Supplemental Digital Content is available in the text.
Keywords: apoptosis, blood platelets, COVID-19, extracellular vesicles, necroptosis, SARS-CoV-2, thrombosis
Rationale:
Coronavirus disease 2019 (COVID-19) is characterized by increased incidence of microthrombosis with hyperactive platelets sporadically containing viral RNA. It is unclear if SARS-CoV-2 (severe acute respiratory syndrome, corona virus-2) directly alters platelet activation or if these changes are a reaction to infection-mediated global inflammatory alterations. Importantly, the direct effect of SARS-CoV-2 on platelets has yet to be studied.
Objective:
To characterize the direct SARS-CoV-2–platelet interactions using in vitro studies with purified infectious virions and samples from infected patients.
Methods and Results:
Platelet RNA analyzed by ARTIC v3 sequencing for SARS-CoV-2 showed presence of fragmented viral genome in all patients with COVID-19. Immunofluorescent imaging of platelets from patients with COVID-19 confirmed presence of SARS-CoV-2 proteins, whereas there was no detection of viral RNA by real-time quantitative polymerase chain reaction. Transmission electron microscopy of platelets incubated with purified SARS-CoV-2 virions demonstrated rapid internalization and digestion leading to distinct morphological changes and resulted in a release of extracellular vesicles. Interactions between SARS-CoV-2 and platelets occurred with or without ACE (angiotensin-converting enzyme) 2 presence as measured by immunofluorescence. Transmission electron microscopy showed that SARS-CoV-2 virions became internalized when they were attached to microparticles, bypassing the need for ACE2. Enrichment analysis of platelet transcriptome from patients with acute COVID-19, compared with those with clinical thrombosis, suggested upregulation of pathways related to virally mediated cell death, specifically necroptosis and apoptosis. Platelets incubated with infectious virus appeared to undergo cell death in 30 minutes postincubation as assessed by transmission electron microscopy and platelets from patients with COVID-19 showed evidence of increased markers of apoptosis and necroptosis by Western blot. Immunofluorescence confirmed colocalization of SARS-CoV-2 with phospho-MLKL (mixed lineage kinase domain-like pseudokinase) and caspase 3 on nonpermeabilized platelets in vitro and in COVID-19 platelets.
Conclusions:
Platelets internalize SARS-CoV-2 virions, directly or attached to microparticles, and viral internalization leads to rapid digestion, programmed cell death, and extracellular vesicle release. During COVID-19, platelets mediate a rapid response to SARS-CoV-2 and this response can contribute to dysregulated immunity and thrombosis.
In This Issue, see p 599
Editorial, see p 647
The coronavirus disease 2019 (COVID-19) pandemic has caused devastation worldwide, and currently, infection with SARS-CoV-2 (severe acute respiratory syndrome, corona virus-2) is the leading cause of morbidity and mortality in the United Statess1 and globally (https://covid19.who.int/). Understanding the wide range of disease manifestations due to COVID-19 is crucial particularly because patients with heart failure, type 2 diabetes, and obesity have increased morbidity and mortality.2,3 COVID-19 is characterized by a dysregulated cytokine profile, dysregulated lymphocyte levels, thrombocytopenia or thrombocytosis, disseminated intravascular coagulation, and severe microthrombosis in the lungs, manifesting as acute thrombotic events such as stroke, venous thromboembolism, and, less commonly, myocardial infarction (MI).4–6 Studies have also suggested that disseminated intravascular coagulation is predictive of death in patients with COVID-19.7,8
Platelets are the second most abundant circulating cell and the major blood component responsible for thrombosis. Platelets are anucleated cell fragments that are produced by their precursor cells, the megakaryocytes.9 In addition to their classical role in hemostasis and thrombosis, platelets have profound immune functions including activation of the innate immune system and involvement in the adaptive immune response by releasing serotonin thereby activating T cells.5,10 Platelets from patients with COVID-19 have been reported to be hyperactive with enhanced potential for aggregation11,12 but reduced procoagulant response.13 In addition, platelets from patients with COVID-19 contain an increased level of inflammatory cytokines related to activation of the immune system.12 The antiviral proteins responsible for inhibition of viral entry into the cytoplasm of the host cell such as IFITM3 (interferon-induced transmembrane protein 3) are also upregulated in platelets from patients with COVID-19.11 SARS-CoV-2 (severe acute respiratory syndrome, corona virus-2) RNA has been reported inconsistently in patients’ platelets11,12 and if/how internalization occurs and whether it directly alters platelet function are unknown.
Similar to influenza virus, SARS-CoV-2 is an RNA virus but, in distinction, carries its genetic information in a positive-strand molecule and does not require a nucleus for viral reproduction.14 SARS-CoV-2 gains access to a cell by interacting with ACE (angiotensin-converting enzyme) 2, such as in alveolar type II epithelial cells in the lower respiratory tract.15 Transmembrane proteases such as TMPRSS2 (transmembrane serine protease 2) are not required for viral entry although their presence allows the virions to gain rapid and direct entry into the cytoplasm.15,16 Other receptors for SARS-CoV-2 may exist, although convincing binding and internalization studies are lacking. Proteoglycans such as heparan sulfate can bind SARS-CoV-2, facilitating and enhancing interaction with ACE2.17 It has also been proposed that sialic acid binds the spike protein of SARS-CoV-2, although how this binding affects viral internalization is unclear.18 Expression of ACE2/TMPRSS2 on platelets has been controversial,5,11,12 and it has been suggested that, in addition to ACE2, cells may interact with SARS-CoV-2 through sialic acid19 and heparan sulfate.20
Platelets are the primary blood cell responsible for thrombosis. Platelets from patients with COVID-19 are hyperactivated with a higher procoagulant potential and elevated cytokine content.11,12 Elevated cytokine content in platelets is associated with increased plasma cytokine levels12 suggesting platelets contribute to the enhanced inflammation seen in COVID-19. The lack of uniform detection of SARS-CoV-2 RNA in platelets has made it difficult to assess if the overall hyperreactive platelet profile is predominantly due to an environmental response or a direct viral effect. In this study, we sought to determine whether direct uptake of SARS-CoV-2 by platelets occurs and if this uptake alters platelet function. We hypothesized that SARS-CoV-2 is internalized by platelets leading to changes in platelets that contribute to a dysregulated immune response and subsequent prothrombotic outcome.
Methods
Data Availability
Data Supplement contains detailed methods of all experiments included in this article. The Data Supplement also contains data, analytic methods, reagents supporting the findings of this study and are available from the corresponding author upon reasonable request.
Results
Fragmented SARS-CoV-2 Genome Is Detected in Platelets From All Patients With COVID-19
Various reports have suggested some detection of SARS-CoV-2 RNA from blood and in platelets.11,12,21 Depending on the primers and SARS-CoV-2 transcript targeted, different levels and inconsistent degrees of expression have been reported but no actual viral presence has been demonstrated in platelets. Similarly, using the 2 primers recommended by Centers for Disease Control and Prevention against the nucleocapsid RNA, we were unable to detect the viral RNA (vRNA) in platelets. To go beyond the limitations of RNA detection by real-time quantitative polymerase chain reaction (RT-qPCR), we resorted to SARS-CoV-2 targeted sequencing using a tiled amplicon approach that spans the entire genome of the virus. To our surprise, the platelets of all 17 patients were positive for SARS-CoV-2 covering 54% to 87% of the entire genome (Figure 1A). The viral genome was fragmented (Figure 1A) similarly to vRNA for structural proteins such as spike, nucleocapsid, and envelope (Figure IA through IC in the Data Supplement). This fragmentation, suggestive of digestion, explains inconsistencies in levels of vRNA detection in previous reports.11,12 To address possible contamination by leukocytes in our platelet preparations we compared lncRNA (long non-coding RNA) from patients’ platelet versus leukocyte (total) samples using individuals screened for SARS-CoV-2 by targeted tiled amplicon approach. lncRNA have distinct signatures in different cells,22 and specific lncRNAs were highly abundant in leukocytes but completely absent in the platelet preparations (See Table III in the Data Supplement). These lncRNA patterns demonstrate differential expression suggesting that the detection of the SARS-CoV-2 genome is specific to platelets and it is not a result of leukocyte contamination.
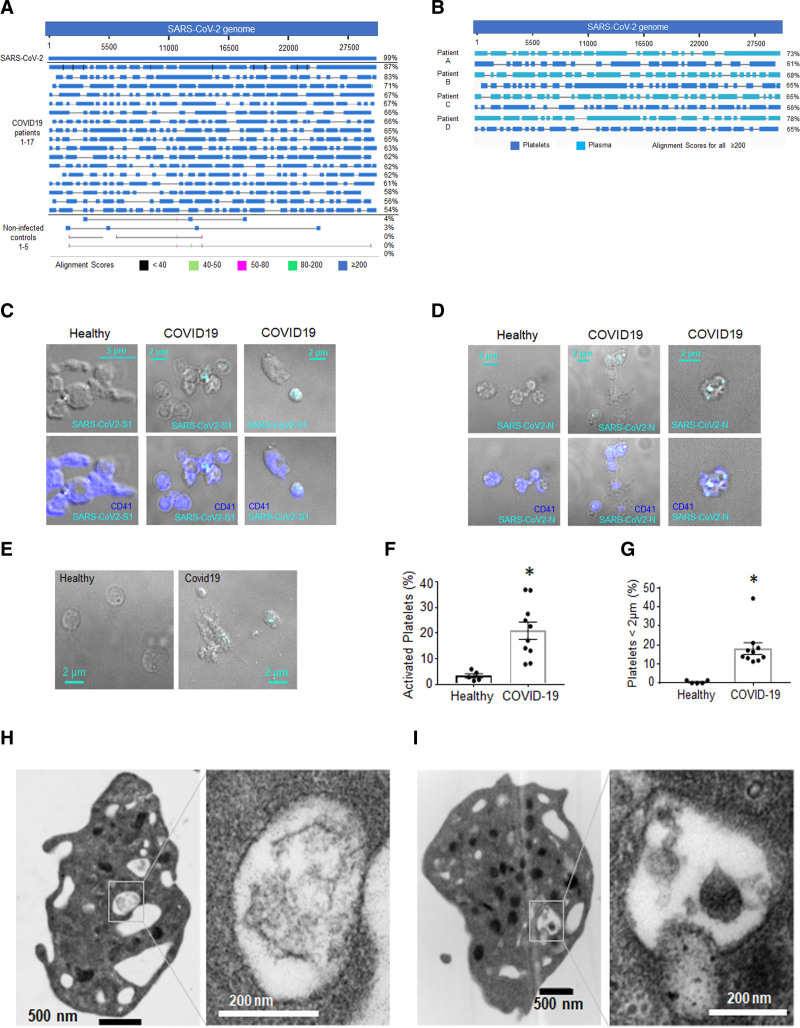
Figure 1.
Detection of SARS-CoV-2 (severe acute respiratory syndrome, corona virus-2) in actively infected patients’ platelets.A and B, RNA from platelets from patients with coronavirus disease 2019 (COVID-19) and noninfected controls was sequenced using Artic V3 sequencing specifically enriching for the SARS-CoV-2 genome. RNA isolated from SARS-CoV-2 (USA-WA1/2020 Strain, BEI) was used as a positive control. Sequencing results were analyzed using Nucleotide Blast National Center for Biotechnology Information server against SARS-CoV-2 NC_045512_Wuhan-Hu-1 genome. A, Alignment of viral RNA (vRNA) in platelets. B, Alignment of vRNA in platelets (included in A) and plasma from the same patients. Whole blood from patients with COVID-19 was fixed and stained with antibodies against (C) SARS-CoV-2 Spike 1 protein and (D) SARS-CoV-2 Nucleocapsid protein with detection by immunofluorescence using spinning disk confocal microscopy and ×100 lens. E, Differential interference contrast (DIC) of healthy and COVID-19 platelets. Quantitation of (F) platelets that show severe activation and (G) platelets that are between 1 and 2 µm using confocal DIC images taken with ×100 (Covid-19: 2 females [F] and 3 males [M]). The healthy donors used here were n=5, 2M (56 y White participant; 44 y Asian); 3F (60 y White participant; 45 y White participant; 44 y White participant). Significance was assessed using a nonparametric test (Mann-Whitney), P=0.0079 for both graphs; data are presented as mean±SEM. H and I, Transmission electron microscopy of washed platelets from 2 different donors (representative of 6) showing structures that could be digested virus.
Sequencing of citrated plasma from the same patients showed viral genome presence in 4 of the 17 patients. Sequencing comparison between the plasma and platelet viral genome of the same patients showed differences in fragmentation (Figure 1B). The vRNA in plasma was also detected by RT-qPCR in 3 of the 4 patients (Table IV in the Data Supplement). Two of the patients who died as a result of COVID-19 were part of this subpopulation. Further studies are necessary to establish whether there is a connection between plasma viral presence and severity of disease.
Using immunofluorescent staining of fixed whole blood from 10 of the 17 actively infected hospitalized patients with COVID-19, we also evaluated the presence of spike and nucleocapsid SARS-CoV-2 proteins in platelets. Whole blood showed the occasional presence of these 2 SARS-CoV-2 proteins associated with CD41-stained platelets (Figure 1C and 1D). Differential interference contrast of platelets from patients with COVID-19 showed the presence of smaller platelets (smaller than 2 µm in diameter) and some consistent with severe activation (Figure 1C through 1G). It is important to note that the blood from patients with COVID-19 also contained platelets bigger than 5 µm (Figure III in the Data Supplement). Transmission electron microscopy (TEM) of platelets isolated from patients with COVID-19 infrequently demonstrated structures consistent with intact virions (Figure 1H and 1I, Figure IF through II in the Data Supplement). Overall, our observations using sequencing of isolated platelets and plasma as well as immunofluorescence of whole blood from patients with COVID-19 suggest that platelets actively take up the virus in infected patients and contribute to the viral fragmentation, without allowing for active replication.
Platelets Internalize SARS-CoV-2 In Vitro
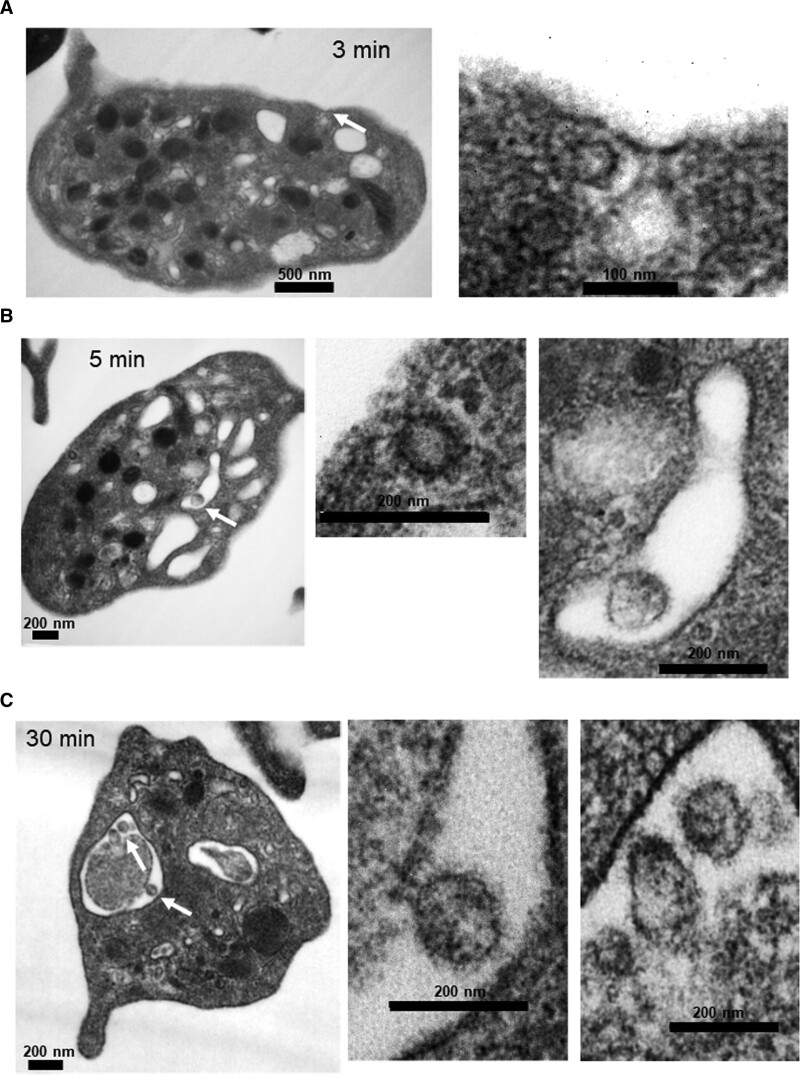
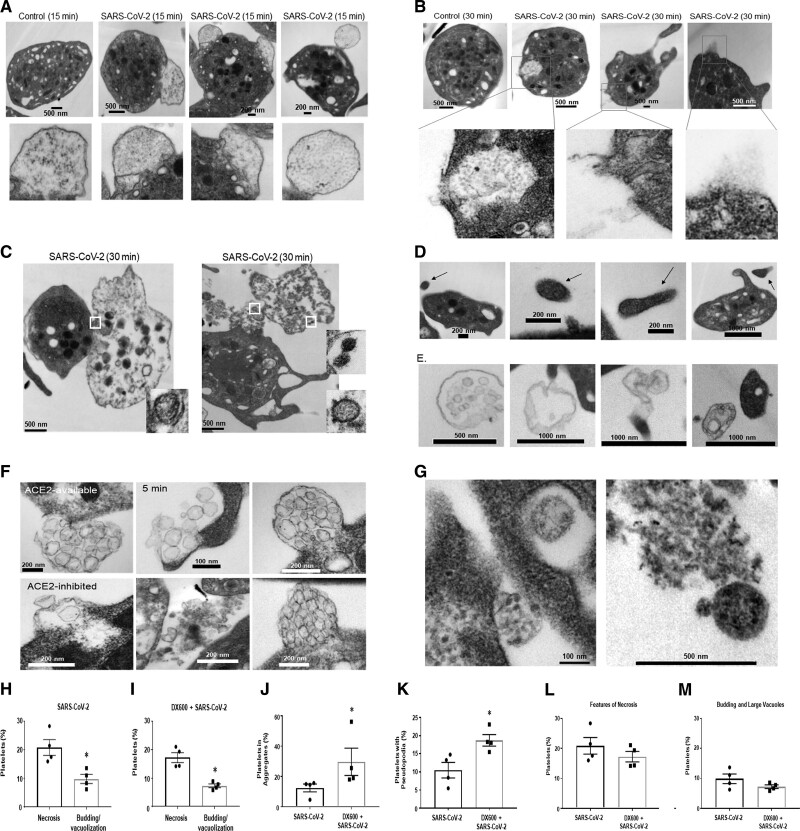
To assess the direct interaction of platelets with SARS-CoV-2, we incubated platelets from healthy donors with infectious SARS-CoV-2 virions. Importantly, the virions used in this study were purified and concentrated (as described in methods) to eliminate procoagulation material found in the media of infected cells used to propagate the virus. As we have previously done for other viral studies,23 platelets were incubated with SARS-CoV-2 (10:1), for 3, 5, 15, and 30 minutes. We compared all time points to platelets incubated with the media in which the virus was resuspended. As compared to control platelets, endocytic vesicles were seen at 3 minutes that appeared to contain the virus lacking spikes and envelope (Figure 2A). At 5 minutes, the virus was also found in large phagocytic vacuoles that we have previously described with influenza virions (Figure 2B). At 15 minutes, the virus appeared to undergo digestion within the large vacuoles (Figure IIA through IIC in the Data Supplement). At 30 minutes, we observed the virus in large vacuoles, but now appearing to be attached to microparticles (Figure 2C). Internalization of platelet-derived microparticles by platelets is not well understood, but it can involve at least partially, receptors such as TLR4 (toll-like receptor 4).24 Of note, microparticles as referred to here are particles between 200 nm and 1 µm that have similar content as the platelet cytoplasm. In healthy individuals, platelets are responsible for ≈80% of the microparticle content in plasma and are vesicles that function as cross-cellular communicators.25 These observations suggest that SARS-CoV-2 can be internalized by platelets by at least 3 different pathways: in endosomes, in phagocytic vacuoles, and by viral attachment to microparticles with subsequent internalization.
Figure 2.
Platelets internalize infectious SARS-CoV-2 (severe acute respiratory syndrome, corona virus-2) in vitro. Infectious SARS-CoV-2 appears to be internalized by platelets via three distinct routes. Washed platelets from healthy human donors were incubated with SARS-CoV-2 virions (10:1) over time. Transmission electron microscopy shows that the virus can locate in (A) endosomes (only part of the virus), (B) in phagosome-like structures by itself (last image). Please note that the image in the middle is the size of a clathrin-coated vesicle and it is not a viral particle, and (C) in phagosome-like structures attached to platelet-derived microparticles.
ACE2 and SARS-CoV-2 Internalization by Platelets
The putative receptor for internalization of SARS-CoV-2 in cells has been determined to be ACE2. Concomitant expression of ACE2 and the protease TMPRSS2 determines if the vRNA or the entire virus enters the cell via the endosomal pathway.16 Previous reports have indicated that platelets do not contain the RNA for ACE2.11,12 Similarly, our RNA-sequencing analysis did not detect ACE2 or TMPRSS2 RNA. RT-qPCR analysis using our preamplified method (and treated with DNAse) detected low ACE2 expression in 10 of 37 patients (COVID-19 and MI) using 3 different Taqman primers. However, all 3 primers for ACE2 in platelets were not detected in any one donor (Table V in the Data Supplement). These observations suggest that ACE2 RNA may be below the level of detection or that there are nucleotide mutations that may interfere with alignment of the primers utilized. To determine if ACE2 protein is present in platelets, we stained whole blood from healthy donors, MI, and patients with COVID-19 with ACE2, CD41, and DAPI. By this method, we were able to observe ACE2 on platelets (Figure 3A), primarily, those consisting of small aggregates or larger platelets above 5 µm in size (Figure III in the Data Supplement). Similarly, staining infected patients’ blood with TMPRSS2 showed expression by immunofluorescence (Figure 3B). Whole blood from most healthy donors expressed variable low levels of ACE2 (0.02%± 0.1 in healthy people, calculated per 100 platelets, n=4). To quantify expression of ACE2 in platelets of healthy individuals, we performed Western blot (WB) analysis of isolated platelets. By this method, we confirmed variable expression of ACE2 and TMPRSS2 (Figure 3C through 3F) in platelets from healthy donors.
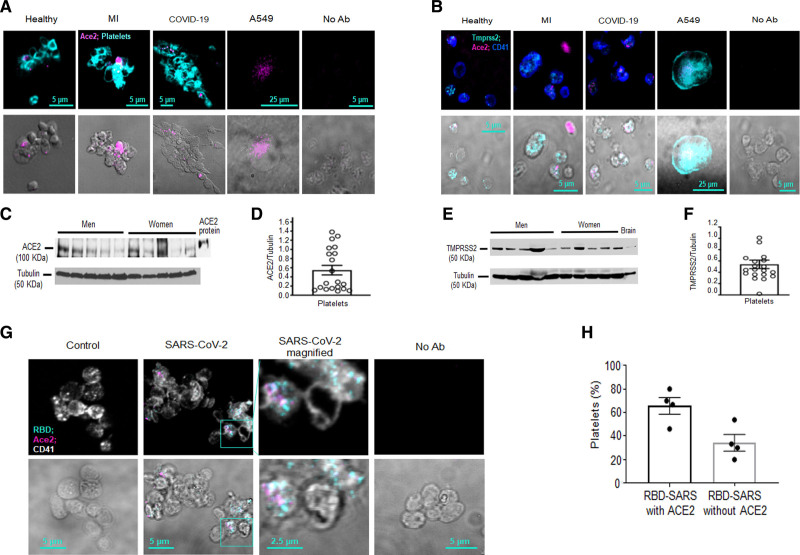
Figure 3.
The binding domain of SARS-CoV-2 (severe acute respiratory syndrome, corona virus-2) does not always associate with ACE (angiotensin-converting enzyme) 2 on platelets.A and B, Platelets from whole blood were stained with ACE2 and CD41 postfixation of whole blood. Human alveolar epithelial cells (A549), transduced to stably overexpress ACE2 and TMPRSS2 (transmembrane serine protease 2), were used as a positive control. No antibody (Ab) control (No Ab) was used to account for autofluorescence of paraformaldehyde-fixed cells. A, Representative images of ACE2 expression in platelets from different donors. Healthy (n=4, 2 females [F] and 2 males [M]), myocardial infarction (MI; n=3, 2M, 1F), and coronavirus disease 2019 (COVID-19; n=6, 4M, 2F) donors were screened. B, ACE2 and TMPRSS2 colocalization in platelets from healthy, MI, and COVID-19 donors (representative images of n=4 healthy, n=3 with MI and n=4 with COVID-19). C–F, ACE2 and TMPRSS2 expression assessed by Western blot analysis of washed platelets from healthy human donors (blots are representative of n=20 healthy donors, 10M and 10F, age 40.1±2.8 y, all White participants); data are presented as mean±SEM. C, Representative Western blots of ACE2 and (D) densitometry of expression. E, TMPRSS2 Western blot and (F) Densitometry of expression. G, SARS-CoV-2 binds ACE2 on the platelet surface after viral incubation. Washed platelets from a healthy human donor were incubated for 5 min with SARS-CoV-2. Platelets were fixed and stained with Abs against ACE2 and the ACE2–receptor-binding domain (RBD) of the SARS-CoV-2 spike protein. H, Quantitation of platelets that have both SARS-CoV-2 and ACE2 vs. platelets that have only SARS-CoV-2 (n=4; 2M and 2F). The healthy donors used here were n=4, 2M (56 y White participant; 44 y Asian participant); 2F (59 y White participant; 46 y White participant). Significance was assessed using a nonparametric t test (Mann-Whitney), P values for all with * is 0.0289; data are presented as mean±SEM. Images and calculations are representative of n=4 different donors (2M and 2F).
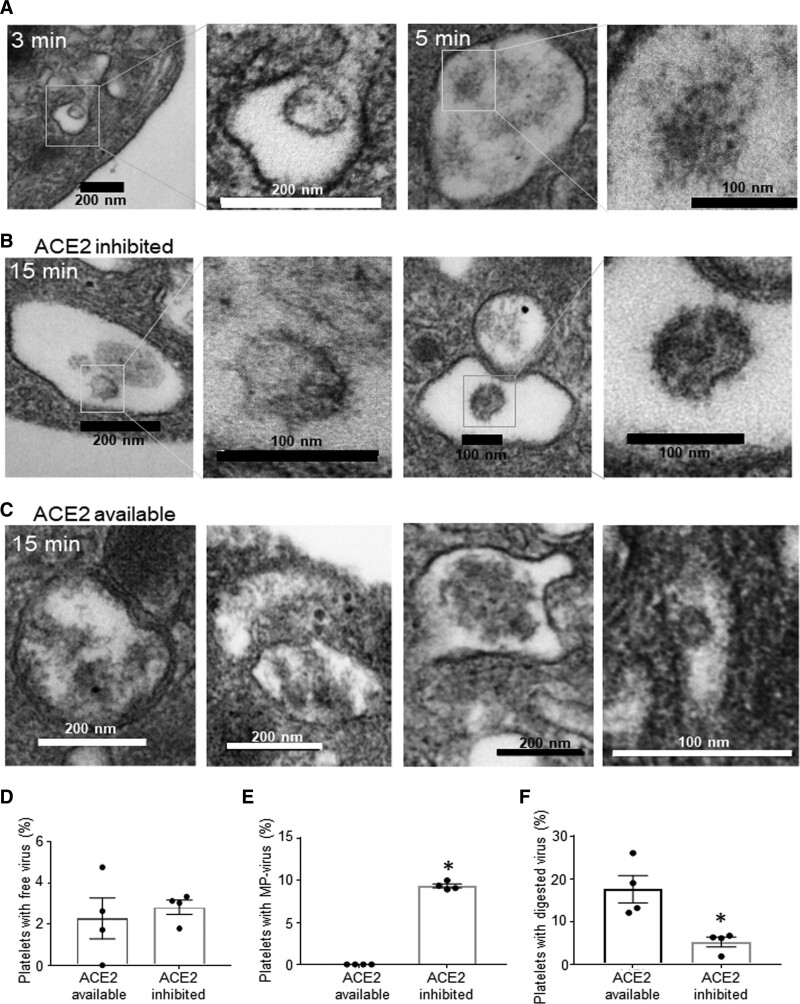
To determine if ACE2 interacts with SARS-CoV-2, we incubated platelets with purified infectious SARS-CoV-2 and stained for the receptor-binding domain in SARS-CoV-2 and ACE2. Immunofluorescence analysis showed that the receptor-binding domain can colocalize with ACE2 on platelets (Figure 3G), although not all platelets that were positive for the Spike–receptor-binding domain had ACE2 on them (Figure 3G through 3H). To further assess if ACE2 activity can affect internalization, we incubated platelets with infectious virus for 15 minutes in the presence and absence of the ACE2 inhibitor DX600. DX600 is a potent specific inhibitor of ACE2 function at 200 nM26; it is stable and nonhydrolysable by ACE2.27 Under these conditions, TEM demonstrated viral internalization; however, virions were primarily located in large vacuoles with microparticle attachment (Figure 4A and 4B; similarly to 30 minutes without ACE2 inhibition, Figure 2C). Lack of DX600 (ACE2 available) led to presence of mostly digested virus at 15 min postincubation (Figure 4A through 4C). Quantification of platelets with free virus versus platelets with virus attached to microparticles is provided in Figure 4D through 4F. These data suggest that platelets can internalize SARS-CoV-2 independent of ACE2 function by attaching to platelet-derived microparticles.
Figure 4.
Platelets internalize SARS-CoV-2 (severe acute respiratory syndrome, corona virus-2) in ACE (angiotensin-converting enzyme) 2–dependent and ACE2-independent ways. Washed platelets from healthy human donors were incubated with SARS-CoV-2, and some were pretreated with DX600 (200 nmol/L) for 10 min. A, Platelets at 3–5 min in the absence of ACE2-inhibitor; platelets in (B) presence of and (C) absence of ACE2 inhibitor (DX600, 200 nmol/L) for 15 min at 37 °C and constant rotation at 1000 rpm (revolutions per minute). Samples were fixed with Karnovsky’s fixative and resolved by transmission electron microscopy. Quantitation of platelets from (B) and (C) showing (D) free virus, (E) virus attached to microparticles (MP), and (F) digested virus. Significance was assessed using a nonparametric t test (Mann-Whitney), P values for all with * is 0.0289; data are presented as mean±SEM. Images and calculations are representative of n=4 different donors (2 males [M] and 2 females [F]). The healthy donors used here were n=4, 2M (56 y White participant; 44 y Asian participant); 2F (59 y White participant; 46 y White participant).
SARS-CoV-2 Mediates Morphological Changes in Platelets and Release of EVs
To assess the effect of SARS-CoV-2 on platelets, we analyzed morphological changes from TEM images of healthy platelets incubated with the virus. When compared with control platelets, we observed platelet budding, not previously seen with influenza23 (Figure 5A). Select platelets had observable cytoplasmic disruptions from which cellular content appears to have been released (Figure 5B). Over time, select platelets became necrotic with visible lack of contents (Figure 5C). Of note, at no point did platelets form 3-dimensional aggregates typical of treatment with platelet prothrombotic agonists.
Figure 5.
SARS-CoV-2 (severe acute respiratory syndrome, corona virus-2) mediates distinct morphological changes in platelets and various forms of extracellular vesicle release. Washed platelets from healthy human donors were incubated with SARS-CoV-2 virions (10:1) for various time points. Transmission electron microscopy shows that SARS-CoV-2 leads to (A) budding (B) content release from broken membrane or channels; presence of degranulates, and (C) necrotic platelets; (D) platelet microparticles; (E) migrasomes; (F) exosome release; (G) intracellular platelet content release of nongranule origin. H–M, Quantification of Transmission electron microscopy images from n=4 healthy donors at 15 min in the presence and absence of ACE (angiotensin-converting enzyme) 2 inhibitor; necrotic features were defined as empty platelets with broken membranes. The healthy donors used here were n=4, 2 males (M; 56 y White participant; 44 y Asian participant); 2 females (F; 60 y White participant; 46 y White participant). Significance was assessed using a nonparametric t test (Mann-Whitney), P values for all with * is 0.0289; data are presented as mean±SEM. SARS-CoV-2 indicates severe acute respiratory syndrome, corona virus 2.
Over time, distinct platelet content was observed externally including microparticles of differing size (Figure 5D) and content (Figure 5D and 5E). Interestingly, distinctive microparticles were observed (Figure 5E) that have been described as migrasomes,28 which are particles that originate from migrating cells and function as local chemoattractants.29,30 Exosomes (Figure 5F) released from platelets were also observed after incubation with SARS-CoV-2, as well as release of other platelet contents (Figure 5G, Figures IID and IV in the Data Supplement), some occurring after platelet membrane loss (Figure 5G, right). Elements of microparticle heterogeneity were also observed in the platelets from patients with COVID-19 (Figure IV in the Data Supplement). Quantification of the TEM images in the presence and absence of ACE2 inhibitor showed that features of SARS-CoV-2 necrosis (emptying of platelet contents by breaking, releasing, and leaking) are elevated when compared with budding and formation of large vacuoles (Figure 5H and 5I). However, ACE2 inhibition seemed to have a similar effect on platelets (Figure 5L and 5M). ACE2-inhibition increased the formation of small platelet aggregates (up to 16) and pseudopodia formation (Figure 5J and 5K). Our observations indicate that platelets both generate microparticles and undergo profound morphological changes with content release.
Platelet Gene Expression From Patients With COVID-19 Compared to Clinical Thrombosis Suggests Programmed Cell Death
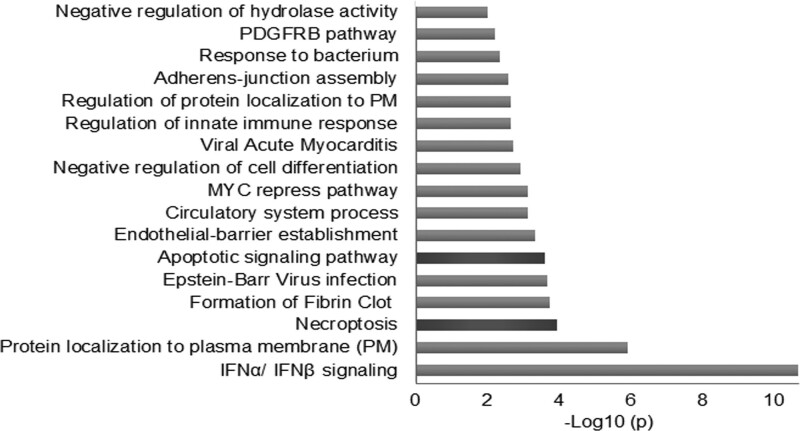
To gain insight into the effect of SARS-CoV-2 on platelets, we performed RNAseq (RNA sequencing) comparing infected and noninfected patients. Previous work has compared the transcriptome of platelets from patients with COVID-19 to healthy platelets11; however, this approach does not account for the basal prothrombotic state seen during infection. Therefore, we compared the transcriptome using platelets from hospitalized patients with acute COVID-19 (n=10) to patients presenting with MI (ST-segment–elevation MI (n=10) and non–ST-segment–elevation MI (n=10; Table I in the Data Supplement). Pathway enrichment analysis of the 40 upregulated and 40 downregulated transcripts in COVID-19 compared with acute MI showed that the most affected transcripts are related to type I interferon signaling (Figure 6, Figure VA in the Data Supplement). Of note, the 40 upregulated and 40 downregulated transcripts were chosen to account for the greatest differences in expression with the highest statistical significance, although guided by biological plausibility these cutoff points were arbitrary. RNAseq also identified changes in pathways related to protein localization to the plasma membrane (GO:0072659) and regulation of that localization (GO:1905475; Figure VB in the Data Supplement). Surprisingly, the pathways with the most significant changes were related to programmed cell death, specifically necroptosis and apoptosis as well as innate immunity. Transcript levels of markers for programmed cell death in platelets associated modestly with plasma levels of IFN (interferon) γ, P-selectin, vWF (von Willebrand factor), and a few other inflammatory cytokines and molecules (Table VI in the Data Supplement). Our approach also uncovered dysregulation in the pathway related to fibrin clot formation consistent with clinical outcomes of COVID-19. With the exception of type I interferon signaling, the pathways that were enriched by our approach were not observed using the same analysis of published data11 comparing COVID-19 platelets to healthy donor platelets (Figure VC in the Data Supplement), suggesting that by eliminating the pathways related to thrombosis, pathways directly related to the effect of the virus on platelets can be uncovered.
Figure 6.
Pathway enrichment analysis of coronavirus disease 2019 (COVID-19) platelet transcriptome suggests programmed cell death. Platelets from patients with COVID-19 (n=10) and patients with myocardial infarction (ST-segment–elevation myocardial infarction [n=10], non–ST-segment–elevation myocardial infarction [n=10]) without COVID-19 were isolated and sequenced. Pathway enrichment analyses, with the respective P values were generated in https://metascape.org/ using the fold changes from the RNAseq (RNA sequencing) data of the highest, statistically significant, 40 upregulated, and 40 downregulated genes. Fold changes of the sequencing results are included in Table VII in the Data Supplement. IFN indicates interferon. MYC indicates myelocytomatosis; PDGFRB, platelet-derived growth factor receptor beta; and PM, plasma membrane.
SARS-CoV-2 Leads to Programmed Cell Death in Platelets
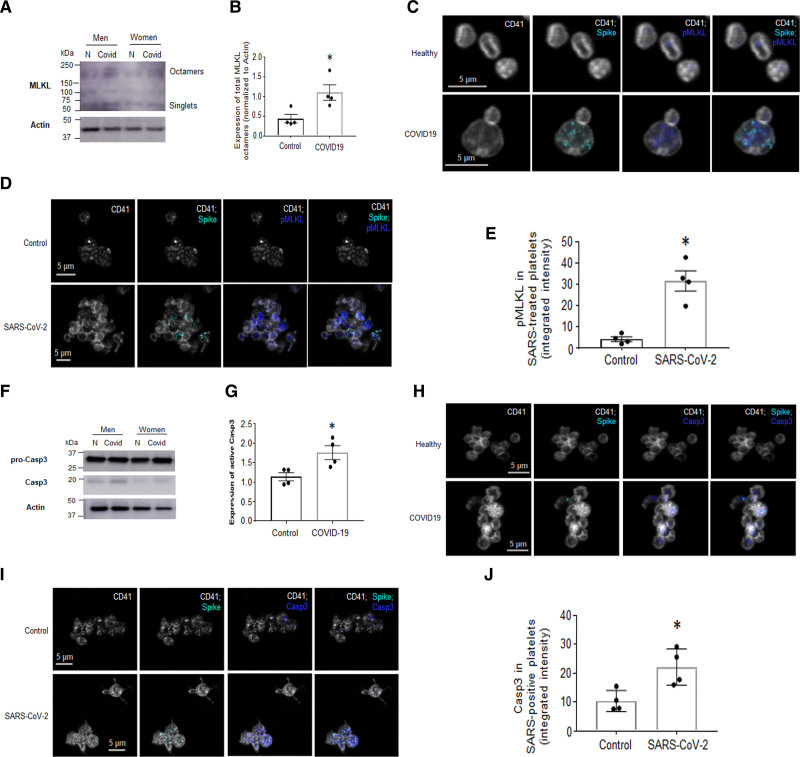
Viral infection of a nucleated cell results in various forms of programmed cell death depending on cell type and function. Our RNAseq results also suggest upregulation of transcripts related to programmed cell death. The specific types of cell death include apoptosis, necroptosis, and pyroptosis. To assess if programmed cell death occurs during COVID-19 and with SARS-CoV-2, we examined markers for cell death in platelets from patients with COVID-19. The observation of platelet content leakage, breaks, and necrosis suggested necroptosis (Figure 5B and 5C, Figure II in the Data Supplement). Thus, we first screened platelets for necroptosis markers from age-, sex-, and race-matched patients with COVID-19. The process of necroptosis ultimately leads to phosphorylation of the MLKL (mixed lineage kinase domain-like pseudokinase) and p-MLKL (phosphorylated MLKL) forms octamer channels that translocate to the surface membrane of dying cells causing release of cellular contents.31 WB analysis of platelets from patients with COVID-19 revealed increased platelet levels of MLKL-octamers in a select number of patients (Figure 7A and 7B), with the exception of one pair (data not shown). Similarly, immunofluorescence of blood of patients with COVID-19 showed the presence of p-MLKL in platelets that stained positive for SARS-CoV-2 spike protein (Figure 7C). To assess if SARS-CoV-2 contributes to p-MLKL-channel formation, platelets from healthy donors after in vitro incubation (30 minutes) with SARS-CoV-2 were stained. To eliminate intracellular staining, cells were not permeabilized. Immunofluorescence of platelets demonstrated p-MLKL on the platelet surface with concomitant detectable levels of SARS-CoV-2 (Figure 7D and 7E).
Figure 7.
Platelets from patients with coronavirus disease 2019 (COVID-19), and platelets from healthy donors treated with SARS-CoV-2 (severe acute respiratory syndrome, corona virus-2), have upregulated markers of programmed cell death. Washed platelets from patients with COVID-19 were subjected to Western blot (WB) analysis. Markers for necroptosis (A) MLKL (mixed lineage kinase domain-like pseudokinase)-octamer formation by WB (representative images of n=4, 3 males [M], 1 female [F]; the images shown here are from sex- and age-matched groups that include a male and a female pair). B, Quantification of A. C and D, Colocalization of SARS-CoV-2 with p-MLKL (phosphorylated MLKL) by immunofluorescence (IF). C, In COVID-19 blood and in D. Platelets postincubation with SARS-CoV-2 for 30 min. E, Quantitation of D using integrated intensity over platelet number (ImageJ). Marker for apoptosis. F, Active Casp3 (caspase 3) expression (representative images of n=4, 3M, 1F; the images shown here are from sex- and age-matched groups that include a male and a female pair) and its (G) quantitation and H and I. Colocalization of SARS-CoV-2 and Casp 3 in platelets in H. Whole blood from patients with COVID-19 and (I) platelets incubated with SARS-CoV-2 for 30 min by IF; (J) quantitation of I as in E. IF was performed on whole blood that was immediately RBC-lysed and fixed with 4% paraformaldehyde and then stained with the indicated markers (platelet marker used is CD41). IF images are representative of n=4 different patients with COVID-19 on n=4 different healthy donors. Significance was assessed using a nonparametric t test (Mann-Whitney), *P values for B and G are 0.0289; *P values for E and J are 0.0286; data are presented as mean±SEM.
Phosphatidyl serine has been previously described on the surface of platelets from patients with COVID-19,12 and our data demonstrated budding as a result of SARS-CoV-2 incubation and presence of smaller platelets in patients with COVID-19 (Figure 1E, Figure ID through IE in the Data Supplement), thus, we examined markers of apoptosis such as Casp3 (Caspase 3) activation. WB analysis revealed an increase in the levels of active Casp3 in sex-, age-, and race-matched patients (Figure 7F and 7G). Similarly, immunofluorescence of platelets stained with Casp3 antibody and SARS-CoV-2 were found on the same platelet (Figure 7H). Incubation of platelets with SARS-CoV-2 virions in vitro also resulted in the presence of Casp3 and SARS-CoV-2 proteins in the same platelets (Figure 7I and 7J). Of note, markers of pyroptosis such as active Caspase 1 or 4 and the N-fragment of GSDMD (gasdermin D) were not detected by WB (Figure VI in the Data Supplement). Our results indicate that platelets undergo necroptosis and apoptosis during COVID-19 and SARS-CoV-2 can directly contribute to these forms of cell death.
Discussion
Patients with COVID-19 have been consistently noted to have enhanced clinical thrombosis presumed to be due to increased coagulation, enhanced platelet function inflammatory cytokines, endothelial damage, and activated immunity. Previous studies of patients with COVID-19 report hyperactive platelets with increased interleukin content.11,12 In this study, we demonstrate the presence of a wide range of fragmented vRNA in platelets from patients with COVID-19, direct platelet uptake, and digestion of SARS-CoV-2 and describe the subsequent platelet response as a result of this internalization. Internalization of the virus does not require ACE2 and entry into platelets appears to occur by viral attachment to microparticles, some of which may be of platelet origin.
Our observations suggest that SARS-CoV-2 leads to programmed cell death of platelets in which markers and morphological changes of apoptosis and necroptosis are evident. Consistent with these morphological changes, pathway enrichment analysis using RNAseq of platelets isolated from COVID-19 as compared to MI patients reveals activation of necroptotic and apoptotic pathways in COVID-19. These data suggest that apoptosis was mediated by Casp3 activation, while necroptosis was mediated by MLKL phosphorylation and formation of octamer channels that are known to puncture the cell membrane. These findings also suggest that ACE2 activity may not be required for apoptosis or for necroptosis to occur, but inhibition of ACE2 activity is associated with enhanced small-aggregate and pseudopodia formation. SARS-CoV-2 also leads to release of extracellular vesicles from platelets such as microparticles, exosomes, and migrasomes. Our findings indicate that SARS-CoV-2 directly affects platelets leading to proinflammatory cell death, extracellular vesicle, and platelet content release.
Infectious SARS-CoV-2 in blood of patients with COVID-19 has not been reported, although blood21,32 and selective blood cells such as platelets11,12 may occasionally test positive for SARS-CoV-2 RNA. In this study, we report that platelets from all hospitalized patients with COVID-19 contained various amounts of the SARS-CoV-2 genome as assessed by SARS-CoV-2 Artic V3 Sequencing; however, RT-qPCR for the nucleocapsid vRNA was negative in platelets. Of note, lack of leukocyte contamination in our platelet preparation was assessed by targeted amplicon sequencing of lncRNA, showing distinct presence of highly abundant leukocyte-specific lncRNAs that were absent in platelets. Citrated plasma was largely devoid of vRNA by RT-qPCR (3 of 17 tested positive) or sequencing (4 of 17 tested positive). Interestingly, plasma and platelet vRNA fragmentation in the same person did not overlap. In both cases, however, the vRNA was fragmented, suggesting a lack of infectivity but not eliminating the possibility for vRNA-mediated inflammation in blood or organs. It is important to note that EDTA activates platelets,33 thus results reporting the presence of vRNA in EDTA-plasma should be viewed with caution as it may be due to platelet contamination. Lastly, by focusing on hospitalized patients, we are not able to assess how long after one first tests positive by qPCR that the SARS-CoV-2 vRNA can be found in platelets. Although more data is necessary to establish when platelets become devoid of SARS-CoV-2 vRNA, lack of the full viral genome suggests that convalescent plasma transfusions should not contain infectious virus.
Using immunofluorescence of whole blood from patients with COVID-19 shows that SARS-CoV-2 spike and nucleocapsid proteins can also be detected in platelets. This suggests that platelets’ active removal of vRNA from the circulation occurs throughout the infection. It is not clear, however, if this process occurs only in people significantly ill with COVID-19 or if it also occurs in asymptomatic individuals. Transmission electron microscopy of platelets incubated with infectious SARS-CoV-2 virus confirmed rapid internalization and digestion. Interestingly, platelets internalized the virus in 3 to 5 minutes, and after 15 minutes, the virus appeared digested. During the first 5 minutes, we found virions in the same phagocytic vesicles as we previously have observed with influenza.23 However, we only observed 1 to 2 virions per platelet in contrast to up to 10 virions with influenza. In addition, at 3 minutes, we were able to observe endocytic vesicles close to the platelet surface that contained virus without the envelope. Both of these types of internalization have been described for SARS-CoV-2 in nucleated cells and depend on ACE2 expression.14
Surprisingly, at 30 minutes of incubation (and when ACE2 activity was inhibited) we detected SARS-CoV-2 in phagocytic vesicles with the virus attached to platelet-generated microparticles. Internalization of platelet-derived microparticles by platelets has been described to be at least partially mediated by TLR4. Regardless of the mechanism of microparticle internalization, the fact that the virus can attach to microparticles and become internalized suggests that SARS-CoV-2 can bypass the requirement for ACE2-mediated mechanisms of internalization. There is also a possibility that platelet-microparticle formation increases the number and surface area of interaction with the viral particles. Finally, because ACE2 can be secreted into the circulation as an enzymatically active ectodomain also known as soluble ACE2 (sACE2),34 there is also a possibility that sACE2 and microparticles collaborate to transfer the virus inside of platelets without the need for ACE2 on the platelet surface. Regardless of the route of internalization, sequestering the virus in platelets may prevent it from reaching other cells in which it can produce progeny.
As discussed, SARS-CoV-2 is a single-stranded RNA virus that contains its genetic information in a positive RNA strand. The positive RNA strand, in turn, does not require nucleus entry and can be translated in the cytoplasm, bypassing additional control mechanisms. Platelets are cell fragments and do not have a nucleus, thus it is unclear if they can synthesize new RNA. Translation in platelets is also limited and it is unclear if all platelets uniformly have endoplasmic reticulum and can translate RNA. Additionally, we have previously shown that once platelets interact with RNA viruses, they form heterotypic aggregates with other leukocytes.10,35 These observations along with our SARS-CoV-2 sequencing results suggest that platelets can internalize the virus but the virus does not reproduce in platelets. As opposed to our in vitro studies, a limitation of our findings from patients was the inability to know the precise timing of infection onset. This, along with the small patient number, did not allow us to compare patients based on days postinfection, limiting our ability to quantify platelet-dependent viral clearance along with the potential contribution of megakaryocytes to infection. The diversity of patient medications at time of blood draw also should be acknowledged as a limitation. However, because both patient groups were on similar medications and SARS-CoV2 treatment caused programmed cell death of platelets from healthy donors, we are confident that the major conclusion of our study is not affected by differences in medications. Despite these limitations, it is interesting to speculate that platelets may serve as a dead-end for viruses such as influenza or SARS-CoV-2, with internalization and rapid digestion serving to halt viral reproduction.
Abrogation of viral replication is supported by the observed features of programmed cell death beginning 15 minutes postincubation with the virus as well as markers of apoptosis and necroptosis seen at 30 minutes. Programmed cell death is an important and necessary response to viral infection.36 Failure to mount a proper cell death response may lead to an increase in viral production, progeny release, and lack of a proper immune reaction.36 Future studies should also address the contribution of the different SARS-CoV-2 variants on programmed cell death and activation of platelets. In general, apoptosis of nucleated cells has been understood to be noninflammatory, whereas necroptosis and pyroptosis are inflammatory programmed cell death. The reasons for these differences are based on sensing, pathway activation, and morphological changes that occur depending on the virus and the infected cell. Morphologically, apoptosis is characterized by nuclear condensation, release of apoptotic bodies, and cell shrinkage, whereas necroptosis is characterized by cell swelling, breakage of the plasma membrane, and release of cytotoxic content into the extracellular space.37 Regardless of the initiation signal, mechanistically, apoptosis leads to activation of effector caspases such as Casp3. These caspases can then cleave a restricted set of target proteins that produce the apoptosis-related morphological and biochemical changes that lead to formation of apoptotic bodies, exposure of surface molecules, such as phosphatidylserine, responsible for the phagocytosis of the intact cell corpses and ultimately cell shrinkage.38
Necroptosis, however, is a caspase-independent cell death that involves activation by phosphorylation of MLKL.37 p-MLKL forms octamers that translocate to the membrane-forming pores that induce cell lysis. This process leads to release of intracellular contents such as ATP, IL (interleukin)-1beta, HMGB1 (high mobility group box 1), and other molecules that have been termed as danger-associated molecular patterns and lead to recruitment of proinflammatory cells.37 Consistently, previous reports have suggested a relationship between changes in platelet and plasma inflammatory interleukin content in patients with COVID-19 and the presence of platelet-specific content in plasma.12 Our results suggest that at least some of the inflammatory contents in plasma could be due to platelet necroptosis. Future studies are necessary to define mechanism and establish association with extent of disease.
Our study revealed that platelets incubated with SARS-CoV-2 undergo either an apoptosis-like process or necroptosis. Since platelets lack a nucleus it is difficult to assess apoptosis by traditional morphological changes such as DNA condensation and apoptotic body formation. Apoptotic platelets, however, have been connected to formation of microparticles.39 Pathway analysis of RNAseq-platelets from patients with COVID-19 showed that transcripts related to both apoptosis and necroptosis are enriched in the top 40 upregulated and top 40 downregulated transcripts. Transmission electron microscopy revealed that there is microparticle formation, budding, and a decrease in platelet size (shrinkage). Phosphatidylserine exposure has also been described as a characteristic of platelets from patients with COVID-19.12 WB analysis revealed that there is increased activation of the effector Casp3 in some patients’ platelets and SARS-CoV-2 and Casp3 colocalized on the same platelet.
Necroptosis of platelets as a programmed cell death has not been previously described. In a nucleated cell, this process takes a significant amount of time and requires viral replication. In platelets, the fascinating morphological changes characterizing necroptosis begin 15 minutes postincubation with SARS-CoV-2. Another striking observation is that platelets from noninfected donors had a basal amount of MLKL in octamer formation that was increased in the COVID-19 platelets. Incubation of platelets with SARS-CoV-2 also led to an observable release of platelet contents, some of which had appeared by TEM to lead to channel formation. Immunofluorescence of platelets in whole blood showed increased MLKL phosphorylation in nonpermeabilized platelets that were positive for SARS-CoV-2. Our study proposes that some platelets respond using necroptosis and others apoptosis; however, the molecular events that control a push toward one versus the other warrant further investigation. Based on our previous reports on influenza and differential expression of TLR7 in platelets,23 it is tempting to speculate that platelets that do not have TLR7, and hence do not activate Akt (protein kinase B)23 (prosurvival), are prone to cell death. Regardless of the initiation signal, platelets seem to be prepared for viral entry and neutralization by programmed cell death, further supporting a lack of internal replication. Dysregulation of these processes suggests why some individuals are more prone to a higher incidence of virally mediated thrombotic effects.
Extracellular vesicle formation of platelet origin has been previously described in the plasma of patients with COVID-19.12 Here, we report that incubation of platelets with SARS-CoV-2 leads to release of extracellular vesicles of various kinds. In addition to microparticles with platelet cytoplasmic content, platelets also release and interact with exosomes. Platelet-derived microparticles are known to modify the function of cells such as neutrophils,40 endothelial cells41 and T cells,42 and even their own precursors, the megakaryocytes,43 enabling them to cross-communicate with many cells during infection. In addition to extracellular vesicles, platelet contents may contribute to inflammatory plasma content elevation by direct lysis and forming tiny semivesicles observed in the TEMs. We also observed another type of microparticle associated with platelets that is suggestive for a migrasome. These recently described microparticles, migrasomes, and migrasome-like structures have been located in blood capillaries28 and detected in serum,44 but the cell of origin has been unclear.44 In migrating cells, migrasomes have been described as a migration-dependent organelle that is composed of cytoplasmic contents released as a result of a process defined as migracytosis.45 Migrasome function is not well characterized. So far, it is clear that migrasomes can serve as chemoattractant molecules29,30 and can be taken up by many cells suggesting a role in mediation of cell-cell communication that platelets may utilize.
Our data show that SARS-CoV-2 can be internalized by platelets, and this internalization leads to a variable platelet response involving cell death or pseudopodia formation. However, the receptors responsible for SARS-CoV-2 internalization in platelets have been debated. We show that some platelets express proteins for the putative receptor-protease axis ACE2-TMPRSS2 by immunofluorescence and immunoblotting. Consistent with our previous studies46 and those of others,11 RNA levels of ACE2 or TMPRSS2 are not detectable by sequencing, suggesting certain limitations. RT-qPCR of preamplified platelet transcripts showed limited detection, particularly in the donors with a higher amount of collected platelets, suggesting the necessity of a critical concentration of low abundant RNA. Similarly, primer efficiency may not be comparable as we were not able to detect ACE2 by all 3 primers used. SARS-CoV-2 internalization, however, also occurred without ACE2 expression and the virions were internalized by attaching to microparticles. These observations suggest that ACE2 expression on microparticles may be more important for viral internalization when platelet ACE2 is limited. Our immunofluorescence images and previous studies12 support ACE2 microparticle expression.
Recently, platelets have been appreciated for their diverse functions related to innate and adaptive immunity. The complex platelet reaction to SARS-CoV-2 presented by these data has further expanded our understanding of platelet immune function during infection and provided an increasingly complex picture of how these cells bridge immunity and thrombosis. Our findings suggest that platelets play an active role during infection by interacting with SARS-CoV-2 and internalizing the virions. Internalization of the virus leads to release of various extracellular vesicles, in addition to initiating apoptosis-like and necroptotic programmed cell death. Cell death may contribute to viral sequestration and neutralization as well as important innate and adaptive immune responses. However, effective viral destruction by platelets could contribute to uncontrolled release of platelet contents followed by dysregulated immunity enhanced prothrombotic function as manifested in COVID-19 microthrombosis. A more complete understanding of these complex processes is required for effective risk assessment and treatment of patients with COVID-19.
Author Contributions
M. Koupenova and J.E. Freedman designed, interpreted the results, and wrote this article. M. Koupenova, M. Somasundaran (viral purification and in vitro treatment of platelets), and H.A. Corkrey executed the experiments; R.W. Finberg and J.P. Wang provided blood from patients with coronavirus disease 2019 (COVID-19) and the propagated infectious virus; S. Soofi collected the blood from all patients with COVID-19 and provided patient data. J. Rade provided blood from ST-segment–elevation myocardial infarction (STEMI) and non–ST-segment–elevation myocardial infarction (NSTEMI) patients; R. Bhandari, M. Godwin, and S.J. Cameron provided the ACE (angiotensin-converting enzyme) 2 immunoblotting. K.M. Parsi, A. Cousineau, and R. Maehr created the A549ACE2+TMPRSS2 clonal cell line. All authors assisted with the preparation and editing of the article. All immunofluorescent work was carried out at the UMass Medical School in the Sanderson Center for Optical Experimentation (SCOPE) light microscopy core facility.
Sources of Funding
This work was supported by University of Massachusetts Medical School internal Clinical and Translational Science Award grant and NIH (R01 HL153235) to M. Koupenova (UL1TR001453); American Heart Association (AHA) coronavirus disease 2019 (COVID-19) rapid response grant to J. E. Freedman; Department of Defense grant to R. Finberg (Peer-Reviewed Medical Research Program (PRMRP) Expansion Award–Funding Level 2), and K08HL128856 and NIH Loan Repayment Program HL120200 to S.J. Cameron. The UMass Electron Microscopy Core Facility is funded in part by Research equipment grants (National Institutes of Health [NIH] S10 OD025113-01) and by UMMS Office of Research.
Disclosures
None.
Supplemental Materials
Expanded Materials and Methods
Online Figures I–IV
Online Tables I–VI
Major Resources Table
Online Tables VII_Data Set
Supplementary Material
Nonstandard Abbreviations and Acronyms
- ACE
- angiotensin-converting enzyme
- Casp3
- caspase 3
- COVID-19
- coronavirus disease 2019
- IFN
- interferon
- IL
- -interleukin
- MI
- myocardial infarction
- MLKL
- mixed lineage kinase domain-like pseudokinase
- p-MLKL
- phosphorylated MLKL
- RNAseq
- RNA sequencing
- RT-qPCR
- real-time quantitative polymerase chain reaction
- SARS-CoV-2
- severe acute respiratory syndrome, corona virus-2
- TEM
- transmission electron microscopy
- vRNA
- viral RNA
- vWF
- von Willebrand factor
- WB
- Western blot
This manuscript was sent to Francesco Violi, Guest Editor, and Jane Leopold, Senior Guest Editor, for review by expert referees, editorial decision, and final disposition.
The Data Supplement is available with this article at https://www.ahajournals.org/doi/suppl/10.1161/CIRCRESAHA.121.319117.
For Sources of Funding and Disclosures, see page 644–645.
Contributor Information
Heather A. Corkrey, Email: heather.corkrey@umassmed.edu.
Olga Vitseva, Email: Olga.Vitseva@umassmed.edu.
Kahraman Tanriverdi, Email: Kahraman.Tanriverdi@umassmed.edu.
Mohan Somasundaran, Email: mohan.somasundaran@umassmed.edu.
Ping Liu, Email: ping.liu@umassmed.edu.
Shaukat Soofi, Email: Shaukat.Soofi@umassmed.edu.
Rohan Bhandari, Email: BHANDAR2@ccf.org.
Matthew Godwin, Email: GODWINM@ccf.org.
Krishna Mohan Parsi, Email: krishnamohan.parsi@umassmed.edu.
Alyssa Cousineau, Email: acousineau95@gmail.com.
René Maehr, Email: Rene.Maehr@umassmed.edu.
Jennifer P. Wang, Email: jennifer.wang@umassmed.edu.
Scott J. Cameron, Email: CAMEROS3@ccf.org.
Jeffrey Rade, Email: jeffrey.rade@umassmed.edu.
Robert W. Finberg, Email: robert.finberg@umassmed.edu.
Jane E. Freedman, Email: jane.freedman@vumc.org.
Novelty and Significance
What Is Known?
Severe coronavirus disease 2019 (COVID-19) is characterized by microthrombosis, increased coagulation, and profound inflammation.
Platelets mediate clinical thrombosis and, in patients with COVID-19, may have an increased thrombotic or inflammatory profile.
Current data from quantitative polymerase chain reaction of platelets has detected SARS-CoV-2 (severe acute respiratory syndrome, corona virus-2) in about 25% of infected patients.
Respiratory viruses such as influenza can be taken up by platelets leading to a platelet-mediated immune-thrombotic response.
What New Information Does This Article Contribute?
By sequencing for SARS-CoV-2, we show that platelets from all patients with COVID-19 contain various levels of viral RNA, but the viral genome is fragmented making it noninfectious.
Platelets can rapidly internalize SARS-CoV-2 either through ACE (angiotensin-converting enzyme) 2 or by taking up virions attached to microparticles.
Internalization of SARS-CoV-2 initiates platelet death programs that lead to release of platelet content and subsequent reduction of their functionality.
COVID-19 is characterized by increased risk for thrombosis, coagulation, and inflammation. Platelets from patients with COVID-19 have increased thrombotic potential but decreased procoagulant response. Although studies have shown that the virus that causes COVID-19, SARS-CoV-2, can be detected in the platelets of one-fourth of patients, it is not clear if the virus is commonly in the circulation of patients or if it has a direct effect on platelets. Using various models, we show that SARS-CoV-2 is present in platelets, but not in plasma, of all patients tested in this study. In platelets, the virus appears fragmented and does not reproduce. After viral uptake, platelets undergo rapid cell death suggesting that the platelet milieu does not permit viral replication and may be protective in the immune response. However, the release of platelet contents during the process of dying can be highly prothrombotic or proinflammatory and can lead to dysregulated immune activation. Our findings suggest that during COVID-19, platelets mediate a rapid response to SARS-CoV-2, and this response, when exaggerated, can contribute to dysregulated immunity and thrombotic outcomes.
References
- 1.Woolf SH, Chapman DA, Lee JH. COVID-19 as the Leading Cause of Death in the United States. JAMA. 2021;325:123–124. doi: 10.1001/jama.2020.24865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petrilli CM, Jones SA, Yang J, Rajagopalan H, O’Donnell L, Chernyak Y, Tobin KA, Cerfolio RJ, Francois F, Horwitz LI. Factors associated with hospital admission and critical illness among 5279 people with coronavirus disease 2019 in New York City: prospective cohort study. BMJ. 2020;369:m1966. doi: 10.1136/bmj.m1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McGurnaghan SJ, Weir A, Bishop J, Kennedy S, Blackbourn LAK, McAllister DA, Hutchinson S, Caparrotta TM, Mellor J, Jeyam A, et al. ; Public Health Scotland COVID-19 Health Protection Study Group; Scottish Diabetes Research Network Epidemiology Group. Risks of and risk factors for COVID-19 disease in people with diabetes: a cohort study of the total population of Scotland. Lancet Diabetes Endocrinol. 2021;9:82–93. doi: 10.1016/S2213-8587(20)30405-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fajgenbaum DC, June CH. Cytokine storm. N Engl J Med. 2020;383:2255–2273. doi: 10.1056/NEJMra2026131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koupenova M, Freedman JE. Platelets and COVID-19: inflammation, hyperactivation and additional questions. Circ Res. 2020;127:1419–1421. doi: 10.1161/CIRCRESAHA.120.318218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elbadawi A, Elgendy IY, Sahai A, Bhandari R, McCarthy M, Gomes M, Bishop GJ, Bartholomew JR, Kapadia S, Cameron SJ. Incidence and outcomes of thrombotic events in symptomatic patients With COVID-19. Arterioscler Thromb Vasc Biol. 2021;41:545–547. doi: 10.1161/ATVBAHA.120.315304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lillicrap D. Disseminated intravascular coagulation in patients with 2019-nCoV pneumonia. J Thromb Haemost. 2020;18:786–787. doi: 10.1111/jth.14781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang N, Li D, Wang X, Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J Thromb Haemost. 2020;18:844–847. doi: 10.1111/jth.14768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Machlus KR, Italiano JE., Jr.The incredible journey: from megakaryocyte development to platelet formation. J Cell Biol. 2013;201:785–796. doi: 10.1083/jcb.201304054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koupenova M. Potential role of platelets in COVID-19: Implications for thrombosis. Res Pract Thromb Haemost. 2020;4:737–740. doi: 10.1002/rth2.12397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manne BK, Denorme F, Middleton EA, Portier I, Rowley JW, Stubben C, Petrey AC, Tolley ND, Guo L, Cody M, et al. Platelet gene expression and function in patients with COVID-19. Blood. 2020;136:1317–1329. doi: 10.1182/blood.2020007214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zaid Y, Puhm F, Allaeys I, Naya A, Oudghiri M, Khalki L, Limami Y, Zaid N, Sadki K, Ben El Haj R, et al. Platelets can associate with sars-cov-2 rna and are hyperactivated in covid-19. Circ Res. 2020;127:1404–1418. doi: 10.1161/CIRCRESAHA.120.317703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denorme F, Manne BK, Portier I, Petrey AC, Middleton EA, Kile BT, Rondina MT, Campbell RA. COVID-19 patients exhibit reduced procoagulant platelet responses. J Thromb Haemost. 2020;18:3067–3073. doi: 10.1111/jth.15107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.V’kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang T, Bidon M, Jaimes JA, Whittaker GR, Daniel S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiviral Res. 2020;178:104792. doi: 10.1016/j.antiviral.2020.104792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clausen TM, Sandoval DR, Spliid CB, Pihl J, Perrett HR, Painter CD, Narayanan A, Majowicz SA, Kwong EM, McVicar RN, et al. SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2. Cell. 2020;183:1043, e15–1057. doi: 10.1016/j.cell.2020.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baker AN, Richards SJ, Guy CS, Congdon TR, Hasan M, Zwetsloot AJ, Gallo A, Lewandowski JR, Stansfeld PJ, Straube A, et al. The SARS-COV-2 Spike Protein Binds Sialic Acids and Enables Rapid Detection in a Lateral Flow Point of Care Diagnostic Device. ACS Cent Sci. 2020;6:2046–2052. doi: 10.1021/acscentsci.0c00855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MADOFF MA, EBBE S, BALDINI M. SIALIC ACID OF HUMAN BLOOD PLATELETS. J Clin Invest. 1964;43:870–877. doi: 10.1172/JCI104972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vögtle T, Sharma S, Mori J, Nagy Z, Semeniak D, Scandola C, Geer MJ, Smith CW, Lane J, Pollack S, et al. Heparan sulfates are critical regulators of the inhibitory megakaryocyte-platelet receptor G6b-B. Elife. 2019;8:e46840. doi: 10.7554/eLife.46840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng L, Liu J, Xu W, Luo Q, Chen D, Lei Z, Huang Z, Li X, Deng K, Lin B, et al. SARS-CoV-2 can be detected in urine, blood, anal swabs, and oropharyngeal swabs specimens. J Med Virol. 2020;92:1676–1680. doi: 10.1002/jmv.25936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Atianand MK, Caffrey DR, Fitzgerald KA. Immunobiology of Long Noncoding RNAs. Annu Rev Immunol. 2017;35:177–198. doi: 10.1146/annurev-immunol-041015-055459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koupenova M, Corkrey HA, Vitseva O, Manni G, Pang CJ, Clancy L, Yao C, Rade J, Levy D, Wang JP, et al. The role of platelets in mediating a response to human influenza infection. Nat Commun. 2019;10:1780. doi: 10.1038/s41467-019-09607-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jerez-Dolz D, Torramade-Moix S, Palomo M, Moreno-Castaño A, Lopez-Vilchez I, Hernandez R, Badimon JJ, Zafar MU, Diaz-Ricart M, Escolar G. Internalization of microparticles by platelets is partially mediated by toll-like receptor 4 and enhances platelet thrombogenicity. Atherosclerosis. 2020;294:17–24. doi: 10.1016/j.atherosclerosis.2019.12.017 [DOI] [PubMed] [Google Scholar]
- 25.Żmigrodzka M, Guzera M, Miśkiewicz A, Jagielski D, Winnicka A. The biology of extracellular vesicles with focus on platelet microparticles and their role in cancer development and progression. Tumour Biol. 2016;37:14391–14401. doi: 10.1007/s13277-016-5358-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joshi S, Balasubramanian N, Vasam G, Jarajapu YP. Angiotensin converting enzyme versus angiotensin converting enzyme-2 selectivity of MLN-4760 and DX600 in human and murine bone marrow-derived cells. Eur J Pharmacol. 2016;774:25–33. doi: 10.1016/j.ejphar.2016.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang L, Sexton DJ, Skogerson K, Devlin M, Smith R, Sanyal I, Parry T, Kent R, Enright J, Wu QL, et al. Novel peptide inhibitors of angiotensin-converting enzyme 2. J Biol Chem. 2003;278:15532–15540. doi: 10.1074/jbc.M212934200 [DOI] [PubMed] [Google Scholar]
- 28.Ma L, Li Y, Peng J, Wu D, Zhao X, Cui Y, Chen L, Yan X, Du Y, Yu L. Discovery of the migrasome, an organelle mediating release of cytoplasmic contents during cell migration. Cell Res. 2015;25:24–38. doi: 10.1038/cr.2014.135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang Y, Zucker B, Zhang S, Elias S, Zhu Y, Chen H, Ding T, Li Y, Sun Y, Lou J, et al. Migrasome formation is mediated by assembly of micron-scale tetraspanin macrodomains. Nat Cell Biol. 2019;21:991–1002. doi: 10.1038/s41556-019-0367-5 [DOI] [PubMed] [Google Scholar]
- 30.Tavano S, Heisenberg CP. Migrasomes take center stage. Nat Cell Biol. 2019;21:918–920. doi: 10.1038/s41556-019-0369-3 [DOI] [PubMed] [Google Scholar]
- 31.Huang D, Zheng X, Wang ZA, Chen X, He WT, Zhang Y, Xu JG, Zhao H, Shi W, Wang X, et al. The mlkl channel in necroptosis is an octamer formed by tetramers in a dyadic process. Mol Cell Biol. 2017;37:e00497–16. doi: 10.1128/MCB.00497-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahnadi CE, Sabrinah Chapman E, Lépine M, Okrongly D, Pujol-Moix N, Hernández A, Boughrassa F, Grant AM. Assessment of platelet activation in several different anticoagulants by the Advia 120 Hematology System, fluorescence flow cytometry, and electron microscopy. Thromb Haemost. 2003;90:940–948. doi: 10.1160/TH03-02-0097 [DOI] [PubMed] [Google Scholar]
- 34.Tipnis SR, Hooper NM, Hyde R, Karran E, Christie G, Turner AJ. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J Biol Chem. 2000;275:33238–33243. doi: 10.1074/jbc.M002615200 [DOI] [PubMed] [Google Scholar]
- 35.Koupenova M, Vitseva O, MacKay CR, Beaulieu LM, Benjamin EJ, Mick E, Kurt-Jones EA, Ravid K, Freedman JE. Platelet-TLR7 mediates host survival and platelet count during viral infection in the absence of platelet-dependent thrombosis. Blood. 2014;124:791–802. doi: 10.1182/blood-2013-11-536003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomson BJ. Viruses and apoptosis. Int J Exp Pathol. 2001;82:65–76. doi: 10.1111/j.1365-2613.2001.iep0082-0065-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orzalli MH, Kagan JC. Apoptosis and necroptosis as host defense strategies to prevent viral infection. Trends Cell Biol. 2017;27:800–809. doi: 10.1016/j.tcb.2017.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fink SL, Cookson BT. Apoptosis, pyroptosis, and necrosis: mechanistic description of dead and dying eukaryotic cells. Infect Immun. 2005;73:1907–1916. doi: 10.1128/IAI.73.4.1907-1916.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vasina EM, Cauwenberghs S, Feijge MA, Heemskerk JW, Weber C, Koenen RR. Microparticles from apoptotic platelets promote resident macrophage differentiation. Cell Death Dis. 2011;2:e211. doi: 10.1038/cddis.2011.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duchez AC, Boudreau LH, Naika GS, Bollinger J, Belleannée C, Cloutier N, Laffont B, Mendoza-Villarroel RE, Lévesque T, Rollet-Labelle E, et al. Platelet microparticles are internalized in neutrophils via the concerted activity of 12-lipoxygenase and secreted phospholipase A2-IIA. Proc Natl Acad Sci USA. 2015;112:E3564–E3573. doi: 10.1073/pnas.1507905112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Faille D, El-Assaad F, Mitchell AJ, Alessi MC, Chimini G, Fusai T, Grau GE, Combes V. Endocytosis and intracellular processing of platelet microparticles by brain endothelial cells. J Cell Mol Med. 2012;16:1731–1738. doi: 10.1111/j.1582-4934.2011.01434.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dinkla S, van Cranenbroek B, van der Heijden WA, He X, Wallbrecher R, Dumitriu IE, van der Ven AJ, Bosman GJ, Koenen HJ, Joosten I. Platelet microparticles inhibit IL-17 production by regulatory T cells through P-selectin. Blood. 2016;127:1976–1986. doi: 10.1182/blood-2015-04-640300 [DOI] [PubMed] [Google Scholar]
- 43.French SL, Butov KR, Allaeys I, Canas J, Morad G, Davenport P, Laroche A, Trubina NM, Italiano JE, Moses MA, et al. Platelet-derived extracellular vesicles infiltrate and modify the bone marrow during inflammation. Blood Adv. 2020;4:3011–3023. doi: 10.1182/bloodadvances.2020001758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao X, Lei Y, Zheng J, Peng J, Li Y, Yu L, Chen Y. Identification of markers for migrasome detection. Cell Discov. 2019;5:27. doi: 10.1038/s41421-019-0093-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, Wang J, Ding Y, Zhang J, Xu Y, Xu J, Zheng S, Yang H. Migrasome and tetraspanins in vascular homeostasis: concept, present, and future. Front Cell Dev Biol. 2020;8:438. doi: 10.3389/fcell.2020.00438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clancy L, Beaulieu LM, Tanriverdi K, Freedman JE. The role of RNA uptake in platelet heterogeneity. Thromb Haemost. 2017;117:948–961. doi: 10.1160/TH16-11-0873 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data Supplement contains detailed methods of all experiments included in this article. The Data Supplement also contains data, analytic methods, reagents supporting the findings of this study and are available from the corresponding author upon reasonable request.