ABSTRACT
The human gut microbiota plays a central role in intestinal health and disease. Yet, many of its bacterial constituents are functionally still largely unexplored. A crucial prerequisite for bacterial survival and proliferation is the creation and/or exploitation of an own niche. For many bacterial species that are linked to human disease, the inner mucus layer was found to be an important niche. Allobaculum mucolyticum is a newly identified, IBD-associated species that is thought be closely associated with the host epithelium. To explore how this bacterium is able to effectively colonize this niche, we screened its genome for factors that may contribute to mucosal colonization. Up to 60 genes encoding putative Carbohydrate Active Enzymes (CAZymes) were identified in the genome of A. mucolyticum. Mass spectrometry revealed 49 CAZymes of which 26 were significantly enriched in its secretome. Functional assays demonstrated the presence of CAZyme activity in A. mucolyticum conditioned medium, degradation of human mucin O-glycans, and utilization of liberated non-terminal monosaccharides for bacterial growth. The results support a model in which sialidases and fucosidases remove terminal O-glycan sugars enabling subsequent degradation and utilization of carbohydrates for A. mucolyticum growth. A. mucolyticum CAZyme secretion may thus facilitate bacterial colonization and degradation of the mucus layer and may pose an interesting target for future therapeutic intervention.
KEYWORDS: Allobaculum mucolyticum, microbiota, intestinal mucin, mucin o-glycans, mucin degradation, cazymes, glycosidase, pathobiont, proteome
Introduction
The intestinal microbiota plays an important role in host health and disease. Due to their close association with the host epithelium, bacteria occupying the intestinal mucosal niche are thought to have a preponderant effect on host health. For example, their close proximity to the intestinal epithelium increases the chances of activating host immune responses, which may drive excessive inflammatory responses as seen in inflammatory bowel disease (IBD).1
In order to provide protection and avoid excessive inflammatory responses, the intestinal epithelial cells are covered by a viscous mucus layer that protects them from direct contact with potentially harmful bacteria. This mucus layer primarily consists of heavily glycosylated mucin proteins, such as MUC2, that are highly decorated with branched O-glycans linked to serine/threonine-rich repeats in the polypeptide backbone. These mucin O-glycans retain large amounts of water (>90% of the mucin weight is due to water), which gives the mucus layer its lubricant properties. Furthermore, mucin glycans also protect the peptide backbone from proteolytic degradation by bacterial and host proteases, thereby safeguarding its main function as a mucosal firewall.
Many enteropathogenic bacteria have developed mechanisms to breach the mucus barrier, e.g. through flagella-driven propulsion. However, in recent years it has also become clear that several commensal bacteria, such as Akkermansia muciniphila and Bacteroides thetaiotamicron, have adopted a mucus-dwelling lifestyle and feed on the monosaccharides present in mucin O-glycans.2–5 To enable this, these bacteria employ one or more mucin O-glycan specific glycoside hydrolases that release the monosaccharides that make up the glycan chain. The glycoside hydrolase families required for mucin O-glycan degradation have previously been described by Tailford et al.6
A decreased mucus layer thickness and increased bacterial invasion into the inner mucus layer are well-known phenomena in IBD pathology.7 Our group has previously demonstrated that IgA-coated bacteria are inclined to encroach toward the intestinal epithelial cells and can cause intestinal inflammation in mouse models of colitis.8 One of these highly IgA-coated bacteria is a newly identified bacterium from the Allobaculum genus: Allobaculum mucolyticum sp. nov. Little is known about bacteria from the Allobaculum genus, which are part of the Erysipelotrichaceae family and inhabitants of the intestinal microbiota. The first reported member of this genus is Allobaculum stercoricanis, a Gram-positive rod shaped bacterium isolated from dog feaces.9 Allobaculum species are mainly identified on the basis of high throughput 16S rRNA gene sequencing, often in studies that focus on changes in gut microbiota composition of mice after dietary interventions. The relative abundance of Allobaculum species in mice and rats has been correlated with aging, high fat diets, and fatty acid metabolism.10–16 Besides correlations with fatty acid metabolism, Herrmann et al. reported Allobaculum to be an active glucose utilizer and producer of lactate and butyrate.13,14 These reports are in line with previous reports about bacteria from the Erysipelotrichaceae family, which are often associated with a western or high fat diet.17,18
Allobaculum species have also been implicated in playing a role in inflammatory processes. For example, Cox et al.16 reported a positive correlation between Allobaculum relative abundance and levels of ileal RORγT and IL-17 levels and protection from metabolic syndrome in mice. Another, more recent report by Miyauchi et al. causatively linked an Allobaculum strain (OTU002) to increased susceptibility to experimental autoimmune encephalitis.19 This was attributed to this bacterium’s ability to adhere to small intestinal epithelial cells which induces the expansion of inflammatory intestinal T helper 17 cells. Finally, A. mucolyticum was originally isolated from an ulcerative colitis patient (UC) based on its high levels of IgA coating. Mice that were colonized with this strain as part of a microbial community also developed IgA responses toward this bacterium and developed much more severe colitis upon exposure to DSS.8 This suggests that A. mucolyticum is immunogenic in vivo and could play an important role in the development of intestinal inflammation.
As the reports about Allobaculum species are increasing in number, and we and others have reported that bacteria within the Allobaculum genus belong to the immunogenic inhabitants of the intestinal mucosal niche, a more detailed characterization of bacteria belonging to this genus is warranted. Here, we subjected A. mucolyticum to a thorough genomic and functional characterization. We show that A. mucolyticum can secrete a wide repertoire of mucin O-glycan targeting carbohydrate active enzymes (CAZymes), which allow it to effectively degrade and feed on intestinal mucins. These potent mucolytic capabilities suggest it can effectively colonize and degrade the protective mucus layer of its host.
Results
Allobaculum mucolyticum utilizes both dietary- and host-derived glycans for growth
Allobaculum species are inhabitants of the intestine and thus encounter multiple dietary- and host-derived nutrients. To investigate the nutrient requirements of A. mucolyticum, we assessed its growth over a period of 72 hours in the presence of a variety of substrates. Robust growth of A. mucolyticum was observed in an enriched gut microbiota medium (GMM), containing multiple mono- and disaccharides, such as glucose, fructose, maltose and cellobiose (Figure 1a). To examine whether other, dietary-derived sources of glycan can also be used for growth, A. mucolyticum was grown in a basal medium supplemented with the plant polysaccharide inulin (Figure S1). The basal medium alone, which lacks all glycans present in GMM, did not support the growth of A. mucolyticum. Supplementation of basal medium with inulin resulted in firm growth, demonstrating the bacterium’s ability to utilize dietary-derived carbon sources.
Figure 1.
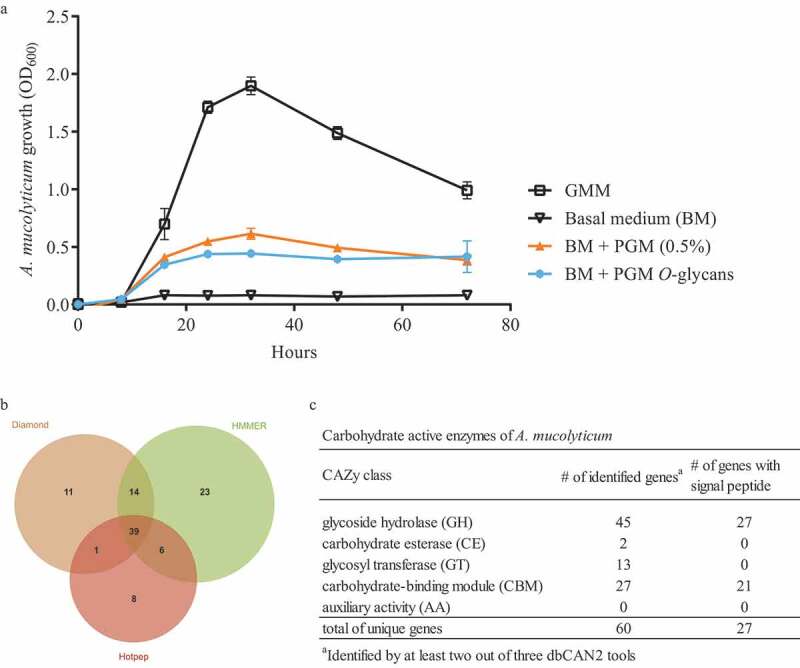
A. mucolyticum growth on mucin and mucin O glycans, and identification of carbohydrate active enzymes (CAZymes) in its genome. (a) A. mucolyticum growth was assessed over a 72 h period by measuring optical density at 600 nm (OD600). Bacteria were grown in Gut Microbiota Medium (GMM), basal medium (BM), or basal medium supplemented with porcine gastric mucin (0.5% w/vol) or purified PGM O-glycans (10 mg/mL). Data represent the mean ± SD of three independent experiments. (b) Proportional Venn diagram showing the number of identified CAZyme encoding genes using the Diamond, HMMER and Hotpep tools that are part of the dbCAN2 metaserver analysis. (c) The table shows the CAZymes and the presence of signal peptides, separated by CAZyme class, that were identified by at least two out of three dbCAN2 tools
As several other Allobaculum species have been reported to be closely associated with the intestinal mucosal niche we next investigated whether A. mucolyticum could also use mucins for growth. Hereto, basal medium was supplemented with porcine gastric mucin (PGM, 0.5% w/vol) and this was sufficient to support A. mucolyticum growth (Figure 1a). To assess whether mucin O-glycans and/or the mucin peptide backbone peptide promoted bacterial growth, bacteria were grown in basal medium supplemented with purified mucin O-glycans (10 mg/mL). After supplementation with these glycans A. mucolyticum was able to grow to a similar optical density as on complete porcine gastric mucin (0.5% w/vol) (Figure 1a). This showed that the O-glycans present on mucin are sufficient to support the growth of A. mucolyticum and indicates that it can, in addition to dietary-derived glycans, also use host-derived mucin as a growth substrate.
The genome of Allobaculum mucolyticum predicts the presence of a wide range of O-glycan-targeting carbohydrate active enzymes (CAZymes)
To investigate how A. mucolyticum can utilize complex O-glycans, we interrogated the available A. mucolyticum genome for the presence of carbohydrate active enzymes (CAZymes) using the dbCAN2 metaserver pipeline for automated CAZyme annotation.20 This pipeline uses a combination of three different annotation tools (Diamond, HMMER and Hotpep) to increase accuracy in predicting and annotating CAZymes within bacterial genomes. This analysis revealed a total of 102 putative ORFs with putative CAZy domains (Figure 1b).
For increased accuracy, we discarded hits that were not identified by at least two out of three tools for further analysis. Among the remaining 60 ORFs multiple types of CAZy domains could be detected, such as glycoside hydrolase (GH), carbohydrate esterase (CE), glycosyl transferase (GT) domains, and carbohydrate-binding modules (CBM), with some genes encoding a combination of multiple domains (Figure 2c). Many of the ORFs, especially those containing glycoside hydrolase domains, also encode signal peptides, suggesting that the proteins may be transferred to the bacterial surface or secreted into the environment. The wide repertoire of secreted and non-secreted CAZymes suggests that A. mucolyticum is highly adapted to living in the glycan-rich mucus niche.
Figure 2.
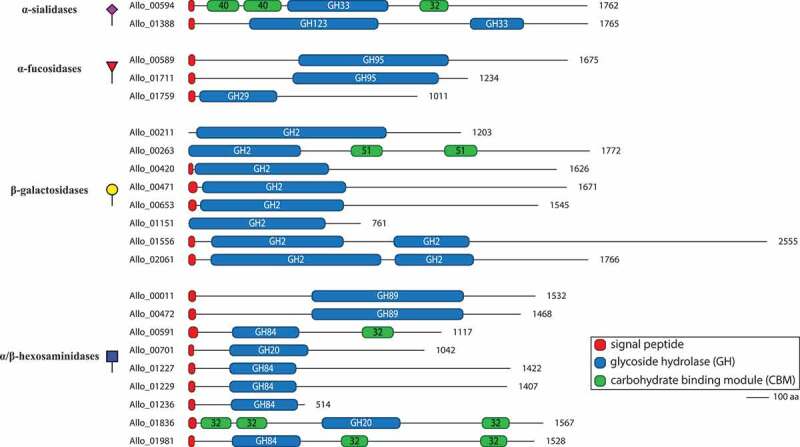
Domain architecture of putative mucin O-glycan targeting CAZymes in genome of A. mucolyticum. CAZymes are divided in groups based upon their predicted enzyme activity: from top to bottom, α-sialidases, α-fucosidases, β-galactosidases and α/β-hexosaminidases. The displayed domains are those identified by HMMER and SignalP 4.0 and are drawn to scale with the size of the polypeptide backbone, in number of amino acids, indicated at the right side of each protein
Closer inspection of the collection of CAZymes identified in the A. mucolyticum genome revealed at least 22 glycoside hydrolases that were predicted to specifically target O-glycans as found on mucins, of which the majority contains a signal peptide (Table 1). Among the 22 glycoside hydrolases are two sialidases (GH33) and three fucosidases (GH29/95), which target the monosaccharides often found at terminal positions of mucin O-glycans, and eight different galactosidases (GH2) and nine hexosaminidases (GH20/GH84/GH89), which are predicted to hydrolyze the underlying glycosidic linkages. The gene structure of the 22 putative enzymes, with the locations of the predicted glycoside hydrolase domains and additional domains (Figure 2), clearly demonstrates the great diversity in domains, domain organization and predicted protein sizes – also between genes predicted to encode the same class of enzymes. Transcription and translation of the entire repertoire of mucin-targeting CAZymes would allow degradation of the wide variety of linkages commonly found within mucin O-glycans.
Table 1.
Putative mucin O-glycan targeting CAZymes of A. mucolyticum.
| Specificity | Gene_ID (putative activity) | CAZy family domainsa | Signal peptide (aa) | Protein size (aa) |
|---|---|---|---|---|
| Sialidase | Allo_00594 (exo-α-sialidase) | CBM40,CBM40,GH33,CBM32 | Y(1-25) | 1762 |
| Allo_01388 (exo-β-N-acetylgalactosaminidase, exo-α-sialidase) | GH123,GH33 | Y(1-25) | 1765 | |
| Fucosidase | Allo_00589 (exo-α-1,2-L-fucosidase) | GH95,CBM51 | Y(1-29) | 1675 |
| Allo_01711 exo-α-1,2-L-fucosidase) | GH95,CBM51 | Y(1-29) | 1234 | |
| Allo_01759 (exo-α-1,3/1,4-L-fucosidase) | GH29 | Y(1-30) | 1011 | |
| Galactosidase | Allo_00211 (exo-β-galactosidase) | GH2,CBM51,CBM67 | N | 1203 |
| Allo_00263 (exo-β-galactosidase) | GH2,CBM51,CBM51 | N | 1772 | |
| Allo_00420 (exo-β-galactosidase) | CBM32,GH2 | Y(1-20) | 1626 | |
| Allo_00471 (exo-β-galactosidase) | GH2,CBM32,CBM67,CBM71 | Y(1-39) | 1671 | |
| Allo_00653 (exo-β-galactosidase) | GH2,CBM32,CBM67,CBM71 | Y(1-35) | 1545 | |
| Allo_01151 (exo-β-galactosidase) | GH2,CBM57 | N | 761 | |
| Allo_01556 (exo-β-galactosidase) | GH2,GH2 | Y(1-28) | 2555 | |
| Allo_02061 (exo-β-galactosidase) | GH2,GH2 | Y(1-31) | 1766 | |
| Hexosaminidase | Allo_00011 (exo-α-N-acetylglucosaminidase) | GH89,CBM32 | Y(1-33) | 1532 |
| Allo_00472 (exo-α-N-acetylglucosaminidase) | GH89 | Y(1-33) | 1468 | |
| Allo_00591 (exo-β-N-acetylglucosaminidase) | GH84,CBM32 | Y(1-42) | 1117 | |
| Allo_00701 (β-N-acetylglucosaminidase) | GH20 | Y(1-24) | 1042 | |
| Allo_01227 (exo-β-N-acetylglucosaminidase) | GH84,CBM32 | Y(1-29) | 1422 | |
| Allo_01229 (exo-β-N-acetylglucosaminidase) | GH84,CBM32 | Y(1-29) | 1407 | |
| Allo_01236 (exo-β-N-acetylglucosaminidase) | GH84,CBM32 | Y(1-29) | 514 | |
| Allo_01836 (β-N-acetylglucosaminidase) | CBM32,CBM32,GH20,CBM32 | Y(1-35) | 1567 | |
| Allo_01981 (β-N-acetylglucosaminidase) | GH84,CBM32,CBM32 | Y(1-27) | 1528 |
aIdentified domains are listed from N to C terminus of the peptide chain
A. mucolyticum secretes a large array of mucin O-glycan-targeting CAZymes
In order to establish which of the predicted mucin-targeting CAZymes is produced and secreted by A. mucolyticum under in vitro culturing conditions, both the bacterial whole-cell lysate and the conditioned medium from bacteria cultured for 24 h in complete GMM were subjected to proteomic analysis using mass spectrometry. In addition, the conditioned media from bacteria grown for 24 h in basal medium or basal medium supplemented with PGM were analyzed to investigate the effect of glycan availability on the regulation or secretion of CAZymes. Mapping of the detected peptide fragments to the A. mucolyticum genome revealed a total of 1005 proteins in the whole-cell lysate and the different conditioned media fractions. Since A. mucolyticum has a predicted 2255 genes, these results show that nearly 45% of all genes are translated under the conditions tested.
Since many CAZymes contain putative signal peptides and are predicted to be secreted, we next analyzed the total A. mucolyticum secretome by focusing on proteins enriched in the conditioned media compared to the whole-cell lysate. This showed that 86 proteins were significantly enriched (>10 fold) in the conditioned medium of at least one of the three different growth conditions (Figure S2). For most proteins, the highest enrichment was detected in the conditioned medium of bacteria grown in GMM. Basal medium supplemented with PGM largely mimicked the protein secretion profile of complete GMM, suggesting that PGM provides similar regulatory cues or secretion signals. Basal medium did not result in a large number of significantly enriched secreted proteins, which correlated with the poor growth in this medium.
A closer inspection of the detected proteins revealed that out of the 60 putative CAZymes identified in the genome, 49 were detected using mass spectrometry of which 26 were significantly enriched in the conditioned medium of at least one of the growth conditions (Figure S3). Eighteen of these were predicted mucin-targeting CAZymes (Figure 3), indicating that over 80% (18 out of 22) of the predicted mucin-targeting CAZymes were produced and secreted into the culture media. Strikingly, the 18 mucin-targeting CAZymes were among the most abundant proteins detected. This analysis demonstrates that A. mucolyticum produces and secretes a large array of putative mucin O-glycan targeting glycoside hydrolases which together are predicted to efficiently deglycosylate mucin proteins.
Figure 3.
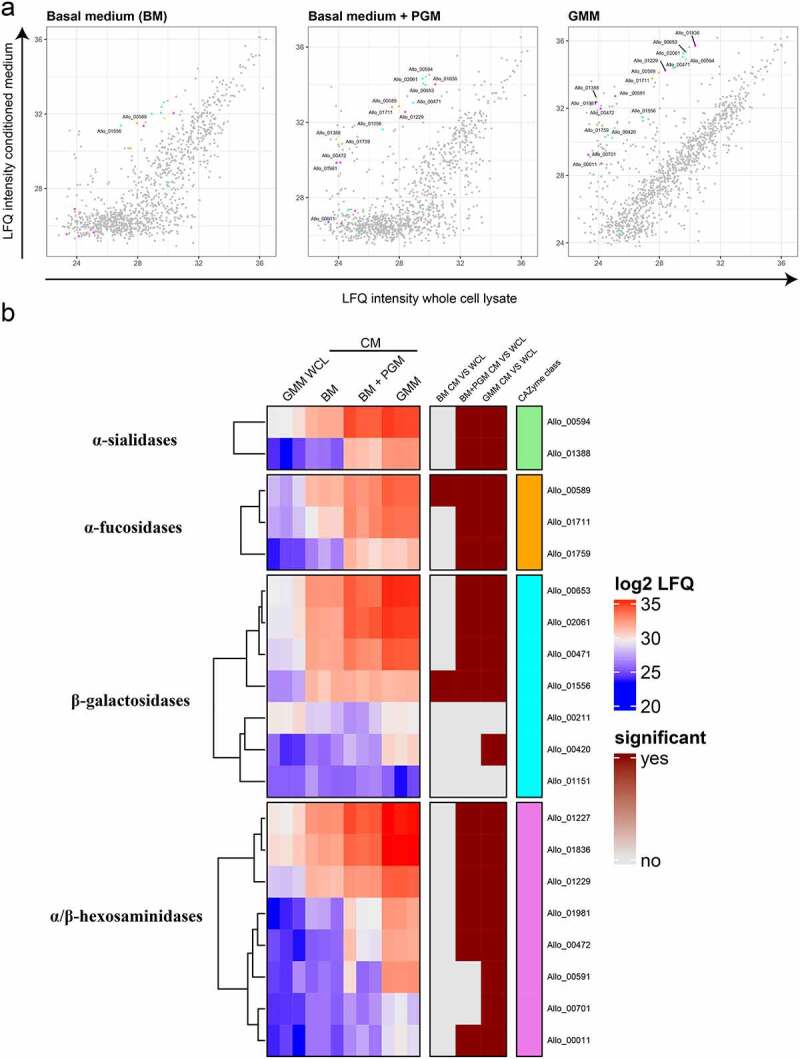
Mass spectrometry analysis of the A. mucolyticum proteome demonstrates the presence of mucin O-glycan targeting CAZymes. Conditioned media (CM) from 24 h cultures in basal medium (BM), basal medium + PGM and Gut Microbiota Medium (GMM) and a whole cell lysate (WCL) from the GMM culture were subjected to mass spectrometry analysis, with three biologically independent replicates per condition. (a) The scatter plots show the log2 LFQ from whole cell lysate of the GMM culture (X-axis) and the secretomes from the three different media (Y-axis). The colored dots indicate the putative O-glycan targeting CAZymes with their color matching the color of their putative CAZyme class shown in the heat map. (b) The red/blue heat map displays the proteins and their relative enrichment in the three different secretomes compared to the GMM WCL. Significance (depicted in the brown heatmap) was calculated using a t-test and indicates a > 10 fold enrichment in the respective secretome versus GMM WCL with an FDR < 0.05
Functional activity of the secreted CAZymes
Mucin O-glycans, such as the typical core 2 mucin O-glycan depicted in figure 5f, are made up of multiple covalently linked monosaccharides. To degrade such glycans the linkages between the different monosaccharides need to be hydrolyzed by CAZymes with the correct linkage specificity. To verify whether the CAZymes secreted by A. mucolyticum are enzymatically active and able to hydrolyze linkages found in mucin O-glycans, conditioned medium or heat-inactivated conditioned medium (30 minutes at 98°C) from a 48 h culture was incubated with five different monovalent fluorescent or chromogenic substrates to detect the presence of sialidase, fucosidase, galactosidase, N-acetyl-glucosaminidase and N-acetyl-galactosaminidase activities. This resulted in clear signals for the presence of α-sialidase, β-galactosidase, β-GlcNAcase and α-GalNAcase activities in A. mucolyticum conditioned medium (Figure 4a-e). Although multiple fucosidases were detected in the secretome during the proteomics analysis, no fucosidase activity could be detected – even after concentrating the culture media 100x. An alternative approach using chemoselective labeling of the fucosidases with an activity-based probe, which can detect fucosidase activity in enzymes with both GH29 and GH95 glycosyl hydrolase domains, also did not detect fucosidase activity. Despite the lack of fucosidase activity, the combination of identified enzymatic activities would likely be able to effectively target and degrade O-linked glycans, such as found on mucins.
Figure 5.
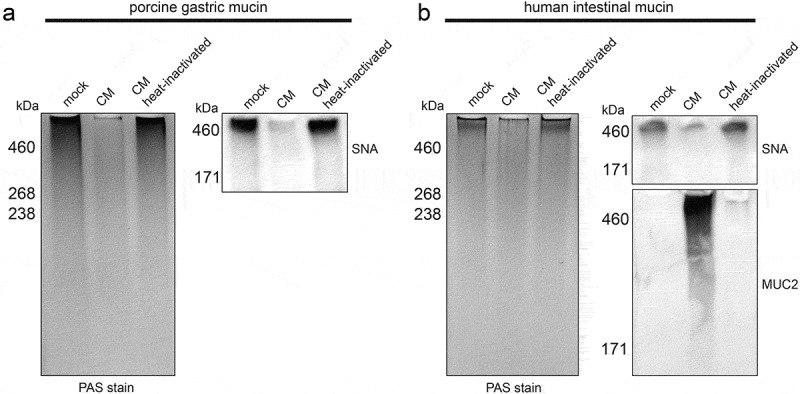
Enzymatic activity within A. mucolyticum conditioned medium degrades porcine and human mucins. (a) Porcine gastric mucin or (b) human intestinal mucin (both 10 mg/mL) were mixed 1:1 with GMM (mock) or active or heat-inactivated A. mucolyticum conditioned medium (CM) from a 48 h culture grown in GMM, and incubated overnight at 37°C. After separation using SDS-PAGE, mucins were either detected in-gel using a Period acid-Schiff (PAS) stain or electroblotted and detected using SNA lectin, and in the case of human intestinal mucin also with an anti-MUC2 antibody. Molecular masses are indicated in kilodaltons (kDa)
Figure 4.
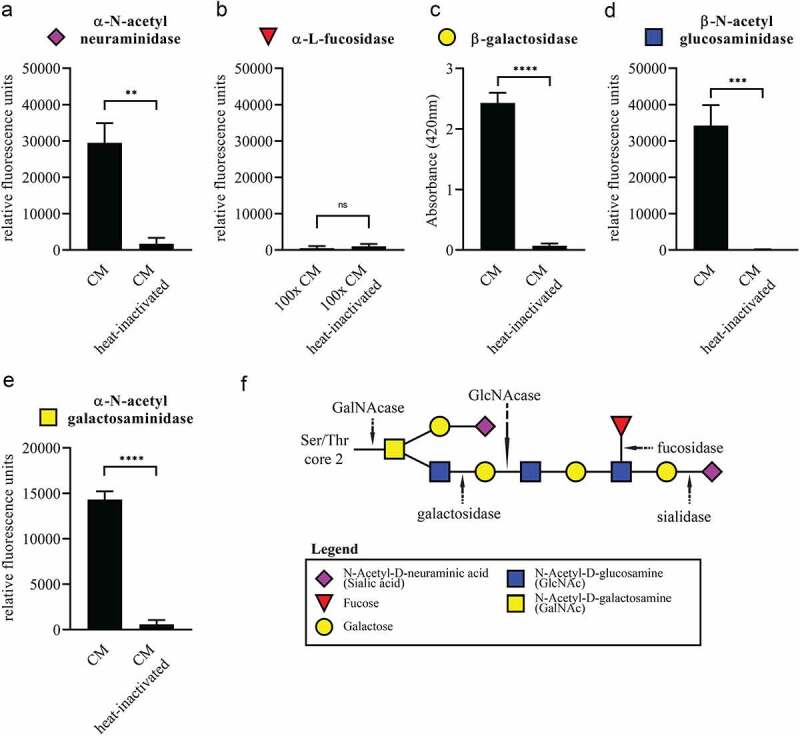
Enzymatic activity of mucin O-glycan-targeting CAZymes in conditioned medium of A. mucolyticum. Filter-sterilized conditioned medium (CM) from a 48 h culture of A. mucolyticum grown in Gut Microbiota Medium was incubated for two hours at 37°C with fluorescent 4-methylumbelliferone linked (a) α-N-acetylneuraminic acid (sialic acid), (b) α-fucose, (d) β-GlcNAc and (e) α-GalNAc or the chromogenic substrate (c) 2-Nitrophenyl β-D-galactopyranoside (ONPG) for the detection of enzymatic activities. A 100x concentrated conditioned medium was used with the α-fucose substrate. (f) Schematic composition of a typical core 2 mucin O-glycan and the target site for each enzyme class. Data represent the mean ± SD of three independent experiments. Statistical significance was determined by an unpaired t test using GraphPad Prism software. **, P < .01; ***, P < .001; ****, P < .0001; ns, not significant
Mucin degradation and utilization by A. mucolyticum
To assess whether secreted CAZymes in the A. mucolyticum conditioned medium are sufficient to degrade mucins, porcine gastric mucin (10 mg/mL) and human intestinal mucin (10 mg/mL) were treated with either control GMM, A. mucolyticum conditioned medium or heat-inactivated conditioned medium (30 min, 98°C). Following incubation (18 h), the mucins were separated using SDS-PAGE and then either stained in-gel with a Periodic acid-Schiff stain (PAS) or electroblotted onto a nitrocellulose membrane and detected with a SNA lectin or an antibody against human MUC2. The PAS-based detection, which detects polysaccharides, clearly demonstrated that both the porcine gastric mucin (Figure 5a) and human intestinal mucin (Figure 5b) were almost entirely degraded by the A. mucolyticum conditioned medium. Heat-inactivation completely abolished the mucin degradation activity. When detecting sialic acids at terminal positions in the glycan chain by probing with the lectin SNA, a clear reduction in signal upon treatment with conditioned medium was observed (Figure 5a-b). Interestingly, human intestinal mucin, which according to the PAS stain barely entered the gel, appeared as a MUC2-positive smear after treatment with the active conditioned medium. The smear pattern indicates degradation of MUC2 and an enhanced ability to be separated by SDS-PAGE.
To assess whether mucin O-glycans products could be used as a substrate for bacterial growth, A. mucolyticum was cultured in a basal medium supplemented with 20 mM of the individual monosaccharides that are typically found within O-glycan chains: sialic acid, fucose, galactose, GlcNAc and GalNAc (Figure 6). The addition of sialic acid or fucose to basal medium did not support growth. In contrast, galactose, GlcNAc and GalNAc did support growth equal to growth levels observed in complete GMM. As sialic acid and fucose normally cap the O-glycans and protect the glycan chain from hydrolysis, this finding supports a model in which A. mucolyticum produces sialidases and fucosidases in order to gain access to and liberate underlying galactose, GlcNAc and GalNAc. Overall these data demonstrate that A. mucolyticum can effectively degrade mucin O-glycans by secreting a wide repertoire of mucin degrading enzymes after which it can use the liberated galactose, GlcNAc and GalNAc as substrates for growth.
Figure 6.
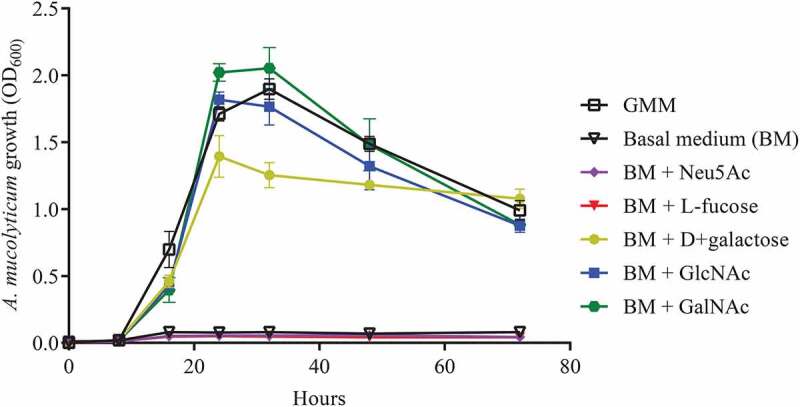
A. mucolyticum uses mucin O-glycan monosaccharides as a substrate for growth. A. mucolyticum growth was assessed over a 72 h period by measuring optical density at 600 nm (OD600). Bacteria were grown in Gut Microbiota Medium (GMM), basal medium (BM) or basal medium supplemented with individually added monosaccharides (20 mM). Graphs depict the mean values ± SD acquired from three independent experiments
Discussion
While the intestinal mucus layer is designed as a barrier against bacterial invasion some bacteria have adapted to living in the mucosal niche by feeding on mucin glycans. Through close proximity to intestinal epithelial cells these bacteria can have a dominant effect on host physiology and intestinal immune responses. Here we provide evidence that the recently classified bacterium Allobaculum mucolyticum produces a large repertoire of mucin O-glycan targeting enzymes that allow it to degrade mucins and forage on host-derived mucin O-glycans.
A search of CAZymes encoded in the A. mucolyticum genome revealed an array of 60 putative CAZymes of which many can target mucin O-glycans. With multiple glycoside hydrolases predicted to target similar glycosidic linkages, there appears to be a considerable redundancy in the A. mucolyticum CAZyme repertoire. For example, there are eight putative β-galactosidases and eight different hexosaminidases. Closer inspection of the proteins revealed that both the domain architecture and organization (Figure 2) and levels of protein production (Figure 3) are markedly different between different CAZymes that are predicted to target the same linkage. This redundancy may be explained by placing it in the perspective of the incredible diversity of glycans and glycan-linkages present in intestinal mucosal niche. Mucin glycans can be branched and contain various structures and modifications, which can greatly affect enzyme-substrate interactions. Having a diverse repertoire of CAZymes therefore allows A. mucolyticum to utilize a wide range of glycan substrates.
Large repertoires of CAZymes are also found in the genomes of other well-known mucolytic bacteria such as Akkermansia muciniphila, Ruminococcus gnavus, Ruminococcus torques and several Bifidobacteria and Bacteroides species, such as Bacteroides thetaiotamicron.6 Glycan generalists such as B. fragilis and B. thetaiotamicron contain a very extensive repertoire of CAZymes, which allows utilization of a wide variety of substrates from both dietary- and host-derived sources. The total repertoire of CAZymes encoded within the A. mucolyticum genome is not as extensive as that of certain Bacteroides species, but A. mucolyticum seems to be highly specialized to target mucin O-glycans, with more than nearly half of its glycoside hydrolases directed to such glycans. This is comparable to the CAZyme repertoire detected in the mucin specialist A. muciniphila (Table S1). Even though the A. mucolyticum CAZyme repertoire is largely directed toward mucin glycans, A. mucolyticum can also use dietary-derived polysaccharide inulin as a substrate for growth (Figure S1). This gives the bacterium the ability to switch between dietary- and host-derived glycans and adapt to fluctuations in nutrient availability as encountered in the dynamic environment of the gastrointestinal tract.
The plasticity of the CAZyme expression in response to changes in the nutritional environment is also evident from our proteomics analysis. This reveals that A. mucolyticum produces a limited set of CAZymes in basal medium, that contains no source of glycans. Under this condition, which allows for minimal growth, four CAZymes were found to be significantly enriched in the conditioned medium as compared to the whole cell lysate. The four enzymes (Allo_589, 898, 1556 & 2010) are a putative α-fucosidase, a carbohydrate esterase, a β-galactosidase and a N-acetyl-β-galactosaminidase, respectively. Although the protein function and exact specificity of CAZymes are difficult to predict based on their amino acid sequence alone, the putative annotation suggests that under nutrient limited conditions A. mucolyticum is able to hydrolyze several host-associated glycans. For example, fucose is known to be abundant on mucin glycans, but also on glycans such as the Lewis antigens, which are known to play a role in regulating inflammation.21 The N-acetyl-β-galactosaminidase (GH123) could be involved in the degradation of glycospingholipids on the membranes of the intestinal epithelial cells.22
When basal medium was supplemented with either mucin or the GMM carbohydrates glucose, fructose and the α- and β-glucans maltose and cellobiose, production and secretion of CAZymes – in particular O-glycan targeting CAZymes – by A. mucolyticum was markedly increased. These results indicate that A. mucolyticum requires the presence of environmental glycans as stimuli to induce expression and secretion of the repertoire of glycosidases. Such regulatory mechanisms are also known from other bacteria that forage on mucin glycans.4 While we have not yet determined in detail the individual components responsible for the induction of the CAZymes, the fact that both mucin and the combination of glucose, fructose, maltose and cellobiose are sufficient for glycosidase expression suggests that a similar glycosidase program is initiated by glycans of both host and dietary origin.
The annotation and detection of secreted O-glycan targeting CAZymes did not in all cases translate to the detection of the predicted enzymatic activity. For example, while three putative fucosidases were identified within the conditioned medium of A. mucolyticum by mass spectrometry, no detectable fucosidase activity was observed in the conditioned medium using the monovalent 4-MU-fucose fluorescent substrate even though the validity of the assay was confirmed with a commercial fucosidase. Nor could fucosidase activity be detected using a novel activity-based probe. Although it is possible that all three fucosidases were annotated incorrectly by the dbCAN2 pipeline, the lack of detectable fucosidase activity is perhaps more likely explained by an incompatibility between the substrate specificity of the A. mucolyticum fucosidases and the monovalent substrates used in this study, as monovalent substrates are structurally different than the complex plant- and host-derived glycans found in the intestine.
In contrast to the lack of detectable levels of fucosidase activity, we were able to detect N-acetyl-α-galactosaminidase activity, which was not expected on basis of the CAZyme annotation (Table 1). This might indicate that A. mucolyticum is capable of hydrolyzing the α-linked GalNAc moiety that makes up the core of O-glycans, thereby enabling the complete degradation of mucin glycans. This hypothesis is supported by the finding that A. mucolyticum can effectively utilize GalNAc for growth (Figure 6). The enzyme(s) responsible for the N-acetyl-α-galactosaminidase activity remain to be identified.
Crost et al.23 have previously demonstrated that the ability of R. gnavus to grow on PGM as a sole carbon source is strain-dependent, and was due to the presence of sialidases and fucosidases. These results highlight the necessity for both types of glycosidases to remove the terminal sialic acid and fucose residues in order to allow degradation and subsequent utilization of the underlying glycans. A. mucolyticum produces multiple sialidases and fucosidases that are suspected to be essential to access mucin glycans for growth. Media supplemented with sialic acid or fucose, however, were not able to support growth (Figure 6), which is in agreement with the absence of core genes in the A. mucolyticum genome normally found in canonical gene clusters for sialic acid or fucose catabolism. Interestingly, a curated blast search of the genome does show the presence of several genes predicted to be involved in the uptake of sialic acid, like genes encoding for sialic acid TRAP transporter SiaQ and sialic acid-binding protein SiaP (Allo_00009 and Allo_00087, respectively).24 Furthermore, there are several putative polysialyltransferases (e.g. Allo_01518 and Allo_02009) with homology to proteins from Neisseria meningitidis known to play a role the sialylation of lipooligosaccharide (LOS). Sialylation of the bacterial LOS or capsule glycans is known to contribute to host colonization and bacterial immune evasion.25,26 When A. mucolyticum does not directly use host glycan-liberated sialic acid or fucose, they could still have a major impact on the ecology of the mucosal niche and host health as free sialic acid and fucose has been shown to boost the expansion of enteric pathogens like Salmonella typhimurium, Clostridium difficile, Campylobacter jejuni and Escherichia coli under certain conditions.27–30 In addition to this, removal of sialic acid from mucin glycans was shown to affect rotavirus adhesion and infection.31,32 Whether A. mucolyticum decorates its surface glycans with endogenous sialic acid or fucose, or affects the microbiota composition indirectly by releasing them into the environment remains to be investigated.
Increased bacterial penetration and degradation of the intestinal mucus layer are major features of IBD. Understanding the mechanisms that drive these processes may thus be key to treating or preventing disease. It has been reported that a diet deprived of dietary fiber enhances the expression of mucin targeting CAZymes of the gut microbiota and promotes the expansion of mucus degrading bacteria such as A. muciniphila and Bacteroides caccae.33 As mucin also seems to be an important nutrient source for A. mucolyticum, it may be that a similar expansion might be observed for this bacterium. Bacterial mucus degradation may cause erosion of the mucus layer and an increased susceptibility to lethal colitis by the enteric pathogen Citrobacter rodentium, as was shown in mice.33 This finding highlights the delicate balance between the microbiota, an intact mucus layer and invading pathogens.
Mucus layer degradation, as observed in IBD patients, is linked to an increased prevalence mucosa-associated mucolytic bacteria, like R. gnavus and R. torques.34 Since A. mucolyticum was isolated from a patient with ulcerative colitis and, as described here, is able to effectively forage on mucins, it is likely that it operates in the same niche as other mucolytic bacteria. It would therefore be of great interest to investigate the interactions with other known mucus-degrading bacteria and to investigate to what extent the mucolytic capacities of A. mucolyticum can contribute to the pathogenesis of IBD.
Materials and methods
General
All commercial chemicals were purchased from Sigma-Aldrich unless stated otherwise. Data visualization was performed using Adobe Illustrator 2021, GraphPad Prism 7, and R.
Carbohydrate-active enzyme (CAZyme) annotation
CAZymes within the Prokka-annotated genome of A. mucolyticum (Genbank accession number: JAHUZH010000000) and A. muciniphila ATCC BAA-835 (NCBI Reference Sequence: NC_010655.1) were annotated using the dbCAN2 metaserver (http://bcb.unl.edu/dbCAN2/) in February 2021.20,35 CAZymes identified by at least two out of three tools were considered for further analysis and are displayed in Figure 1. The putative mucin-targeting glycoside hydrolases, as displayed in Table 1, Figure 2, and supplementary STable 1, were selected based on the mucin targeting CAZyme classes reported by Tailford et al.6
Mucin and mucin O-glycan purification
Porcine gastric mucin (PGM type III, Sigma–Aldrich) was purified by ethanol precipitation as described previously by Ottman et al.36 with minor modifications. Briefly, 10 g of mucin was dissolved in 500 ml 2X PBS with pH 7.8 (NaCl: 274 mM, KCl: 5.4 mM, Na2HPO4: 20 mM, KH2PO4: 3.6 mM) and 100 µL of toluene for 24 h at 4°C, under continuous stirring. After the first hour, the pH was adjusted to 7.2 with 2 M NaOH. After 24 h, the solution was centrifuged at 10,000 x g for 20 min at 4°C, after which the supernatant containing the dissolved mucin was collected, cooled to 0°C and ice-cold (0°C) ethanol was added to a final concentration of 60% (vol/vol). The resulting precipitate was dissolved in 0.1 M NaCl and precipitated again with ice-cold ethanol to 60% (vol/vol). The precipitate was then washed with 100% ice-cold ethanol and dissolved in distilled water (~180 mL). Thereafter, the mucin solution was dialyzed against 5 L of distilled water for 24 h at 4°C, lyophilized, resuspended in Milli-Q ultrapure water, and sterilized by autoclaving (15 min at 121°C).
Porcine gastric mucin O-glycans were purified according to a protocol by Desai et al.33 In short, PGM (Type III, Sigma-Aldrich) was suspended at 2.5% w/v in 100 mM Tris (pH 7.4) and autoclaved immediately (15 min at 121°C). Proteinase K (Sigma) was added to a final concentration of 100 µg/mL and incubated for 16 h at 55°C with slow shaking. The proteolyzed solution was centrifuged at 21,000 x g for 30 min at 4°C to remove insoluble material. Next, NaOH and NaBH4 were added to final concentration of 0.1 M and 1 M, respectively. This solution was incubated for 18 h at 65°C to promote selective release of mucin O-glycans from mucin glycopeptides by alkaline β-elimination. The solution was neutralized to pH 7.0 with 2 M HCl, centrifuged at 21,000 x g for 30 min at 4°C, and filtered with a 0.2 µm filter to remove remaining insoluble material. After extensive dialysis (4 x 4 h with a 100X volume of Milli-Q Ultrapure water, 1 kDa MWCO dialysis tube), the dialyzed mucin O-glycan solution was lyophilized and resuspended in Milli-Q Ultrapure water at desired concentrations.
Human small intestinal mucins were harvested from the urine (~100 mL per batch) of a patient with an orthotopic neobladder reconstruction. Crude mucin was collected by centrifugation (30,000 x g, 4°C, 30 min) and pellets were washed twice with PBS. Washed mucin was purified further using ethanol precipitation, as described for PGM, and redissolved in 5 mL Milli-Q Ultrapure water. Instead of lyophilization, the mucin was directly transferred to a 100 kDa MWCO Amicon Ultra filter (Thermo Fisher Scientific), washed twice with 5 mL Milli-Q and concentrated. Protein concentration was determined using a NanoDrop One UV-Vis Spectrophotometer (Thermo Fisher Scientific) and set to 10 mg/mL in Milli-Q Ultrapure water.
Bacterial culture conditions
A. mucolyticum was routinely cultured at 37°C under anaerobic conditions in a vinyl anaerobic chamber (Coy Labs) with the following gas mix: 85% N2, 10% CO2 and 5% H2. Gut Microbiota Medium (GMM) (Table S2), was the default growth medium and is essentially an enriched Gut Microbiota Medium, as described by Goodman et al.37 that lacks isovaleric acid. The basal medium (BM) is GMM lacking glucose, fructose, maltose and cellobiose. Prior to use for bacterial growth, all media were prereduced in the anaerobic chamber for at least 12 h.
Bacterial growth curves
Gut Microbiota Medium and basal medium were prepared according to recipe in described in Table S2. All media supplements were prepared in Milli-Q Ultrapure water and, if required, solutions were brought to neutral pH using 1 M NaOH. Supplemented media were prepared with supplements at following final concentrations: D+ glucose (2 mg/ml, Sigma), D-fructose (1 mg/mL, Sigma), D+ maltose (1 mg/ml, Sigma), D+ cellobiose (1 mg/ml, Sigma), N-Acetylneuraminic acid (20 mM, Carbosynth), L-Fucose (20 mM, Sigma), D+ Galactose (20 mM, Mikrobiologie), N-acetyl-D-glucosamine (20 mM, Sigma), N-acetyl-D-galactosamine (20 mM, Carbosynth), inulin (10 mg/mL, Sigma), purified porcine gastric mucin (PGM, 0,5% w/vol) or purified PGM O-glycans (10 mg/mL). All solutions, except PGM solution, were filter-sterilized and prereduced in the anaerobic chamber prior to use
A. mucolyticum was grown on GMM agar plates for 72 h at 37°C under anaerobic conditions. Static liquid starter cultures were inoculated with three separate bacterial colonies each and grown for 16 h at 37°C under anaerobic conditions. Bacterial growth was assessed by measuring absorbance (600 nm). Bacteria were collected by centrifugation (3,000 x g, 5 min), gently washed twice and resuspended in 2X basal medium. Washed bacteria were diluted to the desired OD and combined with an equal volume of MilliQ Ultrapure water or medium supplements to a total volume of 200 µL per well with a final starting OD of 0.01. For each time point (0, 8, 16, 24, 32, 48 and 72 h) a 96-Well Multiwell Plate (Corning Costar) was used with duplicate wells per condition. At each time point one plate was removed from the anaerobic chamber and final absorbance values (600 nm) were recorded using a FLUOstar Omega plate reader (BMG Labtech). Values were corrected for background levels in respective negative controls. Experiments were performed in triplicates, using three biologically independent A. mucolyticum starter cultures.
Bacterial conditioned media and whole cell lysate preparation
Bacterial conditioned media and whole-cell lysates for mass spectrometric analysis were harvested from static liquid A. mucolyticum cultures that were grown in Gut Microbiota Medium, basal medium or basal medium supplemented with 0.5% PGM for 24 h at 37°C under anaerobic conditions. For each condition, 4 mL cultures were inoculated from three independent starter cultures and grown as described above. After 24 h, bacteria were harvested by centrifugation at 15,000 x g for 2 min, washed twice with 2 mL of PBS and resuspended in 4 mL of ice-cold lysis buffer (PBS containing cOmplete EDTA-free protease inhibitor cocktail (Roche)). The conditioned media were collected and filter-sterilized using 0.2 µm filter. Bacteria were lysed by sonication (3 x 10 seconds on ice). Large bacterial fragments were removed by centrifugation at 15,000 x g for 1 min, after which cleared lysate was collected. To remove possible media components all bacterial cell lysates and conditioned media were transferred to a 5 kDa MWCO filter (Pierce concentrator, PES, 5 K MWCO, 6 ml, Thermo Fisher Scientific), washed with 5 mL of Milli-Q and concentrated to 1 mL.
The conditioned media used for glycosidase enzymatic activity assays were collected from three independent static liquid A. mucolyticum cultures grown in the Gut Microbiota Medium for 48 h at 37°C under anaerobic conditions. The conditioned media were filter-sterilized using 0.2 µm filter and stored at −20°C prior to use. The 100X concentrated conditioned media used for fucosidase assays were obtained using a 10 kDa MWCO Amicon Ultra filter (Thermo Fisher Scientific).
Mass spectrometry sample preparation
Whole cell lysates and conditioned media were processed using Filter Aided Sample Preparation (FASP), as described by Wiśniewski et al.38 In brief, samples were denatured at 95°C in the presence of DTT, mixed with 8 M urea and loaded onto Centrifugal Filters (Microcon, cat. no MRCF0R030). This was followed by two washes with 8 M urea, treatment with 0.05 M iodoacetamide in 8 M urea, and three 8 M urea washes. After washing out the urea with three washes with 0.05 M ammonium bicarbonate, samples were digested overnight with trypsin at 37°C. The next day, samples were acidified and desalted using Stagetips.39 Half of each of the digested samples was injected into an LTQ-Orbitrap QExactive mass spectrometer (Thermo Fisher Scientific) and measured using a 120-minute gradient.
Mass spectrometry analyses
Thermo Raw files were analyzed using MaxQuant versions 1.5.1.0 using default parameters, with the inclusion of the match between runs and IBAQ features.40,41 Initial analyses were performed in Perseus.42 Proteins flagged as contaminants, reverse or only identified by site were filtered out. Triplicates were grouped and only proteins reproducibly quantified in at least one of the sets of triplicates were retained. Missing values were imputed using default parameters. Differential proteins were determined using a t-test with adjustment for multiple testing (FDR < 0.05). To call proteins enriched in the secreted fractions, they additionally required at least a 10-fold higher LFQ value compared to the whole cell lysates. Data visualization and downstream processing was performed in R.
Glycosidase activity assays
A. mucolyticum conditioned media used for enzymatic assays were collected as described above. The following substrates were used: 4-MU-Neu5Ac (Cayman), 4-MU-Fucose (Carbosynth), 4-MU-GlcNAc (Carbosynth), 4-MU-GalNAc (Carbosynth), and ortho-Nitrophenyl-β-galactoside (ONPG, Sigma). Stocks of the 4-MU linked substrates were diluted to 200 µM in Milli-Q Ultrapure water and ONPG was diluted to 1 mg/mL in Milli-Q Ultrapure water, after which 50 µl of diluted substrates were added to a 96-Well Multiwell flat-bottom Plate (Corning Costar). Next, 50 µL of either the Gut Microbiota Medium (negative control), conditioned medium or heat-inactivated conditioned medium (98°C for 30 min) were added in duplicates. Plates were incubated for 2 h at 37°C shielded from light and transferred to a FLUOstar Omega plate reader (BMG labtech). For the 4-MU linked substrates, signal was detected using a 340 ex./460 em. filter set. For the ONPG substrate, absorbance was measured at a wavelength of 420 nm.
Chemoselective labeling of fucosidases using an activity-based probe was performed as described by Luijkx and Henselijn et al.43
Detection of mucin degradation by Periodic acid-Schiff (PAS) stain and western blotting
PGM or human mucin (10 mg/mL in Milli-Q Ultrapure water) were combined at a 1:1 ratio with medium controls or the same A. mucolyticum conditioned medium that was used for the glycosidase activity assays. This mixture was incubated for 16 h at 37°C after which samples were mixed with one volume of 3x Laemmli sample buffer and boiled for 5 min. Subsequently 25 µL per sample was loaded onto a gel and separated using SDS-PAGE. For SDS-PAGE separation and immunoblotting of large mucin proteins, a Boric acid-Tris gel system was used, as previously described by Li et al,44 with minor modifications. A 5% acrylamide gel (5% Acryl/Bis acryl solution, Bio-Rad 161–0144; 26% 1.5 M Tris pH 8.8; 0.1% SDS; 0.1% ammonium persulfate; 0.1% TEMED) was prepared in a Mini Protean II chamber (Bio-Rad) using 1.5 mm spacer plates. Gels were run in Boric acid-Tris buffer (192 mM Boric acid; 1 mM EDTA; 0.1% SDS, set to pH 7.6 with Tris) at 25 mA per gel for 3 h. To detect glycoproteins, gels were stained using a Periodic acid-Schiff stain (Pierce glycoprotein staining kit, Thermo Fisher Scientific). For detection of MUC2 and sialic acids, mucins were separated as described above and transferred onto nitrocellulose membranes using a wet transfer system with transfer buffer (25 mM Tris; 192 mM glycine; 20% methanol,) for 3 h at 90 V at 4°C. Subsequently, membranes were blocked with 5% bovine serum albumin (BSA, Sigma-Aldrich) in TSMT buffer (20 mM Tris, 150 mM NaCl, 1 mM CaCl2, 2 mM MgCl2, 0.1% Tween 20, adjusted to pH 7 with HCl) overnight at 4°C and then incubated with an anti-human MUC2 rabbit serum (a gift from Dr. Karin Strijbis) at a 1:200 dilution in TSMT containing 1% BSA, or with biotinylated SNA lectin (2 mg/mL stock, Vector Labs) at a dilution of 1:1000 in TSMT containing 1% BSA. After incubation for 1 h at room temperature, blots were washed 4 times with an excess of TSMT and incubated with α-rabbit IgG (A4914, 1:10,000 dilution, Sigma-Aldrich) in TSMT containing 1% BSA for the detection of MUC2, or Streptavidin-HRPO (1 mg/ml stock, 1:10,000 dilution, Jackson ImmunoResearch) in TSMT containing 1% BSA for the detection of sialic acids. Blots were developed with the Clarity Western ECL kit (Bio-Rad) and imaged in a Gel-Doc system (Bio-Rad).
Supplementary Material
Acknowledgments
We would like to kindly thank Dr. Karin Strijbis for providing the anti-human MUC2 rabbit serum.
We would like to kindly thank Dr. Yvette Luijkx and Dr. Tom Wennekes for providing the activity-based probe and advice on experimental setup.
Funding Statement
M.R.d.Z. was supported by a VIDI grant from the Netherlands Organization for Scientific Research (NWO, grant 91715377) and the Utrecht Exposome Hub of Utrecht Life Sciences (www.uu.nl/exposome), funded by the Executive Board of Utrecht University.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD024919.45 Data can be accessed via https://www.ebi.ac.uk/pride/ using dataset identifier above.
The A. mucolyticum Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession JAHUZH000000000. The version described in this paper is version JAHUZH010000000.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website
References
- 1.Honda K, Littman DR.. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535(7610):75–18. doi: 10.1038/nature18848. [DOI] [PubMed] [Google Scholar]
- 2.Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. International Journal of Systematic and Evolutionary Microbiology. 2004;54(5):1469–1476. doi: 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- 3.Van Passel MWJ, Kant R, Zoetendal EG, Plugge CM, Derrien M, Malfatti SA, Chain PSG, Woyke T, Palva A, de Vos WM, et al. The Genome of Akkermansia muciniphila, a Dedicated Intestinal Mucin Degrader, and Its Use in Exploring Intestinal Metagenomes. PLoS ONE 2011;6(3):e16876. doi: 10.1371/journal.pone.0016876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sonnenburg JL. Glycan Foraging in Vivo by an Intestine-Adapted Bacterial Symbiont. Science 2005;307(5717):1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 5.Martens EC, Chiang HC, Gordon JI. Mucosal Glycan Foraging Enhances Fitness and Transmission of a Saccharolytic Human Gut Bacterial Symbiont. Cell Host & Microbe. 2008;4(5):447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tailford LE, Crost EH, Kavanaugh D, Juge N. Mucin glycan foraging in the human gut microbiome. Frontiers in Genetics. 2015;6(FEB). doi: 10.3389/fgene.2015.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johansson MEV, Gustafsson JK, Holmén-Larsson J, Jabbar KS, Xia L, Xu H, Ghishan FK, Carvalho FA, Gewirtz AT, Sjövall H, et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut. 2014;63(2):281–291. doi: 10.1136/gutjnl-2012-303207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, Degnan PH, Hu J, Peter I, Zhang W, et al. Immunoglobulin A Coating Identifies Colitogenic Bacteria in Inflammatory Bowel Disease. Cell. 2014;158(5):1000–1010. doi: 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greetham HL, Gibson GR, Giffard C, Hippe H, Merkhoffer B, Steiner U, Falsen E, Collins MD. Allobaculum stercoricanis gen. nov., sp. nov., isolated from canine feces. Anaerobe. 2004;10(5):301–307. doi: 10.1016/j.anaerobe.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 10.Bárcena C, Valdés-Mas R, Mayoral P, Garabaya C, Durand S, Rodríguez F, Fernández-García MT, Salazar N, Nogacka AM, Garatachea N, et al. Healthspan and lifespan extension by fecal microbiota transplantation into progeroid mice. Nature Medicine. 2019;25(8):1234–1242. doi: 10.1038/s41591-019-0504-5. [DOI] [PubMed] [Google Scholar]
- 11.Thevaranjan N, Puchta A, Schulz C, Naidoo A, Szamosi JC, Verschoor CP, Loukov D, Schenck LP, Jury J, Foley KP, et al. Age-Associated Microbial Dysbiosis Promotes Intestinal Permeability, Systemic Inflammation, and Macrophage Dysfunction. Cell Host and Microbe. 2017;21(4):455–466. e4. doi: 10.1016/j.chom.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ravussin Y, Koren O, Spor A, LeDuc C, Gutman R, Stombaugh J, Knight R, Ley RE, Leibel RL. Responses of Gut Microbiota to Diet Composition and Weight Loss in Lean and Obese Mice. Obesity. 2012;20(4):738–747. doi: 10.1038/oby.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herrmann E, Young W, Rosendale D, Reichert-Grimm V, Riedel CU, Conrad R, Egert M. RNA-Based Stable Isotope Probing Suggests Allobaculum spp. as Particularly Active Glucose Assimilators in a Complex Murine Microbiota Cultured in Vitro. BioMed Research International. 2017:2017. doi: 10.1155/2017/1829685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pujo J, Petitfils C, Le Faouder P, Eeckhaut V, Payros G, Maurel S, Perez-Berezo T, Van Hul M, Barreau F, Blanpied C, et al. Bacteria-derived long chain fatty acid exhibits anti-inflammatory properties in colitis. Gut. 1917;2020. gutjnl-2020-321173. doi: 10.1136/gutjnl-2020-321173. [DOI] [PubMed] [Google Scholar]
- 15.Liu S, Qin P, Wang J. High-fat diet alters the intestinal microbiota in streptozotocin-induced type 2 diabetic mice. Microorganisms. 2019;7(6):176. doi: 10.3390/microorganisms7060176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cox LM, Yamanishi S, Sohn J, A V A, Leung JM, Cho I, Rogers AB, Kim SG, Li H, Gao Z, et al. Altering the Intestinal Microbiota during a Critical Developmental Window Has Lasting Metabolic Consequences. Cell. 2014;158(4):705–721. doi: 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaakoush NO. Insights into the Role of Erysipelotrichaceae in the Human Host. Frontiers in Cellular and Infection Microbiology. 2015;5(November):1–4. doi: 10.3389/fcimb.2015.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Science Translational Medicine. 2009;1(6):1–12. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyauchi E, Kim S, Suda W, Kawasumi M, Onawa S, Taguchi-Atarashi N, Morita H, Taylor TD, Hattori M, Ohno H. Gut microorganisms act together to exacerbate inflammation in spinal cords. Nature. 2020;585(7823):102–106. doi: 10.1038/s41586-020-2634-9. [DOI] [PubMed] [Google Scholar]
- 20.Zhang H, Yohe T, Huang L, Entwistle S, Wu P, Yang Z, Busk PK, Xu Y, Yin Y, Cox L. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Research 2018;46(W1):W95–W101. doi: 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kudelka MR, Stowell SR, Cummings RD, Neish AS. Intestinal epithelial glycosylation in homeostasis and gut microbiota interactions in IBD. Nature Reviews Gastroenterology and Hepatology. 2020;17(10):597–617. doi: 10.1038/s41575-020-0331-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sumida T, Fujimoto K, Ito M. Molecular cloning and catalytic mechanism of a novel glycosphingolipid- degrading β-N-acetylgalactosaminidase from Paenibacillus sp. TS12. Journal of Biological Chemistry. 2011;286(16):14065–14072. doi: 10.1074/jbc.M110.182592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crost EH, Tailford LE, Le Gall G, Fons M, Henrissat B, Juge N. Utilisation of Mucin Glycans by the Human Gut Symbiont Ruminococcus gnavus Is Strain-Dependent. PLoS ONE 2013;8:10. doi: 10.1371/journal.pone.0076341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price MN, Arkin AP, Greene CS. Curated BLAST for Genomes Greene CS, editor. mSystems. 2019;4(2):1–7. doi: 10.1128/mSystems.00072-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carlin AF, Uchiyama S, Chang YC, Lewis AL, Nizet V, Varki A. Molecular mimicry of host sialylated glycans allows a bacterial pathogen to engage neutrophil Siglec-9 and dampen the innate immune response. Blood. 2009;113(14):3333–3336. doi: 10.1182/blood-2008-11-187302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis LA, Gulati S, Burrowes E, Zheng B, Ram S, Rice PA. α-2,3-Sialyltransferase Expression Level Impacts the Kinetics of Lipooligosaccharide Sialylation, Complement Resistance, and the Ability of Neisseria gonorrhoeae to Colonize the Murine Genital Tract. mBio. 2015;6(1):1–11. doi: 10.1128/mBio.02465-14.Editor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502(7469):96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang YL, Chassard C, Hausmann M, Von Itzstein M, Hennet T. Sialic acid catabolism drives intestinal inflammation and microbial dysbiosis in mice. Nature Communications 2015;6:1–11. doi: 10.1038/ncomms9141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pacheco AR, Curtis MM, Ritchie JM, Munera D, Waldor MK, Moreira CG, Sperandio ,V. Fucose sensing regulates bacterial intestinal colonization. Nature. 2012;492(7427):113–117. doi: 10.1038/nature11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luijkx YMCA, Bleumink NMC, Jiang J, Overkleeft HS, Wösten MMSM, Strijbis K, Wennekes T. Bacteroides fragilisfucosidases facilitate growth and invasion of Campylobacter jejuni in the presence of mucins. Cellular Microbiology 2020;22(12):12. doi: 10.1111/cmi.13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Engevik MA, Banks LD, Engevik KA, Chang-Graham AL, Perry JL, Hutchinson DS, Ajami NJ, Petrosino JF, Hyser JM. Rotavirus infection induces glycan availability to promote ileum-specific changes in the microbiome aiding rotavirus virulence. Gut Microbes. 2020;11(5):1324–1347. doi: 10.1080/19490976.2020.1754714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alfajaro MM, Kim J-Y, Barbé L, Cho E-H, Park J-G, Soliman M, Baek Y-B, Kang M-I, Kim SH, Kim G-J, et al. Dual Recognition of Sialic Acid and αGal Epitopes by the VP8* Domains of the Bovine Rotavirus G6P[5] WC3 and of Its Mono-reassortant G4P[5] RotaTeq Vaccine Strains López S, editor. Journal of Virology 2019;93(18):18. doi: 10.1128/JVI.00941-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, Pudlo NA, Kitamoto S, Terrapon N, Muller A, et al. A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell. 2016;167(5):1339–1353.e21. doi: 10.1016/j.cell.2016.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Png CW, Lindén SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, McGuckin MA, Florin THJ. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. American Journal of Gastroenterology. 2010;105(11):2420–2428. doi: 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- 35.Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Research. 2012;40(W1):W445–W451. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ottman N, Davids M, Suarez-Diez M, Boeren S, Schaap PJ, dos Santos VAPM, Smidt H, Belzer C, de Vos WM, Kelly RM. Genome-scale model and omics analysis of metabolic capacities of Akkermansia muciniphila reveal a preferential mucin-degrading lifestyle. Applied and Environmental Microbiology. 2017;83(18):1–15. doi: 10.1128/AEM.01014-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goodman AL, Kallstrom G, Faith JJ, Reyes A, Moore A, Dantas G, Gordon JI. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proceedings of the National Academy of Sciences. 2011;108(15):6252–6257. doi: 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nature Methods. 2009;6(5):359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 39.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nature Protocols. 2007;2(8):1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 40.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature Biotechnology. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 41.Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann MM. Accurate Proteome-wide Label-free Quantification by Delayed Normalization and Maximal Peptide Ratio Extraction, Termed MaxLFQ. Molecular & Cellular Proteomics. 2014;13(9):2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nature Methods. 2016;13(9):731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 43.Luijkx YMCA, Henselijn AJ, Bosman GP, Cramer DAT, Giesbers CAP, Van ’T Veld EM, Boons GJPH, Heck AJR, Reiding KR, Strijbis K, et al. Detection of bacterial α - L -fucosidases with an ortho -quinone methide-based probe and mapping of the probe-protein adducts. Manuscript submitted for publication [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li X, Bleumink-Pluym NMC, Luijkx YMCA, Wubbolts RW, van Putten JPM, Strijbis K. MUC1 is a receptor for the Salmonella SiiE adhesin that enables apical invasion into enterocytes. PLOS Pathogens 2019;15(2):e1007566. doi: 10.1371/journal.ppat.1007566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Research 2019;47(D1):D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD024919.45 Data can be accessed via https://www.ebi.ac.uk/pride/ using dataset identifier above.
The A. mucolyticum Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession JAHUZH000000000. The version described in this paper is version JAHUZH010000000.


