsen C N, Price J S, Wilkinson K D. Substrate binding and catalysis by ubiquitin C-terminal hydrolases: identification of two active site residues. Biochemistry. 1996;35:6735–6744. doi: 10.1021/bi960099f. [DOI] [PubMed] [Google Scholar]
- 28.Lerner A, Clayton L K, Mizoguchi E, Ghendler Y, Van E W, Koyasu S, Bhan A K, Reinherz E L. Cross-linking of T-cell receptors on double-positive thymocytes induces a cytokine-mediated stromal activation process linked to cell death. EMBO J. 1996;15:5876–5887. [PMC free article] [PubMed] [Google Scholar]
- 29.Levinger L, Varshavsky A. Selective arrangement of ubiquitinated and D1 protein-containing nucleosomes within the Drosophila genome. Cell. 1982;28:375–385. doi: 10.1016/0092-8674(82)90355-5. [DOI] [PubMed] [Google Scholar]
- 30.Melton D A, Krieg P A, Rebagliati M R, Maniatis T, Zinn K, Green M R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984;12:7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyers S, Lenny N, Hiebert S W. The t(8;21) fusion protein interferes with AML-1B-dependent transcriptional activation. Mol Cell Biol. 1995;15:1974–1982. doi: 10.1128/mcb.15.4.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller A D, Rosman G J. Improved retroviral vectors for gene transfer and expression. BioTechniques. 1989;7:980–986. , 989. [PMC free article] [PubMed] [Google Scholar]
- 33.Moazed D, Johnson D. A deubiquitinating enzyme interacts with SIR4 and regulates silencing in S. cerevisiae. Cell. 1996;86:667–677. doi: 10.1016/s0092-8674(00)80139-7. [DOI] [PubMed] [Google Scholar]
- 34.Okuda T, Cai Z, Yang S, Lenny N, Lyu C, Van D J, Harada H, Downing J R. Expression of a knocked-in AML1-ETO leukemia gene inhibits the establishment of normal definitive hematopoiesis and directly generates dysplastic hematopoietic progenitors. Blood. 1998;91:3134–3143. [PubMed] [Google Scholar]
- 35.Okuda T, Van D J, Hiebert S W, Grosveld G, Downing J R. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 36.Onno M, Nakamura T, Mariage-Samson R, Hillova J, Hill M. Human TRE17 oncogene is generated from a family of homologous polymorphic sequences by single-base changes. DNA Cell Biol. 1993;12:107–118. doi: 10.1089/dna.1993.12.107. [DOI] [PubMed] [Google Scholar]
- 37.Palis J, Kingsley P D. Differential gene expression during early murine yolk sac development. Mol Reprod Dev. 1995;42:19–27. doi: 10.1002/mrd.1080420104. [DOI] [PubMed] [Google Scholar]
- 38.Papa F R, Hochstrasser M. The yeast DOA4 gene encodes a deubiquitinating enzyme related to a product of the human tre-2 oncogene. Nature. 1993;366:313–319. doi: 10.1038/366313a0. [DOI] [PubMed] [Google Scholar]
- 39.Pardee A B. G1 events and regulation of cell proliferation. Science. 1989;246:603–608. doi: 10.1126/science.2683075. [DOI] [PubMed] [Google Scholar]
- 40.Rhoades K L, Hetherington C J, Rowley J D, Hiebert S W, Nucifora G, Tenen D G, Zhang D E. Synergistic up-regulation of the myeloid-specific promoter for the macrophage colony-stimulating factor receptor by AML1 and the t(8;21) fusion protein may contribute to leukemogenesis. Proc Natl Acad Sci USA. 1996;93:11895–11900. doi: 10.1073/pnas.93.21.11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rock K L, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg A L. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 42.Rowley J D. Recurring chromosome abnormalities in leukemia and lymphoma. Semin Hematol. 1990;27:122–136. [PubMed] [Google Scholar]
- 43.Scheffner M, Huibregtse J M, Vierstra R D, Howley P M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 44.Svedberg H, Chylicki K, Baldetorp B, Rauscher F J, Gullberg U. Constitutive expression of the Wilm’s tumor gene (WT1) in the leukemic cell line U937 blocks parts of the differentiation program. Oncogene. 1998;16:925–932. doi: 10.1038/sj.onc.1201613. [DOI] [PubMed] [Google Scholar]
- 45.Tobias J W, Varshavsky A. Cloning and functional analysis of the ubiquitin-specific protease gene UBP1 of Saccharomyces cerevisiae. J Biol Chem. 1991;266:12021–12028. [PubMed] [Google Scholar]
- 46.Valtieri M, Tweardy D J, Caracciolo D, Johnson K, Mavilio F, Altmann S, Santoli D, Rovera G. Cytokine-dependent granulocytic differentiation. Regulation of proliferative and differentiative responses in a murine progenitor cell line. J Immunol. 1987;138:3829–3835. [PubMed] [Google Scholar]
- 47.Vassilev A P, Rasmussen H H, Christensen E I, Nielsen S, Celis J E. The levels of ubiquitinated histone H2A are highly upregulated in transformed human cells: partial colocalization of uH2A clusters and PCNA/cyclin foci in a fraction of cells in S-phase. J Cell Sci. 1995;108:1205–1215. doi: 10.1242/jcs.108.3.1205. [DOI] [PubMed] [Google Scholar]
- 48.Verma I M, Stevenson J K, Schwartz E M, Van A D, Miyamoto S. Rel/NF-kappa B/I kappa B family: intimate tales of association and dissociation. Genes Dev. 1995;9:2723–2735. doi: 10.1101/gad.9.22.2723. [DOI] [PubMed] [Google Scholar]
- 49.Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe A H, Speck N A. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci USA. 1996;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y, Yeung Y G, Langdon W Y, Stanley E R. c-Cbl is transiently tyrosine-phosphorylated, ubiquitinated, and membrane-targeted following CSF-1 stimulation of macrophages. J Biol Chem. 1996;271:17–20. doi: 10.1074/jbc.271.1.17. [DOI] [PubMed] [Google Scholar]
- 51.Wilkinson K D, Tashayev V L, O’Connor L B, Larsen C N, Kasperek E, Pickart C M. Metabolism of the polyubiquitin degradation signal: structure, mechanism, and role of isopeptidase T. Biochemistry. 1995;34:14535–14546. doi: 10.1021/bi00044a032. [DOI] [PubMed] [Google Scholar]
- 52.Yaglom J, Linskens M H K, Sadis S, Rubin D M, Futcher B, Finley D. p34Cdc28-mediated control of Cln3 cyclin degradation. Mol Cell Biol. 1995;15:731–741. doi: 10.1128/mcb.15.2.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yergeau D A, Hetherington C J, Wang Q, Zhang P, Sharpe A H, Binder M, Marin-Padilla M, Tenen D G, Speck N A, Zhang D E. Embryonic lethality and impairment of haemotopoiesis in mice heterozygous for an AML1-ETO fusion gene. Nat Genet. 1997;15:303–306. doi: 10.1038/ng0397-303. [DOI] [PubMed] [Google Scholar]
- 54.Zhu Y, Carroll M, Papa F R, Hochstrasser M, D’Andrea A D. DUB-1, a deubiquitinating enzyme with growth-suppressing activity. Proc Natl Acad Sci USA. 1996;93:3275–3279. doi: 10.1073/pnas.93.8.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu Y, Lambert K, Corless C, Copeland N G, Gilbert D J, Jenkins N A, D’Andrea A D. DUB-2 is a member of a novel family of cytokine-inducible deubiquitinating enzymes. J Biol Chem. 1997;272:51–57. doi: 10.1074/jbc.272.1.51. [DOI] [PubMed] [Google Scholar]