Supplemental Digital Content is available in the text.
Keywords: cardiovascular disease, glucocorticoid receptor, left ventricular hypertrophy, long noncoding RNA, RNA-binding proteins
Rationale:
RBPs (RNA-binding proteins) play critical roles in human biology and disease. Aberrant RBP expression affects various steps in RNA processing, altering the function of the target RNAs. The RBP SRSF4 (serine/arginine-rich splicing factor 4) has been linked to neuropathies and cancer. However, its role in the heart is completely unknown.
Objective:
To investigate the role of SRSF4 in the heart.
Methods and Results:
Echocardiography of mice specifically lacking SRSF4 in the heart (SRSF4 KO) revealed left ventricular hypertrophy and increased cardiomyocyte area, which led to progressive diastolic dysfunction with age. SRSF4 KO mice showed altered electrophysiological activity under isoproterenol-induced cardiac stress, with a post-QRS depression and a longer QT interval, indicating an elevated risk of sudden cardiac death. RNA-Seq analysis revealed expression changes in several long noncoding RNAs, including GAS5 (growth arrest-specific 5), which we identified as a direct SRSF4 target in cardiomyocytes by individual-nucleotide-resolution cross-linking and immuno-precipitation. GAS5 is a repressor of the GR (glucocorticoid receptor) and was downregulated in SRSF4 KO hearts. This corresponded with elevated GR transcriptional activity in cardiomyocytes, leading to increases in hypertrophy markers and cell size. Furthermore, hypertrophy in SRSF4 KO cardiomyocytes was reduced by overexpressing GAS5.
Conclusions:
Loss of SRSF4 expression results in cardiac hypertrophy, diastolic dysfunction, and abnormal repolarization. The molecular mechanism underlying this effect involves GAS5 downregulation and consequent elevation of GR transcriptional activity. Our findings may help to develop new therapeutic tools for the treatment of cardiac hypertrophy and myocardial pathology in patients with Cushing syndrome.
Meet the First Author, see p 601
RBPs (RNA-binding proteins) regulate protein expression and function by regulating a wide range of posttranscriptional processes, including constitutive and alternative splicing, transport, localization, stability, and translation.1
The human genome encodes >1500 RBPs.2 Aberrant expression of specific RBPs is linked to several human diseases, mostly neurological or neuromuscular disorders such as Alzheimer disease, amyotrophic lateral sclerosis, and frontotemporal lobar dementia.3,4 Altered expression of RBPs is also a common feature of several cancers.5,6 Moreover, recent evidence reveals the involvement of RBPs in a broad spectrum of cardiovascular diseases,7 including dilated cardiomyopathy,8–11 cardiac hypertrophy,12 and electrophysiological alterations.13,14 For example, deletion of MBNL1 (muscleblind-like 1) in mice results in QRS and QTc widening and bundle block,15 whereas RBFox1 deficiency leads to hypertrophic cardiomyopathy.16 Some members of the serine/arginine-rich (SR) family of RBPs show an association with heart disease. A lack of SRSF1 (serine/arginine splicing factor 1) in cardiomyocytes leads to animal death due to severe excitation-contraction coupling defects.17 Similarly, loss of SRSF2 causes dilated cardiomyophaty18 and depletion of SRSF10 causes embryonically lethal cardiac contraction defects.19
The SR family of RBPs is composed of 12 members, each containing one or 2 RNA recognition motifs in its N-terminal region and a C-terminal domain (RS domain) enriched in arginine and serine residues. SR proteins are mainly located in the nucleus, where they participate in mRNA constitutive and alternative splicing, but they also shuttle between the nucleus and the cytoplasm to regulate mRNA export, stability, and translation.20–22
The SR RBP member SRSF4 associates cotranscriptionally and posttranscriptionally with intronless and intron-containing mRNAs and with ncRNAs (noncoding RNAs).23,24 SRSF4 participates in constitutive and alternative splicing and can alter gene expression by regulating detained intron splicing.25 SRSF4 also interacts with RNA classes that are not processed by the spliceosome.24 Moreover, SRSF4 binds to polyadenylated histone mRNAs in the cytoplasm, suggesting that it may be involved in histone mRNA 3’ end formation, export, and translation.24 SRSF4 controls cell cycle and differentiation by regulating hundreds of functionally related mRNAs.26 In the nervous system, SRSF4 is associated with a subset of neurodegenerative diseases called tauopathies through its regulation of the alternative splicing of exon 10 in the tau gene; this results in increased expression of the abnormal tau isoform FTDD-17, which can lead to neurodegeneration.27 Moreover, SRSF4 modulates alternative splicing of genes involved in hematopoietic progenitor cell differentiation28 and SRSF4 downregulation in acute myeloid leukemia causes a splicing change in CASP8 (caspase 8) associated with disease progression.29 These accumulated findings demonstrate the relevance of SRSF4 to neural function and cell proliferation; however, the role of this RBP in the heart is completely unknown.
In this study, we show that loss of SRSF4 in cardiomyocytes produces left ventricular (LV) hypertrophy, diastolic dysfunction, as well as repolarization abnormalities under cardiac stress conditions. Using cross-linking and immuno-precipitation (iCLIP), we found that the lncRNA GAS5 is a direct SRSF4 target in cardiomyocytes whose expression is downregulated in SRSF4 KO hearts. GAS5 is a repressor of the GR (glucocorticoid receptor),30 and we show here that GR transcriptional activity increases in SRSF4 KO cardiomyocytes, resulting in increased cell size and expression of hypertrophy markers. GAS5 overexpression in SRSF4 KO cardiomyocytes reduces their surface area, thereby establishing a causal relationship between SRSF4–GAS5 interaction and cardiac hypertrophy.
Methods
Data Availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Mice
SRSF4 KO mice were originally generated at the Mouse Clinical Institute—Institut Clinique de la Souris, France (EMMA ID: EM: 07637). The neomycin cassette was removed using the FLP-FRT system to obtain a SRSF4 floxed mouse line with LoxP sites flanking SRSF4 exon 2. This line was crossed with C57BL/6 mice to remove the FLP cassette and the resulting SRSF4 floxed mice were crossed with the cardio-specific Cre line Nkx2.5-Cre (C57BL/6 background, provided by Dr José Luis de la Pompa, CNIC, Madrid, Spain). The resulting line was maintained in homozygosis. SRSF4 floxed mice were used as controls.
Experiments were performed with adult males aged from 2 to 14 months. Power calculations were performed to determine the necessary number of animals based on prior data (P=0.05; power=80%). The study was mainly restricted to male mice to avoid the influence of sex differences associated with oestrogen production, which can affect the heart. A group of female mice was also included in the study to confirm the changes observed in male mice. Unless indicated in the figure legend, figures show results in male mice. Mice were housed in an air-conditioned room with a 12-hour light/dark cycle and free access to water and chow. Mice were euthanized in a carbon dioxide chamber. All procedures were approved by the Ethics Committees of the CNIC and the Regional Government of Madrid (PROEX 332-15, PROEX 177-17). Animals were randomly (simple randomization) assigned to groups. Researchers were blinded to the allocations.
Echocardiography
LV wall thickness, cardiac function, and chamber dimensions were analyzed by transthoracic 2-dimensional, M-mode, and Pulse Wave Doppler echocardiography. Scans were performed by operators blinded to genotype and treatment using a 40 MHz linear transducer (Vevo 2100, VisualSonics). Mice were placed on a heating pad under light anesthesia with 0.5% to 2% isoflurane administered with 100% oxygen via nose cone and adjusting the isoflurane delivery trying to maintain the heart rate in 500±50 bpm. LV wall thickness was calculated as the mean of the LV anterior wall thickness in diastole and the LV posterior wall thickness in diastole acquired in short-axis view. Normalized Cardiac Mass is calculated by Vevo 2100 with the following formulas; LV mass=1.053×([LVIDd (left ventricular internal dimension in diastole)+LVPWd (left ventricular posterior wall thickness in diastole)+LVAWd (left ventricular anterior wall thickness in diastole)]3−LVIDd3). LV mass corrected=LV mass×0.8. Normalized cardiac mass=LV mass corrected/body weight. LVIDd (LV internal diameter in diastole). LV ejection fraction and LV volume in diastole were obtained from the long axis view. The mitral inflow pattern was assessed using pulse wave Doppler to evaluate diastolic function from a 4-chamber apical view. Diastolic function was determined by measuring isovolumetric relaxation time and the early and late diastolic peak wave ratio (E/A wave ratio). Images were analyzed off-line by an expert using the Vevo 2100 analysis software (VisualSonics). As with the image acquisition, researchers were blinded to mouse genotype and treatment.
RNA-Seq
Total RNA was extracted from the LV of 2-month-old control and SRSF4 KO hearts using the RNAeasy extraction kit (74104, Qiagen). Libraries were prepared using polyA-positive selection, and RNAs were sequenced using an Illumina-HiSeq apparatus. Raw sequencing data were uploaded to the GEO repository (accession reference GSE131145). RNAs were sequenced by the CNIC Genomics Unit and RNA-Seq analysis was performed by the CNIC Bioinformatics Unit. Fastq files containing the reads for each library were extracted and demultiplexed using the Casava v1.8.2 pipeline. Reads were preprocessed with a pipeline that used cutadapt v1.6 to remove Illumina adaptors and FastQC to perform quality controls after each step. Processed reads were mapped to the Ensembl genebuild 76 reference transcriptome GRCm38 and quantified using RSEM v1.2.3. Data were normalized using TMM (trimmed mean of M values), and genes with at least 1 count per million in at least 3 samples were retained for further analysis. Differential expression was assessed using the edgeR Bioconductor Package.31,32 Changes were considered significant at an adjusted P<0.05. Alternative splicing was analyzed using vast-tools v0.2.0,33 and changes were considered significant when the probability of an absolute difference in the inclusion rate was >95%. The following regular commands were used:
Map reads to the genome, transcriptome and reference built-in library of exon-exon junctions for mouse genome version mm9: vast-tools align --sp Mmu SAMPLE.R1.fastq SAMPLE.R2.fastq
Combine information from all samples: vast-tools combine -sp Mmu
Perform differential splicing analysis: vast-tools diff -a SRSF4_1,SRSF4_2,SRSF4_3 -b Control_1,Control_2,Control_3
Plots summarizing RNA-seq results were generated with custom python scripts using matplotlib and seaborn libraries. The specific scripts used to generate the figures can be found in the public code repository at https://bitbucket.org/cmartiga/srsf4_ko_analysis.
Cross-Linking and Immuno-Precipitation
Wild-type C57BL/6 neonatal cardiomyocytes were isolated at postnatal day 0 or 1.34 A total of 8 million neonatal cardiomyocytes were plated on 10 cm2 plates. The following day, cells were transfected with 15 μg of SRSF4-GFP modRNA using 21 μL of Lipofectamine 2000 (11668027, Thermo Fisher Scientific). At 48-hour posttransfection, cells were irradiated once with 100 mJ UV light (254 nm) and iCLIP was performed as previously described.24 Briefly, crosslinked RNA was digested with RNaseI (Ambion) to produce 80 to 200–nucleotide fragments, and RNA-protein complexes were immunoprecipitated with Protein G Dynabeads (ThermoFisher Scientific) coupled to a goat anti-GFP antibody (D. Drechsel, MPI-CBG, Dresden). RNA fragments were then ligated to preadenylated DNA 3′ adapters (IDT) and reverse-transcribed using barcoded RT-primers and SuperscriptIV (ThermoFisher Scientific). After size-selection, cDNA fragments were circularized using CircLigaseTM (Epicentre) and relinearized with BamHI (NEB). The final cDNA libraries containing 5′ and 3′ adapters were amplified using AccuPrime (ThermoFisher Scientific) and were sequenced in an Illumina HiSeq2000 machine (single-end 75 nucleotide reads, 20 million reads per replicate). Analysis of iCLIP sequencing reads was done using the iCount package (http://icount.biolab.si). Adapters and barcodes were removed, and reads were mapped to the mouse mm9 genome assembly (Ensembl59 annotation) using Bowtie (version 0.10.1). To determine protein-RNA contact sites, only uniquely mapping reads were used. PCR duplicates were removed using the random barcodes in the 3′ adapter. Cross-link (X-link) sites within co-transcribed regions were extracted and randomized. Significant X-links (FDR <0.05) were calculated using normalized numbers of input X-links.35–37 BED files were indexed with tabix,38 and coordinates from significant X-links were intersected with GTF coordinates using pysam39 (https://github.com/pysam-developers/pysam) to characterize the binding preferences of SRSF4 (coding, noncoding, or intronic sequences). Raw sequencing data were uploaded to the GEO repository (accession reference GSE131145). All codes are available at https://github.com/tomazc/iCount.
Statistical Analysis
Data were analyzed for normality using the D´Agostino and Pearson omnibus normality test. If the normality test was not passed, data were analyzed for statistical significance by a Mann-Whitney test for experiments with two conditions and one variable, and by a Kruskal-Wallis test followed by Dunn Multiple comparison test for experiments with >2 conditions and one variable. If the normality test was passed, data were analyzed for statistical significance by Student t test for experiments with 2 conditions and one variable, by 1-way ANOVA followed by Bonferroni posttest for experiments with >2 conditions and one variable, and by 2-way ANOVA followed by Bonferroni posttest for analysis of multiple conditions and 2 variables, as indicated in each figure legend. Fisher test was used to associate 2 qualitative variables. Differences were considered significant at P<0.05. The n in figure legends indicates biological replicates.
For figures that include representative images, the individual sample that best represented the mean of the group was selected. All statistical analyses were performed using Graphpad Prism 7.0.
Results
Cardiac SRSF4 Expression Progressively Decreases Over Time
Cardiac SRSF4 mRNA expression during development and into adulthood was studied by qRT-PCR in hearts isolated from wild-type embryos and mice. SRSF4 mRNA expression peaked in the embryonic heart at embryonic day E13.5, followed by a progressive decline up to 6 months of age (Figure IA in the Data Supplement).
Cardio-Specific SRSF4 Knockout Leads to LV Hypertrophy
To study the role of SRSF4 in the heart, we crossed SRSF4 floxed mice with Nkx2.5-Cre transgenic mice. In the resulting cardio-specific knockout mouse line (SRSF4 KO), the floxed exon in the SRSF4 gene is excised in early heart progenitor cells during embryonic development. Cardiac-specific loss of SRSF4 was confirmed by qRT-PCR and Western blot (Figure IB and IC in the Data Supplement) and had no effect on animal viability. Some mRNA and protein expression was detected in the hearts of KO mice, likely originated in cells that do not derive from cardiac precursors, and in which the Cre recombinase was not expressed. Mice bearing the floxed SRSF4 allele with no Cre recombinase were used as controls.
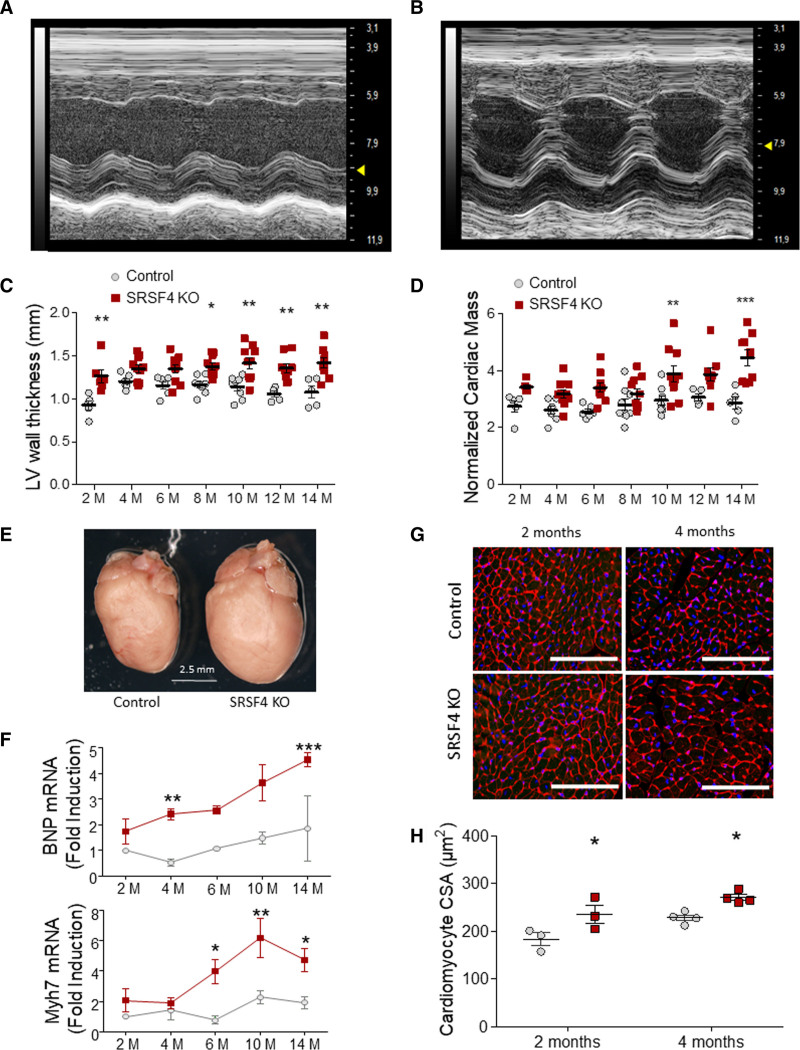
Echocardiography from 2 to 14 months postbirth showed significantly higher values for LV wall thickness and normalized cardiac mass in male SRSF4 KO mice than in controls, together with an increased heart weight to body weight ratio (Figure 1A through 1D, Table II in the Data Supplement). SRSF4 KO mice also showed a significantly thicker LV wall when compared to Nkx2.5-Cre mice without a floxed SRSF4 allele, suggesting that the observed cardiac hypertrophy was not due to nonspecific effects of the Cre recombinase itself (Figure II in the Data Supplement).SRSF4 KO hearts appeared to be enlarged (Figure 1E) and had higher expression than controls of the heart disease markers brain natriuretic peptide (BNP) and MYH7 (myosin heavy chain 7) (Figure 1F). These findings were confirmed in a group of female mice at 4 months of age (Figure III in the Data Supplement). SRSF4 KO mice also showed an increased cardiomyocyte area (Figure 1G and 1H), suggesting that the cardiac hypertrophy observed by echocardiography was due to an increase in the size of individual cardiomyocytes. We did not detect significant increases in the expression of different fibrosis markers by qRT-PCR in SRSF4 KO hearts or in the amount of fibrotic tissue in histological sections (Figure IV in the Data Supplement). We found no significant changes in arterial pressure, suggesting that the observed cardiac hypertrophy was not the result of increased blood pressure (Figure V in the Data Supplement).
Figure 1.
Cardiac-specific loss of SRSF4 (serine/arginine splicing factor 4) leads to left ventricular (LV) hypertrophy.A and B, Representative M-mode short axis view echocardiography of control (A) and SRSF4 KO (B) left ventricles at 10 mo of age. C and D, Echocardiography analysis of LV wall thickness (C) and cardiac mass normalized to body weight (D). Data are shown as mean±SEM. Symbols represent individual animals (biological replicates). E, Representative whole hearts from 10-month-old control and KO mice. F, Quantitative real-time polymerase chain reaction analysis of brain natriuretic peptide (BNP) and Myh7 mRNA in control and SRSF4 KO hearts. n=4 mice per group in all groups except SRSF4 KO mice at 4 mol/L, which was n=5. C, D, and F, Data are shown as mean±SEM. *P<0.05, **P<0.01, ***P<0.001 for SRSF4 KO (red) vs control (gray). Two-way ANOVA followed by Bonferroni correction; M, months. G, Representative myocardial sections from 2- and 4-month-old mice, stained with tetramethylrhodamine-conjugated wheat-germ agglutinin (red) and DAPI (blue). Scale bar: 100 µm. H, Quantification of cardiomyocyte cross-sectional area (CSA). Each symbol represents the CSA average of one mouse. Data are shown as mean±SEM. *P<0.05 SRSF4 KO vs control. Two-way ANOVA followed by Bonferroni posttest. AW indicates anterior wall; and PW, posterior wall.
SRSF4 KO Mice Develop Diastolic Dysfunction
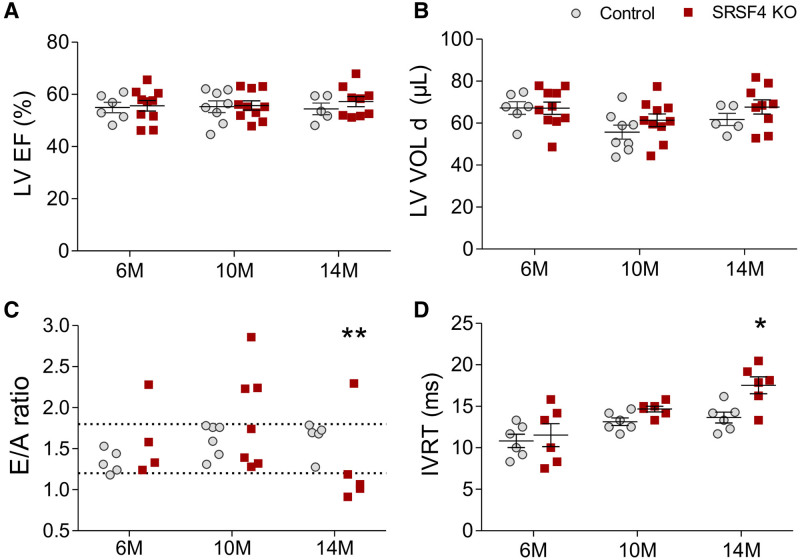
To investigate the effect of LV hypertrophy in SRSF4 KO mice on cardiac function, we analyzed systolic and diastolic function by echocardiography (Table II in the Data Supplement). Loss of SRSF4 had no significant effect on systolic function or LV diastolic volume (Figure 2A and 2 B). However, SRSF4 mice showed a significant and age-dependent decrease in the E/A wave ratio and an elevated isovolumetric relaxation time (Figure 2C and 2D). Thus, although SRSF4 KO mice show preserved contraction, they progressively develop diastolic dysfunction.
Figure 2.
SRSF4 (serine/arginine splicing factor 4) KO mice develop diastolic dysfunction with preserved ejection fraction.A–D, Echocardiography analysis of left ventricular function. A, LV ejection fraction (LVEF). B, LV volume in diastole (LVVOLd). C, E/A wave ratio of mitral flow, with normal mitral flow defined as ranging from 1.2 to 1.8. **P<0.01 normal (1.2–1.8) vs abnormal E/A ratio; Fisher test. M, months. D, Isovolumetric relaxation time (IVRT). Data are shown as mean±SEM. Symbols represent individual animals (ie, biological replicates). A, B, and D, *P<0.05 SRSF4 KO vs control. Two-way ANOVA followed by Bonferroni posttest.
SRSF4 KO Mice Exhibit Electrocardiographic Features of Cardiac Hypertrophy and Abnormal Repolarization
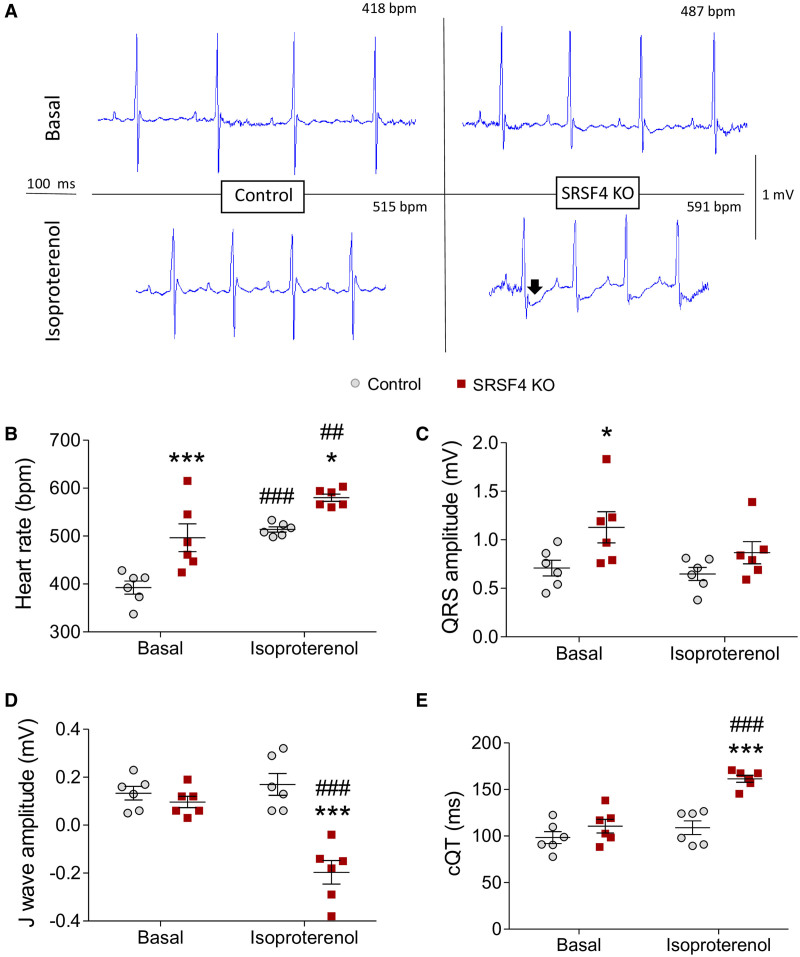
Cardiac hypertrophy is a risk factor for QT-prolongation and sudden cardiac death.40 To study whether the hypertrophy caused by the lack of SRSF4 altered cardiac electrophysiological activity, we performed ECGs under basal and stress conditions (Figure 3A). In basal conditions, KO mice had a significantly higher heart rate than controls and a wider QRS complex amplitude, which is a characteristic of cardiac hypertrophy (Figure 3B and 3C). We also found decreased J wave amplitude and a longer QT interval corrected for heart rate (cQT) in SRSF4 KO mice, although the differences did not reach statistical significance. These repolarization abnormalities were intensified by isoproterenol-induced stress, which further increased the cQT interval, caused ST-segment depression, and produced a negative J wave (Figure 3D and 3E). We found no significant changes in the expression of potassium channel coding genes, suggesting that the electrical abnormalities were the consequence of cardiac hypertrophy, rather than the cause (Figure VI in the Data Supplement).
Figure 3.
SRSF4 (serine/arginine splicing factor 4) KO mice have electrophysiological alterations.A, Representative ECGs of control and SRSF4 KO mice in baseline conditions and after intraperitoneal isoproterenol injection. The arrow marks the depression after the QRS complex. B–E, Analysis of heart rate (B), QRS amplitude (C), J wave amplitude (D), and corrected QT interval (E) in baseline and isoproterenol-stimulated conditions. Data are shown as mean±SEM, and symbols represent individual 6-month-old mice. *P<0.05, ***P<0.001, SRSF4 KO vs control; ##P<0.01, ###P<0.001, isoproterenol vs baseline; 2-way ANOVA followed by the Bonferroni multiple comparison test.
The lncRNA GAS5 Is a Direct SRSF4 Target in Cardiomyocytes, and Its Expression Is Downregulated in SRSF4 KO Hearts
To study the molecular mechanism underlying the development of LV hypertrophy in SRSF4 KO mice, we performed an RNA-Seq analysis of hearts from 2-month-old control and SRSF4 KO mice. Significant expression changes were detected in 858 genes, including many lncRNAs (Figure VIIA and VIIB in the Data Supplement and Table III in the Data Supplement). Gene ontology analysis of significantly increased genes in KO mice showed enrichment in genes associated with sodium transport and vasoconstriction, whereas downregulated genes were associated with action potential regulation (Figure VIIC and VIID in the Data Supplement). Alternative splicing analysis in control and KO hearts, however, revealed few genes with a significant isoform change, suggesting that alternative splicing is not strongly affected by the lack of SRSF4 (Figure VIIIA through VIIIC and Table IV in the Data Supplement).
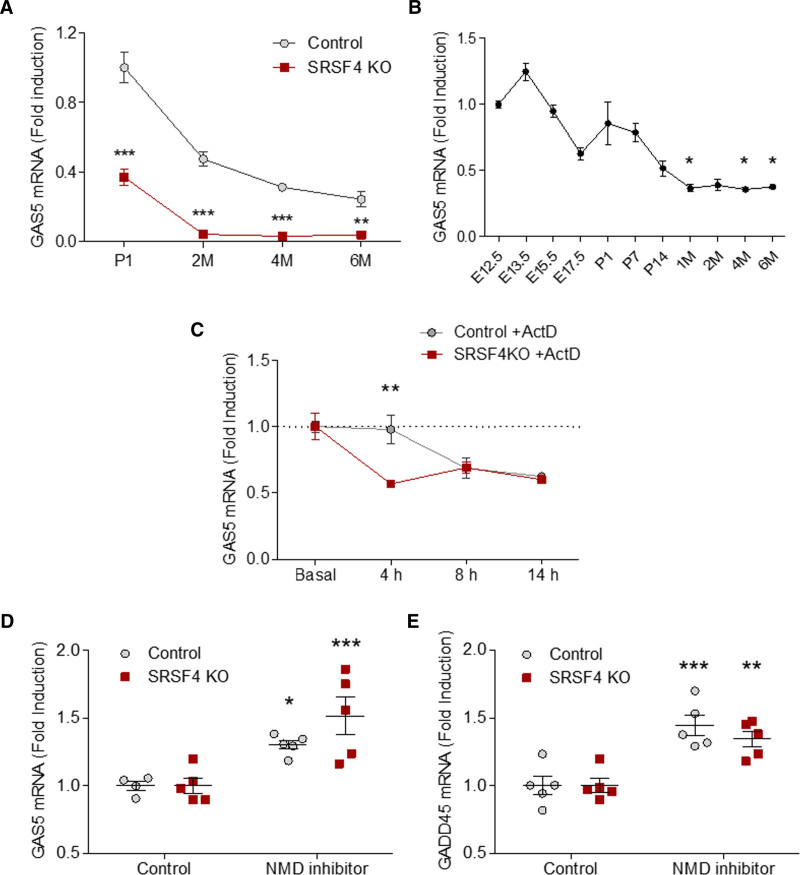
To investigate the direct SRSF4 target genes in cardiomyocytes, we immunoprecipitated SRSF4-bound RNAs from neonatal cardiomyocytes using iCLIP, which enables the identification of RNA-protein interactions.41 The iCLIP results showed specific binding of SRSF4 to 836 RNAs in these cells (Figure IX and Table V in the Data Supplement). To determine which of the differentially expressed genes are direct SRSF4 targets, we intersected RNA-Seq and iCLIP data. The resulting list of 42 genes (Table VI in the Data Supplement) included the lncRNA GAS5. Analysis by qRT-PCR confirmed GAS5 downregulation in SRSF4 KO hearts (Figure 4A), and the GAS5 expression profile in wild-type hearts was very similar to that of SRSF4 (Figure 4B). We next investigated whether GAS5 downregulation in the absence of SRSF4 was due to faster degradation. Transcription in neonatal cardiomyocytes from control and SRSF4 KO mice was blocked with actinomycin D, and GAS5 expression was measured at specific time points posttreatment. At 4-hour posttreatment, GAS5 expression in SRSF4 KO cardiomyocytes was significantly lower than in control cells (Figure 4C).
Figure 4.
Expression of the lncRNA GAS5 decreases in the absence of SRSF4 (serine/arginine splicing factor 4).A, Quantitative real-time polymerase chain reaction (qRT-PCR) analysis of GAS5 expression in the hearts of control and SRSF4 KO mice from birth to adulthood. Data are shown as mean±SEM; n=4 mice per group, except for P1 groups (n=5). **P<0.01, ***P<0.001 SRSF4 KO vs control; 2-way ANOVA followed by Bonferroni posttest. B, Expression profile of GAS5 in the hearts of wild-type mice from embryonic development to adulthood. Data are shown as mean±SEM; n=3 mice per group, except for the E17.5 group (n=4). *P<0.05; Kruskal-Wallis test followed by Dunn Multiple comparison test. C, qRT-PCR analysis of GAS5 expression in control and SRSF4 KO neonatal cardiomyocytes treated with ActD. Values for each treatment time point were normalized to the mean basal (untreated) value and to the mean value for control cells treated with DMSO (dotted line). Data are shown as mean±SEM, n=3 per group. **P<0.01 SRSF4 KO vs control; 2-way ANOVA followed by the Bonferroni posttest. D and E, qRT-PCR analysis of GAS5 (D) and GADD45 (E) expression in control and SRSF4 KO neonatal cardiomyocytes treated with vehicle (DMSO, control) or with nonsense-mediated decay (NMD) inhibitor for7 h. Values were normalized to the mean value of each group treated with DMSO. Data are shown as mean±SEM, and symbols represent individual samples from one experiment. n=5. *P<0.05, **P<0.01, ***P<0.001 for control vs NMD inhibitor. Two-way ANOVA followed by Bonferroni correction.
A previous study has reported that GAS5 is an nonsense-mediated decay (NMD) target.42 To determine whether GAS5 is more degraded by NMD in the absence of SRSF4, we treated control and SRSF4 KO neonatal cardiomyocytes with an NMD inhibitor. Results showed a significant increase in GAS5 expression after blocking NMD in control cells, however, in KO cells, GAS5 levels were even further increased. Such a difference was not seen for the control NMD target GADD45, which suggests that the observed effect is specific. (Figure 4D and 4E). Together, these results suggest that SRSF4 binds the lncRNA GAS5 and protects it from degradation by NMD.
SRSF4–GAS5–Glucocorticoid-Receptor Signaling Controls Cardiomyocyte Hypertrophy
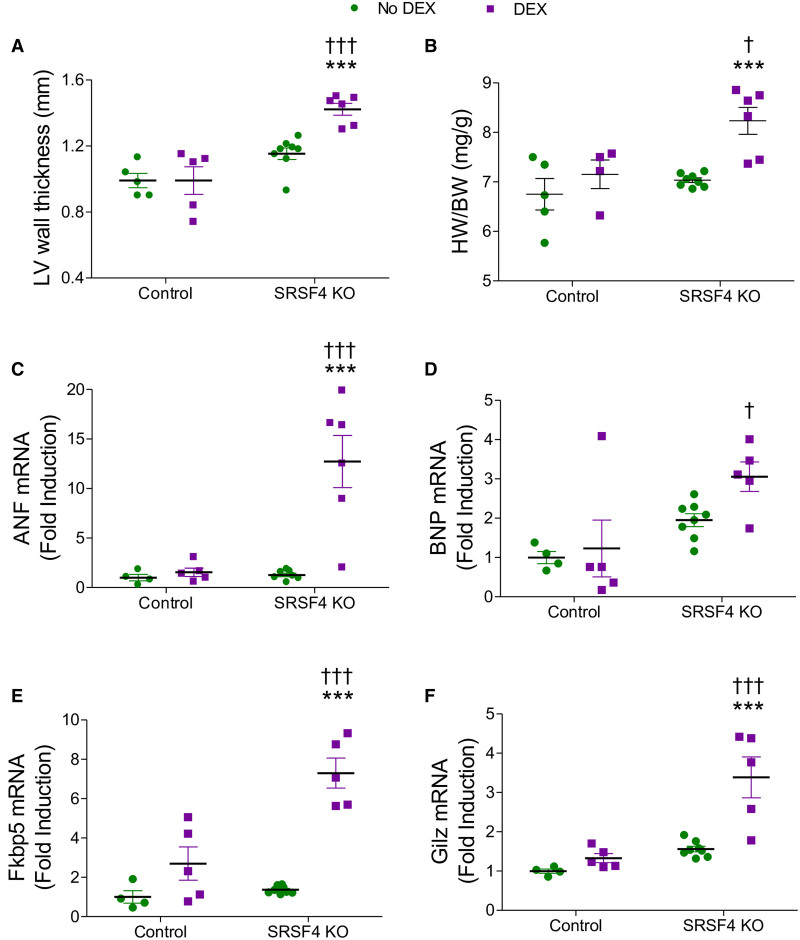
GAS5 represses GR-mediated induction of several genes.30 Moreover, GR activation is known to induce hypertrophic changes in H9C2 cells and primary neonatal rat cardiomyocytes.43 To investigate whether SRSF4 regulates GR activity by controlling GAS5 expression, we first analyzed the localization of GAS5 within the cell and found it was partially located in the nucleus (Figure X in the Data Supplement). We then treated neonatal cardiomyocytes from control and SRSF4 KO mice with dexamethasone to activate the GR. After 72 hours, we determined the expression of hypertrophic markers and several known direct GR targets30,44 by qRT-PCR. Dexamethasone increased the expression of these genes in control and KO cells, but dexamethasone-induced expression of the hypertrophic markers ANF (atrial natriuretic factor) and BNP was significantly higher in SRSF4 KO cells than in controls (Figure XIA and XIB in the Data Supplement). Similarly, SRSF4 KO cells showed significantly higher dexamethasone-induced expression of the direct GR targets Fkbp5 and Gilz (Figure XIC and XID in the Data Supplement). Moreover, dexamethasone-treated SRSF4 KO neonatal cardiomyocytes were significantly larger than both dexamethasone-treated control cells and untreated cells (Figure XIE in the Data Supplement).
To validate these findings in vivo, we treated SRSF4 KO and control mice with dexamethasone and analyzed LV mass by echocardiography. We found a significant increase in LV wall thickness in SRSF4 KO mice compared with control mice. This was paralleled by a significant increase in the heart weight to body weight ratio in mice lacking SRSF4 in the heart (Figure 5A and 5B). In addition, we observed a significant increase in ANF, Fkbp5, and Gilz mRNA expression in SRSF4 KO mice (Figure 5C through 5F). BNP also showed increased expression, although the changes did not reach significance. Together, these results indicate that the hypertrophy observed in SRSF4 KO cardiomyocytes might be due to increased GR transcriptional activity.
Figure 5.
SRSF4 (serine/arginine splicing factor 4) KO mice show an increase in hypertrophy and cardiac expression of hypertrophic markers and known GR (glucocorticoid receptor) targets following DEX treatment.A and B, Echocardiography analysis of left ventricular (LV) wall thickness (A), and heart weight/body weight (B) of SRSF4 KO and control mice following 2 wk treatment with DEX (15 mg/100 g BW) or no treatment. Expression of ANF (C), brain natriuretic peptide (BNP) (D), Fkbp5 (E), and Gliz (F) was analyzed by quantitative real-time polymerase chain reaction. Data are shown as mean±SEM. Symbols represent individual animals. ***P<0.001 DEX (green) vs No DEX (violet). †P<0.05, SRSF4 KO vs control, †††P<0.001 SRSF4 KO vs control. Two-way ANOVA followed by Bonferroni correction.
To rule out that changes in GR expression may underlie the cardiac hypertrophy observed in SRSF4 KO mice, we measured its expression by qRT-PCR and western blot. Results showed no significant changes neither in mRNA nor in protein levels (Figure XII in the Data Supplement).
GAS5 Overexpression Reduces Hypertrophy in SRSF4 KO Cardiomyocytes
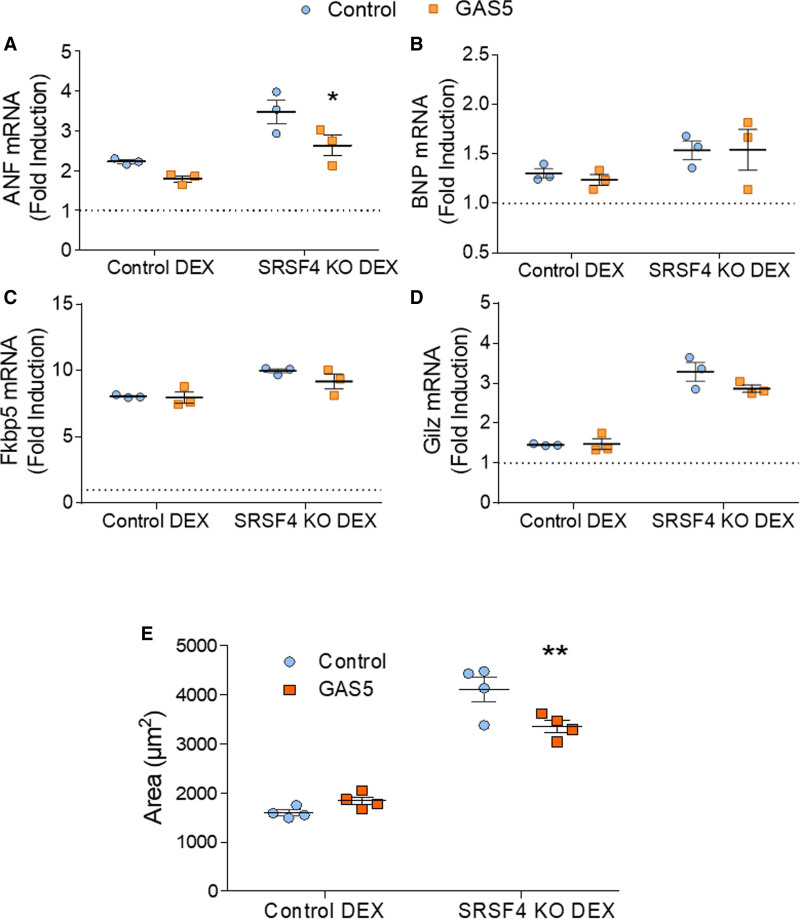
To determine whether the increased cardiomyocyte hypertrophy observed in SRSF4 KO cells is due to decreased GAS5 expression, we overexpressed GAS5 using modRNA in control and SRSF4 KO neonatal cardiomyocytes and stimulated them with dexamethasone. At 72 hours after dexamethasone treatment, we measured the expression of hypertrophic markers and known direct GR targets by qRT-PCR. GAS5 overexpression significantly decreased dexamethasone-induced expression of the hypertrophic marker ANF and partially reduced the GR-regulated gene Gilz (Figure 6A through 6D). GAS5 (growth arrest-specific 5)-overexpression in SRSF4 KO cardiomyocytes also reduced dexamethasone-induced cardiomyocyte hypertrophy (Figure 6E).
Figure 6.
GAS5 (growth arrest-specific 5) overexpression reduces hypertrophy in SRSF4 (serine/arginine splicing factor 4) KO cardiomyocytes.A–D, Quantitative real-time polymerase chain reaction (qRT-PCR) analysis of the hypertrophy markers ANF and brain natriuretic peptide (BNP) (A and B) and the GR (glucocorticoid receptor) targets Fkbp5 and Gliz (C and D) in control and SRSF4 KO neonatal cardiomyocytes treated with 100 nmol/L DEX and transfected with GAS5 or control modRNA. Data are shown as mean±SEM, and symbols represent 3 independent experiments (biological replicates), each of them the average of 2 (control) or 3 (KO) technical replicates. In each independent experiment, values were normalized to the mean value of control samples with no DEX and no GAS5 in each genotype (dotted line). *P<0.05 GAS5 vs control modRNA; 2-way ANOVA followed by the Bonferroni posttest. E, Area of control and SRSF4 KO neonatal cardiomyocytes treated with 100 nm DEX and transfected with GAS5 or control modRNA. Data are shown as mean±SEM, and symbols represent the average of individual cardiomyocytes in each biological replicate. **P<0.01, GAS5 vs control modRNA; 2-way ANOVA followed by the Bonferroni posttest.
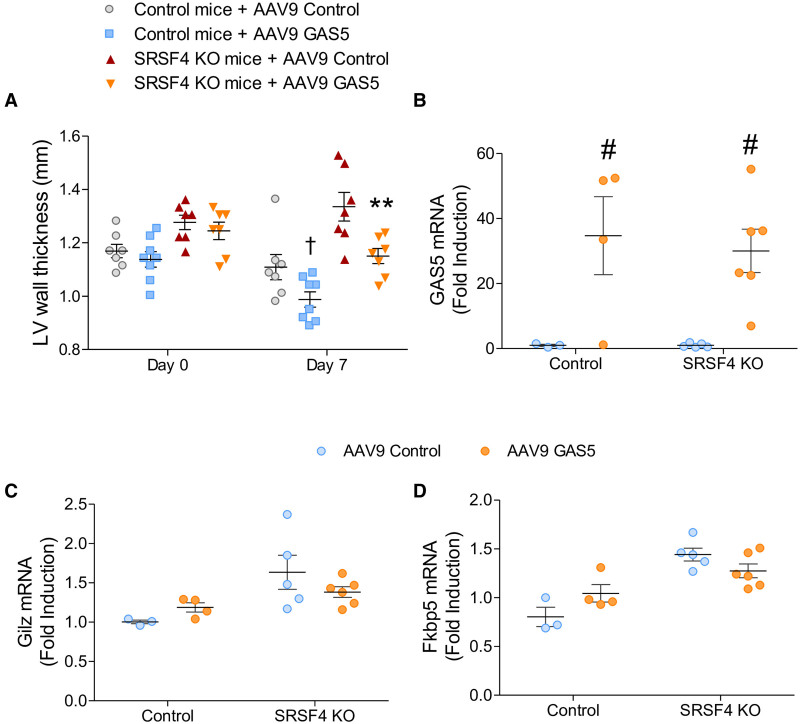
To confirm these results in mice, we overexpressed GAS5 in a cardiac-specific manner using an adeno-associated virus bearing the GAS5 lncRNA under the control of the cardiac troponin promoter. Results showed a significant decrease of the LV wall thickness in SRSF4 KO mice one week after injection of AAV9 (adeno-associated virus type 9)-GAS5, and a partially reduced expression of GR target genes Fkbp5 and Gilz (Figure 7A through 7D). Together these results indicate that the loss of GAS5 in SRSF4 KO hearts thus appears to be at least partly responsible for the hypertrophic response in cardiomyocytes.
Figure 7.
GAS5 (growth arrest-specific 5) overexpression reduces left ventricular hypertrophy in SRSF4 (serine/arginine splicing factor 4) KO mice.A, Echocardiography analysis of left ventricular (LV) wall thickness of control and SRSF4 KO mice injected with AAV9 (adeno-associated virus type 9)-GAS5 or a control AAV9. Data are shown as mean ±SEM. Symbols represent individual mice. **P<0.01, SRSF4 KO mice + AAV9 GAS5 vs SRSF4 KO mice+AAV9 control; †P<0.05, control mice+AAV9 GAS5 vs control mice+AAV9 control; 2-way ANOVA followed by the Bonferroni multiple comparison test. B–D, Quantitative real-time polymerase chain reaction (qRT-PCR) analysis of GAS5 (B), Gilz (C), and Fkbp5 (D) mRNA expression in mice injected with the different AAV9 at 1 wk postinjection. Data are shown as mean±SEM. Symbols represent individual animals, different from A. #P<0.05, AAV9 control vs AAV9 GAS5; 2-way ANOVA followed by the Bonferroni multiple comparison test.
SRSF6 Overexpression Plays a Role in GR Signaling
Since GAS5 overexpression did not completely reverse cardiac hypertrophy in SRSF4 KO cardiomyocytes, we investigated other pathways that might also contribute to the observed phenotype. The RNA-seq analysis had shown an increase in another member of the SR family, SRSF6, in SRSF4 KO hearts, together with changes in other RBPs (Table III in the Data Supplement). While we did not observe major changes in the expression of any of the other RBPs analyzed (Figure XIII in the Data Supplement), we found a significant increase in SRSF6 mRNA in the myocardium of SRSF4 KO mice (Figure XIVA in the Data Supplement). To determine whether the upregulation of SRSF6 could play a role in GR signaling in the absence of SRSF4, we knocked down the expression of SRSF6 in SRSF4 KO cardiomyocytes in vitro, and we measured the expression of the direct GR targets Fkbp5 and Gilz (Figure XIVB in the Data Supplement). Results showed a significant decrease in both GR targets following the reduction in SRSF6, suggesting that this RBP plays a role in the activation of GR signaling observed in the absence of SRSF4.
The Expression of SRSF4 and GAS5 Is Reduced in Patients With Cushing Syndrome
Finally, given the relationship between SRSF4, GAS5, and the GR that we unveiled here, we investigated whether the expression of SRSF4 and GAS5 was downregulated in patients with Cushing syndrome, whose features include elevated blood glucocorticoids and LV hypertrophy. We analyzed previously published RNA-seq data obtained from adipose tissue of patients with Cushing syndrome and controls. We found a downregulation of SRSF4 and GAS5 expression in patients with Cushing syndrome compared with controls (Figure XV in the Data Supplement), suggesting a potential role of this signaling axis in Cushing syndrome.
Discussion
A number of RBPs have recently been implicated in several diseases, including cardiovascular disease. The RBP SRSF4 has been linked to neuropathies and cancer; however, its role in the heart had not been previously explored. In this study, we show that cardiac-specific loss of SRSF4 leads to LV hypertrophy associated with an increase in cardiomyocyte size. Adult SRSF4 KO mice also develop diastolic dysfunction and electrophysiological features associated with an increased risk of sudden cardiac death.
The combination of RNA-Seq and iCLIP analysis identified the lncRNA GAS5 as a direct SRSF4 target in cardiomyocytes. GAS5 expression during development followed the same pattern as that of SRSF4, and loss of SRSF4 resulted in strong downregulation of this lncRNA. GAS5 is degraded through NMD; depletion of the key NMD machinery component UPF1 (up-frameshift 1) induces GAS5 transcript upregulation and consequent downregulation of the apoptosis-related genes cIAP2 and SGK1, whose promoters are regulated by the GR.45 Another study reported that downregulation of the NMD machinery is essential for the differentiation of neural progenitors and is accompanied by increased expression of several NMD targets, including GAS5.46 In addition to mRNAs, NMD targets include miRNAs and lncRNAs such as GAS5.42 Thus, while the precise mechanism by which SRSF4 regulates GAS5 expression remains unknown, our data suggests that it involves mRNA stability or protection from degradation through NMD.
GAS5 is expressed in adult tissues and during embryonic development and is downregulated in several cancers, including breast cancer.47 GAS5 localizes to both the nucleus and the cytoplasm30 and has an estimated half-life of ≈4 hours.48 In lymphocytes, GAS5 regulates apoptosis and the cell cycle.49 Interestingly, GAS5 is decreased in patients with type 2 diabetes; it binds to the insulin receptor promoter to regulate its expression, and its depletion inhibits glucose uptake and insulin signaling.50 The interaction of GAS5 with the GR DNA-binding domain prevents GR binding to the glucocorticoid response element in target genes, thus modulating GR transcriptional activity.30 Our results suggest that SRSF4-GAS5 interaction regulates GR transcriptional activity in the heart, in line with in vivo and in vitro evidence implicating glucocorticoids in the development of cardiac hypertrophy51–53 and the induction by activated GR of hypertrophic changes in H9C2 cells and primary neonatal rat cardiomyocytes.43 Our findings demonstrate that GR transcriptional activity increases in SRSF4 KO cardiomyocytes, in which GAS5 expression is downregulated. We also found that SRSF4 KO cardiomyocytes responded to dexamethasone-mediated GR activation with more pronounced increases in hypertrophy marker expression and cell area than observed in controls. These results were validated in vivo. Moreover, we also found that overexpression of GAS5 reduced hypertrophy in SRSF4 KO cardiomyocytes, suggesting a causal role for the loss of GAS5 expression in the hypertrophic response observed in the absence of SRSF4.
GAS5 overexpression, however, only partially reversed cardiac hypertrophy, suggesting that additional mechanisms mediate cardiac hypertrophy in mice lacking SRSF4. In this regard, we observed that SRSF6 transcripts are increased in SRSF4 KO cardiomyocytes and that reduction of SRSF6 levels results in reduced cardiomyocyte hypertrophy. Although additional investigation is needed to confirm the role of SRSF6 in the heart, these results suggest that the induction of SRSF6 is a prohypertrophic mechanism.
The structural and functional alterations observed in mice lacking SRSF4 in the heart resemble those found in human hypertrophic cardiomyopathy.54 It would be interesting to investigate whether patients with idiopathic cardiac hypertrophy have loss of function mutations in the SRSF4 or GAS5 genes. Moreover, elevated blood glucocorticoids and LV hypertrophy are features of Cushing syndrome and several patients with Cushing syndrome develop LV hypertrophy without hypertension.53 In this regard, we found that SRSF4 and GAS5 expression was downregulated in adipose tissue from patients with Cushing syndrome. Therefore, it would be worth investigating whether these patients have elevated GR activity in the heart due to SRSF4 deficiency.55,56
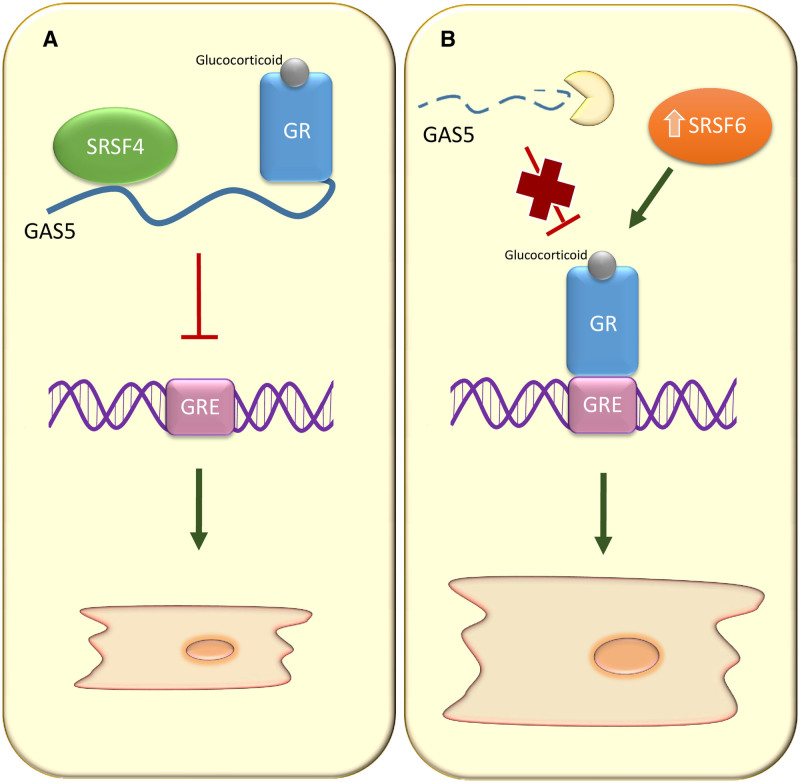
In summary, our study shows that loss of SRSF4 in cardiomyocytes results in faster degradation and subsequent downregulation of GAS5, leading to an increase in the transcriptional activity of the GR, which causes cardiomyocyte hypertrophy (Figure 8). These findings may help to identify new diagnostic and therapeutic tools for the treatment of cardiac hypertrophy and Cushing syndrome.
Figure 8.
Molecular mechanism leading to cardiac hypertrophy in the absence of SRSF4 (serine/arginine splicing factor 4).A, GAS5 (growth arrest-specific 5) is a direct SRSF4 target. When SRSF4 is present, GAS5 binds to the GR (glucocorticoid receptor) and inhibits its binding to the GRE region in its target genes. B, In the absence of SRSF4, GAS5 is downregulated, and the GR induces the expression of target genes involved in cardiomyocyte hypertrophy. Moreover, the absence of SRSF4 produces the upregulation of SRSF6, which further contributes to an increase in GR targets.
Acknowledgments
We thank the CNIC animal facility for mouse work, the Histology Unit for H&E staining, the Viral Vectors Unit for viral production and the Genomics and Bioinformatics Units for RNA-Seq and analysis.
Sources of Funding
This study was supported by grants from the European Union (CardioNeT-ITN-289600 and CardioNext-ITN-608027 to E. Lara-Pezzi), from the Spanish Ministerio de Economía y Competitividad (RTI2018-096961-B-I00, SAF2015-65722-R, and SAF2012-31451 to E. Lara-Pezzi), the Spanish Carlos III Institute of Health (CPII14/00027 to E. Lara-Pezzi, RD12/0042/066 to P. García-Pavía and E. Lara-Pezzi, and RD12/0042/005), the Madrid Regional Government (2010-BMD-2321 Fibroteam to E. Lara-Pezzi). This study was also supported by the Plan Estatal de I+D+I 2013-2016, with funding from the European Regional Development Fund (ERDF) A way to build Europe initiative. The CNIC is supported by the Ministerio de Ciencia, Innovación y Universidades (MCNU) and the Pro-CNIC Foundation, and is a Severo Ochoa Center of Excellence (SEV-2015-0505).
Disclosures
None.
Supplemental Materials
Detailed Methods
Online Figures I–XIV
Online Tables I–VI
Supplementary Material
Nonstandard Abbreviations and Acronyms
- GAS5
- growth arrest-specific 5
- GR
- glucocorticoid receptor
- iCLIP
- individual-nucleotide resolution cross-linking and immunoprecipitation
- LV
- left ventricular
- ncRNA
- noncoding RNA
- NMD
- nonsense-mediated decay
- RBP
- RNA binding protein
- SR
- serine/arginine-rich
- SRSF4
- serine/arginine splicing factor 4
The Data Supplement is available with this article at https://www.ahajournals.org/doi/suppl/10.1161/CIRCRESAHA.120.318577.
For Sources of Funding and Disclosures, see page 682.
Contributor Information
Javier Larrasa-Alonso, Email: jlarrasa@gmail.com.
María Villalba-Orero, Email: maria.villalba@cnic.es.
Carlos Martí-Gómez, Email: carlos.marti@cnic.es.
Paula Ortiz-Sánchez, Email: paula0620@gmail.com.
Marina M. López-Olañeta, Email: mmlopez@cnic.es.
François McNicoll, Email: McNicoll@bio.uni-frankfurt.de.
Michaela Müller-McNicoll, Email: Mueller-McNicoll@bio.uni-frankfurt.de.
Pablo García-Pavía, Email: pablogpavia@yahoo.es.
Novelty and Significance
What Is Known?
RBPs (RNA binding proteins) regulate a wide range of posttranscriptional processes, and their aberrant expression is linked to several human diseases.
Loss of the RBP SRSF4 (serine/arginine-rich splicing factor 4) is associated with neuropathies and cancer; however, its role in the heart is virtually unknown.
What New Information Does This Article Contribute?
Cardiac-specific deletion of SRSF4 results in cardiac hypertrophy and diastolic dysfunction.
Loss of SRSF4 destabilizes the lncRNA GAS5 (growth arrest-specific 5), which results in overactivation of the glucocorticoid receptor.
The SRSF4 is an RBP involved in posttranscriptional regulation in different tissues. Loss of SRSF4 can lead to neurodegeneration or cancer due to changes in alternative splicing of its target mRNAs. Its role in the heart was previously unknown. In this study, we found that cardiac-specific SRSF4 knockout mice develop cardiac hypertrophy, electrophysiological abnormalities, and diastolic dysfunction. In the absence of SRSF4, several lncRNAs were also dysregulated. We observed that SRSF4 binds to and stabilizes the lncRNA GAS5, an inhibitor of the GR (glucocorticoid receptor). In the absence of SRSF4, GAS5 is degraded, and the GR promotes cardiac hypertrophy. This SRSF4-deficient hypertrophic response also upregulated another RBP, SRSF6. Lastly, overexpression of GAS5 using an adeno-associated viral vector reduced cardiac hypertrophy in SRSF4 knockout mice. Our results highlight the novel role of the SRSF4-GAS5-GR axis in the development of cardiac hypertrophy, opening new possibilities for further investigations in its treatment.
References
- 1.Corbett AH. Post-transcriptional regulation of gene expression and human disease. Curr Opin Cell Biol. 2018;52:96–104. doi: 10.1016/j.ceb.2018.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerstberger S, Hafner M, Tuschl T. A census of human RNA-binding proteins. Nat Rev Genet. 2014;15:829–845. doi: 10.1038/nrg3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conlon EG, Manley JL. RNA-binding proteins in neurodegeneration: mechanisms in aggregate. Genes Dev. 2017;31:1509–1528. doi: 10.1101/gad.304055.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maziuk B, Ballance HI, Wolozin B. Dysregulation of RNA binding protein aggregation in neurodegenerative disorders. Front Mol Neurosci. 2017;10:89. doi: 10.3389/fnmol.2017.00089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pereira B, Billaud M, Almeida R. RNA-binding proteins in cancer: old players and new actors. Trends Cancer. 2017;3:506–528. doi: 10.1016/j.trecan.2017.05.003 [DOI] [PubMed] [Google Scholar]
- 6.Wang ZL, Li B, Luo YX, Lin Q, Liu SR, Zhang XQ, Zhou H, Yang JH, Qu LH. Comprehensive genomic characterization of RNA-binding proteins across human cancers. Cell Rep. 2018;22:286–298. doi: 10.1016/j.celrep.2017.12.035 [DOI] [PubMed] [Google Scholar]
- 7.de Bruin RG, Rabelink TJ, van Zonneveld AJ, van der Veer EP. Emerging roles for RNA-binding proteins as effectors and regulators of cardiovascular disease. Eur Heart J. 2017;38:1380–1388. doi: 10.1093/eurheartj/ehw567 [DOI] [PubMed] [Google Scholar]
- 8.Liu J, Kong X, Zhang M, Yang X, Xu X. RNA binding protein 24 deletion disrupts global alternative splicing and causes dilated cardiomyopathy. Protein Cell. 2019;10:405–416. doi: 10.1007/s13238-018-0578-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo W, Schafer S, Greaser ML, Radke MH, Liss M, Govindarajan T, Maatz H, Schulz H, Li S, Parrish AM, Dauksaite V, et al. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat Med. 2012;18:766–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding J, Chen J, Wang Y, Kataoka M, Ma L, Zhou P, Hu X, Lin Z, Nie M, Deng ZL, Pu WT, Wang DZ. Trbp regulates heart function through microRNA-mediated Sox6 repression. Nat Genet. 2015;47:776–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei C, Qiu J, Zhou Y, Xue Y, Hu J, Ouyang K, Banerjee I, Zhang C, Chen B, Li H, et al. Repression of the central splicing regulator RBFox2 is functionally linked to pressure overload-induced heart failure. Cell Rep. 2015;10:1521–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Si Y, Ma N, Mei J. The RNA-binding protein PCBP2 inhibits Ang II-induced hypertrophy of cardiomyocytes though promoting GPR56 mRNA degeneration. Biochem Biophys Res Commun. 2015;464:679–684. doi: 10.1016/j.bbrc.2015.06.139 [DOI] [PubMed] [Google Scholar]
- 13.Li J, Xie D, Huang J, Lv F, Shi D, Liu Y, Lin L, Geng L, Wu Y, Liang D, Chen YH. Cold-inducible RNA-binding protein regulates cardiac repolarization by targeting transient outward potassium channels. Circ Res. 2015;116:1655–1659 [DOI] [PubMed] [Google Scholar]
- 14.Freyermuth F, Rau F, Kokunai Y, Linke T, Sellier C, Nakamori M, Kino Y, Arandel L, Jollet A, Thibault C, et al. Splicing misregulation of SCN5A contributes to cardiac-conduction delay and heart arrhythmia in myotonic dystrophy. Nat Commun. 2016;7:11067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dixon DM, Choi J, El-Ghazali A, Park SY, Roos KP, Jordan MC, Fishbein MC, Comai L, Reddy S. Loss of muscleblind-like 1 results in cardiac pathology and persistence of embryonic splice isoforms. Sci Rep. 2015;5:9042. doi: 10.1038/srep09042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao C, Ren S, Lee J, Qiu J, Chapski DJ, Rau CD, Zhou Y, Abdellatif M, Nakano A, Vondriska TM.RBFox1-mediated RNA splicing regulates cardiac hypertrophy and heart failure. J Clin Invest. 2016;126:195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu X, Yang D, Ding JH, Wang W, Chu PH, Dalton ND, Wang HY, Bermingham JR, Ye Z, Liu F, et al. ASF/SF2-regulated CaMKIIδ alternative splicing temporally reprograms excitation-contraction coupling in cardiac muscle. Cell. 2005;120:59–72 [DOI] [PubMed] [Google Scholar]
- 18.Ding JH, Xu X, Yang D, Chu PH, Dalton ND, Ye Z, Yeakley JM, Cheng H, Xiao RP, Ross J, et al. Dilated cardiomyopathy caused by tissue-specific ablation of SC35 in the heart. EMBO J. 2004;23:885–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng Y, Valley MT, Lazar J, Yang AL, Bronson RT, Firestein S, Coetzee WA, Manley JL. SRp38 regulates alternative splicing and is required for Ca(2+) handling in the embryonic heart. Dev Cell. 2009;16:528–538. doi: 10.1016/j.devcel.2009.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Änkö ML. Regulation of gene expression programmes by serine-arginine rich splicing factors. Semin Cell Dev Biol. 2014;32:11–21. doi: 10.1016/j.semcdb.2014.03.011 [DOI] [PubMed] [Google Scholar]
- 21.Müller-McNicoll M, Botti V, de Jesus Domingues AM, Brandl H, Schwich OD, Steiner MC, Curk T, Poser I, Zarnack K, Neugebauer KM. SR proteins are NXF1 adaptors that link alternative RNA processing to mRNA export. Genes Dev. 2016;30:553–566. doi: 10.1101/gad.276477.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeong S. SR proteins: binders, regulators, and connectors of RNA. Mol Cells. 2017;40:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grishina I, Lorenz M, Pabis M, Poser I, Rollins J, Sapra AK. SR protein family members display diverse activities in the formation of nascent and mature mRNPs In Vivo Mol Cell. 2009179–190 [DOI] [PubMed] [Google Scholar]
- 24.Änkö ML, Müller-McNicoll M, Brandl H, Curk T, Gorup C, Henry I, Ule J, Neugebauer KM. The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol. 2012;13:R17. doi: 10.1186/gb-2012-13-3-r17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boutz PL, Bhutkar A, Sharp PA. Detained introns are a novel, widespread class of post-transcriptionally spliced introns. Genes Dev. 2015;29:63–80. doi: 10.1101/gad.247361.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ankö ML, Morales L, Henry I, Beyer A, Neugebauer KM. Global analysis reveals SRp20- and SRp75-specific mRNPs in cycling and neural cells. Nat Struct Mol Biol. 2010;17:962–970. doi: 10.1038/nsmb.1862 [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, Wang J, Gao L, Stamm S, Andreadis A. An SRp75/hnRNPG complex interacting with hnRNPE2 regulates the 5’ splice site of tau exon 10, whose misregulation causes frontotemporal dementia. Gene. 2011;485:130–138. doi: 10.1016/j.gene.2011.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lucena-Araujo AR, Santana-Lemos BA, Thome CH, Ferreira GA, Ruggero D, Faça VM, Rego EM. Early hematopoietic progenitors of Dkc1 hypomorphic mutant mice display decreased proliferation rate and an impaired control of serine/arginine-rich splicing factor 4 (Srsf4) translation. Blood. 2014124–293725359993 [Google Scholar]
- 29.Liu J, Huang B, Xiao Y, Xiong HM, Li J, Feng DQ, Chen XM, Zhang HB, Wang XZ. Aberrant expression of splicing factors in newly diagnosed acute myeloid leukemia. Onkologie. 2012;35:335–340. doi: 10.1159/000338941 [DOI] [PubMed] [Google Scholar]
- 30.Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Irimia M, Weatheritt RJ, Ellis JD, Parikshak NN, Gonatopoulos-pournatzis T, Tapial J, Raj B, Hanlon DO, Barrios-rodiles M, Babor M, et al. A highly conserved program of neuronal microexons is misregulated in autistic brains Cell. 20141511–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.López-Olañeta MM, Villalba M, Gómez-Salinero JM, Jiménez-Borreguero LJ, Breckenridge R, Ortiz-Sánchez P, García-Pavía P, Ibáñez B, Lara-Pezzi E. Induction of the calcineurin variant CnAβ1 after myocardial infarction reduces post-infarction ventricular remodelling by promoting infarct vascularization. Cardiovasc Res. 2014;102:396–406. doi: 10.1093/cvr/cvu068 [DOI] [PubMed] [Google Scholar]
- 35.Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.König J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17:909–915. doi: 10.1038/nsmb.1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Z, Kayikci M, Briese M, Zarnack K, Luscombe NM, Rot G, Ule J. iCLIP predicts the dual splicing effects of TIA-RNA interactions. PLOS Genet. 2010;8:e1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H. Tabix: fast retrieval of sequence features from generic TAB-delimited files. Bioinformatics. 2011;27:718–719. doi: 10.1093/bioinformatics/btq671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kang YJ. Cardiac hypertrophy: a risk factor for QT-prolongation and cardiac sudden death. Toxicol Pathol. 2006;34:58–66. doi: 10.1080/01926230500419421 [DOI] [PubMed] [Google Scholar]
- 41.Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. iCLIP - transcriptome-wide mapping of protein-RNA interactions with individual nucleotide resolution. J Vis Exp. 20112–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Colombo M, Karousis ED, Bourquin J, Bruggmann R, Mühlemann O. Transcriptome-wide identification of NMD-targeted human mRNAs reveals extensive redundancy between SMG6- and SMG7-mediated degradation pathways. RNA. 2017;23:189–201. doi: 10.1261/rna.059055.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ren R, Oakley RH, Cruz-topete D, Cidlowski JA. Dual role for glucocorticoids in cardiomyocyte hypertrophy and apoptosis. Endocrinology. 2012;153:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.So AY, Chaivorapol C, Bolton EC, Li H, Yamamoto KR. Determinants of cell- and gene-specific transcriptional regulation by the glucocorticoid receptor. PLoS Genet. 2007;3:e94. doi: 10.1371/journal.pgen.0030094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tani H, Torimura M, Akimitsu N. The RNA degradation pathway regulates the function of GAS5 a non-coding RNA in mammalian cells. PLoS One. 2013;8:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karousis ED, Mühlemann O. Nonsense-mediated mRNA decay : novel mechanistic insights and biological impact. WIREs RNA. 2016;7:661–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu X, Li Z. Long non-coding RNA growth arrest-specific transcript 5 in tumor biology. Oncol Lett. 2015;10:1953–1958. doi: 10.3892/ol.2015.3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tani H, Mizutani R, Salam KA, Tano K, Ijiri K, Wakamatsu A, Isogai T, Suzuki Y, Akimitsu N. Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Res. 2012;22:947–956. doi: 10.1101/gr.130559.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28:195–208. doi: 10.1038/onc.2008.373 [DOI] [PubMed] [Google Scholar]
- 50.Shi Y, Parag S, Patel R, Lui A, Murr M, Cai J, Shi Y, Parag S, Patel R, Lui A, et al. Stabilization of lncRNA GAS5 by a small molecule and its implications in diabetic adipocytes article stabilization of lncRNA GAS5 by a small molecule and its implications in diabetic adipocytes. Cell Chem Biol. 2019;26:319–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Vries WB, van der Leij FR, Bakker JM, Kamphuis PJ, van Oosterhout MF, Schipper ME, Smid GB, Bartelds B, van Bel F. Alterations in adult rat heart after neonatal dexamethasone therapy. Pediatr Res. 2002;52:900–906. doi: 10.1203/00006450-200212000-00015 [DOI] [PubMed] [Google Scholar]
- 52.Lister K, Autelitano DJ, Jenkins A, Hannan RD, Sheppard KE. Cross talk between corticosteroids and alpha-adrenergic signalling augments cardiomyocyte hypertrophy: a possible role for SGK1. Cardiovasc Res. 2006;70:555–565. doi: 10.1016/j.cardiores.2006.02.010 [DOI] [PubMed] [Google Scholar]
- 53.Muiesan ML, Lupia M, Salvetti M, Grigoletto C, Sonino N, Boscaro M, Rosei EA, Mantero F. Left ventricular structural and functional characteristics in Cushing’s syndrome. J Am Coll Cardiol. 2003;41:2275–2279 [DOI] [PubMed] [Google Scholar]
- 54.Veselka J, Anavekar NS, Charron P. Hypertrophic obstructive cardiomyopathy. Lancet. 2017;389:1253–1267. doi: 10.1016/S0140-6736(16)31321-6 [DOI] [PubMed] [Google Scholar]
- 55.Avenatti E, Rebellato A, Iannaccone A, Battocchio M. Left ventricular geometry and 24-h blood pressure profile in Cushing’s syndrome. Endocrine. 2017;55:547–554 [DOI] [PubMed] [Google Scholar]
- 56.Tsagarakis DAVS. Cardiac hypertrophy in Cushing’s syndrome : if not hypertension then what? Endocrine. 2017453–455 [DOI] [PubMed] [Google Scholar]
- 57.Gómez-Salinero JM, López-Olañeta MM, Ortiz-Sánchez P, et al. The calcineurin variant CnAβ1 controls mouse embryonic stem cell differentiation by directing mTORC2 membrane localization and activation. Cell Chem Biol. 2016;23:1372–1382 [DOI] [PubMed] [Google Scholar]
- 58.Chen EY, Tan CM, Kou Y, et al. Enrichr : interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kuleshov MV, Jones MR, Rouillard AD, et al. Enrichr : a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:90–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seabold S, Perktold J. Statsmodels : Econometric and Statistical Modeling with Python. SCIPY. 2010;57:61 [Google Scholar]
- 61.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu A, Ibrahim JG, Love MI. Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics. 2019;35:2084–2092. doi: 10.1093/bioinformatics/bty895 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.