Abstract
Aberrant activation of the HOX, MEIS, and PBX homeodomain protein families is associated with leukemias, and retrovirally driven coexpression of HOXA9 and MEIS1 is sufficient to induce myeloid leukemia in mice. Previous studies have demonstrated that HOX-9 and HOX-10 paralog proteins are unique among HOX homeodomain proteins in their capacity to form in vitro cooperative DNA binding complexes with either the PBX or MEIS protein. Furthermore, PBX and MEIS proteins have been shown to form in vivo heterodimeric DNA binding complexes with each other. We now show that in vitro DNA site selection for MEIS1 in the presence of HOXA9 and PBX yields a consensus PBX-HOXA9 site. MEIS1 enhances in vitro HOXA9-PBX protein complex formation in the absence of DNA and forms a trimeric electrophoretic mobility shift assay (EMSA) complex with these proteins on an oligonucleotide containing a PBX-HOXA9 site. Myeloid cell nuclear extracts produce EMSA complexes which appear to contain HOXA9, PBX2, and MEIS1, while immunoprecipitation of HOXA9 from these extracts results in coprecipitation of PBX2 and MEIS1. In myeloid cells, HOXA9, MEIS1, and PBX2 are all strongly expressed in the nucleus, where a portion of their signals are colocalized within nuclear speckles. However, cotransfection of HOXA9 and PBX2 with or without MEIS1 minimally influences transcription of a reporter gene containing multiple PBX-HOXA9 binding sites. Taken together, these data suggest that in myeloid leukemia cells MEIS1 forms trimeric complexes with PBX and HOXA9, which in turn can bind to consensus PBX-HOXA9 DNA targets.
The mechanisms by which the HOX homeodomain proteins regulate tissue patterning during development remain largely unknown. Although they appear to be DNA binding proteins, early observations indicated that many Drosophila and mammalian HOX proteins bind only weakly to DNA by themselves and/or exhibit a high degree of redundancy in binding site specificity (34). More recent studies demonstrated that HOX proteins form heterodimeric DNA binding complexes with members of the EXD/PBX (reviewed in reference 26) or MEIS (41) family of homeodomain proteins. Interactions with PBX provide both increased DNA binding affinity for many HOX proteins (40) and substantial DNA selectivity for HOX proteins from paralog groups 1 to 10 (8). Data from a number of laboratories suggested that EXD/PBX-HOX heterodimers bind to consensus TGAT(T/G)NA(T/C) sites in which EXD or PBX protein binds to the 5′ TGAT site and the HOX proteins bind to the 3′ (T/G)NA(T/C) site. (Throughout the text and in Table 1, PBX consensus binding sites are underlined, HOXA9 consensus sites are double underlined, and MEIS1 consensus binding sites are broken underlined for clarity.) HOX proteins exhibit differential binding specificity through interaction with the variable nucleotide in position 6 (8, 10, 25, 35, 42). In particular, in vitro DNA site selection experiments suggest that HOXA9 and HOXA10 form cooperative binding complexes with PBX on TGATTTAC and TGATTTAT consensus targets (8, 42). For interaction with PBX, the HOX proteins from paralog groups 1 through 8 require a tryptophan within a conserved YPWM motif, while the paralog group 9 and 10 proteins use a tryptophan in a conserved ANW sequence. The ABD-B-like HOX proteins from paralog groups 9 through 13 form cooperative DNA binding complexes with MEIS1 on consensus targets containing a 5′ MEIS1 site (..................TGACAG) followed by a TTA(C/T)GAC HOX protein binding site (41). Thus, HOX proteins from paralog groups 9 and 10, located at the transition between the YPWM-containing proteins from paralog groups 1 through 8 and the ABD-B-like proteins, possess the unique capacity to form DNA binding complexes with either PBX or MEIS on distinct DNA target sequences.
TABLE 1.
DNA site selection in the presence of HOXA9, PBX1A, and MEIS1 yields PBX-HOXA9 sitesa
| Protein precipitated | Content at indicated position
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
| HOXA9 (n = 13) | ||||||||||||
| %G | 33 | 8 | 92 | 8 | 0 | 0 | 8 | 0 | 0 | 48 | 54 | 8 |
| %A | 33 | 0 | 0 | 92 | 0 | 0 | 23 | 100 | 0 | 15 | 23 | 15 |
| %T | 33 | 92 | 8 | 0 | 100 | 100 | 69 | 0 | 85 | 31 | 23 | 46 |
| %C | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 15 | 8 | 0 | 31 |
| Consensus | G/A/T | T | G | A | T | T | T | A | T/C | G/T | G | T/C |
| PBX1 (n = 16) | ||||||||||||
| %G | 43 | 12 | 81 | 0 | 6 | 0 | 0 | 0 | 0 | 38 | 56 | 13 |
| %A | 38 | 12 | 19 | 100 | 0 | 0 | 6 | 100 | 0 | 18 | 38 | 7 |
| %T | 19 | 76 | 0 | 0 | 94 | 100 | 94 | 0 | 75 | 38 | 0 | 27 |
| %C | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 25 | 6 | 6 | 53 |
| Consensus | G/A | T | G | A | T | T | T | A | T/C | G/T | G/A | C/T |
| MEIS1 (n = 12) | ||||||||||||
| %G | 25 | 0 | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 76 | 33 | 33 |
| %A | 75 | 0 | 0 | 100 | 0 | 0 | 0 | 100 | 0 | 8 | 42 | 12 |
| %T | 0 | 100 | 0 | 0 | 100 | 100 | 100 | 0 | 92 | 8 | 8 | 22 |
| %C | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 8 | 17 | 33 |
| Consensus | A/G | T | G | A | T | T | T | A | T/C | G | A/G | C/G |
| HOXA9 + MEIS1 + PBX1 (n = 21) | ||||||||||||
| %G | 30 | 0 | 100 | 0 | 0 | 0 | 5 | 0 | 0 | 62 | 38 | 28 |
| %A | 45 | 10 | 0 | 95 | 0 | 0 | 19 | 100 | 0 | 14 | 19 | 9 |
| %T | 15 | 90 | 0 | 5 | 100 | 100 | 76 | 0 | 81 | 19 | 14 | 9 |
| %C | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 19 | 5 | 19 | 54 |
| Consensus | A/G | T | G | A | T | T | T/A | A | T/C | G | G | C/G |
Combinations of epitope-tagged and untagged proteins were used to select DNA binding sites as described in Materials and Methods.
The existence of cooperative interactions between HOX/HOM-C and EXD proteins in Drosophila was initially surmised from genetic data (33) and later confirmed by in vitro studies (7, 47). In contrast, to date there has been only limited in vivo evidence for HOX-PBX complexes in mammalian cells (14, 36). Several recent studies have provided in vivo data suggesting that in certain nonmyeloid cells, much of the PBX protein in nuclear extracts appears to be bound in tight complexes with MEIS1 (9) or MEIS-like proteins (16). These investigations have used immunoprecipitated PBX proteins from nuclear extracts in subsequent DNA site selection assays to demonstrate a TGAT..................TGACAG consensus binding site for in vivo PBX protein complexes. In these studies, PBX-HOX consensus sites were not detected. Recently, Berthelsen et al. purified a urokinase enhancer binding complex from HeLa cells and demonstrated that it was a mixture of either PBX1 or PBX2 tightly complexed with a novel MEIS1-like protein, Prep1 (5). In Drosophila the MEIS-like protein, homothorax, appears to form protein-protein complexes with EXD and to play a key role in its nuclear localization (32, 38). In contrast, it has been suggested that several Drosophila HOM-C proteins including ABD-B, the homologue of HOXA9, act to prevent the nuclear localization of EXD (2). Data on the subcellular localization of mammalian MEIS or HOX proteins have not been reported, while PBX1 protein is partitioned between the nucleus and cytoplasm along the leg primordium, under the control of Hedgehog signaling (13).
Although Hox genes play an important role in embryonic development (18), these genes are also expressed in lineage- and stage-specific patterns during adult hematopoietic differentiation and in leukemias (reviewed in reference 23). (Throughout this paper, mouse genes appear with an initial capital letter and human genes are completely capitalized.) It is of interest that both PBX (15, 31) and Meis (28) were originally identified because of their involvement in leukemic transformation, and Hoxa9 and Meis1 are coordinately up-regulated by retroviral integration in myeloid leukemias arising in BXH-2 mice (30). We have been studying the role of HOX proteins, and especially HOXA9 and HOXA10, in myeloid blood cell differentiation and leukemogenesis (21, 22, 46). Retrovirus-mediated overexpression of the Hoxa9 or Hoxa10 gene in bone marrow cells produces delayed leukemias in mice (17, 46), while overexpression of Hoxa9 together with Meis1 in hematopoietic progenitor cells results in extremely rapid development of myeloid cell tumors, suggesting that coexpression of these two genes is sufficient for primary transformation (17). Within blood cells, expression of both the HOXA9 and MEIS1 genes appears to be largely restricted to the myeloid cell lineage, and the coexpression of these genes is correlated with myeloid leukemias in human patient samples (21). It should be noted that among the 9 and 10 paralog genes, only the HOXA9 and HOXA10 genes appear to be expressed in myeloid cells. Since PBX genes are also expressed in myeloid cells (27), the HOXA9 and HOXA10 proteins are uniquely positioned among the HOX proteins in being capable of forming DNA binding complexes with both PBX and MEIS proteins in these cells. These observations lead to the question of which DNA-protein binding complexes are physiologically relevant and/or play a role in myeloid leukemic transformation.
In this study, we have demonstrated that MEIS1 forms a triple protein-protein complex with HOXA9 and PBX in the absence of DNA in vitro and also forms trimeric complexes with PBX and HOXA9 on a PBX-HOXA9 DNA binding target. We have extended these findings by using coimmunoprecipitation, electrophoretic mobility shift assays (EMSA), and immunofluoresence localization to demonstrate that a portion of the HOXA9 protein present in myeloid cells is found in the nucleus within ternary complexes with PBX2 and MEIS1.
MATERIALS AND METHODS
Protein expression.
cDNAs encoding full-length murine HOXA9 and the human PBX1a, PBX2, and PBX3 proteins were subcloned into an sp65 vector containing an SP6 promoter (Promega, Madison, Wis.) engineered to express proteins from their respective translational start sites (PBX1a, PBX2, and PBX3) or as fusion proteins containing an N-terminal FLAG epitope sequence (MDYKDDDDK) (PBX1a and HOXA9). HOXA9 and MEIS1a and MEIS1b were also cloned into a pET28 vector (Novagen, Madison, Wis.) containing a T7 promoter, which produces proteins with an N-terminal T7 epitope tag sequence. For use in antibody purification or purification, HOXA9 and HOXA10 were expressed as glutathione S-transferase (GST) fusion proteins in the pGEX vector (Pharmacia, Piscataway, N.J.). For gel shift, in vitro precipitation, and DNA target selection assays, full-length homeodomain proteins were synthesized with the relevant N-terminal epitope tags, using the TNT coupled in vitro transcription-translation system, in parallel reactions in the presence and absence of [35S]methionine. In each case, electrophoresis of the labeled proteins demonstrated synthesis of the appropriate full-length product (Fig. 1B). The identity of each protein was confirmed previously, using specific antisera (41, 42). Using autoradiography and densitometry of the 35S-labeled proteins, and calculating the incorporation of labeled methionine of known specific activity into each protein, we estimated that the relative protein concentrations used were within a twofold range. Each of the epitope-tagged MEIS1, PBX1, and HOXA9 proteins was shown to be functional in DNA site selection or EMSA analyses (see Results) and in previous studies (41, 42).
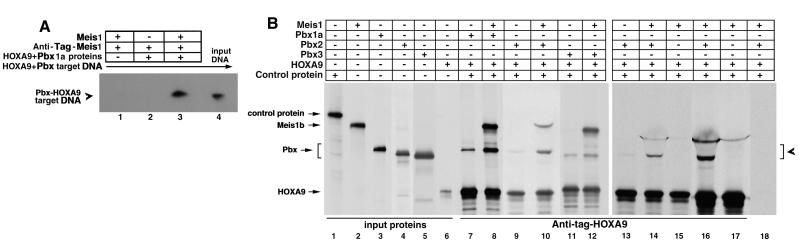
FIG. 1.
MEIS1 forms protein-protein complexes with HOXA9 and PBX1a in the presence and absence of DNA. (A) Immunoprecipitation of target DNA complexed to PBX and HOXA9 proteins is mediated by MEIS1 protein. To demonstrate ternary complex formation on a DNA target, an epitope-tagged MEIS1 protein was mixed with untagged HOXA9 and PBX1a proteins and a 32P-labeled DNA target containing a PBX-HOXA9 consensus binding site. Immunoprecipitation with antiserum to the MEIS1 epitope tag brought down the target DNA (lane 3). The DNA was not precipitated when either the PBX and HOXA9 proteins (lane 1) or the MEIS1 protein (lane 2) was omitted. Lane 4 shows the migration position of the input labeled oligonucleotide. (B) MEIS1 forms a triple complex with PBX and HOXA9 in the absence of DNA. [35S]methionine-labeled, FLAG epitope-tagged HOXA9 protein and untagged PBX1a, PBX2, or PBX3 protein, along with a control luciferase protein, were synthesized in vitro, treated with DNase to remove contaminating DNA, and incubated with or without labeled untagged MEIS1b protein. Lanes 1 to 6 show the migration positions of the input proteins. Lanes 7 through 17 were incubated with antiserum to the epitope tag on the HOXA9 protein to precipitate HOXA9 and associated proteins. Immunoprecipitation of the epitope-tagged HOXA9 protein in the absence of MEIS1b resulted in precipitation of low amounts of PBX1a (lane 7), PBX2 (lane 9), or PBX3 (lane 11). Addition of MEIS1b resulted in a 5- to 10-fold increase in precipitation of the PBX1a protein (lane 8), a 7-fold increase in PBX2 (lane 10), and a 3-fold increase in PBX3 (lane 12). The control protein was not precipitated, demonstrating that the protein-protein interactions were specific. To ensure that contaminating DNAs were not facilitating the observed protein-protein interactions, immunoprecipitation experiments were performed in the presence of EtdBr to prevent protein-DNA interactions (43) (lanes 13 to 18). With EtdBr present, addition of MEIS1b resulted in a 10-fold enhancement of PBX2-HOXA9 binding (lanes 13 and 14). In a similar manner, the binding of MEIS1 to HOXA9 was increased fivefold in the presence of PBX2 (compare lane 15 with lane 14). Lanes 16 and 17 are a longer autoradiographic exposure of lanes 14 and 15, to more clearly demonstrate the previously reported observation that MEIS1 binds to HOXA9 in the absence of PBX proteins (41). The MEIS1b protein reproducibly migrated as a diffuse band in the presence of EtdBr (lanes 14 to 17). When either the epitope-specific antibody (lane 18) or the epitope-tagged HOXA9 protein (not shown) was omitted from the reaction mixture, no proteins were precipitated. The brackets mark the position of the various PBX protein bands coprecipitated with HOXA9.
DNA site selection protocol.
Site selection using the in vitro-synthesized MEIS1b, PBX1a, and HOXA9 proteins was performed according to the basic protocol described by Blackwell and Weintraub (6). In each case, a mixture of T7- and FLAG epitope-tagged proteins was used such that only the desired protein would be immunoprecipitated during the selection assay. The T7- or FLAG epitope-tagged HOXA9, MEIS1b, and PBX1a fusion proteins were synthesized in vitro, and estimated equimolar amounts of each protein were incubated at 4°C for 2 h to overnight with a 63-mer containing a random 24-mer core flanked by arms which contained cloning sites (5′-GCTCGAATTCAAGCTTCTN24CATGGATCCTGCAGAATTCAGT-3′). Bound DNA was immunoprecipitated with an antiserum to the T7 or FLAG tag sequence fused to appropriate HOXA9, PBX1a, or MEIS1b protein. Following extensive washing steps, the DNA was amplified by 15 to 20 cycles of PCR (94°C, 1 min; 54°C, 1 min; 72°C, 1 min), using primers designed against the flanking arms. After four cycles of selection, the amplified DNA was subcloned and sequenced by standard methods. Consensus sequences were determined by visual alignment of sequences from unique clones.
In vitro protein coimmunoprecipitation assays.
Mixtures of 35S-labeled T7-tagged MEIS1b protein, untagged PBX1a, PBX2, or PBX3 protein, and FLAG-tagged HOXA9 protein were incubated at 4°C in binding buffer (75 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 10 mM Tris-HCl [pH 7.5], 1% bovine serum albumin [BSA] 6% glycerol), in the presence of protein G beads which had been preblocked in a solution of 1% BSA in binding buffer, for 60 min prior to addition of anti-FLAG antisera. Reticulocyte lysate containing the appropriate viral polymerase was substituted for specific proteins, omitted from reaction mixtures, to control for possible protein-protein interactions mediated by endogenous lysate factors. Following an additional overnight incubation, the beads were centrifuged and washed extensively (75 mM NaCl, 15 mM Tris-HCl [pH 7.5], 1% BSA, 0.15% Triton X-100). Proteins were solubilized in Laemmli buffer and subjected to polyacrylamide gel electrophoresis (Fig. 1B). Two approaches were used to ensure the absence of DNA interactions in protein-protein reaction mixtures. In most experiments, DNase treatment of the reticulocyte lysate protein synthesis mixture was used to remove contaminating DNA. In some experiments, ethidium bromide (EtdBr) (60 μg/ml) was added to the reaction mixtures to prevent DNA-protein interactions (19). In these experiments, EtdBr was included in all of the precipitation and washing steps. A similar strategy using T7-tagged MEIS1 and FLAG-epitope tagged PBX1a and HOXA9 proteins, in the presence or absence of a 32P-labeled oligonucleotide containing a PBX-HOXA9 binding site, was used for the in vitro DNA precipitation studies in the absence of EtdBr (Fig. 1A).
EMSA.
Complementary oligonucleotides (upper strand shown) containing a previously identified consensus binding site for PBX1a with HOXA9 (ctgcgATGATTTACGACcgc) (42) were synthesized (Operon Technologies, Alameda, Calif.). The standard conditions used were similar to those previously described (41). Briefly, double-stranded, end-labeled DNA (50,000 cpm/binding reaction, 10 nM) was incubated with 2 μl of reticulocyte lysate reaction mixture containing the test HOXA9 and/or PBX protein (1 nM), either in the presence of 2 μl of reticulocyte lysate reaction mixture containing MEIS1 (1 nM) or with 2 μl of the lysate control, in 75 mM NaCl–1 mM EDTA–1 mM dithiothreitol–10 mM Tris-HCl (pH 7.5)–6% glycerol–2 μg of BSA, with 1 μg of dI-dC plus 0.1 μg of single-stranded salmon sperm DNA as nonspecific competitors, in a final reaction volume of 15 μl. Prior to addition of DNA for EMSA, mixtures of the in vitro-synthesized proteins were incubated together for 30 min at 30°C. Labeled DNA targets were then incubated with each protein mixture for 30 min at 4°C. Reaction mixtures were run on a 6% polyacrylamide gel to visualize complex formation by retardation of the 32P-labeled target DNA. In some experiments, polyclonal antisera to the appropriate epitope tag was incubated with aliquots of the reaction mixture for an additional 30 min. The HOXA9 and PBX1a proteins were fused to one epitope tag, while the MEIS1b molecule was fused to a different epitope tag, such that it was possible to use specific antisera to identify the presence of the MEIS1 protein in the complex by supershifting and/or blocking the retarded complex band. Gel electrophoresis was performed in 0.25× Tris-borate-EDTA buffer as described previously (40). For each gel shift reaction, a control containing the reticulocyte lysate and appropriate viral polymerase(s) was used to detect possible DNA binding by endogenous lysate factors.
Nuclear protein extracts were prepared by standard methods (1) from U-937 and KG1 cells grown in continuous log-phase culture. Extracts (1 μl) were incubated with 32P-labeled oligonucleotide probe containing a PBX-HOXA9 binding site and subjected to EMSA as described above. In some cases, specific polyclonal antibodies were preincubated with nuclear extracts for 30 min at 4°C prior to EMSA. To control for nonspecific effects of the various antibodies, gel shift assays in the presence and absence of antibodies were performed with nuclear extracts and two unrelated oligonucleotide probes, PU.1 and C/EBPα, both of which had previously been shown to be shifted by U-937 cell nuclear extracts (43).
In vivo coimmunoprecipitation.
Specific affinity-purified rabbit antipeptide antibodies against PBX1, PBX2, and PBX3 were from Santa Cruz Biochemicals (Santa Cruz, Calif.). A rabbit polyclonal antiserum against the entire PBX1a protein was used together with a mixture of the anti-PBX peptide sera for immunoprecipitation from U-937 cells. Polyclonal chicken antibodies to the full-length murine HOXA9 protein fused to GST were produced by Research Genetics, Inc. (Huntsville, Ala.) and were purified by affinity chromatography on immobilized HOXA9 protein expressed in pET28a. Rabbit antibodies raised against polypeptides N terminal and C terminal to the homeodomain of HOXA9, and rabbit antibodies against a peptide C terminal to the homeodomain of HOXA10, prepared in collaboration with Berkeley Antibodies, Inc. (Richmond, Calif.), were purified by affinity chromatography on immobilized GST-HOXA9 or GST-HOXA10 protein, respectively. Two separate preparations of antibodies to MEIS1 were used to block EMSA band formation with endogenous proteins. Rabbit antibody MEIS1-1, to an N-terminal peptide common to MEIS1a and MEIS1b, was purified by affinity chromatography. Antiserum MEIS1-2, to the whole MEIS1b protein, was raised in guinea pigs and affinity purified on immobilized MEIS1b protein. Equivalent amounts of purified guinea pig immunoglobulin G (IgG) were used as a control for specific guinea pig anti-MEIS antibodies, purified chicken IgY was used as a control for specific chicken anti-HOXA9 antibody, while purified rabbit IgG was used as a control for specific rabbit anti-MEIS, anti-HOXA9, or anti-HOXA10 antibody.
Immunofluorescent localization of proteins.
Cytospins of cells growing in log phase were fixed in 4% formaldehyde (freshly prepared from paraformaldehyde) in phosphate-buffered saline for 24 h. For double-labeling immunohistochemical localization, PBX2 was detected with affinity-purified rabbit antibodies followed by anti-rabbit donkey IgG conjugated to peroxidase (Jackson Immunochemicals, West Grove, Pa.), followed by Cy3-tyramide (NEN, Boston, Mass.). HOXA9 was detected either with biotinylated, affinity-purified rabbit antibodies followed by strepavidin-Alexa488 (Molecular Probes, Eugene, Oreg.) or with affinity-purified chicken antibodies followed by biotinylated donkey anti-chicken IgY (Jackson Immunochemicals) and strepavidin-Alexa488. MEIS1 was detected with affinity-purified guinea pig antibodies followed by anti-guinea pig donkey IgG conjugated to Cy3. For triple labeling, PBX2 was detected with affinity-purified rabbit antibodies followed by anti-rabbit donkey IgG conjugated to 7-amino-4-methylcoumarin-3-acetic acid (AMCA; Jackson Immunochemicals). Some specimens were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) to visualize nuclei. Slides were mounted with Vectashield (Vector, Burlingame, Calif.). Multicolor, quantitative fluorescence imaging was performed with a custom-built quantitative image processing microscope system, remotely controlled by a Sun workstation at the Laboratory for Cell Analysis, UCSF Cancer Center. A 16-bit, gray-level, high-resolution charge-coupled device camera (MicroImager 1400 digital camera; Xillix, Richmond, B.C., Canada) was attached to a Zeiss Axioplan fluorescence microscope equipped with a motorized stage and a focusing drive. The microscope was also fitted with multiband fluorescence emission, excitation, and dichroics filters (Chroma Technologies, Brattleboro, Vt.). The excitation filters were changed by using a computer-controlled filter wheel in the excitation light path. This permitted sequential acquisition of images for each of the fluorochromes used, with all of the images being in proper register to the accuracy with which chromatic aberrations have been corrected. Signals generated by Cy3 and Alexa288 were visualized with a Texas red filter and a fluorescein isothiocyanate FITC filter, respectively, whereas AMCA and DAPI signals were visualized with their corresponding filters. The acquired images were digitally modified (background corrections, contrast adjustments, and magnification calibration) by using ScillImage software, run on a Sun workstation. The final images were assembled and labeled on a Macintosh 8500 PC computer with Adobe Photoshop.
Transcription assays.
A direct repeat of an oligonucleotide containing a 3-mer PBX-HOXA9 binding site, gatcATGATTTACGACgagaattcgaATGATTTACGACcgcctgcgATGATTTACGAC, was inserted into the BglII site of the reporter plasmid pGL3, which contains a simian virus 40 (SV40) minimal promoter (Promega), to create pGL3-(PBX-HOXA9)6. U-937 cells growing in log phase were transfected by electroporation with a gene pulser (960 μFd, 170 V) (Bio-Rad Instruments, Richmond, Calif.). To control for squelching, in each experiment control parent plasmids containing the relevant promoters were substituted when specific DNAs were omitted from the transfection mix such that 10 μg of total DNA was always transfected into 2 × 107 cells. Cells were harvested after growth for 6 h; longer growth periods led to much lower apparent transfection efficiencies. cDNAs encoding the full-length proteins were cloned into standard expression vectors to generate plasmids pPRC/CMV-PBX2, pSV40-VP16-PBX2, pMSCV-MEIS1b, pPRC/CMV-HOXA9, and pRSV-β-gal. pSV40-VP16-PBX2 contains a VP16 activation domain and nuclear localization signal sequence fused N terminal to the PBX2 coding sequence.
RESULTS
DNA site selection with MEIS1, HOXA9, and PBX1a yields a consensus PBX-HOXA9 binding site.
Since we had previously shown that HOXA9, PBX1a, and MEIS1 are capable of forming all three possible heterodimeric complexes on their distinct bipartite DNA targets (41, 42), we first examined whether DNA site selection in the presence of all three proteins would be informative of preferential DNA binding. (In previous studies we demonstrated that the two alternatively spliced forms of MEIS1, which differ in their C termini, behave indistinguishably in protein-protein and protein-DNA interactions [41]. In the present study, MEIS1b was used in all in vitro assays. The antibodies used to detect MEIS1 proteins in vivo recognize both MEIS1a and MEIS1b.) Equal concentrations of the full-length HOXA9, MEIS1b, and PBX1a proteins were incubated with an oligonucleotide mixture containing an N24 core. In each of three site selection experiments, one of the proteins was tagged with a unique epitope, permitting immunoprecipitation of the specific protein, along with the associated proteins and any specifically bound oligonucleotides. In a separate experiment, all of the proteins were fused to the same epitope tag to allow competitive selection with all possible combinations of proteins. Site selection using immunoprecipitation of either HOXA9 alone or PBX1a alone, in the presence of all three proteins, yielded a series of different selected oligonucleotides, each of which contained a highly conserved TGATTTAT/C sequence (Table 1). This binding site was identical within the central core to a previously defined PBX-HOXA9 heterodimeric binding sequence, with lower conservation of a 5′ A in the PBX site and a 3′ GAC within the HOXA9 site (42). Precipitation of HOXA9 also yielded one clone containing both a MEIS-HOXA9 consensus site and a PBX-HOXA9 site, while precipitation of PBX yielded three clones containing partial PBX-MEIS sites in addition to the PBX-HOXA9 sites (Table 2). When epitope-tagged MEIS1 protein was selectively immunoprecipitated during DNA site selection, each unique selected sequence again contained a consensus PBX-HOXA9 site (Table 1). Surprisingly, in this experiment heterodimeric DNA binding sites for MEIS1 with HOXA9 (..................TGACAGTTAT/C) were not detected, while one clone contained a PBX-MEIS1 site (TGAT............TGAC......AG) in addition to a PBX-HOXA9 site. Finally, when all three proteins were epitope tagged and precipitated in the selection assay, each cloned sequence again contained a PBX-HOXA9 consensus site (Table 1). In no case was a triple site detected, even though from previous studies (9, 41), it is theoretically possible to align the three proteins in a PBX-MEIS1-HOXA9 configuration (TGAT...................TGACAGTTAT/C). In each site selection, in addition to the PBX-HOXA9 consensus site, most of the clones contained additional single binding sites for the respective tagged protein arrayed in random orientation and spacing with respect to the PBX-HOXA9 site (Table 2). These secondary sites appear to occur because of enhanced immunoselection of oligonucleotides bound by multiple tagged proteins and demonstrate that the specific epitope-tagged homeodomain protein was being immunoprecipitated in each case. As described below, MEIS1 does not bind to the PBX-HOXA9 consensus oligonucleotide either alone (Fig. 1A, lane 1) or in the presence of either HOXA9 or PBX (Fig. 2, lanes 5 and 6). Thus, the site selection data indicate that in the presence of all three proteins, PBX-HOXA9 binding sites are preferred. Furthermore, HOXA9 forms a cooperative DNA binding complex with PBX, which in turn is bound by the MEIS1 protein, which does not bind to specific DNA sequences in this triple complex.
TABLE 2.
Types of DNA binding sites precipitated with antisera to various homeodomain proteinsa
| Type of site | No. of sites
|
|||
|---|---|---|---|---|
| Anti-HOXA9b | Anti-PBX1 | Anti-MEIS1 | Anti-all three | |
| PBX-HOX | 13 | 16 | 13 | 21 |
| PBX-MEIS | 0 | 3 | 1 | 1 |
| MEIS-HOX | 1 | 0 | 0 | 1 |
| PBX alone | 3 | 5 | 2 | 4 |
| MEIS alone | 2 | 6 | 14 | 8 |
| HOX alone | 11 | 0 | 3 | 4 |
| Total no. of clones sequenced | 13 | 16 | 12 | 21 |
Many selected oligonucleotides contain more than one previously defined consensus recognition site, so that the total number of sites exceeds the number of clones sequenced.
Antiserum used to precipitate protein-DNA complexes. In each case, the antiserum was directed to a unique epitope tag fused to the protein to be specifically precipitated.
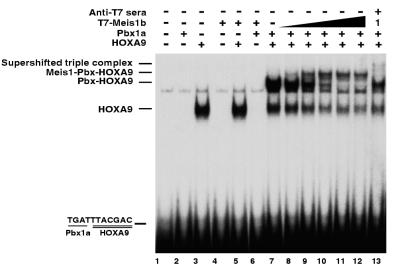
FIG. 2.
Formation of a MEIS1-PBX1a-HOXA9 ternary complex on a PBX-HOXA9 DNA target. EMSA was used to assess DNA binding by in vitro-translated proteins. As previously described, PBX1a and HOXA9 form a dimeric binding complex on their consensus target (lane 7), while HOXA9 alone forms a faster-migrating EMSA band (lane 3). Comparison with a lysate control (lane 1) demonstrates that neither PBX1a alone (lane 2), MEIS1b alone (lane 4), MEIS1b with HOXA9 (lane 5), nor MEIS1b with PBX1a (lane 6) is capable of binding this oligonucleotide. Addition of increasing amounts of MEIS1b to PBX1a and HOXA9 resulted in the formation of a new, slower-migrating band and the disappearance of the dimer band (lanes 8 to 12). Preincubation of the triple protein mixture with antiserum to the epitope tag fused to the MEIS1 protein prevented formation of the slower-migrating band together with the appearance of a weak supershifted band and the reappearance of the dimer band (lane 13). The amount of MEIS1 used in lane 13 was the same as that used in lane 9. We interpret this result to mean that the MEIS1 antiserum supershifts some complex while preventing most of the MEIS1b protein from binding to the PBX-HOXA9 dimeric DNA gel shift complex.
MEIS1 potentiates immunoprecipitation of PBX-HOXA9-bound DNA.
To further investigate the possible formation of a triple protein-DNA complex, we tested whether in vitro immunoprecipitation of epitope-tagged MEIS1 protein could bring down a 32P-labeled oligonucleotide containing the consensus PBX-HOXA9 binding site, in the presence or absence of untagged HOXA9 and PBX1a proteins. MEIS1 alone does not bind to the PBX-HOXA9 target oligonucleotide (Fig. 1A, lane 1). Immunoprecipitation of MEIS1 in the presence of PBX and HOX proteins brought down the labeled DNA (Fig. 1A, lane 3), whereas labeled DNA was not precipitated when the MEIS1 was omitted (lane 2). Mixtures of PBX and MEIS1 or HOXA9 and MEIS1 do not bind cooperatively to this DNA target (Fig. 2, lanes 5 and 6). We interpret these data as support for a model in which MEIS1 interacts with a complex of PBX1a and HOXA9 which are cooperatively bound to a DNA target, by bridging these proteins.
HOXA9 forms a triple complex with MEIS1 and PBX proteins in the absence of DNA.
Previous studies have shown that MEIS1 is capable of forming specific protein-protein complexes with both HOXA9 (41) and PBX1a (9) in the absence of DNA. Since our data suggested that MEIS1 was bound to either HOXA9 and/or PBX1a in the presence of DNA, we wished to study the interactions between these molecules in the absence of a DNA target. We used immunoprecipitation of 35S-labeled proteins to study protein-protein interactions in the absence of DNA. In these studies, the HOXA9 protein was synthesized with an epitope tag so that use of an epitope-specific antibody would precipitate HOXA9 along with untagged associated proteins. As shown in Fig. 1B, when an equimolar amount of PBX1a was incubated with the epitope-tagged HOXA9 protein alone, a modest amount of PBX1a protein coprecipitated with the HOXA9 (lane 7). These data are similar to results of previous studies which show that PBX1a exhibits only minimal interaction with HOXB7 in the absence of DNA (10). When an equimolar concentration of MEIS1b was added to the PBX1a-HOXA9 mixture, there was a sixfold increase in the amount of PBX1a precipitated by antiserum to the epitope-tagged HOXA9 protein (lane 8). A control protein (lane 1) present in each reaction mixture was not precipitated, indicating that the interactions between MEIS1b, PBX1a, and HOXA9 were specific. Specificity was further established by showing that no proteins were precipitated if either the epitope-tagged HOXA9 protein or the antiepitope serum was omitted (lane 18 and data not shown). Since other experiments demonstrated that other members of the PBX family, PBX2 and PBX3, were present in myeloid cell nuclear extracts (below), we repeated these experiments using PBX2 (lanes 9, 10, 13, and 14) and PBX3 (lanes 11 and 12). Addition of MEIS1 increased PBX2 binding to HOXA9 approximately 7- to 10-fold, while MEIS1 produced a 3-fold enhancement of PBX3-HOXA9 interaction. These results indicate that MEIS1 is capable of forming a triple complex with HOXA9 and each of the three known PBX proteins in the absence of DNA. When the complementary experiment was performed, addition of PBX2 increased the coprecipitation of MEIS1 with HOXA9 approximately fivefold (lanes 14 to 17).
MEIS1, PBX, and HOXA9 form an in vitro EMSA triple protein complex on a PBX-HOXA9 target.
We used EMSA to demonstrate formation of an in vitro triple protein complex on an oligonucleotide containing a PBX-HOXA9 consensus binding site (TGATTTACGAC). As shown in Fig. 2, neither PBX1a (lane 2) or MEIS1 (lane 4) alone nor a combination of PBX1a and MEIS1 (lane 6) binds to the oligonucleotide probe. As previously described, HOXA9 alone forms a rapidly migrating band with the target DNA (lane 3), and MEIS1 does not enhance HOXA9 binding to the PBX-HOXA9 target (lane 5). As previously demonstrated in assays using specific antisera (42), HOXA9 and PBX1a together form a heterodimeric complex on their consensus binding target (lane 7, upper band). When increasing amounts of MEIS1 are incubated with the HOXA9 and PBX1a proteins and DNA target, a slower-migrating band appears (lanes 8 to 12, top band). Concomitant with the formation of this new band, the dimeric PBX1a-HOXA9 band disappears. We ascribe the new band to a MEIS1-PBX1a-HOXA9 trimeric complex, because it disappeared upon addition of an antibody to the epitope fusion tag on the MEIS1 protein (lane 13). While a small amount of supershifted band could be visualized, the band representing the dimeric PBX1a-HOXA9 complex reappeared; we hypothesize that the latter is due to the antiserum to the epitope tag fused to the MEIS1 protein preventing MEIS1 binding to the PBX-HOXA9 dimeric complex. Similar experiments with PBX2 showed that this protein was also capable of forming a trimeric DNA binding complex with the HOXA9 and MEIS1 proteins (data not shown). Although a band representing the triple complex was always observed, the intensity of the triple complex band in the EMSA gels was variable in different experiments. In some experiments, the addition of MEIS1 protein to HOXA9-PBX-DNA mixtures resulted in large reductions in the gel shift bands ascribed to PBX-HOXA9 and to HOXA9 alone, without compensatory formation of an equivalent-intensity triple complex band. Since triple complex formation was extremely reproducible by immunoprecipitation, we attribute the variability of the EMSA band intensity to the subtle vagaries of the EMSA technique.
Heterodimers of HOXA9 with MEIS1 and PBX can be detected in myeloid cell nuclear extracts.
To explore the possible interactions between the three proteins in vivo, we examined their expression in myeloid cell lines. We have previously shown that the MEIS1 and HOXA9 genes are expressed in both U-937 and KG1 myeloid leukemia cells (21), while PBX2 and PBX3 gene expression has also been reported to occur in these cells (27). We used affinity-purified antibodies to detect HOXA9 and MEIS1 proteins in both U-937 and KG1 cell nuclear extracts by Western blotting (Fig. 3A). Two bands of endogenous HOXA9 protein migrated with apparent molecular weights of 40,000 and 48,000, while a sample of HOXA9 protein prepared by in vitro translation migrated at 40,000. Thus, the smaller band corresponds in size to the protein derived from a full-length human HOXA9 cDNA clone (accession no. U82759), whose calculated molecular weight was 30,000 and which thus migrates anomalously in this gel system. The larger HOXA9 protein band may represent posttranslationally modified protein (40a) or be derived from one of the alternatively spliced variants previously described for HOXA9 in other species (3, 39). In a similar manner, the 57,000-molecular-weight (57K) and 65K immunoreactive bands detected for MEIS1 protein appear to represent the two alternatively spliced forms of MEIS1 previously predicted from cDNA analysis (28), although each protein migrates somewhat more slowly than the corresponding MEIS1a and MEIS1b protein produced in vitro. Western gel analysis using specific antisera was also used to demonstrate expression of a 55K band of PBX2 protein in nuclear extracts of U-937 or KG1 cells (Fig. 3A). Low-level expression of PBX3 was also detected, but PBX1a was not observed, using a similar approach (data not shown). The endogenous PBX proteins also migrate slightly more slowly than the in vitro-prepared proteins, again suggesting posttranslational modification (40a).
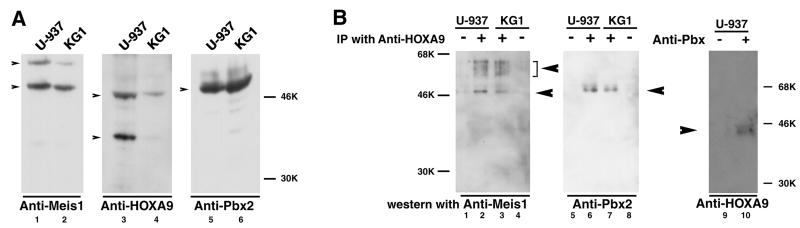
FIG. 3.
Coimmunoprecipitation of HOXA9 and MEIS or PBX from myeloid cell nuclear extracts. (A) HOXA9, MEIS1, and PBX2 are expressed in U-937 and KG1 nuclear cell extracts. Western gel analysis of U-937 and KG1 cell nuclear extracts using affinity-purified specific antibodies showed expression of MEIS1 (lanes 1 and 2), HOXA9 (lanes 3 and 4), and PBX2 (lanes 5 and 6). (B) Coimmunoprecipitation of HOXA9 with PBX2 or HOXA9 with MEIS1. U-937 and KG1 cell nuclear proteins were incubated with affinity-purified chicken antibody to HOXA9 (lanes 2, 3, 6, and 7) or nonimmune IgY (lanes 1, 4, 5, and 8). Following immunoprecipitation, the pellets were assayed by Western blotting using rabbit antibodies to MEIS1 proteins (lanes 1 to 4) or to PBX2 protein (lanes 5 to 8). Immunoprecipitation of HOXA9 brought down MEIS1 (lanes 2 and 3) and PBX2 (lanes 6 and 7). In a similar manner, U-937 cell nuclear proteins were subjected to immunoprecipitation with a mixture of whole rabbit antisera to PBX protein and affinity-purified antibodies to PBX1, PBX2, and PBX3 (lane 10), or control sera (lane 9), and the pellets were analyzed by Western blotting with affinity-purified chicken antibodies to HOXA9 (lanes 9 and 10). Specific HOXA9 bands were detected in proteins precipitated with anti-PBX (lane 10), while immunoprecipitation with a nonspecific serum did not yield HOXA9 immunoreactive protein (lane 9).
To detect the presence of HOXA9 protein complexes in U-937 and KG1 nuclear extracts, we first used immunoprecipitation with affinity-purified chicken antibodies to HOXA9. When the precipitation products were subjected to Western blot analysis with affinity-purified MEIS1-1 antibody bands corresponding to those previously observed in whole nuclear extracts were detected along with several bands of intermediate size which may represent proteolysis products (Fig. 3B, lanes 2 and 3). Precipitation using a nonspecific control IgY did not yield immunoreactive MEIS1 bands (lane 1 and 4). In a similar manner, when a Western blot of proteins which had been immunoprecipitated with anti-HOXA9 was probed with affinity-purified antibody to PBX2, we detected bands which migrated with the same mobility as the bands observed in whole nuclear extracts (lanes 6 and 7). However, no bands were detected when a parallel filter was probed with an antiserum specific to PBX3, suggesting that little of the HOXA9 protein is associated with PBX3. Finally, we performed coimmunoprecipitation using a mixture of antibodies to PBX1, PBX2, and PBX3 proteins. When the precipitated proteins were subjected to Western gel analysis with affinity-purified rabbit antibodies to HOXA9, we detected an immunoreactive protein band which migrated at the same position as the upper HOXA9 band previously visualized in whole nuclear extracts (lane 10).
MEIS1, PBX, and HOXA9 in myeloid cell nuclear extracts can form a DNA binding triple complex.
Following the demonstration that both PBX2 and MEIS1 can be coprecipitated with HOXA9, we used EMSA analysis in conjunction with specific blocking antibodies to demonstrate the presence of a gel shift band containing all three proteins in myeloid cell nuclear extracts. For these studies, a labeled PBX-HOXA9 oligonucleotide target was used for EMSA analysis with nuclear extracts from either U-937 or KG1 cells. EMSA analysis with U-937 nuclear extracts revealed a consistent major band (Fig. 4, lanes 1 to 3, upper arrow), which migrated in a position similar to that of the trimeric MEIS1-PBX-HOXA9-DNA complex observed for in vitro-translated proteins (Fig. 2). In addition, a faster, variable-intensity band was observed (lower arrow). Both bands could be competed with specific oligonucleotide (compare lanes 5 and 6 with lane 4) but not by an unrelated oligonucleotide (lanes 7 and 8). Similar data were obtained for KG1 nuclear extracts (data not shown). We then used specific, affinity-purified blocking antisera to characterize the EMSA gel shift bands. As controls, we first demonstrated that preincubation of each specific antisera with mixtures of in vitro-translated proteins resulted in disruption of specific EMSA bands (lanes 22 and 23 and data not shown). We then used these antisera to demonstrate that the slower-migrating EMSA band in the U-937 cell nuclear extract contains, in part, complexes of MEIS1 associated with HOXA9 and PBX2. Antibodies to MEIS1 (lanes 10 and 17), HOXA9 (lane 11), PBX2 (lane 12), and PBX3 (lane 13) were able to reduce the amount of the slower gel shift band in nuclear extracts, while antibodies to PBX2 and PBX3 together were able to almost completely prevent formation of this complex (lane 14). Consistent with the observation that PBX1 is not expressed in U-937 cell nuclear extracts, antiserum specific to this protein did not reduce the intensity of the slower-migrating EMSA band (lane 15).
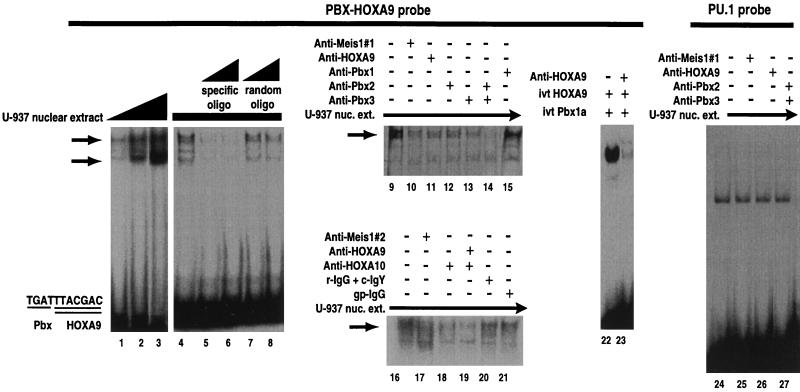
FIG. 4.
MEIS1, PBX, and HOXA9 proteins in U-937 cell nuclear extracts can form a ternary EMSA complex. Increasing amounts of U-937 cell nuclear extract shifted an oligonucleotide (oligo) containing the PBX-HOXA9 consensus binding site to form a highly reproducible upper band and a variable lower band (lanes 1 to 3). Both bands could be competed with cold specific oligonucleotide (lanes 5 and 6 compared to lane 4) but not with a random oligonucleotide (lanes 7 and 8). Two different specific blocking antisera were used to demonstrate the presence of MEIS1 in the upper gel shift band (compare lane 10 with lane 9 and lane 17 with lane 16). In a similar manner, specific antisera were used to demonstrate that HOXA9 (lane 11), PBX2 (lane 12), and PBX3 (lane 13) were present in this band. A combination of antisera to PBX2 plus PBX3 (lane 14) blocked almost all of the slower-migrating EMSA band. In contrast, antiserum to PBX1, which is not present in U-937 cell nuclear extracts, did not diminish the intensity of the upper band (lane 15). Since antiserum to HOXA9 blocked only a fraction of the upper gel shift band, we used specific antiserum against HOXA10 to demonstrate that this protein also contributes to the upper gel shift complex (lanes 18 and 19). Lanes 20 and 21 represent treatment of nuclear extracts (nuc. ext.) with a mixture of normal rabbit IgG and chicken IgY (r-IgG + c-IgY) and nonspecific guinea pig IgG (gp-IgG), respectively, as controls for the specific antibodies used in lanes 17 to 19. To demonstrate that the antiserum to HOXA9 blocked protein-protein interactions, in vitro-synthesized (ivt) HOXA9 and PBX1a proteins were incubated with their consensus DNA target in the absence (lane 22) or presence (lane 23) of affinity-purified antibodies to HOXA9. To demonstrate the specificity of the antibody reduction of the EMSA bands, blocking experiments were repeated using an unrelated PU.1 oligonucleotide probe previously shown to be shifted by U-937 cell extracts (43). None of the specific antibodies were able to block or supershift the EMSA bands formed with the PU.1 probe (lanes 24 to 27).
Since the antiserum to HOXA9 incompletely blocked the slower EMSA band, we considered whether a complex consisting of the related HOXA10 protein bound to MEIS1 and PBX was also a component of this band. We have previously shown that the HOXA10 gene is expressed in these cells and that the HOXA10 protein forms complexes with MEIS1 as well as forming DNA binding complexes with PBX on this target (8, 41, 42). Addition of affinity-purified antisera to HOXA10 alone (Fig. 4, lane 18) and together with the antiserum to HOXA9 (lane 19) substantially diminished the slower EMSA band, suggesting that both HOX proteins form complexes with MEIS1 and PBX in these cells. A combination of control nonspecific chicken IgY for HOXA9 and rabbit IgG for HOXA10 (lane 20) or control nonspecific guinea pig IgG for MEIS1 (lane 21) did not prevent formation of the slower gel shift complexes. Several additional MEIS family members have been identified, some of which are expressed in myeloid cells (29, 44). Preliminary data suggest that these MEIS1-related proteins also form DNA binding complexes with HOXA9 (6a) and thus may also contribute to the gel shift bands observed in the nuclear extracts. Since these proteins would not react with the anti-MEIS1 peptide antibodies, a portion of the EMSA bands would not be diminished by these antibodies. Finally, the disruption of the slower EMSA band by antisera to PBX3 suggests that this protein may be complexed with HOXA10 or other proteins which contribute to this gel shift band. The identity of the faster-migrating EMSA band is uncertain. To demonstrate that the antibody blocking experiments were specific, they were repeated using two unrelated oligonucleotide probes, containing either PU.1 or C/EBPα DNA binding sites, which are shifted by myeloid cell extracts (43). Neither the MEIS1-1 nor HOXA9 antibody nor a mixture of the PBX2 and PBX3 antibodies supershifted EMSA bands formed with the PU.1 probe and U-937 or KG1 cell nuclear extracts (lanes 24 to 27 and data not shown). In a similar manner, none of the antisera were capable of supershifting EMSA bands formed between U-937 or KG1 cell extracts and the C/EBPα probe (data not shown).
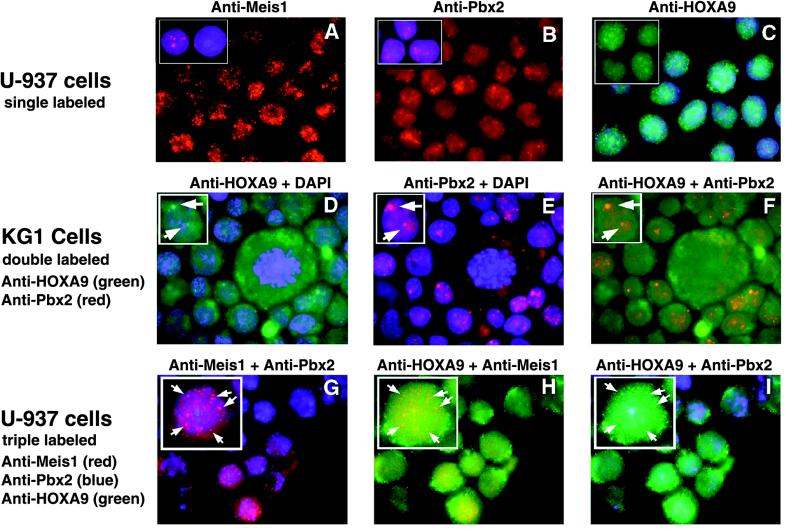
HOXA9 colocalizes with PBX2 and MEIS1 in nuclear speckles in myeloid leukemic cells.
To further explore the in vivo relationship between HOXA9, PBX, and MEIS1 proteins, we used immunohistochemical analysis with specific antibodies to localize proteins in U-937 and KG1 myeloid leukemia cells. For these experiments, HOXA9, PBX2, or MEIS1 were visualized by using affinity-purified primary antibodies which were differentially but simultaneously detected with specific fluorescent secondary reagents. In the initial experiments we used specific antibodies, along with a DAPI stain to identify nuclei, in order to visualize subcellular localization. The majority of the signals for MEIS1, PBX2, and HOXA9 in U-937 cells were localized to the nucleus (Fig. 5A to C). Minor expression of MEIS1 and PBX2 was detected in the cytoplasm, while moderate HOXA9 cytoplasmic expression was observed. In contrast to the relatively constant cell to cell staining intensity seen for HOXA9 and PBX2, we observed a reproducible large variation in MEIS1 signal intensity in both U-937 and KG1 cells. Thus, some cells appeared to express much higher levels of MEIS1 than other cells, but no correlation could be made between MEIS1 levels and cellular phenotype or proliferation state. We next used combinations of two specific antibodies, with DAPI in the third channel, to examine colocalization in KG1 cells (Fig. 5C to F). Alignment of the images for HOXA9 and PBX2 revealed that a portion of the PBX2 signal colocalized with HOXA9 signal within nuclear speckles (Fig. 5F), reminiscent of the expression patterns reported for a number of other transcription factor within the nucleus (reviewed in reference 20). The cytoplasmic HOXA9 expression was more easily visualized in KG1 cells (Fig. 5D). In separate double-labeling experiments, a fraction of the MEIS1 signal also colocalized with HOXA9 and with PBX2 within nuclear speckles in both U-937 and KG1 cells (data not shown). To demonstrate colocalization of all three proteins, we performed a triple-labeling experiment in which each protein was detected with specific antisera in conjunction with a uniquely colored secondary reagent (Fig. 5G to I). Computer-generated coalignment of sets of two images from the same cells revealed that a portion of the HOXA9, PBX2, and MEIS1 signals could be detected in the same nuclear speckles. Colocalization of proteins was not observed in the cytoplasm.
FIG. 5.
Some HOXA9 protein colocalizes with PBX2 and MEIS1 proteins to nuclear speckles in myeloid cells. (A to C) Localization of individual proteins in U-937 cells stained with affinity-purified guinea pig antibodies to MEIS1, rabbit antibodies to PBX2, and chicken antibodies to HOXA9, respectively. The insets in panels A and B show cells counterstained with DAPI to visualize nuclei; panel C shows DAPI counterstaining of cells, and the insert shows HOXA9 signal alone. Strong signals for all three proteins were detected in the nucleus, with enhanced signals within nuclear speckles. Weak staining for MEIS1 and PBX2 and moderate HOXA9 expression were observed in the cytoplasm. For each antibody, virtually all of the signal could be blocked with purified MEIS1, PBX2, or HOXA9 protein or peptide antigen, respectively (data not shown). (D to F) Double labeling with HOXA9 and PBX2 in KG1 cells. Arrows point to the green signal for HOXA9 (D), the red signal for PBX (E), or the combined yellow signal for HOXA9 plus PBX (F). (G to I) Triple labeling of U-937 cells with anti-MEIS1 (red), anti-PBX2 (blue), and anti-HOXA9 (green), showing colocalization of sets of two proteins: purple speckles resulting from the superposition of the MEIS1 and PBX signals (G), yellow speckles resulting from the superposition of the HOXA9 and MEIS1 signals (H), and aqua speckles resulting from the superposition of the HOXA9 and PBX2 signals (I). Arrows point to nuclear speckles in which all three proteins appear to be expressed.
The HOXA9, MEIS1, and PBX proteins do not display transcriptional regulatory activity on a synthetic target site in myeloid cells.
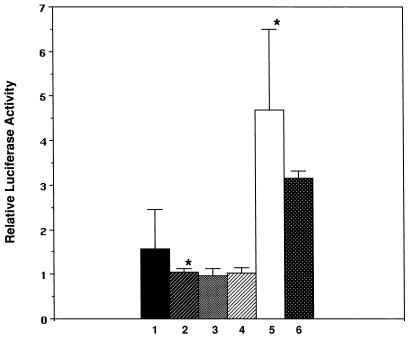
We attempted to demonstrate transcriptional activity in transient transfection assays using a reporter plasmid containing a minimal SV40 promoter and an upstream 6-mer of a TGATTTACGAC sequence representing a consensus PBX-HOXA9 binding site. When this pGL3-(PBX-HOXA9)6 reporter construct was transfected into a series of myeloid cell lines, including U-937, KG1, THP, ML3, and M1, no significant changes in activity were observed relative to the parent pGL3 reporter containing the minimal SV40 promoter alone, while transfection of FDCP1 myeloid cells yielded a twofold increase in activity for the pGL3-(PBX-HOXA9)6 compared to the pGL3 control (Fig. 6 and data not shown). These results demonstrated that endogenous proteins, including HOXA9, MEIS1, and PBX2, which we had detected in these cells were not capable of either strongly activating a reporter containing multiple PBX-HOXA9 sites or repressing the clearly detectable luciferase activity derived from transcription of this plasmid due to the SV40 minimal promoter. To further test for transcriptional activity, we cotransfected combinations of plasmids expressing HOXA9 with PBX2 alone or together with MEIS1 with the pGL3-(PBX-HOXA9)6 reporter construct into U-937, KG1, and FDCP1 cells. For U-937 and KG1 cells, there were no significant changes in reporter gene activity after normalization using a transfection control gene (Fig. 6), while a twofold increase in reporter activity in response to exogenous HOXA9 plus PBX2 was observed in FDCP1 cells (data not shown). In contrast, cotransfection of a plasmid expressing a VP16-PBX2 fusion protein together with HOXA9 resulted in a clearly detectable and reproducible 5- to 7-fold activation in U-937 cells (Fig. 6) and a 15-fold activation in FDCP1 cells. Addition of a MEIS1b expression plasmid to this system did not lead to a further activation in either cell line. These data demonstrated that the reporter system and expression plasmids were functional and suggested that neither the endogenous nor exogenous HOXA9, PBX2, and MEIS1 proteins were capable of substantially influencing the transcription of an artificial gene construct in which the PBX-HOXA9 sites were placed within a region lacking binding sites for possible required cofactors.
FIG. 6.
The HOXA9, MEIS1, and PBX proteins do not display transcriptional regulatory activity on a synthetic target in myeloid cells. U-937 cells were transfected with a pGL3 luciferase reporter plasmid containing a minimal SV40 promoter alone (bar 1) or with a pGL3-(PBX-HOXA9)6 plasmid containing an upstream sixfold repeat of the consensus PBX-HOXA9 binding site (bar 2). The lack of change in activity observed for the pGL3-(PBX-HOXA9)6 construct reflects the apparent inability of the endogenous HOXA9, PBX2, and MEIS1 proteins to activate or repress this reporter. Cotransfection of the pGL3-(PBX-HOXA9)6 reporter together with expression plasmids encoding the HOXA9 plus PBX2 proteins (bar 3), or HOXA9 plus PBX2 with MEIS1b proteins (bar 4), did not produce transcriptional activity. Cotransfection of the pGL3-(PBX-HOXA9)6 reporter together with HOXA9 and VP16-PBX2 expression plasmids yielded activity (bar 5), demonstrating that the reporter system was functional. Cotransfection of an expression plasmid encoding MEIS1b in addition to HOXA9 and VP16-PBX2 did not further change the observed activity (bar 6). In each assay, a cytomegalovirus-LacZ plasmid was used to normalize for transfection efficiency. ∗, P < 0.004.
DISCUSSION
Retrovirally driven coexpression of Hoxa9 and Meis1 in either FDCP cells or mouse primary bone marrow cells leads to rapid myeloid leukemic transformation (17). Since the protein products of these two genes can form heterodimeric DNA binding complexes with each other and each protein can also form heterodimeric DNA binding complexes with PBX proteins, we attempted to analyze the mechanism by which HOXA9 and MEIS1 could transform cells by elucidating their preferential DNA binding partners. We set up a competitive three-way DNA site selection experiment in order to ascertain which proteins would preferentially bind to DNA. Remarkably, in every situation studied, a highly conserved PBX-HOXA9 dimeric protein binding site was selected. We interpreted these data to reflect the existence of a ternary protein complex in which MEIS1 binds to a PBX-HOXA9 dimer bound to DNA. Since we did not detect a contiguous DNA binding site for all three proteins during DNA site selection, we conclude that MEIS1 can form a triple complex without binding to the DNA through its specific recognition site. We cannot exclude the possibility that MEIS1 nonspecifically contacts the DNA or interacts in vivo at a site not accommodated by the N24-containing oligonucleotide used in our site selection studies. Since our in vitro and in vivo data suggest that MEIS1 interacts with DNA binding complexes of HOXA9 with PBX, it is interesting that coexpression of Pbx1a with Hoxa9 does not accelerate leukemic transformation compared to Hoxa9 alone (17). We hypothesize that the relatively high concentrations of PBX proteins found in myeloid cells in the present study may reflect levels of this protein sufficient for interaction with exogenous HOXA9 and MEIS1 in the cell types used by Kroon et al. (17). Alternatively, overexpression of MEIS1 and HOXA9 may change the normal balance of PBX with these proteins, leading to formation of different DNA-protein complexes.
Other studies have used antisera to PBX to precipitate DNA binding complexes from nonmyeloid cells and have identified PBX-MEIS binding sites (9, 16). Although these studies did not detect PBX-HOX sites, the antisera used might not have allowed detection of PBX-HOX proteins bound to DNA, since in our hands, antisera to PBX prevented binding of PBX and HOXA9 to their consensus site. Furthermore, there appears to be more PBX and MEIS1 protein than HOX proteins in the cells that we have studied. If a similar situation exists in the cells previously used for PBX immunoprecipitation, it would be difficult to detect low levels of PBX-HOX complexes. Thus, our demonstration of PBX-HOXA9 complexes in myeloid cell nuclei by coprecipitation and by immunohistochemical localization is not necessarily at variance with the previous studies.
Our model of MEIS1 binding to a cooperative PBX-HOXA9 DNA binding complex is consistent with two recent reports describing evidence for ternary complexes containing an Antennapedia-like homeodomain protein bound to PBX and MEIS-like proteins (4, 45). In both studies, the MEIS-like protein has been proposed to make protein-protein contacts but not to bind directly to the DNA target. In one study, a MEIS-like TALE class homeodomain protein, Prep1, was isolated from HeLa cells in a complex with PBX (5). These workers showed that Prep1 can form a ternary complex in vitro with PBX and HOXB1 on a naturally occurring b1-ARE target, previously shown to be a PBX-HOXB1 binding site (4). In addition, Prep1 can potentiate the activity of HOXB1 and PBX on a b1-ARE reporter construct in transient transfection assays. In the second study, Swift et al. present EMSA data suggesting formation of a triple complex between PDX1, a pancreatic cell-specific homeodomain protein, with PBX2 and MEIS2 (45). Although this report does not present data directly demonstrating protein-protein interactions, the authors propose a model similar to that suggested by our data, in which the MEIS protein binds to the other two proteins, which are bound to the DNA target. Our data also suggest that MEIS proteins, in addition to forming DNA-bound triple complexes with PBX and HOXA9, can disrupt the binding of HOX-PBX heterodimers to DNA (Fig. 2). Thus, the biologic effects of these three proteins probably reflect their relative concentrations within particular cellular milieu.
EXD, the Drosophila homologue of PBX, has been reported to be functional only in the nucleus (12, 37). The Drosophila homologue of MEIS, HTH, forms protein-protein complexes with and is required for EXD transport to the nucleus (32, 38), and the murine MEIS1 protein can at least partially rescue the hth mutant phenotype and induce nuclear localization of EXD (38). Our data are inconclusive with regard to the role of mammalian MEIS proteins on regulation of PBX protein localization to the nucleus. While some of the PBX2 is colocalized with MEIS1 in nuclear speckles, both proteins are present in both the nucleus and cytoplasm in noncongruent patterns. It is of note that the oncogenic fusion protein E2A-PBX1a was localized to nuclear spherical structures following transient transfection (24). However, these bodies did not appear to be associated with transcriptional activity, as has been reported for nuclear speckles (20). In contrast to the E2A fusion protein, PBX1a appeared to be diffusely expressed in the cytoplasm and nucleus (24). These results are consistent with our observation of low diffuse immunohistochemical signals for PBX1a mainly in the cytoplasm of myeloid cells (16a). Furthermore, the PBX3 protein appears in cytoplasmic extracts of 266-6 acinar pancreatic cells although MEIS proteins are found in nuclear extracts of these cells (45). The mechanisms regulating the subcellular distribution of the PBX proteins remain unclear, but they appear to depend on more than the presence of MEIS proteins and may vary according to the cellular context.
Our transfection data indicate that combinations of HOXA9 and PBX2 are not capable of robust regulation of gene transcription, either with or without MEIS1, when an artificial target is transfected into myeloid cells. It is of interest that the Prep1-PBX complex did not exhibit innate transcriptional activity either on a partial b1-ARE site to which it bound (4) or on a fragment derived from its naturally occurring urokinase gene enhancer target (11). These observations support earlier findings that neither coexpressed PBX and HOX proteins (8) nor MEIS and HOX proteins (41) exhibited transcriptional activity on consensus binding site targets incorporated into artificial reporter constructs. We anticipated that the addition of MEIS1 protein might provide transcriptional activity to PBX and HOXA9 on reporter constructs in myeloid cell lines. However, transient transfection assays in a range of myeloid cells did not reveal transcriptional activity for combinations of the three proteins, using a reporter construct containing a 6-mer repeat of a PBX-HOXA9 consensus binding site. In the case of Prep1-PBX, additional cell specific factors, bound to adjacent sites within the natural enhancer, were required for transcriptional activity (5). PDX1 exhibits differential cell-specific transcriptional activity which is dependent on either being free of PBX2 and MEIS2 or being bound to these proteins in the context of additional protein cofactors associated with adjacent sites within a natural promoter region. Unfortunately, gene targets for HOXA9 with PBX have not yet been described; therefore, it remains unclear whether these proteins function as traditional transcription factors when acting on a natural target or perhaps function at other levels within the nucleus such as influencing chromatin folding. Future studies will attempt to address these points.
ACKNOWLEDGMENTS
This work was supported by the Research Service of the Department of Veterans Affairs and by NIH grant DK48642 (H.J.L.). H.J.L. is a Clinical Investigator in the Department of Veterans Affairs. Rabbit antisera to HOXA9 and to HOXA10 were produced under a collaborative research project with Berkeley Antibodies Co., Inc., funded by NIH grant N43-DK-2-2219.
We thank Stephen Fong for technical assistance with cell culture, Mark Kamps for an antiserum to PBX proteins, Linda Shapiro for a gift of U-937 cells and for advice on their transfection, Gerry Krystal for FDCP1 cells, Dan Tenen for the PU.1 and C/EBPα probes, Mike Cleary for PBX1a, PBX2, and PBX3 cDNAs, and Arthur Buchberg and Jeff Montgomery for rabbit antisera to MEIS1 and for providing helpful comments on the manuscript.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology, unit 4.1. New York, N.Y: Green Publishing Associates and Wiley Interscience; 1987. [Google Scholar]
- 2.Azpiazu N, Morata G. Functional and regulatory interactions between Hox and extradenticle genes. Genes Dev. 1998;12:261–273. doi: 10.1101/gad.12.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson G V, Nguyen T-H E, Maas R L. The expression pattern of the murine Hoxa-10 gene and the sequence recognition of its homeodomain reveal specific properties of Abdominal B-like genes. Mol Cell Biol. 1995;15:1591–1601. doi: 10.1128/mcb.15.3.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berthelsen J, Zappavigna V, Ferretti E, Malvilio F, Blasi F. The novel homeoprotein Prep1 modulates Pbx-Hox protein cooperativity. EMBO J. 1998;17:1434–1445. doi: 10.1093/emboj/17.5.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berthelsen J, Zappavigna V, Mavilio F, Blasi F. Prep1, a novel functional partner of Pbx proteins. EMBO J. 1998;17:1423–1433. doi: 10.1093/emboj/17.5.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blackwell T K, Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990;250:1104–1110. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- 6a.Buchberg, A. Personal communication.
- 7.Chan S-K, Jaffe L, Capovilla M, Botas J, Mann R S. The DNA binding specificity of Ultrabithorax is modulated by cooperative interactions with extradenticle, another homeoprotein. Cell. 1994;78:603–615. doi: 10.1016/0092-8674(94)90525-8. [DOI] [PubMed] [Google Scholar]
- 8.Chang C-P, Brocchieri L, Shen W-F, Largman C, Cleary M L. Pbx modulation of Hox homeodomain N-terminal arms establishes a gradient of DNA-binding specificities across the Hox locus. Mol Cell Biol. 1996;16:1734–1745. doi: 10.1128/mcb.16.4.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang C-P, Jacobs Y, Nakamura T, Jenkins N A, Copeland N G, Cleary M L. Meis proteins are major in vivo DNA binding partners for wild-type but not chimeric Pbx proteins. Mol Cell Biol. 1997;17:5679–5687. doi: 10.1128/mcb.17.10.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang C-P, Shen W-F, Rozenfeld S, Lawrence H J, Largman C, Cleary M L. Pbx proteins display hexapeptide-dependent cooperative DNA binding with a subset of Hox proteins. Genes Dev. 1995;9:663–674. doi: 10.1101/gad.9.6.663. [DOI] [PubMed] [Google Scholar]
- 11.De Cesare D, Palazzolo M, Berthelsen J, Blasi F. Characterization of UEF-4, a DNA-binding protein required for transcriptional synergism between two AP-1 sites in the human urokinase enhancer. J Biol Chem. 1997;272:23921–23929. doi: 10.1074/jbc.272.38.23921. [DOI] [PubMed] [Google Scholar]
- 12.Gonzales-Crespo S, Morata G. Control of Drosophila adult pattern by extradenticle. Development. 1995;121:2117–2125. doi: 10.1242/dev.121.7.2117. [DOI] [PubMed] [Google Scholar]
- 13.González-Crespo S, Abu-Shaar M, Torres M, Martínez A C, Mann R S, Morata G. Antagonism between extradenticle function and Hedgehog signalling in the developing limb. Nature. 1998;394:196–200. doi: 10.1038/28197. [DOI] [PubMed] [Google Scholar]
- 14.Gould A, Morrison A, Sproat G, White R A, Krumlauf R. Positive cross-regulation and enhancer sharing: two mechanisms for specifying overlapping Hox expression patterns. Genes Dev. 1997;11:900–913. doi: 10.1101/gad.11.7.900. [DOI] [PubMed] [Google Scholar]
- 15.Kamps M P, Look A T, Baltimore D. The human t(1,19) translocation in pre-B ALL produces multiple nuclear fusion proteins with differing transforming potentials. Genes Dev. 1991;5:358–368. doi: 10.1101/gad.5.3.358. [DOI] [PubMed] [Google Scholar]
- 16.Knoepfler P S, Kamps M P. The highest affinity DNA element bound by Pbx complexes in t(1;19) leukemic cells fails to mediate cooperative DNA-binding or cooperative transactivation by E2a-Pbx1 and class I Hox proteins—evidence for selective targetting of E2a-Pbx1 to a subset of Pbx-recognition elements. Oncogene. 1997;14:2521–2531. doi: 10.1038/sj.onc.1201097. [DOI] [PubMed] [Google Scholar]
- 16a.Kömüves, L. G., and C. Largman. Unpublished observations.
- 17.Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg A M, Sauvageau G. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 1998;17:3714–3725. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 19.Lai J-S, Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc Natl Acad Sci USA. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamond A I, Earnshaw W C. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- 21.Lawrence H J, Rozenfeld S, Cruz C, Matsukuma K, Buchberg A, Largman C. Frequent co-expression of the HOXA9 and MEIS1 homeobox genes in human myeloid leukemias. 1998. Submitted for publication. [DOI] [PubMed] [Google Scholar]
- 22.Lawrence H J, Sauvageau G, Ahmadi A, Lau T, Lopez A R, Le Beau M M, Link M, Largman C. Stage and lineage-specific expression of the HOXA10 homeobox gene in normal and leukemic hematopoietic cells. Exp Hematol. 1995;23:1159–1165. [PubMed] [Google Scholar]
- 23.Lawrence H J, Sauvageau G, Humphries R K, Largman C. The role of HOX genes in normal and leukemic hematopoiesis. Stem Cells. 1996;14:281–290. doi: 10.1002/stem.140281. [DOI] [PubMed] [Google Scholar]
- 24.LeBrun D P, Matthews B P, Feldman B J, Cleary M L. The chimeric oncoproteins E2A-PBX1 and E2A-HLF are concentrated within spherical nuclear domains. Oncogene. 1997;15:2059–2067. doi: 10.1038/sj.onc.1201367. [DOI] [PubMed] [Google Scholar]
- 25.Lu Q, Kamps M P. Structural determinants within Pbx1 that mediate cooperative DNA binding with pentapeptide-containing Hox proteins: proposal for a model of a Pbx1-Hox-DNA complex. Mol Cell Biol. 1996;16:1632–1640. doi: 10.1128/mcb.16.4.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mann R S, Chan S K. Extra specificity from extradenticle: the partnership between HOX and PBX/EXD homeodomain proteins. Trends Genet. 1996;12:258–262. doi: 10.1016/0168-9525(96)10026-3. [DOI] [PubMed] [Google Scholar]
- 27.Monica K, Galili N, Nourse J, Saltman D, Cleary M L. PBX2 and PBX3, new homeobox genes with extensive homology to the human proto-oncogene PBX1. Mol Cell Biol. 1991;11:6149–6157. doi: 10.1128/mcb.11.12.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moskow J J, Bullrich F, Huebner K, Daar I O, Buchberg A M. Meis, a PBX1-related homeobox gene involved in myeloid leukemias in BXH-2 mice. Mol Cell Biol. 1995;15:5434–5443. doi: 10.1128/mcb.15.10.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakamura T, Jenkins N A, Copeland N G. Identification of a new family of Pbx-related homeobox genes. Oncogene. 1996;21:2235–2242. [PubMed] [Google Scholar]
- 30.Nakamura T, Largaespada D A, Shaughnessy J D J, Jenkins N A, Copeland N G. Cooperative activation of Hoxa and Pbx-1-related genes in murine myeloid leukemias. Nat Genet. 1996;12:149–153. doi: 10.1038/ng0296-149. [DOI] [PubMed] [Google Scholar]
- 31.Nourse J, Mellentin J D, Galili N, Wilkinson J, Stanbridge E, Smith S D, Cleary M C. Chromosomal translocation t(1;19) results in synthesis of a homeobox fusion mRNA that codes for a potential chimeric transcription factor. Cell. 1990;60:535–545. doi: 10.1016/0092-8674(90)90657-z. [DOI] [PubMed] [Google Scholar]
- 32.Pai C-Y, Kuo T-S, Jaw T J, Kurant E, Chen C-T, Bessarab D A, Salzberg A, Sun Y H. The homothorax homeoprotein activates the nuclear localization of another homeoprotein, Extradenticle, and suppresses eye development in Drosophila. Genes Dev. 1998;12:435–447. doi: 10.1101/gad.12.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peifer M, Wieschaus W. Mutations in the Drosophila gene extradenticle affect the way specific homeo domain proteins regulate segmental identity. Genes Dev. 1990;4:1209–1223. doi: 10.1101/gad.4.7.1209. [DOI] [PubMed] [Google Scholar]
- 34.Pellerin I, Schnabel C, Catron K M, Abate C. Hox proteins have different affinities for a consensus DNA site that correlate with the positions of their genes on the hox cluster. Mol Cell Biol. 1994;14:4532–4545. doi: 10.1128/mcb.14.7.4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phelan M L, Sadoul R, Featherstone M S. Functional differences between HOX proteins conferred by two residues in the homeodomain N-terminal arm. Mol Cell Biol. 1994;14:5066–5075. doi: 10.1128/mcb.14.8.5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Popperl H, Bienz M, Studer M, Chan S-K, Aparicio S, Brenner S, Mann R S, Krumlauf R. Segmental expression of Hoxb-1 is controlled by a highly conserved autoregulatory loop dependent upon exd/pbx. Cell. 1995;81:1031–1042. doi: 10.1016/s0092-8674(05)80008-x. [DOI] [PubMed] [Google Scholar]
- 37.Rauskolb C, Peifer M, Wieschaus E. Extradenticle determines segment identities throughout Drosophila development. Development. 1995;121:3663–3673. doi: 10.1242/dev.121.11.3663. [DOI] [PubMed] [Google Scholar]
- 38.Rieckhof G E, Casares F, Ryoo H D, Abu-Shaar M, Mann R S. Nuclear translocation of extradenticle requires homothorax, which encodes an extradenticle-related homeodomain protein. Cell. 1997;91:171–183. doi: 10.1016/s0092-8674(00)80400-6. [DOI] [PubMed] [Google Scholar]
- 39.Rubin M R, King W, Toth L E, Sawczuk I S, Levine M S, D’Eustachio P, Nguyen-Huu M C. Murine Hox-1.7 homeobox gene: cloning, chromosomal location, and expression. Mol Cell Biol. 1987;7:3836–3841. doi: 10.1128/mcb.7.10.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen W-F, Chang C-P, Rozenfeld S, Sauvageau G, Humphries R K, Lu M, Lawrence H J, Cleary M L, Largman C. HOX homeodomain proteins exhibit selective complex stabilities with Pbx and DNA. Nucleic Acids Res. 1996;24:898–906. doi: 10.1093/nar/24.5.898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40a.Shen, W.-F., and C. Largman. Unpublished observations.
- 41.Shen W-F, Montgomery J C, Rozenfeld S, Lawrence H J, Buchberg A, Largman C. The Abd-B-like Hox proteins stabilize DNA binding by the Meis1 homeodomain proteins. Mol Cell Biol. 1997;17:6448–6558. doi: 10.1128/mcb.17.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen W-F, Rozenfeld S, Lawrence H J, Largman C. The Abd-B-like Hox homeodomain proteins can be subdivided by the ability to form complexes with Pbx1a on a novel DNA target. J Biol Chem. 1997;272:8198–8206. doi: 10.1074/jbc.272.13.8198. [DOI] [PubMed] [Google Scholar]
- 43.Smith L T, Hohaus S, Gonzalez D A, Dziennis S E, Tenen D G. PU.1 (Spi-1) and C/EBPα regulate the granulocyte colony-stimulating factor receptor promoter in myeloid cells. Blood. 1996;88:1234–1247. [PubMed] [Google Scholar]
- 44.Steelman S, Moskow J J, Muzynski K, North C, Druck T, Montgomery J C, Huebner K, Daar I O, Buchberg A M. Identification of a conserved family of Meis1-related homeobox genes. Genome Res. 1997;7:142–156. doi: 10.1101/gr.7.2.142. [DOI] [PubMed] [Google Scholar]
- 45.Swift G H, Liu Y, Rose S D, Bischof L J, Steelman S, Buchberg A M, Wright C V E, MacDonald R J. An endocrine-exocrine switch in the activity of the pancreatic homeodomain protein PDX1 through formation of a trimeric complex with PBX1b and MRG1 (MEIS2) Mol Cell Biol. 1998;18:5109–5120. doi: 10.1128/mcb.18.9.5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thorsteinsdottir U, Sauvageau G, Hough M R, Dragowska W, Lansdorp P M, Lawrence H J, Largman C, Humphries K R. Overexpression of HOXA10 in murine hematopoietic cells perturbs both myeloid and lymphoid differentiation and leads to acute myeloid leukemia. Mol Cell Biol. 1997;17:495–505. doi: 10.1128/mcb.17.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Dijk M A, Murre C. Extradenticle raises the DNA binding specificity of homeotic selector gene products. Cell. 1994;78:617–624. doi: 10.1016/0092-8674(94)90526-6. [DOI] [PubMed] [Google Scholar]